- 1Department of General Biology and Genetics, Institute of Biochemical Technologies, Ecology and Pharmacy, V.I. Vernadsky Crimean Federal University, Simferopol, Republic of Crimea
- 2Laboratory of Entomology and Phytopathology, Dendrology and Landscape Architecture, Nikita Botanical Gardens—National Scientific Centre of the Russian Academy of Sciences, Yalta, Republic of Crimea
Twenty years ago, it was difficult to imagine the use of nucleic acids in plant protection as insecticides, but today it is a reality. New technologies often work inefficiently and are very expensive; however, qualitative changes occur during their development, making them more accessible and work effectively. Invented in 2008, contact oligonucleotide insecticides (olinscides, or DNA insecticides) based on the CUAD (contact unmodified antisense DNA) platform have been substantially improved and rethought. The main paradigm shift was demonstrating that unmodified antisense DNA can act as a contact insecticide. Key breakthroughs included identifying convenient target genes (rRNA genes), mechanism of action (DNA containment), and discovering insect pests (sternorrhynchans) with high susceptibility to olinscides. Today, the CUAD platform possesses impressive characteristics: low carbon footprint, high safety for non-target organisms, rapid biodegradability, and avoidance of target-site resistance. This next-generation class of insecticides creates opportunities for developing products tailored for specific insect pest populations. The ‘genetic zipper’ method, based on CUAD biotechnology, integrates molecular genetics, bioinformatics, and in vitro nucleic acid synthesis. It serves as a simple and flexible tool for DNA-programmable plant protection using unmodified antisense oligonucleotides targeting pest rRNAs. Aphids, key pests of important agricultural crops, can be effectively controlled by oligonucleotide insecticides at an affordable price, ensuring efficient control with minimal environmental risks. In this article, a low-dose concentration (0.1 ng/µL; 20 mg per hectare in 200 L of water) of the 11 nt long oligonucleotide insecticide Schip-11 shows effectiveness against the aphid Schizolachnus pineti, causing mortality rate of 76.06 ± 7.68 on the 12th day (p<0.05). At a consumption rate of 200 L per hectare, the cost of the required oligonucleotide insecticide is about 0.5 USD/ha using liquid-phase DNA synthesis making them competitive in the market and very affordable for lab investigations. We also show that non-canonical base pairing Golinscide: UrRNA is well tolerated in aphids. Thus, non-canonical base-pairing should be considered not to harm non-target organisms and can be easily solved during the design of oligonucleotide insecticides. The ‘genetic zipper’ method, based on CUAD biotechnology, helps quickly create a plethora of efficient oligonucleotide pesticides against aphids and other pests. Already today, according to our estimations, the ‘genetic zipper’ is potentially capable of effectively controlling 10-15% of all insect pests using a simple and flexible algorithm.
1 Introduction
Unmodified DNA, as a programmable molecule and polymer of natural origin, has always attracted researchers. Unfortunately, for a long time it was believed that unmodified oligonucleotides are toxic to cells and degraded quickly in all eukaryotic cells under the action of nucleases (1), including insects (2). Some articles literally stated that unmodified (phosphodiester) antisense oligonucleotides should not be used for these experiments (3). The mode of action of unmodified antisense DNA and its potential application as contact insecticide have not been investigated on insect pests, and no attempts were made to test this hypothesis until the beginning of the 21st century. An unexpected and surprising turn with unmodified antisense oligonucleotides in plant protection came in 2008, when it was shown that short unmodified antisense DNA has significant insecticidal effect on insect pests (4). For the first time, the equivalence between antisense oligodeoxyribonucleotides and contact insecticides was established in experiments with spongy moth Lymantria dispar, which led to the development of the CUAD (contact unmodified antisense DNA) platform (5–8). The first 18–20 nt long oligonucleotide insecticides, based on anti-apoptotic genes, showed effectiveness on virus-free and nuclear polyhedrosis virus-infected spongy moth caterpillars (9). This discovery opened up an entirely new dimension in insect pest control using nucleic acids as contact insecticides. Scientists studying RNAi also picked up this idea three years later when Wang et al. (10) successfully applied double-stranded RNA fragments as contact insecticides in insect pest control for the first time (10).
In 2019, this innovation was substantially improved and rethought (11). The CUAD biotechnology has a number of features that distinguish it from all modern classes of chemical insecticides and plant protection technologies developing today: unmodified antisense DNA as active substance, DNA containment as mechanism of action, insect pre-rRNA and rRNA as target (12–14). Oligonucleotide insecticides are based on ‘genetic zipper’ method acting through formation of complementary DNA olinscide-rRNA duplex (resembles zipper mechanism) that ‘zips’ target rRNA expression and leads to death of pests. Oligonucleotide insecticides (briefly, olinscides, or DNA insecticides) act on sternorrhynchans through DNA containment mechanism consisting of 2 steps: 1) rRNA and/or pre-rRNA ‘arrest’ and hypercompensation of target rRNA; 2) target rRNA and/or pre-rRNA degradation recruiting RNase H (13–15). Use of insect pest pre-rRNA and mature rRNA as target leads to high efficiency of oligonucleotide insecticides, since pre-rRNA and mature rRNA comprise 80% of all RNA in the cell. The degradation of ribosomal RNA inevitably leads to disruption of protein biosynthesis and the death of insect pests (9). If insecticide resistance occurs, different strategies can be applied. Generally, new and efficient olinscides can be easily re-created displacing target site to the left or to the right from the olinscide-resistant site of target mature rRNA and/or pre-rRNA (11, 13). According to our investigations, contact delivery of unmodified antisense DNA (CUAD) is much more efficient (16) than oral delivery of unmodified antisense DNA (ODUAD) because of active DNases present in the digestive tract of insects (17, 18).
While 1st step of discovered DNA containment mechanism (rRNA ‘arrest’ accompanied with rRNA hypercompensation) was completely unknown before, 2nd step (degradation of rRNA) recruiting RNase H was partially known but not for rRNAs (19, 20). Historically, some principles of the practical application of antisense oligonucleotides (ASOs) for chemical reactions on biopolymers were first formulated in Novosibirsk (Russia) by N. Grineva in 1967. In the case of nucleic acids, it was promising to obtain compounds, containing a reactive group bound to oligonucleotide residue capable of specific base pairing with the complementary nucleotide sequence (21). In 1977, using this antisense approach to modify valine tRNA, N. Grineva and colleagues demonstrated that the method allows alkylation with a reagent bound to the corresponding oligonucleotide at certain points along the valine tRNA (22). These first steps were quite different from the research work currently being carried out using antisense oligonucleotides in agriculture and medicine, but they laid the groundwork for the possibility of regulating gene expression in living organisms using antisense oligonucleotides. Later, in 1978, P. Zamecnik and M. Stephenson showed that modified antisense DNA hinders reproduction of Rous sarcoma virus in chicken embryo fibroblasts in a sequence-specific manner (23). In 1979, H. Donis-Keller presented results showing that RNase H cleaves the RNA strand in RNA–DNA heteroduplexes in a site-specific manner (24). The development of antisense technologies has long been primarily focused on medicine using modified antisense oligonucleotides (25). After 20 years of research with modified antisense oligonucleotides, in 1998, the FDA licensed the first drug, Vitravene (Fomivirsen), based on the 21-mer phosphorothioate oligonucleotide (26). This area of medicine continues to progress and has already seen the registration of other important drugs (27). In any case, it had not been shown until 2008 that DNA can function as insecticide and there were no attempts to investigate precise mechanisms of action of unmodified antisense DNA on insect cells, including rRNA as a target.
To date, contact unmodified antisense DNA (CUAD) biotechnology is the only platform that successfully uses short unmodified antisense DNA for plant protection (11, 13, 14). The CUAD platform works best on hemipterans from suborder Sternorrhyncha (11, 12). The main paradigm shift with unmodified antisense DNA was in showing that it can be a contact insecticide. The main breakthroughs in the development of this approach were in finding the most convenient target genes (rRNA genes), showing mechanism of action (DNA containment) and in searching up insect pests (sternorrhynchans) with high susceptibility to this approach. n the last few years, CUAD biotechnology based on oligonucleotide pesticides has been established as a powerful “genetic zipper” method against soft scale insects, armored scale insects, psyllids, mealybugs, aphids, and mites, opening new frontiers in DNA-programmable plant protection based on contact application of deoxyribonucleic acid (12, 15). Obtained data suggest that short antisense DNA sequences via DNA containment (DNAc) mechanism can participate in regulation of rRNA expression by complementary interaction with cell DNA (direct rDNA master regulation of rRNA expression) and viral DNA (direct rDNA master regulation of rRNA expression by viral DNA, or rRNA switchboard mechanism) (14). Also DNAc can be recruited in innate immunity system (13) against ssDNA viruses for which hemipteran insects serve as major vectors (28, 29) and also against DNA viruses that normally infect them (30).
Because of very efficient and easy algorithm, DNA-guided ‘genetic zipper’ method (CUAD biotechnology) is a unique and very potent alternative to other antisense approaches in insect pest control based on duplexes of unmodified nucleic acids and RNA-guided nucleases: RNA interference and CRISPR/Cas9. Innovative insect pest control technologies (RNAi, CUAD, CRISPR/Cas9) are based on formation of duplexes of unmodified nucleic acids (RNAi: guide RNA-mRNA; CUAD: guide DNA-rRNA; CRISPR/Cas9: guide RNA-genomic DNA) and action of nucleic acid-guided nucleases (RNAi: Argonaute, briefly Ago; CUAD: RNase H; CRISPR/Cas9: CRISPR associated protein 9, briefly Cas9) (31). While RNAi and CRISPR/Cas9 were not discovered on insect pests and initially had fundamental importance rather than practical one, CUAD biotechnology was discovered on insect pests as practical tool and recently fundamental importance of DNA containment mechanism played in rRNA biogenesis was revealed (9, 13, 14, 32). To date, while RNAi and CRISPR/Cas9 are excellent tools for manipulations with unmodified nucleic acids in laboratory, they do not have easy algorithms for creation of end-products for insect pest control; each separate case is special and usually is sorted out using trial and error method.
Oligonucleotide insecticides possess low carbon footprint, high safety for non-target organisms, rapid biodegradability in ecosystems, and avoidance of target-site resistance. While current chemical insecticides require days, months and even years for biodegradation by bacteria and fungi, oligonucleotide insecticides are substantially biodegraded within hours in the presence of nucleases (33). Olinscides have the potential to complement the existing insecticide market and set an eco-precedent for crop protection products where the effectiveness of the insecticide will be determined by its safety for non-target organisms (33). The advantage of using natural oligomers, unmodified antisense oligonucleotides, seems to be the safest way, since the cells of all living organisms contain ubiquitous nucleases that can neutralize them (8). Consequently, for oligonucleotide insecticides there is no need to look for methods of accelerated biodegradation. The principle of using oligonucleotide pesticides is that they must have enough time to act in the right place and on the right organism before their rapid biodegradation (and they successfully do this on sternorrhynchans and other pests). In contrast, due to resistance to biodegradation conventional chemical insecticides have too much time for their action not only in the right place and not only on the right organism (33). Consequently, majority of chemical insecticides were banned in one way or another after decades or years of use in plant protection, when effective competitors with proven or supposed greater safety appeared (34). Recently, the idea of oligonucleotide insecticides attracted the attention of scientists and experts in plant protection (35–40) and to a certain extent the affordability of such insecticides remained in question. If in the near future a balance is found between the effectiveness and cost of such pest control agents, then the insecticide market will be replenished in abundance with species-, genus-, and family-specific oligonucleotide insecticides.
Our research team decided to lower the price of olinscides and find a group of serious insect pests on which oligonucleotide insecticides at low concentrations could have a significant insecticidal effect. Aphids from subfamily Lachninae turned out very susceptible to low-dose concentrations of oligonucleotide insecticides. Generally, aphids (Hemiptera: Aphididae) are significant economic pests that are found globally. Aphids feed on phloem (41) and cause substantial economic losses mainly spreading plant viruses, and producing honeydew (42–44). Among aphids around 100 species are considered to be agricultural pests of a wide range of crops. They are major insect pests of various plants, including alfalfa, wheat, potato, sugar beet and tobacco (45). The damage caused by aphids amounts to hundreds of millions of dollars a year (46). Their management is challenging because the mobility of aphids is extremely high (47). Also these pests reproduce predominantly asexually (48), one female leaves 10-90 offspring in 7-10 days and therefore, theoretically, could produce billions of offspring in one growing season in the absence of mortality factors (49). Chemical insecticides have been used to control aphids, and they quickly develop resistance to various classes of chemical insecticides, including neonicotinoids, carbamates, organophosphates, organochlorines, and pyrethroids (50). This prompts the search for new insecticides with advanced characteristics and multi-decade utility.
In this article we use aphid Schizolachnus pineti for the experiments. S. pineti is a serious pest of Pinus spp., but especially on young Scots pine (Pinus sylvestris) where it forms dense colonies in rows along the previous year’s needles (51). The goal of this article is to show insecticidal effect on S. pineti using low concentrations of olinscides and provide cost-effective solution for aphid control based on the DNA-programmable ‘genetic zipper’ method.
2 Materials and methods
2.1 Origin of material
Individuals of S. pineti were collected from pine forest in Crimea. The experiments with S. pineti (Figure 1) were carried out in laboratory conditions at room temperature (25°C) and 50% humidity on shoots of Pinus sylvestris L. (Coníferae: Pinaceae). Shoots of trees with S. pineti were immersed in water and randomized; 3 shoots were taken for each experimental group; there were 100-120 aphids on each shoot in all experimental groups including water-treated Control (300-360 insect individuals per 3 shoots of P. sylvestris in each replicate for each group of the experiment). Experiments were carried out in triplicate.
2.2 Design, synthesis, and application of oligonucleotide insecticide Schip-11
We designed oligonucleotide insecticide Schip-11 5′-TGT-GTT-CGT-TA-3′ which is almost complementary (at 4th position of olinscide, G instead of A) to the ITS2 region of pre-rRNA of Schizolachnus pineti (Figure 2). We decided to use Schip-11 sequence to find out if non-canonical base pairing Golinscide: UrRNA in aphids is well tolerated as seen in scale insects (13). Also G:U base pairs are among the first-identified and most frequently occurring non-canonical Watson-Crick interactions in structured RNAs (52), thus, non-canonical base-pairing should be taken into consideration during creation of olinscides.

Figure 2. Alignment of the sequenced DNA fragment of S. pineti collected from nature and fragment of ITS2 region of rDNA of M. sanborni (GenBank: OR046519.1) performed using ClustalW 2.0.3.
The sequence of olinscide was synthesized using ASM 800E DNA synthesizer (BIOSSET, Novosibirsk, Russia) according to standard phosphoramidite synthesis procedure. The synthesis was carried out in the direction from the 3′ to the 5′ end. After completion of all cycles of synthesis, the target oligonucleotide was removed from the solid-phase support; the removal of the protective groups was carried out overnight at 55°C in a concentrated ammonia solution (analytical grade, “Vekton”, Saint Petersburg, Russia). Purification of the synthesized olinscide Schip-11 was performed on OPS-12 cartridges used for purification of oligonucleotides (BIOSSET, Novosibirsk, Russia). A BactoSCREEN analyzer based on a MALDI-TOF mass spectrometer was used to determine the quality of produced olinscide Schip-11 (Litech, Moscow, Russia). The ratio of mass-to-charge (m/z) of olinscide Schip-11 was measured as positive ions with 3-hydroxypicolinic acid as a matrix on a LaserToFLT2 Plus device (UK) in a ratio of 2:1. The theoretical m/z ratio was calculated using ChemDraw 18.0 software (ChemDraw, Cambridge Soft, USA) and differed by no >10 units with the resulting m/z ratio. Dilution in nuclease-free water to a required concentration was carried out on NanoDrop Lite spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Concentrations of 200 ng/μL and 0.1 ng/μL of olinscide Schip-11 in nuclease-free water was applied on P. sylvestris leaves using hand sprayer (10 ml of water solution per m2 of leaves). As a control, water-treated group was used. Mortality was recorded every day during 4 days for insects treated with Schip-11 in concentration 200 ng/µL and during 12 days for insects treated with Schip-11 in concentration 0.1 ng/µL. Effectiveness of olinscide Schip-11 was calculated by dividing the number of dead insect individuals by the total number of insect individuals on the shoot and multiplying by 100%, winged insect individuals were excluded from calculations.
2.3 Target gene expression
Isolation of total RNA was carried out according to manufacturer’s instructions using ExtractRNA reagent (Evrogen, Moscow, Russia). RNA extraction was carried out in three replicates. A MMLV RT kit was used to perform first-strand cDNA synthesis (Syntol, Moscow, Russia), following the manufacturer’s instructions. For cDNA synthesis, 10 μl of RNA was taken at a concentration of 20 ng/µl. PCR reactions were carried out on 5 μL of cDNA using 10 μL of SYBR Green Master Reaction Mix reagent (Syntol, Moscow, Russia), 7 μL of ddH2O (Syntol, Moscow, Russia), 1 μL of MgCl2 and 1 μL (80 ng/μL) of each specific primers SP_F 5′-ACG-ACA-ACA-TGC-GTG-TAC-C-3′ and SP_R 5′-GTC-CCA-CAG-TCC-GCT-TCTC-3′. The following PCR program was applied: 10 min of initial denaturation at 95°C, followed by 30 cycles with 10 s of denaturation at 95°C, 15 s of annealing at 62°C, and 20 s of elongation at 72°C was used for amplification on a LightCycler® 96 Real-Time PCR System (Roche, Basel, Switzerland). The expression of the target gene was evaluated on the 1st and 3rd day after treatment with Schip-11.
2.4 DNA sequencing
Primers, forward 5′-CGT-CGT-AAC-CTT-GCC-CTC-TT-3′ and reverse 5′-CGG-GGA-CAT-CGT-GAT-TTT-GG-3′, were used for PCR. PCR reactions were carried out on 5 μL of cDNA using 10 μL of SYBR Green Master Reaction Mix reagent (Syntol, Moscow, Russia), 7 μL of ddH2O (Syntol, Moscow, Russia), 1 μL of MgCl2 and 1 μL (80 ng/μL) of each specific primer. DNA was first denatured for 4 min at 95°C, then 30 cycles of 1 min of denaturation at 94°C, 1 min of hybridization at 62°C, and 1 min of elongation at 72°C, followed by a final elongation step at 72°C for 7 min. PCR products were purified using the Cleanup S-Cap (Evrogen, Moscow, Russia) and the sequencing polymerase reaction was carried out with Big Dye Terminator v 3.1 Cycle Sequencing RR-100 (Applied Biosystems, Vilnius, Lithuania). Polymerase reactions were carried out using 2 μL of purified DNA and 2 μL of primers (12.8 ng/μL). DNA was initially denatured for 1 min at 96°C, followed by 30 cycles of 10 s of denaturation at 96°C, 5 s of hybridization at 50°C, and 4 min of elongation at 60°C. Amplicons were sequenced in both directions using the NANOPHOR-05 capillary DNA sequencer (Syntol, Moscow, Russia). DNA sequences we analyzed using ClustalW 2.0.3 program (53) and BLAST.
2.5 Statistical analysis
The mean and standard error of the mean (SE) were calculated using the Student’s t-test for statistical analysis to evaluate the significance of the difference in mortality and pre-rRNA concentration between water-treated Control and experimental groups. All above-mentioned calculations were preformed using Prism 9 software (GraphPad Soft-ware Inc., Boston, USA).
3 Results
3.1 Mortality of S. pineti after contact application of olinscide Schip-11
After treatment of S. pineti with olinscide Schip-11 in concentration 200 ng/μL mortality of the pest reached 24.32 ± 1.37%, 61.03 ± 2.17%, 76.56 ± 3.67%, and 84.19 ± 3.84% on the 1st, 2nd, 3rd, and 4th day, respectively (Figure 3A) (p<0.05). Of note, the same olinscide (5′-TGT-GTT-CGT-TA-3′; 100 ng/µL) with perfect complementarity to target ITS2 of pre-rRNA also caused significant mortality of closely related species, chrysanthemum aphid Macrosiphoniella sanborni. This olinscide caused 67.15 ± 3.32% mortality rate of the chrysanthemum aphid after a single treatment and 97.38 ± 2.49% mortality rate after a double treatment (with daily interval) on the 7th day (p<0.05) (54).
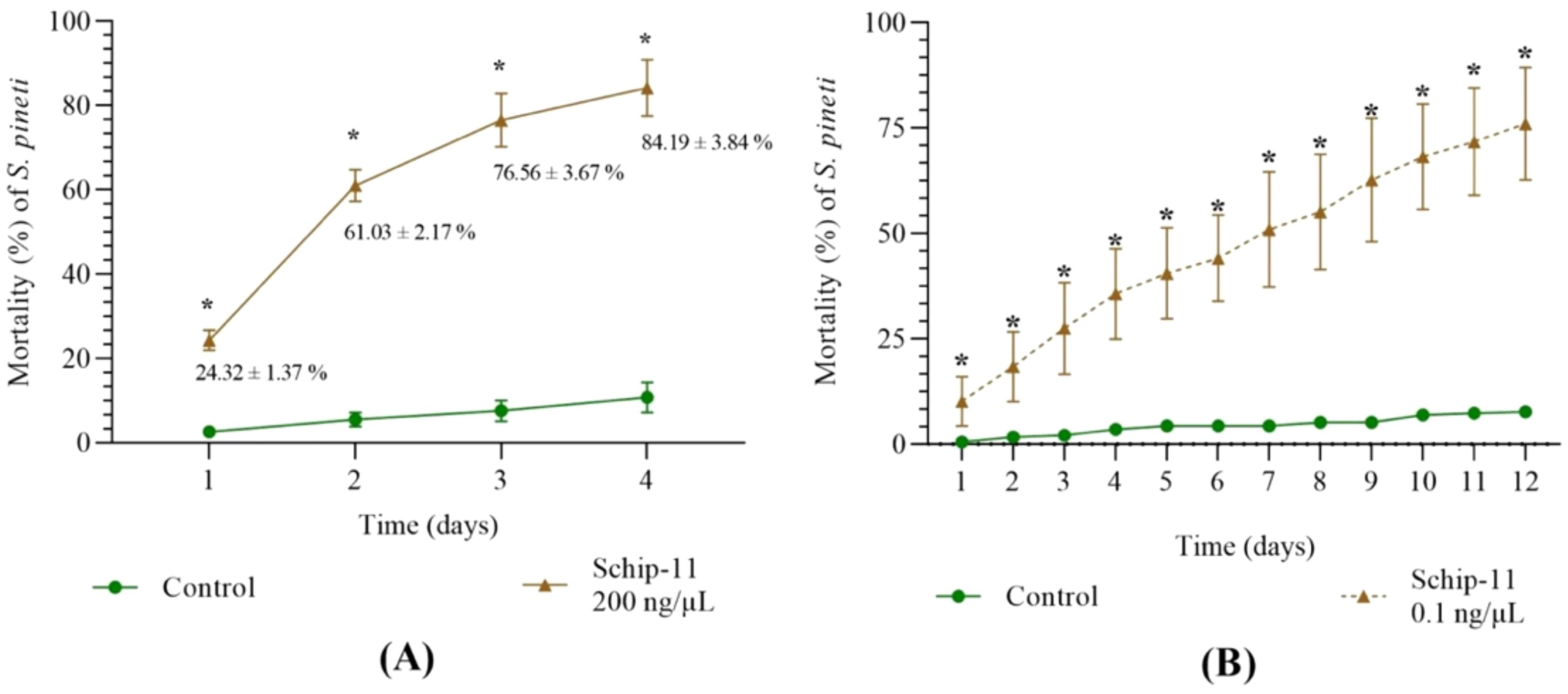
Figure 3. Dynamics of mortality of S. pineti after treatment with water and oligonucleotide insecticide Schip-11 in different concentrations: (A) 200 ng/µL; (B) 0.1 ng/µL; The significance of the difference between Schip-11 group and water-treated Control groups is indicated by *p<0.05.
After treatment of S. pineti with olinscide Schip-11 in concentration 0.1 ng/μL mortality of the pest reached 10.18 ± 3.36%, 18.44 ± 4.79%, 29.51 ± 5.35%, 35.67 ± 6.19%, and 76.06 ± 7.68% on the 1st, 2nd, 3rd, 4th, and 12th day, respectively (p<0.05) (Figure 3B). Interestingly, graph of dynamics of mortality of the pest differs from the standard S-curve which is characteristic for olinscides in concentration 200 ng/µL and 100 ng/µL (13, 32, 55, 56) and represents almost linear graph. It should also be noted that insect mortality occurs more slowly when concentration of olinscide is 0.1 ng/µL. Similar insect mortality (≈76%), obtained on 12th day in the group with a concentration of 0.1 ng/μl, was achieved already on the 3rd day in the group with a concentration of 200 ng/μl of olinscide.
Of note, closely related species of aphids from the same subfamily Lachninae, Cinara pinea and Eulachnus rileyi, also showed high sensitivity to Schip-11 in 0.1 ng/µL concentration. On the 12th day, mortality of C. pinea and E. rileyi comprised 63.66 ± 19.81% and 67.73 ± 9.16%, respectively in comparison with water-treated Controls (9.14 ± 0.83% and 8.31 ± 2.11%, respectively) (p<0.05). It shows high reproducibility of results and perspective of using low-dose concentrations of olinscides on conifer aphids.
3.2 Olinscide Schip-11 significantly decreases concentration of pre-rRNA of S. pineti (investigation carried out on dead insects)
In this article we decided to investigate concentration of pre-rRNA containing target ITS2 region after application of olinscide Schip-11 and used dead individuals of S. pineti. We found significantly decreased concentration of pre-rRNA in dead insects treated with water solutions of olinscides in both concentrations, 200 ng/µL and 0.1 ng/µL, compared to dead insects from water-treated Controls (p<0.05).
On the 1st and 3rd day, concentration of pre-rRNA was 15.62 (p<0.05) and 9.09 (p<0.05) times lower compared to water-treated Control for 200 ng/µL of Schip-11. For 0.1 ng/µL group, on the 1st and 3rd day, concentration of pre-rRNA was 17.85 (p<0.05) and 45.45 (p<0.05) times lower compared to water-treated Control (Figure 4).
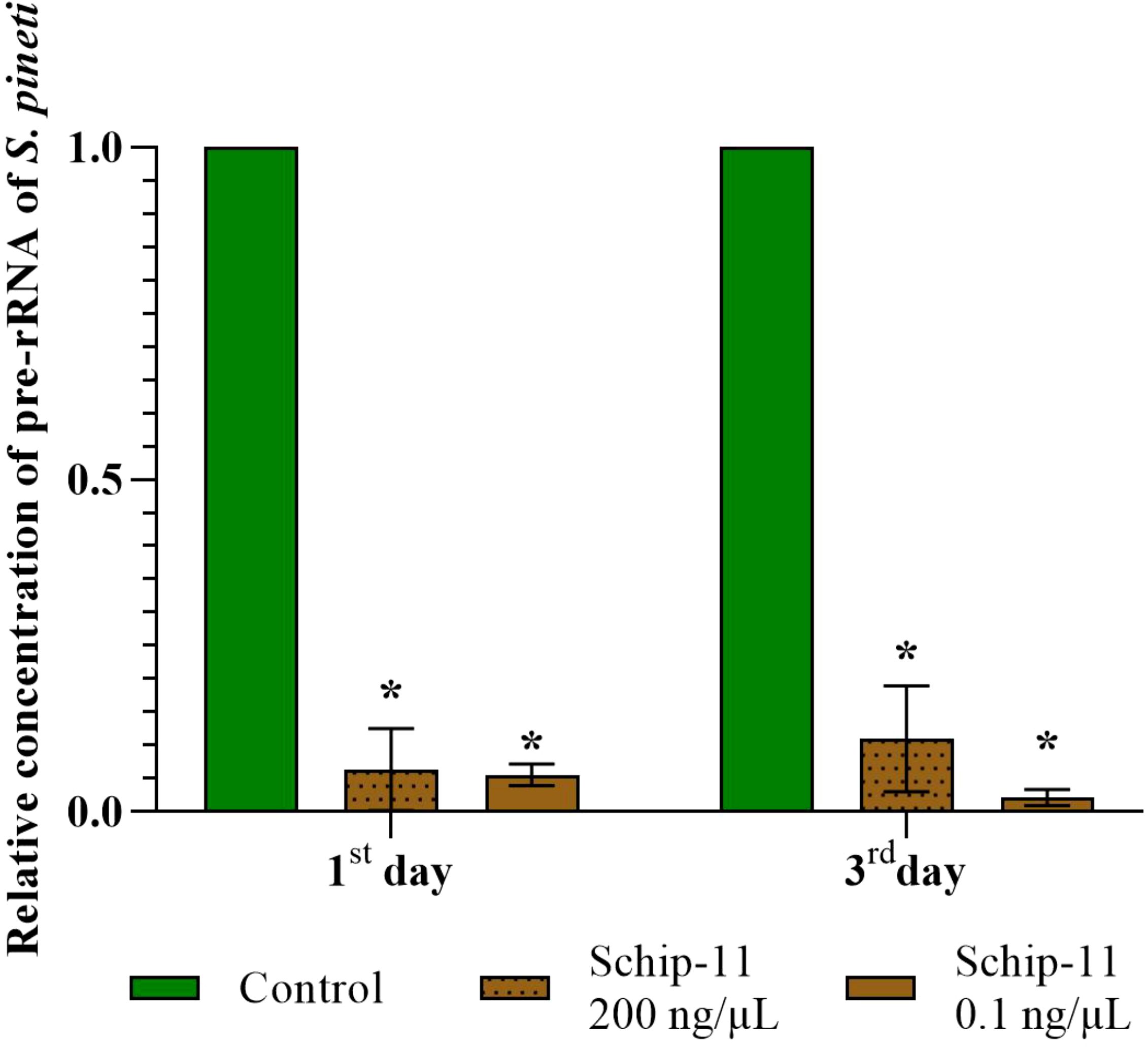
Figure 4. Relative concentration of pre-rRNA of S. pineti after treatment with oligonucleotide insecticide Schip-11 in different concentrations (200 ng/µL; 0.1 ng/µL) on the 1st and 3rd day; water-treated Control was taken as 1 (100%). The significance of differences between Schip–11 group and water-treated Controls is indicated by * at p < 0.05.
Previously, for survived individuals of chrysanthemum aphid M. sanborni we detected hypercompensation of target rRNA and then gradual decrease in 1-3 days after treatment (54) which represents typical reaction of cells of sternorrhynchans on oligonucleotide insecticides through DNA containment mechanism (13, 15). Here we show that dead insect individuals have decreased concentration of target rRNA. It is evident that increased concentration of target rRNA is better than decreased one in survived insects compared to water-treated Control (13), while dead insects are likely to have them decreased (13, 32, 54).
4 Discussion
Obtained results show substantial potential of ITS2 regions of pre-rRNAs as a target for olinscides. Moreover, ITS regions of rRNA genes are more variable (Figure 2) in comparison with sequences of 5.8S, 18S, and 28S rRNAs (57) allowing the creation of plethora unique sequences of oligonucleotide insecticides. The length of an oligonucleotide insecticide ~ 11 nt makes it possible to create selective oligonucleotide insecticides with a uniqueness frequency equal to 1/4.19·106 and is obviously enough to be used in most agrocenoses (32). In the case of ecosystems with increased diversity, such as forests, it is possible to increase the length of oligonucleotide insecticides to 15–20 nt (9). It is important to note that pre-rRNA and rRNA is a convenient target for olinscides, while mRNA, due to much lower concentration, will be much less susceptible to antisense oligonucleotides, even if oligonucleotide insecticides will possess perfect complementarity to it. Pest rRNA comprises 80% of all RNA in the cell (58) and its use as a target for ‘genetic zipper’ helps making this approach very efficient and selective at the same time. Thousands of different mRNAs make up only 5% of all RNA and use of mature rRNA and pre-rRNA for targeting substantially increases signal-to-noise ratio, ca. 105:1 (rRNA vs. random mRNA) (59).
The use of olinscides could solve, or at least improve, the fundamental problem of insecticide selectivity. The results of our previous work with chrysanthemum aphid M. sanborni showed that the change of just one nucleotide at the 1st (5′-end, T to A) and 11th (3′-end, A to T) positions leads to dramatical decrease in biological efficiency of target 11-nucleotides long olinscide Macsan-11 (54). At the same time on scale insects, Dynaspidiotus britannicus and Aonidia lauri, we showed that non-canonical base-pairing, such as A:С (С:A) and Golinscide: UrRNA (52, 60–62) may occur between olinscides and ‘imperfect’ sites of rRNAs of non-target organisms (13). Here for the first time we show that non-canonical base pairing Golinscide: UrRNA between target pre-rRNA and olinscide is also well tolerated in aphids. Definitely, non-canonical base-pairing should be taken into consideration during design of olinscides not to harm non-target organisms. Importantly, on olinscides potential hazard for non-target organisms can be calculated, while for conventional chemical insecticides it is impossible task.
Low concentrations of oligonucleotide insecticides (0.1 ng/µl) showed high insecticidal potential against aphids. The detected high mortality rate indicates an effective and target effect of unmodified antisense DNA on the pest. Of note, Yakubov et al. (63) also reported that for low concentrations of oligonucleotides (<0.5 µM), the uptake efficiency by cells is considerably higher and the average concentration of the oligonucleotide derivatives in mammalian cells exceeds the derivative concentration in the medium. This can be understood by assuming that the cells can absorb a limited amount of oligomer on their surface. This could increase the efficiency of the endocytosis process (absorptive endocytosis). At low oligonucleotide concentrations, the contribution of this process predominates. Undegraded oligodeoxynucleotides were found in cellular nuclei and cytoplasm after penetration to cells (63).
It also should be noted that at a comparable concentration (~0.07 ng/μl) dsRNA also have a significant insecticidal effect on the Colorado potato beetle (64). In parallel with oligonucleotide insecticides, a different class of insecticides based on dsRNA is being developed, the action of which is based on RNAi. RNA biocontrols show the best results on coleopterans (65), and much worse on hemipterans (66). In this context, oligonucleotide insecticides and RNA biocontrols as 2 different next-generation classes of insecticides, are able to complement each other’s action in complex preparations for wide range of pests from different orders, especially against those that have shown resistance to many different compounds from major insecticide classes (67).
Obtained here results show perspective of using olinscides against conifer aphids, including S. pineti, E. rileyi, and C. pinea. Using cold fog generators and big cold fogging machines it is possible to treat vast territories of conifer forests without harm to natural enemies (wasps, mites, beetles, etc.) of insect pests and other non-target organisms. Recent results also demonstrated remarkable specificity of oligonucleotide insecticides in action (8, 68) and showed their safety for several non-target organisms: Quercus robur, Malus domestica (69), Triticum aestivum (70), Manduca sexta, Agrotis ipsilon (71), Galleria mellonella (8). Aphids form an important part of many food chains and can be part of a healthy garden or forest ecosystem. Thus, olinscides can control a distinct pest species while closely related species will stay unharmed. Using unique complementary sequences to target pre-rRNAs and rRNAs of an insect pest it is possible to create well-tailored olinscides with minimal risks to balance of an ecosystem. As a molecule of natural origin, olinscides do not reduce biodiversity, do not impact soil health, and are not accumulated in ecosystems. We can say that olinscides degrade almost immediately after their action recruiting ubiquitous DNases (8, 13, 54). Studies of the nuclease activity of tissue homogenates of target insect pests (Lymantria dispar, Icerya purchasi, Leptinotarsa decemlineata) and their host plants (Quercus pubescens, Pittosporum tobira, Solanum tuberosum) have shown that most of the used olinscides degrade within 24 hours at 27°C (8, 32, 72) and even faster, within 1 hour, recruiting DNases of Macrosiphoniella sanborni (54).
CUAD biotechnology, as well as double-stranded RNA technology, has achieved a significant reduction in the cost of nucleic acid synthesis using innovative methods for production of nucleic acids in vitro (73, 74). СUAD biotechnology has become significantly cheaper due to liquid phase synthesis (72). One of market leaders in liquid phase synthesis, Sumitomo Chemical Co., Ltd. (Tokyo, Japan), offers the synthesis of 1 kg of unmodified oligonucleotides 11 nt long for 25,000 USD (personal communication). In the case of using non-optimized solid-phase DNA synthesis, which is available in many laboratories around the world, including ours, the cost of synthesis of 1 kg of unmodified oligonucleotides (11 nt long) will be ca. 1 million USD. Thus, at a consumption rate of 200 L per hectare, at a concentration of 0.1 mg/L (or 0.1 ng/μL), the price of the required amount of oligonucleotide insecticide will be about 0.5 USD when using liquid-phase DNA synthesis. This price allows to increase the frequency of treatments with oligonucleotide insecticides in real conditions. If non-optimized solid-phase DNA synthesis is used, which is available in many laboratories around the world, including ours, the cost of synthesizing the required amount of oligonucleotide insecticide per hectare will be 20 USD. This price will be very affordable for investigations in the lab.
Of note, today dsRNA-based technology does not have an easy algorithm for creation of pesticides providing selectivity in action and high efficiency like CUAD biotechnology does on several groups of pests (hemipterans and mites) (33). Obviously, long and fragile dsRNAs are more unpredictable in action that is why it is not easy to use them as practical tool for plant protection. In addition to better durability (75) and target specificity, antisense oligonucleotides are easier to synthesize and cheaper than siRNAs or dsRNAs. In the human system, antisense oligonucleotides have been shown to have lower immunoreactivity. For potential applications in edible plants this fact possesses significant importance since accumulation of this kind of means of insect pest control could potentially occur. Importantly, considering potential environmental risks, there are no findings in the numerous human clinical studies that prove, for example, genomic integration events attributable to the use of antisense oligonucleotides (76). While double-stranded RNA insecticides need easy and efficient algorithm and more groups of pests to show their effectiveness on, ‘genetic zipper’ method built on single-stranded DNA insecticides requires only the latter.
Double-stranded RNA biocontrols are perceived as ‘difficult’ insecticides, since they do not have clear and easy algorithm of creation, there is no strategy for RNAi how to avoid target-site resistance in insects, success of their application in the field is unpredictable, affordable production of dsRNA is not publicly available, while publicly available in vitro production is still very expensive (>200 USD/g). The efficiency of a given dsRNA pesticide is largely affected by the selection of the target gene and its targeted region, the size of the dsRNA, the method of dsRNA production and formulation, as well as by the method and dosage of the dsRNA application to crops (77). Undoubtedly, RNAi is an amazing technique for elucidation of gene function but is very fickle tool for insect pest control. On the contrary, CUAD-based oligonucleotide insecticides are considered as ‘easy’ insecticides with simple and efficient algorithm of creation and adaption to potential emergence of target-site resistance (15). Essentially it is about management of minimal risks for the environment and olinscides provide this opportunity, while this idea basically impossible for modern chemical insecticides and long dsRNA, what makes olinscides a unique and perspective approach for plant protection (13). In comparison with dsRNA biocontrols, oligonucleotide pesticides were not only the first contact nucleic acid-based insect pest control agents for plant protection, but also significantly simplified system for creation of efficient and well-tailored insecticides.
The results obtained allow us to look at CUAD biotechnology as a platform capable to occupy a significant part of the insecticide market. Most of the insect pests against which CUAD biotechnology is effective today are representatives from the suborder Sternorrhyncha, which primarily live in the subtropics and tropics, and, to a lesser extent, in the temperate zone (11). Already today, according to our estimations, the ‘genetic zipper’ method is potentially capable of effectively controlling 10-15% of all insect pests using a simple and flexible algorithm of DNA-programmable plant protection. Oligonucleotide insecticides can make a significant contribution to the protection of plants from pests of coffee, cocoa, citrus fruits, cereals, and other important groups of agricultural plants that ensure food security.
5 Conclusion
This article for the first time shows that low concentration of oligonucleotide insecticides (0.1 mg/L) leads to increased mortality of aphids. At a consumption rate of 200 L per hectare, the price of the required amount of oligonucleotide insecticide will be about 0.5 USD when using liquid-phase DNA synthesis. In fact, distribution of olinscides in 20 mg/ha ratio gives huge number of molecules per each mm2 of hectare, ca. 3.6*108/mm2. Thus, each conifer aphid of this size will get enormous amount of contact olinscide molecules. If olinscide molecules have sufficient complementarity (including non-canonical base pairing, like Golinscide: UrRNA) to target rRNA in pest cells, they will be able to cause death of target insect pest. In the case of using non-optimized solid-phase DNA synthesis, the price of the required amount of oligonucleotide insecticide per hectare will be 20 USD. ‘Genetic zipper’ method based on CUAD biotechnology opens up new frontiers for the large-scale implementation of oligonucleotide insecticides as the next generation class of insecticides in plant protection (8, 13, 14, 40). Oligonucleotide insecticides have a low carbon footprint (72), high safety for non-target organisms, rapid biodegradability in ecosystems, and avoidance of target-site resistance (8, 13, 32, 54, 72). Moreover, we are now able to predict the effectiveness of oligonucleotide insecticides on various insect pests based on their effectiveness in closely related species (15).
The CUAD platform is a simple and flexible DNA-programmable biotechnology for creation of oligonucleotide insecticides (15). Investigation of efficiency of low concentrations of oligonucleotide insecticides together with auxiliary substances (spreaders, adhesives, penetrators, etc.) will help discover the most optimal formulations for control of wide range of pests. How far we are from the point in plant protection when crop will contain only crop, without traces of chemical insecticides (organic xenobiotics)? One thing is clear, we are on the way to it.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
VO: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. YP: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. NG: Data curation, Formal analysis, Software, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was funded by a grant from the Russian Science Foundation “Development of oligonucleotide insecticides for plant protection against insect pests from the suborder Sternorrhyncha (order Hemiptera) based on short antisense oligonucleotides of ribosomal RNA genes” (project no. 22-16-20052).
Acknowledgments
We thank our many colleagues, too numerous to name, for the technical advances and lively discussions that prompted us to write this brief research report. We apologize to the many colleagues whose work has not been cited. Experiments were carried out at the Molecular Genetics and Biotechnologies Lab created within the framework of a state assignment V.I. Vernadsky Crimean Federal University for 2024 and the planning period of 2024–2026 No. FZEG-2024–0001. We are very much indebted to all reviewers and our colleagues from the lab on DNA technologies, PCR analysis, and creation of DNA insecticides (V.I. Vernadsky Crimean Federal University, Department of General Biology and Genetics), and OLINSCIDE BIOTECH LLC for valuable comments on our manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Egli M, Manoharan M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. (2023) 51:2529–73. doi: 10.1093/nar/gkad067
2. Peng Y, Wang K, Fu W, Sheng C, Han Z. Biochemical comparison of dsRNA degrading nucleases in four different insects. Front Physiol. (2018) 9:624. doi: 10.3389/fphys.2018.00624
3. Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. (2002) 1:347–55.
4. Oberemok VV. Method of Elimination of Phyllophagous Insects from Order Lepidoptera. Ukraine Patent UA No. 36, 445, 27. (2008).
5. Oberemok VV, Skorokhod OA. Single-stranded DNA fragments of insect-specific nuclear polyhedrosis virus act as selective DNA insecticides for gypsy moth control. Pestic Biochem Physiol. (2014) 113:1–7. doi: 10.1016/j.pestbp.2014.05.005
6. Oberemok VV, Laikova KV, Zaitsev AS, Gushchin VA, Skorokhod OA. The RING for gypsy moth control: Topical application of fragment of its nuclear polyhedrosis virus anti-apoptosis gene as insecticide. Pestic Biochem Physiol. (2016) 131:32–9. doi: 10.1016/j.pestbp.2016.01.006
7. Oberemok VV, Laikova KV, Zaitsev AS, Shumskykh MN, Kasich IN, Gal’chinsky NV, et al. Molecular alliance of lymantria dispar multiple nucleopolyhedrovirus and a short unmodified antisense oligonucleotide of its anti-apoptotic IAP-3 gene: A novel approach for gypsy moth control. Int J Mol Sci. (2017) 18:2446. doi: 10.3390/ijms18112446
8. Oberemok VV, Laikova KV, Gal’chinsky NV, Useinov RZ, Novikov IA, Temirova ZZ, et al. DNA insecticide developed from the Lymantria dispar 5.8S ribosomal RNA gene provides a novel biotechnology for plant protection. Sci Rep. (2019) 9:6197. doi: 10.1038/s41598-019-42688-8
9. Oberemok VV, Laikova KV, Andreeva OA, Gal’chinsky NV. Oligonucleotide insecticides and RNA-based insecticides: 16 years of experience in contact using of the next generation pest control agents. J Plant Dis Prot. (2024). doi: 10.1007/s41348-024-00949-3
10. Wang Y, Zhang H, Li H, Miao X. Second-generation sequencing supply an effective way to screen RNAi targets in large scale for potential application in pest insect control. PloS One. (2011) 6:e18644. doi: 10.1371/journal.pone.0018644
11. Oberemok VV, Laikova KV, Gal’chinsky NV. Contact unmodified antisense DNA (CUAD) biotechnology: list of pest species successfully targeted by oligonucleotide insecticides. Front Agron. (2024) 6:1415314. doi: 10.3389/fagro.2024.1415314
12. Oberemok VV, Gal’chinsky NV, Useinov RZ, Novikov IA, Puzanova YV, Filatov RI, et al. Four most pathogenic superfamilies of insect pests of suborder sternorrhyncha: invisible superplunderers of plant vitality. Insects. (2023) 14:462. doi: 10.3390/insects14050462
13. Gal’chinsky NV, Yatskova EV, Novikov IA, Sharmagiy AK, Plugatar YV, Oberemok VV, et al. Mixed insect pest populations of Diaspididae species under control of oligonucleotide insecticides: 3′-end nucleotide matters. Pesticide Biochem Physiol. (2024) 200:105838. doi: 10.1016/j.pestbp.2024.105838
14. Oberemok VV, Gal’chinsky NV. Oligonucleotide insecticides (contact unmodified antisense DNA biotechnology) and RNA biocontrols (double-stranded RNA technology): newly born fraternal twins in plant protection. biorXiv. (2024) 14. doi: 10.1101/2024.03.13.584797v1
15. Oberemok VV, Novikov IA, Yatskova EV, Bilyk AI, Sharmagiy AK, Gal’chinsky NV. Potent and selective ‘genetic zipper’ method for DNA-programmable plant protection: innovative oligonucleotide insecticides against Trioza alacris Flor. Chem Biol Technol Agric. (2024c) 11:144. doi: 10.1186/s40538-024-00668-9
16. Oberemok VV, Laikova KV, Useinov RZ, Gal’chinsky NV, Novikov IA, Yurchenko KA, et al. Insecticidal activity of three 10–12 nucleotides long antisense sequences from 5.8S ribosomal RNA gene of gypsy moth Lymantria dispar L. against its larvae. J Plant Prot Res. (2019) 59:561–4. doi: 10.24425/jppr.2019.131271
17. Schernthaner JP, Milne RE, Kaplan H. Characterization of a novel insect digestive DNase with a highly alkaline pH optimum. Insect Biochem Mol Biol. (2002) 32:255–63. doi: 10.1016/S0965-1748(01)00084-4
18. Keyel PA. Dnases in health and disease. Dev Biol. (2017) 429:1–11. doi: 10.1016/j.ydbio.2017.06.028
19. Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. (2001) 13:290–301. doi: 10.1016/S0955-0674(00)00211-8
20. Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. (2002) 84:775–90. doi: 10.1016/S0300-9084(02)01402-5
21. Belikova A, Zarytova V, Grineva N. Synthesis of ribonucleosides and diribonucleoside phosphates containing 2-chloroethylamine and nitrogen mustard residues. Tetrahedron Lett. (1967) 37:3557–62. doi: 10.1016/S0040-4039(01)89794-X
22. Knorre DG. Chemical instruments in modern biology (on the example of antisense effects on genetic structures). Soros Educ J. (1998) 12:25–31.
23. Zamecnik PC, Stephenson ML. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci USA. (1978) 75:280–4. doi: 10.1073/pnas.75.1.280
24. Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. (1979) 7:179–92. doi: 10.1093/nar/7.1.179
25. Lin M, Hu X, Chang S, Chang Y, Bian W, Hu R, et al. Advances of antisense oligonucleotide technology in the treatment of hereditary neurodegenerative diseases. Evid Based Complement Alternat Med. (2021) 2021:6678422. doi: 10.1155/2021/6678422
26. Geary RS, Henry SP, Grillone LR. Fomivirsen: clinical pharmacology and potential drug interactions. Clin Pharmacokinet. (2002) 41:255–60. doi: 10.2165/00003088-200241040-00002
27. Thakur S, Sinhari A, Jain P, Jadhav HR. A perspective on oligonucleotide therapy: Approaches to patient customization. Front Pharmacol. (2022) 13:1006304. doi: 10.3389/fphar.2022.1006304
28. Wang XW, Blanc S. Insect transmission of plant single-stranded DNA viruses. Annu Rev Entomol. (2021) 66:389–405. doi: 10.1146/annurev-ento-060920-094531
29. Wu W, Shan HW, Li JM, Zhang CX, Chen JP, Mao Q. Roles of bacterial symbionts in transmission of plant virus by hemipteran vectors. Front Microbiol. (2022) 13:805352. doi: 10.3389/fmicb.2022.805352
30. Guo Y, Ji N, Bai L, Ma J, Li Z. Aphid viruses: a brief view of a long history. Front Insect Sci. (2022) 2:846716. doi: 10.3389/finsc.2022.846716
31. Oberemok VV, Gal’chinsky NV, Novikov IA, Sharmagiy AK, Yatskova EV, Laikova EV, et al. rRNA-specific antisense DNA and dsDNA trigger rRNA biogenesis and cause potent insecticidal effect on insect pest Coccus hesperidum L. biorXiv. (2024). doi: 10.1101/2024.10.15.618468v1
32. Oberemok VV, Useinov RZ, Skorokhod OA, Gal’chinsky NV, Novikov IA, Makalish TP, et al. Oligonucleotide insecticides for green agriculture: regulatory role of contact DNA in plant–insect interactions. Int J Mol Sci. (2022) 23:15681. doi: 10.3390/ijms232415681
33. Oberemok VV, Laikova KV, Andreeva OA, Gal’chinsky NV. Biodegradation of insecticides: oligonucleotide insecticides and double-stranded RNA biocontrols paving the way for eco-innovation. Front Environ Sci. (2024) 12:1430170. doi: 10.3389/fenvs.2024.1430170
34. De Schutter K, Taning CNT, Van Daele L, Van Damme EJM, Dubruel P, Smagghe G. RNAi-based biocontrol products: market status, regulatory aspects, and risk assessment. Front Insect Sci. (2022) 1:818037. doi: 10.3389/finsc.2021.818037
35. Boukouvala MC, Kavallieratos NG, Skourti A, Pons X, Alonso CL, Eizaguirre M, et al. Lymantria dispar (L.) (Lepidoptera: erebidae): current status of biology, ecology, and management in europe with notes from North America. Insects. (2022) 13:854. doi: 10.3390/insects13090854
36. Kumar H, Sharma M, Chandel A. DNA insecticides: future of crop protection(2022). Available online at: http://agrifoodmagazine.co.in/2022/volume-4-issue-6-june-2022/ (Accessed 28 Apr 2024).
37. Manju M, Nirosha V, Tullika T, Mankhanniang G. DNA insecticides: an emerging tool in pest management (2022). Available online at: https://agriallis.com/wp-content/uploads/2022/09/DNA-INSECTICIDES-AN-EMERGING-TOOL-IN-PEST-MANAGEMENT.pdf (Accessed 7 May 2024).
38. Oirdi ME, Yaseen M, Farwa U, Raza MA, Farhan M, Sandhu ZA, et al. Crops and people: the dangers and potential benefits of pesticides. Cogent Food Agric. (2024) 10. doi: 10.1080/23311932.2024.2334096
39. Patil V, Jangra S, Ghosh A. Advances in antisense oligo technology for sustainable crop protection. Crit Rev Plant Sci. (2024) 1–23. doi: 10.1080/07352689.2024.2394001
40. TriLink BioTechnologies. Feasibility of Antisense Oligonucleotides as DNA Insecticides (2024). Available online at: https://www.trilinkbiotech.com/blog/feasibility-of-antisense-oligonucleotides-as-dna-insecticides/ (Accessed 28 Apr 2024).
41. Will T, Vilcinskas A. The structural sheath protein of aphids is required for phloem feeding. Insect Biochem Mol Biol. (2015) 57:34–40. doi: 10.1016/j.ibmb.2014.12.005
42. Brault V, Uzest M, Monsion B, Jacquot E, Blanc S. Aphids as transport devices for plant viruses. C R Biol. (2010) 333:524–38. doi: 10.1016/j.crvi.2010.04.001
43. Donnelly R, Cunniffe NJ, Carr JP, Gilligan CA. Pathogenic modification of plants enhances long-distance dispersal of nonpersistently transmitted viruses to new hosts. Ecology. (2019) 100:e02725. doi: 10.1002/ecy.2725
44. Bass C, Nauen R. The molecular mechanisms of insecticide resistance in aphid crop pests. Insect Biochem Mol Biol. (2023) 156:103937. doi: 10.1016/j.ibmb.2023.103937
45. Khanal N, Vitek C, Kariyat R. The known and unknowns of aphid biotypes, and their role in mediating host plant defenses. Diversity. (2023) 15:186. doi: 10.3390/d15020186
46. Ali J, Bayram A, Mukarram M, Zhou F, Karim MF, Hafez MMA, et al. Peach–potato aphid myzus persicae: current management strategies, challenges, and proposed solutions. Sustainability. (2023) 15:11150. doi: 10.3390/su151411150
47. Roy L, Barrès B, Capderrey C, Mahéo F, Micoud A, Hullé M, et al. Host plants and insecticides shape the evolution of genetic and clonal diversity in a major aphid crop pest. Evol Appl.. (2022) 15(10):1653–69. doi: 10.1111/eva.13417
48. Rubio-Meléndez ME, Barrios-SanMartin J, Pina-Castro FE, Figueroa CC, Ramirez CC. Asexual reproduction of a few genotypes favored the invasion of the cereal aphid Rhopalosiphum padi in Chile. Peer J. (2019) 7:e7366. doi: 10.7717/peerj.7366
49. Loxdale HD, Balog A, Biron DG. Aphids in focus: unravelling their complex ecology and evolution using genetic and molecular approaches. Biol J Linn Society. (2020) 129:507–31. doi: 10.1093/biolinnean/blz194
50. Kaleem Ullah RM, Gao F, Sikandar A, Wu H. Insights into the effects of insecticides on aphids (Hemiptera: aphididae): resistance mechanisms and molecular basis. Int J Mol Sci. (2023) 24:6750. doi: 10.3390/ijms24076750
51. InfluentialPoints.com. Available online at: https://influentialpoints.com/Gallery/Schizolachnus_pineti_grey_waxy_pine_needle_aphid.htm:~:text=Schizolachnus%20pineti%20can%20be%20found,along%20the%20previous%20year's%20needles (Accessed 28 Apr 2024).
52. Sabalette KB, Makarova L, Marcia M. G·U base pairing motifs in long non-coding RNAs. Biochimie. (2023) 214:123–40. doi: 10.1016/j.biochi.2023.06.003
53. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. (1994) 22:4673–80. doi: 10.1093/nar/22.22.4673
54. Puzanova YV, Novikov IA, Bilyk AI, Sharmagiy AK, Plugatar YV, Oberemok VV. Perfect complementarity mechanism for aphid control: oligonucleotide insecticide macsan-11 selectively causes high mortality rate for macrosiphoniella sanborni gillette. Int J Mol Sci. (2023) 24:11690. doi: 10.3390/ijms241411690
55. Gal’chinsky N, Useinov R, Yatskova E, Laikova K, Novikov I, Gorlov M, et al. A breakthrough in the efficiency of contact DNA insecticides: rapid high mortality rates in the sap-sucking insects Dynaspidiotus britannicus Comstock and Unaspis euonymi Newstead. J Plant Prot Res. (2020) 60:220–3. doi: 10.24425/jppr.2020.133315
56. Useinov RZ, Gal’chinsky N, Yatskova E, Novikov I, Puzanova Y, Trikoz N, et al. To bee or not to bee: creating DNA insecticides to replace non-selective organophosphate insecticides for use against the soft scale insect Ceroplastes japonicus Green. J Plant Prot Res. (2020) 60:406–9. doi: 10.24425/jppr.2020.133956
57. Matyášek R, Kuderová A, Kutílková E, Kučera M, Kovařík A. Intragenomic heterogeneity of intergenic ribosomal DNA spacers in Cucurbita moschata is determined by DNA minisatellites with variable potential to form non-canonical DNA conformations. DNA Res. (2019) 26:273–86. doi: 10.1093/dnares/dsz008
58. Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. (1999) 24:437–40. doi: 10.1016/S0968-0004(99)01460-7
59. Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. (2015) 6:2. doi: 10.3389/fgene.2015.00002
60. Fox KR, Allinson SL, Sahagun-Krause H, Brown T. Recognition of GT mismatches by Vsr mismatch endonuclease. Nucleic Acids Res. (2000) 28:2535–40. doi: 10.1093/nar/28.13.2535
61. Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. (2005) 33:1671–7. doi: 10.1093/nar/gki312
62. Luige O, Karalė K, Bose PP, Bollmark M, Tedebark U, Murtola M, et al. Influence of sequence variation on the RNA cleavage activity of Zn2+-dimethyl-dppz-PNA-based artificial enzymes. RSC Adv. (2022) 12:5398–406. doi: 10.1039/d1ra08319h
63. Yakubov LA, Deeva EA, Zarytova VF, Ivanova EM, Ryte AS, Yurchenko LV, et al. Mechanism of oligonucleotide uptake by cells: involvement of specific receptors? Proc Natl Acad Sci USA.. (1989) 86(17):6454–8. doi: 10.1073/pnas.86.17.6454
64. Pallis S, Alyokhin A, Manley B, Rodrigues T, Barnes E, Narva K. Effects of low doses of a novel dsRNA-based biopesticide (Calantha) on the colorado potato beetle. J Econ Entomol. (2023) 116:456–61. doi: 10.1093/jee/toad034
65. Christiaens O, Whyard S, Vélez AM, Smagghe G. Double-stranded RNA technology to control insect pests: current status and challenges. Front Plant Sci. (2020) 11:451. doi: 10.3389/fpls.2020.00451
66. Cooper AM, Silver K, Zhang J, Park Y, Zhu KY. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manage Sci. (2019) 75:18–28. doi: 10.1002/ps.5126
67. Rodrigues TB, Mishra SK, Sridharan K, Barnes ER, Alyokhin A, Tuttle R, et al. First sprayable double-stranded RNA-based biopesticide product targets proteasome subunit beta type-5 in Colorado potato beetle (Leptinotarsa decemlineata). Front Plant Sci. (2021) 12:728652. doi: 10.3389/fpls.2021.728652
68. Oberemok VV, Laikova KV, Repetskaya AI, Kenyo IM, Gorlov MV, Kasich IN, et al. A half-century history of applications of antisense oligonucleotides in medicine, agriculture and forestry: we should continue the journey. Molecules. (2018) 23:1302. doi: 10.3390/molecules23061302
69. Zaitsev A, Omel’chenko O, Nyadar P, Oberemok V. Influence of DNA oligonucleotides used as insecticides on biochemical parameters of Quercus robur and Malus domestica. Bulletin of the Transilvania University of Brasov (2015) 8:37–46.
70. Nyadar PM, Oberemok V, Omelchenko A, Kerimova S, Seidosmanova E, Krasnodubiets A, et al. DNA insecticides: The effect of concentration on non-target plant organisms such as wheat (Triticum aestivum L.). J Plant Prot Res. (2019) 59:60–8. doi: 10.24425/jppr.2019.126038
71. Oberemok VV, Laikova VK, Zaitsev SA, Nyadar MP, Shumskykh NM, Gninenko IY. DNA insecticides based on iap3 gene fragments of cabbage looper and gypsy moth nuclear polyhedrosis viruses show selectivity for non-target insects. Arch Biol Sci. (2015) 67:785–92. doi: 10.2298/ABS141230037O
72. Gal’chinsky NV, Yatskova EV, Novikov IA, Useinov RZ, Kouakou NJ, Kouame KF, et al. Icerya purchasi maskell (Hemiptera: monophlebidae) control using low carbon footprint oligonucleotide insecticides. Int J Mol Sci. (2023) 24:11650. doi: 10.3390/ijms241411650
73. Taning CN, Arpaia S, Christiaens O, Dietz-Pfeilstetter A, Jones H, Mezzetti B, et al. RNA-based biocontrol compounds: current status and perspectives to reach the market. Pest Manage Sci. (2020) 76:841–5. doi: 10.1002/ps.5686
74. Vilkhovoy M, Adhikari A, Vadhin S, Varner JD. The evolution of cell free biomanufacturing. Processes. (2020) 8:675. doi: 10.3390/pr8060675
75. Thorp HH. The importance of being r: greater oxidative stability of RNA compared with DNA. Chem Biol. (2000) 7:33–6. doi: 10.1016/s1074-5521(00)00080-6
76. Gruber C, Gursinsky T, Gago-Zachert S, Pantaleo V, Behrens SE. Effective antiviral application of antisense in plants by exploiting accessible sites in the target RNA. Int J Mol Sci. (2023) 24:17153. doi: 10.3390/ijms242417153
Keywords: ‘genetic zipper’ method, oligonucleotide insecticides, contact unmodified antisense DNA (CUAD) biotechnology, cost-effective aphid control, DNA-programmable plant protection
Citation: Oberemok VV, Puzanova YV and Gal’chinsky NV (2024) The 'genetic zipper' method offers a cost-effective solution for aphid control. Front. Insect Sci. 4:1467221. doi: 10.3389/finsc.2024.1467221
Received: 19 July 2024; Accepted: 11 November 2024;
Published: 11 December 2024.
Edited by:
Frank Chidawanyika, International Centre of Insect Physiology and Ecology (ICIPE), KenyaReviewed by:
Kanakachari Mogilicherla, Indian Institute of Rice Research (ICAR), IndiaNorman Muzhinji, University of the Free State, South Africa
Copyright © 2024 Oberemok, Puzanova and Gal’chinsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nikita V. Gal’chinsky, cGNyLnByb2R1Y3RAZ21haWwuY29t
 Vol V. Oberemok
Vol V. Oberemok Yelizaveta V. Puzanova1
Yelizaveta V. Puzanova1 Nikita V. Gal’chinsky
Nikita V. Gal’chinsky