
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Integr. Neurosci., 15 June 2021
Volume 15 - 2021 | https://doi.org/10.3389/fnint.2021.692948
This article is part of the Research TopicRNA localization and Localized Translation in NeuronsView all 6 articles
Hundreds of messenger RNAs (mRNAs) are transported into neurites to provide templates for the assembly of local protein networks. These networks enable a neuron to configure different cellular domains for specialized functions. According to current evidence, mRNAs are mostly transported in rather small packages of one to three copies, rarely containing different transcripts. This opens up fascinating logistic problems: how are hundreds of different mRNA cargoes sorted into distinct packages and how are they coupled to and released from motor proteins to produce the observed mRNA distributions? Are all mRNAs transported by the same transport machinery, or are there different adaptors or motors for different transcripts or classes of mRNAs? A variety of often indirect evidence exists for the involvement of proteins in mRNA localization, but relatively little is known about the essential activities required for the actual transport process. Here, we summarize the different types of available evidence for interactions that connect mammalian mRNAs to motor proteins to highlight at which point further research is needed to uncover critical missing links. We further argue that a combination of discovery approaches reporting direct interactions, in vitro reconstitution, and fast perturbations in cells is an ideal future strategy to unravel essential interactions and specific functions of proteins in mRNA transport processes.
Neuronal messenger RNA (mRNA) localization and local translation are crucial for a range of processes, such as neuronal development (Yoon et al., 2016; Wong et al., 2017), migration (Preitner et al., 2014), polarization (Ciolli Mattioli et al., 2019), and synaptic plasticity (Miller et al., 2002; Donlin-Asp et al., 2021). mRNAs are transported through the cytoplasm as messenger ribonucleoprotein (mRNP) complexes, with a variety of RNA-binding proteins (RBPs) bound to them to control mRNA stability, localization, and translation (Doyle and Kiebler, 2011; Buxbaum et al., 2015). Most RNA sequences important for mRNA localization are located in their 3'untranslated regions (3'UTRs). In order to establish cytoplasmic mRNA distribution, microtubule-based motor proteins, such as mostly microtubule plus-end-directed kinesins and the minus-end-directed dynein, recognize features of mRNPs that are unknown to a large extent. In this review, we focus specifically on known and potential adaptor proteins that couple mRNAs with motor proteins in mammalian neurons. This focus is motivated by the importance of revealing essential components and the architecture of the mRNA transport machinery, which will allow to disentangle the actual effect of mRNA localization from pleiotropic effects of other components of mRNA transport complexes in the future. To highlight how little is known with certainty and to emphasize potential future research goals, we discuss the value and limitations of methods often used in the past. We further provide a condensed overview on which minimal neuronal mRNA transport complexes were identified to date or can be proposed based on published evidence. We further weigh the strength of evidence for interactions that link mRNAs to motors based on the number of different methods used to show respective interactions. We differentiate explicitly between methods reporting direct and indirect interactions to highlight where further research would be beneficial. We further focus on low-throughput biochemical evidence and regard only those mRNA targets for which the binding site for the motor-coupling RBP is known. Finally, we argue that novel screening approaches detecting direct interactions between RBPs and motor proteins (Yang et al., 2018; Lang et al., 2020), advanced in vitro live-biochemistry assays (Heym et al., 2013; Sladewski et al., 2013; McClintock et al., 2018; Baumann et al., 2020), and fast perturbations in cells (Nishimura et al., 2009; van Bergeijk et al., 2015; Yesbolatova et al., 2020) offer a great potential to not only reveal the essential building blocks of the mammalian mRNA transport machinery, but also to discover their functionality.
Important studies in the past decades have provided valuable information about which factors play a role in neuronal mRNA transport (Figure 1A, Supplementary Table 1). However, because of intrinsic limitations, many methods used cannot determine the exact functionality of a protein and its direct interactions. Results from knockdown or overexpression studies are often difficult to interpret, as it is challenging to disentangle phenotypes caused by the pleiotropic functions of proteins required for mRNA transport, such as RBPs and motor proteins. For instance, several RBPs involved in neuronal mRNA transport, such as Fragile X Mental Retardation Protein (FMRP) (Dictenberg et al., 2008; Goering et al., 2020), are also required for nuclear processes (Shah et al., 2020), nuclear mRNA export (Zhang et al., 2007; Edens et al., 2019), and translation regulation (Feng et al., 1997), whereas an mRNP-transporting motor protein like kinesin-1 KIF5 transports a variety of cargoes (Twelvetrees et al., 2010; Nakajima et al., 2012; Barry et al., 2014; Heisler et al., 2014; Ruane et al., 2016; Henrichs et al., 2020; Serra-Marques et al., 2020). Hence, analyzing the potential role of an RBP or a motor protein in mRNA transport using knockdowns or overexpressions will inevitably affect multiple cellular processes, making the interpretation of results difficult. Further, while imaging provided fascinating insights into mRNP transport dynamics and the mRNA content of neuronal mRNA transport complexes (Buxbaum et al., 2015), co-localization of an RBP and an mRNA can happen by chance unless a sufficient resolution is achieved for imaging (Eliscovich et al., 2017). Another approach, affinity purification–mass spectrometry analysis, provided valuable lists of proteins associated with neuronal mRNA transport complexes (Mallardo et al., 2003; Kanai et al., 2004; Charalambous et al., 2013; Fritzsche et al., 2013; Chu et al., 2019). However, pulldowns come with the risk of losing transiently binding dynamic interactors (Richards et al., 2021) and difficulties to distinguish between protein–protein and protein–RNA–protein interactions, in which an RNA connects two RBPs that are not directly interacting with one another. For instance, many pulled-down neuronal mRNA transport complexes are either sensitive to RNase treatment or pulldowns were done in the presence of RNase inhibitors, which leaves significant uncertainties about direct protein interactions essential for mRNA transport (Mallardo et al., 2003; Kanai et al., 2004; Dictenberg et al., 2008; Chu et al., 2019; Fukuda et al., 2021). Consequently, approaches that can reveal the core engine of mRNA transport complexes, such as biochemical reconstitution, biophysical approaches, or fast perturbations in cells, require knowledge of essential factors, direct interactions, or even minimal interaction interfaces, have been impossible for long.
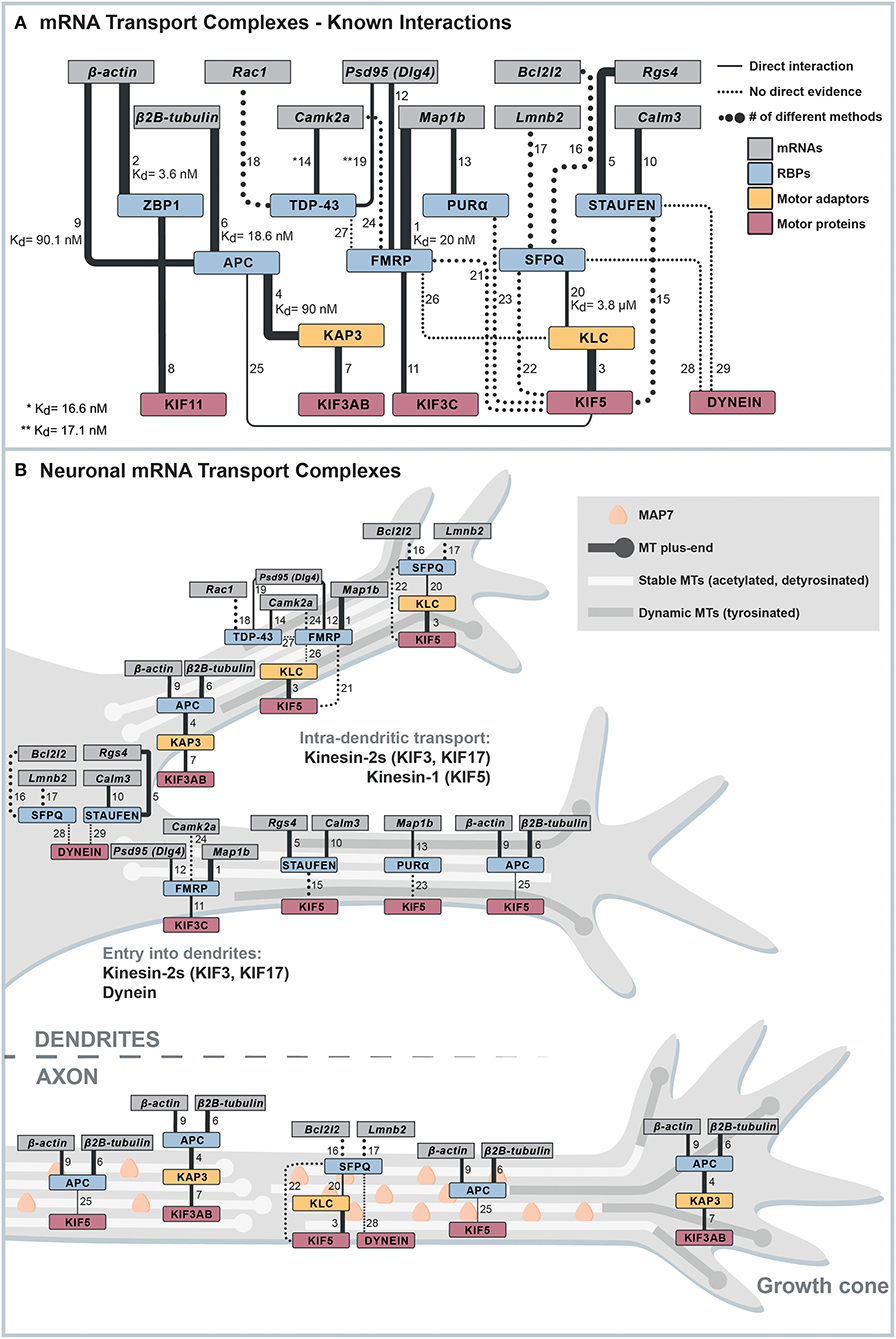
Figure 1. Known interactions linking mRNAs to motor proteins. (A) Cytoscape network of known interactions. The network summarizes and weighs available evidence that allows the establishment of connections between mRNAs and microtubule motor proteins. Only evidence from low-throughput techniques is shown, and RBP-mRNA are only plotted when the mRNA has a known binding motif for the respective RBP. The line width indicates the number of different methods used to show the respective interaction. Dotted lines indicate indirect evidence for interactions, and continuous lines are evidence for direct interactions. All evidence represented in the figure, and further evidence we excluded from the network because of different reasons is listed in Supplementary Table 1. The numbers on the lines connecting two nodes in the network correspond to the interaction number in the first column of Supplementary Table 1. (B) Possible minimal mRNA transport complexes for different transport steps. Combining the interactions shown in (A) and information about motor activity in different zones of neurites, we propose potential minimal mRNA transport complexes for different transport steps.
Most evidence for associations with RBPs and mRNPs was reported for KIF5, a heterotetrameric kinesin comprising two heavy chains (KHCs) and two light chains (KLCs), with methods almost entirely detecting indirect interactions. FMRP was found to interact with either all KIF5 isoforms (Kanai et al., 2004), preferentially the KIF5B isoform (Zhao et al., 2020), or solely with a not specified KIF5 light chain (Dictenberg et al., 2008). Likewise, neuron-specific kinesin-2 KIF3C is a candidate for FMRP transport (Davidovic et al., 2007). While ample direct evidence exists for FMRP directly binding to localized mRNAs, such as Map1b (Brown et al., 2001; Menon et al., 2008) or Psd95 (Zalfa et al., 2007), it is not clear whether FMRP can bind mRNAs and motor proteins simultaneously, which is a requirement for an mRNA-motor adaptor. Similarly, other RBPs such as PUR-alpha/beta (PURα/β) and heterogeneous nuclear ribonucleoprotein U (HNRP-U) are candidates for mRNA-motor adaptors, since they were among the strongest KIF5 tail binders in an early affinity purification study (Kanai et al., 2004). While to the knowledge of the authors there are no known links of HNRP-U and PURβ, respectively, to any localized mRNA targets, PURα is described to directly bind Map1b (Chen et al., 1995; Johnson et al., 2006). Furthermore, the Splicing Factor Proline and Glutamine Rich (SFPQ) immunoprecipitates in an RNase-sensitive manner with dynein and KIF5 (Fukuda et al., 2021). In addition, a fragment of SFPQ binds kinesin-1 light chain KLC1 with an affinity of 3.8 μM independent of RNA. Thus, although SFPQ is a component of RNA granules and regulates Bcl2l2 and Lmnb2 mRNA localization in axons (Cosker et al., 2016), it remains unclear whether KIF5-SFPQ complexes constitute the essential core of an mRNA transport complex and whether the KIF5-SFPQ interaction is sufficiently strong for processive axonal mRNA transport. Finally, another recent study has linked kinesin-1 and dynein to the transport of TDP-43-FMRP-STAU1 complexes carrying Rac1 mRNA (Chu et al., 2019), again, using methods showing solely indirect interactions.
Another well-studied RBP required for mRNA localization is the Zipcode Binding Protein 1 (ZBP-1). Ample evidence exists for its direct binding to the beta-actin mRNA localization sequence (the “zipcode”) (Chao et al., 2010; Patel et al., 2012; Nicastro et al., 2017). Further, co-precipitation experiments show a direct interaction of the kinesin family member 11 (KIF11) with ZBP-1/Insulin Like Growth Factor 2 mRNA Binding Protein 1 (IGF2BP1) (Song et al., 2015). Nevertheless, evidence that this interaction is important for neuronal mRNA transport is lacking. Also, considering that KIF11 is not able to enter dendrites (Lipka et al., 2016), the question of how β-actin mRNA is transported into dendrites remains unanswered.
Another interesting candidate for mRNA-motor linkage is mRNA export factor NXF2 which binds kinesin-2 motor KIF17, KIF9, and a dynein light chain-like protein (Takano et al., 2007). Although NXF2 co-localizes with RNA granule components in hippocampal neurons, it has not been tested yet whether the NXF2-KIF17 interaction is required for neuronal mRNA transport. In addition, KIF9 has no known role in neuronal transport, and it is neither clear whether the dynein light chain-like protein functions as a part of the dynein complex, nor if dynein light chains are involved in cargo recruitment to the latter (Williams et al., 2007). Staufen2 (STAU2) is doubtlessly present in many mRNA transport complexes. It directly binds localized mRNAs (Heber et al., 2019) and affects mRNA transport (Mallardo et al., 2003; Sharangdhar et al., 2017; Bauer et al., 2019; Chu et al., 2019) but to date no direct evidence exists for its interaction with motor proteins.
Currently, the only direct evidence existing for a sufficient set of factors that can processively and selectively transport axonal mRNA packages of one to four mRNAs is based on tumor suppressor Adenomatous Polyposis Coli (APC). Using purified, full-length proteins and 3'UTR fragments, APC was shown to bind KAP3, a kinesin-2 adaptor, and G-rich motif-containing axonal mRNAs (β2B-tubulin and β-actin-mRNA fragments) with lower nanomolar affinity. Biochemical in vitro reconstitutions could then prove that a minimal reconstituted complex consisting of the kinesin-2 KIF3A/B/KAP3 and APC can transport RNAs over tens of micrometers (Baumann et al., 2020). While the involvement of APC in axonal mRNA localization had already been demonstrated beforehand (Preitner et al., 2014), this study underlined the value of in vitro reconstitution to identify essential building blocks of mRNA transport systems and their functions. As shown in Figure 1A, we summarize all known interactions that connect localized neuronal mRNAs to motors using the Cytoscape network visualization software (Cline et al., 2007). The interaction data the network is based on is listed in Supplementary Table 1. The table also contains additional data, which we excluded from the network because of different criteria specified in it.
All examples mentioned until here prove or hint at membrane-free mRNA transport complexes, but neuronal mRNPs can also associate with membrane-bound cargoes. RNAs hitchhike on slowly moving endosomes in Xenopus laevis retinal ganglion cells, a transport mode previously discovered in the filamentous fungus Ustilago maydis (Baumann et al., 2014; Pohlmann et al., 2015). In X. laevis ganglion cells, however, endosomes mostly serve as a translation platform, whereas most of the faster moving mRNAs move independently of endosomes (Cioni et al., 2019). In the future, it will be interesting to understand which proteins couple mRNAs to endosomes and to which extent mRNA-endosome association contributes to the actual mRNA transport process. In a different study, Annexin A11 (ANXA11), with its ordered and disordered domains, was found to function as an adaptor between liquid-like mRNPs and lysosomes (Liao et al., 2019). Even though it is intriguing to consider adaptors that link condensates to motor proteins, it is not clear currently how selectivity for certain localization signal-containing mRNAs can be inferred by such a co-partitioning-based coupling mechanism, unless mRNAs with localization signals preferentially accumulated in the same condensates that the disordered ANXA11 domain partitions into. Furthermore, to the knowledge of the authors, no data exist to date showing the co-localization of lysosomes with endogenous mRNAs. Hence, while mRNP association with endosomes and lysosomes, respectively, is an interesting phenomenon, we believe further research is required to reveal the full picture and clarify what causes mRNPs to choose endosome/lysosome association vs. direct coupling to motors via protein adaptors.
The entry of kinesins and dynein into axons and dendrites is guided by many factors, such as microtubule polarity, selectivity for microtubules with certain post-translational modifications (PTMs) (Janke and Magiera, 2020), or regulation by microtubule-associated proteins (MAPs) with specific neuronal distribution patterns (Monroy et al., 2020). Combining the information of motor-neurite selectivity and possible mRNA-motor adaptors allows postulating some mRNA transport pathways in axons or dendrites, respectively (Figure 1B).
Considering the plus-end out orientation of the microtubule cytoskeleton in axons, anterograde transport both into and inside the axon must be carried out by plus-end directed kinesin motors. Hence, the currently most likely kinesin motors with a known mRNP adaptor are kinesin-1 and kinesin-2 KIF3AB, which both bind to APC (Ruane et al., 2016; Baumann et al., 2020). Regarding dynamic tyrosinated microtubules in the growth cone, KIF3 is again the more suitable motor since MAP7, which KIF5 needs for efficient microtubule recruitment, only slowly populates freshly polymerized microtubules (Monroy et al., 2018, 2020; Hooikaas et al., 2019). Finally, if the KIF11-ZBP-1 interaction (Song et al., 2015) was to be found relevant in neurons, it could mediate axonal β-actin mRNA transport.
In dendrites, the minus-end out orientation of acetylated microtubules inhibits the entrance of kinesin-1 (Tas et al., 2017), whereas dynein and other kinesins can drive mRNA transport into dendrites (Lipka et al., 2016). However, to date, there is no evidence for direct interactions linking mRNAs to either dynein or any of the kinesins classified as strongly dendrite-targeting, such as KIF1A/B/C and KIF21A/B (Lipka et al., 2016). Hence, currently, the only options for postulating an mRNA transport complex for transport into dendrites with a proven direct link to mRNA include two different kinesin-2 motors, KIF3 and KIF17. Both motors can enter dendrites but apparently do so at a lower frequency compared with the above-mentioned kinesin-3 and kinesin-4 motors (Lipka et al., 2016). KIF17 is a special case, as it does not autonomously enter dendrites but instead requires dynein for transport into dendrites (Franker et al., 2016). How KIF17 is coupled to dynein, however, is not understood. While APC directly binds cargo adaptor KAP3 with a 90 nM affinity (Baumann et al., 2020), its interaction with KIF17 has been, so far, only shown with indirect methods (Jaulin and Kreitzer, 2010) and not in neurons. Hence, aside from the possibility that KIF3 also transports APC-mRNPs into dendrites, no other minimal set of factors is known to date that could transport mRNAs into dendrites.
At least for bidirectional mRNA transport along the unipolar axonal microtubule cytoskeleton, it would be important to understand how mammalian mRNPs interact with dynein. Considering that direct links between RBPs and dynein exist, as exemplified by the reconstitution of dynein-based mRNA transport using the Drosophila melanogaster-specific RBP Egalitarian and dynein adaptor BICD2 (McClintock et al., 2018), the existence of a mammalian linker is likely, and its identification would be a significant advancement in the mRNA transport field. A second possibility would be that inactive mRNP-carrying kinesins are transported by dynein. Several indications exist in the literature that kinesins with a demonstrated or likely function in RNP transport, such as KIF3, KIF5, and KIF17, can interact with dynein (Deacon et al., 2003; Ligon et al., 2004; Kodani et al., 2013; Franker et al., 2016; Twelvetrees et al., 2016), but further experiments are required to understand if these interactions contribute to bidirectional mRNA transport.
As recent evidence suggests that 3'UTRs often contain multiple regions that trigger localization to neurites (Mikl et al., 2021), it is possible that a single mRNA is transported by more than one motor. If this was the case, an mRNA could potentially recruit different numbers of adaptor-motor complexes based on the amount of localization motifs encoded in its 3'UTR. mRNAs with longer 3'UTRs, higher numbers of localization motifs (Tushev et al., 2018) and motors attached, would then potentially reach more distal locations, as more motors per cargo increases processivity (Furuta et al., 2013). Binding and unbinding or allosteric regulation of kinesins and dynein, respectively, could then give rise to bidirectional transport; and local cues, such as signal-induced adaptor modification or local adaptor concentrations, could fine-tune direction bias or disassembly of transport complexes, thereby producing the observed mRNA distributions.
We propose that stronger emphasis on the following approaches could help to accelerate progress in the mRNA transport field:
(1) Discovering essential building blocks of the neuronal mRNA transport machinery: while pulldown studies have provided a valuable pool of proteins that are involved in mRNA transport, they might fail to capture important transient interactions and do not report direct interactions. As a complementary technique, high-throughput (HT) screening for direct interactions with proper orthogonal validations could add valuable information to understand how mRNAs, mRNA-motor adaptors, and motor proteins directly interact (Yang et al., 2018; Garriga-Canut et al., 2019; Lang et al., 2020). HT screening can also help to narrow down essential linkers by interaction network analysis and function as a hypothesis-generator for subsequent mechanistic studies.
(2) Acquiring quantitative data about affinities between mRNA transport complex building blocks and their mode of interaction (conventional stereospecific interactions or co-partitioning into condensates because of multivalent transient interactions): neuronal mRNA distributions are likely the result of complex dynamic processes, multiple overlapping reactions, and concentration-dependent competitive mechanisms. Revealing these processes could greatly improve the understanding of how cytoplasmic mRNA distributions are generated. As shown for a variety of cellular processes, bottom-up in vitro reconstitution can provide exceptional clarity and quantitative insights on dynamic processes (Loose et al., 2008; Ohya et al., 2009; Maurer et al., 2012; Sawa-makarska et al., 2020). Also, for microtubule-based mRNA transport, a number of reconstitution studies could already provide a clear picture of essential components and their function (Heym et al., 2013; Sladewski et al., 2013; McClintock et al., 2018; Baumann et al., 2020). In addition, biophysical techniques could also help to identify point mutations in individual factors that specifically impair mRNA-motor coupling. Such mutants would then again be powerful tools for functional studies on mRNA transport in cells.
(3) Developing new approaches to disentangle pleiotropic functions of transport-mRNP building blocks in neuronal models: motor proteins and RBPs have multiple and sometimes redundant functions. After a knockdown, the accumulation of secondary effects and residual activities of the latter, depending on protein stability, can obscure their true functions. Hence, fast depletion and restoration of proteins, e.g., performing auxin-induced degradation (Nishimura et al., 2009; Yesbolatova et al., 2020) or optogenetics (van Bergeijk et al., 2015) in combination with endogenous mRNA transport live imaging (Turner-Bridger et al., 2018; Donlin-Asp et al., 2021) would be ideal to assess how and if a studied factor directly controls mRNA transport. To further avoid artifacts potentially caused by transient (over-)expressions (Rizzo et al., 2009; Gibson et al., 2013), such assays should be carried out using endogenously tagged proteins with, as much as possible, controlled expression levels. Finally, communicating negative results, for example at dedicated sessions during conferences, could contribute to faster progress and economical use of resources.
ECR, JG, SPM, SJB, and NL wrote the manuscript. ECR designed the figure. ECR, JG, and SPM analyzed the literature. All authors contributed to the article and approved the submitted version.
This study was supported by the Spanish Ministry of Economy and Competitiveness (MINECO) [BFU2017-85361-P], the Juan de la Cierva-Incorporación program [IJCI-2015-25994], the Human Frontiers in Science Program (HFSP) [RGY0083/2016], and the Ministerio de Ciencia, Innovación y Universidades and Fondo Social Europeo (FSE) [PRE2018-084501]. ECR was supported by an INPhINIT La Caixa PhD fellowship [LCF/BQ/DI19/11730046]. We further acknowledge the support of the Spanish Ministry of Economy and Competitiveness to the EMBL partnership, Centro de Excelencia Severo Ochoa [SEV-2012-0208] and [SEV-2015-0533], and the CERCA Program/Generalitat de Catalunya.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnint.2021.692948/full#supplementary-material
Barry, J., Gu, Y., Jukkola, P., O'Neill, B., Gu, H., Mohler, P., et al. (2014). Ankyrin-G directly binds to kinesin-1 to transport voltage-gated Na+ channels into axons. Dev. Cell 28, 117–131. doi: 10.1016/j.devcel.2013.11.023
Bauer, K. E., Segura, I., Gaspar, I., Scheuss, V., Illig, C., Ammer, G., et al. (2019). Live cell imaging reveals 3'-UTR dependent mRNA sorting to synapses. Nat. Commun. 10:3178. doi: 10.1038/s41467-019-11123-x
Baumann, S., Komissarov, A., Gili, M., Ruprecht, V., Wieser, S., and Maurer, S. P. (2020). A reconstituted mammalian APC-kinesin complex selectively transports defined packages of axonal mRNAs. Sci. Adv. 6:eaaz1588. doi: 10.1126/sciadv.aaz1588
Baumann, S., König, J., Koepke, J., and Feldbrügge, M. (2014). Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep. 15, 94–102. doi: 10.1002/embr.201338037
Brown, V., Jin, P., Ceman, S., Darnell, J. C., O'Donnell, W. T., Tenenbaum, S. A., et al. (2001). Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107, 477–487. doi: 10.1016/S0092-8674(01)00568-2
Buxbaum, A. R., Yoon, Y. J., Singer, R. H., and Park, H. Y. (2015). Single-molecule insights into mRNA dynamics in neurons. Trends Cell Biol. 25, 468–475. doi: 10.1016/j.tcb.2015.05.005
Chao, J. A., Patskovsky, Y., Patel, V., Levy, M., Almo, S. C., and Singer, R. H. (2010). ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev. 24, 148–158. doi: 10.1101/gad.1862910
Charalambous, D. C., Pasciuto, E., Mercaldo, V., Pilo Boyl, P., Munck, S., Bagni, C., et al. (2013). KIF1Bβ transports dendritically localized mRNPs in neurons and is recruited to synapses in an activity-dependent manner. Cell. Mol. Life Sci. 70, 335–356. doi: 10.1007/s00018-012-1108-0
Chen, N. N., Chang, C. F., Gallia, G. L., Kerr, D. A., Johnson, E. M., Krachmarov, C. P., et al. (1995). Cooperative action of cellular proteins YB-1 and Pur alpha with the tumor antigen of the human JC polyomavirus determines their interaction with the viral lytic control element. Proc. Natl. Acad. Sci. U. S. A. 92, 1087–1091. doi: 10.1073/pnas.92.4.1087
Chu, J.-F., Majumder, P., Chatterjee, B., Huang, S.-L., and Shen, C.-K. J. (2019). TDP-43 regulates coupled dendritic mRNA transport-translation processes in co-operation with FMRP and Staufen1. Cell Rep. 29, 3118–3133.e6. doi: 10.1016/j.celrep.2019.10.061
Ciolli Mattioli, C., Rom, A., Franke, V., Imami, K., Arrey, G., Terne, M., et al. (2019). Alternative 3 UTRs direct localization of functionally diverse protein isoforms in neuronal compartments. Nucl. Acids Res. 47, 2560–2573. doi: 10.1093/nar/gky1270
Cioni, J. M., Lin, J. Q., Holtermann, A. V., Koppers, M., Jakobs, M. A. H., Azizi, A., et al. (2019). Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell 176, 56–72.e15. doi: 10.1016/j.cell.2018.11.030
Cline, M. S., Smoot, M., Cerami, E., Kuchinsky, A., Landys, N., Workman, C., et al. (2007). Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382. doi: 10.1038/nprot.2007.324
Cosker, K. E., Fenstermacher, S. J., Pazyra-Murphy, M. F., Elliott, H. L., and Segal, R. A. (2016). The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat. Neurosci. 19, 690–696. doi: 10.1038/nn.4280
Davidovic, L., Jaglin, X. H., Lepagnol-Bestel, A. M., Tremblay, S., Simonneau, M., Bardoni, B., et al. (2007). The fragile X mental retardation protein is a molecular adaptor between the neurospecific KIF3C kinesin and dendritic RNA granules. Hum. Mol. Genet. 16, 3047–3058. doi: 10.1093/hmg/ddm263
Deacon, S. W., Serpinskaya, A. S., Vaughan, P. S., Lopez Fanarraga, M., Vernos, I., Vaughan, K. T., et al. (2003). Dynactin is required for bidirectional organelle transport. J. Cell Biol. 160, 297–301. doi: 10.1083/jcb.200210066
Dictenberg, J. B., Swanger, S. A., Antar, L. N., Singer, R. H., and Bassell, G. J. (2008). A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 14, 926–939. doi: 10.1016/j.devcel.2008.04.003
Donlin-Asp, P. G., Polisseni, C., Klimek, R., Heckel, A., and Schuman, E. M. (2021). Differential regulation of local mRNA dynamics and translation following long-term potentiation and depression. Proc. Natl. Acad. Sci. U. S. A. 118:e2017578118. doi: 10.1073/pnas.2017578118
Doyle, M., and Kiebler, M. A. (2011). Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 30, 3540–3552. doi: 10.1038/emboj.2011.278
Edens, B. M., Vissers, C., Su, J., Arumugam, S., Xu, Z., Shi, H., et al. (2019). FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 28, 845–854.e5. doi: 10.1016/j.celrep.2019.06.072
Eliscovich, C., Shenoy, S. M., and Singer, R. H. (2017). Imaging mRNA and protein interactions within neurons. Proc. Natl. Acad. Sci. U. S. A. 114, E1875–E1884. doi: 10.1073/pnas.1621440114
Feng, Y., Absher, D., Eberhart, D. E., Brown, V., Malter, H. E., and Warren, S. T. (1997). FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1, 109–118. doi: 10.1016/S1097-2765(00)80012-X
Franker, M. A., Esteves da Silva, M., Tas, R. P., Tortosa, E., Cao, Y., Frias, C. P., et al. (2016). Three-step model for polarized sorting of KIF17 into dendrites. Curr. Biol. 26, 1705–1712. doi: 10.1016/j.cub.2016.04.057
Fritzsche, R., Karra, D., Bennett, K. L., Ang, F., Heraud-Farlow, J. E., Tolino, M., et al. (2013). Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 5, 1749–1762. doi: 10.1016/j.celrep.2013.11.023
Fukuda, Y., Pazyra-Murphy, M. F., Silagi, E. S., Tasdemir-Yilmaz, O. E., Li, Y., Rose, L., et al. (2021). Binding and transport of SFPQ-RNA granules by KIF5A/KLC1 motors promotes axon survival. J. Cell Biol. 220:2020.02.02.931204. doi: 10.1083/jcb.202005051
Furuta, K., Furuta, A., Toyoshima, Y. Y., Amino, M., Oiwa, K., and Kojima, H. (2013). Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc. Natl. Acad. Sci. U. S. A. 110, 501–506. doi: 10.1073/pnas.1201390110
Garriga-Canut, M., Yang, J.-S. S., Preusser, F., Speroni, S., Gili, M., and Maurer, S. P. (2019). rec-Y3H screening allows the detection of simultaneous RNA-protein interface mutations. Methods. 9:2. doi: 10.1016/j.ymeth.2019.09.002
Gibson, T. J., Seiler, M., and Veitia, R. A. (2013). The transience of transient overexpression. Nat. Methods 10, 715–721. doi: 10.1038/nmeth.2534
Goering, R., Hudish, L. I., Guzman, B. B., Raj, N., Bassell, G. J., Russ, H. A., et al. (2020). FMRP promotes RNA localization to neuronal projections through interactions between its RGG domain and G-quadruplex RNA sequences. Elife 9:784728. doi: 10.7554/eLife.52621.sa2
Heber, S., Gáspár, I., Tants, J.-N., Günther, J., Moya, S. M. F., Janowski, R., et al. (2019). Staufen2-mediated RNA recognition and localization requires combinatorial action of multiple domains. Nat. Commun. 10:1659. doi: 10.1038/s41467-019-09655-3
Heisler, F. F., Lee, H. K., Gromova, K. V., Pechmann, Y., Schurek, B., Ruschkies, L., et al. (2014). GRIP1 interlinks N-cadherin and AMPA receptors at vesicles to promote combined cargo transport into dendrites. Proc. Natl. Acad. Sci. U. S. A. 111, 5030–5035. doi: 10.1073/pnas.1304301111
Henrichs, V., Grycova, L., Barinka, C., Nahacka, Z., Neuzil, J., Diez, S., et al. (2020). Mitochondria-adaptor TRAK1 promotes kinesin-1 driven transport in crowded environments. Nat. Commun. 11:3123. doi: 10.1038/s41467-020-16972-5
Heym, R. G., Zimmermann, D., Edelmann, F. T., Israel, L., Ökten, Z., Kovar, D. R., et al. (2013). In vitro reconstitution of an mRNA-transport complex reveals mechanisms of assembly and motor activation. J. Cell Biol. 203, 971–984. doi: 10.1083/jcb.201302095
Hooikaas, P. J., Martin, M., Mühlethaler, T., Kuijntjes, G.-J., Peeters, C. A. E. E., Katrukha, E. A., et al. (2019). MAP7 family proteins regulate kinesin-1 recruitment and activation. J. Cell Biol. 218, 1298–1318. doi: 10.1083/jcb.201808065
Janke, C., and Magiera, M. M. (2020). The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 21, 307–326. doi: 10.1038/s41580-020-0214-3
Jaulin, F., and Kreitzer, G. (2010). KIF17 stabilizes microtubules and contributes to epithelial morphogenesis by acting at MT plus ends with EB1 and APC. J. Cell Biol. 190, 443–460. doi: 10.1083/jcb.201006044
Johnson, E. M., Kinoshita, Y., Weinreb, D. B., Wortman, M. J., Simon, R., Khalili, K., et al. (2006). Role of Purα in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J. Neurosci. Res. 83, 929–943. doi: 10.1002/jnr.20806
Kanai, Y., Dohmae, N., and Hirokawa, N. (2004). Kinesin transports RNA. Neuron 43, 513–525. doi: 10.1016/j.neuron.2004.07.022
Kodani, A., Salomé Sirerol-Piquer, M., Seol, A., Garcia-Verdugo, J. M., and Reiter, J. F. (2013). Kif3a interacts with Dynactin subunit p150 Glued to organize centriole subdistal appendages. EMBO J. 32, 597–607. doi: 10.1038/emboj.2013.3
Lang, B., Yang, J.-S., Garriga-Canut, M., Speroni, S., Gili, M., Tartaglia, G. G., et al. (2020). Matrix screening reveals a vast potential for direct protein-protein interactions among RNA binding proteins. bioRxiv 2020.09.14.296160. doi: 10.1101/2020.09.14.296160
Liao, Y.-C. C., Fernandopulle, M. S., Wang, G., Choi, H., Hao, L., Drerup, C. M., et al. (2019). RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell 179, 147–164.e20. doi: 10.1016/j.cell.2019.08.050
Ligon, L. A., Tokito, M., Finklestein, J. M., Grossman, F. E., and Holzbaur, E. L. F. (2004). A direct interaction between cytoplasmic dynein and kinesin I may coordinate motor activity. J. Biol. Chem. 279, 19201–19208. doi: 10.1074/jbc.M313472200
Lipka, J., Kapitein, L. C., Jaworski, J., and Hoogenraad, C. C. (2016). Microtubule-binding protein doublecortin-like kinase 1 (DCLK1) guides kinesin-3-mediated cargo transport to dendrites. EMBO J. 35, 302–318. doi: 10.15252/embj.201592929
Loose, M., Fischer-Friedrich, E., Ries, J., Kruse, K., and Schwille, P. (2008). Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320, 789–792. doi: 10.1126/science.1154413
Mallardo, M., Deitinghoff, A., Muller, J., Goetze, B., Macchi, P., Peters, C., et al. (2003). Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. U. S. A. 100, 2100–2105. doi: 10.1073/pnas.0334355100
Maurer, S. P., Fourniol, F. J., Bohner, G., Moores, C. A., and Surrey, T. (2012). EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 149, 371–382. doi: 10.1016/j.cell.2012.02.049
McClintock, M. A., Dix, C. I., Johnson, C. M., McLaughlin, S. H., Maizels, R. J., Hoang, H. T., et al. (2018). RNA-directed activation of cytoplasmic dynein-1 in reconstituted transport RNPs. Elife 7:273912. doi: 10.7554/eLife.36312.056
Menon, L., Mader, S. A., and Mihailescu, M.-R. (2008). Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA 14, 1644–1655. doi: 10.1261/rna.1100708
Mikl, M., Eletto, D., Lee, M., Lafzi, A., Mhamedi, F., Sain, S. B., et al. (2021). A massively parallel reporter assay reveals focused and broadly encoded RNA localization signals in neurons. bioRxiv 2021.04.27.441590. doi: 10.1101/2021.04.27.441590
Miller, S., Yasuda, M., Coats, J. K., Jones, Y., Martone, M. E., and Mayford, M. (2002). Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron 36, 507–19. doi: 10.1016/S0896-6273(02)00978-9
Monroy, B. Y., Sawyer, D. L., Ackermann, B. E., Borden, M. M., Tan, T. C., and Ori-Mckenney, K. M. (2018). Competition between microtubule-associated proteins directs motor transport. Nat. Commun. 9, 1–12. doi: 10.1038/s41467-018-03909-2
Monroy, B. Y., Tan, T. C., Oclaman, J. M., Han, J. S., Simó, S., Niwa, S., et al. (2020). A combinatorial MAP code dictates polarized microtubule transport. Dev. Cell 53, 60–72.e4. doi: 10.1016/j.devcel.2020.01.029
Nakajima, K., Yin, X., Takei, Y., Seog, D.-H., Homma, N., and Hirokawa, N. (2012). Molecular motor KIF5A is essential for GABAA receptor transport, and KIF5A deletion causes epilepsy. Neuron 76, 945–961. doi: 10.1016/j.neuron.2012.10.012
Nicastro, G., Candel, A. M., Uhl, M., Oregioni, A., Hollingworth, D., Backofen, R., et al. (2017). Mechanism of β-actin mRNA Recognition by ZBP1. Cell Rep. 18, 1187–1199. doi: 10.1016/j.celrep.2016.12.091
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T., and Kanemaki, M. (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922. doi: 10.1038/nmeth.1401
Ohya, T., Miaczynska, M., Coskun, Ü., Lommer, B., Runge, A., Drechsel, D., et al. (2009). Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature 459, 1091–1097. doi: 10.1038/nature08107
Patel, V. L., Mitra, S., Harris, R., Buxbaum, A. R., Lionnet, T., Brenowitz, M., et al. (2012). Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 26, 43–53. doi: 10.1101/gad.177428.111
Pohlmann, T., Baumann, S., Haag, C., Albrecht, M., and Feldbrügge, M. (2015). A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. Elife 2015:55. doi: 10.7554/eLife.06041.055
Preitner, N., Quan, J., Nowakowski, D. W., Hancock, M. L., Shi, J., Tcherkezian, J., et al. (2014). APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell 158, 368–382. doi: 10.1016/j.cell.2014.05.042
Richards, A. L., Eckhardt, M., and Krogan, N. J. (2021). Mass spectrometry-based protein–protein interaction networks for the study of human diseases. Mol. Syst. Biol. 17, 1–18. doi: 10.15252/msb.20188792
Rizzo, M. A., Davidson, M. W., and Piston, D. W. (2009). Fluorescent protein tracking and detection: applications using fluorescent proteins in living cells. Cold Spring Harb. Protoc. 4:pdb.top64. doi: 10.1101/pdb.top64
Ruane, P. T., Gumy, L. F., Bola, B., Anderson, B., Wozniak, M. J., Hoogenraad, C. C., et al. (2016). Tumour Suppressor Adenomatous Polyposis Coli (APC) localisation is regulated by both Kinesin-1 and Kinesin-2. Sci. Rep. 6:27456. doi: 10.1038/srep27456
Sawa-makarska, J., Baumann, V., Coudevylle, N., Bülow, S., Von Nogellova, V., Abert, C., et al. (2020). Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science 369:aaz7714. doi: 10.1126/science.aaz7714
Serra-Marques, A., Martin, M., Katrukha, E. A., Grigoriev, I., Peeters, C. A. E., Liu, Q., et al. (2020). Concerted action of kinesins kif5b and kif13b promotes efficient secretory vesicle transport to microtubule plus ends. Elife 9, 1–37. doi: 10.7554/eLife.61302
Shah, S., Molinaro, G., Liu, B., Wang, R., Huber, K. M., and Richter, J. D. (2020). FMRP control of ribosome translocation promotes chromatin modifications and alternative splicing of neuronal genes linked to autism. Cell Rep. 30, 4459–4472.e6. doi: 10.1016/j.celrep.2020.02.076
Sharangdhar, T., Sugimoto, Y., Heraud-Farlow, J., Fernández-Moya, S. M., Ehses, J., Ruiz de Los Mozos, I., et al. (2017). A retained intron in the 3'-UTR of Calm3 mRNA mediates its Staufen2- and activity-dependent localization to neuronal dendrites. EMBO Rep. 18, 1762–1774. doi: 10.15252/embr.201744334
Sladewski, T. E., Bookwalter, C. S., Hong, M.-S., and Trybus, K. M. (2013). Single-molecule reconstitution of mRNA transport by a class V myosin. Nat. Struct. Mol. Biol. 20, 952–957. doi: 10.1038/nsmb.2614
Song, T., Zheng, Y., Wang, Y., Katz, Z., Liu, X., Chen, S., et al. (2015). Specific interaction of KIF11 with ZBP1 regulates the transport of β-actin mRNA and cell motility. J. Cell Sci. 128, 1001–1010. doi: 10.1242/jcs.161679
Takano, K., Miki, T., Katahira, J., and Yoneda, Y. (2007). NXF2 is involved in cytoplasmic mRNA dynamics through interactions with motor proteins. Nucl. Acids Res. 35, 2513–2521. doi: 10.1093/nar/gkm125
Tas, R. P., Chazeau, A., Cloin, B. M. C. C., Lambers, M. L. A. A., Hoogenraad, C. C., and Kapitein, L. C. (2017). Differentiation between oppositely oriented microtubules controls polarized neuronal transport. Neuron 96, 1264–1271.e5. doi: 10.1016/j.neuron.2017.11.018
Turner-Bridger, B., Jakobs, M., Muresan, L., Wong, H. H.-W., Franze, K., Harris, W. A., et al. (2018). Single-molecule analysis of endogenous β-actin mRNA trafficking reveals a mechanism for compartmentalized mRNA localization in axons. Proc. Natl. Acad. Sci. U. S. A. 115, E9697–E9706. doi: 10.1073/pnas.1806189115
Tushev, G., Glock, C., Heumüller, M., Biever, A., Jovanovic, M., and Schuman, E. M. (2018). Alternative 3' UTRs modify the localization, regulatory potential, stability, and plasticity of mRNAs in neuronal compartments. Neuron 98, 495–511.e6. doi: 10.1016/j.neuron.2018.03.030
Twelvetrees, A. E., Pernigo, S., Sanger, A., Guedes-Dias, P., Schiavo, G., Steiner, R. A., et al. (2016). The dynamic localization of cytoplasmic dynein in neurons is driven by kinesin-1. Neuron 90, 1000–1015. doi: 10.1016/j.neuron.2016.04.046
Twelvetrees, A. E., Yuen, E. Y., Arancibia-Carcamo, I. L., MacAskill, A. F., Rostaing, P., Lumb, M. J., et al. (2010). Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron 65, 53–65. doi: 10.1016/j.neuron.2009.12.007
van Bergeijk, P., Adrian, M., Hoogenraad, C. C., and Kapitein, L. C. (2015). Optogenetic control of organelle transport and positioning. Nature 518, 111–114. doi: 10.1038/nature14128
Williams, J. C., Roulhac, P. L., Roy, A. G., Vallee, R. B., Fitzgerald, M. C., and Hendrickson, W. A. (2007). Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc. Natl. Acad. Sci. U. S. A. 104, 10028–10033. doi: 10.1073/pnas.0703614104
Wong, H. H.-W., Lin, J. Q., Ströhl, F., Roque, C. G., Cioni, J.-M., Cagnetta, R., et al. (2017). RNA docking and local translation regulate site-specific axon remodeling in vivo. Neuron 95, 852–868.e8. doi: 10.1016/j.neuron.2017.07.016
Yang, J.-S., Garriga-Canut, M., Link, N., Carolis, C., Broadbent, K., Beltran-Sastre, V., et al. (2018). rec-YnH enables simultaneous many-by-many detection of direct protein–protein and protein–RNA interactions. Nat. Commun. 9:3747. doi: 10.1038/s41467-018-06128-x
Yesbolatova, A., Saito, Y., Kitamoto, N., Makino-Itou, H., Ajima, R., Nakano, R., et al. (2020). The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat. Commun. 11:19532. doi: 10.1038/s41467-020-19532-z
Yoon, Y. J., Wu, B., Buxbaum, A. R., Das, S., Tsai, A., English, B. P., et al. (2016). Glutamate-induced RNA localization and translation in neurons. Proc. Natl. Acad. Sci. U. S. A. 113, E6877–E6886. doi: 10.1073/pnas.1614267113
Zalfa, F., Eleuteri, B., Dickson, K. S., Mercaldo, V., De Rubeis, S., di Penta, A., et al. (2007). A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat. Neurosci. 10, 578–587. doi: 10.1038/nn1893
Zhang, M., Wang, Q., and Huang, Y. (2007). Fragile X mental retardation protein FMRP and the RNA export factor NXF2 associate with and destabilize Nxf1 mRNA in neuronal cells. Proc. Natl. Acad. Sci. U. S. A. 104, 10057–10062. doi: 10.1073/pnas.0700169104
Keywords: mRNA trafficking, neurons, kinesin, RNA-binding protein, mRNA localization, dynein
Citation: Rodrigues EC, Grawenhoff J, Baumann SJ, Lorenzon N and Maurer SP (2021) Mammalian Neuronal mRNA Transport Complexes: The Few Knowns and the Many Unknowns. Front. Integr. Neurosci. 15:692948. doi: 10.3389/fnint.2021.692948
Received: 09 April 2021; Accepted: 25 May 2021;
Published: 15 June 2021.
Edited by:
Jean-Michel Cioni, San Raffaele Hospital (IRCCS), ItalyReviewed by:
Pabitra Sahoo, University of South Carolina, United StatesCopyright © 2021 Rodrigues, Grawenhoff, Baumann, Lorenzon and Maurer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastian P. Maurer, c2ViYXN0aWFuLm1hdXJlckBjcmcuZXU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.