
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Insect Sci., 11 March 2025
Sec. Insect Health and Pathology
Volume 5 - 2025 | https://doi.org/10.3389/finsc.2025.1555434
Background: The bacterial diversity of two bee species in the process of honey collection during the flowering season of three different floral sources in the winter was studied. The common bacterium in all samples was Bacillus subtilis.
Methods: In the present study, we collected nectar, honey sacs, and fresh honey during the winter flowering season of Agastache rugosa, Prunus cerasoides, and Brassica rapa. The pure culture method was used to count and analyze the number of bacteria, they were identified using 16S rRNA sequencing, similarities were compared in NCBI, and the common dominant bacterial species B. subtilis in all samples using phylogenetic analysis and intersection analysis were determined to conduct further bacteriostatic experiments.
Results: The results showed that the most abundant quantity of bacteria could be found in the honey sacs, compared to in nectar or fresh honey. At the same time, the highest abundance of bacteria could be found in the honey sacs of A. cerana when collected on Brassica rapa, while the highest abundance of bacteria could be found in the honey sacs of A. mellifera when collected on Prunus cerasoides and Agastache rugosa. A total of 33 bacterial species were isolated, with variations in their distribution across different sample types and sources. The inhibitory effect of 10-1-10-5 on Hafnia alvei by B. subtilis was very significant.
Conclusions: B. subtilis was identified in all sample sources, indicating the potential importance of B. subtilis as a probiotic in the bee gut for honey production, and B. subtilis could promote the disease resistance and health of honeybees in winter.
The gut microbiota of the honeybee has emerged as a key factor influencing the health and vitality of these essential pollinators (1). The symbiotic relationship between bees and their microbial passengers is integral to their ability to digest complex floral nectars and pollen, defend against pathogens, and adapt to environmental stressors (1–3). The composition of the bee gut microbiota is shaped by a variety of factors, including genetic, immunological, social status, and environmental interactions, which ultimately affect the bees’ capacity to perform their ecological roles effectively (4–6).
Among the environmental factors, the choice of floral resources is particularly influential. The nutritional and chemical properties of nectar and pollen can significantly affect the development and stability of the gut microbiota (7, 8). The foraging behavior of bees during different seasons, and their interaction with available flora, introduce temporal dimensions to this microbiota-environment relationship (9, 10).
Seasonal changes in floral availability prompt bees to forage on a variety of plant species, including those that flower in winter. However, only in a few places do researchers have the chance to study the bees foraging on flowers (11). These winter-flowering plants present unique challenges and opportunities for bees, given their distinct biochemical compositions and the bees’ need to adapt to colder temperatures and altered environmental conditions (9). The study of how bees and their microbiota respond to winter nectar sources is therefore critical to understanding the resilience of bee populations in the face of seasonal adversity.
Honey sacs act as a nectar sink and transmitter between floral nectar and freshly downloaded honey, and play a crucial role in filtering the flow of harmful substances to mid-gut or hind-gut with a special valve. The nectar is passed from the dancer bees, who first store the nectar, normally 10-50 uL, in their honey sac, to the next indoor mates during waggle dances (12). The indoor mates, based on the information from the passing nectar and dance, either save the nectar into a honey cell or are triggered by the dancer to fly out for more foraging. The honey sac, as the anterior part of a bee’s digestive system, introduces a microbial flora from the nectar during the process of nectar intake. Additionally, air is also mixed in during this process, so the honey sac is not a strictly anaerobic environment. As the nectar is regurgitated back into the honeycomb, both the nectar and the microorganisms from the honey sac are introduced into the cells. The honey sac, being a relatively open space, can also be colonized by aerobic bacteria such as Bacillus subtilis, which can inhibit pathogenic microorganisms at the front end of the digestive tract (13).
The present study focuses on the honeybee species Apis cerana and A. mellifera, which are known to forage on winter-flowering plants such as Agastache rugosa, Prunus cerasoides, and Brassica rapa (14, 15). These plants provide a vital food source during a period when other floral resources are scarce, especially in Yunnan, China. Unlike previous studies that focused on the diverse microbiome of raw honey, this study focused on diverse sources since this diversity may originate from pollen, nectar, air, the honeybee digestive tract (from mouth parts to the honey sac to the midgut and hindgut), contamination during processing by bees in the hive from fresh honey to ripe honey, and honey extraction by humans (16, 17). The research aims to elucidate the bacterial diversity within the nectar, honey sacs, and fresh honey collected by these bees from the above winter-flowering sources with less contamination from honey processing in the hive and from honey extraction by humans. By doing so, the study seeks to identify the key microbial constituents and their potential implications for bee health and honey quality, with a special focus on the microbiota effect on bee health and digestive effects (18).
Hafnia alvei is one of several Enterobacteriaceae species that are sporadically found in the gut of bees and may represent opportunistic pathogens (19). Indeed, good bee health is clearly linked to the stable functioning of ecosystems and indirectly to human well-being (20). H. alvei is a serious infectious disease that threatens bee populations, but it is mostly reported in A. mellifera. It has the characteristics of rapid spread, severe damage, long disease duration, and contamination of bee products (21). H. alvei is one of the pathogens causing septicemia in adult bees, and research reports that two bacterial members of the bee gut, Gilliamella and Lactobacillus, can clear H. alvei during invasion (22). In addition to isolating common dominant bacteria, this experiment also used B. subtilis to perform bacteriostatic experiments on hive honeydew, further demonstrating the defensive effect of B. subtilis in the honey sac against pathogenic bacteria.
Previous research has shed light on the general composition of bee gut microbiota and its role in honeybee biology. However, there is a dearth of knowledge regarding the specific dynamics of microbial communities as they progress from nectar through the honey production process, particularly during winter months. This study addresses this gap by examining the bacterial diversity at various stages of honey processing, offering a nuanced perspective on the interplay between bees, their microbiota, and the winter floral resources they exploit. Based on the current understanding of bee microbiota and the unique challenges posed by winter foraging, we learned much about the diversity of cultivable bacteria in honeybees during winter foraging, reflecting adaptation to the specific nutritional and chemical properties of winter nectar sources. We are also gaining a better understanding of the resistance of dominant bacteria to pathogens and the promotion of bee health, which may have implications for colony management and improving honey production during the winter season.
Through investigation, it was confirmed that there were no other floral plants that were flowering at the same time. Nectar was collected using a sterile capillary in a field near each apiary. In total, 38,142 A. rugosa (Ar), 297 P. cerasoides (Pc), and 9,810 B. rapa (Br) flowers were artificially collected and 9 mL of each nectar was collected for subsequent analyses. All samples were frozen immediately after collection, stored at –20°C, and screened within 2 months (23) (Figure 1).
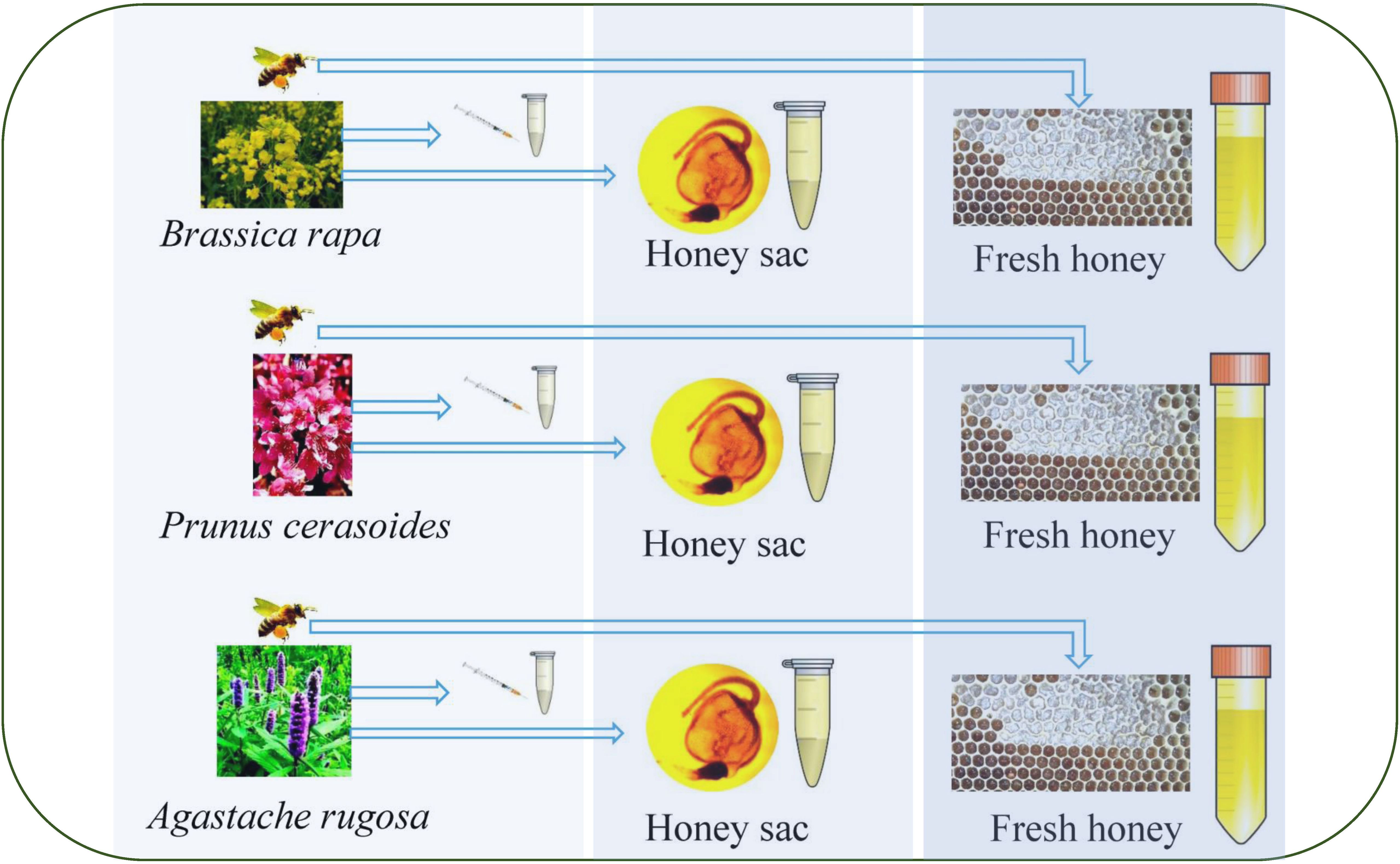
Figure 1. Sampling procedure from nectar to honey sac to fresh honey from the honeycomb. On the left side of the figure is the collection of nectar from different floral sources during the flowering period, in the middle of the figure is the collection of sacs from two honey bee species from three floral source areas, and on the right side of the figure is the collection of honey from two honey bee species from three floral source areas.
We collected honeybees from November to February of the next year, during the full bloom period of A. rugosa, P. cerasoides, and B. rapa, from three apiaries with A. cerana and A. mellifera in Mengzi (N23°29′-N24°22′, E103°18′-E103°33′, Al: 1256-1279m), Kunming (N25°07′, E102°45′ Al: 1958m), and Luoping (N24°49′-N24°57′, E104°18′-E104°22′, Al; 1555-1580m), Yunnan Province, Southwest China.
The colonies were all maintained using standard beekeeping practices. For surface sterilization, honeybees were suspended in 75% ethanol and then incubated in sterile water for 1 min each. All honey sacs were collected by aseptic excision into an aseptic tube. Samples were placed in separate sterile tubes containing 10 mL each of sterile physiological saline (0.9% w/v NaCl, 0.1% w/v Tween 80 and 0.1% w/v peptone) (23).For the two different species, A. cerana and A. mellifera, we collected 30 honey sacs from each colony, 10 of which were filled with a sterile sampling liquid tube. In total, 180 honey sacs were collected (Figure 1).
To obtain fresh honey samples, an empty honeycomb was placed in each of the three colonies of A. cerana and the three colonies of A. mellifera. After 24 hours, fresh honey was collected from each honeycomb placed using a sterile tube. Specifically, 3 mL of honey was collected from each colony. This resulted in a total of 9 mL of fresh honey collected from each honeybee species, a total of 18 mL. The collected honey samples were immediately stored at -20°C to preserve their integrity for subsequent experiments (Figure 1).
The samples were named according to the following abbreviations: LP: Luoping Yunnan province; MZ: Mengzi Yunnan province; KM: Kunming Yunnan province; 30××:Isolation bacterial strain number. Samples from LP were collected during the flowering season of Br. Samples from MZ were collected during the flowering season of Ar. Samples from KM were collected during the flowering season of Pc. All are the same in the text.
Tryptone soy broth (TSB) agar (24) and Man, Rogosa, and Sharpes (MRS) agar were used as culture media (23). Isolates were cultured aerobically in TSB medium at 37°C for 2–3 days or anaerobically in MRS medium at 37°C for 3–4 days using anaerobic flasks with Anaerocult (Merck, Darmstadt, Germany). From each growing surface plate containing 30–300 colonies each, 5–20 colonies of different morphologies were selected and each different colony was then subcultured to obtain pure isolates. A total colony count (CFU) was also performed when selecting the colonies.
The collected bacteria grown on a plate were centrifuged at 10,000 rpm for 30 seconds at 4°C to obtain bacterial thalli according to the kit instructions. This process allowed us to collect 0.1–0.3 g of bacterial thalli. Pure DNA was then extracted from the bacterial thalli using the Tianamp Bacteria DNA Extraction Kit (Tiangen Biotech Co., Beijing, China) according to the manufacturer’s instructions.
PCR amplification of the 16S rRNA gene from each bacterial sample was performed using a thermal cycler (MJ Research, T100TM Thermal Cycler; Bio-Rad Co., Hercules, CA, USA). Each reaction mixture (final volume, 50μL) contained 4μL template DNA, 0.2μL each primer, 25μL 2× TransTaqTM II HiFi PCR SuperMix II (Transgen Co., Beijing, China), and 20.6 μL dH2O.
The universal oligonucleotide primers used to amplify the bacterial 16S rRNA gene were 27F (5′-AGAGTTTGATCCTGGCTC-3′) and 1387R (5′-GGGCGGTGTGTACAAGGC-3′). The PCR conditions included an initial denaturation of the DNA for 5 min at 94°C, then 30 cycles of denaturation of the DNA for 30 s at 94°C, annealing for 1 min at 58°C, and extension for 90 s at 72°C, followed by a final incubation for 7 min at 72°C (25, 26). PCR products were selected for sequencing.
The purified PCR products obtained from the bacterial isolates were sequenced at Sangon Biotech Co. (Shanghai China) using 27F and 1387R primers. To determine the closest known relatives of the partial 16S rRNA gene sequences that we obtained, the sequences were queried against GenBank (National Centre for Biotechnology Information, Rockville Pike, Bethesda, MD, USA) using the Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov/).
We compiled the 16S rRNA sequences from the isolated samples using SeqMan software. Next, the 16S rRNA and test strain sequences were edited using the BioEdit program and aligned using Clustal-W. After the deletion of regions containing ambiguous nucleotides, the distance matrix was calculated using BioEdit. Phylogenetic trees were constructed using the neighbor-joining method. To determine the stability of the phylogenetic tree, the sequence data were sampled 1,000 times for bootstrap analysis using MEGA version 11 with Kimura 2-parameter distances.
We selected three representative dominant strains from three floral source periods as test strains. The test strains were cultured with 0.1 g bacteria and 0.9 g sterile water for gradient dilution 10-1~10-6. Similar to the test strains, the pathogens were able to grow normally on TSB medium. Then, 0.1g of bacteria were collected by centrifugation, diluted with 0.9g of sterile water and 0.1mL were plated on TSB agar medium. The control plate was used as a standard for growth. Simultaneously, the treated test strains were placed on three pieces of 5 mm sterile filter paper equidistant from each plate as shown in Figure 2. The antibacterial distance was measured after 5 days of aerobic cultivation at 37°C (27).
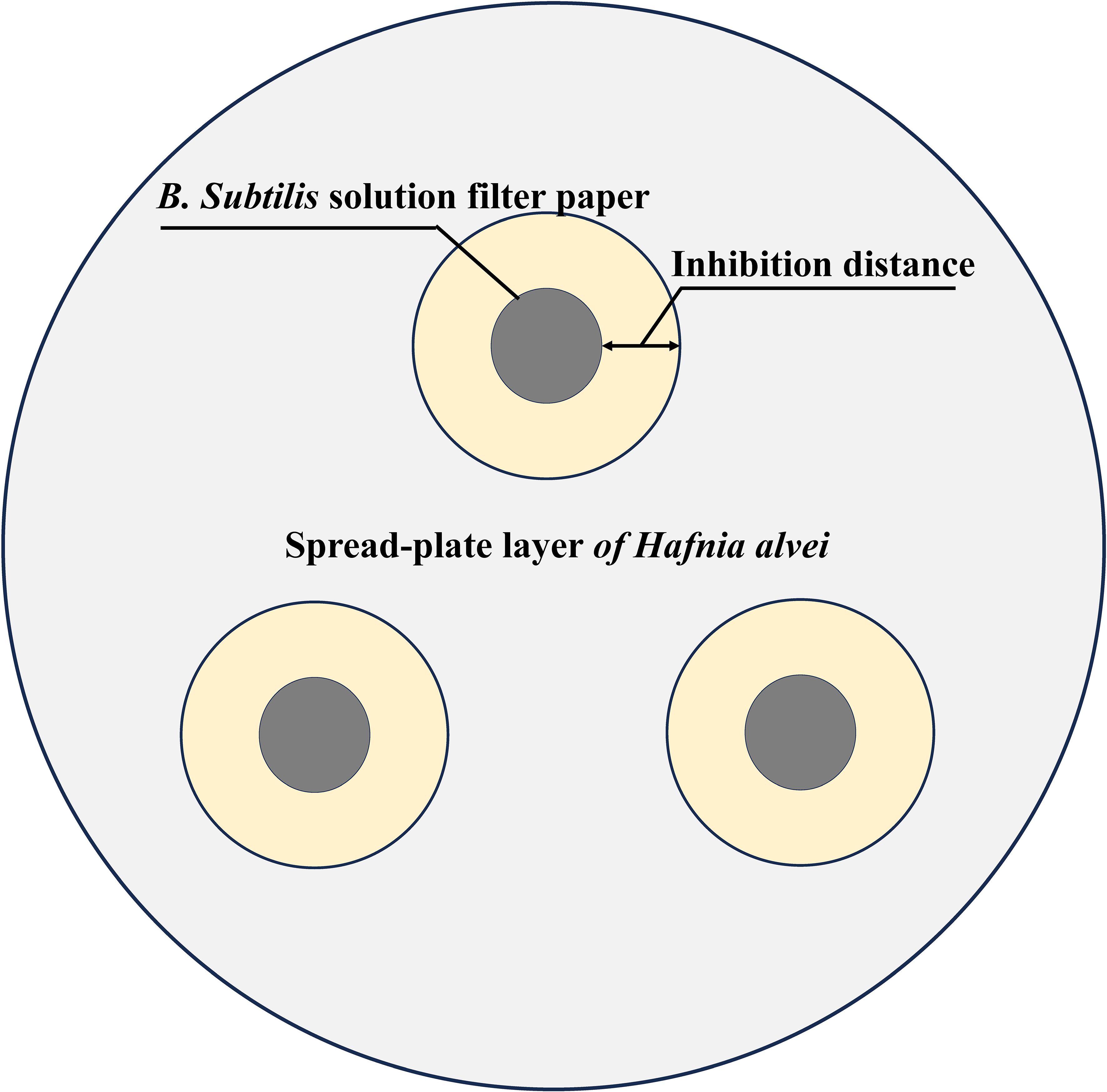
Figure 2. Schematic diagram of B. subtilis inhibiting Hafnia alvei. The gray area is the spread-plate layer of H. alvei, the black area is the B. subtilis filter sheet, and the yellow area is the bacteriostatic distance.
The number of bacteria was determined via two-way ANOVA, with floral sources (A. rugosa, P. cerasoides, and B. rapa) and honey sources (nectar, honey sac honey, and fresh honey) as fixed effects, while honeybee species was set as the random effect. Tukey’s HSD test was used as a post hoc test. All statistic were performed in R Studio (2022.02.3). The results of the antibacterial experiment were determined to be statistically significant using one-way ANOVA.
A sum analysis was performed on all samples and there was more bacteria in the honey sac than in nectar or fresh honey for both bee species (F4,130 = 100.918, P<0.001). The number of B. subtilis was also higher in the honey sac than in nectar and fresh honey (F4,130 = 99.258, P<0.001). Post-hoc comparison results showed that bacterial quantity was highest in honey sacs of A. cerana which was higher than in honey sacs of A. mellifera (Tukey’s HSD, P<0.001) (Figure 3A).
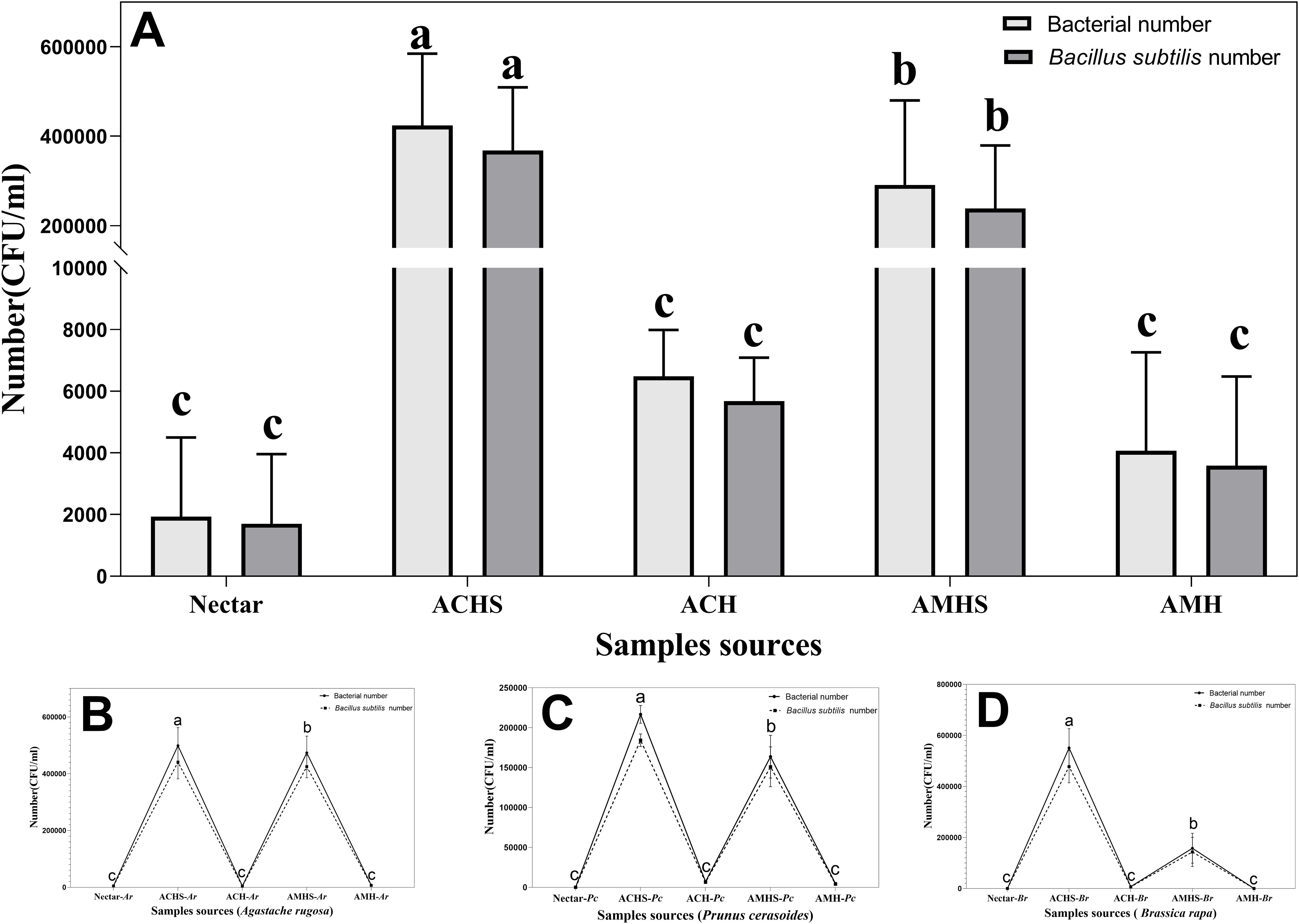
Figure 3. The total number of bacteria and B. subtilis between different sample sources and their interactions. (A) Plot of the number of bacteria in all samples during the winter flowering period of the floral source. Plot of the number of bacteria during the flowering period of Agastache rugosa (B), Prunus cerasoides (C), and Brassica rapa (D). Different letters showed significant differences, P<0.05. ACHS, honey sac of A. cerana; AMHS, honey sac of A. mellifera; ACH, honey of A. cerana; AMH, honey of A. mellifera. Same below.
In the three different winter flowering stages, the number of bacteria in all samples changed in the same way. B. subtilis was the dominant bacteria in the samples and the number and total number of bacteria also changed in the same way. Similar trends of bacterial abundance were found in winter floral sources of Pc (F4,40 = 574.780, P<0.001), Ar (F4,40 = 416.315, P<0.001), and Br (F4,40 = 278.476, P<0.001), while the highest abundance was found in the honey sac samples of A. cerana compared with the honey sac samples of A. mellifera, followed by the nectar and honey samples (Figures 3B–D).
The number of bacteria was significantly different in the three winter flowering sources. The number of bacteria in the honey sacs of A. cerana at Pr anthesis was significantly lower than at Ar and Br anthesis (F2,24 = 89.750, P < 0.001). There was no significant difference between Ar and Br anthesis (P=0.097) (Figure 4A). The number of bacteria in the honey sacs of A. mellifera at Ar anthesis was significantly higher than at Pc and Br anthesis (F2,24 = 115.847, P < 0.001). There was no significant difference between Ar and Br anthesis (P=0.803) (Figure 4B).
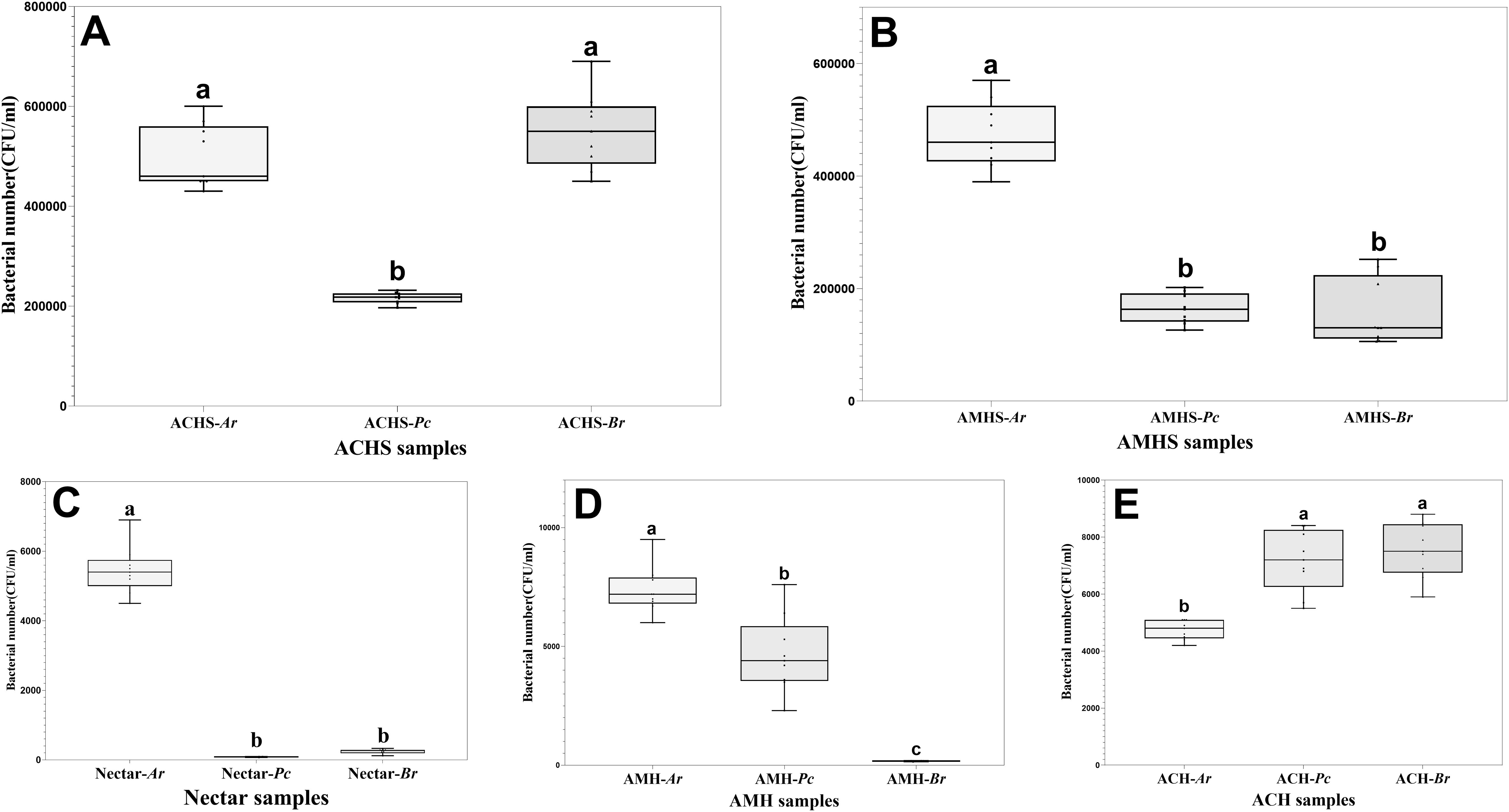
Figure 4. Comparison of bacterial number in honey sacs (A, B), nectar (C), and fresh honey (D, E) from bee colonies during winter flowering of Ar, Pc, and Br, respectively. Different letters show significant differences, P<0.05.
The highest bacterial quantity of nectar was found in Ar compared with the other two winter floral sources (P<0.01), while no statistical difference was found between Br and Pc (P=0.400) (Figure 4C).
The number of bacteria in the honey collected by A. mellifera and A. cerana was the lowest and the difference between the other groups was statistically significant, except that the difference in the honey quantity of A. cerana at the flowering stage of Pc and Br was not significant (Tukey’s HSD, P<0.05) (Figures 4D,E).
Bacterial isolation and identification in the nectar, honey sacs, and honey from the LP (Br), KM (Pc), and MZ (Ar) flowering seasons yielded 66 strains of different bacterial species. The strains were submitted to NCBI, and after comparison, information on similar strains and strain accession numbers from NCBI, and similarity ranges and degrees of similarity were obtained (Table 1). Other similar strains are also listed in Table 1.
In total, 33 bacteria were isolated and identified from the nectar, honey sac, and honey samples from three different flowering sources collected by two bee species. Apart from B. subtilis, which was isolated from all samples, seven bacterial species were isolated and identified from Br. B. amyloliquefaciens was the only bacterial species isolated from the nectar and honey sacs of both bee species. Another seven bacterial species were isolated from only one of the sample sources. Furthermore, 13 bacterial species were isolated and identified from Ar, and other bacterial species were only isolated from one of the sample sources. In total, 22 bacterial species were isolated and identified from Pc and only 4 bacterial species were found in the honey sacs of both A. cerana and A. mellifera species, such as B. amyloliquefaciens and Acetobacteraceae bacteria. Other bacterial species were isolated from only one of the sample sources (Table 1).
Phylogenetic analysis clustered the samples from three different regions with different floral sources into five bacterial clusters (Figure 5). Cluster I: Proteobacteria and Gammaproteobacteria; Cluster II: Proteobacteria, Alphaproteobacteria, Bacteroidetes, and Sphingobacteriia; Cluster III: Actinobacteria and Microbacteriaceae; Cluster IV: Firmicutes and Bacill; Cluster V: Firmicutes, Bacilli, Bacillales, Bacillaceae, and Bacillus. The bacterium Aquifex pyrophilus was set as an outgroup (L37096).
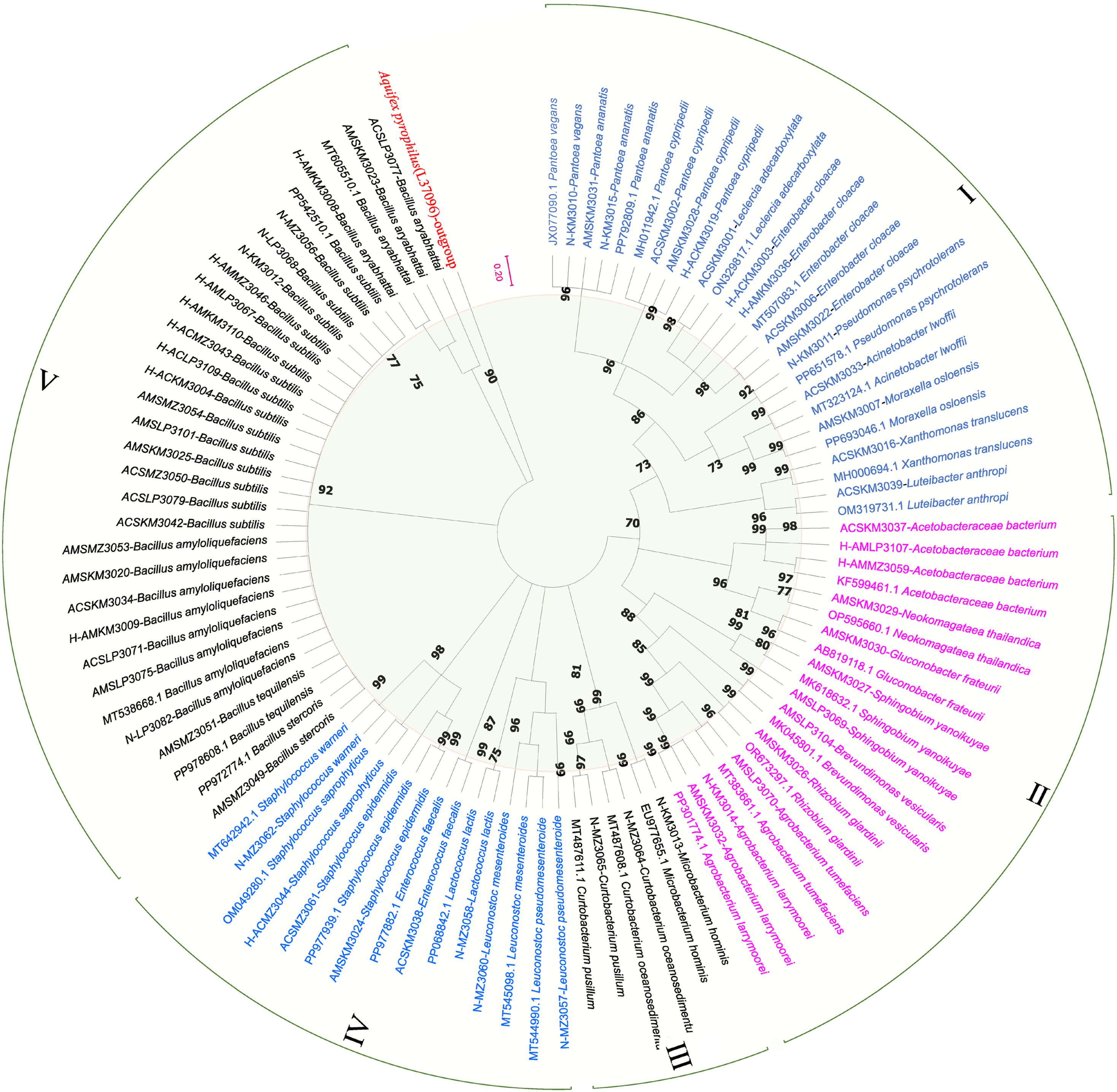
Figure 5. Phylogenetic tree of nectar, honey sac, and honey bacteria from three different nectar sources. The phylogenetic tree is based on a distance matrix analysis of 873 positions in the 16S rRNA gene and was constructed using ClustalW with the neighbor-joining method within the MEGA (11) package. Closely related types and reference strains are shown in parentheses together with accession numbers from GenBank. Bootstrap values based on 1,000 re-samplings display the significance of the interior nodes, and are shown at branch points. As shown by the bifurcation points, all five clusters have a bootstrap value of at least 70, and the scale of 0.2 represents a 20% evolutionary difference. Bacterial samples from different regions of the phylogenetic tree are grouped into five clusters (I-V), distinguished by color. The length of the branch is proportional to the genetic distance, and the longer the branch, the greater the difference between the samples.
This phylogenetic analysis reveals a clear division of the bacterial samples into five well-supported clusters, suggesting distinct evolutionary lineages or ecological adaptations within the sampled regions. The clustering pattern may reflect underlying environmental gradients, geographic separation, or host-specific associations that have shaped the diversification of these bacterial populations.
Figure 6A illustrates the patterns of bacterial distribution, highlighting both regionally specific and widely distributed types. The bacterial cross analysis showed that there were significant differences between the three different types of winter honey sources in the three different regions (LP, KM, and MZ). Only one common species of B. subtilis was present in all the samples, although in different regions. B. amyloliquefaciens was also found in some samples from LP and KM. Proteobacteria and Actinomycetes were found only in the KM region (Pc), but not in other regions.
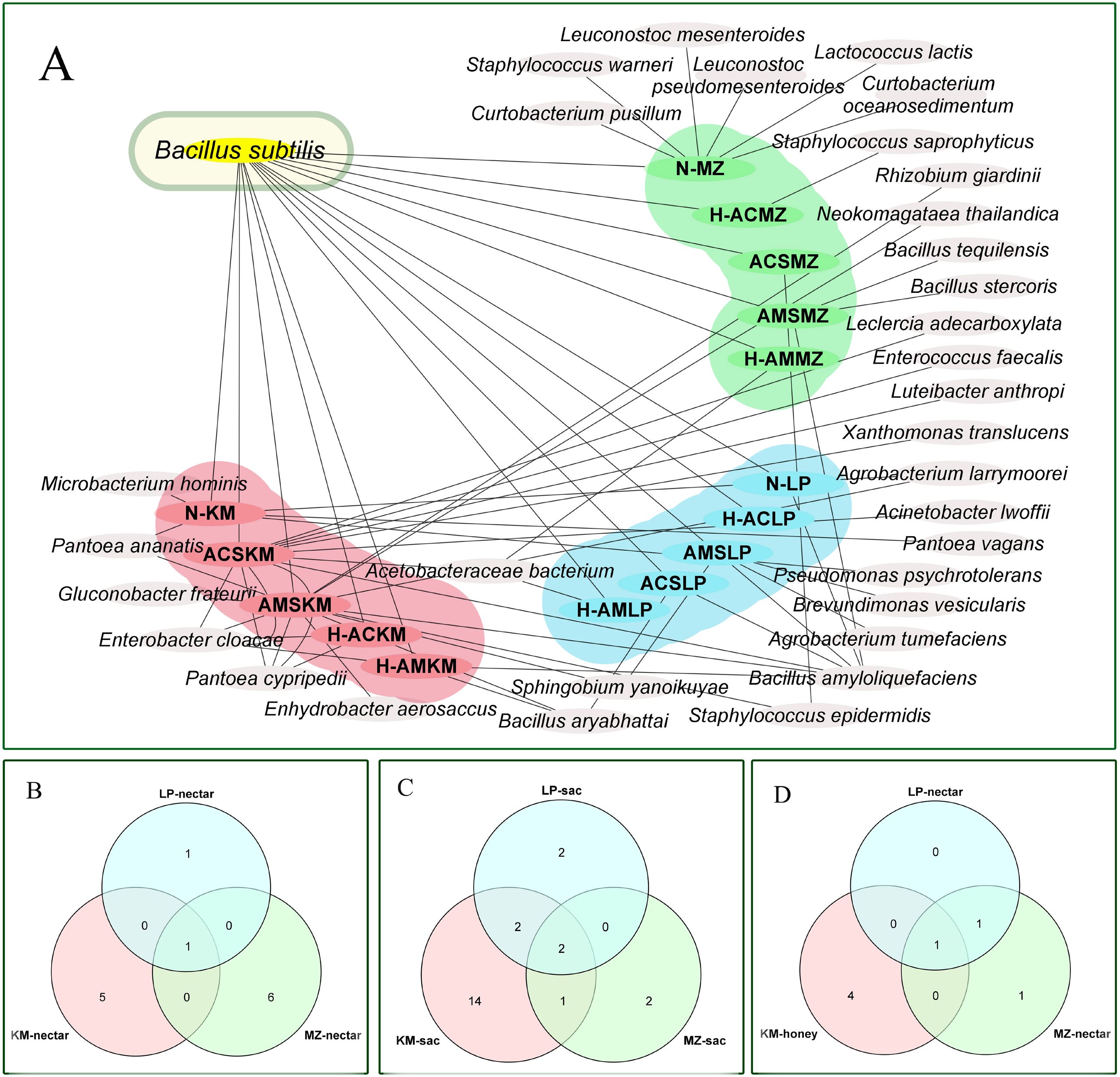
Figure 6. (A) Bacterial overlap map of three types of nectar source samples in three regions. (B) Venn diagram of bacteria in nectar from three regions. (C) Venn diagram of bacteria in honey sacs from three regions. (D) Venn diagram of bacteria in honey from three regions. (LP in blue, MZ in green, and KM in pink).
Figures 6B–D show one type of nectar, two types of honey, and one type of honey that were common to the three regions, respectively.
Six representative B. subtilis strains isolated from the honey sacs of A. cerana and A. mellifera during three winter flowering periods were tested in an inhibition of Hafnia alvei experiment, and the results showed that concentrations of 10-1–10-5 had a significant inhibitory effect on bacteria (P<0.001). The F values are in Figures 7A–F. In contrast to ACSKM3042 with concentrations of 10-1 and 10-2 (P=0.449) and AMSKM3025 with concentrations of 10-1 and 10-3(P=0.355), AMSLP3101, with concentrations of 10-2 and 10-3 (P=0.626), was not significant.
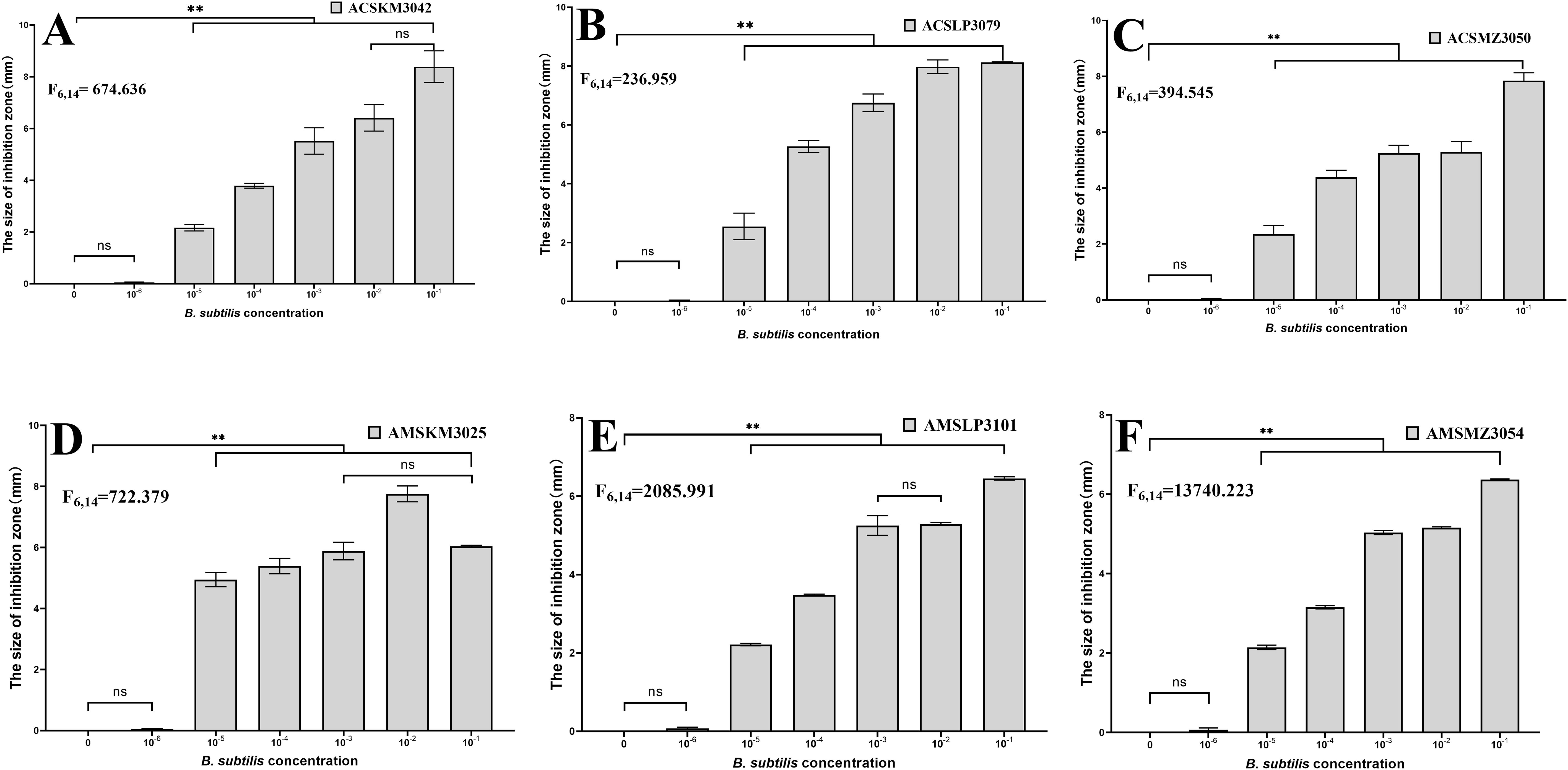
Figure 7. Inhibition of Hafnia alvei CMCC44102 by different dominant B. subtilis isolates from honey sacs. The six representative strains of B. subtilis from (A–F) are numbers ACSKM3042, ACSLP3079, ACSMZ3050, AMSKM3025, AMSLP3101, and AMSMZ3054 respectively. The horizontal axis is the H2O control and the concentration of B. subtilis, and the vertical axis is the inhibition distance. **, significant difference at p ≤ 0.01. ns, indicates no significant difference.
The present study offers a detailed examination of bacterial diversity across the honey production process, from nectar collection to honey sac content and finally to fresh honey, during the winter flowering season of Ar, Pc, and Br. Our findings reveal a dynamic and complex microbial landscape that varies significantly with the bee species and the floral source, highlighting the intricate interplay between bees, their gut microbiota, and the environment.
The isolated and identified bacteria all have a similarity of >97%. It is currently widely accepted that when the homology of the 16S rDNA sequence is higher than 97%, it indicates a relationship within the same genus, and when it is higher than 99%, it indicates a relationship within the same species (28). Therefore, the identified strains have a high degree of reliability at the species and genus levels.
Only 33 bacteria were isolated and identified from the nectar, honey sac, and honey samples which was a relatively low identification rate compared to most other bee gut microbes or honey microbes studies (17). This could simply be a result of culture-based isolation constraints when compared with sequence-only-based identification (Olofsson et al., 2008; 17). This result is also consistent with the low number of bacteria found in honey sacs or mid-gut; while a large number of bacteria can be identified in the hind-gut (29). Due to the low temperature in winter, the growth and reproduction of bacteria would be inhibited, and the number and types of bacteria in the environment would be reduced (30). Therefore, due to the winter environment, nectar, honey sac, and honey bacterial phases will also change accordingly.
The most abundant bacterial quantities were identified in the honey sacs, suggesting that this stage of honey production is a hotspot for microbial activity. This abundance may be attributed to the bees’ regurgitative actions, which facilitate microbial fermentation and enzymatic processing of nectar (18, 23, 31). The differences in bacterial diversity between A. cerana and A. mellifera could be reflective of their distinct foraging behaviors and physiological adaptations to the winter environment (9). A previous study found that A. cerana starts foraging earlier and at lower temperatures than A. mellifera, even in the cold winter in Yunnan (32).
The floral source had a pronounced effect on the bacterial composition within the honey sacs, with Ar, Pc, and Br each contributing to a unique microbial signature. This variability underscores the importance of plant chemistry in shaping the bee gut microbiota and raises questions about how these differences may impact honey quality and bee health. A. rugosa is known for its antifungal, antibacterial, carminative, and antipyretic properties, and has been used as a traditional Chinese herbal medicine (33). It was no surprise that the number of bacteria was lower in Br than in the other two winter flowering flowers. The number of bacteria was higher in the honey sacs of A. cerana than A. mellifera after the bees foraged on Br, which differed from the bees that foraged on Pc or Ar, and could reflect that sympatric A. cerana are more adapted to these nectar sources with more secondary metabolites (34, 35).
The study’s focus on winter flowering plants provides critical insights into bee microbiota during a period of limited floral diversity. As previous studies have shown that the core bacteria are from the Pseudomonas, Paenibacillus, Lonsdalea, Serratia, and Bacillus genera, which are mainly analyzed in seasons other than the winter season (31). Liu et al. (9) analyzed the intestinal bacteria of A. mellifera in the winter season, and found its core bacteria were Gilliamella, Bartonella, Snodgrassella, Lactobacillus, Frischella, Commensalibacter, and Bifidobacterium, which are stable intestinal bacteria (9). Agrobacterium sp. is a bacterium that belongs to the Rhizobiaceae family and is normally associated with plants, not bees. It is found in the honey bee gut and may have been accidentally introduced into the honey sac by environmental factors, including bacteria from the genus Agrobacterium, or by bees during collection. This does not mean that these bacteria play an important role in the bee gut. However, in this study, the bees were sampled inside of the hive overwinter and the bees remained in the hive without any foraging activity. Our results differ from these previous studies either because of the different seasons, or different foraging activities.
The ability of bees to adapt to and process these alternative food sources is vital for their survival and the maintenance of colony health in the off-season. Understanding these adaptations is essential for developing strategies to support bee populations in the face of environmental challenges, such as habitat loss and climate change (36, 37).
The ubiquitous presence of B. subtilis across all sample sources is a noteworthy finding. As a known producer of antimicrobial substances and enzymes, B. subtilis may play a key role in the honey production process, potentially contributing to the stabilization and preservation of honey (18, 38). The similar high osmotic stress of the nectar, honey sac, and fresh honey samples meant that few bacteria could survive in it. B. subtilis existed in all the samples, which is consistent with a previous finding that osmotolerant bacteria can survive and even be transmitted from flower nectar to honey (39, 40).
A previous study showed that B. subtilis isolated from both honey sample and bee gut had high antimicrobial activity against the pathogens Paenibacillus larvae and Ascosphaera apis (19) and also showed antagonistic activity against the chalkbrood pathogen and pesticide degradation (41, 42).
The results showed that the representative B. subtilis in the samples had an inhibitory effect on Hafnia alvei at concentrations above 10-5, which could improve the resistance to disease of overwintering bees in the absence of honey sources and weak colony strength by the dominant B. subtilis, and suggested that B. subtilis enhances resistance against H. alvei infection, thereby contributing to bee health during the winter.
This study provides compelling evidence that the honey production process is significantly influenced by the bee species and the floral sources they exploit during winter months. The observed variations in bacterial diversity and abundance have implications for our understanding of bee health and the quality of honey produced. The findings underscore the need for a nuanced approach to beekeeping practices that consider the health of bees and the biodiversity of their gut microbiota. The significance of this work lies in its potential to inform apicultural practices and conservation strategies. By revealing the impact of winter floral resources on bee gut microbiota, the study offers a basis for selecting floral sources that support robust bee colonies. Additionally, the insights into the microbial dynamics during honey production can guide efforts to improve honey quality and the overall health of honeybee populations. As bees are critical pollinators for both agricultural and natural ecosystems, this research contributes to bee health and biodiversity conservation efforts.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank, accessions PQ158547–PQ158549, PQ151644, PQ160366–PQ160426, PQ160480.
No specific permits were required for the described studies. Collections and experiments were made in the Yunnan Province, China. No special permits were required to be obtained for these locations. Studies involved the European honeybee (A. mellifera) and Easten honeybee (A. cerana), which are neither an endangered nor protected species. The apiaries are not privately owned or protected in any way.
MW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. WZ: Investigation, Visualization, Writing – original draft, Writing – review & editing. DZ: Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. JH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 32060779) to MW and JH.
We would like to thank Mr Shaofang Jian of Luoping County Agriculture Bureau and Mr Teng Li of Huize County Agriculture Bureau for their efforts and company during this study and for their help with beehives, dissection, and feeding. We thank Hongyuan Wang and Zuyan Duan for their assistance with DNA extraction, purification, and sequencing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Motta EVS, Moran NA. The honeybee microbiota and its impact on health and disease. Nat Rev Microbiol. (2024) 22:122–37. doi: 10.1038/s41579-023-00990-3
2. Engel P, Kwong WK, McFrederick Q, Anderson KE, Barribeau SM, Chandler JA, et al. The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. mBio. (2016) 7:e02164–02115. doi: 10.1128/mBio.02164-15
3. Steffan SA, Dharampal PS, Kueneman JG, Keller A, Argueta-Guzmán MP, McFrederick QS, et al. Microbes, the ‘silent third partners’ of bee–angiosperm mutualisms. Trends Ecol Evol. (2024) 39:65–77. doi: 10.1016/j.tree.2023.09.001
4. Ellegaard KM, Engel P. Genomic diversity landscape of the honey bee gut microbiota. Nat Commun. (2019) 10:1–13. doi: 10.1038/s41467-019-08303-0
5. Wu J, Lang H, Mu X, Zhang Z, Su Q, Hu X, et al. Honey bee genetics shape the strain-level structure of gut microbiota in social transmission. Microbiome. (2021) 9:1–19. doi: 10.1186/s40168-021-01174-y
6. Yun JH, Jung MJ, Kim PS, Bae JW. Social status shapes the bacterial and fungal gut communities of the honey bee. Sci Rep. (2018) 8:2019. doi: 10.1038/s41598-018-19860-7
7. Pozo MaríaI, Mariën T, Kemenade Gv, Wäckers F, Jacquemyn &H. Effects of pollen and nectar inoculation by yeasts, bacteria or both on bumblebee colony development. Oecologia. (2021) 195:689–703. doi: 10.1007/s00442-021-04872-4
8. Nicolson SW. Bee food: the chemistry and nutritional value of nectar, pollen and mixtures of the two. Afr Zoology. (2015) 46:197–204. doi: 10.1080/15627020.2011.11407495
9. Liu P, Zhu Y, Ye L, Shi T, Li L, Cao H, et al. Overwintering honeybees maintained dynamic and stable intestinal bacteria. Sci Rep. (2021) 11:22233. doi: 10.1038/s41598-021-01204-7
10. Castelli L, Branchiccela Belén, Romero Héctor, Zunino P, Antúnez &K. Seasonal dynamics of the honey bee gut microbiota in colonies under subtropical climate. Microbial Ecol. (2022) 83:492–500. doi: 10.1007/s00248-021-01756-1
11. Rajagopalan K, DeGrandi–Hofman G, Pruett M, Jones VP, Corby–Harris V, Pireaud J, et al. Warmer autumns and winters could reduce honey bee overwintering survival with potential risks for pollination services. Sci. Rep. (2024) 14:5410. doi: 10.1038/s41598-024-55327-8
12. Nicolson SW, Human H, Pirk CWW. Honey bees save energy in honey processing by dehydrating nectar before returning to the nest. Sci Rep. (2022) 12:16224. doi: 10.1038/s41598-022-20626-5
13. Wueppenhorst K, Alkassab AT, Beims H, Bischoff G, Ernst U, Friedrich E, et al. Nurse honey bees filter fungicide residues to maintain larval health. Curr Biol. (2024) 34:5570–5577.e5511. doi: 10.1016/j.cub.2024.10.008
14. Tiwari P, Tiwari J, Ballabha R. Prunus cerasoides D. Don (Himalayan wild cherry): A boon to hill beekeepers in garhwal Himalaya. Nat Sci. (2009) 7:21–3.
15. Wang M, Xu H, Li Y, Yang L, He S. Changes of amylase value in honey making processing period of Elsholtzia ciliate. J Bee. (2014) 12:11–2.
16. Snowdon JA, Cliver D. Microorganisms in honey. Int J Food Microbiol. (1996) 31:1–26. doi: 10.1016/0168-1605(96)00970-1
17. Xiong ZR, Sogin JH, Worobo RW. Microbiome analysis of raw honey reveals important factors influencing the bacterial and fungal communities. Front Microbiol. (2023) 13:1099522. doi: 10.3389/fmicb.2022.1099522
18. Wang M, Zhao W, Xu H, Wang Z, He S. Bacillus in the guts of honey bees (Apis mellifera; Hymenoptera: Apidae) mediate changes in amylase values. Eur J Entomology. (2015) 112:619–24. doi: 10.14411/eje.2015.095
19. Sabaté DC, Carrillo L, Carina Audisio M. Inhibition of Paenibacillus larvae and Ascosphaera apis by Bacillus subtilis isolated from honeybee gut and honey samples. Res Microbiol. (2009) 160:193–9. doi: 10.1016/j.resmic.2009.03.002
20. Todorov SD, Alves MV, Bueno GCA, Alves VF, Ivanova IV. Bee-associated beneficial microbes—Importance for bees and for humans. Insects. (2024) 15:430. doi: 10.3390/insects15060430
21. Qiu Jy, Zhu X, Zhong Cy, Luo Yy, Hou P, Fan Y, et al. Isolation, identification and drug sensitive test for Hafnia alvei from Apis cerana in Guizhou. China Anim Husbandry Veterinary Med. (2018) 45:1050–8.
22. Lang H, Duan H, Wang J, Zhang W, Guo J, Zhang X, et al. Specific strains of honeybee gut Lactobacillus stimulate host immune system to protect against pathogenic Hafnia alvei. Microbiol Spectr. (2022) 10:e01896–01821. doi: 10.1128/spectrum.01896-21
23. Olofsson T, Vásquez A. Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr Microbiol. (2008) 57:356–63. doi: 10.1007/s00284-008-9202-0
24. Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int J systematic evolutionary Microbiol. (2013) 63:2008–18. doi: 10.1099/ijs.0.044875-0
25. Ibal JC, Pham HQ, Park CE, Shin J-H. Information about variations in multiple copies of bacterial 16S rRNA genes may aid in species identification. PloS One. (2019) 14:e0212090. doi: 10.1371/journal.pone.0212090
26. Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. (1998) 64:795–9. doi: 10.1128/AEM.64.2.795-799.1998
27. Zangeneh M, Khorrami S, Khaleghi M. Bacteriostatic activity and partial characterization of the bacteriocin produced by L. plantarum sp. isolated from traditional sourdough. Food Sci Nutr. (2020) 8:6023–30. doi: 10.1002/fsn3.1890
28. Johnson JS, Spakowicz DJ, Hong B-Y, Petersen LM, Demkowicz P, Chen L, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. (2019) 10:5029. doi: 10.1038/s41467-019-13036-1
29. Martinson VG, Moy J, Moran NA. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol. (2012) 78:2830–40. doi: 10.1128/AEM.07810-11
30. Fang J, Wei S, Shi G, Cheng Y, Zhang X, Zhang F, et al. Potential effects of temperature levels on soil bacterial community structure. E3S Web Conferences. (2021) 292:01008. doi: 10.1051/e3sconf/202129201008
31. Mundo MA, Xiong ZR, Galasong Y, Manns DC, Seeley TD, Vegdahl AC, et al. Diversity, antimicrobial production and seasonal variation of honey bee microbiota isolated from the honey stomachs of the domestic honey bee, Apis mellifera. Front Sustain Food System. (2023) 6:931363. doi: 10.3389/fsufs.2022.931363
32. Tan K, Yang S, Wang Z, Radloff SE, Oldroyd BP. Differences in foraging and broodnest temperature in the honey bees Apis cerana and A. mellifera. Apidologie. (2012) 43:618–23. doi: 10.1007/s13592-012-0136-y
33. Gong H, Li S, He L, Kasimu R. Microscopic identification and in vitrok activity of Agastache rugosa (Fisch. et Mey) from Xinjiang, China. BMC Complementary Altern Med. (2017) 17:1–6. doi: 10.1186/s12906-017-1605-7
34. Zhang J, Wang Z, Klett K, Qu Y, Tan K. Higher toxin tolerance to triptolide, a terpenoid foraged by a sympatric honeybee. J Insect Physiol. (2022) 137:104358. doi: 10.1016/j.jinsphys.2022.104358
35. Zhang J, Wang Z, Wen P, Qu Y, Tan K, C.Nieh J. The reluctant visitor: a terpenoid in toxic nectar can reduce olfactory learning and memory in Asian honey bees. J Exp Biol. (2018) 221:jeb.168344. doi: 10.1242/jeb.168344
36. Kazenel MR, Wright KW, Griswold T, Whitney KD, Rudgers JA. Heat and desiccaton tolerances predict bee abundance under climate change. Nature. (2024) 628:342–7. doi: 10.1038/s41586-024-07241-2
37. Orr MC, Hughes AC, Chesters D, Pickering J, Zhu CD, Ascher JS. Global patterns and drivers of bee distribution. Curr Biol. (2021) 31:451–458.e454. doi: 10.1016/j.cub.2020.10.053
38. Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. (2007) 318:283–7. doi: 10.1126/science.1146498
39. A´lvarez-Pe´rez S, Herrera CM, Vega C. d. Zooming-in on floral nectar a first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol Ecol. (2012) 80:591–602. doi: 10.1111/j.1574-6941.2012.01329.x
40. Fridman S, Izhaki I, Gerchman Y. Bacterial communities in floral nectar. Environ Microbiol Rep. (2012) 4:97–104. doi: 10.1111/j.1758-2229.2011.00309.x
41. Roy N, Moon S, Kim C, Kim J-M, Lee K-S, Shin Y, et al. Probiotic potential of Bacillus subtilis strain I3: antagonistic activity against chalkbrood pathogen and pesticide degradation for enhancing honeybee health. Probiotics Antimicrobial Proteins. (2024) 17:51–61. doi: 10.1007/s12602-024-10248-w
Keywords: honey sac, winter flowering flora, Apis cerana, Apis mellifera, Bacillus subtilis, bacteriostasis
Citation: Wang M, Zhao W, Zhou D and Huang J (2025) Bacterial diversity in the honey sac during bee foraging on winter-flowering flora and dominant Bacillus subtilis inhibits Hafnia alvei. Front. Insect Sci. 5:1555434. doi: 10.3389/finsc.2025.1555434
Received: 04 January 2025; Accepted: 13 February 2025;
Published: 11 March 2025.
Edited by:
Maristella Mastore, University of Insubria, ItalyReviewed by:
Zhenguo Liu, Shandong Agricultural University, ChinaCopyright © 2025 Wang, Zhao, Zhou and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Huang, MjAxMjA2OEB5bmF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.