- 1State Key Laboratory of Marine Environmental Science and Marine Biodiversity and Global Change Research Center, Xiamen University, Xiamen, Fujian, China
- 2Department of Marine Sciences, University of Connecticut, Groton, CT, USA
Phosphorus (P) is an essential nutrient for marine phytoplankton but its preferred form, dissolved inorganic phosphorus (DIP), is often limited in the euphotic zone of the ocean. Many phytoplankton species have developed the ability to utilize dissolved organic phosphorus (DOP) such as phosphoesters or phosphonates. Phosphonates are characterized by the stable C-P bond and its utilization is known to rely on either a C-P lyase pathway or C–P hydrolase pathways in bacteria. In this study by transcriptomic analysis we detected the genes encoding the C-P hydrolase pathway enzymes PhnW and PhnX in Karlodinium veneficum and other dinoflagellates. However, we found that these dinoflagellates are unable to utilize the presumably dominant type of phosphonate in the ocean, 2-aminoethylphosphonic acid (2-AEP), under the antibiotic-treated condition. In accordance, our RT-qPCR and proteomic analyses of K. veneficum grown on 2-AEP showed no up-regulation of PhnW and PhnX at both transcriptional and translational levels. Nevertheless, the genes related to phosphonate biosynthesis and utilization were widely found in dinoflagellates. Taken together our results suggest that the PhnW-PhnX pathway in dinoflagellates may serve for intracellular phosphonate metabolism instead of scavenging environmental phosphonates.
Introduction
Phosphorus (P) is an essential nutrient for the growth of marine phytoplankton and it is considered as the ultimate limiting nutrient of primary production in the global ocean (Tyrrell, 1999). The P reservoir in the ocean comprises dissolved inorganic phosphorus (DIP, mainly orthophosphate or Pi) and dissolved organic phosphorus (DOP). Pi is typically the preferred form (over DOP) of P nutrient and rapidly taken up by phytoplankton while its recycling from the remineralization of sinking organic matters and continental weathering is rather slow (Karl, 2014). Thus, Pi is usually depleted in the euphotic zone and more abundant at greater depths, a vertical trend opposite to that of DOP (Tyrrell, 1999; Paytan and McLaughlin, 2007). Pi limitation has been shown to constrain primary production (Karl, 2014; Lin et al., accepted) and nitrogen fixation (Sañudo-Wilhelmy et al., 2001; Mills et al., 2004) in marine ecosystem. In response, marine phytoplankton have evolved many strategies to cope with Pi-limitation, e.g., maximizing Pi uptake efficiency, mobilizing intracellular P storage such as polyphosphate (Orchard et al., 2009). Phytoplankton can also lower cellular demand of P under Pi-limitation by substituting some of the phospholipids in cell membrane with sulfur- and nitrogen-containing lipids (Snyder et al., 2009; Van Mooy et al., 2009; Wurch et al., 2011). Yet, one of the most important mechanisms to cope with Pi deficiency is to hydrolyze various forms of DOP in the ocean to yield phosphate (Dyhrman et al., 2007). It has been estimated that 30% of the primary production is supported by DOP during the boreal spring in the Northern Atlantic subtropical gyre (Mather et al., 2008).
Phosphoesters (C–O–P bond) and phosphonates (C–P bond) are the two dominant classes of DOP in the ocean and contribute roughly 75 and 25% of the high-molecular-weight DOP reservoir in the ocean respectively (Clark et al., 1998; Kolowith et al., 2001). Hydrolysis of phosphoesters by enzymes such as alkaline phosphatase (AP) has been described in both prokaryotic and eukaryotic lineages (Duhamel et al., 2010; Lin et al., 2012a). However, utilization of phosphonates is currently only known in bacteria (Kononova and Nesmeyanova, 2002; Dyhrman et al., 2006).
Phosphonates are organophosphorus compounds that contain the chemically stable carbon–phosphorus (C–P) bond and are produced in many organisms including marine invertebrates (Quin, 1965) and cyanobacteria (Dyhrman et al., 2009). Anthropogenic phosphonates are also widely introduced into the environment, for instance use of glyphosate herbicide in agriculture (Nowack, 2003; Hove-Jensen et al., 2014). Phosphonate utilization by bacteria has been attributed to either C-P lyase pathway or C–P hydrolase pathways (Quinn et al., 2007; McGrath et al., 2013). C-P lyase pathway regulates the hydrolysis of a broad range of phosphonate substrates, as demonstrated in bacteria (White and Metcalf, 2004; Dyhrman et al., 2006). One of the most important C–P hydrolase pathways so far found in bacteria (Villarreal-Chiu et al., 2012) and some cyanobacteria (Gomez-Garcia et al., 2010) is the phosphonatase pathway (PhnW-PhnX pathway). It acts specifically on 2-aminoethylphosphonic acid (2-AEP), which together with its derivatives, is the predominant phosphonate detected in marine invertebrates, and potentially an important and prevalent source of phosphonates in the ocean (Quin and Quin, 2001; McGrath et al., 2013). Phosphonatase pathway consists of two steps of biochemical reactions (Jiang et al., 1995). The 2-AEP–pyruvate transaminase (PhnW; EC 2.6.1.37) first catalyzes the transamination of 2-AEP to produce alanine and phosphonoacetaldehyde (Kim et al., 2002), and subsequently phosphonoacetaldehyde is transformed to acetaldehyde and phosphate by phosphonoacetaldehyde hydrolase (PhnX; also known as phosphonatase; EC 3.11.1.1; Morais et al., 2004). It has remained unexplored whether eukaryotic phytoplankton are able to utilize phosphonates as a source of P.
In this study, we conducted experiments to examine whether dinoflagellates, one of the most important lineages of eukaryotic phytoplankton, possess the enzyme system to utilize phosphonates. We detected both PhnW and PhnX of the phosphonatase pathway in dinoflagellates, but observed no ability of these algae to scavenge environmental 2-AEP. Our further experiments indicated that bacteria would be an important mediator for dinoflagellates to exploit phosphonates for Pi from the marine environment.
Materials and Methods
Algal Cultures and Phosphorus Treatments
Four species representing four major orders (Gymnodiniales, Prorocentrales, Gonyaulacales, Suessiales) of the dinoflagellate phylum were used in this study. Karlodinium veneficum strain CCMP2778 and Symbiodinium kawagutii strain CCMP2468 were obtained from the Provasoli-Guillard National Center for Marine Algae and Microbiota (NCMA) in Boothbay Harbor, Maine, USA. Prorocentrum donghaiense and Alexandrium pacificum (formerly A. catenella) were provided by Center for Collections of Marine Bacteria and Phytoplankton of Xiamen University. These algal strains were maintained in L1 medium made with seawater (salinity, 28 PSU), which was filtered through 0.22-μm membranes and autoclaved. Cultures were grown at 20°C under a L: D = 14 h: 10 h photocycle with a photon flux of 120 ± 10 μE·m−2·s−1.
The algal cultures were first grown in low-Pi L1 medium until the DIP in the culture were depleted (< 0.5 μM). The P-limited cultures were then inoculated into media with different P nutrient conditions: P-replete (36 μM DIP), P-depleted (< 0.5 μM DIP), and 2-AEP (< 0.5 μM DIP but amended with 36 μM 2-AEP; 2-AEP from Sigma-Aldrich, St. Louis, MO, USA), each in triplicate. In antibiotic-treated cultures, 1 × KAS antibiotics (final concentration 100 μg/L ampicillin, 50 μg/L kanamycin and 50 μg/L streptomycin) were added. Cell counts were carried out using a Sedgwick–Rafter counting chamber (Phycotech, St. Joseph, MI, USA). DIP was measured using the molybdenum blue method (Timothy et al., 1984). Cells were collected by centrifugation and resuspended in 1 mL Trizol reagent (MRC, Cincinnati, OH, USA) and stored at −80°C for subsequent RNA and protein extraction.
Examination of Bacterial Involvement in Phosphonate Utilization
The non-antibiotic-treated dinoflagellates were first acclimated to the 2-AEP condition until significant increase in growth was observed compared to the P-depleted groups. Each of the treatments was then equally split into two portions, one remaining unchanged as the control and the other being gently filtered through autoclaved 3-μm polycarbonate membrane (vacuum < 10 psi) to collect the filtrate, which contained the bacteria from the culture and excluded the algae. For K. veneficum and A. pacificum, both the control and the filtrate were inoculated into fresh medium containing 2-AEP, while the divided portions of P. donghaiense were incubated in the previous media. The DIP concentrations in both the algal and the filtrate (bacterial) cultures were measured. Thus, the increase of DIP concentration in the filtrate cultures would be attributed to the hydrolysis of 2-AEP by the bacteria existing in the cultures.
Amplification of Full-length cDNAs of PhnW and PhnX
RNA of the collected samples was extracted following the instruction of the Trizol, purified using Qiagen RNeasy Mini Kit and treated with DNase RQ1 to eliminate possible genomic DNA contamination (Lin et al., 2010). The first strand cDNA was synthesized using ImProm-II™ Reverse Transcription System (Promega, Madison, WI, USA). Specific primers (Supplementary Table S1) were designed for both 3′- and 5′-RACE based on the partial sequences identified from dinoflagellate-specific spliced leader (DinoSL) based cDNA libraries of K. veneficum (Lin et al., unpublished). The 21-bp highly conserved DinoSL was used as the 5' forward primer for the 5′-RACE (Zhang et al., 2007; Lin et al., 2010).
Computational Characterization of PhnW and PhnX
A set of online programs was used to characterize PhnW and PhnX in K. veneficum deduced from their respective cDNA sequences. The signal peptide was determined by SignalP V4.0 (Petersen et al., 2011). PredGPI (Pierleoni et al., 2008) was used to locate the glycosylphosphatidylinositol (GPI) anchor in the deduced protein sequence. TMpred (Hofmann, 1993) was used to predict transmembrane domains. Four existing computational models, Interpretable Subcellular Localization Prediction (YLoc, Briesemeister et al., 2010), Euk-mPLoc2.0 (Chou and Shen, 2008), WoLF PSORT (Horton et al., 2007), and CELLO (Yu et al., 2006) were adopted to predict subcellular localizations of PhnW and PhnX. The reliability of these programs had previously been verified for alkaline phosphatase (Lin et al., 2012a). The likelihood of the predicted enzyme localization was ranked according to the number of the models that supported the prediction.
Search of Genes Related to Phosphonate Metabolism in Dinoflagellates
In addition to the phosphonatase pathway genes phnW and phnX, phosphoenolpyruvate mutase (pepM) and phosphonopyruvate decarboxylase (ppd), the two major phosphonate synthesis genes were also included in the data mining in dinoflagellates. Sequences of phnW, phnX, pepM, and ppd from K. veneficum were used as queries to blastp against available dinoflagellate genomic and transcriptomic data sets, including the genomes of S. kawagutii (Lin et al., 2015) and Symbiodinium sp. clade B1 (http://marinegenomics.oist.jp/genomes/gallery/), and the transcriptomic datasets in CAMERA (http://camera.crbs.ucsd.edu/ddc/). The threshold E-value was set as e-05. Keyword search was also carried out to get all possible related sequences from CAMERA. All the sequences acquired from those datasets were further confirmed by blastp against NCBI GenBank database.
RT-qPCR Measurement of phnW and phnX Transcript Abundances
Specific primers applicable for all isoforms of each gene (Supplementary Table S1) were designed for RT-qPCR to examine the differential expression of phnW and phnX under different P conditions. Calmodulin (calcium-modulated protein; KM275627) and gapdh (glyceraldehyde-3-phosphate dehydrogenase; KM275628) were used as the reference genes because of relative stable expression previously reported for the dinoflagellate P. donghaiense (Shi et al., 2013). For standard curves, a PCR product for each gene was prepared in ten-fold dilution series (103–107 copies per μL) (Lin et al., 2012b). RT-qPCR was performed using Bio-Rad iQ SYBR Green Supermix kit (Bio-Rad Laboratories, Hercules, CA, USA) with all the reactions set up in triplicate for each gene. Relative transcript levels of these genes were calculated in three ways to facilitate comparison: normalized to the amount of total RNA equivalent to the amount of cDNA used in each reaction, to each cell, and to the expression levels of the two reference genes.
Proteomic Analysis to Identify Differentially Expressed Proteins Under Different P Conditions in K. veneficum
We used iTRAQ (isobaric tags for relative and absolute quantitation) method to study the proteome of K. veneficum collected from the three P conditions as described above. About 3 × 107 cells were harvested from each culture and protein was extracted as previously reported (Li et al., 2013). 100 μg total protein from each sample was used for iTRAQ Labeling. LC-ESI-MS/MS analysis was then performed on TripleTOF 5600 System (AB SCIEX, Concord, ON), followed by protein identification through Mascot search engine (Matrix Science, London, UK; version 2.3.02) against annotated transcriptome data (Lin et al., unpublished).
For protein quantitation, the quantitative protein ratios were weighted and normalized by the median ratio in Mascot. The criteria as fold changes >1.2 and p < 0.05 was adopted to depict significantly differentially expressed proteins. Functional annotations of the proteins were conducted using Blast2GO program against NCBI non-redundant (nr) protein database. The KEGG database (http://www.genome.jp/kegg/) and the COG database (http://www.ncbi.nlm.nih.gov/COG/) were used to classify and group these identified proteins.
Antibody Preparation and Western Blot Analysis of Phnx Abundance
The coding region of phnX was amplified by specific primers (Supplementary Table S1). Purified PCR products were cloned into pEasy-E2 expression vector and transformed into E. coli BL21 (DE3). Positive clones were grown in LB medium with 100 μg/mL ampicillin and 0.5 mM IPTG were added to induce the expression of the PhnX. The cells were harvested by centrifugation and then suspended in Tris buffer (50 mM Tris-Cl, pH 8.0) and homogenized by sonication. After removal of the cell debris by centrifugation, the supernatant was purified using Ni-NTA spin kit column (Transgen, Beijing, China). The protein was then used to immunize a rabbit (Japanese white rabbit) to generate a polyclonal antibody (Proteintech Group Inc., Wuhan, China).
Western blot analysis was conducted essentially following Lin et al. (1994). K. veneficum cells were homogenized by ceramic bead (0.5 mm) beating on FASTPREP-24 instrument (MP, Santa Ana, CA, USA) to obtain the crude total protein. An equal amount of total protein (15 μg) from each sample was used in SDS-PAGE gel electrophoresis. We used PhnX antiserum diluted 1:5000 for the first incubation and anti-rabbit polyclonal antibody diluted 1:10,000 for the second incubation.
Results
Identification and Characterization of PhnW and PhnX in K. veneficum
Full-length cDNAs of the two genes were obtained in this study, with existence of DinoSL at the 5' UTR of these gene transcripts indicating the dinoflagellate origin (Supplementary Figure S1). PhnW is 1221 bp long encoding a deduced protein of 406 amino acid residues giving a molecular mass of 44.5 kDa, while phnX is 717 bp encoding a 25.5 kDa protein with 238 amino acid residues. Blastp analyses showed that these genes matched well-characterized homologs with very strong statistical significance (E-values < e-30; Supplementary Table S2). We did not detect signal peptides, GPI anchor, or transmembrane domains, suggesting that both the PhnW and PhnX are probably not membrane-associated proteins in the dinoflagellates. Further computational analysis of subcellular localizations showed that PhnW is most likely to be located in the mitochondria while PhnX most likely in the cytoplasm (Supplementary Table S3).
Genes Related to Phosphonate Metabolism in Taxonomically Diverse Dinoflagellates
Results from our data mining on PhnW, PhnX, PepM, and Ppd demonstrated the presence and expression in a wide taxonomic range of dinoflagellates (Table 1). The low E-values (< e-20) of blastp (Supplementary Table S2) and good alignments of these protein sequences with representative sequences from bacteria showed that they all share conserved regions (Supplementary Figure S2). Out of the examined 30 species from seven orders (gonyaulacales, gymnodiniales, oxyrrhinales, peridiniales, prorocentrales, suessiales, thoracosphaerales), PepM, Ppd, PhnW, and PhnX were found in 27, 26, 27, 16 species respectively.
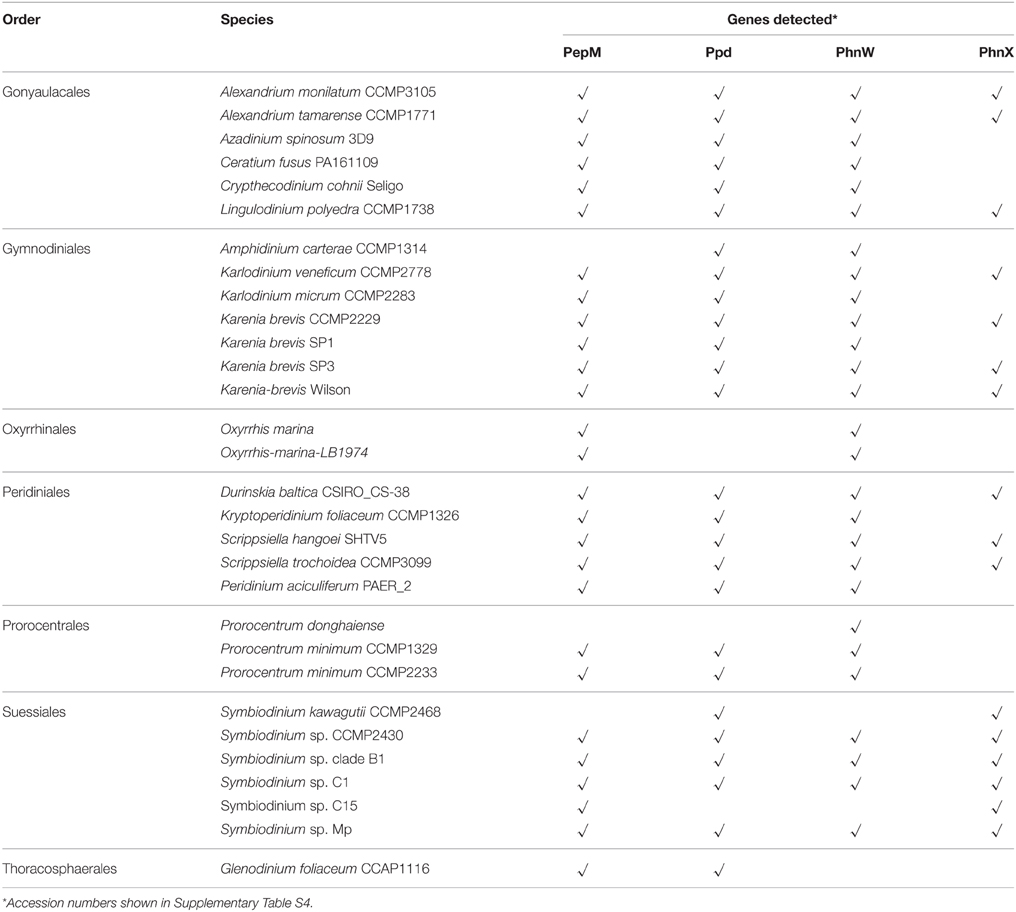
Table 1. Genes related to phosphonate metabolism detected from available transcriptomes in dinoflagellates.
2-AEP Investigated as Sole P Source in Normal Culture of K. veneficum
To determine whether phnW and phnX are able to facilitate the utilization of phosphonate by K. veneficum, 2-AEP was supplied as the sole P source in the culture medium. In the 2-AEP group, only a minimum growth was observed in the first 6 days, but after that there was a remarkable increase of cell concentration compared to that in P-depleted group (Figure 1A), indicating that 2-AEP could be utilized as a sole P source to support the growth of K. veneficum. DIP concentration in the 2-AEP group was the same as that in the P-depleted group initially, but later showed slight but significant increase (Figure 1B; p < 0.05), and DIP accumulation increased remarkably after day 14 (from 0.32 μM in the 14th day to 8.52 μM in the 16th day), suggesting that 2-AEP had been hydrolyzed to release phosphate into the culture.
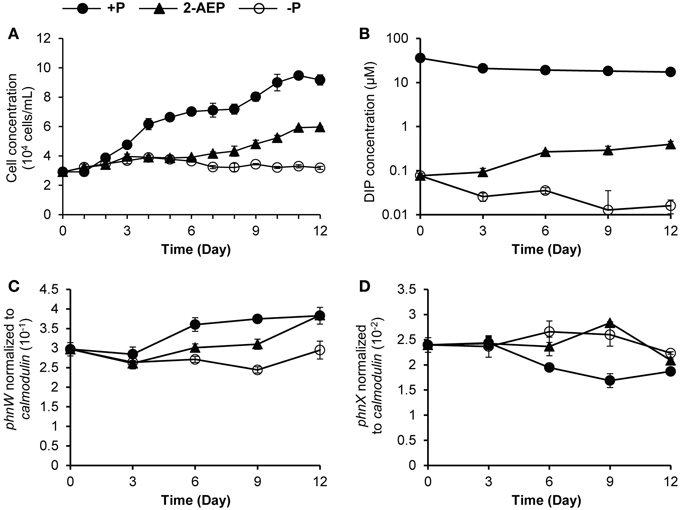
Figure 1. Growth curves (A), DIP concentrations (B), and transcript abundances of phnW and phnX normalized to calmodulin (C,D) in K. veneficum grown under different P conditions. Solid circles, P-replete condition; solid triangles, P-depleted condition amended with 2-AEP; open circles, P-depleted condition.
Transcriptional and Translational Regulation of PhnW and PhnX
When normalized to the same amount of RNA (5 ng) equivalent to the amount of cDNA used in RT-qPCR and normalized to per cell basis, the expression levels of both phnW and phnX showed no up-regulation in the 2-AEP culture compared to P-replete culture (Supplementary Figures S3A–D). When phnW transcript abundance was normalized to calmodulin, the expression level in the 2-AEP culture was between that in P-replete and the P-depleted conditions (p < 0.05; Figure 1C). A different pattern was found in phnX, which showed similar expression levels between the P-depleted group and the 2-AEP group but a higher expression level than that in the P-replete group (p < 0.05; Figure 1D). This trend remained when phnW and phnX were normalized to gapdh (Supplementary Figures S3E,F).
To examine whether the lack of transcriptional up-regulation of PhnW and PhnX genes were because these genes are regulated at the translational level, we further investigated PhnW and PhnX protein abundances. From the iTRAQ proteomic analysis for K. veneficum grown under 2-AEP, P-depleted, and P-replete conditions, 4922 proteins were identified (Supplementary Figure S4), including some proteins related to phosphonate metabolism and phosphate transportation such as Ppd, PhnW, and PhnX (Table 2). Comparative proteomic analysis did not show differential expression of PhnW under different P conditions (Figure 2A). The low expression level of PhnX precluded comparative proteomic analysis on this protein. Western blot analysis showed that PhnX expression was lower in the 2-AEP and P-depleted treatment groups than that in the P-replete group (Figures 2B,C).
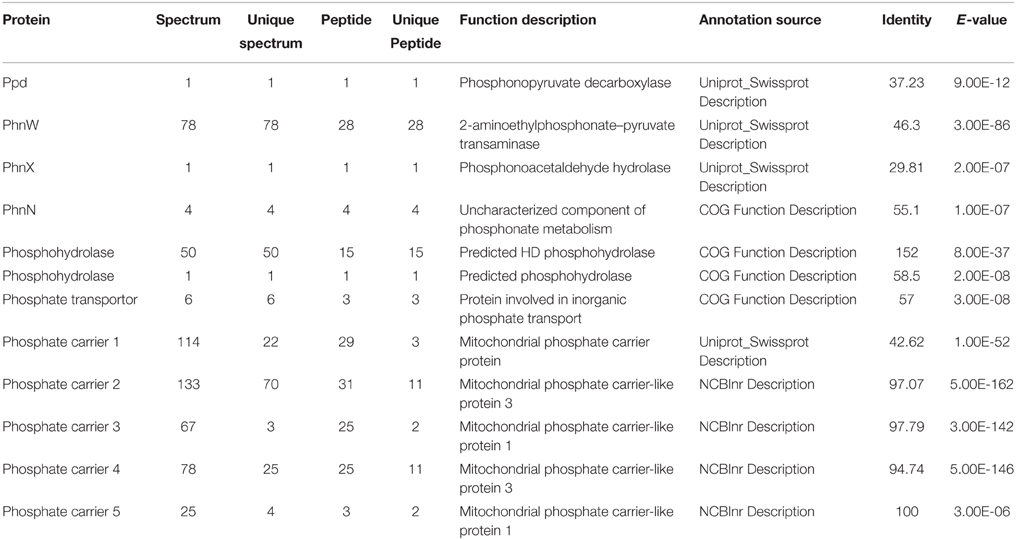
Table 2. LC-ESI-MS/MS analysis results and function description of some proteins related to phosphonate metabolism and phosphate transportation.
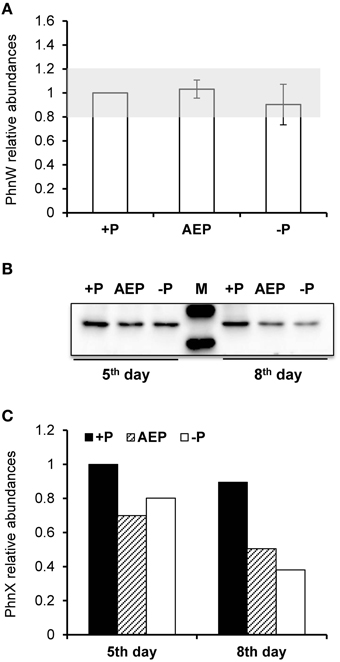
Figure 2. Protein abundances of PhnW and PhnX in K. veneficum grown under different P conditions. (A) Relative abundances of PhnW quantified from iTRAQ analysis using the control culture (+P) as the reference. Gray shading depicts the range within which the fold changes relative to the control are considered as insignificant. (B) Western blot showing abundances of PhnX protein under different P conditions. (C) Relative abundances of PhnX quantified from the western blot using the control culture (+P) as the reference. +P, P-replete condition; AEP, P-depleted condition amended with 2-AEP; −P, P-depleted condition.
Bacterial Involvement in Phosphonate Utilization
To find out whether the delayed dinoflagellate growth promoting effect of 2-AEP was mediated through bacteria, we conducted new experiments to compare the growth in antibiotic-treated cultures with that in the normal (non-antibiotic-treated) cultures, in which 2-AEP were provided as sole P-source. Because K. veneficum was very sensitive to antibiotics-treatment, it was excluded from this experiment, which involved P. donghaiense, A. pacificum, and S. kawagutii. Our results showed that in the normal cultures 2-AEP supported growth of these dinoflagellates, whereas in the antibiotic treatment cultures, hardly any growth was observed in the 2-AEP groups, similar to the P-depleted group (Figure 3). Besides, in the 2-AEP condition of the normal cultures, both K. veneficum (Figure 1A) and P. donghaiense (Figure 3C) showed a lag period of about 6 days before net growth was noticed while in A. pacificum (Figure 3A) and S. kawagutii (Figure 3E) the lag period was within 2 days.
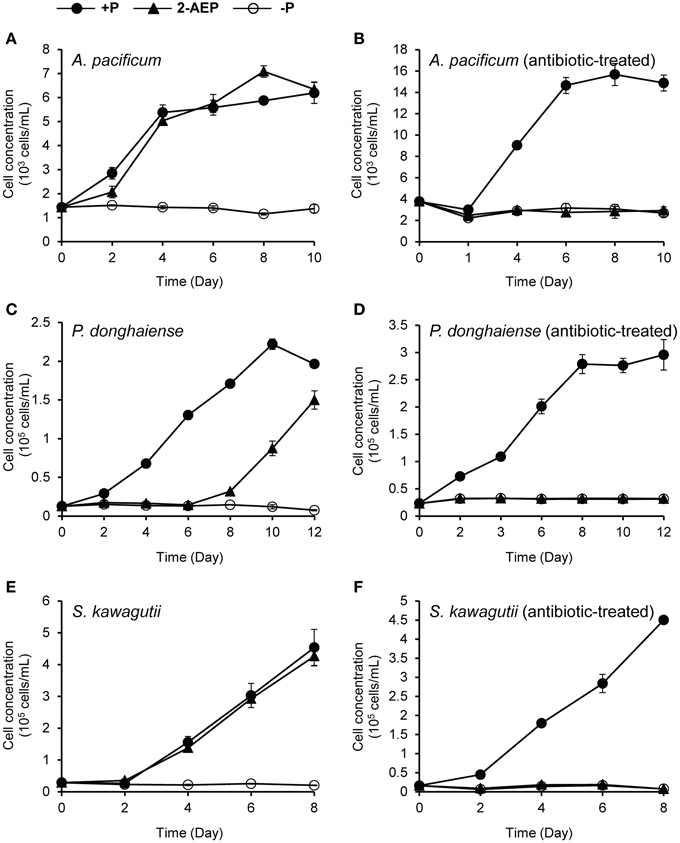
Figure 3. Comparison of the growth curves of different dinoflagellates under different P conditions with vs. without antibiotic treatment. (A,B) Growth curves of A. pacificum cultures without and with antibiotic treatment. (C,D) Growth curves of P. donghaiense cultures without and with antibiotic treatment. (E,F) Growth curves of S. kawagutii cultures without and with antibiotic treatment. Solid circles, P-replete condition; solid triangles, P-depleted condition amended with 2-AEP; open circles, P-depleted condition.
To further confirm that the utilization of 2-AEP was contributed by bacteria, we introduced the 3 μm-filtrate (i.e., bacteria-containing medium) from the 2-AEP cultures of A. pacificum, P. donghaiense, and K. veneficum into two new sets of cultures, one with original dinoflagellate species, and the other just medium without algae, and DIP was measured subsequently. The results showed that both the cultures with or without algae showed an increase in DIP concentration (except for the P. donghaiense cultures with algae, in which the released Pi appeared to be quickly consumed to support the fast growth of algal population; Figure 4), indicating that a substantial amount of 2-AEP was hydrolyzed by bacteria. The DIP concentration increased more rapidly in the cultures without algae than these with algae (p < 0.05), likely due to the consumption of phosphate by the algae.
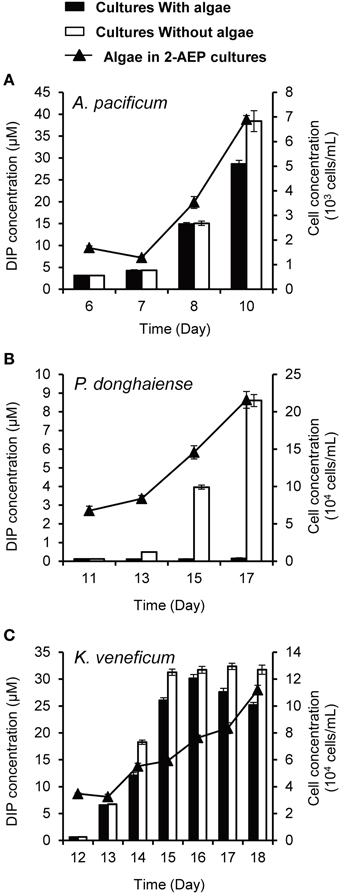
Figure 4. DIP concentrations in the cultures with (solid bars) and without algae (open bars) as well as growth curves of the dinoflagellates A. pacificum (A), P. donghaiense (B), and K. veneficum (C) in the normal cultures grown in 2-AEP amended medium (solid triangles and line).
Discussion
Dinoflagellates constitute one of the most important eukaryotic phytoplankton groups in the marine ecosystem, and they contribute significantly to marine primary production in the coastal ecosystems, coral reef growth (by the genus of Symbiodinium), harmful algal blooms, and marine biotoxins. Previous studies have demonstrated the ability of dinoflagellates to utilize phosphoesters as a source of P in response to Pi limitation (Lin et al., 2011, 2012a,b). It has remained unclear whether phosphonates, the other important kind of DOP in the ocean, could be utilized by dinoflagellates as a sole P source. As the PhnW and PhnX based phosphonatase pathway in bacteria are known to specifically catalyze the cleavage of the presumably dominant kind of biogenic phosphonate in nature, 2-AEP, we set out to explore whether a similar mechanism operates in dinoflagellates. We found these two genes in a number of dinoflagellate species and their encoded proteins in the proteome of K. veneficum. Further experiments were carried out using culture and molecular methods to investigate the availability of 2-AEP as a P source to dinoflagellates. This is the first attempt to explore the bioavailability of phosphonates to any eukaryotic phytoplankton and some interesting insights can be derived from this study.
Absence of the Function to Utilize Phosphonate in Dinoflagellates
It appeared that 2-AEP was utilized as a P source in initial K. veneficum culture experiments, while no induction of PhnW and PhnX by 2-AEP was detected either at the transcriptional or the translational level. Instead, the lower PhnX protein expression levels in 2-AEP condition compared to P-replete condition showed that the intracellular 2-AEP metabolism or related P recycling in K. veneficum was also slowed down by Pi deficiency, despite the substantial amounts of 2-AEP in the extracellular environment. Further culture experiments showed that it was the bacteria instead of the dinoflagellates that were responsible for the release of phosphate from 2-AEP and dinoflagellates could not directly utilize 2-AEP as a P source.
We tried to obtain some insights into why dinoflagellates possess and express PhnW and PhnX genes but these genes are not functional in utilizing 2-AEP in the growth medium. Our computational prediction of subcellular localization showed that both PhnW and PhnX were intracellular enzymes, suggesting that transport of phosphonates into the cell would be required in order for the algae to be able to utilize 2-AEP in the external environment. Yet our search into dinoflagellate molecular datasets yielded none of the phosphonate transporters previously reported in bacteria such as PhnCDE and multicomponent 2-AEP transporter (Jiang et al., 1995; Dyhrman et al., 2006). If any, phosphonate transporters in dinoflagellates must be far divergent from those in bacteria and cyanobacteria that they are currently unrecognizable.
Ecological Significance of Phosphonate Utilization through Bacteria in the Marine Environment
The results of our culture experiments indicated that although dinoflagellates were unable to directly utilize external phosphonates, bacteria present in the cultures effectively made the phosphorus in 2-AEP available to dinoflagellates. It is interesting to note that in the filtrate incubation experiment the DIP concentration continued to increase even when there was a high concentration of added DIP in the culture. This observation indicated that the catabolism of 2-AEP was Pi-independent, which is consistent with early studies showing that 2-AEP catabolism in some bacteria is not restricted by the presence of Pi (Quinn et al., 2007). From the normal cultures we noted that the amount of Pi released from 2-AEP through the action of bacteria was rather high, sufficient to support the growth of dinoflagellates, indicating that these bacteria have quite a strong ability to cleave 2-AEP and release Pi into the environment.
An “acclimation period” for phosphonte utilization has been described in bacteria as well as in cyanobacteria (Kononova and Nesmeyanova, 2002; Gomez-Garcia et al., 2010). Similar results were also observed in our work, and the different lag periods preceding net growth in different dinoflagellate species might be due to differences in the species composition of the microbiomes associated with the dinoflagellate cultures (Figures 1A, 3).
Bacteria that are able to metabolize phosphonates are widely distributed in the marine environment (Villarreal-Chiu et al., 2012). Considering that phosphonates contribute 25% of high-molecular-weight DOP in the ocean, the utilization of natural phosphonates through the mediation of bacteria could supply a substantial amount of Pi to phytoplankton in the euphotic zone of P-limited waters. Further study is needed to gain understanding on whether some of the bacteria are physically associated with the dinoflagellates, as many bacteria have been shown to have intimate association with algae in the algasphere (Seyedsayamdost et al., 2011).
Phosphonate Metabolism in Dinoflagellates
Our search into various datasets showed that genes encoding PhnW and PhnX occur widely in dinoflagellate species (Table 1). It is curious what role PhnW and PhnX may play while they do not facilitate utilization of external 2-AEP. We found that besides PhnW and PhnX, genes encoding PepM and Ppd, the two key enzymes involved in phosphonate biosynthesis in microorganisms (Metcalf and van der Donk, 2009), are also widely distributed in dinoflagellate species. Thus, it is apparent that a phosphonate metabolism pathway including both the biosynthesis and hydrolysis of 2-AEP exist inside dinoflagellate cells. As conceptually shown in Figure 5, the biosynthesis of 2-AEP from phosphoenolpyruvate (PEP) can occur through a pathway consisting of PepM, Ppd, and PhnW, while the PhnW-PhnX pathway can function to metabolize (recycle) the intracellular 2-AEP. Because PepM and PhnW catalyze reversible reactions, PhnW can take part in not only the hydrolysis but also the biosynthesis of 2-AEP. Extracellular 2-AEP cannot be utilized by dinoflagellates apparently due to the absence of 2-AEP transporters in the cell membrane, as initially suggested by the lack of a gene homolog in the genome of S. kawagutii (Lin et al., 2015) and many transcriptomic datasets of other dinoflagellate species.
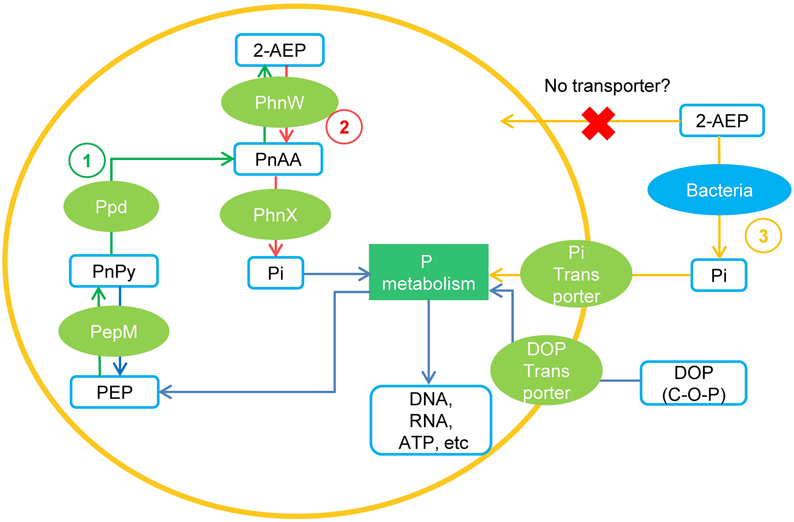
Figure 5. A conceptual model of 2-AEP metabolism in dinoflagelates inferred from current data. (1) Pathway of 2-AEP biosynthesis in the algal cells; (2) Pathway of 2-AEP metabolism (recycling) inside the algal cells. (3) Pathway of dinoflagellates obtaining Pi from environmental 2-AEP through the mediation of bacteria. PEP, phosphoenolpyruvate; PnPy, phosphonopyruvate; PnAA, phosphonoacetaldehyde.
Phosphonates could be a component of membrane phosphonolipids in primitive life forms, and they could increase the rigidity of outer-membrane structure and protect cells from the degradation of enzymes such as phosphatases (Quinn et al., 2007). As such it is no surprise that dinoflagellates, probably other groups of phytoplankton as well, have the genetic potential to synthesize phosphonates. Furthermore, phosphonates might also exist in phytoplankton cells as a form of P storage, which is produced during abundant supply of Pi to sustain phytoplankton growth during Pi-limitation (Ternan et al., 1998). However, the exact function of phosphonates and the PhnW-PhnX pathway in dinoflagellates remains to be further investigated.
Author Contributions
SL conceived the study; SL, YC, and XL designed the research and wrote the paper; YC performed the laboratory work and data analysis; HZ made the K. veneficum cDNA library and assisted with data analysis. LL conducted data mining in search of genes related to phosphonate metabolism in dinoflagellates.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was mainly supported by National Natural Science Foundation of China grant #41176091, #41330959 and explorative research grant # MELRI1203 of the State Key Laboratory of Marine Environmental Sciences at Xiamen University (to SL). The partial sequences of PhnW and PhnX were from a transcriptome dataset generated in a project supported by the National Science Foundation “Microbial Genome Sequence Program” grant EF-0626678 (to SL, HZ, Y.-H. Rogers, T. Gaasterland, and A. Place).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmars.2015.00120
References
Briesemeister, S., Rahnenführer, J., and Kohlbacher, O. (2010). YLoc—an interpretable web server for predicting subcellular localization. Nucleic Acids Res. 38, W497–W502. doi: 10.1093/nar/gkq477
Chou, K. C., and Shen, H. B. (2008). Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3, 153–162. doi: 10.1038/nprot.2007.494
Clark, L. L., Ingall, E. D., and Benner, R. (1998). Marine phosphorus is selectively remineralized. Nature 393, 426–426. doi: 10.1038/30881
Duhamel, S., Dyhrman, S. T., and Karl, D. M. (2010). Alkaline phosphatase activity and regulation in the North Pacific Subtropical Gyre. Limnol. Oceanogr. 55, 1414–1425. doi: 10.4319/lo.2010.55.3.1414
Dyhrman, S. T., Chappell, P. D., Haley, S. T., Moffett, J. W., Orchard, E. D., Waterbury, J. B., et al. (2006). Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439, 68–71. doi: 10.1038/nature04203
Dyhrman, S. N. T., Ammerman, J. W., and Van Mooy, B. A. S. (2007). Microbes and the marine phosphorus cycle. Oceanography 20, 110–116. doi: 10.5670/oceanog.2007.54
Dyhrman, S. T., Benitez-Nelson, C. R., Orchard, E. D., Haley, S. T., and Pellechia, P. J. (2009). A microbial source of phosphonates in oligotrophic marine systems. Nat. Geosci. 2, 696–699. doi: 10.1038/ngeo639
Gomez-Garcia, M. R., Davison, M., Blain-Hartnung, M., Grossman, A. R., and Bhaya, D. (2010). Alternative pathways for phosphonate metabolism in thermophilic cyanobacteria from microbial mats. ISME J. 5, 141–149. doi: 10.1038/ismej.2010.96
Hofmann, K. (1993). TMbase-A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374, 166
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic. Acids. Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Hove-Jensen, B., Zechel, D. L., and Jochimsen, B. (2014). Utilization of glyphosate as phosphate source: biochemistry and genetics of bacterial carbon-phosphorus lyase. Microbiol. Mol. Biol. Rev. 78, 176–197. doi: 10.1128/MMBR.00040-13
Jiang, W., Metcalf, W. W., Lee, K.-S., and Wanner, B. L. (1995). Molecular cloning, mapping, and regulation of Pho regulon genes for phosphonate breakdown by the phosphonatase pathway of Salmonella typhimurium LT2. J. Bacteriol. 177, 6411–6421.
Karl, D. M. (2014). Microbially mediated transformations of phosphorus in the sea: new views of an old cycle. Annu. Rev. Mar. Sci. 6, 279–337. doi: 10.1146/annurev-marine-010213-135046
Kim, A. D., Baker, A. S., Dunaway-Mariano, D., Metcalf, W., Wanner, B., and Martin, B. M. (2002). The 2-aminoethylphosphonate-specific transaminase of the 2-aminoethylphosphonate degradation pathway. J. Bacteriol. 184, 4134–4140. doi: 10.1128/JB.184.15.4134-4140.2002
Kolowith, L. C., Ingall, E. D., and Benner, R. (2001). Composition and cycling of marine organic phosphorus. Limnol. Oceanogr. 46, 309–320. doi: 10.4319/lo.2001.46.2.0309
Kononova, S. V., and Nesmeyanova, M. A. (2002). Phosphonates and their degradation by microorganisms. Biochemistry(Mosc.) 67, 184–195. doi: 10.1023/A:1014409929875
Li, C., Zhang, Y., Xie, Z. X., He, Z. P., Lin, L., and Wang, D. Z. (2013). Quantitative proteomic analysis reveals evolutionary divergence and species-specific peptides in the Alexandrium tamarense complex (Dinophyceae). J. proteomics 86, 85–96. doi: 10.1016/j.jprot.2013.05.007
Lin, S., Chang, J., and Carpenter, E. J. (1994). Detection of proliferating cell nuclear antigen analog in four species of marine phytoplankton. J. Phycol. 30, 449–456. doi: 10.1111/j.0022-3646.1994.00449.x
Lin, S., Cheng, S., Song, B., Zhong, X., Lin, X., Li, W., et al. (2015). The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350, 691–694. doi: 10.1126/science.aad0408
Lin, S., Litaker, R. W., and Sunda, W. G. (accepted). Phosphorus physiological ecology molecular mechanisms in marine phytoplankton. J. Phycol. doi: 10.1111/jpy.12365
Lin, S., Zhang, H., Zhuang, Y., Tran, B., and Gill, J. (2010). Spliced leader–based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc. Natl. Acad. Sci. U.S.A. 107, 20033–20038. doi: 10.1073/pnas.1007246107
Lin, X., Zhang, H., Cui, Y., and Lin, S. (2012a). High sequence variability, diverse subcellular localizations, and ecological implications of alkaline phosphatase in dinoflagellates and other eukaryotic phytoplankton. Front. Microbiol. 3:325. doi: 10.3389/fmicb.2012.00235
Lin, X., Zhang, H., Huang, B., and Lin, S. (2011). Alkaline phosphatase gene sequence and transcriptional regulation by phosphate limitation in Amphidinium carterae (Dinophyceae). J. Phycol. 47, 1110–1120. doi: 10.1111/j.1529-8817.2011.01038.x
Lin, X., Zhang, H., Huang, B., and Lin, S. (2012b). Alkaline phosphatase gene sequence characteristics and transcriptional regulation by phosphate limitation in Karenia brevis (Dinophyceae). Harmful Algae 17, 14–24. doi: 10.1016/j.hal.2012.02.005
Mather, R. L., Reynolds, S. E., Wolff, G. A., Williams, R. G., Torres-Valdes, S., Woodward, E. M. S., et al. (2008). Phosphorus cycling in the North and South Atlantic Ocean subtropical gyres. Nat. Geosci. 1, 439–443. doi: 10.1038/ngeo232
McGrath, J. W., Chin, J. P., and Quinn, J. P. (2013). Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules. Nat. Rev. Microbiol. 11, 412–419. doi: 10.1038/nrmicro3011
Metcalf, W. W., and van der Donk, W. A. (2009). Biosynthesis of phosphonic and phosphinic acid natural products. Annu. Rev. Biochem. 78, 65–94. doi: 10.1146/annurev.biochem.78.091707.100215
Mills, M. M., Ridame, C., Davey, M., La Roche, J., and Geider, R. J. (2004). Iron and phosphorus co-limit nitrogen fixation in the eastern tropical North Atlantic. Nature 429, 292–294. doi: 10.1038/nature02550
Morais, M. C., Zhang, G., Zhang, W., Olsen, D. B., Dunaway-Mariano, D., and Allen, K. N. (2004). X-ray crystallographic and site-directed mutagenesis analysis of the mechanism of Schiff-base formation in phosphonoacetaldehyde hydrolase catalysis. J. Biol. Chem. 279, 9353–9361. doi: 10.1074/jbc.M312345200
Nowack, B. (2003). Environmental chemistry of phosphonates. Water Res. 37, 2533–2546. doi: 10.1016/S0043-1354(03)00079-4
Orchard, E. D., Webb, E. A., and Dyhrman, S. T. (2009). Molecular analysis of the phosphorus starvation response in Trichodesmium spp. Environ. Microbiol. 11, 2400–2411. doi: 10.1111/j.1462-2920.2009.01968.x
Paytan, A., and McLaughlin, K. (2007). The oceanic phosphorus cycle. Chem. Rev. 107, 563–576. doi: 10.1021/cr0503613
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Pierleoni, A., Martelli, P. L., and Casadio, R. (2008). PredGPI: a GPI-anchor predictor. BMC Bioinformatics 9:392. doi: 10.1186/1471-2105-9-392
Quin, L. D. (1965). The prsence of compounds with a carbon-phosphorus bond in some marine invertebrates. Biochemistry 4, 324–330. doi: 10.1021/bi00878a022
Quin, L. D., and Quin, G. S. (2001). Screening for carbon-bound phosphorus in marine animals by high-resolution 31 P-NMR spectroscopy: coastal and hydrothermal vent invertebrates. Comp. Biochem. Phys. B 128, 173–185. doi: 10.1016/S1096-4959(00)00310-9
Quinn, J. P., Kulakova, A. N., Cooley, N. A., and McGrath, J. W. (2007). New ways to break an old bond: the bacterial carbon–phosphorus hydrolases and their role in biogeochemical phosphorus cycling. Environ. Microbiol. 9, 2392–2400. doi: 10.1111/j.1462-2920.2007.01397.x
Sañudo-Wilhelmy, S. A., Kustka, A. B., Gobler, C. J., Hutchins, D. A., Yang, M., Lwiza, K., et al. (2001). Phosphorus limitation of nitrogen fixation by Trichodesmium in the central Atlantic Ocean. Nature 411, 66–69. doi: 10.1038/35075041
Seyedsayamdost, M. R., Carr, G., Kolter, R., and Clardy, J. (2011). Roseobacticides: small molecule modulators of an algal-bacterial symbiosis. J. Am. Chem. Soc. 133, 18343–18349. doi: 10.1021/ja207172s
Shi, X., Zhang, H., and Lin, S. (2013). Tandem repeats, high copy number and remarkable diel expression rhythm of form II RuBisCO in Prorocentrum donghaiense (dinophyceae). PLoS ONE 8:e71232. doi: 10.1371/journal.pone.0071232
Snyder, D. S., Brahamsha, B., Azadi, P., and Palenik, B. (2009). Structure of compositionally simple lipopolysaccharide from marine Synechococcus. J. Bacteriol. 191, 5499–5509. doi: 10.1128/JB.00121-09
Ternan, N. G., McGrath, J. W., McMullan, G., and Quinn, J. P. (1998). Review: organophosphonates: occurrence, synthesis and biodegradation by microorganisms. World J. Microbiol. Biotechnol. 14, 635–647. doi: 10.1023/A:1008848401799
Timothy, R., Yoshiaki, M., and Carol, M. (1984). A Manual of Chemical and Biological Methods for Seawater Analysis. New York, NY: Pergamon Press.
Tyrrell, T. (1999). The relative influences of nitrogen and phosphorus on oceanic primary production. Nature 400, 525–531. doi: 10.1038/22941
Van Mooy, B. A. S., Fredricks, H. F., Pedler, B. E., Dyhrman, S. T., Karl, D. M., Koblížek, M., et al. (2009). Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458, 69–72. doi: 10.1038/nature07659
Villarreal-Chiu, J. F., Quinn, J. P., and McGrath, J. W. (2012). The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front. Microbiol. 3:19. doi: 10.3389/fmicb.2012.00019
White, A. K., and Metcalf, W. W. (2004). Two C-P Lyase operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J. Bacteriol. 186, 4730–4739. doi: 10.1128/JB.186.14.4730-4739.2004
Wurch, L. L., Bertrand, E. M., Saito, M. A., Van Mooy, B. A. S., and Dyhrman, S. T. (2011). Proteome changes driven by phosphorus deficiency and recovery in the brown tide-forming alga Aureococcus anophagefferens. PLoS ONE 6:e28949. doi: 10.1371/journal.pone.0028949
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Keywords: phosphorus, phosphonate, dinoflagellates, PhnW-PhnX pathway, phosphonate metabolism
Citation: Cui Y, Lin X, Zhang H, Lin L and Lin S (2016) PhnW-PhnX Pathway in Dinoflagellates Not Functional to Utilize Extracellular Phosphonates. Front. Mar. Sci. 2:120. doi: 10.3389/fmars.2015.00120
Received: 28 September 2015; Accepted: 21 December 2015;
Published: 11 January 2016.
Edited by:
George S. Bullerjahn, Bowling Green State University, USAReviewed by:
Benjamin Van Mooy, Woods Hole Oceanographic Institution, USAJohn Quinn, Queen's University, UK
Copyright © 2016 Cui, Lin, Zhang, Lin and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Senjie Lin, c2VuamllLmxpbkB1Y29ubi5lZHU=
 Yudong Cui
Yudong Cui Xin Lin
Xin Lin Huan Zhang
Huan Zhang Lingxiao Lin
Lingxiao Lin Senjie Lin
Senjie Lin