- Department of Life and Environmental Sciences, Polytechnic University of Marche, Ancona, Italy
Deep-sea ecosystems represent the largest and most remote biome of the biosphere. They play a fundamental role in global biogeochemical cycles and their functions allow existence of life on our planet. In the last 20 years enormous progress has been made in the investigation of deep-sea microbes, but the knowledge of the microbial ecology of the soft bottoms (representing >90% of the deep-sea floor surface) is still very limited. Deep-sea sediments host the largest fractions of Bacteria, Archaea and viruses on Earth, and potentially, a high diversity. At the same time, available results from metagenomics suggest that a large fraction of microbial taxa is completely unknown to science. Estimating the diversity of deep-sea benthic microbes and understanding their functions are some of the challenges of absolute priority, not only for deep-sea microbial ecology, but also for the entire research field of life sciences. The achievement of these goals, given the importance of the deep-sea microbial life for the functioning of the global biosphere, will open new perspectives for the comprehension of adaptation processes to the impact of global changes.
Deep-Sea Microbial Ecology: an Exciting Challenge in Marine Ecology
Deep-sea environments (the waters and sediments of the ocean interior beneath 200 m depth) are the largest biome of the world, representing more than 65% of the Earth's surface and more than 95% of the global biosphere (Herring, 2001). In the last decades, the deep-sea exploration has resulted in the discovery of tens of previously completely unknown habitats and ecosystems, along with thousands of species of organisms inhabiting these remote systems (Gage and Tyler, 1991; Snelgrove, 1999; Teske and Sørensen, 2008; Rex and Etter, 2010; Danovaro et al., 2014). As a result of the development and improvement of the techniques for observing, mapping and sampling the seabed and all of its features, the deep-sea environment is now defined as a highly dynamic geo- and biosphere. A multiplicity of highly diverse habitats have been recently described, including canyons, seamounts, ridges, deep-water coral reefs, cold seeps, pockmarks, mud volcanoes, carbonate mounds, brine pools, gas hydrates, fractures, and trenches that host rich and highly diversified microbial assemblages (Jørgensen and Boetius, 2007; Bartlett, 2009; Ramirez-Llodra et al., 2010; Danovaro et al., 2014).
Despite the technological advancement, current estimates indicate that only 5% of the deep oceans has been explored in detail so far, and that less than 0.001% has been sampled and described in detail in terms of biodiversity (Snelgrove, 1999; Danovaro, 2010; Danovaro et al., 2014). Even less is known on deep-sea microbes, on their the ecological role and functioning.
Although the scientific discovery of the ubiquitous and enormous abundance of microorganisms dates back to the 1860s, microbes inhabiting the deep sea started to be studied in detail only in the 1950s, along with the appearance of the first historical monographs on marine microbiology (e.g., Benecke, 1933). In the 1950s, deep sea samples that were collected during the Danish Galathea Deep-Sea Expedition from depths of more than 10,000 meters were shown to host millions of viable bacteria per gram of sediment, confirming that pressures up to 1100 atmospheres do not exclude the possibility of life (ZoBell and Morita, 1959). Further investigations carried out in the ‘80 and ‘90 provided evidence of microbial activity in the deep ocean interior (Jannasch and Taylor, 1984; Alongi, 1990; Yayanos, 1995).
Although the “Deep-sea Microbial Ecology” is a relatively young branch of both Ecology and Microbiology, enormous progresses have been made in the last two decades of deep-sea investigations, as indicated by the increase of published articles and citations (Figure 1). Researchers have documented life everywhere in the deep sea, including the deepest trenches with active metabolic life from −2 to >150°C, and microbial life even in sediments at 10,000 m depth and at 1000 m below the seafloor (Jørgensen and Boetius, 2007). In these benthic ecosystems, biomass is generally dominated by microbes belonging to the domains of Bacteria and Archaea, and by unicellular eukaryotes, which are fundamentally important for C cycling and nutrient regeneration and are therefore vital for sustaining oceanic production (Middelburg and Meysman, 2007; Danovaro et al., 2014).
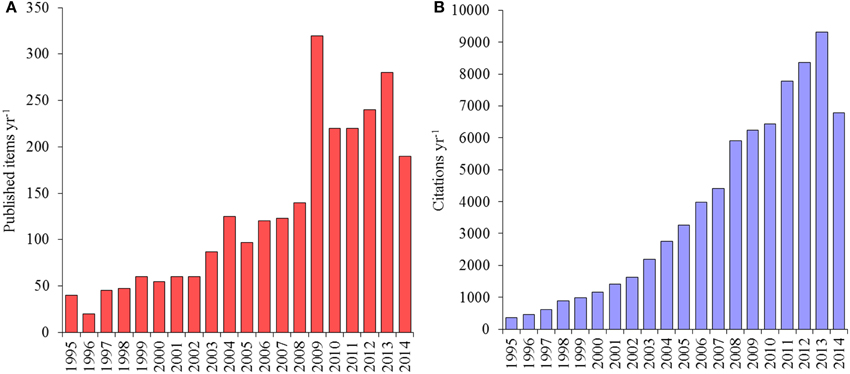
Figure 1. (A) Trend of article publication on deep-sea microbes (B) and relative citations per year from 1995 to 2014 (source Web of Science).
Since most of the deep-sea microorganisms, due to their adaptation to extreme conditions (e.g., oligotrophy, low temperatures, high in situ pressure conditions and in some cases limited oxygen concentrations) are extremely difficult to cultivate with the current technologies, the number of pure cultures that are available is limited to a few hundreds of species, and, only in a limited number of cases, the cultivation of microorganisms under high pressure has been realized (e.g., Kato et al., 1995; Nogi et al., 1998; Mikucki et al., 2003; da Silva et al., 2013). To date, most of the interest from scientists has been focused on habitats of deep-sea environments such as hydrothermal vents, cold seeps, and subsurface sediments where life conditions are even more extreme. However, since such habitats contribute for a likely negligible fraction to entire deep-sea floor (in the order of 0.001% of the seafloor, which is characterized for more than 90% by soft bottom; Danovaro, 2010), this overview will focus on the knowledge on the ecology of benthic bacteria, archaea, and viruses of deep-sea “soft bottoms.”
Recent technological progress in metagenomics is opening new perspectives for investigating microbial life in deep-sea ecosystems. The advent of next-generation sequencing technology allowed us to investigate and to compare the composition of microbial assemblages from different deep-sea habitats, and to provide information even on their metabolic potential (Sogin et al., 2006; DeLong, 2009; Zinger et al., 2011). A number of challenges in deep-sea benthic microbial ecology have to be faced yet, including: the analysis of the composition and functions of the rare biosphere (i.e., low-abundance high-diversity organisms, Sogin et al., 2006), the estimate of microbial (including viral) diversity and its link with the functioning of the dark ocean. The growing interest by the scientific community on the dark portion of the oceans and the technological improvement of tools for exploring it, hint at a bright future for understanding microbial life in the most remote ecosystems of the globe.
Prokaryotic and Viral Abundances in the Deep-Sea Sediments
A global estimate of the number of prokaryotic cells (i.e., here defined as the sum of Bacteria and Archaea) that are present in the oceans, from the surface to subsurface sediments, revealed that deep-sea microbes represent the “hidden majority” of all life forms, comprising between 50 and >80% of the Earth's total microbial biomass and between 10 and 30% of the Earth's total living biomass (Whitman et al., 1998).
In recent years, a much wider portion of the seafloor has been sampled to investigate deep-sea microbes. The analysis of these samples has revealed that at least 2.9 × 1029 prokaryotes inhabit the first few meters of sediment depth (Kallmeyer et al., 2012) suggesting that half of the microbial cells in the Earth's oceans are found in the sediments (Lloyd et al., 2013).
Archaea have been assumed for a long time to be a minor portion of the total microbial community in surface oceans and in seafloor. Information on benthic archaeal assemblage composition and distribution in deep-sea surface sediments is limited and particularly focused on sub-superficial sediments and “extreme” habitats such as hydrothermal vents and cold seeps (Jørgensen and Boetius, 2007). Available investigations indicate that archaeal abundance can equal bacterial abundance or even dominate not only in deep-sea waters beneath 1000 m depth, but also in subsurface sediments (Karner et al., 2001; Biddle et al., 2006; Schippers and Neretin, 2006; Lipp et al., 2008). Recent studies reveal that in deep-sea surface sediments the abundance of archaea can be more relevant than previously thought (ca. 107–108 per gram of sediment dry weigh), contributing in some cases, up to more than 50% of the entire prokaryotic assemblage (Giovannelli et al., 2013; Molari et al., 2013).
In quantitative terms, viruses inhabiting the deep-sea ecosystems are by far more important than they were considered just two decades ago (Danovaro and Serresi, 2000; Danovaro et al., 2008a). Viruses are the most abundant biological entities of the biosphere, with current estimates of the global viral abundance in the order of 1030–1031, outnumbering prokaryotes by at least one order of magnitude (Suttle, 2005, 2007). Data reported here indicate that benthic prokaryotic and viral abundances are high at all depths in the deep sea, from the shelf-break down to abyssal depths (up to >5000-m depth), with values similar to those reported for coastal areas (Danovaro et al., 2008a; Siem-Jørgensen et al., 2008; Figure 2). In deep-sea surface sediments viral abundance is also strongly and positively correlated with prokaryotic abundance (Figure 3A), so that the ratio between the two abundances seems not to be directionally affected by depth nor by pressure. When the abundance of prokaryotes and viruses per m2 of surface sediment is multiplied by the extension of the deep-sea sediments at different bathymetric ranges, the overall abundance of these microbial components in surface sediments shows a marked increase with water depth, with viruses outnumbering prokaryotes also at abyssal depths (Figure 3B).
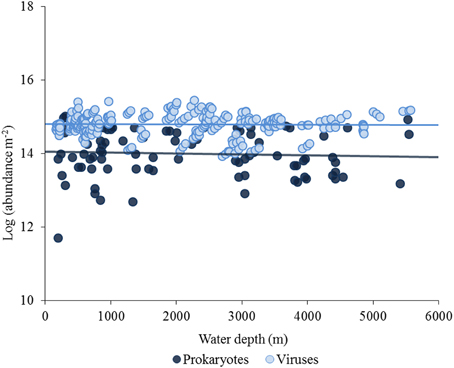
Figure 2. Bathymetric pattern of prokaryotic and viral abundances in the surface sediments (top 1 cm) worldwide (data from Danovaro et al., 2002; Rex et al., 2006; Danovaro et al., 2008b). Abundances were multiplied by the extension (in m2) of the deep-sea sediments at different bathymetric ranges (range of 400 m). Bars represent standard errors.
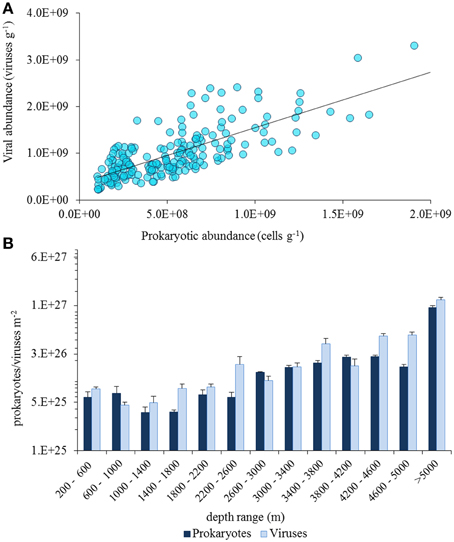
Figure 3. (A) Relationship between viral and prokaryotic abundance in deep-sea surface sediments worldwide (data from Danovaro et al., 2008b). (B) Pattern of total abundance of prokaryotes and viruses in the overall seafloor surface (per bathymetric ranges of 400 m).
A high viral abundance in deep-sea sediments could be due to both the supply of viruses adsorbed onto particles sinking down from the upper water column (Mari et al., 2007) and/or to the high in situ viral production rates (Danovaro et al., 2008a; Siem-Jørgensen et al., 2008). However, the downward flux of viruses associated with settling particles to deep-sea sediments was demonstrated to be extremely low, when compared to in situ abundances (Danovaro et al., 2008b).
The first investigations of global trends in the distribution and abundance of microbial cells showed that the abundance of cells in surface sediments is usually related to the input of fresh detritus, rather than to any other oceanographic parameter (Smith et al., 1997; Boetius and Damm, 1998). Despite recent investigations conducted in different sites of Pacific Oceans in the 1st meter below the sea floor, suggested that the total microbial abundance varies between sites by ca. five orders of magnitude according with mean sedimentation rate and distance from land (Kallmeyer et al., 2012), it is generally accepted that the abundance and biomass of the microbial components are controlled by the quantity and quality of the available organic substrates (Bühring et al., 2006). This applies to most biological components in the deep-sea (e.g., meiofauna macrofauna, megafauna) and, since the availability of food and resources typically decrease with increasing water depth, consequently the biomass of all faunal components decreases exponentially (Rex et al., 2006). However, the analysis of the spatial patterns of the different microbial components reported here, indicates that the distribution of prokaryotes and viruses in deep-sea surface sediments does not show trends similar to those of other benthic components (megafauna, macrofauna, and meiofauna) and does not significantly decrease with increasing water depth (Rex et al., 2006).
In the top 10-cm of deep-sea sediments, prokaryotic biomass, is approximately 160 Pg, which represents ca. 30–45% of the total microbial carbon on Earth (Whitman et al., 1998). The huge, nitrogen- and phosphorus-rich, prokaryotic biomass represents a potentially enormous and high quality food source for benthic consumers in deep-sea ecosystems, but experiments conducted in situ suggest that prokaryotic biomass does not contribute significantly to the food requirements of higher trophic levels (Witte et al., 2003). The high prokaryotic biomass (and in many cases activity) in food-limited deep-sea ecosystems represents a paradox of the deep oceans that needs explanations.
Prokaryotic Activity in Deep-Sea Sediments
The lack of photosynthetic primary production, and the constantly low temperatures (from ca. −2 to 4°C) together with the generally reduced inputs of organic matter contribute to set the limits to benthic microbial activity and secondary production. However, emerging evidence from different studies is confirming that the microbial components inhabiting deep-sea ecosystems are more dynamic than previously thought (Danovaro et al., 2008b; Luna et al., 2012; Molari et al., 2013).
Microbes play a crucial role in deep-sea ecosystems by fundamentally contributing to the total fresh biomass (Danovaro et al., 2014), as prokaryotes represent the largest pool of biomass and biomass production in the world oceans (Whitman et al., 1998). Microbes (including viruses and unicellular eukaryotes) have also key roles in organic matter re-mineralization, in nutrient cycling and in the transfer of energy to higher trophic levels. All of these evidences make these organisms the most important players in the biogeochemical cycles on a global scale (Nealson, 1997; Mason et al., 2009). However, information on the relative importance of bacteria and archaea and their biomass in marine sediments is still limited (Kallmeyer et al., 2012; Xie et al., 2013), and even less is known on the contribution of viruses to the total microbial benthic biomass (Anderson et al., 2013; Engelhardt et al., 2014).
In the surface sediments the main process sustaining the deep-sea benthic heterotrophs is represented by the downward fluxes of organic material (Danovaro et al., 2014). These sediments are generally characterized by a relatively high oxygen content, and since it is consumed slowly free oxygen can diffuse from centimeters to meters down into sediments, representing the dominant terminal electron acceptor for remineralization (Wenzhöfer and Glud, 2002; Fischer et al., 2009).
Little is still known about how prokaryotes respond to changes in the distribution, composition, and flux of organic matter in the oceanic food webs (Azam, 1998), or which microbial species are responsible for most of the carbon, nitrogen and phosphorous cycling in deep waters and sediments (Jørgensen and Boetius, 2007; Arístegui et al., 2009). Approximately 1–2% of the organic carbon supplied from the ocean surface and reaching deep-sea sediments, is oxidized by benthic microorganisms within a few days, but re-mineralization rates slow down if the organic material ages becoming richer in compounds which are refractory and difficult to degrade (Lochte and Turley, 1988; Turley and Lochte, 1990). Recent attempts to derive estimates of carbon cycling in the global ocean suggest that about one-third of the biological production of CO2 in the oceans is due to the microbial activity from the dark ocean interior (meso- bathypelagic waters; Del Giorgio and Duarte, 2002; Arístegui et al., 2005). Heterotrophic C production is potentially high at all depths from ca. 160 m to 5600 m (Figure 4A) and there is evidence that biomass and heterotrophic activity are high even at the deepest depths (down to 11,000 m) of the ocean interior (Danovaro et al., 2003; Glud et al., 2013).
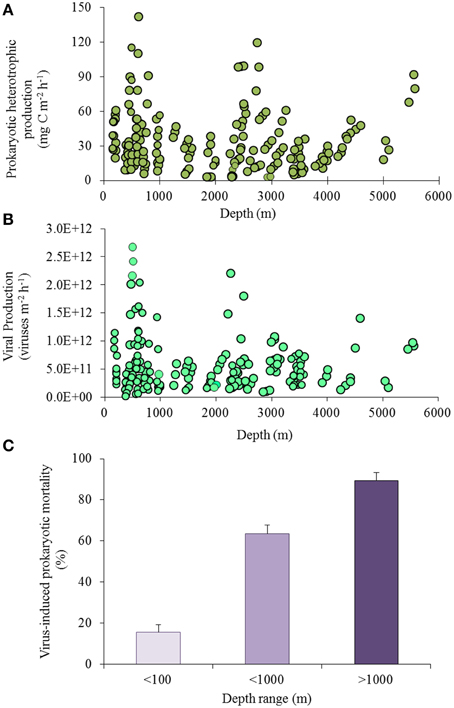
Figure 4. (A) Heterotrophic C production, (B) viral production and (C) virus-induced prokaryotic mortality in deep-sea sediments including sediments at <100-m depth in light purple, <1000-m depth in purple and >1000-m depth in dark purple; data from Mei and Danovaro, 2004; Danovaro et al., 2008b).
The degradation of organic matter pools containing nitrogen is a key process in the oceanic nitrogen cycling of deep-sea sediments (Glud et al., 2009). Nitrification of ammonia to nitrite and nitrate is largely mediated by microorganisms belonging to both the Bacteria (i.e., the β-subgroup and γ-subgroup of Proteobacteria; Swan et al., 2011) and Archaea domains (Könneke et al., 2005). Previous studies revealed that the oxygen consumption by nitrification can account for a significant fraction (21–50%) of the total oxygen demand in organic-rich deep-sea sediments (Christensen and Rowe, 1984), whereas in sediments characterized by high oxygen concentrations that metabolism is limited by the paucity of available organic substrates (Christensen and Rowe, 1984).
Deep-sea sediments are also important for the removal of nitrogen via denitrification (3–12% of the global ocean benthic denitrification, Brunnegård et al., 2004), particularly in organic-rich ecosystems, such as continental slopes (Middelburg et al., 1996). Previous investigations, indeed, suggested that nitrogen released from continental margins is primarily due to prokaryotic denitrification, although anammox processes (i.e., the reduction of nitrites coupled to ammonium oxidation; Thamdrup and Dalsgaard, 2002) can be also important (Glud et al., 2009).
Nitrification is believed to be one of the main processes responsible for dark CO2 fixation in the ocean interior (Herndl et al., 2005; Wuchter et al., 2006; Middelburg, 2011; Swan et al., 2011). In the deep sea chemosynthesis (i.e., chemoautotrophy based on the use of chemical compounds as energy source) is the only source of primary production. Chemosynthetic (also defined, “chemoautotrophic”) organisms produce organic matter through the assimilation of inorganic carbon coupled with the use of reduced chemical compounds as energy source (Swan et al., 2011). Different studies have suggested that chemoautotrophic processes that involve aerobic ammonia oxidation in marine ecosystems are mainly due to archaeal rather than bacterial assemblages (Francis et al., 2005, 2007; Wuchter et al., 2006; Yakimov et al., 2011; Molari et al., 2013). There is evidence, indeed, that non-extremophilic ammonia-oxidizing archaea (AOA) are ubiquitous components of benthic deep-sea ecosystems (Francis et al., 2005). In particular, Thaumarchaeota inhabiting the upper layers of marine sediments have been proposed to contribute significantly to the reservoir of nitrogen oxides of the oceans (Könneke et al., 2005), thus contributing to their productivity and autotrophic carbon fixation (Stahl and de la Torre, 2012).
Information on chemoautotrophic production rates in deep-sea sediments is still very limited, and the overall contribution of chemosynthetic primary production in deep-sea ecosystems could be much higher than previously thought. The chemoautotrophy production estimated for benthic ecosystems is 0.37 PgC year−1, and despite this value can be significantly lower in deep-sea sediments (0.004 PgC year−1; Middelburg, 2011), it could represent an important source of organic carbon for deep-sea benthic consumers. Recent investigations indicated that chemosynthetic primary production by benthic archaea can contribute for up to >20% to the total heterotrophic biomass production and does not show depth-related patterns (Molari et al., 2013).
Recently, benthic archaea belonging to uncultured miscellaneous crenarchaeotal group (MCG) and marine benthic group-D (MBG-D) have been also proposed to contribute to the degradation of sedimentary organic carbon, including proteins (Biddle et al., 2006; Lloyd et al., 2013), but their contribution to the overall heterotrophic metabolism is still largely unknown.
Only one investigation conducted in surface deep-sea sediments reports that benthic archaeal assemblages contribute approximately for 25% to the heterotrophic C production (Molari et al., 2013).
Despite in the last decades important discoveries on the role of Archaea in marine ecosystems have been made, there is still much work ahead for including archaeal chemoautotrophic and heterotrophic production in future global models of deep-sea benthic metabolism and trophodynamics.
Viral Ecology: The New Frontier of Deep-Sea Microbiology
Viruses offer the opportunity to explore a new field of deep-sea ecology so far almost completely neglected, which could be crucial to better understand the functioning of deep-sea ecosystems.
Viruses can likely infect all the known life forms of the oceans, from the largest mammals to the smallest marine microbes. Since the prokaryotes are the predominant life forms in the oceans in terms of biomass, the interactions between viruses and the members of the two domains Bacteria and Archaea are considered one of the most relevant processes driving the ecosystem functioning on the global scale (Proctor and Fuhrman, 1990; Fuhrman, 1999).
Viral infections in deep-sea surface sediments are responsible for the abatement from 40 to up to >80% of the overall heterotrophic carbon production by bacteria and archaea (below 1000-m depth), causing the release of ~0.37–0.63 Gt C year−1 on a global scale (Danovaro et al., 2008b). These finding suggest that viruses can influence global biogeochemical cycles in fundamental ways (Corinaldesi et al., 2014; Danovaro et al., 2014).
By killing their hosts, viruses can transform the living biomass into organic detritus (mostly dissolved organic matter; DOM), which can then be used again by other microbes stimulating their growth. This process defined “viral shunt” (Suttle, 2005, 2007), depending on the balance between the abatement of living cells and the stimulation of the metabolism of the survivors, can either fuel heterotrophic and autotrophic production (Corinaldesi et al., 2012, 2014) and enhancing nutrient regeneration pathways, or it can also decrease the efficiency of the carbon transfer to higher trophic levels (Fuhrman, 1999).
Viral production rates in deep-sea sediments are potentially high at all water depths in surface sediments (Figure 4B). Several studies reported the predominance of the lytic viral cycle (viral replication strategy resulting in the lysis of the infected cells and consequent release of virus progeny and other intracellular components) both in deep-sea sediments and in deep-sea waters (Danovaro et al., 2008b). At the same time, also new replication strategies could be potentially utilized by viruses as it was recently demonstrated for some viruses infecting archaea (Prangishvili et al., 2006; Bize et al., 2009). Analyses of viral and heterotrophic C production in deep-sea sediment samples at in situ, decompressed (at 0.1 MPa) and re-pressurized conditions demonstrated that production rates are not significantly influenced by deep-sea sample recovery (Danovaro et al., 2008b). Unfortunately, only a limited number of studies were conducted under deep-sea in situ conditions so far (Tamburini et al., 2009; Jannasch and Wirsen, 1973), and it is still difficult to draw general conclusions on how pressure and sample recovery affect directly or indirectly, the metabolism and dynamics of microbial communities in deep-sea benthic ecosystems. However, it is clear that deep-sea viruses, and especially viruses in the sediments, are a highly dynamic and active component of deep-sea ecosystems, with high viral turnover, comparable to, or even higher than, that reported for coastal aquatic ecosystems (Danovaro et al., 2008a; Siem-Jørgensen et al., 2008).
Viral production is significantly and positively related with heterotrophic C production. This reflects the theoretical dependence of viral replication on the host abundance and metabolism. Viral lysis of benthic prokaryotes, indeed, causes the abatement of 80% of the total heterotrophic C production in deep-sea sediments. This impact increases with increasing water depth, from values <20% in the sediments at less than 100 m depth up to >80% in deep-sea surface sediments (at depths >3000-m; Figure 4C), indicating that viruses are the main cause of prokaryotic mortality in deep-sea sediments worldwide (Danovaro et al., 2008b).
The shunt of most of the heterotrophic carbon production into organic detritus due to viral lysis is a crucial process for sustaining the high prokaryotic biomass of deep-sea ecosystems and provides an important contribution to prokaryotic metabolism, allowing the deep-sea ecosystems to cope with the severe organic resource limitation (Danovaro et al., 2008b).
These findings could help to explain the paradox of deep-sea ecosystems in which prokaryotic biomass is largely unused by deep-sea fauna, despite severe food limitation (Witte et al., 2003).
In previous investigations the high virus-induced prokaryotic mortality observed in different marine ecosystems (Proctor and Fuhrman, 1990; Suttle et al., 1990) has suggested that viruses can have a major impact on prokaryotic diversity (Fuhrman and Suttle, 1993; Thingstad et al., 1993). This has been theorized in the concept of “killing the winner” (Thingstad and Lignell, 1997), meaning that lytic viruses can keep in check competitive dominants, allowing the co-existence of less competitive taxa and even sustaining bacterial diversity. This model is supported by the finding from isolated phage–host systems that phages propagate as a function of host density and can thus control host abundance (Weinbauer and Rassoulzadegan, 2004). However, results acquired to date, based on manipulative experiments carried out on water, are still equivocal and neither the intensity nor the outcome of this regulation is well-understood (Hewson and Fuhrman, 2006; Bouvier and Del Giorgio, 2007). Some studies reported that viral lysis has a limited effect on the composition of the bacterial assemblages (Hewson and Fuhrman, 2006), whereas others have suggested that rare bacterial groups, being more effective in resource exploitation, can be the most susceptible to viral lysis (Bouvier and Del Giorgio, 2007).
Information on the impact of viruses on microbial diversity in benthic deep-sea ecosystems is practically inexistent. However, understanding if the viral component can represent a driver of the biodiversity and of the functioning in benthic deep-sea ecosystems is a key issue for the comprehension of the functioning of deep-sea ecosystems.
Microbial Diversity
In the last years the application of next generation sequencing technologies revealed that deep-sea ecosystems host highly diversified bacterial, archaeal, and unicellular eukaryotic assemblages and contain novel taxa (Sogin et al., 2006; Jørgensen and Boetius, 2007; Lauro and Bartlett, 2008; Zinger et al., 2011). Studies based on metagenomics, indeed, indicated that the majority of microbial sequences, including those belonging to viruses and unicellular eukaryotes, in deep-sea benthic ecosystems remained unassigned (Scheckenbach et al., 2010; Zinger et al., 2011; Yoshida et al., 2013), suggesting that deep-sea microbial diversity is still largely unknown.
So far, there is evidence that deep-sea surface sediments harbor bacterial assemblages mostly represented by Alpha-, Delta-, and Gammaproteobacteria, Acidobacteria, Actinomycetes, Chloroflexi, and Planctomycetes (Schauer et al., 2010; Orcutt et al., 2011; Zinger et al., 2011; Parkes et al., 2014). The composition of such assemblages appears to be dependent on different environmental factors (e.g., oxygen content, food availability, geographic barriers, past environmental conditions) and their interplay (Ramette and Tiedje, 2007; West et al., 2008; Goffredi and Orphan, 2010; Schauer et al., 2010; Jamieson et al., 2013). Archaeal diversity in oxygenated deep-sea sediments is generally characterized by the dominance of Crenarchaeota over Euryarchaeota (Jørgensen and Boetius, 2007; Corinaldesi et al., 2012; Giovannelli et al., 2013), which instead typically account for the majority in gas-hydrate-bearing and methane-bearing sediments due to the exploitation of the methane released by the seeps (Inagaki et al., 2006; Parkes et al., 2007).
Previous investigations in deep-sea sediments identified phylogenetically distinct lineages of the Marine Group I (MG-I) Thaumarchaeota, which were different from those of the bottom water (Durbin and Teske, 2010). Generally, MG-I Thaumarchaeota (Brochier-Armanet et al., 2008) accounts for the majority of the archaeal assemblages in deep-sea surface benthic ecosystems (Durbin and Teske, 2010, 2011; Jorgensen et al., 2012; Nunoura et al., 2013). However, other archaeal groups, such as the Marine Benthic Group B and E, South African Gold Mine Euryarchaeotal Group, the Miscellaneous Crenarchaeotic Group and the Deep-sea Archaeal Group can provide significant contribution to the archaeal assemblages of such ecosystems (Vetriani et al., 1999; Inagaki et al., 2003; Orcutt et al., 2011; Jorgensen et al., 2012; Wang et al., 2014).
Microbial eukaryotes (protists) have received much less attention, and studies have concentrated mainly on a single taxonomic group, the foraminiferans (Gooday, 1999). Information on eukaryotic microbial diversity in deep-sea surface sediments is particularly focused on specific habitats as hydrothermal vents, cold seeps and hypersaline anoxic basins (Edgcomb et al., 2002, 2009; Takishita et al., 2005; Bernhard et al., 2014). Investigations carried out in three different abyssal plains of the southeastern Atlantic Ocean revealed that the most abundant phylogenetic groups in clone libraries constructed with general eukaryotic primers were Alveolata, Euglenozoa, Heterokonta, and Rhizaria (Scheckenbach et al., 2010). In addition, a high percentage of retrieved clones was affiliated with parasitic species and many others had no close representatives in genetic databases suggesting the presence of novel taxa.
Given the paucity of information, the analysis of micro-eukaryotic diversity and the estimate of its contribution to the entire biodiversity of deep-sea benthic ecosystems represent another key step of future scientific research in deep-sea benthic systems.
Exploring the genetic diversity of viruses in marine ecosystems is complex due to the difficulty in identifying conserved genes in the viral genomes (such as the ribosomal genes of bacteria, archaea, and eukaryotes), and because most of the viral hosts cannot be cultivated (Edwards and Rohwer, 2005; Schoenfeld et al., 2008). The advent of metagenomic approaches has represented a novel opportunity for characterizing the viral diversity as it allow us to capture, to a large extent, the genetic richness of viral assemblages in marine ecosystems (Edwards and Rohwer, 2005; Suttle, 2007; Rosario and Breitbart, 2011). While a number of studies have investigated viral diversity in pelagic ecosystems (e.g., Angly et al., 2006; Duhaime and Sullivan, 2012; Hurwitz and Sullivan, 2013), its exploration in benthic ecosystems is still very scarce. Only very few studies are conducted in deep-sea sediments for investigating viral diversity, but they suggest that these ecosystems could represent a hotspot of viral diversity (Yoshida et al., 2013). Benthic viruses of the deep-sea ecosystems are not only represented by phages, but also by a significant portion of viruses infecting eukaryotes, allowing us to hypothesize a potential role of viruses in regulating the abundance and diversity of (unicellular and multicellular) eukaryotic hosts. Such studies, indeed, showed the presence not only of double-stranded DNA phages mainly belonging to the order Caudovirales (i.e., Myoviridae, Podoviridae, Siphoviridae) believed to infect prokaryotes (Angly et al., 2006; Rosario and Breitbart, 2011; Hurwitz and Sullivan, 2013), but also by ssDNA viruses belonging to Circoviridae infecting eukaryotes (Labonté and Suttle, 2013; Yoshida et al., 2013). Investigating the host range of viruses, their impact on benthic eukaryotes and its ecological and evolutionary implications is another priority for deep-sea microbial ecology in the future.
Future Perspectives
The marine environment is changing rapidly (Hoegh-Guldberg and Bruno, 2010; Doney et al., 2012). For many years, it has been assumed that the deep sea is a stable habitat, buffered from short-term changes in the atmosphere or upper ocean. However, now there is growing evidence that deep-sea ecosystems respond to global climate change over seasonal to annual (Danovaro and Serresi, 2000; Ruhl et al., 2008; Danovaro et al., 2013) and horbital (multi-millennial) timescales (Hunt et al., 2005; Yasuhara and Cronin, 2008; Yasuhara et al., 2012). Despite the extensive research on the potential effects of increasing CO2 concentration and global warming on ecosystems (Hays et al., 2005; Doney et al., 2009), we know very little about the impact of such changes on the deep-sea biodiversity over long time-scales (Yasuhara et al., 2014; Yasuhara and Danovaro, 2014) and even less is known on the effects of climate change on deep benthic microbial assemblages.
During the last 10–15 years, seawater temperatures throughout much of the globe are warmer than ever recorded. A rapid temperature increase has been also reported for the deep ocean interior (Bethoux et al., 1990; Levitus et al., 2005). As viral replication and life cycle are closely linked with host metabolism, temperature changes will likely influence the interactions between viruses and the hosts they infect. For example, it has been argued that as the prokaryotic growth rate increases, the duration of the lytic cycle decreases and burst-size increases (Proctor et al., 1993; Hadas et al., 1997). However, how virus-host interactions respond to temperature shifts in the deep-sea ocean is still unknown and such aspect deserves major attention in the next future.
Since phytoplankton production is also expected to respond to increasing temperatures and enhanced stratification, biogeochemical models predict a global decrease of the oceanic primary productivity (Bopp et al., 2001; Steinacher et al., 2010), and hence a reduction of organic C inputs to the deep seafloor. However, such an effect can be also opposite on regional scales (i.e., increase primary productivity and C export to the deep seafloor; Sarmiento et al., 2004). The consequent changes in the intensity of organic carbon fluxes from the ocean surface down to the ocean interior (Ruhl et al., 2008; Smith et al., 2008) can have downstream consequences on biodiversity and functioning of the benthic deep-sea ecosystems (Ruhl and Smith, 2004; Danovaro et al., 2008c), including microbial diversity and virus-host interactions. Moreover, the viral life strategies could change as a response to shifts in oceanic productivity. An expected pattern is that the relative importance of lytic infections decreases as the proportion of lysogens increases (e.g., in response to lower productivity and lower host metabolic activity; Danovaro et al., 2011).
Ocean acidification is another major effect of global change linked to CO2 emissions. Since the preindustrial era, the pH of the ocean surface has decreased from ca. 8.21 to 8.10 (Raven et al., 2005) and it is expected to decrease by a further 0.3–0.4 pH units by the end of the century (Orr et al., 2005). The direct effects of ocean pH changes on marine viruses are difficult to predict, but the most significant effects could occur on their host organisms (i.e., bacteria, archaea, protists, and metazoan) on which viruses rely.
The potential impact of these global changes (e.g., temperature increase, decrease of pH, deoxygenation of bottom waters) can co-occur in time and space with anthropogenic impacts (e.g., induced by deep-sea drilling for oil extraction, deep-sea mining), which act at local and regional scales (Darling and Côté, 2008; Alsterberg et al., 2014). We cannot predict yet whether climate changes impacts will be intensified by multiple stressors acting on deep-sea benthic bacteria, archaea, and viruses. However, there is increasing evidence that deep-sea microbes interact actively with the present climate change and are a key biotic component that is able to influence the processes regulating the reactions of marine ecosystems to these ongoing changes (Danovaro et al., 2011).
Despite the technological advancements made in the last decades, the remoteness and technological challenges posed by deep-sea ecosystems hamper the development of an intense scientific research, especially in the field of microbial ecology (Danovaro et al., 2014). However, stimulating opportunities are envisaged in the coming years for the exploration and understanding the biodiversity and functioning of the microbial components (including viruses) inhabiting deep-sea sediments. Understanding factors influencing interactions between viruses and bacteria or archaea, their biogeography and functioning, even in the light of a changing ocean, is only a part of the outlooks which could be addressed hereafter.
Deep-sea benthic ecosystems represent the cutting edge of the marine scientific research, as dark oceans potentially harbor mostly gene sequences of yet unknown viruses and unculturable microbes. It can be expected that high-throughput sequencing will provide an unprecedented opportunity to solve the paradoxes and finding to response to the main ecological paradigms proposed for these ecosystems (Danovaro et al., 2014). High throughput sequencing is expected to be a very powerful tool even for deciphering potential functional signals contained within microbial metagenomes, including viromes, which are practically still unknown in deep-sea benthic ecosystems. Certainly, the discovery of new processes and activities mediated by different microbial components could complicate the set-up of ecological models to assess/forecast deep-sea functioning, but this is a challenge that it worth addressing.
It is known, that deep-sea ecosystems have an enormous potential for the discovery and development of new compounds and bioactive molecules of microbial origin (i.e., pharmaceuticals, molecular probes, enzymes, cosmetics, nutritional supplements, and agrichemicals) for technological and industrial purposes (Ohta and Hatada, 2006; Wu et al., 2006; Cavicchioli et al., 2011). Thus, the exploration of the microbial life in deep-sea sediments is needed not only because it is fundamental for understanding its role in the global ecological and biogeochemical processes, but also because provides important goods and services for ameliorating human activities and health. Obviously, the enormous value of deep-sea benthic ecosystems raises the question on which are the terms of their sustainable use in order to preserve them. To date, a legal framework for the management, benefit sharing and resource access in areas beyond national jurisdiction does not exist but this is of fundamental importance to preserve while exploring deep-sea ecosystems. Thus, the actual scientific success of deep-sea microbial ecology will depend on technological and methodological advancements and our ability to promote research projects for the study of deep-sea microbes but also on the ability to identify useful strategies for the conservation of the largest ecosystem on Earth.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the National project EXPLODIVE (FIRB 2008, contract no. I31J10000060001) and the Flagship Project RITMARE—The Italian Research for the Sea—coordinated by the Italian National Research Council and funded by the Italian Ministry of Education, University and Research within the National Research Program 2011–2013. I thank E. Rastelli and A. Dell'Anno for the support during the dataset elaboration.
References
Alongi, D. M. (1990). Bacterial growth rates, production and estimates of detrital carbon utilization in deep-sea sediments of the Solomon and Coral Seas. Deep Sea Res. A. 37, 731–746. doi: 10.1016/0198-0149(90)90002-D
Alsterberg, C., Sundbäck, K., and Gamfeldt, L. (2014). Multiple stressors and multifunctionality: limited effects on an illuminated benthic system. Biol. Lett. 10, 20140640. doi: 10.1098/rsbl.2014.0640
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Anderson, R. E., Brazelton, W. J., and Baross, J. A. (2013). The deep viriosphere: assessing the viral impact on microbial community dynamics in the deep subsurface. Rev. Mineral. Geochem. 75, 649–675. doi: 10.2138/rmg.2013.75.20
Angly, F. E., Felts, B., Breitbart, M., Salamon, P., Edwards, R. A., Carlson, C., et al. (2006). The marine viromes of four oceanic regions. PLoS Biol. 4:e368. doi: 10.1371/journal.pbio.0040368
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Arístegui, J., Duarte, C. M., Gasol, J. M., and Alonso-Sáez, L. (2005). Active mesopelagic prokaryotes support high respiration in the subtropical northeast Atlantic Ocean. Geophys. Res. Lett. 32, L03608. doi: 10.1029/2004GL021863
Arístegui, J., Gasol, J. M., Duarte, C. M., and Herndl, G. J. (2009). Microbial oceanography of the dark ocean's pelagic realm. Limnol. Oceanogr. 54, 1501–1529. doi: 10.4319/lo.2009.54.5.1501
Azam, F. (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 268, 694–695. doi: 10.1126/science.280.5364.694
Bartlett, D. H. (2009). Microbial life in the trenches. Marine Technol. Soc. J. 43, 128–131. doi: 10.1080/10425170400009293
Benecke, W. (1933). Bakteriologie des meeres. Abder. Hand. Biol. Arbeitsmeth. IX Abt. Teil 5, 717–854.
Bernhard, J. M., Konstantinos, K., Pachiadaki, M. G., Rocke, E., Beaudoin, D. J., Morrison, C., et al. (2014). Benthic protists and fungi of Mediterranean deep hypsersaline anoxic basin redoxcline sediments. Front. Microbiol. 5:605. doi: 10.3389/fmicb.2014.00605
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bethoux, J. P., Gentili, B., Raunet, J., and Tailliez, D. (1990). Warming trend in the western Mediterranean deep water. Nature 347, 660–662. doi: 10.1038/347660a0
Biddle, J. F., Lipp, J. S., Lever, M. A., Lloyd, K. G., Sørensen, K. B., Anderson, R., et al. (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U.S.A. 103, 3846–3851. doi: 10.1073/pnas.0600035103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bize, A., Karlsson, E. A., Ekefjärd, K., Quax, T. E. F., Pina, M., Prevost, M. C., et al. (2009). A unique virus release mechanism in the Archaea. Proc. Natl. Acad. Sci. U.S.A. 106, 11306–11311. doi: 10.1073/pnas.0901238106
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Boetius, A., and Damm, E. (1998). Benthic oxygen uptake, hydrolytic potentials and microbial biomass at the Arctic continental slope. Deep Sea Res. Part I Oceanogr. Res. Pap. 45, 239–275. doi: 10.1016/S0967-0637(97)00052-6
Bopp, L., Monfray, P., Aumont, O., Dufresne, J. L., Le Treut, H., Madec, G., et al. (2001). Potential impact of climate change on marine export production. Global Biogeochem. Cycles 15, 81–99. doi: 10.1029/1999GB001256
Bouvier, T., and Del Giorgio, P. A. (2007). Key role of selective viral−induced mortality in determining marine bacterial community composition. Environ. Microbiol. 9, 287–297. doi: 10.1111/j.1462-2920.2006.01137.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brochier-Armanet, C., Boussau, B., Gribaldo, S., and Forterre, P. (2008). Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6, 245–252. doi: 10.1080/10425170400009293
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brunnegård, J., Grandel, S., Ståhl, H., Tengberg, A., and Hall, P. O. J. (2004). Nitrogen cycling in deep-sea sediments of the Porcupine abyssal plain, NE Atlantic. Prog. Oceanogr. 63, 159–181. doi: 10.1016/j.pocean.2004.09.004
Bühring, S. I., Lampadariou, N., Moodley, L., Tselepides, A., and Witte, U. (2006). Benthic microbial and whole-community responses to different amounts of 13C-enriched algae: in situ experiments in the deep Cretan Sea (Eastern Mediterranean). Limnol. Oceanogr. 51, 157–165. doi: 10.4319/lo.2006.51.1.0157
Cavicchioli, R., Charlton, T., Ertan, H., Omar, S. M., Siddiqui, K. S., and Williams, T. J. (2011). Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 4, 449–460. doi: 10.1111/j.1751-7915.2011.00258.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Christensen, J. P., and Rowe, G. T. (1984). Nitrification and oxygen consumption in northwest Atlantic deep-sea sediments. J. Mar. Res. 42, 1099–1116. doi: 10.1357/002224084788520828
Corinaldesi, C., Dell'Anno, A., and Danovaro, R. (2012). Viral infections stimulate the metabolism and shape prokaryotic assemblages in submarine mud volcanoe. ISME J. 6, 1250–1259. doi: 10.1038/ismej.2011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Corinaldesi, C., Tangherlini, M., Luna, G. M., and Dell'Anno, A. (2014). Extracellular DNA can preserve the genetic signatures of present and past viral infection events in deep hypersaline anoxic basins. Proc. Biol. Sci. 281, 20133299. doi: 10.1098/rspb.2013.3299
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Danovaro, R., Manini, E., and Dell'Anno, A. (2002). Higher abundance of bacteria than of viruses in deep Mediterranean sediments. Appl. Environ. Microbiol. 68, 1468–1472. doi: 10.1128/AEM.68.3.1468-1472.2002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Danovaro, R., and Serresi, M. (2000). Viral abundance and virus-to-bacterium ratio in deep-sea sediments of the eastern Mediterranean. Appl. Environ. Microbiol. 66, 1857–1861.
da Silva, M. A. C., Cavalett, A., Spinner, A., Rosa, D. C., Jasper, R. B., Quecine, M. C., et al. (2013). Phylogenetic identification of marine bacteria isolated from deep-sea sediments of the eastern South Atlantic Ocean. Springerplus 2:127. doi: 10.1186/2193-1801-2-127
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Danovaro, R. (2010). Methods for the Study of Deep-Sea Sediments, Their Functioning and Biodiversity. London: CRC press Taylor; Francis Group. 456. doi: 10.1080/10425170400009293
Danovaro, R., and Serresi, M. (2000). Viral density and virus-to-bacterium ratio in deep-sea sediments of the Eastern Mediterranean. Appl. Environ. Microbiol. 66, 1857–1861. doi: 10.1128/AEM.66.5.1857-1861.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Danovaro, R., Carugati, L., Corinaldesi, C., Gambi, C., Guilini, K., Pusceddu, A., et al. (2013). Multiple spatial scale analyses provide new clues on patterns and drivers of deep-sea nematode diversity. Deep-sea Res. II Top. Stud. Oceanogr. 92, 97–106. doi: 10.1016/j.dsr2.2013.03.035
Danovaro, R., Corinaldesi, C., Dell'Anno, A., Fuhrman, J. A., Middelburg, J. J., Noble, R. T., et al. (2011). Marine viruses and global climate change. FEMS Microbiol. Rev. 35, 993–1034. doi: 10.1111/j.1574-6976.2010.00258.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Danovaro, R., Corinaldesi, C., Filippini, M., Fischer, U. R., Gessner, M. O., Jacquet, S., et al. (2008a). Viriobenthos in freshwater and marine sediments: a review. Freshw. Biol. 53, 1186–1213. doi: 10.1111/j.1365-2427.2008.01961.x
Danovaro, R., Dell'Anno, A., Corinaldesi, C., Magagnini, M., Noble, R., Tamburini, C., et al. (2008b). Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454, 1084–1087. doi: 10.1038/nature07268
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Danovaro, R., Della Croce, N., Dell'Anno, A., and Pusceddu, A. (2003). A depocenter of organic matter at 7800-m depth in the SE Pacific Ocean. Deep Sea Res. Part I Oceanogr. Res. Pap. 50, 1411–1420. doi: 10.1016/j.dsr.2003.07.001
Danovaro, R., Gambi, G., Dell'Anno, A., Corinaldesi, C., Fraschetti, S., Vanreusel, A., et al. (2008c). Exponential decline of deep-sea ecosystem functioning linked to benthic biodiversity loss. Curr. Biol. 18, 1–8. doi: 10.1016/j.cub.2007.11.056
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Danovaro, R., Snelgrove, P. V., and Tyler, P. (2014). Challenging the paradigms of deep-sea ecology. Trends Ecol. Evol. 29, 465–475. doi: 10.1016/j.tree.2014.06.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Darling, E. S., and Côté, I. M. (2008). Quantifying the evidence for ecological synergies. Ecol. Lett. 11, 1278–1286. doi: 10.1111/j.1461-0248.2008.01243.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Del Giorgio, P. A., and Duarte, C. M. (2002). Respiration in the open ocean. Nature 420, 379–384. doi: 10.1038/nature01165
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
DeLong, E. F. (2009). The microbial ocean from genomes to biomes. Nature 459, 200–206. doi: 10.1038/nature08059
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Doney, S. C., Fabry, V. J., Feely, R. A., and Kleypas, J. A. (2009). Ocean acidification: the other CO2 problem. Ann. Rev. Mar. Sci. 1, 169–192. doi: 10.1146/annurev.marine.010908.163834
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Doney, S. C., Ruckelshaus, M., Duffy, J. E., Barry, J. P., Chan, F., English, C. A., et al. (2012). Climate change impacts on marine ecosystems. Ann. Rev. Mar. Sci. 4, 11–37. doi: 10.1146/annurev-marine-041911-111611
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Duhaime, M. B., and Sullivan, M. B. (2012). Ocean viruses: rigorously evaluating the metagenomic sample-to-sequence pipeline. Virology 434, 181–186. doi: 10.1016/j.virol.2012.09.036
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Durbin, A. M., and Teske, A. (2010). Sediment-associated microdiversity within the Marine Group I Crenarchaeota. Environ. Microbiol. Rep. 2, 693–703. doi: 10.1111/j.1758-2229.2010.00163.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Durbin, A. M., and Teske, A. (2011). Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ. Microbiol. 13, 3219–3234. doi: 10.1111/j.1462-2920.2011.02544.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Edgcomb, V. P., Kysela, D. T., Teske, A., Gomez, A. D. V., and Sogin, M. L. (2002). Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. U.S.A. 99, 7663–7668. doi: 10.1073/pnas.062186399
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Edgcomb, V., Orsi, W., Leslin, C., Epstein, S. S., Bunge, J., Jeon, S., et al. (2009). Protistan community patterns within the brine and halocline of deep hypersaline anoxic basins (DHABs) in the eastern Mediterranean Sea. Extremophiles 13, 151–167. doi: 10.1007/s00792-008-0206-2
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Edwards, R. A., and Rohwer, F. (2005). Viral metagenomics. Nat. Rev. Microbiol. 3, 504–510. doi: 10.1038/nrmicro1163
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Engelhardt, T., Kallmeyer, J., Cypionka, H., and Engelen, B. (2014). High virus-to-cell ratios indicate on-going production of viruses in deep subsurface sediments. ISME J. 8, 1503–1509.
Fischer, J. P., Ferdelman, T. G., D'Hondt, S., Røy, H., and Wenzhöfer, F. (2009). Oxygen penetration deep into the sediment of the South Pacific gyre. Biogeosciences 6, 1467–1478. doi: 10.5194/bg-6-1467-2009
Francis, C. A., Beman, J. M., and Kuypers, M. M. M. (2007). New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1, 19–27. doi: 10.1038/ismej.2007.8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A. 102, 14683–14688. doi: 10.1073/pnas.0506625102
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fuhrman, J. A. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548. doi: 10.1038/21119
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fuhrman, J. A., and Suttle, C. A. (1993). Viruses in marine planktonic systems. Oceanography 6, 51–63. doi: 10.5670/oceanog.1993.14
Gage, J. D., and Tyler, P. A. (1991). Deep Sea Biology: A Natural History of Organisms at the Deep-Sea Floor. Cambridge: Cambridge University Press. 504. doi: 10.1017/CBO9781139163637
Giovannelli, D., Molari, M., d'Errico, G., Baldrighi, E., Pala, C., and Manini, E. (2013). Large-scale distribution and activity of prokaryotes in deep-sea surface sediments of the Mediterranean Sea and the adjacent Atlantic Ocean. PLoS ONE 8:e72996. doi: 10.1371/journal.pone.0072996
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Glud, R. N., Thamdrup, B., Stahl, H., Wenzhoefer, F., Glud, A., Nomaki, H., et al. (2009). Nitrogen cycling in a deep ocean margin sediment (Sagami Bay, Japan). Limnol. Oceanogr. 54, 723–734. doi: 10.4319/lo.2009.54.3.0723
Glud, R. N., Wenzhöfer, F., Middelboe, M., Oguri, K., Turnewitsch, R., Donald, E., et al. (2013). High rates of microbial carbon turnover in sediments in the deepest oceanic trench on Earth. Nat. Geosci. 6, 284–288. doi: 10.1038/ngeo1773
Goffredi, S. K., and Orphan, V. J. (2010). Bacterial community shifts in taxa and diversity in response to localized organic loading in the deep sea. Environ. Microbiol. 12, 344–363. doi: 10.1111/j.1462-2920.2009.02072.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gooday, A. J. (1999). Biodiversity of foraminifera and other protists in the deep sea: scales and patterns. Belg. J. Zool. 129, 61–80.
Hadas, H., Einav, M., Fishov, I., and Zaritsky, A. (1997). Bacteriophage T4 development depends on the physiology of its host Escherichia coli. Microbiology 143, 179–185. doi: 10.1099/00221287-143-1-179
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hays, G. C., Richardson, A. J., and Robinson, C. (2005). Climate change and marine plankton. Trends Ecol. Evol. 20, 337–344. doi: 10.1016/j.tree.2005.03.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Herndl, G. J., Reinthaler, T., Teira, E., van Aken, H., Veth, C., Pernthaler, A., et al. (2005). Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71, 2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hewson, I., and Fuhrman, J. A. (2006). Viral impacts upon marine bacterioplankton assemblage structure. J. Mar. Biol. Ass. U. K. 86, 577–589. doi: 10.1017/S002531540601349X
Hoegh-Guldberg, O., and Bruno, J. F. (2010). The impact of climate change on the world's marine ecosystems. Science 328, 1523–1528. doi: 10.1126/science.1189930
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hunt, G., Cronin, T. M., and Roy, K. (2005). Species–energy relationship in the deep sea: a test using the quaternary fossil record. Ecol. Lett. 8, 739–747. doi: 10.1111/j.1461-0248.2005.00778.x
Hurwitz, B. L., and Sullivan, M. B. (2013). The Pacific Ocean Virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS ONE 8:e57355. doi: 10.1371/journal.pone.0057355
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Inagaki, F. T., Nunoura, S., Nakagawa, A., Teske, M., Lever, A., Lauer, M., et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U.S.A. 103, 2815–2820. doi: 10.1080/10425170400009293
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Inagaki, F., Suzuki, M., Takai, K., Oida, H., Sakamoto, T., Aoki, K., et al. (2003). Microbial communities associated with geological horizons in coastal subseafloor sediments from the sea of okhotsk. Appl. Environ. Microbiol. 69, 7224–7235. doi: 10.1128/AEM.69.12.7224-7235.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jamieson, R. E., Heywood, J. L., Rogers, A. D., Billett, D. S. M., and Pearce, D. A. (2013). Bacterial biodiversity in deep-sea sediments from two regions of contrasting surface water productivity near the Crozet Islands, Southern Ocean. Deep Sea Res. Part I Oceanogr. Res. Pap. 75, 67–77. doi: 10.1016/j.dsr.2012.12.012
Jannasch, H. W., and Taylor, C. D. (1984). Deep-sea microbiology. Annu. Rev. Microbiol. 38, 487–487. doi: 10.1146/annurev.mi.38.100184.002415
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jannasch, H. W., and Wirsen, C. O. (1973). Deep-sea microorganisms: in situ response to nutrient enrichment. Science 180, 641–643. doi: 10.1126/science.180.4086.641
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jørgensen, B. B., and Boetius, A. (2007). Feast and famine—microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5, 770–781. doi: 10.1038/nrmicro1745
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jorgensen, S. L., Hannisdal, B., Lanzéna, A., Baumberger, T., Flesland, K., Fonseca, R., et al. (2012). Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl. Acad. Sci. U.S.A. 109, E2846–E2855. doi: 10.1073/pnas.1207574109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, E. C., and D'Hondt, S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 109, 16213–16216. doi: 10.1073/pnas.1203849109.
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Karner, M. B., DeLong, E. F., and Karl, D. M. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510. doi: 10.1038/35054051
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kato, C., Sato, T., and Horikoshi, K. (1995). Isolation and properties of barophilic and barotolerant bacteria from deep-sea mud samples. Biodivers. Conserv. 4, 1–9. doi: 10.1007/BF00115311
Könneke, M., Bernhard, A. E., de la Torre, J. R., Walker, C. B., Waterbury, J. B., and Sthal, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Labonté, J. M., and Suttle, C. A. (2013). Previously unknown and highly divergent ssDNA viruses populate the oceans. ISME J. 7, 2169–2177. doi: 10.1038/ismej.2013.110
Lauro, F. M., and Bartlett, D. H. (2008). Prokaryotic lifestyles in deep sea habitats. Extremophiles 12, 15–25. doi: 10.1007/s00792-006-0059-5
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Levitus, S., Antonov, J., and Boyer, T. (2005). Warming of the world ocean, 1955–2003. Geophys. Res. Lett. 32:L02604. doi: 10.1029/2004GL021592
Lipp, J. S., Morono, Y., Inagaki, F., and Hinrichs, K. U. (2008). Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454, 991–994. doi: 10.1038/nature07174
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lloyd, K. G., May, M. K., Kevorkian, R., and Steen, A. D. (2013). Meta analysis of quantification methods shows archaea and bacteria to be similarly abundant in the subseafloor. Appl. Environ. Microbiol. 79, 7790–7799. doi: 10.1128/AEM.02090-13
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lochte, K., and Turley, C. M. (1988). Bacteria and cyanobacteria associated with phytodetritus in the deep-sea. Nature 333, 67–69. doi: 10.1038/333067a0
Luna, G. M., Bianchelli, S., Decembrini, F., De Domenico, E., Danovaro, R., and Dell'Anno, A. (2012). The dark portion of the Mediterranean Sea is a bioreactor of organic matter cycling. Global Biogeochem. Cycles 26, GB2017. doi: 10.1029/2011GB004168
Mari, X., Kerros, M.-E., and Weinbauer, M. G. (2007). Virus attachment to transparent exopolymeric particles along trophic gradients in the Southwestern Lagoon of New Caledonia. Appl. Environ. Microbiol. 73, 5245–5252. doi: 10.1128/AEM.00762-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mason, O. U., Di Meo-Savoie, C. A., Van Nostrand, J. D., Zhou, J., Fisk, M. R., and Giovannoni, S. J. (2009). Prokaryotic diversity, distribution, and insights into their role in biogeochemical cycling in marine basalts. ISME J. 3, 231–242. doi: 10.1038/ismej.2008.92
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mei, M. L., and Danovaro, R. (2004). Virus production and life strategies in aquatic sediments. Limnol. Oceanogr. 49, 459–470. doi: 10.4319/lo.2004.49.2.0459
Middelburg, J. J. (2011). Chemoautotrophy in the ocean. Geophys. Res. Lett. 38:L24604. doi: 10.1029/2011GL049725
Middelburg, J. J., and Meysman, F. J. R. (2007). Burial at sea. Science 316, 1294–1295. doi: 10.1126/science.1144001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Middelburg, J. J., Soetaert, K., Herman, P. M. J., and Heip, C. H. R. (1996). Denitrification in marine sediments: a model study. Global Biogeochem. Cycles 10, 661–673.
Mikucki, J. A., Liu, Y., Delwiche, M., Colwell, F. S., and Boone, D. R. (2003). Isolation of a methanogen from deep marine sediments that contain methane hydrates, and description of Methanoculleus submarinus sp. nov. Appl. Environ. Microbiol. 69, 311–331. doi: 10.1128/AEM.69.6.3311-3316.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Molari, M., Manini, E., and Dell'Anno, A. (2013). Dark inorganic carbon fixation sustains the functioning of benthic deep-sea ecosystems. Global Biogeochem. Cycles 27, 212–221. doi: 10.1002/gbc.20030
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nealson, K. H. (1997). Sediment bacteria: who's there, what are they doing, and what's new? Annu. Rev. Earth Planet. Sci. 25, 403–434. doi: 10.1146/annurev.earth.25.1.403
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nogi, Y., Kato, C., and Horikoshi, K. (1998). Moritella japonica sp. nov., a novel barophilic bacterium isolated from a Japan Trench sediment. J. Gen. Appl. Microbiol. 44, 289–295. doi: 10.2323/jgam.44.289
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nunoura, T., Nishizawa, M., Kikuchi, T., Tsubouchi, T., Hirai, M., Koide, O., et al. (2013). Molecular biological and isotopic biogeochemical prognoses of the nitrification-driven dynamic microbial nitrogen cycle in hadopelagic sediments. Environ. Microbiol. 15, 3087–3107. doi: 10.1111/1462-2920.12152
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ohta, Y., and Hatada, Y. (2006). A novel enzyme, lambda-carrageenase, isolated from a deep-sea bacterium. J. Biochem. 140, 475–481. doi: 10.1093/jb/mvj180
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Orcutt, B. N., Sylvan, J. B., Knab, N. J., and Edwards, K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422. doi: 10.1128/MMBR.00039-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Orr, J. C., Fabry, V. J., Aumont, O., Bopp, L., Doney, S. C., Feely, R. A., et al. (2005). Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 437, 681–686. doi: 10.1038/nature04095
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Parkes, R. J., Cragg, B. A., Banning, N., Brock, F., Webster, G., Fry, J. C., et al. (2007). Biogeochemistry and biodiversity of methane cycling in subsurface marine sediments (Skagerrak, Denmark). Environ. Microbiol. 9, 1146–1161. doi: 10.1111/j.1462-2920.2006.01237.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Parkes, R. J., Cragg, B., Roussel, E., Weightman, A., and Sass, H. (2014). A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere: geosphere interactions. Mar. Geol. 352, 409–425. doi: 10.1016/j.margeo.2014.02.009
Prangishvili, D., Forterre, P., and Garrett, R. A. (2006). Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 4, 837–848. doi: 10.1038/nrmicro1527
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Proctor, L. M., and Fuhrman, J. A. (1990). Viral mortality of marine bacteria and cyanobacteria. Nature 343, 60–62. doi: 10.1038/343060a0
Proctor, L. M., Okubo, A., and Fuhrman, J. A. (1993). Calibrating estimates of phage-induced mortality in marine bacteria: ultrastructural studies of marine bacteriophage development from one-step growth experiments. Microb. Ecol. 25, 161–182. doi: 10.1007/BF00177193
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ramette, A., and Tiedje, J. M. (2007). Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Natl. Acad. Sci. U.S.A. 104, 2761–2766. doi: 10.1073/pnas.0610671104
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ramirez-Llodra, E., Brandt, A., Danovaro, R., De Mol, B., and Escobar, E. (2010). Deep, diverse and definitely different: unique attributes of the world's largest ecosystem. Biogeosciences 7, 2851–2899. doi: 10.5194/bg-7-2851-2010
Raven, J., Caldeira, K., Elderfield, H., Hoegh-Guldberg, O., Liss, P., Riebesell, U., et al. (2005). Ocean Acidification due to Increasing Atmospheric Carbon Dioxide. London: The Royal Society.
Rex, M. A., and Etter, R. J. (2010). Deep-Sea Biodiversity: Pattern and Scale. Cambridge, UK: Harvard University Press.
Rex, M. A., Etter, R. J., Morris, J. S., Crouse, J., McClain, C. R., Johnson, N. A., et al. (2006). Global bathymetric patterns of standing stock and body size in the deep-sea benthos. Mar. Ecol. Prog. Ser. 317, 1–8. doi: 10.3354/meps317001
Rosario, K., and Breitbart, M. (2011). Exploring the viral world through metagenomics. Curr. Opin. Virol. 1, 289–297. doi: 10.1016/j.coviro.2011.06.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ruhl, H. A., and Smith, K. L. (2004). Shifts in deep-sea community structure linked to climate and food supply. Science 305, 513–515. doi: 10.1126/science.1099759
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ruhl, H. A., Ellena, J. A., and Smith, K. L. (2008). Connections between climate, food limitation, and carbon cycling in abyssal sediment communities. Proc. Natl. Acad. Sci. U.S.A. 105, 17006–17011. doi: 10.1073/pnas.0803898105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sarmiento, J. L., Gruber, N., Brzezinski, M. A., and Dunne, J. P. (2004). High-latitude controls of thermocline nutrients and low latitude biological productivity. Nature 427, 56–60. doi: 10.1038/nature02127
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schauer, R., Bienhold, C., Ramette, A., and Harder, J. (2010). Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 4, 159–170. doi: 10.1038/ismej.2009.106
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Scheckenbach, F., Hausmann, K., Wylezich, C., Weitere, M., and Arndt, H. (2010). Large-scale patterns in biodiversity of microbial eukaryotes from the abyssal sea floor. Proc. Natl. Acad. Sci. U.S.A. 107, 115–120. doi: 10.1073/pnas.0908816106
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schippers, A., and Neretin, L. N. (2006). Quantification of microbial communities in near−surface and deeply buried marine sediments on the Peru continental margin using real−time PCR. Environ. Microbiol. 8, 1251–1260. doi: 10.1111/j.1462-2920.2006.01019.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schoenfeld, T., Patterson, M., Richardson, P. M., Wommack, K. E., Young, M., and Mead, D. (2008). Assembly of viral metagenomes from yellowstone hot springs. Appl. Environ. Microbiol. 74, 4164–4174. doi: 10.1128/AEM.02598-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Siem-Jørgensen, M., Glud, R. N., and Middelboe, M. (2008). Viral dynamics in a coastal sediment: seasonal pattern, controlling factors and relations to the pelagicbenthic coupling. Marine Biol. Res. 4, 165–179. doi: 10.1080/17451000801888718
Smith, C. R., Berelson, W., Demaster, D. J., Dobbs, F. C., Hammond, D., Hoover, D. J., et al. (1997). Latitudinal variations in benthic processes in the abyssal equatorial Pacific: control by biogenic particle flux. Deep-sea Res. II Top. Stud. Oceanogr. 44, 2295–2317. doi: 10.1016/S0967-0645(97)00022-2
Smith, C. R., De Leo, F. C., Bernardino, A. F., Sweetmann, A. K., and Arbizu, P. M. (2008). Abyssal food limitation, ecosystem structure and climate change. Trends Ecol. Evol. 23, 518–528. doi: 10.1016/j.tree.2008.05.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Snelgrove, P. V. R. (1999). Getting to the bottom of marine biodiversity: sedimentary habitats. Bioscience 49, 129–138. doi: 10.2307/1313538
Sogin, M. L., Morrison, H. G., Huber, J. A., Welch, D. M., Huse, S. M., Neal, P. R., et al. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120. doi: 10.1073/pnas.0605127103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stahl, D. A., and de la Torre, J. R. (2012). Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66, 83–101. doi: 10.1080/10425170400009293
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Steinacher, M., Joos, F., Frölicher, T. L., Bopp, L., Cadule, P., Cocco, V., et al. (2010). Projected 21st century decrease in marine productivity: a multi-model analysis. Biogeosciences 7, 979–1005. doi: 10.5194/bg-7-979-2010
Suttle, C. A. (2005). Viruses in the sea. Nature 437, 356–361. doi: 10.1038/nature04160
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Suttle, C. A. (2007). Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Suttle, C. A., Chan, A. M., and Cottrell, M. T. (1990). Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347, 467–469. doi: 10.1038/347467a0
Swan, B. K., Martinez-Garcia, M., Preston, C. M., Sczyrba, A., Woyke, T., Lamy, D., et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333, 1296–1300. doi: 10.1126/science.1203690
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Takishita, K., Miyake, H., Kawato, M., and Maruyama, T. (2005). Genetic diversity of microbial eukaryotes in anoxic sediment around fumaroles on a submarine caldera floor based on the small-subunit rDNA phylogeny. Extremophiles 9, 185–196. doi: 10.1007/s00792-005-0432-9
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tamburini, C., Garel, M., Al Ali, B., Mérigot, B., Kriwy, P., Charrière, B., et al. (2009). Distribution and activity of Bacteria and Archaea in the different water masses of the Tyrrhenian Sea. Deep-sea Res. II Top. Stud. Oceanogr. 56, 700–712. doi: 10.1016/j.dsr2.2008.07.021
Teske, A., and Sørensen, B. (2008). Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J. 2, 3–18. doi: 10.1038/ismej.2007.90
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thamdrup, B., and Dalsgaard, T. (2002). Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68, 1312–1318. doi: 10.1128/AEM.68.3.1312-1318.2002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thingstad, T. F., and Lignell, R. (1997). Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13, 19–27. doi: 10.3354/ame013019
Thingstad, T. F., Heldal, M., Bratbak, G., and Dundas, I. (1993). Are viruses important partners in pelagic food webs? Trends Ecol. Evol. 8, 209–213. doi: 10.1016/0169-5347(93)90101-T
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Turley, C. M., and Lochte, K. (1990). Microbial response to the input of fresh detritus to the deep-sea bed. Palaeogeogr. Palaeoclimatol. Palaeoecol. 89, 3–23. doi: 10.1016/0031-0182(90)90048-C
Vetriani, C., Jannasch, H. W., MacGregor, B. J., Stahl, D. A., and Reysenbach, A. L. (1999). Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65, 4375–4384.
Wang, P., Wei, Y., Liac, T., Li, F., Meng, J., and Zhang, C. L. (2014). Archaeal diversity and spatial distribution in the surface sediment of the South China Sea. Geomicrobiol. J. 31, 1–11. doi: 10.1080/01490451.2013.797522
Weinbauer, M. G., and Rassoulzadegan, F. (2004). Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6, 1–11. doi: 10.1046/j.1462-2920.2003.00539.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wenzhöfer, F., and Glud, R. (2002). Benthic carbon mineralization in the Atlantic: a synthesis based on in situ data from the last decade. Deep Sea Res. Part I Oceanogr. Res. Pap. 49, 1255–1279. doi: 10.1016/S0967-0637(02)00025-0
West, N. J., Obernosterer, I., Zemb, O., and Lebaron, P. (2008). Major differences of bacterial diversity and activity inside and outside of a natural iron−fertilized phytoplankton bloom in the Southern Ocean. Environ. Microbiol. 10, 738–756. doi: 10.1111/j.1462-2920.2007.01497.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583. doi: 10.1073/pnas.95.12.6578
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Witte, U., Wenzhöfer, F., Sommer, S., Boetius, A., Heinz, P., Aberle, N., et al. (2003). In situ experimental evidence of the fate of a phytodetritus pulse at the abyssal sea floor. Nature 424, 763–766. doi: 10.1038/nature01799
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wu, S., Liu, B., and Zhang, X. (2006). Characterization of a recombinant thermostable xylanase from deep-sea thermophilic Geobacillus sp. MT-1 in East Pacific. Appl. Microbiol. Biotechnol. 72, 1210–1216. doi: 10.1007/s00253-006-0416-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wuchter, C., Abbas, B., Coolen, M. J. L., Herfort, L., van Bleijswijk, J., Timmers, P., et al. (2006). Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U.S.A. 103, 12317–12322. doi: 10.1073/pnas.0600756103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xie, S., Lipp, J. S., Wegner, G., Ferdelman, T. G., and Hinrichs, K-U. (2013). Turnover of microbial lipids in the deep biosphere and growth of benthic archaeal populations. Proc. Natl. Acad. Sci. U.S.A. 110, 6010–6014. doi: 10.1073/pnas.1218569110
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yakimov, M. M., La Cono, V., Smedile, F., DeLuca, T. H., Juárez, S., Ciordia, S., et al. (2011). Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea). ISME J. 5, 945–961. doi: 10.1038/ismej.2010.197
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yasuhara, M., and Cronin, T. M. (2008). Climatic influences on deep-sea ostracode (Crustacea) diversity for the last three million years. Ecology 89, S53–S65. doi: 10.1890/07-1021.1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yasuhara, M., and Danovaro, R. (2014). Temperature impacts on deep-sea biodiversity. Biol. Rev. doi: 10.1111/brv.12169. [Epub ahead of print].
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yasuhara, M., Hunt, G., Dowsett, H. J., Robinson, M. M., and Stoll, D. K. (2012). Latitudinal species diversity gradient of marine zooplankton for the last three million years. Ecol. Lett. 15, 1174–1179. doi: 10.1111/j.1461-0248.2012.01828.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yasuhara, M., Okahashi, H., Cronin, T. M., Rasmussen, T. L., and Hunt, G. (2014). Response of deep-sea biodiversity to abrupt deglacial and Holocene climate changes in the North Atlantic Ocean. Global Ecol. Biogeogr. 23, 957–967. doi: 10.1111/geb.12178
Yayanos, A. A. (1995). Microbiology to 10,500 meters in the deep sea. Annu. Rev. Microbiol. 49, 777–805. doi: 10.1146/annurev.mi.49.100195.004021
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yoshida, M., Takaki, Y., Eitoku, M., Nunoura, T., and Takai, K. (2013). Metagenomic analysis of viral communities in (hado) pelagic sediments. PLoS ONE 8:e57271. doi: 10.1371/journal.pone.0057271
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zinger, L., Amaral-Zettler, L. A., Fuhrman, J. A., Horner-Devine, M. C., Huse, S. M., Welch, D. B. M., et al. (2011). Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS ONE 6:e24570. doi: 10.1371/journal.pone.0024570
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: prokaryotes, viruses, deep-sea sediments, biodiversity, deep-sea ecosystem functioning
Citation: Corinaldesi C (2015) New perspectives in benthic deep-sea microbial ecology. Front. Mar. Sci. 2:17. doi: 10.3389/fmars.2015.00017
Received: 06 November 2014; Accepted: 22 February 2015;
Published: 11 March 2015.
Edited by:
Anna Metaxas, Dalhousie University, CanadaReviewed by:
Takuro Nunoura, Japan Agency for Marine-Earth Science and Technology (JAMSTEC), JapanBrett J. Baker, University of Texas Austin, USA
Copyright © 2015 Corinaldesi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cinzia Corinaldesi, Department of Life and Environmental Sciences, Polytechnic University of Marche, via Brecce Bianche, 60131 Ancona, ItalyYy5jb3JpbmFsZGVzaUB1bml2cG0uaXQ=
 Cinzia Corinaldesi
Cinzia Corinaldesi