
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ind. Microbiol., 06 January 2025
Sec. Food
Volume 2 - 2024 | https://doi.org/10.3389/finmi.2024.1508079
 Md Asif Hasan Muzumder Niloy1
Md Asif Hasan Muzumder Niloy1 Sharita Islam1
Sharita Islam1 Tasnimul Ferdous1
Tasnimul Ferdous1 Soyabur Rahman2
Soyabur Rahman2 Sabina Yesmin3
Sabina Yesmin3 Shahriar Bin Rasul4
Shahriar Bin Rasul4 Jebunnahar Khandakar1*
Jebunnahar Khandakar1*Introduction: Mold contamination, particularly from green and orange molds, poses a serious threat during the growing stage in mushroom cultivation, exacerbated throughout the hot and humid summer months. Despite extensive studies on green mold, orange mold remains underexplored. Consequently, this study comprehensively investigated orange mold contamination, focusing on identifying the causal agent, assessing its pathogenicity, and exploring potential countermeasures.
Methods: Internal transcribed spacer (ITS) region sequencing was used to confirm the causative entity, while the dual confrontation plate method was employed to assess pathogenicity. Furthermore, control strategies, including plant extract, in vitro media performance, and substrate characteristics, were explored. For estimated substrate qualities, Fourier-transform infrared spectroscopy (FTIR) and scanning electron microscopy (SEM) studies, along with analysis of physicochemical properties including the C:N ratio, carbon, protein, and mineral content were assessed.
Results: The result confirmed Neurospora sitophila as the causal entity. The pathogenicity assessments indicated that this mold impedes the colonization of mushroom mycelium by competing for nutrients and space. The in-vitro studies of media performance demonstrated that Neurospora sitophila growth was inhibited at varying rates in nitrogen supplemented media in the presence of available carbon. Notably, SEM analysis revealed Neurospora sitophila heavily colonized sawdust but not rice straw, attributed to a higher C:N ratio in sawdust.
Discussion: These findings suggest that lower C:N ratio negatively affects orange mold growth, highlighting nitrogen supplementation in sawdust or using rice straw as effective strategies to manage orange mold contamination in mushroom cultivation. This strategy could also be applied to other food industries where Neurospora is used.
Mushrooms are extensively valued for their nutraceutical and therapeutic benefits and serve as key recyclers in the natural ecosystem by decomposing organic substrate. Successful commercial cultivation of edible mushrooms relies heavily on the quality of the growing substrate, including the carbon-nitrogen (C:N) ratio, the amounts of cellulose, hemicellulose, and lignin (Suwannarach et al., 2022; Balan et al., 2022). Environmental conditions, particularly high humidity, and optimal temperature are crucial for the establishment of mushroom mycelium on the substrate (Zhan et al., 2021). However, unwanted intrusions of foreign microorganisms such as various mold, bacteria, insects, and mites pose challenges for mushroom cultivation.
In recent years, the threat of mold contamination in the mushroom industry has intensified due to climate change and rising temperatures. Competitor molds, like green and orange mold significantly hinder the colonization of mushroom mycelium on the substrate. Green mold outbreaks, caused primarily by Trichoderma, and sporadically by other fungi such as Penicillium and Aspergillus, have been a persistent issue in oyster mushroom cultivation across the globe (Allaga et al., 2021; Ahedo-Quero et al., 2024; Šašić Zorić et al., 2023; Cao et al., 2024). Similarly, orange mold, while a more recent concern in Bangladesh, poses a serious threat to the mushroom industry. This issue has been notably problematic during the spawn running stage, where distinctive orange color contamination rapidly overwhelms the farms, often within a week. The problem is exacerbated during the moist summer months, with temperatures exceeding 32°C and relative humidity reaching 70–80%. Moreau in 1956 asserted that orange mold on mushroom beds is caused by the obligatory aerobes Neurospora spp., which naturally thrive in moist tropical or subtropical climates, as their latent ascospores are activated by high temperatures. Despite being a contaminant in the mushroom industry, Neurospora spp. has been utilized in the food industry, particularly as a pigment producer in the traditional Indonesian dish oncom merah (Nout and Aidoo, 2010). Interestingly, no evidence has been obtained that Neurospora is the causal agent of any disease or infection in humans and animals (Perkins and Davis, 2000).
The quick colonization of lignocellulose substrates by orange mold, along with its ability to produce profound spores within a short time at an ambient temperature, underscores the urgency of controlling its spread before causing irreparable damage (Collier et al., 2020). In mushroom cultivation, sterilization, and pasteurization are commonly employed to eliminate competitive mold from substrates; however, these methods do not prevent the reintroduction of new inoculum after treatment (Jaramillo and Albertó, 2013). Chemical control using fungicides is another approach, but this raises environmental and health concerns, especially since mushrooms have a short cropping cycle and are known to bioaccumulate toxic metals (Sharma et al., 2007). Furthermore, the choice of fungicides must be highly selective, given that both molds and mushrooms are fungi. Beyond the substrate treatment methods, mold contamination also depends on substrate qualities including carbon content, protein levels, and the C:N ratio (Osunde et al., 2019). Research has demonstrated that the gene expression pattern of Neurospora crassa during its asexual growth stages is regulated by both external environmental stimuli and internal signals (Wang et al., 2019; Ebbole, 1998; Horowitz et al., 1976). Notably, genes associated with carbon and nitrogen metabolism are particularly sensitive to changes in nutrient availability, adapting their expression to fluctuating conditions (Wang et al., 2019). This interplay between nutritional factors and Neurospora morphogenesis is crucial for developing strategies to optimize mold control in mushroom cultivation systems.
The present study sets out to address the challenges posed by orange mold through a series of multifaced objectives. Accordingly, the research focused on identifying and characterizing the organisms responsible for orange mold and assessing their pathogenicity in relation to Pleurotus ostreatus. In addition, the research explored diverse control measures, including applying natural plant extract, in vitro analysis of different carbon and nitrogen sources, and evaluating substrate properties. The findings revealed the importance of substrate physiochemical characteristics, particularly the C:N ratio, in controlling orange mold. Importantly, this study represents the first comprehensive report on managing orange mold in the mushroom industry, offering valuable insight into controlling Neurospora sitophila, with potential applications extending beyond mushroom cultivation to the broader food industry.
Orange mold-contaminated sawdust mushroom spawn packets were gathered from the local mushroom farm in Savar, Dhaka. A 10 gm of sawdust sample was taken into a 100 mL conical flask containing 90 mL of sterile distilled water, and then shaken briefly to ensure thorough mixing. A 10−3 dilution was prepared from this mixture, and 1mL aliquots were spread onto a Petri dish containing Potato Dextrose Agar (PDA). After incubating the dishes at room temperature for 24 hours, a pure culture of mold was obtained. This pure culture was then subjected to microscopic examination to observe its morphological characteristics. A loopful of the culture was placed on a slide, stained with lactophenol cotton blue, and observed under a light microscope at 40x magnification.
The molecular identification of the fungi was performed using Polymerase Chain Reaction (PCR) with oligonucleotides specific for the internal transcribed spacer (ITS) regions of rDNA. DNA extraction and quantification of DNA followed the protocol provided with the Maxwell Blood DNA extraction kits (Model AS1010, Promega Corp, Madison, WI, USA). The fungal DNA was further purified using The Wizard® Genomic DNA purification Kit (A1120, Promega Corp., Madison, WI, USA). The extracted DNA was quantified at 40.9ng/µL using a NanoDrop 2000c Spectrophotometer. PCR reactions were carried out in a final volume of 25µL using the GoTaq ® Green Master Mix Kit (M7122, Promega Corp, Madison, WI, USA), with a concentration of 1x. The reaction mix included 5µM of the ITS1-Forward primer, 5µM of ITS4-Reverse primer, and 25 ng of genomic DNA. Amplification was performed in a C1000 thermal cycler (Bio-Rad® Germany) under the following conditions: initial denaturation at 95°C for 5 minutes, followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 45 seconds, and extension at 72°C for 45 seconds, with a final extension at 72°C for 10 minutes. The PCR products were separated by agarose gel electrophoresis (2%) at 80V for 40 minutes and visualized using SYBR Gold® (Invitrogen, Carlsbad, CA, USA). DNA fragments of approximately 700 base pairs were selected for purification using ExoSAP-IT (N/P 78200, USB Affymetrix, Inc., Cleveland, OH, USA). The purified fragments were sequenced with the ABI PRISM BigDye® Terminator sequencing kit v3.1 (P/N4336917, Applied Biosystems, Foster City, CA, USA) using the ITS1 forward primer (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4-Reverse primer (5′-TCCTCCGCTTATTGATATGC-3′). Fragment analysis was conducted on a Genetic Analyzer 3130 sequencer (Applied Biosystems® HITACHI Tokyo, Japan). Sequences were assembled using SeqMan software 8 (LaserGene) (DNASTAR®, Madison, WI, USA) and analyzed using the GenBank database (“National Center for Biotechnology Information,” n.d.). The top hit from the BLAST analysis in GenBank was used to identify the fungal species.
A dual culture technique was employed for assessing the aggressiveness of Neurospora sitophila isolates on Pleurotus ostreatus. Initially, 6 mm diameter agar plugs of P. ostreatus mycelium were positioned 1.5 cm from the edge of PDA Petri dishes and incubated at room temperature for 5 days to allow mycelial growth. Notably, P. ostreatus demonstrated a significantly slower growth rate, taking approximately 12 to 15 days to achieve full mycelial coverage. Afterward, similarly prepared mycelial plugs of N. sitophila, were placed on the opposite side of the same plate, also 1.5 cm away from the edge. The plates were then incubated for an additional 24 hours at ambient temperature as N. sitophila exhibited rapid mycelium expansion. The interaction between the N. sitophila and P. ostreatus mycelia was meticulously observed and documented.
An in-vitro experiment was conducted to evaluate the growth rate of Neurospora on various carbon, and nitrogen-enriched media, including N-Acetylglucosamine (GlcNAc), D-glucose, D fructose, Yeast extract, Peptone, and Sodium nitrate. For the assessment of carbon and nitrogen sources, PDA media supplementation with 5% (w/v) of mentioned carbon, and nitrogen sources, while PDA without supplements served as a control. Specifically, a 6mm Neurosopra mycelium plug was inoculated in the center of the Petri dish and subsequently incubated at ambient temperature for 24 hours. Notably, all experiments were performed in triplicate to ensure accuracy and reproducibility. The inhibition percentage was determined by measuring the radial growth of the fungus on both control and experimental plates after 24 and 48 hours of incubation. The calculation followed the formula proposed by (Yazid et al., 2023):
Where: (I%) represents the inhibition percentage of fungal growth on tested media. (R1) denotes the average radial growth in control plates. (R2) represents the average radial growth in experimental plates.
In this study, four locally available wild plants – Ocimum tenuiflorum, Leucas aspera, Persicaria hydropiper, and Helitropium indicum – were selected based on their documented antimicrobial properties from traditional knowledge and literature (Chandini et al., 2022; Rahman and Islam, 2013; Ayaz et al., 2020). The leaves of these plants were thoroughly washed under running water, sterilized for 2 minutes in 2% sodium hypochlorite, rinsed with sterile water, and then dried with absorbent paper. Subsequently, the leaves were dehydrated in an oven at 50°C for three days and ground using a conventional blender. The extracts were obtained by maceration (Hernández-Ceja et al., 2021) using three solvents including ethanol, acetone, and water. For each extraction, 15g of the dry and ground materials were mixed with 100 mL of the respective 100% solvent. Each mixture was placed in separate 200 mL beakers, kept in the dark, and left to stand for 3 days at room temperature. The extracts were then filtered through muslin cloth and centrifuged at 14,000 rpm for 20 min. The resulting supernatant was concentrated at 500°C using a rotary evaporator (model no). All the extracts were re-dissolved in methanol, adjusted to the concentration of 100 mg/mL, and stored at 40°C until further use. A total of 12 extracts were obtained. 5mg/mL of each plant extract stock solution was added to 20 mL of sterilized potato dextrose agar (PDA) in Petri dishes. A 6mm of the actively growing mycelium plug of the orange mold was placed in the center of the dishes and incubated at room temperature. Plates without plant extract served as negative control. Each treatment was conducted in triplicates, and the entire experiment was repeated three times.
The Fourier-transform infrared (FT-IR) spectra of sawdust and rice straw were obtained using an FTIR Spectrometer (model: IRAffinity-1S, Shimadzu Co., Japan) according to (Hamidu et al., 2020). Potassium bromide (KBr) was used as a window material to prepare the sample. The ratio of sample and KBr was 1:100, and the mixture was thoroughly mixed and ground in a porcelain-made mortar and pestle to achieve a homogenous mixture. The mixture was pressed into the pellet die at a high pressure of about 8 Tons to form a transparent pellet. In the FTIR analysis of rice straw and sawdust, a beam of light was directed at the prepared sample pellets to obtain transmission spectra within the wavenumber range of 4000–400 cm−1, facilitating the identification of all functional groups within this region (Ernest, 2015). The procedure involved conducting 45 scans at a resolution of 2 cm−1 to ensure precision and accuracy of the data.
The pH was analyzed in the aqueous extract, prepared by 1: 10 (w/v) fresh substrates and deionized water, using a standard hydrogen electrode, connected with a pH meter (model: pH 211, Hanna Instrument, Italy). The C:N ratio of the substrates were obtained based on the total nitrogen, determined by the Kjeldahl method, and the total carbon content that was determined by the dry ashing method (Kalra, 1997; Sáez-Plaza et al., 2013).
Holocellulose determination was carried out according to the chloride method (Wise and John, 1952). Briefly, 1g oven-dried samples were taken in a 250 mL flask with the addition of a mixture of 3mL of nitric acid and acetic acid (1:10 ratio), diluted with 100 mL distilled water incubated in a water bath at 100°C for 30 min. After centrifuging at 3000 rpm, the supernatant was discarded, and the residual was washed with distilled water. Then, 10 mL of 67% sulphuric acid was added and allowed to react for 1 hour. From this solution, I mL was taken and mixed with 10 mL of anthrone reagent and boiled in a water bath for 10 min until the green color was developed. After cooling, the absorbance was measured with a spectrometer at 630 nm. The cellulose content (%) was then determined relative to the initial full dry weight. About 1 g of powdered sample was placed in a refluxing flask and 10 mL of neutral detergent solution was added. The mixture was mixed with 2 mL of deca-hydro-naphthalene and 0.5 g sodium sulfite and then kept in a water bath at 70°C, refluxed for 60 min and filtered through Gooch crucible. The residue in the crucible was first washed with 100 mL of 8.3% NaOH solution, then with 15 mL of 10% acetic acid and 250 mL of distilled water, finally dried at 100°C, and weighed. Finally, % hemicellulose content was determined relative to oven dried sample.
The substrates were oven-dried at 60 ± 2°C until a constant weight was achieved. The dried samples were ground into a fine powder using a mortar and pestle. For analysis, all samples were mineralized by the wet digestion method. Specifically, 0.5 g of powdered samples were placed into the Teflon vessels with 5mL 65% Nitric Acid (Analar Grade, Merck, Germany) and 2mL 30% Hydrogen peroxide (Merck, Germany). The digestion process was carried out in a Microwave Digester (Model: Ethos One, Milestone, United States) at 180°C for 45 min (Lao et al., 2023). After digestion, the digests were transferred into 50 mL volumetric flasks and made up the volume with Class 1 (18MΩ) deionized water.
The analyses of mineral elements calcium (Ca), and Magnesium (Mg), were conducted using Flame Atomic Absorption Spectrophotometry (FAAS) as described by Brzezicha et al., 2019. The analytical conditions of the FAAS instrument (Model: AA-7000, Shimadzu Co. Japan) including detection limits, wavelength (nm), cathode lamp current (mA) slit width (ranging from 0.2 to 0.7), and air-acetylene flame mixture, were optimized according to established literature guidelines for each metal. The standard recovery percentages of the analytes were within the range of 95 to 105%. The total phosphorus (P) of the digested substrate samples was determined by Visible Spectrophotometric (model: UV-1800, Shimadzu Co. Japan) analysis through Ascorbic Acid reduction method (USEPA Method 365.3).
Fresh sawdust and rice straw were randomly collected, fixated, dehydrated, and covered with gold in an Emscope sputter coater. Subsequently, samples were examined using a scanning electron microscope (ZEISS EVO 18 Model).
To investigate the pathogen responsible for orange mold contamination, fungi were isolated from the contaminated spawn packet (Figure 1A) and cultured on PDA media. The results showed that the young colony of fungi appeared off-white with a fluffy or cottony texture (Figure 1B). As the colony matured, its color transitioned to orange (Figure 1C). Microscopic examination of the isolates revealed distinctive features, including conidia, and conidiophores (Figure 1D). According to Koch’s postulate, the pure culture was inoculated onto a sawdust substrate to verify whether isolated fungi were responsible for orange mold (Byrd and Segre, 2016). The characteristic orange color symptoms subsequently developed in the newly inoculated sawdust packets, indicating that the isolated fungus is a causal organism of orange mold. The fungus starts as an orange-white wisp but rapidly transforms into a bright orange, powdery patch. If allowed to progress, these patches develop into round, lumpy formations. Considering the above characteristics, our isolated fungi presumed Neurospora spp.
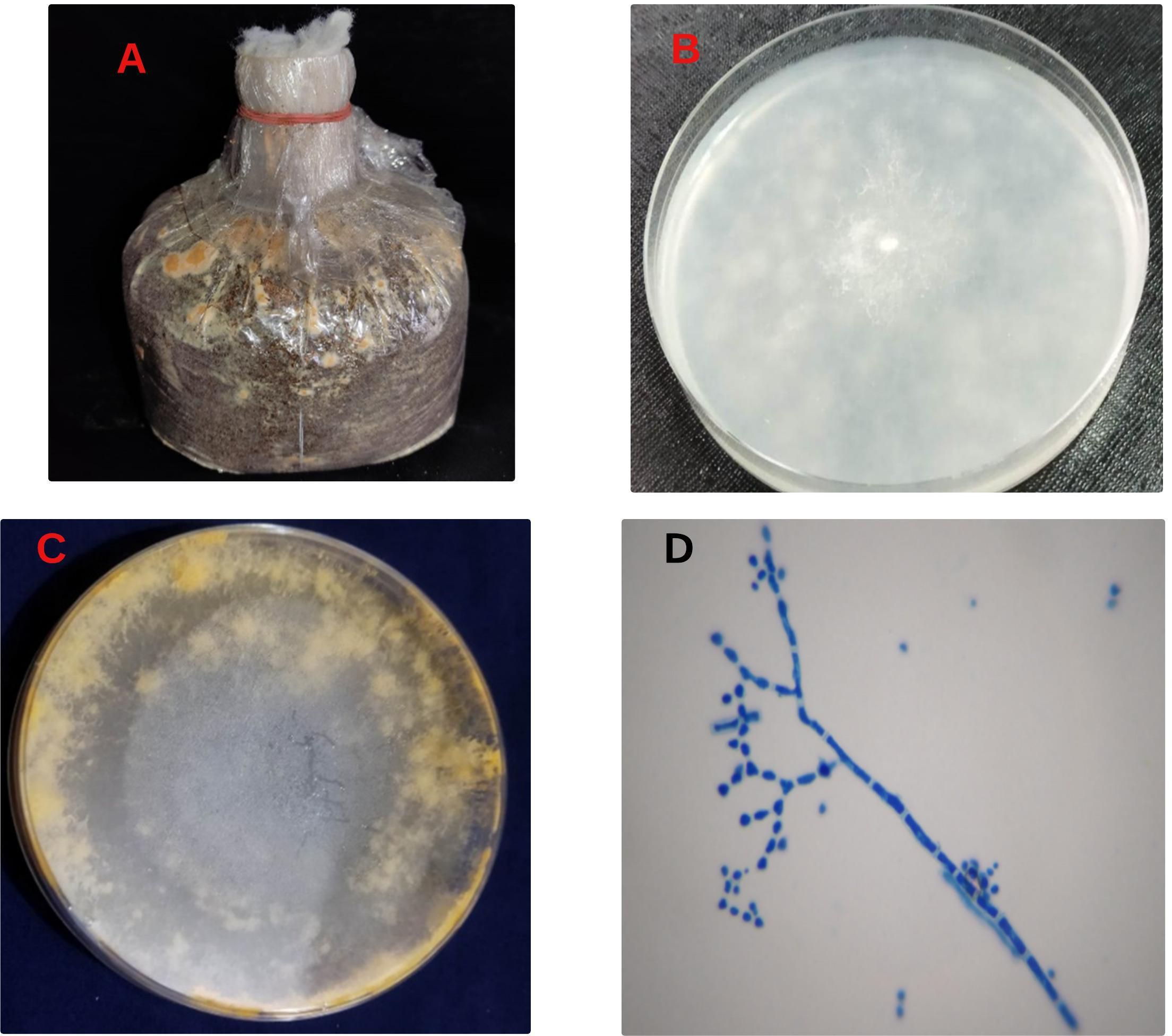
Figure 1. Contamination symptoms, fungal isolation, and microscopic study. Contaminant spawn packet (A), initial colony (B). Mature colony (C) conidia, and conidiophores (D).
Identification based on cultural features was confirmed by sequence analysis of the isolates. The universal primer for fungi identification in the ITS (internal transcribed spacer) region was amplified using PCR and then sequenced. The ITS region is a highly variable genetic marker found in all fungal species, making it an essential tool for distinguishing closely related species (White et al., 1990). The ITS region comprises two variable segments, ITS1 and ITS2, which are separated by the more conserved 5.8S rRNA gene (Schoch et al., 2012). Typically, ITS4 refers to the region that includes ITS2 and occasionally parts of the flanking regions (Gardes and Bruns, 1993). In this study, the ITS region of a presumptive Neurospora species was amplified, and the resultant sequence, designated as Neurospora_ITS_4, was analyzed. Basic Logical Alignment Search Tool (BLAST) results of ITS region in the National Centre for Biotechnology Information (NCBI) database revealed relationships and similarities with reference sequences in GenBank. DNA sequencing of the ITS 4 region showed the highest similarity to Neurospora sitophila (Accession No. ON712132.1) with 100% query coverage and 100% identity match, followed by Neurospora crassa with 100% query coverage and 98% identity match (Accession No. MH790467.1). A phylogenetic tree was constructed using BLAST Tree View provided by the National Center for Biotechnology Information (NCBI) BLAST tool (Altschul et al., 1990), incorporating Neurospora_ITS_4 and other highly similar sequences identified in the BLAST analysis. The phylogenetic tree revealed that Neurospora_ITS_4 clusters closely with Neurospora crassa (MH790549.1) and Neurospora sitophila (OW982620.1), indicating a close evolutionary relationship with these species. This placement suggests that Neurospora_ITS_4 is the Neurospora genus fungal species shown in Figure 2.
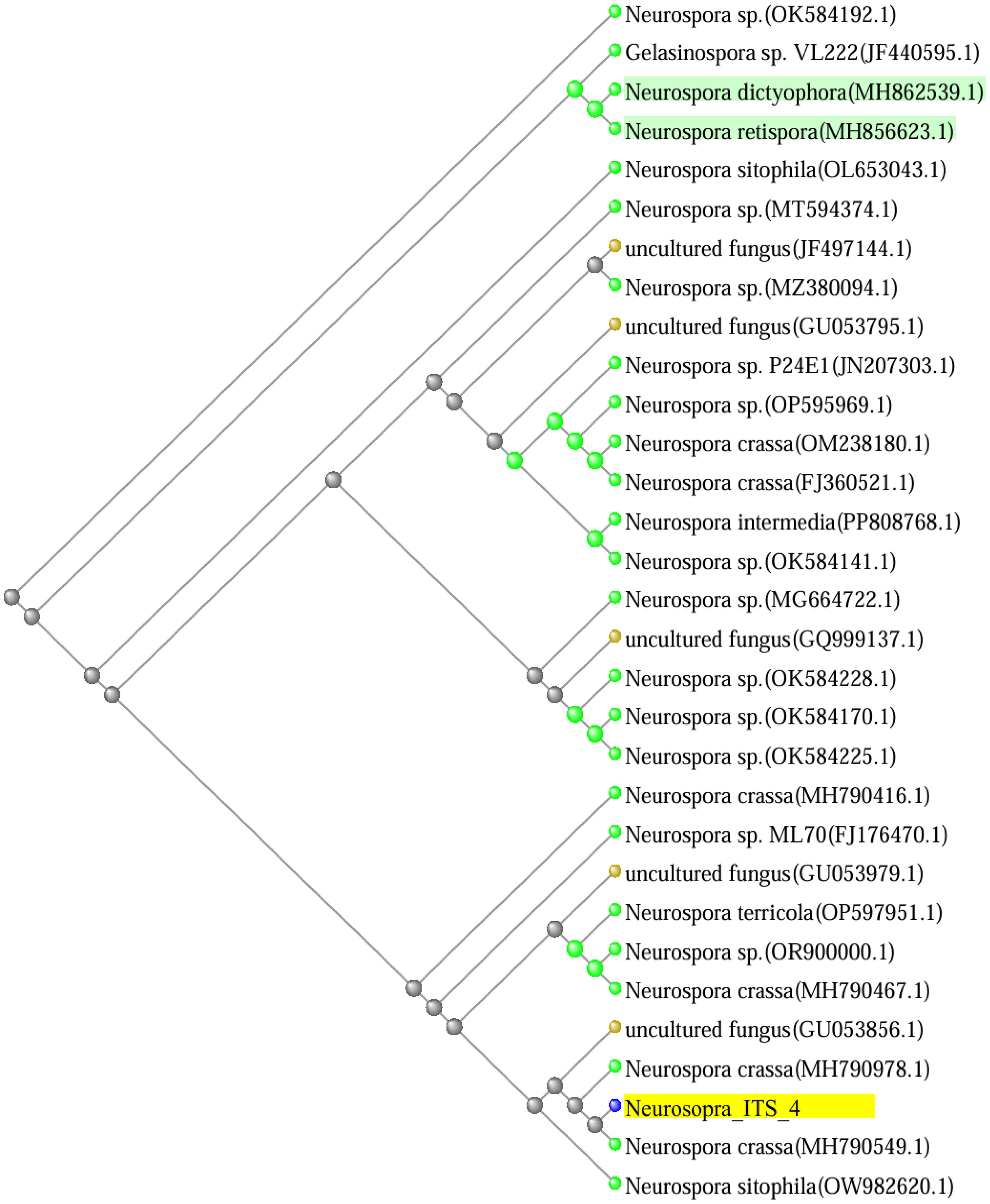
Figure 2. Phylogenetic tree based on ITS gene using BLAST Tree View. Branch length indicates evolutionary distance.
The plate dual culture experiments were conducted to assess the impact of N. sitophila on P. ostreatus. The results revealed that N. sitophila neither inhibited nor exhibited any antagonistic effect on the growth of P. ostreatus (Figure 3). However, within 24 hours, Neurospora mycelium had completely overrun the mushroom mycelium. The mycelium growth of Neurospora notably faster than that of P. ostreatus, eventually producing an irregular cluster of orange-red conidial as it expanded, the same as shown in Figure 1C.
Botanical extracts are widely recognized for their efficacy in controlling fungal pathogens in the food industry, given their safety for human consumption. This study conducted in vitro experiments to find potential botanical extracts that could suppress Neurospora sitophila growth with minimal or no effect on the host mycelium, Pleurotus ostreatus. In our study, we evaluated the leaf extracts of four locally available medicinal plants: Ocimum tenuiflorum, Leucas aspera, Persicaria hydropiper, and Helitropium indicum. The results indicated that the aqueous extracts (Lane 1, Figure 4) from these plants showed no inhibitory effects on N. sitophila. In contrast, acetone extracts (Lane 3, Figure 4) from Ocimum tenuiflorum, and Helitropium indicum, as well as ethanol extracts (Lane 2, Figure 4) from Leucas aspera, and Persicaria hydropiper demonstrated initial antifungal activity against Neurospora, which diminished over time. Importantly, none of the extracts achieved complete inhibition N. sitophila.
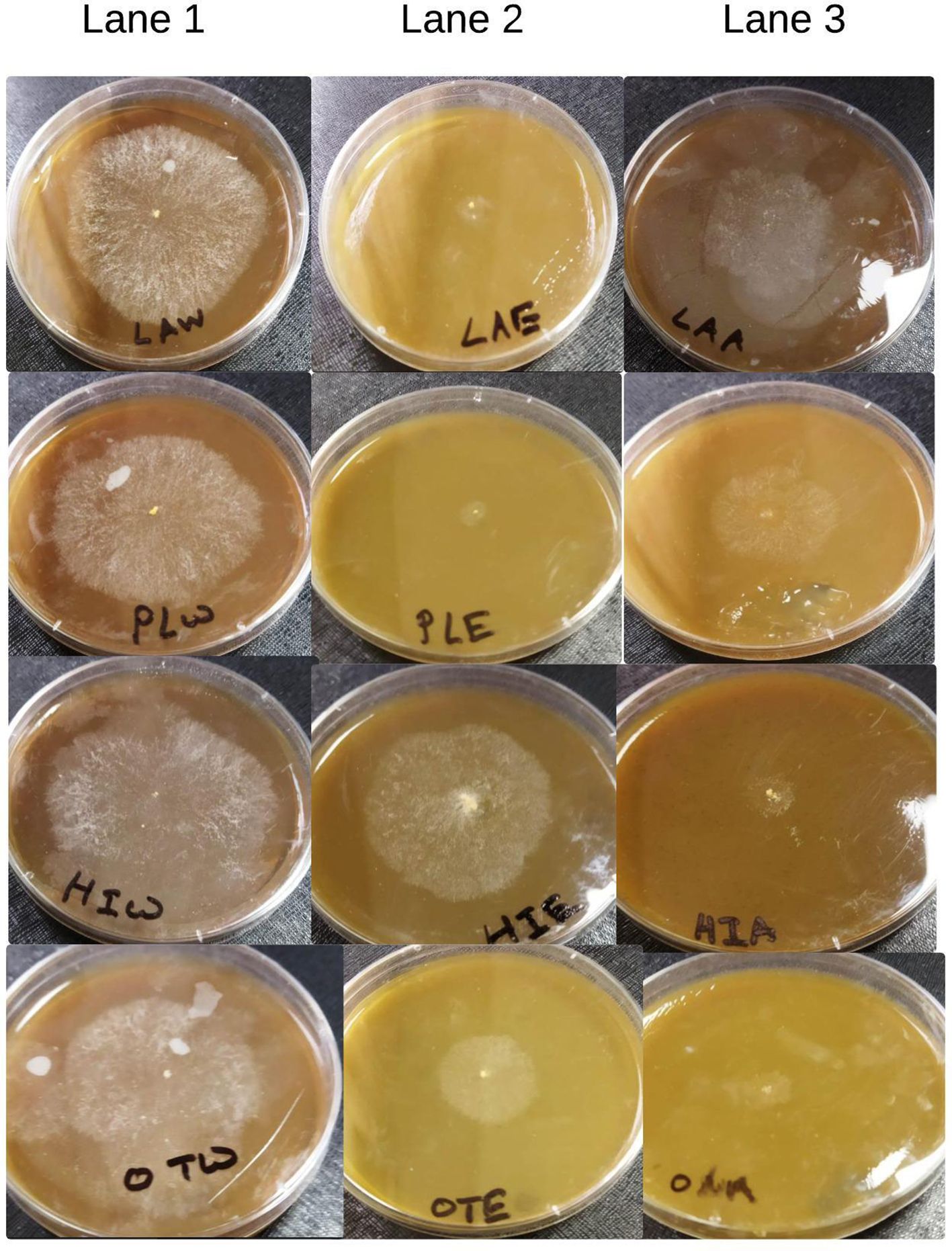
Figure 4. Effect of different botanical extracts on neurospora (Lane 1: aqueous extraction, Lane 2: ethanol extraction, Lane 3: acetone extraction).
In our study, we meticulously evaluated different carbon and nitrogen sources to understand their impact on Neurospora sitophila growth. We tested various carbon forms, including N-Acetylglucosamine (GlcNAc), D-glucose, and D-fructose, along with nitrogen sources such as yeast extract, peptone, and potassium nitrate. Results revealed that D-glucose (control PDA) and D-fructose promoted robust mycelial density without inhibition (Figures 5B, C), whereas GlcNAc-enriched media markedly restricted mycelial development (Figure 5A). Notably, GlcNAc, the second most abundant carbohydrate after cellulose, is a monosaccharide that typically polymerizes linearly through (1,4) β-linkages and potentially serves as both a carbon and nitrogen source. Conversely, in nitrogen-enriched media, Neurospora mycelial growth showed varying levels of restriction: 11.50% with yeast extract, 38.45% with peptone, and complete inhibition (100%) with sodium nitrate (Table 1; Figures 5D–F).
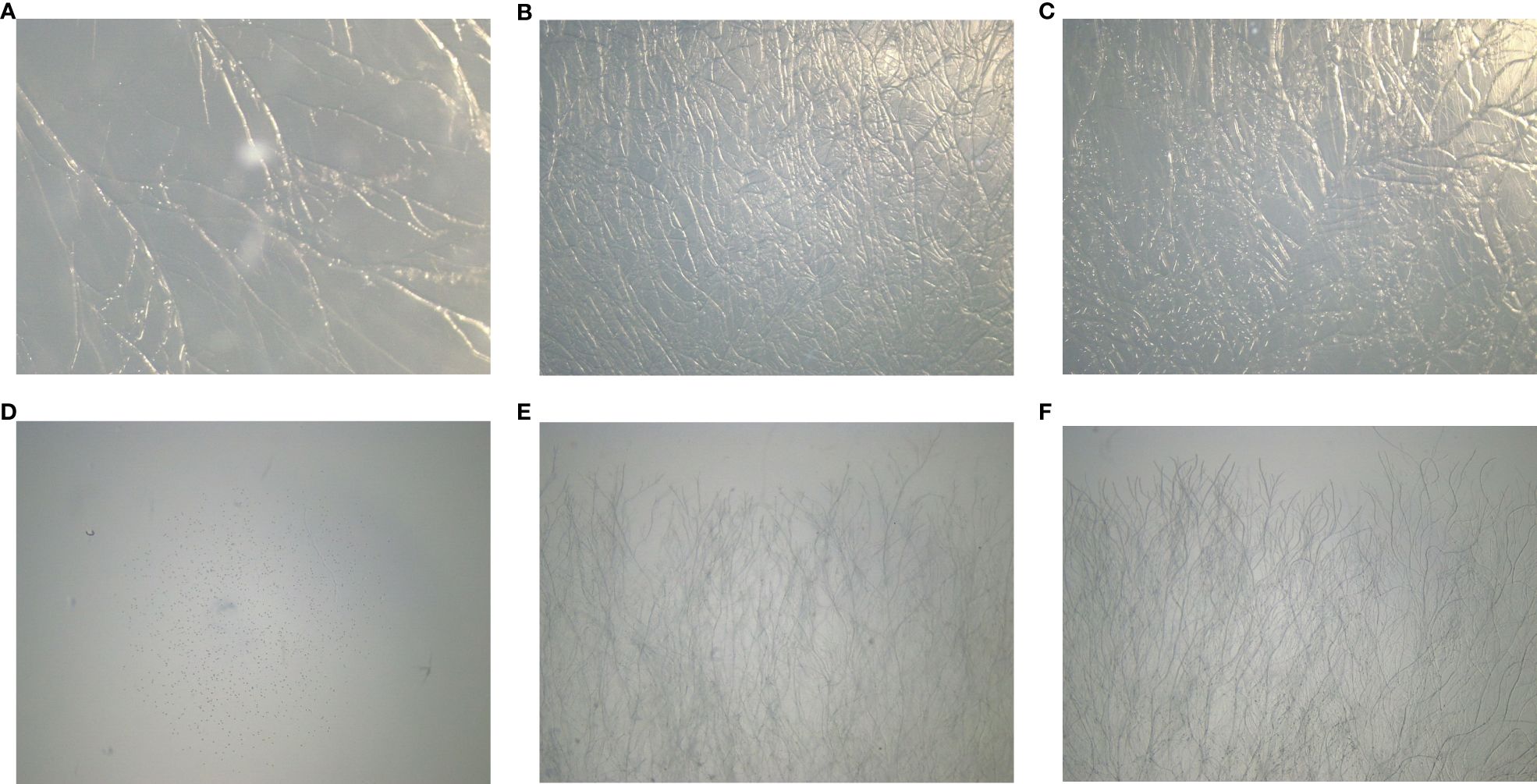
Figure 5. Effect of different carbon and nitrogen mediums on Neurospora sitophila: (A) supplemented with N-Acetylglucosamine (GlcNAc); (B) supplemented with Glucose (Control); (C) supplemented with Fructose; (D) supplemented with Potassium Nitrate; (E) supplemented with Peptone; (F)supplemented with Yeast Extract.
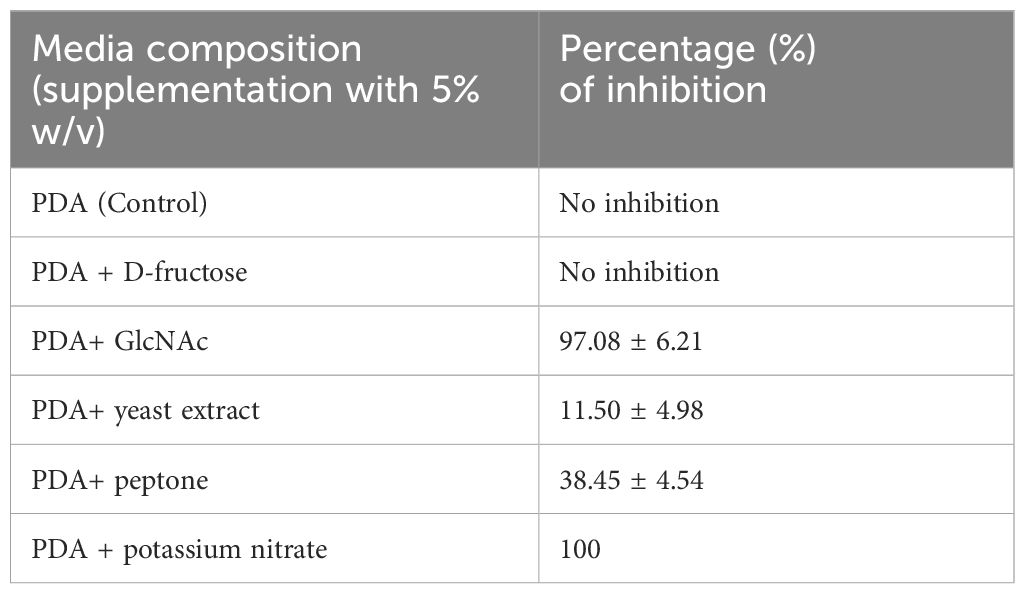
Table 1. Inhibition percentage of Neurospora sitophila in different carbon and nitrogen-enriched media.
The FTIR spectra of both rice straw and sawdust are very similar, though there are subtle differences in the intensity of the transmittance peaks (Figure 6). Rice Straw exhibits distinct peaks at various wavenumbers, noticeable around 3450 cm−1, 2800–2900 cm−1, 1690–1720 cm−1, 1500–1600 cm−1, 1400 cm−1, and 1100 cm−1. Sawdust also shows peaks in similar regions, indicating similar functional groups such as cellulose, hemicellulose, and lignin (Table 2). However, the intensity and position of these peaks in sawdust differ from those in rice straw. Rice straw has a higher transmittance than sawdust, suggesting that these differences can be attributed to the varying chemical composition and structure. Furthermore, it was predicted that rice straw has more complex polysaccharide structures, while sawdust has a higher lignin content.
In addition to carbon and nitrogen, several macronutrients and tress elements, such as phosphorus (P), potassium (K), magnesium (Mg), iron (Fe), zinc (Zn), and manganese (Mn), are crucial for fungal growth and various function (Carrasco et al., 2018). Interestingly, Neurospora can thrive in a medium containing carbon, an inorganic nitrogen source, and several inorganic salts in the presence of vitamin biotin (Beadle and Tatum, 1945). In Bangladesh, farmers primarily cultivate mushrooms on rice straw and sawdust. Therefore, to evaluate the suitability of sawdust and rice straw substrates for Neurospora growth, we carefully assessed their nutrient content. The results showed that rice straw contained 39% cellulose, 24% hemicellulose, and 12% lignin, whereas sawdust had 55% cellulose, 35% hemicellulose, and 25% lignin. Moreover, the C:N ratio of sawdust was notably high at 244:1, compared to the lower ratio of 63:1 in rice straw. Furthermore, rice straw is rich in protein and phosphorus, while sawdust contains higher levels of total carbon, calcium, and magnesium, as presented in Table 3.
The microstructural alterations of rice straw and sawdust during degradation by Neurospora sitophila were analyzed using scanning electron microscopy (SEM), as depicted in Figures 7A–E. Initially, both substrates exhibited uniform, smooth, and compact surfaces, reflecting their structural stability before fungal exposure (Figures 7A, B). However, we observed notable transformation, particularly in the sawdust, after 10–12 days of exposure to Neurospora sitophila, Its surface became more porous, with visible cracks and fissures, indicating the degradation of its structural components (Figure 7D). In contrast, the rice straw remained unchanged before and after treatment, suggesting that it was resistant to degradation by Neurospora sitophila (Figure 7C). However, the cultivation of mushrooms on rice straw resulted in substantial degradation, as Figure 7E illustrates.
This study intended to identify the organism responsible for orange mold, understand its pathogenic effect on Pleurotus ostreatus, and determine effective control measures. Following Koch’s postulates, the isolated pure culture of mold displayed the characteristic orange color when reintroduced to new sawdust packets, confirming its role in causing orange mold. The colony morphology and the presence of macroconidia observed under a bright light microscope suggested that this fungus is likely Neurospora spp (Kuo et al., 2014). It is noted, however, that identifying fungal species solely through morphological and microscopic studies can be challenging without concurrent DNA analysis. To further confirm the identity, we sequenced the internal transcribed spacer-4 (ITS-4) region. The BLAST search against ITS-4 region showed a 100% query coverage and 100% identity match with the Neurospora sitophila (Accession No. ON712132.1) and 100% query coverage and 98% identity match, with Neurospora crassa (Accession No. MH790467.1). Furthermore, molecular phylogenetic analysis revealed that Neurospora_ITS_4 clusters closely with Neurospora crassa (MH790549.1) and Neurospora sitophila (OW982620.1), indicating a close evolutionary relationship with these species. Thus, we concluded that the causal entity of orange mold is Neurospora sitophila.
To examine the nature of Neurospora sitophila pathogenicity with the oyster mushroom Pleurotus ostreatus, an in vitro confrontation assay was meticulously carried out. According to several studies, fungal pathogenicity mechanisms can be broadly categorized into antagonism, inhibition, and competition (Asad, 2022; Zeilinger et al., 2016). Antagonism is characterized by the mycelium growing in the opposite direction, leading to the formation of mycelial cords. Inhibition is marked by the presence of a distinct clear zone, primarily caused by the release of enzymes or metabolites. Competition, however, arises from the rivalry for space and nutrition. The results revealed that the rapidly growing Neurospora mycelium covered the entire Petri dish within 24 hours, forming irregular orange-red conidial clusters over time. These findings indicate that Neurospora significantly impedes mushroom mycelium colonization on lignocellulose substrate by aggressively competing for space and nutrients rather than through antagonism or inhibition. This aggressive competition resulted in the demise of mushroom mycelium growth and the eventual destruction of the mushroom spawn packets. A similar pattern of pathogenicity has also been extensively documented in Trichoderma spp. (Lombardi et al., 2023; Allaga et al., 2021).
Contamination is commonly understood to result from the inoculum’s potential and its capacity for rapid growth in the substrate. As previously stated Neurospora sitophila is a natural contaminant, and its abundant production of airborne powdery conidia leads to rapid growth on the substrate (Gmoser et al., 2017). Therefore, our strategy to control contamination was specifically targeted at inhibiting the proliferation and growth of Neurospora sitophila. Biopesticides, which are based on living microorganisms or natural products, including plant extracts, have great promise in effectively controlling pests and pathogens without decreasing the product quality (Hassan et al., 2021; Kumar et al., 2021). The tested hypothesis assumed that natural plant extracts would suppress mold growth with minimal or no effect on the host mycelium, Pleurotus ostreatus. The results indicated that none of the tested extracts completely inhibited the growth of Neurospora sitophila. Furthermore, the efficacy of the extracts gradually waned as the experiment progressed. The results claimed that this limited spectrum of activity often confines their application to niche situations. However, a noteworthy drawback of this section of our research is its exclusive focus on locally available, low-cost plant species.
A significant step in optimizing fungal growth is adjusting media composition, particularly nitrogen concentration. Consequently, we investigated Neurospora sitophila growth in different carbon and nitrogen media, including GlcNAc, D-glucose, D-fructose, peptone, yeast extract, beef extract, and potassium nitrate. Intriguingly, a notable finding was observed that the growth of Neurospora sitophila was considerably slower on GlcNAc compared to the other carbon media, suggesting that it struggles to utilize GlcNAc as a carbon source. Our results are consistent with the findings of a study (Gaderer et al., 2017) reporting that GlcNAc inhibited Neurospora crassa growth in the presence of other carbon sources. In the context of nitrogen-supplemented media, varying degrees of inhibition were observed. Importantly, beef extract and sodium nitrate supplementation completely inhibit Neurospora sitophila mycelium extension. Taking together it is suggested that Neurospora sitophila cannot efficiently utilize certain nitrogen forms in the presence of an available carbon source. This aligns with reports that Neurospora crassa, exhibits nitrogen metabolite repression, expressing genes necessary for secondary nitrogen sources utilization including nitrate, nitrite, purines, amino acid, and protein in the absence of favored nitrogen sources (ammonia and glutamate) (Marzluf, 1981; Fu and Marzluf, 1987). Genes involved in nitrogen metabolic repression are also integrated with the dynamic balance of asexual and sexual development, which is characteristic of N. crassa. Nitrogen starvation inhibits asexual reproduction (conidiation) and upregulates genes involved in sexual development, resulting in slower dispersal (Park et al., 2008). Conversely, carbon starvation promotes conidiation (Rodriguez-Romero et al., 2010; Ebbole, 1998; Nelson and Metzenberg, 1992). When both carbon and nitrogen are abundant, asexual growth is favored, leading to rapid dispersal. Thus, it is evident that the C:N ratio significantly influences Neurospora growth. Furthermore, saprophytic fungi secrete an array of catabolic enzymes such as protease, into their growth environment to break down the substrate (Hu and St Leger, 2004). The activation of protease is associated with nutrient limitations, including nitrogen, or carbon (Drucker, 1972). Moreover, the addition of different amino acids can suppress the secretion of proteases (Cohen and Drucker, 1977).
Our investigation disclosed a notable disparity in mold growth between sawdust and rice straw substrates. Specifically, contamination was prevalent in sawdust spawn packets, whereas rice straw packets remained unaffected. To assess their susceptibility, we re-inoculated the substrate, and the results were conclusive, sawdust is highly prone to orange mold contamination, while rice straw shows a natural resistance. Scanning electron microscope images provided further validation of the findings. As shown in Figures 7C, D, the sawdust substrate exhibited significant structural changes, becoming porous with visible cracks and fissures following the cultivation of Neurospora sitophila. In contrast, the rice straw substrate remained structurally intact even after 12 days of incubation with Neurospora sitophila. Subsequently, to gain deeper chemical insights, the physical and chemical properties of the substrates, including FTIR analysis were explored. The FTIR spectrum highlighted differences in infrared transmittance between the two substrates, attributable to their distinct chemical compositions and structural characteristics. Remarkably, rice straw exhibited more complex polysaccharide structures, whereas sawdust had a higher lignin content. Additionally, qualitative assays further confirmed these findings. As presented in Table 3, rice straw contains a substantial amount of protein and minerals, and a low amount of carbon including cellulose, hemicellulose, and lignin. Most importantly, rice straw exhibits a lower C:N ratio and calcium (Ca) content compared to sawdust, which contributed to the inhibition of Neurospora growth in rice straw. This finding aligned with the results of invitro media performance in this study and other studies, which have shown that substrates with high levels of carbohydrates and low levels of protein are prone to mold contamination, resulting in poor performances for mushroom growth (Bilal et al., 2014; Girmay et al., 2016; Mandeel et al., 2005). Furthermore, a high cytoplasmic calcium gradient is essential for Neurospora growth, as reported by Silverman-Gavrila and Lew, 2003.
The proliferation of fungal mycelia is primarily driven by the enzymatic breakdown of lignocellulosic biomass like rice straw and sawdust (Saini and Sharma, 2021). Like other saprophytic fungi, Neurospora species are known to secrete key enzymes including cellobiose dehydrogenase, cellulase, β-glycosidase, and xylanase (Østby et al., 2020; Oguntimein et al., 1992; Kanti and Sudiana, 2018). Interestingly, Pedraza-Zapata et al., 2017 demonstrated that the production of the above-mentioned lignocellulolytic enzymes by fungi like Penicillium sp. and Pleurotus ostreatus is significantly influenced by the availability of carbon and nitrogen. Several studies have claimed that substrates with a minimal C:N ratio ranging from 45 to 60/1 are conducive to Oyster mushroom (Pleurotus sp.) hyphal growth, but it depends on the species (Rani et al., 2008; Zhou and Parawira, 2022; Grimm and Wösten, 2018; Chang and Miles, 1989).
However, our study concluded that Neurospora mycelium growth is naturally inhibited in a substrate with a low C:N ratio. Our previous studies demonstrated that supplementing sawdust with nitrogen-enriched waste tea leaves significantly enhanced mushroom yield and increased mineral content compared to using sawdust alone (Ahmed et al., 2024).
The rapid spread of orange mold caused by Neurospora sitophila in the mushroom industry often likened to a wildfire, has garnered less research attention than other molds. Mushrooms are both nutrient-rich foods and hyperaccumulators of metals, contamination prevention strategies must prioritize health and environmental sustainability. Our study demonstrated that controlling orange mold contamination is strongly associated with the carbon-to-nitrogen (C:N) ratio of the substrate, with lower ratios playing a crucial role in its control.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
MAHMN: Writing – original draft, Investigation, Data curation. SI: Writing – original draft, Investigation, Data curation. TF: Writing – review & editing, Investigation, Methodology, Project administration, Supervision. SR: Writing – original draft, Writing – review & editing, Investigation, Data curation. SY: Writing – review & editing, Conceptualization, Formal Analysis, Methodology. SR: Writing – original draft, Data curation, Investigation. JK: Writing – review & editing, Conceptualization, Funding acquisition, Methodology, Project administration, Supervision.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The author(s) declare that this research was funded by the Krishi Gobesona Foundation (KGF), Bangladesh, grant number (CN/FRPP): TF141-C/23. The funding body had no contribution to the methodology, the data analysis, the drafting of the manuscript, or the interpretation of findings.
The corresponding author as the supervisor is grateful to the Mushroom Development Institute (MDI) for providing mushroom germplasm and supported the field experiment, Plasma Plus Laboratory of the Pharmacy Department at Independent University Bangladesh, for conducting mineral analysis, Military Institute of Science and Technology (MIST) for SEM analysis and to the Krishi Gobesona Foundation (KGF) for providing financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahedo-Quero H. O., Aquino-Bolaños T., Ortiz-Hernández Y. D., García-Sánchez E. (2024). Trichoderma diversity in Mexico: A systematic review and meta-analysis. Diversity 16, 685. doi: 10.3390/d16010068
Ahmed R., Niloy Md A. H. M., Islam Md S., Reza Md S., Yesmin S., Rasul S. B., et al. (2024). Optimizing tea waste as a sustainable substrate for oyster mushroom (Pleurotus ostreatus) cultivation: A comprehensive study on biological efficiency and nutritional aspect. Front. Sustain. Food Syst. 7. doi: 10.3389/fsufs.2023.1308053
Allaga H., Zhumakayev A., Büchner R., Kocsubé S., Szűcs A., Vágvölgyi C., et al. (2021). Members of the trichoderma harzianum species complex with mushroom pathogenic potential. Agronomy 11, 24345. doi: 10.3390/agronomy11122434
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arapova O. V., Bondarenko G. N., Chistyakov A. V., Tsodikov M. V. (2017). Vibrational spectroscopy studies of structural changes in lignin under microwave irradiation. Russian J. Phys. Chem. A 91, 1717–1729. doi: 10.1134/S0036024417090059
Ardila N., Daigle F., Heuzey M.-C., Ajji A. (2017). Antibacterial activity of neat chitosan powder and flakes. Molecules 22, 100. doi: 10.3390/molecules22010100
Asad S. A. (2022). Mechanisms of action and biocontrol potential of Trichoderma against fungal plant diseases - A review. Ecol. Complexity 49, 100978. doi: 10.1016/j.ecocom.2021.100978
Ayaz M., Ahmad I., Sadiq A., Ullah F., Ovais M., Khalil A. T., et al. (2020). Persicaria hydropiper (L.) delarbre: A review on traditional uses, bioactive chemical constituents and pharmacological and toxicological activities. J. Ethnopharmacol 251, 112516. doi: 10.1016/j.jep.2019.112516
Balan V., Zhu W., Krishnamoorthy H., Benhaddou D., Mowrer J., Husain H., et al. (2022). Challenges and opportunities in producing high-quality edible mushrooms from lignocellulosic biomass in a small scale. Appl. Microbiol. Biotechnol. 106, 1355–1745. doi: 10.1007/s00253-021-11749-2
Beadle G. W., Tatum E. L. (1945). Neurospora. II. Methods of producing and detecting mutations concerned with nutritional requirements. Am. J. Bot. 32, 678–686. doi: 10.2307/2437625
Bilal S., Mushtaq A., Moinuddin K. (2014). Effect of different grains and alternate substrates on oyster mushroom (Pleurotus ostreatus) production. Afr. J. Microbiol. Res. 8, 1474–1479. doi: 10.5897/AJMR2014.6697
Brzezicha J., Grembecka M., Grochowska I., Falandysz J., Szefer P. (2019). Elemental composition of selected species of mushrooms based on a chemometric evaluation. Ecotoxicol Environ. Saf 173, 353–365. doi: 10.1016/j.ecoenv.2019.02.036
Byrd A., Segre J. (2016). Adapting koch’s postulates. Science 351, 224–226. doi: 10.1126/science.aad6753
Cao Z.-J., Zhao J., Liu Y., Wang S.-X., Zheng S.-Y., Qin W.-T. (2024). Diversity of trichoderma species associated with green mold contaminating substrates of lentinula edodes and their interaction. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1288585
Chandini R., Saranya R., Mohideen K., Nandagopal P., Jayamani L., Jeyakumaran. S. (2022). Anti-candidal effect of ocimum sanctum: A systematic review on microbial studies. Cureus 14, e24749. doi: 10.7759/cureus.24749
Chang S. T., Miles P. G. (1989). Edible Mushrooms and their Cultivation. (Boca Raton, FL, USA; CRC Press).
Cohen B. L., Drucker H. (1977). Regulation of exocellular protease in neurospora crassa: induction and repression under conditions of nitrogen starvation. Arch. Biochem. Biophysics 182, 601–135. doi: 10.1016/0003-9861(77)90541-0
Collier L. A., Ghosh A., Borkovich K. A. (2020). Heterotrimeric G-protein signaling is required for cellulose degradation in neurospora crassa. mBio 11, 105. doi: 10.1128/mbio.02419-20
Carrasco J., Zied D. C., Pardo J. E., Preston G. M., Pardo-Gimenez A. (2018). Supplementation in mushroom crops and its impact on yield and quality. AMB Expr. 8, 146. doi: 10.1186/s13568-018-0678-0
Drucker H. (1972). Regulation of exocellular proteases in neurospora crassa: induction and repression of enzyme synthesis. J. Bacteriol 110, 1041–1049. doi: 10.1128/jb.110.3.1041-1049.1972
Ebbole D. J. (1998). Carbon catabolite repression of gene expression and conidiation in neurospora crassa. Fungal Genet. Biol: FG B 25, 15–21. doi: 10.1006/fgbi.1998.1088
Fu Y. H., Marzluf G. A. (1987). Characterization of nit-2, the major nitrogen regulatory gene of neurospora crassa. Mol. Cell. Biol. 7, 1691–1696. doi: 10.1128/mcb.7.5.1691-1696.1987
Gaderer R., Seidl-Seiboth V., de Vries R.P., Seiboth B., Kappel L. (2017). N-acetylglucosamine, the building block of chitin, inhibits growth of neurospora crassa. Fungal Genet. Biol. 107, 1–11. doi: 10.1016/j.fgb.2017.07.005
Gardes M., Bruns T. D. (1993). ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x
Girmay Z., Gorems W., Birhanu G., Zewdie S. (2016). Growth and yield performance of pleurotus ostreatus (Jacq. Fr.) kumm (Oyster mushroom) on different substrates. AMB Express 6, 875. doi: 10.1186/s13568-016-0265-1
Gmoser R., Ferreira J. A., Lennartsson P. R., Taherzadeh M. J. (2017). Filamentous ascomycetes fungi as a source of natural pigments. Fungal Biol. Biotechnol. 4, 45. doi: 10.1186/s40694-017-0033-2
Grimm D., Wösten H. A. B. (2018). Mushroom cultivation in the circular economy. Appl. Microbiol. Biotechnol. 102, 7795–78035. doi: 10.1007/s00253-018-9226-8
Hamidu L. A. J., Aroke U. O., Osha O. A., Misau M. I. (2020). Fourier transform infrared analysis of sawdust and rice husks waste: A raw material for eco-friendly composite production. Saudi J. Eng. Technol. 5, 343–350. doi: 10.36348/sjet.2020.v05i10.001
Hassan H. S., Mohamed A. A., Feleafel M. N., Salem M. Z.M., Ali H. M., Akrami M., et al. (2021). Natural plant extracts and microbial antagonists to control fungal pathogens and improve the productivity of zucchini (Cucurbita pepo L.) in vitro and in greenhouse. Horticulturae 7, 4705. doi: 10.3390/horticulturae7110470
Hernández-Ceja A., Loeza-Lara P. D., Espinosa-García F. J., García-Rodríguez Y. M., Medina-Medrano J. R., Gutiérrez-Hernández G. F., et al. (2021). In vitro antifungal activity of plant extracts on pathogenic fungi of blueberry (Vaccinium sp.). Plants (Basel Switzerland) 10, 8525. doi: 10.3390/plants10050852
Horowitz N. H., Charlang G., Horn G., Williams N. P. (1976). Isolation and identification of the conidial germination factor of neurospora crassa. J. Bacteriol 127, 135–140. doi: 10.1128/jb.127.1.135-140.1976
Hu G., St Leger R. J. (2004). A phylogenomic approach to reconstructing the diversification of serine proteases in fungi. J. Evolutionary Biol. 17, 1204–1214. doi: 10.1111/j.1420-9101.2004.00786.x
Jaramillo S., Albertó E. (2013). Heat treatment of wheat straw by immersion in hot water decreases mushroom yield in Pleurotus ostreatus. Rev. Iberoam Mico. 30, 125–129. http://hdl.handle.net/11336/24108.
Jiao N., Zhu Y., Li H., Yu Y., Xu Y., Zhu J. (2023). Two-step hydrothermal pretreatments for co-producing xylooligosaccharides and humic-like acid from vinegar residue. Fermentation 9, 5895. doi: 10.3390/fermentation9070589
Kalra Y. (1997). Handbook of Reference Methods for Plant Analysis (Boca Raton, FL, USA: CRC Press). doi: 10.1201/9781420049398
Kanti A., Sudiana I. M. (2018). Production of phytase, amylase and cellulase by Aspergillus, Rhizophus and Neurospora on mixed rice straw powder and soybean curd residue. IOP Conf. Ser.: Earth Environ. Sci. 166, 012010. doi: 10.1088/1755-1315/166/1/012010
Kumar J., Ramlal A., Mallick D., Mishra V. (2021). An overview of some biopesticides and their importance in plant protection for commercial acceptance. Plants 10, 11855. doi: 10.3390/plants10061185
Kuo H.-C., Hui S., Choi J., Asiegbu F. O., Valkonen J. P. T., Lee Y.-H. (2014). Secret lifestyles of neurospora crassa. Sci. Rep. 4, 51355. doi: 10.1038/srep05135
Lao Y. M., Qu C. L., Zhang B., Jin H. (2023). Development and validation of single-step microwave-assisted digestion method for determining heavy metals in aquatic products: health risk assessment. Food Chem. 402, 134500. doi: 10.1016/j.foodchem.2022.134500
Lombardi N., Pironti A., Manganiello G., Marra R., Vinale F., Vitale S., et al. (2023). Trichoderma species problematic to the commercial production of pleurotus in Italy: characterization, identification, and methods of control. Microbiol. Res. 14, 1301–1185. doi: 10.3390/microbiolres14030088
Mandeel Q. A., Al-Laith A. A., Mohamed S. A. (2005). Cultivation of oyster mushrooms (Pleurotus spp.) on various lignocellulosic wastes. World J. Microbiol. Biotechnol. 21, 601–607. doi: 10.1007/s11274-004-3494-4
Marzluf G. A. (1981). Regulation of nitrogen metabolism and gene expression in fungi. Microbiol Rev. 45, 437–461. doi: 10.1128/mr.45.3.437-461.1981
Nelson M. A., Metzenberg R. L. (1992). Sexual development genes of neurospora crassa. Genetics 132, 149–162. doi: 10.1093/genetics/132.1.149
Nout M. J., Aidoo K. (2010). “Asian fungal fermented food.” in Industrial Applications, (2nd edition) (Berlin Heidelberg: Springer), 29–58. doi: 10.1007/978-3-642-11458-8_2
Oguntimein G., Vlach D., Moo-Young M. (1992). Production of cellulolytic enzymes by Neurospora sitophila grown on cellulosic materials. Bioresource Technol. 39, 277–283. doi: 10.1016/0960-8524(92)90217-L
Omole J., Dauda B. (2016). Fourier transform infrared spectroscopy (FTIR) and scanning electron microscopy (SEM) analysis of chemically treated bagasse fibre. Am. Chem. Sci. J. 15, 1–9. doi: 10.9734/ACSJ/2016/24912
Østby H., Hansen L. D., Horn S. J., Eijsink V. G. H., Várnai A. (2020). Enzymatic processing of lignocellulosic biomass: principles, recent advances and perspectives. J. Ind. Microbiol. Biotechnol. 47, 623–657. doi: 10.1007/s10295-020-02301-8
Osunde M., Olayinka A., Fashina C., Torimiro N. (2019). Effect of carbon-nitrogen ratios of lignocellulosic substrates on the yield of mushroom (Pleurotus pulmonarius). OALib 6, 1–8. doi: 10.4236/oalib.1105777
Park G., Pan S., Borkovich K. A. (2008). Mitogen-activated protein kinase cascade required for regulation of development and secondary metabolism in neurospora crassa. Eukaryotic Cell 7, 2113–2225. doi: 10.1128/EC.00466-07
Pedraza-Zapata D. C., Sánchez-Garibello A. M., Quevedo-Hidalgo B., Moreno-Sarmiento N., Gutiérrez-Rojas I. (2017). Promising cellulolytic fungi isolates for rice straw degradation. J. Microbiol. 55, 711–719. doi: 10.1007/s12275-017-6282-1
Perkins D. D., Davis R. H. (2000). Evidence for safety of neurospora species for academic and commercial uses. Appl. Environ. Microbiol. 66, 5107–5195. doi: 10.1128/AEM.66.12.5107-5109.2000
Rahman A., Islam S. (2013). Antioxidant, antibacterial and cytotoxic effects of the phytochemicals of whole leucas aspera extract. Asian Pacific J. Trop. Biomed 3, 273–795. doi: 10.1016/S2221-1691(13)60062-3
Rani P., Kalyani N., Prathiba K. (2008). Evaluation of lignocellulosic wastes for production of edible mushrooms. Appl. Biochem. Biotechnol. 151, 151–159. doi: 10.1007/s12010-008-8162-y
Rodríguez-Lucena P., Lucena J. J., Hernández-Apaolaza L. (2009). Relationship between the Structure of Fe-Lignosulfonate Complexes Determined by FTIR Spectroscopy and Their Reduction by the Leaf Fe Reductase. Available online at: https://escholarship.org/uc/item/9k69q71d (Accessed June 30, 2009).
Rodriguez-Romero J., Hedtke M., Kastner C., Müller S., Fischer R. (2010). Fungi, hidden in soil or up in the air: light makes a difference. Annu. Rev. Microbiol. 64, 585–610. doi: 10.1146/annurev.micro.112408.134000
Sáez-Plaza P., Navas M., Wybraniec S., Michałowski T., Asuero A. G. (2013). An overview of the kjeldahl method of nitrogen determination. Part II. Sample preparation, working scale, instrumental finish, and quality control. Crit. Rev. Analytical Chem. 43, 224-272. doi: 10.1080/10408347.2012.751787
Sahoo S., Chakraborti C., Mishra S., Nanda U., Naik S. (2011a). FTIR and XRD investigations of some fluoroquinolones. Int. J. Pharm. Pharm. Sci. 3, 165–170.
Sahoo S., Chakraborti C., Naik S. (2011b). Analytical characterization of a controlled release polymeric suspension of norfloxacin. Int. J. Pharm. Sci. Nanotechnol 3, 1506–1518.
Saini S., Sharma K. K. (2021). Fungal lignocellulolytic enzymes and lignocellulose: A critical review on their contribution to multiproduct biorefinery and global biofuel research. Int. J. Biol. Macromol 193, 2304–2319. doi: 10.1016/j.ijbiomac.2021.11.063
Schoch C. L., Seifert K. A., Huhndorf S., Robert V., Spouge J. L., Levesque C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. United States America 109, 6241–6246. doi: 10.1073/pnas.1117018109
Suwannarach N., Kumla J., Zhao Y., Kakumyan P. (2022). Impact of cultivation substrate and microbial community on improving mushroom productivity: A review. Biology 11, 5695. doi: 10.3390/biology11040569
Sharma S. R., Kumar S., Sharma V. P. (2007). Diseases and Competitor Moulds of Mushrooms and their Management. Tech. Bulletin. (Chambaghat, Solan, India) pp, 1–43.
Silverman-Gavrila L. B., Lew R. R. (2003). Calcium gradient dependence of Neurospora crassa hyphal growth. Microbiology 149, 2475–2485. doi: 10.1099/mic.0.26302-0
Šašić Zorić L., Janjušević L., Djisalov M., Knežić T., Vunduk J., Milenković I., et al. (2023). Molecular approaches for detection of trichoderma green mold disease in edible mushroom production. Biology 12, 2995. doi: 10.3390/biology12020299
Wang Z., Miguel-Rojas C., Lopez-Giraldez F., Yarden O., Trail F., Townsend J. P. (2019). Metabolism and development during conidial germination in response to a carbon-nitrogen-rich synthetic or a natural source of nutrition in neurospora crassa. mBio 10, e00192–e00195. doi: 10.1128/mBio.00192-19
White T. J., Bruns T., Lee S., Taylor J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Yazid S. N. E., Tajudin N. I., Razman N. A. A., Selamat J., Ismail S. I., Sanny M., et al. (2023). Mycotoxigenic fungal growth inhibition and multi-mycotoxin reduction of potential biological control agents indigenous to grain maize. Mycotoxin Res. 39, 177–925. doi: 10.1007/s12550-023-00484-4
Zeilinger S., Gruber S., Bansal R., Mukherjee P. K. (2016). Secondary metabolism in Trichoderma–chemistry meets genomics. Fungal Biol. Rev. 30, 74–90. doi: 10.1016/j.fbr.2016.05.001
Zhan Z., Xu M., Li Y., Dong M. (2021). The Relationship between Fungal Growth Rate and Temperature and Humidity. Int. J. Eng. Manag. Res. 11. Available online at: https://ssrn.com/abstract=3867229 (Acessed June, 2011).
Keywords: Neurospora sitophila, mushroom, C:N ratio, orange mold, Fourier-transform infrared spectroscopy (FTIR), scanning electron microscopy (SEM)
Citation: Niloy MAHM, Islam S, Ferdous T, Rahman S, Yesmin S, Bin Rasul S and Khandakar J (2025) Deciphering the role of substrate carbon to nitrogen ratio in preventing orange mold contamination caused by Neurospora sitophila in mushroom cultivation. Front. Ind. Microbiol. 2:1508079. doi: 10.3389/finmi.2024.1508079
Received: 08 October 2024; Accepted: 05 December 2024;
Published: 06 January 2025.
Edited by:
Lourdes Agüí, Complutense University of Madrid, SpainReviewed by:
Leyla Nazari, Agricultural Research, Education and Extension Organization (AREEO), IranCopyright © 2025 Niloy, Islam, Ferdous, Rahman, Yesmin, Bin Rasul and Khandakar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jebunnahar Khandakar, ZHIua2hhbmRha2FyQGl1Yi5lZHUuYmQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.