
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
TECHNOLOGY REPORT article
Front. Mar. Sci. , 20 February 2018
Sec. Marine Molecular Biology and Ecology
Volume 5 - 2018 | https://doi.org/10.3389/fmars.2018.00028
 Chuya Shinzato1,2*†
Chuya Shinzato1,2*† Yuna Zayasu2
Yuna Zayasu2 Miyuki Kanda3
Miyuki Kanda3 Mayumi Kawamitsu3
Mayumi Kawamitsu3 Noriyuki Satoh2
Noriyuki Satoh2 Hiroshi Yamashita4†
Hiroshi Yamashita4† Go Suzuki4†
Go Suzuki4†Frequent, high-density coral monitoring is essential to understand coral reef ecosystems. For this purpose, we developed a novel method for simultaneous monitoring of Acropora corals and their symbiont, Symbiodinium, from environmental DNA (eDNA) in seawater using next generation sequencing technology (NGS). We performed a tank experiment with running seawater using 19 Acropora species. Complete mitochondrial genomes of all the Acropora species were assembled to create a database and major types of their Symbiodinium symbionts were identified. Then eDNA was isolated by filtering inlet and outlet seawater from the tanks. Acropora and Symbiodinium DNA were amplified by PCR and sequenced. We detected all of the tested Acropora types from eDNA samples. Proportions and numbers of DNA sequences were both positively correlated with masses of corals in the tanks. In this trial, we detected DNA sequences from as little as 0.04 kg of Acropora colony, suggesting that existence of at least one adult Acropora colony (~30 cm diameter = 1 kg) per m2 at depths < 10 m could be detected using eDNA in the field. In addition, we detected major types of Symbiodinium within host corals from seawater, suggesting that it should be possible to detect major coral symbiont types if Acropora corals exist nearby, and possible free-living state Symbiodinium cells from eDNA in seawater. eDNA abundance of Symbiodinium types did not correlate well with frequencies of major Symbiodinium types in the corals, suggesting that quantification of Symbiodinium is difficult at this stage. Although this is the initial attempt to detect coral and Symbiodinium simultaneously from eDNA in seawater, this method may allow us to perform high-frequency, high-density coral reef monitoring of both corals and their symbionts in the near future.
Coral reefs are estimated to harbor about one-third of all described marine species (Knowlton et al., 2010); however, they face a range of anthropogenic challenges, including ocean acidification and increasing seawater temperatures (Hoegh-Guldberg et al., 2007). It has been reported that tropical storms, predation by crown-of-thorns starfish, and bleaching are major causes of coral reef decline (De'ath et al., 2012). Frequent high-density coral monitoring is essential to understand dramatically changing coral reef environments. In addition, it is also important to develop strategies against many threats and policies for quick rehabilitation and conservation. However, actual coral reef monitoring is generally performed infrequently because it requires direct observation by scuba diving and sampling by hand.
DNA that originates from various sources, such as mucus, metabolic waste, and damaged tissues from multicellular organisms exists in seawater and is called environmental DNA (eDNA) (Bohmann et al., 2014; Kelly et al., 2014). Recently eDNA has begun to be employed in aquatic environmental research (Rees et al., 2014, 2015), and it is also being used to monitor marine biodiversity (Miya et al., 2015; Yamamoto et al., 2017). However, we are unaware of any reports that have attempted to use eDNA from seawater to monitor coral reefs. In this study, we performed tank experiments with running seawater as an initial proof of concept.
The structure of coral reefs is formed by calcium deposition by anthozoan cnidarians known as scleractinian corals. We examined the genus Acropora (Scleractinia, Acroporidae) in this study because it is a keystone reef taxon globally, ranging from the Red Sea through the Indo-Pacific Ocean to the Caribbean. It is also the most speciose coral genus with more than 100 described species (Wallace, 1999). Their widespread distribution notwithstanding, Acropora species are among the most sensitive to increasing water temperatures (Loya et al., 2001); hence, they are expected to decline in the near future (Alvarez-Filip et al., 2013).
Scleractinian corals, including Acropora, form obligate endosymbioses with photosynthetic dinoflagellates of the genus Symbiodinium (Yellowlees et al., 2008; Bourne et al., 2009; Shinzato et al., 2014a). The genus Symbiodinium displays tremendous taxonomic and physiological diversity (Lajeunesse, 2001; Stat et al., 2008). Nine divergent lineages, clades A–I, have been described, based on nuclear ribosomal DNA (rDNA) and chloroplast 23S rDNA (Pochon and Gates, 2010). It has been reported that stress tolerance of Symbiodinium differs among clades or types (Tchernov et al., 2004; Sampayo et al., 2008; Hawkins and Davy, 2012) and Symbiodinium genetic composition may play important roles in coral health and stress tolerance (Jones et al., 2008; Stat et al., 2008; Lajeunesse et al., 2010; Fisher et al., 2012; Keshavmurthy et al., 2012, 2014). Thus, simultaneous investigation of corals and Symbiodinium is probably important to understand coral reef ecosystems.
Corals release massive amounts of soluble mucus, which transfers large amounts of energy and nutrients to the reef substrate (Wild et al., 2004). Due to the symbiont natural rates of increase, corals steadily release Symbiodinium cells into the surrounding environment (Yamashita et al., 2011), suggesting that close to reefs, seawater should contain detectable quantities of DNA from both corals and Symbiodinium. Recently, whole genome sequences of an Acropora coral (Shinzato et al., 2011) and Symbiodinium (Shoguchi et al., 2013; Lin et al., 2015; Aranda et al., 2016) have been published, and next-generation sequencing (NGS) technologies have been used to investigate coral reef biodiversity (Shinzato et al., 2014b, 2015; Combosch and Vollmer, 2015; Bongaerts et al., 2017). In the genus Symbiodinium, each clade contains multiple genetic types, and identification has been performed using ribosomal, mitochondrial, plastid, and nuclear DNA markers (Rowan and Powers, 1991; Wilcox, 1998; Lajeunesse, 2001; Santos et al., 2002; Takabayashi et al., 2004). Recently internal-transcribed spacer 2 (ITS-2) regions have been widely used as genetic markers (Lajeunesse, 2001, 2002, 2005; Pochon et al., 2007; Stat et al., 2011) and databases of Symbiodinium DNA sequences, such as SYM-BLAST (https://www.auburn.edu/~santosr/symblast.htm), SymbioGBR (Tonk et al., 2013), and GeoSymbio (Franklin et al., 2012), are currently available. In this laboratory study, we developed a technique that would enable simultaneous detection and identification of coral and Symbiodinium eDNA in reefs, in order to assess distributions of corals and their symbiont types.
Acropora corals from Sekisei Lagoon, Ishigaki Island, Okinawa Prefecture, Japan were identified and ~20-cm-diameter fragments were collected in May 2016. In an exception, sampling of corals was permitted by the Okinawa Prefectural Government for research use (No. 27-73). In total, we collected 19 species (63 colonies) and put 16 shallow-water (5–10 m) Acropora species in Tank 1 (A. acuminata, A. austera, A. cytherea, A. carduus, A. digitifera, A. florida, A. grandis, A. hyacinthus, A. intermedia, Acropora sp., A. microphthalma, A. muricata, A. nasta, A. selago, A. tenuis) and three deeper-water species (≥15 m) in Tank 2 (A. echinata, Acropora sp., A. awi) respectively at Seikai National Fisheries Research Institute at Ishigaki Island, and kept for 2 days with running seawater (Figure 1, Table 1). Numbers of colonies and coral masses determined in seawater differed among species in order to test detection limits of eDNA in seawater (Table 1). Tank sizes and water flow of the two overflow tanks are as follows; Tank 1: 300 cm length × 80 cm width × 50 cm height (water depth: ~20 cm) and a flow rate of 35 L of seawater/min, Tank 2: 200 cm diameter and 100 cm depth (water depth: ~80 cm) and 10 L/min. Small fragments of each coral colony were isolated and stored in the guanidinium reagent, CHAOS (Fukami et al., 2000), for DNA extraction.
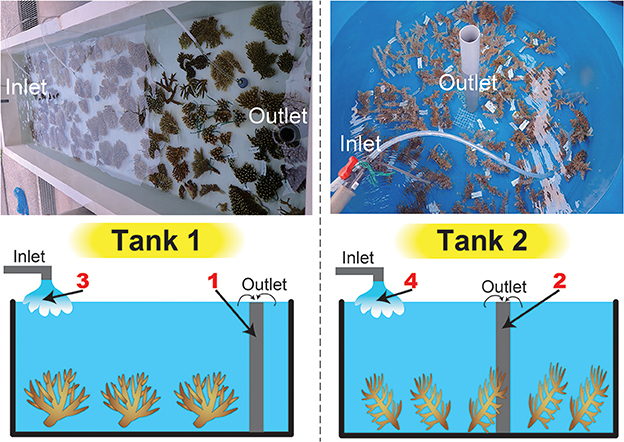
Figure 1. Tanks used in this study and a schematic drawing of water sampling. Photographs of the tanks are shown above, and a schematic drawing of water sampling is shown below. We filtered seawater from inlets and outlets in both tanks (four sample numbers in red).
DNA from each colony was isolated from CHAOS solution using a Maxwell 16 Automated Purification System (Promega). We used 200 ng of DNA from each sample for PCR-free library preparation with KAPA HyperPlus Kits (NIPPON Genetics Co, Ltd) following the manufacturer's instructions. All libraries were multiplexed and sequenced (250-bp paired-end, using a HiSeq 2500 in Rapid mode; Illumina). Raw sequence data were submitted to the DNA Databank of Japan (DDBJ) Sequence Read Archive (DRA) under accession number DRA005695 (Table S1, BioProject PRJDB5633). Sequencing data from each library were assembled with IDBA_UD assembler version 1.1.1 (Peng et al., 2012) with different kmer lengths (60, 80, and 100). Identification of complete circular mitochondrial genomes from assembled contigs was performed by (1) comparing them with the Acropora tenuis complete mitochondrial genome (NCBI accession: AF338425.1) (BLASTN e-value ≤ 1e−100), and (2) confirming that 100 bp of both head and tail DNA sequences of a contig were identical, indicating that the sequence was circular.
After 2 days of incubation, 3 L of raw seawater were collected from outlet pipes of the two tanks (Figure 1) and were filtered through a 0.8 μm pore size polycarbonate filter (47-mmΦ, Advantec Toyo Kaisha, Ltd., Tokyo Japan). In addition, seawater from inlet pipes was also collected to provide control samples (before the seawater was passed over the corals) (Figure 1). Filters were frozen at −20°C for several days, and then boiled in 500 μL of TE buffer. Details of the TE boiling method for DNA extraction are provided in Koike et al. (2007) and Yamashita et al. (2011). A part of the Acropora mitochondrial control region (mtCR, ~800–1,000 bp in length) has been widely used as a molecular marker for Acropora species identification (van Oppen et al., 2002; Suzuki et al., 2008). Thus, eDNA was subjected to PCR amplification using RNS2 (5′-CAGAGTAAGTCGTAACATAG-3′) and GR (5′-AATTCCGGTGTGTGTTCTCT-3′) primers for amplifying mtCR (van Oppen et al., 2002; Suzuki et al., 2008) together with genomic DNA isolated from Acropora echinata as a positive PCR control (expected amplicon size: 935 bp). PCR cycling conditions were 15 min at 95°C, followed by 32 cycles of 30 s at 94°C, 90 s at 58°C, and 60 s at 72°C, with an extension of 30 min at 60°C in the final cycle. To amplify ~600–700 bp of Symbiodinium internal-transcribed spacer regions (both ITS-1 and ITS-2) r18Sf primer (5′-GAAAGTTTCATGAACCTTAT-3′), which binds to an 18S region, and Sym28Sr primer (5′-CTTGTRTGACTTCATGCTA-3′), which binds to the ITS2-flanking 28S region, were used (Yamashita and Koike, 2013), together with genomic DNA isolated from clade C Symbiodinium culture strain CCMP2466 (clade C, type C1; purchased from the Provasoli–Guillard National Center for Culture of Marine Algae and Microbiota, East Boothbay, Maine, USA) as a positive PCR control (expected amplicon size: 676 bp). PCR cycling conditions were 35 cycles of 45 s at 94°C, 45 s at 51°C, and 60 s at 72°C. To check major symbiotic Symbiodinium types in each Acropora colony, DNAs isolated from each colony were also used for PCR amplification using r18Sf and Sym28Sr primers under the same PCR conditions mentioned above. All PCR products were cleaned with a QIAquick PCR Purification Kit (Qiagen).
Sequencing libraries of cleaned PCR products (ten samples, DNA amounts used from each sample are shown in Figure S1) were prepared using a KAPA Hyper Prep Kit (NIPPON Genetics Co, Ltd) without fragmentation. Libraries were multiplexed and 300-bp paired-end reads were sequenced with a MiSeq platform (Illumina) with PhiX control (Illumina). Raw sequence data were submitted to the DDBJ DRA under accession number DRA005680 (BioProject PRJDB5633). Read pairs that both PCR primers of Acropora (RNS2 and GR) or Symbiodinium (r18Sf and Sym28Sr) recognized were selected using custom perl scripts (Supplementary Materials) and subjected to quality filtering. Low quality bases (Phred quality score <30) were trimmed using SolexaQA (Cox et al., 2010) and high-quality sequences longer than 200 bp were retained for subsequent analyses.
To identify Acropora species from eDNA, we used mtCR sequences based on the complete, assembled, mitochondrial genomes of corals used in this study (see above) and on A. digitifera sequencing data from five individuals previously collected at Kabira, Ishigaki, Japan (Shinzato et al., 2015). Although mtCR is highly variable within Acropora mitochondrial genomes (Figure S2), percent identities of mtCR sequences between species are high (88.5–100%, Table S2). Lengths of mtCR exceeding paired-end sequencing length (300-bp paired-end) and 300-bp sequences flanking the RNS2 primer are more variable than those of the GR primer (Tables S3, S4). Accordingly, 300 bp flanking the RNS2 primer sequence in the mtCR from each colony were extracted and exactly the same sequences of clustered using CDHIT-EST (Li and Godzik, 2006), and the clustered sequences (14 types) comprised a mtCR database in this study (Table S3). Quality trimmed reads having the RNS2 primer sequence were identified and chimera filtering based on the mtCR database was performed using the UCHIME2 algorithm (Edgar, in review). We aligned these filtered sequences to the database using BLASTN (Camacho et al., 2009) with e-value cutoff of 1e−100. Output files were stringently filtered and numbers of valid alignments that ensure all of the following criteria were counted (details are available on Supplementary Materials); (1) percent identity = 100% as mitochondrial genome sequences of all colonies were in the database; (2) the BLASTN bit score of the best alignment must be greater than the second-best alignment. In order to determine the correlation between coral wet masses and the number of valid alignments, regression analysis was performed using R, version 3.1.3 (R Core Team, 2015).
To develop a Symbiodinium ITS-2 database, we first clustered exactly the same DNA sequences in a database of ITS-2 gene sequences of most reported Symbiodinium types, GeoSymbio (410 ITS-2 sequences) (Franklin et al., 2012) using CDHIT-EST (Li and Godzik, 2006) with option “–c 1.” This resulted in 391 sequences (different ITS-2 sequences sharing same DNA sequences in the original GeoSymbio database were annotated using a concatenation of the original GeoSymbio names with “_”). Then we downloaded Symbiodinium DNA sequences containing the term “internal transcribed spacer 2” from NCBI in October 2017, and Blasted (Camacho et al., 2009) these against the clustered GeoSymbio (BLASTN, e-value < 1e−5). Aligned sequences longer than 200 bp were extracted and annotated with the names of BLASTN top-hit sequences in the clustered GeoSymbio together with their NCBI accession numbers. Finally, exactly the same DNA sequences were clustered using CDHIT-EST (Li and Godzik, 2006) with option “–c 1.” For identification of Symbiodinium types from eDNA and each Acropora colony, ITS-2 gene sequences flanking the Sym28Sr primer were subjected to chimera filtering (Edgar, in review) using the database and aligned to the Symbiodinium ITS-2 database using BLASTN (Camacho et al., 2009) with an e-value cutoff of 1e−100. Output files were stringently filtered and the number of valid alignments that ensure all of the following criteria were counted (details are available on Supplementary Materials); (1) percent identity ≥ 98% as there might be Symbiodinium types that are not included in the NCBI database; (2) the BLASTN bit score of the best alignment must be greater than the second-best alignment.
We isolated genomic DNA from tissues taken from each of 63 colonies, containing DNA from both Acropora and Symbiodinium, except for A. digitifera, from which we failed to isolate useful DNA. We successfully assembled complete (circular) mitochondrial genomes of all species (accession numbers: LC201813—LC201870) (Table S1). We identified putative mtCR in the assembled mitochondrial genomes and used these to create a database comprised 14 mtCR types (Table S3).
For development of a Symbiodinium ITS-2 database, 5,962 DNA sequences of possible Symbiodinium ITS-2 were downloaded from NCBI, annotated using GeoSymbio database, and exactly the same DNA sequences were clustered. Then, we prepared a Symbiodinium ITS-2 database containing 2,352 non-redundant sequences. Using the latter database, we identified major types of Symbiodinium from each colony (Figure 2). Most colonies exclusively harbored clade C (99.96% of total valid alignments, 4533872 out of 4535626, Figure 2 and Supplementary Material), but clade D was also a major symbiont in some colonies (e.g., D1 and D1a in A. tenuis ten3 and ten12 colonies, Figure 2). Interestingly, C50 is the most major in most colonies in shallow-water Acropora in Tank 1, although C3d_C21 occurred in half of Tank 2 Acropora colonies (Figure 2).
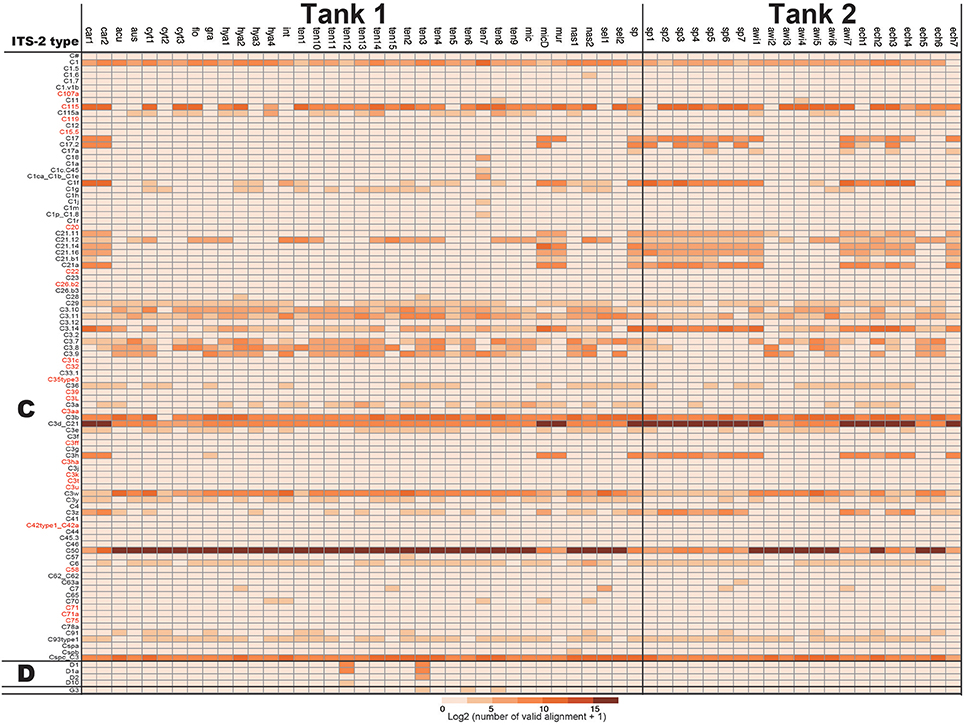
Figure 2. Numbers of ITS-2 types detected by PCR using r18Sf and Sym28Sr primers for identifying symbiotic Symbiodinium types in each Acropora colony. ITS-2 types that share the same GeoSymbio names in the database were summed (raw numbers of valid alignments are available in Supplementary Material). Numbers of valid alignment were shown at log2 scale and colored. ITS-2 types belonging to the same clade (C or D) are separated. ITS-2 types in red were not detected from eDNA (see Figure 4).
After culturing corals for 2 days, we isolated eDNA from both inlet and outlet seawater. We performed PCR to amplify Acropora mtCR and the Symbiodinium ITS-2 region (Figure S1). Although all PCR products using Symbiodinium primers could be identified, some PCR products of Acropora mtCR were not visible by electrophoresis. However, we did isolate DNA from all Acropora mtCR PCR products (Tank 1 outlet: 8.4 ng/μL, Tank 2 outlet: 4.3 ng/μL, Tank 1 inlet: 4.6 ng/μL, Tank 2 inlet: 4.5 ng/μL, respectively) and successfully prepared Illumina sequencing libraries for all samples (Figure S1). These ten amplicon samples, including two positive controls (A. echinata and Symbiodinium culture strain CCMP2466), were multiplexed and sequenced (Table 2).
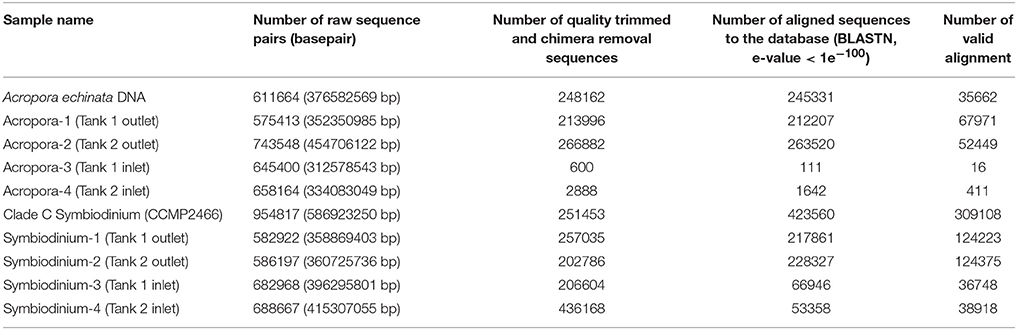
Table 2. Summary of sequencing of PCR amplicons from eDNA in culturing tanks (Tank 1 and Tank 2) and positive controls for Acropora and Symbiodinium (isolated DNAs of Acropora echinata and cultured clade C Symbiodinium CCMP2466, respectively).
As mtCR sequences are highly conserved among Acropora corals (Tables S2–S4), most sequences mapped to more than two sequences in the databases. Thus, we subsequently filtered mapping output so as to count only valid alignments to the database. In the positive control of A. echinata DNA PCR amplicon, 99.1% of valid alignments (35339 of 35662) were correctly assigned to Acropora sp. (Figure 3), indicating that our filtering method worked properly. As expected, few valid alignments from inlet water of Tank 1 and Tank 2 were detected (Table S5). We identified all 14 mtCR types in the database, although only a single valid alignment for car1 (A. carduus) was detected (Figure 3A). The most abundant mtCR types in Tank 1 included A. tenuis (Figure 3A), probably reflecting 15 A. tenuis colonies in the tank (Table 1). In Tank 2, mtCR sequences from Acropora sp. and A. echinata were detected more frequently than A. awi (Figure 3). However, although coral species in Tank 1 and Tank 2 were different, a small proportion of alignments to some mtCR types of coral species that did not exist in each Tank were detected in outlet water of both tanks (Tank 1: 0.05%; 40 out of 67971, Tank 2: 0.8%; 440 out of 52449, Figure 3 in red colors). In inlet water of Tanks 1 and 2, few valid alignments were observed (Figure 3). The proportions of valid alignments of each mtCR types to the total numbers of valid alignments and proportions of weights of corals (Figure 3B, Table S5) were positively correlated in both tanks (regression analysis, R2 = 0.7793, p-value < 0.001), in Tank 1 (regression analysis, R2 = 0.9807, p-value < 0.001) and in Tank 2 (regression analysis, R2 = 0.5985, p-value < 0.01), respectively, suggesting that proportions of DNA sequences reflect the proportions of coral types in surrounding water. In addition, the number of sequences and weights of corals (Figure 3C, Table S5) were positively, but not strongly correlated in both tanks (regression analysis, R2 = 0.6371, p-value < 0.01), in Tank 1 (regression analysis, R2 = 0.6923, p-value < 0.01), and in Tank 2 (regression analysis, R2 = 0.7109, p-value < 0.01), respectively. We detected coral colonies heavier than 0.04 kg (e.g., A. microphthalma deeper-morph: 0.04 kg in Tank 1, Table 1). Overall, the calculated average number of valid alignments per kg (Table S5) was 6145 ± 7058 (R2 = 0.7962, p < 0.001), while for Acropora species in Tank 1 it was 3389 ± 2895 (R2 = 0.987, p < 0.001) and in Tank 2 it was 13492 ± 10411 (R2 = 0.5658, p < 0.05).
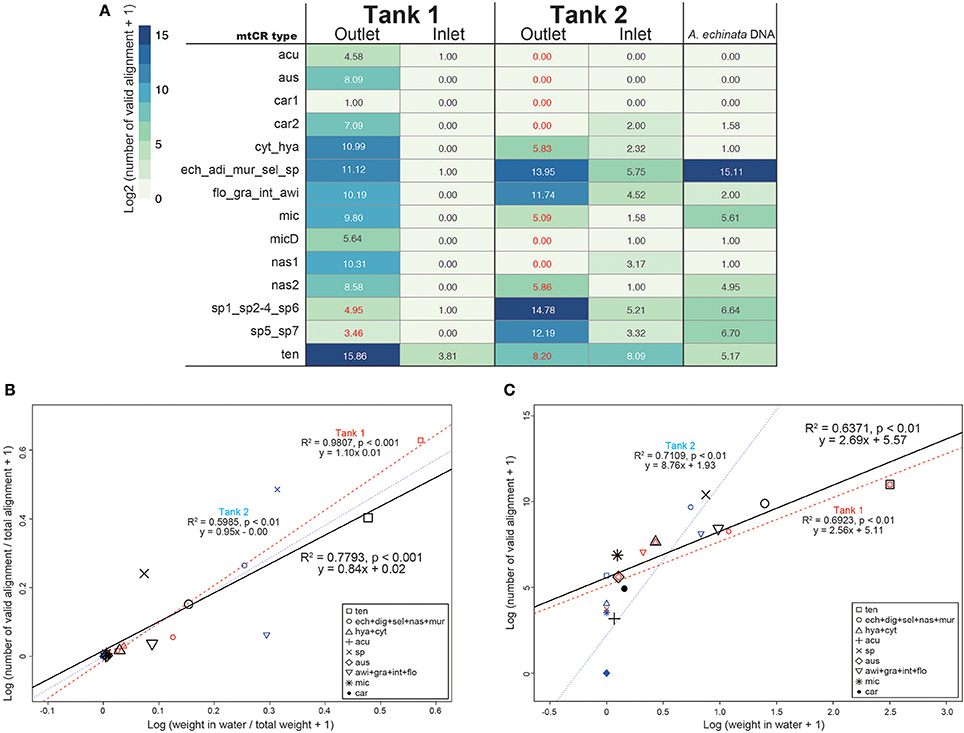
Figure 3. (A) The number of Acropora mtCR types detected from eDNA and Acropora echinata DNA as a positive control. Numbers of valid alignment are shown at log2 scale and colored. IDs of the 300 bp flanking the RNS2 primer sequence in the mtCR are identical are concatenated. Numbers of mtCR types that did not exist in either Tank 1 or Tank 2 outlet are shown in red. (B) Log-transformed proportions of coral masses of each mtCR types to the total weights of the all corals in seawater in kg (Table 1) and proportions of numbers of valid alignments from each mtCR type to the total numbers of alignments in both tanks, Tanks 1 and 2 shown in a scatter plot. (C) Log-transformed coral masses determined in seawater in kg (Table 1) and numbers of sequence reads identified from each mtCR type in both tanks, Tanks 1 and 2 shown in a scatter plot. Dots and regression lines of both tanks in the scatter plot are shown in black, red, and blue, respectively. mtCR types from the same species or different species sharing same mtCR sequences are clustered.
In the positive control of Symbiodinium CCMP2466 (clade C, type C1) PCR amplicon, 97.23% of valid aliments were correctly assigned to C1 types (C1 and C1ca_C1b_C1e, Figure 4), indicating that our bioinformatics filtering worked properly for Symbiodinium as well. Clade C, the major symbiotic Symbiodinium clade in the corals used (Figure 2), was detected in both tanks (Figure 4). The most abundant clade C type detected was C50 in Tank 1 and C3d_C21 in Tank 2, respectively, possibly reflecting abundant clade C types of the Acropora corals in each tank (Tank 1: C50, Tank 2: C3d_C21, Figure 2). We were not able to detect some clade C types that were identified from the corals; however, the total number of valid alignments of these types comprised only ~1% of total alignments (46314 out of 4535626) of symbiotic Symbiodinium in the Acropora corals (Figure 2, Supplementary Materials), indicating that these were minor types of clade C symbiont, which could not be detected in seawater. Although clade C types were exclusively detected from Acropora colonies, those in outlet water were low (Tank 1: 35.6%; 44178 out of 36748, Tank 2: 89.3%; 111107 out of 124375). We detected considerable number of clade D types in outlet water of both Tanks 1 and 2 (Figure 4). Within clade D, D1 was the most abundant in both tanks, possibly reflecting more D1 symbionts in the corals (Figure 2). We also detected other clade D and clade A types, which were not detected in the Acropora corals used in this study (Figure 2), from both outlet and inlet seawater. In outlet water, only small numbers of valid alignments originated from ITS-2 types that were not detected in Acropora corals (Figure 2 and Supplementary Materials, Tank 1: 1%; 1003 out of 124223, Tank 2: 3.2%; 4026 out of 124375), albeit in inlet water, large proportions of valid alignments originated from ITS-2 types that were not detected in the corals (Figure 2 and Supplementary Materials, Tank 1: 51.7%; 19005 out of 36748, Tank 2: 99.8%; 38821 out of 38918). In inlet water detected ITS-2 types were considerably different between Tanks 1 and 2 (Figure 4). For example, some abundant types in Tank 1 inlet water (A2-ISS-C2-Sy and D5) were not detected in Tank 2 inlet water, whereas A3 and F4.4b were abundant types in Tank 2 inlet water (Figure 4).
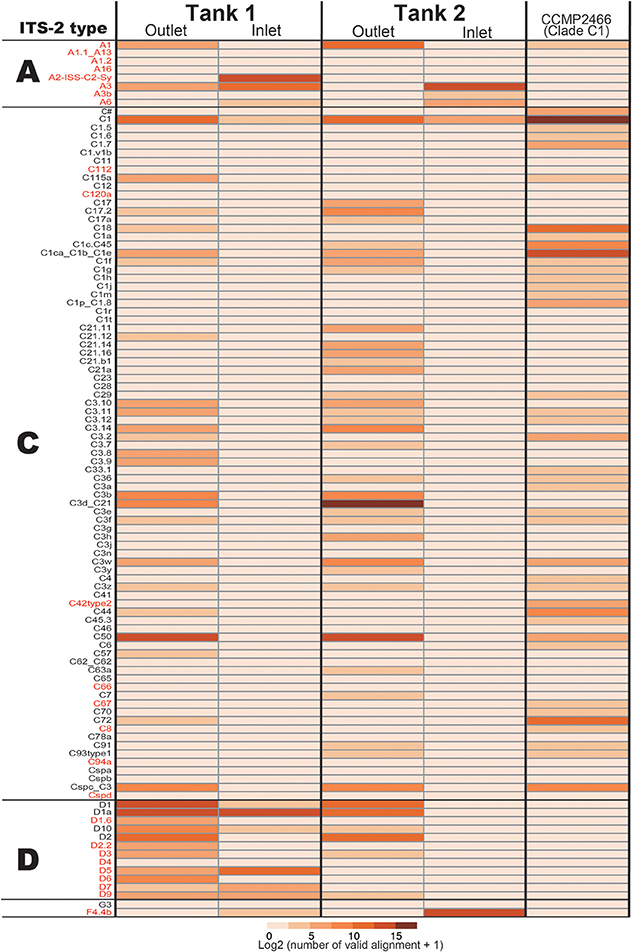
Figure 4. The number of Symbiodinium ITS-2 types detected by eDNA PCR amplicon sequencing and CCMP2466 as a positive control. ITS-2 types that share the same GeoSymbio names in the database were summed (raw numbers of valid alignments are available in Supplementary Materials). ITS-2 types belonging to the same clade (A, C, or D) are separated and ITS-2 types in red indicate that these were not detected in the Acropora colonies used in this study (Figure 2).
In this study, we succeeded in simultaneously detecting both Acropora corals and Symbiodinium from eDNA isolated from aquaria with running seawater. Considering that this was the first attempt to detect Acropora coral eDNA from seawater, these results suggest a possible new technique for estimation of coral cover and species composition by analyzing small amounts of seawater in the field. In addition, the simultaneous detection of eDNA of Symbiodinium suggested that this technique might be applicable to monitoring not only coral distributions, but also their symbionts at the same time.
Successful detection of Acropora colonies with masses ≥0.04 kg in this study suggests that this method might be able to detect an Acropora colony ≥1 kg/12,000 L in seawater (0.04 kg/480 L of seawater in Tank 1). This means that existence of at least one adult Acropora colony (~30 cm diameter = 1 kg) per m2 in water shallower than 10 m could be detected by eDNA in the field. However, the sensitivity of Acropora coral detection from eDNA could vary between species. Although there were almost identical masses for each of the three species in Tank 2 (A. echinata, Acropora sp., and A. awi), the number of valid alignments for A. awi was smaller than other two species (Figure 4, Table S5). Mucus production rates of corals vary greatly (Nakajima and Tanaka, 2014) and this might affect the sensitivity toward each Acropora species. Differences in the calculated average number of valid alignments per kg (Table S5) between the two tanks indicate that estimation of Acropora corals masses from eDNA might be possible, but accuracy depends on each Acropora species.
Acropora sequences were detected from eDNA isolated from inlet water (Figure 3A), suggesting that these might originate from neighboring coral reefs. Running seawater at Seikai National Fisheries Research Institute comes from Urasoko Bay, where Acropora corals once maintained 20–60% coverage (Suzuki et al., 2012a), but were seriously damaged due to an outbreak of crown-of-thorns starfish, Acanthaster planci, in 2012 (Suzuki et al., 2012b). Therefore, the amount of Acropora eDNA contained in flowing seawater could be small. Acropora corals observed around the marine station were A. selago, A. muricata, and A. nasuta and their coverage was less than 1% (Suzuki et al., 2012a). Although some of the minor reads mapping to the mtCR database from inlet pipes might have originated from minor contaminants or artifacts from other samples, all of the observed Acropora species naturally occurring near the research institute were detected from inlet pipes (Figure 3), suggesting that these reads possibly originated from outer reefs and this technology could detect small amounts of Acropora DNA in seawater. There are still some difficulties in applying eDNA techniques to monitoring coral reefs, e.g., lack of suitable DNA markers for corals and reference DNA databases. In this study, it was not clear that we could distinguish DNA sequences from all colonies due to the high conservation and slow evolution of mitochondrial DNA in anthozoan cnidarians, including scleractinian corals (Shearer et al., 2002). For example, mtCR sequences of A. grandis, A. intermedia, and A. awi are same (Table S2). Thus, additional markers for nuclear DNA will be needed to reliably identify Acropora species using eDNA, specifically markers that are more variable than mitochondrial DNA (Ladner and Palumbi, 2012; Suzuki et al., 2016), although nuclear DNA of marine organisms is usually present at low concentrations and is rapidly degraded in seawater (Rees et al., 2014). However, the success of using a nuclear DNA marker (e.g., internal transcribed spacer 1) to detect common carp from eDNA has recently been reported (Minamoto et al., 2016), implying that it might be possible to employ nuclear markers for identifying corals from seawater. An additional problem is that species that are not in the database cannot be detected. Species identification from eDNA depends upon completeness of the reference database and enhancement of a coral DNA database will be required for more precise coral species detection and identification.
In a previous study, detection of Symbiodinium DNA from seawater was complicated by the low primer specificity (Pochon et al., 2010). In this study, we used r18Sf and Sym28Sr primers (Yamashita and Koike, 2013) that were used for amplifying Symbiodinium DNA from environmental samples (Yamashita et al., 2013, 2014). This primer set could be used not only for analyzing Symbiodinium within host animals, but also for detecting free-living state Symbiodinium in coral reef environments (Yamashita and Koike, 2013; Yamashita et al., 2013, 2014). As hoped, major types of Symbiodinium symbionts in corals used in this study were detected from eDNA in outlet water. Most valid alignments in outlet water were assigned to ITS-2 types detected from Acropora corals (Figures 2, 4, Supplementary Materials, Tank 1: 99%, Tank 2: 96.7%), suggesting that it would be possible to detect most coral symbiont types from seawater, if Acropora corals exist nearby. However, eDNA abundance did not correlate well with frequencies of the major Symbiodinium types of the corals used in this study. For example, clade C types were exclusively detected from Acropora colonies, but not from outlet water of both Tanks 1 and 2, indicating that quantification of Symbiodinium types is presently difficult. It might be due to a failure to detect minor symbiotic Symbiodinium types in Acropora corals (Figure 2) or to significant copy number differences of the ITS-2 gene among Symbiodinium types. Indeed, it is reported that there are thousands of copies of ITS regions in Symbiodinium genomes and copy numbers of ITS1 in clade D are ~3x higher than in clade C (Mieog et al., 2007). These might be major factors accounting for the inconsistency between sequence reads and major Symbiodinium types.
The primer set r18Sf and Sym28Sr can detect even relatively rare Symbiodinium clades in coral reef environments (Yamashita and Koike, 2013; Yamashita et al., 2013, 2014). Thus, the primer set was powerful enough to amplify the ITS-2 gene from low concentrations of Symbiodinium cells. In our experiments, clade A was detected in both inlets and clade D types that were not major coral symbionts in the coral used in this study (Figure 2) were identified in the Tank 1 inlet. Likewise, clade F was detected in the Tank 2 inlet (Figure 4). Clade F is a common foraminiferan symbiont in the tropical and subtropical Indo-Pacific and Caribbean Oceans (Pochon and Pawlowski, 2006), but was previously detected in corals (Lien et al., 2012). The detection of different Symbiodinium types in inlet water for the two tanks probably stems from different cell types between two inlets and the great sensitivity of the technique. Chance events involving the distribution of individual Symbiodinium cells to the two tanks were potentially able to bias the results. Therefore, it should be taken into account that both free-living state Symbiodinium, as well as coral symbionts are thoroughly mixed in the water column and regular, continuous observation of symbiotic Symbiodinium in corals under different environmental conditions would be necessary for assessing diverse coral reef environments.
In this study, we demonstrate that detection of nearby coral species and their symbiotic algae is feasible in coral reef environments using eDNA isolated from seawater. With high-quality molecular markers and an extensive database, we could perform more precise, detailed monitoring of coral reef ecosystems, including corals, symbionts, and other organisms. Coral damage and recovery after bleaching or severe storms varies even at small spatial scales, depending on reef environmental conditions, such as reef topology and seawater temperature. It is expected that high-frequency, high-density monitoring will enable us to establish effective conservation strategies. We hope that eDNA technology may become a powerful tool for conservation management of coral reefs in the future.
CS, HY, and GS: conceptualization and commencement of the study. YZ and GS: Animal collection and Acropora species identification. HY and GS: Coral culturing, environmental DNA isolation, and PCR amplification. MiK and MaK: Preparation of sequencing libraries and producing sequencing data. NS provide sequencing reagents and facilities. CS analyzed the data and wrote the main manuscript text, and all authors edited and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported by JSPS KAKENHI grants (17KT0027, 17K07949 and 26290065 for CS, 17K15179 for YZ, 26291094 for HY, 15H04538 for GS). We thank Dr. Steven D. Aird (OIST) for editing the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00028/full#supplementary-material
Supplementary Materials: Five files and directories (Supplementary_Fig-Table.pdf, Perl_scripts, Database, Commands.doc and Sym_num-valid-alignments.xlsx) are included.
Explanation of files:
Supplementary_Fig-Table.pdf: Supplementary Figures and Tables shown in PDF format.
Commands.doc: bioinformatics commands that we used in this study.
Sym_num-valid-alignments.xlsx: Actual numbers of valid alignments used in Figures 2, 4.
Perl_scripts: custom perl scripts used in this study.
Database: Fasta files used for BLASTN databases (Acropora: RNS2_acropora.fa, Symbiodinium: sym-ITS-2_NCBI-GeoSym.fa) in this study.
Figure S1. Electrophoresis of PCR amplicons from eDNA. These were used for library preparation (total amounts of DNA used are shown) and sequenced with a MiSeq (300-bp paired-end libraries). A 100-bp DNA Ladder (TaKaRa, code 3422) was used to determine DNA marker sizes. Amplicon sizes of positive controls (Acropora ehinata and cultured clade C Symbiodinium CCMP2466) are 935 and 676 bp, respectively.
Figure S2. Rate of identical nucleotides in 60-bp sliding window of aligned mitochondrial genomes of Acropora corals used in this study. mtCR and RNS2 and GR primer flanking sequences are shown.
Table S1. Shotgun sequencing data (HiSeq2500, 250-bp paired-end libraries) from coral colonies used in this study. Abbreviations are same as Table 1.
Table S2. Percent identity of mtCR sequences to the most similar sequence within the database used in this study. Abbreviations are same as Table 1.
Table S3. Percent identities of 300 bp flanking the RNS2 primer to the most similar mtCR sequence in the corals used in this study.
Table S4. Percent identities of 300 bp flanking the GR primer to the most similar mtCR sequence in the corals used in this study.
Table S5. Weight in water and number of valid alignments of Acropora species used in this study.
Alvarez-Filip, L., Carricart-Ganivet, J. P., Horta-Puga, G., and Iglesias-Prieto, R. (2013). Shifts in coral-assemblage composition do not ensure persistence of reef functionality. Sci. Rep. 3:3486. doi: 10.1038/srep03486
Aranda, M., Li, Y., Liew, Y. J., Baumgarten, S., Simakov, O., Wilson, M. C., et al. (2016). Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6:39734. doi: 10.1038/srep39734
Bohmann, K., Evans, A., Gilbert, M. T., Carvalho, G. R., Creer, S., Knapp, M., et al. (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 29, 358–367. doi: 10.1016/j.tree.2014.04.003
Bongaerts, P., Riginos, C., Brunner, R., Englebert, N., Smith, S. R., and Hoegh-Guldberg, O. (2017). Deep reefs are not universal refuges: reseeding potential varies among coral species. Sci. Adv. 3:e1602373. doi: 10.1126/sciadv.1602373
Bourne, D. G., Garren, M., Work, T. M., Rosenberg, E., Smith, G. W., and Harvell, C. D. (2009). Microbial disease and the coral holobiont. Trends Microbiol. 17, 554–562. doi: 10.1016/j.tim.2009.09.004
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Combosch, D. J., and Vollmer, S. V. (2015). Trans-Pacific RAD-Seq population genomics confirms introgressive hybridization in Eastern Pacific Pocillopora corals. Mol. Phylogenet. Evol. 88, 154–162. doi: 10.1016/j.ympev.2015.03.022
Cox, M. P., Peterson, D. A., and Biggs, P. J. (2010). SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485. doi: 10.1186/1471-2105-11-485
De'ath, G., Fabricius, K. E., Sweatman, H., and Puotinen, M. (2012). The 27-year decline of coral cover on the Great Barrier Reef and its causes. Proc. Natl. Acad. Sci. U.S.A. 109, 17995–17999. doi: 10.1073/pnas.1208909109
Fisher, P. L., Malme, M. K., and Dove, S. (2012). The effect of temperature stress on coral-Symbiodinium associations containing distinct symbiont types. Coral Reefs 31, 473–485. doi: 10.1007/s00338-011-0853-0
Franklin, E. C., Stat, M., Pochon, X., Putnam, H. M., and Gates, R. D. (2012). GeoSymbio: a hybrid, cloud-based web application of global geospatial bioinformatics and ecoinformatics for Symbiodinium-host symbioses. Mol. Ecol. Resour. 12, 369–373. doi: 10.1111/j.1755-0998.2011.03081.x
Fukami, H., Omori, M., and Hatta, M. (2000). Phylogenetic relationships in the coral family Acroporidae, reassessed by inference from mitochondrial genes. Zool. Sci. 17, 689–696. doi: 10.2108/zsj.17.689
Hawkins, T. D., and Davy, S. K. (2012). Nitric oxide production and tolerance differ among Symbiodinium types exposed to heat stress. Plant Cell Physiol. 53, 1889–1898. doi: 10.1093/pcp/pcs127
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Jones, A. M., Berkelmans, R., Van Oppen, M. J., Mieog, J. C., and Sinclair, W. (2008). A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. Proc. Biol. Sci. 275, 1359–1365. doi: 10.1098/rspb.2008.0069
Kelly, R. P., Port, J. A., Yamahara, K. M., Martone, R. G., Lowell, N., Thomsen, P. F., et al. (2014). Environmental monitoring. Harnessing DNA to improve environmental management. Science 344, 1455–1456. doi: 10.1126/science.1251156
Keshavmurthy, S., Hsu, C. M., Kuo, C. Y., Meng, P. J., Wang, J. T., and Chen, C. A. (2012). Symbiont communities and host genetic structure of the brain coral Platygyra verweyi, at the outlet of a nuclear power plant and adjacent areas. Mol. Ecol. 21, 4393–4407. doi: 10.1111/j.1365-294X.2012.05704.x
Keshavmurthy, S., Meng, P. J., Wang, J. T., Kuo, C. Y., Yang, S. Y., Hsu, C. M., et al. (2014). Can resistant coral-Symbiodinium associations enable coral communities to survive climate change? A study of a site exposed to long-term hot water input. PeerJ 2:e327. doi: 10.7717/peerj.327
Knowlton, N., Brainard, R. E., Fisher, R., Moews, M., Plaisance, L., and Caley, M. J. (2010). “Coral reef biodiversity,” in Life in the World's Oceans: Diversity, Distribution, and Abundance, ed A. D. McIntyre (Chichester: Wiley-Blackwell), 65–79.
Koike, K., Yamashita, H., Oh-Uchi, A., Tamaki, M., and Hayashibara, T. (2007). A quantitative real-time PCR method for monitoring Symbiodinium in the water column. J. Jpn. Coral Reef Soc. 9, 1–12. doi: 10.3755/jcrs.9.1
Ladner, J. T., and Palumbi, S. R. (2012). Extensive sympatry, cryptic diversity and introgression throughout the geographic distribution of two coral species complexes. Mol. Ecol. 21, 2224–2238. doi: 10.1111/j.1365-294X.2012.05528.x
Lajeunesse, T. C. (2001). Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the its region: In search of a “species” level marker. J. Phycol. 37, 866–880. doi: 10.1046/j.1529-8817.2001.01031.x
Lajeunesse, T. C. (2002). Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Mar. Biol. 141, 387–400. doi: 10.1007/s00227-002-0829-2
Lajeunesse, T. C. (2005). “Species” radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene transition. Mol. Biol. Evol. 22, 570–581. doi: 10.1093/molbev/msi042
Lajeunesse, T. C., Smith, R., Walther, M., Pinzón, J., Pettay, D. T., Mcginley, M., et al. (2010). Host-symbiont recombination versus natural selection in the response of coral-dinoflagellate symbioses to environmental disturbance. Proc. Biol. Sci. 277, 2925–2934. doi: 10.1098/rspb.2010.0385
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Lien, Y. T., Fukami, H., and Yamashita, Y. (2012). Symbiodinium clade C dominates zooxanthellate corals (Scleractinia) in the temperate region of Japan. Zool. Sci. 29, 173–180. doi: 10.2108/zsj.29.173
Lin, S., Cheng, S., Song, B., Zhong, X., Lin, X., Li, W., et al. (2015). The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350, 691–694. doi: 10.1126/science.aad0408
Loya, Y., Sakai, K., Yamazato, K., Nakano, Y., Sambali, H., and Van Woesik, R. (2001). Coral bleaching: the winners and the losers. Ecol. Lett. 4, 122–131. doi: 10.1046/j.1461-0248.2001.00203.x
Mieog, J. C., Van Oppen, M. J. H., Cantin, N. E., Stam, W. T., and Olsen, J. L. (2007). Real-time PCR reveals a high incidence of Symbiodinium clade D at low levels in four scleractinian corals across the Great Barrier Reef: implications for symbiont shuffling. Coral Reefs 26, 449–457. doi: 10.1007/s00338-007-0244-8
Minamoto, T., Uchii, K., Takahara, T., Kitayoshi, T., Tsuji, S., Yamanaka, H., et al. (2016). Nuclear internal transcribed spacer-1 as a sensitive genetic marker for environmental DNA studies in common CARP Cyprinus carpio. Mol. Ecol. Resour. 17, 324–333. doi: 10.1111/1755-0998.12586
Miya, M., Sato, Y., Fukunaga, T., Sado, T., Poulsen, J. Y., Sato, K., et al. (2015). MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2:150088. doi: 10.1098/rsos.150088
Nakajima, R., and Tanaka, Y. (2014). The role of coral mucus in the material cycle in reef ecosystems: biogeochemical and ecological perspectives. J. Jpn. Coral Reef Soc. 16, 3–27. doi: 10.3755/jcrs.16.3
Peng, Y., Leung, H. C., Yiu, S. M., and Chin, F. Y. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428. doi: 10.1093/bioinformatics/bts174
Pochon, X., Garcia-Cuetos, L., Baker, A. C., Castella, E., and Pawlowski, J. (2007). One-year survey of a single Micronesian reef reveals extraordinarily rich diversity of Symbiodinium types in soritid foraminifera. Coral Reefs 26, 867–882. doi: 10.1007/s00338-007-0279-x
Pochon, X., and Gates, R. D. (2010). A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai'i. Mol. Phylogenet. Evol. 56, 492–497. doi: 10.1016/j.ympev.2010.03.040
Pochon, X., and Pawlowski, J. (2006). Evolution of the soritids-Symbiodinium symbiosis. Symbiosis 42, 77–88. Available online at: http://www.cawthron.org.nz/media_new/publications/pdf/2017_01/PochonPawlowski_Symbiosis2006.pdf
Pochon, X., Stat, M., Takabayashi, M., Chasqui, L., Chauka, L. J., Logan, D. D. K., et al. (2010). Comparison of endosymbiotic and free-living Symbiodinium (Dinophyceae) diversity in a hawaiian reef environment. J. Phycol. 46, 53–65. doi: 10.1111/j.1529-8817.2009.00797.x
Rees, H. C., Gough, K. C., Middleditch, D. J., Patmore, J. R. M., and Maddison, B. C. (2015). Applications and limitations of measuring environmental DNA as indicators of the presence of aquatic animals. J. Appl. Ecol. 52, 827–831. doi: 10.1111/1365-2664.12467
Rees, H. C., Maddison, B. C., Middleditch, D. J., Patmore, J. R. M., and Gough, K. C. (2014). REVIEW The detection of aquatic animal species using environmental DNA - a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 51, 1450–1459. doi: 10.1111/1365-2664.12306
Rowan, R., and Powers, D. A. (1991). A molecular genetic classification of zooxanthellae and the evolution of animal-algal symbioses. Science 251, 1348–1351. doi: 10.1126/science.251.4999.1348
Sampayo, E. M., Ridgway, T., Bongaerts, P., and Hoegh-Guldberg, O. (2008). Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. Proc. Natl. Acad. Sci. U.S.A. 105, 10444–10449. doi: 10.1073/pnas.0708049105
Santos, S. R., Taylor, D. J., Kinzie, R. A. III., Hidaka, M., Sakai, K., and Coffroth, M. A. (2002). Molecular phylogeny of symbiotic dinoflagellates inferred from partial chloroplast large subunit (23S)-rDNA sequences. Mol. Phylogenet. Evol. 23, 97–111. doi: 10.1016/S1055-7903(02)00010-6
Shearer, T. L., Van Oppen, M. J., Romano, S. L., and Wörheide, G. (2002). Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Mol. Ecol. 11, 2475–2487. doi: 10.1046/j.1365-294X.2002.01652.x
Shinzato, C., Inoue, M., and Kusakabe, M. (2014a). A snapshot of a coral “holobiont”: a transcriptome assembly of the scleractinian coral, Porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS ONE 9:e85182. doi: 10.1371/journal.pone.0085182
Shinzato, C., Mungpakdee, S., Arakaki, N., and Satoh, N. (2015). Genome-wide SNP analysis explains coral diversity and recovery in the Ryukyu Archipelago. Sci. Rep. 5:18211. doi: 10.1038/srep18211
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. doi: 10.1038/nature10249
Shinzato, C., Yasuoka, Y., Mungpakdee, S., Arakaki, N., Fujie, M., Nakajima, Y., et al. (2014b). Development of novel, cross-species microsatellite markers for Acropora corals using next-generation sequencing technology. Front. Mar. Sci. 1:11. doi: 10.3389/fmars.2014.00011
Shoguchi, E., Shinzato, C., Kawashima, T., Gyoja, F., Mungpakdee, S., Koyanagi, R., et al. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399-1408. doi: 10.1016/j.cub.2013.05.062
Stat, M., Bird, C. E., Pochon, X., Chasqui, L., Chauka, L. J., Concepcion, G. T., et al. (2011). Variation in Symbiodinium ITS2 sequence assemblages among coral colonies. PLoS ONE 6:e15854. doi: 10.1371/journal.pone.0015854
Stat, M., Morris, E., and Gates, R. D. (2008). Functional diversity in coral-dinoflagellate symbiosis. Proc. Natl. Acad. Sci. U.S.A. 105, 9256–9261. doi: 10.1073/pnas.0801328105
Suzuki, G., Arakaki, S., Kai, S., and Hayashibara, T. (2012a). Habitat differentiation in the early life stages of simultaneously mass-spawning corals. Coral Reefs 31, 535–545. doi: 10.1007/s00338-011-0865-9
Suzuki, G., Hayashibara, T., Shirayama, Y., and Fukami, H. (2008). Evidence of species-specific habitat selectivity of Acropora corals based on identification of new recruits by two molecular markers. Mar. Ecol. Prog. Ser. 355, 149–159. doi: 10.3354/meps07253
Suzuki, G., Kai, S., and Yamashita, H. (2012b). Mass stranding of crown-of-thorns starfish. Coral Reefs 31, 821–821. doi: 10.1007/s00338-012-0906-z
Suzuki, G., Keshavmurthy, S., Hayashibara, T., Wallace, C. C., Shirayama, Y., Chen, C. A., et al. (2016). Genetic evidence of peripheral isolation and low diversity in marginal populations of the Acropora hyacinthus complex. Coral Reefs 35, 1419–1432. doi: 10.1007/s00338-016-1484-2
Takabayashi, M., Santos, S. R., and Cook, C. B. (2004). Mitochondrial DNA phylogeny of the symbiotic dinoflagellates (Symbiodinium, Dinophyta). J. Phycol. 40, 160–164. doi: 10.1111/j.0022-3646.2003.03-097.x
Tchernov, D., Gorbunov, M. Y., De Vargas, C., Narayan Yadav, S., Milligan, A. J., Häggblom, M., et al. (2004). Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals. Proc. Natl. Acad. Sci. U.S.A. 101, 13531–13535. doi: 10.1073/pnas.0402907101
Tonk, L., Bongaerts, P., Sampayo, E. M., and Hoegh-Guldberg, O. (2013). SymbioGBR: a web-based database of Symbiodinium associated with cnidarian hosts on the Great Barrier Reef. BMC Ecol. 13:7. doi: 10.1186/1472-6785-13-7
van Oppen, M. J., Catmull, J., Mcdonald, B. J., Hislop, N. R., Hagerman, P. J., and Miller, D. J. (2002). The mitochondrial genome of Acropora tenuis (Cnidaria; Scleractinia) contains a large group I intron and a candidate control region. J. Mol. Evol. 55, 1–13. doi: 10.1007/s00239-001-0075-0
Wallace, C. (1999). Staghorn Corals of the World: A Revision of the Genus Acropora. Collingwood, VIC: CSIRO PUBLISHING
Wilcox, T. P. (1998). Large-subunit ribosomal RNA systematics of symbiotic dinoflagellates: morphology does not recapitulate phylogeny. Mol. Phylogenet. Evol. 10, 436–448. doi: 10.1006/mpev.1998.0546
Wild, C., Huettel, M., Klueter, A., Kremb, S. G., Rasheed, M. Y., and Jørgensen, B. B. (2004). Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428, 66–70. doi: 10.1038/nature02344
Yamamoto, S., Masuda, R., Sato, Y., Sado, T., Araki, H., Kondoh, M., et al. (2017). Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci. Rep. 7:40368. doi: 10.1038/srep40368
Yamashita, H., and Koike, K. (2013). Genetic identity of free-living Symbiodinium obtained over a broad latitudinal range in the Japanese coast. Phycol. Res. 61, 68–80. doi: 10.1111/pre.12004
Yamashita, H., Suzuki, G., Hayashibara, T., and Koike, K. (2011). Do corals select zooxanthellae by alternative discharge? Mar. Biol. 158, 87–100. doi: 10.1007/s00227-010-1544-z
Yamashita, H., Suzuki, G., Hayashibara, T., and Koike, K. (2013). Acropora recruits harbor “rare” Symbiodinium in the environmental pool. Coral Reefs 32, 355–366. doi: 10.1007/s00338-012-0980-2
Yamashita, H., Suzuki, G., Kai, S., Hayashibara, T., and Koike, K. (2014). Establishment of coral-algal symbiosis requires attraction and selection. PLoS ONE 9:e97003. doi: 10.1371/journal.pone.0097003
Keywords: scleractinian coral, Acropora, Symbiodinium, environmental DNA, illumina next-generation sequencing
Citation: Shinzato C, Zayasu Y, Kanda M, Kawamitsu M, Satoh N, Yamashita H and Suzuki G (2018) Using Seawater to Document Coral-Zoothanthella Diversity: A New Approach to Coral Reef Monitoring Using Environmental DNA. Front. Mar. Sci. 5:28. doi: 10.3389/fmars.2018.00028
Received: 09 September 2017; Accepted: 19 January 2018;
Published: 20 February 2018.
Edited by:
Iliana B. Baums, Pennsylvania State University, United StatesReviewed by:
Mikhail V. Matz, University of Texas at Austin, United StatesCopyright © 2018 Shinzato, Zayasu, Kanda, Kawamitsu, Satoh, Yamashita and Suzuki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuya Shinzato, Yy5zaGluemF0b0Bhb3JpLnUtdG9reW8uYWMuanA=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.