
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 20 February 2025
Sec. Alloimmunity and Transplantation
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1554876
Diabetes mellitus (DM) is a global health crisis affecting millions, with islet transplantation emerging as a promising treatment strategy to restore insulin production. This review synthesizes the current research on single-cell and spatial transcriptomics in the context of islet transplantation, highlighting their potential to revolutionize DM management. Single-cell RNA sequencing, offers a detailed look into the diversity and functionality within islet grafts, identifying specific cell types and states that influence graft acceptance and function. Spatial transcriptomics complements this by mapping gene expression within the tissue’s spatial context, crucial for understanding the microenvironment surrounding transplanted islets and their interactions with host tissues. The integration of these technologies offers a comprehensive view of cellular interactions and microenvironments, elucidating mechanisms underlying islet function, survival, and rejection. This understanding is instrumental in developing targeted therapies to enhance graft performance and patient outcomes. The review emphasizes the significance of these research avenues in informing clinical practices and improving outcomes for patients with DM through more effective islet transplantation strategies. Future research directions include the application of these technologies in personalized medicine, developmental biology, and regenerative medicine, with the potential to predict disease progression and treatment responses. Addressing ethical and technical challenges will be crucial for the successful implementation of these integrated approaches in research and clinical practice, ultimately enhancing our ability to manage DM and improve patient quality of life.
Diabetes mellitus (DM) is a burgeoning global health crisis, with the International Diabetes Federation (IDF) estimating that over 537 million adults are currently affected, a figure expected to grow to 643 million by 2030 (1). This increase, particularly in developing countries, exacerbates the strain on healthcare systems, necessitating innovative treatments like islet transplantation (2). Islet transplantation holds particular promise for DM management by restoring insulin production and metabolic control (3). However, the procedure faces challenges such as a lack of donor organs, lifelong immunosuppression requirements, and the risk of islet loss due to autoimmune or inflammatory responses (4). To overcome these, research into biomaterials and encapsulation techniques aims to improve patient outcomes (5, 6).
Recent advancements in single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics (ST) are transforming our comprehension of complex biological processes, particularly in the context of islet transplantation and DM management (7, 8). ScRNA-seq allows for the analysis of gene expression patterns on a per-cell basis, shedding light on the heterogeneity of cell populations within islets and their responses to transplantation (8). This technology is crucial for identifying specific cell types, their states, and interactions within the graft microenvironment, which can inform strategies for enhancing transplant outcomes and improving patient management. ST, on the other hand, complements scRNA-seq by mapping gene expression within the spatial context of tissues. This is essential for understanding the microenvironment surrounding transplanted islets and their interactions with host tissues. The spatial information provided by this technology helps to reveal how different cell types and their functions are organized within the tissue, which is critical for the success of islet transplantation.
The integration of these technologies enables researchers to explore cellular heterogeneity and spatial organization simultaneously, leading to a more holistic view of cellular interactions and microenvironments. This integrative approach is crucial for elucidating complex biological processes and disease mechanisms, and recent studies have demonstrated the efficacy of computational tools, such as the iSpatial algorithm. This facilitates the analysis of high-dimensional datasets, paving the way for novel discoveries in tissue biology and pathology (9).
The integration of scRNA-seq and ST provides a comprehensive view of cellular interactions and microenvironments, crucial for understanding complex biological processes (9). Recent studies, including high-plex protein and transcriptome co-mapping with spatial CITE-seq (10), multimodal tri-omics mapping of brain development and neuroinflammation (11), and spatially resolved in vivo CRISPR screening via Perturb-DBiT (12), demonstrate the efficacy of these technologies in revealing cellular heterogeneity and spatial organization. These advancements offer new insights into gene function and regulation, enhancing our ability to develop targeted therapies for islet transplantation and DM management.
This review aims to provide an overview of research on single-cell and ST in islet transplantation for DM treatment. By integrating these cutting-edge technologies into the study of islet transplantation, researchers can enhance the efficacy of this promising therapeutic approach and improve outcomes for patients suffering from DM through more effective islet transplantation strategies.
ScRNA-seq has emerged as a transformative method in islet transplantation, offering unprecedented insights into cellular heterogeneity and functionality (13). This technology allows researchers to dissect the complex cellular composition of islet grafts, enhancing our understanding of graft acceptance, function, and failure. The application of scRNA-seq is pivotal for identifying specific cell types, their states, and interactions within the graft microenvironment, which can inform strategies for enhancing transplant outcomes and improving patient management (14).
ScRNA-seq has revolutionized the study of cellular heterogeneity by quantifying unique gene expression profiles for each cell, providing a detailed view of the transcriptomic landscape (12). Recent advancements have improved resolution and throughput, enabling the analysis of thousands of cells simultaneously and facilitating the identification of specific cell types and their activation states (15). This technology is particularly advantageous in islet transplantation, where understanding distinct cellular populations and their functional states is crucial. Genomic medicine’s ongoing evolution is set to enhance clinical applications and expand genomics’ role in practice (16, 17). Best practices for scRNA-seq have been compiled to offer a comprehensive guide for beginners and update advanced users on the latest methods (18).
Recent advancements in scRNA-seq have greatly expanded our comprehension of islet cell dysfunction in DM (19, 20). Integration of chromatin accessibility, gene expression, and functional data with genetic associations has identified disease-causal gene regulatory changes, revealing two subtypes of β cell subtypes that shift in prevalence as type 2 DM progresses (21). A single-cell atlas of mouse islets (MIA) (22) and dynamic scRNA-seq of human pancreatic slices (HPSs) (23) have provided comprehensive resources for exploring pancreatic cell states and tissue plasticity. These studies have defined transcriptional signatures under endoplasmic reticulum and inflammatory stress, identified unique cell type responses, and highlighted the heterogeneity of T cell responses to insulin-derived epitopes (24). Additionally, human vascularized macrophage-islet organoids have modeled immune-mediated beta cell pyroptosis following viral infections, uncovering mechanisms of beta cell damage (25). Together, these discoveries highlight the complexity of immune responses and cellular heterogeneity in islet transplantation and DM, offering new avenues for targeted therapeutic interventions and improved treatment strategies.
The application of scRNA-seq has unveiled significant cellular heterogeneity within islet grafts, which is crucial for understanding transplant outcomes. Islet cells are not homogeneous, consisting of various cell types, each playing distinct roles in glucose homeostasis (26). ScRNA-seq allows researchers to categorize these cells based on their transcriptional profiles, revealing subpopulations that may respond differently to transplant conditions. For instance, certain beta cell subtypes have been shown to exhibit greater resilience to stress and inflammation (27), which are key factors in islet graft survival. Furthermore, scRNA-seq enables the exploration of intercellular communication networks within the graft, highlighting how different cell types interact and influence each other’s functions. This understanding of cellular heterogeneity is essential for enhancing graft function and longevity, ultimately improving the management of DM in transplant recipients. Recent scRNA-seq studies have shed new light on the immune context of islet transplants, identifying T lymphocytes and myeloid cells as key post-transplant immune elements, particularly in allografts (8, 13). Significantly, allogeneic islet cells can adopt antigen-presenting cell-like properties, potentially enhancing interactions with CD8+ T cells and contributing to allograft destruction (7). These findings underscore the immune response in islet transplantation and the potential for targeted immunomodulatory strategies to enhance graft survival.
ScRNA-seq has profound implications for the recovery of islet function following transplantation. By elucidating the cellular mechanisms that govern islet viability and function, researchers can identify potential therapeutic targets to enhance graft performance. For example, single-cell analyses have demonstrated that specific signaling pathways and gene expression profiles are associated with successful islet engraftment and function restoration (13). Moreover, understanding the dynamic changes in the islet cellular landscape post-transplantation can inform strategies to mitigate the effects of inflammation and immune rejection, which are significant barriers to long-term graft survival (28). The interventions aimed at modulating the immune response or enhancing the regenerative capacity of beta cells can lead to improved outcomes for transplant recipients (29). Thus, the insights gained from scRNA-seq are instrumental in advancing therapeutic strategies that promote islet recovery, ultimately contributing to better glycemic control and quality of life for patients with type 1 DM.
ST is a rapidly evolving field that integrates transcriptomic data with spatial information from tissue samples, allowing researchers to explore the complex cellular architecture of tissues in their native environments (30). This innovative technology overcomes the limitations of traditional scRNA-seq, which lacks spatial context (31). ST provides insights into cellular interactions and the microenvironment, which are crucial for understanding various biological processes and disease mechanisms. Recent advancements in ST techniques, such as multiplexed barcoding and high-resolution imaging, have significantly improved the ability to analyze large tissue sections, enabling the study of heterogeneous tissues, including tumors and organs affected by various diseases (32). The continuous development of ST provides a more comprehensive understanding of tissue biology.
ST technology, which enables the direct mapping of gene expression profiles onto tissue sections, has revolutionized our ability to visualize the spatial distribution of RNA molecules, thereby revealing the complex cellular organization and interactions within tissues (33). This approach has been further enhanced by the integration with scRNA-seq, allowing for the identification of distinct cell populations and their functional states within the tissue microenvironment (33). The technology has shown tremendous potential in disease understanding and drug discovery, providing new perspectives for drug target identification and pharmacological research model building (34). Moreover, spatially resolved transcriptomic technologies have accelerated discovery in fields, offering insights into cell-cell interactions and changes in response to injury (35). As the field progresses, the challenges and opportunities in translating high-resolution ST into clinical practice are being addressed, with the potential to identify disease mechanisms and guide personalized therapies (36). Overall, ST stands as a pivotal tool in advancing our understanding of tissue biology and driving the development of precision medicine.
The pancreatic islet microenvironment is critical for preserving the function and survival of islet cells in DM and islet transplantation (37). Recent studies have utilized ST to construct a detailed spatial map of the islet microenvironment, highlighting the interactions between different cell types (38). This spatial analysis reveals how the local cellular context influences islet function and response to metabolic challenges. Furthermore, dynamic monitoring of the islet microenvironment using ST allows researchers to observe shifts in gene expression profiles over time, yielding insights into the causes of islet dysfunction and the effects of therapeutic interventions. For instance, studies have shown that alterations in the spatial arrangement of immune cells in the islet can contribute to autoimmune attacks in T1D (39). By elucidating the spatial dynamics of the islet microenvironment, ST holds promise for improving strategies in islet transplantation and developing targeted therapies for DM.
ST holds the potential to significantly advance our understanding of the graft microenvironment in transplantation biology in the future. This technology, when applied to islet transplantation, could offer a spatially detailed perspective on gene expression in graft tissues, potentially enabling the detection of molecular patterns that correlate with successful graft integration and function. While current research has not yet fully explored this area, the technology’s promise suggests it could reveal the heterogeneity of immune cell populations surrounding the graft, which is crucial for understanding immune responses and potential rejection mechanisms. For instance, understanding the spatial distribution of T cells and regulatory cells within the graft could influence the outcome of transplantation. Additionally, ST could facilitate the exploration of interactions between graft cells and the surrounding microenvironment, including the extracellular matrix and signaling molecules. This comprehensive analysis could inform the development of novel immunomodulatory strategies aimed at enhancing graft survival and function. As the field progresses, integrating ST with other omics technologies may further enhance our ability to dissect the complex biological processes governing graft success and failure.
Recent advancements in scRNA-seq and ST offer exciting new avenues for understanding the complexities of islet transplantation and DM management. ScRNA-seq enables cell-level gene expression analysis, revealing cell populations within islets and their response to transplantation. ST, on the other hand, enables the mapping of gene expression within tissues, which is crucial for understanding the microenvironment surrounding transplanted islets and their interactions with host tissues. These technologies hold immense potential for elucidating the mechanisms underlying islet function, survival, and rejection, opening up new avenues for improved therapeutic strategies in DM management. Combining single-cell multi-omics with existing knowledge allows for functional insights into gene regulation and cell interactions, which is particularly relevant in immunology (40). ST has become a key instrument in disease understanding and drug discovery, offering new perspectives into how drugs interact with targets and their mechanisms of action (34). ScRNA-seq and spatial multi-omics is rapidly evolving, with methods such as ST playing a significant role in uncovering spatial heterogeneity and creating comprehensive spatial maps (41), which is particularly relevant in the context of islet transplantation and DM management. The application of spatial multi-omics in unraveling the synergy between different omics data highlights the importance of these approaches in advancing our understanding of complex diseases and their treatments (42). Furthermore, the potential of single-cell and spatial resolution in deciphering drug targets and actions cannot be overlooked (43), which is crucial for the development of personalized therapeutic strategies in DM.
The future of research in the integration of scRNA-seq and ST is promising, with several potential directions for exploration. One significant area of interest is the application of these technologies in personalized medicine, where understanding the unique cellular composition and gene expression profiles of individual patients can lead to tailored therapeutic strategies (34, 44). Furthermore, the integration of single-cell and spatial transcriptomic data with clinical outcomes could enhance our ability to predict disease progression and treatment responses (45).
To fully harness the potential of scRNA-seq and ST, the development of integrative analysis methods is essential (46). These methods address the challenges associated with the high dimensionality and complexity of the data generated by these technologies. Various computational frameworks have been proposed to facilitate the synthesis of multi-omics data, allowing researchers to analyze gene expression, protein levels, and epigenetic modifications concurrently (47). For instance, recent advancements in machine learning and statistical modeling have enabled the concurrent analysis of scRNA-seq and spatial transcriptomic data, providing insights into the interplay between cellular states and their spatial contexts (48). Additionally, the application of integrative methods, such as multi-dimensional scaling and clustering algorithms, aids in the identification of distinct cell populations and their spatial distributions within tissues (49). The computational tools used for integrating scRNA-seq and ST data published in high-impact journals after 2023 were summarized in Table 1. These advancements and tools highlight the progress in multi-modal single-cell omics and computational integration, providing researchers with a more comprehensive and robust framework for analyzing complex biological systems. By incorporating these cutting-edge methodologies, the field of islet transplantation and DM management can benefit from a deeper understanding of cellular heterogeneity and spatial organization, leading to more effective therapeutic strategies.
While the potential clinical applications of scRNA-seq and ST are significant, several barriers must be addressed to facilitate their widespread use in clinical settings. Cost is a major factor, with the high expense of these technologies currently limiting their use in routine clinical diagnostics. However, as technology advances and the market expand, costs are expected to decrease, making these tools more accessible. The need for specialized equipment also poses a challenge for many medical institutions. Research is ongoing to develop more convenient and cost-effective devices, reducing the dependence on high-end equipment. Additionally, handling large numbers of samples is a current limitation. Optimizing experimental processes and data analysis methods, as well as improvements in computational algorithms and data management systems, can enhance the scalability of these technologies, making them more feasible for clinical use.
Despite the exciting prospects of integrating scRNA-seq and ST, several ethical challenges must be addressed. Ethically, the collection and use of biological samples raise significant concerns regarding informed consent, privacy, and data security, particularly when dealing with sensitive patient information. The potential for misuse of genetic data necessitates robust ethical guidelines and regulatory frameworks to protect individuals’ rights. Ensuring that patients are fully informed about the purposes, risks, and benefits of the samples being used in research is crucial. Additionally, maintaining the confidentiality and security of genetic data is essential to prevent unauthorized access and potential discrimination. Addressing these ethical challenges will be crucial for the successful implementation of integrated single-cell and spatial transcriptomic approaches in both research and clinical practice.
Pre-transplant, scRNA-seq can identify optimal donor-recipient matches by analyzing islet cell subsets, as demonstrated in highly purified human islet cell studies. ST can monitor disease progression, aiding in timing transplant decisions. Post-transplant, scRNA-seq can detect early signs of graft rejection, while ST assesses graft integration. Collaboratively, bioinformaticians and clinicians can use tools like SpatialScope to integrate these data, enhancing our understanding of the transplant microenvironment. Standardization and benchmarking, as well as advancements in permeabilization and single-cell resolution technologies, are crucial for improving these methods. These integrations and collaborations will lead to more effective therapeutic strategies and improved patient outcomes.
ScRNA-seq provides high-resolution identification of cell types and their states but lacks the ability to pinpoint their spatial distribution or capture local cell–cell interactions and the ligands and receptors that mediate these interactions. This limitation hinders our ability to fully explore the islet microenvironment and the complexity of cell–cell interactions therein. ST, while providing spatial context, often has limitations in spatial resolution. For example, the Visium platform, despite its widespread use, has a spot size of 55 μm, which often contains multiple cells, limiting its ability to resolve detailed tissue structure and characterize cellular communications.
While imaging-based ST has a longer history and a collaborative benchmarking effort has been initiated with the SpaceTX consortium, a systematic benchmarking study has not been done for ST. This lack of benchmarking complicates the establishment of universal evaluation standards. Moreover, ST technologies diverge notably in aspects such as spatial resolution and the preparation of spatially barcoded oligo arrays. This variability introduces challenges in method selection and complicates the establishment of universal evaluation standards.
Unexpected gene-capturing biases were existed in the polyA-based Visium platform, with marker genes consistently captured by other technologies not showing up in the Visium (polyA) data. Considering Visium is the most widely used commercial platform, it is important to further verify its gene-capturing bias on other tissues. Besides, the spot size has become an important metric as a surrogate of the resolution for each method. However, diffusion is a key factor that affects the actual resolution. Varying the permeabilization time has a substantial impact on diffusion, and different technologies exhibited varied diffusion profiles across various tissue types.
To address the limitations of current methods in integrating scRNA-seq and ST data, we propose a multi-faceted approach. First, we suggest using deep generative models like SpatialScope to integrate scRNA-seq and ST data, achieving single-cell resolution and a comprehensive understanding of the transcriptome’s spatial distribution. Additionally, developing accurate statistical and computational methods, such as multi-dimensional scaling and clustering algorithms, is crucial for handling the data’s complexity. Second, we recommend establishing a standardized benchmarking framework and generating cross-platform datasets to evaluate ST methods systematically. Finally, optimizing permeabilization and developing technologies for true single-cell resolution, such as Slide-tag, will improve sensitivity and provide more detailed spatial information. These advancements will enhance the utility of scRNA-seq and ST in islet transplantation and DM management, leading to more effective therapeutic strategies.
Single-cell omics and ST have revolutionized islet transplantation research, providing detailed insights into cellular diversity and spatial organization (Figure 1). These technologies not only reveal the complex interactions within the transplant microenvironment but also identify key regulatory pathways and therapeutic targets to enhance graft survival and function. The integration of these approaches has significant translational potential, offering a more comprehensive framework for improving clinical outcomes in patients with DM. Future research should focus on longitudinal studies and interdisciplinary collaborations to fully realize this potential, ensuring that emerging discoveries translate effectively into clinical practice.
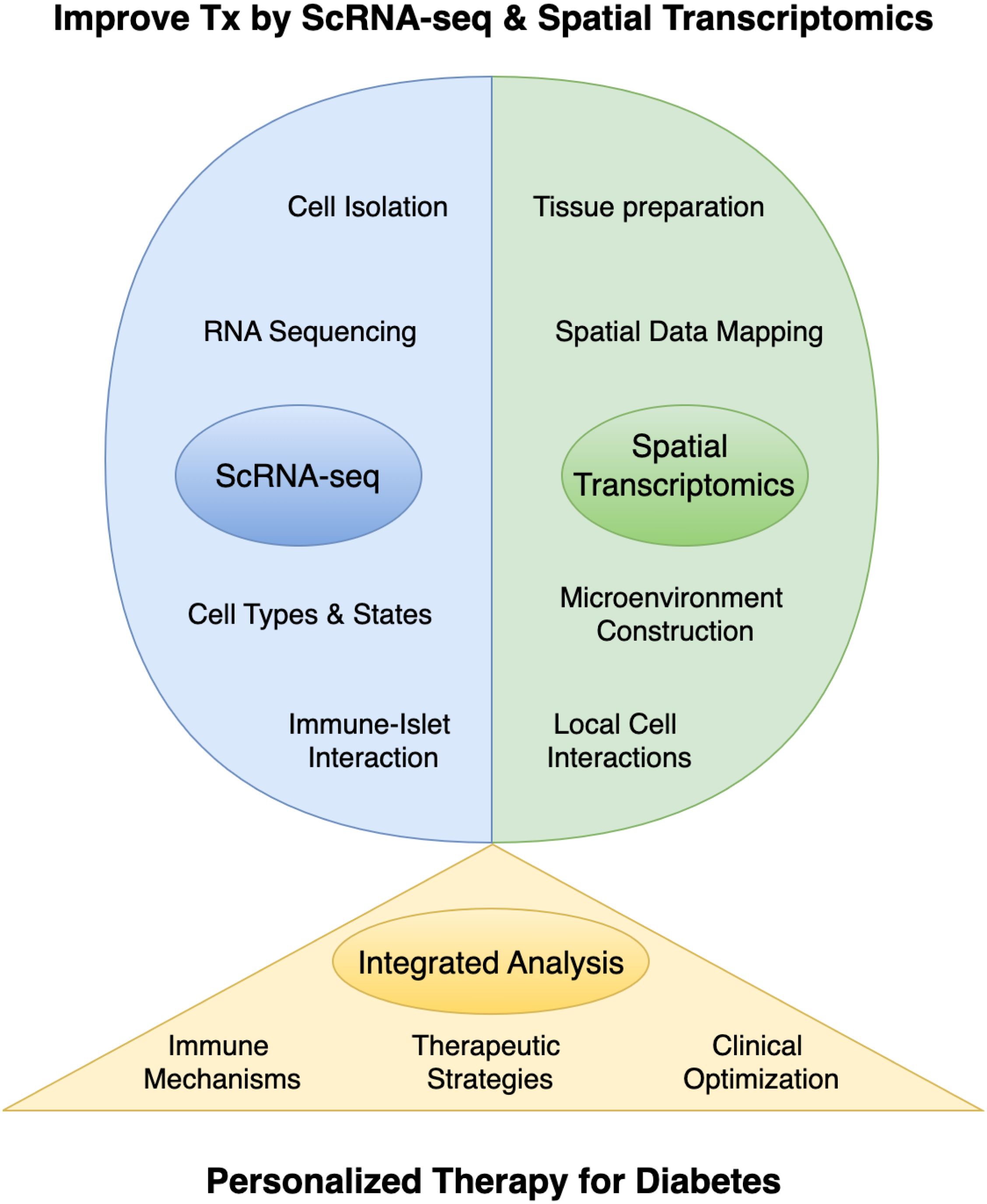
Figure 1. The application of single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics in islet transplantation research. Tx, Islet Transplantation.
LM: Writing – original draft, Writing – review & editing, Funding acquisition. TW: Writing – review & editing. YC: Writing – review & editing. ZL: Writing – review & editing. XW: Writing – review & editing, Funding acquisition. ZP: Writing – review & editing, Funding acquisition.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported in part by the Shenzhen Science and Technology Program (JCYJ20220818102001003, JCYJ20230807115107015, GJHZ20240218114714027 and GCZX2015043017281705) and Team-based Medical Science Research Program (2024YZZ04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Ogurtsova K, Guariguata L, Barengo NC, Ruiz PL-D, Sacre JW, Karuranga S, et al. IDF diabetes Atlas: Global estimates of undiagnosed diabetes in adults for 2021. Diabetes Res Clin Pract. (2022) 183:109118. doi: 10.1016/j.diabres.2021.109118
2. Stabler CL, Russ HA. Regulatory approval of islet transplantation for treatment of type 1 diabetes: Implications and what is on the horizon. Mol Ther. (2023) 31:3107–8. doi: 10.1016/j.ymthe.2023.10.011
3. Chetboun M, Drumez E, Ballou C, Maanaoui M, Payne E, Barton F, et al. Association between primary graft function and 5-year outcomes of islet allogeneic transplantation in type 1 diabetes: a retrospective, multicentre, observational cohort study in 1210 patients from the Collaborative Islet Transplant Registry. Lancet Diabetes Endocrinol. (2023) 11:391–401. doi: 10.1016/S2213-8587(23)00082-7
4. Ramzy A, Belmonte PJ, Braam MJS, Ida S, Wilts EM, Levings MK, et al. A century-long journey from the discovery of insulin to the implantation of stem cell–derived islets. Endocr Rev. (2023) 44:222–53. doi: 10.1210/endrev/bnac021
5. Kioulaphides S, García AJ. Encapsulation and immune protection for type 1 diabetes cell therapy. Advanced Drug Delivery Rev. (2024) 207:115205. doi: 10.1016/j.addr.2024.115205
6. Zhang Q, Gonelle-Gispert C, Li Y, Geng Z, Gerber-Lemaire S, Wang Y, et al. Islet encapsulation: new developments for the treatment of type 1 diabetes. Front Immunol. (2022) 13:869984. doi: 10.3389/fimmu.2022.869984
7. Chen P, Yao F, Lu Y, Peng Y, Zhu S, Deng J, et al. Single-cell landscape of mouse islet allograft and syngeneic graft. Front Immunol. (2022) 13:853349. doi: 10.3389/fimmu.2022.853349
8. Zhou H, Pu Z, Lu Y, Zheng P, Yu H, Mou L. Elucidating T cell dynamics and molecular mechanisms in syngeneic and allogeneic islet transplantation through single-cell RNA sequencing. Front Immunol. (2024) 15:1429205. doi: 10.3389/fimmu.2024.1429205
9. Zhang C, Chen R, Zhang Y. Accurate inference of genome-wide spatial expression with iSpatial. Sci Adv. (2022) 8:eabq0990. doi: 10.1126/sciadv.abq0990
10. Liu Y, DiStasio M, Su G, Asashima H, Enninful A, Qin X, et al. High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat Biotechnol. (2023) 41:1405–9. doi: 10.1038/s41587-023-01676-0
11. Zhang D, Rubio-Rodríguez-Kirby LA, Lin Y, Song M, Wang L, Wang L, et al. Spatial dynamics of mammalian brain development and neuroinflammation by multimodal tri-omics mapping. bioRxiv. (2024). doi: 10.1101/2024.07.28.605493
12. Baysoy A, Tian X, Zhang F, Renauer P, Bai Z, Shi H, et al. Spatially Resolvedin vivoCRISPR Screen Sequencing via Perturb-DBiT. bioRxiv. (2024). doi: 10.1101/2024.11.18.624106
13. Pu Z, Chen S, Lu Y, Wu Z, Cai Z, Mou L. Exploring the molecular mechanisms of macrophages in islet transplantation using single-cell analysis. Front Immunol. (2024) 15:1407118. doi: 10.3389/fimmu.2024.1407118
14. Qu H-Q, Kao C, Hakonarson H. Single-cell RNA sequencing technology landscape in 2023. Stem Cells. (2024) 42:1–12. doi: 10.1093/stmcls/sxad077
15. Baysoy A, Bai Z, Satija R, Fan R. The technological landscape and applications of single-cell multi-omics. Nat Rev Mol Cell Biol. (2023) 24:695–713. doi: 10.1038/s41580-023-00615-w
16. Lim J, Chin V, Fairfax K, Moutinho C, Suan D, Ji H, et al. Transitioning single-cell genomics into the clinic. Nat Rev Genet. (2023) 24:573–84. doi: 10.1038/s41576-023-00613-w
17. Cuomo ASE, Nathan A, Raychaudhuri S, MacArthur DG, Powell JE. Single-cell genomics meets human genetics. Nat Rev Genet. (2023) 24:535–49. doi: 10.1038/s41576-023-00599-5
18. Heumos L, Schaar AC, Lance C, Litinetskaya A, Drost F, Zappia L, et al. Best practices for single-cell analysis across modalities. Nat Rev Genet. (2023) 24:550–72. doi: 10.1038/s41576-023-00586-w
19. Mitchell JS, Spanier JA, Dwyer AJ, Knutson TP, Alkhatib MH, Qian G, et al. CD4+ T cells reactive to a hybrid peptide from insulin-chromogranin A adopt a distinct effector fate and are pathogenic in autoimmune diabetes. Immunity. (2024) 57:2399–2415.e8. doi: 10.1016/j.immuni.2024.07.024
20. Wang L, Wu J, Sramek M, Obayomi SMB, Gao P, Li Y, et al. Heterogeneous enhancer states orchestrate β cell responses to metabolic stress. Nat Commun. (2024) 15:9361. doi: 10.1038/s41467-024-53717-0
21. Wang G, Chiou J, Zeng C, Miller M, Matta I, Han JY, et al. Integrating genetics with single-cell multiomic measurements across disease states identifies mechanisms of beta cell dysfunction in type 2 diabetes. Nat Genet. (2023) 55:984–94. doi: 10.1038/s41588-023-01397-9
22. Hrovatin K, Bastidas-Ponce A, Bakhti M, Zappia L, Büttner M, Salinno C, et al. Delineating mouse β-cell identity during lifetime and in diabetes with a single cell atlas. Nat Metab. (2023) 5:1615–37. doi: 10.1038/s42255-023-00876-x
23. Doke M, Álvarez-Cubela S, Klein D, Altilio I, Schulz J, Mateus Gonçalves L, et al. Dynamic scRNA-seq of live human pancreatic slices reveals functional endocrine cell neogenesis through an intermediate ducto-acinar stage. Cell Metab. (2023) 35:1944–1960.e7. doi: 10.1016/j.cmet.2023.10.001
24. Maestas MM, Ishahak M, Augsornworawat P, Veronese-Paniagua DA, Maxwell KG, Velazco-Cruz L, et al. Identification of unique cell type responses in pancreatic islets to stress. Nat Commun. (2024) 15:5567. doi: 10.1038/s41467-024-49724-w
25. Yang L, Han Y, Zhang T, Dong X, Ge J, Roy A, et al. Human vascularized macrophage-islet organoids to model immune-mediated pancreatic β cell pyroptosis upon viral infection. Cell Stem Cell. (2024) 31:1612–1629.e8. doi: 10.1016/j.stem.2024.08.007
26. Li J, Klughammer J, Farlik M, Penz T, Spittler A, Barbieux C, et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. (2016) 17:178–87. doi: 10.15252/embr.201540946
27. Benninger RKP, Dorrell C, Hodson DJ, Rutter GA. The impact of pancreatic beta cell heterogeneity on type 1 diabetes pathogenesis. Curr Diabetes Rep. (2018) 18:112. doi: 10.1007/s11892-018-1085-2
28. Wang Q, Huang Y, Liu L, Zhao X, Sun Y, Mao X, et al. Pancreatic islet transplantation: current advances and challenges. Front Immunol. (2024) 15:1391504. doi: 10.3389/fimmu.2024.1391504
29. Knoedler L, Dean J, Diatta F, Thompson N, Knoedler S, Rhys R, et al. Immune modulation in transplant medicine: a comprehensive review of cell therapy applications and future directions. Front Immunol. (2024) 15:1372862. doi: 10.3389/fimmu.2024.1372862
30. Qiu X, Zhu DY, Lu Y, Yao J, Jing Z, Min KH, et al. Spatiotemporal modeling of molecular holograms. Cell. (2024) 187:7351–7373.e61. doi: 10.1016/j.cell.2024.10.011
31. Zhou R, Yang G, Zhang Y, Wang Y. Spatial transcriptomics in development and disease. Mol BioMed. (2023) 4:32. doi: 10.1186/s43556-023-00144-0
32. Gulati GS, D’Silva JP, Liu Y, Wang L, Newman AM. Profiling cell identity and tissue architecture with single-cell and spatial transcriptomics. Nat Rev Mol Cell Biol. (2025) 26:11–31. doi: 10.1038/s41580-024-00768-2
33. Tian L, Chen F, Macosko EZ. The expanding vistas of spatial transcriptomics. Nat Biotechnol. (2023) 41:773–82. doi: 10.1038/s41587-022-01448-2
34. Cao J, Li C, Cui Z, Deng S, Lei T, Liu W, et al. Spatial transcriptomics: A powerful tool in disease understanding and drug discovery. Theranostics. (2024) 14:2946–68. doi: 10.7150/thno.95908
35. Jain S, Eadon MT. Spatial transcriptomics in health and disease. Nat Rev Nephrol. (2024) 20:659–71. doi: 10.1038/s41581-024-00841-1
36. Pentimalli TM, Karaiskos N, Rajewsky N. Challenges and opportunities in the clinical translation of high-resolution spatial transcriptomics. Annu Rev Pathol. (2024) 20:405–32. doi: 10.1146/annurev-pathmechdis-111523-023417
37. Yan L, Ye L, Chen Y, He S, Zhang C, Mao X, et al. The influence of microenvironment on survival of intraportal transplanted islets. Front Immunol. (2022) 13:849580. doi: 10.3389/fimmu.2022.849580
38. Olaniru OE, Kadolsky U, Kannambath S, Vaikkinen H, Fung K, Dhami P, et al. Single-cell transcriptomic and spatial landscapes of the developing human pancreas. Cell Metab. (2023) 35:184–199.e5. doi: 10.1016/j.cmet.2022.11.009
39. James EA, Joglekar AV, Linnemann AK, Russ HA, Kent SC. The beta cell-immune cell interface in type 1 diabetes (T1D). Mol Metab. (2023) 78:101809. doi: 10.1016/j.molmet.2023.101809
40. Schäfer PSL, Dimitrov D, Villablanca EJ, Saez-Rodriguez J. Integrating single-cell multi-omics and prior biological knowledge for a functional characterization of the immune system. Nat Immunol. (2024) 25:405–17. doi: 10.1038/s41590-024-01768-2
41. Vandereyken K, Sifrim A, Thienpont B, Voet T. Methods and applications for single-cell and spatial multi-omics. Nat Rev Genet. (2023) 24:494–515. doi: 10.1038/s41576-023-00580-2
42. Liu X, Peng T, Xu M, Lin S, Hu B, Chu T, et al. Spatial multi-omics: deciphering technological landscape of integration of multi-omics and its applications. J Hematol Oncol. (2024) 17:72. doi: 10.1186/s13045-024-01596-9
43. Pang Z, Cravatt BF, Ye L. Deciphering drug targets and actions with single-cell and spatial resolution. Annu Rev Pharmacol Toxicol. (2024) 64:507–26. doi: 10.1146/annurev-pharmtox-033123-123610
44. Wiedmeier JE, Noel P, Lin W, Von Hoff DD, Han H. Single-cell sequencing in precision medicine. Cancer Treat Res. (2019) 178:237–52. doi: 10.1007/978-3-030-16391-4_9
45. Longo SK, Guo MG, Ji AL, Khavari PA. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat Rev Genet. (2021) 22:627–44. doi: 10.1038/s41576-021-00370-8
46. Wan X, Xiao J, Tam SST, Cai M, Sugimura R, Wang Y, et al. Integrating spatial and single-cell transcriptomics data using deep generative models with SpatialScope. Nat Commun. (2023) 14:7848. doi: 10.1038/s41467-023-43629-w
47. Du J, Yang Y-C, An Z-J, Zhang M-H, Fu X-H, Huang Z-F, et al. Advances in spatial transcriptomics and related data analysis strategies. J Transl Med. (2023) 21:330. doi: 10.1186/s12967-023-04150-2
48. Zeng Z, Li Y, Li Y, Luo Y. Statistical and machine learning methods for spatially resolved transcriptomics data analysis. Genome Biol. (2022) 23:83. doi: 10.1186/s13059-022-02653-7
49. Chu L-X, Wang W-J, Gu X-P, Wu P, Gao C, Zhang Q, et al. Spatiotemporal multi-omics: exploring molecular landscapes in aging and regenerative medicine. Military Med Res. (2024) 11:31. doi: 10.1186/s40779-024-00537-4
50. Fan Z, Luo Y, Lu H, Wang T, Feng Y, Zhao W, et al. SPASCER: spatial transcriptomics annotation at single-cell resolution. Nucleic Acids Res. (2023) 51:D1138–49. doi: 10.1093/nar/gkac889
51. Zhu B, Chen S, Bai Y, Chen H, Liao G, Mukherjee N, et al. Robust single-cell matching and multimodal analysis using shared and distinct features. Nat Methods. (2023) 20:304–15. doi: 10.1038/s41592-022-01709-7
52. Chidester B, Zhou T, Alam S, Ma J. SpiceMix enables integrative single-cell spatial modeling of cell identity. Nat Genet. (2023) 55:78–88. doi: 10.1038/s41588-022-01256-z
53. Liu W, Liao X, Luo Z, Yang Y, Lau MC, Jiao Y, et al. Probabilistic embedding, clustering, and alignment for integrating spatial transcriptomics data with PRECAST. Nat Commun. (2023) 14:296. doi: 10.1038/s41467-023-35947-w
54. Long Y, Ang KS, Li M, Chong KLK, Sethi R, Zhong C, et al. Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST. Nat Commun. (2023) 14:1155. doi: 10.1038/s41467-023-36796-3
55. Bae S, Choi H, Lee DS. spSeudoMap: cell type mapping of spatial transcriptomics using unmatched single-cell RNA-seq data. Genome Med. (2023) 15:19. doi: 10.1186/s13073-023-01168-5
56. Qian J, Liao J, Liu Z, Chi Y, Fang Y, Zheng Y, et al. Reconstruction of the cell pseudo-space from single-cell RNA sequencing data with scSpace. Nat Commun. (2023) 14:2484. doi: 10.1038/s41467-023-38121-4
57. Tang X, Zhang J, He Y, Zhang X, Lin Z, Partarrieu S, et al. Explainable multi-task learning for multi-modality biological data analysis. Nat Commun. (2023) 14:2546. doi: 10.1038/s41467-023-37477-x
58. Villemin J-P, Bassaganyas L, Pourquier D, Boissière F, Cabello-Aguilar S, Crapez E, et al. Inferring ligand-receptor cellular networks from bulk and spatial transcriptomic datasets with BulkSignalR. Nucleic Acids Res. (2023) 51:4726–44. doi: 10.1093/nar/gkad352
59. Govek KW, Nicodemus P, Lin Y, Crawford J, Saturnino AB, Cui H, et al. CAJAL enables analysis and integration of single-cell morphological data using metric geometry. Nat Commun. (2023) 14:3672. doi: 10.1038/s41467-023-39424-2
60. Chen S, Zhu B, Huang S, Hickey JW, Lin KZ, Snyder M, et al. Integration of spatial and single-cell data across modalities with weakly linked features. Nat Biotechnol. (2024) 42:1096–106. doi: 10.1038/s41587-023-01935-0
61. Tang Z, Li Z, Hou T, Zhang T, Yang B, Su J, et al. SiGra: single-cell spatial elucidation through an image-augmented graph transformer. Nat Commun. (2023) 14:5618. doi: 10.1038/s41467-023-41437-w
62. Su J, Reynier J-B, Fu X, Zhong G, Jiang J, Escalante RS, et al. Smoother: a unified and modular framework for incorporating structural dependency in spatial omics data. Genome Biol. (2023) 24:291. doi: 10.1186/s13059-023-03138-x
63. Yang P, Jin L, Liao J, Jin K, Shao X, Li C, et al. Revealing spatial multimodal heterogeneity in tissues with SpaTrio. Cell Genomics. (2023) 3:100446. doi: 10.1016/j.xgen.2023.100446
64. Zhang D, Schroeder A, Yan H, Yang H, Hu J, Lee MYY, et al. Inferring super-resolution tissue architecture by integrating spatial transcriptomics with histology. Nat Biotechnol. (2024) 42:1372–7. doi: 10.1038/s41587-023-02019-9
65. Rahimikollu J, Xiao H, Rosengart A, Rosen ABI, Tabib T, Zdinak PM, et al. SLIDE: Significant Latent Factor Interaction Discovery and Exploration across biological domains. Nat Methods. (2024) 21:835–45. doi: 10.1038/s41592-024-02175-z
66. Duan B, Chen S, Cheng X, Liu Q. Multi-slice spatial transcriptome domain analysis with SpaDo. Genome Biol. (2024) 25:73. doi: 10.1186/s13059-024-03213-x
67. Ding J, Liu R, Wen H, Tang W, Li Z, Venegas J, et al. DANCE: a deep learning library and benchmark platform for single-cell analysis. Genome Biol. (2024) 25:72. doi: 10.1186/s13059-024-03211-z
68. He S, Jin Y, Nazaret A, Shi L, Chen X, Rampersaud S, et al. Starfysh integrates spatial transcriptomic and histologic data to reveal heterogeneous tumor–immune hubs. Nat Biotechnol. (2024). doi: 10.1038/s41587-024-02173-8
69. Mah CK, Ahmed N, Lopez NA, Lam DC, Pong A, Monell A, et al. Bento: a toolkit for subcellular analysis of spatial transcriptomics data. Genome Biol. (2024) 25:82. doi: 10.1186/s13059-024-03217-7
70. Rahimi A, Vale-Silva LA, Fälth Savitski M, Tanevski J, Saez-Rodriguez J. DOT: a flexible multi-objective optimization framework for transferring features across single-cell and spatial omics. Nat Commun. (2024) 15:4994. doi: 10.1038/s41467-024-48868-z
71. Sethi R, Ang KS, Li M, Long Y, Ling J, Chen J. ezSingleCell: an integrated one-stop single-cell and spatial omics analysis platform for bench scientists. Nat Commun. (2024) 15:5600. doi: 10.1038/s41467-024-48188-2
72. Curion F, Rich-Griffin C, Agarwal D, Ouologuem S, Rue-Albrecht K, May L, et al. Panpipes: a pipeline for multiomic single-cell and spatial transcriptomic data analysis. Genome Biol. (2024) 25:181. doi: 10.1186/s13059-024-03322-7
73. Li J, Shyr Y, Liu Q. aKNNO: single-cell and spatial transcriptomics clustering with an optimized adaptive k-nearest neighbor graph. Genome Biol. (2024) 25:203. doi: 10.1186/s13059-024-03339-y
74. Li Y, Luo Y. STdGCN: spatial transcriptomic cell-type deconvolution using graph convolutional networks. Genome Biol. (2024) 25:206. doi: 10.1186/s13059-024-03353-0
75. Stassen SV, Kobashi M, Lam EY, Huang Y, Ho JWK, Tsia KK. StaVia: spatially and temporally aware cartography with higher-order random walks for cell atlases. Genome Biol. (2024) 25:224. doi: 10.1186/s13059-024-03347-y
76. Yang W, Wang P, Xu S, Wang T, Luo M, Cai Y, et al. Deciphering cell–cell communication at single-cell resolution for spatial transcriptomics with subgraph-based graph attention network. Nat Commun. (2024) 15:7101. doi: 10.1038/s41467-024-51329-2
77. Jiang J, Liu Y, Qin J, Chen J, Wu J, Pizzi MP, et al. METI: deep profiling of tumor ecosystems by integrating cell morphology and spatial transcriptomics. Nat Commun. (2024) 15:7312. doi: 10.1038/s41467-024-51708-9
78. Samaran J, Peyré G, Cantini L. scConfluence: single-cell diagonal integration with regularized Inverse Optimal Transport on weakly connected features. Nat Commun. (2024) 15:7762. doi: 10.1038/s41467-024-51382-x
79. Tang Z, Luo S, Zeng H, Huang J, Sui X, Wu M, et al. Search and match across spatial omics samples at single-cell resolution. Nat Methods. (2024) 21:1818–29. doi: 10.1038/s41592-024-02410-7
80. Zhang Y, Yu B, Ming W, Zhou X, Wang J, Chen D. SpaTopic: A statistical learning framework for exploring tumor spatial architecture from spatially resolved transcriptomic data. Sci Adv. (2024) 10:eadp4942. doi: 10.1126/sciadv.adp4942
81. Keller MS, Gold I, McCallum C, Manz T, Kharchenko PV, Gehlenborg N. Vitessce: integrative visualization of multimodal and spatially resolved single-cell data. Nat Methods. (2025) 22:63–7. doi: 10.1038/s41592-024-02436-x
82. Wang M-G, Chen L, Zhang X-F. Dual decoding of cell types and gene expression in spatial transcriptomics with PANDA. Nucleic Acids Res. (2024) 52:12173–90. doi: 10.1093/nar/gkae876
83. Nguyen ND, Rosas L, Khaliullin T, Jiang P, Hasanaj E, Ovando-Ricardez JA, et al. scDOT: optimal transport for mapping senescent cells in spatial transcriptomics. Genome Biol. (2024) 25:288. doi: 10.1186/s13059-024-03426-0
Keywords: diabetes, ScRNA-seq, spatial transcriptomics, islet transplantation, islet graft, microenvironment
Citation: Mou L, Wang TB, Chen Y, Luo Z, Wang X and Pu Z (2025) Single-cell genomics and spatial transcriptomics in islet transplantation for diabetes treatment: advancing towards personalized therapies. Front. Immunol. 16:1554876. doi: 10.3389/fimmu.2025.1554876
Received: 03 January 2025; Accepted: 21 January 2025;
Published: 20 February 2025.
Edited by:
Yi Wang, Sichuan Academy of Medical Sciences and Sichuan Provincial People’s Hospital, ChinaCopyright © 2025 Mou, Wang, Chen, Luo, Wang and Pu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisha Mou, bGlzaGFtb3VAZW1haWwuc3p1LmVkdS5jbg==; Zuhui Pu, enVodWlwdUBlbWFpbC5zenUuZWR1LmNu; Xinyu Wang, d3h5aG9yc2VAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.