
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 06 January 2017
Sec. Marine Megafauna
Volume 3 - 2016 | https://doi.org/10.3389/fmars.2016.00272
In the US, marine mammals are protected by the Endangered Species Act (ESA) and the Marine Mammal Protection Act (MMPA). Most of these species are listed by Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) and thus international trade in their products is restricted. Therefore, commercial sale of unfossilized marine mammal bone is largely prohibited. Sale of Steller's sea cow (Hydrodamalis gigas) bone is legal, however, since the animals have been extinct since 1768. The current study outlines a simple test that can identify bone which is bona fide Steller's sea cow—and thus legal to sell. The test uses a segment of the D-loop of the mitochondrion, which has the power to exclude samples which are not specifically H. gigas or a Sirenian relative. The test also includes a reliable method to extract DNA from bone and amplify it using Polymerase Chain Reaction (PCR). Extracted DNA was sequenced to verify that only manatees, dugongs, elephants, and their relatives produced a positive result. Using this test, products being sold commercially as legal “mermaid ivory” (Steller sea cow bone) were found to actually come from gray whale (Eschrichtius robustus), pantropical spotted dolphin (Stenella attenuata), and white-beaked dolphin (Lagenorhynchus albirostris) bone. This finding indicates that government agencies should monitor bones being sold as “mermaid ivory” because protected species are being illegally traded under the guise of being legal Steller's sea cow bone.
With many animal species threatened by illegal trade in wildlife products, it is important to be able to distinguish between products that are legal to sell and those that are not. All of the marine mammals in the US are protected by the Endangered Species Act (ESA) of 1973 and the Marine Mammal Protection Act (MMPA) of 1972 which have provisions related to controlling trade of marine mammal products. The Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) of 1973 further governs and technically limits the international trade in these products. Sale of unfossilized bone from marine mammals is generally prohibited. There is an exception that allows extinct marine mammal bone, often worked into products by Alaskan native artisans, to be sold in the US. These carved products are either sold directly to visitors to St. Lawrence Island or resold by dealers. One such dealer is Alaskan Ivory Tower, which works with customers through Ebay. The sale of Steller's sea cow (Hydrodamalis gigas) bone is legal because the animals were driven to extinction in 1768 via over-exploitation by Russian fur hunters (Stejneger, 1887; Domning, 1978).
In recent years, there has been an active trade in bone samples advertised as Steller's sea cow from St. Lawrence Island, Alaska (63°3′ North 173°2′ West). These bones are sometimes sold under the epithet “mermaid ivory.” There does not seem to be regulation of this trade by authorities. The raw bones are sold to dealers who come to St. Lawrence Island. The Alaskan Native Claims Settlement Act (ANCSA) of 1971 gave the Yupik title to the land and the legal right to sell ivory and other artifacts from the island (43 USC Ch. 33: ALASKA NATIVE CLAIMS SETTLEMENT). Due to the fact that H. gigas bone is pachyosteosclerotic (formed entirely of compact rather than cancellous bone), this material is prized among artisans who make decorative knife handles and carved pieces. Thus, most of the bone fragments coming from St. Lawrence Island are not large enough to positively identify them to species by their morphology. Regulation of this trade is really necessary as some of the bone being sold from the island is not legal to sell. We have developed a simple, successful test that allows for differentiation of Steller's sea cow bone, which is legal to sell, from that of other North Pacific marine mammals.
In order to determine whether bone being sold as Steller's sea cow was actually from a sea cow, 158 bone pieces of carved statuettes were collected from several sources. The vendors of this bone can be found on the Internet, on Ebay, and at large knife shows around the US.
The verification test used a segment of the displacement loop (D-loop) of the mitochondrion that has the power to exclude samples that are not H. gigas or a tethytherian relative. Tethytherians include dugongs (Dugong dugon), manatees (Trichechus spp.), elephants (Elephas maximus and Loxodonta spp.), mammoths (Mammuthus primigenius), and mastodons (Mammut americanum). Since all other extant Sirenians are warm-water species, it was hypothesized that bone from St. Lawrence Island yielding a positive result most likely would be either Steller's sea cow, mammoth, or mastodon bone. Since mammoth and mastodon bones are truly ancient, DNA extraction from these bones is more difficult and unlikely to work using this method, reducing the likelihood of a positive identification via this test. For the unknown samples from St. Lawrence Island, there was the greatest sequence homology with the initial reference sequence for the Steller's sea cow deposited by Ozawa et al. (1997). While there was sequence homology between the unknown samples and the mammoth and mastodon reference sequences, it was much lower than even homology of the unknowns with manatee and dugong reference sequences.
Three fragments of mitochondrial DNA (mtDNA) were extracted, amplified using Polymerase Chain Reaction (PCR) and sequenced. The first two pieces of mtDNA are segments of the D-loop and the third is a small segment of the cytochrome b gene. None of these fragments overlap. It was hypothesized that only the sea cow, mammoth or mastodon would produce a positive result in this test. Sequencing the DNA would identify the species. This test uses simple electrophoresis and agarose gel visualization to determine whether a bone was H. gigas and the test does not require DNA sequencing.
In order to verify the identity of bones sold as Steller's sea cow, over 150 bones or pieces of bone were collected (LDC). Some bones were carved into decorative statuettes by artisans in Alaska (Figure 1). The bone samples were purported to come from St. Lawrence Island, which was verified for a sub-set of samples via referral to shipping manifests. Radioisotope analysis identified the St. Lawrence bone as coming from a population distinct from Bering Island samples (Crerar et al., 2014). A sample with provenance as coming from Bering Island was used as a positive control for the test.
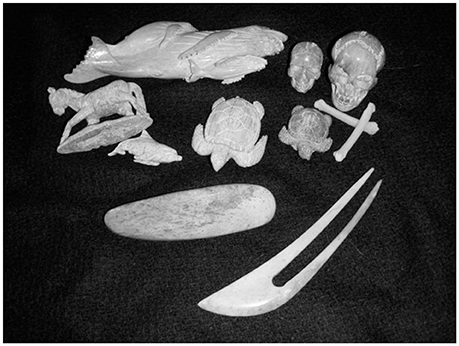
Figure 1. Examples of samples obtained from Alaska Ivory Tower, Fairbanks, AK (photo by Lorelei Crerar).
Samples include 11 carved pieces that were purchased from Alaska Ivory Tower (Fairbanks, AK) (Figure 1). Most of the remaining samples (N = 147) were purchased from David Boone at Boone Trading Company (Brinnon, WA 98320) (N = 51), Jerry Kochheiser at Alaskan Fossil Ivory (Mansfield, OH 44904) (N = 46), Meriam Linder at Converging Traditions (N = 7), Yinan Wang (N = 6), and Damian O'Halloran (N = 1). Dealers bought bone in the town of Gambell on St. Lawrence Island, AK (63.7° N, 171.7° W) from local people who collect the bone from middens located within 10 km of Gambell and the dates of visits were documented using airline receipts. A verified H. gigas rib was loaned from the National Museum of Natural History (Washington, DC) (USNM#593920) and there were 24 other samples extracted from shards in the drawers of various specimens at the US NMNH. Extraction was also attempted on 11 samples of mammoth bone from Webb Knives and Jewelry (Seabeck, WA) but no DNA was retrieved from these samples. Manatee blood used as a positive control was provided by Dr. Bob Bonde at the U.S. Geological Survey Sirenia Project (Gainesville, FL). DNA was also extracted from a known blue whale (Balaenoptera musculus) (USNM#124326) for comparison. At this point in time, all of these presumptive sea cow samples have been sequenced for several sections of the DNA, multiple times (Table 1).
All of the bone samples were cleaned using 30% sodium hypochlorite as recommended by Rohland and Hofreiter (2007). After cleaning and drying, the samples were drilled with a Proxxon Micromot 50/E and 2.78 mm tungsten vanadium drilling bits (No. 28722). Drilling the bone produces a fine powder suited to DNA extraction. After collecting each sample, the lab bench was cleaned with 30% sodium hypochlorite and the fume hood was cleaned between each extraction or PCR session.
Care was always taken to eliminate cross contamination between samples. In order to prevent contamination, DNA extraction of bone and blood were carried out on different days and the extraction area was always carefully cleaned before DNA extraction. Extraction blanks for the DNA extractions never produced a positive result after PCR. DNA was also extracted in a clean fume hood. All work was carried out in a lab that does not process DNA.
The method for DNA extraction was a modification of a standard protocol for the Qiagen DNeasy Kit (Product Number 69504). Samples over 100 g were placed into a 50 mL Falcon conical tube with 40 mL of 0.5 M EDTA at pH 8.0 in a 37°C rocking water bath. Samples under 100 g were extracted in 2 mL tubes using 1 mL of 0.5 M EDTA at pH 8.0 in a 37°C rocking water bath. After 3 days, the samples were washed with HPLC water and 50 mg of the resulting pellet was removed from the tube and weighed on a Mettler Toledo NewClassic MF balance (MS 4035) with a disposable spatula (USA Scientific 9101-7211) and then placed into a new 2 mL tube. To digest the remaining proteins, 40 μL proteinase K was added to the pellet with 360 μL of Qiagen ATL tissue lysis buffer (Product Number 939016). This mixture was returned to a 56°C water bath for 3 h. The rest of the DNA extraction protocol followed the user developed protocol for extracting DNA from bones and teeth published by Qiagen for the DNeasy Kit.
Primers from Ozawa et al. (1997) were used to obtain the cytochrome b gene sequence. The primers extract a segment of the cytochrome b gene that is 230 bp in length. The sequences for these primers are:
fragment 3: L15306 5′-CGATTCTTCGCTTTCCACTTCATCCTACCATT-3′
H15494 5′-TAGTTGTCCGGGTCTCCTAG-3′
The D-loop primers used in the study were created using Lasergene® PrimerSelect™(DNA Star). A consensus sequence was created using published sequences from a manatee (Trichechus manatus latirostris; GenBank accession number 59799930) and a dugong (Dugong dugon; GenBank accession number 17981678). This sequence was then used to create the D-loop consensus primers:
Consensus_Dloop1_1: 5′-CCTGGCATCTGGTTCTTT-3′
Consensus_Dloop1_2: 5′-CGGCCTAGTTGAGTCGKT-3′
Consensus_Dloop2_1: 5′-ATCCTGTACTTCTCCATCATCCTC-3′
Consensus_Dloop2_2: 5′-AGTAGAATTTCAGCTTTGGGTGTT-3′
Primers were ordered from Invitrogen. The Qiagen TopTaq DNA Polymerase Kit (Product Number 200201) was used to amplify DNA. The mixture used a 10 mM primer concentration in a 20 μL reaction mixture. The reaction contained 6 μL TopTaq buffer (Qiagen #200201), 0.5 μL dNTPs (Qiagen #201900), 1 μL of the forward and reverse primers, 8.375 μL of HPLC grade water, and 0.125 μL of TopTaq polymerase (Qiagen #200201).
DNA was amplified using an initial heating step of 95°C for 5 min to activate the polymerase, a denaturation step of 94°C for 30 s, an annealing step of 57–53°C for 1 min, an extension step at 72°C for 1 min and a repetition of this program for 40 cycles. An additional extension step at 72°C for 10 min was added. This program was run on a PTC-200 Thermocycler.
After amplification, DNA was visualized on a GelPilot Small Fragment 3% agarose gel (Qiagen Product Number 129832). A Gel Pilot 100 bp ladder (Qiagen Product Number 239035) was used to monitor the size of the fragments. Manatee blood was used as a positive control and for each PCR reaction a negative control was used. If the negative controls produced a product, the reaction was discarded and run again.
The DNA was cleaned up using ExoSAP-IT® (USB® Product number 78200/01/05/50). A reaction mixture of 5 μL of DNA and 2 μL of ExoSAP-IT® was heated to 37°C for 60 min to dephosphorylate the DNA and then heated to 80°C for 20 min to inactivate the enzyme.
The DNA was sent to Eton Bioscience, Inc. (104 T.W. Alexander Drive, Building 7, Research Triangle Park, NC 27709) in 0.2 mL strip PCR tubes. DNA samples were sequenced using ABI automated sequencers and compared to H. gigas voucher sequences in the GenBank database (Ozawa et al., 1997) using DNA Baser™.
The Dloop_2 segment of DNA was sufficient to determine identity of the samples. The only samples that provided a bright band on the agarose gel were either confirmed or presumed Hydrodamalis gigas or a tethytherian relative. Samples that were not sirenian did not produce a positive result on the gels. A bone from Bering Island was used as positive identification for the DNA. When the proposed sea cow fragments were sequenced, a 100% match for H. gigas was obtained from GenBank. Figure 2 shows an example of an electrophoresis gel with positive results for known and presumed H. gigas samples and a 100 bp ladder running in the extreme left lane. The known H. gigas DNA was extracted from a rib housed in the USNM (#593920) collection. The final three samples were all taken from that same rib at different locations on the bone. The electrophoresis gel shown in Figure 3 demonstrates negative results arising from samples that were not H. gigas. The sample in lane 2 was extracted from bone known to be from the blue whale for comparative purposes. The sample in lane 4 (a negative result) was sold as H. gigas, but when the DNA sequence was uploaded into the BLAST database, it was identified as coming from a white-beaked dolphin (Lagenorhynchus albirostris). The test demonstrated here that the white-beaked dolphin caused a negative result in the gel.
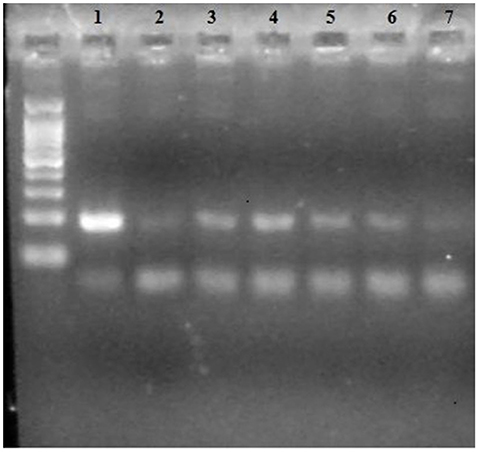
Figure 2. Electrophoresis gel of samples. Lane 1: T. m. latirostris blood (positive control); Lane 2: Dugong dugon (USNM#28495); Lanes 3 and 4: H. gigas from St. Lawrence Island (GMU#00003 and 00004); Lanes 5, 6, 7: known H. gigas DNA from three different areas of a rib (USNM#593920).
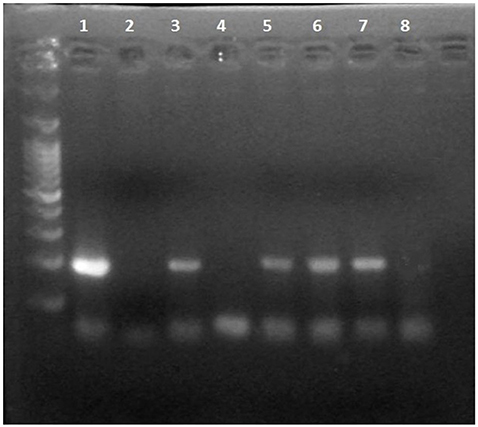
Figure 3. Electrophoresis gel of samples. Lane 1: T. m. latirostris blood (positive control); Lane 2: blue whale (USNM# 124326); Lane 3: elephant (USNM#20922); Lane 4: GMU#00006; Lane 5 and 6: elephant (USNM#49489 and 16323); Lane 7: dugong (USNM#28495); and Lane 8 is a PCR negative.
Sequencing data were used to create a multiple alignment in Sequencher® 5.1 (Gene Codes Corporation in Ann Arbor, MI). Many DNA sequences from the bones were an exact match for each other. This was expected from such a small section of DNA. Only six of the bones have been examined using radiocarbon data to suggest an origin for the bones other than Bering Island (Crerar et al., 2014). Four more samples were examined in 2016 and were all consistent with having come from St. Lawrence Island (unpublished). As the sea cow is a large animal, it is possible that some of the samples could have come from the same animal. Work to identify the number of individuals in the current sample is on-going. GenBank reference D-loop sequences for the Amazonian manatee (T. inunguis) and dugong (D. dugon) were used to identify unknown samples. An example of a match between sample USNM#593920 (Sbjct 716) and the reference sequence for H. gigas is shown in Figure 4. The diagram seen in Figure 4 shows the sequence homology between the submitted sequence, from our sample, and the sequence deposited in GenBank for a sea cow. Missing vertical lines indicate a mismatch in the sequence. Sequencing confirmed that there were samples from a gray whale (Eschrichtius robustus) (Figure 5), a pantropical spotted dolphin (Stenella attenuata) (Data not shown), and a white-beaked dolphin (L. albirostris) (Data not shown) which were sold as H. gigas. It can easily be seen in Figure 5 that our sample was a 95% match with the reference sequence for the gray whale. A list of the samples and their identification is summarized in Table 1.

Figure 4. Data from GenBank for USNM#593920 sequence BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
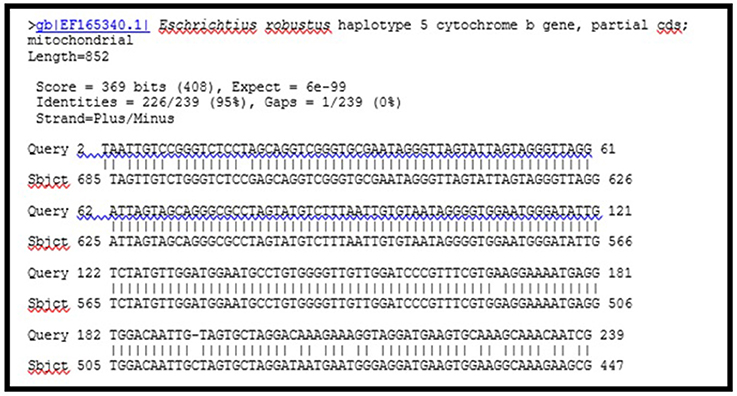
Figure 5. Data from GenBank for GMU#00006 sequence BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Electrophoresis gels visualizing DNA were sufficient to identify a bone as sea cow. Samples sold as Steller's sea cow which did not produce a positive result were then sequenced. Sequencing the DNA confirmed that there were three samples from protected species sold as Steller's sea cow.
The gray whale is listed on CITES Appendix I, and trade of products from this species is highly controlled. The International Union for the Conservation of Nature (IUCN) Red List classifies the gray whale as either of “least concern” for the eastern Pacific population, or “critically endangered” for the western Pacific population. The western population is listed as “endangered” under the ESA and “depleted” under the MMPA. Gray whales occur in the North Pacific, with sightings as far north as 71°N (Rugh and Fraker, 1981), which is north of St. Lawrence Island. Hypothetically, the gray whale bone could have been from an animal stranded on St. Lawrence Island, and was mistakenly sold as a Steller's sea cow bone or it could have been from an animal killed in the past. In order to determine which of these is true, radiocarbon dating would need to be performed on the sample.
Both the pantropical spotted dolphin and white-beaked dolphin are listed under CITES Appendix II, and as of “least concern” by the IUCN. Under the MMPA, the Pacific northeastern offshore stock of the spotted dolphin is listed as “depleted.” The pantropical spotted dolphin has a tropical habitat and thus is unlikely to have been stranded or washed ashore on St. Lawrence Island. The white-beaked dolphin is only found in the North Atlantic, and this bone is also unlikely to have come from an animal that stranded on St. Lawrence Island. Therefore, illegal transport of dolphin bone is a valid conclusion.
Genetic verification of samples to determine levels of illegal trade has been conducted for many marine mammals. Baker et al. (1996) purchased whale meat from commercial sources in Japan. Using DNA extraction, PCR and sequencing, several species of protected whales were identified in meat, including fin (Balaenoptera physalus) and humpback whales (Megaptera novaeangliae). It was also found that dolphin meat was being sold as whale meat (Baker et al., 1996). Palumbi and Cipriano (1998) also identified protected species for sale in Asian markets using mtDNA analyses, including endangered Bryde's whales (Balaenoptera edeni).
Lee et al. (2009) and Vogel et al. (1990) used mtDNA to identify sources of ivory and verify samples that were legal to sell. As international trade in elephant products is controlled by CITES, a method for identification of illegal ivory is important. Lee et al. (2009) used mtDNA to track ivory back to the source population. Vogel et al. (1990) also located the environment in which animals lived using mtDNA from ivory.
Identifying the source of bone carvings has also been undertaken previously. Using a segment of D-loop, Gupta et al. (2011) extracted DNA from two ivory idols, the first study to accomplish this from ivory statuettes, and found that these were from Asian elephant (E. maximus) ivory. Similar work was carried out in this current study, extracting DNA from carved statuettes of Steller's sea cow bone (shown in Figure 1). All of the carvings were subsequently positively identified as Steller's sea cow. As the number of carved samples is so much smaller (N = 11) than the raw bone (N = 147), it could simply be chance that all of the carved bone was sea cow. It could also be that artisans carving the bone prefer to work with “mermaid ivory” and are able to distinguish that bone from other species using morphological properties of the bone.
Genetic analysis is very useful for forensic identification of illegal wildlife trafficking (Gupta, 2012). For example, animal products more than five years old were used to identify tiger-derived products and bring the poacher to justice (Gupta, 2012). However, a major problem with DNA analysis for forensic identification is that there are few sequences published in GenBank that are useful for sample identification (Gupta, 2012). This was the case in the current study as it was the first time a D-loop sequence was obtained from Steller's sea cow bone.
Using cytochrome b and the newly documented D-loop sequences, it was possible to determine whether a bone was a bona fide Steller's sea cow product. The new H. gigas D-loop sequences can also be used to verify the identity of ancient bones that are fragmented or badly weathered, such as ones from archeological sites. The relatively simple method of identification used in this study could be used on bone pieces or statuettes that cannot be readily identified using morphological analysis, with minimal damage to the art pieces (i.e., a small hole drilled in the base). Given the sample collected for this study, it seems that most of the bone (93%) coming from St. Lawrence Island is in pieces too small to be identified morphologically, so this test could be quite useful as a general screening for bone being sold from the island. It may be more useful to test bone that is already in the hands of dealers. However, policing this trade may be quite difficult.
The findings of this study support the idea that government agencies, such as the US Fish and Wildlife Service and the National Marine Fisheries Service, should monitor the sale of “mermaid ivory”. Even though only three samples were not Steller sea cow products, the samples from the gray whale, pantropical spotted dolphin and white-beaked dolphin were effectively being traded illegally, with the latter two items being derived from a location far removed from St Lawrence Island, and thus transport and trade of these items were potentially a violation of CITES. Unfortunately, the dealer of these particular bones was unable to provide information on specifically where the bones were purchased, so it is difficult to determine how or when the animals came to St. Lawrence Island. Information of this type could be determined using stable isotope analysis, which is more difficult and expensive than the scope of this test allowed.
The animals are extinct. Although bone was used, no animals were harmed to conduct this research.
LC conceived the experimental design and completed the sample collection and processing. LC and EP interpreted the data and prepared the manuscript. All authors critically revised the manuscript and approved of the final submission.
This work was funded by George Mason University and Andrew P. Crerar. Publication costs of this article were kindly funded for LC, EF and EP by the George Mason University Libraries Open Access Publishing Fund.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
LC gratefully recognizes the help of Andrew Crerar (who funded this work) and Dan Horner (who prepared the bone), the Gillevet Lab (GMU), and lab assistants Chris Broadhurst, Chris Monk, J. Stuart Pate, Henry Johnson, Michael Harpole, Jeffery Warner, and Caroline Benzel. We all thank the wonderful reviewers and editors of this manuscript.
Baker, C., Cipriano, F., and Palumbi, S. (1996). Molecular genetic identification of whale and dolphin products from commercial markets in Korea and Japan. Mol. Ecol. 5, 671–685. doi: 10.1111/j.1365-294X.1996.tb00362.x
Crerar, L. D., Crerar, A. P., Domning, D. P., and Parsons, E. C. (2014). Rewriting the history of an extinction—was a population of Steller's sea cows (Hydrodamalis gigas) at St. Lawrence Island also driven to extinction? Biol. Lett. 10:20140878. doi: 10.1098/rsbl.2014.0878
Domning, D. P. (1978). Sirenian evolution in the North Pacific Ocean. Univ. Calif. Publ. Geol. Sci. 118, 1–176.
Gupta, S. K. (2012). DNA wildlife forensics: present and future. J. Forensic Res. 3:e103. doi: 10.4172/2157-7145.1000e103
Gupta, S. K., Thangaraj, K., and Singh, L. (2011). Identification of the source of ivory idol by DNA analysis. J. Forensic Sci. 56, 1343–1345. doi: 10.1111/j.1556-4029.2011.01750.x
Lee, J. C., Hsieh, H. M., Huang, L. H., Kuo, Y. C., Wu, J. H., Chin, S. C., et al. (2009). Ivory identification by DNA profiling of cytochrome b gene. Int. J. Legal Med. 123, 117–121. doi: 10.1007/s00414-008-0264-0
Ozawa, T., Hayashi, S., and Mikhelson, V. M. (1997). Phylogenetic position of mammoth and Steller's sea cow within Tethytheria demonstrated by mitochondrial DNA sequences. J. Mol. Evol. 44, 406–413. doi: 10.1007/PL00006160
Palumbi, S. R., and Cipriano, F. (1998). Species identification using genetic tools: the value of nuclear and mitochondrial gene sequences in whale conservation. J. Heredity 89:459. doi: 10.1093/jhered/89.5.459
Rohland, N., and Hofreiter, M. (2007). Ancient DNA extraction from bones and teeth. Nat. Protoc. 2, 1756–1762. doi: 10.1038/nprot.2007.247
Rugh, D. J., and Fraker, M. A. (1981). Gray whale (Eschrichtius robustus) sightings in eastern Beaufort Sea. Arctic 34, 186–187. doi: 10.14430/arctic2521
Stejneger, L. (1887). How the great northern sea-cow (Rytina) became exterminated. Am. Naturalist 21, 1047–1054. doi: 10.1086/274607
Keywords: Steller's sea cow, illegal trade, DNA, Hydrodamalis gigas, wildlife products, Sirenian
Citation: Crerar LD, Freeman EW, Domning DP and Parsons ECM (2017) Illegal Trade of Marine Mammal Bone Exposed: Simple Test Identifies Bones of “Mermaid Ivory” or Steller's Sea Cow (Hydrodamalis gigas). Front. Mar. Sci. 3:272. doi: 10.3389/fmars.2016.00272
Received: 26 July 2016; Accepted: 08 December 2016;
Published: 06 January 2017.
Edited by:
Mark Meekan, Australian Institute of Marine Science, AustraliaReviewed by:
Rebecca Ruth McIntosh, Phillip Island Nature Parks, AustraliaCopyright © 2017 Crerar, Freeman, Domning and Parsons. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorelei D. Crerar, bGNyZXJhckBnbXUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.