- 1Pivot Bio, Inc., Berkeley, CA, United States
- 2Institute of Biotechnology, Department of Chemical Engineering, University of Granada, Granada, Spain
- 3Department of Chemical Sciences, Benemérita Autonomous University of Puebla, Puebla, Mexico
Adopting a risk assessment paradigm that is contextualized and strain-specific will be critical to enabling the continued development and safe use of microbes, particularly bacteria, in numerous industries and applications. An overly simplistic approach of labeling bacterial species as either harmful or beneficial is ill-suited for the complexities of their interactions with hosts and other microbes, where the lines between friend, foe, and innocent bystander are often unclear. Many such nuanced relationships have been described in human microbiome studies, illustrating the inherent challenges of defining bacterial safety. Any effective risk assessment framework must take into account bacterial niche and environment, fitness, host health, route and extent of exposure, and strain characterization. Klebsiella variicola, a diazotrophic soil bacterium isolated around the world, has been the subject of increasing interest on both environmental and clinical fronts, and has been used commercially as a biofertilizer on millions of farm acres. Here we review its population structure, relevance in clinical and environmental settings, and use as a biofertilizer in light of the risk assessment framework described.
Introduction
As tiny living factories, bacteria have provided a host of applications in industries ranging from food to fuels (Akoijam et al., 2022). They hold the potential to make tremendous contributions toward a more sustainable future (Robbins, 2021), but evaluating their safe use remains a challenge. One of the pitfalls of assessing bacterial safety is creating a false dichotomy wherein species are considered categorically harmful or beneficial. In reality, the relationship between bacteria and their hosts is incredibly complex and dynamic, and the concept of bacterial safety must be viewed along a spectrum that is highly niche-dependent, strain-specific, and never static. The need for this conceptual shift in safety assessment has been illustrated and informed largely by human microbiome research, and its relevance to other domains such as industry and agriculture is deeply evident (Sansinenea, 2022).
Reframing the concept of bacterial risk
Bacteria are a pervasive presence in the human body. The average adult carries roughly 1 kilogram of bacteria in their gut, many of which are uncultivable, and it is estimated that less than 1% of all microbes are considered human pathogens (Microbiology by numbers, 2011). Many bacteria are beneficial – even vital – to our systemic health, yet the pathogenic few have devastated human populations throughout history, and millions still die every year of infectious diseases, despite tremendous strides made with the use of antibiotics. Given our confounding relationship with bacteria, the desire to identify and eliminate bad actors is not surprising. However, it is becoming increasingly apparent that many members of the human microbiota cannot easily be classified as friend or foe, and attempts to label them as such are often shortsighted (Jochum and Stecher, 2020). Human microbiomes bring to light multifaceted, often surprising associations, wherein the same bacterial species can be considered commensal, mutualistic, or pathogenic, depending on its niche access, environmental conditions and resources, and specific strain genotype.
For example, Helicobacter pylori has colonized the human stomach for millennia, has been considered a gastric pathogen for the last few decades, and more recently appears to be protective against certain diseases, including childhood-onset asthma (Chen and Blaser, 2008; Blaser, 2010). Attempting to reconcile these observations raises numerous questions. Is H. pylori a driver in development or progression of gastric disease, or simply a passenger? Is pathogenicity only linked to certain strains or genotypes of this species? Might eliminating it from its long-held niche have unintended consequences? Additional research is needed to answer these questions, but what is clear is that humankind’s relationship with this microbe is more complex than previously thought.
Enterococcus faecalis is a ubiquitous human and animal gut microbe that can behave as a commensal, probiotic, or pathogen, depending on the strains and environment (Franz et al., 2011; Hanchi et al., 2018; Braïek and Smaoui, 2019; Krawczyk et al., 2021). It is widely used as a starter culture for fermented dairy products, can have protective effects against foodborne pathogens, and has been evaluated as an adjuvant therapy for sinusitis in children (Kitz et al., 2012; Hanchi et al., 2018). However, outside of its normal food and gut-associated niches, it is a frequent cause of hospital-acquired bacteremia, urinary tract infections, and infectious endocarditis, posing an increased risk to immunocompromised patients with comorbidities, catheters, and history of broad-spectrum antibiotic use (Arias and Murray, 2012; Dubin and Pamer, 2017). Several studies have been carried out to determine the differences between clinical and food-derived isolates, especially with respect to genes associated with virulence and antibiotic resistance (Eaton and Gasson, 2001; Franz et al., 2001; Semedo et al., 2003). While such genes were more frequently found in clinical isolates, they were also present in the genomes of nonpathogenic food isolates, and some (e.g., gelE) were phenotypically silent under similar conditions. In short, the species defies a clear-cut risk designation, and the safety of E. faecalis must be evaluated by individual strain.
Escherichia coli is yet another species in which diverse roles are observed due to strain-to-strain genetic variation. Certain E. coli serotypes introduced during vaginal delivery typically colonize a newborn’s gastrointestinal tract within hours of birth, are recognized as critical symbionts for healthy microbiome development, and persist throughout life (Blount, 2015). One particular strain, marketed under the name Symbioflor® 2, has been approved as a treatment for irritable bowel syndrome (IBS) in the European Union (EMA recommends that Symbioflor 2 can continue to be used for irritable bowel syndrome, 2017). Other serotypes, such as the enterohemorrhagic colitis causing O157:H7, are highly pathogenic, possessing specific combinations of virulence-related factors that render them capable of causing potentially fatal kidney damage (Kaper et al., 2004). Once again, the line between friend and foe is nuanced, and attempting to apply a single risk designation for the entire E. coli species is futile.
Other human gut bacteria are widely viewed as categorically safe, and have long been used as probiotics and food additives. The global probiotic market is valued at over $50 billion USD, and more than 1,000 clinical trials have been conducted to study the benefits of probiotics in over 700 conditions and diseases (Dronkers et al., 2020; Probiotics Market Size, 2021). Lactobacillus and Bifidobacterium are the most commonly used genera in probiotics and fermented foods, with well-documented health benefits (Probiotics - Health Professional Fact Sheet, 2023). Lactobacilli dominate healthy vaginal microbiota, and their depletion is associated with several disease states, including infection, pelvic inflammatory disease, and infertility (Ravel et al., 2011; Kroon et al., 2018; Younes et al., 2018). Bifidobacteria species are among the most dominant gut colonizers of breastfed infants, and their presence has a profound positive impact on health throughout life, with inverse associations observed for obesity, metabolic disease, allergy, and infection (Bäckhed et al., 2015; Arboleya et al., 2016; Turroni et al., 2020; Stuivenberg et al., 2022). Yet even Lactobacillus and Bifidobacterium species can become pathogenic when introduced outside of their normal niches, especially in individuals with comorbidities or risk factors such as age (specifically neonates and the elderly), and history of broad-spectrum antibiotic use (Pathak et al., 2014; Butta et al., 2017; Esaiassen et al., 2017; Rossi et al., 2019).
When considering these and a myriad of similar examples from plant, animal and soil microbiomes, the picture of bacterial risk that begins to emerge is one with a great deal of nuance. From it, we can make several observations:
1) The importance of bacterial niche specificity and environmental stimuli cannot be overstated. What causes disease in one context may provide an essential benefit in another. Symbiotic bacteria are highly niche-specific, living in delicately balanced communities that have co-evolved with their hosts. Perturbations to this balance – known as “dysbiosis” – can have deleterious effects, and the potential risks of indiscriminately eradicating a species that evolved to inhabit a specific niche are clear. In such communities, fine-tuning, rather than wholesale elimination of a species, should be the prevailing approach.
2) In a sense, all microbes are opportunists. A host experiencing disease, dysbiosis, or weakened immune function provides an environment where even its closest bacterial allies can become enemies. In such an environment, it is also possible for passengers in a disease to be mistaken for drivers. Considering overall host health and unraveling specific roles in disease are difficult but necessary steps.
3) Different strains of the same species can vary widely in their genotypic and phenotypic profiles, and as a result, may have remarkably different behaviors. Therefore, risks must by necessity be evaluated at the individual strain level.
4) Given the constant evolution of bacteria in response to environmental cues and pressures, ongoing surveillance is necessary.
Clearly, bacterial behavior and impact in any given habitat are the results of a delicate balance of environmental, host health, and strain-specific factors (Figure 1). In such a complex, ever-shifting landscape, defining which bacteria are considered safe and under which circumstances requires moving beyond simplistic labels of “good” or “bad.” Accurately evaluating bacterial risk requires a wide-angle lens, and context is critical. This paradigm shift is gaining increasing recognition (Wiles and Guillemin, 2019; Jochum and Stecher, 2020), and its widespread adoption will create a path forward for bringing countless new microbial products and technologies to life.
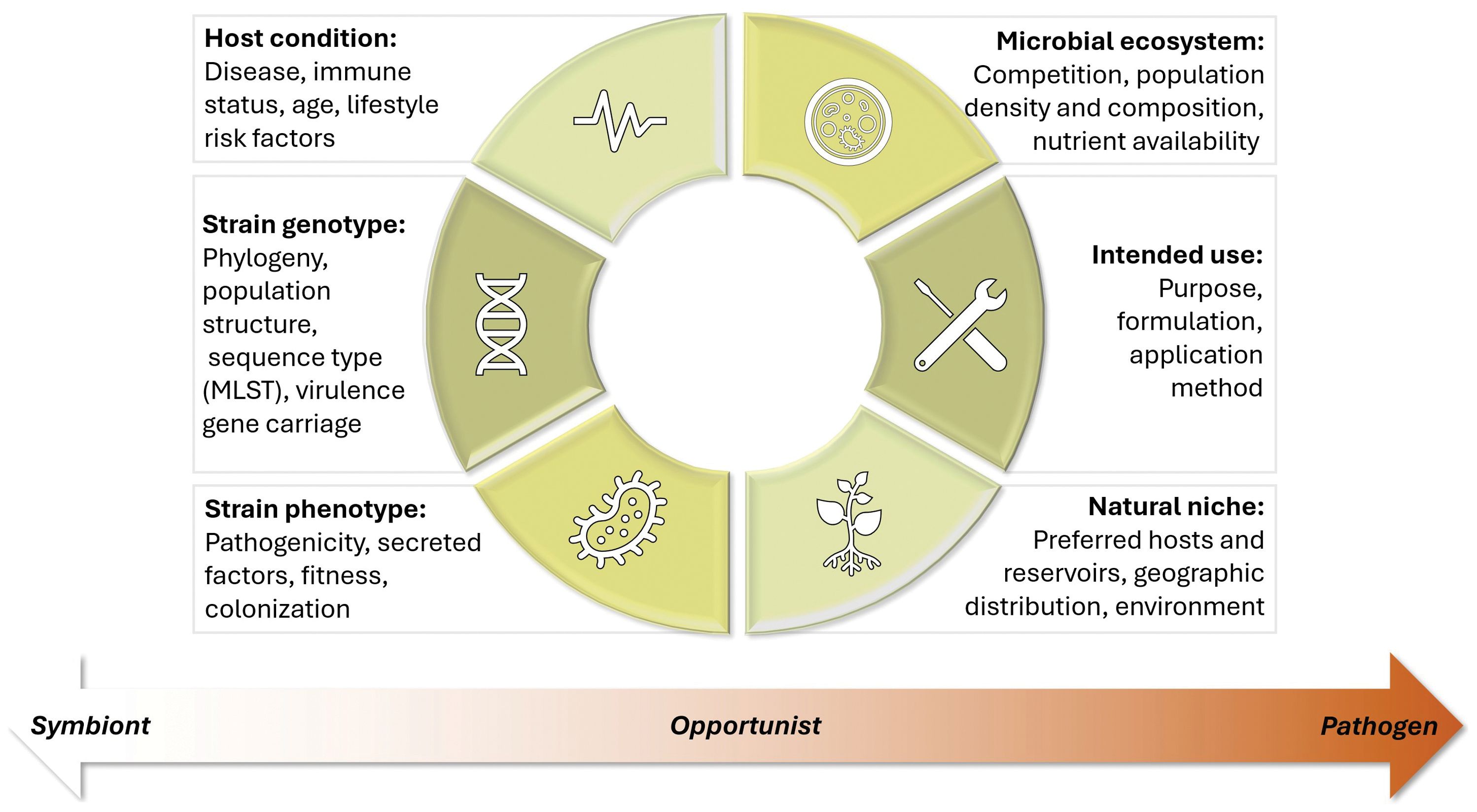
Figure 1 Factors for consideration when assessing bacterial safety. Bacterial safety should be viewed along a continuum between symbiont and pathogen. Determination of where a given bacterial strain falls on this continuum cannot be made without considering a number of factors, including host condition, strain phenotype and genotype, microbial ecosystem, natural niche, and intended use.
Klebsiella variicola
An illustration of this contextualized approach to safety evaluation in the agricultural arena can be drawn from the diazotrophic bacterium Klebsiella variicola, a member of the Klebsiella pneumoniae species complex (KpSC) which has been successfully developed for use as a biofertilizer. First isolated from healthy banana plants and identified as a novel nitrogen-fixing species in 2004, K. variicola has gained increasing attention in recent years as a bacterial species of both clinical and environmental interest (Rosenblueth, 2004; Rodríguez-Medina et al., 2019; Duran-Bedolla et al., 2021). Like other members of the Klebsiella genus, K. variicola is a gram-negative, facultative anaerobic non-spore-forming bacterium, with plants considered to be its natural reservoir (Rosenblueth, 2004). It has widespread geographic distribution and can colonize a wide range of diverse niches. Strains of K. variicola have been isolated from many sources, including leaf-cutter ant fungus gardens (Pinto-Tomas, 2009), fecal samples of healthy breastfed infants (Rosales-Bravo et al., 2017; Gómez et al., 2021), the traditional fermented Mayan beverage pozol (Rizo et al., 2020), and industrial effluents (Afzal et al., 2017). Gut carriage of K. variicola and other KpSC member species has also been confirmed in healthy adults (Raffelsberger et al., 2021). Furthermore, in-house sequence-based screening of the publicly available Human Microbiome Project database (Human Microbiome Project Data Portal, 2013) for the presence of KpSC members in healthy human subjects revealed frequent detection of these microbes across different body sites, particularly oral (Figure 2).
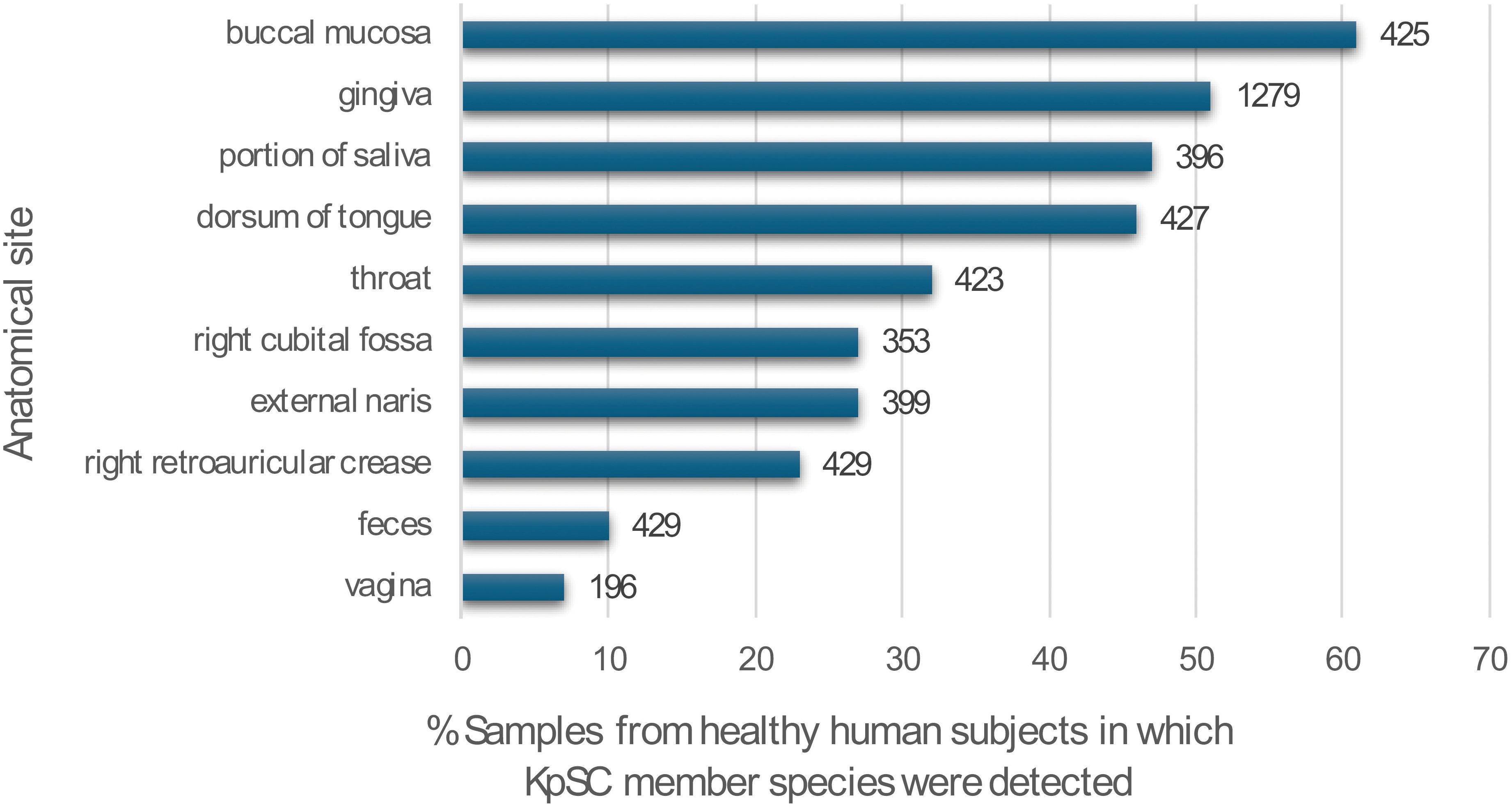
Figure 2 Prevalence of KpSC member species detected across different anatomical sites in 5,906 sample sequences collected from healthy adults and deposited in the Human Microbiome Project database (Human Microbiome Project Data Portal, 2013). Bars labeled with sample size from each site. Screening methods can be found in Supplementary File 1. Sample metadata can be accessed in Supplementary File 2.
The ability to inhabit these different niches stems from the species’ remarkably high genetic diversity. Molecular genotyping of K. variicola has identified over 150 sequence types to date, with some strictly isolated from one niche (plant, animal, or human), and others identified across kingdoms (Guo et al., 2018; Barrios-Camacho et al., 2019; Morgado et al., 2022). Based on their biosafety profiles, K. variicola strains have been identified with containment level assignments of BSL1 and BSL2 – for example, strains ATCC F2R9 and LMG 23571, respectively (ATCC F2R9, 2001; LMG 23571, 2004).
Genomic differences can significantly alter the behavior and niche of different K. variicola strains, but attempting to make associations quickly becomes problematic. For example, a phylogenetic core genome study of 145 primarily clinical K. variicola isolates showed two distinct lineages, one with two genomes, and the other with 143 genomes (Potter et al., 2018). The genomes in the second lineage formed 26 clusters, with variations in virulence and infection severity between different strains, but no correlation was observed between cluster and geography, niche, or virulence level. A mosaic distribution of virulence-related genes has been observed among clinical and plant-associated K. variicola strains (Martínez-Romero et al., 2017b), but when considering the possibility that some genes may be phenotypically silent, it is difficult to draw meaningful conclusions without sufficient in vitro and in vivo data.
The behavior and niche of these microbes are also impacted by their metabolic profiles. Nitrogen fixation, for example, is a metabolic capability that strongly affects niche preference. In contrast to K. pneumoniae, where nitrogen fixation genes are located in the accessory genome and encountered infrequently, nitrogen fixation is a hallmark of K. variicola, with the requisite nif genes contained in its highly conserved core genome (Martin and Bachman, 2018). Indeed, of 294 K. pneumoniae genomes analyzed in a global survey, only one contained the nif operon (Holt et al., 2015), and it has been proposed that the absence of nitrogen-fixation capacity contributed to K. pneumoniae’s evolution toward human hosts and its increased pathogenicity compared to its consistently nitrogen-fixing plant-associated relative, K. variicola (Fouts et al., 2008; Brisse et al., 2014). One K. variicola outlier strain, NL49/58, acquired the capability to ferment adonitol – a metabolic characteristic typically present in other Klebsiella species such as K. aerogenes, K. planticola, K. oxytoca, and K. pneumoniae (Brisse and Verhoef, 2001; Davidson et al., 2015). This trait likely rendered NL49/58 capable of colonizing milk, underlying its reported link to cases of bovine mastitis, which had previously been associated with K. pneumoniae and K. oxytoca (Podder et al., 2014; Masse et al., 2020).
Taken together, these studies highlight the importance of delineating genotypic and phenotypic differences not only between K. variicola and other Klebsiella species, but also between different K. variicola strains. This is a critical step in the process of evaluating the safety of this highly heterogenous species, and should include not only in silico screening, but also in vitro and in vivo characterization of pathogenicity potential in plants and animals.
Beneficial environmental Klebsiella variicola strains
Environmental K. variicola strains are of special interest for their potential uses in sustainable agriculture and industry (Duran-Bedolla et al., 2021). Drawn to nutrient-rich root exudates (Feng et al., 2021), plant-associated strains exist as endophytic or rhizospheric nitrogen-fixing symbionts, and provide their hosts with a bioavailable source of nitrogen and increased resilience toward environmental stressors (Souza et al., 2015; Vives-Peris et al., 2020). Examples of such strains abound in the literature, and K. variicola has been isolated from and shown to support the growth of numerous crops, including sugarcane, soybeans, maize, tomatoes, and peanuts (Wei et al., 2013; Kim et al., 2017; Yang and Yang, 2020; Guerrieri et al., 2021; Qin et al., 2022; Wang, 2022; Zhang et al., 2022).
The reported effects of K. variicola on its plant hosts are notable; a greenhouse study in sugarcane reported a 79% increase in whole-plant nitrogen content and 51% greater whole-plant dry biomass with K. variicola inoculation (Wei et al., 2013), and studies in tomato plants showed significant increases in yield and above-ground biomass (Guerrieri et al., 2021; Nava-Faustino et al., 2022). It has been reported to be a highly conserved core member of the nitrogen-fixing microbial community in maize plant xylem – a finding that was consistent across substantially different soil types, fertilization regimes, and soil microbial communities analyzed in the study (Zhang et al., 2022). It has also been shown to mitigate flood stress in soybeans (Kim et al., 2017), and improves salinity stress tolerance in wheat, maize, and tuberose (Ghazi et al., 2021; Kusale et al., 2021a, 2021b). Isolated instances of plant pathogenicity have also been reported, primarily in tropical monsoon climates: banana rhizome rot, carrot soft rot, and sorghum leaf streak in India (Chandrashekar et al., 2018; Loganathan et al., 2021; Malik, 2023), banana soft rot in China (Fan, 2016), and plantain soft rot in Haiti (Fulton et al., 2020). However, it should be noted that these plant pathogenicity reports rely on intrinsically limited culture-based isolation and detection methods. Additional field trials are needed to identify all relevant species, both cultivable and uncultivable, to recapitulate pathogenicity under conditions consistent with the natural environments of these crops, and to validate plant beneficial activity and elucidate mechanisms. Meanwhile, selected strains have been successfully harnessed as biofertilizers for cereal crops, and K. variicola’s utility in providing a stable, safe, and sustainable nitrogen source has now been demonstrated commercially on millions of farm acres (Wen et al., 2021; Arbaugh et al., 2022; Temme, 2022).
K. variicola soil isolates have also been identified and studied for use in the remediation of soil and industrial waste. Several strains tolerant to toxic halides, heavy metals, and aromatic compounds show efficacy in the treatment of wastewater, industrial effluents, and contaminated soil. For example, a strain isolated in Bangladesh effectively reduced highly toxic chromium (VI) produced by leather tanning, a major source of industrial pollution, and showed a high tolerance to cadmium, zinc, nickel, and mercury (Kabir et al., 2018). When loaded onto biochar, another strain removed up to 96% of chromium (VI) from groundwater samples collected near a leather tannery in China (Yu et al., 2021). Another biochar system, in this case loaded with a bacterial consortium including K. variicola and two Enterobacter species, reduced cadmium levels in contaminated soil, reduced its levels in bok choy plants by up to 50%, and simultaneously increased dry shoot biomass of the plants by an average of 240% (Wei et al., 2022). Strains with bioremediation activity against toxic aromatic compounds (Liu et al., 2016), lead (Das and Osborne, 2017), red azo dye (Eslami et al., 2019), and atrazine (Pan et al., 2023) have also been reported. Future studies are needed to further validate these findings, but reports to date clearly indicate K. variicola’s potential for the remediation of a range of toxic waste and pollutants, provided the right strains are selected, optimized, and applied to the specific niche and context in which they’re intended to function.
Clinical strains of Klebsiella variicola: direct drivers of infection, or opportunistic passengers?
The emergence of enhanced diagnostic techniques has enabled several large-scale population structure studies of the KpSC (Berry et al., 2015; Long et al., 2017; Martínez-Romero et al., 2017a; Ohama, 2022). These studies show that (1) K. variicola accounts for only 2-6% of KpSC-positive human samples, and (2) pathogenicity is not clearly associated with lineage for these species, but rather linked to specific virulence factors, which are found with consistently lower frequency in K. variicola (Holt et al., 2015; Lam et al., 2021; Dereeper et al., 2022; Morgado et al., 2022). Phylogenetic analyses of gyrA, parC, and rpoB gene sequences have categorized K. variicola as a distinct phylogroup of KpSC, specifically KpIII (Brisse and Verhoef, 2001; Brisse et al., 2004; Garza-Ramos et al., 2015), while K. pneumoniae (KpI) is by far the dominant agent of Klebsiella-associated human infections (Pendleton et al., 2013; Lam et al., 2021). Furthermore, a global population analysis of over 11,000 Klebsiella genomes showed an extremely low prevalence of both virulence and antimicrobial resistance factors for K. variicola, with the exception of β-lactamase resistance, which is a chromosomally encoded feature of all KpSC species (Lam et al., 2021). K. variicola isolates associated with human infection, primarily isolated from blood, urine, and sputum, have been reported in immunosuppressed (e.g., cancer and trauma patients) and immunocompromised (e.g., diabetics, the elderly, neonates) populations (Maatallah et al., 2014; Long et al., 2017; Farzana et al., 2019; Imai et al., 2019; Piepenbrock et al., 2020; Kiley et al., 2021; Watanabe et al., 2022). Both retrospective and case studies underscore the clear opportunistic nature of this commensal bacterium, describing the infection of individuals with weakened immunity or comorbidities. Several studies also show evidence that a major source of KpSC infections in hospitalized patients may in fact be the patients’ own microbiota opportunistically spreading or overgrowing under conditions of weakened immunity (Martin et al., 2016; Gorrie et al., 2017, 2018).
One retrospective analysis of bloodstream infections (BSI) in Norway has suggested that K. variicola carries a higher 30-day mortality rate compared to K. pneumoniae (Maatallah et al., 2014); however, this conclusion is based on a single study with considerable limitations. The study was limited to one site, had a relatively small sample size (139 KpSC cases, 34 of which were K. variicola infections), and lacked sufficient biological and clinical relevance to inform practice or draw solid conclusions. BSI are generally considered to be a marker of severe primary disease irrespective of the causative agent, and present a poor prognosis and high mortality rate (Goto and Al-Hasan, 2013). Among the BSI patients included in this study, K. pneumoniae remained the most frequent causative agent (69%). Of the 10 K. variicola infected patients suffering 30-day mortality, all 10 presented comorbidities, and their average age was 75. Moreover, mortalities in the K. variicola group were not linked to any known virulence factors. While this study does not provide sufficient evidence to make meaningful risk determinations or impact patient treatments or outcomes, it does highlight, yet again, the prerequisite of weakened immunity for successful K. variicola infection.
A contextualized risk assessment paradigm
The demand for safer and more sustainable agricultural inputs has increased tremendously in recent years, and industry is responding accordingly. The global biofertilizer market, valued at $1.8 billion USD in 2021, is projected to grow to $4.4 billion by 2029 (Biofertilizer Market Size, 2021). Significant technological advances have enabled agricultural companies to develop a wide array of microbial products, all of which require appropriate safety evaluation. Given the recognized nuance and complexity of host-microbe interactions and the significant differences between strains within a species, strains must be evaluated individually, with a focus on their intended application and usage parameters. As described above, multiple beneficial activities of Klebsiella variicola in environmental settings have been reported. Evaluation of this highly heterogeneous species could encompass several key considerations, including, but not limited to, a strain’s genotype and phenotype (including virulence and antimicrobial resistance factors), in vivo toxicity, virulence and non-target effects, and potential routes of human and environmental exposure based upon intended application methods.
In the case of K. variicola strains developed for use as biofertilizers, the intended context is exclusively environmental, by way of application to crops. Revisiting the question of safety in light of this context leads to several observations. First, studies to date do not indicate infection risks in healthy humans, and virulence and antibiotic resistance factors, already found infrequently in K. variicola, would not encounter the acquisition pressures found in a hospital or community environment when applied in the field. Furthermore, these strains are not intended for direct human exposure, and application methods are designed to minimize exposure risk. Strains selected for field use were well characterized in terms of mechanism of action, genotype, and phenotype, and did not show toxicity or pathogenicity in a standard battery of in vivo biosafety tests (Wen et al., 2021). Finally, given that K. variicola is a ubiquitous soil microbe and plant symbiont found around the world, its presence in agricultural environments is not novel.
Conclusion
The concept of bacterial risk is nuanced, dynamic, and highly dependent on context. As evidenced increasingly in human microbiome studies, the lines between commensal, mutualistic, and pathogenic bacteria are often hazy. Whether a host-microbe interaction is beneficial, detrimental, or neutral depends on numerous factors, including a bacterial strain’s genotype, fitness, and niche, as well as host health, degree of exposure, and the prevailing microbial environment. Klebsiella variicola is a near ubiquitous diazotroph found primarily in soil and plants, and has tremendous potential as a biofertilizer. It is also a frequent resident of the human microbiome, and clinical reports do not indicate a risk of pathogenicity in healthy individuals. Moreover, K. variicola strains developed for biofertilizer use are intended strictly for targeted application in select crops, and are chosen and optimized for the selective colonization of plants over other hosts. With these considerations in mind, we find no indication that the judicious application of K. variicola strains optimized for use as biofertilizers in select crops presents undue safety concerns to healthy humans or the environment. The context-specific approach presented herein illustrates a critical paradigm shift in the microbial risk evaluation process – a necessary step in creating a path forward for the development, commercialization, and safe use of microbial products across multiple industries.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ChM: Writing – review & editing, Writing – original draft, Visualization, Conceptualization. FaS: Writing – review & editing, Supervision, Conceptualization. CaM: Writing – review & editing, Conceptualization. FoS: Writing – review & editing, Supervision, Conceptualization. NH: Writing – review & editing, Supervision, Conceptualization, Project administration, Resources. NR: Writing – original draft, Data curation, Methodology, Visualization. NV: Writing – review & editing. ES: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We are grateful to Nicholas Colaianni for providing technical guidance on screening the Human Microbiome Project database, Chris Cornelius for formatting figures, and Sophie Kowaliczko for assistance with reviewing literature. We also thank Allen Kuo for providing legal insight and contract preparation, and Mitchell Craft, Wendy Watkins, and Mary Burich for manuscript review.
Conflict of interest
Authors ChM, FaS, CaM, FoS, NH and NR were employed by the company Pivot Bio. Pivot Bio has filed patent applications and is developing and commercializing products based on microbial strains presented in this manuscript.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/finmi.2024.1412302/full#supplementary-material
References
Afzal A. M., Rasool M. H., Waseem M., Aslam B. (2017). Assessment of heavy metal tolerance and biosorptive potential of Klebsiella variicola isolated from industrial effluents. AMB Express 7, 184. doi: 10.1186/s13568-017-0482-2
Akoijam N., Kalita D., Joshi S. R. (2022). Bacteria and their industrial importance. Ind. Microbiol. Biotechnol. 63–79. doi: 10.1007/978-981-16-5214-1_2
Arbaugh B. M., Rezaei F., Mohiti-Asli M., Pena S., Scher H. B., Jeoh T. (2022). A strategy for stable, on-seed application of a nitrogen-fixing microbial inoculant by microencapsulation in spray-dried cross-linked alginates. ACS Agric. Sci. Technol. 2, 950–959. doi: 10.1021/acsagscitech.2c00107
Arboleya S., Watkins C., Stanton C., Ross R. P. (2016). Gut bifidobacteria populations in human health and aging. Front. Microbiol. 7 1204. doi: 10.3389/fmicb.2016.01204
Arias C. A., Murray B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
ATCC F2R9 (2001). Available online at: https://www.atcc.org/products/baa-830 (Accessed March 26, 2024).
Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703. doi: 10.1016/j.chom.2015.04.004
Barrios-Camacho H., Aguilar-Vera A., Beltran-Rojel M., Aguilar-Vera E., Duran-Bedolla J., Rodriguez-Medina N., et al. (2019). Molecular epidemiology of Klebsiella variicola obtained from different sources. Sci. Rep. 9, 10610. doi: 10.1038/s41598-019-46998-9
Berry G. J., Loeffelholz M. J., Williams-Bouyer N. (2015). An Investigation into Laboratory Misidentification of a Bloodstream Klebsiella variicola Infection. J. Clin. Microbiol. 53, 2793–2794. doi: 10.1128/JCM.00841-15
Biofertilizer Market Size (2021). Available online at: https://www.fortunebusinessinsights.com/industry-reports/biofertilizers-market-100413.
Blaser M. J. (2010). Helicobacter pylori and esophageal disease: wake-up call? Gastroenterology 139, 1819–1822. doi: 10.1053/j.gastro.2010.10.037
Blount Z. D. (2015). The unexhausted potential of E. coli. eLife 4, e05826. doi: 10.7554/elife.05826
Braïek O. B., Smaoui S. (2019). Enterococci: between emerging pathogens and potential probiotics. BioMed. Res. Int. 2019, 1–13. doi: 10.1155/2019/5938210
Brisse S., Himbergen T. V., Kusters K., Verhoef J. (2004). Development of a rapid identification method for Klebsiella pneumoniae phylogenetic groups and analysis of 420 clinical isolates. Clin. Microbiol. Infect. 10, 942–945. doi: 10.1111/j.1469-0691.2004.00973.x
Brisse S., Passet V., Grimont P. A. D. (2014). Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella Singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int. J. Syst. Evol. Microbiol. 64, 3146–3152. doi: 10.1099/ijs.0.062737-0
Brisse S., Verhoef J. (2001). Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int. J. Syst. Evol. Microbiol. 51, 915–924. doi: 10.1099/00207713-51-3-915
Butta H., Sardana R., Vaishya R., Singh K. N., Mendiratta L. (2017). Bifidobacterium: an emerging clinically significant metronidazole-resistant anaerobe of mixed pyogenic infections. Cureus 9, e1134. doi: 10.7759/cureus.1134
Chandrashekar B. S., Prasannakumar M. K., Puneeth M. E., Teli K., Priyanka K., Mahesh H. B., et al. (2018). First report of bacterial soft rot of carrot caused by Klebsiella variicola in India. N. Dis. Rep. 37, 21–21. doi: 10.5197/j.2044-0588.2018.037.021
Chen Y., Blaser J. ,. M. (2008). Helicobacter pylori colonization is inversely associated with childhood asthma. J. Infect. Dis. 198, 553–560. doi: 10.1086/590158
Das A., Osborne J. W. (2017). Enhanced bioremoval of lead by earthworm–Lumbricus terrestris co-cultivated with bacteria–Klebsiella variicola. J. Photochem. Photobiol. B: Biol. 175, 65–72. doi: 10.1016/j.jphotobiol.2017.08.031
Davidson F. W., Whitney H. G., Tahlan K. (2015). Genome sequences of klebsiella variicola isolates from dairy animals with bovine mastitis from Newfoundland, Canada. Genome Announc. 3, e00938–e00915. doi: 10.1128/genomeA.00938-15
Dereeper A., Gruel G., Pot M., Couvin D., Barbier E., Bastian S., et al. (2022). Limited Transmission of Klebsiella pneumoniae among Humans, Animals, and the Environment in a Caribbean Island, Guadeloupe (French West Indies). Microbiol. Spectr. 10, e01242–e01222. doi: 10.1128/spectrum.01242-22
Dronkers T. M. G., Ouwehand A. C., Rijkers G. T. (2020). Global analysis of clinical trials with probiotics. Heliyon 6, e04467. doi: 10.1016/j.heliyon.2020.e04467
Dubin K., Pamer E. G. (2017). Enterococci and their interactions with the intestinal microbiome. Microbiol. Spectr. 5, 309–330. doi: 10.1128/microbiolspec.BAD-0014-2016
Duran-Bedolla J., Garza-Ramos U., Rodriguez-Medina N., Vera A. A., Barrios-Camacho H. (2021). Exploring the environmental traits and applications of Klebsiella variicola. Braz. J. Microbiol. 52, 2233–2245. doi: 10.1007/s42770-021-00630-z
Eaton T. J., Gasson M. J. (2001). Molecular screening of enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67, 1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001
EMA recommends that Symbioflor 2 can continue to be used for irritable bowel syndrome (2017). Available online at: https://www.ema.europa.eu/en/medicines/human/referrals/symbioflor-2.
Esaiassen E., Hjerde E., Cavanagh J. P., Simonsen G. S., Klingenberg C. (2017). Bifidobacterium bacteremia: clinical characteristics and a genomic approach to assess pathogenicity. J. Clin. Microbiol. 55, 2234–2248. doi: 10.1128/JCM.00150-17
Eslami H., Shariatifar A., Rafiee E., Shiranian M., Salehi F., Hosseini S. S., et al. (2019). Decolorization and biodegradation of reactive Red 198 Azo dye by a new Enterococcus faecalis–Klebsiella variicola bacterial consortium isolated from textile wastewater sludge. World J. Microbiol. Biotechnol. 35, 38. doi: 10.1007/s11274-019-2608-y
Fan H. C. (2016). First report of banana soft rot caused by klebsiella variicola in China. Plant Dis. 100, 517–517. doi: 10.1094/PDIS-05-15-0586-PDN
Farzana R., Jones L. S., Rahman M. A., Andrey D. O., Sands K., Portal E., et al. (2019). Outbreak of hypervirulent multidrug-resistant klebsiella variicola causing high mortality in neonates in Bangladesh. Clin. Infect. Dis. 68, 1225–1227. doi: 10.1093/cid/ciy778
Feng H., Fu R., Hou X., Lv Y., Zhang N., Liu Y., et al. (2021). Chemotaxis of beneficial rhizobacteria to root exudates: the first step towards root-microbe rhizosphere interactions. Int. J. Mol. Sci. 22, 6655. doi: 10.3390/ijms22136655
Fouts D. E., Tyler H. L., Deboy R. T., Daugherty S., Ren Q., Badger J. H., et al. (2008). Complete genome sequence of the N2-fixing broad host range endophyte klebsiella pneumoniae 342 and virulence predictions verified in mice. PloS Genet. 4, e1000141. doi: 10.1371/journal.pgen.1000141
Franz C. M., Huch M., Abriouel H., Holzapfel W., Galvez A. (2011). Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 151, 125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014
Franz C. M. A. P., Muscholl-Silberhorn A. B., Yousif N. M. K., Vancanneyt M., Swings J., Holzapfel W. H. (2001). Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl. Environ. Microbiol. 67, 4385–4389. doi: 10.1128/AEM.67.9.4385-4389.2001
Fulton J. C., Klein J. M., Bec S., Fayette J., Garrett K. A., Jones J. B., et al. (2020). Draft genome sequences of plant-pathogenic klebsiella variicola strains isolated from plantain in Haiti. Microbiol. Resour. Announc. 9, e00336–e00320. doi: 10.1128/MRA.00336-20
Garza-Ramos U., Silva-Sánchez J., Martínez-Romero E., Tinoco P., Pina-Gonzales M., Barrios H., et al. (2015). Development of a Multiplex-PCR probe system for the proper identification of Klebsiella variicola. BMC Microbiol. 15, 64. doi: 10.1186/s12866-015-0396-6
Ghazi A., Atia E., Elsakhawy T. (2021). Evaluation of an endophytic plant growth-promoting bacterium, Klebsiella variicola, in mitigation of salt stress in tuberose (Polianthes tuberosa L.). J. Hortic. Sci. Biotechnol. 96, 770–782. doi: 10.1080/14620316.2021.1926343
Gómez M., Valverde A., Campo R. D., Rodríguez J. M., Maldonado-Barragán A. (2021). Phenotypic and molecular characterization of commensal, community-acquired and nosocomial klebsiella spp. Microorganisms 9, 2344. doi: 10.3390/microorganisms9112344
Gorrie C. L., Mirceta M., Wick R. R., Edwards D. J., Thomson N. R., Strugnell R. A., et al. (2017). Gastrointestinal carriage is a major reservoir of klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208–215. doi: 10.1093/cid/cix270
Gorrie C. L., Mirceta M., Wick R. R., Judd L. M., Wyres K. L., Thomson N. R., et al. (2018). Antimicrobial-resistant klebsiella pneumoniae carriage and infection in specialized geriatric care wards linked to acquisition in the referring hospital. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc Am. 67, 161–170. doi: 10.1093/cid/ciy027
Goto M., Al-Hasan M. N. (2013). Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin. Microbiol. Infect. 19, 501–509. doi: 10.1111/1469-0691.12195
Guerrieri M. C., Fiorini A., Fanfoni E., Tabaglio V., Cocconcelli P. S., Trevisan M., et al. (2021). Integrated genomic and greenhouse assessment of a novel plant growth-promoting rhizobacterium for tomato plant. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.660620
Guo Y., Zhai Y., Zhang Z., Li D., Wang Z., Li J., et al. (2018). Complete genomic analysis of a kingdom-crossing klebsiella variicola isolate. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02428
Hanchi H., Mottawea W., Sebei K., Hammami R. (2018). The genus enterococcus: between probiotic potential and safety concerns-an update. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01791
Holt K. E., Wertheim H., Zadoks R. N., Baker S., Whitehouse C. A., Dance D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Human Microbiome Project Data Portal (2013). Available online at: https://portal.hmpdacc.org/.
Imai K., Ishibashi N., Kodana M., Tarumoto N., Sakai J., Kawamura T., et al. (2019). Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study, Japan 2014–2017. BMC Infect. Dis. 19, 946. doi: 10.1186/s12879-019-4498-x
Jochum L., Stecher B. (2020). Label or concept – what is a pathobiont? Trends Microbiol. 28, 789–792. doi: 10.1016/j.tim.2020.04.011
Kabir M. M., Fakhruddin A. N. M., Chowdhury M. A. Z., Pramanik M. K., Fardous Z. (2018). Isolation and characterization of chromium(VI)-reducing bacteria from tannery effluents and solid wastes. World J. Microbiol. Biotechnol. 34, 126. doi: 10.1007/s11274-018-2510-z
Kaper J. B., Nataro J. P., Mobley H. L. T. (2004). Pathogenic escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Kiley J. L., Mende K., Beckius M. L., Kaiser S. J., Carson M. L., Lu D., et al. (2021). Resistance patterns and clinical outcomes of Klebsiella pneumoniae and invasive Klebsiella variicola in trauma patients. PloS One 16, e0255636. doi: 10.1371/journal.pone.0255636
Kim A.-Y., Shahzad R., Kang S.-M., Seo C.-W., Park Y.-G., Park H.-J., et al. (2017). IAA-producing Klebsiella variicola AY13 reprograms soybean growth during flooding stress. J. Crop Sci. Biotechnol. 20, 235–242. doi: 10.1007/s12892-017-0041-0
Kitz R., Martens U., Zieseniß E., Enck P., Rose M. (2012). Probiotic E.faecalis — adjuvant therapy in children with recurrent rhinosinusitis. Cent. Eur. J. Med. 7, 362–365. doi: 10.2478/s11536-011-0160-8
Krawczyk B., Wityk P., Gałęcka M., Michalik M. (2021). The many faces of enterococcus spp.—Commensal, probiotic and opportunistic pathogen. Microorganisms 9, 1900. doi: 10.3390/microorganisms9091900
Kroon S. J., Ravel J., Huston W. M. (2018). Cervicovaginal microbiota, women’s health, and reproductive outcomes. Fertil. Steril. 110, 327–336. doi: 10.1016/j.fertnstert.2018.06.036
Kusale S. P., Attar Y. C., Sayyed R. Z., Enshasy H. E., Hanapi S. Z., Ilyas N., et al. (2021a). Inoculation of klebsiella variicola alleviated salt stress and improved growth and nutrients in wheat and maize. Agronomy 11, 927. doi: 10.3390/agronomy11050927
Kusale S. P., Attar Y. C., Sayyed R. Z., Malek R. A., Ilyas N., Suriani N. L., et al. (2021b). Production of Plant Beneficial and Antioxidants Metabolites by Klebsiella variicola under Salinity Stress. Molecules 26, 1894. doi: 10.3390/molecules26071894
Lam M. M. C., Wick R. R., Watts S. C., Cerdeira L. T., Wyres K. L., Holt K. E. (2021). A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12 4188. doi: 10.1038/s41467-021-24448-3
Liu W., Wang Q., Hou J., Tu C., Luo Y., Christie P. (2016). Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci. Rep. 6, 26710. doi: 10.1038/srep26710
LMG 23571 (2004). Available online at: https://bccm.belspo.be/catalogues/lmg-strain-details?NUM=23571&COLTYPE=&LIST1=STRNUM&TEXT1=23571&LIST2=SPECIES&TEXT2=&LIST3=ORIGSUBST&TEXT3=&LIST4=ORIGIN&TEXT4=&LIST5=ALL%20FIELDS&TEXT5=&CONJ=OR&RANGE=20 (Accessed March 26, 2024).
Loganathan M., Thangavelu R., Pushpakanth P., Kalimuthu M., Ramesh R., Selvarajan R., et al. (2021). First report of rhizome rot of banana caused by Klebsiella variicola in India. Plant Dis. 105, 2011. doi: 10.1094/PDIS-10-20-2316-PDN
Long S. W., Linson S. E., Saavedra M. O., Cantu C., Davis J. J., Brettin T., et al. (2017). Whole-Genome Sequencing of Human Clinical Klebsiella pneumoniae Isolates Reveals Misidentification and Misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2, e00290–e00217. doi: 10.1128/mSphereDirect.00290-17
Maatallah M., Vading M., Kabir M. H., Bakhrouf A., Kalin M., Nauclér P., et al. (2014). Klebsiella variicola Is a Frequent Cause of Bloodstream Infection in the Stockholm Area, and Associated with Higher Mortality Compared to K. pneumoniae. PloS One 9, e113539. doi: 10.1371/journal.pone.0113539
Malik V. (2023). First report of Klebsiella Leaf Streak on Sorghum Caused by Klebsiella variicola in Haryana, India. Plant Dis. 107, 2215. doi: 10.1094/PDIS-09-22-2200-PDN
Martin R. M., Bachman M. A. (2018). Colonization, infection, and the accessory genome of klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00004
Martin R. M., Cao J., Brisse S., Passet V., Wu W., Zhao L., et al. (2016). Molecular epidemiology of colonizing and infecting isolates of klebsiella pneumoniae. mSphere 1, e00261–e00216. doi: 10.1128/mSphere.00261-16
Martínez-Romero E., Rodríguez-Medina N., Beltrán-Rojel M., Silva-Sánchez J., Barrios-Camacho H., Pérez-Rueda E., et al. (2017a). Genome misclassification of Klebsiella variicola and Klebsiella quasipneumoniae isolated from plants, animals and humans. Salud Pública México 60, 56. doi: 10.21149/8149
Martínez-Romero E., Rodríguez-Medina N., Beltrán-Rojel M., Toribio-Jiménez J., Garza-Ramos U. (2017b). Klebsiella variicola and Klebsiella quasipneumoniae with capacity to adapt to clinical and plant settings. Salud Pública México 60, 29. doi: 10.21149/8156
Masse J., Dufour S., Archambault M. (2020). Characterization of Klebsiella isolates obtained from clinical mastitis cases in dairy cattle. J. Dairy Sci. 103, 3392–3400. doi: 10.3168/jds.2019-17324
Morgado S., Fonseca E., Vicente A. C. (2022). Genomics of klebsiella pneumoniae species complex reveals the circulation of high-risk multidrug-resistant pandemic clones in human, animal, and environmental sources. Microorganisms 10, 2281. doi: 10.3390/microorganisms10112281
Nava-Faustino G., Ramírez-Rojas S., Francisco-Palemón A., Díaz D. O., Forero-Forero Á.V., Toribio-Jiménez J. (2022). In vivo translocation of Klebsiella variicola PB02 and Klebsiella quasipneumoniae HPA43 in fruits of Solanum lycopersicum cultivar DT-22. Rev. Mex. Cienc. Agrícolas 13, 799–811. doi: 10.29312/remexca.v13i5.2880
Ohama Y. (2022). Accurate identification of klebsiella variicola by MALDI-TOF mass spectrometry in clinical microbiology laboratories. Microbiol. Spectr. 10, e02844–e02822. doi: 10.1128/spectrum.02844-22
Pan Z., Wu Y., Zhai Q., Tang Y., Liu X., Xu X., et al. (2023). Immobilization of bacterial mixture of Klebsiella variicola FH-1 and Arthrobacter sp. NJ-1 enhances the bioremediation of atrazine-polluted soil environments. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1056264
Pathak P., Trilligan C., Rapose A. (2014). Bifidobacterium–friend or foe? A case of urinary tract infection with Bifidobacterium species. BMJ Case Rep. 2014, bcr2014205122–b. doi: 10.1136/bcr-2014-205122
Pendleton J. N., Gorman S. P., Gilmore B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 11, 297–308. doi: 10.1586/eri.13.12
Piepenbrock E., Higgins P. G., Wille J., Xanthopoulou K., Zweigner J., Jahn P., et al. (2020). Klebsiella variicola causing nosocomial transmission among neonates – an emerging pathogen? J. Méd. Microbiol. 69, 396–401. doi: 10.1099/jmm.0.001143
Pinto-Tomas A. A. (2009). Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326, 1120–1123. doi: 10.1126/science.1173036
Podder M. P., Rogers L., Daley P. K., Keefe G. P., Whitney H. G., Tahlan K. (2014). Klebsiella species associated with bovine mastitis in Newfoundland. PloS One 9, e106518. doi: 10.1371/journal.pone.0106518
Potter R. F., Lainhart W., Twentyman J., Wallace M. A., Wang B., Burnham C. A., et al. (2018). Population structure, antibiotic resistance, and uropathogenicity of klebsiella variicola. mBio 9, e02481–e02418. doi: 10.1128/mBio.02481-18
Probiotics - Health Professional Fact Sheet (2023). Available online at: https://ods.od.nih.gov/factsheets/Probiotics-HealthProfessional/#:~:text=The%20seven%20core%20genera%20of,Enterococcus%2C%20Escherichia%2C%20and%20Bacillus.
Probiotics Market Size (2021). Available online at: https://www.grandviewresearch.com/industry-analysis/probiotics-market.
Qin Y., Xie X. Q., Khan Q., Wei J. L., Sun A. N., Su Y. M., et al. (2022). Endophytic nitrogen-fixing bacteria DX120E inoculation altered the carbon and nitrogen metabolism in sugarcane. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1000033
Raffelsberger N., Hetland M. A. K., Svendsen K., Småbrekke L., Löhr I. H., Andreassen L. L. E., et al. (2021). Gastrointestinal carriage of Klebsiella pneumoniae in a general adult population: a cross-sectional study of risk factors and bacterial genomic diversity. Gut Microbes 13, 1939599. doi: 10.1080/19490976.2021.1939599
Ravel J., Gajer P., Abdo Z., Schneider G. M., Koenig S. S. K., Mcculle S. L., et al. (2011). Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. 108, 4680–4687. doi: 10.1073/pnas.1002611107
Rizo J., Rogel M. A., Guillen D., Wacher C., Martinez-Romero E., Encarnacion S., et al. (2020). Nitrogen fixation in pozol, a traditional fermented beverage. Appl. Environ. Microbiol. 86. doi: 10.1128/AEM.00588-20
Robbins A. M. (2021). “The Role of Microbiology in Sustainable Development,” in American Society for Microbiology. Available at: https://asm.org/Articles/2021/October/The-Role-of-Microbiology-in-Sustainable-Developmen.
Rodríguez-Medina N., Barrios-Camacho H., Duran-Bedolla J., Garza-Ramos U. (2019). Klebsiella variicola: an emerging pathogen in humans. Emerg. Microbes Infect. 8, 973–988. doi: 10.1080/22221751.2019.1634981
Rosales-Bravo H., Morales-Torres H. C., Vázquez-Martínez J., Molina-Torres J., Olalde-Portugal V., Partida-Martínez L. P. (2017). Novel consortium of Klebsiella variicola and Lactobacillus species enhances the functional potential of fermented dairy products by increasing the availability of branched-chain amino acids and the amount of distinctive volatiles. J. Appl. Microbiol. 123, 1237–1250. doi: 10.1111/jam.2017.123.issue-5
Rosenblueth M. (2004). Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst. Appl. Microbiol. 27, 27–35. doi: 10.1078/0723-2020-00261
Rossi F., Amadoro C., Colavita G. (2019). Members of the lactobacillus genus complex (LGC) as opportunistic pathogens: A review. Microorganisms 7, 126. doi: 10.3390/microorganisms7050126
Sansinenea E. (2022). Microorganisms: a problem or a solution in our current life? Futur. Microbiol. 17, 1433–1435. doi: 10.2217/fmb-2021-0294
Semedo T., Santos M. A., Martins P., Lopes M. F. S., Marques J. J. F., Tenreiro R., et al. (2003). Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J. Clin. Microbiol. 41, 2569–2576. doi: 10.1128/JCM.41.6.2569-2576.2003
Souza R. D., Ambrosini A., Passaglia L. M. P. (2015). Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 38, 401–419. doi: 10.1590/S1415-475738420150053
Stuivenberg G. A., Burton J. P., Bron P. A., Reid G. (2022). Why are bifidobacteria important for infants? Microorganisms 10, 278. doi: 10.3390/microorganisms10020278
Temme K. (2022). History in the making: A reflection of innovation, growth and endless opportunities. Available online at: https://blog.pivotbio.com/history-in-the-making#:~:text=Exceeding%20%2450%20million%20in%20sales,a%20replacement%20for%20synthetic%20nitrogen.
Turroni F., Milani C., Duranti S., Lugli G. A., Bernasconi S., Margolles A., et al. (2020). The infant gut microbiome as a microbial organ influencing host well-being. Ital. J. Pediatr. 46, 16. doi: 10.1186/s13052-020-0781-0
Vives-Peris V., Ollas C., Gomez-Cadenas A., Perez-Clemente R. M. (2020). Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17. doi: 10.1007/s00299-019-02447-5
Wang D. (2022). Insights into Genomic Evolution and the Potential Genetic Basis of Klebsiella variicola subsp. variicola ZH07 Reveal Its Potential for Plant Growth Promotion and Autotoxin Degradation. Microbiol. Spectr. 10, e00846–e00822. doi: 10.1128/spectrum.00846-22
Watanabe N., Watari T., Otsuka Y., Yamagata K., Fujioka M. (2022). Clinical characteristics and antimicrobial susceptibility of Klebsiella pneumoniae, Klebsiella variicola and Klebsiella quasipneumoniae isolated from human urine in Japan. J. Méd. Microbiol. 71. doi: 10.1099/jmm.0.001546
Wei T., Li X., Li H., Gao H., Guo J., Li Y., et al. (2022). The potential effectiveness of mixed bacteria-loaded biochar/activated carbon to remediate Cd, Pb co-contaminated soil and improve the performance of pakchoi plants. J. Hazard. Mater. 435, 129006. doi: 10.1016/j.jhazmat.2022.129006
Wei C.-Y., Lin L., Luo L.-J., Xing Y.-X., Hu C.-J., Yang L.-T., et al. (2013). Endophytic nitrogen-fixing Klebsiella variicola strain DX120E promotes sugarcane growth. Biol. Fertil. Soils 50, 657–666. doi: 10.1007/s00374-013-0878-3
Wen A., Havens K. L., Bloch S. E., Shah N., Higgins D. A., Davis-Richardson A. G., et al. (2021). Enabling biological nitrogen fixation for cereal crops in fertilized fields. ACS Synth. Biol. 10, 3264–3277. doi: 10.1021/acssynbio.1c00049
Wiles T. J., Guillemin K. (2019). The other side of the coin: what beneficial microbes can teach us about pathogenic potential. J. Mol. Biol. 431, 2946–2956. doi: 10.1016/j.jmb.2019.05.001
Yang L., Yang K. (2020). Biological function of Klebsiella variicola and its effect on the rhizosphere soil of maize seedlings. PeerJ 8, e9894. doi: 10.7717/peerj.9894
Younes J. A., Lievens E., Hummelen R., van der Westen R., Reid G., Petrova M. I. (2018). Women and their microbes: the unexpected friendship. Trends Microbiol. 26, 16–32. doi: 10.1016/j.tim.2017.07.008
Yu R., Man M., Yu Z., Wu X., Shen L., Liu Y., et al. (2021). A high-efficiency Klebsiella variicola H12-CMC-FeS@biochar for chromium removal from aqueous solution. Sci. Rep. 11, 6611. doi: 10.1038/s41598-021-85975-z
Keywords: safety assessment, commensal microbiota, biotechnology, microbiome dynamics, diazotroph, biofertilizer, sustainable agriculture, Klebsiella variicola
Citation: Marian C, Sanjar F, Maxwell C, Sabitu F, Hubbard N, Ratib N, Vassilev N and Sansinenea E (2024) A paradigm for the contextual safety assessment of agricultural microbes: a closer look at Klebsiella variicola. Front. Ind. Microbiol. 2:1412302. doi: 10.3389/finmi.2024.1412302
Received: 13 April 2024; Accepted: 12 July 2024;
Published: 30 July 2024.
Edited by:
Boonfei Tan, Singapore-MIT Alliance for Research and Technology (SMART), SingaporeReviewed by:
Leonardo Magalhães Cruz, Federal University of Paraná, BrazilCopyright © 2024 Marian, Sanjar, Maxwell, Sabitu, Hubbard, Ratib, Vassilev and Sansinenea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christina Marian, Y21hcmlhbkBwaXZvdGJpby5jb20=
†These authors share first authorship
‡ORCID: Christina Marian, orcid.org/0009-0002-4783-374X
Fatemeh Sanjar, orcid.org/0000-0002-8749-9910
Nikolay Vassilev, orcid.org/0000-0002-6144-2940
Estibaliz Sansinenea, orcid.org/0000-0002-1593-1890
 Christina Marian
Christina Marian Fatemeh Sanjar1†‡
Fatemeh Sanjar1†‡ Nikolay Vassilev
Nikolay Vassilev Estibaliz Sansinenea
Estibaliz Sansinenea