- CERMAV, Centre National de la Recherche Scientifique, Grenoble Alpes Université, Grenoble, France
Polysaccharides are the most abundant and the most complex organic molecules in the ocean. In contrast to land polysaccharides, many marine polysaccharides are highly sulfated; in particular, the cell walls of macroalgae harbor a high diversity of sulfated polysaccharides (carrageenans, agarans, fucoidans, ulvans, etc.). These sulfated polysaccharides, biosynthesized by macroalgal primary producers, represent an important food source for heterotrophic organisms. Their biodegradation requires a set of enzymes that can cleave the glycosidic linkages of the carbohydrate backbone (called glycoside hydrolases) and the sulfate ester groups (called polysaccharide sulfatases). This review first provides on overview of the current state of knowledge on the classification and mechanisms of sulfatases in general. Then, based on an exploration of marine genomic and metagenomics data that reveals the diversity of carbohydrate sulfatases, it focuses on strategies to predict these sulfatases. In particular, the modularity of sulfatases and their location in marine polysaccharide utilization loci (PUL) provide clues as to their potential substrates and can drive future functional assays. Finally, the review underscores the low number of currently biochemically characterized marine carbohydrate sulfatases (e.g., agarases, carrageenanases, and fucose sulfatases). Bottlenecks encountered in studies on sulfatases likely lie in the difficulties in purifying them and producing them in heterologous systems.
Introduction
After carbon, oxygen, and nitrogen, sulfur is the most abundant element in living organisms. Sulfur participates in the composition of cysteine and methionine, which are key amino acids for protein folding (e.g., disulfide bonds) and essential for the reactivity of many enzymes as catalytic amino acids or complexing cofactors (e.g., iron-sulfur cofactors). Sulfur is also found in vitamins, steroids, and many carbohydrates including polysaccharides, proteoglycans, and glycolipids. Sulfur is assimilated by living organisms in the form of inorganic sulfate ions, which are the second most abundant anions in seawater. The concentration of sulfate ions in the oceans ranges from 25 to 28 mM; these values are very high compared with those found in freshwater and soil (10–50 μM) (Wright and Colling, 1995; Friedlander, 2001; Bochenek et al., 2013).
The abundance of sulfate ions and its ubiquity has resulted in a wide distribution of sulfated molecules and more especially, sulfated polysaccharides in marine organisms. In contrast to land polysaccharides, a huge number marine polysaccharides are decorated with sulfate ester groups. Except for sulfated glycosaminoglycans such as chondroitin sulfate or herapan sulfate found in the matrix of animal cells, anionic polysaccharides found in terrestrial organisms carry uronic groups (i.e., pectins, hyaluronic acid, polysialic acids). The occurrence of sulfated polysaccharides in macroalgae, marine angiosperms, and seagrasses has been proposed to be an adaptation to marine environments (Aquino et al., 2005; Olsen et al., 2016), suggesting that the sulfation of polysaccharides is a marker of marine origins. Nevertheless, in all cases, the biosynthesis of sulfated polysaccharides requires the activation of sulfate in PAPS (3′-phosphoadenosine-5′-phosphosulfate), catalyzed by PAPS synthase, and then the grafting of a sulfate ester group onto the carbohydrate via a specific sulfotransferase (Klaassen and Boles, 1997; Zhang et al., 1998).
The most studied sulfated marine polysaccharides are those extracted from the cell walls of macroalgae due to the industrial interest in their hydrocolloid properties. Agarans and carrageenans are the most exploited polysaccharides from red algae and the wide diversity of red algae polysaccharides has been reviewed recently (Usov, 2011). Similarly, the sulfated polysaccharides extracted from brown and green algae also show high diversity. Several reviews have highlighted their structural complexity and their potential applications (see, for example, Kloareg and Quatrano, 1988; Berteau and Mulloy, 2003; Lahaye and Robic, 2007; Pomin and Mourão, 2008). The diversity of sulfated polysaccharides biosynthesized by marine microorganisms, such as bacteria and microalgae, has been less explored so far. However, sulfate groups were found to decorate the carbohydrate backbone of several secreted polysaccharides and matrix lipo-polysaccharides biosynthesized by marine bacteria (Mancuso Nichols et al., 2005; Nazarenko et al., 2011). Composition analyses attested also the structural diversity of sulfated polysaccharides biosynthesized in microalgae but only a few polysaccharide structures have been resolved (Hoagland et al., 1993; Gügi et al., 2015). For example, studies and structural analyses of the main cell-wall polysaccharide in diatoms—a group of ecologically important marine organisms—suggest that it is a sulfated glucuronmannan (Percival and McDowell, 1967; Willis et al., 2013).
Marine polysaccharides make up a large part of algal biomass and are a food source for heterotrophic organisms. This reservoir of sulfated polysaccharides are produced by concomitant fixation of sulfur and carbon by photosynthetic organisms. Enzymatic degradation of complex sulfated polysaccharides requires a set of enzymes that can cleave the glycosidic bond and remove any decorations, such as sulfate groups, from the carbohydrate backbone. Specific glycoside hydrolases or polysaccharide lyases depolymerize marine polysaccharides break the glycosidic bonds via hydrolysis and β-elimination mechanisms, respectively. Concerted action with sulfatases can result in the production of neutral mono- and oligosaccharides then used for energy consumption.
The avalanche of gene sequences produced during the numerous marine genomics and metagenomics programs have revealed the important role of sulfate in marine biology (Glöckner et al., 2003; Teeling et al., 2012). Genomes of polysaccharide-degrading bacteria reveal the co-occurrence of glycoside hydrolases and polysaccharide lyases with sulfatases. More interestingly, in the bacterial genus Bacteroidetes known to include many polysaccharide-degrading strains, all the genes coding for polysaccharide-degrading enzymes leading to the saccharification of sulfated polysaccharides are clustered in the genome in so-called “polysaccharide utilization loci” (PUL) (Sonnenburg et al., 2006; Flint et al., 2008; White et al., 2014). More recently, the discovery of marine polysaccharide-degrading enzymes in Bacteroidetes strains found in the microbiota of the human gut—likely acquired by gene transfer from marine organisms associated with seafood (Hehemann et al., 2010)—suggest that the investigation of the enzymatic degradation of marine polysaccharides goes beyond the ecological understanding of the ocean cycle, and also involves human health. Likewise, genomic analyses of human microbiota have revealed numerous PULs with sulfatase genes involved in the degradation of many sulfated polysaccharides, probably of marine origin.
Therefore, the purpose of this review is to summarize the work dealing with marine polysaccharide sulfatases starting with a short introduction to sulfatase classification, followed by a highlight on the diversity of marine polysaccharides sulfatases revealed by genomic data mining and then a description of the marine polysaccharide sulfatases that have been biochemically characterized to date. Glycosaminoglycan sulfatases will not be considered here, although several chondroitin or heparin sulfatases have been identified in marine prokaryotes (Han et al., 2014; Wang et al., 2015a).
Mechanism and Structure of Sulfatases
Sulfatases are grouped into four classes based on sequence homology, crystallographic structure and mechanisms. Type I sulfatase—or formylglycine-dependent sulfatase—encompasses the vast majority of the known sulfatases and to date, all the biochemically characterized carbohydrate sulfatases (Hanson et al., 2004). These sulfatases require post-translational conversion of a cysteine or a serine into a formylglycine amino acid residue essential for catalysis. The oxidation of amino acids is catalyzed by a formylglycine-generating enzyme (FGE), also named sulfatase-modifying factor 1 (SUMF1), which recognizes the consensus amino acid sequence C/S-X-P-X-R (Cosma et al., 2003; Dierks et al., 2003; Sardiello et al., 2005; Bojarová and Williams, 2008).
In anaerobic bacteria, cysteine and serine residues can be modified by another family of enzymes called anaerobic sulfatase-maturating enzymes (anSMEs) (Berteau et al., 2006). Crystallographic structure of FGE (Bond et al., 1997; von Bülow et al., 2001) and anSME (Goldman et al., 2013) demonstrate that their mechanisms differ in the use or non-use of oxygen molecules, respectively. A recent review covers the state of knowledge on these enzymes as well as their potential biotechnological applications (Appel and Bertozzi, 2015).
Based on inhibition experiments and the analysis of the crystal structure of sulfatase complexes, two mechanisms of desulfation have been proposed, differing essentially in the hydration state of the formylglycine residue at the beginning of the reaction (Bond et al., 1997; Lukatela et al., 1998). However, both mechanisms generally agree that the last steps of the reaction involve a covalent bond between the sulfate anion and the formylglycine amino acid, and the regeneration of formylglycine by hydrolysis (Figure 1). The reaction results in the retention of the carbon configuration, the chirality of the carbon center being unaffected by the desulfation catalysis (Williams et al., 2014; Figure 2).
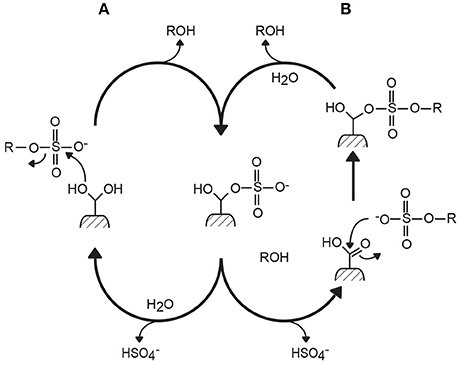
Figure 1. Two mechanisms of action proposed for formylglycine-dependent sulfatases (the type I sulfatase family). Mechanisms (A,B) differ by the hydration state at the beginning of the reaction. However, they both lead to the formation of a covalent intermediate between the sulfate ester group and formylglycine amino acid (Bond et al., 1997; Lukatela et al., 1998).
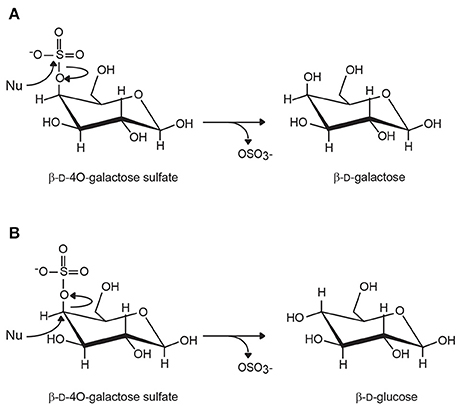
Figure 2. Inverting and retaining carbohydrate sulfatases. (A) Retaining sulfatases cleave the S-O bonds without inverting the configuration of the carbon carrying the sulfate ester group. In the case of 4O-β-D-galactose sulfate, the removal of the sulfate leads to the production of β-D-galactose. (B) In inverting sulfatases, the C-O bond is broken and the stereochemical inversion of the configuration of the ring carbon results in the epimerization of the residue. Although this situation has never been observed, the removal of sulfate of 4O-β-D-galactose would result in the formation of β-D-glucose. (Nu, nucleophilic attack).
Type II sulfatase encompasses Fe(II) α-ketoglutarate-dependent alkylsulfatases, which belong to the dioxygenase superfamily (Müller et al., 2004). The catalysis requires molecular oxygen (O2) and α-keto-glutarate as a co-substrate and leads to the oxidation of the primary sulfate ester group in the aldehyde and concomitantly to the oxidative decarboxylation of the α-keto-glutarate into succinate. Type III sulfatases are represented by enzymes related to Zn2+- or Mn2+-dependent metallo-β-lactamases. The cleavage of the C-OS bond occurs via a nucleophilic attack of the carbon by a molecule of water activated by the binuclear metal ion complex located in the active site (Hagelueken et al., 2006). The loss of the sulfate group results in the inversion of the configuration of the carbon (Figure 2). No carbohydrate sulfatases fall in the type II or type III sulfatase categories.
Two other sets of enzymes represent another potential sulfatase families. One sulfatase belonging to the superfamily of amidohydrolases has been purified from the marine bacteria Pseudoalteromonas carrageenovora; the recombinantly expressed enzyme is active on a synthetic substrate (i.e., methylemberliferyl sulfate), but showed less activity than the native purified enzyme (Genicot et al., 2014). The galactose-6-sulfurylase (discussed below in Section Predicted marine polysaccharide sulfatases) catalyzes the formation of anhydrogalactose concomitantly to the release of the sulfate group (Rees, 1961a,b).
Predicted Marine Polysaccharide Sulfatases
The massive compilation of gene sequences through the numerous genome and metagenome sequencing programs has led to an exponential increase in the number of putative proteins whose functions have, for the most part, not yet been determined (Hanson et al., 2010). Despite the development of bioinformatics tools, prediction of protein function requires experimental validation, including a comprehensive overview of metabolic pathways and functional characterization of the proteins. Like other classes of proteins, sulfatases have seen the number of predicted genes increase exponentially in databases, particularly owing to the massive sequencing of marine organisms. The precise function of the sulfatases, such as the nature of their sulfated substrates (carbohydrate, lipids, metabolites, etc.), cannot be predicted based on sequence comparison with biochemically characterized sulfatases due to the lack of data. Based on the surge in sulfatase gene sequences and their genomic environment, probable function and preferred substrates of carbohydrate sulfatases can be suspected.
Modular Sulfatases
Most sulfatases are made of one catalytic module; however, in rare cases, two sulfatase modules are found in the same protein, both having the predicted catalytic amino acid residues. More interestingly, some proteins are multi-modular, containing one sulfatase catalytic module and one glycoside hydrolase catalytic module. For example, the protein found in the bacterium Nonlabens ulvanivorans, combines two catalytic modules, one attributed to a sulfatase, and the other to a glycoside hydrolase belonging to family GH78 (rhamnosidases). This organization reflects a genomic rearrangement adapted to the degradation of ulvan, the sulfated cell-wall matrix polysaccharide found in green algae of the genera Ulva and Enteromorpha (Kopel et al., 2014). Ulvan, composed of 3O-sulfate-rhamnose, is likely more efficiently degraded by a bi-modular protein. Similarly, the bi-modular protein found in Flammeovirga sp. SJP92 genome (GenBank accession no. PRJNA306821), composed of a xylosidase (GH10) linked to a sulfatase, suggests evolution for optimized degradation of sulfated xylan, found especially in the cell walls of red algae.
A rapid survey of the Uniprot (The UniProt Consortium, 2015) and CAZy databases (Terrapon et al., 2015) revealed several modular sulfatases (Figure 3), suggesting that a thorough analysis of database should multiply examples of modular sulfatases. Observations of these modular proteins indicate that the preferred substrate of these sulfatases is a sulfated carbohydrate and provide strong clues as to the sugar residues involved.
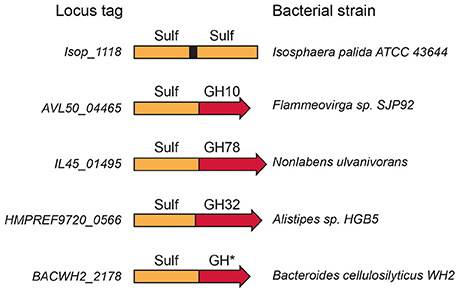
Figure 3. Modular sulfatases. Several examples of modular sulfatases retrieved from databases. The catalytic sulfatase module (Sulf) is linked to a catalytic module attributed to glycoside hydrolase (GH). The GH number refers to the CAZy family name.
Sulfated Polysaccharide Utilization Loci
PULs are clusters of genes co-localized in the genome that encode proteins involved in the same polysaccharide-degradation pathway, starting from the detection of the polysaccharide to its depolymerization and uptake of the degradation products (Flint et al., 2008; White et al., 2014; Terrapon et al., 2015). PULs are found in Bacteroidetes, which are recognized as important polysaccharide degraders given the number polysaccharide-degrading enzymes found in their genomes. The starch utilization loci of the human gut symbiont Bacteroidetes thetaiotaomicron was the first PUL to be described (Anderson and Salyers, 1989). Starch is first bound by the outer membrane SusD protein, and then undergoes partial hydrolysis by an amylase, SusG. The malto-oligosaccharides are then imported into the cell by SusC, which is a TonB-dependent transporter. The complete degradation of malto-oligosaccharides is ensured by SusA and SusB, located in the periplasm. This gene organization is used as paradigm for the prediction of other PULs and always include proteins that are homologous to SusC and SusD and a series of polysaccharide-degrading enzymes.
Because protein expression of PUL is co-regulated, transcriptomic methods can monitor the expression level of proteins of the targeted PUL when the bacteria are grown in presence of selected polysaccharides, thereby identifying the function of the identified PUL (Martens et al., 2008, 2011; McNulty et al., 2013; Despres et al., 2016). Co-expression of proteins located in the same PUL by carbohydrate inducers validate the PUL organization and give some indication as to the putative polysaccharides metabolized in vivo. PULs found in Bacteroidetes of the human microbiota have been recorded in the CAZy database (http://www.cazy.org/PULDB/) and experimentally analyzed PULs are also included (Terrapon et al., 2015). Many PULs, including the experimentally analyzed ones, contain sulfatases and glycoside hydrolases (or polysaccharide lyases), suggesting that these sulfatases are active on carbohydrates.
The first documented marine PUL includes proteins involves in the degradation of porphyran (the biosynthetic precursor of agarose; Figure 4). It contains several glycoside hydrolases, sulfatases, and a set of proteins of unknown function. This PUL has been transferred laterally from marine Bacteroidetes to human microbiota Bacteroidetes (Hehemann et al., 2010). Many other PULs dedicated to the degradation of sulfated polysaccharides have been observed in several bacteria genomes or metagenomes (Gómez-Pereira et al., 2012; Mann et al., 2013; Kabisch et al., 2014; Hahnke et al., 2015; Xing et al., 2015; Panschin et al., 2016; Sun et al., 2016). The functions of PULs are hypothesized based on sequence homology of glycoside hydrolases found in the PUL with characterized enzymes. Then, the presumed enzyme activities encountered in the PUL are compared with the literature available on the composition or structure of marine polysaccharides to attribute the putative metabolized polysaccharide to the PUL. Additional information, such as the location where the Bacteroidetes strains have been isolated or sequenced (e.g., surface of an algae, microalgae bloom) may also help refine the inference.
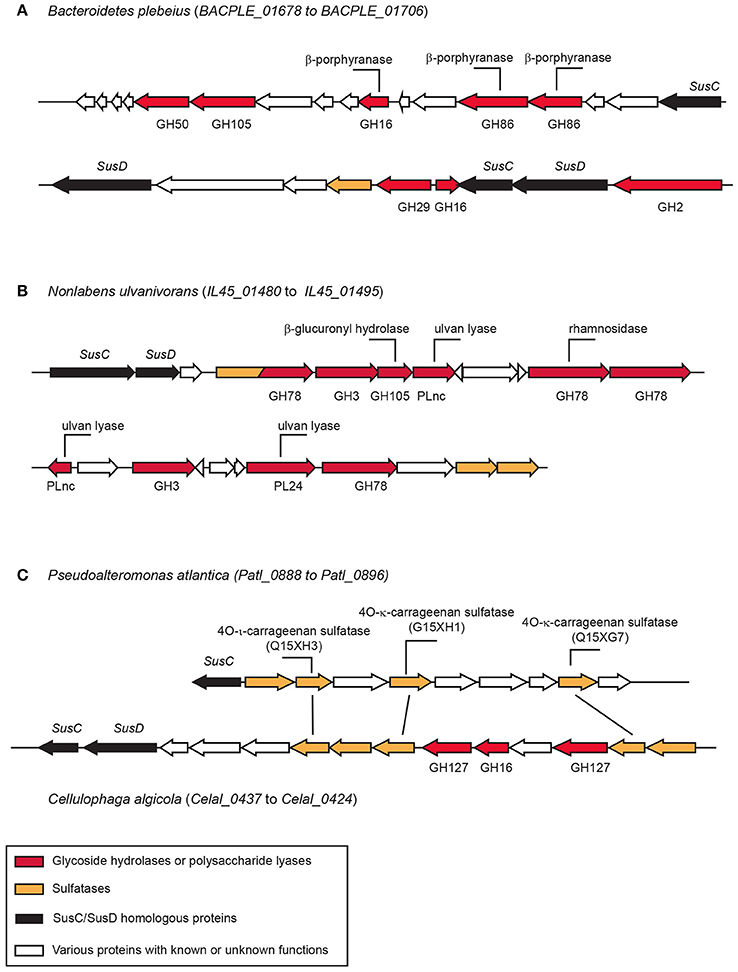
Figure 4. Examples of polysaccharide utilization loci (PUL) containing sulfatases. (A) The porphyranase PUL in Bacteroidetes plebeius found in the human microbiota (Hehemann et al., 2012). (B) Ulva degradation PUL observed in Nonlabens ulvanivorans isolated from the feces of Aplysia sp. (Kopel et al., 2014). (C) Carrageenan PUL in Cellulophaga algicola. The sulfatases found in this PUL have strong homologies with the biochemically characterized sulfatase of Pseudolateromonas atlantica T6c (Préchoux et al., 2013, 2016). The function of the biochemically characterized enzymes are indicated on their corresponding genes.
Biochemically Characterized Marine Polysaccharide Sulfatases
Agaran and Carrageenan Sulfatases
Agarans and carrageenans are found in the cell wall of many red algae (Rhodophyta). They are sulfated galactans made of D-galactose alternately linked by α- and β- glycosidic bonds. In agarans, the α-linked galactose belongs to the L series. These polysaccharides are classified according the position and number of sulfate ester groups and the occurrence of α-anhydrogalactose. The best-known agaran is agarose, appreciated for its gelling properties. Other agarans, such as the less-studied porphyran or funoran, are found in many species of red algae. The diversity of carrageenans is very high, based on the high number of different repetition moieties that have been reported; however, κ- (kappa-), ι- (iota-), hybrid κ-/ι- (also called κ2-carrageenan), and λ- (lambda-) carrageenans are the most frequent marine polysaccharides in industry, employed as gelling, or thickening ingredients (Usov, 2011).
Enzymatic degradation of red algal polysaccharides by glycoside hydrolases which cleave the glycosidic bonds have been observed in a wide diversity of marine bacteria (Michel et al., 2006). Porphyranases have also been found in the genome in human gut microbiota Bacteroidetes strains. Surprisingly, agaran- and carrageenan-degrading enzymes (i.e., agarases, porphyranases, or carrageenases) are not predicted from the genome sequence analysis of agarophyte and carrageenophyte algae (Bhattacharya et al., 2013; Collén et al., 2013).
Despite the high number of described agaran- and carrageenan-depolymerizing enzymes, only a few sulfatases have been biochemically characterized. They are all observed in marine bacteria and as for agarases and carrageenases, no sulfatases have been predicted from the analysis of red algae genomes (Ho, 2015).
The pioneering work of Weigl and Yaphe (1966) demonstrated the first carrageenan sulfatase activity in protein extracts prepared from the marine bacterium P. carrageenovora. Ten years later, this enzyme—a 4O-κ-carrabiose sulfatase of about 55 kDa—was purified and NMR revealed an exo-type mode of action. The sulfate ester group located at the non-reducing end of oligo-κ-carrageenans produced by P. carrageenovora κ-carrageenase are specifically eliminated, suggesting that the sulfatase intervenes after enzymatic depolymerization of κ-carrageenan (McLean and Williamson, 1979, 1981).
More recently, a similar approach was undertaken with the carrageenolytic strain Pseudoalteromonas atlantica T6c whose genome has been sequenced (Copeland et al., 2006). An endo-ι-carrageenan-sulfatase active on ι-carrageenan was isolated and biochemically characterized. The enzymes catalyzes the specific removal of the sulfate at position 4 of the β-linked galactose resulting in the conversion of ι-carrabiose into α- (alpha-) carrabiose repetition units (Figure 5) (Préchoux et al., 2013; Préchoux and Helbert, 2014). The gene coding for the sulfatase has been cloned and expressed recombinantly in Escherichia coli. These experiments demonstrate that the endo-4O-ι-carrageenan-sulfatase is a type I sulfatase, a formylglycine-dependent sulfatase.
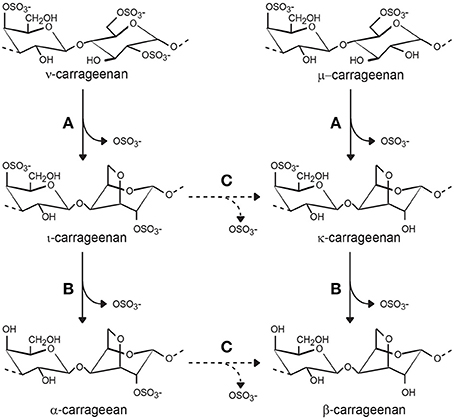
Figure 5. Carrageenan sulfatases observed during biosynthesis and biodegradation of carrageenan. (A) Sulfatases (named galactose-6-sulfurylases) catalyze the conversion of ν- (nu) and μ-(mu-) carrabiose into ι-(iota-) and κ-(kappa-)-carrabioses, respectively. (B) Several endo- and exo-sulfatases have been biochemically characterized. They all catalyze the removal of the sulfate ester group at position 4 (solid line arrows). (C) Desulfation of the group located at position 2 is hypothetical, but necessary for complete degradation of carrageenan (dashed arrows).
Another carrageenan-sulfatase was isolated from P. atlantica T6c, but it catalyzes the desulfation of κ-carrabiose, resulting in a neutral β-carrabiose repetition moiety (Figure 5). Overexpression of this endo-4O-κ-carrageenan sulfatase in E. coli also validated its grouping in the type I sulfatase family (Préchoux et al., 2016). Interestingly, in contrast to the first carrageenan sulfatase investigations, these studies revealed endo-acting carrageenan sulfatases that are efficient on polymers instead of oligosaccharides, suggesting that degradation of carrageenan may follow different pathways. One pathway involves the depolymerization of carrageenan prior to desulfation of oligo-carrageenans and another pathway supposes the desulfation of the polysaccharides before the depolymerization of α- or β-carrageenans by yet-to-be-discovered α- and β-carrageenases.
Desulfation of agars has also been reported in several marine bacteria strains: P. carrageenovora (Kim et al., 2005), Pyrococcus furiosus (Jung et al., 2012), Thermotoga marina (Lee et al., 2013), Marinomonas sp. FW-1 (Wang et al., 2015b), and Flammeovirga pacifica (Gao et al., 2015). In all cases, the modalities of desulfation were not demonstrated, notably, the position of the eliminated sulfate ester group. Heterologous expression of T. marina sulfatase (Lee et al., 2013) demonstrates that it belongs to the type I family. Interestingly, P. carrageenonvora sulfatase has homology with type III β-lactamase-like sulfatase. The gene was originally identified in neighboring genes involved in κ-carrageenan degradation, suggesting potential involvement in sulfated galactan metabolism (Barbeyron et al., 1995).
The galactose-6-sulfurylases, which intervene at the last step in the biosynthesis of agarans and carrageenans, catalyze the removal of sulfate ester groups at position 6 in galactose. The hydroxyl group in position 3 of the galactose residue attacks the carbon, resulting in the loss of the sulfate group. This nucleophilic substitution leads to the inversion of the carbon configuration, the formation of 3,6 anhydrogalactose and inversion of the 4C1 chair conformation to 1C4 (Figure 5). In this reaction, the molecule of water required for hydrolysis is replaced by an endogenous hydroxyl group; therefore, the term galactose-6-sulfurylase seems inappropriate (Usov, 2011).
The first evidence for the formation of an anhydro-ring was obtained by Rees (1961a,b) using protein extracts from Porphyra sp. incubated on porphyran purified from the same algae. Similar approaches conducted with carrageenophyte algae also lead to the formation of an anhydro-ring in κ- and ι-carrageenan (Wong and Craigie, 1978; Zinoun et al., 1997; Genicot-Joncour et al., 2009). The pure enzyme was sequenced and expression of the corresponding gene was attempted in E. coli without success. The function of the gene could not be validated experimentally, but its occurrence in recently sequenced red algal genomes confirm its algal origin and may explain the difficulty of expressing a eukaryotic protein in a prokaryotic cell (Collén et al., 2013).
Fucoidan Sulfatases and Others
Fucoidans represent a class of sulfated polysaccharides composed mainly of α-L-fucose found in the cell walls of brown algae and in marine invertebrates (i.e., worms, sea urchins). The structure of fucoidans depends on their biological origin and differs among algal species. Fucoidans are co-products of the alginate industry, which make them attractive as bioactive compounds and several potential applications have been examined (Berteau and Mulloy, 2003; Pomin and Mourão, 2008). Several marine strains have been shown to produce the enzymatic arsenal to degrade brown algal fucoidans (Michel and Czjzek, 2013). Fucanolytic enzymes include glycoside hydrolases and sulfatases. Gene sequences of fucanases have been validated by overexpression of active enzymes (Colin et al., 2006). However, although fucoidan sulfatase activity has been demonstrated in several bacterial strains, its sequence is still unknown and as for agarases and carrageenases in red algae, no sulfatase genes are predicted in brown algal genome (Cock et al., 2010). Sulfatases active on polymeric substrates or specific to oligosaccharide chain ends have been reported, hinting at the unexplored diversity of fucoidan sulfatases (Daniel et al., 2001).
Desulfation of marine sulfated glycosylated metabolites is grossly unexplored: one report demonstrates a 4O-xylose sulfatase in a protein extract of the liver of the marine mollusk Littorina kurila (Kusaykin et al., 2006). The enzyme is active on the sulfated xylose residue of holostan triterpene glycoside (Kusaykin et al., 2006).
Outlook
Sequencing of marine life has shed light on the diversity of carbohydrate sulfatases. The modularity of sulfatases and the genome organization of Bacteroidetes (i.e., PUL organization) have the potential to drive future functional characterization of the large amount of sulfatase sequences. Ultimately, the comprehensive classification of sulfatases based on sequence homology, allowing prediction of function is a challenging task for future work, calling for bioinformatics analyses (Wegner et al., 2013; Barbeyron et al., 2016) and crystallographic description of catalytic sites to identify the key amino acids involved in substrate recognition.
Functional characterization can benefit from genetic tools, such as gene mutation, to inactivate encoded proteins, and from more classic approaches, such as biochemical analyses of the enzymes purified from the organism in which they have been detected or heterologous expression in E. coli. However, only very few carbohydrate sulfatases have been biochemically characterized, underlining the difficulties in preparing them to purity. Recombinant marine bacterial sulfatases have been obtained, but always with a low level of activity. This low activity can be explained by the low rate of post-translational maturation of sulfatases in classic proteins expression systems that are naturally unable to convert cysteine and serine into formylglycine amino acid. Co-expression of sulfatases with known maturating enzymes (i.e., FGE, AnSME) can result in the production of a low amount of active enzymes, but better knowledge of the sulfatase maturating system—which probably involves several yet-to-be-discovered proteins—is a bottleneck to the production of active recombinant sulfatases.
The diversity of sulfatases lies in the diversity of sulfated polysaccharides present in the oceans. The composition and structure are simply unknown for most of them. Therefore, a comprehensive analysis of sulfatase function requires, in parallel, novel data on marine polysaccharides. Macro- and micro-algae, the primary producers of the oceans, harbor an unexplored and massive source of sulfated polysaccharides that contain both carbon and inorganic sulfate ions. Deciphering the turnover of marine polysaccharides involving sulfatases and other enzyme partners, will enhance our molecular understanding of the biogeochemistry of the ocean.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the French National Research Agency (ANR) under the national research grant ANR-14-CE05-0043.
References
Anderson, K. L., and Salyers, A. A. (1989). Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J. Bacteriol. 171, 3192–3198. doi: 10.1128/jb.171.6.3192-3198.1989
Appel, M. J., and Bertozzi, C. R. (2015). Formylglycine, a post-transitionally generated residue with unique catalytic capabilities and biotechnology applications. ACS Chem. Biol. 10, 72–84. doi: 10.1021/cb500897w
Aquino, R. S., Landeira-Fernandez, A. M., Valente, A. P., Andrade, L. R., and Mourão, P. A. (2005). Occurrence of sulfated galactans in marine angiosperms: evolutionary implications. Glycobiology 15, 11–20. doi: 10.1093/glycob/cwh138
Barbeyron, T., Brillet-Guéguen, L., Carré, W., Carrière, C., Caron, C., Czjzek, M., et al. (2016). Matching the diversity of sulfated biomolecules: creation of a classification database for sulfatases reflecting their substrate specificity. PLoS ONE 11:e0164846. doi: 10.1371/journal.pone.0164846
Barbeyron, T., Potin, P., Richard, C., Collin, O., and Kloareg, B. (1995). Arylsulphatase from Alteromonas carrageenovora. Microbiology 141, 2897–2904. doi: 10.1099/13500872-141-11-2897
Berteau, O., Guillot, A., Benjdia, A., and Rabot, S. (2006). A new type of bacterial sulfatase reveals a novel maturation pathway in prokaryotes. J. Biol. Chem. 281, 22464–22470. doi: 10.1074/jbc.m602504200
Berteau, O., and Mulloy, B. (2003). Sulfated fucans, fresh perspectives: structures, functions, and biological properties of sulfated fucans and an overview of enzymes active toward this class of polysaccharide. Glycobiology 13, 29–40. doi: 10.1093/glycob/cwg058
Bhattacharya, D., Price, D. C., Chan, C. X., Qiu, H., Rose, N., Ball, S., et al. (2013). Genome of the red alga Porphyridium purpureum. Nat. Commun. 4:1941. doi: 10.1038/ncomms2931
Bochenek, M., Etherington, G. J., Koprivova, A., Mugford, S. T., Bell, T. G., Malin, G., et al. (2013). Transcriptomic analysis of the sulfate deficiency response in the marine microalga Emiliania huxleyi. New Phytol. 199, 650–662. doi: 10.1111/nph.12303
Bojarová, P., and Williams, S. J. (2008). Sulfotransferases, sulfatases and formylglycine-generating enzymes: a sulfation fascination. Curr. Opin. Chem. Biol. 12, 573–581. doi: 10.1016/j.cbpa.2008.06.018
Bond, C. S., Clements, P. R., Ashby, S. J., Collyer, C. A., Harrop, S. J., Hopwood, J. J., et al. (1997). Structure of a human lysosomal sulfatase. Structure 5, 277–289. doi: 10.1016/s0969-2126(97)00185-8
Cock, J. M., Sterck, L., Rouzé, P., Scornet, D., Allen, A. E., Amoutzias, G., et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465, 617–621. doi: 10.1038/nature09016
Colin, S., Deniaud, E., Jam, M., Descamps, V., Chevolot, Y., Kervarec, N., et al. (2006). Cloning and biochemical characterization of the fucanase FcnA: definition of a novel glycoside hydrolase family specific for sulfated fucans. Glycobiology 16, 1021–1032. doi: 10.1093/glycob/cwl029
Collén, J., Porcel, B., Carré, W., Ball, S. G., Chaparro, C., Tonon, T., et al. (2013). Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on the evolution of the Archaeplastida. Proc. Natl. Acad. Sci. U.S.A. 110, 5247–5252. doi: 10.1073/pnas.1221259110
Copeland, A., Lucas, S., Lapidus, A., Barry, K., Detter, J. C., Glavina del Rio, T., et al. (2006). Complete Sequence of Pseudoalteromonas Atlantica T6c. Walnut Creek, CA: US DOE Joint Genome Institute.
Cosma, M. P., Pepe, S., Annunziata, I., Newbold, R. F., Grompe, M., Parenti, G., et al. (2003). The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell 113, 445–456. doi: 10.1016/s0092-8674(03)00348-9
Daniel, R., Berteau, O., Chevolot, L., Varenne, A., Gareil, P., and Goasdoue, N. (2001). Regioselective desulfation of sulfated L-fucopyranoside by a new sulfoesterase from the marine mollusk Pecten maximus - application to the structural study of algal fucoidan (Ascophyllum nodosum). Eur. J. Biochem. 268, 5617–5626. doi: 10.1046/j.1432-1033.2001.02497.x
Despres, J., Forano, E., Lepercq, P., Comtet-Marre, S., Jubelin, G., Yeoman, C. J., et al. (2016). Unraveling the pectinolytic function of Bacteroides xylanisolvens using a RNA-seq approach and mutagenesis. BMC Genomics 17:147. doi: 10.1186/s12864-016-2472-1
Dierks, T., Schmidt, B., Borissenko, L. V., Peng, J., Preusser, A., Mariappan, M., et al. (2003). Multiple sulfatase deficiency is caused by mutations in the gene encoding the human Cα-formylglycine generating enzyme. Cell 113, 435–444. doi: 10.1016/s0092-8674(03)00347-7
Flint, H. J., Bayer, E. A., Rincon, M. T., Lamed, R., and White, B. A. (2008). Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6, 121–131. doi: 10.1038/nrmicro1817
Friedlander, M. (2001). Inorganic nutrition in pond cultivated Gracilaria conferta (Rhodophyta): nitrogen, phosphate and sulfate. J. Appl. Phycol. 13, 278–296. doi: 10.1023/A:1011139329415
Gao, C., Jin, M., Yi, Z., and Zeng, R. (2015). Characterization of a recombinant thermostable arylsulfatase from deep-sea bacterium Flammeovirga pacifica. J. Microbiol. Biotech. 25, 1894–1901. doi: 10.4014/jmb.1504.04028
Genicot, S. M., Groisillier, A., Rogniaux, H., Meslet-Cladière, L., Barbeyron, T., and Helbert, W. (2014). Discovery of a novel iota-carrageenan sulfatase isolated from the marine bacterium Pseudoalteromonas carrageenovora. Front. Chem. 2:67. doi: 10.3389/fchem.2014.00067
Genicot-Joncour, S., Poinas, A., Richard, O., Potin, P., Rudolph, B., Kloareg, B., et al. (2009). The cyclization of the 3,6-anhydro ring of iota-carrageenan is catalyzed by two D-galactose-2,6-sulfurylases in the red alga Chondrus crispus. Plant Physiol. 151, 1609–1616. doi: 10.1104/pp.109.144329
Glöckner, F. O., Kube, M., Bauer, M., Teeling, H., Lombardot, T., Ludwig, W., et al. (2003). Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Nat. Acad. Sci. U.S.A. 100, 8298–8303. doi: 10.1073/pnas.1431443100
Goldman, P. J., Grove, T. L., Sites, L. A., McLaughlin, M. I., Booker, S. J., and Drennan, C. L. (2013). X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc. Natl. Acad. Sci. U.S.A. 110, 8519–8524. doi: 10.1073/pnas.1302417110
Gómez-Pereira, P. R., Schüler, M., Fuchs, B. M., Bennke, C., Teeling, H., Waldmann, J., et al. (2012). Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environ. Microbiol. 14, 52–66. doi: 10.1111/j.1462-2920.2011.02555.x
Gügi, B., Le Costaouec, T., Burel, C., Lerouge, P., Helbert, W., and Bardor, M. (2015). Oligosaccharides and polysaccharides structures in diatoms unravel their biosynthetic capabilities. Mar. Drugs 13, 5993–6018. doi: 10.3390/md13095993
Hagelueken, G., Adams, T. M., Wiehlmann, L., Widow, U., Kolmar, H., Tümmler, B., et al. (2006). The crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, defines a third class of sulfatases. Proc. Nat. Acad. Sci. U.S.A. 103, 7631–7636. doi: 10.1073/pnas.0510501103
Hahnke, R. L., Stackebrandt, E., Meier-Kolthoff, J. P., Tindall, B. J., Huang, S., Rohde, M., et al. (2015). High quality draft genome sequence of Flavobacterium rivuli type strain WB 3.3-2T (DSM 21788T), a valuable source of polysaccharide decomposing enzymes. Stand. Genomic Sci. 10, 46 doi: 10.1186/s40793-015-0032-y
Han, W., Wang, W., Zhao, M., Sugahara, K., Li, F. (2014). A novel eliminase from a marine bacterium that degrades hyaluronan and chondroitin sulfate. J. Biol. Chem. 289, 27886–27898. doi: 10.1074/jbc.M114.590752
Hanson, A. D., Pribat, A., Waller, J. C., and de Crécy-Lagard, V. (2010). “Unknown” proteins and “orphan” enzymes: the missing half of the engineering parts list - and how to find it. Biochem. J. 425, 1–11. doi: 10.1042/BJ20091328
Hanson, S. R., Best, M. D., and Wong, C.-H. (2004). Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew. Chem. Int. Ed. 43, 5736–5763. doi: 10.1002/anie.200300632
Hehemann, J. H., Correc, G., Barbeyron, T., Helbert, W., Czjzek, M., and Michel, G. (2010). Seaweed diet enabled transfer of CAZymes from marine bacteria to Japanese gut bacteria. Nature 464, 908–912. doi: 10.1038/nature08937
Hehemann, J. H., Kelly, A. G., Pudlo, N. A., Martens, E. C., and Boraston, A. B. (2012). Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc. Nat. Acad. Sci. U.S.A. 109, 19786–19791. doi: 10.1073/pnas.1211002109
Ho, C.-L. (2015). Phylogeny of algal sequences encoding carbohydrate sulfotransferase, formylglycine-dependent sulfatases, and putative sulfatase modifying factors. Front. Plant. Sci. 6:1057. doi: 10.3389/fpls.2015.01057
Hoagland, K. D., Rosowski, J. R., Gretz, M. R., and Roemer, S. C. (1993). Diatom extracellular polymeric substances: function, fine structure, chemistry, and physiology. J. Phycol. 29, 537–566. doi: 10.1111/j.0022-3646.1993.00537.x
Jung, K. T., Kim, H. W., You, D. J., Nam, S. W., Kim, B. W., and Jeon, S. J. (2012). Identification of the first archaeal arylsulfatase from Pyrococcus furiosus and its application to desulfatation of agar. biotech. Bioproc. Eng. 17, 1140–1146. doi: 10.1007/s12257-012-0228-6
Kabisch, A., Otto, A., König, S., Becher, D., Albrecht, D., Schüler, M., et al. (2014). Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes “Gramella forsetii” KT0803. ISME J. 8, 1492–1502. doi: 10.1038/ismej.2014.4
Kim, D. E., Kim, K. H., Bae, Y. J., Lee, J. H., Jang, Y. H., and Nam, S. W. (2005). Purification and characterization of the recombinant arylsulfatase cloned from Pseudoalteromonas carrageenovora. Prot. Expr. Purif. 39, 107–115. doi: 10.1016/j.pep.2004.09.007
Klaassen, C. D., and Boles, J. W. (1997). Sulfation and sulfotransferases.5. The importance of 3'-phosphoadenosine 5'-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 11, 404–418.
Kloareg, B., and Quatrano, R. S. (1988). Structure of the cell-walls of marine-algae and ecophysiological functions of the matrix polysaccharides. Oceanogr. Mar. Biol. 26, 259–315.
Kopel, M., Helbert, W., Henrissat, B., Doniger, T., and Banin, E. (2014). Draft genome sequence of Nonlabens ulvanivorans, an ulvan degrading bacterium. Genome Announc. 2:e00793–14. doi: 10.1128/genomea.00793-14
Kusaykin, M. I., Pesentseva, M. S., Sil'chenko, A. S., Avilov, S. A., Sova, V. V., Zvyagintseva, T. N., et al. (2006). Aryl sulfatase of unusual specificity from the liver of marine mollusk Littorina kutila. Russ. J. Bioorg. Chem. 32, 63–70. doi: 10.1134/S1068162006010067
Lahaye, M., and Robic, A. (2007). Structure and functional properties of ulvan, a polysaccharide from green seaweeds. Biomacromolecules 8, 1765–1774. doi: 10.1021/bm061185q
Lee, D. G., Shin, J. G., Jeon, M. J., and Lee, S. H. (2013). Heterologous expression and characterization of a recombinant thermophilic arylsulfatase from Thermotoga maritima. Biotech. Bioproc. Eng. 18, 897–902. doi: 10.1007/s12257-013-0094-x
Lukatela, G., Krauss, N., Theis, K., Selmer, T., Gieselmann, V., von Figura, K., et al. (1998). Crystal structure of human arylsulfatase A: the aldehyde function and the metal ion at the active site suggest a novel mechanism for sulfate ester hydrolysis. Biochemistry 37, 3654𢀓3664. doi: 10.1021/bi9714924
Mancuso Nichols, C. A., Guezennec, J., and Bowman, J. P. (2005). Bacterial exopolysaccharides from extreme marine environments with special consideration of the southern ocean, sea ice, and deep-sea hydrothermal vents: a review. Mar. Biotech. 7, 253–271. doi: 10.1007/s10126-004-5118-2
Mann, A. J., Hahnke, R. L., Huang, S., Werner, J., Xing, P., Barbeyron, T., et al. (2013). the genome of the alga-associated marine Flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl. Environ. Microbiol. 79, 6813–6822. doi: 10.1128/AEM.01937-13
Martens, E. C., Chiang, H. C., and Gordon, J. I. (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457. doi: 10.1016/j.chom.2008.09.007
Martens, E. C., Lowe, E. C., Chiang, H., Pudlo, N. A., Wu, M., McNulty, N. P., et al. (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9:e1001221. doi: 10.1371/journal.pbio.1001221
McLean, M. W., and Williamson, F. B. (1979). Glycosulphatase from Pseudomonas carrageenovora. Purification and some properties. Eur. J. Biochem. 101, 497–505. doi: 10.1111/j.1432-1033.1979.tb19744.x
McLean, M. W., and Williamson, F. B. (1981). Neocarratetraose 4-O-monosulfate β-hydrolase from Pseudomonas carrageenovora. Eur. J. Biochem. 113, 447–456. doi: 10.1111/j.1432-1033.1981.tb05084.x
McNulty, N. P., Wu, M., Erickson, A. R., Pan, C., Erickson, B. K., Martens, E. C., et al. (2013). Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 11:e1001637. doi: 10.1371/journal.pbio.1001637
Michel, G., and Czjzek, M. (2013). “16. Polysaccharide-degrading enzymes from marine bacteria,” in Marine Enzymes for Biocatalysis: Sources, Biocatalytic Characteristics and Bioprocesses of Marine Enzymes, ed A. Trincone (Cambridge: Wood Publishing Series in Biomedicine Number 38), 429–464.
Michel, G., Nyvall-Collen, P., Barbeyron, T., Czjzek, M., and Helbert, W. (2006). Bioconversion of red seaweed galactans: a focus on bacterial agarases and carrageenases. Appl. Microbiol. Biotechnol. 71, 23–33. doi: 10.1007/s00253-006-0377-7
Müller, I., Kahnert, A., Pape, T., Sheldrick, G. M., Meyer-Klaucke, W., Dierks, T., et al. (2004). Crystal structure of the alkylsulfatase AtsK: Insights into the catalytic mechanism of the Fe(II) alpha-ketoglutarate-dependent dioxygenase superfamily. Biochemistry 43, 3075–3088. doi: 10.1021/bi035752v
Nazarenko, E. L., Crawford, R. J., and Ivanova, E. P. (2011). The structural diversity of carbohydrate antigens of selected gram-negative marine bacteria. Mar. Drugs 9, 1914–1954. doi: 10.3390/md9101914
Olsen, J. L., Rouzé, P., Verhelst, B., Lin, Y. C., Bayer, T., Collen, J., et al. (2016). The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530, 331–335. doi: 10.1038/nature16548
Panschin, I., Huang, S., Meier-Kolthoff, J. P., Tindall, B. J., Rohde, M., Verbarg, S., et al. (2016). Comparing polysaccharide decomposition between the type strains Gramella echinicola KMM 6050(T) (DSM 19838(T)) and Gramella portivictoriae UST040801-001(T) (DSM 23547(T)), and emended description of Gramella echinicola Nedashkovskaya et al. 2005 emend. Shahina et al. 2014 and Gramella portivictoriae Lau et al. (2005). Stand. Genomic Sci. 11, 37. doi: 10.1186/s40793-016-0163-9
Percival, E., and McDowell, R. H. (1967). “Polysaccharides containing uronic acid and ester sulfates,” in Chemistry and Enzymology of Marine Algal Polysaccharides (London: London Academic Press Inc. Ltd.), 186–189.
Pomin, V. H., and Mourão, P. A. (2008). Structure, biology, evolution, and medical importance of sulfated fucans and galactans. Glycobiology 18, 1016–1027. doi: 10.1093/glycob/cwn085
Préchoux, A., Genicot, S., Rogniaux, H., and Helbert, W. (2013). Controlling carrageenan structure using a formylglycine-dependent sulfatase, an endo-4S-iota-carrageenan sulfatase. Mar. Biotech. 15, 265–274. doi: 10.1007/s10126-012-9483-y
Préchoux, A., Genicot, S., Rogniaux, H., and Helbert, W. (2016). Enzyme-assisted preparation of furcellaran-like κ–/β-carrageenan. Mar. Biotech. 18, 133–143. doi: 10.1007/s10126-015-9675-3
Préchoux, A., and Helbert, W. (2014). Preparation and detailed NMR analyses of a series of oligo α-carrageenans. Carbohydr. Polym. 101, 864–870. doi: 10.1016/j.carbpol.2013.10.007
Rees, D. A. (1961a). Enzymatic desulphation of porphyran. Biochem. J. 80, 449–453. doi: 10.1042/bj0800449
Rees, D. A. (1961b). Enzymic synthesis of 3:6-anhydro-L-galactose within porphyran from L-galactose 6-sulphate units. Biochem. J. 81, 347–352. doi: 10.1042/bj0810347
Sardiello, M., Annunziata, I., Roma, G., and Ballabio, A. (2005). Sulfatases and sulfatase modifying factors: an exclusive and promiscuous relationship. Hum. Mol. Genet. 14, 3203–3217. doi: 10.1093/hmg/ddi351
Sonnenburg, E. D., Sonnenburg, J. L., Manchester, J. K., Hansen, E. E., Chiang, H. C., and Gordon, J. I. (2006). A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc. Nat. Acad. Sci. U.S.A. 103, 8834–8839. doi: 10.1073/pnas.0603249103
Sun, C., Fu, G. Y., Zhang, C. Y., Hu, J., Xu, L., Wang, R. J., et al. (2016). Isolation and complete genome sequence of Algibacter alginolytica sp. nov., a novel seaweed-degrading Bacteroidetes bacterium with diverse putative polysaccharide utilization loci. Appl. Environ. Microbiol. 82, 2975–2987. doi: 10.1128/AEM.00204-16
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Terrapon, N., Lombard, V., Gilbert, H. J., and Henrissat, B. (2015). Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics 31, 647–655. doi: 10.1093/bioinformatics/btu716
The UniProt Consortium (2015). UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212. doi: 10.1093/nar/gku989
Usov, A. I. (2011). “Polysaccharides of the Red Algae,” in Advance Carbohydrate Chemistry Biochemtry, Vol. 65, ed D. Horton (Burlington, NJ: Academic Press). 115–217.
von Bülow, R., Schmidt, B., Dierks, T., von Figura, K., and Usón, I. (2001). Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis. J. Mol. Biol. 305, 269–277. doi: 10.1006/jmbi.2000.4297
Wang, W. S., Han, W., Cai, X., Zheng, X., Sugahara, K., and Li, F. (2015a). Cloning and characterization of a novel chondroitin sulfate/dermatan sulfate 4-O-endosulfatase from a marine bacterium. J. Biol. Chem. 290, 7823–7832. doi: 10.1074/jbc.M114.629154
Wang, X., Duan, D., Xu, J. C., Gao, X., and Fu, X. (2015b). Characterization of a novel alkaline arylsulfatase from Marinomonas sp FW-1 and its application in the desulfation of red seaweed agar. J. Indus. Microbio. Biotech. 42, 1353–1362. doi: 10.1007/s10295-015-1625-6
Wegner, C. E., Richter-Heitmann, T., Klindworth, A., Klockow, C., Richter, M., Achstetter, T., et al. (2013). Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar. Genomics 9, 51–61. doi: 10.1016/j.margen.2012.12.001
Weigl, J., and Yaphe, W. (1966). Glycosulfatase of Pseudomonas carrageenovora: desulfation of disaccharide from κ- carrageenan. Can. J. Microbiol. 12, 874–876. doi: 10.1139/m66-118
White, B. A., Lamed, R., Bayer, E. A., and Flint, H. J. (2014). Biomass Utilization by Gut Microbiomes. Ann. Rev. Microbiol. 68, 279–296. doi: 10.1146/annurev-micro-092412-155618
Williams, S. J., Denehy, E., and Krenske, E. H. (2014). Experimental and theoretical insights into the mechanisms of sulfate and sulfamate ester hydrolysis and the end products of type i sulfatase inactivation by aryl sulfamates. J. Org. Chem. 79, 1995–2005. doi: 10.1021/jo4026513
Willis, A., Chiovitti, A., Dugdale, T. M., and Wetherbee, R. (2013). Characterization of the extracellular matrix of Phaeodactylum tricornutum (Bacillariophyceae): structure, composition, and adhesive characteristics. J. Phycol. 49, 937–949. doi: 10.1111/jpy.12103
Wong, K. F., and Craigie, J. S. (1978). Sulfohydrolase activity and carrageenan biosynthesis in Chondrus crispus (Rhodophyceae). Plant Physiol. 61, 663–666. doi: 10.1104/pp.61.4.663
Wright, O., and Colling, A. (1995). “Chapter 3 - Salinity in the oceans,” in Seawater: its Composition, Properties and Behaviour, 2nd Edn., ed G. Bearman (Pergamon: ISBN 9780080425184), 29–38.
Xing, P., Hahnke, R. L., Unfried, F., Markert, S., Huang, S., Barbeyron, T., et al. (2015). Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J. 9, 1410–1422. doi: 10.1038/ismej.2014.225
Zhang, H., Varmalova, O., Vargas, F. M., Falany, C. N., and Leyh, T. S. (1998). Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. J. Biol. Chem. 273, 10888–10892. doi: 10.1074/jbc.273.18.10888
Keywords: sulfatases, polysaccharides, polysaccharide utilization loci, module, carrageenan, agars, genomic
Citation: Helbert W (2017) Marine Polysaccharide Sulfatases. Front. Mar. Sci. 4:6. doi: 10.3389/fmars.2017.00006
Received: 18 October 2016; Accepted: 09 January 2017;
Published: 26 January 2017.
Edited by:
Antonio Trincone, Istituto di Chimica Biomolecolare (CNR), ItalyReviewed by:
Irina Bakunina, G.B. Elyakov Pacific Institute of Bioorganic Chemistry (FEB RAS), RussiaVitor Hugo Pomin, Federal University of Rio de Janeiro, Brazil
Copyright © 2017 Helbert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: William Helbert, d2lsbGlhbS5oZWxiZXJ0QGNlcm1hdi5jbnJzLmZy
 William Helbert
William Helbert