
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Immunol., 03 April 2025
Sec. Vaccines and Molecular Therapeutics
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1569251
This article is part of the Research TopicVaccines and Breakthrough InfectionsView all 6 articles
 Lauren M. Schwartz1
Lauren M. Schwartz1 Jose Vila-Belda1
Jose Vila-Belda1 Jerome Carless1
Jerome Carless1 Sadish Dhakal1
Sadish Dhakal1 Koen Hostyn1
Koen Hostyn1 Trina Gorman1
Trina Gorman1 Deborah Ogbeni2
Deborah Ogbeni2 Gathoni Kamuyu2
Gathoni Kamuyu2 Mark Manak3
Mark Manak3 Valentina Bernasconi4
Valentina Bernasconi4 Ali Azizi5*
Ali Azizi5*The CEPI-Centralized Laboratory Network (CLN) has significantly contributed to the development of several approved SARS-CoV-2 vaccines by conducting over 70,000 clinical samples for testing from various vaccine developers. A centralized data management system was developed to track, review, store and share immunological clinical results generated from sample testing. The data system ensures the completeness and accuracy of submitted results and checks the set criteria in controls for each assay. Each testing facility within the network submits their results to a secure storage system using report forms with embedded data quality checks. Upon submission, a statistical program runs additional checks to identify errors in completeness and uniqueness. Any discrepancies or errors are shared with the testing facility to rectify. Reports are further reviewed by CEPI-CLN experts before releasing to the vaccine developer. Study results are then consolidated into an internal relational database management system, enabling CEPI to analyze the data through an interactive dashboard that visualizes control trends and sample results across all studies. This analysis facilitates the harmonization of immunological data and helps to inform CEPI’s programmatic and strategic decision making. Given the success of this approach with SARS-CoV-2 vaccines, the system will be adopted for new pathogens and assay types currently under development at CEPI-CLN.
As of May 2024, the CEPI-Centralized Laboratory Network (CLN) has significantly contributed to the development of several approved SARS-CoV-2 vaccines by conducting over 120,000 assay runs (over 70,000 clinical samples) for testing from various vaccine developers worldwide (1–4). In summary six SARS-CoV-2 immunological assays have been developed, validated or qualified, and transferred to the network using the same materials, key reagents, and protocols: three binding assays (S-, RBD, and N-ELISA), a microneutralization assay (MNA), a pseudotyped virus-based neutralization assay (PNA), and an IFN-γ T-cell ELISpot assay. Inter-lab studies using replicate assays, as well as revalidation in receiving facilities, have shown that results are highly reproducible, allowing for direct comparison of different vaccines throughout the network (2). Reliability of clinical sample testing is assured through the implementation of an internal centralized system designed to store sensitive and proprietary data and perform data quality checks on the immunological clinical results, ensuring the integrity and consistency of data collected. Additionally, the system enables trend analysis of reference standards and controls, generated by the Medicines and Healthcare Products Regulatory Agency (MHRA, formerly NIBSC), allowing CEPI-CLN to harmonize results across laboratories and to identify any potential issues or anomalies in the data that indicate of loss of consistency between facilities. This centralized system plays a crucial role in maintaining the quality and integrity of clinical sample testing processes.
The automated process by which submitted clinical data is checked for consistency, consolidated, cleaned, and stored in a database to enable analysis, is called the data pipeline (Figure 1). All steps in the data pipeline were developed by CEPI-CLN and for internal use only. The data pipeline is managed by Apache Airflow (5), an open-source platform that initiates and tracks each dependent step. Further, isolated and encrypted environments on both Amazon Web Services (AWS) and Heroku are used to house Apache Airflow and the database, respectively. Compliance with international data privacy laws (General Data Protection Regulation [GDPR]) (6) and ISO 27001 is achieved through multiple technical and organizational measures that have been deployed throughout the data pipeline including: pseudonymized specimen identifiers, regulated and restricted data access to all software and database storage systems through the principle of least privilege, weekly database and systems backups, and the use of isolated and encrypted cloud environments. The data pipeline was first built within a testing space to ensure a valid and secure pipeline before moving to a production space where regular audits and vulnerability assessments are conducted.
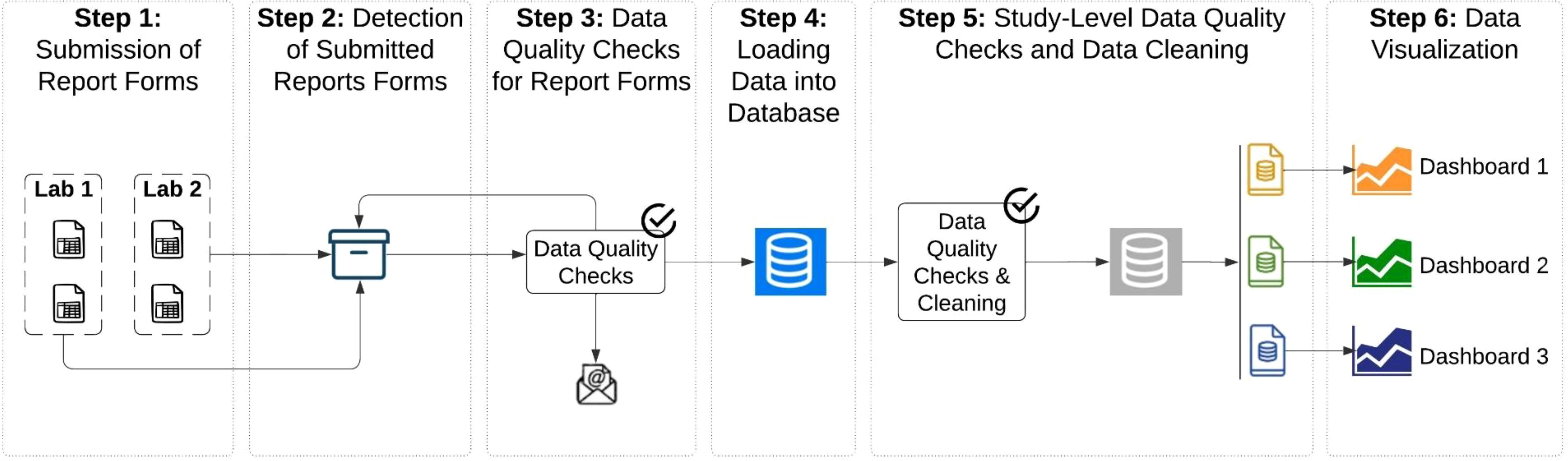
Figure 1. Design of the data pipeline from report submission to visualization. Image shows the different steps of the data pipeline including labs submitting report forms (Step 1), detection of report forms in the centralized storage repository (Step 2), automated data quality checks on the report forms (Step 3), loading (Step 4) and further cleaning of the data in the database (Step 5) and finally visualizations in dashboards (Step 6).
Testing facilities within the network have quality management systems in place, employing quality control procedures throughout the analytical process. From sample receipt through to reporting, facilities are responsible for assuring the integrity of the results they generate.
Assay results are entered into a standardized report form at each facility. All facilities participate in virtual training sessions to review consistent data entry protocols. Each report may contain all or a subset of sample testing results for each clinical study with only information relevant for immunologic testing including sample and participant unique identifiers, study time point, data of collection, plate control identifier, and assay result. Additional information collected during the course of a clinical trial, such as participant-level demographic variables, is not shared with the facilities. The report form contains data validation rules, drop down menus, formulas, and conditional formatting (Table 1) as a first step in ensuring the completeness and consistency of results. For example, cells change color if any information related to a test sample or plate control is missing. This allows facility staff and reviewers to scan the report to identify missing critical data before submission. Additionally, to complement facility quality control procedures, warnings appear when plate control values fall outside of pre-defined acceptance ranges, signaling samples that need to be retested. The report form is locked, and password protected so that the facility staff cannot accidentally change the embedded functionality. Within each facility, all report forms are approved by a quality control manager or designated expert before being uploaded onto a central encrypted file storage system that is compliant with GDPR (6).
Following data submission, sample and control immunological results enter CEPI’s automated data pipeline, where each subsequent step is initiated by Apache Airflow (5). The file storage system is automatically checked every hour to detect if new report forms have been submitted. Once a new report form is detected, a python program downloads the file to a temporary directory, triggering a series of automated data quality checks, as described in Step 3.
A statistical program designed in Stata is run to ensure each report form retains embedded functionality and calculations, is complete, and passes a series of other quality checks, such as that dates are valid and that plate control acceptance criteria, based on the reagent lot, is calculated correctly (Table 1). Results of these checks are saved in a spreadsheet that is uploaded to the central file storage system. An email is sent to relevant CEPI-CLN staff informing them of the result of these checks. The CEPI-CLN team reviews the spreadsheet, identifying and communicating any data quality errors to the facility if needed. Subsequently, the facility may submit a revised report form, which supersedes the previous version, thereby avoiding duplicates in the central file storage system. Report forms approved by CEPI are shared with vaccine developers through the central file storage system. Interpretation of clinical testing results and their relevance to efficacy of the vaccine are the responsibility of the vaccine developer.
All rows from the submitted report form with complete sample and plate control data are bulk inserted into a single relational PostgreSQL database. The database is located in a dedicated and isolated environment designed for storing sensitive data. PostgresSQL is inherently ACID compliant. Specifically, the psycopg2 package (7, 8) handles Atomicity and Isolation by managing transactions using connection objects, commits, and rollbacks. Consistency is maintained by enforcing unique constraints across all tables in the database. Durability is achieved through a combination of postgres’ internal Write-Ahead Logging (9) and hourly backup snapshots managed by Heroku (10).
Data is first moved to staging tables which contain all pre-processed data. This provides a historical snapshot of the clinical trial data across each batch. Data are loaded into separate tables for clinical sample results, plate control results, and data quality check summaries. Key identifiers are maintained across all tables and include facility name, assay type, report name, study ID and load date. To maintain idempotency, records matching the bulk insert load date are removed before each insert, ensuring no unexpected duplication.
A second round of data quality checks are performed within the database across all reports submitted for each study, to ensure all key indicators (sample identifiers, assay type, facility name, study ID, dates) are complete and properly recorded and time points are consistent and valid (Table 1). Discrepant results are again reviewed by the CEPI team and, if needed, the facility resubmits revised reports to correct issues. The quality checks are run automatically as each new report is submitted, but it may take weeks until all samples are tested and for final data quality checks to be performed. In the final production tables, version control for revised report forms is maintained through tracking the submission time of each report form and upsert queries ensure that only unique data are inserted. Various data cleaning processes are performed including calculating international standard unit conversions and aligning study visit time point variables across studies (e.g. ‘day1’ and ‘D01’ cleaned to ‘Day 1’). Materialized views are created from the final production tables that aggregate, combine, or reshape data as needed for the visualizations.
Using an encrypted connection to the materialized views in the database, an interactive dashboard of data visualizations enables CEPI to continually analyze and monitor submitted results. The dashboard visualizes aspects such as 1) the number and status of reports that have been submitted to support scheduling and inventory control, 2) trends in immunological results by assay and study (Figure 2A), and 3) plate control trends by assay, lot, and lab over time (Figure 2B). As of May 2024, over 300 reports have been uploaded and quality checked since the start of the data pipeline in 2022. Importantly, control results are used to track trends by facility and across time to ensure consistency and identify any possible quality issues.
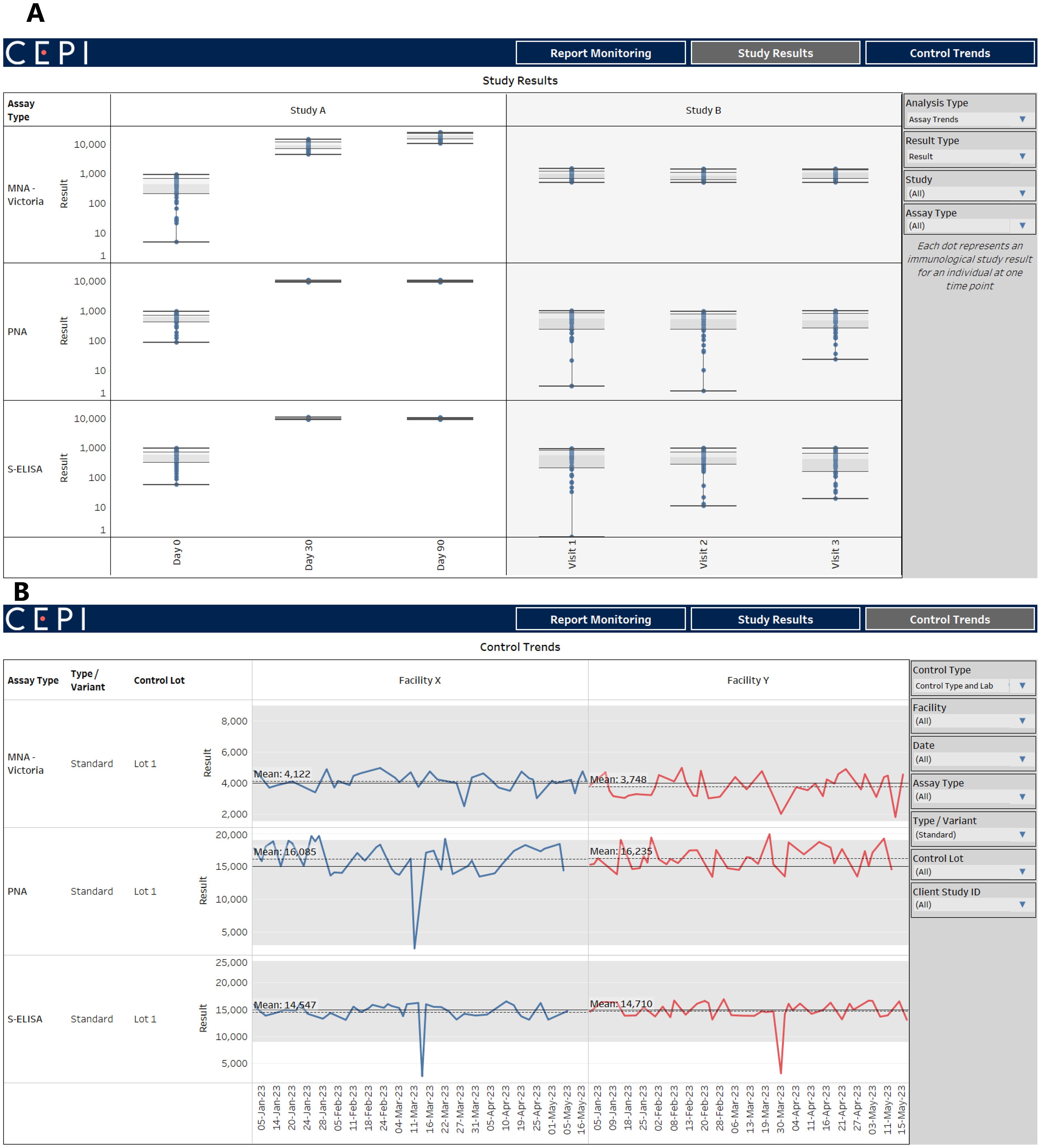
Figure 2. Tableau dashboard with with assay results and control trends. (A) shows example clinical trials results on a dashboard connected to CEPI-CLN database. (B) shows example control results which can be used to track trends by facility and across time to ensure consistency and identify any possible quality issues.
The CEPI-CLN currently includes 18 facilities across the world. This network relies on a series of processes to ensure the consistency, completeness, and reliability of vaccine test sample results across all facilities. These processes include standardized and harmonized assay procedures, regular proficiency testing after the post-technology transfer, data quality checks, and ongoing communication and collaboration among the network. By implementing these rigorous processes, the CEPI-CLN aims to maintain high standards of quality assurance and control, ultimately contributing to the development of safe and effective vaccines against emerging infectious diseases. The data quality checks and ongoing analysis provide additional confidence in the data, which is both shared with vaccine developers and used to inform CEPI’s programmatic and strategic decisions. Using the resulting data and visualizations, CEPI can facilitate rapid evaluation and dissemination of the most effective vaccine candidates. Additionally, CEPI can also obtain a better understanding of aspects such as the correlation and duration of protection across multiple SARS-CoV-2 vaccine clinical trials, as well as identifying which vaccine platforms require support towards licensure. Given the success of this approach with COVID-19 vaccines, the system is currently being adopted for new pathogens and assay types currently under development at CEPI-CLN (11). The database may also start to leverage machine learning or Artificial Intelligence (AI) tools to supplement quality control systems. Since 2023, CEPI has made significant investments and partnered with several private companies and recognized academic institutions to incorporate AI-driven tools in various areas to support CEPI’s 100 Days Mission: to quickly make safe and effective vaccines against any viral pandemic threat. Additionally, we plan to incorporate study-level demographic information in the database to support high-level analyses related to vaccine response in different populations. In summary, these processes not only maintain high-quality standards but also strengthen global preparedness, reinforcing CEPI’s commitment to equitable access to vaccines against rare pathogens.
The datasets presented in this article are not readily available because of privacy concerns. Requests to access the datasets should be directed to author AA, YWxpLmF6aXppQGNlcGkubmV0.
LS: Conceptualization, Data Curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. JV-B: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. JC: Data curation, Formal analysis, Investigation, Software, Writing – original draft, Writing – review & editing. SD: Data curation, Formal analysis, Investigation, Software, Writing – original draft, Writing – review & editing. KH: Methodology, Software, Writing – original draft, Writing – review & editing. TG: Methodology, Writing – original draft, Writing – review & editing. DO: Investigation, Writing – original draft, Writing – review & editing. GK: Investigation, Writing – original draft, Writing – review & editing. MM: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. VB: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. AA: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. All work discussed here in was funded by CEPI under contractual agreements with the respective laboratories. The authors do not receive any royalties, licenses, stock options, or other financial benefit.
We would like to thank CEPI-CLN facilities for their support during the development of the database.
Authors LS, JV-B, JC, SD, KH and TG were employed by company Gorman Consulting. Author MM was employed by company Turesol Consulting.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Azizi A, Manak M, Bernasconi V. The CEPI centralized laboratory network for COVID-19 will help prepare for future outbreaks. Nat Med. (2023) 29:2684–5. doi: 10.1038/s41591-023-02534-x
2. Manak M, Gagnon L, Phay-Tran S, Levesque-Damphousse P, Fabie A, Daugan M, et al. Standardised quantitative assays for anti-SARS-CoV-2 immune response used in vaccine clinical trials by the CEPI Centralized Laboratory Network: a qualification analysis. Lancet Microbe. (2024) 5:e216–25. doi: 10.1016/S2666-5247(23)00324-5
3. Azizi A, Bernasconi V. Unifying global efforts by CEPI’s centralized laboratory network. Front Immunol. (2024) 15:1404309. doi: 10.3389/fimmu.2024.1404309
4. Azizi A, Kamuyu G, Ogbeni D, Levesque-Damphousse P, Knott D, Gagnon L, et al. Driving consistency: CEPI-Centralized Laboratory Network’s conversion factor initiative for SARS-CoV-2 clinical assays used for efficacy assessment of COVID vaccines. Hum Vaccines Immunother. (2024) 20:2344249. doi: 10.1080/21645515.2024.2344249
5. Apache Airflow. Home (2024). Available online at: https://airflow.apache.org/ (Accessed June 24, 2024).
6. GDPR.eu. GDPR compliance checklist (2024). Available online at: https://gdpr.eu/checklist/ (Accessed June 24, 2024).
7. The connection class — Psycopg 2.9.10 documentation (2024). Available online at: https://www.psycopg.org/docs/connection.html (Accessed June 24, 2024).
8. Basic module usage — Psycopg 2.9.10 documentation (2024). Available online at: https://www.psycopg.org/docs/usage.html (Accessed June 24, 2024).
9. PostgreSQL Documentation. 28.3. Write-Ahead Logging (WAL) (2024). Available online at: https://www.postgresql.org/docs/17/wal-intro.html (Accessed June 24, 2024).
10. Heroku PGBackups | Heroku Dev Center . Available online at: https://devcenter.heroku.com/articles/heroku-postgres-backups (Accessed June 24, 2024).
Keywords: vaccine results, data management, quality, trial monitoring, database
Citation: Schwartz LM, Vila-Belda J, Carless J, Dhakal S, Hostyn K, Gorman T, Ogbeni D, Kamuyu G, Manak M, Bernasconi V and Azizi A (2025) Monitoring immunological COVID-19 vaccine clinical testing across the CEPI Centralized Laboratory Network. Front. Immunol. 16:1569251. doi: 10.3389/fimmu.2025.1569251
Received: 31 January 2025; Accepted: 18 March 2025;
Published: 03 April 2025.
Edited by:
Sonia Jangra, The Rockefeller University, United StatesReviewed by:
Willy A. Valdivia-Granda, Orion Integrated Biosciences, United StatesCopyright © 2025 Schwartz, Vila-Belda, Carless, Dhakal, Hostyn, Gorman, Ogbeni, Kamuyu, Manak, Bernasconi and Azizi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali Azizi, YWxpLmF6aXppQGNlcGkubmV0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.