- Department of Entomology, Martin-Gatton College of Agriculture, Food and Environment, University of Kentucky, Lexington, KY, United States
Since its discovery in 1998, RNA interference (RNAi), a Nobel prize-winning technology, made significant contributions to advances in biology because of its ability to mediate the knockdown of specific target genes. RNAi applications in medicine and agriculture have been explored with mixed success. The past 25 years of research on RNAi resulted in advances in our understanding of the mechanisms of its action, target specificity, and differential efficiency among animals and plants. RNAi played a major role in advances in insect biology. Did RNAi technology fully meet insect pest and disease vector management expectations? This review will discuss recent advances in the mechanisms of RNAi and its contributions to insect science. The remaining challenges, including delivery to the target site, differential efficiency, potential resistance development and possible solutions for the widespread use of this technology in insect management.
1 Introduction
The target-specific interference of mRNA transcription, stability, and translation by complexes of Argonaute (Ago) family proteins and small RNAs such as small interfering RNA (siRNA) and microRNA (miRNA) resulting in decreased levels of gene products and their function is referred to as RNA interference (RNAi) (1, 2). Twenty-five years of research on methods development, mechanisms of action, and development of applications in animals, humans, and plants led to its widespread use in basic and applied research (3, 4). RNAi methods contributed significantly to advances in all areas of biology. RNAi-based therapeutics have been developed and are being used for the treatment of human diseases. Also, RNAi-based methods have been developed for pest and disease control and crop improvement in agriculture. Science Journal declared RNAi as the technology of the year in 2002. The Nobel Prize in Medicine for 2006 was awarded to Fire and Mello for discovering RNAi. Since its discovery, RNAi has played an important role in advances in insect science. Initial research on RNAi in insects was conducted in the model insect, Drosophila melanogaster. During the next few years, RNAi function was discovered in many insect species, including some economically important insects such as pests, disease vectors, and beneficial insects. RNAi works efficiently in some groups of insects, such as beetles but is inefficient in other insects, such as moths and butterflies. The differential efficiency of RNAi among pests and the potential development of resistance to RNAi are some of the major hurdles to the widespread use of RNAi in insect pest and disease vector management. There are many reviews published on RNAi; rather than adding one more to this list of reviews on this subject, I will focus my discussion on the critical evaluation of contributions of RNAi over the past 25 years along with challenges and possible solutions for widespread use of this technology for the management of pests and disease vectors.
2 RNAi discovery
Gene silencing research started with an unexpected phenotype detected in petunia plants. When a gene coding for an enzyme to enhance purple color was introduced into petunia plants, the transgenic plants showed a decrease rather than an increase in purple color (5). Further research revealed co-suppression of the endogenous gene coding for the enzyme involved in the production of purple color (6). Co-suppression of the endogenous gene was reported for virus resistance in plants and animals. La Crosse (LA) virus gene introduced into an infectious Sindbis virus expression vector caused interference to its replication (7). Also, the expression of virus genes interfered with the replication of the California serogroup virus in mosquito cells and mosquitoes (8). The silencing of genes was reported in the fungus, Nuerospora crassa (9). However, in these studies, the trigger of silencing and the mechanism of silencing were not identified. A landmark publication in RNAi by Fire and Mello (2) in 1998, for the first time, identified double-stranded RNA (dsRNA) as the molecule that triggered RNA silencing and coined the term RNA interference (RNAi). This publication also reported on the mechanism of action of dsRNA to achieve target gene silencing. Subsequent research during the next few years reported RNAi in nematodes, insects, plants, and trypanosomes (10–18). The functioning of RNAi was reported in mammalian cells in 2001 (19) and mice in 2002 (20). Research on RNAi during the past 25 years resulted in development of many applications in medicine and agriculture (21–28).
3 RNAi in insects
The first insect to demonstrate RNAi function was the fruit fly, Drosophila melanogaster (12, 29). Fruit fly genetic and genomic tools were employed to study the functions of genes using transgenic flies (30). Transgenic fly lines expressing RNAi triggers for almost all genes identified in the Drosophila genome are available from Drosophila stock centers. These UAS transgenic flies can be crossed with GAL4 lines expressing GAL4 in a tissue and stage-specific manner, allowing the knockdown of target genes in specific tissues and stages. The addition of RNAi to the Drosophila toolbox revolutionized functional genomics in this model insect and facilitated studies on the function of genes involved in almost every aspect of insect life (3). High throughput screening assays were developed using Drosophila cell lines and transgenic flies and used to identify genes and their function in developmental and physiological processes in flies (31–57). During the next few years functioning of RNAi was demonstrated in other insects such as Anopheles gambiae (30), Tribolium castaneum (18), Anopheles stephensi (22), Blattella germanica (58), Manduca sexta (59), Apis mellifera (60–62), Aedes aegypti (63), Locusta migratoria (64), Leptinotarsa decemliata (65) and Nilaparvata lugens (66).
4 RNAi mechanism
Double-stranded RNAs in the cytoplasm of the cells are recognized by dicer enzymes and digest them into small interference RNA (siRNA) of 18-21 base pair length (20, 67, 68). The RNA-Induced Silencing Complex (RISC) containing argonaute proteins (69, 70) facilitates the binding of siRNA to complementary DNA/mRNA blocking replication, transcription and translation (71). This results in reduced levels of target gene products hence the knockdown or knockout effect of the target gene function. In mammalian cells, dsRNAs induce interferon response (72). Most of the the critical players including, dicers and argonautes involved in RNAi response are conserved in insects. However proteins involved in dsRNA uptake and transport into cytoplasm vary among insect species contributing to differences in RNAi response among insect species (more discussion on this in section 6) However, the interferon response in insect cells is not robust. Therefore, long dsRNAs could be used as RNAi triggers in insects. The exogenously applied dsRNA is processed to multiple siRNAs by insect cells bypassing the need for optimization of siRNA for each gene normally required for efficient silencing of target genes in vertebrates. Long dsRNAs (200-400 bp) are delivered to insect cell lines in vitro or injected into insects in vivo to induce RNAi response. This makes it easier to implement RNAi technology in laboratories without access to bioinformatics tools. This may be one of the reasons for the extensive use of RNAi in entomology laboratories all over the world. RNAi technology contributed to advances in our knowledge of pest biology, insecticide targets and resistance development to insecticides. Most of this research on non-model insects during the past 25 years would not have been possible without the use of RNAi technology.
5 Applications of RNAi in basic science and pest control
RNAi made significant contributions to advances in our understanding of insect biology. A combination of genetics and genomic tools and the availability of genome sequences in model insects such as the fruit fly, silk moth and red flour beetle facilitated research in functional genomics to determine functions of genes involved in insect development, reproduction and behavior. RNAi also played an important role in advancing the biology of non-model insects that include pests, disease vectors and beneficial insects. Research on the identification and characterization of insecticide target sites and mode of action of insecticides and resistance development against insecticides by insects is aided by the use of RNAi methods. Functional genomics studies aided by RNAi in destructive pests such as the brown plant hopper, locust and disease vectors such as the yellow fever mosquito advanced our knowledge on development, reproduction and immune response in these insects.
RNAi works well and is used extensively in the red flour beetle, Tribolium castaneum to study the function of genes involved in growth, development, reproduction and insecticide resistance (73–100). The western corn rootworm, Diabrotica virgifera virgifera is another coleopteran insect where RNAi helped to identify targets for insecticide development (101, 102).
Expression of RNAi triggers in plants results in the knockdown of target genes in insects that feed on transgenic plants resulting in their mortality (101, 103). These results showed the potential of RNAi technology in pest management and attracted investment from both public and private sectors for the development of RNAi-based methods for controlling pests and disease vectors. The utility of RNAi technology to protect crops and trees from insect damage was tested in multiple systems, including cotton (104–106), rice (107), potato (108, 109), tobacco (110–113), poplar (114) and wheat (115). Surprisingly, the plant-mediated delivery of RNAi triggers was tested only in a few crop-pest systems. Difficulty in producing transgenic plants for economically important crops and hesitancy in public acceptance of food derived from genetically modified crops may have contributed to the slow progress of research in this area. Despite 20 years of efforts, only one plant-mediated RNAi product, corn seed for protection against corn rootworm has been commercialized.
In insects such as the Colorado potato beetle, tephritid fruit fly (116) and beet armyworm (117), feeding dsRNA produced in bacteria induces efficient knockdown of target genes and mortality. dsRNA produced in yeast (118) and algae (119–121) was also reported to trigger RNAi in mosquitoes. In vitro synthesized dsRNA also induces efficient RNAi in coleopteran insects such as the Colorado potato beetle and flea beetle, and these products are under development for registration to control these pests. RNAi is also being developed to protect beneficial insects from pathogens such as the Israeli Acute Paralysis Virus that infects honey bees (122). Remebee-1, a dsRNA product, was developed to control this disease of honey bees (60). Transgenic silkworms expressing dsRNA targeting lef-1 gene of Bombyx mori nucleopolyhedrovirus (BmNPV) were developed to control this viral disease (123, 124). Another application of RNAi is for blocking the transmission of human and plant pathogens by insect vectors. dsRNA targeting the defensin gene in Anopheles gambiae adults reduced antimicrobial defense against Gram-positive bacteria (30). Also, knockdown of a complement-like protein 1, TEP1, gene in An. gambiae increased the number of developing parasites in the susceptible strain but abolished melanotic refractoriness in the refractory strain (125). RNAi played a critical role in understanding host-parasite interactions and immune responses in mosquitoes (126–140).
6 Roadblocks to widespread use of dsRNA pesticides
After 25 years of research on RNAi, why do we have only a few RNAi-based pesticides registered for pest management? The differential efficiency of RNAi among insects and its lower efficiency in major pests, potential resistance development and public hesitancy in accepting food from genetically modified crops are the major hurdles to the widespread use of RNAi in pest management. After discovering the functioning of RNAi in many insects, it became clear that RNAi works with variable efficiency among insect species tested. In beetles belonging to Tenebrionidae, Chrysomelidae and other families, the RNAi is efficient and systemic (141–147). Injection of dsRNA induces efficient RNAi in hemipteran insects, the milkweed bug (Oncopeltus fasciatus) (148), the brown marmorated stink bug (Halyomorpha halys) (149), the bed bug (Cimex lectularius) (150) and also in other insects such as B. germanica (58), S. gregaria and L. migratoria (151, 152) belonging to other orders. In contrast, RNAi is variable and inefficient in insects from most of the other orders, such as Lepidoptera (153). Interestingly, RNAi works well in leafhoppers from the order Hemiptera but is inefficient in aphids belonging to the same order. In most insects, injection works better than feeding for delivering dsRNA. In Drosophila, transgenic expression of hairpin dsRNA works more efficiently compared to feeding or injection of dsRNA. In locust and a few other insects only injection of dsRNA triggers RNAi; feeding dsRNA in these insects does not result in the knockdown of target genes (151). RNAi efficiency is also variable in different tissues and stages of an insect. RNAi efficiency also depends on the target gene and its expression levels (154). Constitutively and highly expressed target genes are knocked down much more efficiently than those expressed at lower levels and restricted developmental stages and tissues (154). These results suggest that degradation of RNA in the alimentary canal, delivery of dsRNA to cytoplasm for digestion by dicer enzymes and recruitment by RISC complex and expression levels of genes involved in RNAi might play important roles in determining RNAi efficiency.
The first roadblocks encountered by the dsRNA en route to its site of action are double-stranded ribonucleases (dsRNases) (155–160) (Figure 1). In Locusta migratoria, injection but not oral delivery of dsRNA induces RNAi (151). dsRNA digestion by nucleases is a major contributor to inefficient RNAi (161). Silencing of dsRNase genes improves RNAi efficiency (156, 162–164). dsRNases are more active in the tobacco budworm (Heliothis virescens, inefficient RNAi) than those in L. decemlineata (165). dsRNA added to the medium of Sf9 cells is degraded by dsRNases secreted by these cells; silencing dsRNases improve RNAi efficiency in these cells (166). RNAi efficiency-related nuclease (REase) identified in lepidopterans has been proposed as a main factor contributing to inefficient RNAi in these insects (157). Formulations that protect dsRNA from nucleases might improve RNAi efficiency in insects that are refractive to dsRNA.
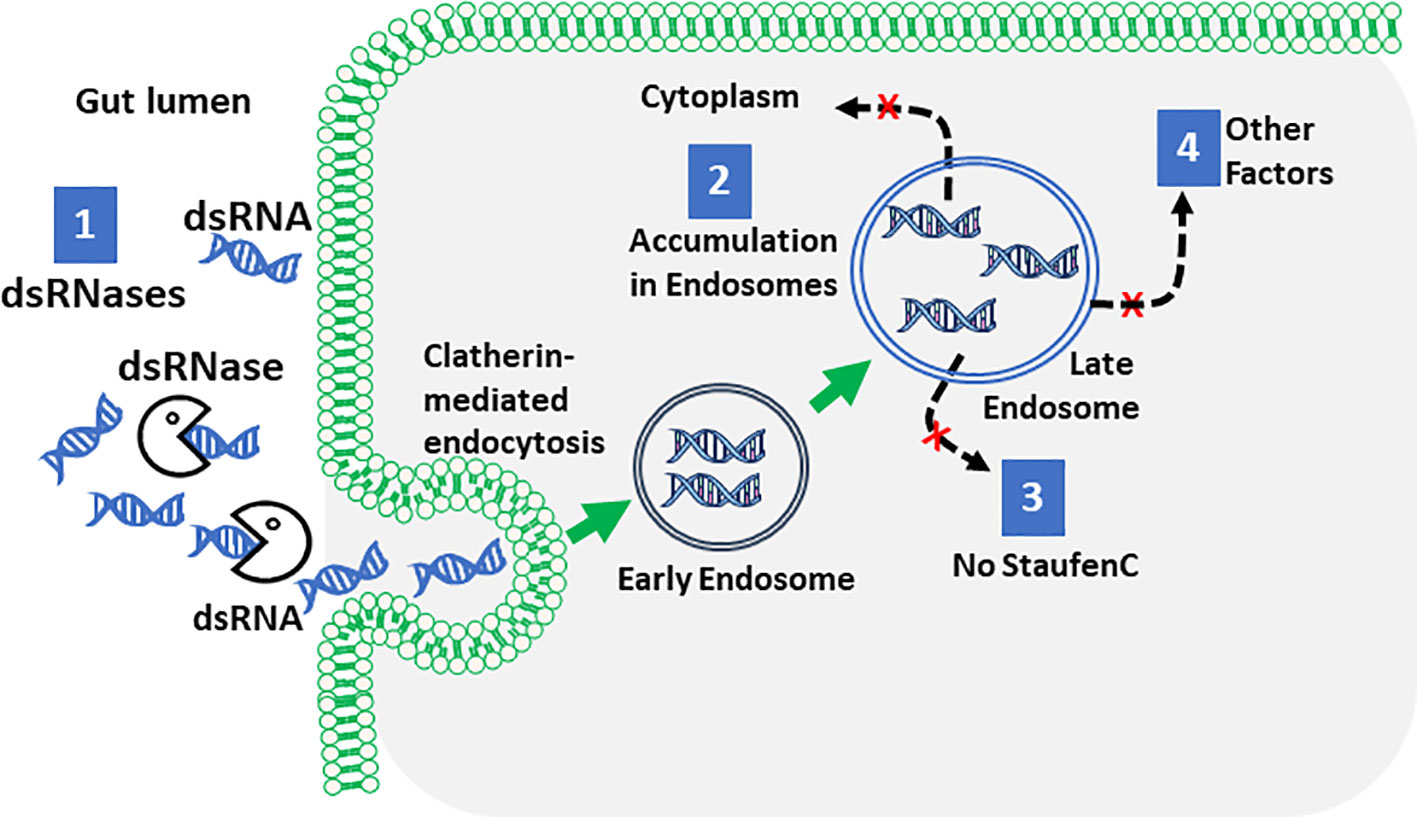
Figure 1 The major roadblocks to efficient RNAi in insects. The cartoon illustrates dsRNA degradation by dsRNases, endosomal entrapment and lack of key players in RNAi such as StaufenC as the major reasons for inefficient RNAi Lepidopterans and other insects.
The second roadblock dsRNA encounters while reaching its site of action in the cytoplasm of the cells is crossing the cell membrane and intracellular transport (Figure 1). Clatherin-mediated endocytosis was identified as a major pathway for dsRNA uptake (167–174). Macropinocytosis was identified as the major route of dsRNA uptake in the boll weevil (Anthonomus grandis) (175). Depending on the concentration and size, dsRNA may enter cells through multiple mechanisms (171). In L. migratoria, dsRNA does not induce RNAi in follicle cells and oocytes because these tissues do not take up dsRNA (176). In coleopterans, intracellular transport is efficient in delivering dsRNA to the cytoplasm (165). In contrast, in lepidopterans, the intracellular transport is inefficient and most of the dsRNA is trapped in the endosomes (165, 177).
Differences in the expression levels of key players involved in the intracellular transport of dsRNA and its processing to siRNA and recruitment to RISC complex influence RNAi efficiency. Differences in the processing of dsRNA to siRNA were detected between L. decemlineata and S. frugiperda (165). In insects belonging to orders Lepidoptera, Orthoptera, Hemiptera and Diptera, the processing of dsRNA to siRNA is not as efficient as in coleopterans (178). Differences in the expression levels of Dicer enzymes and their activity could contribute to the differences observed in dsRNA to siRNA processing. The number of Argonaut genes identified, as well as participation of these in siRNA, miRNA and piRNA pathways, vary among insects (172, 179–185). Transgenic expression of Ago2 in B. mori improved RNAi efficiency, suggesting that this protein may be one of the limiting factors responsible for the refractiveness of lepidopterans to RNAi (186).
A double-stranded RNA binding protein (StaufenC) was identified as a key player for efficient RNAi in coleopterans, L. decemlineata (65) and T. castaneum (187). Interestingly, this gene has been identified only in coleopterans and is required for processing dsRNA to siRNA in these insects. L. decemlineata, Lepd-SL1 RNAi resistant cells selected by exposure to dsRNA trigger for multiple generations express lower levels of StaufenC than RNAi susceptible cells. These studies showed that StaufenC is a key player in efficient RNAi coleopterans and a potential target for resistance development to RNAi-based pesticides (187). Recent studies showed that StaufenC functions in coleopterans like Loquacious in D. melanogaster which is involved in processing dsRNA to siRNA (188). The nematode C. elegans requires systemic RNA interference defective protein 1 (CeSid1) for successful RNAi (189). The expression of CeSid1 in insect cell lines improves RNAi (190–192). However, the requirement of SID1 homologs for RNAi function varies among insects tested (193, 194). Expression of CeSid1 in two S. frugiperda cell lines, ovarian-derived Sf9 and midgut-derived Sf17 cells, showed that CeSid1 increases RNAi efficiency in Sf9 but not in Sf17 cells (195). In Sf9 cells expressing CeSid1, decreased accumulation of dsRNA in late endosomes and increased processing dsRNA to siRNA was observed. The Verson’s glands showed the greatest improvement, while the midgut showed the least improvement in RNAi efficiency in CeSID1-expressing transgenic insects. These data point to the variability of RNAi machinery among cell and tissue types (195).
7 Strategies to improve RNAi efficiency in major pests and disease vectors
Delivery of dsRNA to the site of action in the cytoplasm of the cell will likely improve RNAi efficiency in major pests and disease vectors that are refractive to RNAi. As with any other insecticide-active ingredients, the formulation is key to the successful delivery of dsRNA to the site of action. Several materials were developed to formulate dsRNA with the goal of improving RNAi efficiency (Figure 2). In the mosquito, Anopheles gambiae, larvae, chitosan and dsRNA formulations improved RNAi efficiency (196). Several other nanoformulations of dsRNA, including guanidine-containing polymers, nanocarrier/dsRNA/detergent formulation, guanylated polymers and branched amphiphilic peptide bilayer conjugated gold nanoparticles were also shown to improve RNAi (175, 197–215). The nanoformulations of dsRNA help protect dsRNA from dsRNases (210–214), facilitate dsRNA penetration through the body wall (216), and improve the cellular uptake and endosomal escape of dsRNA (210–214, 217, 218), ultimately resulting in increased RNAi efficiency (Figure 2). Short (23 nt) RNAs with partially closed ends (pcRNAs) were shown to enter cells through a clathrin-independent pathway and improve knockdown of target genes (219). The pcRNAs might help in overcoming some of the hurdles faced by dsRNA reaching the cytoplasm in insects that are recalcitrant to RNAi. dsRNA expressed in microorganisms such as bacteria (117, 220–223), yeast (118, 224–226), algae (119–121), Bacillus thuringiensis bacteria (227, 228) and plant viruses (229) were shown to induce knockdown of target genes in insects that ingest these microorganisms expressing dsRNA. Plant-mediated delivery of dsRNA is another option. Indeed, the first RNAi commercial product is based on this method; transgenic maize expressing dsRNA targeting the SNF7 gene of corm rootworm protects plants from its damage (101). In-planta expression of dsRNA has been demonstrated to protect crops from pests (104, 109, 111, 230–232).
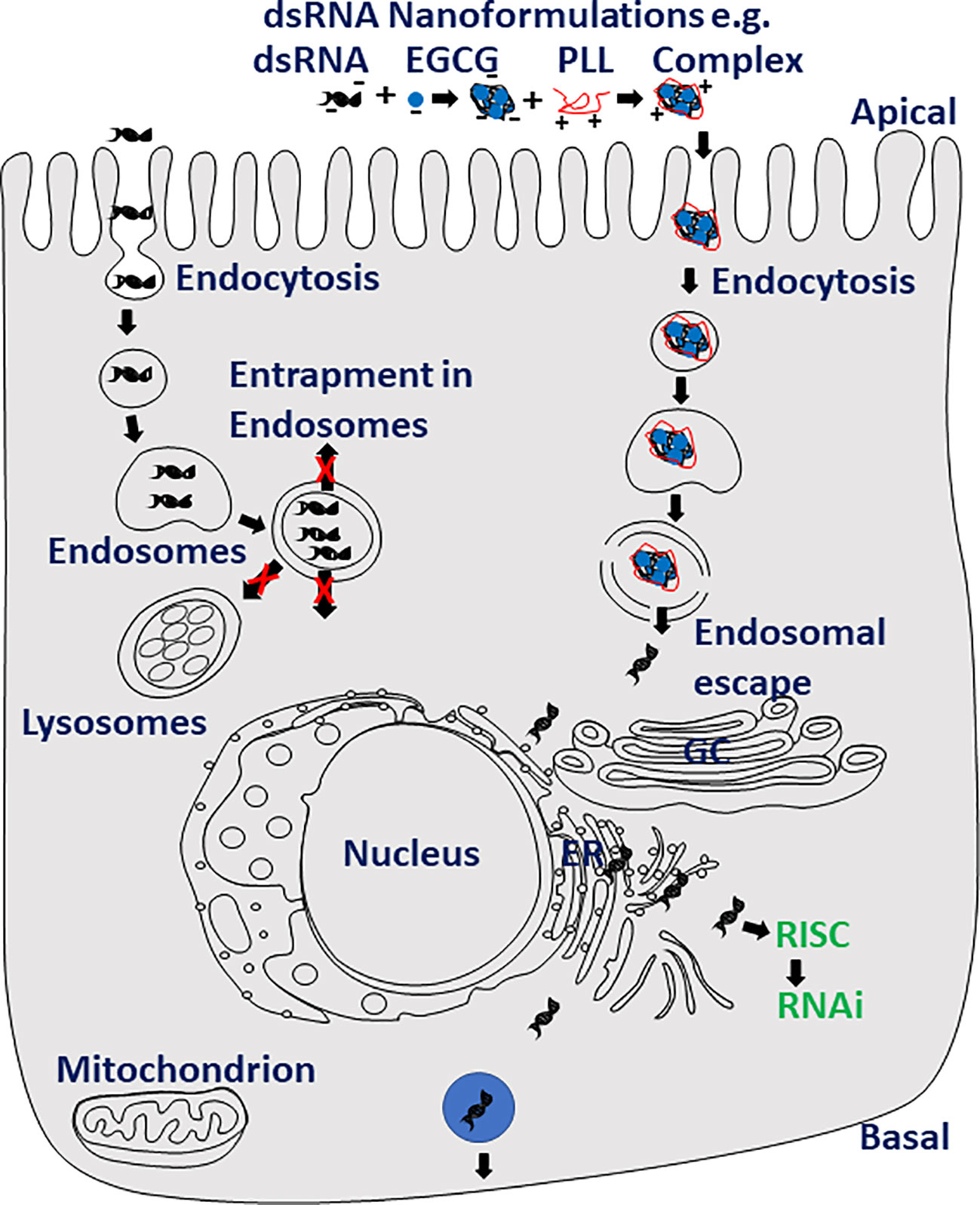
Figure 2 Nanotechnology may help increasing RNAi efficiency in major pests. Nanoformulations of dsRNA such as dsRNA, polyphenol (−)-epigallocatechin-3-O-gallate (EGCG) and poly-l-lysine (PLL) protect dsRNA from degradation by dsRNases, improve uptake, intracellular transport and endosomal escape of dsRNA resulting in increased RNAi efficiency.
8 Conclusions and future perspectives
Over the past 25 years, RNAi made irrefutable contributions to advances in our understanding of insect structure and function. However, advances in developing RNAi methods for pest and disease vector control are not that encouraging; only one commercial product and a few additional products are in the pipeline. One of the major roadblocks to the widespread use of RNAi technology in insect control is its variable efficiency among major insect pests and disease vectors. Also, the availability of transgenic crop plants expressing B. thuringiensis toxins that are effective against Lepidopteran pests and inefficient RNAi in major sucking pests slowed the development of RNAi-based products. The potential development of pest resistance to RNAi products is also a major concern. Nanoformulations of dsRNA could help deliver dsRNA to the site of action and improve RNAi efficiency. Advances in methods for producing and delivering dsRNA through microorganisms are encouraging. The research on RNAi during the next 25 years will likely find solutions to these problems and promote widespread use of this technology in insect control and other fields.
Author contributions
SP confirms being the sole contributor of this work and has approved it for publication.
Funding
Research in the Palli laboratory is supported by grants from the National Institutes of Health (GM070559 and 1R21AI131427–01), the National Science Foundation (Industry/University Cooperative Research Centers, the Center for Arthropod Management Technologies under grant IIP-1821936), and the USDA/NIFA (under Hatch Project 2351177000). The content is solely the responsibility of the author and does not necessarily represent the official views of granting agencies.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author SP declared that he is an editorial board member of Frontiers, at the time of submission.This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet (2013) 14(7):447–59. doi: 10.1038/nrg3462
2. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature (1998) 391(6669):806–11. doi: 10.1038/35888
3. Perrimon N, Ni J-Q, Perkins L. In vivo RNAi: today and tomorrow. Cold Spring Harbor Perspect Biol (2010) 2(8):a003640. doi: 10.1101/cshperspect.a003640
4. Belles X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annual review of entomology. Annu Rev Entomol (2010) 55:111–28. doi: 10.1146/annurev-ento-112408-085301
5. Napoli C, Lemieux C, Jorgensen R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell (1990) 2(4):279–89. doi: 10.1105/tpc.2.4.279
6. Jorgensen R. Silencing of plant genes by homologous transgenes. Agbiotech News Inf (1992) 4(9):265N–73N. doi: 10.1104/pp.108.117275
7. Powers AM, Olson KE, Higgs S, Carlson JO, Beaty BJ. Insertion of a La Crosse virus gene into an infectious Sindbis virus expression vector and its potential to induce interference. Am J Trop Med Hygiene (1992) 47(4 SUPPL.):137. doi: 10.1016/0168-1702(94)90061-2
8. Powers AM, Kamrud KI, Olson KE, Higgs S, Carlson JO, Beaty BJ. Molecularly engineered resistance to California serogroup virus replication in mosquito cells and mosquitoes. Proc Natl Acad Sci United States America (1996) 93(9):4187–91. doi: 10.1073/pnas.93.9.4187
9. Cogoni C, Irelan JT, Schumacher M, Schmidhauser TJ, Selker EU, Macino G. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J (1996) 15(12):3153–63. doi: 10.1002/j.1460-2075.1996.tb00678.x
10. Montgomery MK, Fire A. Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genetics: TIG (1998) 14(7):255–8. doi: 10.1016/S0168-9525(98)01510-8
11. Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci United States America (1998) 95(26):15502–7. doi: 10.1073/pnas.95.26.15502
12. Misquitta L, Paterson BM. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): A role for nautilus in embryonic somatic muscle formation. Proc Natl Acad Sci United States America (1999) 96(4):1451–6. doi: 10.1073/pnas.96.4.1451
13. Marie B, Bacon JP, Blagburn JM. Double-stranded RNA interference shows that Engrailed controls the synaptic specificity of identified sensory neurons. Curr Biol (2000) 10(5):289–92. doi: 10.1016/S0960-9822(00)00361-4
14. Shippy TD, Guo JH, Brown SJ, Beeman RW, Denell RE. Analysis of maxillopedia expression pattern and larval cuticular phenotype in wild-type and mutant tribolium. Genetics (2000) 155(2):721–31. doi: 10.1093/genetics/155.2.721
15. Adelman ZN, Blair CD, Carlson JO, Beaty BJ, Olson KE. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol Biol (2001) 10(3):265–73. doi: 10.1046/j.1365-2583.2001.00267.x
16. Moss EG. RNA interference: It’s a small RNA world. Curr Biol (2001) 11(19):R772–R5. doi: 10.1016/S0960-9822(01)00467-5
17. Adelman ZN, Sanchez-Vargas I, Travanty EA, Carlson JO, Beaty BJ, Blair CD, et al. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J Virol (2002) 76(24):12925–33. doi: 10.1128/JVI.76.24.12925-12933.2002
18. Bucher G, Scholten J, Klingler M. Parental RNAi in tribolium (Coleoptera). Curr Biol (2002) 12(3):R85–R6. doi: 10.1016/S0960-9822(02)00666-8
19. Hope IA. RNAi surges on: application to cultured mammalian cells. Trends Genetics: TIG (2001) 17(8):440. doi: 10.1016/S0168-9525(01)02385-X
20. McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature (2002) 418(6893):38–9. doi: 10.1038/418038a
21. Worby CA, Simonson-Leff N, Dixon JE. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci STKE: Signal Trans Knowledge Environ (2001) 2001(95):pl1. doi: 10.1126/stke.2001.95.pl1
22. Brown AE, Bugeon L, Crisanti A, Catteruccia F. Stable and heritable gene silencing in the malaria vector Anopheles stephensi. Nucleic Acids Res (2003) 31(15):1–6. doi: 10.1093/nar/gng085
23. Hoa NT, Keene KM, Olson KE, Zheng L. Characterization of RNA interference in an Anopheles Gambiae cell line. Insect Biochem Mol Biol (2003) 33(9):949–57. doi: 10.1016/S0965-1748(03)00101-2
24. Hong YS, Ton LQ, Collins FH. On methods of germ-line transformation and RNA interference in African malaria mosquito, Anopheles Gambiae. J Asia-Pacific Entomol (2003) 6(2):111–7. doi: 10.1016/S1226-8615(08)60176-4
25. Means JC, Muro I, Clem RJ. Silencing of the baculovirus Op-iap3 gene by RNA interference reveals that it is required for prevention of apoptosis during Orgyia pseudotsugata M nucleopolyhedrovirus infection of Ld652Y cells. J Virol (2003) 77(8):4481–8. doi: 10.1128/JVI.77.8.4481-4488.2003
26. Schroder R. The genes orthodenticle and hunchback substitute for bicoid in the beetle Tribolium. Nature (2003) 422(6932):621–5. doi: 10.1038/nature01536
27. Tomoyasu Y, Denell RE. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol (2004) 214(11):575–8. doi: 10.1007/s00427-004-0434-0
28. Ding SW. RNAi: Mechanisms, biology and applications. FEBS Letters. (2005) 579(26):5821–. doi: 10.1016/j.febslet.2005.08.036
29. Yang D, Lu H, Erickson JW. Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr Biol (2000) 10(19):1191–200. doi: 10.1016/S0960-9822(00)00732-6
30. Blandin S, Moita LF, Kacher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles Gambiae: Targeted disruption of the Defensin gene. EMBO Rep (2002) 3(9):852–6. doi: 10.1093/embo-reports/kvf180
31. Wang J, Zhou X, Bradley PL, Chang SF, Perrimon N, Wong STC. Cellular phenotype recognition for high-content RNA interference genome-wide screening. J Biomol Screening (2008) 13(1):29–39. doi: 10.1177/1087057107311223
32. Sepp KJ, Hong P, Lizarraga SB, Liu JS, Mejia LA, Walsh CA, et al. Identification of neural outgrowth genes using genome-wide RNAi. PloS Genet (2008) 4(7):e1000111. doi: 10.1371/journal.pgen.1000111
33. Bai JW, Binari R, Ni JQ, Vijayakanthan M, Li HS, Perrimon N. RNA interference screening in Drosophila primary cells for genes involved in muscle assembly and maintenance. Development (2008) 135(8):1439–49. doi: 10.1242/dev.012849
34. Ramadan N, Flockhart I, Booker M, Perrimon N, Mathey-Prevot B. Design and implementation of high-throughput RNAi screens in cultured Drosophila cells. Nat Protoc (2007) 2(9):2245–64. doi: 10.1038/nprot.2007.250
35. Perrimon N, Mathey-Prevot B. Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics (2007) 175(1):7–16. doi: 10.1534/genetics.106.069963
36. Perrimon N, Mathey-Prevot B. Matter arising - Off targets and genome scale RNAi screens in Drosophila. Fly (2007) 1(1):1–5. doi: 10.4161/fly.3601
37. Perrimon N, Friedman A, Mathey-Prevot B, Eggert US. Drug-target identification in Drosophila cells: combining high-throughout RNAi and small-molecule screens. Drug Discovery Today (2007) 12(1-2):28–33. doi: 10.1016/j.drudis.2006.10.006
38. Lu JR, Ruhf ML, Perrimon N, Leder P. A genome-wide RNA interference screen identifies putative chromatin regulators essential for E2F repression. Proc Natl Acad Sci United States America (2007) 104(22):9381–6. doi: 10.1073/pnas.0610279104
39. DasGupta R, Nybakken K, Booker M, Mathey-Prevot B, Gonsalves F, Changkakoty B, et al. A case study of the reproducibility of transcriptional reporter cell-based RNAi screens in Drosophila. Genome Biol (2007) 8(9):R203. doi: 10.1186/gb-2007-8-9-r203
40. Bai J, Hartwig JH, Perrimon N. SALS, a WH2-domain-containing protein, promotes sarcomeric actin filament elongation from pointed ends during Drosophila muscle growth. Dev Cell (2007) 13(6):828–42. doi: 10.1016/j.devcel.2007.10.003
41. Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila (vol 19, pg 1861, 2005). Genes Dev (2007) 21(7):875–7. doi: 10.1101/gad.1320705
42. Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong PY, Perrimon N, et al. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods (2006) 3(10):833–8. doi: 10.1038/nmeth935
43. Friedman A, Perrimon N. High-throughput approaches to dissecting MAPK signaling pathways. Methods (2006) 40(3):262–71. doi: 10.1016/j.ymeth.2006.05.002
44. Flockhart I, Booker M, Kiger A, Boutros M, Armknecht S, Ramadan N, et al. FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res (2006) 34:D489–D94. doi: 10.1093/nar/gkj114
45. Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user’s guide. Nat Rev Genet (2006) 7(5):373–84. doi: 10.1038/nrg1836
46. DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-Wingless signaling pathway. Science (2005) 308(5723):826–33. doi: 10.1126/science.1109374
47. Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, et al. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev (2005) 19(4):445–52. doi: 10.1101/gad.1267905
48. Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev (2005) 19(16):1861–70. doi: 10.1101/gad.1320705
49. Armknecht S, Boutros M, Kiger A, Nybakken K, Mathey-Prevot B, Perrimon N. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol (2005) 392:55–73. doi: 10.1016/S0076-6879(04)92004-6
50. Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science (2005) 309(5738):1248–51. doi: 10.1126/science.1116008
51. Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, et al. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PloS Biol (2004) 2(12):2135–43. doi: 10.1371/journal.pbio.0020379
52. Dasgupta R, Perrimon N. Using RNAi to catch Drosophila genes in a web of interactions: insights into cancer research. Oncogene (2004) 23(51):8359–65. doi: 10.1038/sj.onc.1208028
53. Wang J, Zhou XB, Li FH, Bradley PL, Chang SF, Perrimon N, et al. An image score inference system for RNAi genome-wide screening based on fuzzy mixture regression modeling. J Biomed Informatics (2009) 42(1):32–40. doi: 10.1016/j.jbi.2008.04.007
54. Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics (2009) 182(4):1089–100. doi: 10.1534/genetics.109.103630
55. Kondo S, Booker M, Perrimon N. Cross-species RNAi rescue platform in drosophila melanogaster. Genetics (2009) 183(3):1165–73. doi: 10.1534/genetics.109.106567
56. Bai JW, Sepp KJ, Perrimon N. Culture of Drosophila primary cells dissociated from gastrula embryos and their use in RNAi screening. Nat Protoc (2009) 4(10):1502–12. doi: 10.1038/nprot.2009.147
57. Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science (2004) 303(5659):832–5. doi: 10.1126/science.1091266
58. Ciudad L, Piulachs MD, Belles X. Systemic RNAi of the cockroach vitellogenin receptor results in a phenotype similar to that of the Drosophila yolkless mutant. FEBS J (2006) 273(2):325–35. doi: 10.1111/j.1742-4658.2005.05066.x
59. Eleftherianos I, Millichap PJ. ffrench-Constant RH, Reynolds SE. RNAi suppression of recognition protein mediated immune responses in the tobacco hornworm Manduca sexta causes increased susceptibility to the insect pathogen Photorhabdus. Dev Comp Immunol (2006) 30(12):1099–107. doi: 10.1016/j.dci.2006.02.008
60. Hunter W, Ellis J, Vanengelsdorp D, Hayes J, Westervelt D, Glick E, et al. Large-scale field application of rnai technology reducing Israeli acute paralysis virus disease in Honey Bees (Apis mellifera, Hymenoptera: Apidae). PloS Pathog (2010) 6(12):e1001160. doi: 10.1371/journal.ppat.1001160
61. Desai SD, Eu YJ, Whyard S, Currie RW. Reduction in deformed wing virus infection in larval and adult honey bees (Apis mellifera L.) by double-stranded RNA ingestion. Insect Mol Biol (2012) 21(4):446–55. doi: 10.1111/j.1365-2583.2012.01150.x
62. Aronstein KK PT, Saldivar E. SID-1 is implicated in systemic gene silencing in the honey bee. J Apicult Res (2006) 45:20–4. doi: 10.1080/00218839.2006.11101307
63. Pridgeon JW, Zhao L, Becnel JJ, Strickman DA, Clark GG, Linthicum KJ. Topically applied AaeIAP1 double-stranded RNA kills female adults of Aedes aEgypti. J Med Entomol (2008) 45(3):414–20. doi: 10.1093/jmedent/45.3.414
64. Zhang JZ, Liu XJ, Zhang JQ, Li DQ, Sun Y, Guo YP, et al. Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust, Locusta migratoria manilensis (Meyen). Insect Biochem Mol Biol (2010) 40(11):824–33. doi: 10.1016/j.ibmb.2010.08.001
65. Zhu F, Xu JJ, Palli R, Ferguson J, Palli SR. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manage Sci (2011) 67(2):175–82. doi: 10.1002/ps.2048
66. Bao YY, Wang Y, Wu WJ, Zhao D, Xue J, Zhang BQ, et al. De novo intestine-specific transcriptome of the brown planthopper Nilaparvata lugens revealed potential functions in digestion, detoxification and immune response. Genomics (2012) 99(4):256–64. doi: 10.1016/j.ygeno.2012.02.002
67. Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol (2003) 4(6):457–67. doi: 10.1038/nrm1129
69. Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell (2001) 107(3):309–21. doi: 10.1016/S0092-8674(01)00547-5
70. Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev (2001) 15(2):188–200. doi: 10.1101/gad.862301
71. Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J (2001) 20(23):6877–88. doi: 10.1093/emboj/20.23.6877
72. Gantier MP, Williams BR. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev (2007) 18(5-6):363–71. doi: 10.1016/j.cytogfr.2007.06.016
73. Minakuchi C, Namiki T, Yoshiyama M, Shinoda T. RNAi-mediated knockdown of juvenile hormone acid O-methyltransferase gene causes precocious metamorphosis in the red flour beetle Tribolium castaneum. FEBS J (2008) 275(11):2919–31. doi: 10.1111/j.1742-4658.2008.06428.x
74. Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proc Natl Acad Sci United States America (2005) 102(32):11337–42. doi: 10.1073/pnas.0504982102
75. Arakane Y, Muthukrishnan S, Kramer KJ, Specht CA, Tomoyasu Y, Lorenzen MD, et al. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol Biol (2005) 14(5):453–63. doi: 10.1111/j.1365-2583.2005.00576.x
76. van der Zee M, Berns N, Roth S. Distinct functions of the Tribolium zerknullt genes in serosa specification and dorsal closure. Curr Biol (2005) 15(7):624–36. doi: 10.1016/j.cub.2005.02.057
77. Choe CP, Miller SC, Brown SJ. A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc Natl Acad Sci United States America (2006) 103(17):6560–4. doi: 10.1073/pnas.0510440103
78. Ober KA, Jockusch EL. The roles of wingless and decapentaplegic in axis and appendage development in the red flour beetle, Tribolium castaneum. Dev Biol (2006) 294(2):391–405. doi: 10.1016/j.ydbio.2006.02.053
79. Choe CP, Brown SJ. Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev Biol (2007) 302(1):281–94. doi: 10.1016/j.ydbio.2006.09.037
80. Aikins MJ, Schooley DA, Begum K, Detheux M, Beeman RW, Park Y. Vasopressin-like peptide and its receptor function in an indirect diuretic signaling pathway in the red flour beetle. Insect Biochem Mol Biol (2008) 38(7):740–8. doi: 10.1016/j.ibmb.2008.04.006
81. Arakane Y, Specht CA, Kramer KJ, Muthukrishnan S, Beeman RW. Chitin synthases are required for survival, fecundity and egg hatch in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol (2008) 38(10):959–62. doi: 10.1016/j.ibmb.2008.07.006
82. Aranda M, Marques-Souza H, Bayer T, Tautz D. The role of the segmentation gene hairy in Tribolium. Dev Genes Evol (2008) 218(9):465–77. doi: 10.1007/s00427-008-0240-1
83. Cerny AC, Grossmann D, Bucher G, Klingler M. The Tribolium ortholog of knirps and knirps-related is crucial for head segmentation but plays a minor role during abdominal patterning. Dev Biol (2008) 321(1):284–94. doi: 10.1016/j.ydbio.2008.05.527
84. Farzana L, Brown SJ. Hedgehog signaling pathway function conserved in Tribolium segmentation. Dev Genes Evol (2008) 218(3-4):181–92. doi: 10.1007/s00427-008-0207-2
85. Niu BL, Shen WF, Liu Y, Weng HB, He LH, Mu JJ, et al. Cloning and RNAi-mediated functional characterization of MaLac2 of the pine sawyer, Monochamus alternatus. Insect Mol Biol (2008) 17(3):303–12. doi: 10.1111/j.1365-2583.2008.00803.x
86. Parthasarathy R, Palli SR. Proliferation and differentiation of intestinal stem cells during metamorphosis of the red flour beetle, Tribolium castaneum. Dev Dynamics (2008) 237(4):893–908. doi: 10.1002/dvdy.21475
87. Parthasarathy R, Tan A, Bai H, Palli SR. Transcription factor broad suppresses precocious development of adult structures during larval-pupal metamorphosis in the red flour beetle, Tribolium castaneum. Mech Dev (2008) 125(3-4):299–313. doi: 10.1016/j.mod.2007.11.001
88. Tan A, Palli SR. Identification and characterization of nuclear receptors from the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol (2008) 38(4):430–9. doi: 10.1016/j.ibmb.2007.09.012
89. Tan A, Palli SR. Edysone receptor isoforms play distinct roles in controlling molting and metamorphosis in the red flour beetle, Tribolium castaneum. Mol Cell Endocrinol (2008) 291(1-2):42–9. doi: 10.1016/j.mce.2008.05.006
90. Zhu Q, Arakane Y, Beeman RW, Kramer KJ, Muthukrishnan S. Functional specialization among insect chitinase family genes revealed by RNA interference. Proc Natl Acad Sci United States America (2008) 105(18):6650–5. doi: 10.1073/pnas.0800739105
91. Bitra K, Tan A, Dowling A, Palli SR. Functional characterization of PAS and HES family bHLH transcription factors during the metamorphosis of the red flour beetle, Tribolium castaneum. Gene (2009) 448(1):74–87. doi: 10.1016/j.gene.2009.08.003
92. Minakuchi C, Namiki T, Shinoda T. Kruppel homolog 1, an early juvenile hormone-response gene downstream of Methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev Biol (2009) 325(2):341–50. doi: 10.1016/j.ydbio.2008.10.016
93. Bai H, Palli SR. Functional characterization of bursicon receptor and genome-wide analysis for identification of genes affected by bursicon receptor RNAi. Dev Biol (2010) 344(1):248–58. doi: 10.1016/j.ydbio.2010.05.003
94. Bitra K, Palli SR. The members of bHLH transcription factor superfamily are required for female reproduction in the red flour beetle, Tribolium castaneum. J Insect Physiol (2010) 56(10, Sp. Iss. SI):1481–9. doi: 10.1016/j.jinsphys.2010.03.005
95. Parthasarathy R, Palli SR. Molecular analysis of nutritional and hormonal regulation of female reproduction in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol (2010) 41(5):294–305. doi: 10.1016/j.ibmb.2011.01.006
96. Parthasarathy R, Sheng Z, Sun Z, Palli SR. Ecdysteroid regulation of ovarian growth and oocyte maturation in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol (2010) 40(6):429–39. doi: 10.1016/j.ibmb.2010.04.002
97. Parthasarathy R, Sun Z, Bai H, Palli SR. Juvenile hormone regulation of vitellogenin synthesis in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol (2010) 40(5):405–14. doi: 10.1016/j.ibmb.2010.03.006
98. Xu J, Tan A, Palli SR. The function of nuclear receptors in regulation of female reproduction and embryogenesis in the red flour beetle, Tribolium castaneum. J Insect Physiol (2010) 56(10):1471–80. doi: 10.1016/j.jinsphys.2010.04.004
99. Zhu F, Parthasarathy R, Bai H, Woithe K, Kaussmann M, Nauen R, et al. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc Natl Acad Sci U.S.A. (2010) 107(19):8557–62. doi: 10.1073/pnas.1000059107
100. Bai H, Zhu F, Shah K, Palli S. Large-scale RNAi screen of G protein-coupled receptors involved in larval growth, molting and metamorphosis in the red flour beetle. BMC Genomics (2011) 12(1):388. doi: 10.1186/1471-2164-12-388
101. Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol (2007) 25(11):1322–6. doi: 10.1038/nbt1359
102. Alves AP, Lorenzen MD, Beeman RW, Foster JE, Siegfried BD. RNA interference as a method for target-site screening in the western corn rootworm, Diabrotica virgifera virgifera. J Insect Sci (2010) 10:2061. doi: 10.1673/031.010.14122
103. Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol (2007) 25(11):1307–13. doi: 10.1038/nbt1352
104. Mao Y-B, Tao X-Y, Xue X-Y, Wang L-J, Chen X-Y. Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Res (2011) 20(3):665–73. doi: 10.1007/s11248-010-9450-1
105. Mao YB, Xue XY, Tao XY, Yang CQ, Wang LJ, Chen XY. Cysteine protease enhances plant-mediated bollworm RNA interference. Plant Mol Biol (2013) 83(1-2):119–29. doi: 10.1007/s11103-013-0030-7
106. Tian G, Cheng LL, Qi XW, Ge ZH, Niu CY, Zhang XL, et al. Transgenic cotton plants expressing double-stranded RNAs target HMG-coA reductase (HMGR) gene inhibits the growth, development and survival of cotton bollworms. Int J Biol Sci (2015) 11(11):1296–305. doi: 10.7150/ijbs.12463
107. Zha WJ, Peng XX, Chen RZ, Du B, Zhu LL, He GC. Knockdown of Midgut Genes by dsRNA-Transgenic Plant-Mediated RNA Interference in the Hemipteran Insect Nilaparvata lugens. PloS One (2010) 6(5):e20504. doi: 10.1371/journal.pone.0020504
108. Zhang J, Khan SA, Hasse C, Ruf S, Heckel DG, Bock R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science (2015) 347(6225):991–4. doi: 10.1126/science.1261680
109. Murtaza S, Tabassum B, Tariq M, Riaz S, Yousaf I, Jabbar B, et al. Silencing a myzus persicae macrophage inhibitory factor by plant-mediated RNAi induces enhanced aphid mortality coupled with boosted RNAi efficacy in transgenic potato lines. Mol Biotechnol (2022) 64(10):1152–63. doi: 10.1007/s12033-022-00498-w
110. Xiong YH, Zeng HM, Zhang YL, Xu DW, Qiu DW. Silencing the HaHR3 Gene by Transgenic Plant-mediated RNAi to disrupt Helicoverpa armigera development. Int J Biol Sci (2013) 9(4):370–81. doi: 10.7150/ijbs.5929
111. Mao JJ, Zeng FR. Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae. Transgenic Res (2014) 23(1):145–52. doi: 10.1007/s11248-013-9739-y
112. Zhu JQ, Liu SM, Ma Y, Zhang JQ, Qi HS, Wei ZJ, et al. Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene ecR. PloS One (2012) 7(6):e38572. doi: 10.1371/journal.pone.0038572
113. Thakur N, Upadhyay SK, Verma PC, Chandrashekar K, Tuli R, Singh PK. Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase A gene. PloS One (2014) 9(3):9. doi: 10.1371/journal.pone.0087235
114. Sun LL, Gao Y, Zhang QH, Lv YT, Cao CAW. Resistance to Lymantria dispar larvae in transgenic poplar plants expressing CYP6B53 double-stranded RNA. Ann Appl Biol (2022) 181(1):40–7. doi: 10.1111/aab.12752
115. Zhang JH, Li HY, Zhong X, Tian JF, Segers A, Xia LQ, et al. Silencing an aphid-specific gene SmDSR33 for aphid control through plant-mediated RNAi in wheat. Front Plant Sci (2023) 13. doi: 10.3389/fpls.2022.1100394
116. Li XX, Zhang MY, Zhang HY. RNA Interference of Four Genes in Adult Bactrocera dorsalis by Feeding Their dsRNAs. PloS One (2011) 6(3):e17788. doi: 10.1371/journal.pone.0017788
117. Tian HG, Peng H, Yao Q, Chen HX, Xie Q, Tang B, et al. Developmental Control of a Lepidopteran Pest Spodoptera exigua by Ingestion of Bacteria Expressing dsRNA of a Non-Midgut Gene. PloS One (2009) 4(7):e6225. doi: 10.1371/journal.pone.0006225
118. Murphy KA, Tabuloc CA, Cervantes KR, Chiu JC. Ingestion of genetically modified yeast symbiont reduces fitness of an insect pest via RNA interference. Sci Rep (2016) 6:22587. doi: 10.1038/srep22587
119. Fei XW, Zhang Y, Ding LL, Xiao S, Xie XQ, Li YJ, et al. Development of an RNAi-based microalgal larvicide for the control of Aedes aEgypti. Parasites Vectors (2021) 14(1):387. doi: 10.1186/s13071-021-04885-1
120. Fei XW, Xiao S, Huang XD, Li ZJ, Li XH, He CH, et al. Control of Aedes mosquito populations using recombinant microalgae expressing short hairpin RNAs and their effect on plankton. PloS Negl Trop Dis (2023) 17(1):e0011109. doi: 10.1371/journal.pntd.0011109
121. Fei XW, Zhang Y, Ding LL, Li YJ, Deng XD. Controlling the development of the dengue vector Aedes aegypti using HR3 RNAi transgenic Chlamydomonas. PloS One (2020) 15(10):e0240223. doi: 10.1371/journal.pone.0240223
122. Maori E, Paldi N, Shafir S, Kalev H, Tsur E, Glick E, et al. IAPV, a bee-affecting virus associated with Colony Collapse Disorder can be silenced by dsRNA ingestion. Insect Mol Biol (2009) 18(1):55–60. doi: 10.1111/j.1365-2583.2009.00847.x
123. Isobe R, Kojima K, Matsuyama T, Quan GX, Kanda T, Tamura T, et al. Use of RNAi technology to confer enhanced resistance to BmNPV on transgenic silkworms. Arch Virol (2004) 149(10):1931–40. doi: 10.1111/j.1365-2583.2009.00847.x
124. Kanginakudru S, Royer C, Edupalli SV, Jalabert A, Mauchamp B, Chandrashekaraiah, et al. Targeting ie-1 gene by RNAi induces baculoviral resistance in lepidopteran cell lines and in transgenic silkworms. Insect Mol Biol (2007) 16(5):635–44. doi: 10.1111/j.1365-2583.2007.00753.x
125. Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, et al. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles Gambiae. Cell (2004) 116(5):661–70. doi: 10.1016/S0092-8674(04)00173-4
126. Abraham EG, Pinto SB, Ghosh A, Vanlandingham DL, Budd A, Higgs S, et al. An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proc Natl Acad Sci United States America (2005) 102(45):16327–32. doi: 10.1073/pnas.0508335102
127. Blandin SA, Levashina EA. Phagocytosis in mosquito immune responses. Immunol Rev (2007) 219:8–16. doi: 10.1111/j.1600-065X.2007.00553.x
128. Blandin SA, Wang-Sattler R, Lamacchia M, Gagneur J, Lycett G, Ning Y, et al. Dissecting the genetic basis of resistance to malaria parasites in Anopheles Gambiae. Sci (Washington) (2009) 326(5949):147–50. doi: 10.1126/science.1175241
129. Kato N, Mueller CR, Fuchs JF, McElroy K, Wessely V, Higgs S, et al. Evaluation of the function of a type i peritrophic matrix as a physical barrier for midgut epithelium invasion by mosquito-borne pathogens in Aedes aEgypti. Vector-Borne Zoonotic Dis (2008) 8(5):701–12. doi: 10.1089/vbz.2007.0270
130. Brackney DE, Foy BD, Olson KE. The effects of midgut serine proteases on dengue virus type 2 infectivity of Aedes aEgypti. Am J Trop Med Hygiene (2008) 79(2):267–74. doi: 10.4269/ajtmh.2008.79.267
131. Chen Y, Ling E, Weng Z. Functional characterization of PGRP-LC1 of Anopheles Gambiae through deletion and RNA interference. Insect Sci (2009) 16(6):443–53. doi: 10.1111/j.1744-7917.2009.01296.x
132. Cheon HM, Shin SW, Bian GW, Park JH, Raikhel AS. Regulation of lipid metabolism genes, lipid carrier protein lipophorin, and its receptor during immune challenge in the mosquito Aedes aEgypti. J Biol Chem (2006) 281(13):8426–35. doi: 10.1074/jbc.M510957200
133. Huang CY, Christensen BM, Chen CC. Role of dopachrome conversion enzyme in the melanization of filarial worms in mosquitoes. Insect Mol Biol (2005) 14(6):675–82. doi: 10.1111/j.1365-2583.2005.00597.x
134. Meister S, Kanzok SM, Zheng XL, Luna C, Li TR, Hoa NT, et al. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles Gambiae. Proc Natl Acad Sci United States America (2005) 102(32):11420–5. doi: 10.1073/pnas.0504950102
135. Shin SW, Bian GW, Raikhel AS. A toll receptor and a cytokine, Toll5A and Spz1C, are involved in toll antifungal immune signaling in the mosquito Aedes aEgypti. J Biol Chem (2006) 281(51):39388–95. doi: 10.1074/jbc.M608912200
136. Shin SW, Kokoza V, Bian GW, Cheon HM, Kim YJ, Raikhel AS. REL1, a homologue of Drosophila dorsal, regulates Toll antifungal immune pathway in the female mosquito Aedes aEgypti. J Biol Chem (2005) 280(16):16499–507. doi: 10.1074/jbc.M500711200
137. Shin SW, Kokoza VA, Raikhel AS. Transgenesis and reverse genetics of mosquito innate immunity. J Exp Biol (2003) 206(21):3835–43. doi: 10.1242/jeb.00640
138. Volz J, Mueller H-M, Zdanowicz A, Kafatos FC, Osta MA. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol (2006) 8(9):1392–405. doi: 10.1111/j.1462-5822.2006.00718.x
139. Volz J, Osta MA, Kafatos FC, Muller HM. The roles of two clip domain serine proteases in innate immune responses of the malaria vector Anopheles Gambiae. J Biol Chem (2005) 280(48):40161–8. doi: 10.1074/jbc.M506191200
140. Wang Y, Hao H, Qiu Z-W, Xu W-Y, Zhang J, Zhou T-L, et al. Involvement of prophenoloxidases in the suppression of Plasmodium yoelii development by Anopheles dirus. Exp Parasitol (2009) 123(1):6–10. doi: 10.1016/j.exppara.2009.05.017
141. Rodrigues TB, Dhandapani RK, Duan JJ, Palli SR. RNA interference in the asian longhorned beetle: identification of key RNAi genes and reference genes for RT-qPCR. Sci Rep (2017) 7:8913. doi: 10.1038/s41598-017-08813-1
142. Laudani F, Strano CP, Edwards MG, Malacrino A, Campolo O, Abd El Halim HM, et al. RNAi-mediated gene silencing in Rhynchophorus ferrugineus (Oliver) (Coleoptera: Curculionidae). Open Life Sci (2017) 12(1):214–22. doi: 10.1515/biol-2017-0025
143. Prentice K, Christiaens O, Pertry I, Bailey A, Niblett C, Ghislain M, et al. RNAi-based gene silencing through dsRNA injection or ingestion against the African sweet potato weevil Cylas puncticollis (Coleoptera: Brentidae). Pest Manag Sci (2017) 73(1):44–52. doi: 10.1002/ps.4337
144. Rodrigues TB, Rieske LK, Duan JJ, Mogilicherla K, Palli SR. Development of RNAi method for screening candidate genes to control emerald ash borer, Agrilus planipennis. Sci Rep (2017) 7:7379. doi: 10.1038/s41598-017-07605-x
145. Powell ME, Bradish HM, Gatehouse JA, Fitches EC. Systemic RNAi in the small hive beetle (Aethina tumida Murray, Coleoptera: Nitidulidae), a serious pest of the European honey bee (Apis mellifera). Pest Manag Sci (2017) 73(1):53–63. doi: 10.1002/ps.4365
146. Bodemann RR, Rahfeld P, Stock M, Kunert M, Wielsch N, Groth M, et al. Precise RNAi-mediated silencing of metabolically active proteins in the defence secretions of juvenile leaf beetles. Proc Biol Sci (2012) 279(1745):4126–34. doi: 10.1098/rspb.2012.1342
147. Macedo LLP, Antonino de Souza Junior JD, Coelho RR, Fonseca FCA, Firmino AAP, Silva MCM, et al. Knocking down chitin synthase 2 by RNAi is lethal to the cotton boll weevil. Biotechnol Res Innovation (2017) 1(1):72–86. doi: 10.1016/j.biori.2017.04.001
148. Hughes CL, Kaufman TC. RNAi analysis of Deformed, proboscipedia and Sex combs reduced in the milkweed bug Oncopeltus fasciatus: novel roles for Hox genes in the hemipteran head. Development (2000) 127(17):3683–94. doi: 10.1242/dev.127.17.3683
149. Mogilicherla K, Howell JL, Palli SR. Improving RNAi in the Brown Marmorated Stink Bug: Identification of target genes and reference genes for RT-qPCR. Sci Rep-Uk (2018) 8:3720. doi: 10.1038/s41598-018-22035-z
150. Zhu F, Gujar H, Gordon JR, Haynes KF, Potter MF, Palli SR. Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci Rep (2013) 3:1456. doi: 10.1038/srep01456
151. Luo Y, Wang X, Wang X, Yu D, Chen B, Kang L. Differential responses of migratory locusts to systemic RNA interference via double-stranded RNA injection and feeding. Insect Mol Biol (2013) 22(5):574–83. doi: 10.1111/imb.12046
152. Wynant N, Verlinden H, Breugelmans B, Simonet G, Vanden Broeck J. Tissue-dependence and sensitivity of the systemic RNA interference response in the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol (2012) 42(12):911–7. doi: 10.1016/j.ibmb.2012.09.004
153. Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Kanginakudru S, et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol (2011) 57(2):231–45. doi: 10.1016/j.jinsphys.2010.11.006
154. Chen JS, Peng YC, Zhang HN, Wang KX, Tang YJ, Gao J, et al. Transcript level is a key factor affecting RNAi efficiency. Pesticide Biochem Physiol (2021) 176:104872. doi: 10.1016/j.pestbp.2021.104872
155. Arimatsu Y, Kotani E, Sugimura Y, Furusawa T. Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori. Insect Biochem Mol Biol (2007) 37(2):176–83. doi: 10.1016/j.ibmb.2006.11.004
156. Wynant N, Santos D, Verdonck R, Spit J, Van Wielendaele P, Vanden Broeck J. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol (2014) 46:1–8. doi: 10.1016/j.ibmb.2013.12.008
157. Song HF, Zhang JQ, Li DQ, Cooper AMW, Silver K, Li T, et al. A double-stranded RNA degrading enzyme reduces the efficiency of oral RNA interference in migratory locust. Insect Biochem Mol Biol (2017) 86:68–80. doi: 10.1016/j.ibmb.2017.05.008
158. Guan RB, Li HC, Fan YJ, Hu SR, Christiaens O, Smagghe G, et al. A nuclease specific to lepidopteran insects suppresses RNAi. J Biol Chem (2018) 293(16):6011–21. doi: 10.1074/jbc.RA117.001553
159. Yan S, Yin H, Li N, Chen Y, Ji CD, Jiang QH, et al. Combination of a nanocarrier delivery system with genetic manipulation further improves pesticide efficiency: a case study with chlorfenapyr. Environ Science-Nano (2022) 9(6):2020–31. doi: 10.1039/D2EN00126H
160. Li MS, Ma ZZ, Peng M, Li L, Yin MZ, Yan S, et al. A gene and drug co-delivery application helps to solve the short life disadvantage of RNA drug. Nano Today (2022) 43:2020–31. doi: 10.1016/j.nantod.2022.101452
161. Christiaens O, Swevers L, Smagghe G. DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides (2014) 53:307–14. doi: 10.1016/j.peptides.2013.12.014
162. Spit J, Philips A, Wynant N, Santos D, Plaetinck G, Broeck JV. Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol (2017) 81:103–16. doi: 10.1016/j.ibmb.2017.01.004
163. Zhang YH, Ma ZZ, Zhou H, Chao ZJ, Yan S, Shen J. Nanocarrier-delivered dsRNA suppresses wing development of green peach aphids. Insect Sci (2022) 29(3):669–82. doi: 10.1111/1744-7917.12953
164. Guo W, Guo MJ, Yang CX, Liu ZQ, Chen SM, Lu J, et al. RNA interference-mediated silencing of vATPase subunits A and E affect survival and development of the 28-spotted ladybeetle, Henosepilachna vigintioctopunctata. Insect Sci (2021) 28(6):1664–76. doi: 10.1111/1744-7917.12899
165. Shukla JN, Kalsi M, Sethi A, Narva KE, Fishilevich E, Singh S, et al. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol (2016) 13(7):656–69. doi: 10.1080/15476286.2016.1191728
166. Koo JP SR. dsRNase1 contribution to dsRNA degradation activity in the Sf9 cells conditioned medium. Front Insect Sci (2023) 3:1118775. doi: 10.3389/finsc.2023.1118775
167. Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol (2006) 8(8):793–802. doi: 10.1038/ncb1439
168. Wynant N, Santos D, Vanden Broeck J. Biological mechanisms determining the success of RNA interference in insects. Int Rev Cell Mol Biol (2014) 312:139–67. doi: 10.1016/B978-0-12-800178-3.00005-1
169. Xiao D, Gao XW, Xu JP, Liang X, Li QQ, Yao JX, et al. Clathrin-dependent endocytosis plays a predominant role in cellular uptake of double-stranded RNA in the red flour beetle. Insect Biochem Mol Biol (2015) 60:68–77. doi: 10.1016/j.ibmb.2015.03.009
170. Li XX, Dong XL, Zou C, Zhang HY. Endocytic pathway mediates refractoriness of insect Bactrocera dorsalis to RNA interference. Sci Rep-Uk (2015) 5:8700. doi: 10.1038/srep08700
171. Cappelle K, de Oliveira CFR, Van Eynde B, Christiaens O, Smagghe G. The involvement of clathrin-mediated endocytosis and two Sid-1-like transmembrane proteins in doublestranded RNA uptake in the Colorado potato beetle midgut. Insect Mol Biol (2016) 25(3):315–23. doi: 10.1111/imb.12222
172. Yoon JS, Shukla JN, Gong ZJ, Mogilicherla K, Palli SR. RNA interference in the Colorado potato beetle, Leptinotarsa decemlineata: Identification of key contributors. Insect Biochem Mol Biol (2016) 78:78–88. doi: 10.1016/j.ibmb.2016.09.002
173. Pinheiro DH, Velez AM, Fishilevich E, Wang HC, Carneiro NP, Valencia-Jimenez A, et al. Clathrin-dependent endocytosis is associated with RNAi response in the western corn rootworm, Diabrotica virgifera virgifera LeConte. PloS One (2018) 13(8):e0201849. doi: 10.1371/journal.pone.0201849
174. Wynant N, Santos D, Van Wielendaele P, Vanden Broeck J. Scavenger receptor-mediated endocytosis facilitates RNA interference in the desert locust, Schistocerca gregaria. Insect Mol Biol (2014) 23(3):320–9. doi: 10.1111/imb.12083
175. Gillet FX, Garcia RA, Macedo LLP, Albuquerque EVS, Silva MCM, Grossi-de-Sa MF. Investigating engineered ribonucleoprotein particles to improve oral RNAi delivery in crop insect pests. Front Physiol (2017) 8. doi: 10.3389/fphys.2017.00256
176. Ren D, Cai Z, Song J, Wu Z, Zhou S. dsRNA uptake and persistence account for tissue-dependent susceptibility to RNA interference in the migratory locust, Locusta migratoria. Insect Mol Biol (2014) 23(2):175–84. doi: 10.1111/imb.12074
177. Yoon JS, Gurusamy D, Palli SR. Accumulation of dsRNA in endosomes contributes to inefficient RNA interference in the fall armyworm, Spodoptera frugiperda. Insect Biochem Mol Biol (2017) 90:53–60. doi: 10.1016/j.ibmb.2017.09.011
178. Singh IK, Singh S, Mogilicherla K, Shukla JN, Palli SR. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci Rep (2017) 7:17059. doi: 10.1038/s41598-017-17134-2
179. Wang G, Jiang L, Zhu L, Cheng T, Niu W, Yan Y, et al. Characterization of Argonaute family members in the silkworm, Bombyx mori. Insect Sci (2013) 20(1):78–91. doi: 10.1111/j.1744-7917.2012.01555.x
180. Lewis SH, Salmela H, Obbard DJ. Duplication and diversification of dipteran argonaute genes, and the evolutionary divergence of Piwi and Aubergine. Genome Biol Evol (2016) 8(3):507–18. doi: 10.1093/gbe/evw018
181. Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, Koonin EV, et al. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol (2014) 21:743. doi: 10.1038/nsmb.2879
182. Hartig JV, Tomari Y, Förstemann K. piRNAs—the ancient hunters of genome invaders. Genes Dev (2007) 21(14):1707–13. doi: 10.1101/gad.1567007
183. Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol (2011) 12(4):246–58. doi: 10.1038/nrm3089
184. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell (2009) 136(4):642–55. doi: 10.1016/j.cell.2009.01.035
185. Rubio M, Maestro JL, Piulachs M-D, Belles X. Conserved association of Argonaute 1 and 2 proteins with miRNA and siRNA pathways throughout insect evolution, from cockroaches to flies. Biochim Biophysics Acta Gene Regul Mechanisms (2018) 1861(6):554–60. doi: 10.1016/j.bbagrm.2018.04.001
186. Li Z, Zeng B, Ling L, Xu J, You L, Aslam AFM, et al. Enhancement of larval RNAi efficiency by over-expressing Argonaute2 in Bombyx mori. Int J Biol Sci (2015) 11(2):176–85. doi: 10.7150/ijbs.10235
187. Yoon JS, Mogilicherla K, Gurusamy D, Chen X, Chereddy S, Palli SR. Double-stranded RNA binding protein, Staufen, is required for the initiation of RNAi in coleopteran insects. Proc Natl Acad Sci United States America (2018) 115(33):8334–9. doi: 10.1073/pnas.1809381115
188. Kim K, Koo J, Yoon JS, Reddy Palli S. Coleopteran-specific StaufenC functions like Drosophila melanogaster Loquacious-PD in dsRNA processing. RNA Biol (2021) 18(sup1):467–77. doi: 10.1080/15476286.2021.1960687
189. Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C-elegans requires the putative transmembrane protein SID-1. Science (2002) 295(5564):2456–9. doi: 10.1126/science.1068836
190. Kobayashi I, Tsukioka H, Komoto N, Uchino K, Sezutsu H, Tamura T, et al. SID-1 protein of Caenorhabditis elegans mediates uptake of dsRNA into Bombyx cells. Insect Biochem Mol Biol (2012) 42(2):148–54. doi: 10.1016/j.ibmb.2011.11.007
191. Mon H, Kobayashi I, Ohkubo S, Tomita S, Lee JM, Sezutsu H, et al. Effective RNA interference in cultured silkworm cells mediated by overexpression of Caenorhabditis elegans SID-1. RNA Biol (2012) 9(1):40–6. doi: 10.4161/rna.9.1.18084
192. Xu J, Nagata Y, Mon H, Li ZQ, Zhu L, Iiyama K, et al. Soaking RNAi-mediated modification of Sf9 cells for baculovirus expression system by ectopic expression of Caenorhabditis elegans SID-1. Appl Microbiol Biotechnol (2013) 97(13):5921–31. doi: 10.1007/s00253-013-4785-1
193. Luo Y, Wang XH, Yu D, Kang L. The SID-1 double-stranded RNA transporter is not required for systemic RNAi in the migratory locust. RNA Biol (2012) 9(5):663–71. doi: 10.4161/rna.19986
194. Ye C, An X, Xie BQ, Ding BY, Niu J, Wang JJ. The involvement of systemic RNA interference deficient-1-like (SIL1) in cellular dsRNA uptake in Acyrthosiphon pisum. Insect Sci (2022) 10:13167. doi: 10.1111/1744-7917.13167
195. Chen X, Koo J, Gurusamy D, Mogilicherla K, Palli SR. Caenorhabditis elegans systemic RNA interference defective protein 1 enhances RNAi efficiency in a lepidopteran insect, the fall armyworm, in a tissue-specific manner. RNA Biol (2021) 18(9):1291–9. doi: 10.1080/15476286.2020.1842632
196. Zhang X, Zhang J, Zhu KY. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles Gambiae). Insect Mol Biol (2010) 19(5):683–93. doi: 10.1111/j.1365-2583.2010.01029.x
197. Christiaens O, Tardajos MG, Martinez Reyna ZL, Dash M, Dubruel P, Smagghe G. Increased RNAi Efficacy in Spodoptera exigua via the Formulation of dsRNA With Guanylated Polymers. Front Physiol (2018) 9. doi: 10.3389/fphys.2018.00316
198. Zhang X, Mysore K, Flannery E, Michel K, Severson DW, Zhu KY, et al. Chitosan/interfering RNA nanoparticle mediated gene silencing in disease vector mosquito larvae. J Visual Experiments (2015) 97:e52523. doi: 10.3791/52523.3791/52523
199. Ramesh Kumar D, Saravana Kumar P, Gandhi MR, Al-Dhabi NA, Paulraj MG, Ignacimuthu S. Delivery of chitosan/dsRNA nanoparticles for silencing of wing development vestigial (vg) gene in Aedes aEgypti mosquitoes. Int J Biol Macromol (2016) 86:89–95. doi: 10.1016/j.ijbiomac.2016.01.030
200. Das S, Debnath N, Cui Y, Unrine J, Palli SR. Chitosan, Carbon quantum dot, and silica nanoparticle mediated dsrna delivery for gene silencing in Aedes aEgypti: A comparative analysis. ACS Appl Mater Interfaces (2015) 7(35):19530–5. doi: 10.1021/acsami.5b05232
201. Taning CNT, Christiaens O, Li XX, Swevers L, Casteels H, Maes M, et al. Engineered Flock House Virus for Targeted Gene Suppression Through RNAi in Fruit Flies (Drosophila melanogaster) in vitro and in Vivo. Front Physiol (2018) 9. doi: 10.3389/fphys.2018.00805
202. Mysore K, Andrews E, Li P, Duman-Scheel M. Chitosan/siRNA nanoparticle targeting demonstrates a requirement for single-minded during larval and pupal olfactory system development of the vector mosquito Aedes aEgypti. BMC Dev Biol (2014) 14:9. doi: 10.1186/1471-213X-14-9
203. Albishi NM, Palli SR. Autophagy genes AMBRA1 and ATG8 play key roles in midgut remodeling of the yellow fever mosquito, Aedes aEgypti. Front. Insect Sci (2023) 3:1113871. doi: 10.3389/finsc.2023.1113871
204. Castellanos NL, Smagghe G, Sharma R, Oliveira EE, Christiaens O. Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest Manage Sci (2019) 75(2):537–48. doi: 10.1002/ps.5167
205. Baddar Z, Dhandapani G, Laisney J, Tripathi P, Palli S, Unrine JM. Polymer-coated hydroxyapatite nanocarrier for double-stranded RNA delivery. J Agric Food Chem (2020) 68(25):6811–8. doi: 10.1021/acs.jafc.0c02182
206. Edwards CH, Christie CR, Masotti A, Celluzzi A, Caporali A, Campbell EM. Dendrimer-coated carbon nanotubes deliver dsRNA and increase the efficacy of gene knockdown in the red flour beetle Tribolium castaneum. Sci Rep (2020) 10(1):69068. doi: 10.1038/s41598-020-69068-x
207. Dhandapani RK, Gurusamy D, Palli SR. Protamine-lipid-dsRNA nanoparticles improve RNAi efficiency in the fall armyworm, spodoptera frugiperda. J Agric Food Chem (2022) 70(22):6634–43. doi: 10.1021/acs.jafc.2c00901
208. Niu L, Yan HX, Sun YJ, Zhang DL, Ma WH, Lin YJ. Nanoparticle facilitated stacked-dsRNA improves suppression of the Lepidoperan pest Chilo suppresallis. Pesticide Biochem Physiol (2022) 187:105183. doi: 10.1016/j.pestbp.2022.105183
209. Geng KX, Zhang Y, Zhao X, Zhang WL, Guo XH, He L, et al. Fluorescent nanoparticle-rnai-mediated silencing of sterol carrier protein-2 gene expression suppresses the growth, development, and reproduction of Helicoverpa armigera. Nanomaterials (2023) 13(2):13020245. doi: 10.3390/nano13020245
210. Dhandapani RK, Gurusamy D, Howell JL, Palli SR. Development of CS-TPP-dsRNA nanoparticles to enhance RNAi efficiency in the yellow fever mosquito, Aedes aEgypti. Sci Rep (2019) 9:45019. doi: 10.1038/s41598-019-45019-z
211. Gurusamy D, Mogilicherla K, Palli SR. Chitosan nanoparticles help double-stranded RNA escape from endosomes and improve RNA interference in the fall armyworm, Spodoptera frugiperda. Arch Insect Biochem Physiol (2020) 104(4):21677. doi: 10.1002/arch.21677
212. Gurusamy D, Mogilicherla K, Shukla JN, Palli SR. Lipids help double-stranded RNA in endosomal escape and improve RNA interference in the fall armyworm, Spodoptera frugiperda. Arch Insect Biochem Physiol (2020) 104(4):21678. doi: 10.1002/arch.21678
213. Laisney J, Gurusamy D, Baddar ZE, Palli SR, Unrine JM. RNAi in Spodoptera frugiperda Sf9 Cells via Nanomaterial Mediated Delivery of dsRNA: A Comparison of Poly-L-arginine Polyplexes and Poly-L-arginine-Functionalized Au Nanoparticles. ACS Appl Mater Interfaces (2020) 12(23):25645–57. doi: 10.1021/acsami.0c06234
214. Dhandapani RK, Gurusamy D, Palli SR. Development of catechin, poly-L-lysine, and double-stranded RNA nanoparticles. ACS Appl Bio Mater (2021) 4(5):4310–8. doi: 10.1021/acsabm.1c00109
215. Avila LA, Chandrasekar R, Wilkinson KE, Balthazor J, Heerman M, Bechard J, et al. Delivery of lethal dsRNAs in insect diets by branched amphiphilic peptide capsules. J Controlled Release (2018) 273:139–46. doi: 10.1016/j.jconrel.2018.01.010
216. Zheng Y, Hu Y, Yan S, Zhou H, Songm D, Yin M, et al. A polymer/detergent formulation improves dsRNA penetration through the body wall and RNAi-induced mortality in the soybean aphid Aphis glycines. Pest Manag Sci (2019) 75(7):1993-99. doi: 10.1002/ps.5313
217. Natarajan P, Sukthankar P, Changstrom J, Holland CS, Barry S, Hunter WB, et al. Synthesis and characterization of multifunctional branched amphiphilic peptide bilayer conjugated gold nanoparticles. ACS Omega (2018) 3(9):11071–83. doi: 10.1021/acsomega.8b01633
218. Kolge H, Kadam K, Ghormade V. Chitosan nanocarriers mediated dsRNA delivery in gene silencing for Helicoverpa armigera biocontrol. Pesticide Biochem Physiol (2023) 189:105292. doi: 10.1016/j.pestbp.2022.105292
219. Abbasi R, Heschuk D, Kim B, Whyard S. A novel paperclip double-stranded RNA structure demonstrates clathrin-independent uptake in the mosquito Aedes aEgypti. Insect Biochem Mol Biol (2020) 127:103492. doi: 10.1016/j.ibmb.2020.103492
220. Ganbaatar O, Cao BD, Zhang YA, Bao DR, Bao WH, Wuriyanghan H. Knockdown of Mythimna separata chitinase genes via bacterial expression and oral delivery of RNAi effectors. BMC Biotechnol (2017) 17:9. doi: 10.1186/s12896-017-0328-7
221. Al Baki A, Jung JK, Kim Y. Alteration of insulin signaling to control insect pest by using transformed bacteria expressing dsRNA. Pest Manage Sci (2020) 76(3):1020–30. doi: 10.1002/ps.5612
222. Bento FMM, Marques RN, Campana FB, Demetrio CGB, Leandro RA, Parra JRP, et al. Gene silencing by RNAi via oral delivery of dsRNA by bacteria in the South American tomato pinworm, Tuta absoluta. Pest Manage Sci (2020) 76(1):287–95. doi: 10.1002/ps.5513
223. Ma ZZ, Zhang YH, Li MS, Chao ZJ, Du XG, Yan S, et al. A first greenhouse application of bacteria-expressed and nanocarrier-delivered RNA pesticide for Myzus persicae control. J Pest Sci (2023) 96(1):181–93. doi: 10.1007/s10340-022-01485-5
224. Duman-Scheel M. Saccharomyces cerevisiae (Baker’s yeast) as an interfering RNA expression and delivery system. Curr Drug Targets (2019) 20(9):942–52. doi: 10.2174/1389450120666181126123538
225. Mysore K, Hapairai LK, Wei N, Realey JS, Scheel ND, Severson DW, et al. Preparation and use of a yeast shRNA delivery system for gene silencing in mosquito larvae. In: Brown SJ, Pfrender ME, editors. Insect Genomics: Methods and Protocols. Methods in Molecular Biology. p. 213–31.
226. Mysore K, Sun LH, Hapairai LK, Wang CW, Igiede J, Roethele JB, et al. A Yeast RNA-Interference pesticide targeting the irx gene functions as a broad-based mosquito larvicide and adulticide. Insects (2021) 12(11):12110986. doi: 10.3390/insects12110986
227. Caccia S, Astarita F, Barra E, Di Lelio I, Varricchio P, Pennacchio F. Enhancement of Bacillus thuringiensis toxicity by feeding Spodoptera littoralis larvae with bacteria expressing immune suppressive dsRNA. J Pest Sci (2020) 93(1):303–14. doi: 10.1007/s10340-019-01140-6
228. Jiang YX, Chen JZ, Li MW, Zha BH, Huang PR, Chu XM, et al. The Combination of Bacillus thuringiensis and Its engineered strain expressing dsrna increases the toxicity against Plutella Xylostella. Int J Mol Sci (2022) 23(1):3010444. doi: 10.3390/ijms23010444
229. Kolliopoulou A, Kontogiannatos D, Swevers L. The use of engineered plant viruses in aTrans-kingdom silencing strategy against their insect vectors. Front Plant Sci (2020) 11. doi: 10.3389/fpls.2020.00917
230. Zha WJ, Peng XX, Chen RZ, Du B, Zhu LL, He GC. Knockdown of Midgut Genes by dsRNA-Transgenic Plant-Mediated RNA Interference in the Hemipteran Insect Nilaparvata lugens. PloS One (2011) 6(5):e20504. doi: 10.1371/journal.pone.0020504
231. Malik HJ, Raza A, Amin I, Scheffler JA, Scheffler BE, Brown JK, et al. RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci Rep (2016) 6:38469. doi: 10.1038/srep38469
Keywords: RNAi, dsRNA, endosomes, insects, pests, nanoparticles
Citation: Palli SR (2023) RNAi turns 25:contributions and challenges in insect science. Front. Insect Sci. 3:1209478. doi: 10.3389/finsc.2023.1209478
Received: 20 April 2023; Accepted: 26 May 2023;
Published: 04 October 2023.
Edited by:
Aaron D. Gross, Virginia Tech, United StatesReviewed by:
Mohammad Mehrabadi, Tarbiat Modares University, IranKris Silver, Kansas State University, United States
Copyright © 2023 Palli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subba Reddy Palli, cnBhbGxpQHVreS5lZHU=
 Subba Reddy Palli
Subba Reddy Palli