- Department of Biology, School of Life Sciences, Xiamen University, Xiamen, China
A small flagellate alga was isolated from the phycosphere of a toxic red tide dinoflagellate Alexandrium tamarense. Phylogenetic analysis and ultrastructural observations demonstrated that the small flagellate alga is a species belong to Ochrophyte Ochromonas sp. The process of ingesting bacteria by Ochromonas sp. was recorded by a time lapse capture under a light microscope. Through the use of different assemblages in the co-culture experiment, the species interactions in this phycosphere microenvironment were analyzed. We demonstrated that the growth of Ochromonas sp. was supported by bacteria. Three strains of bacteria ingested by Ochromonas sp. were isolated and identified to belong to α-, δ- and γ-Proteobacteria. The growth of A. tamarense was suppressed when co-cultured with bacteria. In contrast, Ochromonas sp. triggered the growth of A. tamarense by inhibiting the growth of algicidal bacteria. This result firstly demonstrated a positive effect of a flagellate on a dinoflagellate in the phycosphere of A. tamarense. Combined with other negative effects between dinoflagellates and bacteria or bacteria and flagellates, this study showed a series of clear interactions among dinoflagellate, bacterium, and flagellate in the dinoflagellate microenvironment.
Introduction
Quantitative studies about the interactions among species in microecosystems are critical because these interactions control the structure and function of the ecosystem as a whole (Paine, 1966; Wootton and Emmerson, 2005). Algal-bacterial interactions are a typical model in microecosystem studies (Doucette et al., 1998; Amaro et al., 2005), in part because bacteria transform energy and regenerate inorganic nutrient in marine ecosystems (Azam et al., 1983; Su et al., 2011). Alexandrium tamarense is a model dinoflagellate alga that is globally distributed and known for producing harmful algal blooms (HABs) that cause paralytic shellfish poisoning (Anderson et al., 1996). A. tamarense growth is inhibited by certain marine bacteria, including Brevibacterium (Bai et al., 2011), Thalassobius, Alteromonoas, Rhodobacteracea (Wang et al., 2010), Pseudoalteromonas (Su et al., 2007b; Wang et al., 2012), Vibrio and Halomonas (Su et al., 2011). The mechanism is allelopathy, a natural phenomenon whereby a chemical released by one organism serve as a signal affecting the interaction among other organisms (Zhou et al., 2007). The allelopathy of algicidal bacteria could be an effective way to manage HABs (Manage et al., 2001; Li et al., 2014). The allelochemicals released by A. tamarense tend to negatively affect competing algae (Tillmann et al., 2008; Tillmann and Hansen, 2009), though it is possible that they may positive effects on other organisms.
Aside from bacteria, dinoflagellates are also known to associate with flagellates, which sometimes consume bacteria in marine and freshwater habitats (Fenchel, 1982; Sieburth and Davis, 1982). Ochromonas are marine, brackish, and freshwater flagellates that contain one or two chloroplasts (Doddema and Van Der Veer, 1983; Andersson et al., 1985). However, the photosynthetic apparatus of Ochromonas is less efficient than that of other algae (Myers and Graham, 1956). Ultrastructural research revealed that Ochromonas possess highly developed phagotrophic organs, which enables efficient phagotrophy of the bacteria (Bouck, 1971; Aaronson, 1974). These characteristics suggest that Ochromonas plays an important role in moderating bacterial abundance through direct ingestion (Pringsheim, 1952). Additionally, some Ochromonas excrete antibiotic compounds (Hanse, 1973; Blom and Pernthaler, 2010).
The “phycosphere” is an environmental region extending outward from an algal cell where bacterial growth is stimulated by algal extracellular products (Bell and Mitchell, 1972). Bacteria in the phycosphere may be free-living (Blackburn et al., 1998), attached to the algal surface (Vaqué et al., 1990). The interactions, or “microbial loop” (Azam, 1998), between the algae and bacteria is intimate (Wang et al., 2010). The high level of organic compounds released by microalgae stimulate nearby bacteria, which transform these organic compounds into inorganic nutrients (Azam and Ammerman, 1984). The biomass of bacteria increase dramatically during the decline of the algal bloom, suggesting that lytic bacteria play an important role in the control and elimination of the algal bloom (Mayali and Azam, 2004; Zheng et al., 2005). Bacteria in the phycosphere can inhibit the growth of algae or even lyse algal cells in a short time (Lee et al., 2000; Mayali and Azam, 2004; Su et al., 2007a).
Because of the complex relationships between these organisms, little is known about the interactions between different algae, bacteria, and other members of the phycosphere. To characterize and quantify these interactions, we set up a time lapse microscopy recording system and used culturing techniques to check for the presence (and measure the performance) of other algal and bacteria species within the A. tamarense phycosphere. We found evidence for complex interactions between A. tamarense, Ochromonas, and several bacteria.
Materials and Methods
Algal Cultures
A. tamarense (strain no.ATGD98-006) was obtained from the Algal Culture Collection, Institute of Hydrobiology, Jinan University (Guangzhou China). The alga (containing A. tamarense and its phycosphere microorganisms) was cultured in f/2 medium (75 mg NaNO3, 5 mg NaH2PO4.H2O, 4.36 mg Na2EDTA, 3.15 mg FeCl3.6H2O, 0.01 mg CuSO4.5H2O, 0.022 mg ZnSO4.7H2O, 0.01 mg CoCl2.6H2O, 0.18 mg MnCl2.4H2O, 0.006 mg Na2MoO4.2H2O, 0.1 mg thiamine HCl, 0.5 μg biotin, and 0.5 μg vitamin B12, in 1 L filtered seawater, Guillard, 1975) at 20±1 °C under a 12:12 (light/dark) cycle with an illumination of 50 μE/m2 s.
Isolation of A. tamarense, Flagellates and Bacteria
At mid-exponential phase, 100 mL of the algal culture was filtered through a 5-μm Isopore membrane filter. The algal cells of A. tamarense on the filter were immediately suspended in 50 mL of sterile f/2 medium and then purified according to a procedure for removing bacteria (Su et al., 2007a).
Flagellates in the supernatant were cultured in f/2 medium using the same conditions as mentioned above. The nutrition of mixotroph flagellate cells was maintained with bacteria in f/2 medium. Axenic cultures of the flagellates were produced by adding a mixture of antibiotics (chloramphenicol, streptomycin, and gentamycin; ratio 1: 1: 0.5) (Corno and Jürgens, 2006), followed by culture for 5 days.
To obtain bacteria cultures, 10 mL of mid-exponential phase algal cultures (containing A. tamarense and its phycosphere microorganisms) were filtered through a 1-μm Isopore membrane filter. The supernatant was added into Erlenmeyer flasks with 25 mL of 2216E medium (peptone, 5 g; yeast extraction, 1 g; ferric phosphorous acid, 0.1 g; agar, 10 g; pH 7.6–7.8, fixed capacity to 1 L using natural sea water), followed by incubation for 24 h at 28°C with shaking at 120 rpm.
Analysis of the Symbiotic Relationship Between Algae and Bacteria
To analyze interactions in the phycosphere, we recombined A. tamarense, flagellates and bacteria in a series of co-culture treatments (Table 1). In the co-culture experiment, the growth of each species was monitored. The initial density of A. tamarense, flagellates and bacteria were approximately 1.0 × 104 cell/mL, 5.0 × 104 cell/mL and 5.0 × 106 cell/mL, respectively. To determine whether Ochromonas can ingest dead cells of bacteria, we added cells heat-killed at 70°C for 2 h (treatment 5). Each co-culture experiment was conducted in triplicate. Samples for algal density measurements were collected every day and fixed with Lugol's iodine and counted using microscopy with a hemocytometer. Bacterial abundance was estimated by microscopic cell counting following the protocol of Patel et al. (2007).
Results were analyzed using Microsoft Excel (Microsoft Corporation, Redmond, Washington). Averaged results were presented as mean ±SEM taken from the number of experiments indicated. Statistical significance was evaluated using Student's t-test. All data were from three independent experiments.
Cells of A. tamarense in the exponential growth phase were assigned to 24-well plates and incubated with bacteria (1.0 × 109 cell/mL). The lysis process in cells was observed by an inverted microscope equipped with a CCD camera.
Identification of Flagellates and Ingested Bacteria
Flagellates and attached bacteria were isolated from A. tamarense cultures by filtering through a 5-μm Isopore membrane filter. The genomic DNA of the flagellate was extracted using the DNeasy Plant Mini Kit (Qiagen Valencia, CA, Catalog no. 69104), according to the manufacturer's suggested protocols. 18S rRNA genes were amplified using the forward primer SSU-F (5′-ACCTGGTTGATCCTGCCAGT-3′) and the reverse primer SSU-R (5′-TCACCTACGGAAACCTTGT-3′). PCR was conducted in a 50-μL reaction mixture containing 4 μL of genomic DNA, 2 μL of each primer, 25 μL of GOTaq-Colorless Master Mix, 2 × and 12 μL of ddH2O. Cyclic sequencing reactions included an initial denaturation for 10 min at 94°C, then 32 cycles of 30 s at 94°C, 45 s at 55°C, and 90 s at 72°C, followed by a final extension step of 10 min at 72°C. The PCR product was cloned into the pGEM-T Easy (Promega) vector, and the resulting plasmid was sequenced. Isolation and 16S rRNA sequencing protocols (Oh et al., 2011) were used to identify the ingested bacteria that remained within the flagellate.
Microscopy Observation
The cultured cells were centrifuged (3000 r/min, 5 min), and the medium was removed and replaced by 0.1 M PBS (pH 7.4). Cells were transferred onto a coverslip presoaked with 5% HNO3. The slide was then stored in a specific container and 2.5% glutaraldehyde in 0.1 M PBS (pH 7.4) was added to fix the flagellates for 24 h. The glutaraldehyde solution was replaced with 0.1 M PBS (pH 7.4). After three buffer exchanges, each lasting for 15 min, the slides were taken out and naturally dried in a clean vessel. The slides were mounted on a specimen holder and dried at 30°C in a dry chamber and then sputtered with gold-palladium in a SCD Sputter Coater before examination with a JMS6390LV scanning electron microscope (JEOL, Japan).
The collected cells were fixed in 2.5% glutaraldehyde in 0.1 M PBS (pH 7.4) for 2 h at 4°C and were then embedded in 2% agarose (Reize and Melkonian, 1989) and fixed in 2.5% glutaraldehyde in 0.1 M PBS (pH 7.4) over night. The glutaraldehyde solution was replaced with 0.1 M PBS (pH 7.4). After being rinsed three times for 15 min in 0.1 M PBS (pH 7.4), cells in agarose were post-fixed in 1% OsO4 for 2–3 h at room temperature. The cells were dehydrated through a graded series of ethanol and propylene oxide, and then embedded in an Epon812 Embedding medium (3 g of DDSA, 7 g of MNA, and 10 g of Epon812 in 0.32 mL of DMP). Ultrathin sections were made using a PowerTomo-XL ultramicrotome (RMC, U.S.A.), collected on 200-mesh copper grids, and post-stained with 1% aqueous uranylacetate and lead citrate (Reynolds, 1963) before viewing with a JEM2100HC electron microscope (JEOL, Japan).
Because the small size of marine bacteria makes micrography under a light microscopy unsuitable, the samples were stained by SYBR Green I (Molecular Probes–Invitrogen) before being photographed. Fluorescence and bright field images were taken for the same vision by using a fluorescence microscope (Olympus BX-41, Olympus, Inc.).
Results
Biological Characteristic of the Ochromonas
Light microscopy and electron microscopy (TEM and SEM) were used to investigate the morphology and ultrastructure of the flagellates. Under light microscopy, the cells swam continuously in the anterior direction and displayed a spiral path, while the long flagellum showed a curved wave, and the posterior flagellum showed no active motion but may have acted like a rudder to change direction. The cells ingesting food were non-motile because the flagellum is an important tool in capturing of particles (see Supplementary Video 1).
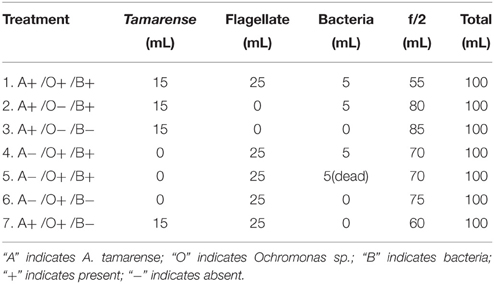
As shown in Figure 1, the flagellate cells were spherical, naked and only covered by the plasmalemma. The fluorescence microscopy images indicate that each flagellate cell contains two chloroplasts (Figure 1B). The cell measured 2–3 μm, and two unequal flagella arose from the anterior face of the cell. The long anterior flagellum (approximately 3–5 times the length of the cell), which bore mastigoneme (Figure 1D), is directed forwards. The short smooth posterior flagellum (equal to the body length), which was not easily observed under light microscopy, emerges laterally. The TEM image shows that the flagella are approximately 250 nm in diameter and that the flagellar fibers form the usual “9+2” pattern in the cross section (Figure 1F). The nucleus and chloroplast are enclosed by the chloroplast endoplasmatic reticulum, which may be continuous with the nuclear envelope. The chloroplast (Figure 1E) is bound by a double chloroplast envelope, and inside, there are arranged lamellae composed of three adpressed thylakoids. Three to four pigment granules were observed to be located on the face of the lamellae. Over half of the cell volume is occupied by the nucleus and chloroplast.
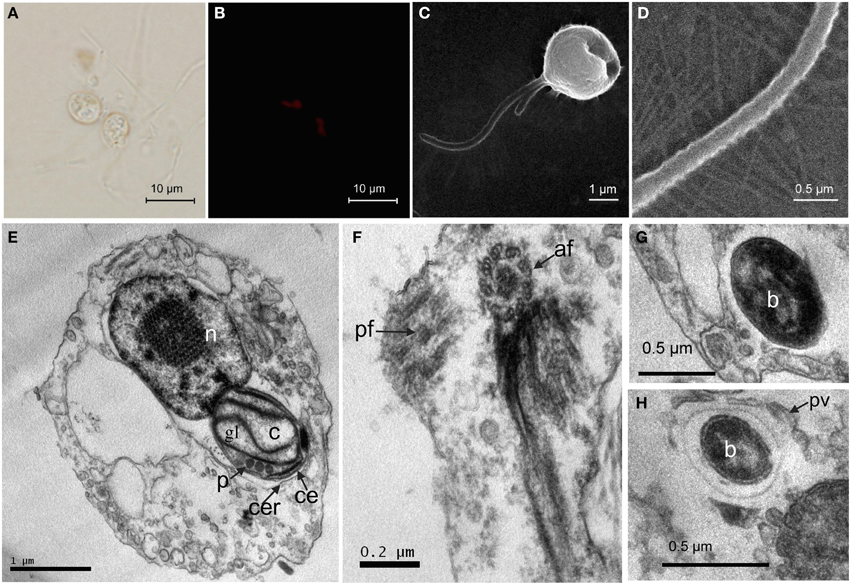
Figure 1. Morphology and ultrastructure of the flagellate. (A) Bright-field image of the flagellate; (B) Autofluorescence of a chloroplast under the fluorescence microscopy; (C) SEM image showing the anterior flagellum (AF) and the posterior flagellum (PF); (D) High magnification view of the mastigoneme bearing AF; (E) Flagellate cell containing a single nucleus (n), a chloroplast (c). The chloroplast (c) is surrounded by the chloroplast endoplasmic reticulum (cer), which is connected to the nuclear envelope. The girdle lamella (gl) lies parallel to the double layers of the chloroplasts envelope (ce), and three pigment granules (p) are within the chloroplast; (F) Cross section of a posterior flagellum (pf), anterior flagellum (af); (G) Bacteria (b) within the flagellate. (H) Protoplasmic vesicle (pv) surrounding a bacterium (b).
To obtain the phylogenetic information of the flagellate, 18S rRNA sequencing was performed and the 1700-bp gene fragments were compared. According to the blast results of the 18S rRNA gene sequence (GenBank accession numbers KT877396) on NCBI, 18 strains of algae from different locations with a similar sequence to the flagellate were selected to construct the phylogenetic tree (Figure 2). Combined with the morphology and ultrastructure result, the flagellate in this study belonged to the Ochromonas.
Figure 2. Phylogenetic tree of the flagellate based on 18S rRNA gene sequence. Phylogenetic tree based on comparison of the 18S rRNA gene sequences indicating that this flagellate (in bold) belongs to Ochromonas. The phylogenetic tree was generated using the neighbor-joining method. Bootstrap values, expressed as percentages of 1000 replications, are given at the branching points. The scale bar indicates substitutions per nucleotide position.
Inhibitory Activity of Ochromonas sp. on Bacteria
The process of Ochromonas sp. capturing bacteria was observed (Supplementary Video 1). First, sessile flagellate cells are observed to cause the bacteria and other particle approaching them to spin by using the long anterior flagellum. Once the bacteria move into the reach of the long flagellum, they lie within a clear vesicle and are rapidly taken into the algal cell body. The Ochromonas sp. can capture 6–8 bacteria, and the captured bacteria moving in the cell of Ochromonas sp. also can be observed.
To determine whether the Ochromonas sp. was supported by bacteria, growth of Ochromonas sp. in different treatments was monitored, as is illustrated in Figure 3. Ochromonas sp. appeared to have no increase in cell density after day 1 in axenic cultures compared with cultures fed with heat-killed bacteria, which showed a slight increase in cell density of 4.93 × 104 cell/mL. In contrast, Ochromonas sp. co-cultured with live bacteria showed a rapid increase in growth and reached a maximum density of 2.74 × 105 cell/mL on the 15th day. In the treatment with Ochromonas sp. co-cultured with A. tamarense but no bacteria, the growth of Ochromonas sp. was similar to the treatment of axenic cultures. After the addition of both bacteria and A. tamarense, growth of Ochromonas sp. was suppressed at first. Then, there was a sudden growth increase on the 7th day. Moreover, it reached a maximum density of 3.26 × 105 cell/mL on the 15th day.
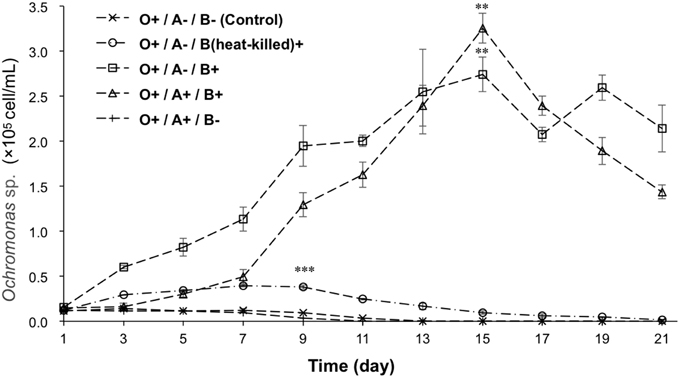
Figure 3. The growth curves of Ochromonas sp. under the effects of bacteria and A. tamarense. “A” indicates A. tamarense; “O” indicates Ochromonas sp.; “B” indicates bacteria; “+” indicates present; “−” indicates absent. The asterisks **, and *** denoted statistical significance with p-values less than 0.01, and 0.001, respectively. All error bars indicate the SE of the three biological replicates.
To investigate the impact of Ochromonas sp. on the bacteria, the bacterial abundance in both treatments where Ochromonas sp. was present and absent were estimated. The result showed bacterial growth was inhibited by Ochromonas sp. (Figure 4).
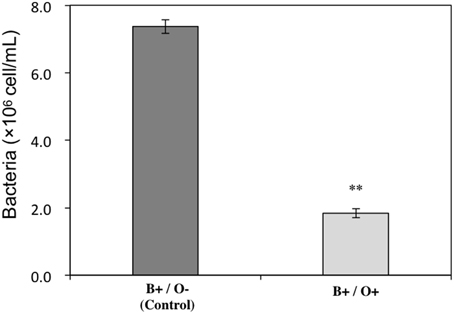
Figure 4. Inhibitory effect of Ochromonas sp. on the bacteria cocultured with A. tamarense. “O” indicates Ochromonas sp.; “B” indicates bacteria; “+” indicates present; “−” indicates absent. The asterisks ** denoted statistical significance with p-values less than 0.01. All error bars indicate the SE of the three biological replicates.
Identification of Ingested Bacteria of Ochromonas sp.
To obtain the phylogenetic information of the bacteria ingested by this Ochromonas sp., 16S rRNA sequencing and phylogenetic analysis was performed. As shown in Figure 5, the sequencing results indicate that isolate 1 (GenBank accession numbers KT877397) belongs to the δ-subdivisions of Proteobacteria, isolate 2 (GenBank accession numbers KT877398) exhibits 100% identity to Roseobacter sp. JL-351 and isolate 3 (GenBank accession numbers KT877399) shows 100% identity to Maribacter sp. E-8-2, which belongs to Alteromonadaceae.
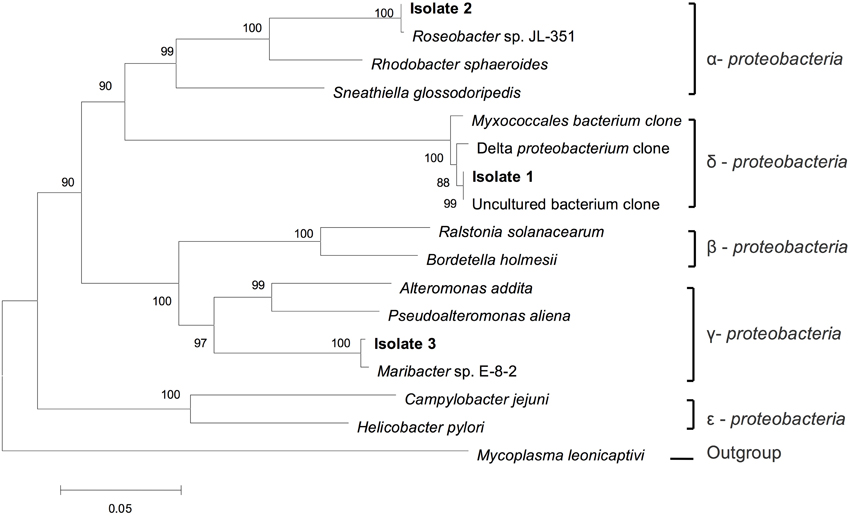
Figure 5. Phylogenetic tree of the ingested bacteria. Phylogenetic tree based on a comparison of the 16S rRNA gene sequences, indicating the phylogenetic position of the strains isolated (isolation 1, 2, and 3) from flagellate culture. The phylogenetic tree was generated using the neighbor-joining method. Bootstrap values, expressed as percentages of 1000 replications, are given at the branching points. The scale bar indicates substitutions per nucleotide position.
Algicidal Effects of Bacteria on the Growth of A. tamarense
The effects of bacteria on the lysis of A. tamarense cells in the absence of Ochromonas sp. were observed continuously for 24 h under light microscopy. Figure 6 shows the morphological changes of the A. tamarense cells, which were incubated with bacteria at a density of 109 cell/mL. Figure 6A shows the normal cells with the integrity of the cell membrane and cell wall when the bacteria were just added. After 10 min of treatment, some cells began to break and many velum vesicles appeared on the girdle and sulcus of the cell wall and some cells escaped from their cell wall. With the exposure time increased to 30 min, the cells lysed, and the decomposed components were released from the cell (Figure 6C). After 24 h, most of the cells were degraded into debris and covered by a layer of bacteria.
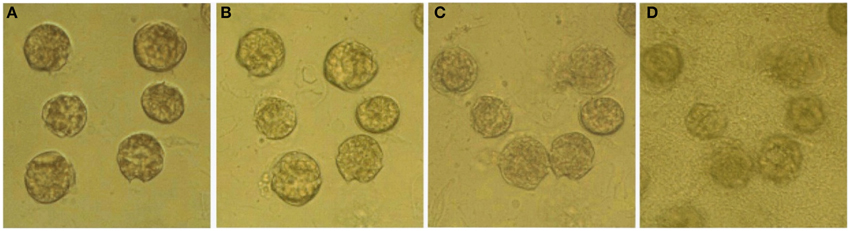
Figure 6. Variation of algal structure after bacteria were added to A. tamarense cultures. (A–D) respectively present cells treated with bacteria for 0, 10, 30 min and 24 h.
Effects of Ochromonas sp. and Bacteria on the Growth of A. tamarense
To analyze the relationship among A. tamarense, Ochromonas sp., and bacteria, the growth of A. tamarense was investigated. As shown in Figure 7, the axenic culture of A. tamarense increased steadily and reached a maximum density of 3.13 × 104 cell/mL on the 19th day. In the treatment that A. tamarense co-cultured with Ochromonas sp. only, the growth of A. tamarense was similar to the treatment of axenic cultures. However, when co-cultured with bacteria only, a significant growth-inhibiting effect was showed on A. tamarense, which resulted in a lower maximum density of 1.27 × 104 cell/mL compared with the axenic culture. After adding both bacteria and Ochromonas sp., the growth of A. tamarense was similar to the axenic culture and the inhibiting effects of bacteria decreased. In these mixed cultures, Ochromonas sp. was observed to live close to resting A. tamarense cells or within the debris. Fluorescence imaging (Figure 8B) showed that the bacteria spread throughout the entire debris field of A. tamarense.
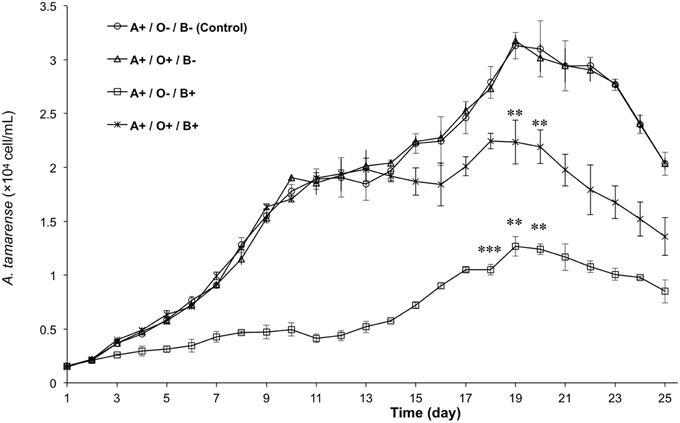
Figure 7. The growth curves of A. tamarense under different effects of Ochromonas sp. and bacteria. “A” indicates A. tamarense; “O” indicates Ochromonas sp.; “B” indicates bacteria; “+” indicates present; “−” indicates absent. The asterisks **, and *** denoted statistical significance with p-values less than 0.01, and 0.001, respectively. All error bars indicate the SE of the three biological replicates.
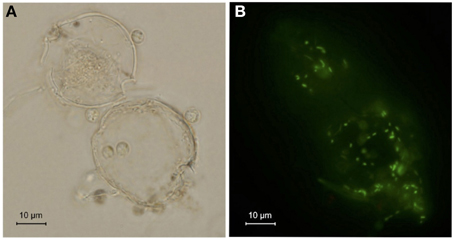
Figure 8. Debris of A. tamarense attached with flagellate and bacteria. (A) Bright-field image. (B) Fluorescence image of bacteria after staining with SYBR Green I.
Discussion
A diagnostic characteristic of the members of genus Ochromonas is that they are mixotrophic (Andersson et al., 1989; Zubkov et al., 2001). Heterotrophy is the major mechanism to support the growth of Ochromonas, and the photosynthetic apparatus is only a survival strategy during poor phototrophic conditions (Andersson et al., 1989). Some studies indicate that axenically pregrown flagellates did not significantly increase in cell number when incubated in inorganic media (Estep et al., 1986; Andersson et al., 1989). Our results showed (Figure 3) that the cell density of axenically cultured flagellate continuously decreased, and after 13 days, no flagellate cell was observed. By adding a certain amount of heat-killed bacteria, the number of flagellate cells slowly increased and started to decline on the 7th day (at a cell density of 3.93 × 104 cell/mL). By contrast, the group that was provided live bacteria showed a rapid increase and reached a maximum density of 2.74 × 105 cell/mL on the 15th day. This is because the heat-killed bacteria provide nutrition to the flagellate in the short term, but living bacteria proliferate and steadily feed the flagellate. The results demonstrate that the autotrophic growth of the flagellate was poor.
The main nutrient strategy of this heterotrophic organism was to phagocytose small particles. Aaronson (1974) studied the phagocytic process of Ochromonas danica and described the vesicles and secretion as “flypaper.” The ingestion of bacteria by Ochromonas cells is mainly attributed to vesicles secreted materials (Aaronson, 1971, 1973). As shown in the TEM images, bacteria were surrounded by vesicles with a double membrane released by the flagellate (Figure 1H), and then they were taken into the cell body and the vesicles were withdrawn into the cytoplasm and formed a food vacuole. Protoplasm near the anterior end of the cells tends to form several small outgrowths, as observed in Figure 1H, where ingestion takes place. A single flagellate cell can continuously take in numerous particles, whatever the nutrient state (Pringsheim, 1952). The whole process was first captured (Supplementary Video 1) by a CCD camera. Sessile Ochromonas sp. cells cause a small bacterium to approach the front of the cell by the long flagellum, and then, the bacterium suddenly stops moving and is observed to lie within a vesicle. Gavaudan (1931) describes a protoplasmic cylinder that closes over the particle and becomes a vesicle surrounding it. However, this structure was not observed in the TEM image, and the formation of the vesicle was too rapid to be seen. The vesicle carried the bacterium inside the cell body, which like a bubble, was formed at the surface of the body. This process only lasted for several seconds from the capture to the phagocytosis. The ingested bacteria kept moving within the Ochromonas sp. cell for a period of time, which suggests that the efficiency of digestion was low.
Grazing by small algae proved to be a major mortality factor for aquatic bacteria (Sherr and Sherr, 2002; Corno and Jürgens, 2006; Zubkov and Tarran, 2008). The Ochromonas diet has been described in previous studies and includes Esherichia coli, Proteus mirabilis, Aerobacter aerogenes, Bacillus megaterium, Sarcina lutea, and Serratia marcescens (Aaronson, 1974), as well as some blue-green alga (Daley et al., 1973; Li et al., 2010) and starch grains, casein, and oil droplets (Pringsheim, 1952). As a mixotrophic bacterivore, Ochromonas sp. showed a significant inhibiting effect on the growth of bacteria that live in the phycosphere of A. tamarense (Figure 4). To establish what types of bacteria are ingested, free-living bacteria were removed, and those that were ingested and remained within the cells of Ochromonas sp. were isolated. Phylogenetic analysis revealed that the three isolates belong to α-, δ-, and γ-Proteobacteria. Many of the α- and γ-Proteobacteria have been shown to be the phycosphere bacteria of A. tamarense (Mayali and Azam, 2004; Bai et al., 2011). The Pseudomonas, alteromonas, Pseudoalteromonas, and Roseobacter group, which show a high identity to our isolations, were well known to be key algicidal bacteria of A. tamarense and many other harmful algae (Lovejoy et al., 1998; Mayali and Azam, 2004; Amaro et al., 2005; Wang et al., 2010; Oh et al., 2011), and this finding suggests that bacteria might be useful for regulating HABs.
In natural communities, species have been found to affect each other both directly and indirectly through negative or positive interactions; thus, the study of species interactions is one of the most fundamental fields in ecology (Callaway et al., 2002). Previous reports generally focused on how negative interactions, including competition, predation and disturbance, affected the community's structure. Over the past two decades, many types of bacteria have been reported to kill algal cells by direct attack (Mayali and Azam, 2004) or produce algicidal substances (Pokrzywinski et al., 2012). These algicidal bacteria sometimes concurrently increase in abundance following the peak of some algal blooms, suggesting that negative interactions may affect algal bloom dynamics (Mayali and Azam, 2004; Pokrzywinski et al., 2012). In our experiment, the growth curves of A. tamarense (Figure 7) showed that axenic cultures of A. tamarense grew quickly and reached a maximum density of 3.13 × 104 cell/mL. By contrast, the growth of cells co-cultured with bacteria (at a density of 106 cell / mL) only reached a maximum density of 1.27 × 104 cell/mL. When treated by high-density bacteria (109 cell/mL), the cells of A. tamarense were lysed and degraded into debris within 24 h (Figure 6). However, some reports indicated that the algicidal activity of bacteria can be influenced by other coexisting organisms, such as predation by heterotrophic protists and competition from non-algicidal bacteria (Mayali and Doucette, 2002; Mayali and Azam, 2004). In our research, the growth of algicidal bacteria was inhibited though the predation of Ochromonas sp. This result revealed that there are two types of negative interactions in the phycosphere of A. tamarense: growth inhibition by algicidal bacteria and predation on bacteria by Ochromonas sp.
After adding Ochromonas sp. to the co-culture system, the inhibition of algal growth was relaxed (Figure 7) and the growth of A. tamarense was controlled at a maximum density of 2.24 × 104 cell/mL. Combined with phylogenetic analysis results, Ochromonas sp. in this study was shown to facilitate the growth of A. tamarense by ingesting algicidal bacteria in the phycosphere of A. tamarense. During our observation, cells of Ochromonas sp. tended to gather around the cell that was attacked by bacteria or even the debris of A. tamarense (Figure 8). Bacteria were attracted to the extracellular products or the A. tamarense cell itself and gathered with a high density. Quickly swimming Ochromonas cells do not seem to ingest food, because the flagellum plays an important role in the capture of particles and, if used in swimming, it would be inefficient for this purpose (Pringsheim, 1946, 1952). Thus, A. tamarense provides suitable habitat for Ochromonas sp. In the early period, the growth of A. tamarense was rapid, and a large amount of nutriments consumed by A. tamarense cells may disturbed bacterial growth, which indirectly affected the growth of Ochromonas sp. With the reduction of nutrition and accumulation of secondary products, the cell vitality of A. tamarense is likely weaker and more easily attacked by bacteria, while the growth of Ochromonas sp. also accelerated. This finding may explain the reason for the slow growth of Ochromonas sp. (Figure 3) at the early period of co-culture with both bacteria and A. tamarense, and the rapid growth from the 7th to the 15th days, leading to a maximum density of 3.26 × 105 cell/mL. The fact that both parties benefit suggested a mutualistic interaction in the phycosphere of A. tamarense. In the treatment where bacteria were absent, A. tamarense and Ochromonas sp. both had no effect to the each other's growth. That suggested there was no direct interaction between A. tamarense and Ochromonas sp. Considering that there are negative interactions of bacteria on A. tamarense and negative interactions of Ochromonas sp. on bacteria, a positive interaction of Ochromonas sp. on A. tamarense could be recognized as emergent effect. That is to say, the growth of A. tamarense was controlled by the bacteria and the bacteria were controlled by Ochromonas sp. In general, the results of species interactions are the final outcome of a balance between negative and positive interactions, and consideration of the role played by species interactions in communities could contribute to maintaining community diversity and stability (Mulder et al., 2001; Cardinale et al., 2002; Butterfield, 2009; Cavieres and Badano, 2009).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (40776082, 31371444).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmars.2015.00100
References
Aaronson, S. (1971). The synthesis of extracellular macromolecules and membranes by a population of the phytoflagellate Ochromonas danica. Limnol. Oceanogr. 16, 1–9. doi: 10.4319/lo.1971.16.1.0001
Aaronson, S. (1973). Particle aggregation and phagotrophy by Ochromonas. Arch. Mikrobiol. 92, 39–44. doi: 10.1007/BF00409509
Aaronson, S. (1974). The biology and ultrastructure of phagotrophy in Ochromonas danica (Chrysophyceae: Chrysomdnadida). J. Gen. Microbiol. 83, 21–29. doi: 10.1099/00221287-83-1-21
Amaro, A. M., Fuentes, M. S., Ogalde, S. R., Venegas, J. A., and Suárez-Isla, B. A. (2005). Identification and characterization of potentially algal-lytic marine bacteria strongly associated with the toxic dinoflagellate Alexandrium catenella. J. Eukaryot. Microbiol. 52, 191–200. doi: 10.1111/j.1550-7408.2005.00031.x
Anderson, D. M., Kulis, D. M., Qi, Y.-Z., Zheng, L., Lu, S., and Lin, Y.-T. (1996). Paralytic shellfish poisoning in Southern China. Toxicon 34, 579–590. doi: 10.1016/0041-0101(95)00158-1
Andersson, A., Falk, S., Samuelsson, G., and Hagström, A. (1989). Nutritional characteristics of a mixotrophic nanoflagellate, Ochromonas sp. Microb. Ecol. 17, 251–262. doi: 10.1007/BF02012838
Andersson, A., Lee, C., Azam, F., and Hagström, A. (1985). Release of amino acids and inorganic nutrients by heterotropic marine microflagellates. Mar. Ecol. Prog. Ser. 23, 99–106. doi: 10.3354/meps023099
Azam, F. (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280, 690–694. doi: 10.1126/science.280.5364.694
Azam, F., and Ammerman, J. W. (1984). Cycling of organic matter by bacterioplankton in pelagic marine ecosystems: microenvironmental considerations. Flows Energy Mater. Mar. Ecosyst. 13, 345–360. doi: 10.1007/978-1-4757-0387-0_14
Azam, F., Fenchel, T., Field, J. G., Gray, J., Meyer-Reil, L., and Thingstad, F. (1983). The ecological role of water-column microbes in the sea. Estuaries 50, 257–263. doi: 10.3354/meps010257
Bai, S. J., Huang, L. P., Su, J. Q., Tian, Y., and Zheng, T. L. (2011). Algicidal effects of a novel marine actinomycete on the toxic dinoflagellate Alexandrium tamarense. Curr. Microbiol. 62, 1774–1781. doi: 10.1007/s00284-011-9927-z
Bell, W., and Mitchell, R. (1972). Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143, 265–277. doi: 10.2307/1540052
Blackburn, N., Fenchel, T., and Mitchell, J. (1998). Microscale nutrient patches in planktonic habitats shown by Chemotactic bacteria. Science 282, 2254–2256. doi: 10.1126/science.282.5397.2254
Blom, J. F., and Pernthaler, J. (2010). Antibiotic effects of three strains of chrysophytes (Ochromonas, Poterioochromonas) on freshwater bacterial isolates. FEMS Microbiol. Ecol. 71, 281–290. doi: 10.1111/j.1574-6941.2009.00800.x
Bouck, G. B. (1971). The structure, origin, isolation, and composition of the tubular mastigonemes of the Ochromonas flagellum. J. Cell Biol. 50, 362–384. doi: 10.1083/jcb.50.2.362
Butterfield, B. J. (2009). Effects of facilitation on community stability and dynamics: synthesis and future directions. J. Ecol. 97, 1192–1201. doi: 10.1111/j.1365-2745.2009.01569.x
Callaway, R. M., Brooker, R., Choler, P., Kikvidze, Z., Lortie, C. J., Michalet, R., et al. (2002). Positive interactions among alpine plants increase with stress. Nature 417, 844–848. doi: 10.1038/nature00812
Cardinale, B. J., Palmer, M. A., and Collins, S. L. (2002). Species diversity enhances ecosystem functioning through interspecific facilitation. Nature 415, 426–429. doi: 10.1038/415426a
Cavieres, L. A., and Badano, E. I. (2009). Do facilitative interactions increase species richness at the entire community level? J. Ecol. 97, 1181–1191. doi: 10.1111/j.1365-2745.2009.01579.x
Corno, G., and Jürgens, K. (2006). Direct and indirect effects of protist predation on population size structure of a bacterial strain with high phenotypic plasticity. Appl. Environ. Microbiol. 72, 78–86. doi: 10.1128/AEM.72.1.78-86.2006
Daley, R. J., Morris, G., and Brown, S. (1973). Phagotrophic ingestion of a blue-green alga by Ochromonas. J. Protozool. 20, 58–61. doi: 10.1111/j.1550-7408.1973.tb06001.x
Doddema, H., and Van Der Veer, J. (1983). Ochromonas monocis sp. nov., a particle feeder with bacterial endosymbionts. Cryptogam. Algol. 4, 89–97.
Doucette, G., Kodama, M., Franca, S., and Gallacher, S. (1998). Bacterial interactions with harmful algal bloom species: bloom ecology, toxigenesis, and cytology. NATO ASI Ser. G Ecol. Sci. 41, 619–648.
Estep, K. W., Davis, P. G., Keller, M. D., and Sieburth, J. M. (1986). How important are oceanic algal nanoflagellates in bacterivory? 1. Limnol. Oceanogr. 31, 646–650. doi: 10.4319/lo.1986.31.3.0646
Fenchel, T. (1982). Ecology of heterotrophic microflagellates. IV. Quantitative occurrence and importance as bacterial consumers. Mar. Ecol. Prog. Ser. 9, 35–42. doi: 10.3354/meps009035
Gavaudan, P. (1931). Quelques Remarques Sur Chlorochromonas Polymorpha, Spec. Nov. Paris: Botaniste. 277–300.
Guillard, R. R. (1975). “Culture of phytoplankton for feeding marine invertebrates,” in Culture of Marine Invertebrate Animals, eds W. L. Smith and M. H. Chanley (New York, NY: Springer), 29–60.
Hanse, J. A. (1973). Antibiotic activity of the chrysophyte Ochromonas malhamensis. Physiol. Plant. 29, 234–238. doi: 10.1111/j.1399-3054.1973.tb03098.x
Lee, S.-O., Kato, J., Takiguchi, N., Kuroda, A., Ikeda, T., Mitsutani, A., et al. (2000). Involvement of an extracellular protease in algicidal activity of the marine bacterium pseudoalteromonassp. strain A28. Appl. Environ. Microbiol. 66, 4334–4339. doi: 10.1128/AEM.66.10.4334-4339.2000
Li, F. C., Wang, F. Q., Zhang, F. H., Cao, W. R., and Kang, X. J. (2010). Grazing and trophic strategy of Ochromonas sp. on Chlorella pyrenoidosa. Chin. J. Ecol. 3, 539–542.
Li, Y., Zhu, H., Guan, C., Zhang, H., Guo, J., Chen, Z., et al. (2014). Towards molecular, physiological, and biochemical understanding of photosynthetic inhibition and oxidative stress in the toxic Alexandrium tamarense induced by a marine bacterium. Appl. Microbiol. Biotechnol. 98, 4637–4652. doi: 10.1007/s00253-014-5578-x
Lovejoy, C., Bowman, J. P., and Hallegraeff, G. M. (1998). Algicidal effects of a novel marinepseudoalteromonas isolate (Class Proteobacteria, Gamma Subdivision) on harmful algal bloom species of the generachattonella, gymnodinium, andheterosigma. Appl. Environ. Microbiol. 64, 2806–2813.
Manage, P. M., Kawabata, Z. I., and Nakano, S.-I. (2001). Dynamics of cyanophage-like particles and algicidal bacteria causing Microcystis aeruginosa mortality. Limnology 2, 73–78. doi: 10.1007/s102010170002
Mayali, X., and Azam, F. (2004). Algicidal bacteria in the sea and their impact on algal blooms1. J. Eukaryot. Microbiol. 51, 139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x
Mayali, X., and Doucette, G. J. (2002). Microbial community interactions and population dynamics of an algicidal bacterium active against Karenia brevis (Dinophyceae). Harmful Algae 1, 277–293. doi: 10.1016/S1568-9883(02)00032-X
Mulder, C., Uliassi, D., and Doak, D. (2001). Physical stress and diversity-productivity relationships: the role of positive interactions. Proc. Natl. Acad. Sci. U.S.A. 98, 6704–6708. doi: 10.1073/pnas.111055298
Myers, J., and Graham, J. R. (1956). The role of photosynthesis in the physiology of Ochromonas. J. Cell. Comp. Physiol. 47, 397–414. doi: 10.1002/jcp.1030470307
Oh, J.-I., Kim, M.-J., Lee, J.-Y., Ko, I.-J., Kim, W., and Kim, S. W. (2011). Isolation and characterization of algicidal bacteria from Cochlodinium polykrikoides culture. Biotechnol. Bioprocess Eng. 16, 1124–1133. doi: 10.1007/s12257-011-0232-2
Paine, R. T. (1966). Food web complexity and species diversity. Am. Nat. 100, 65–75. doi: 10.1086/282400
Patel, A., Noble, R. T., Steele, J. A., Schwalbach, M. S., Hewson, I., and Fuhrman, J. A. (2007). Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nat. Protoc. 2, 269–276. doi: 10.1038/nprot.2007.6
Pokrzywinski, K. L., Place, A. R., Warner, M. E., and Coyne, K. J. (2012). Investigation of the algicidal exudate produced by Shewanella sp. IRI-160 and its effect on dinoflagellates. Harmful Algae 19, 23–29. doi: 10.1016/j.hal.2012.05.002
Pringsheim, E. G. (1946). Pure Cultures of Algae: Their Preparation and Maintenance. London: CUP Archive.
Reize, I. B., and Melkonian, M. (1989). A new way to investigate living flagellated/ciliated cells in the light microscope: immobilization of cells in agarose. Bot. Acta 102, 145–151. doi: 10.1111/j.1438-8677.1989.tb00083.x
Reynolds, E. S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17, 208–212. doi: 10.1083/jcb.17.1.208
Sherr, E. B., and Sherr, B. F. (2002). Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81, 293–308. doi: 10.1023/A:1020591307260
Sieburth, J., and Davis, P. (1982). The role of heterotrophic nanoplankton in the grazing and nurturing of planktonic bacteria in the Sargasso and Caribbean Seas. Ann. Inst. Oceanogr. 58, 285–296.
Su, J. Q., Yang, X. R., Zheng, T. L., Tian, Y., Jiao, N. Z., Cai, L. Z., et al. (2007b). Isolation and characterization of a marine algicidal bacterium against the toxic dinoflagellate Alexandrium tamarense. Harmful Algae 6, 799–810. doi: 10.1016/j.hal.2007.04.004
Su, J., Yang, X., Zheng, T., and Hong, H. (2007a). An efficient method to obtain axenic cultures of Alexandrium tamarense—a PSP-producing dinoflagellate. J. Microbiol. Methods 69, 425–430. doi: 10.1016/j.mimet.2006.07.005
Su, J., Yang, X., Zhou, Y., and Zheng, T. (2011). Marine bacteria antagonistic to the harmful algal bloom species Alexandrium tamarense (Dinophyceae). Biol. Control 56, 132–138. doi: 10.1016/j.biocontrol.2010.10.004
Tillmann, U., Alpermann, T., John, U., and Cembella, A. (2008). Allelochemical interactions and short-term effects of the dinoflagellate Alexandrium on selected photoautotrophic and heterotrophic protists. Harmful Algae 7, 52–64. doi: 10.1016/j.hal.2007.05.009
Tillmann, U., and Hansen, P. J. (2009). Allelopathic effects of Alexandrium tamarense on other algae: evidence from mixed growth experiments. Aquat. Microb. Ecol. 57, 101. doi: 10.3354/ame01329
Vaqué, D., Duarte, C., and Marrasé, C. (1990). Influence of algal population-dynamics on phytoplankton colonization by bacteria- evidence from 2 diatom species. Inter Res. 65, 201–203. doi: 10.3354/meps065201
Wang, B., Yang, X., Lu, J., Zhou, Y., Su, J., Tian, Y., et al. (2012). A marine bacterium producing protein with algicidal activity against Alexandrium tamarense. Harmful Algae 13, 83–88. doi: 10.1016/j.hal.2011.10.006
Wang, X., Li, Z., Su, J., Tian, Y., Ning, X., Hong, H., et al. (2010). Lysis of a red-tide causing alga, Alexandrium tamarense, caused by bacteria from its phycosphere. Biol. Control 52, 123–130. doi: 10.1016/j.biocontrol.2009.10.004
Wootton, J. T., and Emmerson, M. (2005). Measurement of interaction strength in nature. Annu. Rev. Ecol. Evol. Syst. 36, 419–444. doi: 10.1146/annurev.ecolsys.36.091704.175535
Zheng, T., Su, J., Maskaoui, K., Yu, Z., Hu, Z., Xu, J., et al. (2005). Microbial modulation in the biomass and toxin production of a red-tide causing alga. Mar. Pollut. Bull. 51, 1018–1025. doi: 10.1016/j.marpolbul.2005.02.039
Zhou, L., Zheng, T., Wang, X., Ye, J., Tian, Y., and Hong, H. (2007). Effect of five Chinese traditional medicines on the biological activity of a red-tide causing alga—Alexandrium tamarense. Harmful Algae 6, 354–360. doi: 10.1016/j.hal.2006.10.002
Zubkov, M. V., and Tarran, G. A. (2008). High bacterivory by the smallest phytoplankton in the North Atlantic Ocean. Nature 455, 224–226. doi: 10.1038/nature07236
Keywords: Alexandrium tamarense, Ochromonas sp., bacteria, phycosphere, species interactions
Citation: Hu L, Peng X, Zhou J, Zhang Y, Xu S, Mao X, Gao Y and Liang J (2015) Characterizing the Interactions Among a Dinoflagellate, Flagellate and Bacteria in the Phycosphere of Alexandrium tamarense (Dinophyta). Front. Mar. Sci. 2:100. doi: 10.3389/fmars.2015.00100
Received: 01 July 2015; Accepted: 06 November 2015;
Published: 26 November 2015.
Edited by:
Russell T. Hill, University of Maryland Center for Environmental Science, USAReviewed by:
Kimberly B. Ritchie, Mote Marine Laboratory, USAJohn Everett Parkinson, University of the Ryukyus, Japan
Copyright © 2015 Hu, Peng, Zhou, Zhang, Xu, Mao, Gao and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingyue Peng, eHlwZW5nQHhtdS5lZHUuY24=;
Yahui Gao, Z2FveWhAeG11LmVkdS5jbg==
 Lidan Hu
Lidan Hu Xingyue Peng*
Xingyue Peng*