
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ind. Microbiol. , 20 September 2023
Sec. Food
Volume 1 - 2023 | https://doi.org/10.3389/finmi.2023.1257483
This article is part of the Research Topic Fermented Foods: Characterization of the Autochthonous Microbiota View all 10 articles
Introduction: Several human health benefits have been ascribed to probiotics, while traditional fermented foods have been acknowledged as rather important sources of these microorganisms. The objective of this study was the isolation of microorganisms from fermented food products of both animal and plant origin and the evaluation of their probiotic potential.
Methods: Microbial isolation was performed from milk kefir and table olives, while an olive mill waste sample also was analyzed given its anticipated association with the autochthonous microbiota of olive drupes. Among the 16 macroscopically distinct recovered microorganisms, 14 microbial isolates were identified as presumptive lactic acid bacteria (LAB), whereas two isolates corresponded to yeasts. The microbial isolates exhibiting a reproducibly robust growth profile in appropriate culture broth media (11 out of the 16 isolates) were assessed for their probiotic potential based on a set of in vitro assays: resistance to low pH; autoaggregation; biofilm formation; antioxidant activity; and safety assessment through evaluation of hemolytic activity.
Results and discussion: Based on the collective evaluation of the results of the abovementioned assays, five presumptive LAB as well as the two yeast isolates were identified as exhibiting desirable in vitro probiotic traits. Hence, these microbial isolates could be regarded as good candidates for inclusion in further studies aiming, ultimately, at their potential utilization in novel functional food products.
Food fermentation is a long-lasting food processing technology which has traditionally allowed for enhanced preservation and considerable shelf-life extension of various perishable foods of both plant and animal origin. Fermented foods and beverages, defined as “foods made through desired microbial growth and enzymatic conversions of food components” (Marco et al., 2021), have been consumed for thousands of years (Liu et al., 2019). Nonetheless, the claimed health benefits of their consumption have resulted in significant research interest and increasing popularity among consumers the last decades (Xiang et al., 2019). The functional character of fermented foods has been associated with the potential probiotic effect of their constituent microorganisms, the production (during fermentation) of bioactive compounds (e.g., short-chain fatty acids, peptides, and polyamines), as well as with the reduction of toxic compounds and anti-nutrients in the end products (Dimidi et al., 2019; Xiang et al., 2019; Annunziata et al., 2020). Moreover, assessment of the potential impact of fermented foods on human cognitive function has also attracted the interest of researchers recently (Casertano et al., 2022).
The functional attributes of fermented foods are regarded as being essentially delineated by their ample content in microorganisms with probiotic traits. According to the definition provided by the Food and Agriculture Organization (FAO) of the United Nations and the World Health Organization (WHO), probiotics are “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (FAO/WHO, 2002; Hill et al., 2014). The contribution of probiotics to human health protection has been linked to various modes of action including: (i) modification of the gut microbiota (and its metabolic activity) contributing, among others, to the improvement of lactose fermentation (Ibrahim et al., 2021; Ma et al., 2023); (ii) reduction of the bowel’s pH and prevention of its colonization by pathogenic organisms (Marco et al., 2006); (iii) prevention of gastric ulcer due to their activity against the bacterium Helicobacter pylori (Boltin, 2016); (iv) modulation of the host’s immune responses (Yan and Polk, 2011); (v) protection against allergens (Lopez-Santamarina et al., 2021); and (vi) anti-carcinogenic activity (Legesse Bedada et al., 2020). Given their well-established benefits for human health, probiotics have been increasingly utilized as nutraceuticals in the production of food supplements and as ingredients of food products, with the latter being either fermented foods or non-fermented products enriched with probiotics. Numerous microbial strains have been identified as having probiotic traits, with most of them corresponding to species of the genus Bifidobacterium, lactic acid bacteria (LAB) species of the genera Lactobacillus (recently emended and reclassified into 25 genera), Enterococcus, Streptococcus, Pediococcus and Leuconostoc, as well as to the yeast species Saccharomyces cerevisiae (Fijan, 2014; Zheng et al., 2020; Soemarie et al., 2021; Staniszewski and Kordowska-Wiater, 2021).
Assessment of the putative association of probiotics with the health benefits of traditional fermented foods is a research objective of enduring value. Such association is strongly supported by the profuse participation of the former microorganisms in these products’ microbiota. Indeed, fermented foods constitute valuable natural reservoirs of microorganisms with probiotic traits (Rezac et al., 2018; Soemarie et al., 2021), with the latter being evaluated in vitro, ex vivo and/or in the context of clinical trials (Nuraida, 2015; Kim and Park, 2018). Fermented foods and beverages which have been evaluated as important sources of microorganisms with probiotic potential are various dairy products (e.g., yogurt, kefir, sour cream, ripened cheeses), meat products (e.g., salami, pepperoni), produce commodities (e.g., kimchi, sauerkraut, table olives, tempeh, tofu) and herbal extracts (e.g., kombucha) (Marco et al., 2017; Bonatsou et al., 2018; Kok and Hutkins, 2018; Marco et al., 2021; Soemarie et al., 2021). Isolation of microorganisms with probiotic potential from fermented foods is anticipated to provide a competitive advantage in their effective utilization as functional microbial cultures (starter or adjunct cultures) in the manufacture of the so-called “probiotic fermented foods” (Marco et al., 2021). Such microorganisms are highly likely to retain their probiotic traits in situ (i.e., in the food matrix) during the manufacturing process, and thus, to result in food products of enhanced functional quality throughout their shelf life. Furthermore, microorganisms with probiotic traits can be used for the functional supplementation of various products, even non-fermented ones, as well as for the development of nutritional supplements.
Given the above, the objective of the present study was the isolation of microorganisms from selected fermented food products of both animal and plant origin and the in vitro screening of their probiotic potential. The fermented foods used for microbial isolation were milk kefir and table olives, while an olive mill waste sample also was analyzed, given its anticipated association with the autochthonous microbiota of olive drupes.
The food products used in the present study for microbial isolation purposes are compiled in Table 1 and included: (i) four kefir products (three industrially manufactured commercial products purchased from local retail stores and one homemade product), and (ii) natural black cv. Kalamata olives. Regarding kefir products, they were all made from cow’s milk, while one of the industrial products (Kefir 3), was flavored and sweetened with steviol glycosides (Table 1). With reference to table olives, two distinct batches of fermented olives were provided by a local manufacturer, and microbial isolation was performed both from the olive drupes and the corresponding brines. Finally, an olive mill waste (OMW) sample provided by a local olive oil extraction plant also was analyzed, given its anticipated microbial association with the autochthonous microbiota of olive drupes.
The aforementioned samples were subjected to microbiological analyses aiming at the isolation of microorganisms present at high concentration levels and as such, contributing appreciably to these products’ microbiota. For this purpose, 1-mL aliquots of the kefir products were transferred aseptically to 9 mL of sterilized quarter strength Ringers solution (Neogen®, Lansing, MI, USA), and appropriate serial decimal dilutions in the same solution were pour/surface plated in/on selected agar media. The same procedure was followed for the microbiological analyses of the olive brines and the OMW. Regarding the isolation of microorganisms from olive drupes, three randomly selected drupes from each batch were rinsed with sterilized Ringers solution (10 mL per drupe), to remove brine and loosely attached microbial cells, and were depitted with sterile scalpel and forceps under aseptic conditions. The olive drupes’ flesh was then transferred aseptically in 400-mL sterile stomacher bags (BagLight® Polysilk 400, Interscience, St Nom, France), was 1:10 diluted with Ringers solution and was homogenized in a Stomacher apparatus (BagMixer® 400, Interscience) for 120 s. Appropriate serial decimal dilutions of the homogenates were then pour/surface plated in/on selected agar media.
Overall, the microbiological analyses conducted and the agar media used in the present study were the following: (i) surface plating on Standard Methods Agar-PCA (Condalab, Madrid, Spain) for the total viable count (TVC) after incubation of plates at 28°C for 72 h; (ii) pour plating in de Man, Rogosa and Sharpe (MRS) agar (Condalab) along with medium’s overlay for the determination of presumptive lactic acid bacteria (LAB) after incubation of plates at 28°C for 72–96 h; (iii) surface plating on potato dextrose agar (PDA, Condalab) supplemented with 0.05 g/L chloramphenicol (Sigma-Aldrich, Seelze, Germany) for the determination of yeasts and molds after incubation of plates at 28°C for 5 days; and (iv) pour plating in violet red bile glucose (VRBG) agar (Condalab) along with medium’s overlay for the assessment of the presence (at detectable levels) of bacteria of the Enterobacteriaceae family after incubation of plates at 37°C for 24 h. Upon completion of the microbiological analyses, the pH values of the samples also were measured using a digital pH meter (Orion Model 420A, ATI Orion, Boston, MA, USA) with a glass electrode (HANNA® Instruments, Athens, Greece).
Morphologically distinct colonies growing in/on the different agar media (in the dilutions allowing for their accurate enumeration) were streaked on appropriate media aiming at the isolation of presumably different microorganisms. The isolated colonies were then used to inoculate different culture broth media, which were then incubated at different temperatures (depending on the medium) and for different time periods (24–48 h) in order to identify the incubation conditions better supporting their growth. Specifically, the microbial colonies recovered from PDA were grown in potato dextrose broth (PDB, Himedia, Mumbai, India) at 28°C, whereas the microbial colonies retrieved from MRS agar were grown in MRS broth (Condalab), M-17 broth (Himedia) and Columbia broth (Condalab) at 28, 37 and 45°C. The microbial isolates were phenotypically characterized based on microscopic observation using an optical microscope equipped with a digital camera (BioBlue BB.4267, Euromex Microscopen BV, Arnhem, The Netherlands), and in the case if bacterial isolates using Gram stain as well as the results of catalase and oxidase tests.
All microbial isolates were maintained as frozen (−80°C) stock cultures in MRS broth supplemented with 20% glycerol (presumptive LAB isolates) or PDB broth supplemented with 30% glycerol (yeast isolates). Working cultures were stored refrigerated (4°C) on slants of appropriate agar media (MRS agar and PDA for presumptive LAB and yeast isolates, respectively) and were renewed bimonthly. The probiotic potential of the microbial isolates was assessed based on a series of in vitro assays (described in detail in the following sections): resistance to low pH; autoaggregation capability; biofilm formation; antioxidant activity; and safety assessment through evaluation of hemolytic activity. The microbial cultures used in these assays were prepared by transferring a loopful from the working cultures (agar slants) in 10 mL of culture broth (MRS broth or PDB) and incubating at 28 or 37°C for 24 or 48 h, depending on the isolate. Beyond the isolated microorganisms, the Saccharomyces cerevisiae strain AXAZ-1, originally isolated from a Greek vineyard plantation (Argiriou et al., 1992) and maintained at the cultures collection of the laboratory of Microbiology (Department of Biology, University of Patras), also was evaluated in the present study.
The resistance of the microbial isolates to low pH was evaluated according to previously described procedures with some modifications (Argyri et al., 2013; Pavli et al., 2016). Specifically, cultures of the microbial isolates were centrifuged (Heraeus, Biofuge Stratos, Thermo Scientific, Osterode, Germany) at 10,000× g for 5 min at 4°C. The harvested cells were washed twice with phosphate buffered saline (PBS, pH 7.2) by centrifugation under the same conditions, and were finally re-suspended in 10 mL of PBS solution with pH adjusted to 2.5 using HCl (min. 37%; Sigma-Aldrich). The pH adjustment of the PBS solution was performed using a digital pH meter with a glass electrode, and its pH value was also measured (and confirmed not to be considerably different) after autoclaving. The microbial suspensions were incubated at 37°C with constant stirring at medium amplitude in a water bath (Memmert WB22, Schwabach, Germany) for a total period of 3 h. At 0 and 3 h of the applied acid challenge, samples were taken from each microbial suspension to determine the initial and surviving population, respectively. The populations of presumptive LAB isolates were enumerated by pour-plating of appropriate serial decimal dilutions (in Ringers solution) in MRS agar, while the populations of the yeast isolates by surface plating on PDA. Microbial colonies were counted after incubation of plates at 28°C for a total of 120 h (colonies were counted at 72 h and the counts were confirmed at 96 and 120 h). The acid resistance of each isolate was assessed in three independent experiments (n=3).
The autoaggregation assay was performed based on previously described protocols (Ogunremi et al., 2015; Bonatsou et al., 2018). In brief, cultures of the microbial isolates were centrifuged as described above, washed once with PBS (pH 7.2) and resuspended in 10 mL of the same solution. The microbial suspensions were vortexed for 10 s and incubated at 37°C for 24 h. At 0, 2, 4 and 24 h of incubation, 1-mL aliquots of the supernatant of each isolate were carefully transferred in plastic cuvettes and their optical density (OD) was measured at 600 nm using a UV spectrophotometer (UV-1800, Shimadzu Europa GmbH, Duisburg, Germany). The autoaggregation capability was quantified based on the following equation:
where Atand A0 is the OD measurement at time t and zero, respectively.
The autoaggregation assay was performed twice (independent assays), with the OD at 2 and 4 h, however, being recorded once.
The biofilm-forming ability of the microbial isolates was evaluated in vitro using a colorimetric microtiter plate method, frequently used for the indirect quantification of biofilm formation. The biofilm formation assay applied herein is based on the measurement of the OD of the biofilm mass in microtiter plate wells after crystal violet staining and was performed according to previously described procedures (Lianou and Koutsoumanis, 2012; Pachla et al., 2021) with some modifications. More specifically, 20-μL aliquots of each microbial isolate’s culture were added to 180 μL of appropriate culture broth, namely MRS broth for the presumptive LAB isolates and PDB for the yeast isolates, dispensed in 96-well polystyrene microtiter plates (CytoOne®, STARLAB International GmbH, Hamburg, Germany). Each microbial isolate was tested in eight replicate wells, while negative control wells (containing only 200 μL of culture broth) also were included. The microplates were incubated statically at 37°C for 72 h. The content of the microplates was then discarded, and the wells were rinsed with 200 μL of Ringers solution, aiming at the removal of the planktonic or reversibly attached cells. During the applied rinsing step, the microplates were agitated on a rocking platform (Stuart STR9 3D Rocking Platform, Stuart Scientific, Staffordshire, UK) at 40 rpm for 5 min. Next, the adherent bacterial cells were fixed with 200 μL of methanol (min. 99.8%; Fisher Scientific, Loughborough, UK) per well for 15 min, the wells were emptied by inversion of the microplates and the latter were air dried for 20 min. Afterward, the biofilm mass was stained with crystal violet (Gram’s crystal violet solution, Sigma-Aldrich) for 20 min, after which the excess stain was rinsed off (at least three times) through filling the microplates’ wells with deionized water and emptying them by inversion. Then, the microplates were vigorously tapped on absorbent paper and air dried for 1 h. The bound to the formed biofilm stain was solubilized in 200 μL of ethanol (min. 99.8%; Fisher Scientific) per well and the OD of each well was measured at 595 nm using a microplate reader (MRX Microplate Absorbance Reader, Dynex Technologies, Chantilly, VA, USA). The average value of the OD measurements of the control wells was subtracted from the OD of each test well, and this difference, referred to as ΔOD595 nm, was used as an index of the biofilm-forming ability of the microbial isolates. Two independent biofilm formation assays were performed for each microbial isolate, with eight replicate wells per treatment (isolates and negative control) being included in each assay (n=16).
The antioxidant activity was assessed as the percentage of reduction of 2,2-diphenyl-1-picrylhydrazyl (DPPH) according to previously described protocols (Gil-Rodríguez et al., 2015; Cho et al., 2018) with some modifications. One-milliliter aliquots of each microbial culture were centrifuged (LabNet Hermle Z 233 MK-2 microcentrifuge, Hudson, MA, USA) at 12,000 rpm for 5 min at 4°C, and the harvested cells were washed twice with Ringers solution and finally resuspended in 1 mL of the same solution. One-milliliter aliquots of freshly prepared DPPH (Sigma-Aldrich) solution (0.2 mM in methanol) were then added to the cell suspensions, and the mix was vortexed and incubated at 37°C in darkness for 30 min. Afterward, the reaction tubes were centrifuged under the same conditions, 200 μL of the mixtures’ supernatants were transferred into 96-well polystyrene microtiter plates, and absorbance measurements were recorded at 490 nm using a microplate reader (MRX Microplate Absorbance Reader). The scavenged DPPH was calculated using the following equation:
where Asample is the absorbance of the DPPH solution with the sample and Ablank is the absorbance of the DPPH solution and Ringers solution. The absorbance values of pure DPPH solution as well as of ascorbic acid (Sigma-Aldrich) solution (0.2 mM in methanol), serving as positive control of the applied assay, were also recorded.
The antioxidant activity of each microbial isolate (and the positive control) was evaluated in four replicate microplate wells (n=4). Given the anticipated directly proportional association of the scavenging activity of each microbial isolate with the cell concentration of the tested microbial suspension, the cell concentration of the used microbial cultures was determined by pour/surface plating (as described in Section 2.3), and the attained antioxidant activity results were standardized for a microbial concentration of 107 CFU/mL.
For assessing the safety of the isolated microorganisms, fresh microbial cultures were streaked on duplicate plates of Columbia agar containing 5% (v/v) sheep blood (Bioprepare® Microbiology, Keratea, Attica, Greece). The plates were incubated at 28°C for 48–72 h and were examined for signs of hemolytic activity. Specifically, green-hued zones around colonies are regarded as sign of α-hemolysis, clear zones around colonies as sign of β-hemolysis, whereas absence of zones around the colonies is indicative of no hemolytic activity also referred to as γ-hemolysis.
The experimental data derived from the acid resistance, autoaggregation capability, biofilm formation and antioxidant activity assays were evaluated by analysis of variance using the general linear model procedure of the software IBM® SPSS® Statistics 27 (IBM Corp., Armonk, NY, USA). Means were separated using the Tukey HSD post-hoc test at a significance level of α=0.05.
The total mesophilic populations and presumptive LAB constituting the autochthonous microbiota of the fermented food samples and the OMW sample tested herein, along with the samples’ pH values are compiled in Table 2. Bacteria belonging to the Enterobacteriaceae family were not recovered at detectable levels from none of the tested samples. Aiming at the recovery of microorganisms present at high concentration levels in the tested samples and as such, contributing appreciably to their microbiota, microbial isolation was conducted at the dilutions of the tested samples allowing for their enumeration. The microorganisms isolated from the selected fermented food products and the OMW are presented in Table 3, while the results of the in vitro trials aiming at the evaluation of their probiotic traits are illustrated in Figures 1–5. The characterization of the probiotic potential of the isolated microorganisms was based on the collective evaluation of the compiled data, allowing for the selection of isolates that would be of interest for further investigation and utilization in industrial applications.
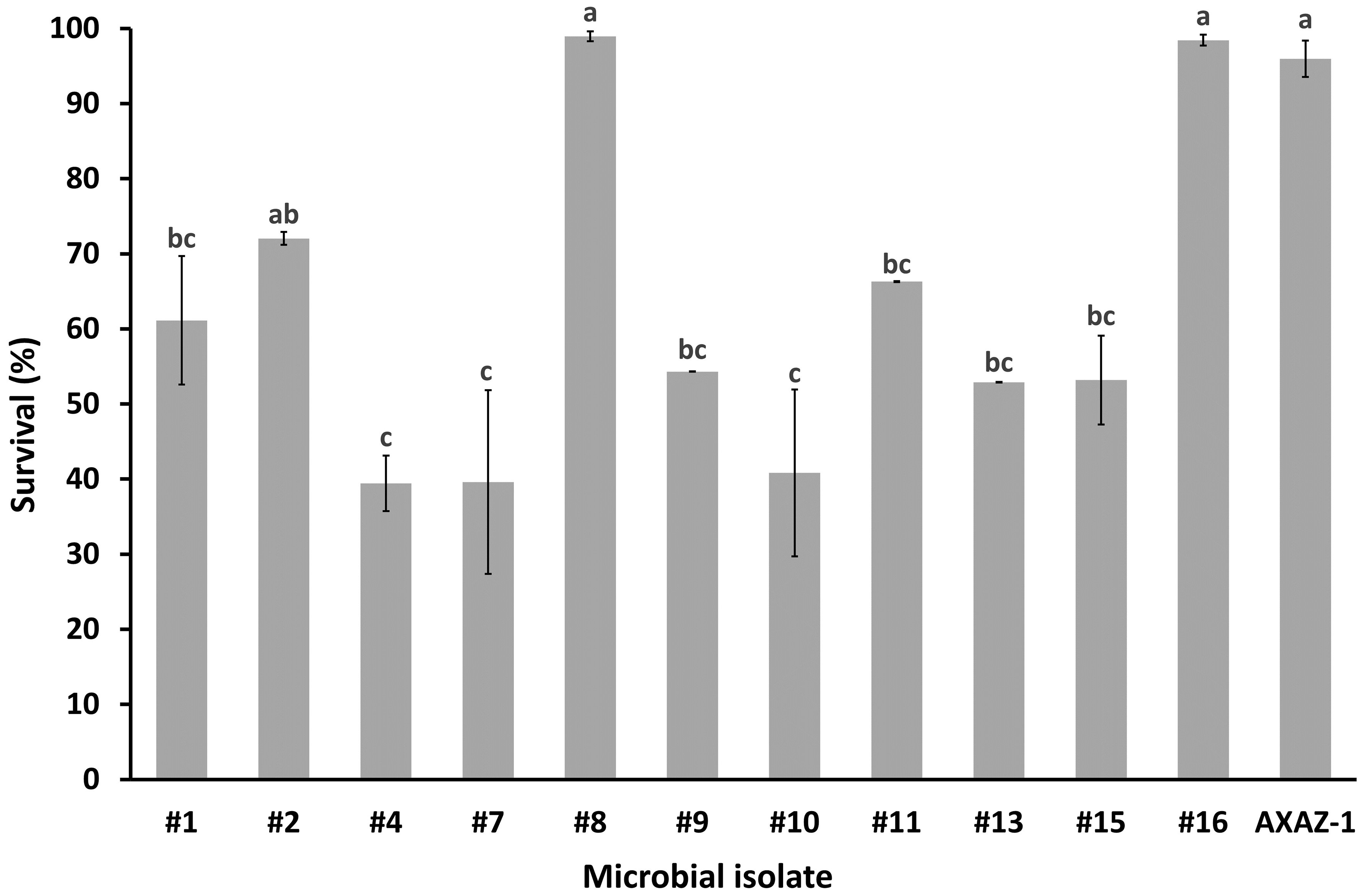
Figure 1 Survival percentage of microbial populations after 3 h of exposure to pH 2.5 in phosphate buffered saline. Values are means ± standard errors (n=3). Means lacking a common letter are significantly different (P<0.05).
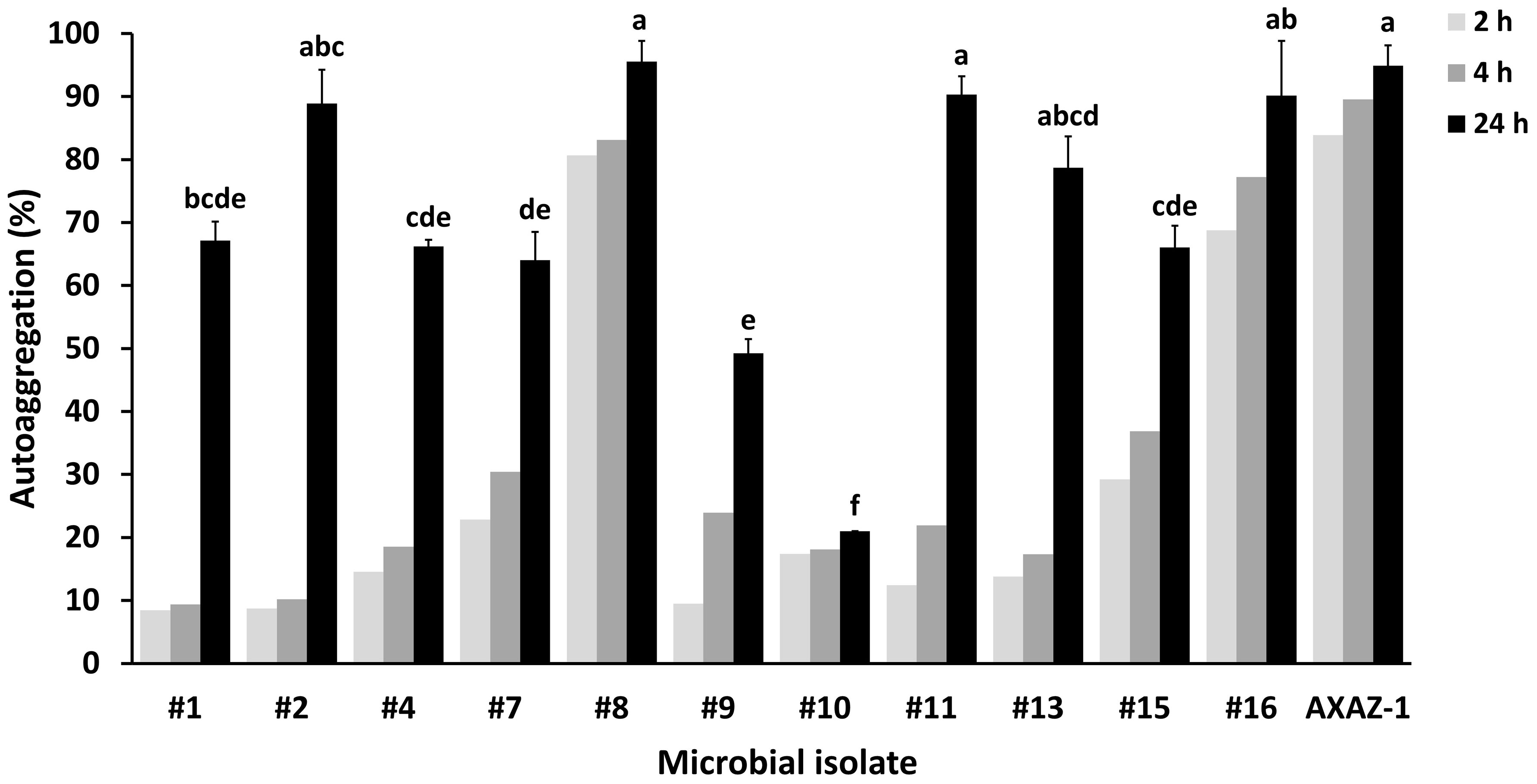
Figure 2 Autoaggregation capability (as percentage) of the microbial isolates after incubation in phosphate buffered saline for 2, 4 and 24 h and optical density measurements at 600 nm. The values corresponding to 24 h are means ± standard errors (n=2). Means lacking a common letter are significantly different (P<0.05).
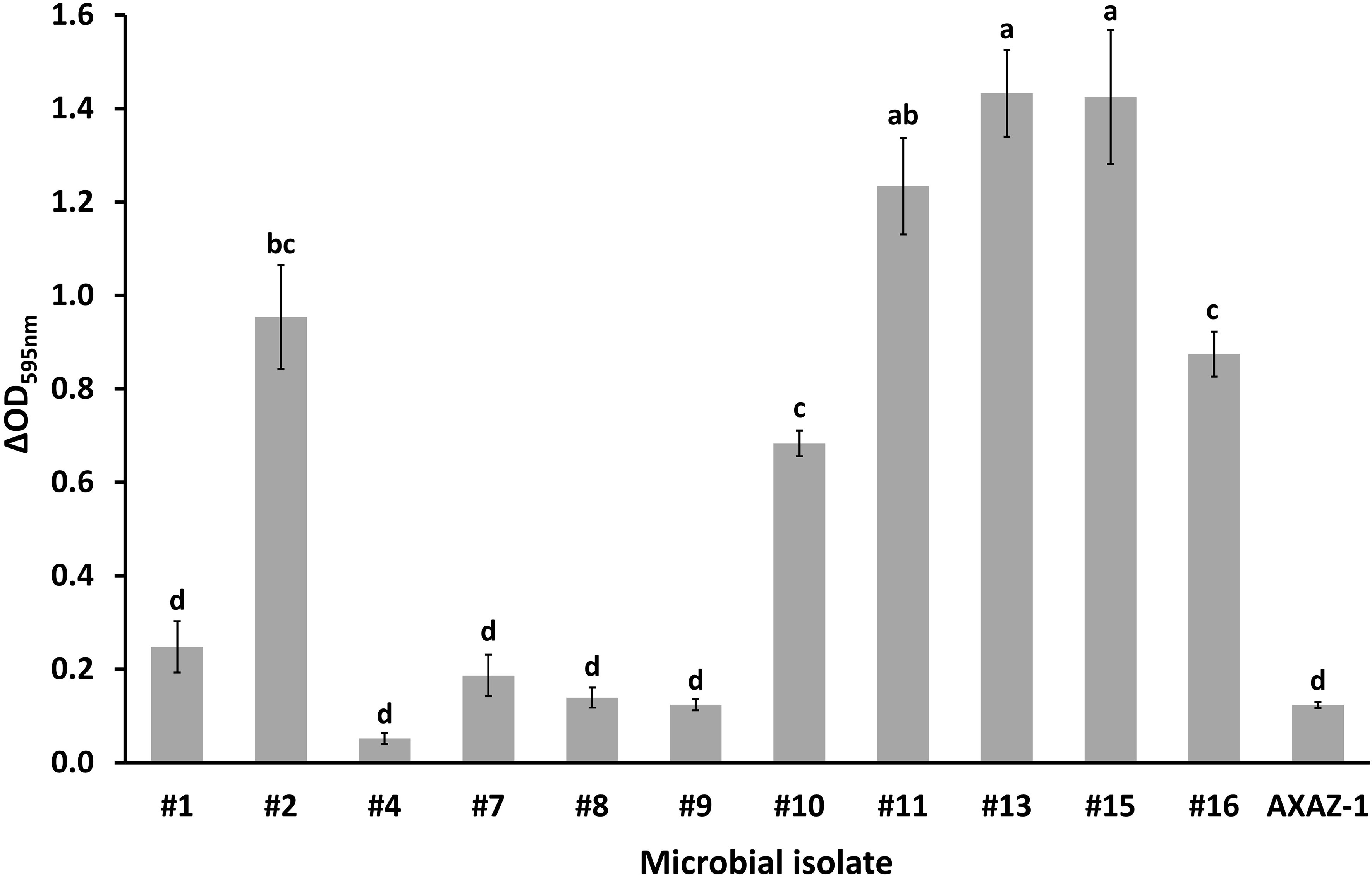
Figure 3 Biofilm formation in polystyrene microtiter plates by the microbial isolates, based on crystal violet staining and the difference between the optical density measurements of the test and negative control samples (ΔOD595nm). Values are means ± standard errors (n=16). Means lacking a common letter are significantly different (P<0.05).
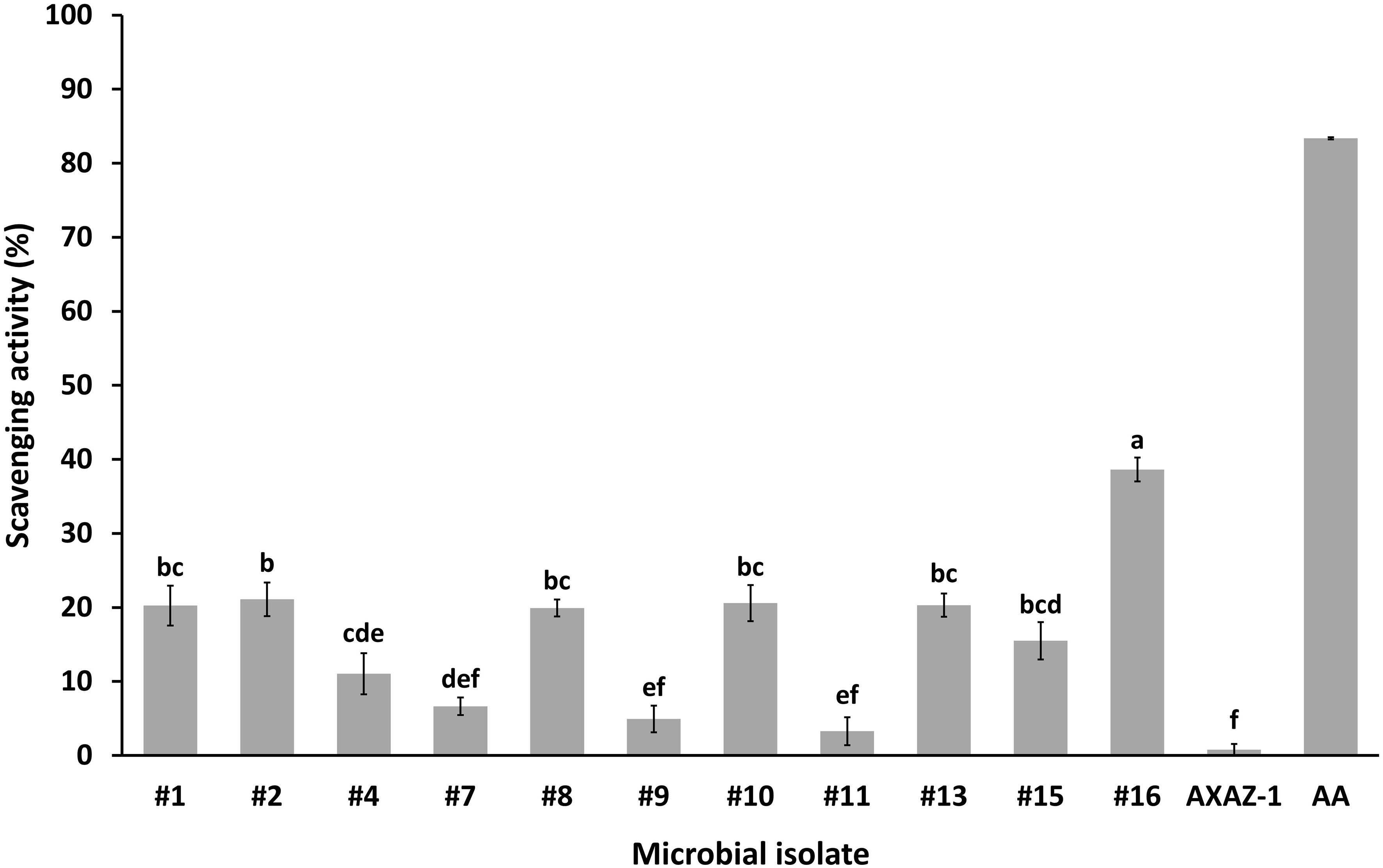
Figure 4 Antioxidant effects of intact cells of the microbial isolates on DPPH (1,1-diphenyl-2-picrylhydrazyl) radical-scavenging activity. Values are means ± standard errors (n=4). Means lacking a common letter are significantly different (P<0.05). AA: ascorbic acid (positive control).
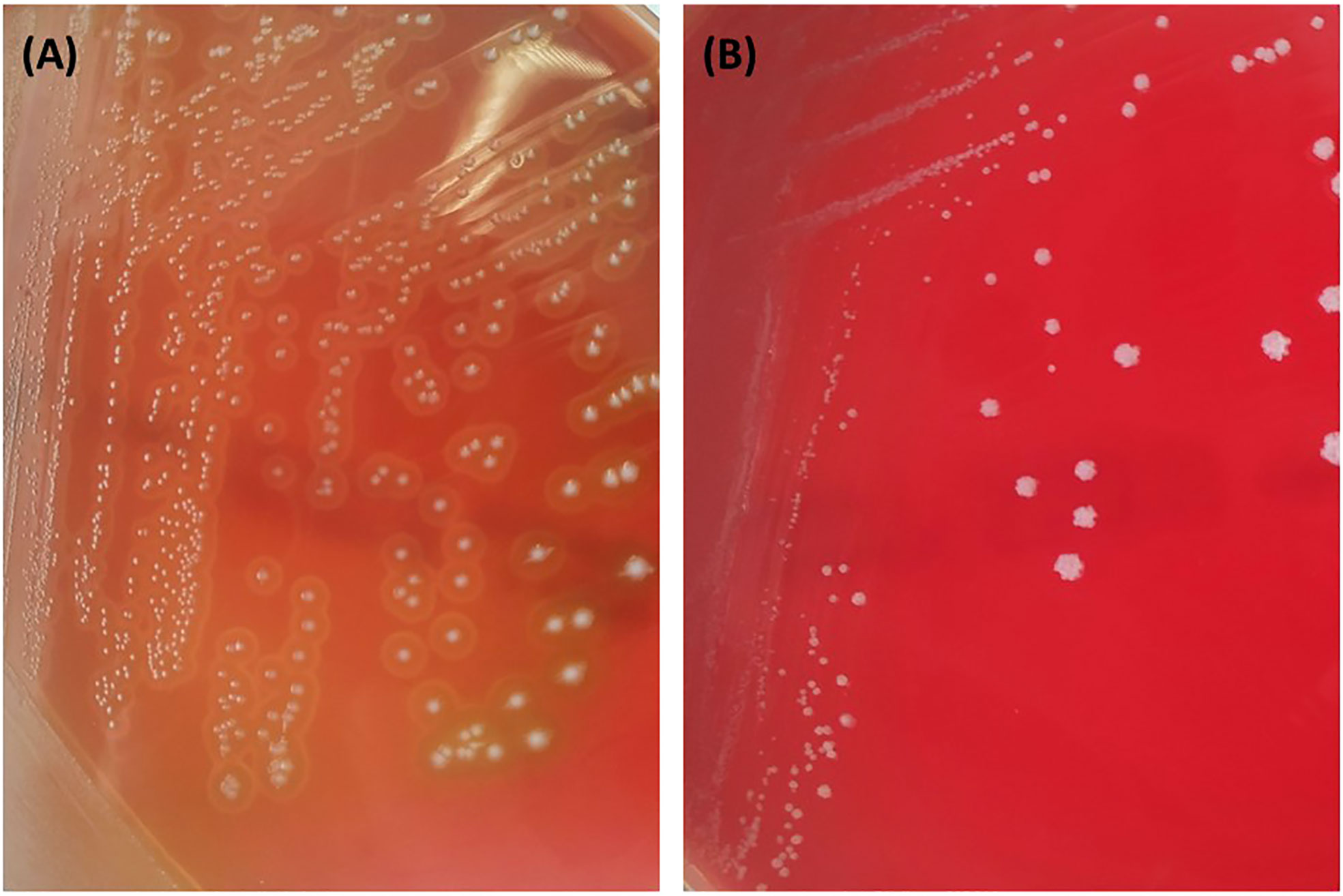
Figure 5 Observation of α-hemolysis (partial hemolysis, (A)) and γ-hemolysis (absence of hemolysis, (B)) for the microbial isolate #1 and #8, respectively, on Columbia agar containing 5% (v/v) sheep blood.
In total, 16 microbial isolates with distinct macroscopic characteristics were recovered from the tested food/OMW samples. The macroscopic distinctiveness (i.e., evidently different colony morphology) of the isolated microorganisms was aimed to maximize the likelihood of isolating different microbial strains from each tested sample. Based on their phenotypic characteristics, the majority of microbial isolates (14/16) were recovered from MRS agar and were identified as presumptive LAB (Gram-positive, catalase- and oxidase-negative bacteria), while two isolates were recovered from PDA with chloramphenicol and were identified as yeasts. From the isolated microorganisms, 11 exhibited reproducibly robust growth in culture broth under the conditions of this study and were therefore, selected to be further investigated for their probiotic potential. The phenotypic and culture characteristics of the isolated microorganisms, as well as the isolates selected for further study are presented in Table 3. Microscopy images of the 11 selected microbial isolates (nine bacteria and two yeasts) are illustrated in Supplementary Figures 1, 2. The S. cerevisiae strain AXAZ-1 (maintained at the laboratory’s cultures collection) also was included in all conducted trials.
When the microbial isolates were assessed regarding their resistance to low pH, the microorganisms which were noted for their acid tolerance were the isolates #1, #2, #8, #11, #16 as well as the strain AXAZ-1, with the estimated survival (%) after 3 h of acid challenge (pH 2.5) being 61.1, 72.0, 98.9, 66.3, 98.4 and 96.0, respectively. Nonetheless, sufficient acid resistance was also recorded for the rest of the microbial isolates, for which the survival percentages varied from 39.4 to 54.3% (Figure 1).
The abovementioned isolates also exhibited considerable autoaggregation capability. Particularly, the isolates #2, #8, #11 and #16, along with the yeast strain AXAZ-1, were capable of autoaggregating at levels exceeding 80% after 24 h (Figure 2). Appreciable autoaggregation was also demonstrated by the microbial isolates #1, #4, #7, #13 and #15, with the estimated percentages after 24 h varying from 64.0 to 78.7%. The yeasts investigated in the context of this study, namely the isolates #8 and #16 and the strain AXAZ-1, were evidently prompter to form cell aggregates than the bacterial isolates, since high autoaggregation percentages were noted even within the first 2 h of incubation (Figure 2). On the other hand, most of the bacterial isolates exhibited either a similarly low autoaggregation capability at 2 and 4 h followed by a considerably higher capability at 24 h of incubation (e.g., isolates #1 and #2), or a gradually increasing autoaggregation as a function of time (e.g., isolates #9 and #15). The microbial isolate exhibiting the lowest (P<0.05) autoaggregation capability was the bacterial isolate #10 (Figure 2).
With reference to the biofilm-forming ability of the microbial isolates, which was quantified in vitro via a colorimetric microtiter plate method, the collected ΔOD595nm data are illustrated in Figure 3. According to these data, which are indicative of the total biofilm mass produced under the conditions of this study by each one of the tested microorganisms, the bacterial isolates #2, #10, #11, #13 and #15, as well as the yeast isolate #16, could be characterized as strong biofilm formers. Significantly (P<0.05) poorer biofilm formation was recorded for the rest of the tested microorganisms, including the yeast AXAZ-1, with the bacterial isolate #4 being evaluated as the weakest biofilm former (Figure 3). Since the autoaggregation capability of microorganisms constitutes a putatively important parameter for their initial adhesion on surfaces, it is worth and relevant for someone to comparatively evaluate this capability with the corresponding biofilm formation of the tested isolates. Indeed, important capability of both autoaggregation and biofilm formation was recorded for all the aforementioned isolates, except for the bacterial isolate #10, which, albeit demonstrating considerable biofilm formation, did not exhibit remarkable autoaggregation (Figures 2, 3).
The antioxidant activity of the microbial isolates, expressed as the percent reduction of DPPH, was evaluated as rather variable (Figure 4). The highest (P<0.05) antioxidant activity was exhibited by the yeast isolate #16, with the estimated scavenging activity being 38.6%. The minimum value of the estimated scavenging activity was 3.3% (isolate #11), while the yeast strain AXAZ-1 demonstrated negligible antioxidant activity. The suitability of the wavelength of 490 nm for the determination of the microbial isolates’ scavenging activity in the context of the applied methodology (section 2.6) was assessed and confirmed in preliminary experimental trials; according to the results of these trials, the absorbance of DPPH at 490 nm was maximized to 1.35 ± 0.08 (mean ± standard deviation, n=4) compared to the significantly lower absorbance values recorded at the wavelengths of 450 nm, 550 nm, 595 nm and 630 nm at which the microplate reader could operate.
Finally, the vast majority of the microbial isolates investigated in the present study were evaluated as safe in terms of hemolysis, namely as γ-hemolytic. The sole exception to this was the isolate #1 (bacterial isolate from kefir), which exhibited α-hemolysis (partial hemolysis) with green-hued zones being formed around the colonies of the isolated microorganism on Columbia agar containing sheep blood (Figure 5).
The presence at high populations of microorganisms, being characterized as presumptive LAB and/or yeasts in fermented food products was anticipated, justifying the selection of such products in the search of potential probiotics in the first place. Indeed, the fermented milk beverage of kefir, originating from the Caucasus region, is manufactured through the addition to milk of a symbiotic starter culture referred to as “kefir grains” (Lopitz-Otsoa et al., 2003). This symbiotic culture typically consists of lactic and acetic acid-producing bacteria and yeasts (lactose-fermenting and/or non-lactose fermenting), being housed and interacting within kefiran, a polysaccharide and protein matrix (Lopitz-Otsoa et al., 2003; Dimidi et al., 2019). Among the eight macroscopically different microorganisms isolated from the kefir products tested in this study, seven were identified as presumptive LAB and one as yeast, with five of them being ultimately selected to be further investigated. Although a wide range of microbial species have been identified in kefir grains, the microorganisms most recovered from the final product are LAB species belonging to the genera Lactobacillus and Lactococcus, acetic acid bacteria of the genus Acetobacter, and the yeast species Kluyveromyces lactis and Saccharomyces cerevisiae (Dimidi et al., 2019). Furthermore, it has been shown that even though the abundance of yeasts in kefir grains may be relatively low compared to that of bacteria, their contribution to the fermentation process and the characteristics of the final product (e.g., acid production and aroma) can be significant (Chen et al., 2021). In the case of the kefir products investigated herein, the sole yeast isolate (isolate #8) originated from the homemade product, while only bacterial isolates were recovered from the industrial products, probably reflecting corresponding differences in the microbial consortia used as starter cultures.
Table olives are regarded as one of the oldest fermented vegetables in the Mediterranean area, constituting an important element of the economy of several countries, with the main producers being Spain, Greece, Italy, and Portugal (IOC, 2022). Olive fermentation, which still adheres to traditional processes, is based on the enzymatic activity of various members of the autochthonous microbiota of olive drupes. This indigenous microbiota is mainly composed of LAB and yeasts; the most identified bacteria are Lactobacillus spp., while yeast species of the genera Candida, Pichia and Saccharomyces have also been isolated (Argyri et al., 2020; Torres et al., 2020). As is the case for kefir, fermented table olives also exhibit a favorable health profile which, beyond their high content in bioactive compounds (e.g., unsaturated fatty acids and phenolic compounds), expands to their putative role as food vehicles for probiotics’ delivery (Bonatsou et al., 2017; Perpetuini et al., 2020). The specific microbial species that will shape both the technological characteristics (e.g., texture, flavor) and functional traits of the final product seem to depend largely on the applied fermentation process. Specifically, in natural black olives which were one of the microbial isolation sources in this study, the fermentation is principally driven by lactobacilli, with Lactobacillus pentosus and Lb. plantarum being identified as the main LAB species involved, while Leuconostoc mesenteroides has also been recovered from the final product (Doulgeraki et al., 2013; Bonatsou et al., 2017; Perpetuini et al., 2020). The contribution of yeasts is also undeniably important, with this microbial group playing a decisive role in the development of the organoleptic characteristics of the final product, while at the same time enhancing LAB growth through the release of nutritive compounds (Nisiotou et al., 2010; Perpetuini et al., 2020). However, certain yeast species can cause significant product deterioration due to spoilage (Nisiotou et al., 2010), justifying to some extent why the microbiota of successfully fermented table olives is predominantly populated by LAB species. The outgrowth and prevalence of LAB over yeasts during olive fermentation results in the acidification of brine (due to the production of lactic acid) which, in turn, ensures the microbiological stability of the final product (Perpetuini et al., 2020). Consistent with the anticipated dominance of LAB, six macroscopically distinct bacterial isolates were recovered from table olives in the present investigation (three isolates from drupes and three isolates from brine samples), while no yeast was isolated, not at least at the sample dilution levels required for TVC determination. On the other hand, the isolation of yeast #16 from OMW demonstrates that even if not present in the final product, yeasts are likely to participate in the indigenous microbiota of olive drupes, contributing mainly to the first stages of the fermentation process. From the table olives’ isolates, four (two from drupes and two from brines) were selected for further investigation. In quest of probiotics originating from table olives, the ones recovered from the surface of drupes may have a competitive advantage over microorganisms recovered from the brine. Such advantage is mainly associated with the indigenous ability of these microorganisms to adhere, an ability which is highly likely to allow for enhanced maintenance of microbial cell integrity and stability during transit in the gastrointestinal tract and ultimately, to favorably contribute to microbial adhesion to the intestinal epithelium (Larsen et al., 2009; Peres et al., 2015).
Based on the results of this study, the microbial isolates exhibiting the highest acid resistance were the bacteria #1 and #2 originating from kefir, the bacterial isolate #11 recovered from olive brine, and both isolated yeasts, namely the isolates #8 (from kefir) and #16 (from OMW). High survival rates under low pH conditions have been commonly recorded among LAB and yeasts isolated from fermented foods in studies assessing the probiotic potential of these organisms (Argyri et al., 2013; Pavli et al., 2016; Oliveira et al., 2017; Bonatsou et al., 2018). The resistance of microorganisms to low pH is an important prerequisite for their putative probiotic label, since in the absence of such inherent acid resistance they will not be able to survive the transit through gastric fluids and thus, successfully colonize the intestinal epithelium. Although most of the tested isolates (8/11) exhibited survival exceeding 50% after 3-h exposure to the rather low pH of 2.5, the acid survival recorded for the two yeast isolates was well above 90%, and a similarly high survival was also noted for the yeast strain AXAZ-1. The remarkable acid resistance of yeasts has been well identified and documented in the scientific literature, constituting essentially one of their most important traits for their value as putative probiotics (Oliveira et al., 2017; Bonatsou et al., 2018). The collected in this study resistance data would be useful to be further substantiated in future research through additional experimental trials involving either exposure of the microbial isolates to simulated gastric fluid or utilization of gastrointestinal model systems.
Most of the tested in this study microbial isolates exhibited considerable autoaggregation capability. Indeed, 10 out of the 12 tested microorganisms demonstrated autoaggregation equal to or higher than 60% after 24 h, with five of them (i.e., isolates #2, #8, #11, #16 and the yeast strain AXAZ-1) demonstrating a rather high autoaggregation capability (>80%). The remarkable autoaggregation demonstrated by the yeasts (isolates #8 and #16 as well as AXAZ-1), even within the first 2 h of incubation, is most likely associated with the larger size of yeast cells compared to that of bacterial cells, facilitating the precipitation of the formed cell aggregates. The autoaggregation capability of microorganisms is regarded as indicative, to some extent, of their ability to adhere on biotic and/or abiotic surfaces, including the intestinal epithelium, an apparently desirable trait for microorganisms evaluated for their putative probiotic traits (Javanshir et al., 2021). Nonetheless, additional parameters may play an important role, such as the hydrophobicity and co-aggregation of microbial cells, while autoaggregation is commonly evaluated in conjunction with the biofilm-forming ability of microorganisms (Ogunremi et al., 2015; Bonatsou et al., 2018). As demonstrated by the biofilm formation data generated in the present study, the bacterial isolates #2, #10, #11, #13 and #15, as well as the yeast isolate #16, could be regarded as strong biofilm formers, and all these isolates, excluding #10, also demonstrated a high autoaggregation capability. It is also noteworthy that when the isolation source is considered, it seems that table olives may be a more propitious reservoir of microorganisms with high biofilm-forming ability than kefir. Only one of the kefir isolates (out of the five that were tested) was a strong biofilm former (isolate #2), while the rest of the isolates evaluated as such originated from fermented table olives (#10, #11, #13) and OMW (#15, #16). The high biofilm-forming ability of both LAB and yeast isolates originating from table olives has been also reported by other researchers (Grounta et al., 2016; León-Romero et al., 2016; Bonatsou et al., 2018; Vergara Alvarez et al., 2023), while findings relevant to the important probiotic properties of microorganisms isolated from table olive biofilms are certainly very interesting (Benítez-Cabello et al., 2020). Extracellular polymeric substances (EPS), being composed primarily of polysaccharides, are a vital structural and functional component of biofilms (Flemming, 2016). As supported by research findings, EPS production enhances the attachment of probiotics to the intestinal epithelium, inhibiting in this manner its colonization by foodborne pathogenic bacteria (Tatsaporn and Kornkanok, 2020). The biofilm-forming ability of the microbial isolates tested in this study was assessed via an indirect quantification methodology which refers to the total biofilm mass, not allowing the distinction among viable cells, dead cells, and EPS matrix components. Thus, a further research step could be the compositional characterization of the biofilms formed by the isolates identified herein as strong biofilm formers. Moreover, the attained in vitro results should be also ascertained in situ and ultimately ex vivo/in vivo, since biofilm formation on abiotic surfaces is not necessarily associated with microbial adhesivity on biotic surfaces (Peres et al., 2015).
The antioxidant activity of microbial metabolites constitutes an important field of study in Microbiology and Microbial Biochemistry, since the formation of reactive oxygen species (ROS) and the resulting oxidative stress is an important etiologic factor for many human diseases including diabetes, rheumatoid arthritis, and cardiovascular diseases (Forman and Zhang, 2021). Probiotics and their fermented metabolites, referred to as “postbiotics” have been shown to exhibit antioxidative activities and as such, are rather promising for the regulation of oxidative stress and eventually the protection of cells from oxidative damage (Hoffmann et al., 2021; Lin et al., 2022). Regarding the microorganisms investigated herein, the greatest antioxidant activity was exhibited by the isolate #16, corresponding to yeast isolated from OMW. The rest of the microbial isolates, being in their vast majority presumptive LAB, were evaluated as having variable antioxidant activity, with the estimated scavenging activity (expressed as percent reduction of DPPH) varying from 0.8 to 21.1%. These observations are generally in good agreement with the findings of previous studies assessing, among other probiotic traits, the antioxidant activity of LAB (Chen et al., 2014) and yeasts (Gil-Rodríguez et al., 2015). Although the mechanisms of their antioxidant action have not been fully elucidated, it has been proposed that probiotics may modulate the redox status of the host via their metal ion chelating ability, antioxidant enzyme systems (e.g., superoxide dismutase and catalase), antioxidant metabolites (e.g., glutathione, butyrate, and folate), as well as through regulation of signaling pathways, enzymes producing ROS and the intestinal microbiota (Wang et al., 2017).
For microorganisms with beneficial activity to be utilized by the food, pharmaceutical and/or cosmetic industries, their safety for humans must be explicitly demonstrated. The hemolytic activity of microbial strains is among the safety criteria that are commonly assessed since its absence is indicative of their safe use in consumer products’ applications (FAO/WHO, 2002). Hemolytic activity is regarded as an indication of potential pathogenicity, and thus, microbial isolates exhibiting β-hemolysis cannot be considered as potential probiotics. Excluding potentially isolate #1, which exhibited α-hemolysis (partial hemolysis), the rest of the microbial isolates investigated in the present study were evaluated as safe in terms of hemolysis. Alpha hemolysis is mainly caused by the microbial production of hydrogen peroxide, oxidizing oxy-hemoglobin to met-hemoglobin (McDevitt et al., 2020). In any case, however, observations based on in vitro trials should be confirmed through virulence assays in cell lines and subsequently, in the context of in vivo investigations.
Based on the collective evaluation of the results of the in vitro assays carried out in this study, five presumptive LAB (#2, #10, #11, #13, #15) as well as two yeast isolates (#8, #16) were identified as exhibiting desirable in vitro probiotic traits. Hence, these microbial isolates could be regarded as good candidates for inclusion in further studies aiming, ultimately, at their potential utilization in novel functional food products. The S. cerevisiae strain AXAZ-1 is an alcohol-resistant and psychrotolerant strain previously isolated from the agricultural area of North Achaia, Greece (Argiriou et al., 1992). Given its biotechnological interest, related mainly to the valorization of agro-industrial residues for the production of high-added value products such as ethanol (Kallis et al., 2019; Dourou et al., 2021), and the fact that S. cerevisiae strains have been commonly identified as having probiotic potential (Staniszewski and Kordowska-Wiater, 2021), it was thought as worthy and interesting to also evaluate the probiotic traits of this strain. Nevertheless, a probiotic potential was not plainly conveyed for AXAZ-1 under the conditions of this study; despite its high acid resistance and autoaggregation capability, this yeast strain was evaluated as a poor biofilm former with negligible antioxidant activity.
Microbial strains with functional properties, such as probiotic potential, may be used in various food product development applications. Such applications involve utilization of such strains as starter cultures (provided of course that they also possess the required technological properties) to produce probiotic fermented foods, or as adjunct cultures in various novel multi-functional food products of both enhanced health benefits and microbiological safety (Blana et al., 2014; Botta et al., 2015; Peres et al., 2015). The seven microbial isolates that were identified herein as exhibiting the most desirable in vitro probiotic traits, namely isolates #2, #8, #10, #11, #13, #15 and #16, could be regarded as good candidates for inclusion in further studies. Such studies should include: (i) molecular characterization of the identified isolates; (ii) assessment of additional probiotic properties (e.g., antimicrobial activity) and safety criteria (e.g., antibiotic resistance); (iii) in situ assessment of their performance as starter/adjunct cultures in selected food products; and (iv) ex vivo evaluation of both their safety and potential health benefits. The data collected in the present preliminary study provide the information needed for the targeted genetic characterization of the microbial isolates exhibiting probiotic potential, a characterization which is of even greater importance in the light of the recently emended taxonomy of LAB (Zheng et al., 2020). The genetic characterization of the identified microbial isolates, and the assessment of their behavior and performance in situ constitute our imminent research goals.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
NN: Formal Analysis, Investigation, Writing – original draft. AL: Conceptualization, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing.
The present work was supported by the University of Patras.
The authors would like to thank PROJECT 140 I.K.E. GREEK OLIVE COMPANY (138th KM National Road Athens – Lamia, Atalanti, GR 35200 https://www.project140.gr/) for kindly providing the table olives in brine used in the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author Alexandra Lianou declared that she was an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/finmi.2023.1257483/full#supplementary-material
Annunziata G., Arnone A., Ciampaglia R., Tenore G. C., Novellino E. (2020). Fermentation of foods and beverages as a tool for increasing availability of bioactive compounds. Focus on short-chain fatty acids. Foods 9 (8), 999. doi: 10.3390/foods9080999
Argiriou T., Kalliafas A., Psarianos C., Kana K., Kanellaki M., Koutinas A. A. (1992). New alcohol resistant strains of Saccharomyces cerevisiae species for potable alcohol production using molasse. Appl. Biochem. Biotechnol. 36, 153–161. doi: 10.1007/bf02921775
Argyri K., Doulgeraki A. I., Manthou E., Grounta A., Argyri A. A., Nychas G.-G. E., et al. (2020). Microbial diversity of fermented Greek table olives of Halkidiki and Konservolia varieties from different regions as revealed by metagenomic analysis. Microorganisms 8 (8), 1241. doi: 10.3390/microorganisms8081241
Argyri A. A., Zoumpopoulou G., Karatzas K. A., Tsakalidou E., Nychas G.-J., Panagou E. Z., et al. (2013). Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 33 (2), 282–291. doi: 10.1016/j.fm.2012.10.005
Benítez-Cabello A., Torres-Maravilla E., Bermúdez-Humarán L., Langella P., Martín R., Jiménez-Díaz R., et al. (2020). Probiotic properties of Lactobacillus strains isolated from table olive biofilms. Probiotics Antimicrob. Proteins 12 (3), 1071–1082. doi: 10.1007/s12602-019-09604-y
Blana V. A., Grounta A., Tassou C. C., Nychas G.-J. E., Panagou E. Z. (2014). Inoculated fermentation of green olives with potential probiotic Lactobacillus pentosus and Lactobacillus plantarum starter cultures isolated from industrially fermented olives. Food Microbiol. 38, 208–218. doi: 10.1016/j.fm.2013.09.007
Boltin D. (2016). Probiotics in Helicobacter pylori-induced peptic ulcer disease. Best Pract. Res. Clin. Gastroenterol. 30 (1), 99–109. doi: 10.1016/j.bpg.2015.12.003
Bonatsou S., Karamouza M., Zoumpopoulou G., Mavrogonatou E., Kletsas D., Papadimitriou K., et al. (2018). Evaluating the probiotic potential and technological characteristics of yeasts implicated in cv. Kalamata natural black olive fermentation. Int. J. Food Microbiol. 271, 48–59. doi: 10.1016/j.ijfoodmicro.2018.02.018
Bonatsou S., Tassou C. C., Panagou E. Z., Nychas G.-J. E. (2017). Table olive fermentation using starter cultures with multifunctional potential. Microorganisms 5 (2), 30. doi: 10.3390/microorganisms5020030
Botta C., Bertolino M., Zeppa G., Cocolin L. (2015). Evaluation of Toma Piemontese PDO cheese as a carrier of putative probiotics from table olive fermentations. J. Funct. Foods 18, Part A, 106–116. doi: 10.1016/j.jff.2015.06.063
Casertano M., Fogliano V., Ercolini D. (2022). Psychobiotics, gut microbiota and fermented foods can help preserving mental health. Food Res. Int. 152, 110892. doi: 10.1016/j.foodres.2021.110892
Chen Z., Liu T., Ye T., Yang X., Xue Y., Shen Y., et al. (2021). Effect of lactic acid bacteria and yeasts on the structure and fermentation properties of Tibetan kefir grains. Int. Dairy J. 114, 104943. doi: 10.1016/j.idairyj.2020.104943
Chen P., Zhang Q., Dang H., Liu X., Tian F., Zhao J., et al. (2014). Screening for potential new probiotic based on probiotic properties and α-glucosidase inhibitory activity. Food Control 35 (1), 65–72. doi: 10.1016/j.foodcont.2013.06.027
Cho Y. J., Kim D. H., Jeong D., Seo K. H., Jeong H. S., Lee H. G., et al. (2018). Characterization of yeasts isolated from kefir as a probiotic and its synergic interaction with the wine byproduct grape seed flour/extract. LWT-Food Sci. Technol. 90, 535–539. doi: 10.1016/j.lwt.2018.01.010
Dimidi E., Cox S. R., Rossi M., Whelan K. (2019). Fermented foods: Definitions and characteristics, impact on the gut microbiota and effects on gastrointestinal health and disease. Nutrients 11 (8), 1806. doi: 10.3390/nu11081806
Doulgeraki A. I., Pramateftaki P., Argyri A. A., Nychas G.-J. E., Tassou C. C., Panagou E. Z. (2013). Molecular characterization of lactic acid bacteria isolated from industrially fermented Greek table olives. LWT-Food Sci. Technol. 50 (1), 353–356. doi: 10.1016/j.lwt.2012.07.003
Dourou M., Economou C. N., Aggeli L., Janák M., Valdés G., Elezi N., et al. (2021). Bioconversion of pomegranate residues into biofuels and bioactive lipids. J. Clean. Prod. 323, 129193. doi: 10.1016/j.jclepro.2021.129193
FAO/WHO (2002) Joint FAO/WHO working group report on drafting guidelines for the evaluation of probiotics in food. Available at: https://www.fao.org/3/a0512e/a0512e.pdf (Accessed July 11, 2023).
Fijan S. (2014). Microorganisms with claimed probiotic properties: an overview of recent literature. Int. J. Environ. Res. 11 (5), 4745–4767. doi: 10.3390/ijerph110504745. Public Health.
Flemming H. C. (2016). EPS-then and now. Microorganisms 4 (4), 41. doi: 10.3390/microorganisms4040041
Forman H. J., Zhang H. (2021). Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat. Rev. Drug Discovery 20 (9), 689–709. doi: 10.1038/s41573-021-00233-1. Erratum in: Nat. Rev. Drug. Discov. 20(8), 652.
Gil-Rodríguez A. M., Carrascosa A. V., Requena T. (2015). Yeasts in foods and beverages: In vitro characterisation of probiotic traits. LWT-Food Sci. Technol. 64 (2), 1156–1162. doi: 10.1016/j.lwt.2015.07.042
Grounta A., Doulgeraki A. I., Nychas G.-J. E., Panagou E. Z. (2016). Biofilm formation on Conservolea natural black olives during single and combined inoculation with a functional Lactobacillus pentosus starter culture. Food Microbiol. 56, 35–44. doi: 10.1016/j.fm.2015.12.002
Hill C., Guarner F., Reid G., Gibson G. R., Merenstein D. J., Pot B., et al. (2014). Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11 (8), 506–514. doi: 10.1038/nrgastro.2014.66
Hoffmann A., Kleniewska P., Pawliczak R. (2021). Antioxidative activity of probiotics. Arch. Med. Sci. 17 (3), 792–804. doi: 10.5114/aoms.2019.89894
Ibrahim S. A., Gyawali R., Awaisheh S. S., Ayivi R. D., Silva R. C., Subedi K., et al. (2021). Fermented foods and probiotics: An approach to lactose intolerance. J. Dairy Res. 88 (3), 357–365. doi: 10.1017/S0022029921000625
IOC (2022) World olive oil and table olive figures. Available at: https://www.internationaloliveoil.org/what-we-do/economic-affairs-promotion-unit/#figures (Accessed July 11, 2023).
Javanshir N., Hosseini G. N. G., Sadeghi M., Esmaeili R., Satarikia F., Ahmadian G., et al. (2021). Evaluation of the function of probiotics, emphasizing the role of their binding to the intestinal epithelium in the stability and their effects on the immune system. Biol. Proced. 23 (1), 23. doi: 10.1186/s12575-021-00160-w
Kallis M., Sideris K., Kopsahelis N., Bosnea L., Kourkoutas Y., Terpou A., et al. (2019). Pistacia terebinthus resin as yeast immobilization support for alcoholic fermentation. Foods 8, 127. doi: 10.3390/foods8040127
Kim H.-Y., Park K.-Y. (2018). Clinical trials of kimchi intakes on the regulation of metabolic parameters and colon health in healthy Korean young adults. J. Funct. Foods 47, 325–333. doi: 10.1016/j.jff.2018.05.052
Kok C. R., Hutkins R. (2018). Yogurt and other fermented foods as sources of health-promoting bacteria. Nutr. Rev. 76 (Suppl 1), 4–15. doi: 10.1093/nutrit/nuy056
Larsen N., Michaelsen K. F., Pærregaard A., Vogensen F. K., Jakobsen M. (2009). A comparative study on adhesion and recovery of potential probiotic strains of Lactobacillus spp. by in vitro assay and analysis of human colon biopsies. Microb. Ecol. Health Dis. 21 (2), 95–99. doi: 10.1080/08910600902907632
Legesse Bedada T., Feto T. K., Awoke K. S., Garedew A. D., Yifat F. T., Birri D. J. (2020). Probiotics for cancer alternative prevention and treatment. BioMed. Pharmacother. 129, 110409. doi: 10.1016/j.biopha.2020.110409
León-Romero Á., Domínguez-Manzano J., Garrido-Fernández A., Arroyo-López F. N., Jiménez-Díaz R. (2016). Formation of in vitro mixed-species biofilms by Lactobacillus pentosus and yeasts isolated from Spanish-style green table olive fermentations. Appl. Environ. Microbiol. 82 (2), 689–695. doi: 10.1128/AEM.02727-15
Lianou A., Koutsoumanis K. P. (2012). Strain variability of the biofilm-forming ability of Salmonella enterica under various environmental conditions. Int. J. Food Microbiol. 160 (2), 171–178. doi: 10.1016/j.ijfoodmicro.2012.10.002
Lin W. Y., Lin J. H., Kuo Y. W., Chiang P. R., Ho H. H. (2022). Probiotics and their metabolites reduce oxidative stress in middle-aged mice. Curr. Microbiol. 79 (4), 104. doi: 10.1007/s00284-022-02783-y
Liu L., Wang J., Levin M. J., Sinnott-Armstrong N., Zhao H., Zhao Y., et al. (2019). The origins of specialized pottery and diverse alcohol fermentation techniques in Early Neolithic China. Proc. Natl. Acad. Sci. U.S.A. 116 (26), 12767–12774. doi: 10.1073/pnas.1902668116
Lopez-Santamarina A., Gonzalez E. G., Lamas A., Mondragon A. D. C., Regal P., MIrand J. M. (2021). Probiotics as a possible strategy for the prevention and treatment of allergies. A narrative review. Foods. 10 (4), 701. doi: 10.3390/foods10040701
Lopitz-Otsoa F., Rementeria A., Elguezabal N., Garaizar J. (2003). Kefir: a symbiotic yeasts-bacteria community with alleged healthy capabilities. Rev. Iberoam Micol. 23 (2), 67–74. doi: 10.1016/s1130-1406(06)70016-x
Ma T., Shen X., Shi X., Sakandar H. A., Quan K., Li Y., et al. (2023). Targeting gut microbiota and metabolism as the major probiotic mechanism - An evidence-based review. Trends Food Sci. Technol. 138, 178–198. doi: 10.1016/j.tifs.2023.06.013
Marco M. L., Heeney D., Binda S., Cifelli C. J., Cotter P. D., Foligné. B. (2017). Health benefits of fermented foods: microbiota and beyond. Curr. Opin. Biotechnol. 44, 94–102. doi: 10.1016/j.copbio.2016.11.010
Marco M. L., Pavan S., Kleerebezem M. (2006). Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 17 (2), 204–210. doi: 10.1016/j.copbio.2006.02.005
Marco M. L., Sanders M. E., Gänzle M., Arrieta M. C., Cotter P. D., De Vuyst L., et al. (2021). The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat. Rev. Gastroenterol. Hepatol. 18 (3), 196–208. doi: 10.1038/s41575-020-00390-5
McDevitt E., Khan F., Scasny A., Thompson C. D., Eichenbaum Z., McDaniel L. S., et al. (2020). Hydrogen peroxide production by Streptococcus pneumoniae results in alpha-hemolysis by oxidation of oxy-hemoglobin to met-hemoglobin. mSphere 5 (6), e01117–e01120. doi: 10.1128/mSphere.01117-20
Nisiotou A. A., Chorianopoulos N., Nychas G.-J. E., Panagou E. Z. (2010). Yeast heterogeneity during spontaneous fermentation of black Conservolea olives in different brine solutions. J. Appl. Microbiol. 108 (2), 396–405. doi: 10.1111/j.1365-2672.2009.04424.x
Nuraida L. (2015). A review: Health promoting lactic acid bacteria in traditional Indonesian fermented foods. Food Sci. Hum. Wellness 4 (2), 47–55. doi: 10.1016/j.fshw.2015.06.001
Ogunremi O. R., Sanni A. I., Agrawal R. (2015). Probiotic potentials of yeasts isolated from some cereal‐based Nigerian traditional fermented food products. J. Appl. Microbiol. 119 (3), 797–808. doi: 10.1111/jam.12875
Oliveira T., Ramalhosa E., Nunes L., Pereira J. A., Colla E., Pereira E. L. (2017). Probiotic potential of indigenous yeasts isolated during the fermentation of table olives from Northeast of Portugal. Innov. Food Sci. Emerg. Technol. 44, 167–172. doi: 10.1016/j.ifset.2017.06.003
Pachla A., Ptaszyńska A. A., Wicha M., Kunat M., Wydrych J., Oleńska E., et al. (2021). Insight into probiotic properties of lactic acid bacterial endosymbionts of Apis mellifera L. derived from the Polish apiary. Saudi J. Biol. Sci. 28 (3), 1890–1899. doi: 10.1016/j.sjbs.2020.12.040
Pavli F. G., Argyri A. A., Papadopoulou O. S., Nychas G.-J. E., Chorianopoulos N. G., Tassou C. C. (2016). Probiotic potential of lactic acid bacteria from traditional fermented dairy and meat products: Assessment by in vitro tests and molecular characterization. J. Prob. Health 4 (3), 1000157. doi: 10.4172/2329-8901.1000157
Peres C. M., Hernandez-Mendonza A., Bronze M. R., Peres C., Xavier Malcata F. (2015). Synergy of olive bioactive phytochemicals and probiotic strain in control of Escherichia coli. LWT-Food Sci.Technol. 64 (2), 938–945. doi: 10.1016/j.lwt.2015.06.074
Perpetuini G., Prete R., Garcia-Gonzalez N., Khairul Alam M., Corsetti A. (2020). Table olives more than a fermented food. Foods 9 (2), 178. doi: 10.3390/foods9020178
Rezac S., Kok C. R., Heermann M., Hutkins R. (2018). Fermented foods as a dietary source of live organisms. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01785
Soemarie Y. B., Milanda T., Barliana M. I. (2021). Fermented foods as probiotics: A Review. J. Adv. Pharm. Technol. Res. 12 (4), 335–339. doi: 10.4103/japtr.japtr_116_21
Staniszewski A., Kordowska-Wiater M. (2021). Probiotic and potentially probiotic yeasts-Characteristics and food application. Foods 10 (6), 1306. doi: 10.3390/foods10061306
Tatsaporn T., Kornkanok K. (2020). Using potential lactic acid bacteria biofilms and their compounds to control biofilms of foodborne pathogens. Biotechnol. Rep. (Amst). 26, e00477. doi: 10.1016/j.btre.2020.e00477
Torres S., Verón H., Contreras L., Isla M. I. (2020). An overview of plant-autochthonous microorganisms and fermented vegetable foods. Food Sci. Hum. Wellness 9 (2), 112–123. doi: 10.1016/j.fshw.2020.02.006
Vergara Alvarez S. C., Leiva Alaniz M. J., Mestre Furlani V. M., Vazquez F., Mancha Agresti P., Nally C. M., et al. (2023). Bioprospecting of the probiotic potential of yeasts isolated from a wine environment. Fungal Genet. Biol. 164, 103767. doi: 10.1016/j.fgb.2022.103767
Wang Y., Wu Y., Wang Y., Xu H., Mei X., Yu D., et al. (2017). Antioxidant properties of probiotic bacteria. Nutrients 9 (5), 521. doi: 10.3390/nu9050521
Xiang H., Sun-Waterhouse D., Waterhouse G. I., Cui C., Ruan Z. (2019). Fermentation-enabled wellness foods: A fresh perspective. Food Sci. Hum. Wellness 8 (3), 203–243. doi: 10.1016/j.fshw.2019.08.003
Yan F., Polk B. D. (2011). Probiotics and immune health. Curr. Opin. Gastroenterol. 27 (6), 496–501. doi: 10.1097/MOG.0b013e32834baa4d
Zheng J., Wittouck S., Salvetti E., Franz C. M. A. P., Harris H. M. B., Mattarelli P., et al. (2020). A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70 (4), 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: fermented foods, probiotic potential, milk kefir, table olives, lactic acid bacteria, yeasts
Citation: Ntiantiasi N and Lianou A (2023) Isolation and in vitro screening of the probiotic potential of microorganisms from fermented food products. Front. Ind. Microbiol. 1:1257483. doi: 10.3389/finmi.2023.1257483
Received: 12 July 2023; Accepted: 21 August 2023;
Published: 20 September 2023.
Edited by:
Marta Laranjo, University of Evora, PortugalReviewed by:
Katarzyna Pobiega, Warsaw University of Life Sciences, PolandCopyright © 2023 Ntiantiasi and Lianou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandra Lianou, YWxpYW5vdUB1cGF0cmFzLmdy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.