
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Malar., 09 April 2025
Sec. Case Management
Volume 3 - 2025 | https://doi.org/10.3389/fmala.2025.1497613
 Elizabeth Kibwana1†
Elizabeth Kibwana1† Domtila Kimani1†
Domtila Kimani1† Nick J. Edwards2
Nick J. Edwards2 Kelvias Keter1
Kelvias Keter1 Agnes Mutiso1
Agnes Mutiso1 Lydia Nyamako1
Lydia Nyamako1 Rose Gatheru1
Rose Gatheru1 Adrian V. S. Hill2
Adrian V. S. Hill2 Philip Bejon1,3
Philip Bejon1,3 Katie J. Ewer2
Katie J. Ewer2 Melissa C. Kapulu1,3*
Melissa C. Kapulu1,3*Background: Controlled human malaria infection (CHMI) studies are considered a powerful tool for assessing the efficacy of malaria vaccines and investigating immunity against infection. The monitoring of infection has historically been carried out by microscopy and/or more recently sensitive quantitative polymerase chain reaction (qPCR) based on the 18S ribosomal RNA (rRNA) gene. Here we describe the validation of a molecular assay previously developed to quantify malaria parasites in CHMI.
Methods: We used primers and probes for the 18S rRNA Plasmodium falciparum gene. The validation of the assay was performed using cultured 3D7 parasites to generate standards of known quantities of parasites. We determined the specificity, accuracy, precision, lower limit of detection, linearity, and robustness. We also evaluated the effect of using different volumes of whole blood for DNA extraction on the assay performance.
Results: The validation revealed: (1) specificity of 100% (n=5 independent experiments); (2) linearity with R2 values ≥ 0.98, a slope of −3.8 to −3.1, and efficiency 89–100%; (3) lower limit of detection of 0.3 parasites/microliter and lower limit of quantification of 2.6 parasites/microliter; (4) precision with both inter-assay repeatability and intra-assay reproducibility with coefficients of variation (CV) values of <10%; and (5) accuracy and good extraction efficiency of >90%. The use of large blood volumes for extraction had an adverse effect on precision.
Conclusion: We show that this qPCR method for P. falciparum parasite quantification from whole blood is specific, precise, sensitive, accurate, and robust. However, our whole blood qPCR method is not suitable when DNA extraction is from large blood volumes, where using a larger extraction volume of 1000 µl has a considerable effect on the robustness, reproducibility, and repeatability of qPCR, highlighting the importance of standardized protocols in DNA extraction. Other qPCR methods should be considered where high volumes of blood are required.
Malaria remains a significant public health burden in tropical and subtropical regions, with an estimated 249 million cases occurring globally in 2022 (WHO, 2023). Recent recommendations and roll out of malaria vaccines could be complementary tool to existing control measures to reduce the global burden of malaria (malERA Consultative Group on Vaccines, 2011; Duffy and Patrick Gorres, 2020). The controlled human malaria infection (CHMI) model is a well-established, powerful system that assesses the efficacy of anti-malarial drugs and early-stage vaccines. In this model, healthy volunteers are infected with malaria parasites either via the bites of infected mosquitoes or by needle-based administration of a known amount of cryopreserved sporozoites or infected red blood cells (Verhage et al., 2005; Epstein et al., 2007; Duncan and Draper, 2012; Billingsley et al., 2014; Mordmüller et al., 2015). The current gold standard for monitoring infection is by thick blood smears at regular time points, antimalarial treatment is then administered once parasites have been detected by microscopy. The detection limit by blood smear is ∼4–20 parasites/microliter (p/µl) depending on the number of fields evaluated and the expertise of the microscopist (Seilie et al., 2019). However, blood smear detection has its challenges, including the need for expert personnel and poor sensitivity in detecting low parasite levels. To overcome these challenges, various nucleic acid tests (NATs) have been developed to detect and quantify patent blood stage parasitemia as well as provide definite species identification (Roth et al., 2016). The most common NAT target of Plasmodium is the 18S (small subunit) ribosomal RNA gene (18S rRNA) (Kimura et al., 1997; Murphy et al., 2012) that can be detected by quantitative PCR (qPCR) (Hermsen et al., 2001), nested polymerase chain reaction (PCR) (Kimura et al., 1997), nucleic acid sequence-based amplification (NASBA) (Schoone et al., 2000), and quantitative reverse transcription PCR (qRT-PCR) (Kamau et al., 2011; Kamau et al., 2014). NAT are significantly more sensitive, specific, and reliable in detecting low levels of parasitemia than microscopy and work well with large sample sets and larger blood volumes (i.e. defined as ≥ 200 μl to 2 ml in previous studies) (Imwong et al., 2014; Hodgson et al., 2015). The objective of this study was to validate a previously developed quantitative PCR method by Hermsen et al. (2001) (Ogwang et al., 2015) that was transferred to Oxford (Andrews et al., 2005; Hodgson et al., 2015) and adapted for use in Kilifi (Ogwang et al., 2015) for the quantification of malaria parasite densities from whole blood in CHMI studies. We previously used this assay in a CHMI study (Kapulu et al., 2021) and here validate it to be able to analyze samples in further CHMI studies particularly in the context of intervention testing.
The P. falciparum 3D7 parasites were cultured in vitro in human red blood cell (RBC) suspensions using RPMI 1640 medium (Gibco/Invitrogen, Germany) supplemented with 2-mM L-glutamine (Gibco/Invitrogen, Germany), 37.5-mM HEPES (Gibco/Invitrogen, Germany), 25-µg/ml Gentamicin (Sigma, Germany), 0.2% glucose (Sigma, Germany) and 50.4-µg/ml Hypoxanthine (Sigma, Germany) and maintained at 37°C under 5% CO2. The growth of the parasite was followed by examination of the thin blood smear stained with Giemsa by microscopy and maintained at >10% parasitemia. Cultures were synchronized at the ring stage by treating cultured parasites with 5% sorbitol and subsequently reintroducing them back into culture. Asexual stage parasite cultures were harvested by centrifugation and counted by microscopy and flow cytometry. The parasites were frozen at −80°C until needed.
Five microliters of harvested P. falciparum-infected RBCs were stained with 1 ml of 1:1000 SYBR green (Molecular Probes, Lithuania) for 30 min at room temperature, washed twice in 1 ml phosphate buffered saline (PBS), and resuspended in 1 ml of PBS (Sigma, Germany) and analyzed by flow cytometry using BD FACS Canto (BD Biosciences, Germany) with an acquisition of 1,000,000 events per sample. Initial gating was performed using uninfected red blood cells.
Donor whole blood from O positive healthy donors was screened for the presence of P. falciparum as determined by rapid diagnostic test (RDT) and later confirmed by Taqman 18S rRNA qPCR. These donor blood samples were then spiked with an in vitro cultured and counted asexual stage frozen P. falciparum parasites. A two-fold serial dilution of spiked whole blood was made resulting in a parasite count range of 0.25 to 2,500 p/μl.
DNA was extracted from whole blood spiked with P. falciparum using the QIAsymphony machine (Qiagen, Germany) for automated DNA extraction and purification (Qiagen, Germany) according to the manufacturer’s instructions. DNA extraction was performed on four different sample volumes (200 μl, 400 μl, 500 μl and 1000 μl) to determine whether the sensitivity of the assay would be increased when using larger blood extraction volumes. Each extraction was also performed with a known positive and negative control sample. DNA was eluted in 50 µl (for extraction volumes of 200 µl) and 100 µl of elution buffer (for 400, 500 and 1000 µl extraction volume). From this 10 µl of eluted DNA was used to amplify the 18S ribosomal RNA gene by qPCR in a TaqMan assay using primers and probes on an Applied Biosystems 7500 real-time PCR system by Applied Biosystems 7500 quantification software (Thermo Fisher Scientific, USA). Final concentrations of 1× universal PCR Master Mix, 10pmol/µl of each primer, and 10µmol of Non-Florescent Quencher with Minor Groove Binder moiety (NFQ-MGB) probe were used in a 50µl reaction. Primers were based on the multicopy (four to eight per parasite) 18S (small subunit) ribosomal RNA genes of the Pf malaria parasite (NCBI nucleotide database accession number: M19173) developed by Hermsen et al. (2001) (Hermsen et al., 2001). The probes were based on the Oxford qPCR method as previously described (Sheehy et al., 2012). The primers and probes were synthesized by Eurofins Genomics (Germany).
18S Pf Forward-5’GTAATTGGAATGATAGGAATTTACAAGGT 3’
18S Pf Reverse-5’ TCAACTACGAACGTTTTAACTGCAAC 3’
18S Pf MGB -5’ FAM- AACAATTGGAGGGCAAG-NFQ-MGB 3’
The thermal profile used for qPCR is as follows: 10 min at 95°C; 45 cycles of 15 sec at 95°C; 1 min at 60°C. Included in all qPCR runs were known cultured parasite standards comprising of seven serial dilutions of extracted DNA run in triplicates, the unknown samples also in triplicate, negative control (uninfected red blood cells) in triplicate, and three replicates of the Non-Template Control [NTC (water)].
To generate the standard curve for relative quantification by qPCR, highly synchronized 3D7 ring stage parasites were cultured as previously described, serially diluted in fresh red blood cells (ranging from 117500 to 2.5 p/µl) and DNA was extracted as above using the QIAsymphony machine. Samples were run in triplicate in each qPCR plate. Standard curves were generated by plotting the mean cycle CT values versus the logarithmic parasite concentration. The parasite concentration (p/µl) was based on the relative quantified parasites for each dilution using flow cytometry.
The assays were validated in accordance with the ICH Harmonized Tripartite Guideline Part II: Validation of analytical procedures (ICH Harmonised Tripartite Guideline, 2005). The guidelines for assay validation define the following parameters that must be investigated to validate a bioanalytical assay: (1) specificity (2), accuracy (3), precision (repeatability, intermediate precision, reproducibility) (4), sensitivity (lower limit of detection and lower limit of quantification) (5), linearity (6), range, and (7) robustness. The specificity of the qPCR assay was determined using the WHO external quality assurance (EQA). The EQA scheme offers an independent means for research laboratories to verify the quality of their NAT diagnostic assays. This is achieved by providing well-characterized blinded controls consisting of mixed positive and negative Plasmodium samples (WHO, 2018). DNA was extracted from 500 µl of the EQA samples followed by qPCR as described above. Sensitivity [lower limit of detection (LLOD)] was defined as the ability of the assay to detect very low parasite densities. This corresponded to the lowest detected parasite density in a sample that was consistently detected at a 95% confidence level in all replicate wells, but not necessarily quantified as an exact value (Almeida-De-Oliveira et al., 2019). Limit of quantification (LOQ) was defined as the lowest concentration that can be quantified and has acceptable assay precision and accuracy and at which the coefficient of variation (CV) is ≤20%. Linearity was used as an indication that the relationship between parasite concentration and cycle threshold (Ct) values is proportional, allowing for accurate quantification. Linearity was evaluated by calculating the coefficient of determination (R2) from the standard curves. Precision (inter-assay repeatability, intra-assay reproducibility) was used to determine the repeatability and reproducibility of the assay, DNA was extracted from 2 to 4 replicates of a high, medium, and low cultured quantified 3D7 parasite followed by qPCR. The coefficient of variation (CV) was used as a measure of reproducibility. Reproducibility (inter-lab comparison) was used to determine how our assay performed against an independent laboratory. Standard curve samples were obtained from the Jenner Institute, University of Oxford, and the qPCR assay was performed. The standards were extracted Plasmodium DNA from 400 µl of spiked whole blood and eluted in 100 µl quantified by using previously prepared plasmid DNA standards of known concentration by qPCR (quantified as 100,000 parasites/ml) (Murphy et al., 2014). We then quantified the Oxford standard curve samples, and unknown spiked samples made in-house, as described in the Methods section. In addition, we used the Oxford standard curve to quantify our standard curve and the same unknown spiked in-house samples. Accuracy was defined as the degree of closeness of a measured value to the true value. Precision was determined by the recovery percentage (( x100).
No amplification occurred in any of the nontarget Plasmodium species tested (Figure 1). The amplification reaction did not show any Ct value in the non-template control or the internal negative controls (uninfected whole blood), indicating the absence of any non-specific amplification and/or contamination. Only samples containing P. falciparum were successfully amplified (100% specificity based on n=9) (Figure 1). Also included in the run was the internal positive control (3D7 cultured parasites), which was positively amplified.
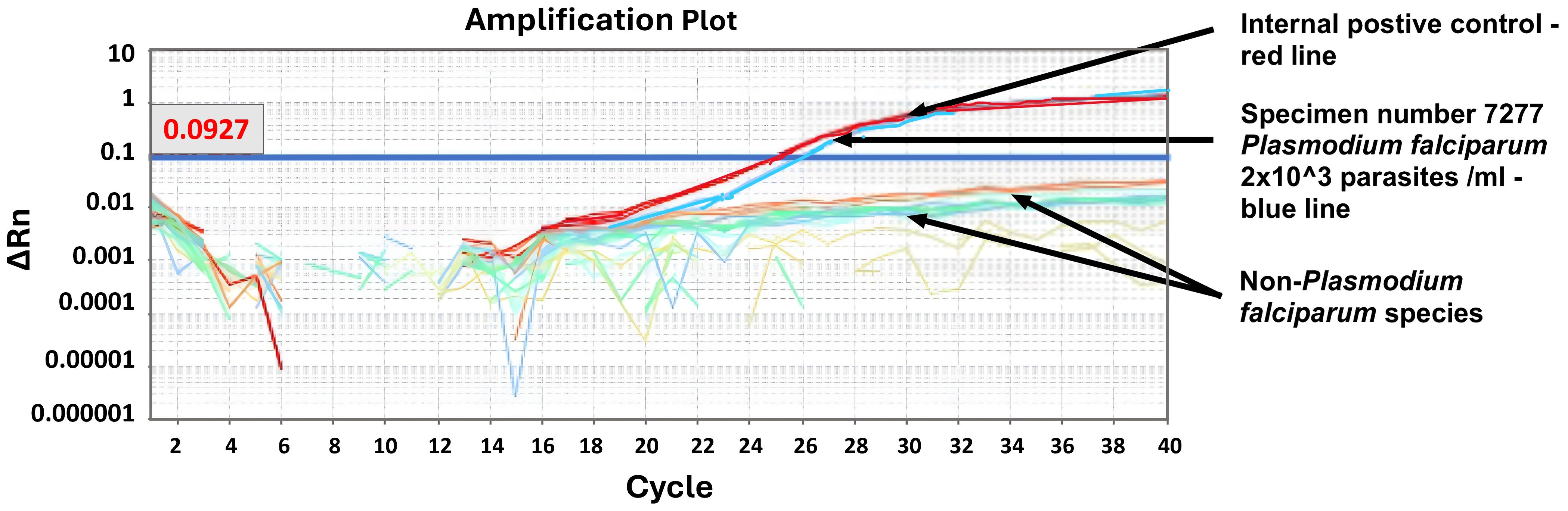
Figure 1. Specificity test. Machine derived qPCR amplification plot for samples run following WHO NAAT distribution of 6th June 2022. Only samples containing P. falciparum showed an amplification curve (WHO sample 7277, blue line). The positive control was an in-house known positive control (red line).
The standard curve was created by plotting the Ct values against the logarithm of the DNA concentration, which in this case was previously determined by flow cytometry. The standard curve is typically a linear relationship (y = mx + b), with y as the Ct value and x as the logarithmic transformation of the DNA concentration. The slope of the standard curve is also used to calculate the efficiency of PCR using the following equation:
Standard curves were linear, and the correlation coefficients (R2) were ≥0.994 and efficiency >90% on all days (Figure 2). We concluded that the acceptable criteria for linearity for this assay are the following. R2 ≥ 0.98| slope −3.8 to −3.1| efficiency 89–100%.
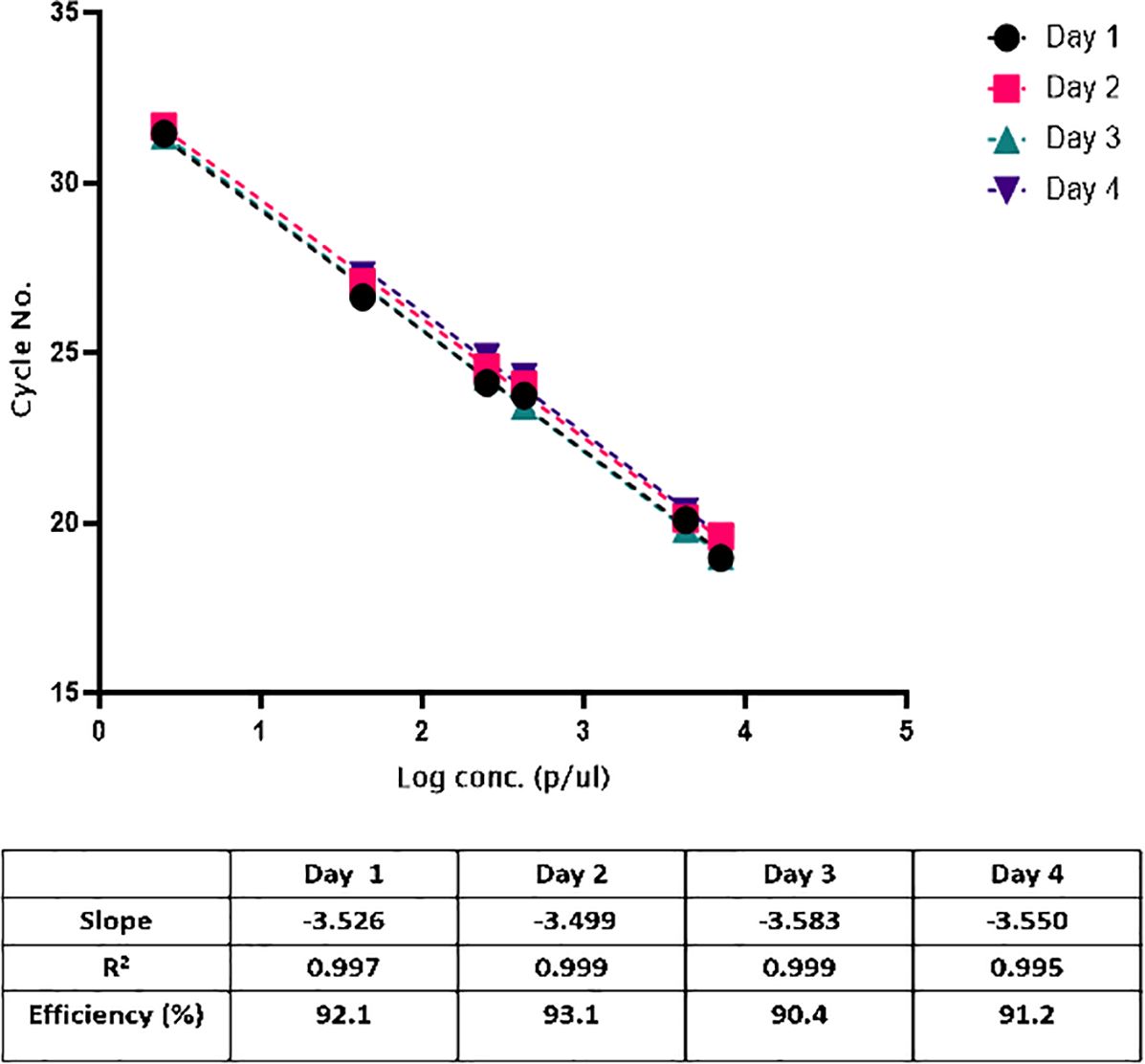
Figure 2. Construction of the standard curve for quantifying parasitemia. Serial six-fold dilution of cultured 3D7 parasites (n=3 for each point), ranging from 7000 to 2.5 p/μl, showed a linearity (R2>0.99).
LOQ: To determine the LOQ, counted, spiked positive samples were serially diluted 5-fold, covering a range of 2500 to 0.25 P. falciparum 3D7 ring-stage parasites/μl and the qPCR ran. The experimental results determined that the LOQ for this assay was 2.6 p/μl. The experiment was carried out on three different days with five replicates. With a treatment threshold for the CHMI studies conducted in our setting at KEMRI-Wellcome Trust Programme being 500 p/µl (Kapulu et al., 2021), there is thus confidence that the qPCR assay can readily identify positive samples above this threshold 100% of the time.
LOD: LOD was determined as the lowest concentration of cultured parasites that consistently produced a positive result in 95% of the samples tested in all replicate wells (Almeida-De-Oliveira et al., 2019). The lowest concentration detected was 0.3 p/µl (Standard deviation [SD], 0.1) with an average Ct of 34.52. This implies that when the assay is performed on the 3D7 parasite strain, there is a 95% chance of detecting parasites as low as 0.3 p/µl.
Accuracy: Four different concentrations of (1300, 130, 13 and 6.5 p/µl) were obtained by spiking different concentrations of frozen culture parasites into fresh whole blood. The accuracy was evaluated by calculating the recovery rate. The average percentage recovery was 89.8–110.2% (Table 1) with a CV of< 20%. These results also demonstrate that the DNA extraction method has a good extraction efficiency.
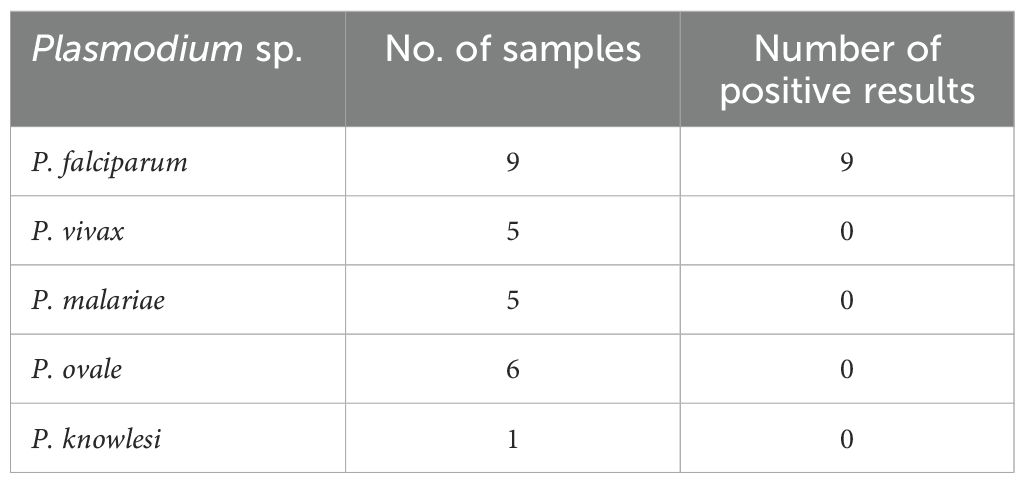
Table 1. Test of the specificity of the qPCR method on relevant Plasmodium species using blinded samples provided by the UK NEQAS (tested between 2021 and 2022).
Robustness: Five confirmed parasite-negative blood samples were spiked with 250 p/µl and 25 p/µl of cultured 3D7 ring-stage parasites (n=10). DNA was extracted using the automated QIAsymphony machine for DNA extraction and purification followed by qPCR. Control samples of non-infected blood samples (negative) were processed along (n=5). All negative control samples were negative (no amplification), whereas all spiked samples were positive. Indicating a robustness of >99%.
Repeatability was analyzed among triplicates of an assay (in-run), while reproducibility (between runs) was determined in three repeated day assays performed by different operators for the four different parasite quantities (high, medium, low and LLOD). The variation in both inter-assay and intra-assay was low with CV<10% (Tables 2, 3).
The results showed good agreement in quantification, specificity, reproducibility, and linearity (data shown is an average of 3 independent experiments). No significant differences were observed between the observed quantities when using Kilifi or Oxford standards to quantify spiked samples (Figures 3, 4).
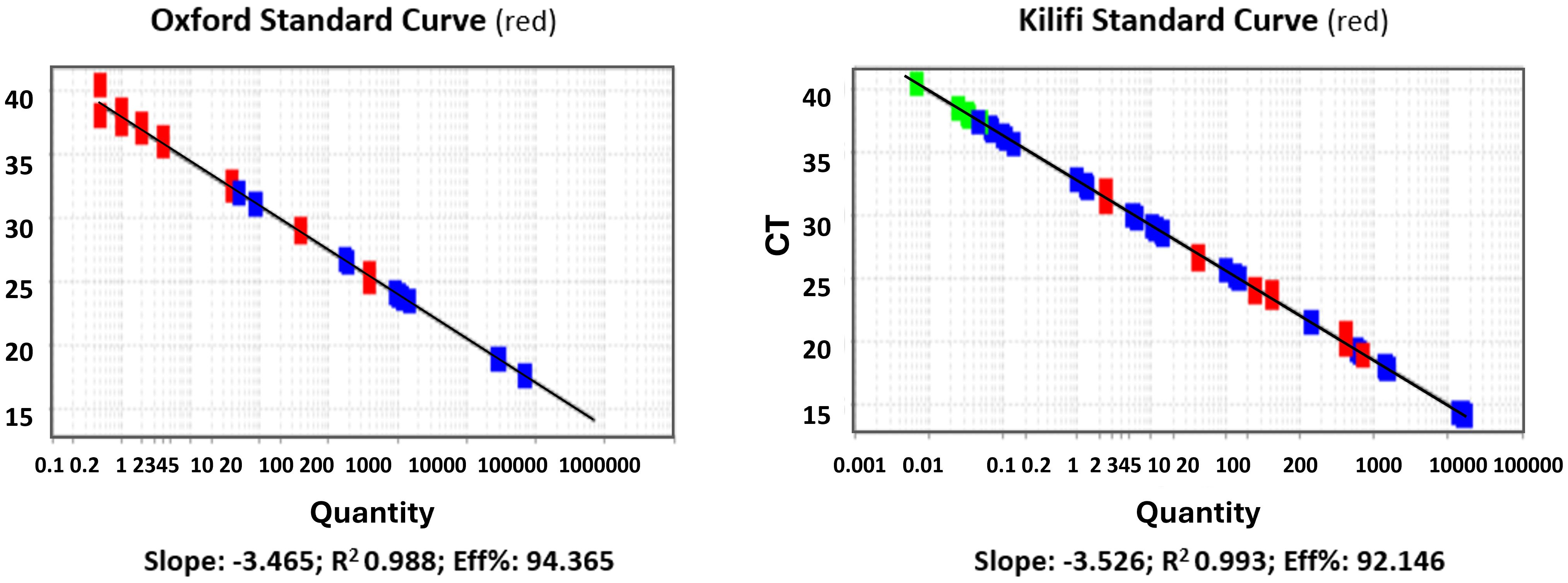
Figure 3. Linearity of the standard curves (log10 p/μl vs Ct). Standard curve from the Jenner Institute, Oxford, performed at the KEMRI-Wellcome Trust, Kilifi (left panel). Standard curve used in the Kilifi qPCR assay (right panel). red denotes standard samples from either Oxford (left panel) or Kilifi (right panel); blue denotes test samples being quantified by the respective standards; green denotes samples at the lower extreme end of detection.
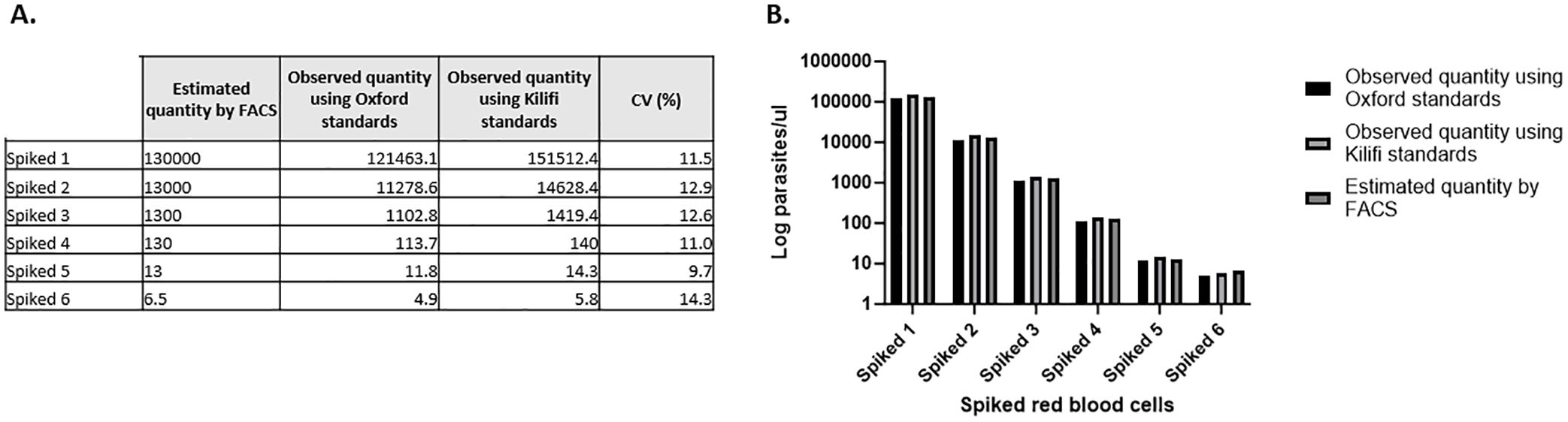
Figure 4. Quantification of spiked red blood cells in parasites per microliter. (A) Absolute quantities determined using two independent standard curves. (B) Graphical representation of the data showing no difference between the three different parasitemia measured. Showing the average of 3 experiments.
DNA was extracted from four different sample volumes 200, 400, 500, and 1000µl at different parasite concentrations followed by qPCR analysis as described above. The results of this experiment are shown in Figure 5. Using a larger extraction volume of 1000 µl has a considerable effect on the robustness, reproducibility, and repeatability of the assay.
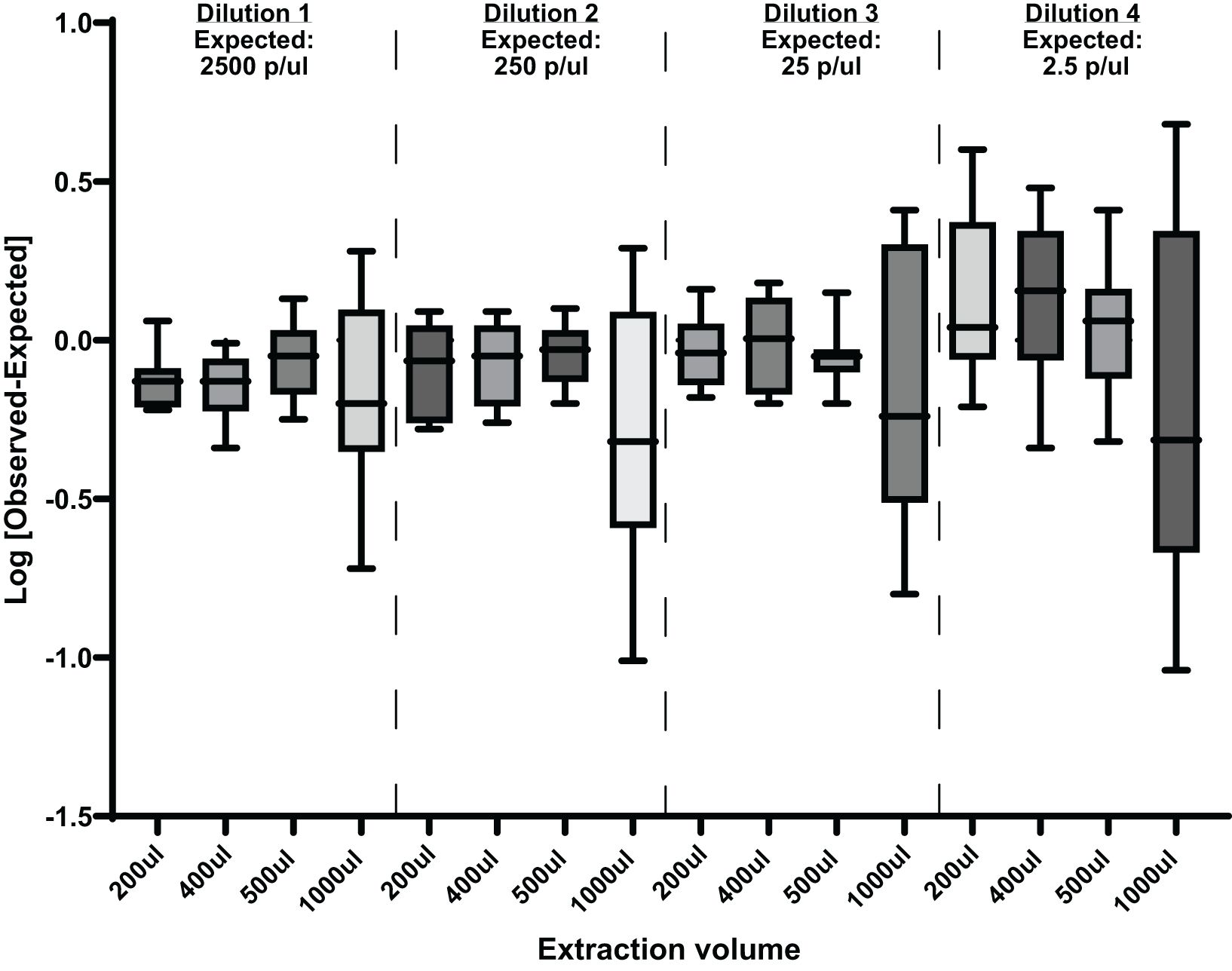
Figure 5. Effects of different extraction volumes at different parasite concentrations on this qPCR assay. Showing [Log Observed-Log Expected] mean with ranges. p/µl, parasites per µl.
Assay validation is essential for the monitoring of clinical trials, a high degree of confidence is needed in the data obtained, and assays must be fully validated to provide this assurance, particularly when used to determine vaccine or drug efficacy. In CHMI vaccine studies, volunteers generally develop submicroscopic parasitemia for several days before they become thick smear positive (Hermsen et al., 2001; Andrews et al., 2005). The pre-patency period is crucial for estimating vaccine efficacy, and early treatment mitigates the clinical risks posed to the participants. Walk et al. (2016) have further shown that utilization of the more sensitive qPCR method instead of microscopy in CHMI, has the potential to decrease pre-patent period leading the less severe clinical outcomes for volunteers. Therefore, the use of well validated and qualified qPCR is highly beneficial for CHMI outcome as a primary diagnostic tool. Validation of the effectiveness of increasingly sensitive qPCR in clinical care would require larger sample sizes and clinical samples. Here, we describe the validation of a qPCR method for the detection and quantitation of malaria parasites from whole blood. The assay was highly specific, linear, and precise, and was optimized, using primers targeting the P. falciparum 18S RNA gene in a wide range of parasitemia. The assessment of specificity here was limited (i.e., n=9 samples). The LLOD of the qPCR assay was 3 parasites/μl and the assay has good sensitivity whereas other reported similar qPCR assays have reported much lower quantities of malaria parasites (Roestenberg et al., 2012; Kamau et al., 2014). The treatment diagnostic criteria in a recent CHMI study conducted at the KEMRI-Wellcome Trust Programme was based on individuals having a parasite threshold of 500 parasites/μl and/or clinical symptoms with parasitemia (Kapulu et al., 2021). This assay easily captures parasitemia above the threshold with high confidence. Thus, the assay is able capture and detect a range of very high-density parasites as well as low density parasitemia to a limit lower than 3 parasites/μl. In the spiked recovery experiments (Table 4), the experimental results of the assay showed that the precision ranged from 89.8% to 100%. The spiked recovery experiments also demonstrated the efficiency of the DNA extraction process.
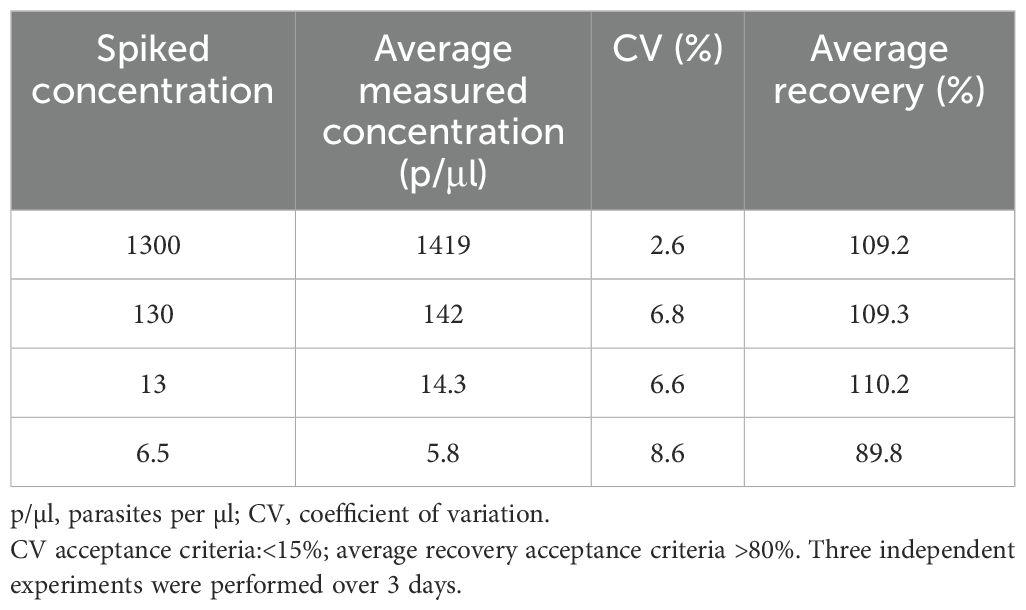
Table 4. Measurement of accuracy of qPCR assay – tested by measuring the recovery of spiked red blood cells.
We also conducted an independent comparison to verify our assay fitness using standard curve samples provided by the Jenner Institute, Oxford, as used in the Jenner’s assay that had previously been subjected to a similar validation process for their CHMI studies. Based on experimental evidence, the general agreement between the two standard curves was high, with R2>0.98 and PCR efficiency of >92% for both curves. No significant differences were observed when using Kilifi or Oxford standard curves to quantify spiked blood samples. The assay demonstrated high reproducibility, repeatability, and precision when different standard curves were used for quantification.
We also investigated the effect of using a larger DNA extraction volume on the sensitivity of the assay sensitivity. It has been demonstrated that increasing the extraction sample volume accelerated early diagnosis and/or increased sensitivity when using the qRT-PCR assay (Imwong et al., 2014; Hodgson et al., 2015). However, these studies used different blood-processing and DNA extraction methods. The data generated here show that increasing the DNA extraction volume to 1 ml does not increase the sensitivity of this assay. In theory, a larger volume would increase the sensitivity of the assay by extracting more parasite infected blood, but in practice, the assay becomes less reproducible and therefore less robust at 1 ml volumes. We did not observe a similar deterioration in reproducibility when using either 200 µl, 400 µl, or 500 µl as extraction volumes. As expected at lower parasite dilutions of 0.25p/µl, there is substantial variability at all extraction volumes (Figure 5), since 0.25p/µl is lower than the established LLOD. When the sample volume is increased to 1 ml, this qPCR assay is no longer reliable. In this set of experiments, the use of a 1 ml sample volume had a significant effect on the precision and consistency of the assay, as demonstrated by the wide spread of the data shown in Figure 5. We speculate that this is due to PCR inhibitors. As the volume of the sample increases, so do any PCR inhibitory substances. Studies have shown that heme, IgG, leukocyte DNA, and anticoagulants (EDTA and/or heparin) can play a role in PCR inhibition by either affecting DNA polymerase activity, binding to genomic DNA, or binding to fluorescent dyes, causing fluorescence quenching (Al-soud and Radstrom, 2001; Sidstedt et al., 2018). This could be the reason for the inconsistency observed when using the 1 ml extraction volume. Our assay acceptance criteria were as follows: linearity of the standard curve of R2 0.98, efficiency 87–100%, accuracy 90–100%, intra-assay precision<5%, and inter-assay precision<10%.
In summary, the data presented here demonstrate that this malaria qPCR assay is suitable for use in clinical studies and/or other investigations to quantify P. falciparum parasites from human whole blood from field and clinical CHMI studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The datasets presented in this study can be found at the online repository for KEMRI-Wellcome Trust Research Programme: Harvard Dataverse.
EK: Formal Analysis, Writing – original draft. DK: Formal Analysis, Writing – review & editing. NE: Writing – review & editing. KK: Formal Analysis, Writing – review & editing. AM: Formal Analysis, Writing – review & editing. LN: Formal Analysis, Writing – review & editing. RG: Formal Analysis, Writing – review & editing. AH: Supervision, Writing – review & editing. PB: Supervision, Writing – review & editing. KE: Supervision, Writing – review & editing. MK: Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded as part of the EDCTP2 programme supported by the European Union (grant number RIA2016V-1649-MMVC) and by a Wellcome Trust grant (grant number 107499/Z/15/Z).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are grateful to all the KEMRI-Wellcome Trust laboratory team for support. This manuscript is published with permission/approval from KEMRI Director.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Almeida-De-Oliveira N. K., Moreira O. C., de Lavigne A. R., Mendonça-Lima L., Werneck G. L., Daniel-Ribeiro C. T., et al. (2019). Analytical validation of real-time quantitative PCR assays for optimum diagnosis of vivax malaria. Mem. Inst. Oswaldo Cruz 114, e180350. doi: 10.1590/0074-02760180350
Al-soud W. A., Radstrom P. (2001). Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 29, 485–493. doi: 10.1186/1743-8454-4-1
Andrews L., Andersen R. F., Webster D., Dunachie S., Walther R. M., Bejon P., et al. (2005). Quantitative real-time polymerase chain reaction for malaria diagnosis and its use in malaria vaccine clinical trials. Am. J. Trop. Med. Hyg. 73, 191–198. doi: 10.4269/AJTMH.2005.73.191
Billingsley P., Sim B. K. L., Bijker E., Roestenberg M., Lyke K., Laurens M., et al. (2014). Controlled human malaria infections using aseptic, purified cryopreserved Plasmodium falciparum sporozoites administered by needle and syringe. Malar. J. 13 (Supp 2), 12. doi: 10.1186/1475-2875-13-s1-p12
Duffy P. E., Patrick Gorres J. (2020). Malaria vaccines since 2000: progress, priorities, products. NPJ Vaccines 5, 1–9. doi: 10.1038/s41541-020-0196-3
Duncan C. J. A., Draper S. J. (2012). Review: Controlled human blood stage malaria infection: Current status and potential applications. Am. J. Trop. Med. Hygiene 86, 561–565. doi: 10.4269/ajtmh.2012.11-0504
Epstein J. E., Rao S., Williams F., Freilich D., Luke T., Sedegah M., et al. (2007). Safety and clinical outcome of experimental challenge of human volunteers with plasmodium falciparum –infected mosquitoes: an update. J. Infect. Dis. 196, 145–154. doi: 10.1086/518510
Hermsen C. C., Telgt D. S. C., Linders E. H. P., Van De Locht L. A. T. F., Eling W. M. C., Mensink E. J. B. M., et al. (2001). Detection of Plasmodium falciparum malaria parasites in vivo by real-time quantitative PCR. Mol. Biochem. Parasitol. 118, 247–251. doi: 10.1016/S0166-6851(01)00379-6
Hodgson S. H., Douglas A. D., Edwards N. J., Kimani D., Elias S. C., Chang M., et al. (2015). Increased sample volume and use of quantitative reverse-transcription PCR can improve prediction of liver-to-blood inoculum size in controlled human malaria infection studies. Malar. J. 14 (33), 1–9. doi: 10.1186/s12936-015-0541-6
ICH Harmonised Tripartite Guideline (2005). Validation of Analytical Procedures: Text and Methodology Q2(R1). International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use, Geneva, 1–13.
Imwong M., Hanchana S., Malleret B., Rénia L., Day N. P. J., Dondorp A., et al. (2014). High-throughput ultrasensitive molecular techniques for quantifying low-density malaria parasitemias. J. Clin. Microbiol. 52, 3303–3309. doi: 10.1128/JCM.01057-14
Kamau E., Alemayehu S., Feghali K. C., Komisar J., Regules J., Cowden J., et al. (2014). Measurement of parasitological data by quantitative real-time PCR from controlled human malaria infection trials at the Walter Reed Army Institute of Research. Malar. J. 13, 288. doi: 10.1186/1475-2875-13-288
Kamau E., Tolbert L. S., Kortepeter L., Pratt M., Nyakoe N., Muringo L., et al. (2011). Development of a highly sensitive genus-specific quantitative reverse transcriptase real-time PCR assay for detection and quantitation of plasmodium by amplifying RNA and DNA of the 18S rRNA genes. J. Clin. Microbiol. 49, 2946–2953. doi: 10.1128/JCM.00276-11
Kapulu M. C., Njuguna P., Hamaluba M., Kimani D., Ngoi J. M., Musembi J., et al. (2021). Safety and PCR monitoring in 161 semi-immune Kenyan adults following controlled human malaria infection. JCI Insight 6 (17), 1–13. doi: 10.1172/jci.insight.146443
Kimura M., Kaneko O., Liu Q., Zhou M., Fumihiko K., Wataya Y., et al. (1997). Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitol. Int. 46, 91–95. doi: 10.1016/S1383-5769(97)00013-5
malERA Consultative Group on Vaccines. (2011). A research agenda for malaria eradication: vaccines. PloS Med. 8 (1), e1000398. doi: 10.1371/journal.pmed.1000398
Mordmüller B., Supan C., Sim K. L., Gómez-Pérez G. P., Ospina Salazar C. L., Held J., et al. (2015). Direct venous inoculation of Plasmodium falciparum sporozoites for controlled human malaria infection: A dose-finding trial in two centres. Malar. J. 14, 0–10. doi: 10.1186/s12936-015-0628-0
Murphy S. C., Hermsen C. C., Douglas A. D., Edwards N. J., Petersen I., Fahle G. A., et al. (2014). External quality assurance of malaria nucleic acid testing for clinical trials and eradication surveillance. PloS One 9, e97398. doi: 10.1371/JOURNAL.PONE.0097398
Murphy S. C., Prentice J. L., Williamson K., Wallis C. K., Fang F. C., Fried M., et al. (2012). Real-time quantitative reverse transcription PCR for monitoring of blood-stage Plasmodium falciparum infections in malaria human challenge trials. Am. J. Trop. Med. Hygiene 86, 383–394. doi: 10.4269/ajtmh.2012.10-0658
Ogwang C., Kimani D., Edwards N. J., Roberts R., Mwacharo J., Bowyer G., et al. (2015). Prime-boost vaccination with chimpanzee adenovirus and modified vaccinia Ankara encoding TRAP provides partial protection against Plasmodium falciparum infection in Kenyan adults. Sci. Transl. Med. 7, 286re5. doi: 10.1126/SCITRANSLMED.AAA2373
Roestenberg M., O’Hara G. A., Duncan C. J. A., Epstein J. E., Edwards N. J., Scholzen A., et al. (2012). Comparison of clinical and parasitological data from controlled human malaria infection trials. PloS One 7, 1–8. doi: 10.1371/journal.pone.0038434
Roth J. M., Korevaar D. A., Leeflang M. M. G., Mens P. F. (2016). Molecular malaria diagnostics: A systematic review and meta-analysis. Crit. Rev. Clin. Lab. Sci. 53 (2), 87–105. doi: 10.3109/10408363.2015.108499
Schoone G. J., Oskam L., Kroon N. C. M., Schallig H. D. F. H., Omar S. A. (2000). Detection and quantification of Plasmodium falciparum in blood samples using quantitative nucleic acid sequence-based amplification. J. Clin. Microbiol. 38, 4072–4075. doi: 10.1128/jcm.38.11.4072-4075.2000
Seilie A. M., Chang M., Hanron A. E., Billman Z. P., Stone B. C., Zhou K., et al. (2019). Beyond blood smears: Qualification of plasmodium 18s rrna as a biomarker for controlled human malaria infections. Am. J. Trop. Med. Hygiene 100, 1466–1476. doi: 10.4269/ajtmh.19-0094
Sheehy S. H., Duncan C. J., Elias S. C., Choudhary P., Biswas S., Halstead F. D., et al. (2012). ChAd63-MVA-vectored blood-stage malaria vaccines targeting MSP1 and AMA1: assessment of efficacy against mosquito bite challenge in humans. Mol. Ther. 20, 2355–2368. doi: 10.1038/mt.2012.223
Sidstedt M., Hedman J., Romsos E. L., Waitara L., Wadsö L., Steffen C. R., et al. (2018). Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal. Bioanal Chem. 410, 2569–2583. doi: 10.1007/s00216-018-0931-z
Verhage D. F., Telgt D. S. C., Bousema J. T., Hermsen C. C., van Gemert G. A., van der Meer J. W. M., et al. (2005). Clinical outcome of experimental human malaria induced by Plasmodium falciparum-infected mosquitoes. Netherlands J. Med. 63, 52–58.
Walk J., Schats R., Langenberg M. C. C., Reuling I. J., Teelen K., Roestenberg M., et al. (2016). Diagnosis and treatment based on quantitative PCR after controlled human malaria infection. Malar. J. 15 (1), 398.#. doi: 10.1186/s12936-016-1434-z
WHO (2018). WHO external quality assurance scheme for malaria nucleic acid amplification testing (NAAT EQA). Geneva: World Health Organization. Available online at: https://www.who.int/malaria/areas/diagnosis/faq-nucleic-acid-amplification-tests/en/ (Accessed February 8, 2021).
WHO (2023). World malaria report 2023. Geneva: World Health Organization. Available online at: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2023 (Accessed April 11, 2024).
Keywords: CHMI, P. falciparum, 18S qPCR, whole blood, parasite quantification, assay validation
Citation: Kibwana E, Kimani D, Edwards NJ, Keter K, Mutiso A, Nyamako L, Gatheru R, Hill AVS, Bejon P, Ewer KJ and Kapulu MC (2025) Quantification of Plasmodium falciparum: validation of quantitative polymerase chain reaction assays for detection of parasites in controlled human malaria infection studies. Front. Malar. 3:1497613. doi: 10.3389/fmala.2025.1497613
Received: 17 September 2024; Accepted: 04 March 2025;
Published: 09 April 2025.
Edited by:
Christian Nsanzabana, Swiss Tropical and Public Health Institute (Swiss TPH), SwitzerlandReviewed by:
Erasto Vitus Mbugi, Muhimbili University of Health and Allied Sciences, TanzaniaCopyright © 2025 Kibwana, Kimani, Edwards, Keter, Mutiso, Nyamako, Gatheru, Hill, Bejon, Ewer and Kapulu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melissa C. Kapulu, bWthcHVsdUBrZW1yaS13ZWxsY29tZS5vcmc=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.