
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Insect Sci. , 07 April 2025
Sec. Invasive Insect Species
Volume 5 - 2025 | https://doi.org/10.3389/finsc.2025.1527130
The purpose of this research was to examine the Solenopsis invicta venom 2 protein and transcript among Solenopsis invicta fire ants exhibiting an unusual response to antibody interrogation of this protein. Sequence and phylogenetic analyses combined with Western blotting and lateral flow immunoassay were employed to examine the venom proteins from these fire ants. Genotypic variation was discovered in the Solenopsis invicta venom 2 gene. Many of these unique genotypes exhibited strong identity to the Solenopsis richteri venom 2 ortholog from the congener, Solenopsis richteri. Phylogenetic analysis of these sequences revealed a significant evolutionary relationship with Solenopsis richteri despite being obtained from Solenopsis invicta. A unique, truncated, Solenopsis invicta venom 2-like protein was also discovered in these colonies originating from a unique locus on chromosome 10 where multiple duplication events have apparently copied this gene. These results suggest the possible presence of a cryptic species.
Solenopsis richteri Forel and Solenopsis invicta Buren were introduced accidentally from South America into the United States at the Mobile, Alabama, port area around 1918 and 1933, respectively (1). S. richteri is a more temperate species from southern Argentina and S. invicta has subtropical origins in Brazil and northern Argentina. Interestingly, the two species intermate and produce a reproductively viable hybrid (2, 3). Since their introductions, S. invicta has greatly expanded its range to include areas from Florida to Virginia and west into California (4), and the species have segregated roughly based on their cold tolerance (S. richteri > hybrid > S. invicta). Currently, S. richteri is largely confined to a contiguous region in northern Mississippi (5), Alabama (6), and Tennessee (7). The hybrid occupies a large band between S. richteri to the north and S. invicta to the south and is currently the dominant form in Tennessee (7, 8; Oliver et al., In Press)1.
Both species and the hybrid possess a potent venom that poses a significant human health risk (9, 10). Among those stung, 5% require medical attention and 2% exhibit serious allergic responses, including anaphylaxis (9, 11, 12). The venom is comprised of a mix of alkaloids (>95%) and more than 46 proteins (0.01%) (13, 14). Four of these proteins have been studied extensively and are well characterized, namely Solenopsis (invicta or richteri depending on the species) venom 1 protein, venom 2 protein, venom 3 protein, and venom 4 protein (3, 15) with the remaining proteins playing undefined roles (13). Solenopsis venom protein 2 comprises the largest proportion (~67%) of these four proteins (10) and each of the parent invasive fire ant species, S. invicta and S. richteri, expresses an ortholog with 80% identity between the protein sequences (15, 16).
Lateral flow immunoassay tests (InvictDetect™ and InvictDetect Plus™, Agdia, Inc., Elkhart, IN) were developed and commercialized for rapid field identification of S. invicta, S. richteri, and their hybrid, targeting the Solenopsis venom 2 protein (16, 17). Field evaluations with InvictDetect™ and InvictDetect Plus™ revealed false negative responses among a small number of S. invicta colony samples. The objective was to examine the Solenopsis invicta venom 2 protein (henceforth Soli2) in these false negative-yielding S. invicta colonies to understand the reason for the failed responses.
Field evaluations of S. invicta fire ants with InvictDetect™ or InvictDetect Plus™ immunostrips were conducted in Gainesville, Florida, and Atlanta, Georgia, from 2015 through to 2020. Colonies yielding three consecutive false negative InvictDetect™ or InvictDetect Plus™ immunostrip responses were taken to the laboratory for additional analysis, including venom purification and Soli2 transcript sequencing (Table 1). Two normally responding (to InvictDetect™ or InvictDetect Plus™) ant colonies were included and served as controls, Soli_WtA and Soli_WtB (Table 1). These are referred to as “wildtype” S. invicta. Venom was partially purified using a modified method of Paterson Fox et al. (18). Briefly, 5 g of worker ants were placed into a 50 ml conical tube containing 5 µl of 12.1 M HCl on the inside cap of the tube for 15 minutes. The acid vapors irritated the ants, inducing the release of their venom. Deionized water (5 ml) and hexane (5 ml) were added to the tube to extract and separate the venom protein components from the alkaloids. The tube was rocked for 5 minutes, centrifuged at 3,000 RPM, and the hexane phase was removed and discarded. Fresh hexane (5 ml) was added to the tube, and the process was repeated two additional times. The aqueous phase was frozen and lyophilized overnight to dry.
Soli2 transcripts were amplified by reverse transcription polymerase chain reaction (RT-PCR) with oligonucleotide primers specific to the conserved 5’ and 3’ termini of the mature Soli2 transcripts (19). Total RNA was extracted from a pooled group (n = 10) of worker ants using the TRIzol method (Thermo Fisher Scientific) according to the manufacturer’s directions. RNA was treated with DNAse and used as a template for two-step RT-PCR.
Briefly, 0.5 µl (10-50 ng) of the DNAse-treated total RNA was mixed with 10 mM dNTPs and 1 µM reverse oligonucleotide primer CS5 (5’ TTATATGCACAATATTTTATTGTTAACCCAACAC), heated to 65°C for 5 minutes, and then placed on ice for 1 minute. To this mixture, 5x first strand buffer and Superscript Reverse Transcriptase (RT, Life Technologies) were added, and the reaction was incubated at 55°C for 30 minutes. The RT reaction was terminated by incubation at 70°C for 15 minutes. The cDNA was used as a template for PCR with oligonucleotide primers 4CS (5’ GTTAAATGAATAAAGTCACTCATACAACTTCTCTA) and 5CS under the following cycling conditions: 94°C for 2 minutes; 35 cycles of 94°C for 15 seconds and 55°C for 15 seconds; and 68°C for 1.5 minutes and 1 cycle of 68°C for 5 minutes. Amplicons were purified on 0.75% Agarose gels and visualized with SYBR Safe (Life Technologies). Amplicons were ligated into the pCR4-TOPO vector (Life Technologies) and transformed into TOP10 E. coli competent cells (Life Technologies). Bacterial colonies were picked and inoculated in Luria broth with ampicillin (75µg/ml), incubated at 37°C while shaking at 225 rpm, and positively determined for the presence of the insert by PCR with the original oligonucleotide primers.
Insert-positive plasmid DNA was purified using the QIAprep Miniprep Spin kit (Qiagen) and clones (a minimum of 5 per colony) were sequenced by the Sanger method. Consensus sequences for each clone were used in subsequent analyses.
5’ Random amplification of cDNA ends (RACE) was conducted on RNA isolated from several colonies expressing the Soli2 truncated venom transcript to confirm the size of the transcript. cDNA was synthesized with a gene-specific oligonucleotide primer (5CS), and the PCR was subsequently completed with a nested gene-specific primer (8CS, 5’ GGCCTTGAGTCTCTCTATCTACACATCTAGAA) and the GeneRacer Abridged Anchor Primer. The anticipated size of the amplicon was compared with one empirically derived.
Colony social form (monogyne and polygyne) was determined by detection of the Gp-9 alleles as previously described (20).
The mitochondrial cytochrome oxidase I gene from a subset of the S. invicta colonies (Soli_3, Soli_4, Soli_7, Soli_10, Soli_11, and Soli_WtA) was amplified, cloned, and sequenced. The PCR was conducted using total DNA extracted from a pooled group of 10 worker ants as a template. The universal oligonucleotide primers LCO1490 (5’ggtcaacaaatcataaagatattgg) and HCO2198 (5’taaacttcagggtgaccaaaaaaatca) reported by Folmer et al. (21) were used. Cloning and Sanger sequencing were completed as described above.
The mitochondrial genomes of colonies Soli_3, Soli_7, Soli_WtA, and Soli_WtB were sequenced using nanopore R10 sequencing chemistry. Total DNA from each colony was end prepped and repaired using NEBNext FFPE Repair Mix and Ultra2 End Prep Mix (New England Biolabs) following the genomic DNA ligation sequencing protocol (SQK-NBD114.96, Protocol: NBE_0171_v114_revO_15Sep2022, Oxford Nanopore Technologies, Oxford, UK). After the initial end prep, subsequent steps were conducted using the amplicon sequencing protocol (SQK-NBD114.96, Protocol: NBA_9170_v144_revM_15Sep2022, Oxford Nanopore Technologies). Barcoded samples were sequenced on a MinION Mk1B with an R10.4.1 flow cell (Oxford Nanopore Technologies) using MinKNOW software. The sequencing was conducted for 72 hr. Reads were binned and basecalled automatically by the MinKNOW software using a minimum quality cutoff of 10. Raw sequencing data for each sample has been deposited in NCBI under BioProject: PRJNA1179381.
Assembly and indexing of each mitochondrial genome were conducted by mapping to the S. invicta mitochondrial genome (NCBI Accession: NC014672.2) using Minimap2 (22). The resulting binary assembly files were displayed in IGV to examine the coverage and the consensus option was used to generate a final consensus for each sample (23).
To determine whether the truncated transcript identified by molecular analysis was being translated into protein, Western analysis was conducted against the purified worker ant venom from ant colonies Soli_4, Soli_11, Soli_WtA, and Soli_WtB. Venom proteins were applied (7.5 µg) and separated on a 4%–20% gradient SDS-PAGE gel. Gel-separated proteins were electroblotted onto PVDF membranes and blocked in TBS (tris buffered saline; 20 mM Tris-HCl, 500 mM NaCl, pH 7.5) + 1% BSA (bovine serum albumin) for 2 hours. Blots were probed with monoclonal antibody (mAb) 6A8D7 [prepared to a 15 amino acid sequence (NKELKIIRKDVAECL) corresponding to positions 2–16 of the Soli2 venom protein], mAb 3H6B9 [corresponding to a fourteen amino acid sequence (IEAQRVLRKDIAEC) at positions 2–15 of the Soli2 venom protein] (16), or mAb 7D7F7 [corresponding to a 17 amino acid sequence (NPDPAVIKERSMKMCTK) at positions 48–64 of the Soli2 venom protein]. Monoclonal antibodies were produced by ProMab Biotechnologies Incorporated (Richmond, CA) using the hybridoma method. Monoclonal antibody in concentrations normalized to S. invicta (6A8D7 = 1.6 µg/ml; 3H6B9 = 1.8 µg/ml; 7D7F7 = 0.5 µg/ml) was added to the TBS + 1% BSA solution containing the blot for 2 hours at room temperature with shaking (40 rpm). Membranes were rinsed twice with TTBS (TBS + 0.05% Tween 20), probed with secondary antibody (0.1 μg/ml), goat anti-mouse conjugated with alkaline phosphatase (Sigma, St. Louis, MO) for 1 hour, and rinsed twice with TTBS. The membrane was incubated for several minutes with BCIP (5-bromo-4-chloro-3-indolyl-phosphate) and NBT (nitro blue tetrazolium) for the colorimetric detection of alkaline phosphatase activity. Once bands were detected on the blot, the reaction was terminated by rinsing the membrane with deionized water.
Worker ants from S. invicta colonies Soli_9, Soli_11, and Soli_13 were qualitatively analyzed for their venom alkaloids and compared with a standard S. invicta colony. Chromatograms were obtained from an Agilent Intuvo 9000 GC system (Santa Clara, CA) equipped with an HP-5 MS ultra-inert nonpolar column, 30 m × 0.25 m i.d. column, coupled to a 5977 B mass spectral detector and a MassHunter Data Acquisition Workstation version 10.0.368 (Santa Clara, CA). The injector temperature was set at 250°C. The oven temperature was programmed at 40°C for 2 min and then to 285°C at 5°C/min, followed by a 10 min hold at 285°C.
The piperidine alkaloids were identified by their base peak fragment at m/z 98. The alkaloid composition of S. invicta has been known for decades due to its importance as an invasive species throughout the southeastern U.S.A. The four major S. invicta worker alkaloids are trans-piperidines: 2-methyl-6-tridecyl-piperidine (compound 2), 2-methyl-6-tridecenyl-piperidine (compound 3), 2-methyl-6-pentadecyl-piperidine (compound 4), and 2-methyl-6-pentadecenyl-piperidine (compound 5). In addition, there are minor amounts of 2-methyl-6-undecyl-piperidine (compound 1) and 2-methyl-6-heptadecenyl-piperidine (compound 6). The component peak heights are proportional to the relative amounts of each component. The variation in peak ratios in the samples was within normal observed variation in S. invicta samples.
Evolutionary relationships among Soli2 venom protein sequences and S. invicta cytochrome oxidase I sequences were inferred using the maximum likelihood method and JTT matrix-based model (24). Phylogenetic trees with the highest log likelihood were used for the analysis. The percentage of trees in which the associated sequences clustered together was calculated with 500 bootstrap iterations. The initial trees for the heuristic search were obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model, and then selecting the topology with the superior log likelihood value. Trees are drawn to scale, with branch lengths measured in the number of substitutions per site. Fourteen Soli2 sequences with a total of 138 positions comprised the final dataset for the Soli2 venom proteins and eight sequences with a total of 449 positions comprised the final dataset for cytochrome oxidase I. Evolutionary analyses were conducted in MEGA11 (25). A similar evolutionary analysis was conducted on the mitochondrial genome sequences but with the use of the Kimura Two Parameter model on a matrix of 5 sequences by 15,549 positions (26).
Soli2 venom protein transcript sequences were aligned against the Solenopsis invicta reference genome (UNIL_SINV_3.0, GCA_016802725.1) using the Splign alignment tool (27) to identify the locus, expressed (exons), and intervening sequences (introns). Locus assignment was made based on the highest exon identity observed for a given sequence. Introns were expressed as the number of nucleotides comprising the sequence.
The open reading frames (ORFs) predicted for the Soli2 and Soli2-like translated sequences were aligned using ClustalW in the Vector NTI Advance 11.5 (AlignX) software package.
The sequences obtained throughout the study were deposited into GenBank. Accession numbers assigned to Solenopsis venom 2 proteins from S. invicta, S. richteri, and S. geminata are listed in Table 2. Cytochrome oxidase sequence accession numbers include Si_COI_WtA (PQ038572); Si_COI_3 (PQ038575); Si_COI_4 (PQ038574); Si_COI_7 (PQ038573); Si_COI_10 (PQ038577); and Si_COI_11 (PQ038576). Nanopore sequences were deposited into GenBank under Bioproject PRJNA1179381 (Soli_WtA: SAMN44490061; Soli_WtB: SAMN44490062; Soli_3: SAMN44490063; Soli_7: SAMN44490064).
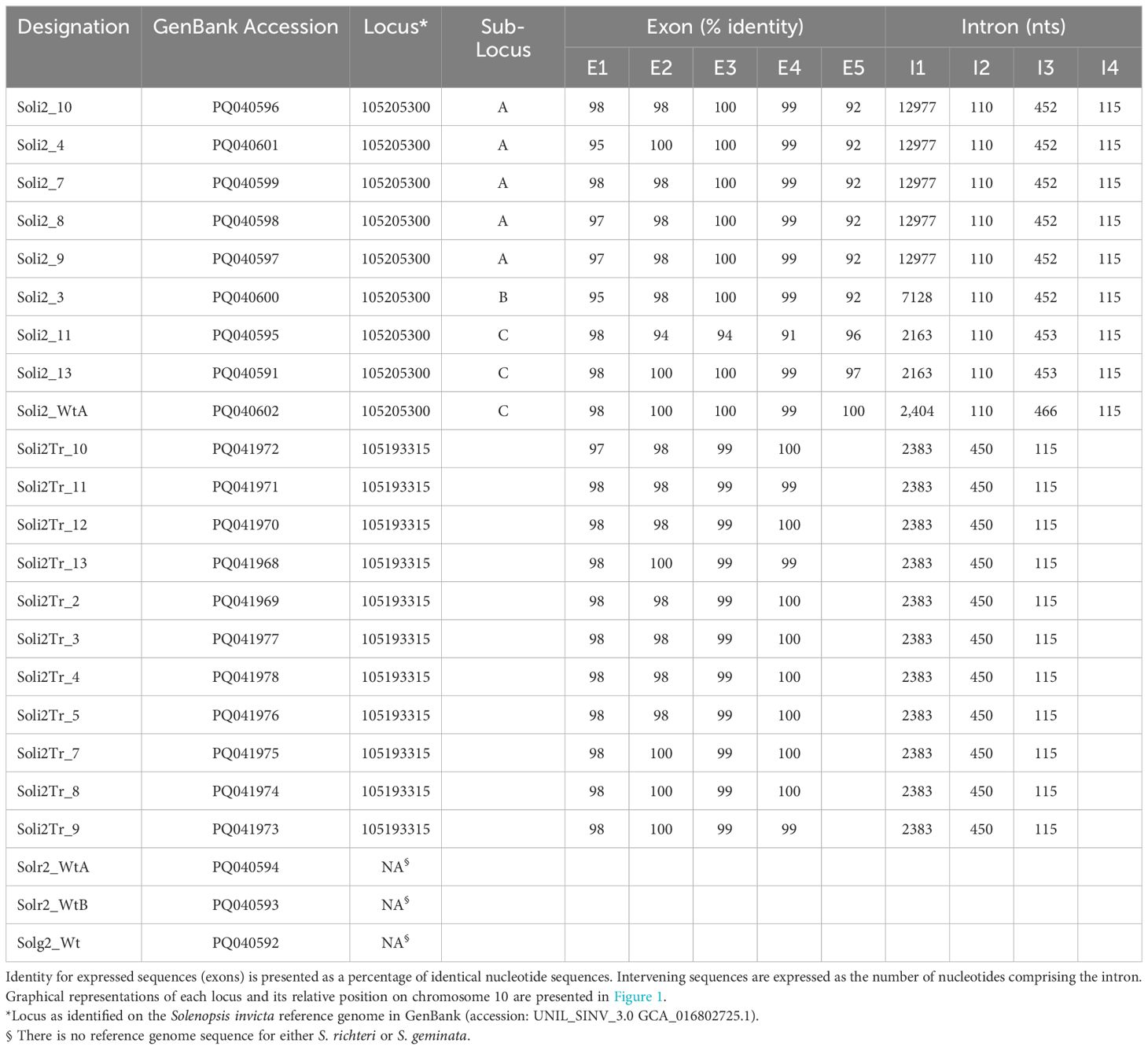
Table 2. Solenopsis invicta venom 2 protein loci identified by alignment of the transcript sequence against the Solenopsis invicta reference genome (UNIL_SINV_3.0, GCA_016802725.1) using the Splign alignment tool (27).
Among the 861 evaluations of verified Solenopsis invicta colonies with the InvictDetect™ and/or InvictDetect Plus™ lateral flow immunoassay kits, 12 false negatives (1.39%) for S. invicta occurred that did not resolve despite repeated evaluations and increasing the number of pooled worker ants used in the assays. Worker ants from 11 of these colonies were collected and evaluated by RT-PCR for the presence of the Soli2 venom protein transcript. Eight colonies (i.e., Soli_3, _4, _7, _8, _9, _10, _11, _and 13) produced two amplicons [one of 496 nts (short transcript) and one of 596 nts (long transcript)]. Three colonies (Soli_2, _5, and _12) produced a single 496 nt amplicon (short transcript). BLAST analysis (28) of the short and long amplicons revealed similarity to the Soli2 gene (Genbank accession L09560.1). When the nucleotide sequences were aligned to the Solenopsis invicta reference genome, they mapped to a 43,627-nucleotide region of the minus strand of chromosome 10 where Soli2 resides, specifically the region encompassed by nucleotides 9,600,319 to 9,643,946 (Figure 1). Within this region, multiple copies and variations of Soli2 are present; six Soli2 loci occupy this region of chromosome 10 (Figure 1), including loci 105206474, 120358770, 113003936, 105193335, 10520533, and 105193315. The long transcripts (596 nt) all mapped to locus 105205300 (Table 2), but the exons are spread across this locus in multiple copies and locations. Thus, this locus can be thought of as being comprised of three “sub” loci, which we refer to as A, B, and C (Figure 1). Five exons comprise the open reading frame (ORF) of the long transcript. Sub-loci A and B share positions for exons 2, 3, 4, and 5, but multiple copies of exon 1 are found upstream. Sub-locus C is found near the 5′ end of the locus, and exons 2, 3, 4, and 5 have different positions and sequences from sub-loci A and B. Another difference between the transcripts generated at each sub-locus was the size of intron 1, which ranged from 2,163 to 12,977 nts (Table 2). The remaining introns (2, 3, and 4) were consistent in size (Table 1). The original published nucleotide sequence for Soli2 (Genbank accession L09560.1) was identical to the transcript from colony Soli_WtA (Genbank accession PQ040591), which was collected in 2015 (Table 1) and produced a positive response with InvictDetect™.
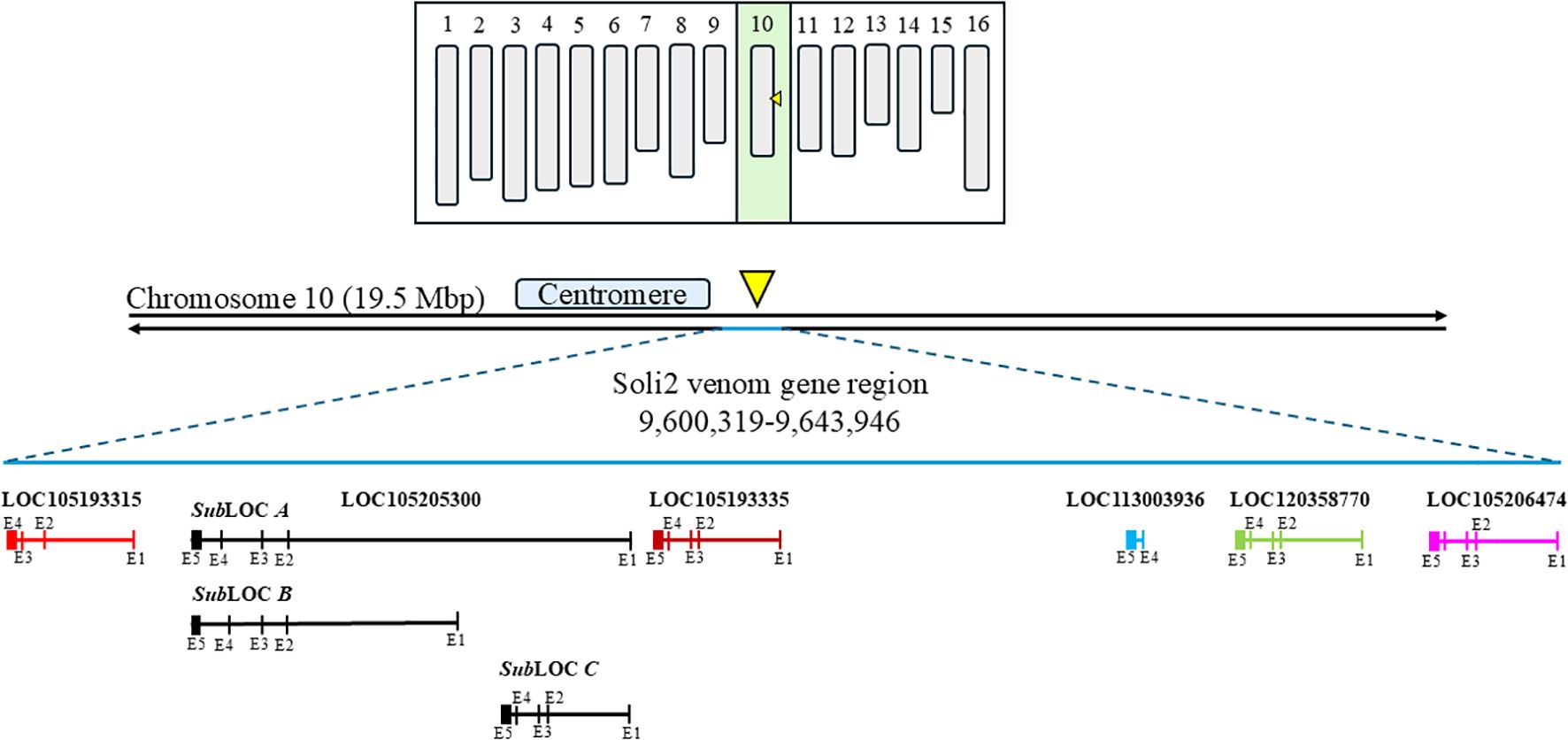
Figure 1. Diagrammatic karyotype of Solenopsis invicta identifying the contiguous region on chromosome 10 where Solenopsis invicta venom 2 protein genes reside (yellow arrowhead). The two black horizontal lines represent chromosome 10. The approximate location of the centromere is shown and the Solenopsis invicta venom 2 protein gene region and loci are identified in bold font. Solenopsis invicta venom 2 protein genes are represented by various colored horizontal lines. Exons are approximated and illustrated by vertical lines through the identified locus.
The short transcripts (496 nts) all mapped to locus 105193315 and were comprised of only four exons. The sequences of exons 1, 2, 3, and 4 of locus 105193315 exhibited sequence similarity with exons 1, 3, 4, and 5 of locus 105205300, respectively. The sequence of exon 2 of locus 105205300 is absent in locus 105193315. Loss of exon 2 (100 nts) in the duplicated locus 105193315 results in a shift of the start codon of the nascent transcript it generates, yielding a 5′-proximally truncated ORF [as compared with the ORF generated from the other (5-exon-containing) Soli2 paralogs].
Amino acid sequences from the long transcript group of ant colonies that failed to be detected with the Invictdetect™ kit [596 nts, 138 amino acids (aa)] were unique and distinct from the Soli2 venom protein published previously (Genbank accession L09560.1 and P35775) and from our S. invicta control colonies (Soli_WtA and Soli_WtB) (29). Alignments of the translated ORF from the long transcript show significant divergence from Soli2 (P35775) in sequence identity (Figure 2), which is illustrated in the phylogenetic analysis (Figure 3). Two sequences, Soli2_11 and Soli2_13, from ant colonies Soli_11 and Soli_13, respectively, associated most closely with the original Soli2 sequence (P35775). Conversely, sequences Soli2_3, _4, _7, _8, _9, and _10 formed a unique clade most closely aligned with Solenopsis richteri venom 2 protein (Solr2; GenBank accession P35776) from S. richteri (Figure 3). Notice that the region near the N-terminus of the venom 2 protein sequences is identical to Solr2 (i.e., IEAQRVL) and distinct from Soli2 (NKELKII). Sequences from S. invicta colonies Soli2_3, _4, _7, _8, _9, and _10 mapped to sub-locus A and B (LOC105205300), while those with the invicta-like NKELKII N-proximal sequence mapped to sub-locus C (Figure 2). This divergence was mirrored in a phylogenetic analysis of the cytochrome oxidase I sequences from a limited number of these colonies (Figure 4). Notice that cytochrome oxidase I sequences from colonies Soli_3, _4, _7, and _10 (richteri-like sequence IEAQRVL) similarly formed a distinct grouping separate from the S. invicta (DQ353293) cytochrome oxidase I sequence.
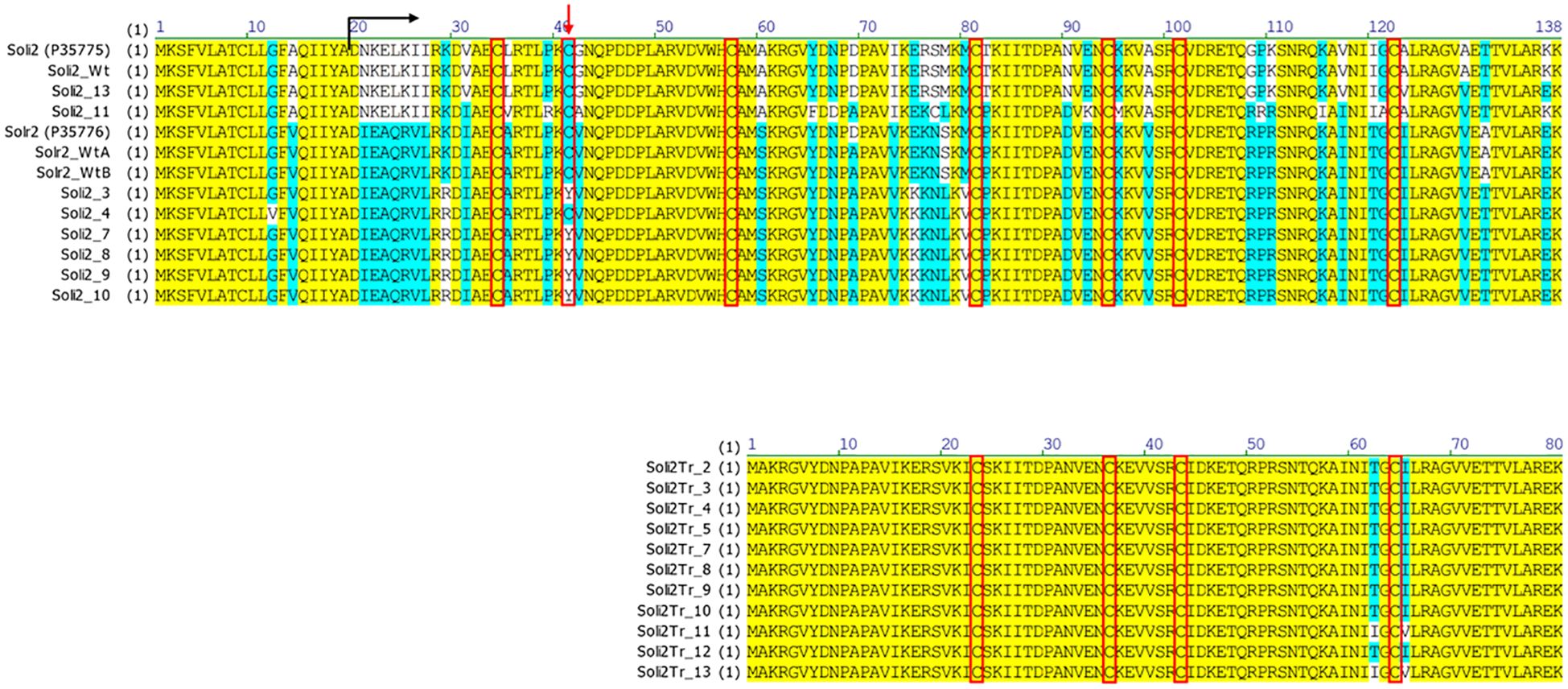
Figure 2. Predicted protein sequences of long (upper) and short (lower) nascent transcripts from S. invicta colonies failing an InvictDetect™ assay {compared with normal, or wildtype, Solenopsis invicta venom 2 protein [Soli2 (P35775) and Soli2_Wt] and Solenopsis richteri venom 2 protein [Solr2 (P35776), Solr2_WtA, and Solr2_WtB]}. The black arrow indicates the position of the mature protein and the red arrow indicates the position of the cysteine residue involved in the dimerization of the monomers. Red boxes call out the cysteine positions.
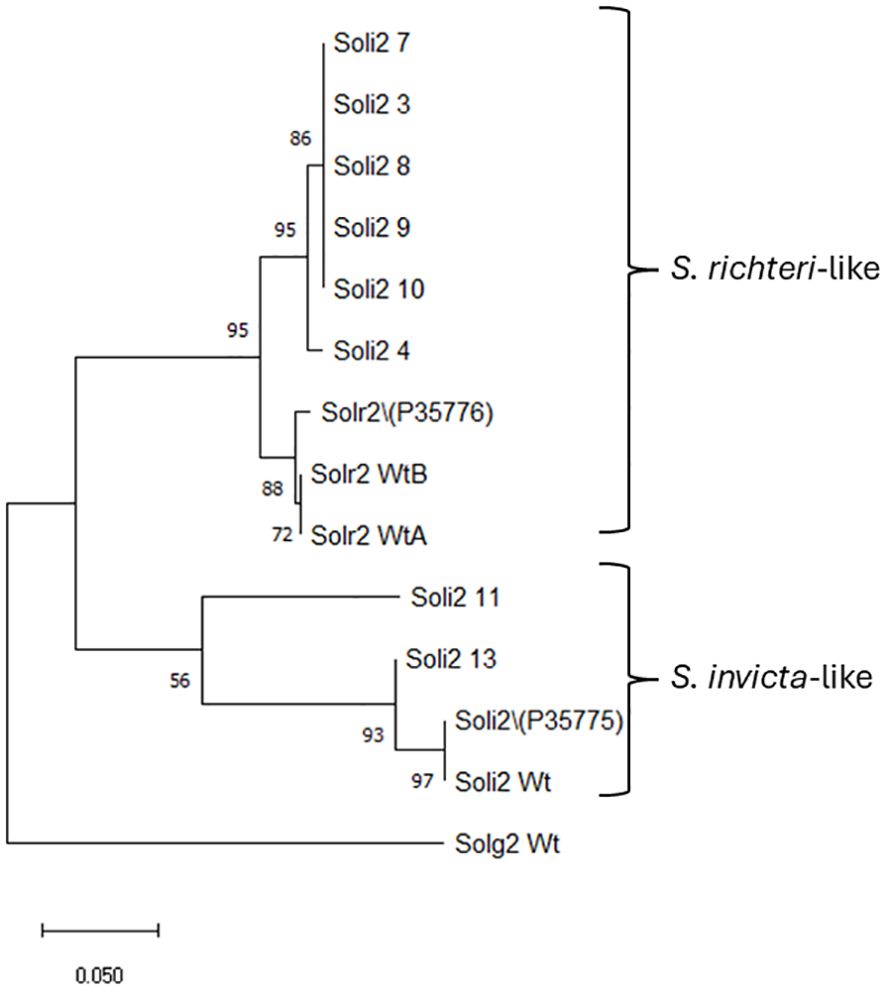
Figure 3. Phylogenetic analysis of Solenopsis venom 2 protein sequences (long sequence) by the maximum likelihood method. Trees are drawn to scale, with branch lengths measured in the number of substitutions per site, 500 bootstrap iterations, and branch support provided as a percentage. Fourteen Soli2 sequences with a total of 138 positions comprised the final dataset for the Soli2 venom proteins. Sample names are provided in Table 2.
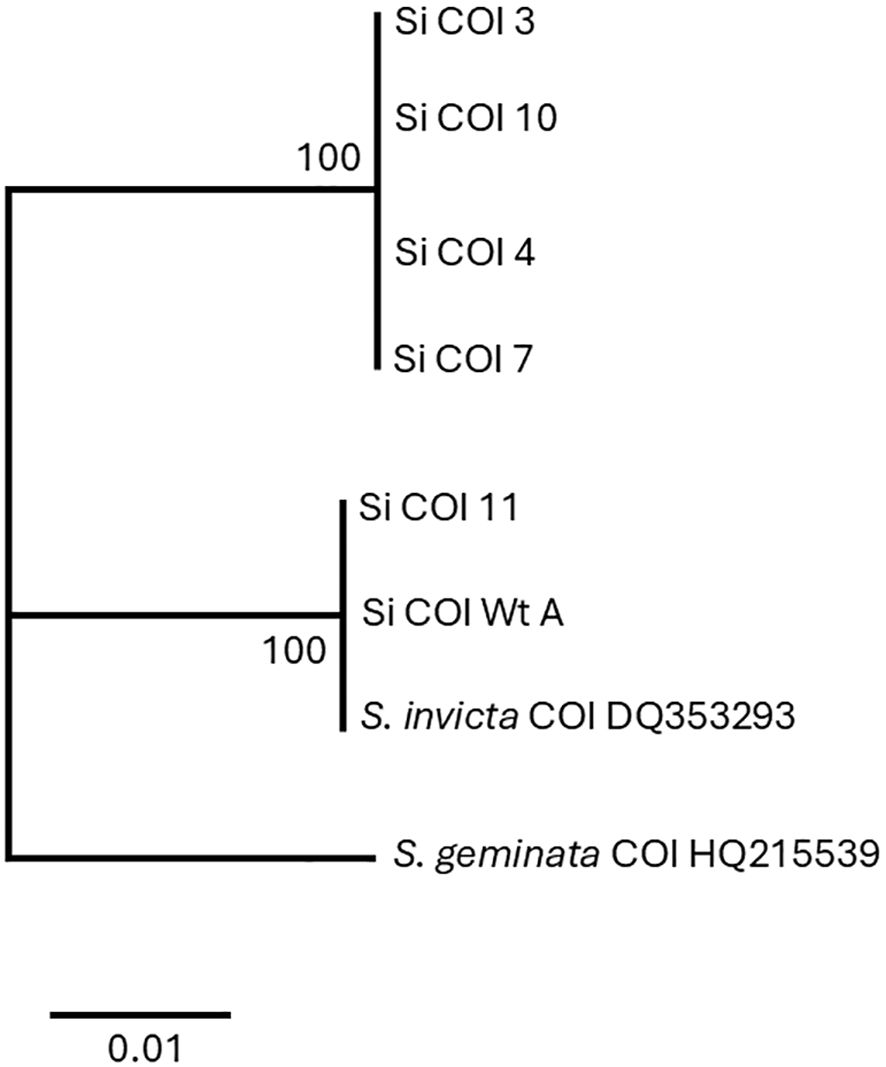
Figure 4. Phylogenetic analysis of cytochrome oxidase I nucleotide sequences by the maximum likelihood method. Trees are drawn to scale, with branch lengths measured in the number of substitutions per site, 500 bootstrap iterations, and branch support provided as a percentage. Eight sequences with a total of 449 positions comprised the final dataset.
Full mitochondrial genomes were assembled after nanopore sequencing of Soli_WtA, Soli_WtB, Soli_3, and Soli_7. Comparison to the S. invicta mitochondrial genome (NC014672.2) revealed greater than 99.99% nucleotide identity for Soli_WtA and Soli_WtB (Figure 5). In contrast, Soli_3 and Soli_7 had approximately 97.3% nucleotide identity. Phylogenetic reconstruction showed topology similar to that recovered from the COI analysis (Figures 4, 5).
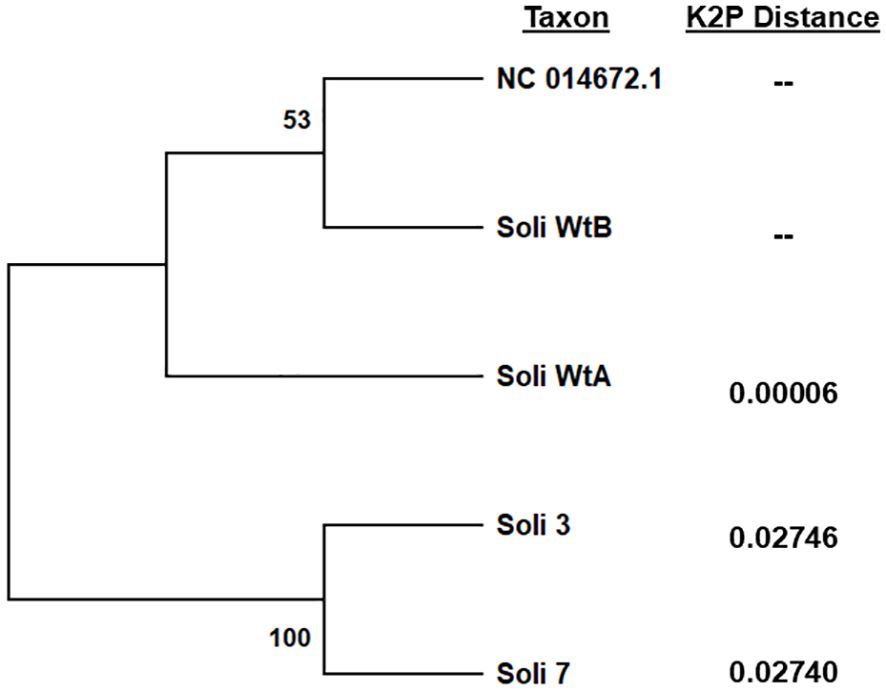
Figure 5. Phylogenetic analysis of complete mitochondrial genome sequences by the maximum likelihood method. Trees are drawn to scale, with branch lengths measured in the number of substitutions per site, 500 bootstrap iterations, and branch support provided as a percentage. Kimura 2-parameter distances were calculated relative to S. invicta mitochondrial genome accession NC014672.2. Five sequences with a total of 15,549 positions comprised the final dataset.
Amino acid sequence alignments of the short transcript ORFs (Figure 2) showed complete conservation among sequences Soli2Tr_2, Soli2Tr_3, Soli2Tr_4, Soli2Tr_5, Soli2Tr_7, Soli2Tr_8, Soli2Tr_9, Soli2Tr_10, and Soli2Tr_12. Two amino acid differences from this group were observed in Soli2Tr_11 and Soli2Tr_13 (Figure 2). Translation of these truncated proteins was confirmed by Western blotting of colonies Soli_4 and Soli_11, which was not observed among S. invicta ant colonies, yielding a positive result for InvictDetect™ (i.e., Soli_WtA & B; Figure 6). Soli2Tr proteins have a predicted molecular weight of 8.8 kDa (Figure 2). In purified venom protein preparations from ant colonies Soli_4 and Soli_11 exhibiting the truncated transcript, an 8.5 kDa protein was detected (Figure 6).
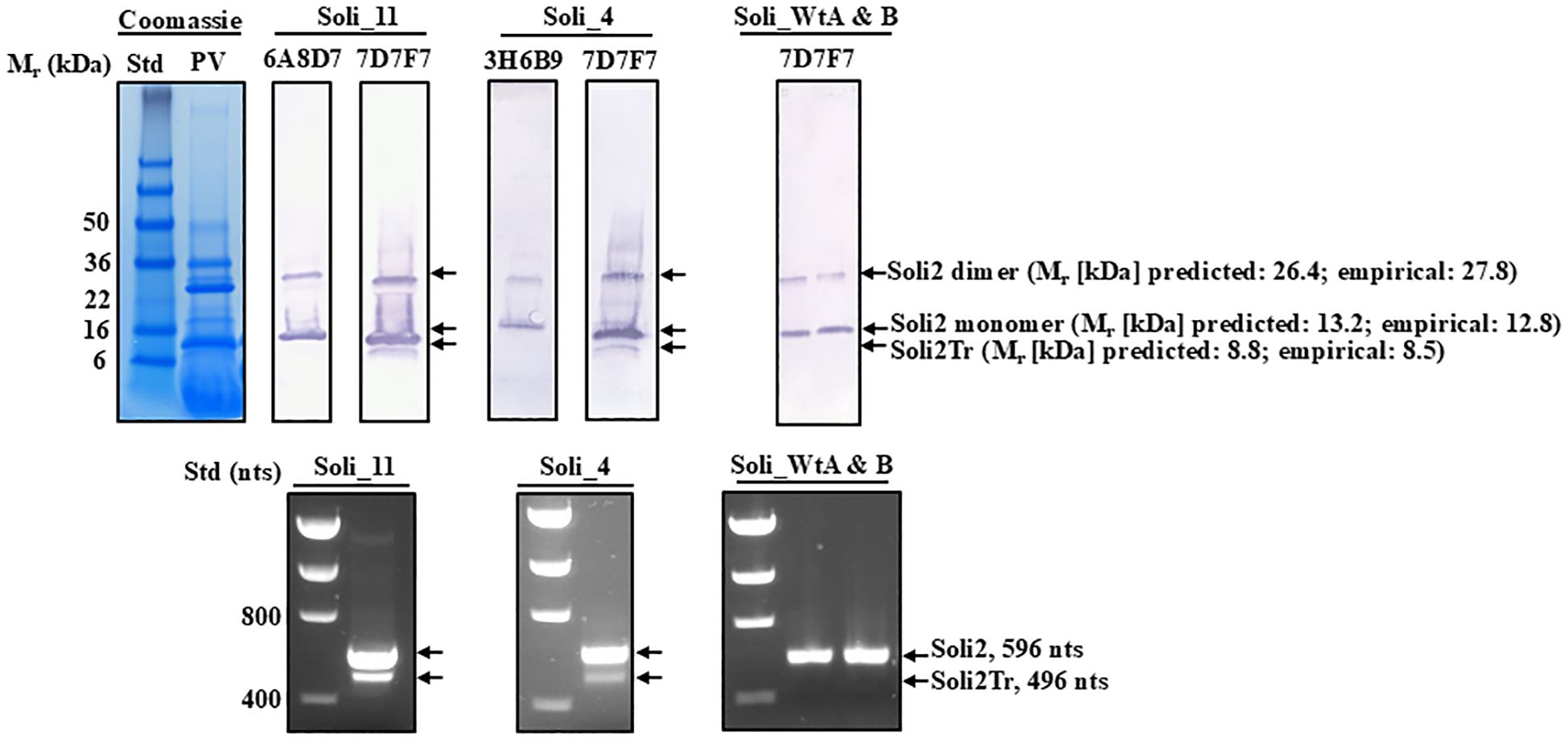
Figure 6. Western blot analysis of purified preparations of venom proteins from worker ants from S. invicta colonies Soli_4, Soli_11, Soli_WtA, and Soli_WtB probed with different monoclonal antibody preparations to specific regions of Soli2 and Solr2. Positions and molecular weights of the monomer and dimer of Soli2 and truncated versions of Soli2-like venom proteins are provided on the right. Molecular weight standards (kDa) are shown on the left. The lower panel set shows the amplicons produced for each of the colonies identified in the Western blot directly above. Molecular standards are on the left side and indicate the number of nucleotides.
Venom alkaloid chemical analysis of Soli_9, Soli_11, and Soli_13 revealed a variation in peak ratios consistent with S. invicta (Figure 7).
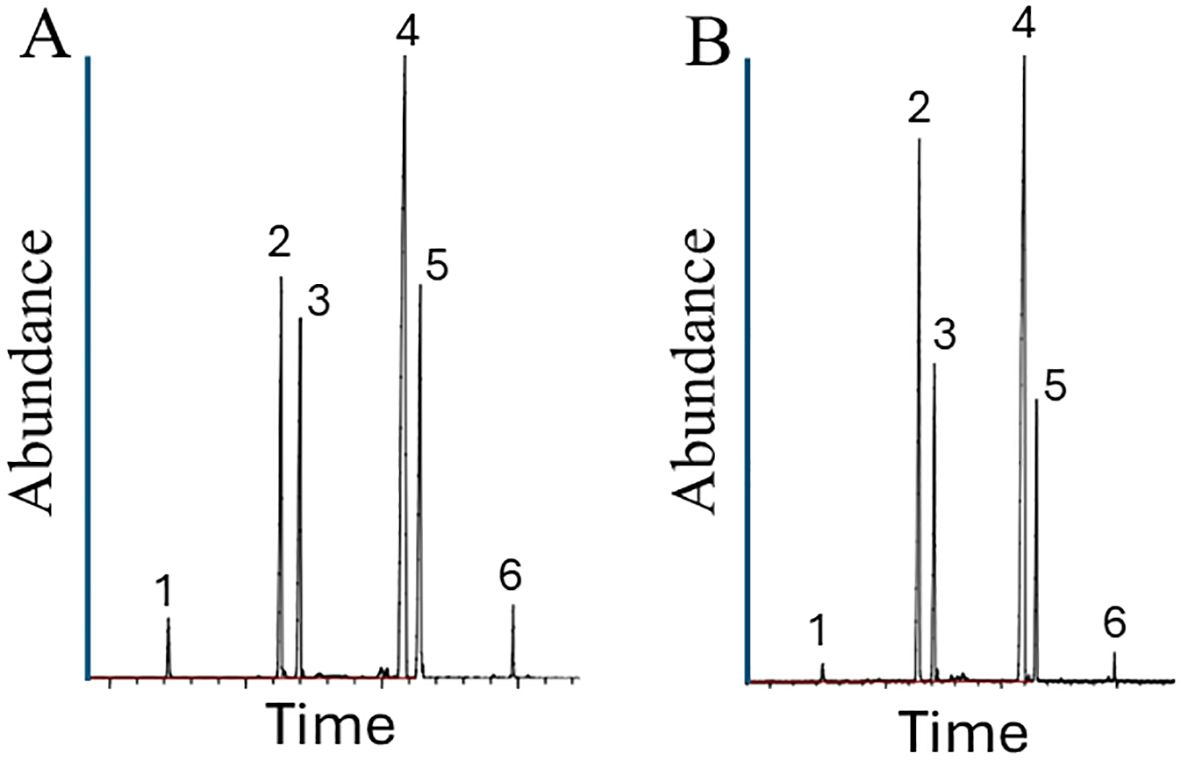
Figure 7. Venom alkaloid analysis of worker ants from S. invicta colonies Soli_9 (A) and Soli_11 (B). The four major S. invicta worker alkaloids are trans-piperidines: 2-methyl-6-tridecyl-piperidine (compound 2), 2-methyl-6-tridecenyl-piperidine (compound 3), 2-methyl-6-pentadecyl-piperidine (compound 4), and 2-methyl-6-pentadecenyl-piperidine (compound 5). In addition, there are minor amounts of 2-methyl-6-undecyl-piperidine (compound 1) and 2-methyl-6-heptadecenyl-piperidine (compound 6).
The lateral flow immunoassays to discriminate and identify the invasive fire ant species, Solenopsis invicta and Solenopsis richteri (and their hybrids), target the Solenopsis (invicta and richteri) venom 2 protein (16, 17). Monoclonal antibodies are used in tandem to capture and report the presence of these proteins from each ant species. Extensive testing of InvictDetect™ and InvictDetect Plus™ against S. invicta nests identified a very small proportion of colonies (1.39%) that yielded unexplained false negative results; the ants were taxonomically identified as S. invicta (30) but failed to give a response from the venom-specific antibodies. Nucleotide sequences obtained from Soli2 venom protein transcripts from these ant colonies revealed unique sequences distinct from the sequences used in the development of the immunoassay and originally published decades ago (29, 31, 32). Additionally, a newly discovered truncated Soli2-like protein (Soli2Tr) was identified in these InvictDetect™-non-responding colonies. Initially, we considered the possibility that the truncated Soli2-like transcripts were generated by alternative splicing at locus LOC105205300 on the S. invicta reference genome (Genbank accession UNIL_Sinv_3.0). However, this does not appear to be the case. The Soli2 truncated transcripts are not alternatively spliced versions but are transcribed at a unique locus (LOC105193315) upstream of LOC105205300 on chromosome 10, where Soli2 has obviously undergone multiple duplication events. Thus, the InvictDetect™ false negative results may be explained by 1) alteration of the carboxyl-proximal epitope (mAb reporting location) as in colony Sinv_11; 2) alteration of the amino-proximal epitope (mAb capture location) as in colonies Sinv_3, Sinv_4, Sinv_7, Sinv_8, Sinv_9, Sinv_10, and Sinv_11; and 3) complete elimination of the amino-proximal epitope (truncated Soli2-like venom protein) as in colonies Sinv_2, Sinv_5, and Sinv_12. Colony Sinv_13 produced two transcripts, one truncated (Soli2Tr_13) and one identical to the normal, or wildtype, Soli2 (P35775) sequence from LOC105205300. InvictDetect™ failure against this colony may have occurred because of a low expression of Soli2_13 (below InvictDetect™ detection levels) and/or antibody competition with Soli2Tr_13. While the sequence recognized by the reporting antibody in the carboxyl-proximal region on the truncated Soli2 protein is intact, the sequence in the amino-proximal region recognized by the reporting antibody is completely missing, which renders the InvictDetect™ and InvictDetect Plus™ immunoassay ineffective because the missing region prevents the capture of the protein from occurring. Translation of the truncated Soli2 transcript does appear to take place, as a protein of the predicted size was detected by Western analysis in colonies Soli_4 and Soli_11 (Figure 6). However, wildtype ant colonies do not appear to express the truncated version from locus 105193315. Thus, multiple variants of Soli2 have been identified, and a new, truncated Soli2-like protein has been discovered. A truncated version of venom 2 protein was also reported from the S. invicta x S. richteri hybrid ants in Tennessee (19), which was described as Solh2Tr97. In contrast to the S. invicta truncated versions of Soli2, Solh2Tr97 is an alternatively spliced version (19).
An interesting, and perplexing, aspect of these sequence variations for the Soli2 long transcript is the evolutionary relationship between the two invasive species of ants, S. invicta and S. richteri. All of the colonies discovered by false negative immunoassay tests in this study were identified morphologically as S. invicta (30), collected from areas known to be populated exclusively by S. invicta (1), and exhibited venom alkaloid profiles consistent with S. invicta (2, 33). However, phylogenetic analysis of the venom 2 protein sequence predicted from six of these S. invicta colonies appeared more closely related to the venom 2 ortholog from S. richteri (i.e. Solr2 P35776) than S. invicta (Soli2 P35775) (Figure 3). Indeed, the specificity of the InvictDetect™/InvictDetect Plus™ to capture monoclonal antibodies for each species resides in the unique amino-proximal sequence of the venom protein of each species and was chosen for the immunoassay because of this significant difference. Note that sequence NKELKII (aa position 21) was originally described from S. invicta (P35775) and amino acid sequence IEAQRVI (also at aa position 21) was described from S. richteri (P35776) (Figure 2). Until the present study, the IEAQRVI sequence was exclusive to S. richteri. Why is this apparently Solr2 phenotype detected in the S. invicta population and at such a low incidence? Could this represent a cryptic species within the S. invicta range? While a possibility, it is more likely that inter-species gene flow occurred during the incipient introduction period for both species into the U.S.A. S. richteri and S. invicta were introduced through the Mobile, Alabama, port around 1918 and 1935, respectively (1), and would have been afforded an opportunity to hybridize. Indeed, we suggest that because a fertile hybrid forms readily between S. invicta and S. richteri in the U.S.A (2), they are functionally a single species in North America. Evidence to support this notion is found in Tennessee, where the two parent species were sympatric decades ago but are almost entirely hybrid today (8; Oliver et al., In Press)1. Gene flow is not restricted or dead-ended once the hybrid forms. The hybrid form breeds equally successfully with other hybrids and both “parent” species (34). Cohen and Privman (34) offer several possibilities for the ready hybridization of S. invicta and S. richteri observed in the U.S.A., including sympatry in the U.S.A. The specific ants introduced into the U.S.A. were genetically compatible with each other, and the introduced ants were released from various exogenous factors that prevented hybridization in the native range [e.g., diet changes, parasite infection (like Wolbachia)], but most likely geographic separation was removed as a species separation variable. S. richteri occupies the wetlands northeast of Buenos Aires, Argentina, and eastward into Uruguay, whereas S. invicta is native to the Pantanal area of Brazil (type specimens from Cuiaba, Brazil) and south into northern Argentina (it is found 2,690 km from Cuiaba, Brazil, in Buenos Aires, Argentina).
Interestingly, a class of compound called tyramides have been found in males from all ant species thus far examined in the Myrmicinae (35). These compounds are transferred from males to females during mating and function to initiate reproductive development in the newly mated queens (36). This transfer is critical for successful colony formation and may also act as a mechanism to maintain species integrity. One would, therefore, expect that in their native ranges where the two species are allopatric, the tyramide structure would not experience selective pressure to maintain species separation. Indeed, chemical analyses have shown this to be the case (36, 37).
Finally, phylogenetic analysis of the cytochrome oxidase I gene and the mitochondrial genome sequence data were consistent with the unusual phylogeny of the long transcript Soli2 protein sequences obtained from the InvictDetect™-failed colonies. As with the long Soli2 transcript data, cytochrome oxidase I of colonies Soli_3, Soli_4, Soli_7, and Soli_10 assorted independently of S. invicta and S. richteri but have strong sequence identity with S. richteri. We admit that proposing the InvictaDetect™-failed ant colonies could represent a cryptic species is not sufficiently supported by our scant data. However, one could consider the possibility because these ant colonies fit the definition of cryptic. Specifically, they are classified as a single species (S. invicta), morphologically indistinguishable, recently diverged, separable only with chemical/molecular data, and occur in sympatry (38).
Six Soli2 paralogs occupy a contiguous 43,627-nucleotide region of the minus strand of chromosome 10, specifically the region encompassed by nucleotides 9,600,319 to 9,643,946. These paralogs appear to be derived from duplication events, which play a major role in the rapid evolution of new biological functions (39, 40). Interestingly, gene duplication is significantly (6–7-fold in humans) higher in chromosome regions near the centromere. These pericentromeric areas are considered rapidly evolving regions of the genome (41). The S. invicta chromosome 10 exhibits an unusually large (6–10 Mbp) submetacentric centromere with the Soli2 region occupying an area just downstream and adjacent to the centromere of this chromosome (42). Thus, the duplication of Soli2 supports the notion that peri-centromeric regions of chromosomes exhibit rapid evolution.
Ohno (40) hypothesized that gene duplication frees one form of the gene from selective pressure, permitting the accumulation of neutral mutations that may lead to a new function (43). This hypothesis has since been validated repeatedly and among varied organisms, including invertebrates responding to various environmental pressures (44–46). Paralogs have been shown to evolve more rapidly than unduplicated genes and, if they become fixed in a population, increase fitness, which is especially noted for secreted proteins (39) such as the venom 2 protein of Solenopsis. Indeed, this mechanism has been reported to increase the diversity and functions of snake venoms (47). Among the small group of ant colonies collected for our study, nucleotide changes of the long Soli2 venom gene were significant, resulting in drastic amino acid changes of the protein, most notably the IEAQRVL S. richteri-like amino-proximal sequence (aa position 21) and the loss of the cysteine at position 41, which is changed to tyrosine in colonies Soli_3, Soli_7, Soli_8, Soli_9, and Soli_10. This cysteine is the site where dimerization occurs between monomers of the Soli2 venom protein (48).
Finally, the expression pattern for Soli2 and Soli2Tr appears to be unique in the InvictDetect™-failing ant colonies. The Soli2Tr transcript and protein were produced in these colonies from locus 105193315, which did not appear to be expressed in 98.6% of the ant colonies (based on a positive InvictDetect™ response). Soli2Tr was accompanied by Soli2 from locus 105205300 most of the time. However, in the three ant colonies, only Soli2Tr was detected, Soli_2, Soli_5, and Soli_12. Additional investigation will be required to understand the basis for the differential expression patterns and why such a small number of colonies produce this truncated protein despite the gene’s presence in the genome of S. invicta.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The manuscript presents research on animals that do not require ethical approval for their study.
SV: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. RV: Conceptualization, Formal analysis, Investigation, Resources, Writing – review & editing. AE: Conceptualization, Data curation, Formal analysis, Investigation, Resources, Writing – review & editing.
The author(s) declare that no financial support was received for the research and/or publication of this article.
We thank Roxie White (USDA) and Charles Strong (deceased) for technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The use of trade, firm, or corporation names in this publication is for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the United States Department of Agriculture or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable.
2. Vander Meer RK, Lofgren CS, Alvarez FM. Biochemical evidence for hybridization in fire ants. Fla Entomol. (1985) 68:501–6. doi: 10.2307/3495147
3. Ross KG, Vander Meer RK, Fletcher DJC, Vargo EL. Biochemical phenotypic and genetic studies of two introduced fire ants and their hybrid (Hymenoptera, Formicidae). Evolution. (1987) 41:280–93. doi: 10.2307/2409138
4. Callcott AMA, Collins HL. Invasion and range expansion of imported fire ants (Hymenoptera: Formicidae) in North America from 1918-1995. Fla Entomol. (1996) 79:240–51. doi: 10.2307/3495821
5. Streett DA, Freeland TB, Vander Meer RK. Survey of imported fire ant (Hymenoptera: Formicidae) populations in Mississippi. Fla Entomol. (2006) 89:91–2. doi: 10.1653/0015-4040
6. Bertagnolli D, Graham LC, Vander Meer RK. Ouch! Who bit me? IFA in Alabama. In: 2007 annual imported fire ant conference (2007) (Tuscaloosa, Alabama: University of Alabama). p. 112–5.
7. Oliver JB, Vander Meer RK, Ochieng SA, Youssef NN, Pantaleoni E, Mrema FA, et al. Statewide survey of imported fire ant (Hymenoptera: Formicidae) populations in Tennessee. J Entomol Sci. (2009) 44:149–57. doi: 10.18474/0749-8004-44.2.149
8. Pandey M, Addesso KM, Archer RS, Valles SM, Baysal-Gurel F, Ganter PF, et al. Worker size, geographical distribution, and introgressive hybridization of invasive Solenopsis invicta and Solenopsis richteri (Hymenoptera: Formicidae) in Tennessee. Environ Entomol. (2019) 48:727–32. doi: 10.1093/ee/nvz023
9. Stafford CT, Hoffman DR, Rhoades RB. Allergy to imported fire ants. South Med J. (1989) 82:1520–7. doi: 10.1097/00007611-198912000-00015
10. Hoffman DR. Fire ant venom allergy. Allergy. (1995) 50:535–44. doi: 10.1111/j.1398-9995.1995.tb01196.x
11. deShazo RD, Butcher BT, Banks WA. Reactions to the stings of the imported fire ant. New Engl J Med. (1990) 323:462–6. doi: 10.1056/NEJM199008163230707
12. Rhoades RB, Stafford CT, James FK. Survey of fatal anaphylactic reactions to imported fire ant stings. J Allergy Clin Immun. (1989) 84:159–62. doi: 10.1016/0091-6749(89)90319-9
13. dos Santos Pinto JR, Fox EG, Saidemberg DM, Santos LD, da Silva Menegasso AR, Costa-Manso E, et al. Proteomic view of the venom from the fire ant Solenopsis invicta Buren. J Prot Res. (2012) 11:4643–53. doi: 10.1021/pr300451g
14. Fox EGP. Venom toxins of fire ants. In: Gopalakrishnakone P, Calvete JJ, editors. Venom genomics and proteomics. Springer Netherlands, Dordrecht (2016). p. 149–67.
15. Hoffman DR. Ant venoms. Curr Opin Allergy Clin Immunol. (2010) 10:342–6. doi: 10.1097/Aci.0b013e328339f325
16. Valles SM, Strong CA, Callcott A-MA. Development of a lateral flow immunoassay for rapid field detection of the red imported fire ant, Solenopsis invicta (Hymenoptera: Formicidae). Anal Bioanal Chem. (2016) 408:4693–703. doi: 10.1007/s00216-016-9553-5
17. Valles SM, Strong CA, Callcott AMA. Multiplexed lateral flow immunoassay to discriminate Solenopsis invicta, Solenopsis richteri, and Solenopsis invicta x richteri hybrids. Insect Soc. (2018) 65:493–501. doi: 10.1007/s00040-018-0637-4
18. Fox EGP, Solis DR, dos Santos LD, Pinto JRAD, Menegasso ARD, Silva RCMC, et al. A simple, rapid method for the extraction of whole fire ant venom (Insecta: Formicidae). Toxicon. (2013) 65:5–8. doi: 10.1016/j.toxicon.2012.12.009
19. Valles SM, Oliver JB, Addesso KM, Perera OP. Unique venom proteins from Solenopsis invicta x Solenopsis richteri hybrid fire ants. Toxicon X. (2021) 9-10:100065. doi: 10.1016/j.toxcx.2021.100065
20. Valles SM, Porter SD. Identification of polygyne and monogyne fire ant colonies (Solenopsis invicta) by multiplex PCR of Gp-9 alleles. Insect Soc. (2003) 50:199–200. doi: 10.1007/s00040-003-0662-8
21. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech. (1994) 3:294–9.
22. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. (2018) 34:3094–100. doi: 10.1093/bioinformatics/bty191
23. Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinf. (2013) 14:178–92. doi: 10.1093/bib/bbs017
24. Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. (1992) 8:275–82. doi: 10.1093/bioinformatics/8.3.275
25. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
26. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. (1980) 16:111–20. doi: 10.1007/BF01731581
27. Kapustin Y, Souvorov A, Tatusova T, Lipman D. Splign: algorithms for computing spliced alignments with identification of paralogs. Biol Direct. (2008) 3:20. doi: 10.1186/1745-6150-3-20
28. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. (1997) 25:3389–402. doi: 10.1093/nar/25.17.3389
29. Hoffman DR, Dove DE, Jacobson RS. Isolation of allergens from imported fire ant (Solenopsis invicta) venom. J Allergy Clin Immun. (1988) 81:203–3. doi: 10.1016/0091-6749(88)90376-4
30. Pitts JP. A cladisitic analysis of the Solenopsis saevissima species-group (Hymenoptera: Formicidae). Athens: University of Georgia (2002).
31. Hoffman DR, Dove DE, Jacobson RS. Allergens in hymenoptera venom. XX. Isolation four allergens imported fire ant (Solenopsis invicta) venom.(1988) 82:818–27. doi: 10.1016/0091-6749(88)90084-X
32. Hoffman DR, Dove DE, Moffitt JE, Stafford CT. Allergens in hymenoptera venom.21. Cross-reactivity and multiple reactivity between fire ant venom and bee and wasp venoms. J Allergy Clin Immun. (1988) 82:828–34. doi: 10.1016/0091-6749(88)90085-1
33. Vander Meer RK, Lofgren CS. Biochemical and behavioral evidence foe hybridization between fire ants,Solenopsis invicta andSolenopsis richteri (Hymenoptera: Formicidae). J Chem Ecol. (1989) 15:1757–65. doi: 10.1007/BF01012263
34. Cohen P, Privman E. Speciation and hybridization in invasive fire ants. BMC Evol Biol. (2019) 19:111. doi: 10.1186/s12862-019-1437-9
35. Jones TH, Chinta SP, Vander Meer RK, Cartwright KC. Branched tyramides from males of the harvester ant. Pogonomyrmex badius. (2023) 110:57. doi: 10.1007/s00114-023-01885-2
36. Vander Meer RK, Chinta SP, Jones TH, O’Reilly EE, Adams RMM. Male fire ant neurotransmitter precursors trigger reproductive development in females after mating. Comm Biol. (2021) 4:1400. doi: 10.1038/s42003-021-02921-5
37. Chen J, Grodowitz MJ. Tyramides in male alates of black imported fire ants Solenopsis richteri. Insect Sci. (2017) 24:169–72. doi: 10.1111/1744-7917.12304
38. Bickford D, Lohman DJ, Sodhi NS, Ng PK, Meier R, Winker K, et al. Cryptic species as a window on diversity and conservation. Trends Ecol Evol. (2007) 22:148–55. doi: 10.1016/j.tree.2006.11.004
39. Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. (2002) 3:RESEARCH0008. doi: 10.1186/gb-2002-3-2-research0008
41. Pich IRO, Kondrashov FA. Long-term asymmetrical acceleration of protein evolution after gene duplication. Genome Biol Evol. (2014) 6:1949–55. doi: 10.1093/gbe/evu159
42. Huang YC, Lee CC, Kao CY, Chang NC, Lin CC, Shoemaker D, et al. Evolution of long centromeres in fire ants. (2016) 16:189. doi: 10.1186/s12862-016-0760-7
43. Kimura M, King JL. Fixation of a deleterious allele at one of two “duplicate” loci by mutation pressure and random drift. (1979) 76:2858–61. doi: 10.1073/pnas.76.6.2858
44. Tabashnik BE. Implications of gene amplification for evolution and management of insecticide resistance. (1990) 83:1170–6. doi: 10.1093/jee/83.4.1170
45. Guillemaud T, Raymond M, Tsagkarakou A, Bernard C, Rochard P, Pasteur N. Quantitative variation and selection of esterase gene amplification in Culex pipiens. (1999) 83:87–99. doi: 10.1038/sj.hdy.6885370
46. Otto E, Young JE, Maroni G. Structure and expression of a tandem duplication of the Drosophila metallothionein gene. (1986) 83:6025–9. doi: 10.1073/pnas.83.16.6025
47. Lynch VJ. Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol Biol. (2007) 7:2. doi: 10.1186/1471-2148-7-2
Keywords: Solenopsis invicta, Solenopsis richteri, venom, alkaloids, Formicidae
Citation: Valles SM, Vander Meer RK and Estep AS (2025) A lateral flow immunoassay-based survey reveals a low-frequency truncated Solenopsis invicta venom 2-like protein and unique Solenopsis invicta venom 2 protein genotypes in Solenopsis invicta. Front. Insect Sci. 5:1527130. doi: 10.3389/finsc.2025.1527130
Received: 12 November 2024; Accepted: 06 March 2025;
Published: 07 April 2025.
Edited by:
Francesco Nugnes, National Research Council (CNR), ItalyReviewed by:
Satya Chinta, United States Department of Agriculture (USDA), United StatesCopyright © 2025 Valles, Vander Meer and Estep. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven M. Valles, c3RldmVuLnZhbGxlc0B1c2RhLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.