
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mamm. Sci., 31 May 2024
Sec. Evolution, Anatomy and the Paleosciences
Volume 3 - 2024 | https://doi.org/10.3389/fmamm.2024.1399358
 Mariya Antonosyan1*†
Mariya Antonosyan1*† Eden Hill1†
Eden Hill1† Margaret Jodry2
Margaret Jodry2 Noel Amano1
Noel Amano1 Samantha Brown3
Samantha Brown3 Torben Rick2
Torben Rick2 Nicole Boivin1,4,5
Nicole Boivin1,4,5Museum legacy collections, often derived from large-scale archaeological excavations, can serve as paleoenvironmental archives of Late Pleistocene megafaunal composition and dynamics. Many of these collections, however, contain large quantities of highly fragmented and morphologically indistinct bones that cannot be identified to a specific taxon and are therefore of limited use to paleoenvironmental and archaeological analyses. Here, we explore the potential of Zooarchaeology by Mass Spectrometry (ZooMS) to identify fossil bone fragments and complement morphological identifications in legacy collections housed at the Smithsonian’s National Museum of Natural History. To undertake this work, we collected fragmented bone specimens of Late Pleistocene megafauna from six archaeological sites in Colorado that are currently housed in the Department of Anthropology, and then performed pilot ZooMS screening. Our analysis successfully retrieved taxonomic information from 80% of the analyzed material, highlighting the potential of future ZooMS studies on museum collections to investigate human-megafaunal interactions in late Pleistocene North America.
Collections of Late Pleistocene fauna from archaeological and fossil sites are currently housed in museums and other repositories around the world. These collections constitute an invaluable source of information about past megafauna, including many now extinct species Cleland et al., 2016; Sanchez et al., 2018; St. Amand et al., 2020). For example, they provide an important reservoir of information about changes in megafaunal diversity and distribution, as well as the chronologies of particular species. Their detailed study accordingly holds the potential to shed further light on species loss and its broader causes and impacts, including long standing questions about the causes of Late Quaternary megafaunal extinctions (Brook and Bowman, 2002; Barnosky et al., 2004; Firestone et al., 2007; Gill et al., 2009; Ripple and Van Valkenburgh, 2010; Faith, 2011; Lorenzen et al., 2011; Cooper et al., 2015; Grayson and Meltzer, 2015; Bartlett et al., 2016; Broughton and Weitzel, 2018; Haynes, 2018; Meltzer, 2020; Seersholm et al., 2020; Monteath et al., 2021; Stewart et al., 2021; Bergman et al., 2023; O’Keefe et al., 2023).
Collection-based studies, however, can be complicated by the differential excavation, recovery, and curation histories of museum collections and the fragmentary nature of fossil assemblages. A significant component of legacy collections include highly fragmentary, undiagnostic fossil and subfossil material remains. Recent developments in biomolecular approaches, including ancient DNA (aDNA), and proteomic tools, provide researchers with new opportunities to address some of these limitations, and to exploit the potential of biological markers preserved in museum fossil and subfossil collections. With the advance of molecular techniques, fossil collections housed in museums have been revisited to reevaluate taxonomic identifications and retrieve in-depth information concerning the evolutionary and demographic histories of prehistoric animals and humans (Cleland et al., 2016; McGuire et al., 2018; Wagner et al., 2020; Hahn et al., 2022).
At the same time, long-archived museum specimens present numerous and substantial challenges to molecular analyses. Museum specimens are commonly stored at room temperature as dry or fluid preserved collections, which has been shown to promote degradation and subsequent loss of DNA over time (Lehmann and Kreipe, 2001; Zimmermann et al., 2008; Raxworthy and Smith, 2021). Another challenge is the high rate of exogenous contamination of samples with microbial, parasitic, and bacterial DNA as well as the potential for cross contamination between specimens stored in close proximity. The application of consolidants in museum conservation practice may result in the unintentional introduction of modern proteins, such as animal collagens in glues (Prüfer et al., 2021). Growing evidence also indicates that biomolecules begin to degrade as soon as skeletal remains are removed from the burial environment (Pruvost et al., 2007; Bollongino et al., 2008). To ensure the continued advancement of molecular applications to museum collections, further assessment of the effects of museum storage on sample preservation is required.
Recent aDNA studies on museum specimens have allowed for the reconstruction of evolutionary processes (Holmes et al., 2016; Lamichhaney et al., 2019), inference of demographic history (White et al., 2018), investigation of epigenetic effects (Rubi et al., 2020), resolution of taxonomic composition (Seersholm et al., 2021), and clarification of taxonomic confusion (Vershinina et al., 2020; Kehlmaier et al., 2021). Proteomics tools have also been extensively applied to museum collections, often from regions and time periods in which aDNA is unlikely to survive, providing insights into phylogeny (e.g. Buckley et al., 2019; Cappellini et al., 2019) and past diets and subsistence (e.g. Bleasdale et al., 2021; Wilkin et al., 2021), as well as improving understanding of past ecosystems and environments (e.g. Brown et al., 2021a; Antonosyan et al., 2024). One key method in palaeoproteomic research is Zooarchaeology by Mass Spectrometry (ZooMS), a form of peptide mass fingerprinting used to taxonomically identify ambiguous animal specimens. Compared with other biomolecular identification methods such as aDNA analyses, ZooMS allows for a more rapid, minimally destructive, and cost-effective approach, that could provide results on heavily fragmented, processed, and degraded material (Buckley, 2018; Richter et al., 2022). The method has been applied to bone assemblages from museum collections as a complement to or even substitute technique for morphology-based identification of fragmented and worked bones (Desmond et al., 2018; Martisius et al., 2020; Dekker et al., 2021). ZooMS has also been applied to various archaeological materials other than bone, including ivory (Coutu et al., 2016), eggshell (Stewart et al., 2013), antler (Ashby et al., 2015), baleen (O'Connor et al., 2015) and leather objects (Ebsen et al., 2019), demonstrating the broad applicability of the method.
In North America, a number of important megafaunal collections are housed in museums and repositories, and finding new ways to analyze these collections affords a prime opportunity to reevaluate the causes and consequences of the continent’s Late Quaternary megafaunal extinctions. These include extensive collections housed in the Smithsonian’s National Museum of Natural History (NMNH), and particularly materials stored in the Departments of Anthropology and Paleobiology from several important archaeological and paleontological sites, as well as a broad range of palaeobiological reference specimens. These collections were obtained during the late 19th through to the 21st centuries, and provide excellent resources for morphometric studies, radiocarbon dating, stable isotope analyses, genetics, and proteomics (see Graham, 1981; Stanford et al., 1981; Rick et al., 2019; Knell and Lee, 2020). To explore the potential of ZooMS analysis for the study of museum-housed North American megafauna, we conducted a pilot ZooMS investigation of bone fragments from six Late Pleistocene archaeological sites held in the collections of the NMNH. Applying ZooMS for the first time to these collections, we focused on sites with purported evidence for hunting and/or scavenging of bison and other megafauna, and sites where the relationships and associations between artifacts and megafauna remain unclear. A number of these sites were suggested to be megafaunal ‘kill sites’, meaning primary sites where megafauna were dispatched by ancient hunters, although this identification has remained contested. All six of the sites investigated are in Colorado, in a region characterized by Clovis and Folsom, and in some cases perhaps pre-Clovis, cultural occupation (see Pitblado and Brunswig, 2007). These traditions represent early cultural phenomena in North American prehistory, often centered around distinctive projectile point technologies used by Paleoindians. Clovis groups (ca. 13,000-12,600 years ago) were efficient hunters often targeting large game such as mammoths and mastodons in addition to other resources. Following the Clovis tradition, the Folsom groups (ca. 12,600-11,500 years ago) are associated with the remains of bison, suggesting a shift in subsistence strategy towards more specialized hunting.
Megafaunal diversity in the Colorado region is high, with previously identified taxa including mammoth, bison, camelids, equids, felines, canids and other fauna identified from fossil localities and/or archaeological sites. Excavated between 1934 and 1980, the sites offer a range of excavation histories, time frames, and degrees of preservation against which we test the potential of ZooMS. By exploring the degree of collagen type I (COL1) preservation, the clarity of the spectra produced, and the resolution of taxonomic identification that ZooMS could provide, the study aimed to clarify the extent to which faunal remains in museum collections in North America and beyond offer a potential untapped reservoir of information for assessing patterns of megafaunal presence, persistence and disappearance from the Late Quaternary landscape.
ZooMS analyses were conducted on fragmented megafaunal remains recovered from six archaeological sites excavated between 1934 and 1980 and deposited at the NMNH (Figure 1): Lamb Spring, Selby, Dutton, Lindenmeier, Linger and Zapata Mammoth Site (see Supplementary Materials for detailed description of the sites). The assemblages analyzed were recovered from purported Clovis and Folsom animal kill and/or carcass processing sites, two of which (Lamb Spring and Selby-Dutton) may have been occupied by pre-Clovis people, though this is not widely accepted (Stanford et al., 1981; Jodry, 1999). Although diagnostic specimens are present in these collections (Wilmsen and Roberts, 1978: Graham, 1981; Rancier et al., 1981; Figure 1; Supplementary Table 1), and these enabled the taxonomic identification of fauna at all of the sites (Figure 1), much of the recovered material was highly fragmented and did not retain any morphologically diagnostic features. No anthropic modifications were observed on any of the specimens analyzed.
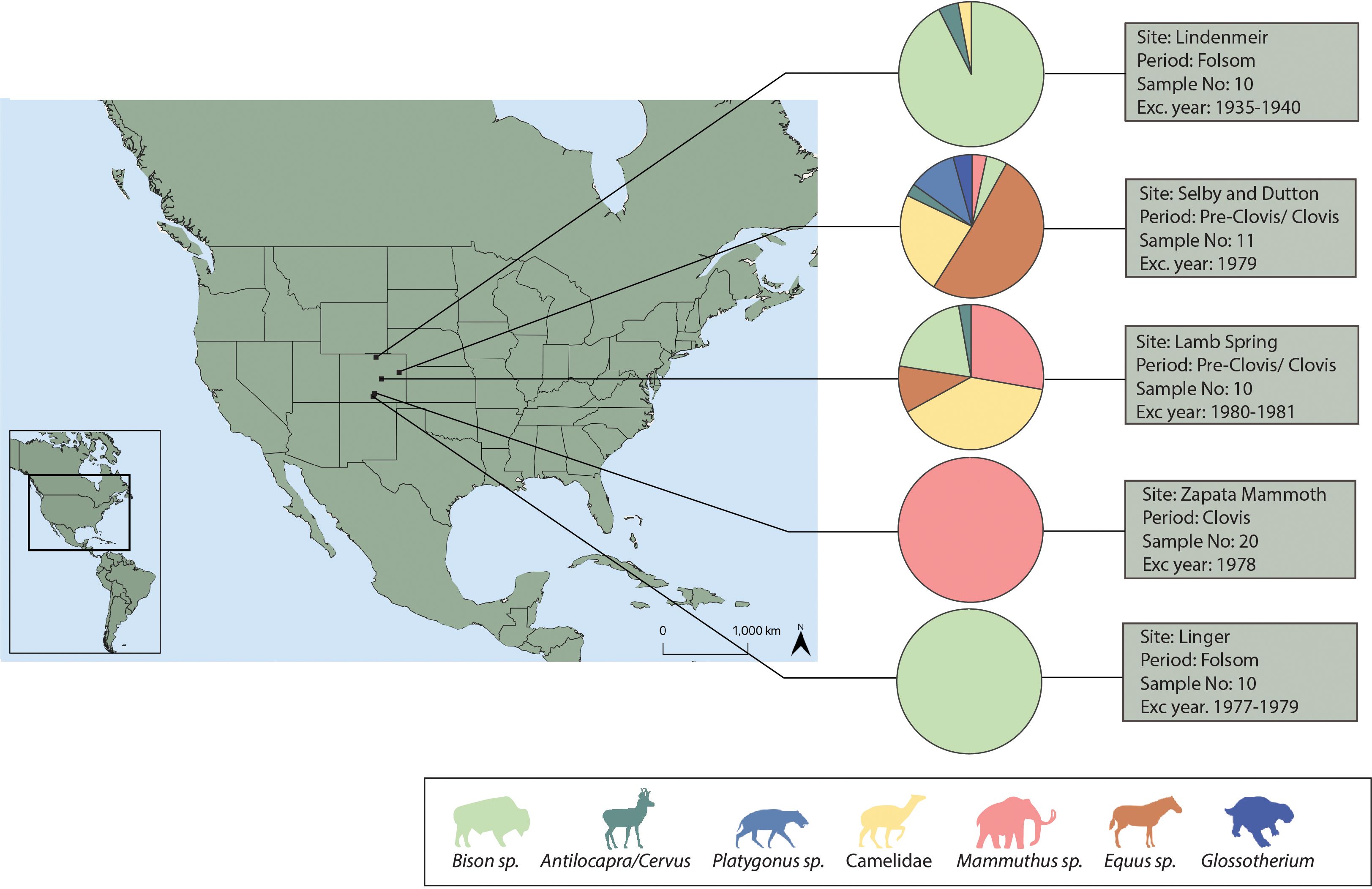
Figure 1 Site locations and published zooarchaeological data. For each site pie chart indicates the percentage of number of identified specimens (%NISP) of morphologically identified taxa (the published zooarchaeological counts can be found in Supplementary Table 1).
After excavation, the materials were transported for analysis and ultimately housed at the NMNH’s Museum Support Center (MSC) in Suitland, Maryland. Details on the handling and treatment of these specimens varies, but all appear to have been washed/cleaned of sediment at some point and none are known to have been treated with any pesticides or other materials. During the early portion of their storage some of these collections may for a time have been in non-climate-controlled conditions, handled by researchers without gloves, and occasionally exposed to ambient light. For the last decade or so, these materials have been stored at the MSC, a state of the art, climate-controlled, and secure facility where all of the assemblages are stored in their own cabinets by site.
To evaluate the efficacy of ZooMS for the investigation of Late Pleistocene faunal assemblages housed in the NMNH, and to complement morphological identifications, samples that displayed minimal to no diagnostic features and a high degree of fragmentation were primarily selected for analysis. In particular, sixty-one bone fragments were randomly sampled for ZooMS screening from the six archaeological sites. Most of the samples were represented by morphologically indistinct small fragments, only eight specimens from Lindenmeier and three from Lamb Spring were previously taxonomically identified based on morphological characteristics (Supplementary Table 2). All samples were obtained from the Department of Anthropology, NMNH and sent to the Max Planck Institute for the Science of Human History (now Max Planck Institute of Geoanthropology) in Jena, Germany for analysis in a dedicated palaeoproteomics laboratory. For each specimen, bone fragments of approximately 20 mg were removed, demineralized and processed following previously published protocols (Buckley et al., 2009; Brown et al., 2020). The samples were demineralized overnight in (4°C) 0.6 M hydrochloric acid (HCI), and then rinsed in 50 mM ammonium bicarbonate (AmBic) three times. Humic acids were removed by adding 0.1 M sodium hydroxide (NaOH), followed by 5 minutes incubation at room temperature. The NaOH was discarded, and a neutral pH restored by rinsing the sample in 50 mM AmBic. Samples were incubated at 65°C for one hour. 0.4 µg/µl trypsin solution (ThermoFisher Pierce™) was added, and samples were incubated for 18 hours at 37°C. Trypsin digestion was halted with 0.1 µl of 5% trifluoroacetic acid (TFA). Peptides were concentrated and desalted using C18 ZipTips (EMD Millipore) and eluted in a final solution of 50% acetonitrile and 0.1% TFA. 0.5 μl of α-cyano-4-hydroxycinnamic acid matrix solution was mixed with 0.5 μl of the eluted final solution and spotted in triplicate on a ground steel 385 spot target plate. All samples were run on a Bruker Autoflex Speed Matrix-Assisted Laser Desorption/Ionization - Time of Flight (MALDI-TOF) mass spectrometer (Bruker Daltonics).
Taxonomic identification was performed manually on MALDI-generated spectra using FlexAnalysis v. 3.4 (Bruker Daltonics), comparing identified collagen peptides with established reference library markers (Buckley et al., 2009; Buckley and Kansa, 2011; Buckley et al., 2011; Rybczynski et al., 2013; Welker et al., 2016; Brown et al., 2021b). No collagen peptides were detected in blanks and calibration standards performed as expected. Low-quality spectra were subsequently diluted (1:10 and 1:20) and rerun on the MALDI-TOF mass spectrometer.
The specificity of ZooMS classification relies on the evolutionary distance of taxonomic groups. It is possible to distinguish between families and often genera with high authenticity; however, separation between closely related species can be challenging, due to low variability of peptide sequences. For example, Bos sp. and Bison sp. can be distinguished from other artiodactyls based on combination of m/z values of 1192.6, 1208.6 (COL1ɑ2 978 - 990), 1427.7 (COL1ɑ2 484 - 498), 2853.4 (COL1ɑ1 586 - 618), and 3033.4 (COL1ɑ2 757 - 789). However, the reference markers for Bison are identical to those for Bos and identification, where possible, relies on consideration of the current range of the fauna and their past range as indicated by archaeological records from the study region. In our case, the attribution to Bos sp. can be excluded since this genus has not been registered amongst the fauna recovered for Late Pleistocene North America.
Likewise, members of the Elephantidae family share similar peptide marker series, making it impossible to discern between a number of possible genera (Elephas, Loxodonta, Mammuthus and Palaeoloxodon) using ZooMS alone. Our identification was guided by knowledge regarding the range of closely related species present in North American archaeological and palaeobiological assemblages. In light of the geographical location of our samples, we can infer that generated Elephantidae markers are indicative of Mammuthus sp. (mammoths). At the same time, mammoths share most of their COL1 peptide markers with mastodons (Mammut sp.), which were also present in Late Pleistocene North America. Mastodons can be separated from mammoths with only the COL1ɑ2 757–789 peptide marker, often hampering the differentiation between the two genera due to poor collagen preservation. This peptide for mastodon is present at m/z 2985.4/3001.4 and for mammoths at 2999.4/3015.4.
Glutamine deamidation level has been suggested as an indicator of collagen preservation variability in ZooMS samples (Welker et al., 2017) and was evaluated here to assess faunal preservation patterns. Deamidation of glutamine residues into glutamic acid is a common post-translational modification that results in a +0.98402 Da mass shift. In order to address protein degradation and collagen preservation, glutamine deamidation ratios were measured on all samples for the COL1ɑ1 508–519 peptide following using Q2E package (Wilson et al., 2012). The deamidation ratios were recorded ranging from 1 (entirely non-deamidated) to 0 (completely deamidated) and visualized via dot and box plots.
Our analysis resulted in successful ZooMS identifications for 80% of the total bone assemblage, with varying success rates observed across the archaeological sites. Of the 61 total samples, only one specimen from Linger (14A) yielded a full set of 12 markers, while 8 samples displayed 11-8 markers and 39 specimens generated 8-4 diagnostic markers sufficient for taxonomic identification. At the same time, 12 samples failed to preserve sufficient collagen for taxonomic identification (Figure 2; Supplementary Table 3).
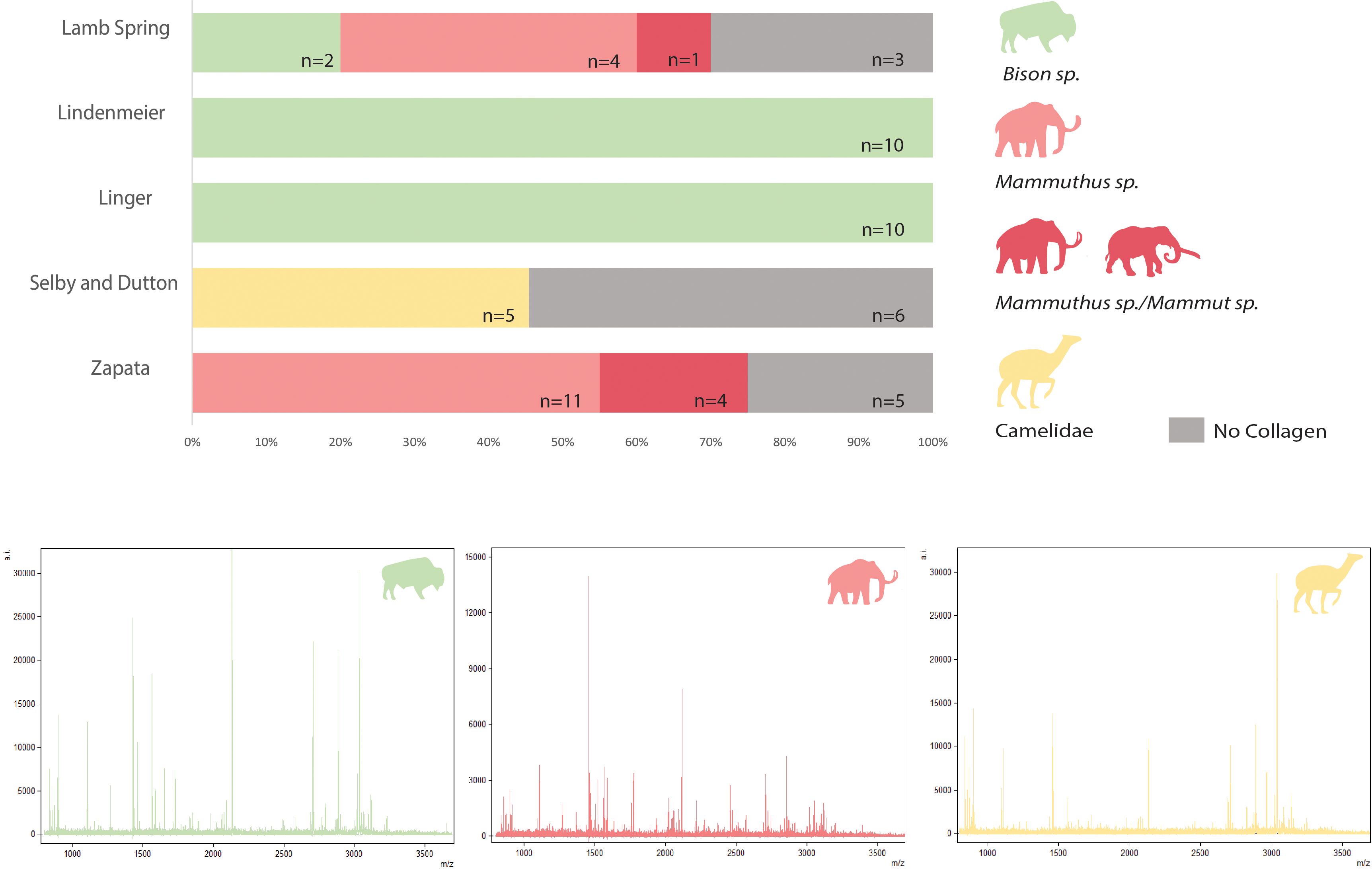
Figure 2 Taxa identified via ZooMS for each site (NISP) and example spectra (samples 14A for Bison sp., A591469-0 for Mammut sp. and 591314c for Camelidae).
From the 48 specimens that yielded sufficient collagen, 35 (72.9%) could be identified to genus level, whereas identification was restricted to family level for 13 specimens. The identified taxa include 22 specimens of Bison sp., 15 specimens of Mammuthus sp., 9 specimens of Mammuthus sp./Mammut sp., and 5 specimens from the family Camelidae. Further taxonomic determination of the camelids into species/genus was hampered by the lack of a securely established set of reference peptide markers for North American taxa. The taxonomic composition varied across the different sites investigated, and agreed with previous osteomorphological identifications that revealed the same set of taxa.
In total, 46 specimens were analyzed for glutamine deamidation of the COL1ɑ1 508–519 peptide (this was not measured for specimens with no or low collagen preservation). On the whole, all samples display relatively low deamidation (>0.6 for the majority of specimens, where 0 indicates fully deamidated and 1 not deamidated), with no extreme variations in the deamidation measurements amongst the archeological sites studied (Figure 3; Supplementary Table 4). The samples from Lindenmeier and Selby-Dutton were the best preserved, displaying the highest mean deamidation (0.75) while those from Zapata and Lamb Spring were the least well preserved (0.62). Unfortunately, due to an absence of recent excavation at the sites, we are unable to compare the deamidation values of recently acquired samples with those preserved in the museum for decades. As a result, we cannot assess the direct impact of museum storage on collagen preservation. However, the remarkably low deamidation values indicate favorable preservation in Late Pleistocene North American contexts. This suggests that successful outcomes can be obtained from museum-held specimens originally recovered from the region, even those collected in the early 20th century.
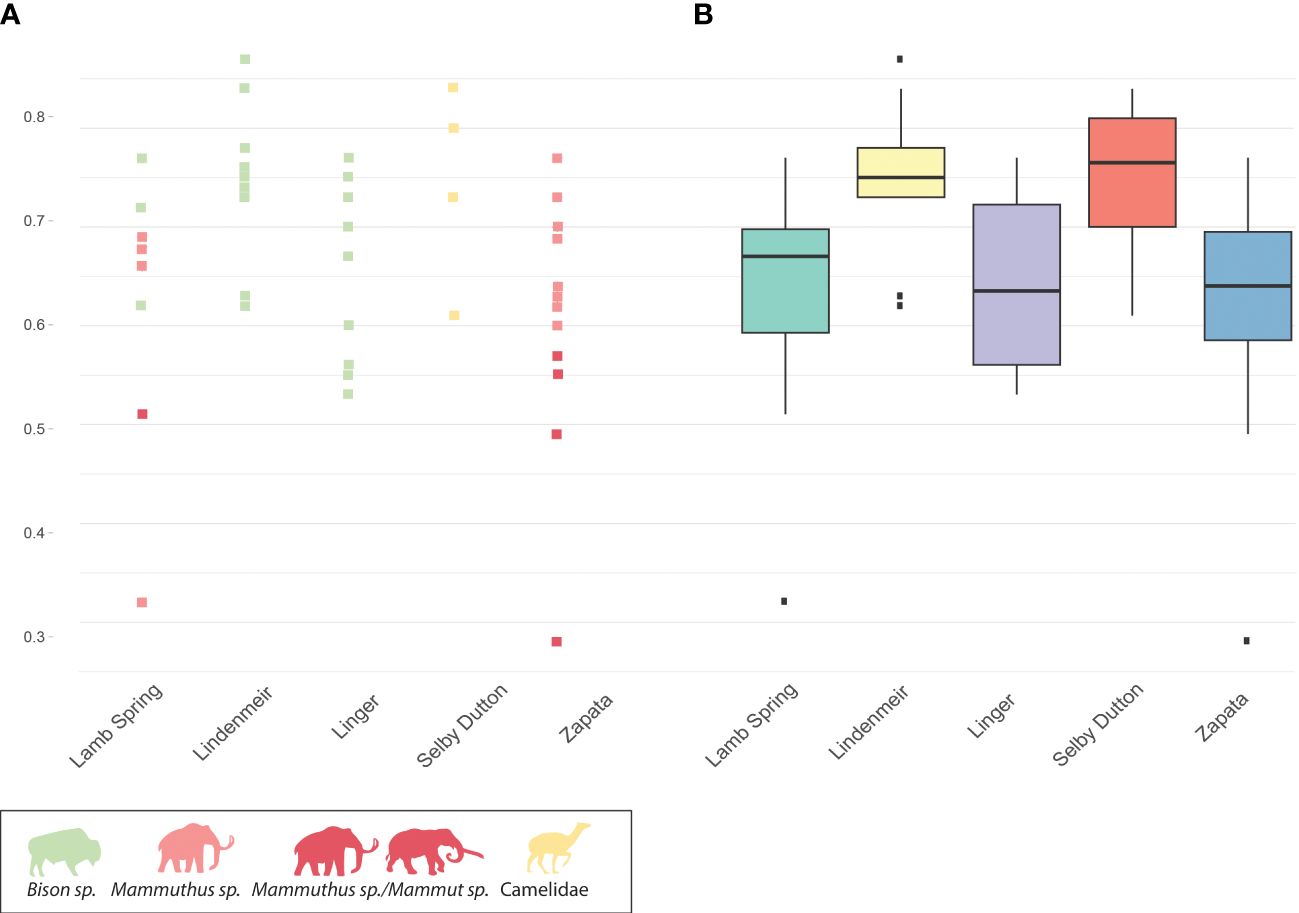
Figure 3 Results of glutamine deamidation measurements. (A) Individual bone measurements of glutamine deamidation per taxon. (B) Box plots for all measurements per site.
Ten unidentified bone fragments from Lamb Spring were analyzed via ZooMS, of which three were identified morphologically as Mammoth sp. ZooMS screening allowed for identification of seven specimens, three samples did not retain enough collagen for taxonomic identification. The samples exhibit varying degrees of molecular preservation, with the number of preserved diagnostic collagen peaks fluctuating from three to nine. In total, eight specimens from Lamb Spring were analyzed for glutamine deamidation. A large variation in deamidation values was observed at the site ranging between 0.32 to 0.77, with mean deamidation value 0.6. The box plot indicates presence of an outlier towards 0.
Regardless of the relatively poor preservation of the materials, it was possible to identify most of the samples to genus level (six specimens) backed up with the knowledge on the geographic distribution of taxa. ZooMS screening revealed the presence of two bison specimens. Four specimens were identified as mammoth and one specimen did not preserve enough collagen to allow for distinction between mastodon and mammoth, restricting identification to Mammuthus sp./Mammut sp. These results confirm the taxonomic identifications from Lamb Spring provided by Rancier et al. (1981), and broadly agree with its identification as a mammoth kill site, though the number of specimens analyzed is insufficient to enable any further insight into the issue.
Ten specimens from Lindenmeier were targeted for ZooMS screening, of which eight were identified morphologically as bison. The ZooMS screening confirmed the previous osteomorphological identifications, with all ten specimens generating spectra diagnostic of Bos sp./Bison sp. Glutamine deamidation of nine samples from the site was measured, with resulting values ranging from 0.62 to 0.87 (mean 0.75). The box plot indicates presence of tree outliers.
All ten morphologically indistinct small fragments from Linger were assigned to a single taxon based on ZooMS: Bison sp., confirming the faunal lists previously published for this site (Wilmsen and Roberts, 1978) and the fauna recorded in the NMNH Anthropology Online Catalogue. Notably, sample 14A from Linger presented an exceptionally well-preserved Bison sp. collagen spectrum (Figure 2), retaining 12 out of 12 published markers (Buckley et al., 2009) and five specimens preserve 10 or 11 markers, highlighting the relatively high rate of molecular preservation at the site. The deamidation analyses confirms relatively good preservation of the specimens from the site, displaying values between 0.53-0.77 and a mean of 0.64.
Of the 11 samples from Selby and Dutton, five samples were identified to family level and the remaining six samples did not retain enough collagen for identification. The reconstructed taxonomic composition is represented by a single taxonomic group: Camelidae. These identifications are in accordance with the fauna recorded by Graham (1981). Currently, reference markers have been reconstructed for four species of Camelids that share a number of peptide markers: Bactrian camel, dromedary camel, alpaca and vicuña (Buckley et al., 2009; Rybczynski et al., 2013). The best-preserved specimens from Selby and Dutton displayed the presence of six published peptide markers that overlap with those identified for Camelus dromedarius (collagen peaks at m/z 1105, m/z 1221, m/z 1453, m/z 2131, m/z 2820, m/z 2883), however unlike Camelus, which carries m/z 2975.4/2991.4 for the COL1ɑ2 757 – 789 marker, the specimens from Selby and Dutton displayed m/z 3017.4/3033.4, which has previously been shown to be distinctive for extinct Camelops (Buckley et al., 2019). However, lack of securely identified markers for North American camelids restricts further interpretation of these specimens. Only four specimens from the sites contained sufficient collagen for deamidation analyses, displaying values ranging from 0.61 to 0.84 (mean 0.75).
Fifteen of the 20 samples from Zapata Mammoth were identified as Proboscideans, while five did not retain enough collagen to determine taxonomic affiliation. The identification revealed eleven specimens of mammoth, however for the remaining four specimens, poor collagen preservation restricted the identification to mammoth/mastodon. This taxonomic determination falls within the faunal composition previously identified at the site, which recorded the presence of mammoth only. The number of diagnostic markers varied from 4 to 6. Samples from Zapata Mammoth displayed the lowest deamidation values among the analyzed sites with large inter-site variation, ranging from 0.28 to 0.77 (mean 0.61). The box plot indicates the presence of an outlier towards 0.
Our preliminary ZooMS screening has demonstrated the rich potential of peptide mass fingerprinting for the analysis of Late Pleistocene megafaunal assemblages housed as legacy collections in museums in North America. Despite the fact that collections were excavated in some cases more than a century ago, and that specimens selected for analysis were often fragmentary and highly weathered, ZooMS success rates and collagen preservation were relatively high across all sites, with an overall success rate of 80%, and a mean deamidation value 0.66. Long-term storage in the NMNH and millennia of exposure at the archaeological sites does not appear to have been detrimental to peptide preservation.
ZooMS analysis of 61 bone fragments across the 5 sites investigated allowed for identification of 4 taxonomic groups: Bison sp., Mammuthus sp., Mammuthus sp./Mammut sp. and Camelidae. The taxa identified are all megafauna representatives that have been found in the region and were previously recorded at each respective site (Wilmsen and Roberts, 1978; Graham, 1981; Rancier et al., 1981). As described above, Delby and Sutton, Lamb Springs, and Lindenmeier were reported to contain a range of different taxa such as mammoths, horses, bison, camelids, and other species, while previous zooarchaeological efforts at the Zapata Mammoth site revealed the presence of only mammoth, and at Linger only bison. Although the consistency of our ZooMS results with data obtained from the morphological analyses allowed us to increase the NISP, it did not reveal the presence of previously unrecorded taxonomic groups for the sites (Figure 4). A larger sample of ZooMS specimens could help further expand taxonomic richness at each site.
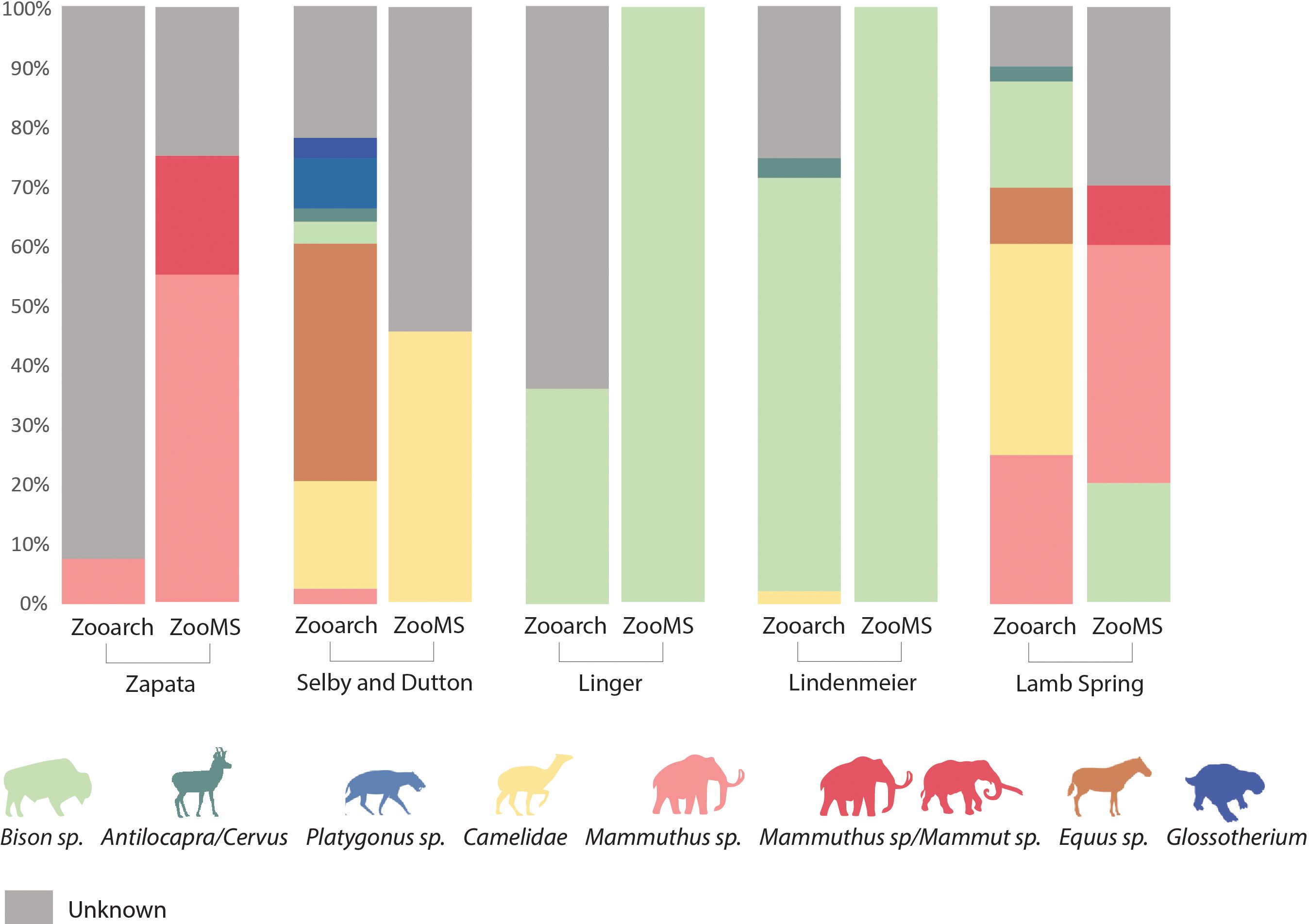
Figure 4 Comparison of percentage of number of identified specimens (%NISP) of traditional Zooarchaeological and ZooMS taxa determinations across sites.
The sites show remarkable variation in terms of faunal composition, with the ZooMS-based identifications of the majority limited to a single taxon. In particular, samples from Linger and Lindenmeier generated collagen fingerprints associated solely with Bison sp., Zapata Mammoth samples revealed the presence of mammoths, while Sutton and Delby samples returned collagen spectra diagnostic of Camelids. Meanwhile, the identifications of Lamb Springs samples revealed the presence of more than one taxonomic group (mammoth and/or mastodon, and bison). In contrast, the morphologically identified set of taxa from the sites are generally more diverse (Figure 4). This limitation can be explained by the small sample size and the fact that the carcasses of larger animals tend to generate higher numbers of fragments, creating a bias in taxonomic representation. These biases could be addressed by increasing the number of specimens sampled, while also strategically sampling a broad and representative range of the diversity of bone types and animal sizes observed in the assemblage. Inflation in the NISP for large species is a recognized issue in ZooMS studies, and researchers in the field are currently exploring ways to improve the statistical significance of faunal spectra and the abundance of each taxon with tailored sampling strategies (Bouchard et al., 2020; Silvestrini et al., 2022).
The utility of ZooMS for a particular study is strongly dependent on whether an appropriate collagen fingerprint reference library is available for relevant species in the geographic region of interest. In contrast to other regions where the method has been used successfully, such as southwestern Asia and Europe (e.g., Buckley and Kansa, 2011; Sinet-Mathiot et al., 2019; Ruebens et al., 2023), North America so far has not seen the creation of comprehensive regional reference libraries, especially for extinct taxa, limiting the applicability of ZooMS. For instance, the dearth of published reference markers for camelid species imposed a significant limitation on our analysis, precluding in-depth taxonomic identification of camelid specimens, and enabling their identification only to family level. This highlights the need for construction of open access ZooMS marker reference libraries for extinct and extant North American species. Such development will enable reinvestigation of the fragmented assemblages in legacy collections from Late Pleistocene kill sites, allowing for more precise and accurate identification of fossil fauna and fruitful exploitation of megafaunal remains held on the continent.
Traditional zooarchaeological analyses of fragmented faunal remains from many Paleoindian sites have been hampered by an absence of taxonomically significant morphological features (Cannon and Meltzer, 2004; Hill, 2007; Hofman et al., 2018), and a number of questions remain about the composition of the Late Pleistocene fauna recovered from the six sites we have revisited with ZooMS. However, our work confirms past identifications and presents a clear roadmap for expanding reference marker libraries and performing additional ZooMS analyses to address ongoing debates about these sites and the link between megafaunal remains and human activity. ZooMS will also be able to help clarify whether or not confirmed North American Pleistocene kill sites are exclusively characterized by proboscidean, bovid, camelid, and equid remains, as the archaeological record appears to indicate (Grayson and Meltzer, 2015), or whether a more diverse array of fauna were targeted.
Our study used glutamine deamidation as a proxy for biomolecular preservation. Deamidation ratios could be calculated for 46 specimens displaying relatively high levels of preservation (mean: 0.62-0.75). Analysis of glutamine deamidation rates has been incorporated into ZooMS studies at various Pleistocene sites (Welker et al., 2017; Sinet-Mathiot et al., 2019; Brown et al., 2021c; Ruebens et al., 2022; Silvestrini et al., 2022; Ruebens et al., 2023) and has the potential to provide additional insights into differing patterns of sample preservation. This potential becomes particularly clear when our rates are compared to available glutamine deamidation values from other Late Pleistocene sites across Europe, such as Uluzzo C, Roccia San Sebastiano, and Riparo del Broion sites in Italy (with a mean deamidation value of 0.33), Quincay in France (mean: 0.5) and Grotte du Renne in France (mean: 0.15). Our samples show relatively lower deamidation rates on average than those from such European Palaeolithic sites for which data is available. This may reflect the older age of European samples, the preservation conditions of samples, or any number of other variables, but they do support the utility of revisiting North American legacy collections using ZooMS.
We did not observe any correlation between collagen preservation and age of excavation. Indeed, the specimens excavated from Lindenmeier in the late 1930s exhibit better preservation than those obtained during the late 1970s and early 1980s. At the same time, some collections displayed substantial variability of deamidation values across samples from a single site, which can perhaps best be explained by localized geological or environmental conditions or different diagenetic processes (Scotchler and Robinson, 1974). These collections have had similar histories of curation and records of handling and storage over the years, but variation in exposure to taphonomic processes was nonetheless not fully controlled. At times, some of the collections were handled without gloves, perhaps exposed to ambient light, and kept in variable climatic conditions in indoor storage prior to being stored at the MSC starting 10 years ago in climate/humidity controlled, secure conditions, away from direct or ambient light.
Nonetheless, it is challenging to suggest a clear explanation for varying rates of preservation across different sites, since the effects of long-term museum storage on proteins are not well understood. While molecular fragmentation is a time-dependent process, the rate at which this occurs is influenced by environmental factors such as temperature, pH, humidity, and the chemical composition of the surroundings. It is therefore impossible to derive a simple correlation between preservation and sample age across different preservation environments (Dal Sasso et al., 2016; Schroeter and Cleland, 2016; Kistler et al., 2017; Brown et al., 2021c). In future, more detailed assessments of the effects of museum storage on sample preservation will be important for improving our understanding of both the molecular potential of particular samples, as well as the factors that support optimal biomolecular preservation of museum remains more broadly.
Archaeological collections of megafauna exist in repositories around the world. Higher resolution investigation of these collections using novel biomolecular tools like ZooMS will foster new insights into local and regional links between climate, environment, human behavior and megafaunal composition, offering new perspectives on the timing and extent of past biodiversity change and the causes and consequences of megafaunal extinction. Our pilot study has revealed that remarkable collagen preservation is exhibited by the fragmented assemblages recovered from Late Quaternary/Paleoindian sites housed at the NMNH, demonstrating the rich potential of ZooMS for examining legacy collections of megafauna, including in North America. Through successful ZooMS screening, we were able to both evaluate the molecular preservation of these materials and obtain valuable taxonomic information from 80% of our samples, reflecting continuity between the ZooMS identifications and previous morphological assessments.
While our study is not able to resolve long-standing debates about the cause of megafaunal extinctions in North America, it does point the way towards a novel approach to existing collections that might offer new insights into old questions. However, there are still challenges that will require more focused research. In particular, a better understanding of the factors influencing biomolecular preservation in museum collections is vital. Future research is needed to explore the effects of environmental conditions, storage practices, and sample age on collagen degradation processes. Additionally, expansion and refinement of ZooMS reference libraries to include a wider range of species, particularly those from regions beyond Europe, is a critical next step.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. The generated ZooMS spectra for this study can be found at https://zenodo.org/records/10798233 an open-source online data repository hosted at Zenodo.
MA: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. EH: Investigation, Methodology, Writing – review & editing. MJ: Conceptualization, Validation, Writing – review & editing. NA: Resources, Supervision, Writing – review & editing. SB: Methodology, Software, Supervision, Writing – review & editing. TR: Conceptualization, Resources, Validation, Writing – review & editing. NB: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was funded by the Max Planck Society.
We are grateful to the Smithsonian NMNH staff for access to and information about collections, especially Amanda Lawrence and Esther Rimer in the Department of Anthropology and Kay Behrensmeyer, Dave Bohaska, and Amanda Millhouse in the Department of Paleobiology. We thank Dennis Stanford for his thoughts on the early phases of this project and the sites he excavated or studied that we report on here. All analyses were reviewed and approved by the NMNH Anthropology Sampling Committee.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmamm.2024.1399358/full#supplementary-material
Antonosyan M., Roberts P., Aspaturyan N., Mkrtchyan S., Lucas M., Boxleitner K., et al. (2024). Multiproxy evidence for environmental stability in the Lesser Caucasus during the Late Pleistocene. Quaternary Sci. Rev. doi: 10.1016/j.quascirev.2024.108559
Ashby S. P., Coutu A. N., Sindbæk S. M. (2015). Urban networks and Arctic outlands: craft specialists and reindeer antler in Viking towns. Eur. J. Archaeology 18, 679–704. doi: 10.1179/1461957115Y.0000000003
Barnosky A. D., Koch P. L., Feranec R. S., Wing S. L., Shabel A. B. (2004). Assessing the causes of late Pleistocene extinctions on the continents. science 306, 70–75. doi: 10.1126/science.1101476
Bartlett L. J., Williams D. R., Prescott G. W., Balmford A., Green R. E., Eriksson A., et al. (2016). Robustness despite uncertainty: regional climate data reveal the dominant role of humans in explaining global extinctions of Late Quaternary megafauna. Ecography 39, 152–161. doi: 10.1111/ecog.01566
Bergman J., Pedersen R.Ø., Lundgren E. J., Lemoine R. T., Monsarrat S., Pearce E. A., et al. (2023). Worldwide Late Pleistocene and Early Holocene population declines in extant megafauna are associated with Homo sapiens expansion rather than climate change. Nat. Commun. 14, 7679. doi: 10.1038/s41467-023-43426-5
Bleasdale M., Richter K. K., Janzen A., Brown S., Scott A., Zech J., et al. (2021). Ancient proteins provide evidence of dairy consumption in eastern Africa. Nat. Commun. 12, 632. doi: 10.1038/s41467-020-20682-3
Bollongino R., Tresset A., Vigne J. D. (2008). Environment and excavation: Pre-lab impacts on ancient DNA analyses. Comptes Rendus Palevol 7, 91–98. doi: 10.1016/j.crpv.2008.02.002
Bouchard G. P., Riel-Salvatore J., Negrino F., Buckley M. (2020). Archaeozoological, taphonomic and ZooMS insights into The Protoaurignacian faunal record from Riparo Bombrini. Quaternary Int. 551, 243–263. doi: 10.1016/j.quaint.2020.01.007
Brook B. W., Bowman D. M. (2002). Explaining the Pleistocene megafaunal extinctions: models, chronologies, and assumptions. Proc. Natl. Acad. Sci. 99, 14624–14627. doi: 10.1073/pnas.232126899
Broughton J. M., Weitzel E. M. (2018). Population reconstructions for humans and megafauna suggest mixed causes for North American Pleistocene extinctions. Nat. Commun. 9, 5441. doi: 10.1038/s41467-018-07897-1
Brown S., Douka K., Collins M. J., Richter K. K. (2021b). On the standardization of ZooMS nomenclature. J. Proteomics 235, 104041. doi: 10.1016/j.jprot.2020.104041
Brown S., Hebestreit S., Wang N., Boivin N., Douka K., Richter K. (2020). Zooarchaeology by Mass Spectrometry (ZooMS) for bone material-Acid insoluble protocol. doi: 10.17504/protocols.io.bf43jqyn
Brown S., Kozlikin M., Shunkov M., Derevianko A., Higham T., Douka K., et al. (2021c). Examining collagen preservation through glutamine deamidation at Denisova Cave. J. Archaeological Sci. 133, 105454. doi: 10.1016/j.jas.2021.105454
Brown S., Wang N., Oertle A., Kozlikin M. B., Shunkov M. V., Derevianko A. P., et al. (2021a). Zooarchaeology through the lens of collagen fingerprinting at Denisova Cave. Sci. Rep. 11, 15457. doi: 10.1038/s41598-021-94731-2
Buckley M. (2018). “Zooarchaeology by mass spectrometry (ZooMS) collagen fingerprinting for the species identification of archaeological bone fragments,” in Zooarchaeology in practice: case studies in methodology and interpretation in archaeofaunal analysis, pp.227–pp.247.
Buckley M., Collins M., Thomas-Oates J., Wilson J. C. (2009). Species identification by analysis of bone collagen using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrometry: Int. J. Devoted to Rapid Dissemination Up-to-the-Minute Res. Mass Spectrometry 23, 3843–3854.
Buckley M., Kansa S. W. (2011). Collagen fingerprinting of archaeological bone and teeth remains from Domuztepe, South Eastern Turkey. Archaeological Anthropological Sci. 3, 271–280. doi: 10.1007/s12520-011-0066-z
Buckley M., Larkin N., Collins M. (2011). Mammoth and Mastodon collagen sequences; survival and utility. Geochimica Cosmochimica Acta 75, 2007–2016. doi: 10.1016/j.gca.2011.01.022
Buckley M., Lawless C., Rybczynski N. (2019). Collagen sequence analysis of fossil camels, Camelops and cf Paracamelus, from the Arctic and sub-Arctic of Plio-Pleistocene North America. J. Proteomics 194, 218–225. doi: 10.1016/j.jprot.2018.11.014
Cannon M. D., Meltzer D. J. (2004). Early PaleoIndian foraging: examining the faunal evidence for large mammal specialization and regional variability in prey choice. Quaternary Sci. Rev. 23, 1955–1987. doi: 10.1016/j.quascirev.2004.03.011
Cappellini E., Welker F., Pandolfi L., Ramos-Madrigal J., Samodova D., Rüther P. L., et al. (2019). Early Pleistocene enamel proteome from Dmanisi resolves Stephanorhinus phylogeny. NATURE 574, 103–107. doi: 10.1038/s41586-019-1555-y
Cleland T. P., Schroeter E. R., Feranec R. S., Vashishth D. (2016). Peptide sequences from the first Castoroides ohioensis skull and the utility of old museum collections for palaeoproteomics. Proc. R. Soc. B: Biol. Sci. 283, 20160593.
Cooper A., Turney C., Hughen K. A., Brook B. W., McDonald H. G., Bradshaw C. J. (2015). Abrupt warming events drove Late Pleistocene Holarctic megafaunal turnover. Science 349, 602–606. doi: 10.1126/science.aac4315
Coutu A. N., Whitelaw G., Le Roux P., Sealy J. (2016). Earliest evidence for the ivory trade in southern Africa: Isotopic and ZooMS analysis of seventh–tenth century AD ivory from KwaZulu-Natal. Afr. Archaeological Rev. 33, 411–435. doi: 10.1007/s10437-016-9232-0
Dal Sasso G., Lebon M., Angelini I., Maritan L., Usai D., Artioli G., et al. (2016). Bone diagenesis variability among multiple burial phases at Al Khiday (Sudan) investigated by ATR-FTIR spectroscopy. Palaeogeography, Palaeoclimatology, Palaeoecology 463, 168-179
Dekker J., Sinet-Mathiot V., Spithoven M., Smit B., Wilcke A., Welker F., et al. (2021). Human and cervid osseous materials used for barbed point manufacture in Mesolithic Doggerland. J. Archaeological Science: Rep. 35, 102678.
Desmond A., Barton N., Bouzouggar A., Douka K., Fernandez P., Humphrey L., et al. (2018). ZooMS identification of bone tools from the North African Later Stone Age. J. Archaeological Sci. 98, 149–157. doi: 10.1016/j.jas.2018.08.012
Ebsen J. A., Haase K., Larsen R., Sommer D. V.P., Brandt L. Ø. (2019). Identifying archaeological leather–discussing the potential of grain pattern analysis and zooarchaeology by mass spectrometry (ZooMS) through a case study involving medieval shoe parts from Denmark. Journal of cultural heritage 39, 21–31.
Faith J. T. (2011). Late Pleistocene climate change, nutrient cycling, and the megafaunal extinctions in North America. Quaternary Sci. Rev. 30, 1675–1680. doi: 10.1016/j.quascirev.2011.03.011
Firestone R. B., West A., Kennett J. P., Becker L., Bunch T. E., Revay Z. S., et al. (2007). Evidence for an extraterrestrial impact 12,900 years ago that contributed to the megafaunal extinctions and the Younger Dryas cooling. Proc. Natl. Acad. Sci. 104, 16016–16021. doi: 10.1073/pnas.0706977104
Gill J. L., Williams J. W., Jackson S. T., Lininger K. B., Robinson G. S. (2009). Pleistocene megafaunal collapse, novel plant communities, and enhanced fire regimes in North America. Science 326, 1100–1103. doi: 10.1126/science.1179504
Graham R. W. (1981). “Preliminary report on late Pleistocene vertebrates from the Selby and Dutton archeological/paleontological sites, Yuma County, Colorado,” in Contributions to Geology, vol. 20. (University of Wyoming, Laramie), pp.33–pp.56.
Grayson D. K., Meltzer D. J. (2015). Revisiting PaleoIndian exploitation of extinct North American mammals. J. Archaeological Sci. 56, 177–193. doi: 10.1016/j.jas.2015.02.009
Hahn E. E., Alexander M. R., Grealy A., Stiller J., Gardiner D. M., Holleley C. E. (2022). Unlocking inaccessible historical genomes preserved in formalin. Mol. Ecol. Resour. 22, 2130–2147. doi: 10.1111/1755-0998.13505
Hill M. E. (2007). A moveable feast: variation in faunal resource use among central and western North American PaleoIndian sites. Am. Antiquity 72, 417–438. doi: 10.2307/40035854
Hofman C. A., Rick T. C., Erlandson J. M., Reeder-Myers L., Welch A. J., Buckley M. (2018). Collagen fingerprinting and the earliest marine mammal hunting in North America. Sci. Rep. 8, 10014. doi: 10.1038/s41598-018-28224-0
Holmes M. W., Hammond T. T., Wogan G. O., Walsh R. E., LaBarbera K., Wommack E. A., et al. (2016). Natural history collections as windows on evolutionary processes. Mol. Ecol. 25, 864–881. doi: 10.1111/mec.13529
Jodry M. A. B. (1999). Folsom technological and socioeconomic strategies: views from Stewart's Cattle Guard and the Upper Rio Grande Basin, Colorado (American University).
Kehlmaier C., López-Jurado L. F., Hernández-Acosta N., Mateo-Miras A., Fritz U. (2021). “Ancient DNA“ reveals that the scientific name for an extinct tortoise from Cape Verde refers to an extant South American species. Sci. Rep. 11, 17537. doi: 10.1038/s41598-021-97064-2
Kistler L., Ware R., Smith O., Collins M., Allaby R. G.. (2017). A new model for ancient DNA decay based on paleogenomic meta-analysis. Nucleic Acids Research 45 (11), 6310-6320
Knell E. J., Lee C. M. (2020). New radiocarbon dates from the late paleoIndian cody complex component at the lamb spring site, Douglas county, Colorado. PaleoAmerica 6, 374–378. doi: 10.1080/20555563.2020.1762408
Lamichhaney S., Card D. C., Grayson P., Tonini J. F., Bravo G. A., Näpflin K., et al. (2019). Integrating natural history collections and comparative genomics to study the genetic architecture of convergent evolution. Philos. Trans. R. Soc. B 374, 20180248. doi: 10.1098/rstb.2018.0248
Lehmann U., Kreipe H. (2001). Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods 25, 409–418. doi: 10.1006/meth.2001.1263
Lorenzen E. D., Nogués-Bravo D., Orlando L., Weinstock J., Binladen J., Marske K. A., et al. (2011). Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479, 359–364. doi: 10.1038/nature10574
Martisius N. L., Welker F., Dogandžić T., Grote M. N., Rendu W., Sinet-Mathiot V., et al. (2020). Non-destructive ZooMS identification reveals strategic bone tool raw material selection by Neandertals. Sci. Rep. 10, 7746. doi: 10.1038/s41598-020-64358-w
McGuire J. A., Cotoras D. D., O’Connell B., Lawalata S. Z., Wang-Claypool C. Y., Stubbs A., et al. (2018). Squeezing water from a stone: high-throughput sequencing from a 145-year old holotype resolves (barely) a cryptic species problem in flying lizards. PeerJ 6, e4470. doi: 10.7717/peerj.4470
Meltzer D. J. (2020). Overkill, glacial history, and the extinction of North America’s Ice Age megafauna. Proc. Natl. Acad. Sci. 117, 28555–28563. doi: 10.1073/pnas.2015032117
Monteath A. J., Gaglioti B. V., Edwards M. E., Froese D. (2021). Late Pleistocene shrub expansion preceded megafauna turnover and extinctions in eastern Beringia. Proc. Natl. Acad. Sci. 118, e2107977118. doi: 10.1073/pnas.2107977118
O'Connor S., Solazzo C., Collins M. (2015). Advances in identifying archaeological traces of horn and other keratinous hard tissues. Stud. Conserv. 60, 393–417. doi: 10.1179/2047058414Y.0000000134
O’Keefe F. R., Dunn R. E., Weitzel E. M., Waters M. R., Martinez L. N., Binder W. J., et al. (2023). Pre–Younger Dryas megafaunal extirpation at Rancho La Brea linked to fire-driven state shift. Science 381, eabo3594.
Pitblado B. L., Brunswig R. H. (2007). Frontiers in Colorado paleoIndian archaeology: from the Dent Site to the Rocky Mountains (University Press of Colorado).
Prüfer K., Posth C., Yu H., Stoessel A., Spyrou M. A., Deviese T., et al. (2021). A genome sequence from a modern human skull over 45,000 years old from Zlatý kůň in Czechia. Nat. Ecol. Evol. 5, 820–825.
Pruvost M., Schwarz R., Correia V. B., Champlot S., Braguier S., Morel N., et al. (2007). Freshly excavated fossil bones are best for amplification of ancient DNA. Proc. Natl. Acad. Sci. 104, 739–744. doi: 10.1073/pnas.0610257104
Rancier J., Haynes G., Stanford D. (1981). “Investigations of lamb spring,” in Southwestern Lore, vol. 48. , pp.1–pp17.
Raxworthy C. J., Smith B. T. (2021). Mining museums for historical DNA: advances and challenges in museomics. Trends Ecol. Evol. 36, 1049–1060. doi: 10.1016/j.tree.2021.07.009
Richter K. K., Codlin M. C., Seabrook M., Warinner C. (2022). A primer for ZooMS applications in archaeology. Proc. Natl. Acad. Sci. 119, e2109323119. doi: 10.1073/pnas.2109323119
Rick T., Harvey V. L., Buckley M. (2019). Collagen fingerprinting and the chumash billfish fishery, Santa Barbara Channel, California, USA. Archaeological Anthropological Sci. 11, 6639–6648. doi: 10.1007/s12520-019-00930-4
Ripple W. J., Van Valkenburgh B. (2010). Linking top-down forces to the Pleistocene megafaunal extinctions. BioScience 60, 516–526. doi: 10.1525/bio.2010.60.7.7
Rubi T. L., Knowles L. L., Dantzer B. (2020). Museum epigenomics: Characterizing cytosine methylation in historic museum specimens. Mol. Ecol. Resour. 20, 1161–1170. doi: 10.1111/1755-0998.13115
Ruebens K., Sinet-Mathiot V., Talamo S., Smith G. M., Welker F., Hublin J. J., et al. (2022). The late Middle Palaeolithic occupation of Abri du Maras (layer 1, Neronian, southeast France): integrating lithic analyses, ZooMS and radiocarbon dating to reconstruct Neanderthal hunting behaviour. J. Paleolithic Archaeology 5, 4. doi: 10.1007/s41982-022-00113-z
Ruebens K., Smith G. M., Fewlass H., Sinet-Mathiot V., Hublin J. J., Welker F. (2023). Neanderthal subsistence, taphonomy and chronology at Salzgitter-Lebenstedt (Germany): a multifaceted analysis of morphologically unidentifiable bone. J. Quaternary Sci. 38, 471–487. doi: 10.1002/jqs.3499
Rybczynski N., Gosse J. C., Richard Harington C., Wogelius R. A., Hidy A. J., Buckley M. (2013). Mid-Pliocene warm-period deposits in the High Arctic yield insight into camel evolution. Nat. Commun. 4, 1550. doi: 10.1038/ncomms2516
Sanchez G. M., Rick T. C., Culleton B. J., Kennett D. J., Buckley M., Erlandson J. M., et al. (2018). Radiocarbon dating legacy collections: A Bayesian analysis of high-precision AMS 14C dates from the Par-Tee site, Oregon. J. Archaeological Science: Rep. 21, 833–848. doi: 10.1016/j.jasrep.2018.08.033
Schroeter E. R., Cleland T. P. (2016). Glutamine deamidation: an indicator of antiquity, or preservational quality? Rapid Commun. mass spectrometry 30, 251–255.
Scotchler J. W., Robinson A. B. (1974). Deamidation of glutaminyl residues: dependence on pH, temperature, and ionic strength. Analytical Biochem. 59, 319–322. doi: 10.1016/0003-2697(74)90040-2
Seersholm F. V., Hansen K. L., Heydenrych M., Hansen A. J., Bunce M., Allentoft M. E. (2021). Ancient DNA preserved in small bone fragments from the PW Lund collection. Ecol. Evol. 11, 2064–2071. doi: 10.1002/ece3.7162
Seersholm F. V., Werndly D. J., Grealy A., Johnson T., Keenan Early E. M., Lundelius E. L. Jr., et al. (2020). Rapid range shifts and megafaunal extinctions associated with late Pleistocene climate change. Nat. Commun. 11, 2770. doi: 10.1038/s41467-020-16502-3
Silvestrini S., Lugli F., Romandini M., Real C., Sommella E., Salviati E., et al. (2022). Integrating ZooMS and zooarchaeology: New data from the Uluzzian levels of Uluzzo C Rock Shelter, Roccia San Sebastiano cave and Riparo del Broion. PLoS One 17, e0275614. doi: 10.1371/journal.pone.0275614
Sinet-Mathiot V., Smith G. M., Romandini M., Wilcke A., Peresani M., Hublin J. J., et al. (2019). Combining ZooMS and zooarchaeology to study Late Pleistocene hominin behaviour at Fumane (Italy). Sci. Rep. 9, 12350. doi: 10.1038/s41598-019-48706-z
St. Amand F., Childs S. T., Reitz E. J., Heller S., Newsom B., Rick T. C., et al. (2020). Leveraging legacy archaeological collections as proxies for climate and environmental research. Proc. Natl. Acad. Sci. 117, 8287–8294. doi: 10.1073/pnas.1914154117
Stanford D., Wedel W. R., Scott G. R. (1981). “Archaeological investigations of the Lamb Spring site,” in Southwestern Lore, vol. 47. , pp.14–pp.27.
Stewart J. R., Allen R. B., Jones A. K., Penkman K. E., Collins M. J. (2013). ZooMS: making eggshell visible in the archaeological record. J. Archaeological Sci. 40, 1797–1804. doi: 10.1016/j.jas.2012.11.007
Stewart M., Carleton W. C., Groucutt H. S. (2021). Climate change, not human population growth, correlates with Late Quaternary megafauna declines in North America. Nat. Commun. 12, 965. doi: 10.1038/s41467-021-21201-8
Vershinina A. O., Kapp J. D., Baryshnikov G. F., Shapiro B. (2020). The case of an arctic wild ass highlights the utility of ancient DNA for validating problematic identifications in museum collections. Mol. Ecol. Resour. 20, 1182–1190. doi: 10.1111/1755-0998.13130
Wagner A., Richter K. K., Ludes E., Arbogast R. M., Carita D., Guidez A., et al. (2020). Whale bone puzzles: Reconstructing and identifying historical whale skeletons using archive records, osteology, and zooarchaeology by Mass Spectrometry (ZooMS). J. Conserv. museum Stud. 18, 1–12. doi: 10.5334/jcms.196
Welker F., Hajdinjak M., Talamo S., Jaouen K., Dannemann M., David F., et al. (2016). Palaeoproteomic evidence identifies archaic hominins associated with the Châtelperronian at the Grotte du Renne. Proc. Natl. Acad. Sci. 113, 11162–11167. doi: 10.1073/pnas.1605834113
Welker F., Soressi M. A., Roussel M., van Riemsdijk I., Hublin J. J., Collins M. J. (2017). Variations in glutamine deamidation for a Châtelperronian bone assemblage as measured by peptide mass fingerprinting of collagen. STAR: Sci. Technol. Archaeological Res. 3, 15–27.
White L. C., Mitchell K. J., Austin J. J. (2018). Ancient mitochondrial genomes reveal the demographic history and phylogeography of the extinct, enigmatic thylacine (Thylacinus cynocephalus). J. Biogeography 45, 1–13. doi: 10.1111/jbi.13101
Wilkin S., Ventresca Miller A., Fernandes R., Spengler R., Taylor W. T. T., Brown D. R., et al. (2021). Dairying enabled early bronze age yamnaya steppe expansions. Nature 598, 629–633. doi: 10.1038/s41586-021-03798-4
Wilmsen E. N., Roberts F. H. Jr. (1978). Lindenmeier 1934-1974: Concluding report on investigations. doi: 10.5479/si.00810223.24.1
Wilson J., van Doorn N. L., Collins M. J. (2012). Assessing the extent of bone degradation using glutamine deamidation in collagen. Analytical Chem. 84, 9041–9048. doi: 10.1021/ac301333t
Keywords: museum collections, Pleistocene, collagen peptide fingerprinting, megafauna, North America
Citation: Antonosyan M, Hill E, Jodry M, Amano N, Brown S, Rick T and Boivin N (2024) A new legacy: potential of zooarchaeology by mass spectrometry in the analysis of North American megafaunal remains. Front. Mamm. Sci. 3:1399358. doi: 10.3389/fmamm.2024.1399358
Received: 11 March 2024; Accepted: 24 April 2024;
Published: 31 May 2024.
Edited by:
S. Ivan Perez, National Scientific and Technical Research Council (CONICET), ArgentinaReviewed by:
Luciano Prates, Museo de La Plata, ArgentinaCopyright © 2024 Antonosyan, Hill, Jodry, Amano, Brown, Rick and Boivin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mariya Antonosyan, YW50b25vc3lhbkBnZWEubXBnLmRl
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.