
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci. , 22 July 2016
Sec. Aquatic Microbiology
Volume 3 - 2016 | https://doi.org/10.3389/fmars.2016.00130
This article is part of the Research Topic Aeromonas: a Jack of All Trades View all 26 articles
Aeromonas salmonicida subsp. salmonicida is an important pathogen in salmonid aquaculture and is responsible for furunculosis, a common infectious disease in salmon, trout and char. The type-three secretion system (T3SS) is considered as the major virulence attribute of A. salmonicida. It is used by the bacterium to secrete and translocate a large number of toxins and effector proteins into the host cell. Some of these factors such as the bi-functional ADP ribosylating—GTPase activating protein AexT have been shown to have a detrimental impact on the integrity of the cell cytoskeleton, and hence contribute to impair phagocytosis. Other effector proteins that are injected to the host cell such as AopP act by inhibiting the NF-κB signaling pathway blocking the translocation of NF-κB (p65) into the nucleus, thus influencing the host's inflammatory response. Several additional effectors that are secreted and translocated via the T3SS including Ati2, AopN, and ExsE have been suggested to modulate the host's immune response in particular by down regulating the inflammatory reaction. The analysis of the immune response in rainbow trout (Oncorhynchus mykiss) infected with virulent, T3SS harboring A. salmonicida subsp. salmonicida revealed that the infection leads to a rapid and strong downregulation of several immune-relevant markers affecting both the innate and the adaptive immune response, leading to mortality of the infected fish. These findings show that T3SS-delivered effector molecules and toxins of A. salmonicida impair the host's cytoskeleton thus damaging cell physiology and phagocytosis, but also affect the host's immune defense.
Aeromonas salmonicida subsp. salmonicida (hereafter referred to A. salmonicida) is the etiologic agent of the disease furunculosis in Salmonidae fish (mostly salmon, trout, and char), a major health problem for the growing salmonid aquaculture throughout the world. The disease is known for over a century and results in significant economic and ecologic losses. It still leads to a large consumption of antibiotics. The introduction of vaccination of salmon against furunculosis by classical oil-adjuvant bactrin vaccines, based on particular empirically selected A. salmonicida strains has significantly reduced the use of antibiotics (http://www.who.int/features/2015/antibiotics-norway/en/#). However, frequent outbreaks of furunculosis caused by A. salmonicida persistently occur in fish farms. Profound knowledge on the virulence attributes of A. salmonicida is required to understand its pathogenicity mechanisms in order to develop novel and improved preventive and therapeutic approaches. Several putative virulence factors of A. salmonicida such as the A-layer protein, extracellular proteases, aerolysin, and extracellular hemolysins failed to explain the pathogenicity of A. salmonicida (Ellis et al., 1988; Vipond et al., 1998). Currently, a large body of evidence exists that the type-three secretion system (T3SS) and the T3SS-related toxins and effector proteins are responsible for the pathogenic phenotype of strains of the genus Aeromonas and represent the main virulence system in A. salmonicida. This review gives a concise overview on the highly developed T3SS in A. salmonicida that consists of a proper bacterial membrane located secretion complex coupled to an extracellular nano-syringe to inject toxins and effector proteins into host cells. The role of the different toxins and effector proteins of A. salmonicida T3SS that enables the pathogen to efficiently infect the host by impairing its early warning system and immune defense is discussed. A detailed molecular description of the individual reactions of the various components of the A. salmonicida T3SS found in the extensive review by Vanden Bergh and Frey (2014).
The type III secretion system of A. salmonicida consists of three major structures, (i) the proper secretion apparatus that enables the delivery of protein toxins and effectors across the inner and outer bacterial membrane, (ii) the injection needle that can bridge the environmental gap between bacterium and host cell and transport the substances to the host, and (iii) the translocation apparatus that enables the translocation of the toxins and effectors from the needle across the cellular host membrane into the host cell (Figure 1). Comparison of the individual components or their genetic bases of the T3SS in A. salmonicida with the well described analogs found in Yersinia and Salmonella show strong conservation in particular among the structural proteins of the secretion- and translocation apparatus. Therefore, the morphological information of the A. salmonicida could be derived to a large extent from the archetype T3SS in Yersinia and Salmonella that have been analyzed extensively (Hodgkinson et al., 2009; Cornelis, 2010; Diepold et al., 2011, 2012; Abrusci et al., 2013; Dewoody et al., 2013; Galan et al., 2014). The T3SS is a sophisticated nano-syringe device as it is used by several pathogenic prokaryotes to deliver effector proteins to eukaryotic cells. The T3SS has been proposed to have evolved from the flagellar apparatus (Pallen and Wren, 2007), which through distinct evolutionary passages would have first lost its motility and then acquired needle-like organelles that provide the ability to secrete polypeptides (Galan et al., 2014). T3SS in A. salmonicida is composed by more than 20 proteins including the Asc (Aeromonas secretion) proteins that build the blocks of the secretion channel, the translocation needle and the infectisome, and the Aop (Aeromonas outer proteins) that are released by the bacterium and injected into the host cell. Studies in Salmonella and in Yersinia revealed that the base of the T3SS consists of several rings which span the bacterial inner and outer membranes and the periplasmic space (Figure 1). A large number of AscD and AscJ proteins (24 each in Yersinia) and AscV build up the ring of the inner membrane and AscC mostly the outer membrane channel. A special domain in the center of the inner membrane rings is formed by AscR, AscS, AscT, AscU, which together with AscV are responsible for the secretion mechanisms, as deduced from Yersinia and Salmonella as well as from work done on components of T3SS of A. salmonicida (Burr et al., 2005; Hodgkinson et al., 2009; Cornelis, 2010; Galan et al., 2014) (Table 1). AscV seems to constitute one of the central protein in the secretion mechanisms and is highly conserved among all bacterial T3SS. The translocation needle itself is composed of more than 100 units of the AscF protein that is topped by a ring of AcrV units that connect to the translocon composed of AopB and AopD, which itself integrates as a pore into the host cell membrane (Figure 1). The AopB/AopD pore is expected to strongly determine host-specificity. Hence, AopB and AopD are among the least conserved proteins shared by bacterial T3SS. In analogy to the T3SS of Yersinia enterocolitica (Mota et al., 2005), the needle of the T3SS of A. salmonicida is assumed to be built up of polymerized multimers of AscF. The polymerization of the AscF subunits is performed by AscH. The control of the length of the needle, as determined by the number of AscF subunits, is assumed to be controlled by AscP. After bridging AscV with AopB/ApoD, secretion and translocation of the effectors is initiated orchestrated by the control proteins AscK, AscL, AscQ, and the ATPase AscN (Broz et al., 2007). The functionality of AscV from A. salmonicida has been demonstrated by deletion mutagenesis in A. salmonicida and in Yersinia by a LcrV/AcrV hybrid, which constituted a functional tip of the translocation needle (Burr et al., 2003a; Broz et al., 2007). During the building up of the syringe and the subsequent secretion and translocation of effector molecules to the host cell, several regulators and chaperons stabilize and control the building process of the formation of the needle, the translocon, and the secretion. This regulation is strongly influenced by host cell contact and by the concentration of free Ca2+ ions in the growth medium of A. salmonicida (Burr et al., 2003a).
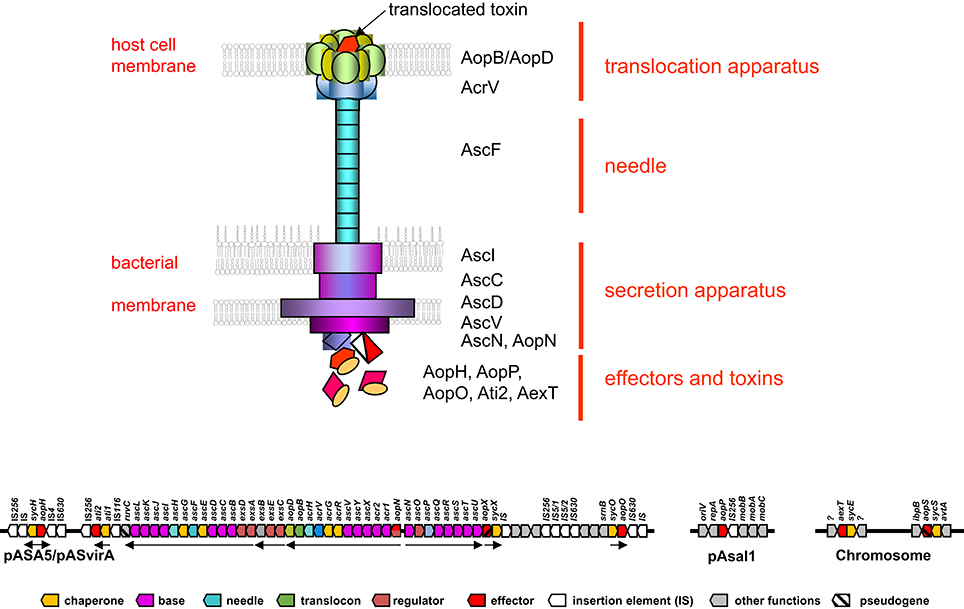
Figure 1. Schematic overview of a Type III secretion apparatus (top) and its genetic arrangement in Aeromonas salmonicida subsp. salmonicida (bottom). The colored arrows represent the individual genes and are labeled with analogous colors of the constituents of the T3SS apparatus drawn on top. Black arrows indicate the major operons. Note only the prominent Asc proteins are indicated in the drawing on the top part of the figure. Data taken from: (Burr et al., 2003a; Reith et al., 2008; Cornelis, 2010; Galan et al., 2014).
The genes encoding the structural components of the T3SS in A. salmonicida are encoded on a large conjugative plasmid of 150 kbp named pASvirA in strain JF2267 and pASA5 in strain A449 (Stuber et al., 2003a; Reith et al., 2008) where they are arranged in five major polycistronic operons (Burr et al., 2003a) (Figure 1). The full genome sequence of the virulent A. salmonicida strain A449 has been determined and confirmed the five structural T3SS operons containing the genes that build up the secretion and translocation apparatus (i) exsA,D,ascB,C,D,E,F,G,H,I,J,K,L; (ii) exsC,E,B; (iii) aopN,acr12,ascX,Y,V,acrR,G,V,H, aopB,D; (iv) ascN,O,P,Q,R,S,T,U, and (v) aopX,sycX (Figure 1). They are flanked on both sides by smaller operons containing the genes for various effector proteins and their corresponding chaperones (ati1, ati2; aopH, sycH; aopO, sycO), interspaced by several insertion elements belonging to IS116, IS256, IS630 (Reith et al., 2008; Vanden Bergh and Frey, 2014) (Figure 1). T3SS genes in A. salmonicida are unstable. In strain JF2267, isolated from an arctic char (Savelinus alpinus) with furunculosis, the T3SS virulence plasmid pASvirA is thermo-labile and rapidly lost if the strain is grown at temperatures above 20°C, although such temperatures (22–28°C) are generally recommended for growth of A. salmonicida by reference literature such as Bergey's Manual of Systematic Bacteriology and other guidelines (Stuber et al., 2003a). Furthermore, stressful conditions such as growth at or exposure to temperatures above 20°C also can lead to loss of the T3SS operons by homologous recombination between the two IS256 copies that flank the T3SS gene cluster (Daher et al., 2011; Tanaka et al., 2012) or by smaller rearrangement events by IS256 and other ISs such as IS630 (Studer et al., 2013; Tanaka et al., 2013; Emond-Rheault et al., 2015a,b). IS elements are also expected to be involved in the integration of the T3SS gene cluster into the chromosome as this has been observed in Aeromonas hydrophila strain SSU and Aeromonas veronii strain AER39. Furthermore, the T3SS effector aopH and its corresponding chaperon sycH gene have also been found on a smaller plasmid pASA6 (18.6 kb) which might be a derivative of the virulence plasmid pASA5. Interestingly the gene of the adenosine diphosphate (ADP) ribosylating toxin AexT a main toxin of A. salmonicida, as well of a second ADP-riboslyase (AopS) that however seems to be inactive in this species, are found chromosomally located on both strains A449 and JF2267 (Braun et al., 2002; Reith et al., 2008).
In general the basic genetic structure of A. salmonicida seems to be well conserved worldwide and over a relatively long time period and is generally linked with geographical origins as recently shown by Studer et al. (2013) and Emond-Rheault et al. (2015a). However, the cluster of virulence genes related to T3SS seems to be continuously exposed to local genetic rearrangements and deletions through internal insertion elements, thermosensitive plasmids and gene exchanges by horizontal transfer among environmental bacteria (Stuber et al., 2003b; Burr and Frey, 2007; Tanaka et al., 2012). An important consequence of gene deletions is the progressive loss of virulence genes observed with A. salmonicida under laboratory conditions where the selective pressure for virulence is absent. This has led to a total loss of all structural T3SS genes in the type strain of A. salmonicida ATCC 33658T and that has completely lost its virulence for fish (Burr and Frey, 2009) and hence is unsuitable for studies of A. salmonicida pathogenicity.
Furthermore, genomic investigations combined with PCR genotyping of a wild type A. salmonicida strain isolated from a brown trout with furunculosis revealed that this strain also is sensitive to genetic instability when exposed to 25°C, leading to insertion sequence-dependent rearrangement of the locus on the pAsa5 plasmid that encodes a T3SS, deleting an area which is essential for the virulence of the bacterium, including the genes exsD, ascV, and ascU (Emond-Rheault et al., 2015b). Hence molecular epidemiological assessments of A. salmonicida require the confirmation of the presence the major T3SS genes in order to be relevant for the occurrence of the disease.
While several virulence factors such as aerolysin AerA, surface layer protein VapA, serine protease Ahe2, and the LPS activated CGAT lipase SatA (Salte et al., 1992, 1993) had been characterized since a long time, only the T3SS could be determined to have a major effect on virulence. Independent studies with isogenic A. salmonicida strains containing deletion mutations for genes of structural proteins of the T3SS proved to be non-virulent both in cellular systems and in Amoebae, as well as in vivo in the natural host, the trout (Burr et al., 2002, 2003b, 2005; Dacanay et al., 2006, 2010; Froquet et al., 2007; Daher et al., 2011). In a recent study it was shown that intraperitoneal injection of 500 colony forming units (cfu) of A. salmonicida wild-type strain JF2267 resulted in 80% mortality after 7 days, whereas the isogenic secretion-deficient ΔascV derivative showed no mortality at a 200 times higher dose (105 cfu/fish) (Burr et al., 2005) and injection of a 200,000 times higher dose (108 cfu/fish), which is far from any natural infection, caused only 20% mortality (Vanden Bergh and Frey, 2014).
The T3SS effectors of A. salmonicida are mainly composed of AexT, AopH, Ati2, AopP, AopO, AopN as identified by mass spectrometry (Vanden Bergh et al., 2013a) (Table 1). From these proteins, AexT, AopP, and Ati2 have been investigated in detail with regard to cytotoxicity and showed functional homology with the analogs genes in Salmonella and Yersinia, while the others were deduced by similarity with T3SS effectors from other bacteria. They will be described shortly individually below. Furthermore, the regulator ExsE and the chaperone Ati1 have been found to be secreted by T3SS (Vanden Bergh et al., 2013a) but their function as effectors is not known.
AexT is a potent bifunctional toxin and possesses a GTPase-activating domain that acts on small monomeric GTPases of the Rho family (Rho, Rac, and Cdc42) and an ADP ribosylating domain that ribosylates in particular non-muscular but also muscular actin (Braun et al., 2002; Burr et al., 2003a; Fehr et al., 2007). Both enzymatic domains on AexT are able to depolymerize actin and cause cell rounding independently (Fehr et al., 2007). AexT is considered as one of the main toxins of A. salmonicida. However, deletion of the aexT gene leads only to a partial attenuation as demonstrated by a delayed cytotoxicity in rainbow trout gonad (RTG) and epithelioma papulosum cyprinid (EPC) cells (Fehr et al., 2007).
AopP was shown to inhibit the translocation of the p50/p65 protein complex (NFκB1/RelA) of the NFκB signaling pathway into the nucleus of the target cells. AopP inhibits IκB kinase β (IKKβ) phosphorylation and blocks IκBα phosphorylation revealing that the AopP mediated inhibition of the NFκB pathway occurs upstream of IκB phosphorylation (Fehr et al., 2006). Inhibition of the NFκB pathway by AopP after stimulation with tumor necrosis factor α (TNFα) induces a strong pro-apoptotic reaction in the host cell (Fehr et al., 2006; Jones et al., 2012).
AopH is an analog to the Yersinia phosphotyrosine phosphatase YopH that dephosphorylates tyrosine residues of multiple proteins at the cellular membrane of the host cell that are involved in focal adhesion complexes. This results in the loss of local adhesion alteration of the actin cytoskeleton structure and leads to loss of the phagocytosis of the affected phagocytes. In Y. enterocolitica, it was shown that the protein YopH, downregulates the respiratory burst while blocking T cell signaling activation and initiation of the adaptive immune response (Cornelis, 2002; Broberg and Orth, 2010). The role of AopH in A. salmonicida has not yet been studied by itself. However, it is thought to have a prominent activity in disturbing cell signaling that would be responsible for the initiation of the adaptive immune response (Vanden Bergh et al., 2013b; Vanden Bergh and Frey, 2014).
AopO is an analog to the Yersinia YopO, a serine/threonine kinase that disturbs the normal distribution of actin in the host cell as shown by Nejedlik et al. (2004). It inactivates Gαq signaling pathways by phosphorylation affecting multiple downstream targets involved in the actin cytoskeleton assembly. In A. salmonicida deletion mutants of the aopO gene in connection with ΔaexT and ΔaopH mutations were shown to affect leukocyte activation and downstream immune responses in Atlantic salmon (Fast et al., 2009). The specific role of AopO in infections by A. salmonicida has not yet been analyzed in detail. However, AopO was shown to undergo a particularly strong induction upon infection of fish compared to growth in axenic culture medium and hence seems to be required for the infections process (Menanteau-Ledouble and El-Matbouli, 2016).
AopN is involved in controlling the secretion of translocator proteins as a gate keeper inside the bacterium. However, it is also secreted by the T3SS in virulent A. salmonicida like effector proteins (Vanden Bergh et al., 2013b). Secretion of AopN, however occurred at a somewhat lower extent than the other effectors described (Bergh et al., 2013). AopN homologs in other bacteria are described as T3SS effectors which play a role in virulence and can have a dual role: controlling the secretion of translocator proteins inside bacteria and suppressing immunity (Vanden Bergh et al., 2013b). Hence, AopN like AopH is assumed to contribute to the strong inhibition of basic immune mediators such as interferon γ (INFγ) particularly in the beginning of infection with virulent A. salmonicida that harbors a functional T3SS.
Ati2 was identified in Vibrio parahaemolyticus as a phosphatidylinositol phosphatase involved in detachment of actin binding proteins from the plasma membrane leading to the destabilization of the host cell and its cytolysis (Broberg et al., 2010; Dallaire-Dufresne et al., 2013).
Fish challenged with virulent, T3SS+, A. salmonicida show a typical dissemination of bacteria that starts 12 h post challenge and is heavily detected 3–4 days in kidneys (Farto et al., 2011; Vanden Bergh and Frey, 2014). Subsequently A. salmonicida colonizes spleen, liver, and terminally the cardiac and skeletal muscles (Burr et al., 2005; Wahli et al., 2005; Farto et al., 2011). Virulent A. salmonicida with a functional T3SS (strain JF2267) was found to provoke a relatively modest Toll-like receptor (TLR) response mainly inducing the TLR11 family. In contrast a marked trafficking of myeloid cells as an immunophysiological outcome of inflammation in whitefish after an infection with A. salmonicida was observed (Altmann et al., 2016). Similarly, infection of rainbow trout with the same A. salmonicida wild-type showed to only moderately induce the expression of factors contributing to TLR signaling, but strongly activated the innate and adaptive cellular response. In fact, a massive migration of macrophages and granulocytes into the coelomic cavity was observed together with emigration of B lymphocytes from, and immigration of T lymphocytes into the head kidney (Brietzke et al., 2015).
A major effect of the T3SS toxins that are translocated to the host cells is actin depolymerization and destabilization of the actin cytoskeleton caused by the ADP ribosylase—GTPase activating toxin AexT, the serine/threonine kinase AopO and the phosphotyrosine phosphatase AopH. Potentially the phosphatidylinositol phosphatase Ati2 shares a similar activity that leads to cell rounding in cultivated fish cells to survival in phagocytic host cells causing spread of the pathogen and colonization of the host in vivo respectively, as a ΔaexT—ΔaopO—ΔaopH triple mutant did not fully abolish destabilization of actin cytoskeleton (Dacanay et al., 2006). Colonization of the host and survival in the macrophage must be considered as a special impairment of the macrophages in their task as immune cells to clear infections.
The immune suppressive effect induced by A. salmonicida was postulated already in 1935 (Scotland. Furunculosis committee and Mackie, 1935). However, until the discovery of T3SS as a major virulence attribute and its instability in A. salmonicida (Burr et al., 2002, 2003a,b, 2005; Stuber et al., 2003a) the variable effects shown in different experiments could not be explained. Furthermore, studies using the type strain of A. salmonicida, that was shown to be non-pathogenic due to the loss of the T3SS-containing virulence plasmid pASA5/pASvirA (Burr and Frey, 2009), resulted in erroneous results with regards to virulence mechanisms. Hence, any data in relation with virulence and immune-suppression or immune activation reported from infection studies with A. salmonicida needs to be interpreted with care and only if clear data concerning the presence respectively absence of the T3SS are stated.
In many fish species, filaments of cells connecting directly the thymus with the head kidney are expected to be used for lymphocyte migration from the thymus to the head kidney (Bowden et al., 2005). Interestingly, thymectomised fish challenged with A. salmonicida reveal higher antibody titres than intact controls. The reconstitution of thymectomised fish with preserved thymocytes depresses the immunoglobulinemia to levels similar to those of the control group suggesting that A. salmonicida promotes thymocytes mediated suppression of B-cell activation or plasma cell secretion (Findlay and Tatner, 1996). Moreover, in vitro, virulent A. salmonicida elicits a significant increase in IL-10 expression by fish leukocytes from the head kidney; IL-10 is a cytokine produced by various cell populations which downregulates the cellular immune response and contributes to Treg-mediated suppression in association with other cytokines. In contrast, deletion of T3SS genes significantly decreases the expression of this cytokine (Fast et al., 2009). Furthermore, in the 3 days following a challenge with A. salmonicida, the expression of genes associated with the following immunosuppressive Treg response are enhanced in the head kidney of fish: fibroleukin Fgl2, Es1 (HES1/KNPI), and serum amyloid A (Millan et al., 2011). Current data show, that virulent A. salmonicida use the whole arsenal of the T3SS and its effectors to neutralize the fish's immune response immediately after infection and lasting at least 5 days post infection. Rainbow trout infected with a fully virulent A. salmonicida that harbors a functional T3SS was invariably associated with a remarkable downregulation or even complete shut off of the expression of specific immune markers affecting both the innate and the adaptive immune response and caused mortality of the infected fish. Hence, translocation of T3SS effectors of A. salmonicida is expected to translate into a concerted blockade of relevant immune players (Vanden Bergh et al., 2013a,b; Vanden Bergh and Frey, 2014).
Altogether, T3SS-delivered toxins and effector molecules of A. salmonicida do not only impair the host's cytoskeleton destabilization of the actin cytoskeleton and thus damaging cell physiology and phagocytosis, but also directly affect the induction of critical immune markers including the shut-down of important warning signals to neighboring cells of the host in order recognize infection and induce immune defense.
Growth of A. salmonicida in inanimate conditions such as in axenic growth medium does not require the presence of a functional T3SS. Indeed T3SS genes remain silent in standard growth media and induction of the T3SS genes is regulated by contact with fish cells or under conditions of low levels of free calcium ions which is prevalent e.g., intracellularly (Burr et al., 2003a). Hence the presence of the whole genetic T3SS background is expected to represent an unnecessary fitness cost to A. salmonicida replicating or surviving outside the infected host, such as in open aquatic environment. However, the presence of such a sophisticated system as T3SS, found in A. salmonicida and other environmental bacteria, which is directed to disrupt the host cytoskeleton of an eukaryote lets speculate that these virulence strategies are the results of selective pressure of these bacteria to survive in the aquatic environment by escaping from the engulfment and killing of feral phagocytes such as amoebas. Hence, the actin cytoskeleton destabilizing mechanisms not only serve to tie up bacteriophageous protozoans and escape phagocytosis, but also to use concurrently the potential predator to survive and replicate inside, and finally to lyse the raider, escape and spread to the environment. Virulent A. salmonicida JF2267 have been demonstrated to be lethal for the amoeba Dictyostelium discoideum while the isogenic T3SS deficient ΔascV mutant (JF2747) was a-virulent in this host. In addition, trans-complementation of the ΔascV mutation restored the virulent phenotype for Dictyostelium (Froquet et al., 2007). It is therefore speculated that wild phagocytes such as Dictyostelium species might serve as reservoir and multiplier for virulent A. salmonicida, thus keeping the T3SS harboring strains in the environment, particularly in fecal and food waste enriched sediment, as it is found in proximity of fish farms. This explains why the high prevalence of protistan hosts in water with fecal contamination or plankton is considered as a source of virulent A. salmonidica and origin of furunculosis outbreaks (King and Shotts, 1988; Nese and Enger, 1993; Vanden Bergh and Frey, 2014).
The T3SS of A. salmonicida constitutes its major virulence which enables this pathogen to impair the host's cytoskeleton thus damaging cell physiology and phagocytosis and concurrently to directly affect the induction of critical immune markers of the innate and adaptive immune system shutting off completely the host's important warning signals to recognize infection and prevent onset of disease. Moreover, the T3SS enables the host to survive phagocytosis by protozoans in the environment using them as an alternative host to multiply and preserve their large genetic background of this complex virulence apparatus.
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was funded by the Swiss National Science Foundation (SNF) grant nr. 31003A-135808.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abrusci, P., Vergara-Irigaray, M., Johnson, S., Beeby, M. D., Hendrixson, D. R., Roversi, P., et al. (2013). Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 20, 99–104. doi: 10.1038/nsmb.2452
Altmann, S., Korytar, T., Kaczmarzyk, D., Nipkow, M., Kuhn, C., Goldammer, T., et al. (2016). Toll-like receptors in maraena whitefish: evolutionary relationship among salmonid fishes and patterns of response to Aeromonas salmonicida. Fish Shellfish Immunol. 54, 391–401. doi: 10.1016/j.fsi.2016.04.125
Bergh, P. V., Burr, S. E., Benedicenti, O., von Siebenthal, B., Frey, J., and Wahli, T. (2013). Antigens of the type-three secretion system of Aeromonas salmonicida subsp. salmonicida prevent protective immunity in rainbow trout. Vaccine 31, 5256–5261. doi: 10.1016/j.vaccine.2013.08.057
Bowden, T. J., Cook, P., and Rombout, J. H. (2005). Development and function of the thymus in teleosts. Fish Shellfish Immunol. 19, 413–427. doi: 10.1016/j.fsi.2005.02.003
Braun, M., Stuber, K., Schlatter, Y., Wahli, T., Kuhnert, P., and Frey, J. (2002). Characterization of an ADP-ribosyltransferase toxin (AexT) from Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184, 1851–1858. doi: 10.1128/JB.184.7.1851-1858.2002
Brietzke, A., Korytar, T., Jaros, J., Kollner, B., Goldammer, T., Seyfert, H. M., et al. (2015). Aeromonas salmonicida infection only moderately regulates expression of factors contributing to toll-like receptor signaling but massively activates the cellular and humoral branches of innate immunity in rainbow trout (Oncorhynchus mykiss). J. Immunol. Res. 2015:901015. doi: 10.1155/2015/901015
Broberg, C. A., and Orth, K. (2010). Tipping the balance by manipulating post-translational modifications. Curr. Opin. Microbiol. 13, 34–40. doi: 10.1016/j.mib.2009.12.004
Broberg, C. A., Zhang, L., Gonzalez, H., Laskowski-Arce, M. A., and Orth, K. (2010). A Vibrio effector protein is an inositol phosphatase and disrupts host cell membrane integrity. Science 329, 1660–1662. doi: 10.1126/science.1192850
Broz, P., Mueller, C. A., Muller, S. A., Philippsen, A., Sorg, I., Engel, A., et al. (2007). Function and molecular architecture of the Yersinia injectisome tip complex. Mol. Microbiol. 65, 1311–1320. doi: 10.1111/j.1365-2958.2007.05871.x
Burr, S. E., and Frey, J. (2007). Analysis of type III effector genes in typical and atypical Aeromonas salmonicida. J. Fish Dis. 30, 711–714. doi: 10.1111/j.1365-2761.2007.00859.x
Burr, S. E., and Frey, J. (2009). Aeromonas salmonicida subsp. salmonicida type strain does not possess a type III secretion system. J. Clin. Microbiol. 47, 3062–3063. doi: 10.1128/JCM.00749-09
Burr, S. E., Pugovkin, D., Wahli, T., Segner, H., and Frey, J. (2005). Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbow trout model. Microbiology 151, 2111–2118. doi: 10.1099/mic.0.27926-0
Burr, S. E., Stuber, K., and Frey, J. (2003a). The ADP-ribosylating toxin, AexT, from Aeromonas salmonicida subsp. salmonicida is translocated via a type III secretion pathway. J. Bacteriol. 185, 6583–6591. doi: 10.1128/JB.185.22.6583-6591.2003
Burr, S. E., Stuber, K., Wahli, T., and Frey, J. (2002). Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184, 5966–5970. doi: 10.1128/JB.184.21.5966-5970.2002
Burr, S. E., Wahli, T., Segner, H., Pugovkin, D., and Frey, J. (2003b). Association of Type III secretion genes with virulence of Aeromonas salmonicida subsp. salmonicida. Dis. Aquat.Org. 57, 167–171. doi: 10.3354/dao057167
Cornelis, G. R. (2002). The Yersinia Ysc-Yop ‘type III’ weaponry. Nat. Rev. Mol. Cell Biol. 3, 742–752. doi: 10.1038/nrm932
Cornelis, G. R. (2010). The type III secretion injectisome, a complex nanomachine for intracellular ‘toxin’ delivery. Biol. Chem. 391, 745–751. doi: 10.1515/bc.2010.079
Dacanay, A., Boyd, J. M., Fast, M. D., Knickle, L. C., and Reith, M. E. (2010). Aeromonas salmonicida Type I pilus system contributes to host colonization but not invasion. Dis. Aquat. Org. 88, 199–206. doi: 10.3354/dao02157
Dacanay, A., Knickle, L., Solanky, K. S., Boyd, J. M., Walter, J. A., Brown, L. L., et al. (2006). Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology 152(Pt 6), 1847–1856. doi: 10.1099/mic.0.28768-0
Daher, R. K., Filion, G., Tan, S. G., Dallaire-Dufresne, S., Paquet, V. E., and Charette, S. J. (2011). Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet. Microbiol. 152, 353–360. doi: 10.1016/j.vetmic.2011.04.034
Dallaire-Dufresne, S., Barbeau, X., Sarty, D., Tanaka, K. H., Denoncourt, A. M., Lague, P., et al. (2013). Aeromonas salmonicida Ati2 is an effector protein of the type three secretion system. Microbiology 159(Pt 9), 1937–1945. doi: 10.1099/mic.0.067959-0
Dewoody, R. S., Merritt, P. M., and Marketon, M. M. (2013). Regulation of the Yersinia type III secretion system: traffic control. Front. Cell. Infect. Microbiol. 3:4. doi: 10.3389/fcimb.2013.00004
Diepold, A., Wiesand, U., Amstutz, M., and Cornelis, G. R. (2012). Assembly of the Yersinia injectisome: the missing pieces. Mol. Microbiol. 85, 878–892. doi: 10.1111/j.1365-2958.2012.08146.x
Diepold, A., Wiesand, U., and Cornelis, G. R. (2011). The assembly of the export apparatus (YscR,S,T,U,V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol. Microbiol. 82, 502–514. doi: 10.1111/j.1365-2958.2011.07830.x
Ellis, A. E., Burrows, A. S., and Stapleton, K. J. (1988). Lack of relationship between virulence of Aeromonas salmonicida and the putative virulence factors: A-layer, extracellular proteyses and extracellular haemolysins. J. Fish Dis. 11, 309–323. doi: 10.1111/j.1365-2761.1988.tb01227.x
Emond-Rheault, J. G., Vincent, A. T., Trudel, M. V., Brochu, F., Boyle, B., Tanaka, K. H., et al. (2015a). Variants of a genomic island in Aeromonas salmonicida subsp. salmonicida link isolates with their geographical origins. Vet. Microbiol. 175, 68–76. doi: 10.1016/j.vetmic.2014.11.014
Emond-Rheault, J. G., Vincent, A. T., Trudel, M. V., Frey, J., Frenette, M., and Charette, S. J. (2015b). AsaGEI2b: a new variant of a genomic island identified in the Aeromonas salmonicida subsp. salmonicida JF3224 strain isolated from a wild fish in Switzerland. FEMS Microbiol. Lett. 362:fnv093. doi: 10.1093/femsle/fnv093
Farto, R., Milton, D. L., Bermudez, M. B., and Nieto, T. P. (2011). Colonization of turbot tissues by virulent and avirulent Aeromonas salmonicida subsp. salmonicida strains during infection. Dis. Aquat. Organ. 95, 167–173. doi: 10.3354/dao02342
Fast, M. D., Tse, B., Boyd, J. M., and Johnson, S. C. (2009). Mutations in the Aeromonas salmonicida subsp salmonicida type III secretion system affect Atlantic salmon leucocyte activation and downstream immune responses. Fish Shellfish Immunol. 27, 721–728. doi: 10.1016/j.fsi.2009.09.009
Fehr, D., Burr, S. E., Gibert, M., d'Alayer, J., Frey, J., and Popoff, M. R. (2007). Aeromonas exoenzyme T of Aeromonas salmonicida is a bifunctional protein that targets the host cytoskeleton. J. Biol. Chem. 282, 28843–28852. doi: 10.1074/jbc.M704797200
Fehr, D., Casanova, C., Liverman, A., Blazkova, H., Orth, K., Dobbelaere, D., et al. (2006). AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-kappaB signalling pathway. Microbiology 152(Pt 9), 2809–2818. doi: 10.1099/mic.0.28889-0
Findlay, C., and Tatner, M. F. (1996). The effect of reconstitution with cryopreserved thymocytes on the in vivo antibody response to sheep red blood cells, Aeromonas salmonicida, and DNP-KLH, in adult long term thymectomised rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 6, 371–381. doi: 10.1006/fsim.1996.0037
Froquet, R., Cherix, N., Burr, S. E., Frey, J., Vilches, S., Tomas, J. M., et al. (2007). Alternative host model to evaluate Aeromonas virulence. Appl. Environ. Microbiol. 73, 5657–5659. doi: 10.1128/AEM.00908-07
Galan, J. E., Lara-Tejero, M., Marlovits, T. C., and Wagner, S. (2014). Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438. doi: 10.1146/annurev-micro-092412-155725
Hodgkinson, J. L., Horsley, A., Stabat, D., Simon, M., Johnson, S., da Fonseca, P. C., et al. (2009). Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat. Struct. Mol. Biol. 16, 477–485. doi: 10.1038/nsmb.1599
Jones, R. M., Luo, L., and Moberg, K. H. (2012). Aeromonas salmonicida-secreted protein AopP is a potent inducer of apoptosis in a mammalian and a Drosophila model. Cell. Microbiol. 14, 274–285. doi: 10.1111/j.1462-5822.2011.01717.x
King, C. H., and Shotts, E. B. (1988). Enhancement of Edwardsiella tarda and Aeromonas salmonicida through ingestion by the ciliated protozoan Tetrahymena pyriformis. FEMS Microbiol. Lett. 51, 95–99. doi: 10.1111/j.1574-6968.1988.tb02976.x
Menanteau-Ledouble, S., and El-Matbouli, M. (2016). Antigens of Aeromonas salmonicida subsp. salmonicida specifically induced in vivo in Oncorhynchus mykiss. J. Fish Dis. 39, 1015–1019. doi: 10.1111/jfd.12430
Millan, A., Gomez-Tato, A., Pardo, B. G., Fernandez, C., Bouza, C., Vera, M., et al. (2011). Gene expression profiles of the spleen, liver, and head kidney in turbot (Scophthalmus maximus) along the infection process with Aeromonas salmonicida using an immune-enriched oligo-microarray. Mar. Biotechnol. (NY) 13, 1099–1114. doi: 10.1007/s10126-011-9374-7
Mota, L. J., Journet, L., Sorg, I., Agrain, C., and Cornelis, G. R. (2005). Bacterial injectisomes: needle length does matter. Science 307, 1278. doi: 10.1126/science.1107679
Nejedlik, L., Pierfelice, T., and Geiser, J. R. (2004). Actin distribution is disrupted upon expression of Yersinia YopO/YpkA in yeast. Yeast 21, 759–768. doi: 10.1002/yea.1135
Nese, L., and Enger, O. (1993). Isolation of Aeromonas salmonicida from salmon lice Lepeophtheirus salmonis and marine plankton. Dis. Aquat. Org. 16, 79–81. doi: 10.3354/dao016079
Pallen, M. J., and Wren, B. W. (2007). Bacterial pathogenomics. Nature 449, 835–842. doi: 10.1038/nature06248
Reith, M. E., Singh, R. K., Curtis, B., Boyd, J. M., Bouevitch, A., Kimball, J., et al. (2008). The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9:427. doi: 10.1186/1471-2164-9-427
Salte, R., Norberg, K., Arnesen, J. A., Odegaard, O. R., and Eggset, G. (1992). Serine protease and glycerophospholipid - cholesterol acyltransferase of Aeromonas salmonicida work in concert in thrombus formation - In vitro the process is counteracted by plasma antithrombin and alpha-2-macroglobulin. J. Fish Dis. 15, 215–227. doi: 10.1111/j.1365-2761.1992.tb00658.x
Salte, R., Norberg, K., Odegaard, O. R., Arnesen, J. A., and Olli, J. J. (1993). Exotoxin-induced consumptive coagulopathy in Atlantic salmon, Salmo salar I - Inhibitory effects of exogenous antithrombin and alpha(2)-macroglobulin on Aeromonas salmonicida serine protease. J. Fish Dis. 16, 425–435. doi: 10.1111/j.1365-2761.1993.tb00876.x
Scotland. Furunculosis committee Mackie, T. J. (1935). Final report (July 1935) of the Furunculosis Committee. Edinburgh: H. M. Stationery off. (printed by J. & J. Gray).
Stuber, K., Burr, S. E., Braun, M., Wahli, T., and Frey, J. (2003a). Type III secretion genes in Aeromonas salmonicida subsp. salmonicida are located on a large thermolabile virulence plasmid. J. Clin. Microbiol. 41, 3854–3856. doi: 10.1128/JCM.41.8.3854-3856.2003
Stuber, K., Frey, J., Burnens, A. P., and Kuhnert, P. (2003b). Detection of type III secretion genes as a general indicator of bacterial virulence. Mol. Cell. Probes 17, 25–32. doi: 10.1016/S0890-8508(02)00108-1
Studer, N., Frey, J., and Vanden Bergh, P. (2013). Clustering subspecies of Aeromonas salmonicida using IS630 typing. BMC Microbiol. 13:36. doi: 10.1186/1471-2180-13-36
Tanaka, K. H., Dallaire-Dufresne, S., Daher, R. K., Frenette, M., and Charette, S. J. (2012). An insertion sequence-dependent plasmid rearrangement in Aeromonas salmonicida causes the loss of the type three secretion system. PLoS ONE 7:e33725. doi: 10.1371/journal.pone.0033725
Tanaka, K. H., Frenette, M., and Charette, S. J. (2013). IS-mediated loss of virulence by Aeromonas salmonicida: a tangible piece of an evolutionary puzzle. Mob. Genet. Elements 3:e23498. doi: 10.4161/mge.23498
Vanden Bergh, P., and Frey, J. (2014). Aeromonas salmonicida subsp. salmonicida in the light of its type-three secretion system. Microb. Biotechnol. 7, 381–400. doi: 10.1111/1751-7915.12091
Vanden Bergh, P., Heller, M., Braga-Lagache, S., and Frey, J. (2013a). The Aeromonas salmonicida subsp. salmonicida exoproteome: determination of the complete repertoire of type-three secretion system effectors and identification of other virulence factors. Proteome Sci. 11:42. doi: 10.1186/1477-5956-11-42
Vanden Bergh, P., Heller, M., Braga-Lagache, S., and Frey, J. (2013b). The Aeromonas salmonicida subsp. salmonicida exoproteome: global analysis, moonlighting proteins and putative antigens for vaccination against furunculosis. Proteome Sci. 11:44. doi: 10.1186/1477-5956-11-44
Vipond, R., Bricknell, I. R., Durant, E., Bowden, T. J., Ellis, A. E., Smith, M., et al. (1998). Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect. Immun. 66, 1990–1998.
Keywords: furunculosis, virulence, type three secretion system, toxins, immunosuppression, fish, infectious disease
Citation: Frey J and Origgi FC (2016) Type III Secretion System of Aeromonas salmonicida Undermining the Host's Immune Response. Front. Mar. Sci. 3:130. doi: 10.3389/fmars.2016.00130
Received: 10 June 2016; Accepted: 07 July 2016;
Published: 22 July 2016.
Edited by:
Brigitte Lamy, University of Montpellier, FranceReviewed by:
Susana Merino, University of Barcelona, SpainCopyright © 2016 Frey and Origgi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joachim Frey, am9hY2hpbS5mcmV5QHZldHN1aXNzZS51bmliZS5jaA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.