- JT Biohistory Research Hall, Takatsuki, Japan
Lepidopteran insects are mostly monophagous or oligophagous. Female butterflies distinguish their host plants by detecting a combination of specific phytochemicals through the gustatory sensilla densely distributed on their foreleg tarsi, thereby ensuring oviposition on appropriate host plants. In this study, to gain insight into the molecular mechanism underlying host plant recognition by the gustatory sensilla, using Asian swallowtail, Papilio xuthus, we focused on a family of small soluble ligand-binding molecules, odorant-binding proteins (OBPs), and found that three OBP genes showed enriched expression in the foreleg tarsus. Multicolor fluorescence in situ hybridization analyses demonstrated the coexpression of these three OBP genes at the bases of the foreleg gustatory sensilla. Further analyses on other appendages revealed that PxutOBP3 was exclusively expressed in the tissues which could have direct contact with the leaf surface, suggesting that this OBP gene specifically plays an important role in phytochemicals perception.
Introduction
Most phytophagous (plant-feeding) insects utilize a limited range of host plants (1, 2). For these monophagous and oligophagous insect species, precise discrimination of right host plants is fundamentally important (3, 4). In Lepidoptera, because of low mobility of larvae, host plant selection is mainly executed by female adults during oviposition (5). Before egg laying, female butterflies quickly drum their forelegs on the leaf surface to examine whether a plant is the suitable host species for larvae by detecting chemicals secreted from the leaves through chemosensilla densely distributed on the ventral surface of the foreleg tarsi (6, 7).
Asian swallowtail, Papilio xuthus, belongs to the family of Papilionidae. P. xuthus larvae feed exclusively on Rutaceae plants, whereas adults suck floral nectar and no longer feed on leaves (8, 9). A complete set of oviposition stimulants for P. xuthus, composed of 10 compounds in the Rutaceae leaves, has been identified (10). We previously demonstrated that the foreleg gustatory sensilla housed at least three types of gustatory receptor neurons which were tuned for five oviposition stimulants (11), and identified PxutGr1 as a receptor gene for one of the oviposition stimulants, synephrine (12). Importantly, however, it has been argued that not only chemoreceptors but also some soluble ligand-binding proteins cooperatively play a crucial role in chemoreception (i.e., olfaction and gustation) (13, 14).
The family of odorant-binding proteins (OBPs) is a representative of the soluble ligand-binding molecules (15–17). Insect OBPs are small (~15 kDa) proteins and generally consist of 130–150 amino acids (15). A structural feature of OBPs is a set of six cysteine residues that form three disulfide bridges providing stable globular conformations (15). According to the conserved cysteine patterns, OBPs are categorized into three classes: classic, minus-C, and plus-C OBPs (15–17). Most of the OBP genes are expressed in support cells located at the bases of chemosensory sensilla, especially olfactory sensilla, and synthesized proteins are localized in the sensillar lymph (16, 17). Functional analysis proposed several roles of OBPs in chemoreception, for example, transport of hydrophobic chemicals through the sensillar lymph to chemoreceptors, protection of ligands from degradative enzymes, and filtering chemicals (16, 17). Some OBPs of Drosophila melanogaster were reported to contribute to taste perception (18–20). Notably, altered expression level of OBPs in the leg gustatory sensilla affected to taste perception and host plant preference in a close relative of D. melanogaster (21–23).
There has been limited information on P. xuthus OBPs. We previously constructed EST libraries from P. xuthus females but obtained only three OBP sequences (24). In this study, we performed a genome-wide search of P. xuthus OBP genes, and then identified additional 41 OBP genes, and found that 3 out of in total 44 OBP genes showed enriched expression patterns in the foreleg tarsus. Subsequent multicolor fluorescence in situ hybridization (FISH) analyses revealed coexpression of these three OBP genes in identical support cells at the bases of the foreleg gustatory sensilla.
Materials and Methods
Animals
P. xuthus used in this study were laboratory-raised summer forms. Adult females were collected at Takatsuki, Osaka, Japan, and allowed to lay eggs on leaves of Citrus unshu in the laboratory at 25°C under light/dark photoperiods of 16 and 8 h, respectively. Larvae were reared on artificial diet (mixture of Insecta F-Ii (Nosan, Kanagawa, Japan) and powder of dried Zanthoxylum ailanthoides leaves).
RNA-Sequencing and De novo Assembly
We collected three pairs of forelegs from 0-, 1-, 3-, and 5-day-old females. Also, a pair of antennae from a 0-day-old female was also collected. Total RNA samples were extracted using QuickGene RNA tissue kit SII (Kurabo, Osaka, Japan). cDNA library preparation was carried out using TruSeq RNA Sample Prep Kit v2 (Illumina, CA, USA) following the manufacturer's protocol with a modification of the incubation time for RNA fragmentation. To obtain longer fragments, we set the incubation time to 1 min. RNA-sequencing runs were performed using MiSeq system with MiSeq sequencing kit v3 (Illumina). Raw RNA-sequencing data have been deposited in the DNA Data Bank of Japan (DDBJ) Sequence Data Archive under accession numbers DRA011862 and DRA011865. Sequenced reads were assembled using Trinity 2.0.6 and 2.8.4 for antenna samples and foreleg samples, respectively, with Quality Trimming Options by Trimmomatic (LEADING: 10; TRAILING: 10; SLIDINGWINDOW: 4:20; MINLEN: 150). Kmer_size parameter was set at 32. Coding regions and amino acid sequences of the assembled contigs were predicted using TransDecoder.
Identification of PxutOBP Genes
To obtain candidate OBP genes, we performed BLASTp searches in both the genome database of P. xuthus and our assembled contigs (e-value > 0.01), using each of the full set of individual Danaus plexippus, Heliconius melpomene, and Manduca sexta OBPs reported previously (25) as queries (Supplementary Data Sheet 1). We also performed a HMMER search in our assembled contigs with the Pfam database and collected contigs that could encode the proteins classified as PBP_GOBP (e-value > 1e-10). Possible OBP-encoding contigs which found only by HMMER search were evaluated through further BLASTp searches against the NCBI non-redundant protein database (nr) and those homologous to OBPs of other insect species (e-value > 0.05) were additionally considered as candidate OBP genes. Subsequently, the deduced amino acid sequences of the candidate OBP genes were aligned by MUSCLE program in MEGA6 software (26) and confirmed the position of conserved cysteines.
Phylogenetic Analysis
To examine the phylogenetic relationship between PxutOBPs and other lepidopteran OBPs, we performed a phylogenetic tree construction using amino acid sequences of OBPs identified in D. plexippus, H. melpomene, M. sexta, Bombyx mori, and Vanessa cardui (Supplementary Data Sheet 1). OBP sequences of V. cardui was kindly provided by Mr. Hiromu C. Suzuki (27). After the multiple alignments, the neighbor-joining (NJ) tree was generated using MEGA with 1,000 rounds of bootstrapping (p-distance, pairwise deletion; otherwise default settings).
Quantitative RT-PCR
After eclosion, each adult female was individually kept in a translucent plastic cup under ad libitum feeding on Pocari Sweat (Otsuka Pharmaceutical, Tokyo, Japan). If needed, mating was performed artificially (hand-pairing) a day after eclosion. Note that all of the adult females had never touched any plant leaves. A previous electrophysiological study reported essentially no differences in response patterns of the foreleg gustatory sensilla varying in age, at least from 0 to 8 days after eclosion (11). To avoid the deficit of appendages during keeping in a small cup, we mainly used adults 0 day after eclosion. Appendages were collected by forceps into 1.5 ml tubes floating on liquid nitrogen and stored at −80°C until use. Total RNA samples were isolated using RNeasy Micro Kit (Qiagen, The Netherland) and then reverse-transcribed using PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Shiga, Japan), which eliminated potentially contaminated genomic DNA. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using SYBR Premix Ex Taq II (Tli RNase H Plus) (Takara) and Thermal Cycler Dice Real Time System II (Takara) in accordance with the manufacturer's protocol and with gene-specific primers listed in Supplementary Table 1. All primer sets were designed to amplify 100–150 bp fragments, and standard curves were prepared using six points with progressive quantities of PCR amplicons (1 × 10−1 to 1 × 10−6 pg/μl). Relative expression levels of each gene were calculated using the expression value of ribosomal protein L32 (Rpl32). Statistical analyses were conducted using Student's t-test, Tukey–Kramer's test, or Dunnett's test with STATCEL2 (OMS, Saitama, Japan).
Fluorescence in situ Hybridization
Riboprobes were prepared by in vitro transcription using PCR products as a template (Supplementary Table 2). Digoxigenin (DIG)- and fluorescein (FLU)-labeled riboprobes were synthesized using RNA labeling kits (Roche, Basel, Switzerland). To synthesize 2,4-dinitrophenyl (DNP)-labeled probes, we prepared a 10× solution [3.5 mM DNP-11-UTP (PerkinElmer, MA, USA), 6.5 mM UTP, 10 mM ATP, CTP, and GTP] and performed in vitro transcription as well as the other probes. Synthesized products were purified by LiCl precipitation.
Tissues were embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek, Tokyo, Japan), immediately frozen in a deep freezer, and stored at −80°C until use. 10 μm fresh-frozen sections were prepared using a cryostat (OT/FAS/EC/MR/Z, Bright Instrument Company, UK) set at −18°C. Sections were collected on APS-coated microscope slides (Matsunami Glass Ind, Osaka, Japan) using wooden toothpicks. After overnight air-drying, sections were fixed in 4% paraformaldehyde in 0.1 M phosphate buffer (PB) overnight at 4°C, treated with 12.5 μg/ml proteinase K (29442-14, Nacalai, Kyoto, Japan) for 15 min and then with 0.2 M HCl for 10 min, followed by acetylation solution [0.25% acetic anhydride, 0.1 M pH 8.0 Triethanolamine hydrochloride (T1502, Sigma-Aldrich, MO, USA)] for 10 min at room temperature. Slides were rinsed with PB between each step. After dehydration through a series of ethanol solutions (70, 80, 90, and 100%), sections were hybridized with the riboprobes overnight at 60°C. The riboprobes were diluted in hybridization buffer (50% formamide, 10 mM Tris-HCl (pH 7.6), 200 μg/ml yeast tRNA (15401-011, Thermo Fisher Scientific, MA, USA), 50 μg/ml heparin, 1× Denhardt's solution, 100 mg/ml sodium dextran sulfate, 0.6 M NaCl, 0.25% SDS, 1 mM EDTA) at a concentration of 1 μl/ml, heat-denatured at 85°C for 5 min, and then added to each slide. A strip of Parafilm (Bemis, IL, USA) was placed on top and slides were incubated in a box moisturized with 50% formamide at 60°C overnight. After hybridization, slides were washed in a wash solution [50% formamide, 2× standard sodium citrate (SSC)] at 60°C for 30 min, treated with 6.25 μg/ml RNase A (30142-04, Nacalai) in Tris-NaCl-EDTA buffer (10 mM Tris-HCl (pH7.6), 1 mM EDTA, 0.5 M NaCl) at 37°C for 30 min, and washed at 60°C in 2× SSC for 20 min and twice in 0.2× SSC for 20 min. Slides were then blocked with Tris-NaCl-Blocking (TNB) buffer [0.1 M Tris-HCl (pH 7.6), 0.15 M NaCl, 5 mg/ml blocking reagent (FP1020, PerkinElmer)] for 1 h at room temperature.
Signals were detected immunocytochemically by a combination of peroxidase (POD)-conjugated antibody and tyramide signal amplification (TSA) system. To obtain clear signals, we used the “enhanced POD-TSA reaction” protocol (28, 29), instead of the manufacturer's standard TSA protocol. After blocking, slides were incubated with anti-DIG-POD (11207733910, Roche) diluted in TNB buffer (1:500) one or two overnight at 4°C, washed three times in Tris-NaCl-Tween20 (TNT) buffer [0.1 M Tris-HCl (pH 7.6), 0.15 M NaCl, 0.05% Tween20] for 5 min, further rinsed three times in borate buffer [0.1 M H3BO3 (pH 8.5), 0.1% Tween20], and incubated with TSA reaction solution (20 mg/ml sodium dextran sulfate, 0.3 mg/ml 4-iodophenol, 0.003% H2O2 in borate buffer) containing TSA Plus Cy3 Reagent (1:200; TS-000202, Akoya Biosciences, MA, USA) for 30 min at room temperature without shaking. After final washing in TNT buffer, sections were counterstained using 4',6-diamidino- 2-phenylindole (DAPI) and mounted in Fluoromount/Plus (K048, Diagnostic BioSystems, CA, USA). Fluorescent images were captured using a TCS SPE confocal system (Leica Microsystems, Germany).
When applied to multicolor staining, after the first TSA reaction, slides were washed in 0.1% Tween20 containing PB (PBT) and incubated in 3% H2O2 containing PBT for 10 min at room temperature to inactivate POD, followed by three times washing in TNT buffer. Slides were then incubated with another POD conjugated antibody, and we performed the above-mentioned signal detection process repeatedly. FLU- and DNP-labeled riboprobes were detected in combination with 5-(and-6)-carboxyfluorescein tyramide (29) (kindly provided by Dr. Yasuko Akiyama-Oda) and TSA Plus Cy5 Reagent (TS-000203, Akoya Biosciences), respectively. In our protocol, the order of detection greatly affected the signal intensity (signals of the first detected genes were stronger than those detected later). In addition, DIG-labeled riboprobes tended to provide better signals than FLU- and DNP-labeled probes. Therefore, in multicolor detection, we used DIG-labeled probes for a gene whose expression level was lower than the other target genes.
Results
Identification and Phylogenetic Analysis of OBP Genes in P. xuthus
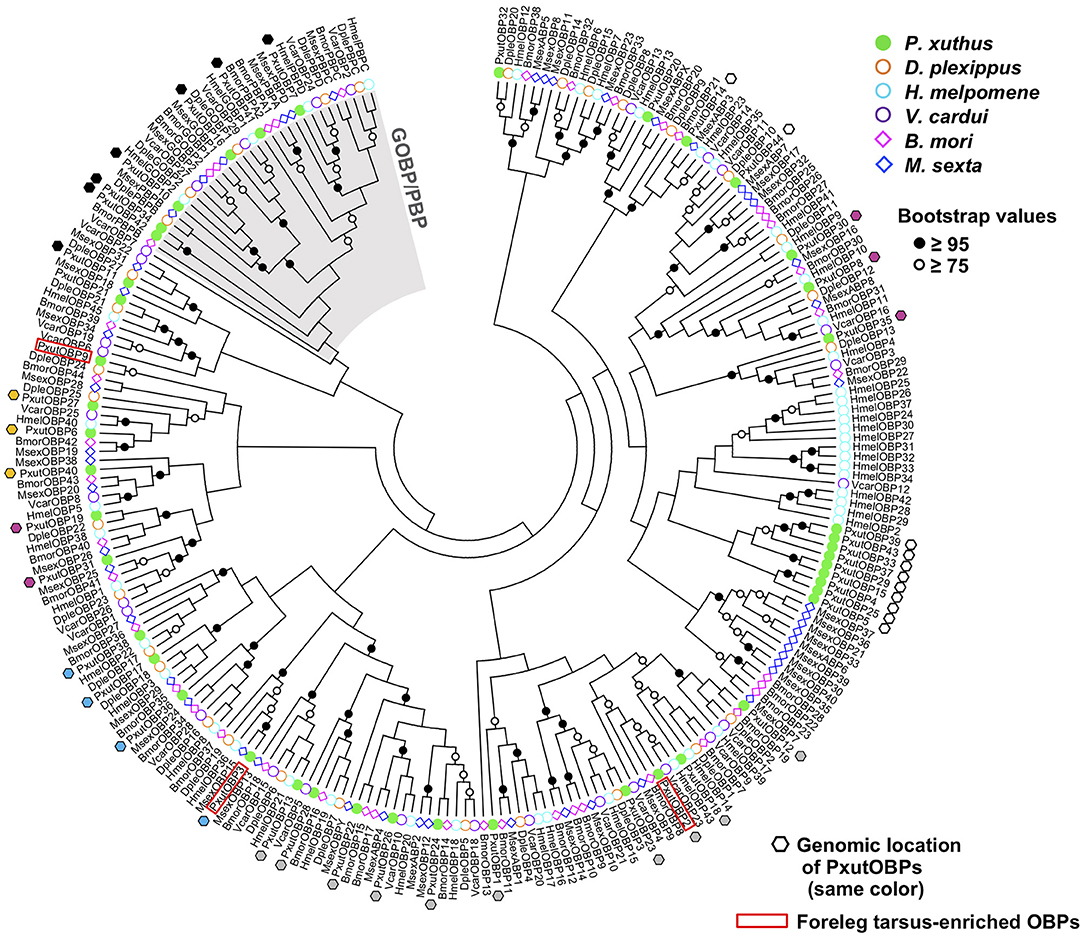
By searching the genomic database of P. xuthus and our RNA-sequencing assemblies, we found in total 44 OBP genes, including three previously reported genes (PxutOBP1, 2, and 3). Most of the OBP genes were clustered on the genome; 38 genes resided on only six scaffolds, each of which had 4–11 OBP genes (Figure 1, Supplementary Data Sheet 2, and Supplementary Figure 1). Based on the cysteine patterns of translated proteins, 21, 16, and 7 genes were classified as the classic, minus-C, and plus-C OBPs, respectively (Supplementary Figure 1). The names of newly characterized 41 OBP genes were designated according to the order of expression levels in the foreleg tarsi (see below). Regarding the overall profile of OBPs, the NJ tree indicated that P. xuthus has GOBP/PBP complex, which is monophyletic OBP genes specific to Lepidoptera, and genes locating on the same scaffolds tend to be phylogenetically close, consistent with previously characterized lepidopteran species (25, 30) (Figure 1).
Expression Profiles in Appendages
We first compared the expression levels of 44 OBP genes between the tarsus of forelegs and that of other legs (Figure 2A) by qRT-PCR analysis. Eleven OBP genes (PxutOBP1, 2, 3, 9, 10, 12, 13, 15, 18, 23, and 28) showed significantly higher expression levels or tended to be preferentially expressed (>five-fold) in the foreleg tarsus (Figure 2B). Most of the insect OBP genes are known to be expressed in the antennal olfactory sensilla (16, 17). We further examined expression levels of these 11 OBP genes among 6 appendages (foreleg tarsus, midleg tarsus, hindleg tarsus, antenna, proboscis, and ovipositor) (Figure 2A). qRT-PCR and statistical analyses revealed that PxutOBP10, 13, 15, and 28 were expressed predominantly in the antenna, and PxutOBP12, 18, and 23 were highly expressed in the antenna as well as the foreleg tarsus (PxutOBP12 and 18 showed the highest expression levels in the ovipositor) (Figure 2C). Thus, we focused on the remainder (PxutOBP1, 2, 3, and 9) in subsequent experiments.
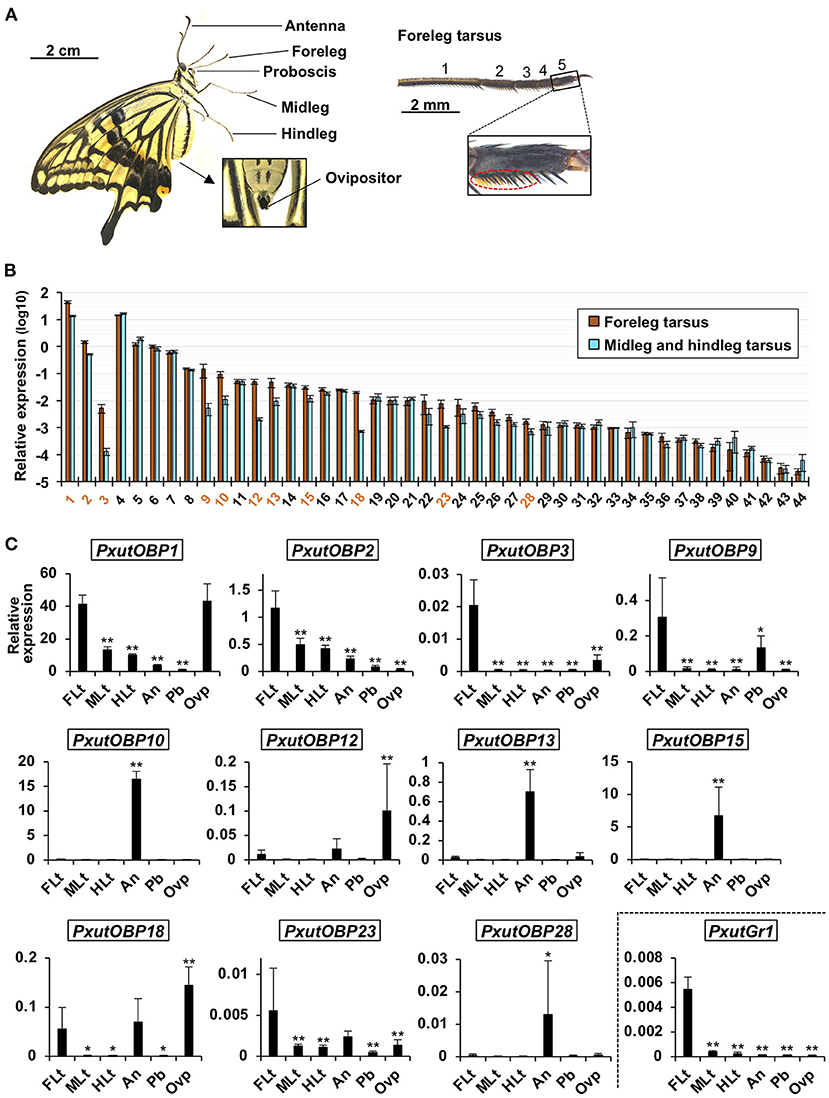
Figure 2. Expression levels of identified PxutOBPs. (A) Appendages examined for gene expression with a magnified view of the foreleg tarsus. Orange brushes in the dotted ellipse are the gustatory sensilla. As few gustatory sensilla are distributed in the first tarsomere (11), we collected the other four tarsomeres as “leg tarsus” samples. (B) Comparison of expression levels of OBPs between the tarsus of the forelegs and that of the other legs sampled from 0-day-old adult females performed by qRT-PCR. Three individuals were used per lot. All data are shown as the means ± SEM (n = 5, each leg). Student's t-test was conducted. OBP genes expressed significantly higher in foreleg tarsus (p < 0.05) or exhibiting prominent average fold change (the ratio of expression levels in foreleg tarsus to those in the other leg tarsus > 5) were indicated by orange-colored numbers. (C) Comparison of expression levels of OBPs among the foreleg tarsus (FLt), midleg tarsus (MLt), hindleg tarsus (HLt), antenna (An), proboscis (Pb), and ovipositor (Ovp) sampled from 0-day-old adult females performed by qRT-PCR. Two individuals were used per lot. All data are shown as the means ± SD (n = 6, each appendage). Multiple comparisons were performed using Dunnett's test. Asterisks indicate significant differences compared to expression levels of the foreleg tarsus (*, p < 0.05; **, p < 0.01).
Visualization of OBP Expression in the Foreleg Tarsus
Under our laboratory conditions (25°C, 16L8D), it took 9 days for P. xuthus to complete pupal metamorphosis. Adult-like legs could be observed 3 days before eclosion (−3 days). We sampled foreleg tarsi of −3-, −2-, −1-, 0-, and 3-day-old females and quantified the expression levels of PxutOBP2, 3, and 9. Significant upregulation of OBP genes was detected at −1 day, and transcription continued after adult eclosion, although PxutOBP3 expression was decreased at relatively low levels (Figure 3A and Supplementary Figure 2).
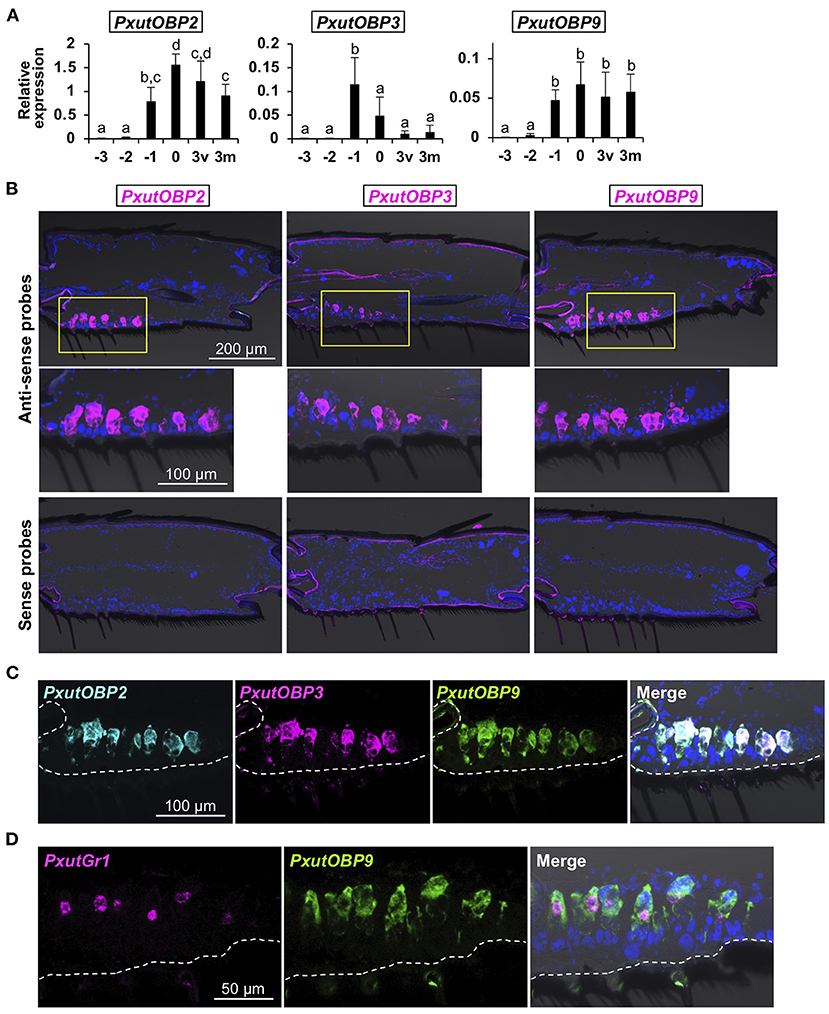
Figure 3. Visualization of PxutOBP2, 3, and 9 expression in the foreleg tarsus. (A) Developmental time-course of the expression of PxutOBP2, 3, and 9 in the female foreleg tarsus analyzed by qRT-PCR. −3, −2, −1: 3, 2, and 1 day before eclosion, respectively; 0: the day of eclosion; 3v: 3-day-old virgin female; 3m: 3-day-old mated female. Two individuals were used per lot. All data are shown as the means ± SD (n = 5, each developmental stage). Multiple comparisons were performed using Tukey-Kramer's test. Different letters indicate significant differences (p < 0.05). (B) Spatial distribution of OBP-expressing cells in the female fifth tarsomere visualized by FISH. As shown in (A), PxutOBP3 expression was peaked on 1 day before eclosion. Therefore, −1-day-old females were used for detection of PxutOBP3 transcripts while 0-day-old females were analyzed for the visualization of PxutOBP2 and 9 expression. (C) Simultaneous detection of PxutOBP2, 3, and 9 expression in the female fifth tarsomere by triple-color FISH. −1-day-old females were used. DNP-, DIG-, and FLU-labeled riboprobes were synthesized for PxutOBP2, 3, and 9, respectively. (D) Simultaneous detection of PxutGr1 and PxutOBP9 expression in the female fifth tarsomere. 0-day-old females were analyzed. DIG- and FLU-labeled riboprobes were synthesized for PxutGr1 and PxutOBP9, respectively.
Next, to examine spatial expression patterns, we performed fluorescence in situ hybridization (FISH) analyses with fresh-frozen sections. Since the gustatory sensilla are most abundant in the fifth tarsomere (11, 31), subsequent analyses were performed using the fifth tarsomeres of female forelegs. PxutOBP2, 3, and 9 were expressed at the bases of the gustatory sensilla (Figure 3B). In contrast, PxutOBP1 signals were broadly detected in the tissues along the ventral cuticle (Supplementary Figure 2), suggesting non-chemosensory function of this gene, and thus we excluded PxutOBP1 from the subject of further FISH analysis.
Triple-color staining successfully detected co-localized signals of PxutOBP2, 3, and 9 (Figure 3C). Because the amounts of transcripts of insect gustatory receptor (Gr) genes are generally quite low (32, 33) (Figure 2C), visualization of Grs expression by in situ hybridization has been thought to be difficult (34). Preliminary experiments showed that our FISH protocol (see section Materials and Methods) was effective to visualize the expression of a previously identified synephrine (one of the oviposition stimulants) receptor gene PxutGr1 (12) (Supplementary Figure 3). Then, we next performed simultaneous detection of PxutGr1 and PxutOBP9. PxutGr1-expressing small cells (gustatory receptor neurons) were observed in close proximity to PxutOBP9-expressing large support cells (Figure 3D and Supplementary Figure 3). These results indicated the coexpression of all the foreleg tarsus-enriched OBP genes in the oviposition stimulants-responsive sensilla. Similar localization patterns were also observed in males (Supplementary Figure 4). This is in agreement with a previous finding of no sexual dimorphism in the electrophysiological response of foreleg gustatory sensilla to the oviposition stimulants (11), although ecological significance has been unknown. Lower expression levels in males than in females (Supplementary Figure 4) possibly resulted from a smaller number of gustatory sensilla on the foreleg tarsus of males (11, 31).
OBP Expression in Other Appendages
Insects can taste chemicals by various body parts (35, 36). FISH analyses revealed that PxutOBP2, 3, and 9 were also coexpressed in the other legs and ovipositor (Figure 4 and Supplementary Figure 5). On the other hand, no PxutOBP3 signal was detected in the proboscis, whereas co-localized signals of PxutOBP2 and 9 were again observed (Figure 5A). We only found PxutOBP2-expressing cells in the antenna without any signals of PxutOBP3 and 9 (Figure 5B).
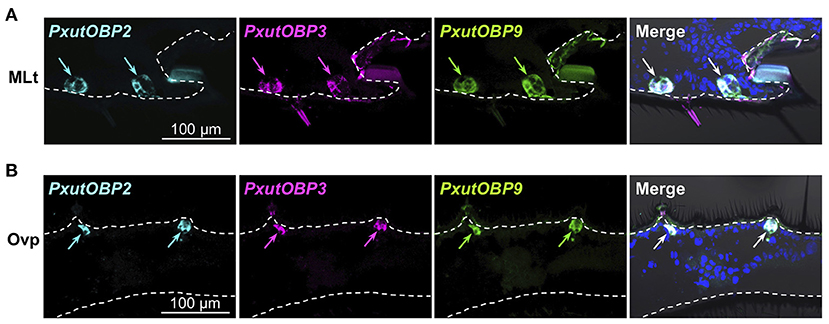
Figure 4. Coexpression of PxutOBP2, 3, and 9 in the midleg tarsus (A) and ovipositor (papilla analis) (B). −1-day-old females were analyzed. DNP-, DIG-, and FLU-labeled riboprobes were synthesized for PxutOBP2, 3, and 9, respectively. Arrows indicate signals of OBPs expression.
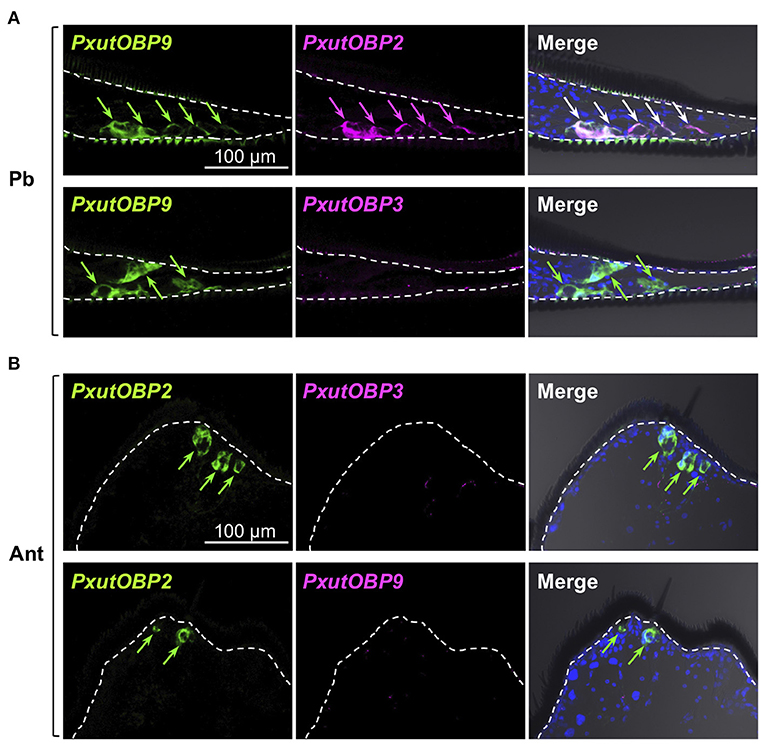
Figure 5. Visualization of PxutOBP2, 3, and 9 expression in the proboscis and antenna. Simultaneous detection of two different pairs of OBP genes was performed. −1-day-old females were analyzed. Arrows indicate signals of OBPs expression. (A) Longitudinal sections of the proboscis. FLU-labeled riboprobe was synthesized for PxutOBP9. For detection of PxutOBP2 or 3 transcripts, DIG-labeled riboprobes were used. (B) Longitudinal sections of the tip of antenna. FLU-labeled riboprobe was synthesized for PxutOBP2. For detection of PxutOBP3 or 9 transcripts, DIG-labeled riboprobes were used.
OBP Expression in Larval Gustatory Organs
Unlike adults, the vast majority of lepidopteran larvae feed on plant tissues (4, 37). The major gustatory organ of larvae, the maxilla, mainly consists of two parts called the maxillary galea (MG) and the maxillary palp (MP) (36) (Figure 6A), and chemosensory neurons housed in MG and MP contribute to food choice (38, 39). We investigated the expression patterns of PxutOBP2, 3, and 9 in the MG and MP of P. xuthus fifth instar larvae. In the MG, similar to the adult proboscis, co-localized signals of PxutOBP2 and 9 were observed, whereas PxutOBP3 signal was undetected (Figure 6B). In the MP, PxutOBP3 with PxutOBP2 and 9 signals were observed, although PxutOBP3-expressing cells differed from PxutOBP2 and 9 coexpressing cells (Figure 6C).
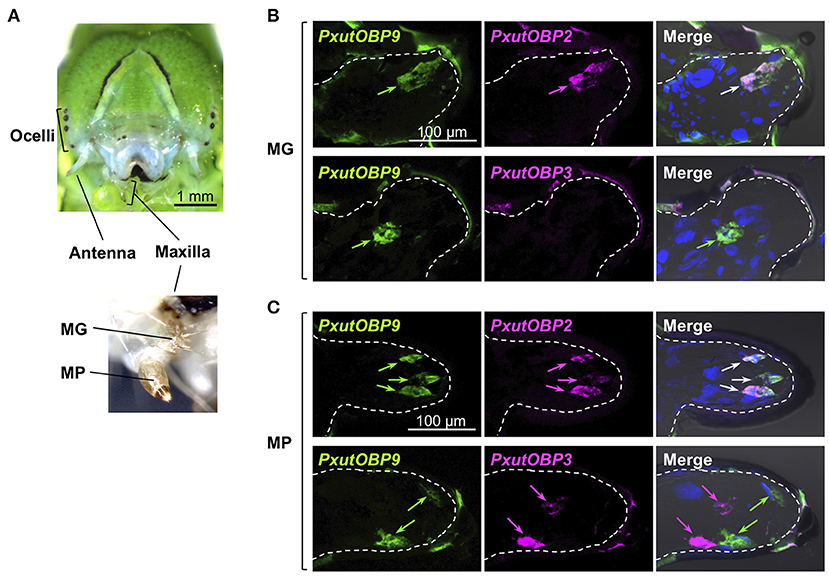
Figure 6. Visualization of PxutOBP2, 3, and 9 expression in the larval gustatory system. (A) Frontal view of a head of a final instar larva with a magnified view of the larval maxilla. (B,C) Simultaneous detection of two different pairs of OBP genes in the maxillary galea (MG) (B) and the maxillary palp (MP) (C). FLU-labeled riboprobe was synthesized for PxutOBP9. For detection of PxutOBP2 or 3 transcripts, DIG-labeled riboprobes were used. Arrows indicate signals of OBPs expression.
Discussion
In the present study, we performed genome-wide searching and found 41 OBP genes (Figure 1), besides the previously identified PxutOBP1, 2, and 3 from EST analyses of female foreleg tarsi (24). Vogt et al. summarized the OBP genes found in the four lepidopteran genomes (D. plexippus, H. melpomene, M. sexta, and B. mori) (25). The numbers of OBP genes in these species are ~30–50, similar to that of our identified PxutOBPs. Three OBP genes, PxutOBP2, 3, and 9 showed enriched expression in the foreleg tarsus (Figure 2). By applying multicolor FISH protocols, we demonstrated the coexpression of these three OBP genes at the bases of the gustatory sensilla of the foreleg tarsus (Figure 3).
Among the adult appendages which we investigated (Table 1), PxutOBP2 and 9 were commonly coexpressed in the same cells, except for the antenna where only PxutOBP2 signals were detected (Figures 4, 5). In the proboscis, PxutOBP3 signal was never observed (Figure 5). By contrast, besides the foreleg tarsus, co-localized signals of PxutOBP2, 3, and 9 were also observed in other legs and the ovipositor, both of which could have direct contact with the leaf surface during the drumming and egg-laying behavior. A small but considerable amount of PxutGr1 transcript was detected in the midleg and hindleg by our qRT-PCR analysis (Figure 2C). These legs may serve an ancillary role in host plant recognition. Some insects employ gustatory information received by the ovipositor to evaluate the oviposition substrate (40, 41). Not only photoreception and mechanosensation (42) but taste perception on the ovipositor could affect the final decision on egg deposition in P. xuthus. PxutOBP2, 3, and 9 were also expressed in the larval MP, whereas no PxutOBP3 signal was observed in the MG (Figure 6 and Table 1). Since the proboscis in adults is developmentally derived from the MG in larvae, the similarity in the undetectable expression of PxutOBP3 is interesting. Recently, in B. mori, Tsuneto and colleagues reported that gustatory neurons housed in MP were tuned into feeding stimulants derived from mulberry leaves (host plant of B. mori), whereas those in MG responded to sucrose (39). Based on these findings, they proposed fascinating model; the MP contributes to host selection via the detection of unique combinations of phytochemicals, and the MG participates in nutritional evaluation (39). Taken together, it is plausible that PxutOBP2 and 9 are basic OBPs of gustatory systems, and cooperation with PxutOBP3 enables P. xuthus larvae and adults to detect the cue phytochemicals leading to make appropriate decisions.
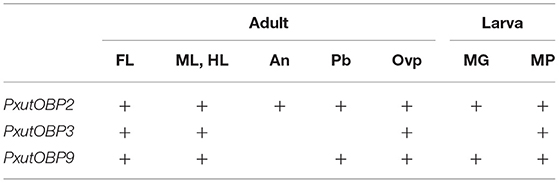
Table 1. Topographic expression patterns of PxutOBP2, 3, and 9 among the appendages based on the results of FISH experiments.
In the case of P. xuthus, 10 and 11 compounds have been identified as oviposition stimulants (10) and larval feeding stimulants (43), respectively. Interestingly, the components are quite different in the two sets of stimulants, and only stachydrine is commonly included, raising the possibility that PxutOBP3 interacts with stachydrine. It is also possible that PxutOBP3 is involved in avoiding the unpreferable plant species. Hydroxybenzoic acid derivatives have been known as deterrents to both oviposition and larval feeding (44). These compounds are contained in Orixa japonica, which is rutaceous plants but always rejected by P. xuthus. Alternatively, because insect OBPs show highly diverse binding affinity and selectivity [i.e., some OBPs bind to specific ligands, but others interact with a variety of compounds (15, 17, 45), it is not surprising that PxutOBP3 would interact with a broader spectrum of phytochemicals. To identify the ligand for PxutOBP3, future biochemical study is required. Intriguingly, a previous study showed that OBPs which coexpressed within the same olfactory sensilla in the antennae of Anopheles mosquitoes can form heterodimers with novel ligand specificities leading to amplify the number of volatiles that can be perceived (46). To uncover the role of PxutOBP3, future identification of the ligand by biochemical analysis and loss of function experiment with careful consideration of the cooperative interactions between coexpressing-PxutOBP2 or 9 is particularly important.
Phylogenetic analysis indicated that OBP genes orthologous to the foreleg tarsus-enriched PxutOBP2, 3, and 9 were conserved in other lepidopteran species (Figure 1). Comparative analyses of these orthologous genes, particularly PxutOBP3 orthologs, across a broad range of species would provide insight into the functional significance of OBPs in host plant selection in Lepidoptera. Furthermore, although we focused on OBP genes preferentially expressed in the foreleg tarsus in this study, longer-distance olfactory information also plays an important role in the process of host plant recognition (5, 7, 47). Therefore, the OBPs highly expressed in the antenna are important targets for future analysis.
Insects utilize diverse chemical signals for survival, and thus, the study of the chemosensory-related molecules is a fundamental topic in the field of insect science. Recent progress in the next-generation sequencing techniques leads to the comprehensive identification of chemosensory genes in various insects, including “non-model species” (17, 27, 48–52). Because chemosensory organs have many sensilla housing multiple sensory neurons each of which responds to different stimuli in general (35, 53, 54), investigation of cellular-level expression patterns of chemosensory genes is fundamental for understanding the molecular and neural mechanisms of chemosensation. In the non-model insects, however, spatial distribution pattern of chemosensory-related molecules has not been well characterized (46, 48, 49). The multicolor FISH analysis applied in this study allowed us to clearly visualize the low-level transcripts of Gr1 and the coexpression of three OBP genes in P. xuthus (Figures 3, 4). We expect that our protocol is an effective approach to gain spatial and combinatorial expression patterns of the chemosensory genes in other non-model insects.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: DDBJ [accession: PRJDB11535, PRJDB11545].
Author Contributions
AU and KO conceived and designed the study and performed the bioinformatic analyses. AU carried out the molecular lab work, analyzed the data, and wrote the manuscript. All authors gave final approval for publication.
Funding
This work was supported in part by JSPS KAKENHI Grant Numbers JP18K14478 (Grant-in-Aid for Early-Career Scientist, for AU) and JP18K06374 (Grant-in-Aid for Scientific Research (C), for KO).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Drs. Yasuko Akiyama-Oda and Hiroki Oda for providing technical advice on the enhanced POD-TSA reaction. We would also like to thank Ms. Yurie Hirosaki for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/finsc.2021.696179/full#supplementary-material
References
1. Bernays E, Graham M. On the evolution of host specificity in phytophagous arthropods. Ecology. (1988) 69:886–92. doi: 10.2307/1941237
2. Wiens JJ, Lapoint RT, Whiteman NK. Herbivory increases diversification across insect clades. Nat Comm. (2015) 6:8370. doi: 10.1038/ncomms9370
3. Gatehouse JA. Plant resistance towards insect herbivores: a dynamic interaction. New Phytol. (2002) 156:145–69. doi: 10.1046/j.1469-8137.2002.00519.x
4. Muto-Fujita A, Takemoto K, Kanaya S, Nakazato T, Tokimatsu T, Matsumoto N, et al. Data integration aids understanding of butterfly–host plant networks. Sci Rep. (2017) 7:43368. doi: 10.1038/srep43368
5. Renwick JAA, Chew FS. Oviposition behavior in Lepidoptera. Annu Rev Entomol. (1994) 39:377–400. doi: 10.1146/annurev.en.39.010194.002113
6. Ilse D. New observations on responses to colours in egg-laying butterflies. Nature. (1937) 140:544–5. doi: 10.1038/140544a0
7. Feeny P, Rosenberg L, Carter M. Chemical aspects of oviposition behavior in butterflies. In: Ahmad S, editor. Herbivorous Insects: Host-Seeking Behavior and Mechanisms. Cambridge, MA: Academic Press (1983) p. 27–76.
8. Honda K, Hayashi N. Chemical factors in Rutaceous plants regulating host selection by two swallowtail butterflies, Papilio protenor and P. xuthus (Lepidoptera: Papilionidae). Appl Entomol Zool. (1995) 30:327–34. doi: 10.1303/aez.30.327
9. Inoue TA, Asaoka K, Seta K, Imaeda D, Ozaki M. Sugar receptor response of the food-canal taste sensilla in a nectar-feeding swallowtail butterfly, Papilio xuthus. Naturwissenschaften. (2009) 96:355–63. doi: 10.1007/s00114-008-0483-8
10. Ohsugi T, Nishida R, Fukami H. Multi-component system of oviposition stimulants for a Rutaceae-feeding swallowtail butterfly, Papilio xuthus. Appl Entomol Zool. (1991) 26:29–40. doi: 10.1303/aez.26.29
11. Ryuda M, Calas-List D, Yamada A, Marion-Poll F, Yoshikawa H, Tanimura T, et al. Gustatory sensing mechanism coding for multiple oviposition stimulants in the swallowtail butterfly, Papilio xuthus. J Neurosci. (2013) 33:914–24. doi: 10.1523/JNEUROSCI.1405-12.2013
12. Ozaki K, Ryuda M, Yamada A, Utoguchi A, Ishimoto H, Calas D, et al. A gustatory receptor involved in host plant recognition for oviposition of a swallowtail butterfly. Nat Comm. (2011) 2:542. doi: 10.1038/ncomms1548
13. Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. (2006) 63:1658–76. doi: 10.1007/s00018-005-5607-0
14. Leal WS. Odorant reception in insects: roles of receptors, binding proteins, degrading enzymes. Annu Rev Entomol. (2013) 58:373–91. doi: 10.1146/annurev-ento-120811-153635
15. Brito NF, Moreira MF, Melo ACA. A look inside odorant-binding proteins in insect chemoreception. J Insect Physiol. (2016) 95:51–65. doi: 10.1016/j.jinsphys.2016.09.008
16. Sun JS, Xiao S, Carlson JR. The diverse small proteins called odorant-binding proteins. Open Biol. (2018) 8:180208. doi: 10.1098/rsob.180208
17. Rihani K, Ferveur JF, Briand L. The 40-year mistery of insect odorant-binding proteins. Biomolecules. (2021) 11:509. doi: 10.3390/biom11040509
18. Jeong YT, Shim J, Oh SR, Yoon HI, Kim CH, Moon SJ, et al. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron. (2013) 79:725–37. doi: 10.1016/j.neuron.2013.06.025
19. Swarup S, Morozova TV, Sridhar S, Nokes M, Anholt RRH. Modulation of feeding behavior by odorant-binding proteins in Drosophila melanogaster. Chem Senses. (2014) 39:125–32. doi: 10.1093/chemse/bjt061
20. Rihani K, Fraichard S, Chauvel I, Poirier N, Delompré T, Neiers F, et al. A conserved odorant binding protein is required for essential amino acid detection in Drosophila. Comm Biol. (2019) 2:425. doi: 10.1038/s42003-019-0673-2
21. Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama T. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. (2007) 5:e118. doi: 10.1371/journal.pbio.0050118
22. Yasukawa J, Tomioka S, Aigaki T, Matsuo T. Evolution of expression patterns of two odorant-binding protein genes, Obp57d and Obp57e, in Drosophila. Gene. (2010) 467:25–34. doi: 10.1016/j.gene.2010.07.006
23. Anholt RRH. Chemosensation and evolution of Drosophila host plant selection. iScience. (2020) 23:100799. doi: 10.1016/j.isci.2019.100799
24. Ozaki K, Utoguchi A, Yamada A, Yoshikawa H. Identification and genomic structure of chemosensory proteins (CSP) and odorant binding proteins (OBP) genes expressed in foreleg tarsi of the swallowtail butterfly Papilio xuthus. Insect Biochem Mol Biol. (2008) 38:969–76. doi: 10.1016/j.ibmb.2008.07.010
25. Vogt RG, Große-Wilde E, Zhou JJ. The lepidoptera odorant binding protein gene family: gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem Mol Biol. (2015) 62:142–53. doi: 10.1016/j.ibmb.2015.03.003
26. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum par- simony methods. Mol Biol Evol. (2011) 28:2731–9. doi: 10.1093/molbev/msr121
27. Suzuki HC, Ozaki K, Makino T, Uchiyama H, Yajima S, Kawata M. Evolution of gustatory receptor gene family provides insights into adaptation to diverse host plants in nymphalid butterflies. Genome Biol Evol. (2018) 10:1351–62. doi: 10.1093/gbe/evy093
28. Lauter G, Söll I, Hauptmann G. Multicolor fluorescent in situ hybridization to define abutting and overlapping gene expression in the embryonic zebrafish brain. Neural Dev. (2011) 6:10. doi: 10.1186/1749-8104-6-10
29. Akiyama-Oda Y, Oda H. Multi-color FISH facilitates analysis of cell-type diversification and developmental gene regulation in the Parasteatoda spider embryo. Develop Growth Differ. (2016) 58:215–24. doi: 10.1111/dgd.12263
30. Li MY, Jiang XY, Qi YZ, Huang YJ, Li GS, Liu S. Identification and expression profiles of 14 odorant-binding protein genes from Pieris rapae (Lepidoptera: Pieridae). J Insect Sci. (2020) 20:1–10. doi: 10.1093/jisesa/ieaa087
31. Inoue T. Morphology of foretarsal ventral surfaces of Japanese Papilio butterflies and relations between these morphology, phylogeny and hostplant preferring hierarchy. Zool Sci. (2006) 23:169–89. doi: 10.2108/zsj.23.169
32. Scott K, Brady R, Jr., Cravchik A, Morozov P, Rzhetsky A, Zuker C, et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. (2001) 104:661–73. doi: 10.1016/S0092-8674(01)00263-X
33. Xu W, Papanicolaou A, Zhang HJ, Anderson A. Expansion of a bitter taste receptor family in a polyphagous insect herbivore. Sci Rep. (2016) 6:23666. doi: 10.1038/srep23666
34. Ling F, Dahanukar A, Weiss LA, Kwon JY, Carlson JR. The molecular and cellular basis of taste coding in the legs of Drosophila. J Neurosci. (2014) 34:7148–64. doi: 10.1523/JNEUROSCI.0649-14.2014
35. Liman ER, Zhang YV, Montell C. Peripheral coding of taste. Neuron. (2014) 81:984–1000. doi: 10.1016/j.neuron.2014.02.022
36. Xu W. How do moth and butterfly taste? —Molecular basis of gustatory receptors in Lepidoptera. Insect Sci. (2020) 27:1148–57. doi: 10.1111/1744-7917.12718
37. Strong DR, Lawton JH, Southwood TRE. Insects on Plants: Community Patterns and Mechanisms. Cambridge, MA: Harvard University Press (1984).
38. Chapman RF. Contact chemoreception in feeding by phytophagous insects. Annu Rev Entomol. (2003) 48:455–84. doi: 10.1146/annurev.ento.48.091801.112629
39. Tsuneto K, Endo H, Shii F, Sasaki K, Nagata S, Sato R. Diet choice: the two-factor host acceptance system of silkworm larvae. PLoS Biol. (2020) 18:e3000828. doi: 10.1371/journal.pbio.3000828
40. Tousson E, Hustert R. Central projections from contact chemoreceptors of the locust ovipositor and adjacent cuticle. Cell Tissue Res. (2000) 302:285–94. doi: 10.1007/s004410000276
41. Ruschioni S, van Loon JJA, Smid HM, van Lenteren JC. Insects can count: sensory basis of host discrimination in parasitoid wasps revealed. PLoS ONE. (2015) 10:e0138045. doi: 10.1371/journal.pone.0138045
42. Arikawa K, Takagi N. Genital photoreceptors have crucial role in oviposition in Japanese yellow swallowtail butterfly, Papilio xuthus. Zool Sci. (2001) 18:175–9. doi: 10.2108/zsj.18.175
43. Murata T, Mori N, Nishida R. Larval feeding stimulants for a Rutaceae-feeding swallowtail butterfly, Papilio xuthus L. in Citrus unshiu leaves. J Chem Ecol. (2011) 37:1099–109. doi: 10.1007/s10886-011-0022-5
44. Ono H, Kuwahara Y, Nishida R. Hydroxybenzoic acid derivatives in a nonhost rutaceous plant, Orixa japonica, deter both oviposition and larval feeding in a rutaceae-feeding swallowtail butterfly, Papilio xuthus L. J Chem Ecol. (2004) 30:287–301. doi: 10.1023/B:JOEC.0000017978.73061.a0
45. Sandler BH, Nikonova L, Leal WS, Clardy J. Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex. Chem Biol. (2000) 7:143–51. doi: 10.1016/S1074-5521(00)00078-8
46. Qiao H, He X, Schymura D, Ban L, Field L, Dani FR, et al. Cooperative interactions between odorant-binding proteins of Anopheles gambiae. Cell Mol Life Sci. (2011) 68:1799–813. doi: 10.1007/s00018-010-0539-8
47. Yoshida M, Itoh Y, Ômura H, Arikawa K, Kinoshita M. Plant scents modify innate colour preference in foraging swallowtail butterflies. Biol Lett. (2015) 11:20150390. doi: 10.1098/rsbl.2015.0390
48. Hojo MK, Ishii K, Sakura M, Yamaguchi K, Shigenobu S, Ozaki M. Antennal RNA-sequencing analysis reveals evolutionary aspects of chemosensory proteins in the carpenter ant, Camponotus japonicus. Sci Rep. (2015) 5:13541. doi: 10.1038/srep13541
49. Jiang X, Krieger J, Breer H, Pregitzer P. Distinct subfamilies of odorant binding proteins in locust (Orthoptera, Acrididae): molecular evolution, structural variation, sensilla-specific expression. Front Physiol. (2017) 8:734. doi: 10.3389/fphys.2017.00734
50. Liu G, Xuan N, Rajashekar B, Arnaud P, Offmann B, Picimbon JF. Comprehensive history of CSP genes: evolution, phylogenetic distribution and functions. Genes. (2020) 11:413. doi: 10.3390/genes11040413
51. Yan H, Jafari S, Pask G, Zhou X, Reinberg D, Desplan C. Evolution, developmental expression and function of odorant receptors in insects. J Exp Biol. (2020) 223:jeb208215. doi: 10.1242/jeb.208215
52. Cassau S, Krieger J. The role of SNMPs in insect olfaction. Cell Tissue Res. (2021) 383:21–33. doi: 10.1007/s00441-020-03336-0
53. Isono K, Morita H. Molecular and cellular designs of insect taste receptor system. Front Cell Neurosci. (2010) 4:20. doi: 10.3389/fncel.2010.00020
Keywords: butterfly, Papilio xuthus, host plant selection, odorant-binding protein, gustatory sensilla, fluorescence in situ hybridization
Citation: Ugajin A and Ozaki K (2021) Coexpression of Three Odorant-Binding Protein Genes in the Foreleg Gustatory Sensilla of Swallowtail Butterfly Visualized by Multicolor FISH Analysis. Front. Insect Sci. 1:696179. doi: 10.3389/finsc.2021.696179
Received: 16 April 2021; Accepted: 06 July 2021;
Published: 30 July 2021.
Edited by:
Axel Brockmann, National Centre for Biological Sciences, IndiaCopyright © 2021 Ugajin and Ozaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Atsushi Ugajin, YXRzdXNoaS51Z2FqaW5AYnJoLmNvLmpw
 Atsushi Ugajin
Atsushi Ugajin Katsuhisa Ozaki
Katsuhisa Ozaki