
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 08 September 2017
Sec. Marine Biotechnology and Bioproducts
Volume 4 - 2017 | https://doi.org/10.3389/fmars.2017.00287
 Erik Borchert1
Erik Borchert1 Joseph Selvin2
Joseph Selvin2 Seghal G. Kiran3
Seghal G. Kiran3 Stephen A. Jackson1
Stephen A. Jackson1 Fergal O'Gara1,4,5
Fergal O'Gara1,4,5 Alan D. W. Dobson1,6*
Alan D. W. Dobson1,6*Esterases catalyze the hydrolysis of ester bonds in fatty acid esters with short-chain acyl groups. Due to the widespread applications of lipolytic enzymes in various industrial applications, there continues to be an interest in novel esterases with unique properties. Marine ecosystems have long been acknowledged as a significant reservoir of microbial biodiversity and in particular of bacterial enzymes with desirable characteristics for industrial use, such as for example cold adaptation and activity in the alkaline pH range. We employed a functional metagenomic approach to exploit the enzymatic potential of one particular marine ecosystem, namely the microbiome of the deep sea sponge Stelletta normani. Screening of a metagenomics library from this sponge resulted in the identification of a number of lipolytic active clones. One of these encoded a highly, cold-active esterase 7N9, and the recombinant esterase was subsequently heterologously expressed in Escherichia coli. The esterase was classified as a type IV lipolytic enzyme, belonging to the GDSAG subfamily of hormone sensitive lipases. Furthermore, the recombinant 7N9 esterase was biochemically characterized and was found to be most active at alkaline pH (8.0) and displays salt tolerance over a wide range of concentrations. In silico docking studies confirmed the enzyme's activity toward short-chain fatty acids while also highlighting the specificity toward certain inhibitors. Furthermore, structural differences to a closely related mesophilic E40 esterase isolated from a marine sediment metagenomics library are discussed.
Metagenomic based approaches have proven extremely useful as a means of discovering microbial enzymes with entirely new biochemical properties, thereby exploiting the microbial diversity of a variety of different environmental ecosystems (Kennedy et al., 2011). These approaches are typically employed to help overcome the problem of cultivating bacteria from different environments, where typically only 0.001 to 1% of bacterial isolates can be recovered and grown under standard laboratory conditions (Bernard et al., 2000). Bioprospecting for novel enzymes with interesting biotechnological applications using metagenomics based approaches particularly from extreme environments such as acidic, cold, hot, and hypersaline environments has proven to be particularly successful (Mirete et al., 2016). Nevertheless, it is clear that relative to the number of metagenomic sampling sites that have been reported to date that up until now we have largely under sampled many of these sites with respect to enzyme discovery (Ferrer et al., 2016). Thus, a large part of the microbial biodiversity present in the earth's biosphere has yet to be explored or exploited for novel enzymes (Alcaide et al., 2015b).
The impetus to explore novel environments for new industrial enzymes comes from the need to meet the ongoing global demand for these enzymes which in 2014 was estimated to have a value of around $4.2 billion, and which is expected to reach nearly $6.2 billion by 2020 (Singh et al., 2016). The deep oceans are one part of the earth's biosphere which have to date received little attention. With mean depths of 3,800 m and 50% of the oceans being deeper than 3000 m, the “deep sea” constitutes not only a potential large resource from a microbial biodiversity perspective, but also a very unique environment; with temperatures ranging from 2 to 3°C and a salinity of about 3.5% together with hundreds of bars of hydrostatic pressure (Wirsen and Molyneaux, 1999). Thus, microbial communities which have adapted to these extremes of temperature, salinity, pressure, and low levels of light are likely to possess novel biochemistry; and have enzymes that may be uniquely suited to many industrial processes (Alcaide et al., 2015a). In addition seawater samples are an extremely rich source of potential biocatalytic biodiversity when one considers that with bacteria capable of achieving densities of up to 106 per milliliter of seawater (Azam, 1998), and assuming that there are ~3,000 genes in a single genome and that 40% of these genes have catalytic activity then there may be as many as 3 × 109 genes mediating up to 1·2 × 109 putative reactions in a milliliter of seawater (Dinsdale et al., 2008; Vieites et al., 2009). Thus, although the deep sea is likely to be a rich source of microbial biocatalytic biodiversity, very few studies have to date attempted to access or exploit this biodiversity; most likely due to both the technical difficulties and costs associated with sampling at lower depths.
Lipolytic enzymes can be classified into eight different families and numerous subfamilies (Arpigny and Jaeger, 1999). The overall three-dimensional structure of all lipases and esterases is defined by a characteristic α/β-hydrolase fold (Ollis et al., 1992), with “true lipases,” members of family I; also having a characteristic lid and possessing characteristic interfacial activation properties (Arpigny and Jaeger, 1999). Furthermore, lipolytic enzymes can be categorized as either lipases (triacylglycerol hydrolases, EC 3.1.1.3) or esterases (EC 3.1.1.1) corresponding to their specific hydrolytic activity, where lipases hydrolyze long-chain acyl groups to fatty acids and acylglycerols and esterases hydrolyze ester bonds of fatty acid esters with short-chain acyl groups (Verger, 1997). Structural analyses has revealed that microbial esterases typically consist of two domains, a CAP domain and a catalytic domain. The CAP domain consists of two N-terminal alpha helices and participates in substrate binding (Mandrich et al., 2005). The catalytic domain contains a functional serine at the active site, which is usually located within the G-X-S-X-G consensus sequence. This serine, along with an acidic residue such as aspartate or glutamate and a histidine, to which the acidic residue is hydrogen-bonded, form a highly conserved catalytic triad (Bornscheuer, 2002). Esterases typically exhibit broad pH, temperature and substrate spectra, together with tolerance to metal ions, solvents and salts to varying extents. This coupled with their region-, chemo-, and stero-selectivity make them an important group of industrial biocatalysts (Singh and Mukhopadhyay, 2012).
The industrial applications of lipolytic enzymes are wide ranging and include applications in the detergent industry, biodiesel production, food industry, pulp and paper industry, fats and oils production via transesterification, as well as environmental applications for the degradation of lipid wastes (Panda and Gowrishankar, 2005; Jegannathan and Nielsen, 2013; Sharma and Kanwar, 2014; Sasso et al., 2016; Ramnath et al., 2017; Rao et al., 2017). Lipolytic enzymes from Burkholderia are for example employed in biodiesel production, as they can be used for transesterification of waste oils with short chain alcohols in the presence of high levels of methanol (Sasso et al., 2016). Furthermore, lipolytic enzymes can be used for bioremediation of environmental hazards such as oil spills, which is important in the context of exploitation for new and remote sources of oils, especially in cold environments (Yang et al., 2009).
We have previously studied the microbial biodiversity of a number of deep sea sponges sampled at depths between 760 and 2,900 m below sea level, and the sponge species Stelletta normani in particular (Jackson et al., 2013). S. normani appears to possess a very diverse microbial community, comparable to high microbial abundance sponges from shallow water habitats (Jackson et al., 2013; Kennedy et al., 2014). Furthermore, the microbial community structures of deep sea sponges appear to possess a huge potential secondary metabolite biodiversity (Borchert et al., 2016). With this in mind we set out to assess the biocatalytic potential of the metagenome of the deep sea sponge S. normani using a functional metagenomic based approach. The S. normani metagenomic fosmid library was found to express a large number of lipolytic activities, from which we subsequently characterized a cold-active esterase from the hormone sensitive lipase family IV. Cold-active enzymes possess unique biochemical properties that are of particular interest for industrial biocatalysis. These include low substrate affinity, thermolability, and high specific activity at low temperatures, which can together help achieve saving in energy costs and in reducing undesirable chemical side reactions; as well as allowing rapid thermal inactivation (Cavicchioli et al., 2002; Santiago et al., 2016). Other “cold-active” lipolytic enzymes from family IV have previously been described, but these usually possess higher optimal temperatures (35–50°C; Hårdeman and Sjöling, 2007; Fu et al., 2011), whereas our esterase has a high activity at 4–40°C, identifying it as truly “cold-active.” In addition this work also broadens the description of members of the lipolytic enzyme family IV, as thermophilic and mesophilic enzymes of this family have to date been already described (Rhee et al., 2005).
The sponge S. normani was sampled in Irish territorial waters off the west coast of Ireland (Latitude 53.9861, Longitude −12.6100) at a depth of 760 m with the help of the remotely operated vehicle (ROV) Holland I abroad the R.V. Celtic Explorer during a Biodiscovery cruise in 2013. The sponge sample was rinsed with sterile artificial seawater [3.33% (w/v) Instant Ocean, Aquarium Systems] and stored at −80°C until further processing.
Total metagenomic DNA was extracted as described in Kennedy et al. (2008). In brief, the sponge tissue was ground under liquid nitrogen using a sterile pestle and mortar. The obtained sample was suspended in lysis buffer [100 mM Tris, 100 mM EDTA, 1.5 M NaCl (w/v), 1% CTAB (w/v), 2% SDS (w/v)] in a 1:5 ratio and then incubated for 2 h at 70°C. This solution was then centrifuged until a clear solution was obtained, which was subsequently used to precipitate the dissolved metagenomic DNA with 0.7 volumes of isopropanol for 30 min at room temperature. The precipitation mixture was centrifuged at 6,000 × g for 30 min, followed by a washing step with 70% ethanol and the obtained DNA pellet was air dried, before resuspending in a suitable amount of Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 8.0).
The metagenomic DNA was size-separated using pulse field-gel-electrophoresis and the size fraction of ~40 kb was cloned into the fosmid Copy Control pCCERI (derivative of pCC1FOS™) vector for metagenomic library construction. The insert harboring fosmids were packed into lambda phages and used to transfect E. coli TransforMax™ EPI300™ cells. In total a library of ~14,000 clones was generated from the metagenomic DNA and this library was plated onto agar Q-Tray plates containing 1% tributyrin. Positive clones were selected for sequencing on the PacBio platform (Beckman Coulter Genomics).
The lipase harboring fosmid was sequenced on the PacBio platform, by Beckman Coulter Genomics; and was assembled manually from the quality filtered and preassembled reads according to overlapping regions. The assembled fosmid was annotated using the BASys online pipeline (Van Domselaar et al., 2005) and all gene annotations were confirmed by BLAST searches (BLASTX) against the NCBI non redundant protein sequence database. The sequence of the esterase 7N9 has been submitted to the NCBI database and the accession is MF521837.
The full length 7N9 esterase gene was amplified using primers incorporating enzyme restriction sites allowing in-frame cloning into the pBAD/mycHIS-A overexpression vector (Invitrogen). Forward primer f7N9 (TATATACCATGGCTAGTCCTGAGCTCGATACGG) incorporates an NcoI restriction site (underlined) at the start codon (italics) of the gene. Reverse primer r7N9 (ATATATAAGCTTGCCAGTGTGCTTTTTAATGAACTCC) incorporates a HindIII restriction site replacing the stop codon of the esterase gene. The amplified PCR product and pBAD/mycHis-A were digested with NcoI and HindIII and subsequently an overnight ligation was carried out at 4°C at a 10:1 ratio of insert to plasmid. Two microliters of the ligation reaction were added to 50 μl TOP10 chemically competent cells (ThermoScientific) and the transformation was carried out according to the manufacturer's guidelines. The transformation mixture was plated in different amounts on LB plates containing 50 μg/ml ampicillin, 0.02% arabinose, and 1% tributyrin.
Pre inoculum was prepared by inoculating a loop full of culture (E. coli TOP10 bearing the pBAD/mycHIS-A vector with esterase insert) in 3 ml of LB broth supplemented with 50 μg/ml ampicillin and incubated at 37°C with constant shaking at 250 rpm on a shaking incubator overnight. Next day 10 ml of LB broth was inoculated with 100 μl of the pre inoculum and 50 μg/ml of ampicillin following incubation at 37°C with constant shaking at 250 rpm on a shaking incubator until it reached the mid log phase of growth (0.6 OD at 600 nm). Then the culture was induced with 0.02% arabinose and was further incubated overnight under the same conditions. Protein expression was confirmed by performing SDS-PAGE.
The esterase enzyme was purified from the overnight culture using the Ni-NTA spin column (Qiagen). Enzyme purification steps were followed as described in the Ni-NTA spin kit hand book (Under Native condition). Five milliliters of overnight culture was used and centrifuged at 4,000 × g for 15 min at 4°C. The pellet was re suspended in 630 μl of lysis buffer (NPI-10) (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0) and 70 μl of lysozyme (10 mg/ml) was added and kept on ice for 30 min. After this the lysate was centrifuged at 12,000 × g for 30 min at 4°C and the supernatant was collected. The Ni-NTA column was equilibrated with 600 μl of NPI-10 buffer, centrifuged at 890 × g for 2 min at 4°C. Six hundred microliters of the supernatant from the previous step was loaded onto the pre-equilibrated Ni-NTA spin column and then centrifuged at 270 × g for 5 min at 4°C, the flow through was collected. The column was washed twice with 600 μl of NPI-20 (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) buffer by centrifuging the column at 270 × g for 2 min at 4°C. Protein was eluted in two fractions by adding 300 μl of NPI-500 (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole, pH 8.0) buffer twice to the column and centrifuged at 890 × g for 2 min at 4°C. The eluted fractions were then checked on SDS-PAGE.
The effect of pH on esterase activity was evaluated at different pH values ranging from pH5 to pH10. The buffers used were glycine-HCl (pH 5.0–pH 6.0), 2.5 mM sodium phosphate (pH 6.0–pH 8.0), 2.5 mM Tris-HCl (pH 8.0–pH 9.0), and glycine-NaOH (pH 9.0–pH 10.0). Cell free supernatant solution and pH buffer were added together in a 1:1 ratio and incubated at 37°C for 1 h. The effect of varying temperature on esterase activity was analyzed by subjecting the cell free supernatant to different temperatures (4°, 20°, 25°, 30°, 37°, and 40°C) for 1 h. The halotolerance was assessed at sodium chloride concentration ranging from 1 to 24%. The enzyme activity was then measured spectrophotometrically. For this, substrate solutions were prepared comprising of solutions A and B, A comprised of 40 mg p-nitrophenyl plamitate dissolved in 12 ml of isopropanol and B comprising 0.1 g of gum Arabic, 0.2 g Sodium deoxycholate, 500 μl Triton X-100 dissolved in 90 ml 50 mM Tris-HCl buffer pH 8. Solutions A and B were mixed in a 1:20 ratio. For each assay 100 μl substrate solution, 50 μl Glycine-NaOH buffer and 10 μl enzyme sample were mixed and pipetted into a microtiter plate, incubated for 45 min at 37°C. The absorbance at 410 nm was then measured and plotted against a p-nitrophenyl standard curve (Mobarak-Qamsari et al., 2011). The enzyme assays were performed in replicates and values presented with standard deviation.
The effect of different metal ions (Ag+, Cu2+, K+, Co2+, Mg2+, and Ba2+, as well as the heavy metal ions Hg+ and Pb2+) on the enzyme activity was tested by adding different concentrations (1, 2, 3, 4, and 5 mM) of the metal ions to the cell free supernatant following incubation for 1 h at room temperature and subsequently measuring the esterase activity as described above in Section Biochemical Characterization of Recombinant Esterase.
The esterase sequence obtained from the PacBio whole fosmid sequencing was subjected to BLAST searches at NCBI and a query cover of 99% of the sequence with 66% identity to the bacterial hormone sensitive lipase E40 (PDB ID: 4xvc) was obtained. The E40 crystal structure was used as a template for homology modeling using Modeller 9.10 (Webb and Sali, 2016) and five models were generated. All the models were stereo chemically optimized by Ramachandran plot and one model was selected for further docking studies (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php). Quality control of the obtained model was performed by using ERRAT (Colovos and Yeates, 1993) and VERITY-3D (Bowie et al., 1991; Lüthy et al., 1992) from the SAVeS 4.0 software (https://services.mbi.ucla.edu/SAVES/). The known inhibitors and substrates were docked against the esterase model in silico by using Ligprep and Glide (Friesner et al., 2004) from the Maestro Schrödinger software package (Maestro, 2016).
Different concentrations of various substrates, pNPP (p-nitrophenyl palmitate), pNPM (p-nitrophenyl myristate), pNPL (p-nitrophenyl laurate), pNPC (p-nitrophenyl caprate), pNPB (p-nitrophenyl butyrate), and pNPA (p-nitrophenyl acetate) from 0.1 to 2.0 mM were added to the Ni-NTA column purified esterase enzyme sample. Based on these values from microplate-readings at 410 nm, Vmax and Km values were calculated and Michaelis–Menten plots were generated.
A metagenomic library was constructed from the marine sponge S. normani. The sponge had been collected by an ROV from a depth of 760 m. Metagenomic DNA was extracted and size selected for ~40 kb DNA fragments following pulsed-field gel electrophoresis and subsequently concentrated using an Amicon centrifugal concentrator. The library which was constructed using the fosmid vector pCCERI (Selvin et al., 2012) contained ~14,000 clones which were screened for lipase activity (Figure 1A). High throughput plate screening using 1% tributyrin resulted in the initial identification of 31 positive clones (data not shown). From amongst the 20 most highly active clones, the 7N9 fosmid was chosen as it displayed the highest level of activity and it was subsequently sequenced using the PacBio system.
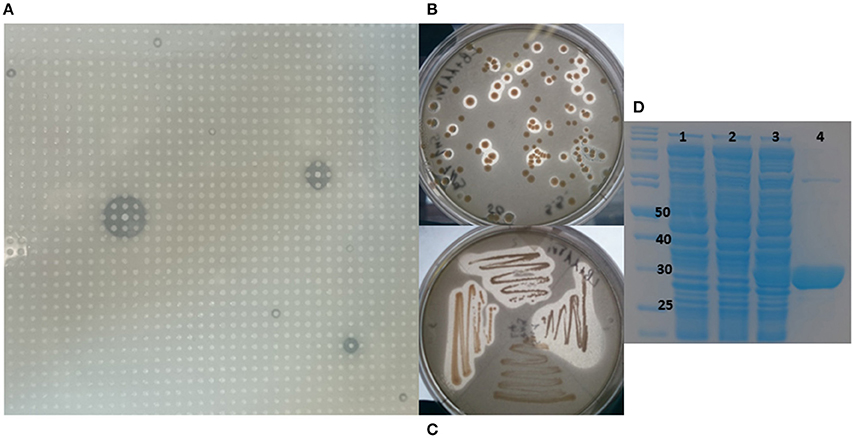
Figure 1. Metagenomic screening, cloning, and purification of 7N9 esterase. (A) Metagenomic library of Stelletta normani plated onto 1% tributyrin agar. (B) Lipase activity of E. coli clones carrying the 7N9 esterase gene on pBAD plasmid. (C) Restreak of active clones, (D) SDS-PAGE analysis of the expression and purification of 7N9 esterase, first lane marker, lane 1 (0.02% arabinose) E. coli culture with empty pBAD expression vector, lane 2 uninduced E. coli culture with pBAD harboring 7N9, lane 3 induced (0.02% arabinose) E. coli with pBAD harboring 7N9, lane 4 partial purification of 7N9 esterase using Ni-NTA resin.
The sequenced fosmid comprised of 41,407 bp and contained 65 coding DNA sequences of which 31 were annotated by BASys (Supplementary Table 1). A contig (contig 30,107–30,997, bah, Supplementary Table 1) was identified as containing an ORF encoding a gene with putative esterase function. The bah contig description refers to a N-acetyl hydrolase named bah in the bialaphos antibiotic production gene cluster (bap) in Streptomyces hygroscopicus. The putative esterase ORF, named 7N9, was found to comprise 296 amino acids, with a GC content of 60.5% and was annotated as an acetyl-hydrolase. BLASTX comparison subsequently classified the esterase as being part of the alpha/beta hydrolase family. The enzyme showed highest homology (66%) to an esterase of the bacterial hormone sensitive lipase family (E40), which had itself been isolated from a marine sediment metagenomics library (Li P. Y. et al., 2015). The E40 esterase is part of the GDSAG motif subfamily within the lipase family IV; phylogenetic comparisons (Figure 2) and multiple sequence alignments (Figure 3) indicate that the 7N9 esterase is part of the same subfamily of lipolytic enzymes as it also contains the characteristic GDSAG motif (hormone sensitive family, Hsl). Furthermore, the esterase also contains the highly conserved His-Gly-Gly (HGG) motif, which together with the GDSAG motif is involved in oxyanion hole formation (Mohamed et al., 2013; Ramnath et al., 2017).
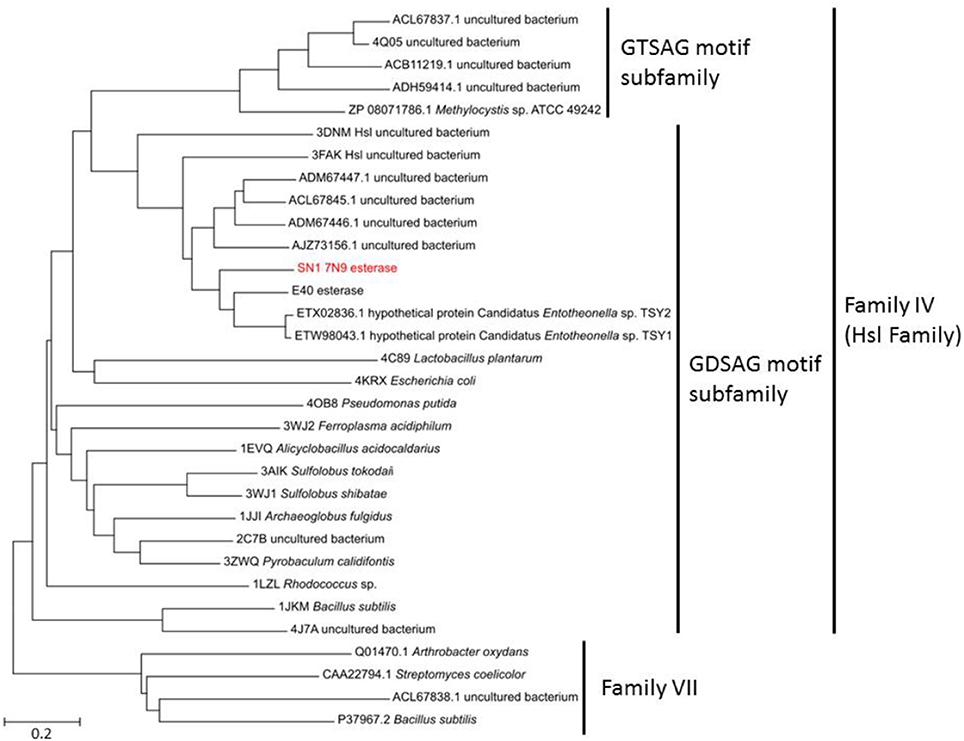
Figure 2. Phylogenetic comparison of the 7N9 esterase and other representative sequences of different lipase families. The phylogenetic tree was built using the neighbor joining method and bootstrap analysis with 500 replicates was conducted, reference sequences from lipase family VII were used as outgroups.
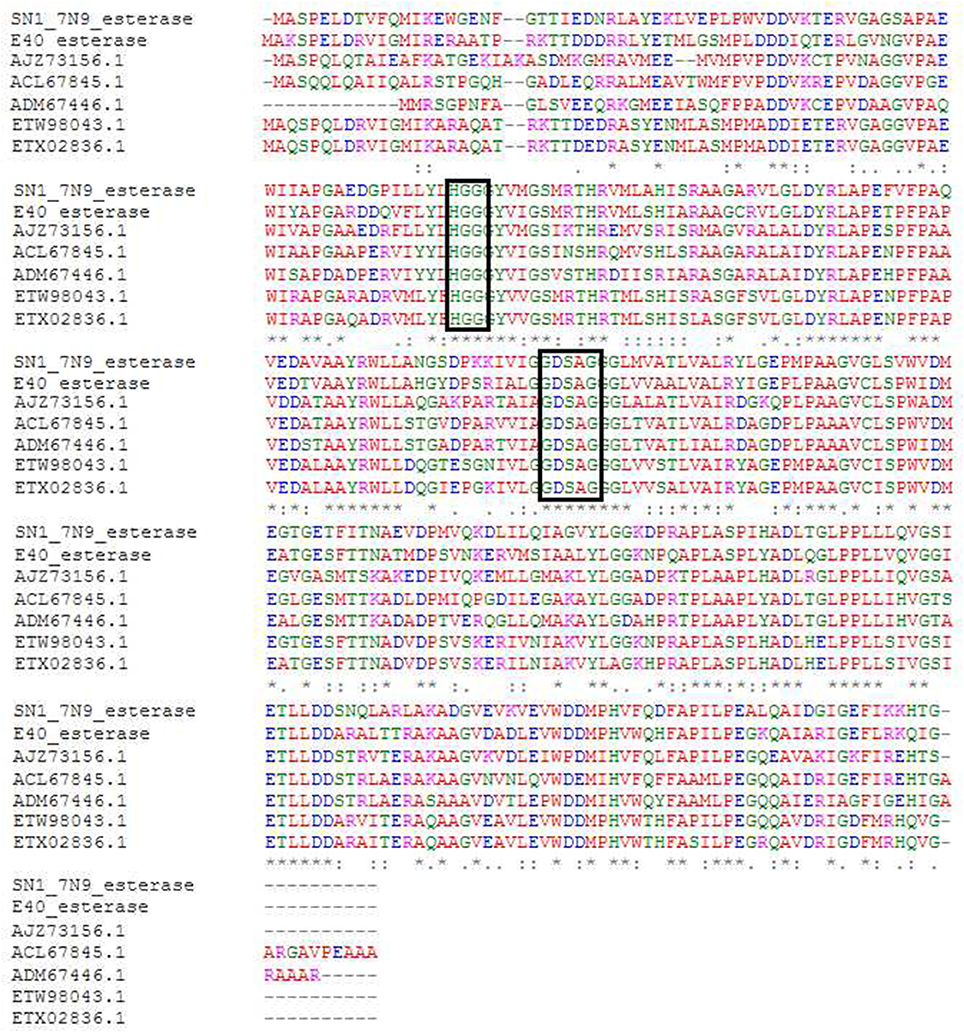
Figure 3. Multiple sequence alignment of most closely related esterase sequences. The conserved GDSAG (GXSXG) and HGG motifs are shown in the black boxes. Red indicates small hydrophobic and aromatic amino acids, blue indicates acidic amino acids, magenta indicates basic amino acids, and green indicates hydroxyl, sulfhydryl amino acids, and glycine. (*) Indicates a fully conserved residue, (:) indicates a group of strongly similar residues, and (.) indicates conservation of a group of weakly similar residues. The alignment was produced by Clustal Omega (Sievers et al., 2011; McWilliam et al., 2013; Li W. et al., 2015) and MEGA6 (Tamura et al., 2013).
The 7N9 esterase gene was PCR amplified, cloned into the pBAD/mycHIS-A vector, transformed into TOP10 E. coli cells and transformants were tested for esterase activity on 1% tributyrin plates (Figures 1B,C). Purification of the protein was performed by using the His-tag and a Ni-NTA resin column approach, with the tag being fused to the protein while transforming it into the expression vector. The ORF encoding the esterase resulted in a protein of a calculated mass of 31.7 kDa with a theoretical pI of 4.59 and ~34 kDa including the fused His-tag and myc-epitope (Figure 1D).
The model of the E40 esterase was used as a template to generate a 3D model of the 7N9 esterase and stereo chemical optimization was performed using Ramachandran plotting. When comparing to the template (E40; Pdb id: 4xvc) there is a slight variation in our models CAP and catalytic domain. The template (Pdb id: 4xvc) contains a CAP domain at Met1–Ile45 and a catalytic domain at Gln46–Gly297. Residues Gly76 and Gly77 within the conserved HGG motif comprise the oxyanion hole that is involved in substrate binding for HSL esterases. The catalytic triad composed of residues Ser145, Glu239, and His269 is below the oxyanion hole. In contrast in our model the CAP domain is located at Met1–Lys45 and the catalytic domain at Thr46–Gly296 and the catalytic triad is composing of the residues Ser144, Glu238, and His268 located below the oxyanion hole (Figure 4). The model was subsequently used to dock different substrates and inhibitors (Supplementary File 1.1). Docking scores indicate a high specificity for the substrate pNPA (Figure 5) and the inhibitor Phenylmethansulfonic acid (Table 1). Phenylmethansulfonic acid is also able to covalently bind to the nucleophilic Ser145 of E40. In the Supplementary Files 1.2, 1.3 3D binding models of the esterase and the different substrates and inhibitors can be found.
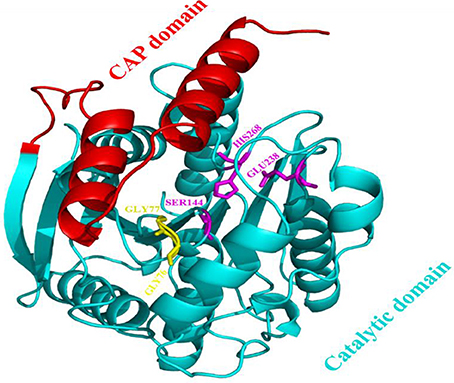
Figure 4. Domain architecture of the 7N9 esterase. The CAP domain is highlighted in red, the catalytic triad Ser144, Glu238, and His268 is highlighted in magenta, the oxyanion hole comprising of residues Gly76 and Gly77 is highlighted in yellow, the catalytic domain is highlighted in light blue.
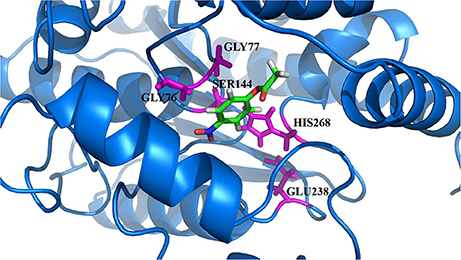
Figure 5. 3D docking model of the preferred substrate, 4-nitrophenol acetate. The catalytic site residues of 7N9 are highlighted in magenta with the substrate placed in the center.
The 7N9 esterase was found to have a higher specificity toward short chain fatty acids (Table 2). Fatty acid substrates ranging in carbon chain length from 16 (pNPP) to two (pNPA) carbon atoms were assessed; with a Vmax for pNPP being 1.507 mM/ml/min and a Km 0.6275 mM, while pNPA had a Vmax of 2.731 mM/ml/min and a Km 0.1674 mM.
The activity of the enzyme was assessed at different temperatures ranging from 4° to 60°C (Figure 6). The enzyme displayed the highest activity between 4° and 20°C with activity declining slightly thereafter. High levels of activity were observed up to 40°C, with no activity being observed at 60°C, identifying it as a cold-adapted type of hormone sensitive esterase.
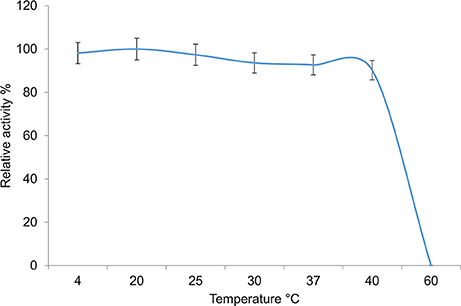
Figure 6. Temperature dependency of the 7N9 esterase. The temperature dependency of the 7N9 esterase was tested at 4°, 20°, 25°, 30°, 37°, and 40°C. The values displayed are the means and standard deviations of triplicate measurements.
The pH dependency of the esterase was tested at different pH ranging from 5 to 10. Optimal activity was achieved at pH 8.0, with higher and lower pH values leading to a decline in activity. Nonetheless activity was observed at all the pH values investigated, with 80% activity being observed at alkaline pH values of pH 9 and 10 (Figure 7). Interestingly the optimal observed pH is in line with normal pH conditions encountered in seawater, where the pH ranges from 7.5 to 8.4 (Chester and Jickells, 2012).
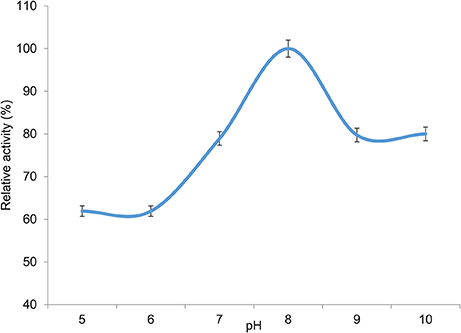
Figure 7. pH dependency of 7N9 esterase. The pH dependency of the 7N9 esterase was assessed at pH 5, pH 6, pH 7, pH 8, pH 9, and pH 10. The values displayed are the means and standard deviations of triplicate measurements.
The effect of different concentrations (1–5 mM) of various metal ions (Ag+, Cu2+, K+, Co2+, Mg2+, and Ba2+, as well as the heavy metal ions Hg+ and Pb2+) on enzyme activity was assessed. Increasing activity was observed with increasing concentrations of Cu2+, Ag+, and Ba2+; while a decrease in activity was observed for K+, Mg2+, Co2+, and the heavy metals Hg2+ and Pb2+ (Figure 8). The increase in Pb2+ concentration had the most detrimental effect on esterase activity, with only residual activity remaining at elevated levels of this heavy metal ion.
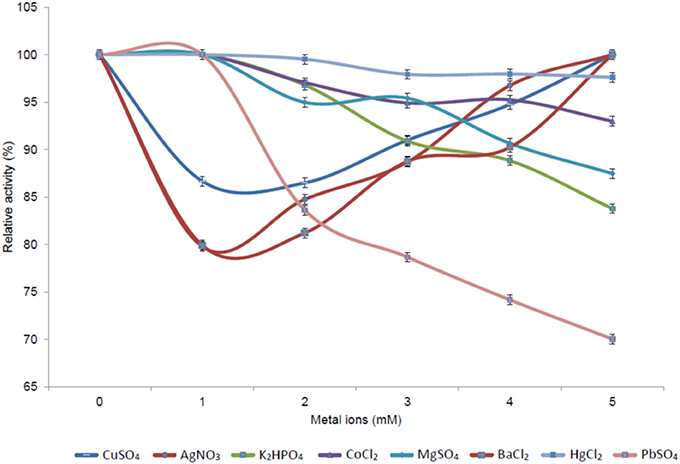
Figure 8. Effect of different metal ion concentrations on the activity of the 7N9 esterase. The effect of various metals (Ag+, Cu2+, K+, Co2+, Mg2+, and Ba2+) and heavy metals (Hg+ and Pb2+) at concentrations ranging from 1 to 5 mM on 7N9 esterase activity was assessed. The values displayed are the means and standard deviations of triplicate measurements.
The halotolerance of the esterase activity in 7N9 was then investigated, by measuring activity at different percentages of sodium chloride, ranging from 1 to 24% (Figure 9). Good levels of activity were observed over the range of sodium chloride concentrations up to 16%, with 87% of relative activity remaining at that concentration; with a more rapid decline thereafter. The overall salt concentration of sea water is typically around 3.5% (Chester and Jickells, 2012) and therefore falls within the range of optimal activity of the enzyme.
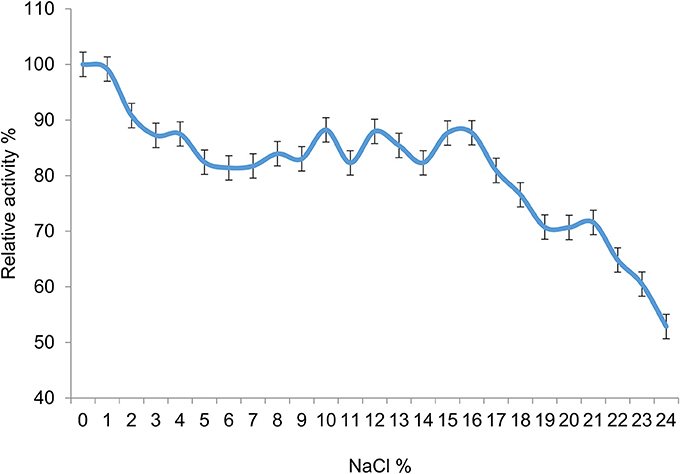
Figure 9. Halotolerance of the 7N9 esterase activity. Sodium chloride concentrations ranging from 1 to 24% were tested for their effect on the activity of the 7N9 esterase. The values displayed are the means and standard deviations of triplicate measurements.
The ever increasing demand for novel biocatalysts, has resulted in the development of a range of different approaches to explore and exploit the genetic resources in various environmental ecosystems. One approach which has been successfully employed to this end is metagenomics which helps facilitate access to genetic resources from uncultured microorganism (Kennedy et al., 2011; Baweja et al., 2016; Parages et al., 2016). Lipolytic enzymes such as esterases and lipases which catalyze the hydrolysis and synthesis of ester substrates are widely used in the agrochemical, cosmetics, dairy, detergent, food, paper processing, and pharmaceutical industries amongst others (Jaeger and Eggert, 2002). In many of these industrial processes, lipolytic enzymes are often subjected to harsh conditions such as high salt content, therefore it is important to obtain halotolerant esterases and lipases for such industrial applications (Ferrer et al., 2007). Despite this there have been only a few reports of the identification of halotolerant or halophilic esterases in the literature. These include three halotolerant esterases EstKT4, EstKT7, and EstKT9 from a tidal flat sediment metagenomic library (Jeon et al., 2012), a halotolerant esterase (PE10) from the halophilic archaea Pelagibacterium halotolerans B2T (Jiang et al., 2012), a halophilic lipase (LipBL) from the moderately halophilic bacterium Marinobacter lipolyticus SM19 (Perez et al., 2011), and a halophilic esterase (Hm EST) from Haloacrcula marismortui (Muller-Santos et al., 2009).
Marine environments in particular are proving particularly interesting as a source for novel microbial biodiversity, with numerous examples of metagenomics based approaches being employed to identify novel biocatalysts with potential biotechnological applications (Kodzius and Gojobori, 2015; Popovic et al., 2015). Marine esterases are known to play an important role in marine carbon degradation and cycling, and given that a functional metagenomics based approach had previously been successfully employed to identify the E40 esterase from sediment from the South China Sea (Li et al., 2012; Li P. Y. et al., 2015), and that marine sponge and sediment metagenomics libraries had resulted in the discovery of a variety of novel lipases including the recent reports of a cold-active salt tolerant esterase from artic sediment (De Santi et al., 2016); and a high organic solvent tolerant and thermostable esterase from marine mud (Gao et al., 2016); we targeted the metagenome of the deep sea sponge S. normani in an attempt to increase the repertoire of marine derived halophilic esterases. To this end we succeeded in identifying a gene encoding a novel psychrophilic esterase (7N9) from the hormone sensitive lipase (Hsl) family IV following the functional screening of a S. normani metagenomic library.
The esterase was heterologously expressed in Escherichia coli and the recombinant 7N9 protein was subsequently biochemically characterized. The esterase was found to be closely related to the aforementioned E40 esterase (66% amino acid homology), which was itself isolated from metagenomic DNA from a depth of 154 m Sea (Li et al., 2012; Li P. Y. et al., 2015). 7N9 and E40 both possess the two highly conserved GDSAG and HGG motifs which group them into the correspondent subfamily of lipase family IV (Mohamed et al., 2013; Ramnath et al., 2017; Figure 3). In contrast however the 7N9 esterase has a much lower optimal temperature (20°C) than the E40 esterase (45°C) and is therefore the first truly cold-adapted esterase in this lipase subfamily. As both enzymes were retrieved from metagenomic libraries one cannot say with certainty from what type of microorganism these esterases may have originated, but phylogenetic comparison and protein homology suggest a close relatedness to hypothetical proteins from the marine symbiont Candidatus genus Entotheonella (Figures 2, 3). Entotheonella is a filamentous marine sponge symbiont which produces almost all the known bioactive compounds derived from the sponge Theonella swinhoei (Wilson et al., 2014). Genome mining of two “Entotheonella” genomes has recently uncovered features of endospore formation and it has been speculated that endospore formation strengthens the resistance of “Entotheonella” to environmental stress and increase its ability to thrive in a diverse range of niches; making it highly resilient to harsh conditions. In addition it appears to possess the potential to secrete extracellular hydrolases (Liu et al., 2016). Interestingly a novel carboxylesterase Est06, isolated from a forest soil metagenomics library has also recently been reported to share 61% similarity with a hypothetical protein from Candidatus Entotheonella sp. TSY1 (Dukunde et al., 2017).
The 3D model of the 7N9 esterase was calculated using the 3D crystal structure of the closely related E40 esterase (Li P. Y. et al., 2015) as a template and subsequently in silico docking studies with different substrates and inhibitors were performed. Similar types of molecular docking studies have previously been employed to predict the enantioselectivity of various lipases (Juhl et al., 2009; Ji et al., 2012) and for the substrate specificity of feruloyl esterases (Gupta Udatha et al., 2012). Esterase 7N9 was found to have subtle differences in its CAP and catalytic domain when compared to E40 esterase. This structural difference may contribute toward the different substrate specificities and the different temperature activity profiles which we have observed. Our 7N9 esterase was most catalytically active with pNPA (p-nitrophenylacetate) as a substrate, whereas E40 was found to be more active with pNPB (p-nitrophenylbutyrate). The in silico docking studies confirmed the high specificity toward short chain fatty acids (Figure 5), as well as toward the inhibitor phenylmethanesulfonic acid, which is most likely able to bind covalently to a serine residue (Table 1).
The recombinant 7N9 esterase was biochemically characterized with respect to its temperature and pH activity profiles, together with its halotolerance and the effect of metal ions on activity was also assessed. The enzyme can be classified as cold-active and slightly alkaliphilic (Colak et al., 2005), as highest activity was observed in the range of 4–20°C and at pH 8.0 (Figures 6, 7). Metal ions were found to have a marked effect on the activity of the enzyme (Figure 8). Mechanisms involved in metal ion resistance in esterases are not well established, with different effects on enzyme activity being observed with some metal ions. For example while the Est06 esterase has been shown to be resistant to Cu2+ (Dukunde et al., 2017), we found Cu2+ to have a positive effect on 7N9 esterase activity as it also has on esterase activity in a cold adapted enzyme isolated from Shewanella sp. (Hang et al., 2016). It has also been shown however to have the opposite effect on an alkalophilic lipase from an Antarctic Pseudomonas strain (Ganasen et al., 2016). Other metal ions including Ag+ and BA2+ were also found to have a positive effect on enzyme activity at concentrations ranging from 1 to 5 mM. The ability to withstand meta ions is an important factor for enzymes used in the bioremediation of environmental waste, for example environmental increases in metal ions like Cu2+ and Pb2+ are known to be associated with oil spills (Moreno et al., 2011) and are therefore may be of particular interest in evaluating the potential use of this esterase in oil spill bioremediation applications. On the one hand Cu2+ ions increase the enzyme activity, but conversely Pb2+ is detrimental to activity. Furthermore, oil spills in cold environments are becoming increasingly common due to the increased industrial exploitation of these environment; thus specialized bioremediation strategies will be required to treat these spills in the future (Yang et al., 2009). In addition with respect to oil spills, for saline industry wastewaters a certain halotolerance is also beneficial, as those wastewaters can contain up to 14% (w/v) sodium chloride (Margesin and Schinner, 2001). The esterase was found to be halotolerant with activity being observed over a wide range of sodium chloride concentrations. Highest relative activity was observed up to 16% salt concentration; with <50% relative activity still being observed at a 34% salt concentration (Figure 9). The E40 esterase was previously reported to be quite unstable at 0.3M NaCl with both kcat and substrate affinity being significantly reduced in 0.5M NaCl (Li P. Y. et al., 2015). The halotolerance of E40 was however subsequently improved by introducing hydrophobic residues in the α4 and α5 helices of the CAP domain (Luan et al., 2016). Interestingly following the introduction of hydrophobic residues into the mutant E40 esterase, the amino acid sequences in both mutant esterases is almost identical in this CAP region; with the LVVAALVAL amino acid sequence present in both halotolerant mutants being almost identical to the LMVATLVAL present in our esterase. Subsequent work has shown that this increased halotolerance is likely to be due to increased intra-protein hydrophobic interactions in the mutant E40 esterases making them more rigid and more stable at higher salt concentrations (Li et al., 2017). This may explain why our 7N9 esterase is inherently halotolerant.
Thus, in conclusion we describe here a novel truly cold active esterase of the GDSAG subfamily of the hormone sensitive lipase family IV. The recombinant esterase is most active against short chain fatty acids such as p-nitrophenylacetate. It displays close structural relatedness to a previously described esterase (E40) isolated from a marine sediment sample, despite its different physicochemical properties. Optimal enzyme activity is achieved at low temperatures (4–20°C), at an alkaline pH (pH 8.0) and salt concentrations only have a minor influence on activity levels, resembling native physiological conditions of the environment from which the initial deep sea metagenomic sample was retrieved. The combined properties of the 7N9 esterase including its halotolerance coupled with its activity at low temperatures and responsiveness to metal ions, places it amongst novel biocatalysts that have potential bioremediation applications as well as industrial related low temperature applications such as in the manufacture of food ingredients, thermolabile pharmaceutical products together with the production of cold-wash detergents.
The study was conceived and designed by SJ, JS, FO, and AD. EB performed the metagenomic experiments, the fosmid sequencing, subcloned the esterase, and data analysis. JS and SK performed the biochemical characterization and the in silico docking studies. The manuscript was written by EB, SJ, and AD.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
EB acknowledges the financial support from the BluePharmTrain project (http://www.benthis.eu/en/bluepharmtrain.htm) funded through the MC-ITN scheme (contract no. 607786). The authors AD, FO, and SJ acknowledge the financial support from the PharmaSea project (http://www.pharma-sea.eu/) funded by the EU Seventh Framework Programme (contract no. 312184) and by the Beaufort Marine Research Award, part of the Sea Change Strategy and the Strategy for Science Technology and Innovation (2006–2012), with the support of The Marine Institute under the Marine Research Sub-Programme of the National Development Plan 2007–2013.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmars.2017.00287/full#supplementary-material
Alcaide, M., Stogios, P. J., Lafraya, Á., Tchigvintsev, A., Flick, R., Bargiela, R., et al. (2015a). Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats. Environ. Microbiol. 17, 332–345. doi: 10.1111/1462-2920.12660
Alcaide, M., Tchigvintsev, A., Martínez-Martínez, M., Popovic, A., Reva, O. N., Lafraya, Á., et al. (2015b). Identification and characterization of carboxyl esterases of gill chamber-associated microbiota in the deep-sea shrimp Rimicaris exoculata by using functional metagenomics. Appl. Environ. Microbiol. 81, 2125–2136. doi: 10.1128/AEM.03387-14
Arpigny, J. L., and Jaeger, K. E. (1999). Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343(Pt 1), 177–183.
Azam, F. (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280, 694–696. doi: 10.1126/science.280.5364.694
Baweja, M., Nain, L., Kawarabayasi, Y., and Shukla, P. (2016). Current technological improvements in enzymes toward their biotechnological applications. Front. Microbiol. 7:965. doi: 10.3389/fmicb.2016.00965
Bernard, L., Schaefer, H., Joux, F., Courties, C., Muyzer, G., and Lebaron, P. (2000). Genetic diversity of total, active and culturable marine bacteria in coastal seawater. Aquat. Microb. Ecol. 23, 1–11. doi: 10.3354/ame023001
Borchert, E., Jackson, S. A., O'Gara, F., and Dobson, A. D. (2016). Diversity of natural product biosynthetic genes in the microbiome of the deep sea sponges Inflatella pellicula, Poecillastra compressa, and Stelletta normani. Front. Microbiol. 7:1027. doi: 10.3389/fmicb.2016.01027
Bornscheuer, U. T. (2002). Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 26, 73–81. doi: 10.1111/j.1574-6976.2002.tb00599.x
Bowie, J. U., Lüthy, R., and Eisenberg, D. (1991). A method to identify protein sequences that fold into a known three-dimensional structure. Science 253, 164–170. doi: 10.1126/science.1853201
Cavicchioli, R., Siddiqui, K. S., Andrews, D., and Sowers, K. R. (2002). Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 13, 253–261. doi: 10.1016/S0958-1669(02)00317-8
Chester, R., and Jickells, T. (2012). Marine Geochemistry 3rd Edn. New Jersey, NJ: Blackwell Publishing; John Wiley & Sons.
Colak, A., Sişik, D., Saglam, N., Güner, S., Canakçi, S., and Beldüz, A. O. (2005). Characterization of a thermoalkalophilic esterase from a novel thermophilic bacterium, Anoxybacillus gonensis G2. Bioresour. Technol. 96, 625–631. doi: 10.1016/j.biortech.2004.06.003
Colovos, C., and Yeates, T. O. (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519. doi: 10.1002/pro.5560020916
De Santi, C., Altermark, B., Pierechod, M. M., Ambrosino, L., de Pascale, D., and Willassen, N. P. (2016). Characterization of a cold-active and salt tolerant esterase identified by functional screening of Arctic metagenomic libraries. BMC Biochem. 17:1. doi: 10.1186/s12858-016-0057-x
Dinsdale, E. A., Edwards, R. A., Hall, D., Angly, F., Breitbart, M., Brulc, J. M., et al. (2008). Functional metagenomic profiling of nine biomes. Nature 452, 629–632. doi: 10.1038/nature06810
Dukunde, A., Schneider, D., Lu, M., Brady, S., and Daniel, R. (2017). A novel, versatile family IV carboxylesterase exhibits high stability and activity in a broad pH spectrum. Biotechnol. Lett. 39, 577–587. doi: 10.1007/s10529-016-2282-1
Ferrer, M., Martínez-Martínez, M., Bargiela, R., Streit, W. R., Golyshina, O. V., and Golyshin, P. N. (2016). Estimating the success of enzyme bioprospecting through metagenomics: current status and future trends. Microb. Biotechnol. 9, 22–34. doi: 10.1111/1751-7915.12309
Ferrer, M., Golyshins, O., Beloqui, A., and Golyshin, P. N. (2007). Mining enzymes from extreme environments. Curr. Opin. Microbiol. 10, 207–214. doi: 10.1016/j.mib.2007.05.004
Friesner, R. A., Banks, J. L., Murphy, R. B., Halgren, T. A., Klicic, J. J., Mainz, D. T., et al. (2004). Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749. doi: 10.1021/jm0306430
Fu, C., Hu, Y., Xie, F., Guo, H., Ashforth, E. J., Polyak, S. W., et al. (2011). Molecular cloning and characterization of a new cold-active esterase from a deep-sea metagenomic library. Appl. Microbiol. Biotechnol. 90, 961–970. doi: 10.1007/s00253-010-3079-0
Ganasen, M., Yaacob, N., Rahman, R. N., Leow, A. T., Basri, M., Salleh, A. B., et al. (2016). Cold-adapted ornaic solvent tolerant alkalophilic family 1.3 lipase from an Antarctic Pseudomonas. Int. J. Biol. Macromol. 92, 1266–1276. doi: 10.1016/j.ijbiomac.2016.06.095
Gao, W., Wu, K., Chen, L., Fan, H., Zhao, Z., Gao, B., et al. (2016). A novel esterase from a marine mud metagenomic library for biocatalytic synthesis of short-chain flavor esters. Microb. Cell Fact. 15, 41. doi: 10.1186/s12934-016-0435-5
Gupta Udatha, D. B. R. K., Sugaya, N., Olsson, L., and Panagiotou, G. (2012). How well do the substrates KISS the enzyme? Molecular docking program selection for feruloyl esterases. Sci. Rep. 2:323. doi: 10.1038/srep00323
Hang, Y., Ran, S., Wang, X., Jiao, J., Wang, S., and Liu, Z. (2016). Mutational analysis and stability characterization of a novel esterase of lipolytic enzyme family VI from Shewanella sp. Int. J. Biol. Macromol. 93, 655–664. doi: 10.1016/j.ijbiomac.2016.09.032
Hårdeman, F., and Sjöling, S. (2007). Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol. Ecol. 59, 524–534. doi: 10.1111/j.1574-6941.2006.00206.x
Jackson, S. A., Flemer, B., McCann, A., Kennedy, J., Morrissey, J. P., O'Gara, F., et al. (2013). Archaea appear to dominate the microbiome of Inflatella pellicula deep sea sponges. PLoS ONE 8:e84438. doi: 10.1371/journal.pone.0084438
Jaeger, K. E., and Eggert, T. (2002). Lipases for biotechnology. Curr. Opin. Biotechnol. 13, 390–397. doi: 10.1016/S0958-1669(02)00341-5
Jegannathan, K. R., and Nielsen, P. H. (2013). Environmental assessment of enzyme use in industrial production - a literature review. J. Clean. Prod. 42, 228–240. doi: 10.1016/j.jclepro.2012.11.005
Jeon, H. J., Lee, H. S., Kim, J. T., Kim, S.-J., Choi, S. H., Kang, S. G., et al. (2012). Identification of a new subfamily of salt-tolerant esterases from a metagenomics library of tidal flat sediment. Appl. Microbiol. Biotechnol. 93, 623–631. doi: 10.1007/s00253-011-3433-x
Ji, L., Xiaoling, T., and Hongwei, Y. (2012). Prediction of the enantioselectivity of lipases and esterases by molecular docking method with modified force field parameters. Biotechnol. Bioeng. 105, 687–696. doi: 10.1002/bit.22596
Jiang, X., Huo, Y., Cheng, H., Zhang, X., Zhu, X., and Wu, M. (2012). Cloning, expression and characterization of a halotolerant esterase from the marine bacterium Pelagibacterium halotolerans B2T. Extremophiles 16, 427–435. doi: 10.1007/s00792-012-0442-3
Juhl, P. B., Trodler, P., Tyagi, T., and Pleiss, J. (2009). Modelling substrate specificity and enantioselectivity for lipases and esterases by substrate-imprinted docking. BMC Struct. Biol. 9:39. doi: 10.1186/1472-6807-9-39
Kennedy, J., Codling, C. E., Jones, B. V., Dobson, A. D., and Marchesi, J. R. (2008). Diversity of microbes associated with the marine sponge, Haliclona simulans, isolated from Irish waters and identification of polyketide synthase genes from the sponge metagenome. Environ. Microbiol. 10, 1888–1902. doi: 10.1111/j.1462-2920.2008.01614.x
Kennedy, J., Flemer, B., Jackson, S. A., Morrissey, J. P., O'Gara, F., and Dobson, A. D. (2014). Evidence of a putative deep sea specific microbiome in marine sponges. PLoS ONE 9:e91092. doi: 10.1371/journal.pone.0091092
Kennedy, J., O'Leary, N. D., Kiran, G. S., Morrissey, J. P., O'Gara, F., Selvin, J., et al. (2011). Functional metagenomic strategies for the discovery of novel enzymes and biosurfactants with biotechnological applications from marine ecosystems. J. Appl. Microbiol. 111, 787–799. doi: 10.1111/j.1365-2672.2011.05106.x
Kodzius, R., and Gojobori, T. (2015). Marine metagenomics as a source for bioprospecting. Mar. Genomics 24(Pt 1), 21–30. doi: 10.1016/j.margen.2015.07.001
Li, P. Y., Chen, X. L., Ji, P., Li, C. Y., Wang, P., Zhang, Y., et al. (2015). Interdomain hydrophobic interactions modulate the thermostability of microbial esterases from the hormone-sensitive lipase family. J. Biol. Chem. 290, 11188–11198. doi: 10.1074/jbc.M115.646182
Li, P. Y., Xie, B. B., Zhang, X. Y., Qin, Q. L., Dang, H. Y., Wang, X. M., et al. (2012). Genetic structure of three fosmid-fragments encoding 16S rRNA genes of the Miscellaneous Crenarchaeotic Group (MCG): implications for physiology and evolution of marine sedimentary archaea. Environ. Microbiol. 14, 467–479. doi: 10.1111/j.1462-2920.2011.02637.x
Li, P. Y., Zhang, Y., Xie, B. B., Zhang, Y. Q., Hao, J., Wang, Y., et al. (2017). Structural and mechanistic insights into the improvement of halotolerant ability of a marine microbial esterase by increasing intra- and inter- domain hydrophobic interactions. Appl. Environ. Microbiol. doi: 10.1128/AEM.01286-17. [Epub ahead of print].
Li, W., Cowley, A., Uludag, M., Gur, T., McWilliam, H., Squizzato, S., et al. (2015). The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584. doi: 10.1093/nar/gkv279
Liu, F., Li, J., Feng, G., and Zhiyong, L. (2016). New genomic insights into Entotheonella symbionts in Theonella swinhoei: mixotrophy, anaerobic adaptatio, resilence and interaction. Front. Microbiol. 7:1333. doi: 10.3389/fmicb.2016.01333
Luan, Z. J., Yu, H. L., Ma, B. D., Qi, Y. K., Chen, Q., and Xu, J.-H. (2016). Dramatically improved performance of an esterase for cilastatin synthesis by cap doamin engineering. Ind. Eng. Chem. Res. 55, 12167–12172. doi: 10.1021/acs.iecr.6b02440
Lüthy, R., Bowie, J. U., and Eisenberg, D. (1992). Assessment of protein models with three-dimensional profiles. Nature 356, 83–85. doi: 10.1038/356083a0
Mandrich, L., Merone, L., Pezzullo, M., Cipolla, L., Nicotra, F., Rossi, M., et al. (2005). Role of the N terminus in enzyme activity, stability and specificity in thermophilic esterases belonging to the HSL family. J. Mol. Biol. 345, 501–512. doi: 10.1016/j.jmb.2004.10.035
Margesin, R., and Schinner, F. (2001). Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 5, 73–83. doi: 10.1007/s007920100184
McWilliam, H., Li, W., Uludag, M., Squizzato, S., Park, Y. M., Buso, N., et al. (2013). Analysis Tool web services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600. doi: 10.1093/nar/gkt376
Mirete, S., Morgante, V., and González-Pastor, J. E. (2016). Functional metagenomics of extreme environments. Curr. Opin. Biotechnol. 38, 143–149. doi: 10.1016/j.copbio.2016.01.017
Mobarak-Qamsari, E., Kasra-Kermanshahi, R., and Moosavi-Nejad, Z. (2011). Isolation and identification of a novel, lipase-producing bacterium, Pseudomnas aeruginosa KM110. Iran. J. Microbiol. 3, 92–98.
Mohamed, Y. M., Ghazy, M. A., Sayed, A., Ouf, A., El-Dorry, H., and Siam, R. (2013). Isolation and characterization of a heavy metal-resistant, thermophilic esterase from a Red Sea brine pool. Sci. Rep. 3:3358. doi: 10.1038/srep03358
Moreno, R., Jover, L., Diez, C., and Sanpera, C. (2011). Seabird feathers as monitors of the levels and persistence of heavy metal pollution after the Prestige oil spill. Environ. Pollut. 159, 2454–2460. doi: 10.1016/j.envpol.2011.06.033
Muller-Santos, M., de Souza, E. M., Pedrosa Fde, O., Mitchell, D. A., Longhi, S., Carriere, F., Canaan, S., et al. (2009). First evidence for the salt-dependent folding and activity of an esterase from the halophilic archaea Haloarcula marismortui. Biochim. Biophys. Acta 1791, 719–729. doi: 10.1016/j.bbalip.2009.03.006
Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., et al. (1992). The alpha/beta hydrolase fold. Protein Eng. 5, 197–211. doi: 10.1093/protein/5.3.197
Panda, T., and Gowrishankar, B. S. (2005). Production and applications of esterases. Appl. Microbiol. Biotechnol. 67, 160–169. doi: 10.1007/s00253-004-1840-y
Parages, M. L., Gutiérrez-Barranquero, J. A., Reen, F. J., Dobson, A. D., and O'Gara, F. (2016). Integrated (Meta) genomic and synthetic biology approaches to develop new biocatalysts. Mar. Drugs 14:62. doi: 10.3390/md14030062
Perez, D., Martin, S., Fernandez-Lorente, G., Filice, M., Guisan, J. M., Ventosa, A., et al. (2011). A novel halophilic lipase, LipBL, showing high efficiency in the production of eicosapentaenoic acid (EPA). PLoS ONE 6:e23325. doi: 10.1371/journal.pone.0023325
Popovic, A., Tchigvintsev, A., Tran, H., Chernikova, T. N., Golyshina, O. V., Yakimov, M. M., et al. (2015). Metagenomics as a tool for enzyme discovery: hydrolytic enzymes from marine-related metagenomes. Adv. Exp. Med. Biol. 883, 1–20. doi: 10.1007/978-3-319-23603-2_1
Ramnath, L., Sithole, B., and Govinden, R. (2017). Classification of lipolytic enzymes and their biotechnological applications in the pulping industry. Can. J. Microbiol. 63, 179–192. doi: 10.1139/cjm-2016-0447
Rao, T. E., Imchen, M., and Kumavath, R. (2017). Marine enzymes: production and applications for human health. Adv. Food Nutr. Res. 80, 149–163. doi: 10.1016/bs.afnr.2016.11.006
Rhee, J. K., Ahn, D. G., Kim, Y. G., and Oh, J. W. (2005). New thermophilic and thermostable esterase with sequence similarity to the hormone-sensitive lipase family, cloned from a metagenomic library. Appl. Environ. Microbiol. 71, 817–825. doi: 10.1128/AEM.71.2.817-825.2005
Santiago, M., Ramírez-Sarmiento, C. A., Zamora, R. A., and Parra, L. P. (2016). Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front. Microbiol. 7:1408. doi: 10.3389/fmicb.2016.01408
Sasso, F., Natalello, A., Castoldi, S., Lotti, M., Santambrogio, C., and Grandori, R. (2016). Burkholderia cepacia lipase is a promising biocatalyst for biofuel production. Biotechnol. J. 11, 954–960. doi: 10.1002/biot.201500305
Selvin, J., Kennedy, J., Lejon, D. P., Kiran, G. S., and Dobson, A. D. (2012). Isolation identification and biochemical characterization of a novel halo-tolerant lipase from the metagenome of the marine sponge Haliclona simulans. Microb. Cell Fact. 11:72. doi: 10.1186/1475-2859-11-72
Sharma, S., and Kanwar, S. S. (2014). Organic solvent tolerant lipases and applications. ScientificWorldJournal 2014:625258. doi: 10.1155/2014/625258
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Singh, A. K., and Mukhopadhyay, M. (2012). Overview of fungal lipase: a review. Appl. Biochem. Biotechnol. 166, 486–520. doi: 10.1007/s12010-011-9444-3
Singh, R., Kumar, M., Mittal, A., and Mehta, P. K. (2016). Microbial enzymes: industrial process in 21st century. 3 Biotech 6:174. doi: 10.1007/s13205-016-0485-8
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Van Domselaar, G. H., Stothard, P., Shrivastava, S., Cruz, J. A., Guo, A., Dong, X., et al. (2005). BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 33, W455–W459. doi: 10.1093/nar/gki593
Verger, R. (1997) ‘Interfacial activation’ of lipases: facts and artifacts. Trends Biotechnol. 15, 32–38.
Vieites, J. M., Guazzaroni, M. E., Beloqui, A., Golyshin, P. N., and Ferrer, M. (2009). Metagenomics approaches in systems microbiology. FEMS Microbiol. Rev. 33, 236–255. doi: 10.1111/j.1574-6976.2008.00152.x
Webb, B., and Sali, A. (2016). Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 86, 2.9.1–2.9.37. doi: 10.1002/cpps.20
Wilson, M. C., Mori, T., Rückert, C., Uria, A. R., Helf, M. J., Takada, K., Gernert, C., et al. (2014). An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62. doi: 10.1038/nature12959
Wirsen, C. O., and Molyneaux, S. J. (1999). A study of deep-sea natural microbial populations and barophilic pure cultures using a high-pressure chemostat. Appl. Environ. Microbiol. 65, 5314–5321.
Keywords: cold active esterase, metagenomic library, deep sea sponge
Citation: Borchert E, Selvin J, Kiran SG, Jackson SA, O'Gara F and Dobson ADW (2017) A Novel Cold Active Esterase from a Deep Sea Sponge Stelletta normani Metagenomic Library. Front. Mar. Sci. 4:287. doi: 10.3389/fmars.2017.00287
Received: 18 May 2017; Accepted: 23 August 2017;
Published: 08 September 2017.
Edited by:
Antonio Trincone, Consiglio Nazionale Delle Ricerche (CNR), ItalyReviewed by:
Jochen Schmid, Technische Universität München, GermanyCopyright © 2017 Borchert, Selvin, Kiran, Jackson, O'Gara and Dobson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alan D. W. Dobson, YS5kb3Nib25AdWNjLmll
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.