- 1Department of Life, Health and Environmental Sciences, University of L’Aquila, L’Aquila, Italy
- 2SPAA Srl, Città Sant’Angelo, Italy
- 3Agroqualis Srl, Academic Spin off of University of L’Aquila, L’Aquila, Italy
Microbial-based inoculants for agricultural use consist of different strains. The consortia production process involves growing pure cultures separately and combining them in the appropriate ratio. However, the inclusion of multiple strains in the formulation increases production costs. By developing co-culture fermentations, it is possible to create consortia with the correct bacterial charge. This study aims to develop a cost-effective co-culture approach for producing an inoculum that includes the appropriate ratio of four Gram-negative bacteria, i.e., Azospirillum brasilense, Burkholderia ambifaria, Gluconacetobacter diazotrophicus, and Herbaspirillum seropedicae. The specific growth rates of strains were studied using the T4 medium, previously optimized for their culture. The co-fermentation process was optimized in 500 mL flasks to attain an equivalent density of 9.7-10 Log CFU mL-1. Then, it was successfully scaled up to a 5 L bioreactor, obtaining an equivalent density of 9.7-9.9 CFU mL-1. This first co-formulation of a four multistrain consortium formed by Gram-negative plant growth-promoting bacteria pave the road for future evaluations of other products useful for sustainable agriculture.
Introduction
Using bacterial consortia is a sustainable and promising tool to improve the sustainability of fertile land while achieving high production rates (Duncker et al., 2021; Seenivasagan and Babalola, 2021). Combining two or more strains with plant growth-promoting (PGP) traits to enhance the inoculum’s efficacy is a proven method (Deter and Lu, 2022). The bacterial consortium consisting of Herbaspirillum seropedicae Z67, Gluconacetobacter diazotrophicus Pal5, Azospirillum brasilense Cd, and Burkholderia ambifaria PHP7 has been shown to have positive biostimulating effects on various crops, including tomato, carrots, hemp, Apennines genepì, and ancient Triticum genotypes (Del Gallo et al., 2010; Botta et al., 2013; Pagnani et al., 2018; Pagnani et al., 2020; Pellegrini et al., 2020a; Pellegrini et al., 2021a; Pellegrini et al., 2021b).
The favorable results achieved with this consortium are due to the synergy of various plant-growth-promoting qualities found in these strains. A. brasilense is a well-known species of PGPB and has great potential for biofertilization (Bashan and De-Bashan, 2010). Azospirillum spp. and Gluconacetobacter spp. are known to enhance plant growth through various processes. Azospirillum spp. are involved in phytohormone synthesis, N2-fixation, mineral mobilization, synthesis of small molecules and enzymes, root system proliferation, increased membrane activity, and mineral and water uptake (Bashan, 1986; James et al., 2001; Logeshwarn et al., 2011; Brígido et al., 2015; Suleman et al., 2018; Silva et al., 2019). Gluconacetobacter spp. are also linked to N2-fixation, synthesis of siderophores and phytohormones, and the solubilization of inorganic forms of zinc and phosphorus (James et al., 2001). Herbaspirillum spp. and Burkholderia species assist in plant growth by producing phytohormones, synthesizing siderophores, and N2-fixing (Olivares et al., 1997; Tawfik et al., 2010; Xu et al., 2020). They also reduce stressful ethylene levels through 1-aminocyclopropane-1-carboxylate (ACC) deaminase. Furthermore, the biocontrol potential against bacterial and fungal phytopathogens has been documented for each strain (Zahir et al., 2009; Djebaili et al., 2021).
The manufacturing process of this consortium involves growing pure cultures separately and then combining them in the appropriate ratio (Bashan et al., 2014). Researchers and enterprises commonly use this technique. However, this procedure increases operational and production costs by considering multiple strains in the formulation and their individual fermentations. Application of multi-strain fermentation techniques has gained relevance in recent years in an effort to lower total bioprocess costs (Jangra et al., 2016). The co-culture systems could overcome the limitations of monocultures or consortia with the added advantages of exploring allelopathic interactions in several biotechnological applications due to their versatility, robustness, and complexity (Hays et al., 2015). Furthermore, several studies underlined how the complexity of co-culturing fermentation could increase its resilience against contamination (Hathi et al., 2021). By creating a co-culture fermentation, costs can be saved by eliminating the need for several processes. Creating multistrain inoculants with the appropriate ratios of each strain is a complex process that requires extensive research (Hussain et al., 2022; Sobhi et al., 2023). Previous attempts at mixed-culture bioprocesses have utilized this technique to reduce production costs (Kapoore et al., 2022). However, microbial strains interact in various ways, such as mutualism, competition, parasitism, commensalism, and predation (Wu et al., 2023). Therefore, understanding the population dynamics and interactions between microbial strains during growth is crucial for co-culture fermentation development.
This work aimed to develop a biomass-producing process that incorporates a co-culture of the four mentioned PGPB strains. The co-fermentation was developed on the T4 medium, previously designed to maximize the four-strain growth. The co-fermentation procedure was studied in 500 mL flasks. Subsequently, the fermentation process was successfully scaled up to a 5-L bioreactor.
Method
Microbial strains and inoculum preparation
The four bacteria studied are G. diazotrophicus strain Pal5, H. seropedicae strain Z67, B. ambifaria gv VII strain PHP7, and A. brasilense strain Cd. These strains are part of the collection of the Agricultural Microbiology Laboratory of the University of L’Aquila. The bacteria were cultured on T4 medium at 30°C previously designed by Botta et al (Botta et al., 2013). The density of the strain was established by Densimat Densitometer (Biomérieux).
Co-culture design
Pure cultures for each strain were obtained by adding 2% w/v of the cell biomass to 250 mL of T4 media in 500 mL flasks and by shaking at 130 rpm at 30°C. The growth of pure cultures was studied for 60 hours with periodic sampling (14 samplings in total). Samples were centrifuged for 15 minutes at 20°C at 8000 g, washed, and resuspended in saline solution (0.9%) to establish bacterial density. The number of colony-forming units per milliliter (CFU mL-1) was determined using the plate counting method, with samples being serially diluted and plated on modified Okon medium - OK (Martinez-Drets et al., 1984), Jensen’s Nitrogen-Fixing bacteria - J-NFb (Döbereiner, 1992), Pseudomonas cepacia, azelaic acid, tryptamine - PCAT (Burbage and Sasser, 1982), and Liquid Glucose Ivo Pernambuco - LGI-P (Baldani et al., 2014), specific for A. brasilense, H. seropedicae, B. ambifaria, and G. diazotrophicus, respectively. The results were graphically displayed, and the growth curves were compared. Different scalar addition times were hypothesized to inoculate the strains and tested in 500 mL flasks. Experiment A was carried out to test the possibility of co-culturing the same strains together for 48 hours. Experiment B was carried out with inoculations at 0 (for G. diazotrophicus), 24 (for A. brasilense), 48 (for H. seropedicae), and 60 hours (B. ambifaria). Experiment C was set with inoculations at 0 (for G. diazotrophicus), 24 (for A. brasilense and H. seropedicae), and 60 hours (B. ambifaria). Experiment D was performed with inoculations at 0 (for G. diazotrophicus), 48 (for A. brasilense and H. seropedicae), and 60 hours (B. ambifaria). All inoculations were performed with standardized cultures to obtain a final density of 6 Log CFU mL-1.
5 L bioreactor scale up
Batch fermentation was conducted using a 5 L bioreactor (Biostat® B). The fermentation was obtained with an agitation speed of 150 rpm, continuous air-sparging, and at 30°C using a T4 medium volume of 2.5 L. The pH was maintained at 6.8 ± 0.2 with the pH controller unit of the bioreactor and NaOH and HCl 0.1 M solutions. A peristaltic pump was used to perform inoculations as described in Experiment D (for which the best results were obtained). Samples were regularly taken to evaluate CFU mL-1 as previously described.
Statistical analysis
Trails and evaluations were repeated five times (n = 5 independent experiments) and values were expressed as mean ± standard deviations. After collection, the dataset was analyzed for outlier’s presence and distribution. Results of the densities obtained from the co-culture experiments, with no normal distribution, were analyzed by Kruskal-Wallis, followed by Conovan-Iman multiple comparisons of the mean. 5L bioreactor data, with a normal distribution, were analyzed by ANOVA, followed by Tukey’s post-hoc test. Data processing was carried out using XLSTAT 2016 software.
Results
The four-strain growth behavior in the T4 medium was studied to keep the goal of a combined process. Figure 1 displays the growth cultures obtained for each strain. A first comparison shows that the growth curves for A. brasilense and H. seropedicae have very similar kinetics, whereas B. ambifaria has a more rapid growth. For G. diazotrophicus the growth is slower. In particular, A. brasilense and H. seropediace start the exponential phase after a lag phase of 5 hours, which lasts up to 24 hours. B. ambifaria starts the exponential phase after 2 hours and lasts up to 12 hours. G. diazotrophicus has a slow adaptation phase and starts the exponential phase after 12 hours, that lasts up to 48 hours. For all the strains, the maximum density recorded was 10 Log CFU mL-1.
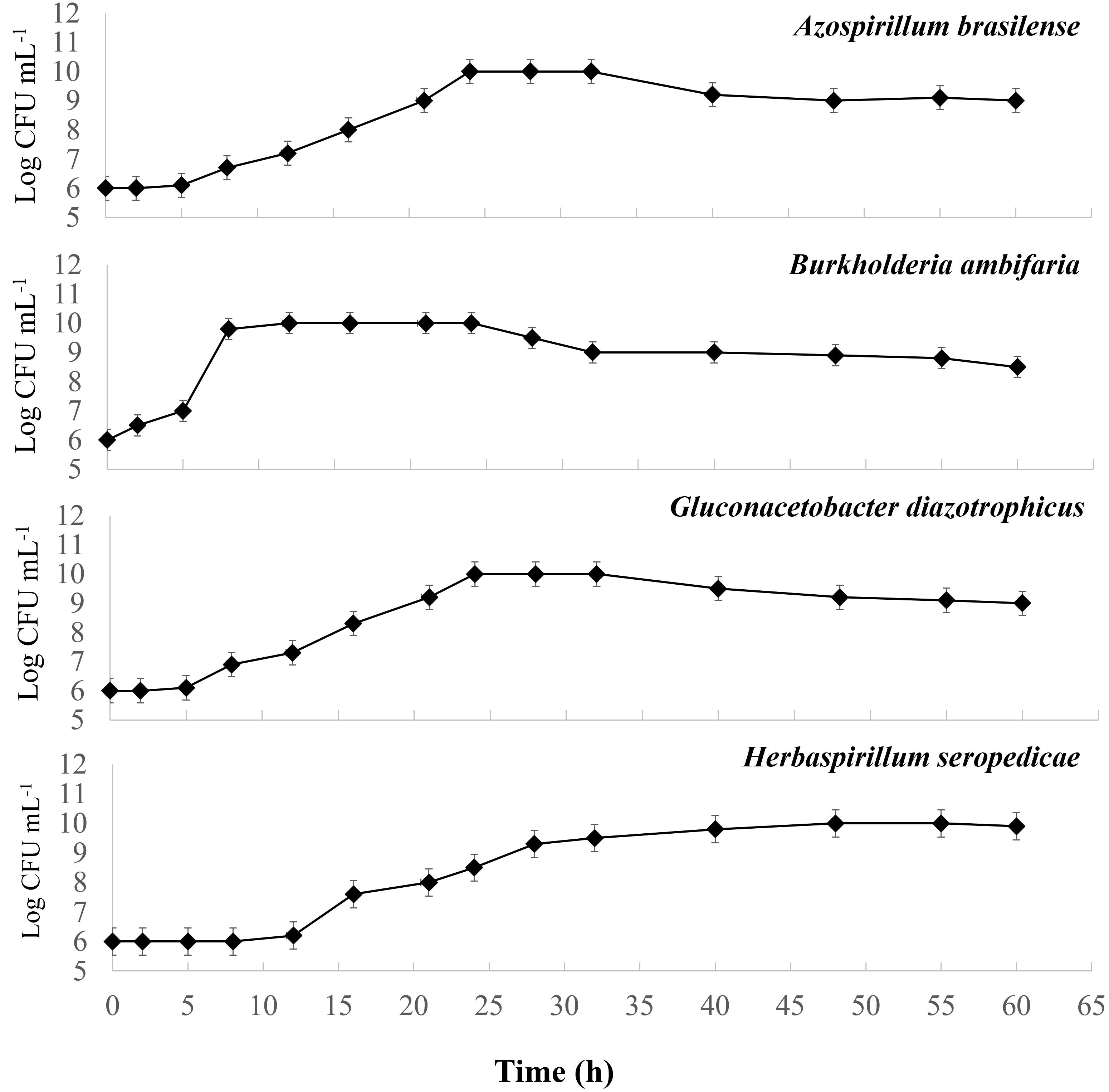
Figure 1 Growth curves of pure cultures of Azospirillum brasilense, Burkholderia ambifaria, Gluconacetobacter diazotrophicus, and Herbaspirillum seropedicae obtained in T4 medium for 60 hours.
Analyzing this behavior, it was hypothesized that co-cultures could only be grown by scalar inoculations. Therefore, three different experiments of scalar additions were set up (Experiments B-D). A control co-culture was established as control by adding all strains at time 0 (Experiment A). Figure 2 shows the results obtained for each experiment and the statistical analysis performed for comparison. From the counts performed for each strain, it was observed that the control culture only allowed the growth of the B. ambifaria strain up to 9.5 Log CFU mL-1 (after 12 hours), which appeared to significantly inhibit the growth of the other three strains (which obtained a maximum density of 107 CFU mL-1). In Experiment B and Experiment C good densities were obtained for G. diazotrophicus and B. ambifaria (9 Log CFU mL-1). However, the other two strains were negatively influenced by these set up. The best results were obtained for Experiment D and at the end of the co-fermentation each strain obtained a final density of 10 Log CFU mL-1 on average.
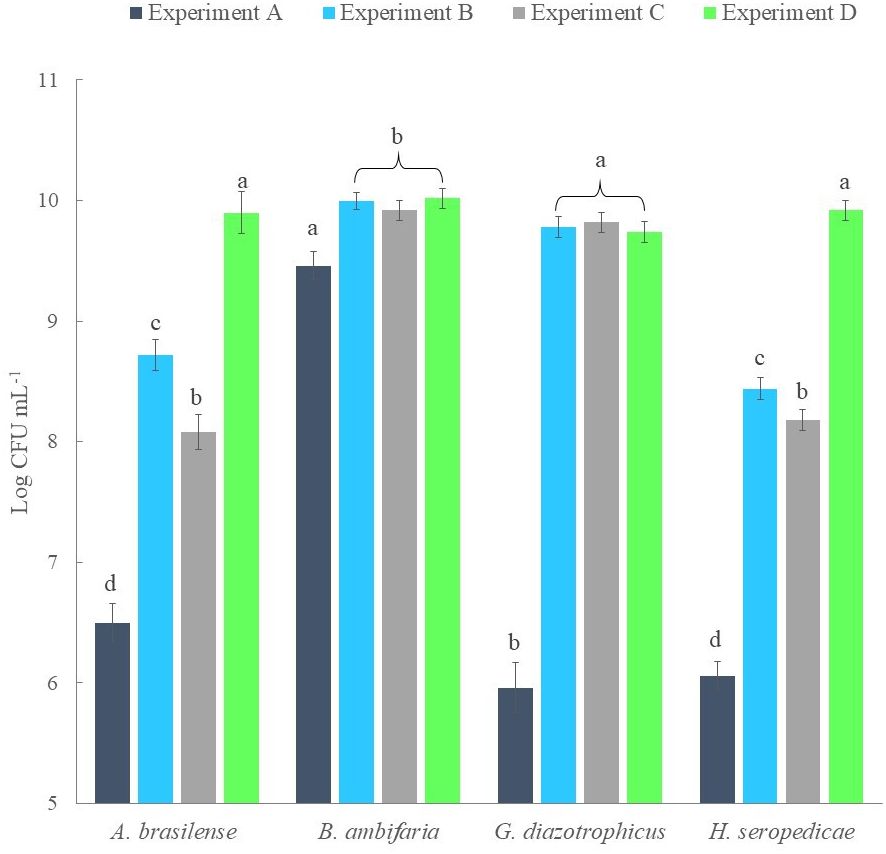
Figure 2 Comparison of the strain bacterial density obtained from co-culture experiments and established at the end of each flask scale fermentation.
Experiment D was also scaled up to a 5 L bioreactor. Figure 3 shows the strain’s densities obtained after 80 hours of co-culture fermentation. The strains achieved a similar final density, comparable to that obtained with the 500 mL flask scale (9.8 Log CFU mL-1). No significant changes in the culture medium were registered during the fermentations.
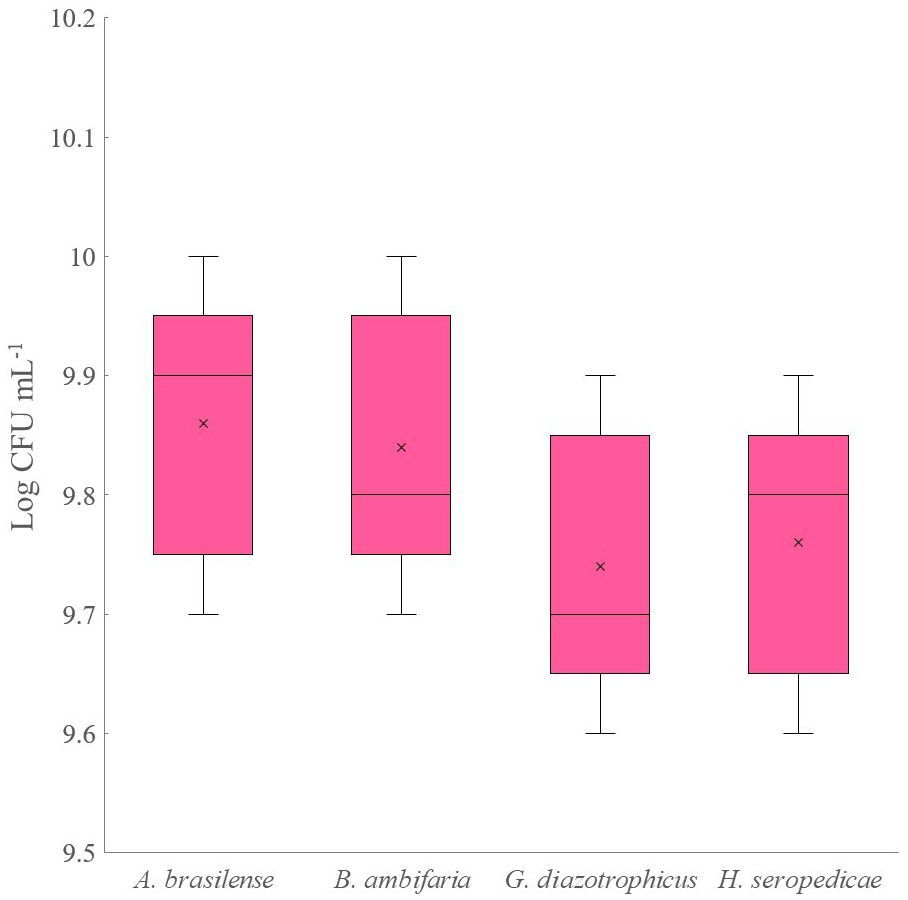
Figure 3 Four strains bacterial density obtained from co-culture experiment on 5-L scale bioreactor.
Discussion
In the first part of this work, we explored the growth behavior of four Gram-negative bacteria. The observed growth rates are in line with the nature of each bacteria. Studying a single fermentation process is crucial to comprehensively characterize the strain’s bacteriological characteristics and metabolic demands. The strains analyzed in this work have been the subject of numerous studies over the years, which have determined their metabolic demands for optimal development (Eskew et al., 1977; Baldani et al., 1986; Stephan et al., 1991; Coenye et al., 2001; Canini et al., 2006). In addition, Botta et al (Botta et al., 2013). developed a culture medium suitable for all four bacteria. These findings are of great importance for experimental research with potential industrial applications. These aspects may influence a company’s decision to scale up a formulation for industrial use.
After completing the initial stage, it is recommended to carefully analyze the growth process and consider potential bioprocesses that may help reduce production costs. This analysis involves evaluating nutrient levels and the duration of the bioprocess. Moreover, the possible interactions among bacteria must be considered (Wu et al., 2023). Given the growth rates and interactions of these strains with other living organisms, adding the strains in a scalar manner was suggested, beginning with the slowest strain, G. diazotrophicus, and concluding with the one with the highest competitive potential, B. ambifaria. The slow growth of G. diazotrophicus is usually reported. Compared with other bacteria with similar sugar assimilation pathways, it is usually described as slower (Restrepo et al., 2023). In fact, when the cultures were added during G. diazotrophicus growth, this resulted in lower charges, probably given to competition behaviors established among strains. B. ambifaria, instead, is a fast-growing bacterium with competitive behavior by secondary metabolites production (e.g., antibiotics and other secondary metabolites) (Pal et al., 2022). This aspect could be considered as one of the most relevant limits of co-culturing fermentation, mainly related to the competition for nutrients and the production of secondary compounds that interfere with the growth of other organisms (Santos et al., 2014). The different trials showed a good compatibility of A. brasilense and H. seropedicae, which grew together without any interference. This compatibility is probably due to the A. brasilense ability of to easily adapt its metabolism and organization of the cells to respond to environmental changes (Bible et al., 2015). This ability could be essential to facilitate the creation of effective synergism and growth rated of the strains during the early exponential phase. Thus, understanding the ideal combination of growth medium, culture condition, and fermentation technique is essential for promoting development, stability, and preservation of functional activity through downstream procedures and storage (Rodrigues et al., 2006).
Microorganisms interact with each other in various ways. These interactions can be positive, negative, or neutral in simple ecological models. Any close, long-term biological interaction between two different biological organisms, either positive or negative, is called symbiosis (Hector et al., 2022). In both symbioses and transitional associations, interactions between two units can be classified as mutualism, commensalism, amensalism, competition, and predation/parasitism, depending on the directionality of the interaction (Wu et al., 2023). Interactions can become more complex and highly dynamic in systems with multiple units due to interspecific, intraspecific, and environmental factors. As the number of entities involved increases, community complexity also increases due to the combined growth in the number and type of pairwise interactions (Duncker et al., 2021). A third unit can alter the strength or nature of a pairwise association, which a fourth unit can further modify. These interactions, along with their types and intensity, can significantly impact a community’s stability and can be used to predict community composition over time (Bairey et al., 2016).
It is widely recognized that co-culturing microorganisms with different metabolic capabilities can have additive or synergistic interactions, with beneficial effects on the microbial community and, directly or indirectly, on plant growth and protection. For example, Pagnussat et al. (2016) found that the amount of biofilm produced by Azospirillum brasilense Sp245 significantly increased in co-culture with Pseudomonas protegens CHA0 (Pagnussat et al., 2016). Few authors focused on optimizing growth conditions of PGPBs strains in co-culture. Bagheri et al. (2022) studied the interactions between Azospirillum oryzae NBT506 and Bacillus velezensis UTB96 in a co-culture system. Using the adjusted population of each strain (8 Log CFU mL-1 of A. oryzae NBT506 and 6 Log CFU mL-1 of B. velezensis UTB96), they observed the maximum growth efficiency (11 Log CFU mL-1) based on different preculture concentrations for both strains (Bagheri et al., 2022). Similar results were obtained by Masciarelli and collaborators, who tested Bradyrhizobium japonicum E109 strain and Bacillus amyloliquefaciens in a co-fermentation to enhance soybean nodulation (Masciarelli et al., 2014). These findings underline the bacterial activity and ability to increase their growth in an optimized co-culture bioreaction. Creating multi-microbial formulations can be valuable, providing economic and industrial benefits for microbial-based inoculants. This strategy can lead to a reduction in the number of bioprocesses required, resulting in lower material and energy costs and increased efficiency.
The selection of a co-culture is an important factor in optimizing economic resources for product development. It might be useful to enhance biomass yield in several biotechnological processes such as fermentation, biofuels, bioremediation, nutraceutical, and chemical production (Kapoore et al., 2022). Controlled co-culturing enables the synergistic utilization of metabolic pathways of participating microorganisms under industrial, reproducible, and controlled conditions. Significant energy and resource savings can be achieved during industrial production by defining optimal values for process parameters, substrate, and product contents (Bader et al., 2010). The ability to produce multiple strains in a single bioreaction process can result in lower process waste production and limit the need for multiple culture broths to obtain biomass. This approach can lead to less economic effort in management and a lower environmental impact of industrial production. By using fewer resources and producing less waste, higher environmental sustainability for the farm can be achieved (Rosero-Chasoy et al., 2021).
Our results provide a solid basis for developing a product that includes these microorganisms and pave the way for further research. As far as we know, no other studies have co-formulated a four multistrain consortium formed by Gram-negative PGPB. Given the large global market for these products and their increasing use, research in this area is very important (Pellegrini et al., 2020b; Chaudhary et al., 2023). Microbial fermentation is of utmost importance in the development of next-generation bioinoculants. Recent research breakthroughs have provided effective methods for optimizing the fermentation of endophytic microbes, from fundamental to scaling-up production. This progress represents cutting-edge agricultural, food, and nutrition technology, ushering in a new scientific era (Ganeshan et al., 2021). However, there is a lack of integration between scientific results and the industrial sector (Singh et al., 2024). Therefore, such studies should be encouraged. This initial research report sets the stage for future evaluations of any product on an industrial and commercial level, including bioprocessing and technoeconomic feasibility analysis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
BF: Data curation, Formal Analysis, Investigation, Visualization, Writing – original draft. FP: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft. RD: Formal Analysis, Investigation, Writing – original draft. DS: Conceptualization, Methodology, Validation, Writing – original draft. MD: Methodology, Validation, Writing – review & editing. MP: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research received private research funding from SPAA Srl.
Acknowledgments
We gratefully acknowledge the support of Dr. Filippo Vitulli and SPAA Srl for our research activities.
Conflict of interest
Author FP is employed by SPAA Srl., Authors MP CEO and DS Consultant are part of Agroqualis Srl - University of L’Aquila Academic Spin off.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constitute as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bader J., Mast-Gerlach E., Popović M. K., Bajpai R., Stahl U. (2010). Relevance of microbial coculture fermentations in biotechnology. J. Appl. Microbiol. 109, 371–387. doi: 10.1111/j.1365-2672.2009.04659.x
Bagheri N., Ahmadzadeh M., Mariotte P., Jouzani G. S. (2022). Behavior and interactions of the plant growth-promoting bacteria Azospirillum oryzae NBT506 and Bacillus velezensis UTB96 in a co-culture system. World J. Microbiol. Biotechnol. 38, 101. doi: 10.1007/s11274-022-03283-8
Bairey E., Kelsic E. D., Kishony R. (2016). High-order species interactions shape ecosystem diversity. Nat. Commun. 7, 12285. doi: 10.1038/ncomms12285
Baldani J. I., Baldani V. L. D., Seldin L., Dobereiner J. (1986). Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a Root-Associated Nitrogen-Fixing Bacterium. Int. J. Syst. Bacteriol 36, 86–93. doi: 10.1099/00207713-36-1-86
Baldani J. I., Reis V. M., Videira S. S., Boddey L. H., Baldani V. L. D. (2014). The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: a practical guide for microbiologists. Plant Soil 384, 413–431. doi: 10.1007/s11104-014-2186-6
Bashan Y. (1986). Migration of the Rhizosphere Bacteria Azospirillum brusilense and Pseudomonas fluorescens Towards Wheat Roots in the Soil. Microbiol. (N Y) 132, 3407–3414. doi: 10.1099/00221287-132-12-3407
Bashan Y., De-Bashan L. E. (2010). ““How the plant growth-promoting bacterium azospirillum promotes plant growth—A critical assessment,”,” in Advances in Agronomy. Ed. Sparks D. L. (Cambridge, MA, USA: Academic Press) 77–136. doi: 10.1016/S0065-2113(10)08002-8
Bashan Y., De-Bashan L. E., Prabhu S. R., Hernandez J.-P. (2014). Advances in plant growth-promoting bacterial inoculant technology: formulations and practical perspectives, (1998–2013). Plant Soil 378, 1–33. doi: 10.1007/s11104-013-1956-x
Bible A. N., Khalsa-Moyers G. K., Mukherjee T., Green C. S., Mishra P., Purcell A., et al. (2015). Metabolic adaptations of azospirillum brasilense to oxygen stress by cell-to-cell clumping and flocculation. Appl. Environ. Microbiol. 81, 8346–8357. doi: 10.1128/AEM.02782-15
Botta A. L., Santacecilia A., Ercole C., Cacchio P., Del Gallo M. (2013). In vitro and in vivo inoculation of four endophytic bacteria on Lycopersicon esculentum. N Biotechnol. 30, 666–674. doi: 10.1016/j.nbt.2013.01.001
Brígido C., Duan J., Glick B. R. (2015). ““Methods to study 1-aminocyclopropane-1-carboxylate (ACC) deaminase in plant growth-promoting bacteria,”,” in Handbook for Azospirillum (Springer International Publishing, Cham), 287–305. doi: 10.1007/978-3-319-06542-7_16
Burbage D. A., Sasser M. (1982). “A medium selective for Pseudomonas-cepacia,” in Phytopathology, vol. 706. (AMER PHYTOPATHOLOGICAL SOC 3340 PILOT KNOB ROAD, ST PAUL, MN 55121).
Canini A., Canuti L., Grilli Caiola M., Del Gallo M. (2006). Immunocytochemical characterisation of endophytic bacteriaAzospirillum brasilense, Herbaspirillum seropedicae, Burkholderia ambifaria andGluconacetobacter diazotrophicus. Ann. Microbiol. 56, 393–398. doi: 10.1007/BF03175039
Chaudhary P., Xu M., Ahamad L., Chaudhary A., Kumar G., Adeleke B. S., et al. (2023). Application of synthetic consortia for improvement of soil fertility, pollution remediation, and agricultural productivity: A review. Agronomy 13, 643. doi: 10.3390/agronomy13030643
Coenye T., Mahenthiralingam E., Henry D., LiPuma J. J., Laevens S., Gillis M., et al. (2001). Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51, 1481–1490. doi: 10.1099/00207713-51-4-1481
Del Gallo M., Santacecilia A., Bozzelli P., Centi V., Cacchio P., Ercole C., et al. (2010). Inoculation of four endophytic bacteria on lycopersicon esculentum and their antagonism towards some pathogenic fungus. J. Biotechnol. 150, 494–494. doi: 10.1016/j.jbiotec.2010.09.763
Deter H. S., Lu T. (2022). Engineering microbial consortia with rationally designed cellular interactions. Curr. Opin. Biotechnol. 76, 102730. doi: 10.1016/j.copbio.2022.102730
Djebaili R., Pellegrini M., Ercole C., Farda B., Kitouni M., Del Gallo M. (2021). Biocontrol of Soil-Borne Pathogens of Solanum lycopersicum L. and Daucus carota L. by Plant Growth-Promoting Actinomycetes: In Vitro and In Planta Antagonistic Activity. Pathogens 10, 1305. doi: 10.3390/pathogens10101305
Duncker K. E., Holmes Z. A., You L. (2021). Engineered microbial consortia: strategies and applications. Microb. Cell Fact 20, 211. doi: 10.1186/s12934-021-01699-9
Eskew D. L., Focht D. D., Ting I. P. (1977). Nitrogen fixation, denitrification, and pleomorphic growth in a highly pigmented Spirillum lipoferum. Appl. Environ. Microbiol. 34, 582–585. doi: 10.1128/aem.34.5.582-585.1977
Ganeshan S., Kim S. H., Vujanovic V. (2021). Scaling-up production of plant endophytes in bioreactors: concepts, challenges and perspectives. Bioresour Bioprocess 8, 63. doi: 10.1186/s40643-021-00417-y
Hathi Z., Mettu S., Priya A., Athukoralalage S., Lam T. N., Choudhury N. R., et al. (2021). Methodological advances and challenges in probiotic bacteria production: Ongoing strategies and future perspectives. Biochem. Eng. J. 176, 108199. doi: 10.1016/j.bej.2021.108199
Hays S. G., Patrick W. G., Ziesack M., Oxman N., Silver P. A. (2015). Better together: engineering and application of microbial symbioses. Curr. Opin. Biotechnol. 36, 40–49. doi: 10.1016/j.copbio.2015.08.008
Hector T. E., Hoang K. L., Li J., King K. C. (2022). Symbiosis and host responses to heating. Trends Ecol. Evol. 37, 611–624. doi: 10.1016/j.tree.2022.03.011
Hussain M. H., Mohsin M. Z., Zaman W. Q., Yu J., Zhao X., Wei Y., et al. (2022). Multiscale engineering of microbial cell factories: A step forward towards sustainable natural products industry. Synth Syst. Biotechnol. 7, 586–601. doi: 10.1016/j.synbio.2021.12.012
James E. K., Olivares F. L., De Oliveira A. L. M., Dos Reis F. B., Da Silva L. G., Reis V. M. (2001). Further observations on the interaction between sugar cane and Gluconacetobacter diazotrophicus under laboratory and greenhouse conditions. J. Exp. Bot. 52, 747–760. doi: 10.1093/jexbot/52.357.747
Jangra M., Belur P. D., Oriabinska L. B., Dugan O. M. (2016). Multistrain probiotic production by co-culture fermentation in a lab-scale bioreactor. Eng. Life Sci. 16, 247–253. doi: 10.1002/elsc.201500069
Kapoore R. V., Padmaperuma G., Maneein S., Vaidyanathan S. (2022). Co-culturing microbial consortia: approaches for applications in biomanufacturing and bioprocessing. Crit. Rev. Biotechnol. 42, 46–72. doi: 10.1080/07388551.2021.1921691
Logeshwarn P., Thangaraju M., Rajasundari K. (2011). Antagonistic potential of Gluconacetobacter diazotrophicus against Fusarium oxysporum in sweet potato (Ipomea batatus) Arch. Phytopathol. Pflanzenschutz. 44, 216–223. doi: 10.1080/03235400902952707
Martinez-Drets G., Del Gallo M., Burpee C., Burris R. H. (1984). Catabolism of carbohydrates and organic acids and expression of nitrogenase by azospirilla. J. Bacteriol 159, 80–85. doi: 10.1128/jb.159.1.80-85.1984
Masciarelli O., Llanes A., Luna V. (2014). A new PGPR co-inoculated with Bradyrhizobium japonicum enhances soybean nodulation. Microbiol. Res. 169, 609–615. doi: 10.1016/j.micres.2013.10.001
Olivares F. L., James E. K., Baldani J. I., Dobereiner J. (1997). Infection of mottled stripe disease-susceptible and resistant sugar cane varieties by the endophytic diazotroph Herbaspirillum. New Phytol. 135, 723–737. doi: 10.1046/j.1469-8137.1997.00684.x
Pagnani G., Galieni A., Stagnari F., Pellegrini M., Del Gallo M., Pisante M. (2020). Open field inoculation with PGPR as a strategy to manage fertilization of ancient Triticum genotypes. Biol. Fertil Soils 56, 111–124. doi: 10.1007/s00374-019-01407-1
Pagnani G., Pellegrini M., Galieni A., D’Egidio S., Matteucci F., Ricci A., et al. (2018). Plant growth-promoting rhizobacteria (PGPR) in Cannabis sativa ‘Finola’ cultivation: An alternative fertilization strategy to improve plant growth and quality characteristics. Ind. Crops Prod 123, 75–83. doi: 10.1016/j.indcrop.2018.06.033
Pagnussat L. A., Salcedo F., Maroniche G., Keel C., Valverde C., Creus C. M. (2016). Interspecific cooperation: enhanced growth, attachment and strain-specific distribution in biofilms through Azospirillum brasilense-Pseudomonas protegens co-cultivation. FEMS Microbiol. Lett. 363, fnw238. doi: 10.1093/femsle/fnw238
Pal G., Saxena S., Kumar K., Verma A., Sahu P. K., Pandey A., et al. (2022). Endophytic Burkholderia: Multifunctional roles in plant growth promotion and stress tolerance. Microbiol. Res. 265, 127201. doi: 10.1016/j.micres.2022.127201
Pellegrini M., Ercole C., Di Zio C., Matteucci F., Pace L., Del Gallo M. (2020a). In vitro and in planta antagonistic effects of plant growth-promoting rhizobacteria consortium against soilborne plant pathogens of Solanum tuberosum and Solanum lycopersicum. FEMS Microbiol. Lett. 367, fnaa099. doi: 10.1093/femsle/fnaa099
Pellegrini M., Ercole C., Gianchino C., Bernardi M., Pace L., Del Gallo M. (2021a). Fusarium Oxysporum f. sp. Cannabis Isolated from Cannabis Sativa L.: In Vitro and In Planta Biocontrol by a Plant Growth Promoting-Bacteria Consortium. Plants 10, 2436. doi: 10.3390/plants10112436
Pellegrini M., Pagnani G., Bernardi M., Mattedi A., Spera D. M., Del Gallo M. (2020b). Cell-free supernatants of plant growth-promoting bacteria: A review of their use as biostimulant and microbial biocontrol agents in sustainable agriculture. Sustainability 12, 9917. doi: 10.3390/su12239917
Pellegrini M., Spera D. M., Ercole C., Del Gallo M. (2021b). Allium cepa L. Inoculation with a consortium of plant growth-promoting bacteria: effects on plants, soil, and the autochthonous microbial community. Microorganisms 9, 639. doi: 10.3390/microorganisms9030639
Restrepo G. M., Rincón A., Sánchez Ó.J. (2023). Kinetic Analysis of Gluconacetobacter diazotrophicus Cultivated on a Bench Scale: Modeling the Effect of pH and Design of a Sucrose-Based Medium. Fermentation 9, 705. doi: 10.3390/fermentation9080705
Rodrigues L. R., Teixeira J. A., Oliveira R. (2006). Low-cost fermentative medium for biosurfactant production by probiotic bacteria. Biochem. Eng. J. 32, 135–142. doi: 10.1016/j.bej.2006.09.012
Rosero-Chasoy G., Rodríguez-Jasso R. M., Aguilar C. N., Buitrón G., Chairez I., Ruiz H. A. (2021). Microbial co-culturing strategies for the production high value compounds, a reliable framework towards sustainable biorefinery implementation – an overview. Bioresour Technol. 321, 124458. doi: 10.1016/j.biortech.2020.124458
Santos C.C.A.d., Libeck B.da S., Schwan R. F. (2014). Co-culture fermentation of peanut-soy milk for the development of a novel functional beverage. Int. J. Food Microbiol. 186, 32–41. doi: 10.1016/j.ijfoodmicro.2014.06.011
Seenivasagan R., Babalola O. O. (2021). Utilization of microbial consortia as biofertilizers and biopesticides for the production of feasible agricultural product. Biol. (Basel) 10, 1111. doi: 10.3390/biology10111111
Silva E. R., Zoz J., Oliveira C. E. S., Zuffo A. M., Steiner F., Zoz T., et al. (2019). Can co-inoculation of Bradyrhizobium and Azospirillum alleviate adverse effects of drought stress on soybean (Glycine max L. Merrill.)? Arch. Microbiol. 201, 325–335. doi: 10.1007/s00203-018-01617-5
Singh R., Kaur S., Bhullar S. S., Singh H., Sharma L. K. (2024). Bacterial biostimulants for climate smart agriculture practices: Mode of action, effect on plant growth and roadmap for commercial products. J. Sustain. Agric. Environ. 3, e12085. doi: 10.1002/sae2.12085
Sobhi M., Zakaria E., Zhu F., Liu W., Aboagye D., Hu X., et al. (2023). Advanced microbial protein technologies are promising for supporting global food-feed supply chains with positive environmental impacts. Sci. Total Environ. 894, 165044. doi: 10.1016/j.scitotenv.2023.165044
Stephan M. P., Oliveira M., Teixeira K. R. S., Martinez-Drets G., Döbereiner J. (1991). Physiology and dinitrogen fixation of Acetobacter diazotrophicus. FEMS Microbiol. Lett. 77, 67–72. doi: 10.1111/fml.1991.77.issue-1
Suleman M., Yasmin S., Rasul M., Yahya M., Atta B. M., Mirza M. S. (2018). Phosphate solubilizing bacteria with glucose dehydrogenase gene for phosphorus uptake and beneficial effects on wheat. PloS One 13, e0204408. doi: 10.1371/journal.pone.0204408
Tawfik K. A., Jeffs P., Bray B., Dubay G., Falkinham J. O., Mesbah M., et al. (2010). Burkholdines 1097 and 1229, potent antifungal peptides from Burkholderia ambifaria 2.2N. Org Lett. 12, 664–666. doi: 10.1021/ol9029269
Wu D., Wang W., Yao Y., Li H., Wang Q., Niu B. (2023). Microbial interactions within beneficial consortia promote soil health. Sci. Total Environ. 900, 165801. doi: 10.1016/j.scitotenv.2023.165801
Xu X., Cheng W., Liu X., You H., Wu G., Ding K., et al. (2020). Selenate reduction and selenium enrichment of tea by the endophytic herbaspirillum sp. Strain WT00C. Curr. Microbiol. 77, 588–601. doi: 10.1007/s00284-019-01682-z
Zahir Z. A., Ghani U., Naveed M., Nadeem S. M., Asghar H. N. (2009). Comparative effectiveness of Pseudomonas and Serratia sp. containing ACC-deaminase for improving growth and yield of wheat (Triticum aestivum L.) under salt-stressed conditions. Arch. Microbiol. 191, 415–424. doi: 10.1007/s00203-009-0466-y
Keywords: fermentation, multi-strain consortium, sustainability, biostimulants, biopesticides
Citation: Farda B, Pasquarelli F, Djebaili R, Spera DM, Del Gallo M and Pellegrini M (2024) Co-culturing a multistrain Gram-negative inoculant useful in sustainable agriculture. Front. Ind. Microbiol. 2:1380037. doi: 10.3389/finmi.2024.1380037
Received: 31 January 2024; Accepted: 15 March 2024;
Published: 26 March 2024.
Edited by:
Ivo Oliveira, University of Trás-os-Montes and Alto Douro, PortugalReviewed by:
Travis Robert Glare, Lincoln University, New ZealandWilgince Apollon, National Polytechnic Institute (IPN), Mexico
Copyright © 2024 Farda, Pasquarelli, Djebaili, Spera, Del Gallo and Pellegrini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beatrice Farda, YmVhdHJpY2UuZmFyZGFAZ3JhZHVhdGUudW5pdmFxLml0
 Beatrice Farda
Beatrice Farda Fabrizia Pasquarelli
Fabrizia Pasquarelli Rihab Djebaili
Rihab Djebaili Daniela M. Spera
Daniela M. Spera Maddalena Del Gallo
Maddalena Del Gallo Marika Pellegrini
Marika Pellegrini