- 1Marine Genomics Unit, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan
- 2DNA Sequencing Section, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan
- 3Marine Biophysics Unit, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan
The genus Acropora (Scleractinia, Acroporidae) is one of the most widespread coral genera, comprising the largest number of extant species among scleractinian (reef-building) corals. Molecular phylogenetic studies have suggested that A. tenuis belongs to the most basal clade (clade I) while A. digitifera belongs to a derived clade (clade IV). In order to develop microsatellite markers that would be useful for most Acropora species, we sequenced the genomic DNA of A. tenuis, using a next generation sequencer (Illumina MiSeq), and designed primer sets that amplify microsatellite loci. Afterward we selected primer pairs with perfectly matched nucleotide sequences from which at least one primer was uniquely mapped to the A. digitifera genome. Fourteen microsatellite markers showed non-significant departure from Hardy–Weinberg equilibrium (HWE) in both A. tenuis and A. digitifera. Thus these markers could be used for wide range of species and may provide powerful tools for population genetics studies and conservation of Acropora corals.
Introduction
Acropora (Scleractinia, Acroporidae) is a common, emblematic genus of reef-building corals. It is also one of the most widespread coral genera, ranging from the Red Sea through the Indo-Pacific Ocean to the Caribbean, and has the largest number of extant species (113) (Wallace, 1999). The fossil record suggests that the genus probably originated about 50 million years ago (MYR) (Veron, 1995). There are two distinct groups of Acropora corals: mass spawning acroporids and “early spawners,” that spawn 1.5–3 h earlier than other mass-spawning species (Hatta et al., 1999; Fukami et al., 2000). The two groups are believed to have diverged 6.6 MYA (Fukami et al., 2000). Major diversification within these groups has occurred more recently (2 MYR) (Veron, 1995). In addition, molecular phylogenetic analyses using a single copy gene PaxC intron and mitochondrial markers show that Acropora corals can be divided into four major clades (Van Oppen et al., 2001; Marquez et al., 2002) (Figure 1). Early spawning species, including A. tenuis, belong to the most basal clade (Clade I) (Fukami et al., 2000; Van Oppen et al., 2001), while most mass-spawning species belong to Clade III. Clade IV has relatively small number of species, including A. digitifera (Van Oppen et al., 2001).
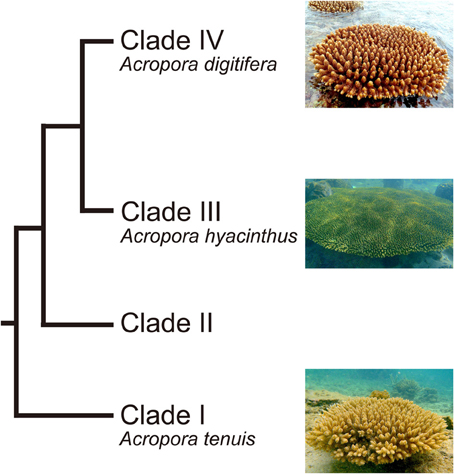
Figure 1. Phylogenetic relationship of Acropora corals inferred from molecular markers. Acropora species used in this study (A. tenuis, A. digitifera, and A. hyacinthus) are shown. Modified from Van Oppen et al. (2001) and Marquez et al. (2002).
Coral reefs are estimated to harbor around one-third of all described marine species (Knowlton et al., 2010); however, they face a range of anthropogenic challenges, including ocean acidification and increasing seawater temperatures (e.g., Hoegh-Guldberg et al., 2007). Although Acropora species are major components of coral reefs worldwide, they are the most sensitive to increased water temperatures (Loya et al., 2001) and are expected to decline in the near future (Alvarez-Filip et al., 2013). For proper maintenance and conservation of Acropora corals, it is important to understand genetic diversity and connectivity among populations. High-resolution genetic markers, such as microsatellites, are essential for such studies. Previous studies have succeeded in developing microsatellite markers specific to several Acropora species, e.g., A. palmata (Baums et al., 2005), A. millepora (Van Oppen et al., 2007), A. cytherea (Concepcion et al., 2010), and Acropora sp1 and A. digitifera (Nakajima et al., 2009). However, cross-species amplification was confirmed for several markers (Nakajima et al., 2009). Because microsatellite markers are currently available for only about 5 of the 113 Acropora species, an increased number of “universal” Acropora microsatellite markers would be extremely useful. In this study we developed cross-species microsatellite markers that can be applied to a variety of Acropora species. To achieve this we used two Acropora species that belong to taxonomically distant clades (I: A. tenuis, IV: A. digitifera) and we took advantage of next-generation sequencing technology to design novel microsatellite primer pairs that can be used for both species.
Materials and Methods
Genomic DNA was isolated from an A. tenuis colony collected at Sesoko Island, Okinawa, Japan, under Okinawa prefectural permit (Number: 24–48), using the guanidinium reagent, CHAOS (Fukami et al., 2000). We sequenced 250 bp paired end reads using a MiSeq sequencer (Illumina) according to manufacturer's instructions. Low quality bases (Phred quality value, QV ≥ 20) were trimmed from raw data and read pairs of at least 80 bp were retained using SolexaQA (Cox et al., 2010). We used PAL_FINDER (Castoe et al., 2012) for detection of simple sequence repeats (SSRs) and PCR primer design from paired end sequencing data. In order to select microsatellite loci that may be highly variable, we selected primer pairs amplifying longer repeat stretches (thresholds: 2 mer; 15 repeats more, 3 mer; 10, 4 mer; 7, 5 mer; 5 and 6 mer; 4, respectively). To remove primers originating from DNA of the symbiotic Symbiodinium, nucleotide sequences of both primers in each pair were mapped to the recently decoded A. digitifera genome (Shinzato et al., 2011), using Symbiodinium-free sperm DNA, and employing BLASTN software (Altschul et al., 1990). In addition, primer pairs from which at least one primer was mapped uniquely to the A. digitifera genome were selected in order to avoid selecting primer pairs that could produce nonspecific PCR amplification.
For fragment analyses, 30 colonies of A. tenuis were collected at Sesoko Island, Okinawa, Japan, and 45 colonies of A. digitifera were collected in the Kerama Islands, Okinawa, Japan, respectively (Okinawa prefecture permit number: 24–48). To avoid multiple collections of colonies that could have been produced through asexual fragmentation or propagation, only colonies that were physically distinct and at least 2 m from other colonies were sampled. Genomic DNA was extracted using a DNeasy kit (QIAGEN). The reaction mixture (10 μ L) contained template DNA (<1 ng/ L), AmpliTaq Gold 360 Master Mix (Qiagen), and three primers for each locus: a non-tailed reverse primer (0.1 μ M), a forward primer with an M13 Reverse (5′- CAGGAAACAGCTATGAC-3′) sequence tail (0.5 μ M), and an M13 Reverse primer (0.5 μ M) fluorescently labeled with FAM, based on the method of Schuelke (2000). PCR cycling conditions were 15 min at 95°C, followed by 32 cycles of 30 s at 94°C, 90 s at 58°C (all loci), and 60 s at 72°C, with an extension of 30 min at 60°C in the final cycle. In addition to A. tenuis and A. digitifera, we also used A. hyacinthus (Clade III) (Figure 1) (Marquez et al., 2002) genomic DNA to confirm PCR amplification (data not shown). PCR products from A. tenuis and A. digitifera were identified and analyzed with the ABI 3130 capillary sequencer (Applied Biosystems) and GeneMapper v4.1 (Applied Biosystems). The number of alleles and observed and expected heterozygosities were calculated and the probability of deviation from Hardy–Weinberg equilibrium (HWE) was tested for each locus and species, using GenAlEx ver. 6.5 (Peakall and Smouse, 2012). Linkage disequilibrium between the loci was tested after Bonferroni correction (P < 0.05) using Genepop v4.2 at http://genepop.curtin.edu.au/index.html (Raymond and Rousset, 1995; Rousset, 2008).
Results and Discussion
We obtained 6,327,391,737 bp (12,802,836 read pairs) of raw sequence data from A. tenuis genomic DNA. From those we selected high quality 2,534,049,158 bp (6,783,510 read pairs), which were used for microsatellite detection and primer design. Primer pairs (7,200) were produced by PAL_FINDER. In order to eliminate primer pairs that could have produced non-specific PCR amplification and that originated from symbiotic Symbiodinium, we selected pairs from which at least one primer sequence was unique to the A. digitifera genome sequence. Subsequently 141 primer pairs were selected. Among those, we confirmed that 74 pairs could produce PCR amplicons in three Acropora species (A. tenuis, A. digitifera, and A. hyacinthus) and performed fragment analyses. We identified 14 polymorphic nuclear microsatellite DNA makers that did not show significant deviation from HWE after applying Bonferroni correction (P < 0.05) in both A. tenuis and A. digitifera (Table 1). Four markers (9079m3, 11192m4, 8010m6, and 12198m3) showed significant deviation from HWE in A. digitifera, but not in A. tenuis, and one marker (7805m4) showed significant deviation from HWE in A. tenuis, but not in A. digitifera (Table 1). We confirmed that no previously reported Acropora microsatellite primer sequences (Baums et al., 2005; Van Oppen et al., 2007; Nakajima et al., 2009; Concepcion et al., 2010) were detected in the PCR amplicon sequences, indicating that all microsatellite loci identified in this study are novel. The number of alleles per locus ranged from 3 to 14 in A. tenuis and 2 to 13 in A. digitifera, respectively (Table 1). Observed and expected heterozygosities ranged from 0.192 to 0.933 and 0.341 to 0.892 in A. tenuis and 0.200 to 0.911 and 0.241 to 0.862 in A. digitifera, respectively (Table 1). Although only the linkage disequilibrium between 11401m4 and 11745m3 was significant in A. digitifera, linkage disequilibrium between 11 loci combinations (11401m4-11745m3, 11401m4-11543m5, 11401m4-7203m5, 11401m4-12406m3, 11745m3-12406m3, 12406m3-10366m5, 441m6-12406m3, 530m4-11543m5, 530m4-8346m3, 7203m5-11745m3, and 8346m3-11543m5) was significant in A. tenuis. Alignment of the A. tenuis nucleotide sequences to the A. digitifera genome revealed high nucleotide conservation between A. tenuis and A. digitifera (about 93%, BLASTN, 1e-5, alignment length longer than 100 bp), indicating high genomic similarity between the two species. In addition, all microsatellite loci are located in different scaffold sequences in the A. digitifera genome (Table 1), suggesting that the loci are evenly distributed across these Acropora genomes. Since genome structures of A. digitifera and A. tenuis should be similar, this may reflect the significant A. tenuis population decrease after a massive 1998 bleaching event around Sesoko island (Loya et al., 2001). Thus, the population used in this study was new and recently recruited, possibly resulting in more loci combinations with significant linkage disequilibrium. Although not tested on a large number of Acropora species, 14 primer pairs showing no significant deviation from HWE in two phylogenetically distant species. These can be used for a variety of Acropora species and may provide powerful tools for Acropora population genetic studies. We hope that they will contribute to establishment of reef conservation guidelines and coral reef transplantation and restoration.
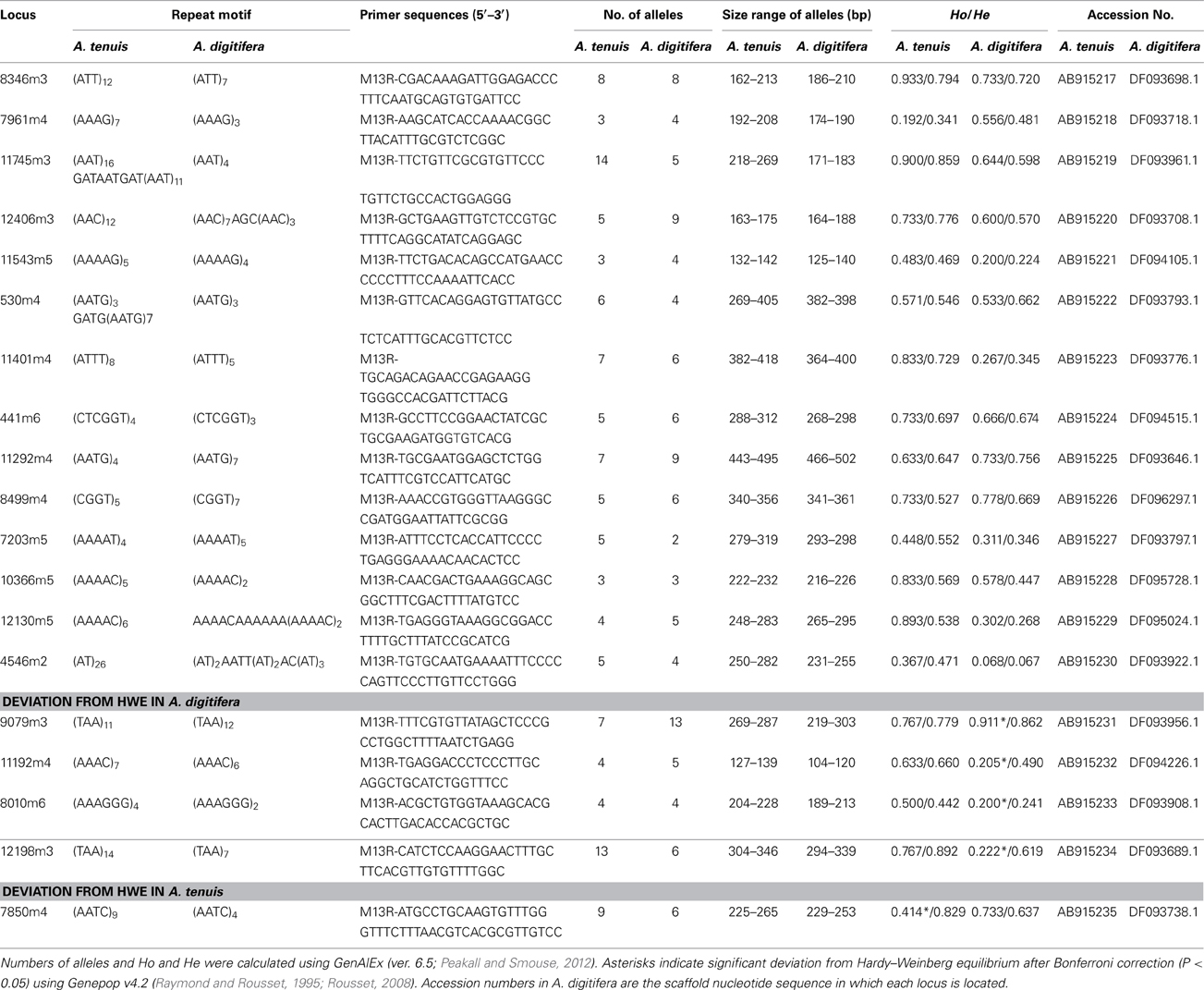
Table 1. Characteristics of the 19 developed polymorphic microsatellite loci from A. tenuis and A. digitifera: locus name, repeat motif, primers sequence, number of alleles, size range, observed (Ho) and expected (He) heterozygotes and GenBank accession number.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was conducted as a part of the Coral Reef Conservation and Restoration Project by Okinawa prefecture, Japan. This study was also supported in part by JSPS KAKENHI (24241071 to Nori Satoh, 25660172 and 26290065 to Chuya Shinzato, 25840152 to Yuichi Nakajima). We thank IDEA Consultants, Inc., Japan, for helping with the collection of A. digitifera specimens from the Kerama Islands. Drs. Kazuhiko Sakai (Sesoko Station, University of the Ryukyus) and Akira Iguchi (Okinawa National College of Technology) provided A. tenuis genomic DNA for fragment analysis. We thank Dr. Steven D. Aird (OIST) for editing the manuscript.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alvarez-Filip, L., Carricart-Ganivet, J. P., Horta-Puga, G., and Iglesias-Prieto, R. (2013). Shifts in coral-assemblage composition do not ensure persistence of reef functionality. Sci. Rep. 3:3486. doi: 10.1038/srep03486
Baums, I. B., Hughes, C. R., and Hellberg, M. E. (2005). Mendelian microsatellite loci for the Caribbean coral Acropora palmata. Mar. Ecol. Prog. Ser. 288, 115–127. doi: 10.3354/meps288115
Castoe, T. A., Poole, A. W., De Koning, A. P., Jones, K. L., Tomback, D. F., Oyler-Mccance, S. J., et al. (2012). Rapid microsatellite identification from Illumina paired-end genomic sequencing in two birds and a snake. PLoS ONE 7:e30953. doi: 10.1371/journal.pone.0030953
Concepcion, G. T., Polato, N. R., Baums, I. B., and Toonen, R. J. (2010). Development of microsatellite markers from four Hawaiian corals: Acropora cytherea, Fungia scutaria, Montipora capitata and Porites lobata. Conserv. Genet. Resour. 2, 11–15. doi: 10.1007/s12686-009-9118-4
Cox, M. P., Peterson, D. A., and Biggs, P. J. (2010). SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485. doi: 10.1186/1471-2105-11-485
Fukami, H., Omori, M., and Hatta, M. (2000). Phylogenetic relationships in the coral family acroporidae, reassessed by inference from mitochondrial genes. Zool. Sci. 17, 689–696. doi: 10.2108/zsj.17.689
Hatta, M., Fukami, H., Wang, W., Omori, M., Shimoike, K., Hayashibara, T., et al. (1999). Reproductive and genetic evidence for a reticulate evolutionary history of mass-spawning corals. Mol. Biol. Evol. 16, 1607–1613. doi: 10.1093/oxfordjournals.molbev.a026073
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Knowlton, N., Brainard, R. E., Fisher, R., Moews, M., Plaisance, L., and Caley, M. J. (2010). “Coral reef biodiversity,” in Life in the World's Oceans: Diversity, Distribution, and Abundance, ed A. D. McIntyre (Chichester: Wiley-Blackwell), 65–79.
Loya, Y., Sakai, K., Yamazato, K., Nakano, Y., Sambali, H., and Van Woesik, R. (2001). Coral bleaching: the winners and the losers. Ecol. Lett. 4, 122–131. doi: 10.1046/j.1461-0248.2001.00203.x
Marquez, L. M., Van Oppen, M. J., Willis, B. L., Reyes, A., and Miller, D. J. (2002). The highly cross-fertile coral species, Acropora hyacinthus and Acropora cytherea, constitute statistically distinguishable lineages. Mol. Ecol. 11, 1339–1349. doi: 10.1046/j.1365-294X.2002.01526.x
Nakajima, Y., Nishikawa, A., Iguchi, A., and Sakai, K. (2009). Novel and cross-species amplifiable microsatellite markers in two Acropora species. Plankton Benthos Res. 4, 38–41. doi: 10.3800/pbr.4.38
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Raymond, M., and Rousset, F. (1995). Genepop (Version-1.2) - population-genetics software for exact tests and Ecumenicism. J. Heredity 86, 248–249.
Rousset, F. (2008). genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Schuelke, M. (2000). An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18, 233–234. doi: 10.1038/72708
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. doi: 10.1038/nature10249
Van Oppen, M. J. H., Underwood, J. N., Muirhead, A. N., and Peplow, L. (2007). Ten microsatellite loci for the reef-building coral Acropora millepora (Cnidaria, Scleractinia) from the Great Barrier Reef, Australia. Mol. Ecol. Notes 7, 436–438. doi: 10.1111/j.1471-8286.2006.01610.x
Van Oppen, M. J., Mcdonald, B. J., Willis, B., and Miller, D. J. (2001). The evolutionary history of the coral genus Acropora (Scleractinia, Cnidaria) based on a mitochondrial and a nuclear marker: reticulation, incomplete lineage sorting, or morphological convergence? Mol. Biol. Evol. 18, 1315–1329. doi: 10.1093/oxfordjournals.molbev.a003916
Keywords: scleractinian coral, Acropora, microsatellite, population genetics, cross-species, Illumina next-generation sequencing
Citation: Shinzato C, Yasuoka Y, Mungpakdee S, Arakaki N, Fujie M, Nakajima Y and Satoh N (2014) Development of novel, cross-species microsatellite markers for Acropora corals using next-generation sequencing technology. Front. Mar. Sci. 1:11. doi: 10.3389/fmars.2014.00011
Received: 04 March 2014; Accepted: 08 May 2014;
Published online: 23 May 2014.
Edited by:
Irene Wagner-Doebler, Helmholtz Centre for Infection Research, GermanyReviewed by:
Carmelo Fruciano, University of Konstanz, GermanySusanna López-Legentil, University of North Carolina Wilmington, USA
Copyright © 2014 Shinzato, Yasuoka, Mungpakdee, Arakaki, Fujie, Nakajima and Satoh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuya Shinzato and Nori Satoh, 1919-1, Tancha, Onna, Okinawa, Japan e-mail: c.shinzato@oist.jp; norisky@oist.jp
 Chuya Shinzato
Chuya Shinzato Yuki Yasuoka1
Yuki Yasuoka1 Yuichi Nakajima
Yuichi Nakajima Nori Satoh
Nori Satoh