
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Insect Sci. , 03 December 2021
Sec. Insect Health and Pathology
Volume 1 - 2021 | https://doi.org/10.3389/finsc.2021.756886
This article is part of the Research Topic Obstacles, Advantages and Recent Progress in Honey Bee Virus Research View all 5 articles
As a phoretic parasite and virus vector, the mite Varroa destructor and the associated Deformed wing virus (DWV) form a lethal combination to the honey bee, Apis mellifera. Routine acaricide treatment has been reported to reduce the diversity of mites and select for tolerance against these treatments. Further, different DWV strains face selective pressures when transmitted via mites. In this study, the haplotypes of Varroa mites and associated DWV variants were quantified using long reads. A single haplotype dominated the mite mitochondrial gene cytochrome oxidase subunit I, reflecting an ancient bottleneck. However, highly polymorphic genes were present across the mite genome, suggesting the diversity of mites could be actively maintained at a regional level. DWV detected in both mites and honey bees show a dominant variant with only a few low-frequency alternate haplotypes. The relative abundances of DWV haplotypes isolated from honey bees and mites were highly consistent, suggesting that some variants are favored by ongoing selection.
Varroa destructor is a an ectoparasitic mite of honey bees, which feeds on honey bee fat bodies and hemocytes (1). During the phoretic stage, mites parasitize adult honey bees and disperse among host individuals and colonies through body contact and host drifting (2, 3). The reproductive phase starts with locating a suitable larval cell. The female mite hides in the brood cell just before it is capped with wax and initiates vitellogenin production. Within the capped cell, the female mite first lays an unfertilized haploid egg that develops into a male, followed by a few fertilized diploid eggs that develop into females (4). The female and male offspring mate within the capped cell, substantially reducing heterozygosity (5). Crossing of mite lineages only occurs when the colony is heavily infected and two or more female mites reproduce within the same brood cell (6). Thus far, the mite mitochondria gene cytochrome c oxidase subunit 1 (COX1) was used to identify haplotypes, where only two were found in European honey bees (7).
In addition to the destructive feeding on fat bodies (the most important immune-response tissues in honey bees), mites also transmit several RNA viruses. Among these viruses, Deformed wing virus (DWV), when combined with the mites, critically proliferates in all developmental stages of honey bees (8, 9). DWV originates from European honey bees, but was selected for and vectored by the mite V. destructor (10). Currently, the dispersal and titer of DWV in honey bee colonies heavily depends on V. destructor. The presence of mites has altered the landscape of DWV strains, in some cases leading to the emergence of dominant strains (11). Acaricides are widely used to control mites, selecting for resistant mite strains and arguably leading to founder effects that reduce mite diversity (12). The aim of this study is to quantify the current diversity of mites and associated DWV. In addition, highly polymorphic loci are selected for haplotype construction. Then the relative abundances of DWV haplotypes are quantified in mites and honey bees to test for competition during the proliferation in a controlled infestation assay.
Mites (Varroa destructor) were collected in four honey bee colonies (Apis mellifera), from the experimental apiary in Jiangxi Agricultural University. All colonies were maintained within one apiary. The colonies were 1 year old and had been treated with Amitraz during the summer. Twenty mites were collected from each of four honey bee colonies using the powdered sugar method in Spring (13). The body surface of the mites was rinsed with distilled water and twenty mites of each colony were pooled for RNA extraction using TRIzol. One Pacbio Iso-Seq library was prepared for each RNA pool to sequence in a SMRT cell. In total, four Iso-Seq libraries were prepared and sequenced in four SMRT cells.
The long reads were aligned to the V. destructor genome assembly (version V_des 3.0) using Minimap2 with default transcript parameters (14). Single nucleotide variants (SNVs) were identified using Longshot with minimal coverage of 20 (15). The SNVs were further annotated with SNPeff (16). SNVs along the contigs were analyzed with Pearson's Chi-Squared test. To analyze the haplotypes of the transcripts, the reads were aligned to the protein coding sequences of the V. destructor genome using Minimap2 with default parameters. The SNVs shared in all four colonies were concatenated to form haplotypes using SAM4WebLogo and summarized using Whatshap (15, 17, 18).
RNA reads that could not be aligned to the mite genome were retrieved using Samtools and assigned to microbes using Kraken2 (16, 19). The results were viewed with Krona (20). The reads assigned to Deformed wing virus (strain A) were extracted and aligned to a reference DWV genome (NCBI Genome version GCF000852585.1) with Minimap2. SNVs were predicted using Longshot and the nucleotides at SNV positions were clipped with SAM4WebLogo (13, 17, 21), which were concatenated to form a haplotype for each long read. The distribution of haplotypes among the colonies was analyzed with Pearson's Chi-Squared test, using R (22). Additionally, the reads were aligned to DWV-A (GCF000852585.1), DWV-B (AY251269.2) and DWV-C (CEND01000001.1) in parallel to quantify the relative abundance of DWV variants associated with the mites using Minimap2 (13, 23, 24).
To further study the competition of DWV haplotypes in mites and bees, a controlled mite infestation assay was performed. Second-instar larvae were collected from honey bee frames randomly selected from the colonies in the apiary. The larvae were artificially reared to the pupal stage in 48-well microtiter plates maintained in an incubator, with a temperature and humidity setting of 34°C, 70% RH, respectively (25). V. destructor mites were collected from capped cells to minimize any physical damage. During the pupation stage, one mite was transplanted onto each pupa to form a paired infection group (N = 10). Pupae without mites served as a control group to assess mortality due to artificial rearing (N = 10). Pupae, and their associated mites, were collected 4 days after infestation (Npupa = 5, Nmite = 5), as well as the emerged adults (Npupa = 5, Nmite = 5). Total RNA was extracted from individual bees and mites with TransZol Up Plus RNA kit (Transgen). cDNA was synthesized using PrimeScriptTM RT reagent Kit (Takara), with 1 μl gDNA Eraser L, 2 μl 5 × gDNA Eraser buffer, 5 μl RNase Free H2O and 2 μl total RNA incubating at 42°C for 2 min. Additionally, 1 μl PrimeScript RT Enzyme Mix I, 4 μl 5 × PrimeScript Buffer, 1 μl RT Primer Mix and 4 μl RNase Free H2O were added to the reaction for 37°C, 15 min, 85°C 5 min. The cDNA was stored at −20°C.
Based on the above SNV analysis of the long reads, the PCR fragments containing the highly dense and variable loci at the 218, 325, 555, and 567th nucleotides were amplified in both mites and bees using customized primers (DWV_h_5': ACGCGCGCGATAATGAGT, DWV_h_3′: GATCTCTGGTTTTGCCTGCAC). A 20 μl amplification reaction system was as follows: 1 μl 5′ primer, 1 μl 3′ primer, 10 μl 2x Tag PCR StarMix, 3 μl DEPC water, 5 μl RNA template. The reaction program consisted of 95°C 5 min, 95°C 10 s, 58°C 30 s, for 40 cycles. The 443 bp PCR product was purified and recovered from an agarose gel using a column DNA back kit (TIANDZ). PCR products were sequenced in both directions using the Illumina Nova 6,000 platform. These paired reads were joined and then aligned to the DWV CDS region using Minimap2 (13). The nucleotides at SNV positions (at 218, 325, 555, and 567th nucleotides) was clipped with SAM4WebLogo to form haplotypes (17). The distribution of haplotypes were analyzed with Pearson's Chi-square test, using R (22). The transmission of haplotypes was visualized using Circlize package in R (26).
On average, over 75 million long reads were generated from a SMRT cell, with a mean length of 3.5Kbp (over 150x of the mite transcriptome) (Supplementary Table 1). In total, 4,633, 5,683, 4,214, and 4,803 biallelic SNVs were identified from four honey bee colonies, out of which 892 SNVs were shared (Figure 1). The number of synonymous and non-synonymous SNVs was not significantly different among the four honey bee colonies (Pearson's Chi-square test, P > 0.05). On average, 5.25 SNVs were phased (assigned variants to the maternal or paternal chromosome) in a haplotype block (Supplementary Table 2). The mitochondrial genes cytochrome c oxidase subunit I (NP_758874.1), cytochrome c oxidase subunit III (NP_758878.1), and cytochrome b (NP_758884.1) were highly expressed. However, SNVs were not found in either of the above genes, nor for the mitochondrial gene cytochrome c (XP_022658338.1). The transcript activating signal cointegrator 1 complex subunit 3-like (ASCC3, XP_022662619.1) showed the highest number of SNVs (146 SNVs) in all four colonies. As 20 mites were pooled, a maximum of 40 haplotypes could be identified from this gene in each colony. In theory, 6 biallelic SNVs (26 = 64) were sufficient to distinguish between the 40 haplotypes. Two SNVs at the 3' end of the transcript XP_022662619.1, with the highest coverage, were used as an anchor to construct haplotypes. One SNV was added at a time, until the observed number of haplotypes was less than the expected ones. In total, 30 haplotypes were identified using 5 SNVs and 12 haplotypes were shared among the colonies (Supplementary Figure 1). Surprisingly, a dominant haplotype (VD-1) was found in all four colonies (Figure 2A).
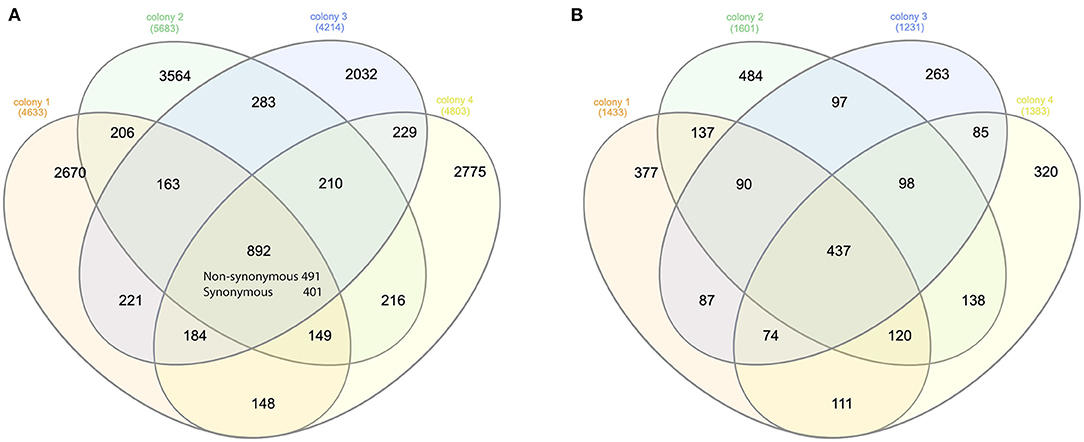
Figure 1. Venn diagram of the SNVs and genes. (A) Venn diagram of the synonymous and non-synonymous SNVs among the mites of four honey bee colonies. Overall, 892 SNVs were shared among the four colonies. The number of synonymous SNV was significantly higher than random for each honey bee colony (Pearson's Chi-square test, P < 0.001). (B) Venn diagram of the genes with non-synonymous SNVs among the mites of four honey bee colonies. Overall, 437 genes were shared in all four colonies, which was again significantly higher than random (Pearson's Chi-square test, P < 0.001).
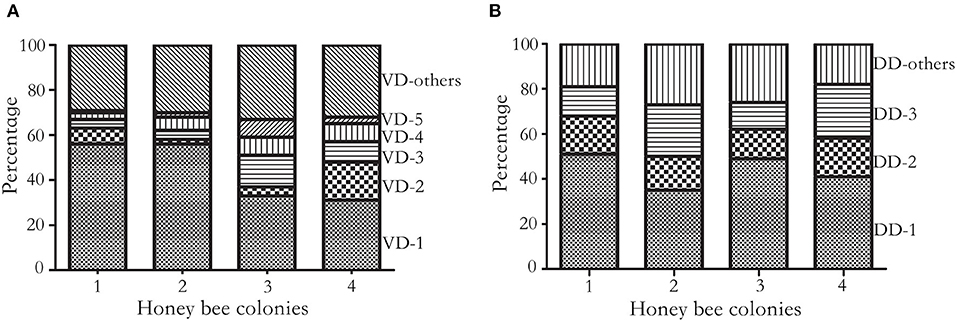
Figure 2. Cumulative frequency histogram of haplotypes. (A) Frequency of V. destructor haplotypes. Haplotypes were constructed on five SNVs of the most diversified transcript XP_22662619.1 at 4920, 4947, 4957, 4980, and 5076th nucleotide. In total, 30 haplotypes were identified, and 25 haplotypes were shared at least in two colonies. The four colonies shared dominant haplotypes (VD-1 to VD-5) and a few low frequency haplotypes (VD-others). VD represents V. destructor dominant haplotype. (B) Frequency of DWV haplotypes. Haplotypes were constructed on three SNVs at the 1809, 1938, and 1977th nucleotide. In total, 11 haplotypes were identified, and 8 haplotypes were shared at least in two colonies. Three haplotypes were shared among the four honey bee colonies, which accounted for 70% of total haplotypes. DD represents DWV dominant haplotype.
On average, 525,922 reads aligned to microbes associated with mites. Among the DWV variants, 95.41 ± 2.02% (Mean ± SD) reads were assigned to DWV-A, which was orders of magnitude higher than DWV-B (4.58 ± 2.01%) and DWV-C (0.02 ± 0.01%). In total, 381 SNVs were identified from DWV, among which 11 SNVs were commonly shared among the four colonies (at 218, 325, 555, 567, 615, 693, 780, 1809, 1938, 1977, and 8676th nucleotide, respectively) and 266 SNVs were colony specific. The proportion of shared SNVs was significantly higher in mites compared with DWV along the genome (Pearson's Chi-square test, P < 0.05). Three commonly shared SNVs at the 1809, 1938, and 1977th nucleotides were used to construct the haplotypes. In total, 11 haplotypes were identified, and 3 haplotypes dominated over 70% of total haplotype counts in all four colonies (Figure 2B; Supplementary Figure 2).
To determine whether the dominant DWV haplotypes in bees originated from high virus abundance in mites, or by viral competition during proliferation, a controlled mite infestation assay was performed. As noted above, for this experiment larvae were reared in a laboratory incubatory until pupation. All larvae survived this incubation. Then, each experimental pupal sample was paired with a Varroa mite. In the control group (no mite), DWV was not detected. In the treatment groups, 2.2 million PCR fragments were sequenced. The 218, 325, 555, and 567th nucleotides were identified as having the highest diversity, on average, in the DWV genome. Surprisingly, two haplotypes accounted for 95% of DWV in both the mites and the bees. By analyzing the paired pupae and mite, the relative abundance of the DWV haplotypes in mites and bees was consistent, a result that deviated significantly from random (Fisher's exact test, P < 0.001) (Figure 3).
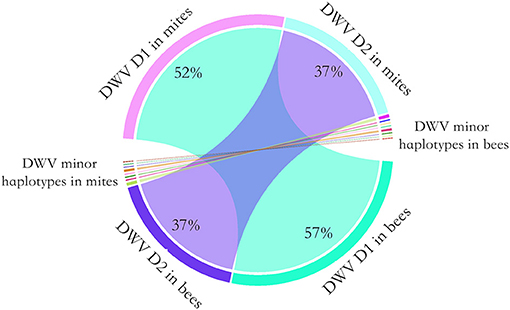
Figure 3. Chord diagram of the DWV haplotype transmission from the mites to honey bees. To quantify the transmission of DWV haplotypes from mites to bees, a controlled mite infestation assay was performed. The larvae were artificially reared in 48-well microtiter plates and a female mite was transplanted to each of the larvae. Then DWV haplotypes was quantified for each bee-mite pair. Two DWV haplotypes dominated in both bee and mites. The dominant haplotype 1 (D1) accounted for 52% of observed haplotypes in mite, which accounted for 57% of observed haplotypes in bees. The dominant haplotype 2 (D2) accounted for 37% of observed haplotypes in mite, which also accounted for 37% of observed haplotypes in bees. A few minor haplotypes were also observed in bees and mites. Overall, the relative haplotype abundance of DWV in bees was highly consistent with the mites (Fisher's exact test, P < 0.001).
V. destructor is a native ectoparasite of the Asian honey bee that has successfully established infestation in European honey bee colonies. The impact of V. destructor infestation on the native host is minor, as the bees can effectively inhibit mite reproduction (27). However, the novel host lacks this acquired defense and the number of the mites infesting colonies can increase by orders of magnitudes, often leading to the collapse of the colony (28–30). Previously, 6 haplotypes of V. destructor were identified based on a 458 bp DNA fragment of cytochrome c oxidase subunit 1 (COX1), with one haplotype per geographic location (7). Two (Korea and Japan haplotypes) out of these 6 haplotypes were found parasitizing the novel host honey bees and the remaining 4 haplotypes were confined to the native host honey bees (7). However, within the Korea and Japan haplotypes, mite haplotypes cannot be further refined using the COX1 gene. Instead, a highly polymorphic nuclear gene is required. In our data, SNVs were not found in the COX1 gene. Further, the COX1 gene perfectly aligned to the Korean haplotype sequence, suggesting all 80 studied mites originated from this lineage (31). This haplotype has spread to, and thrived in, novel host honey bees while also maintaining reproductive potential in the native host honey bees (27, 32). In our study, we identified a highly polymorphic gene, ASCC3, which can serve as an alternate candidate gene for fine-scale population genetic studies in these mites.
Although current diversity in mite populations is low, variation among colonies and apiaries has been reported (33, 34). In our data, the small number of SNVs suggests low standing genetic diversity. Colony-level variation exists and the overall selective pressure on mites remains similar at the colony level, as suggested by the number of synonymous and non-synonymous SNVs. Additionally, the Korea haplotype may have switched back to the native host, boosting mite diversity and facilitating the exchange of viruses (33, 35, 36). DWV is transmitted from queens to offspring, by feeding on contaminated food, and via feeding by mites. In the absence of mites, infection levels of DWV were generally low and didn't present the indicative symptom of deformed wings (37). In natural conditions, DWV was found in pollen, feces and flowers, facilitating horizontal transmission among pollinators, as well as to insect predators (38–42). Spillover of DWV among pollinators could enhance mutation and recombination events. Multiple transmission routes could lead to increased DWV diversity (43–45), perhaps helping to explain why observed DWV diversity is higher than that of mites. In our data, several bee viruses were associated with V. destructor but DWV was the most abundant of these. Using amplified fragments of DWV, haplotype variance was found among bee colonies (46, 47). The diversity between individuals remained high and 286 divergent nucleotides were identified from DWV isolated from bees (48). In our data, 381 SNVs were identified from DWV isolated from mites and numerous haplotypes were found from each colony. Less than 5% of the SNVs were commonly shared among colonies and the distribution of haplotypes was significantly different among the colonies. Interestingly, we found the relative abundance of DWV was highly consistent in paired mites and bees. Even though the transmission of DWV between honey bees and mites is reciprocal and the mites may contain bee cells, the impacts on haplotype structure are minor as long as the dominant haplotypes are circulating in local population. Overall, our results suggest minor competition among the haplotypes during the proliferation within the selected mite—honey bee pairs. Our study is limited by using randomly collected phoretic mites, different gene expression patterns may be identified using other life stages.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: NCBI [accession: PRJNA726354].
WB conducted controlled mite infestation analysis. QH conceived the project. WB, ZGL, LZZ, WYY, JE, and QH wrote the manuscript. All authors contributed to the article and approved the submitted version.
This project was funded by Science and Technology Program Project of Jiangxi Province 20202BBFL63028 (QH), initiation package of Jiangxi Agricultural University 050014/923230722 (QH), National Natural Science Foundation of China 31902220 (ZGL), and the earmarked fund for Jiangxi Agriculture Research System JXARS-14 (WYY). Natural Science Foundation of Jiangxi Province 20192BAB214022 (LZZ). Supported by USDA National Institute of Food and Agriculture (NIFA) grant 2017-06481 (JE).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We appreciate the constructive comments from Prof. Robert Paxton.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/finsc.2021.756886/full#supplementary-material
1. Ramsey SD, Ochoa R, Bauchan G, Gulbronson C, Mowery JD, Cohen A, et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc Natl Acad Sci. (2019) 116:1792–801. doi: 10.1073/pnas.1818371116
2. DeGrandi-Hoffman G, Ahumada F, Graham H. Are dispersal mechanisms changing the host-parasite relationship and ncreasing the virulence of Varroa destructor (Mesostigmata: Varroidae) in managed honey bee (Hymenoptera: Apidae) colonies? Environ Entomol. (2017) 46:737–46. doi: 10.1093/ee/nvx077
3. Peck DT, Smith ML, Seeley TD. Varroa destructor mites can nimbly climb from flowers onto foraging honey bees. PLoS ONE. (2016) 11:e0167798. doi: 10.1371/journal.pone.0167798
4. Rosenkranz P, Aumeier P, Ziegelmann B. Biology and control of Varroa destructor. J Invertebr Pathol. (2010) 103:96–119. doi: 10.1016/j.jip.2009.07.016
5. Evans JD, Cook SC. Genetics and physiology of varroa mites. Curr Opin Insect Sci. (2018) 26:130–5. doi: 10.1016/j.cois.2018.02.005
6. Beaurepaire AL, Krieger KJ, Moritz RFA. Seasonal cycle of inbreeding and recombination of the parasitic mite Varroa destructor in honeybee colonies and its implications for the selection of acaricide resistance. Infect Genet Evol. (2017) 50:49–54. doi: 10.1016/j.meegid.2017.02.011
7. Anderson DL, Trueman JWH. Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp Appl Acarol. (2000) 24:165–89. doi: 10.1023/A:1006456720416
8. de Miranda JR, Genersch E. Deformed wing virus. J Invertebr Pathol. (2010) 103:48–61. doi: 10.1016/j.jip.2009.06.012
9. Ryabov EV, Childers AK, Chen Y, Madella S, Nessa A, VanEngelsdorp D, et al. Recent spread of varroa destructor virus-1, a honey bee pathogen, in the United States. Sci Rep. (2017) 7:17447. doi: 10.1038/s41598-017-17802-3
10. Wilfert L, Long G, Leggett HC, Schmid-Hempel P, Butlin R, Martin SJM, et al. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science. (2016) 351:594–7. doi: 10.1126/science.aac9976
11. Martin SJ, Highfield AC, Brettell L, Villalobos EM, Budge GE, Powell M, et al. Global honey bee viral landscape altered by a parasitic mite. Science. (2012) 336:1304–6. doi: 10.1126/science.1220941
12. González-Cabrera J, Bumann H, Rodríguez-Vargas S, Kennedy PJ, Krieger K, Altreuther G, et al. A single mutation is driving resistance to pyrethroids in European populations of the parasitic mite, Varroa destructor. J Pest Sci. (2004) (2018) 91:1137–1144. doi: 10.1007/s10340-018-0968-y
13. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. (2018) 34:3094–100. doi: 10.1093/bioinformatics/bty191
14. Dietemann V, Nazzi F, Martin SJ, Anderson DL, Locke B, Delaplane KS, et al. Standard methods for Varroa research. J Apic Res. (2013) 52:1–54. doi: 10.3896/IBRA.1.52.1.09
15. Edge P, Bansal V. Longshot enables accurate variant calling in diploid genomes from single-molecule long read sequencing. Nat Commun. (2019) 10:4660. doi: 10.1038/s41467-019-12493-y
16. Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. (2012) 6:80–92. doi: 10.4161/fly.19695
17. Lindenbaum P. JVarkit: java-based utilities for Bioinformatics. Figshare J Contrib. (2015). doi: 10.6084/m9.figshare.1425030.v1. [Epub ahead of print].
18. Patterson M, Marschall T, Pisanti N, van Iersel L, Stougie L, Klau GW, et al. WhatsHap: weighted haplotype assembly for future-generation sequencing reads. J Comput Biol. (2015) 22:498–509. doi: 10.1089/cmb.2014.0157
19. Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. (2019) 20. doi: 10.1186/s13059-019-1891-0
20. Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinform. (2011) 12:385. doi: 10.1186/1471-2105-12-385
21. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. (2003) 19. doi: 10.1093/bioinformatics/btg180
22. R Core Team. R: A Language Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing (2018). Available online at: https://www.R-project.org/
23. Mordecai GJ, Wilfert L, Martin SJ, Jones IM, Schroeder DC. Diversity in a honey bee pathogen: first report of a third master variant of the deformed wing virus quasispecies. ISME. (2016) 10:1264–73. doi: 10.1038/ismej.2015.178
24. Ongus JR, Peters D, Bonmatin J-M, Bengsch E, Vlak JM, Overs MM. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J Gen Virol. (2004) 85:3747–55. doi: 10.1099/vir.0.80470-0
25. Crailsheim K, Brodschneider R, Aupinel P, Behrens D, Genersch E, Vollmann J, et al. Standard methods for artificial rearing of Apis mellifera larvae. J Apic Res. (2013) 52:1–16. doi: 10.3896/IBRA.1.52.1.05
26. Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. (2014) 30:2811–2. doi: 10.1093/bioinformatics/btu393
27. Li W, Wang C, Huang ZY, Chen Y, Han R. Reproduction of distinct Varroa destructor genotypes on honey bee worker brood. Insects. (2019) 10. doi: 10.3390/insects10110372
28. Nazzi F, Le Conte Y. Ecology of Varroa destructor, the major ectoparasite of the western honey bee, Apis mellifera. Annu Rev Entomol. (2016) 61:417–32. doi: 10.1146/annurev-ento-010715-023731
29. Neumann P, Carreck NL. Honey bee colony losses. J Apic Res. (2010) 49:1–6. doi: 10.3896/IBRA.1.49.1.01
30. Strauss U, Pirk CWW, Crewe RM, Human H, Dietemann V. Impact of Varroa destructor on honeybee (Apis mellifera scutellata) colony development in South Africa. Exp Appl Acarol. (2015) 65:89–106. doi: 10.1007/s10493-014-9842-7
31. Techer MA, Rane RV, Grau ML, Roberts JMK, Sullivan ST, Liachko I, et al. Divergent evolutionary trajectories following speciation in two ectoparasitic honey bee mites. Commun Biol. (2019) 2:357. doi: 10.1038/s42003-019-0606-0
32. Lin Z, Qin Y, Page P, Wang S, Li L, Wen Z, et al. Reproduction of parasitic mites Varroa destructor in original and new honeybee hosts. Ecol Evol. (2018) 8:2135–45. doi: 10.1002/ece3.3802
33. Dynes TL, De Roode JC, Lyons JI, Berry JA, Delaplane KS, Brosi BJ. Fine scale population genetic structure of Varroa destructor, an ectoparasitic mite of the honey bee (Apis mellifera). Apidologie. (2016) 2016:1–9. doi: 10.1007/s13592-016-0453-7
34. Solignac M, Cornuet J.-M, Vautrin D, Le Conte Y, Anderson D, et al. The invasive Korea and Japan types of Varroa destructor, ectoparasitic mites of the Western honeybee (Apis mellifera), are two partly isolated clones. Proc Biol Sci. (2005) 272:411–9. doi: 10.1098/rspb.2004.2853
35. Dietemann V, Beaurepaire A, Page P, Yañez O, Buawangpong N, Chantawannakul P, et al. Population genetics of ectoparasitic mites Varroa spp. In Eastern and Western honey bees. Parasitology. (2019) 146:1429–39. doi: 10.1017/S003118201900091X
36. Lin Z, Wang S, Neumann P, Chen G, Page P, Li L, et al. Population genetics and host specificity of Varroa destructor mites infesting eastern and western honeybees. J Pest Sci. (2021) 94:1487–504. doi: 10.1007/s10340-020-01322-7
37. Shen M, Yang X, Cox-Foster D, Cui L. The role of Varroa mites in infections of Kashmir bee virus (KBV) and deformed wing virus (DWV) in honey bees. Virology. (2005) 342:141–9. doi: 10.1016/j.virol.2005.07.012
38. Chen YP, Pettis JS, Collins A, Feldlaufer MF. Prevalence and transmission of honeybee viruses. Appl Environ Microbiol. (2006) 72:606–11. doi: 10.1128/AEM.72.1.606-611.2006
39. Fürst MA, McMahon DP, Osborne JL, Paxton RJ, Brown MJF. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature. (2014) 506:364–6. doi: 10.1038/nature12977
40. Gusachenko ON, Woodford L, Balbirnie-Cumming K, Ryabov EV, Evans DJ. Evidence for and against deformed wing virus spillover from honey bees to bumble bees: a reverse genetic analysis. Sci Rep. (2020) 10:16847. doi: 10.1038/s41598-020-73809-3
41. Singh R, Levitt AL, Rajotte EG, Holmes EC, Ostiguy N, Vanengelsdorp D, et al. RNA viruses in hymenopteran pollinators: evidence of inter-Taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE. (2010) 5:e14357. doi: 10.1371/journal.pone.0014357
42. Yang S, Gayral P, Zhao H, Wu Y, Jiang X, Wu Y, et al. Occurrence and molecular phylogeny of honey bee viruses in vespids. Viruses. (2019) 12. doi: 10.3390/v12010006
43. Acevedo MA, Dillemuth FP, Flick AJ, Faldyn MJ, Elderd BD. Virulence-driven trade-offs in disease transmission: a meta-analysis. Evolution. (2019) 73:636–47. doi: 10.1111/evo.13692
44. Elliot SL. The virulence-transmission trade-off and virulence management. Trends Microbiol. (2003) 11:205–9. doi: 10.1016/S0966-842X(03)00071-4
45. Greenspoon PB, M'Gonigle LK. The evolution of mutation rate in an antagonistic coevolutionary model with maternal transmission of parasites. Proc Biol Sci. (2013) 280:20130647. doi: 10.1098/rspb.2013.0647
46. Berényi O, Bakonyi T, Derakhshifar I, Köglberger H, Topolska G, Ritter W, et al. Phylogenetic analysis of deformed wing virus genotypes from diverse geographic origins indicates recent global distribution of the virus. Appl Environ Microbiol. (2007) 73:3605–11. doi: 10.1128/AEM.00696-07
47. Jamnikar-Ciglenecki U, Pislak Ocepek M, Toplak I. Genetic diversity of deformed wing virus from Apis mellifera carnica (Hymenoptera: Apidae) and Varroa mite (Mesostigmata: Varroidae). J Econ Entomol. (2019) 112:11–9. doi: 10.1093/jee/toy312
Keywords: haplotype, honey bee, mite, virus, selection, long reads
Citation: Bai WF, Lin ZG, Yan WY, Zhang LZ, Evans JD and Huang Q (2021) Haplotype Analysis of Varroa destructor and Deformed Wing Virus Using Long Reads. Front. Insect Sci. 1:756886. doi: 10.3389/finsc.2021.756886
Received: 11 August 2021; Accepted: 03 November 2021;
Published: 03 December 2021.
Edited by:
Wei-Fone Huang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Chuleui Jung, Andong National University, South KoreaCopyright © 2021 Bai, Lin, Yan, Zhang, Evans and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Huang, cWlhbmctaHVhbmdAbGl2ZS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.