- 1Center for Interdisciplinary Research in Animal Health (CIISA) – Center for Interdisciplinary Research in Animal Health, Faculty of Veterinary Medicine, University of Lisbon, Lisbon, Portugal
- 2Associate Laboratory for Animal and Veterinary Sciences (AL4AnimalS), Faculty of Veterinary Medicine, University of Lisbon, Lisbon, Portugal
- 3BioISI – Biosystems & Integrative Sciences Institute, Faculty of Sciences, University of Lisbon, Lisbon, Portugal
The microbial communities inhabiting Portuguese traditional cheeses play a fundamental role in shaping their unique flavor, texture, and safety characteristics. This comprehensive review synthesizes findings from both conventional microbiological studies and advanced OMICs analyses to provide a deeper understanding of the microbiota dynamics in these cheeses. We explore the microbial composition, diversity, and functional roles of bacteria, yeasts, and molds across various Protected Designation of Origin (PDO) cheeses, highlighting their contributions to cheese ripening, flavor development, and safety. Additionally, we discuss the potential of OMICs technologies, namely metagenomics, in unraveling the complex microbial ecosystems of Portuguese traditional cheeses. Through this integrative approach, we aim to shed light on the intricate interplay between microorganisms and cheese matrices, unveiling the secrets behind the rich heritage and distinctiveness of Portuguese traditional cheeses.
1 Traditional cheeses with Protected Designation of Origin: preserving heritage
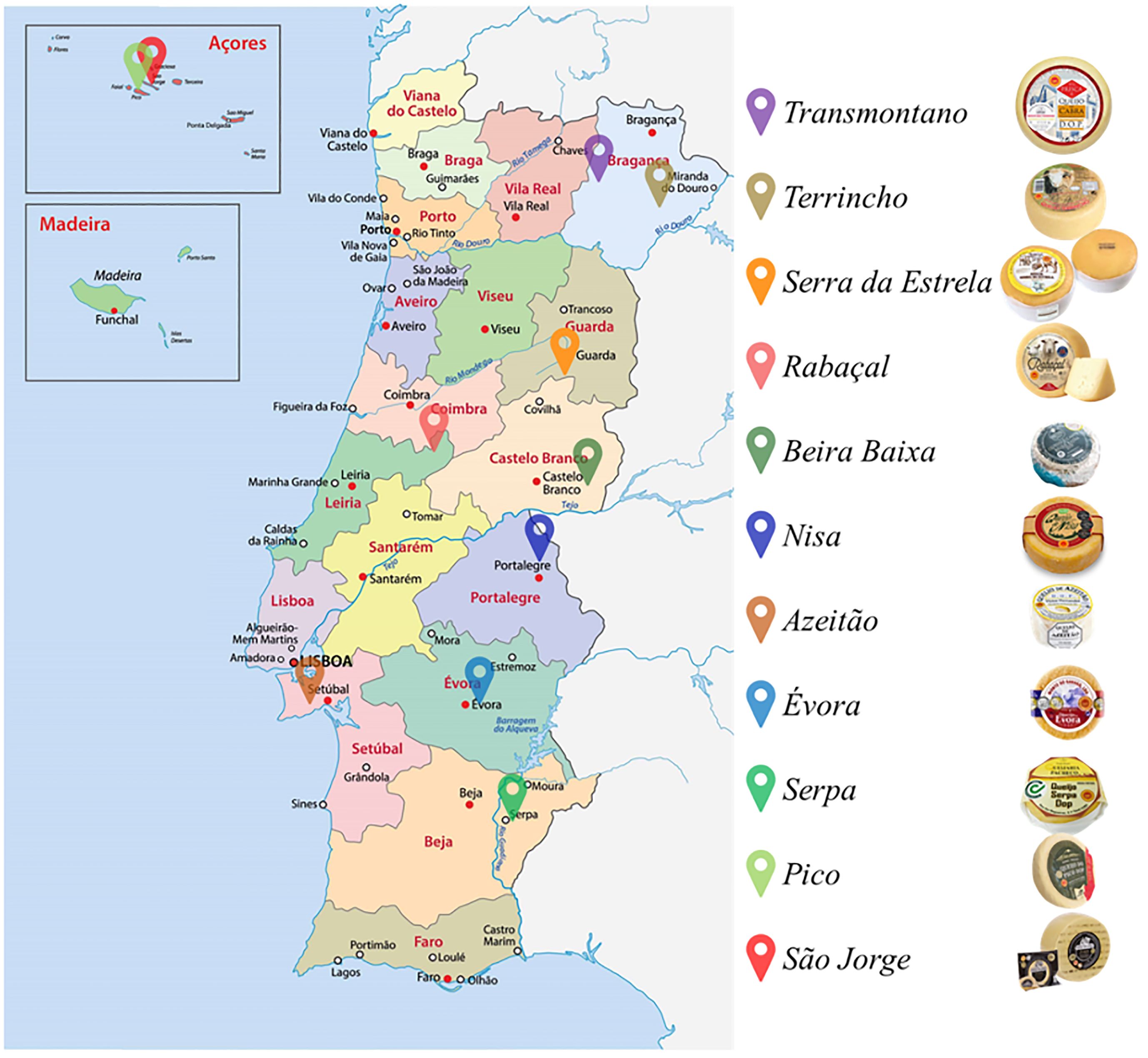
Artisanal food products constitute an important cultural heritage. In Mediterranean countries, such as Portugal, a wide variety of traditional cheeses is manufactured, harboring distinct flavor, texture and overall composition (see Figure 1). These dairies hold significant importance, not only in social and cultural contexts, preserving history and tradition, but also in terms of economic impact, as they are regarded as value-added products (Guiné and Florença, 2020). The majority of Portuguese traditional cheeses, like those in many other countries, are manufactured in rural regions by small, multi-generational enterprises. Consequently, the sale of these products provides essential income to local communities, dependent on cattle farming and/or cheese production as their livelihood (Reis and Malcata, 2011). These products are manufactured using ancient traditions and expertise, which must be safeguarded to maintain their distinctiveness. In Portugal, the art of cheesemaking traces its origins to the Roman era, with raw milk and its byproducts, including cheese, being integral components of the European diet ever since (Araújo-Rodrigues et al., 2020). In those early periods, cheese production represented a pioneering approach to preserving milk products in a more stable form (Salque et al, 2013). Furthermore, during that era, cheese held such significant value that it was frequently utilized in commercial transactions (Freitas and Malcata, 2000). Today, PDO cheeses are manufactured in small-scale industrial dairies, preserving the rich tradition of their production.
Recognizing the significance of traditional products to their countries of origin, the European Union (EU) introduced geographical indication schemes for agricultural products and foodstuffs, including Protected Designation of Origin (PDO), Protected Geographical Indication (PGI), and Traditional Specialty Guaranteed (TSG) (EU, 2022). These designations serve to aid consumers in identifying traditional products, while also safeguarding and promoting their unique qualities, which are intricately tied to geographical origin and manufacturing expertise (Dias and Mendes, 2018).
According to Regulation (EU) No 1151/2012 (European Commission, 2012) and Commission Implementing Regulation (EU) No 668/2014 (European Comission, 2015), implemented by EU, in PDO products “Every part of the production, processing and preparation process must take place in the specific region”, while in PGI “at least one of the stages of production, processing or preparation takes place in the region” and TSG “highlights the traditional aspects, such as the way the product is made or its composition, without being linked to a specific geographical area” (EU, 2022). In this review manuscript, our focus will be exclusively on Portuguese PDO cheeses and their associated microbiota.
While manufacturing techniques may vary depending on the region, certain practices are consistent across all PDO cheese-making facilities. For example, these cheeses are exclusively made with raw milk, never heated above 40°C, and employ coagulating agents or rennet along with salt, limited to a maximum of 25 g/L. Additionally, no starter or non-starter microbiota is added during the manufacturing process (Freitas and Malcata, 2000). For each PDO cheese, a specification book outlines all mandatory production details, encompassing the type of milk utilized and any treatments applied, specified animal breeds (when applicable), coagulating agents, ripening temperatures, humidity levels, cheese dimensions, and labeling requirements.
In traditional cheese manufacturing, one of the pivotal steps is milk clotting, typically achieved using a coagulating agent such as animal, plant, or microbial rennet (Arbita et al., 2020). The choice of rennet not only impacts milk coagulation, but also influences the development of organoleptic characteristics, primarily attributed to various enzymatic activities (Andrén, 2021). For example, cheeses may acquire a bitter flavor if the rennet used exhibits high non-specific proteolysis activity, a trait often disfavored by consumers (Arbita et al., 2020; Faccia et al., 2020; Andrén, 2021).
Since the beginning of cheese production, calf rennet has been the primary animal coagulating agent utilized, specifically an extract derived from the abomasum of suckling calves. The abomasum of young calves produces caseinolytic enzymes, notably chymosin and pepsin (Arbita et al., 2020; Andrén, 2021). Additionally, chymosin, renowned for its high milk-clotting activity and low proteolysis, is one of the most frequently employed enzymes in cheese production (Mohanty et al., 1999; Andrén, 2021).
In Portuguese traditional cheeses bearing the PDO label, the use of thistle (Cynara cardunculus L.), as a coagulating agent, is prevalent. C. cardunculus L., commonly known as cardoon, is an edible flower native to the Mediterranean region, characterized by its large heads and purple flowers (Gostin and Waisundara, 2019; Folgado et al., 2020). The significance of C. cardunculus L. in cheese manufacturing lies in its enzymes with proteolytic activity, which target milk proteins (Folgado et al., 2020). Among these enzymes, cardosins, particularly cardosin A, play a pivotal role in milk clotting. Cardosin A exhibits proteolytic activity similar to chymosin, specifically targeting κ-casein while also cleaving α and β-caseins, contributing to a softer texture and flavor in the cheese (Folgado et al., 2020; Barracosa et al., 2021).
Overall, the selection of coagulating agents in Portuguese traditional PDO cheeses can vary depending on geographical location and the type of milk utilized. Undoubtedly, these agents play a crucial role in shaping the microbial ecosystem and significantly impact the organoleptic characteristics of the final product. Besides playing an effective role in the coagulation process, coagulating agents also contribute to the products microbiota (Cruciata et al., 2014). Associated enzymes take part in the proteolytic process during cheese manufacture, contributing to organoleptic characteristics through volatile compound formation (Pereira et al., 2008). Moreover, a previous study on cheese produced from animal rennet, characterized the microbial load of the coagulating agent and found several genera, including LAB, Enterococcus and lower counts of E.coli, coliforms and Staphylococcus aureus (Voidarou et al., 2011).
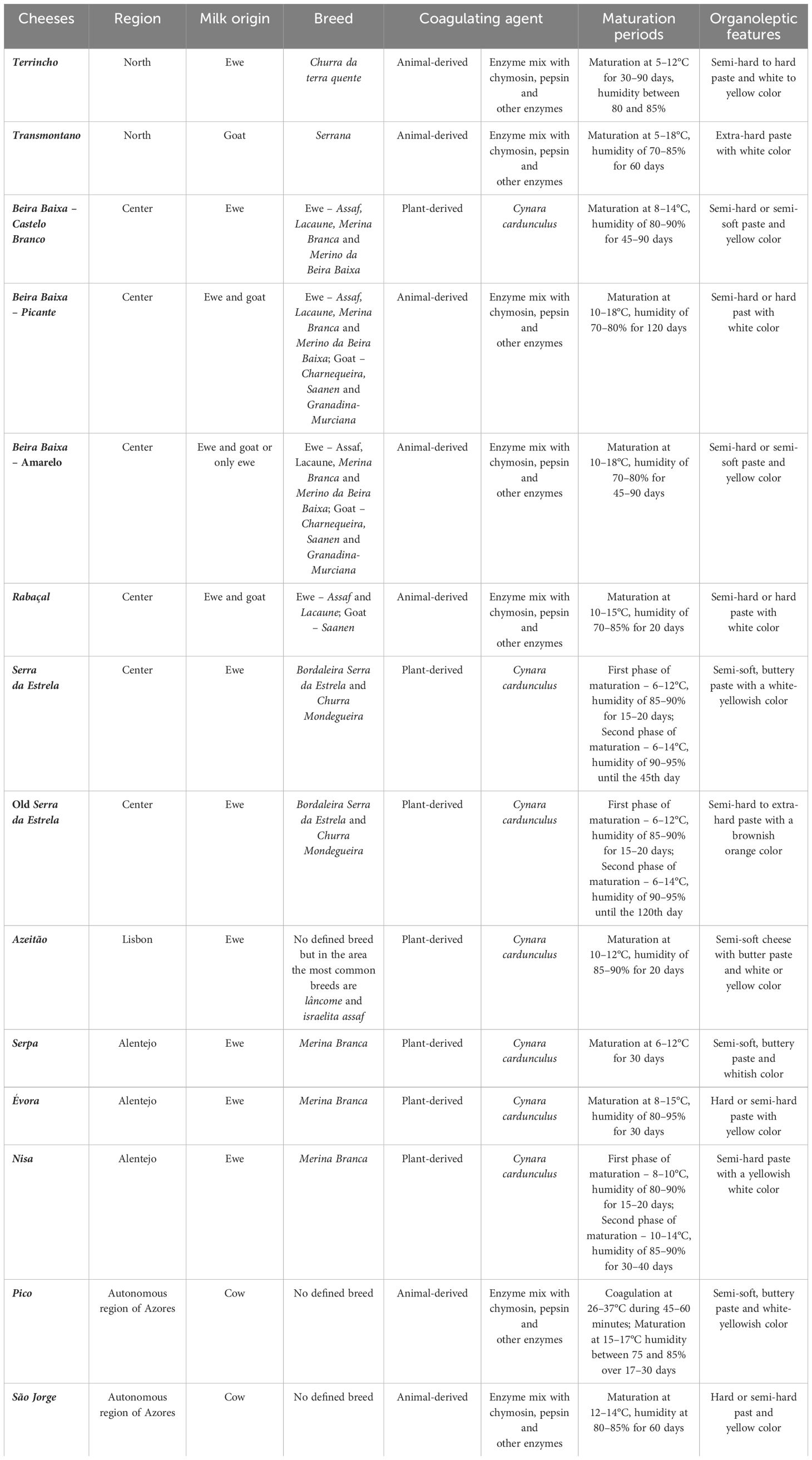
In the rich landscape of Portuguese traditional PDO cheeses, beginning from the northern region of Portugal, two prominent PDO cheeses stand out: Terrincho and Transmontano. Terrincho cheese is manufactured using Churra da Terra Quente ewes milk and animal rennet. This cheese comes in two variants, one aged for a minimum of 30 days and another matured for 90 days (known as old Terrincho cheese), both marketed under the Terrincho PDO designation. The extended maturation period endows old Terrincho with a firmer texture and intensifies its flavor and aroma. In contrast, Transmontano goat cheese, produced from Serrana breed goats milk and animal rennet, features an exceptionally hard paste, along with a robust aroma and spicy flavor profile (Despacho 7822/2011, 2011).
In central Portugal, several PDO cheeses hold prominence: Beira Baixa, Rabaçal, Serra da Estrela, and Azeitão cheeses. The Beira Baixa PDO encompasses three variants: Castelo Branco, Picante from Beira Baixa, and Queijo Amarelo (Direção-Geral de Agricultura e Desenvolvimento Rural, 2022). Castelo Branco cheese is manufactures from ewes milk using Cynara cardunculus L. as a coagulant, resulting in a semi-hard or semi-soft paste with intense flavor and aroma. An aged variant, old Castelo Branco cheese, matures for a minimum of 90 days, yielding a harder paste and a spicy flavor profile (Despacho 9633/2016, 2016). Picante from Beira Baixa, made from a blend of ewe and goat milk with animal rennet, boasts a semi-hard or semi-soft paste with an intense aroma and spicy flavor. Queijo Amarelo, produced from a mixture of ewe and goats milk or solely ewes milk, also employs animal rennet, resulting in a semi-hard to hard paste with intense aroma and a slightly acidic and spicy flavor (Despacho 4185/2011, 2011). Rabaçal cheese, produced from a blend of ewe and goats milk with animal rennet, features a semi-soft or semi-hard paste with a distinctive flavor, imparted by the presence of Santa Maria thyme in the grazing pasture (Despacho 6400/2003, 2003). Moving to Serra da Estrela cheeses, they are made from ewes milk and Cynara cardunculus L., resulting in a semi-soft buttery paste with a mild aroma and slightly acidic flavor. Serra da Estrela cheese can undergo extended ripening, yielding Serra da Estrela old cheese, which presents a semi-hard to extra-hard paste, a robust aroma, and a slightly spicy and salty flavor. Lastly, in the Lisbon region, Azeitão PDO cheese is produced using ewes milk and Cynara cardunculus L., resulting in a cheese with a semi-soft and buttery paste, a yellow hue, and a slightly spicy and acidic flavor (Despacho 6400/2003, 2003).
In the region of Alentejo, three PDO cheeses are produced: Serpa, Évora, and Nisa. These cheeses are all manufactured using ewes milk sourced from the Merina branca breed and employ plant-based rennet Cynara cardunculus L. Serpa cheese boasts a cured, buttery, and semi-soft paste, characterized by a strong odor and a slightly spicy flavor (Despachon.5511/2020, 2020). Évora cheese features a semi-hard to hard paste with a robust aroma and a slightly spicy and acidic flavor profile (Despacho 8601-N/2005, 2005). Lastly, Nisa cheese presents a semi-hard paste with an acidic flavor and a strong odor (Direção-Geral de Agricultura e Desenvolvimento Rural, 2022).
Moving to the island of Azores, two PDO cheeses are manufactured: Pico and São Jorge cheeses. Both cheeses are made from cows milk without a defined breed, using animal rennet (Direção-Geral de Agricultura e Desenvolvimento Rural, 2022). Pico cheese is cured with a soft paste, offering a salty flavor and intense aroma (Despacho32/1996/SRAP, 1996). São Jorge cheese, on the other hand, features a semi-hard or hard paste with a slightly spicy flavor and intense aroma (DespachoSRAP/94/1, 1994).
All the aforementioned characteristics of the PDO cheeses are summarized in Table 1 for easier comprehension and comparison.
2 Microbial impact on cheeses’ organoleptic characteristics
Cheese hosts diverse and intricate microbial communities, which evolve over time and differ based on the cheese type, especially traditional varieties that dont depend on starter cultures for fermentation, as well as regions. Microorganisms in cheese originate not only from milk but also from the production environment. The intricate interplay between microbes and growth substrates, such as milk proteins and fatty acids, significantly impacts the quality and safety of the end product (Afshari et al., 2020).
Due to the diversity of cheese-LAB, these microorganisms harbor both starter and non-starter features. Starter LAB (SLAB) arise in the first stages of manufacture, being crucial participants in the production of lactic acid inducing an acidic pH environment and consequentially curd formation. SLAB can originate from the autochthonous raw milk microbiota, or be added during production, although the latter is not the case in Portuguese PDO cheeses (Coelho et al., 2022). In contrast, non-starter LAB (NSLAB) have a more significant role in the subsequent stages of cheese manufacture (Settanni and Moschetti, 2010), being mostly associated with volatile production, and consequentially odor and flavor development, due to proteolytic activities, as further discussed below. NSLAB can also originate from the raw milk microbiota, or may be introduced from cheesemaking settings, equipment or operators (Irlinger et al., 2017; Coelho et al., 2022).
Considering that raw milk is nutrient rich matrix, the probability of microbial contaminations cannot be overlooked, and may occur by contact with the animals teat surface, milking machinery or collection and storage containers. Due to inadequate hygiene practices, contamination may also occur by staff handling and associated settings, such as bedding, feed or water. Additionally, at cheese production stage, contamination can be associated with manufacturing facilities (storage rooms and shelfs), as well as working staff (Quigley et al., 2013; OSullivan and Cotter, 2017).
In cheese manufacturing, a series of biochemical reactions occur, including glycolysis, proteolysis, and lipolysis, which are influenced by microorganisms, environmental conditions, and the type of coagulant used (Tavaria et al., 2002; Pereira et al., 2008). These reactions shape the microbial composition and organoleptic characteristics of the final cheese, with volatile compounds playing a significant role in flavor development. Lactic acid bacteria (LAB) are key players in these processes. LAB, including genera like Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, or Enterococcus, are non-motile, microaerophilic Gram-positive bacteria capable of tolerating environmental stresses such as high temperature, low pH, or high salt concentrations (Tavaria et al., 2002). Their ability to metabolize various carbon sources and produce antimicrobial compounds makes them indispensable in the food industry.
These microorganisms act as biopreservatives due to their fermentation and acidification capabilities. Milk acidification begins with LAB utilizing lactose as a carbon source, leading to lactate production. This decrease in pH causes the coagulation of casein, the main protein in milk, resulting in the formation of cheese curd, which contains protein, fat, and whey (the liquid portion containing serum proteins, lactose, minerals, and vitamins). Furthermore, the reduction in pH helps prevent the growth of undesirable microorganisms, including foodborne pathogens and spoilage microbes (Favaro et al., 2015). Additionally, the production of organic acids such as lactic, acetic, formic, or propionic acids contributes to extending the shelf life and enhancing the safety of the food product (Favaro et al., 2015). Apart from alterations in pH, another safety strategy employed by these bacteria is the production of bacteriocins. LAB are known to produce various types of bacteriocins such as nisin, reuterin, reutericyclin, pediocin, lacticin, enterocin, among others, as well as bacteriocin-like inhibitor substances (BLIS) (Favaro et al., 2015).
During cheese ripening, LAB also play a central role in the formation of aroma compounds due to their enzymatic activity, which involves the catabolism of aromatic compounds. Additionally, LAB contribute to the breakdown of peptides, which impacts the aroma and texture of the cheese (Cardinali et al., 2022). One group of proteins that is particularly important in this process is the cell-envelope proteinases. These proteins degrade caseins into oligopeptides, which are then transported across the bacterial membrane and further degraded into shorter peptides and amino acids that are essential for LABs survival (Ji et al., 2021). The action of these proteinases generates unique hydrolysates and peptides with distinctive sensory and bioactive properties, which contribute to the overall characteristics of the cheese. Moreover, the degree of proteolysis and the different catalytic properties of these proteases may vary among LAB strains, leading to further diversification of the organoleptic features appreciated by consumers (Ji et al., 2021).
In PDO Portuguese traditional cheeses, the main bacteria found in the final product are Lactococcus spp., Leuconostoc spp., Lactobacillus spp. and, in some cheeses, Enterococcus spp., as it will be discussed in the following sections. All these genera belong to LAB and contribute to cheese maturation in different stages and levels, either by acidification of the milk, proteolytic activity, or production of different bacteriocins active against foodborne pathogens (i.e., nisin produced by Lactococcus lactis against Listeria monocytogenes) (Cotter and Beresford, 2017; Lee et al., 2020; Afrin et al., 2021).
The genus Lactococcus spp. has emerged as the predominant LAB group in all PDO Portuguese traditional cheeses studied to date (Allers et al., 2004; Ayrapetyan et al., 2015a, Ayrapetyan et al., 2015b; Baptista, 2018; Abbasi and Emtiazi, 2020; Câmara et al., 2020; Beltrán-Espinoza et al., 2021). Extensively researched for its influence on cheese manufacture and its applicability across various industries, Lactococcus spp. are starter LAB alongside certain species of Lactobacillus (Lactobacillus delbrueckii and Lactobacillus helveticus). During the initial stages of ripening, these bacteria produce lactic acid, leading to milk acidification (Ruggirello et al., 2014; Cotter and Beresford, 2017). Lactococcus lactis, one of the main species found in Portuguese PDO cheeses, plays a crucial role not only in ensuring the safety of the cheese but also in flavor development. In terms of safety, Lactococcus lactis is known to produce over 40 types of bacteriocins, ribosomally synthesized proteins with antimicrobial activity against other bacteria. These bacteriocins, belonging to class I and II, help control undesirable microorganisms, including potentially pathogenic bacteria (Takala et al., 2023).
Furthermore, apart from their role in acidification, Lactococcus lactis and related species can convert amino acids into aroma compounds through aminotransferase activity. They also contribute to cheese properties by producing exopolysaccharides (Van De Bunt et al., 2014; Cardinali et al., 2022). Given these characteristics, Lactococcus lactis is widely employed as a starter culture, either alone or in combination with other cultures, in the production of various dairy products (OSullivan and Cotter, 2017).
In addition to Lactococcus spp., other LAB genera such as Leuconostoc, Lactobacillus, and Enterococcus are also prominent in Portuguese PDO cheeses (Tavaria and Malcata, 1998; Dahl et al., 2000; Domingos-Lopes et al., 2017; Câmara et al., 2019; Rocha et al., 2021; Rampanti et al., 2023; Rocha et al., 2023). These microorganisms become more noticeable during the later stages of ripening and are often found at the core of the cheeses. They contribute significantly to the flavor, texture, and safety of the final product (Montel et al., 2014). Leuconostoc, much like certain species of Lactobacillus, possesses the capability to metabolize lactose into lactic acid. Thus, alongside Lactococcus, Leuconostoc plays a pivotal role in initiating the maturation process of cheese (OSullivan and Cotter, 2017).
Indeed, Leuconostoc spp. also exhibits several important technological aspects for cheese ripening. For instance, it contributes to the production of aromatic compounds such as diacetyl and acetoin through the degradation of citrate. Additionally, Leuconostoc spp. is also capable of producing gas and dextrans. Dextrans are homopolysaccharides of D-glucose known for their viscosifying, emulsifying, texturizing, and stabilizing attributes in food applications. Therefore, they have the potential to serve as substitutes for commercial hydrocolloids commonly used in the food industry for the same purposes. These glucans are synthesized by extracellular dextransucrase enzymes released by certain LAB, including Leuconostoc species like Leuconostoc mesenteroides, as well as other genera such as Lactobacillus, Streptococcus, Weissella, and Pediococcus (Morelli and von Wright, 2019).
Regarding Lactobacillus spp., this genus is highly diverse within LAB and is commonly found in dairy products, including L. delbrueckii subsp. bulgaricus, L. helveticus, and Lacticaseibacillus casei. Additionally, various Lactobacillus species are also found in the human gastrointestinal tract, such as L. acidophilus, L. gasseri, and Lacticaseibacillus rhamnosus (Morelli and von Wright, 2019).
As mentioned earlier, some species of Lactobacillus have starter activity. However, lactobacilli also play a crucial role in non-starter activities, significantly impacting flavor development and ensuring the quality and durability of cheeses. Lactobacillus (and related genera, according to the new taxonomy; Zheng et al., 2020) is one of the major contributors of non-starter LAB during cheese ripening (Morelli and von Wright, 2019). Some of the species commonly found at this stage of ripening include L. casei, Lacticaseibacillus paracasei, and Lacticaseibacillus rhamnosus. L. casei is a ubiquitous microorganism found in various niches, exhibiting great genetic versatility. In a study by Cai et al. (2009), it was observed that cheese-isolated L. casei displayed a significant number of genes related to carbohydrate metabolism, transcriptional regulation, and signal transduction compared to L. casei from other environments (Cai et al., 2009).
Moreover, besides its proteolytic activity and role in flavor development in cheeses, L. casei can also act as a protective microorganism by detoxifying biogenic amines (BA). These toxic compounds are formed through microbial degradation of amino acids and, if present in food products, may cause symptoms of intoxication such as headaches, itchy skin rashes, heart palpitations, or diarrhea (Linares et al., 2011). Cheese is a fermented food in which BA may be present, with tyramine, histamine and putrescine being the most commonly found (Linares et al., 2011; Herrero-Fresno et al., 2012; Renes et al., 2014). In a genomic study conducted with L. casei, genes for methyltransferases and oxidoreductases related to BA degradation were identified (Ladero et al., 2014). These adaptabilities of L. casei make it an appealing candidate for potential applications in food industry environments and products, as well as a possible source of probiotics (Morelli and von Wright, 2019).
As for Enterococcus spp., these bacteria are constituents of the native microbiota of several traditional products from the Mediterranean area and are used as sanitary indicators (Freitas and Malcata, 2000). However, due to safety concerns regarding specific species or strains with virulence factors that could pose a risk to consumers, the European Food Safety Authority (EFSA) has created a list of microorganisms with Qualified Presumption of Safety (QPS) for use in the food industry. For the Enterococcus genus specifically, since it is not included in this directory, safety assessments are performed on a case-by-case basis, including screenings for species or strain virulence factors (Câmara et al., 2020). Nevertheless, in Portuguese traditional cheeses, Enterococcus spp. are typically present in the raw milk microbiota used for cheese manufacture, resulting in their presence in the cheese itself. Their proteolytic and lipolytic activities have a significant impact on the development of the cheeses flavor characteristics, making this genus essential for cheese ripening (Dias et al., 2021). To date, no outbreaks related to the consumption of any Portuguese PDO cheese containing these microorganisms have been reported, further highlighting their importance in traditional Portuguese PDO cheese production (Rocha et al., 2023).
Regarding the fungal community in Portuguese PDO cheeses, the main genera encountered include Candida spp., Debaryomyces spp., Yarrowia spp., and Kluyveromyces spp. These microorganisms are typically found in the cheese rind, but they can also be present in the cheese matrix (OSullivan and Cotter, 2017). In the case of Candida and Debaryomyces, some studies suggest that these yeasts may enter the cheese through the salt used in traditional cheese manufacture (Cotter and Beresford, 2017). Yeasts influence all stages of cheese ripening, as they have the ability to ferment lactose, contribute to milk acidification, and perform lipolytic and proteolytic activities for texture and flavor development, as well as produce aromatic compounds for scent and flavor (Gonçalves Dos Santos et al., 2017). The influence of molds on cheese properties is less understood. While various molds are found in some types of surface-ripened cheeses like Brie or Camembert (Irlinger et al., 2017), in Portuguese cheeses, the presence of molds is mainly due to contamination.
Overall, apart from the diversity of the microbiota present in the milk, variations in manufacturing practices (e.g., ripening duration, temperature, and humidity conditions) also induce differences in the final product properties (Araújo-Rodrigues et al., 2022). Moreover, the dominance of LAB in the cheese microbiota plays a pivotal role in the organoleptic characteristics of cheeses regardless of the region or milk used for their production.
3 Integrating conventional microbiology and OMICs
To fully capture the intricate microbial ecosystem of traditional cheeses, it is crucial to employ both conventional microbiology and OMIC technologies. These complementary approaches provide a comprehensive understanding of the diverse microbial communities present in these cheeses.
3.1 Conventional microbiology
Conventional microbiological procedures encompass culture-dependent methods, which involve the growth and isolation of microbial populations utilizing selective media. Briefly, after collection the samples are processed by homogenization using an isotonic buffer on a stomacher blender, serial dilutions are prepared and plated in appropriate media. Media selection varies according to the targeted microbial groups, the most commonly used being Man, Rogosa and Sharpe (MRS) agar for total LAB, M17 for lactococci, Slanetz Bartley Agar (SBA) for enterococci, Mayeux, Sandine and Elliker Agar (MSE) for Leuconostoc spp., Rogosa Agar for lactobacilli, potato dextrose agar or Rose Bengal Chloramphenicol (RBC) for yeast and molds, Baird Parker agar for Staphylococcus spp. and Violet Red Bile Glucose Agar (VRBGA) for Enterobacteriaceae (Pinho et al., 2004; Câmara et al., 2017; Gonçalves et al., 2018; Dias, 2021; Rocha et al., 2023; Salamandane et al., 2024).
Subsequent identification relies on phenotypic characteristics, such as morphology and biochemical traits, as well as genotypic techniques such as species-specific PCR (Anastasiou et al., 2022) (Anastasiou et al., 2022). Over the years, numerous studies have been conducted to investigate the microbiota of PDO cheeses, as microbial communities undergo changes during various stages of ripening. Previous research has identified several groups of microorganisms, including bacteria, filamentous fungi (molds), and yeasts. Among these, LAB were found to be the most prevalent group of microorganisms (Freitas and Malcata, 2000), namely in Terrincho (Pinho et al., 2004), Picante (Freitas et al., 1996; Freitas and Malcata, 2000), Serra da Estrela (Rocha et al., 2023), Azeitão, Serpa (Gonçalves et al., 2018; Araújo-Rodrigues et al., 2020), Évora, Pico and São Jorge cheeses (Kongo et al., 2008).
Terrincho cheese has not been extensively studied however, some authors have shown that LAB are the most representative group of microorganisms, based on counts of colony forming units (CFUs), it was verified that Lactobacillus spp. and Lactococcus spp. showed levels of 109 CFU/g followed by Enterococcus spp. with numbers ranging from 107 to 108 CFU/g (Pinho et al., 2004; Pintado et al., 2008). In the study performed by Pinho et al (2004), Terrincho cheese microbiota was studied during ripening time, and the presence of yeasts and molds, Pseudomonas spp., coliforms and Staphylococcus spp. was assessed, with the latest ranging up to 104 CFU/g at the end of the ripening process. Coliform abundance was also studied, with numbers reaching 106 CFU/g after 60 days of ripening (Pinho et al., 2004). Pintado et al (2008) also assessed Terrincho cheese microbiota, and, once again, LAB were the predominant group. Lactococcus spp. and Lactobacillus spp., presented counts of around 109 CFU/g, while Enterococcus counts were lower, with numbers of ca 107 CFU/g. The presence of yeast and molds (~104–106 CFU/g), Pseudomonas spp., and Staphylococcus spp., were equally studied, and the numbers of the latest were concordant with the previous study, ranging from 104 to 105 CFU/g. Enterobacteriaceae were also in the order of 106 CFU/g (Pintado et al., 2008).
In the case of Beira Baixa cheese, which encompasses Amarelo, Picante, and Castelo Branco PDO cheese types, there have been only a few studies on their microbiota to date, to the best of our knowledge. Cardinali et al (2022) investigated the microbiota of Castelo Branco cheese and identified Lactococcus spp. and Lactobacillus spp. as the predominant microorganisms with CFUs counts ranging from 106–109 to 108–109, respectively (Cardinali et al., 2022). In the same study, Enterococcus spp. presence was also determined (104–106 CFU/g), as well as Enterobacteriaceae (10–104 CFU/g) yeasts and molds (10–104 CFU/g). In the study by Freitas et al. (1996), Picante cheese was examined, and once again, LAB was identified as the predominant group. Additionally, the presence of Enterobacteriaceae, Staphylococcus spp., yeasts, and molds was evaluated, with no molds found in the studied cheeses (Freitas et al., 1996).
Due to the limited availability of published papers on some traditional Portuguese cheeses, masters theses have been utilized over the years to characterize their indigenous microbiota. For Amarelo cheese, a masters dissertation conducted by Rodrigues (2023) reported similar results to those found in other Beira Baixa cheeses, with a predominance of LAB (ca 108 CFU/g). In terms of different genera, as this was a preliminary study with some limitations, the authors did not provide detailed information on that matter. Yeasts and molds were also detected, albeit in smaller quantities (ranging from 10 to 104 CFU/g) (Rodrigues, 2023). Potentially pathogenic microorganisms were also investigated: Salmonella spp. or Listeria monocytogenes were not found, but Pseudomonas spp., on the other hand, were present at levels ranging from 105 to 106 CFU/g (Rodrigues, 2023).
Rabaçal cheese is one of the lesser-studied PDO cheeses, and similarly to Amarelo from Beira Baixa, our review was based on masters dissertations. The study conducted by Dias et al. (2021) assessed the microbiota of Portuguese PDO cheeses, including the examination of Rabaçal microbiota in both the cheeses crust/rind and interior. The results align with studies conducted on other cheeses, with LAB being the predominant group of bacteria, with counts of approximately 108 CFU/g. Enterobacteriaceae, coliforms, and yeasts were also isolated in both the cheeses rind and interior, with counts of around 104 CFU/g. Staphylococcus spp. and molds were also detected, although in lesser quantities (ranging from 10 to 102 CFU/g) (Dias et al., 2021).
Serra da Estrela cheese is often regarded as a hallmark of Portuguese traditional PDO cheeses and has been the subject of numerous studies, employing both conventional microbiology and metagenomic approaches. Tavaria and Malcata have contributed with various articles on Serra da Estrela cheese. In a study conducted in 1998, the cheeses microbiota was analyzed, revealing that LAB counts increased throughout ripening and remained predominant until the end of the ripening period (60 days), representing 55.1% of the total microbiota (Tavaria and Malcata, 1998). Enterobacteriaceae and yeasts were the second most abundant groups, each representing 20.4% of the cheeses microbiota, with Staphylococcus spp. also being present, even though in lower percentages (4.1%) (Tavaria and Malcata, 1998). Another study by Tavaria and Malcata (2000) focused on evaluating the microbiota of Serra da Estrela cheeses manufactured in different years and geographic areas. LAB were identified as the main group of bacteria, with other less abundant groups such as yeasts and Staphylococcus spp. also isolated. Enterobacteriaceae were present, and their numbers were observed to decrease throughout ripening. This study also highlighted the influence of geographical location on the cheese microbiota (Freitas and Malcata, 2000). In another study by Dahl et al. (2000), the dominance of LAB throughout ripening was once again confirmed, even in longer ripening periods (duration assessed ranged from 60 to 180 days). Conversely, Enterobacteriaceae and yeast species showed a significant decrease throughout ripening (Dahl et al., 2000). More recent studies on Serra da Estrela cheese attest the prevalence of LAB over other groups of microorganisms with yeasts and molds being the less represented groups (Rampanti et al., 2023; Rocha et al., 2023). A 2024 study performed by Salamandane et al (2024), including PDO and non-PDO Serra da Estrela cheeses, showed a richer microbiota in PDO cheeses, with count numbers ranging from 106 to 109 CFU/g for Lactococcus and Enterococcus, ca 109 for Lactobacillus. E. coli and Staphylococcus were also found in counts around 103–105 and 104–105 respectively. Listeria monocytogenes and Salmonella were not found in any of the samples analyzed. These results highlight the diversity of microorganisms in fermented foods, particularly those with no selection of initial microbiota, as Portuguese PDO cheeses (Salamandane et al., 2024).
Studies on Azeitão and Nisa cheeses microbiota have also been conducted, primarily through masters dissertations. Baptista (2018) characterized the microbiota of these cheeses by analyzing cheeses manufactured in different dairies and different years. They found LAB counts ranging from 107 to 1010 CFU/g for Azeitão cheese and from 106 to 108 CFU/g for Nisa cheese. Specific counts of Lactococcus and Enterococcus were also assessed using selective media. In Azeitão cheeses, Lactococcus numbers ranged from approximately 107 to 1011 CFU/g, and Enterococcus counts ranged from 104 to 107 CFU/g. Nisa cheeses had Lactococcus counts of about 106–1012 CFU/g and Enterococcus counts of 104–105 CFU/g (Baptista, 2018). Another study conducted by MaChado (2020) assessed the presence of coagulase-positive Staphylococcus and Escherichia coli in Azeitão cheese. They found that unlike Staphylococcus counts, which decreased throughout the ripening period, E. coli counts increased (MaChado, 2020).
Regarding Évora cheese, a study by Dias et al. (2021) evaluated the cheese microbiota throughout ripening. Similarly to other cheeses, the majority of the microorganisms belong to the LAB group. At the end of ripening time (25 days), LAB counts were in the order of 107–108 CFU/g, while Enterococcus spp. showed levels of around 105 CFU/g and Enterobacteriaceae and yeasts were found in the same approximate range of counts 104–106 CFU/g (Dias et al., 2021).
Serpa cheese has undergone some studies regarding its microbial content (Roseiro et al., 2003; Gonçalves Dos Santos et al., 2017; Gonçalves et al., 2018). A study performed by Roseiro et al. (2003) identified LAB as the predominant group throughout ripening, with count numbers reaching 109 CFU/g. Molds, Listeria monocytogenes and coagulase positive Staphylococcus were not encountered, contrarily to E. coli and coliforms, that were found in numbers of around 105–106 CFU/g, having increased during ripening. The presence of yeasts was also acknowledged, reaching 103 CFU/g (Roseiro et al., 2003). Posterior studies, assessed the microbiological community, namely yeasts and bacteria, in different manufactures and seasons (winter and spring). Yeasts counts reached 106 CFU/g and were found to be higher in winter compared to spring (Gonçalves Dos Santos et al., 2017; Gonçalves et al., 2018). Regarding bacterial communities, concordantly to the study by Roseiro et al. (2003), LAB were the major constituents, with the following genus represented: Lactobacillus spp., Lactococcus spp., Leuconostoc spp. and Enterococcus spp. and found in numbers up to 1010 CFU/g. Enterobacteriaceae were shown to be in high numbers in winter (105–107CFU/g), but rather low in spring (10–104). E. coli was also present, as well as Staphylococcus spp. but in lower numbers (Staphylococcus spp. present only in spring) (Gonçalves Dos Santos et al., 2017; Gonçalves et al., 2018).
Moving to the Azores, Domingos-Lopes et al. (2017) performed the phenotypic identification of bacteria present in Pico cheese to the genus and species level: 56.1% of the total isolates were Lactobacillus spp., while 30.7% were identified as Enterococcus spp., 4.4% as Lactococcus spp., 3.5% as Leuconostoc spp. and 0.9% as Streptococcus spp (Domingos-Lopes et al., 2017). On the other hand, on a study performed by Câmara et al. (2017), Lactococcus spp. were the most abundant group at the end of the cheese ripening time (21 days), followed by Leuconostoc spp., Lactobacillus spp. and Enterococcus spp. Staphylococcus spp., and yeasts were also found, but no molds were detected (Câmara et al., 2017).
Kongo and colleagues performed some studies with São Jorge cheese throughout the years (Kongo et al., 2007, Kongo et al, 2008, Kongo et al, 2009). The main finding of these studies was that Lactobacillus is the predominant genera. The study performed in 2007, reveals that besides Lactobacillus, in the end of the cheese ripening time, Enterococcus spp. was the second most abundant group of microorganisms, followed by Pediococcus and Leuconostoc spp. Lactococcus species were also found in São Jorge cheese, but only in early stages of ripening (Kongo et al., 2007). A subsequent study in 2008 assessed the hygienic safety of this cheese by evaluating the presence of Enterobacteriaceae and Micrococcaceae. Some Klebsiella species were found, as well as E. coli and Staphyloccocus, while no Salmonella species were detected (Kongo et al., 2008). In another study performed in 2009, the second most prevalent group of microorganisms, after Lactobacillus spp. (107–108 CFU/g) found in this cheese were Lactococcus spp., with viable counts around 106–107 CFU/g, followed by Enterococcus spp. with 105–106 CFU/g. Yeasts and molds were also present in levels around 104 and 105 CFU/g, respectively. Moreover, the presence of Enterobacteriaceae was also assessed and observed in small quantities, around 102 CFU/g (Kongo et al., 2009).
In conclusion, lactic acid bacteria are undoubtedly the predominant group in all cheese types, although the most abundant genera may vary from cheese to cheese. Significant variations in cheese microbiota among dairies and seasons are also evident. All of the aforementioned information is summarized in Table 2.
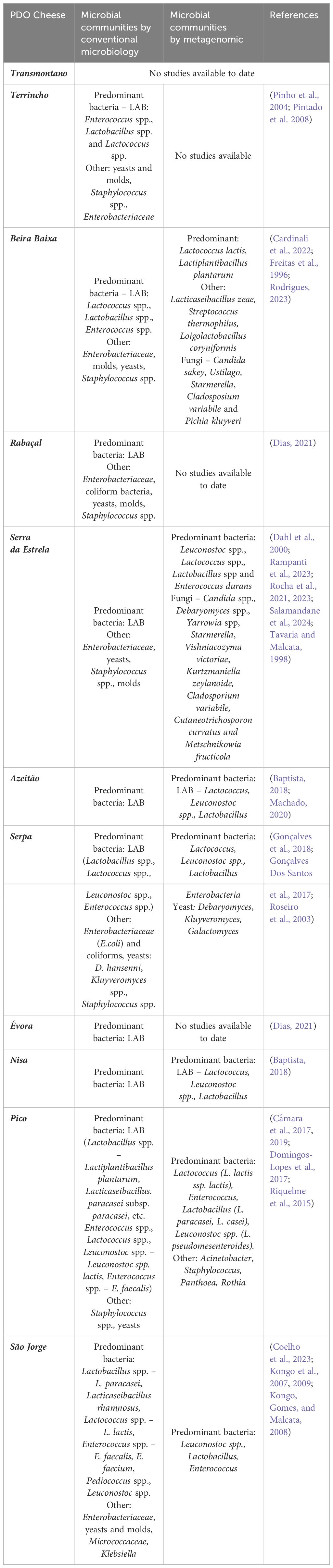
Table 2 Microbial communities of Portuguese PDO cheeses assessed by conventional microbiology and metagenomic approach.
3.2 OMIC technologies
While conventional microbiology remains fundamental and should be conducted concurrently, it presents several drawbacks, primarily due to its time-consuming nature, challenges in selecting suitable growth media and conditions, and the potential oversight of less abundant microorganisms that may be overshadowed by predominant ones. Additionally, the presence of viable but non-culturable (VBNC) microorganisms poses a significant challenge when employing culture-dependent techniques (Anastasiou et al., 2022; Araújo-Rodrigues et al., 2022). In contemporary times, the emergence of OMIC technologies has revolutionized the fields of food science and microbial ecology (Afshari et al., 2020). This new generation of culture independent techniques, like metagenomics, proteomics, metabolomics or transcriptomics, used individually or in integrative analysis, will undoubtedly shed light on the complex microbial ecology of traditional PDO cheeses.
Several reviews have described the importance and interest of studying the cheese microbiota, aiming to correlate microbial interactions with the quality and flavor of the final product (Sattin et al., 2016; Papademas et al., 2019; Afshari et al., 2020; Jiang et al., 2023). However, only a limited number of OMIC studies has been conducted on Portuguese traditional PDO cheeses, including lipidomics, volatilomics (Reis Lima et al., 2020; Inácio et al., 2023), and metagenomics (Riquelme et al., 2015; Rocha et al., 2021; Araújo-Rodrigues et al., 2022; Coelho et al., 2023).
Regarding the metagenomic analysis, to our knowledge, the following Portuguese PDO cheese have been studied using targeted metagenomics, directed to the 16S rDNA and/or 26S rDNA amplicons: Beira Baixa, Serra da Estrela, Azeitão, Serpa, Nisa, Pico and São Jorge cheeses. Moreover, a recently published report featuring Serra da Estrela cheese applied shotgun metagenomics (Salamandane et al., 2024).
In Beira Baixa cheese, specifically Castelo Branco, a survey of the microbial community was performed by Cardinali et al. (2022). The authors sequenced the V3–V4 regions of the 16S rDNA for bacterial and the 26S rDNA for fugal analysis, respectively (Cardinali et al., 2022). In brief, the authors tested three producers and found no significant differences the in terms of microbial content being the most prevalent species Lactococcus lactis and Lactiplantibacillus plantarum. In lower prevalence there was also Lacticaseibacillus zeae, Streptococcus thermophilus and Loigolactobacillus coryniformis (Cardinali et al., 2022). As for the fungal community, there were some differences detected within the different producers where Candida sake and Ustilago were the most prevalent in producer 1, Starmerella and Cladosposium variabile in producer 2 and C. variabile and Pichia kluyveri in producer 3 (Cardinali et al., 2022).
For Serra da Estrela PDO cheese, Rocha et al (2021), conducted a similar study of the cheeses microbial communities. The sequencing strategy was similar to the one applied to Beira Baixa cheese, so for the 16S rDNA the regions V3–V4 were selected and for the fungi community the Internal Transcribed Spacer 2 (ITS-2). In summary, from the 500 taxa identified, the authors appointed 30 as core taxa, present in all samples tested, including genus like Leuconostoc spp. and Lactococcus spp. for bacteria, and Candida spp., Debaryomyces spp. and Yarrowia spp. for fungi (Rocha et al., 2021). Another study by Rampanti et al. (2023) has also resorted to 16S rDNA sequencing of the region V3–V4 to analyze the microbial content of four producers of PDO Serra da Estrela cheese. Their metataxonomic analysis showed, once again, Lactococcus lactis as one of the most prevalent species, being detected in all cheese samples analyzed. Moreover, some species of Leuconcostoc were also identified (i.e. Leuconostoc mesenteroides and Leuconostoc sakei) as well as Enterococcus spp (Rampanti et al., 2023). The authors also conducted a survey on the cheeses mycobiota, verifying that this community of microorganisms could be dependent on each producer. They observed a wide variation in the fungal content of cheeses from different producers, likely due to different manufacturing techniques. Nevertheless, the authors performed detection of major and minor taxa. Among the major taxa, Debaryomyces hansenii, Starmerella, Vishniacozyma victoriae and Kurtzmaniella zeylanoides were detected. As for minor taxa it was detected Cladosporium variabile, Cutaneotrichosporon curvatus and Metschnikowia fructicola (Rampanti et al., 2023).
Still in Serra da Estrela cheese, a shotgun metagenomic approach was used to evaluate the microbiome, resistome and virulome, of both PDO and non-PDO cheeses (Salamandane et al., 2024). Briefly, the authors explored four different producers, two PDO producers (QG1/QG2 and QI1/QI2) and two non-PDO producers (QL1/QL2 and QT1/QT2). Regarding the cheeses microbiota, the authors observed a clear predominance of Leuconostoc mesenteroides throughout the different samples. As for the PDO specific microbiota, samples from both producers harbored Enterococcus durans, Kocuria salsicia, Glutamicibacter ardleyensis, Lactococcus lactis, Raoultella ornithinolytica, Lactobacillus coryniformis and Lactiplantibacillus plantarum. Contrarily, non-PDO cheeses showed higher diversity of associated species, even within samples from the same producer. Briefly, in QL1 and QL2 there was a predominance of Enterococcus durans while in QL2 the predominant species was Lactobacillus paraplantarum. Moreover, increased disparities were found in QT producers, with three predominant species, namely Lactococcus lactis, Lacticaseibacillus rhamnosus and Leuonostoc. Mesenteroides being associated with QT1, while QT2 samples showed a set of five predominant species: Lacticaseibacillus paracasei, Lacticaseibacillus rhamnosus, Lactiplantibacillus plantarum, Leuconostoc mesenteroides and Lactococcus lactis (Salamandane et al., 2024). Overall, this study further highlights the importance of using metagenomic approaches for the in depth clarification of the complex microbial ecosystem associated with traditional cheeses, either PDO or non-PDO.
In the case of Azeitão and Nisa cheeses, owing to the scarcity of metagenomic studies on the microbiotas communities, we turned to a masters dissertation where a metagenomic study was conducted to characterize the bacterial community (Baptista, 2018).
In brief, the authors studied PDO cheeses from 2016 to 2018 produced in these two regions, by sequencing the regions V1–V3 of the 16S rDNA for bacterial identification. In the analyzed cheese samples, Baptista (2018) identified over 22 different genera, mostly belonging to the LAB group. Throughout the different producers in both regions, the most prevalent genus was Lactococcus followed by Leuconostoc spp. and Lactobacillus. Moreover, concerning the microbial diversity of each producer in different years, some differences were observed in the relative abundance of the most prevalent genera. Additionally, in the less abundant genera, there was higher diversity, with some genera not being present in different years (Baptista, 2018).
PDO cheeses produced in Serpa have also been studied. In a study by Gonçalves Dos Santos et al. (2017), an analysis of the yeast community of Serpa PDO and non-PDO cheeses was conducted. In brief, sequencing of the 26S rDNA was performed for the ITS region and D1/D2 to enable identification at the species level. The authors assessed the diversity of the fungal community during different seasons (winter and spring) and verified that the most common fungal genera were Debaryomyces and Kluyveromyces. Moreover, the genus Galactomyces was also found, however its abundance varied within the different cheese factories (Gonçalves Dos Santos et al., 2017). Another study, also from the same research group, focused on the 16S rRNA sequencing of the V3–V4 regions, verifying a prevalence of the Lactococcus followed by Leuconostoc spp., Lactobacillus genus and Enterobacteriaceae family (Gonçalves et al., 2018).
To our knowledge, only one study has focused on the analysis of microbial communities in Pico cheese using metagenomics, specifically targeting the 16S rRNA sequencing of the V3–V4 regions. Riquelme et al. (2015), verified that the most prevalent microorganisms, (included in the dominant category defined by the authors), as core bacteria of Pico cheese were: Lactococcus, Streptococcus, Acinetobacter, Enterococcus, Lactobacillus, Leuconostoc spp., Staphylococcus, Panthoea and Rothia (Riquelme et al., 2015). Another study, also from the same research group used 16S rRNA gene sequencing for identification of autochthonous LAB from the Pico cheese, aiming to assess their technological potential. The most interesting species in terms of technological potential were Lactococcus lactis ssp. lactis, Lacticaseibacillus paracasei, Leuconostoc pseudomesenteroides and L. casei (Câmara et al., 2019).
Regarding the São Jorge PDO cheese a metagenomic study was performed by Coelho et al. (2023) focusing on next-generation sequencing (NGS) as a tool to distinguish PDO from non-PDO cheeses. The authors observed that certified cheeses were richer in Leuconostoc, Lactobacillus, and Enterococcus, whereas in the non-certified cheeses, there was a prevalence of Streptococcus followed by Lactococcus (Coelho et al., 2023).
Regarding the PDO cheeses Rabaçal, Terrincho, Évora, and Transmontano, to our knowledge, there is still no metagenomic data available.
Overall, the most commonly used strategy for metagenomic studies involves sequencing the 16S rDNA of the V3–V4 regions for bacterial characterization and the 26S rDNA of the ITS or D1/D2 regions for fungal characterization. From the information gathered in this section and Section 3.1, it is evident that an OMIC approach, combined with culture-dependent techniques, provides several beneficial aspects for studying PDO cheese microbiota. This approach not only helps assess the quality of the cheeses but also aids in distinguishing certified cheeses. However, to our knowledge, research in this field, particularly regarding Portuguese cheeses, is still limited. All of the above information is summarized in Table 2.
4 Conclusions
In conclusion, our review underscores the pivotal role of microbial communities in shaping the distinct characteristics of Portuguese traditional cheeses, including their flavor, texture, and safety profiles. By synthesizing findings from conventional microbiological studies and cutting-edge OMICs analyses, we have gained valuable insights into the complex dynamics of cheese microbiota. Our exploration of microbial composition, diversity, and functional roles across various PDO cheeses has revealed the significant contributions of the microbiota to cheese ripening, flavor development, and safety assurance. Moreover, our discussion highlights the potential of OMICs technologies, particularly metagenomics, in elucidating the intricate microbial ecosystems of these cheeses.
Through this integrative approach, we have attempted to unveil the secrets behind the rich heritage and distinctiveness of Portuguese traditional cheeses. By furthering our understanding of the interplay between microorganisms and cheese matrices, we pave the way for continued advancements in cheese production, quality assurance, and preservation of cultural heritage.
Author contributions
TS-L: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. SS: Writing – original draft, Writing – review & editing. SM: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by FCT – Fundação para a Ciência e Tecnologia IP Portugal, through projects PTDC/OCE-ETA/1785/2020 (EMOTION), UIDB/00276/2020 (CIISA – Centre for Interdisciplinary Research in Animal Health, Faculty of Veterinary Medicine, University of Lisbon) and LA/P/0059/2020-AL4ANIMALS (AL4AnimalS). SS holds a PhD fellowship supported by national funds through FCT – UI/BD/153073/2022 and SM a research fellowship under project EMOTION.
Acknowledgments
The authors wish to thank all the authors of manuscripts and other reports on traditional cheeses for sharing their knowledge with the scientific community.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbasi S., Emtiazi G. (2020). MALDI-TOF analysis of a novel extremophile peptide purified from Halarchaeum acidiphilum ASDL78 with antiarchaeal and antibacterial activities. J. Basic Microbiol. 60, 920–930. doi: 10.1002/jobm.202000392
Afrin S., Hoque M. A., Sarker A. K., Satter M. A., Bhuiyan M. N. I. (2021). Characterization and profiling of bacteriocin-like substances produced by lactic acid bacteria from cheese samples. Access Microbiol. 3, 1–9. doi: 10.1099/acmi.0.000234
Afshari R., Pillidge C. J., Dias D. A., Osborn A. M., Gill H. (2020). Cheesomics: the future pathway to understanding cheese flavour and quality. Crit. Rev. Food Sci. Nutr. 60, 33–47. doi: 10.1080/10408398.2018.1512471
Allers T., Ngo H. P., Mevarech M., Lloyd R. G. (2004). Development of Additional Selectable Markers for the Halophilic Archaeon Haloferax volcanii Based on the leuB and trpA Genes. Appl. Environ. Microbiol. 70, 943–953. doi: 10.1128/AEM.70.2.943-953.2004
Anastasiou R., Kazou M., Georgalaki M., Aktypis A., Zoumpopoulou G., Tsakalidou E. (2022). Omics approaches to assess flavor development in cheese. Foods 11, 188. doi: 10.3390/foods11020188
Andrén A. (2021). “Milk-clotting enzymes,” in Agents of change - Enzymes in Milk and Dairy producers (Cham: Springer), 349–362. doi: 10.1007/978–3-030–55482-8_14
Araújo-Rodrigues H., Tavaria F. K., dos Santos M. T. P., Alvarenga N., Pintado M. M. (2020). A review on microbiological and technological aspects of Serpa PDO cheese: An ovine raw milk cheese. Int. Dairy J. 100, 104561. doi: 10.1016/j.idairyj.2019.104561
Araújo-Rodrigues H., Martins A. P., Tavaria F. K., Santos M. T. G., Carvalho M. J., Dias J., et al. (2022). Organoleptic chemical markers of serpa PDO cheese specificity. Foods 11, 1–25. doi: 10.3390/foods11131898
Arbita A. A., Williams T. C., Baxter R., Oliver J. D. (2020). Extraction, partial purification and characterization of proteases from the red seaweed Gracilaria edulis with similar cleavage sites on κ-casein as calf rennet. Food Chem. 330, 127324. doi: 10.1016/j.foodchem.2020.127324
Ayrapetyan M., Williams T. C., Baxter R., Oliver J. D. (2015a). Viable but nonculturable and persister cells coexist stochastically and are induced by human serum. Infection Immun. 83, 4194–4203. doi: 10.1128/IAI.00404-15
Ayrapetyan M., Williams T. C., Oliver J. D. (2015b). Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 23, 7–13. doi: 10.1016/j.tim.2014.09.004
Baptista M. (2018). Caracterização do microbioma de queijos tradicionais Portugueses com DOP. Faculdade de Ciências (Universidade de Lisboa). http://hdl.handle.net/10451/36484.
Barracosa P., Simões I., Martins A. P., Barros M., Pires E. (2021). Biochemical diversity of cardoon flowers (Cynara cardunculus L.): Predicting PDO Mediterranean cheese textures. Food Bioscience 39, 100805. doi: 10.1016/j.fbio.2020.100805
Beltrán-Espinoza J. A., Domínguez-Lujan B., Gutiérrez-Méndez N., Chávez-Garay D. R., Nájera-Domínguez C., Leal-Ramos M.Y. (2021). The impact of chymosin and plant-derived proteases on the acid-induced gelation of milk. Int. J. Dairy Technol. 74, 297–306. doi: 10.1111/1471–0307.12760
Cai H., Thompson R., Budinich M. F., Broadbent J. R., Steele J. L. (2009). Genome Sequence and Comparative Genome Analysis of Lactobacillus casei: Insights into Their Niche-Associated Evolution. Genome Biol. Evol. 1, 239–257. doi: 10.1093/gbe/evp019
Câmara S. P. A., Dapkevicius A., Rosa H. J., Silva C. C., Malcata F. X., Enes Dapkevicius M. L. (2017). Physicochemical, biochemical, microbiological and safety aspects of Pico cheese: Assessment throughout maturation and on the final product. Int. J. Dairy Technol. 70, 542–555. doi: 10.1111/1471-0307.12424
Câmara S. P., Dapkevicius A., Riquelme C., Elias R. B., Silva C. C. G., Malcata F. X., et al. (2019). Potential of lactic acid bacteria from Pico cheese for starter culture development. Food Sci. Technol. Int. 25, 303–317. doi: 10.1177/1082013218823129
Câmara S. P. A., Dapkevicius A., Silva C. C. G., Malcata F. X., Enes Dapkevicius L. N. M. (2020). Artisanal Pico cheese as reservoir of Enterococcus species possessing virulence and antibiotic resistance properties: implications for food safety. Food Biotechnol. 34, 25–41. doi: 10.1080/08905436.2019.1710844
Cardinali F., Foligni R., Ferrocino I., Harasym J., Orkusz A., Franciosa I., et al. (2022). Microbial diversity, morpho-textural characterization, and volatilome profile of the Portuguese thistle-curdled cheese Queijo da Beira Baixa PDO. Food Res. Int. 157, 111481. doi: 10.1016/j.foodres.2022.111481
Coelho M. C., Malcata F. X., Silva C. C. G. (2022). Lactic acid bacteria in raw-milk cheeses: from starter cultures to probiotic functions. Foods 11, 2276. doi: 10.3390/foods11152276
Coelho M. C., Malcata F. X., Silva C. C. G. (2023). Distinct bacterial communities in São Jorge cheese with Protected Designation of Origin (PDO). Foods 12, 990. doi: 10.3390/foods12050990
Cotter P. D., Beresford T. P. (2017). “Microbiome changes during ripening,” in Cheese: chemistry, physics and microbiology: fourth edition, 4th ed. (Elsevier Ltd. Academic press). doi: 10.1016/B978-0-12-417012-4.00015-6
Cruciata M., Sannino C., Ercolini D., Scatassa M. L., De Filippis F., Mancuso I., et al. (2014). Animal rennets as sources of dairy lactic acid bacteria. Appl. Environ. Microbiol. 80, 2050–2061. doi: 10.1128/AEM.03837-13
Dahl S., Tavaria F. K., Xavier Malcata F. (2000). Relationships between flavour and microbiological profiles in Serra da Estrela cheese throughout ripening. Int. Dairy J. 10, 255–262. doi: 10.1016/S0958-6946(00)00042-X
Despacho32/1996/SRAP (1996)Available online at: https://eur-lex.europa.eu/legal-content/PT/TXT/?uri=uriserv:OJ.L_.1998.175.01.0007.01.POR.
Despacho4185/2011 (2011)Available online at: https://files.diariodarepublica.pt/2s/2011/03/045000000/1078210784.pdf.
Despacho6400/2003 (2003)Available online at: https://diariodarepublica.pt/dr/detalhe/despacho/6400–2003-3039924.
Despacho7822/2011 (2011)Available online at: https://diariodarepublica.pt/dr/detalhe/despacho/7822–2011-828956.
Despacho8601-N/2005 (2005)Available online at: https://files.diariodarepublica.pt/2s/2005/04/076000001/0010100101.pdf.
Despacho9633/2016 (2016)Available online at: https://files.diariodarepublica.pt/2s/2016/07/143000000/2331523316.pdf.
Despachon.5511/2020 (2020)Available online at: https://diariodarepublica.pt/dr/detalhe/despacho/5511–2020-133614823.
DespachoSRAP/94/1 (1994)Available online at: https://eur-lex.europa.eu/legal-content/PT/TXT/PDF/?uri=CELEX:31996R1107R%2801%29&qid=1438340783658&from=EN.
Dias J. (2021). Microbiota de leite cru e de Queijos DOP da Região Centro: Beira Baixa (Rabaçal e Serra da Estrela). http://hdl.handle.net/10400.6/11840.
Dias J. M., Lage P., Alvarenga N., Garcia J., Borrega J., Santos M. T., et al. (2021). Impact of environmental conditions on the ripening of Queijo de Évora PDO cheese. J. Food Sci. Technol. 58, 3942–3952. doi: 10.1007/s13197-020-04856-x
Dias C., Mendes L. (2018). Protected Designation of Origin (PDO), Protected Geographical Indication (PGI) and Traditional Speciality Guaranteed (TSG): A bibiliometric analysis. Food Res. Int. 103, 492–508. doi: 10.1016/j.foodres.2017.09.059
Direção-Geral de Agricultura e Desenvolvimento Rural (2022). Portuguese traditional products. Available online at: https://tradicional.dgadr.gov.pt/en/.
Domingos-Lopes M. F. P., Stanton C., Ross P. R., Dapkevicius M. L.E., Silva C.C. G. (2017). Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal Pico cheese. Food Microbiol. 63, 178–190. doi: 10.1016/j.fm.2016.11.014
EU (2022). “Geographical indications and quality schemes explained,” in European Commission website. Available at: https://agriculture.ec.europa.eu/farming/geographical-indications-and-quality-schemes/geographical-indications-and-quality-schemes-explained_en.
European Comission (2015). Commission implementing regulation (EU). Off. J. Eur. Union. 15, 21. https://eur-lex.europa.eu/legal-content/GA/TXT/?uri=CELEX%3A32014R0668.
European Commission (2012). Regulation (EU) no 1151/2012. Off. J. Eur. Union. https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32012R1151&from=EN.
Faccia M., D’Alessandro A. G., Summer A., Hailu Y. (2020). Milk products from minor dairy species: A review. Animals 10, 1–25. doi: 10.3390/ani10081260
Favaro L., Barretto Penna A. L., Todorov S. D. (2015). Bacteriocinogenic LAB from cheeses - Application in biopreservation? Trends Food Sci. Technol. 41, 37–48. doi: 10.1016/j.tifs.2014.09.001
Folgado A., Pires A. S., Figueiredo A. C., Pimentel C., Abranches R. (2020). Toward alternative sources of milk coagulants for cheese manufacturing: establishment of hairy roots culture and protease characterization from Cynara cardunculus L. Plant Cell Rep. 39, 89–100. doi: 10.1007/s00299-019-02475-1
Freitas A. C., Pais C., Malcata F. X., Hogg T. A. (1996). Microbiological characterization of Picante da Beira Baixa cheese. J. Food Prot. 59, 155–160. doi: 10.4315/0362-028X-59.2.155
Freitas C., Malcata F. X. (2000). Microbiology and biochemistry of cheeses with appélation dorigine protegée and manufactured in the Iberian Peninsula from ovine and caprine milks. J. Dairy Sci. 83, 584–602. doi: 10.3168/jds.S0022-0302(00)74918-6
Gonçalves M. T. P., Benito M. J., Córdoba M. D.G., Egas C., Merchán A. V., Galván A. I., et al. (2018). Bacterial communities in serpa cheese by culture dependent techniques, 16S rRNA gene sequencing and high-throughput sequencing analysis. J. Food Sci. 83, 1333–1341. doi: 10.1111/1750–3841.14141
Gonçalves Dos Santos M. T. P., Benito M. J., de Guía Córdoba M., Alvarenga N., de Herrera S. R.M.S. (2017). Yeast community in traditional Portuguese Serpa cheese by culture-dependent and -independent DNA approaches. Int. J. Food Microbiol. 262, 63–70. doi: 10.1016/j.ijfoodmicro.2017.09.013
Gostin A. I., Waisundara V. Y. (2019). Edible flowers as functional food: A review on artichoke (Cynara cardunculus L.). Trends Food Sci. Technol. 86, 381–391. doi: 10.1016/j.tifs.2019.02.015
Guiné R. P. F., Florença S. G. (2020). The science behind traditional products: the case of portuguese cheeses. ETP Int. J. Food Eng. 6, 45–51. doi: 10.18178/ijfe.6.2.45-51
Herrero-Fresno A., Martínez N., Sánchez-Llana E., Díaz M., Fernández M., Martin M. C., et al. (2012). Lactobacillus casei strains isolated from cheese reduce biogenic amine accumulation in an experimental model. Int. J. Food Microbiol. 157, 297–304. doi: 10.1016/j.ijfoodmicro.2012.06.002
Inácio R. S., Rodríguez-Alcalá L. M., Pimentel L. L., Saraiva J. A., Gomes A. M. (2023). Evolution of Qualitative and Quantitative Lipid Profiles of High-Pressure-Processed Serra da Estrela Cheese throughout Storage. Appl. Sci. (Switzerland) 13, 5927. doi: 10.3390/app13105927
Irlinger F., Helinck S., Jany J. L. (2017)econdary and adjunct cultures,” in Cheese (Elsevier, Academic press), 273–300. doi: 10.1016/B978-0-12-417012-4.00011-9
Ji D., Ma J., Xu M., Agyei D. (2021). Cell-envelope proteinases from lactic acid bacteria: Biochemical features and biotechnological applications. Compr. Rev. Food Sci. Food Saf. 20, 369–400. doi: 10.1111/1541-4337.12676
Jiang N., Wu R., Wu C., Wang R., Wu J., Shi H. (2023). Multi-omics approaches to elucidate the role of interactions between microbial communities in cheese flavor and quality. Food Rev. Int. 39, 5446–5458. doi: 10.1080/87559129.2022.2070199
Kongo J. M., Ho A. J., Malcata F. X., Wiedmann M. (2007). Characterization of dominant lactic acid bacteria isolated from São Jorge cheese, using biochemical and ribotyping methods. J. Appl. Microbiol. 103, 1838–1844. doi: 10.1111/j.1365-2672.2007.03423.x
Kongo J. M., Gomes A. M., Malcata F. X., McSweeney P. L. H. (2009). Microbiological, biochemical and compositional changes during ripening of São Jorge – a raw milk cheese from the Azores (Portugal). Food Chem. 112, 131–138. doi: 10.1016/j.foodchem.2008.05.067
Kongo J. M., Gomes A. P., Malcata F. X. (2008). Monitoring and identification of bacteria associated with safety concerns in the manufacture of São Jorge, a Portuguese traditional cheese from raw cows milk. J. Food Prot. 71, 986–992. doi: 10.4315/0362-028X-71.5.986
Ladero V., Herrero-Fresno A., Martinez N., Del Río B., Linares D. M., Fernández M., et al. (2014). Genome sequence analysis of the biogenic amine-degrading strain lactobacillus casei 5b. Genome Announcements 2, 2. doi: 10.1128/genomeA.01199-13
Lee J., Seo Y., Ha J., Kim S., Choi Y., Oh H., et al. (2020). Influence of milk microbiota on Listeria monocytogenes survival during cheese ripening. Food Sci. Nutr. 8, 5071–5076. doi: 10.1002/fsn3.1806
Linares D. M., Martín M., Ladero V., Alvarez M. A., Fernandez M. (2011). Biogenic amines in dairy products. Crit. Rev. Food Sci. Nutr. 51, 691–703. doi: 10.1080/10408398.2011.582813
MaChado A. B. (2020). Avaliação do ph e verificação da sua relação com o desenvolvimento de microrganismos no queijo de azeitão ao longo do processo de maturação. Faculdade de Medicina Veterinária (Universidade de Lisboa). http://hdl.handle.net/10437/11938.
Mohanty A. K., Mukhopadhyay U. K., Grover S., Batish V. K. (1999). Bovine chymosin: Production by rDNA technology and application in cheese manufacture. Biotechnol. Adv. 17, 205–217. doi: 10.1016/S0734-9750(99)00010-5
Montel M. C., Buchin S., Mallet A., Delbes-Paus C., Vuitton D. A., Desmasures N., et al. (2014). Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 177, 136–154. doi: 10.1016/j.ijfoodmicro.2014.02.019
Morelli L., von Wright A. (2019). Genetics of lactic acid bacteria. Fourth edi, lactic acid bacteria: microbiological and functional aspects. 4th ed. (Boca Raton: Elsevier Ltd). doi: 10.1201/9780429057465–2
OSullivan O., Cotter P. D. (2017). Microbiota of raw milk and raw milk cheeses. Fourth edi, cheese: chemistry, physics and microbiology: fourth edition. 4th ed. (Elsevier Ltd). doi: 10.1016/B978-0-12-417012-4.00012-0
Papademas P., Aspri M., Mariou M., Dowd S. E., Kazou M., Tsakalidou E. (2019). Conventional and omics approaches shed light on Halitzia cheese, a long-forgotten white-brined cheese from Cyprus. Int. Dairy J. 98, 72–83. doi: 10.1016/j.idairyj.2019.06.010
Pereira C. I., Gomes E. O., Gomes A. M., Malcata F. X. (2008). Proteolysis in model Portuguese cheeses: Effects of rennet and starter culture. Food Chem. 108, 862–868. doi: 10.1016/j.foodchem.2007.11.050
Pinho O., Pintado A. I., Gomes A. M., Pintado M. M. E., Malcata F. X., Ferreira I. M. (2004). Interrelationships among microbiological, physicochemical, and biochemical properties of terrincho cheese, with emphasis on biogenic amines. J. Food Prot. 67, 2779–2785. doi: 10.4315/0362-028X-67.12.2779
Pintado A. I. E., Pinho O., Ferreira I. M., Pintado M. M.E., Gomes A. M., Malcata F.X. (2008). Microbiological, biochemical and biogenic amine profiles of Terrincho cheese manufactured in several dairy farms. Int. Dairy J. 18, 631–640. doi: 10.1016/j.idairyj.2007.11.021
Quigley L., O'Sullivan O., Stanton C., Beresford T. P., Ross R. P., Fitzgerald G. F., et al. (2013). The complex microbiota of raw milk. FEMS Microbiol. Rev. 37, 664–698. doi: 10.1111/1574-6976.12030
Rampanti G., Ferrocino I., Harasym J., Foligni R., Cardinali F., Orkusz A., et al. (2023). Queijo Serra da Estrela PDO Cheese: Investigation into Its Morpho-Textural Traits, Microbiota, and Volatilome. Foods 12, 1–24. doi: 10.3390/foods12010169
Reis P. J. M., Malcata F. X. (2011). Current state of Portuguese dairy products from ovine and caprine milks. Small Ruminant Res. 101, 122–133. doi: 10.1016/j.smallrumres.2011.09.032
Reis Lima M. J., Santos A. O., Falcão S., Fontes L., Teixeira-Lemos E., Vilas-Boas M., et al. (2020). Dataset on free amino acids contents of Serra da Estrela PDO cheeses determined by UPLC-DAD-MS/MS. Data Brief 28, 417–432. doi: 10.1016/j.dib.2019.104908
Renes E., Diezhandino I., Fernandez D., Ferrazza R. E., Tornadijo M. E., Fresno J. M. (2014). Effect of autochthonous starter cultures on the biogenic amine content of ewes milk cheese throughout ripening. Food Microbiol. 44, 271–277. doi: 10.1016/j.fm.2014.06.001
Riquelme C., Câmara S., Maria de Lurdes N., Vinuesa P., da Silva C. C.G., Malcata F. X., et al. (2015). Characterization of the bacterial biodiversity in Pico cheese (an artisanal Azorean food). Int. J. Food Microbiol. 192, 86–94. doi: 10.1016/j.ijfoodmicro.2014.09.031
Rocha R., Vaz Velho M., Santos J., Fernandes P. (2021). Serra da estrela pdo cheese microbiome as revealed by next generation sequencing. Microorganisms 9, 2007. doi: 10.3390/microorganisms9102007
Rocha R., Couto N., Pinto R. P., Vaz-Velho M., Fernandes P., Santos J. (2023). Microbiological characterization of protected designation of origin serra da estrela cheese. Foods 12, 2008. doi: 10.3390/foods12102008
Rodrigues A. S. (2023). Qualidade do Queijo Amarelo da Beira Baixa DOP e efeito da época do ano na qualidade microbiológica. Instituto Politécnico de Castelo Branco (Escola Superior Agrária). http://hdl.handle.net/10400.11/8484.
Roseiro L. B., Andrew Wilbey R., Barbosa M. (2003). Serpa Cheese: Technological, biochemical and microbiological characterisation of a PDO ewes milk cheese coagulated with Cynara cardunculus L. Le Lait 83, 469–481. doi: 10.1051/lait:2003026
Ruggirello M., Dolci P., Cocolin L. (2014). Detection and viability of Lactococcus lactis throughout cheese ripening. PloS One 9, 1–14. doi: 10.1371/journal.pone.0114280
Salamandane A., Leech J., Almeida R., Silva C., Crispie F., Cotter P. D., et al. (2024). Metagenomic analysis of the bacterial microbiome, resistome and virulome distinguishes Portuguese Serra da Estrela PDO cheeses from similar non-PDO cheeses: An exploratory approach. Food Res. Int. 189, 114556. doi: 10.1016/j.foodres.2024.114556
Salque M., Bogucki P. I., Pyzel J., Sobkowiak-Tabaka I., Grygiel R., Szmyt M., et al. (2013). Earliest evidence for cheese making in the sixth millennium bc in northern Europe. Nature 493, 522–525. doi: 10.1038/nature11698
Sattin E., Andreani N. A., Carraro L., Lucchini R., Fasolato L., Telatin A., et al. (2016). A multi-omics approach to evaluate the quality of milk whey used in ricotta cheese production. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01272
Settanni L., Moschetti G. (2010). Non-starter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiol. 27, 691–697. doi: 10.1016/j.fm.2010.05.023
Takala T. M., Mokhtari S., Ahonen S. L., Wan X., Saris P.E. (2023). Wild-type Lactococcus lactis producing bacteriocin-like prophage lysins. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1219723
Tavaria F. K., Dahl S., Carballo F. J., Malcata F. X. (2002). Amino acid catabolism and generation of volatiles by lactic acid bacteria. J. Dairy Sci. 85, 2462–2470. doi: 10.3168/jds.S0022-0302(02)74328-2
Tavaria F. K., Malcata F. X. (1998). Microbiological Characterization of Serra da Estrela Cheese throughout Its Appellation dOrigine Protégée Region. J. Food Prot. 61, 601–607. doi: 10.4315/0362-028X-61.5.601
Tavaria F. K., Malcata F. X. (2000). On the microbiology of Serra da Estrela cheese: geographical and chronological considerations. Food Microbiol. 17, 293–304. doi: 10.1006/fmic.1999
Van De Bunt B., Bron P. A., Sijtsma L., de Vos W. M., Hugenholtz J. (2014). Use of non-growing Lactococcus lactis cell suspensions for production of volatile metabolites with direct relevance for flavour formation during dairy fermentations. Microbial Cell Factories 13, 1–9. doi: 10.1186/s12934-014-0176-2
Voidarou C., Tzora A., Malamou O., Akrida-Demertzi K., Demertzis P. G., Vassos D., et al. (2011). Chemical and microbiological characterization of artisan inoculants used for the fermentation of traditional dairy products in Epirus area (Greece). Anaerobe 17, 354–357. doi: 10.1016/j.anaerobe.2011.07.004
Zheng J., Wittouck S., Salvetti E., Franz C. M., Harris H. M., Mattarelli P., et al. (2020). A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Systematic Evolutionary Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: Portuguese traditional cheeses, microbiota, PDO cheeses, conventional microbiology, OMICs analyses, metagenomics, flavor development, cheese ripening
Citation: Serrano S, Morais S and Semedo-Lemsaddek T (2024) Tradition unveiled: a comprehensive review of microbiological studies on Portuguese traditional cheeses, merging conventional and OMICs analyses. Front. Ind. Microbiol. 2:1420042. doi: 10.3389/finmi.2024.1420042
Received: 19 April 2024; Accepted: 24 June 2024;
Published: 16 July 2024.
Edited by:
Alessia Levante, University of Parma, ItalyReviewed by:
Flora Valeria Romeo, Council for Agricultural and Economics Research (CREA), ItalyLaura Espina, University of Zaragoza, Spain
Copyright © 2024 Serrano, Morais and Semedo-Lemsaddek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teresa Semedo-Lemsaddek, dGxlbXNhZGRla0BmbXYudWxpc2JvYS5wdA==
 Susana Serrano
Susana Serrano Susana Morais
Susana Morais Teresa Semedo-Lemsaddek
Teresa Semedo-Lemsaddek