
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 02 April 2025
Sec. Comparative Immunology
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1552151
 Hongyu Li1,2†*
Hongyu Li1,2†* Yijie Zhang1†
Yijie Zhang1† Yunhuan Zhu1
Yunhuan Zhu1 Qingzhi Zhao1
Qingzhi Zhao1 Jialu Xu1
Jialu Xu1 Xianwei Li1
Xianwei Li1 Ling Zhao1
Ling Zhao1 Hairun Li1
Hairun Li1 Mingcheng Liu1
Mingcheng Liu1 Yuncheng Qian1
Yuncheng Qian1 Xiaofen Zhang1
Xiaofen Zhang1 Keda Chen1*
Keda Chen1*The Immunoglobulin Superfamily (IgSF) represents a vital protein family widely distributed in animal genomes, encompassing multifunctional proteins with immunoglobulin-like domains, including immunoglobulins. These proteins play pivotal roles in various biological processes, such as development, differentiation, adhesion, activation, regulation, and signal transduction. While the functions of IgSF in vertebrates are relatively well understood, their roles in invertebrates remain underexplored. This review aims to comprehensively summarize the functions and mechanisms of IgSF in invertebrates, focusing on arthropods, mollusks, and other primitive phyla. In arthropods, research on IgSF has primarily emphasized its roles in the nervous system, especially in axonal and synaptic regulation, and its critical functions in the immune system. Studies in mollusks have predominantly highlighted the immunological functions of IgSF in pathogen recognition, clearance responses, and signal transduction. In contrast, research on protozoa and platyhelminths has mainly focused on identifying IgSF molecules, with relatively limited insights into their functional roles. In sponges, IgSF is primarily associated with cell adhesion and intercellular recognition. By exploring the genetic and protein structural diversity of IgSF in invertebrates, this review reveals their multifunctionality and complexity in biological systems. It not only enhances our understanding of the roles of IgSF in invertebrates but also lays the groundwork for future studies on their potential applications in evolutionary biology and disease models.
Approximately 450 million years ago, the evolution of upper and lower jaws marked a pivotal advancement in early vertebrates, greatly enhancing their hunting efficiency and adaptability to diverse environments. Molecular clock analyses suggest that the origin of gnathostomes can be traced back to the late Ordovician period, underscoring the significance of this evolutionary milestone in vertebrate history (1, 2). The emergence of jawed vertebrates was accompanied by the evolution of a highly developed adaptive immune system (3). This system comprises B lymphocytes and T lymphocytes, which possess the ability to specifically recognize and eliminate pathogens and other harmful substances, serving as the cornerstone of the body’s immune defense. Furthermore, various components of the vertebrate body contribute distinctively to immune protection. For instance, the first line of innate immunity, consisting of physical and chemical barriers such as the skin, mucous membranes, and sweat, plays a critical role in preventing the entry of pathogens and foreign substances (4).
In contrast, invertebrates have existed on Earth far earlier than vertebrates. Their habitats are fraught with threats from diverse pathogenic microorganisms (5). As the oldest and most diverse group of animals on Earth, invertebrates include species that exhibit both the longest lifespans and the simplest anatomical structures. Examples include Porifera and Cnidaria, which are classified as basal metazoans. Despite their comparatively simple physiological organization, the immune systems of invertebrates have demonstrated remarkable efficacy, enabling these organisms to endure and thrive through prolonged evolutionary challenges and competition (6).
From an evolutionary perspective, invertebrates have undergone a transition from simple unicellular organisms to complex multicellular systems. These organisms are classified into multiple phyla, including Protozoa, Placozoa, Porifera, Cnidaria, Platyhelminthes, Nematomorpha, Annelida, Mollusca, Arthropoda, and Echinodermata. Each phylum exhibits distinct biological characteristics and evolutionary trajectories. Throughout their extensive evolutionary history, invertebrates have adapted to diverse ecological environments, encountering a wide variety of complex pathogens. From the deep sea (e.g., Mesozoic sea anemones Amphianthus inornata (7), Gracilechinus affinis (8), Neoaulaxinia zingiberadix (9)) to forests (e.g., Clitellata and Collembola (10)), and from polar to tropical regions, these extreme and variable habitats pose significant challenges to invertebrate immune systems. Traditionally, invertebrates were thought to rely primarily on innate immune systems to defend against pathogenic invasions (11). While jawed vertebrates rely on the immunoglobulin (Ig) family to facilitate adaptive immunity, invertebrates have independently evolved members of the IgSF to adapt to their complex ecological environments. Although the functional mechanisms differ between these groups, this evolutionary development underscores the sophisticated and diverse immune strategies that invertebrates have cultivated to meet their unique ecological challenges (12).
Ig is a specialized class of proteins primarily secreted by B cells in vertebrates, responsible for recognizing and neutralizing foreign antigens such as bacteria and viruses. The IgSF represents a broader family of proteins, encompassing not only immunoglobulins but also numerous other proteins with Ig-like domains (13, 14). Through ligand-receptor interactions, IgSF molecules are involved in diverse biological processes, including development, differentiation, activation, cell adhesion, signal transduction, and immune regulation (15, 16). In Figure 1, we depict how IgSF plays a role in pathogen recognition and mediates the clearance of pathogens through phagocytosis under certain external pressures, including changes in temperature, trauma, and pathogen invasion. Additionally, IgSF plays an important role in tissue repair and the processes of intercellular recognition and adhesion.
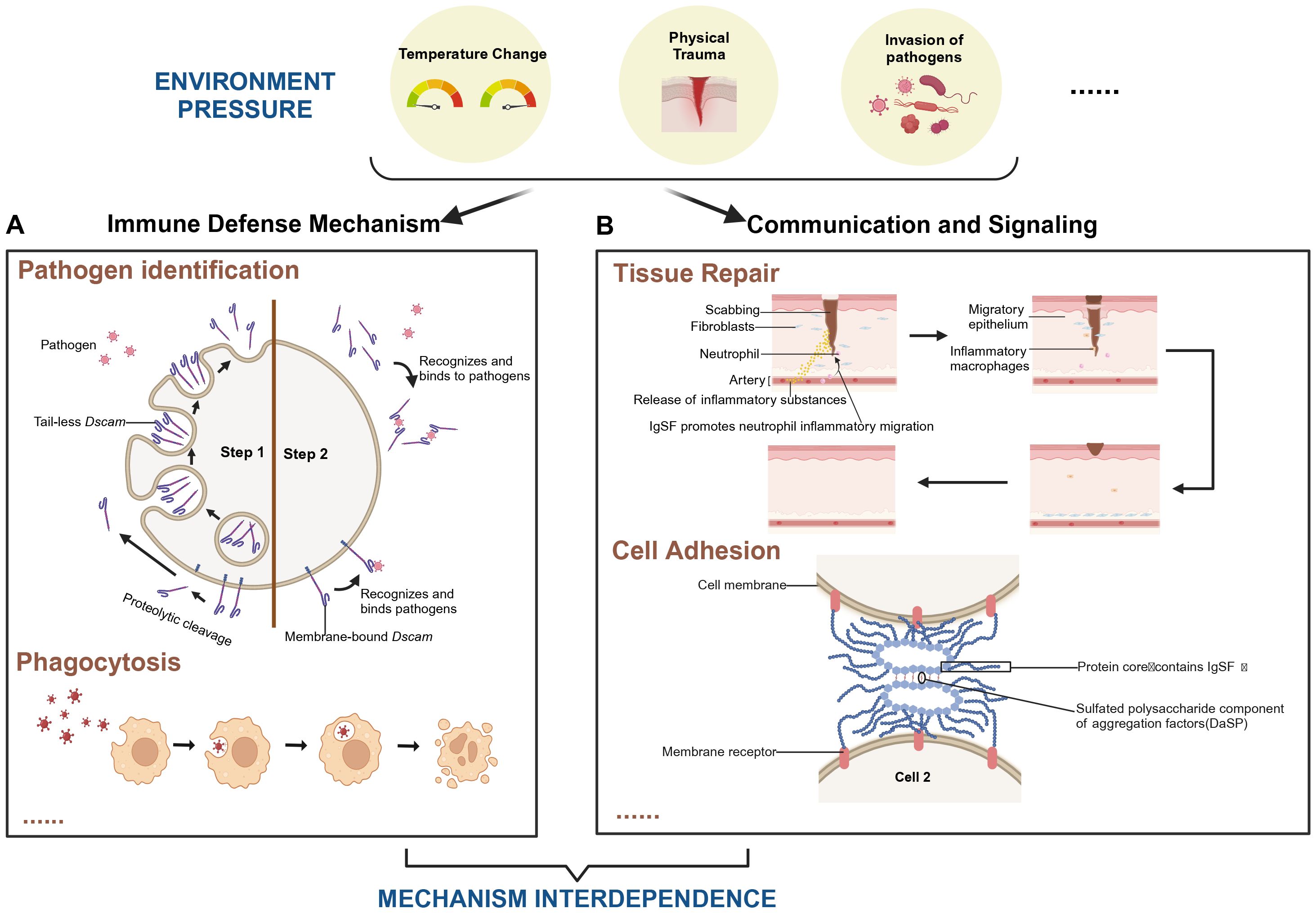
Figure 1. Role of IgSF-mediated immune mechanisms in tissue repair and pathogen clearance under environmental stress. The coordination between immune defense mechanisms and intercellular signaling pathways in supporting tissue repair and pathogen clearance under various environmental stresses, including temperature fluctuations, physical trauma, and pathogen invasion. (A) The “Pathogen Identification” module exemplifies the role of Dscam in pathogen recognition within arthropods. Step 1: Upon pathogen invasion of arthropod cells, intracellular tail-less Dscam is released via exocytosis. Alternatively, proteolytic cleavage of membrane-bound Dscam can lead to the shedding of its extracellular domain. Step 2: The released Dscam selectively recognizes and binds to specific pathogens, initiating the adhesion process required for subsequent phagocytosis (106). The “Phagocytosis” module illustrates how cells engulf and digest external particles by forming phagosomes. (B) The “Tissue Repair” module highlights IgSF’s role in promoting neutrophil migration and initiating inflammation. This accelerates wound healing by attracting fibroblasts and macrophages, which contribute to skin regeneration. The “Cell Adhesion” module focuses on how IgSF-containing proteins, in conjunction with sulfated polysaccharides present in cell aggregation factors, ensure stable cell adhesion, maintaining tissue integrity. Together, these mechanisms underscore the interdependence of immune defense and cellular communicat ion in responding to environmental stresses. They illustrate IgSF’s multifaceted roles in immunity, cell adhesion, and tissue repair, emphasizing its importance in maintaining organismal homeostasis under challenging conditions. Created in https://BioRender.com.
The Ig-like domain exhibits high diversity while maintaining remarkable conservation. Its characteristic structure consists of a sandwich-like fold composed of two β-sheets, commonly referred to as the Ig fold (17). For example, cell adhesion molecules (CAMs), which are rich in Ig-like domains, play critical roles in cell-cell interactions, particularly between neurons and between neurons and other cell types (18). In the nervous system, IgSF molecules regulate intercellular interactions, contributing to neural development, synapse formation, and the construction and function of neural circuits. The structural diversity and functional properties of IgSF molecules provide key insights into the mechanisms of cell-cell interactions in the nervous system (19).
This paper first explores the diverse characteristics of the IgSF, which are reflected not only in genetic diversity but also in structural diversity trends. Among invertebrates, we focus specifically on two representative groups: arthropods and mollusks. In arthropods, the functions of IgSF are primarily associated with the nervous and immune systems. In the nervous system, IgSF plays a critical role in synaptic signal transmission, while in the immune system, numerous molecules within the IgSF family have been identified as key players in immune responses. In mollusks, the immune functions of IgSF are more pronounced, involving pathogen recognition, immune response activation, and signal reconstruction both inside and outside the cell. Additionally, IgSF plays a significant role in the activation of immune-related genes. Beyond arthropods and mollusks, IgSF also functions in other animal phyla, such as protozoa, plankton, and sponges, where its molecules have been identified and are involved in cell adhesion processes. Our review summarizes current research findings on IgSF in invertebrates, highlighting its diverse functions across different invertebrate phyla, particularly its critical roles in immune defense and intercellular communication. These findings offer new perspectives on the immune mechanisms of these organisms and lay a solid foundation for further studies.
The IgSF is one of the largest and most diverse protein families in the human body, encompassing over 765 members (15). This diversity is evident not only in the vast number of members but also in the complexity of their gene structures and functional variety.
To illustrate the genetic diversity and the process that generates it, let’s consider the mechanism of Ig gene rearrangement, a critical step in the development of B cells. The gene rearrangement process in both heavy and light chains involves precise joining of variable (V), diversity (D), and joining (J) gene segments through somatic recombination. This process is fundamental to the generation of a vast array of antibodies with different specificities.
During the early stages of B-cell development, V(D)J recombination gradually establishes precursor cells expressing Ig heavy chains (IgH) (20, 21). For instance, in fish, clusters of activation-induced cytidine deaminase-positive (Aicda+) cells surrounding “melanomacrophages” have shown evidence of B-cell clonal expansion and frequent somatic mutations in VDJ segments (22). Following gene rearrangement, B cells undergo somatic hypermutation (SHM) and class switch recombination (CSR), enhancing antibody affinity and altering their biological properties while retaining high specificity for antigens (23). In certain species, such as chickens, immune responses are further optimized through immunoglobulin gene conversion (23).
Inter-species genetic diversity also underscores the functional variability of IgSF. For example, in mammals, natural killer cell protein 30 (NKp30) acts as an activating receptor for natural killer cells and features a single exon-encoded VJ-type IgSF domain with a transmembrane region (24). Furthermore, paralogous homologs of the natural cytotoxicity receptor 3 homolog (NCR3H) are present in most vertebrates but absent in mammals, suggesting that these gene complexes formed early during whole-genome duplications in jawed vertebrates (24). In humans, Ig gene polymorphisms are also widespread; for instance, the constant heavy chain regions encoded by immunoglobulin heavy chain gamma 1 (IGHG1), immunoglobulin heavy chain gamma 2 (IGHG2), and immunoglobulin heavy chain gamma 3 (IGHG3) exhibit significant individual variation, potentially impacting antibody functions (25).
The mechanisms underlying the diversity of the IgSF share both similarities with and differences from the gene rearrangement processes observed in Ig. As a broad molecular family, IgSF exhibits diverse mechanisms of variation, which differ among its members.
In the adaptive immune system, Ig and T-cell receptors (TCRs) primarily rely on V(D)J recombination to generate diversity, enabling the recognition of a wide range of antigens. For instance, during early B-cell development, V(D)J rearrangement sequentially gives rise to precursor cells expressing the IgH (20, 21). However, not all IgSF members rely on gene rearrangement for diversity. Certain non-immune-related IgSF molecules, such as the Down syndrome cell adhesion molecule (Dscam) in arthropods, achieve extensive variability through alternative splicing. This mechanism enables the generation of thousands of distinct isoforms (26), which play crucial roles in both neural development and immune recognition (27).
Beyond genetic mechanisms, the diversity of IgSF members is further shaped by post-translational modifications (PTMs). Modifications such as glycosylation and dephosphorylation (28) can alter protein conformation and function, thereby expanding their biological properties.
Additionally, Gene duplication also plays a significant role in immune diversity among invertebrates. For example, fibrinogen-related proteins (FREPs) in Biomphalaria glabrata exhibit high variability through point mutations and recombination (29).
Invertebrates also display IgSF gene diversity. Huene et al. identified 41 members of the Allorecognition (Alr) gene family within the Hydractinia symbiolongicarpus allorecognition complex, highlighting the remarkable diversity of IgSF genes (14). Their unique structural features provide new insights into the evolution of IgSF genes in invertebrates. For another example, Drosophila neurons express over 100 different cell adhesion molecule (CAM) genes during development, highlighting the critical role of genetic diversity in nervous system development (30). This extensive genetic diversity forms the foundation for the wide-ranging functions of IgSF, demonstrating its significance in immune systems and other biological processes.
The structural diversity of IgSF members is central to their functional versatility (31). This diversity manifests across genomic and domain levels. For instance, in the Aplysia californica, two distinct Aplysia californica Fibrinogen-related protein (AcFREP) proteins were identified: AcFREP1 lacks introns entirely, whereas AcFREP2 is encoded by four exons (32). Such differences in gene structure directly influence protein structure and function, showcasing IgSF’s remarkable genomic flexibility.
Ig structural diversity arises from complex genetic and molecular mechanisms. This diversity is initially reflected in Ig classification, including five types of heavy chain constant region gene segments (α, β, δ, μ, and ϵ) and two types of light chain constant region gene segments (κ and λ), which divide Ig into five classes (IgM, IgD, IgG, IgA, and IgE) and two types (κ and λ) (33). Moreover, variations in certain amino acid residues in the constant regions of heavy and light chains further lead to subclasses and subtypes (34).
Structurally, greater diversity is derived from variations in the amino acid residues of the variable regions of heavy and light chains, particularly in the complementarity-determining regions (CDRs), with CDR3 being a key site for diversity (33, 35). These changes result in the high diversity of the IgFab fragment, enhancing antibody specificity in antigen binding (36).
Unlike mammalian immunoglobulins, which are classified into five major types based on their function and structure, members of the IgSF in invertebrates exhibit a much broader diversity. Nevertheless, based on their functional and structural characteristics, they can still be categorized into several subfamilies. Below are some typical subfamilies along with representative members.
One of the most prominent subfamilies is the immunoglobulin cell adhesion molecules (IgCAMs). These proteins typically contain multiple Ig-like domains and primarily mediate cell-cell adhesion, playing crucial roles in neural development, synapse formation, and immune response (37). The presence of IgCAMs has been extensively reported across various invertebrate species (30, 38–42). To date, more than 100 IgCAMs have been identified in both vertebrates and invertebrates (43). For instance, N-CAM has been shown to play significant biological roles in Drosophila (38) and Aplysia (39).
Another key subfamily includes the Ig-like receptors, whose members typically contain Ig-like domains and are involved in immune responses, cell-cell recognition, and pathogen detection. Additionally, the IgSF subfamily associated with Toll-like receptors (TLRs) plays a pivotal role in the immune systems of invertebrates (44). By recognizing pathogen-associated molecular patterns (PAMPs), TLRs initiate immune responses (45). In cnidarians such as Hydra, despite the absence of mobile phagocytic cells, immune responses rely predominantly on epithelial cells, which utilize non-conventional TLR signaling pathways (46). Moreover, IgSF members like DSCAM (47–49) share homology with components of the vertebrate adaptive immune system, providing compelling evidence for evolutionary links between immune mechanisms.
Self-recognition molecules represent another crucial subfamily within invertebrate IgSF. For example, in the cnidarian Hydractinia symbiolongicarpus, allorecognition proteins such as Allorecognition 1 (Alr1) and Allorecognition 2 (Alr2) play key roles in distinguishing self from non-self (14, 50), highlighting the complexity of invertebrate immune systems. These proteins belong to a 41-gene family within the allorecognition complex and contain unique V-set and I-set Ig domains (14, 50).
Immunoeffector molecules also constitute a significant IgSF subfamily. For instance, the Molluscan Defence Molecule (MDM) identified in Lymnaea stagnalis contains multiple Ig-like domains and plays a crucial role in modulating immune defense responses during parasitic infections (51).
Finally, receptor modulatory molecules represent another important subfamily within the invertebrate IgSF. These proteins are involved in fine-tuning receptor-mediated signaling pathways, further expanding the functional diversity of IgSF members in invertebrate immunity.
The IgSF family includes various domain types, such as the V-set, C1-set, C2-set, and I-set, each with unique functional and evolutionary characteristics (31). The core structure consists of four β-strands (b, c, e, and f) forming an antiparallel β-sheet sandwich. This stable core underpins the fundamental functionality of IgSF (52). However, IgSF also exhibits significant structural flexibility. Variations in the topology of edge strands (e.g., a, c1, c2, d, and g) and alterations in inter-sheet angles ensure adaptability across diverse environments (52). Additionally, the high mutation rate of Ig-like domains and variability in disulfide bonds further enhance this flexibility, enabling IgSF to meet various biological demands (53).
The IgSF family encompasses not only immunoglobulins but also numerous critical biomolecules, such as major histocompatibility complex (MHC) molecules, TCRs, myelin protein P0, β2-microglobulin, and Thy-1 protein (54, 55). This member diversity emphasizes the structural and functional complexity of IgSF and underscores its pivotal roles in immune systems and other biological processes.
Furthermore, the various subfamilies of the IgSF not only share structural similarities but also exhibit functional overlap and interconnections. Despite differences in their specific roles, all these families contain Ig-like domains, a common feature that contributes to their morphological and functional resemblance (17). Among the most prominent shared functions across IgSF subfamilies are cell adhesion and signal transduction. IgCAMs exemplify this functional versatility. Proteins such as NCAM and ICAM are crucial for cell adhesion in the nervous and immune systems (38, 39), whereas Drosophila melanogaster Dscam, with its alternative splicing domains, guides axon growth and neuronal recognition during neural development. For instance, Dscam1 demonstrates remarkable specificity in neuronal differentiation and connectivity, with its splice variants playing distinct roles in neural circuit formation (26). Although Dscam exhibits a diverse splicing pattern, it shares functional similarities with IgCAMs in mediating cell adhesion and recognition (56–58), highlighting common mechanisms within the IgSF. However, the precise organizational and assembly mechanisms of Dscam at cell adhesion interfaces require further investigation.
Arthropods, the most diverse and morphologically varied animal group, play a key role in global ecosystems and were likely early terrestrial and aerial inhabitants. Today, arthropods are widely distributed across nearly all types of ecosystems (59, 60). Within this diverse group, the IgSF not only plays a pivotal role in the nervous system but also contributes significantly to immune responses.
Certain members of the IgSF, such as Fasciclin, Neuroglial, and Amalgam, are found in insects (61–63), indicating that the diversification of IgSF domains began early in the evolution of metazoans. Through gene duplication and subsequent divergence, these domains have evolved into multifunctional units with distinct roles. Fasciclin is renowned for its role in axon guidance and synapse formation, facilitating proper neuronal connections (61). Another CAM, Neuroglian, is crucial for maintaining nervous system integrity and is closely linked to the regulation of neuronal growth and differentiation (41). Amalgam interacts with Fasciclin and Neuroglian, suggesting its synergistic role in establishing and maintaining synaptic connections (62).
In the nervous system of Drosophila, epithelial IgCAMs play a vital role (37, 42). IgCAMs are involved in axonal growth, fasciculation, neuronal migration, survival, synaptic plasticity, and post-injury regeneration (42). As depicted in Figure 2A, IgCAMs interact through their extracellular domains to promote adhesion between axons and guide their growth along neural pathways (64). Notably, at sites of axon-axon interactions, CAMs on growth cones use a specific “zip code” system to ensure precise axonal interactions, forming complex and stable neural network architectures (64).
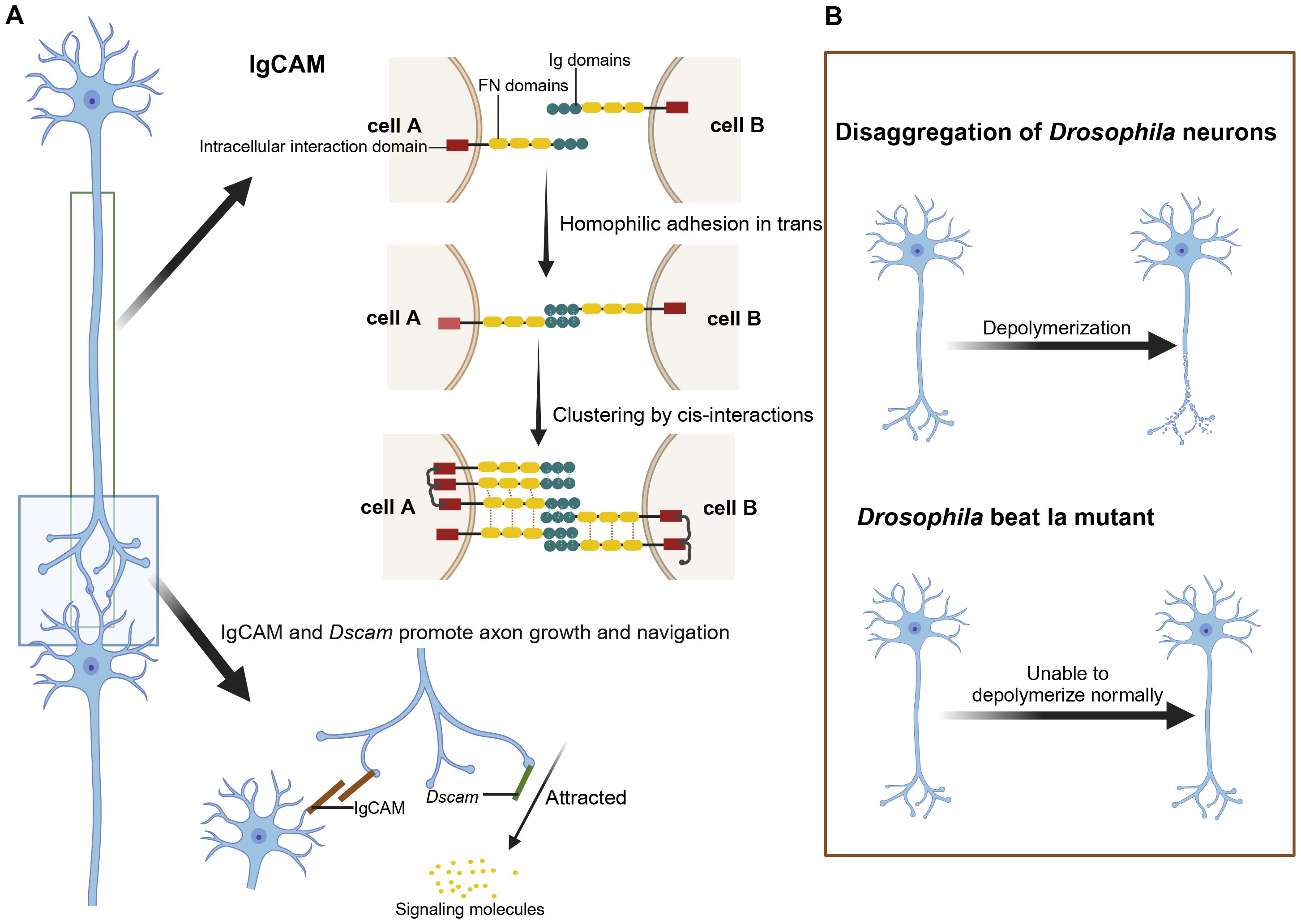
Figure 2. The role of IgCAM and Dscam in axon growth and navigation and the effects of their mutations. (A) Role of IgCAM and Dscam in neuronal connectivity and axon guidance. IgCAM mediates cell-cell adhesion through trans (homoadhesion) and cis interactions, forming stable clusters that bridge cell A and cell B by connecting their Ig and FN domains. Together, IgCAM and Dscam support axon extension and directed growth toward signaling molecules. (B) Depolymerization in Drosophila neurons: Wild-type neurons undergo controlled depolymerization, while Beat Ia mutants exhibit a failure to properly depolymerize axons. Created in https://BioRender.com.
The Dscam, a critical member of the IgSF, is essential for neuronal connectivity in Drosophila and some chelicerates (47). Through alternative splicing, Dscam generates up to 38,016 isoforms that mediate axonal sorting in the Drosophila mushroom body and regulate interactions between adjacent axons (48, 49). In certain arthropods, such as mites and spiders, non-classical forms of Dscam proteins lacking typical immunoglobulin and fibronectin III domains have been discovered, potentially having unique functions in neural development (65). Brites et al. defined Dscam genes with selective exon arrays as Dscam-hv (27). Studies have shown that in Drosophila, Dscam-hv acts as an axon guidance receptor, ensuring the precise transmission of olfactory receptor neuron signals to specific olfactory regions in the brain (66, 67). Additionally, Dscam-hv genes were first identified outside insects in organisms like Daphnia, where they contain variable cytoplasmic tail regions. Further research revealed that Dscam-hv genes in Daphnia are expressed in both brain tissue and hemocytes, with variable expression of exons 4, 6, and 11 reflecting their diverse roles in the nervous and immune systems (27).
Through genetic screening in Drosophila, researchers identified the beaten path Ia (beat Ia) gene, the first member characterized among the 14-beat gene families in Drosophila (68). The proteins encoded by Beats genes exhibit homology ranging from 24% to 72% in their primary amino acid sequences, primarily concentrated within their Ig domains, with higher homology at the N-terminus that gradually decreases toward the C-terminus. All Beats genes are expressed in the adult Drosophila head (68).
Studies indicate that Beat proteins may have anti-adhesive functions (69). In beat Ia mutants, axons display abnormal hyper-adhesion, preventing proper defasciculation, as illustrated in Figure 2B, which shows the disaggregation of Drosophila neurons in a beat Ia mutant (68). Conversely, beaten path Ic (beat Ic) promotes axonal bundling, while beat Ia inhibits this process. Their synergistic actions are critical for axon guidance and neural network formation (68).
The IgSF member Sidekick (Sdk) encodes a large transmembrane protein primarily responsible for neuronal positioning, dynamic cell shape adjustments, and precise axon targeting in the visual system of Drosophila (70).
Furthermore, Yu et al. used Mosaic Analysis with a Repressible Cell Marker (MARCM) to screen for cell-autonomous proteins required for axon pruning in Drosophila mushroom body (MB) γ neurons (71). They identified a Plexin-D1 like protein with unique domains (Plum), an IgSF transmembrane protein, which is closely associated with intracellular signaling and intercellular interactions during axon pruning. Notably, Plum’s role is confined to axon pruning within MB γ neurons (71).
IgSF surface proteins play critical roles in synaptic specificity in neuromuscular circuits, visual circuits, and the pathfinding of olfactory neuronal axons (72). Research reveals that Drosophila Plexin Receptors (Dprs) and Drosophila Interacting Proteins (DIPs) are two major subfamilies of IgSF proteins, offering new perspectives on synaptic specificity regulation (73).
Dprs exhibit specific expression patterns across neuronal clusters and bind to DIP proteins expressed selectively at synapses. The precise expression of specific cell surface proteins (CSPs) is essential for stable and accurate synaptic connections (74). For instance, DIP-α is selectively expressed in two known motor neurons, decisively influencing the correct presynaptic connection of MNISN-1 (73). Synaptic specificity refers to the high precision of neurons in selecting synaptic partners and determining synapse locations (75).
In Drosophila leg neuromuscular systems, the transmembrane IgSF proteins DIP-α and Dpr10, located in motor neurons (MN) and target muscles, respectively, are crucial for several developmental stages, including axonal fasciculation, synaptic specificity, and proper synapse formation. Similarly, these proteins participate in synaptic connections within the adult Drosophila olfactory and visual systems, as well as larval body-wall neuromuscular junctions (76).
Xu et al. identified a Dpr-ome interaction network comprising 21 Dpr proteins and 11 DIP proteins through an in vitro interactome screening. This network exhibits complex interaction patterns (30). During the pupal stage in the optic lobe (OL), neurons expressing specific DIPs often correspond to postsynaptic neurons expressing binding Dprs in vitro. Loss of function in DIPs or Dprs results in altered synaptic connections and neuronal death, underscoring their essential roles in synaptic specificity regulation (30).
In Drosophila neuromuscular and visual systems, Dpr-DIP interaction networks, determined primarily by shape complementarity, are involved in synaptic connectivity, terminal development, and neuronal survival (77). Single-cell RNA sequencing of embryonic Drosophila motor neurons, combined with imaging techniques and genetic perturbation, revealed the importance of homeodomain transcription factors (TFs) and IgSF proteins in synaptic wiring within neuromuscular systems (78).
Certain invertebrate proteins with Ig-like domains in the IgSF play crucial roles in host (e.g., insect) immune defenses (79).
Through transcriptomic analysis of Scylla paramamosain, researchers identified a novel IgSF member, Leucine-Rich Repeats, and Leucine-rich repeat and immunoglobulin-like domain-containing protein 1 (Lrig-1) (80). This gene is expressed across various tissues and is significantly upregulated in hemocytes during primary and secondary infections with Vibrio parahaemolyticus (80). Experiments showed that RNA interference (RNAi) knockdown of Lrig-1 markedly reduced antimicrobial peptide expression, demonstrating its critical role in host immune responses (80).
Another typical IgSF member is Hemolin, encoded by a single gene in the Hyalophora cecropia. This gene contains introns located within and between Ig-like domains, exhibiting unique genomic organization. As an immunoglobulin-like protein, Hemolin plays a vital role in insect immune defense by recognizing and binding pathogen surface molecules, thereby promoting pathogen aggregation and clearance (81).
Mollusks is one of the bilaterally symmetrical eumetazoan phyla, second only to arthropods in biodiversity, with approximately 76,000 extant species currently identified (82–84). This phylum is typically divided into 7 or 8 classes, two of which are entirely extinct. Mollusks exhibit a rich diversity of species and morphological variations, generally characterized by soft bodies protected by hard shells (85). Despite their relatively simple tissue and organ structures, the immune system of mollusks demonstrates a high level of complexity and diversity, especially highlighted in studies of IgSF molecules.
In gastropod mollusks such as Biomphalaria glabrata, multiple IgSF proteins have been identified, among which fibrinogen-related protein (BgFREP) plays a crucial role in immune defense. The N-terminal of BgFREP contains two tandem IgSF domains, while its C-terminal features a fibrinogen domain. During parasitic infections, BgFREP levels significantly increase, indicating its pivotal role in immune responses (86, 87). Studies have shown that BgFREP, through its polymorphism and ability to form multimers, binds specifically to parasitic antigens such as SmPoMucs, influencing host-parasite compatibility dynamics and serving as a core component of immune defense (88). This mechanism enhances the snail’s ability to recognize and respond to various parasitic antigens, effectively improving its immune defense efficiency.
Additionally, in another freshwater snail, Lymnaea stagnalis, an immune-related molecule known as MDM, which contains five C2-like Ig-like domains, was identified. This molecule endows snails with specific immune recognition and regulatory functions and plays a vital role in immune defense during parasitic infections (51). Experiments have also revealed that upon exposure to digenetic trematodes such as Echinostoma paraensei, Biomphalaria glabrata produces large amounts of hemolymph lectins containing IgSF and fibrinogen domains, aiding in the clearance of invading pathogens (89).
The functional diversity of BgFREP arises from mechanisms such as exon loss and somatic mutations, enabling it to bind to various parasitic antigens and regulate host-parasite compatibility and resistance (88, 90). Its structure comprises upstream IgSF domains and downstream fibrinogen-related domains (FreD), functioning in pathogen clearance during parasitic infections (91).
Moreover, research indicates that parasitic infections lead to a reduction in MDM levels, thereby weakening the host’s immune recognition capacity. This may serve as a critical strategy for parasites to evade host immune systems (51). In Sinonovacula constricta, two fibrinogen-related proteins, ScFREP-1 and ScFREP-2, have been identified. These proteins can recognize and bind to Gram-positive bacteria, Gram-negative bacteria, and pathogen-associated molecular patterns (PAMPs), inducing bacterial agglutination in the presence of calcium ions and ultimately clearing invading pathogens (92). Thus, ScFREP-1 and ScFREP-2 play crucial roles in the innate immunity of clams, offering new perspectives for studying Molluscan immunity (92).
In Crassostrea gigas, a novel IgSF member, CgIgIT2, has been identified (93–95). This protein is predominantly expressed in hemolymph and comprises five extracellular Ig domains, a transmembrane (TM) domain, and a classic immunoreceptor tyrosine-based activation motif (ITAM) (93). CgIgIT2 recognizes various microorganisms in the hemolymph, such as Vibrio splendidus and lipopolysaccharide (LPS), and activates intracellular signal transduction by forming dimers. Specifically, the signaling mechanism involves inducing the production of defensive proteins, such as CgICP, through the ERK phosphorylation pathway (95).
Further studies reveal that defensive proteins CgCAICP-1, CgICP-2, and CgLRRIG-1, which contain transmembrane domains, are distributed on the surface of hemocytes. They can recognize and phagocytose V. splendidus, subsequently digesting it within lysosomes. These findings demonstrate that through dimer formation and activation of the ERK phosphorylation signaling pathway, CgIgIT2 plays a vital role in the immune defense of Crassostrea gigas (95).
Inhibitory leukocyte immunoglobulin-like receptors (LILRBs), a class of single-pass transmembrane glycoproteins (Figure 3), were first named in 2001 (63). This family’s extracellular region contains Ig-like domains, while the intracellular region carries immunoreceptor tyrosine-based inhibitory motif (ITIM) (96). The LILRB family includes five members (LILRB1–LILRB5, also known as CD85J, CD85D, CD85A, CD85K, and CD85C) and a related member, LAIR1. As a novel immune checkpoint protein, LILRB’s ITIM structure, similar to CTLA-4 and PD-1, can be directly expressed on the surface of tumor cells, participating in tumor cell growth and survival. This characteristic positions LILRB as a significant target in tumor immunotherapy drug development (97).
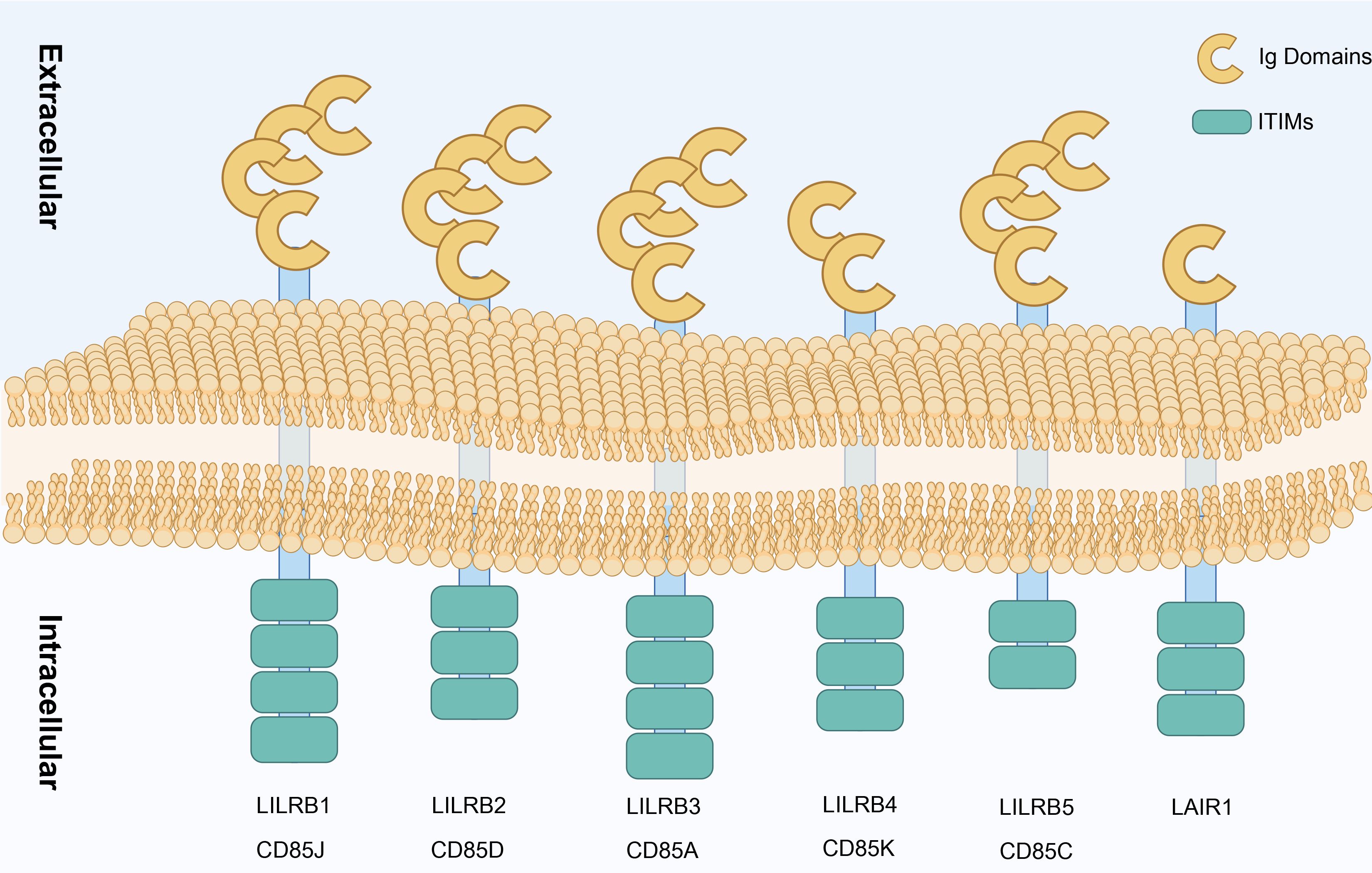
Figure 3. Types of LILRB. Structural representation of LILRB family receptors (LILRB1–LILRB5) and LAIR1 showing their extracellular Ig domains and ITIMs. Each receptor spans the cell membrane, with multiple Ig domains in the extracellular region facilitating ligand binding, while the ITIM in the intracellular region participates in downstream inhibitory signaling, contributing to immune regulation. Created in https://BioRender.com.
In the Aplysia californica, two fibrinogen-related protein genes, AcFREP1 and AcFREP2, have been discovered (32). Although the specific immune responses of AcFREPs have not yet been observed, their homology with the agglutinin functions of Biomphalaria suggests that these genes may play similar roles in immune defense (32). However, the functional differences and precise mechanisms of AcFREP1 and AcFREP2 remain subjects for further experimental research.
Protozoa are the most primitive and simplest unicellular organisms, often referred to as unicellular animals. Despite their structural simplicity, studies have identified molecules resembling the IgSF in protozoa. For example, receptor tyrosine kinases with Ig-like RTKs have been identified in protozoa (98), suggesting that early invertebrates already possessed fundamental immune and signal transduction systems.
Placozoa represents another evolutionarily significant group of animals. In 1883, the German zoologist F.E. Schulze first described Trichoplax adhaerens, a tiny marine organism (99). In 1971, K. Grell adopted the “placula” hypothesis, positioning Placozoa as potential common ancestors of sponges and cnidarians and as the origin of the lineage leading to all higher metazoans (100).
Further studies revealed the presence of a primitive major histocompatibility complex (MHC) in T. adhaerens, which is involved in innate immune processes within cells (101). Although no typical markers of adaptive immunity (e.g., MHC class I and II molecules) have been found in Placozoa, these findings provide critical insights into the early evolution of the animal immune system. They highlight the pivotal role of Placozoa in the evolutionary trajectory of immunity and their influence on subsequent higher organisms.
The sponges, also known as Phylum Porifera, represents one of the most basal groups of extant metazoans. Despite their relatively simple body structure and lack of true organ systems, with tissue complexity limited to the cellular level, sponges hold significant importance in the study of invertebrate evolution and biology. For years, sponges have been extensively used as model systems to study cellular adhesion (102, 103).
In sponges, CAMs are surface glycoproteins that play a crucial role in development and tissue stability. These molecules often contain one or more Ig-like repeat sequences (103), which mediate cell-to-cell adhesion and interaction. For example, sponge cell reaggregation depends on an extracellular proteoglycan complex called the aggregation factor. This factor is essential for whole-body regeneration (WBR) through cell reorganization in sponges (104).
Further research has identified glyconectins (GNs), a novel class of proteoglycan-like cell adhesion and recognition molecules, which are widely distributed across various sponges. Researchers Y. Guerardel and G.N. Misevic have highlighted that the functional mechanism of GNs exhibits evolutionary similarities with selective mechanisms in the IgSF, where carbohydrate domains play a crucial role in cell adhesion and recognition (105).
Additionally, in the ancient sponge Geodia cydonium, often referred to as a living fossil, researchers J. Kubrycht and J. Borecký identified a specific adhesion molecule, GSMS, which contains Ig-like domain sequences. This discovery provides new insights into the origins of early animals immunity and cell adhesion functions (105).
To provide a comprehensive overview of the roles and distributions of the IgSF members across invertebrates, we have compiled a detailed table (Table 1). This table encapsulates the diverse functions of IgSF molecules, ranging from immune defense and neural development to synaptic plasticity and axon guidance.
The IgSF has maintained remarkable functional conservation throughout evolution, especially in cell recognition, adhesion, and signal transduction. These functions are critical across species, with IgSF-mediated interactions playing key roles in nervous system development and immune defense. In the nervous system, CAMs, a major IgSF subgroup, are essential for neural development. In Drosophila, IgSF members such as Fasciclin (61) and Neuroglian (41) regulate axon guidance and synapse formation (41, 61, 62). Similarly, in Porifera, CAMs mediate intercellular adhesion, contributing to tissue stability (103, 104). Despite the conservation of core functions, significant functional diversification has emerged across species. In Drosophila, Dscam undergoes alternative splicing to produce 38,016 isoforms, allowing precise axon sorting (48, 49). Non-canonical Dscam variants in arthropods, like mites and spiders, have distinct neural roles, reflecting functional remodeling for unique neural architectures (65). In the immune system, IgSF diversification has led to more complex regulatory mechanis ms. In mollusks, BgFREP (86–88) and ScFREP (92) not only recognize pathogens but also regulate host-parasite compatibility through polymorphism (86–88, 92). In Crassostrea gigas, CgIgIT2 enhances immune defense by activating the ERK phosphorylation pathway (95). Thus, while IgSF retains its core functions in pathogen recognition, its diversification supports species-specific adaptations in both nervous and immune systems, highlighting its versatility.
This review highlights the remarkable diversity of the IgSF in invertebrates and their pivotal roles in various biological processes. IgSF proteins are integral to key cellular functions such as recognition, adhesion, and signal transduction, with significant involvement in both the immune and nervous systems. Through their structural diversity and specificity, these proteins have laid the foundation for the emergence of adaptive immunity, enabling organisms to efficiently recognize and counteract a wide range of pathogens. Simultaneously, IgSF proteins contribute to critical neurodevelopmental processes, including axon guidance, synapse formation, and the functional maintenance of neural networks, underscoring their essential role in nervous system development.
The review emphasizes the role of IgSF in innate immunity among invertebrates, highlighting how diverse IgSF members utilize molecular recognition mechanisms to detect and neutralize various pathogens in mollusks, demonstrating both specificity and functional complexity. In arthropods, IgSF proteins play crucial roles in neural development, synaptic connectivity, and the precise transmission of neuronal signals, illustrating their multifunctionality across different biological processes.
By comparing the structure and function of IgSF across various organisms, this review traces their evolutionary trajectories, revealing how these proteins have adapted to changing environmental pressures and biological needs. These studies demonstrate that the functional diversification and specialization of IgSF proteins represent a dynamic evolutionary response to external challenges, offering new insights into the complexity of immune and nervous systems.
Advancements in IgSF research will deepen our understanding of their molecular mechanisms in immune defense and neuronal functions, opening the door to potential breakthroughs in disease prevention and neuroscience. Furthermore, studying the evolution of IgSF not only brings insight into their adaptive mechanisms across diverse organisms but also offers innovative solutions in fields such as biomedicine and ecological conservation. These findings enhance our understanding of biological evolution and provide a scientific foundation for developing strategic applications to address environmental challenges and related diseases.
HL: Supervision, Writing – original draft, Writing – review & editing. YZ: Writing – original draft, Writing – review & editing. YHZ: Supervision, Writing – review & editing. QZ: Investigation, Supervision, Writing – review & editing. JX: Investigation, Supervision, Writing – review & editing. XL: Investigation, Writing – review & editing. LZ: Investigation, Writing – review & editing. HL: Investigation, Writing – review & editing. ML: Investigation, Supervision, Writing – review & editing. YQ: Investigation, Supervision, Writing – review & editing. XZ: Supervision, Writing – review & editing. KC: Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China (Grant No: 32000293), Guangxi Natural Science Foundation (Grant Nos: 2020JJA130077 and 2018JJB140423), the University Level Scientific Research Project of Zhejiang Shuren University (Grant No: 2022R064), and Zhejiang Shuren University Basic Scientific Research Special Funds (Grant No: 2024XZ014), the National Innovation and Entrepreneurship Training Program for College Students in 2023 (Grant No: 202311842052X), Zhejiang Shuren University School-level Scientific Research Project (2023A11001).
The authors sincerely thank HL for her invaluable guidance and insightful suggestions during the preparation of this review. The authors also express their gratitude to her team for their support in literature collection and critical discussions, which greatly enriched the content and perspectives of this work. The authors gratefully acknowledge the generous financial support. The authors sincerely appreciate BioRender for providing an intuitive and efficient platform that greatly facilitated the creation of scientific illustrations.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Irisarri I, Baurain D, Brinkmann H, Delsuc F, Sire JY, Kupfer A, et al. Phylotranscriptomic consolidation of the jawed vertebrate timetree. Nat Ecol Evol. (2017) 1:1370–8. doi: 10.1038/s41559-017-0240-5
2. Zhu YA, Li Q, Lu J, Chen Y, Wang J, Gai Z, et al. The oldest complete jawed vertebrates from the early Silurian of China. Nature. (2022) 609:954–8. doi: 10.1038/s41586-022-05136-8
3. Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. (2006) 124:815–22. doi: 10.1016/j.cell.2006.02.001
4. Riera Romo M, Pérez-Martínez D, Castillo Ferrer C. Innate immunity in vertebrates: an overview. Immunology. (2016) 148:125–39. doi: 10.1111/imm.2016.148.issue-2
5. Pan G, Bao J, Ma Z, Song Y, Han B, Ran M, et al. Invertebrate host responses to microsporidia infections. Dev Comp Immunol. (2018) 83:104–13. doi: 10.1016/j.dci.2018.02.004
6. Petralia RS, Mattson MP, Yao PJ. Aging and longevity in the simplest animals and the quest for immortality. Ageing Res Rev. (2014) 16:66–82. doi: 10.1016/j.arr.2014.05.003
7. Bronsdon S, Rogers AD, Tyler PA, Rice AL, Gage JD. Genetic study of the extent and consequences of sexual and asexual reproduction in the deep-sea epizoic anemones Amphianthus inornata and Kadosactis commensalis (Cnidaria: Anthozoa). Mar Biol. (1997) 128:231–9. doi: 10.1007/s002270050087
8. Gooch JL, Schopf TJ. Genetic variability in the deep sea: relation to environmental variability. Evolution. (1972) p:545–52. doi: 10.2307/2407051
9. Ekins M, Erpenbeck D, Wörheide G, Hooper JNA. Staying well connected–Lithistid sponges on seamounts. J Mar Biol Assoc United Kingdom. (2016) 96:437–51. doi: 10.1017/S0025315415000831
10. McGee KM, Porter TM, Wright M, Hajibabaei M. Drivers of tropical soil invertebrate community composition and richness across tropical secondary forests using DNA metasystematics. Sci Rep. (2020) 10:18429. doi: 10.1038/s41598-020-75452-4
11. Iwanaga S, Lee BL. Recent advances in the innate immunity of invertebrate animals. J Biochem Mol Biol. (2005) 38:128–50. doi: 10.5483/BMBRep.2005.38.2.128
12. Zárate-Potes A, Ocampo ID, Cadavid LF. The putative immune recognition repertoire of the model cnidarian Hydractinia symbiolongicarpus is large and diverse. Gene. (2019) 684:104–17. doi: 10.1016/j.gene.2018.10.068
13. Zinn K, Özkan E. Neural immunoglobulin superfamily interaction networks. Curr Opin Neurobiol. (2017) 45:99–105. doi: 10.1016/j.conb.2017.05.010
14. Huene AL, Sanders SM, Ma Z, Nicotra ML. A family of unusual immunoglobulin superfamily genes in an invertebrate histocompatibility complex. Proc Natl Acad Sci U.S.A. (2022) 119:e2207374119. doi: 10.1073/pnas.2207374119
15. Srinivasan M, Roeske RW. Immunomodulatory peptides from IgSF proteins: a review. Curr Protein Pept Sci. (2005) 6:185–96. doi: 10.2174/1389203053545426
16. Kaas Q, Ehrenmann F, Lefranc MP. IG, TR and IgSF, MHC and MhcSF: what do we learn from the IMGT Colliers de Perles? Brief Funct Genomic Proteomic. (2007) 6:253–64. doi: 10.1093/bfgp/elm032
17. Halaby DM, Mornon JP. The immunoglobulin superfamily: an insight on its tissular, species, and functional diversity. . J Mol Evol. (1998) 46:389–400. doi: 10.1007/PL00006318
18. Tan RPA, Leshchyns’ka I, Sytnyk V. Glycosylphosphatidylinositol-anchored immunoglobulin superfamily cell adhesion molecules and their role in neuronal development and synapse regulation. Front Mol Neurosci. (2017) 10:378. doi: 10.3389/fnmol.2017.00378
19. Yoshihara Y, Oka S, Ikeda J, Mori K. Immunoglobulin superfamily molecules in the nervous system. Neurosci Res. (1991) 10:83–105. doi: 10.1016/0168-0102(91)90033-U
20. Li Y, Xue Y, Peng Z, Zhang L. Immune diversity in lophotrochozoans, with a focus on recognition and effector systems. Comput Struct Biotechnol J. (2023) 21:2262–75. doi: 10.1016/j.csbj.2023.03.031
21. Melchers F, Yamagami T, Rolink A, Andersson J. Rules for the rearrangement events at the L chain gene loci of the mouse. Adv Exp Med Biol. (2007) 596:63–70. doi: 10.1007/0-387-46530-8_6
22. Waly D, Muthupandian A, Fan CW, Anzinger H, Magor BG. Immunoglobulin VDJ repertoires reveal hallmarks of germinal centers in unique cell clusters isolated from zebrafish (Danio rerio) lymphoid tissues. Front Immunol. (2022) 13:1058877. doi: 10.3389/fimmu.2022.1058877
23. Chi X, Li Y, Qiu X. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: mechanism and regulation. Immunology. (2020) 160:233–47. doi: 10.1111/imm.v160.3
24. Janes ME, Kinlein A, Flajnik MF, Du Pasquier L, Ohta Y. Genomic view of the origins of cell-mediated immunity. Immunogenetics. (2023) 75:479–93. doi: 10.1007/s00251-023-01319-3
25. Bashirova AA, Zheng W, Akdag M, Augusto DG, Vince N, Dong KL, et al. Population-specific diversity of the immunoglobulin constant heavy G chain (IGHG) genes. Genes Immun. (2021) 22:327–34. doi: 10.1038/s41435-021-00156-2
26. Dong H, Yang X, Wu L, Zhang S, Zhang J, Guo P, et al. A systematic CRISPR screen reveals redundant and specific roles for Dscam1 isoform diversity in neuronal wiring. PloS Biol. (2023) 21:e3002197. doi: 10.1371/journal.pbio.3002197
27. Brites D, McTaggart S, Morris K, Anderson J, Thomas K, Colson I, et al. The Dscam homologue of the crustacean Daphnia is diversified by alternative splicing like in insects. Mol Biol Evol. (2008) 25:1429–39. doi: 10.1093/molbev/msn087
28. Itoh K, Shimono K, Lemmon V. Dephosphorylation and internalization of cell adhesion molecule L1 induced by theta burst stimulation in rat hippocampus. Mol Cell Neurosci. (2005) 29:245–9. doi: 10.1016/j.mcn.2005.02.014
29. Zhang SM, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. (2004) 305:251–4. doi: 10.1126/science.1088069
30. Xu S, Sergeeva AP, Katsamba PS, Mannepalli S, Bahna F, Bimela J, et al. Affinity requirements for control of synaptic targeting and neuronal cell survival by heterophilic IgSF cell adhesion molecules. Cell Rep. (2022) 39:110618. doi: 10.1016/j.celrep.2022.110618
31. Williams AF, Barclay AN. The immunoglobulin superfamily–domains for cell surface recognition. Annu Rev Immunol. (1988) 6:381–405. doi: 10.1146/annurev.iy.06.040188.002121
32. Gorbushin AM, Panchin YV, Iakovleva NV. In search of the origin of FREPs: characterization of Aplysia californica fibrinogen-related proteins. Dev Comp Immunol. (2010) 34:465–73. doi: 10.1016/j.dci.2009.12.007
33. Bengtén E, Wilson M, Miller N, Clem LW, Pilström L, Warr GW. Immunoglobulin isotypes: structure, function, and genetics. Curr Top Microbiol Immunol. (2000) 248:189–219. doi: 10.1007/978-3-642-59674-2_9
34. Diesterbeck US, Aboelhassan DM, Stein SK, Czerny CP. Detection of new allotypic variants of bovine λ-light chain constant regions in different cattle breeds. Dev Comp Immunol. (2012) 36:130–9. doi: 10.1016/j.dci.2011.06.011
35. Guloglu B, Deane CM. Specific attributes of the V(L) domain influence both the structure and structural variability of CDR-H3 through steric effects. Front Immunol. (2023) 14:1223802. doi: 10.3389/fimmu.2023.1223802
36. Sankar K, Hoi KH, Hötzel I. Dynamics of heavy chain junctional length biases in antibody repertoires. Commun Biol. (2020) 3:207. doi: 10.1038/s42003-020-0931-3
37. Finegan TM, Bergstralh DT. Neuronal immunoglobulin superfamily cell adhesion molecules in epithelial morphogenesis: insights from Drosophila. Philos Trans R Soc Lond B Biol Sci. (2020) 375:20190553. doi: 10.1098/rstb.2019.0553
38. Budnik V, Zhong Y, Wu CF. Morphological plasticity of motor axons in Drosophila mutants with altered excitability. J Neurosci. (1990) 10:3754–68. doi: 10.1523/JNEUROSCI.10-11-03754.1990
39. Mayford M, Barzilai A, Keller F, Schacher S, Kandel ER. Modulation of an NCAM-related adhesion molecule with long-term synaptic plasticity in Aplysia. Science. (1992) 256:638–44. doi: 10.1126/science.1585176
40. Zhong Y, Budnik V, Wu CF. Synaptic plasticity in Drosophila memory and hyperexcitable mutants: role of cAMP cascade. J Neurosci. (1992) 12:644–51. doi: 10.1523/JNEUROSCI.12-02-00644.1992
41. Liu W, Li Q. Single-cell transcriptomics dissecting the development and evolution of nervous system in insects. Curr Opin Insect Sci. (2024) p:101201. doi: 10.1016/j.cois.2024.101201
42. Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. (2007) 10:19–26. doi: 10.1038/nn1827
43. Stoeckli ET, Kilinc D, Kunz B, Kunz S, Lee GU, Martines E, et al. Analysis of cell-cell contact mediated by Ig superfamily cell adhesion molecules. Curr Protoc Cell Biol. (2013) 61:9.5.1–9.5.85. doi: 10.1002/0471143030.2013.61.issue-1
44. Satake H, Sekiguchi T. Toll-like receptors of deuterostome invertebrates. Front Immunol. (2012) 3:34. doi: 10.3389/fimmu.2012.00034
45. Zheng L, Zhang L, Lin H, McIntosh MT, Malacrida AR. Toll-like receptors in invertebrate innate immunity. Invertebrate Survival J. (2005) 2:105–13.
46. Bosch TCG, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, et al. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. (2009) 33:559–69. doi: 10.1016/j.dci.2008.10.004
47. Yue Y, Meng Y, Ma H, Hou S, Cao G, Hong W, et al. A large family of Dscam genes with tandemly arrayed 5’ cassettes in Chelicerata. Nat Commun. (2016) 7:11252. doi: 10.1038/ncomms11252
48. Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, et al. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. (2004) 43:673–86. doi: 10.1016/j.neuron.2004.07.020
49. Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. (2004) 118:619–33. doi: 10.1016/j.cell.2004.08.021
50. Huene AL, Sanders SM, Ma Z, Nguyen AD, Koren S, Michaca MH, et al. The Hydractinia Allorecognition Complex encodes a novel family of immunoglobulin superfamily. Neural Networks. (2019) 37:420–3. doi: 10.1101/2022.03.04.482883
51. Hoek RM, Smit AB, Frings H, Vink JM, Jong-Brink M, Geraerts WPM. A new Ig-superfamily member, molluscan defence molecule (MDM) from Lymnaea stagnalis, is down-regulated during parasitosis. Eur J Immunol. (1996) 26:939–44. doi: 10.1002/eji.1830260433
52. Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. (1994) 242:309–20. doi: 10.1016/S0022-2836(84)71582-8
53. Levin SD, Evans LS, Bort S, Rickel E, Lewis KE, Wu RP, et al. Novel immunomodulatory proteins generated via directed evolution of variant igSF domains. Front Immunol. (2019) 10:3086. doi: 10.3389/fimmu.2019.03086
54. Galaktionov VG. Evolutionary development of the immunoglobulins super family. Izv Akad Nauk Ser Biol. (2004) 2):133–45.
55. Zhai R, Wang Q. Phylogenetic analysis provides insight into the molecular evolution of nociception and pain-related proteins. Evol Bioinform Online. (2023) 19:11769343231216914. doi: 10.1177/11769343231216914
56. Guo L, Wu Y, Chang H, He Y. Structure of cell-cell adhesion mediated by the Down syndrome cell adhesion molecule. Proc Natl Acad Sci U.S.A. (2021) 118(39). doi: 10.1073/pnas.2022442118
57. Jin L, Zhou W. Progress and prospect of dscam gene in drosophila immunity. Int J Biol Life Sci. (2024) 5:17–9. doi: 10.54097/t7tcq290
58. Hizawa K, Sasaki T, Arimura N. A comparative overview of DSCAM and its multifunctional roles in Drosophila and vertebrates. Neurosci Res. (2024) 202:1–7. doi: 10.1016/j.neures.2023.12.005
59. Damborenea C, Rogers DC, Thorp JH. Thorp and covich’s freshwater invertebrates: volume 5: keys to neotropical and antarctic fauna. Cambridge, MA, USA: Academic Press (2020).
60. Aria C. The origin and early evolution of arthropods. Biol Rev Camb Philos Soc. (2022) 97:1786–809. doi: 10.1111/brv.12864
61. Garcia-Alonso LA. Fasciclin 2 functions as an expression-level switch on EGFR to control organ shape and size in Drosophila. bioRxiv. (2024) 2024:05. 02.592138. doi: 10.1371/journal.pone.0309891
62. Yokoi K, Kato Y, Suzuki M, Miura K. Molecular cloning and functional analyses of an adhesion molecule, neuroglian, in Mythimna separata (Lepidoptera: Noctuidae). Eur J Entomology. (2018) 115:157–66. doi: 10.14411/eje.2018.015
63. Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT. Leukocyte Ig-like receptor complex (LRC) in mice and men. Trends Immunol. (2002) 23:81–8. doi: 10.1016/S1471-4906(01)02155-X
64. Kamiguchi H. The role of cell adhesion molecules in axon growth and guidance. Adv Exp Med Biol. (2007) 621:95–103. doi: 10.1007/978-0-387-76715-4_7
65. Cao G, Shi Y, Zhang J, Ma H, Hou S, Dong H. A chelicerate-specific burst of nonclassical Dscam diversity. BMC Genomics. (2018) 19:66. doi: 10.1186/s12864-017-4420-0
66. Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, Zipursky SL. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron. (2003) 37:221–31. doi: 10.1016/S0896-6273(02)01183-2
67. Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. (2000) 101:671–84. doi: 10.1016/S0092-8674(00)80878-8
68. Pipes GC, Lin Q, Riley SE, Goodman CS. The Beat generation: a multigene family encoding IgSF proteins related to the Beat axon guidance molecule in Drosophila. Development. (2001) 128:4545–52. doi: 10.1242/dev.128.22.4545
69. Fambrough D, Goodman CS. The Drosophila beaten path gene encodes a novel secreted protein that regulates defasciculation at motor axon choice points. Cell. (1996) 87:1049–58. doi: 10.1016/S0092-8674(00)81799-7
70. Yamagata M. Structure and functions of sidekicks. Front Mol Neurosci. (2020) 13:139. doi: 10.3389/fnmol.2020.00139
71. Yu XM, Gutman I, Mosca TJ, Iram T, Özkan E, Garcia KC, et al. Plum, an immunoglobulin superfamily protein, regulates axon pruning by facilitating TGF-β signaling. Neuron. (2013) 78:456–68. doi: 10.1016/j.neuron.2013.03.004
72. Lobb-Rabe M, DeLong K, Salazar RJ, Zhang R, Wang Y, Carrillo RA. Dpr10 and Nocte are required for Drosophila motor axon pathfinding. Neural Dev. (2022) 17:10. doi: 10.1186/s13064-022-00165-5
73. Ashley J, Sorrentino V, Lobb-Rabe M, Nagarkar-Jaiswal S, Tan L, Xu S, et al. Transsynaptic interactions between IgSF proteins DIP-α and Dpr10 are required for motor neuron targeting specificity. Elife. (2019) 8. doi: 10.7554/eLife.42690
74. Sanes JR, Zipursky SL. Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell. (2020) 181:536–56. doi: 10.1016/j.cell.2020.04.008
75. Margeta MA, Shen K. Molecular mechanisms of synaptic. specificity. Mol Cell Neurosci. (2010) 43:261–7. doi: 10.1016/j.mcn.2009.11.009
76. Venkatasubramanian L, Guo Z, Xu S, Tan L, Xiao Q, Nagarkar-Jaiswal S, et al. Stereotyped terminal axon branching of leg motor neurons mediated by IgSF proteins DIP-α and Dpr10. Elife. (2019) 8. doi: 10.7554/eLife.42692
77. Carrillo RA, Özkan E, Menon KP, Nagarkar-Jaiswal S, Lee PT, Jeon M, et al. Control of synaptic connectivity by a network of drosophila igSF cell surface proteins. Cell. (2015) 163:1770–82. doi: 10.1016/j.cell.2015.11.022
78. Velten J, Gao X, Van Nierop y Sanchez P, Domsch K, Agarwal R, Bognar L, et al. Single-cell RNA sequencing of motoneurons identifies regulators of synaptic wiring in Drosophila embryos. Mol Syst Biol. (2022) 18:e10255. doi: 10.15252/msb.202110255
79. Watson FL, Püttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. (2005) 309:1874–8. doi: 10.1126/science.1116887
80. Sheng Y, Wan H, Xie Y, Zhang X, Zou P, Zhang Z, et al. A member of the immunoglobulin superfamily lrig-1 might be involved in the immune priming of Scylla paramamosain in response to the infection and re-infection by Vibrio parahaemolyticus. Dev Comp Immunol. (2023) 147:104757. doi: 10.1016/j.dci.2023.104757
81. Wojda I, Cytryńska M, Zdybicka-Barabas A, Kordaczuk J. Insect defense proteins and peptides. Subcell Biochem. (2020) 94:81–121. doi: 10.1007/978-3-030-41769-7_4
82. Rosenberg G. A new critical estimate of named species-level diversity of the recent Mollusca. Am Malacological Bull. (2014) 32:308–22. doi: 10.4003/006.032.0204
83. Ponder WF, Lindberg DR, Ponder JM. Biology and evolution of the mollusca, volume 1. Boca Raton, FL, USA: CRC Press (2019).
86. Zhang SM, Léonard PM, Adema CM, Loker ES. Parasite-responsive IgSF members in the snail Biomphalaria glabrata: characterization of novel genes with tandemly arranged IgSF domains and a fibrinogen domain. Immunogenetics. (2001) 53:684–94. doi: 10.1007/s00251-001-0386-8
87. Wu XJ, Dinguirard N, Sabat G, Lui HD, Gonzalez L, Gehring M, et al. Proteomic analysis of Biomphalaria glabrata plasma proteins with binding affinity to those expressed by early developing larval Schistosoma mansoni. PloS Pathog. (2017) 13:e1006081. doi: 10.1371/journal.ppat.1006081
88. Portet A, Pinaud S, Tetreau G, Galinier R, Cosseau C, Duval D, et al. Integrated multi-omic analyses in Biomphalaria-Schistosoma dialogue reveal the immunobiological significance of FREP-SmPoMuc interaction. Dev Comp Immunol. (2017) 75:16–27. doi: 10.1016/j.dci.2017.02.025
89. Léonard PM, Adema CM, Zhang SM, Loker ES. Structure of two FREP genes that combine IgSF and fibrinogen domains, with comments on diversity of the FREP gene family in the snail Biomphalaria glabrata. Gene. (2001) 269:155–65. doi: 10.1016/S0378-1119(01)00444-9
90. Adema CM. Fibrinogen-related proteins (FREPs) in mollusks. Results Probl Cell Differ. (2015) 57:111–29. doi: 10.1007/978-3-319-20819-0_5
91. Dheilly NM, Duval D, Mouahid G, Emans R, Allienne J-F, Galinier R, et al. A family of variable immunoglobulin and lectin domain containing molecules in the snail Biomphalaria glabrata. Dev Comp Immunol. (2015) 48:234–43. doi: 10.1016/j.dci.2014.10.009
92. Wu Y, Zheng Y, Li Y, Li Y, Niu D. Two fibrinogen-related proteins (FREPs) in the razor clam (Sinonovacula constricta) with a broad recognition spectrum and bacteria agglutination activity. Dev Comp Immunol. (2021) 121:104075. doi: 10.1016/j.dci.2021.104075
93. Yan X, Sun J, Yang W, Li X, Yang Q, Li Y, et al. An immunoglobulin superfamily member (CgIgIT2) functions as immune inhibitory receptor to inhibit the inflammatory cytokine expressions in Crassostrea gigas. Dev Comp Immunol. (2023) 144:104708. doi: 10.1016/j.dci.2023.104708
94. Sun J, Wang L, Yang W, Wang L, Fu Q, Song L. IgIT-mediated signaling inhibits the antimicrobial immune response in oyster hemocytes. J Immunol. (2020) 205:2402–13. doi: 10.4049/jimmunol.2000294
95. Sun J, Wang L, Yang W, Wang L, Fu Q, Song L. An ancient BCR-like signaling promotes ICP production and hemocyte phagocytosis in oyster. iScience. (2020) 23:100834. doi: 10.1016/j.isci.2020.100834
96. Deng M, Chen H, Liu X, Huang R, He Y, Yoo B. Leukocyte immunoglobulin-like receptor subfamily B: therapeutic targets in cancer. Antib Ther. (2021) 4:16–33. doi: 10.1093/abt/tbab002
97. Wang H, Kaur G, Sankin AI, Chen F, Guan F, Zang X. Immune checkpoint blockade and CAR-T cell therapy in hematologic Malignancies. J Hematol Oncol. (2019) 12:59. doi: 10.1186/s13045-019-0746-1
98. Grassot J, Gouy M, Perrière G, Mouchiroud G. Origin and molecular evolution of receptor tyrosine kinases with immunoglobulin-like domains. Mol Biol Evol. (2006) 23:1232–41. doi: 10.1093/molbev/msk007
99. Grell K. Trichoplax adhaerens FE Schulze und die Entstehung der Metazoen. Naturwiss. Rundschau. (1971) 24:160–1.
100. Schierwater B, DeSalle R. Placozoa. Curr Biol. (2018) 28:R97–r98. doi: 10.1016/j.cub.2017.11.042
101. Ohta Y, Kasahara M, O’Connor TD, Flajnik MF. Inferring the “primordial immune complex”: origins of MHC class I and antigen receptors revealed by comparative genomics. J Immunol. (2019) 203:1882–96. doi: 10.4049/jimmunol.1900597
102. Thorp JH, Covich AP. Ecology and classification of North American freshwater invertebrates. San Diego, CA, USA: Academic Press (2009).
103. Fernàndez-Busquets X, Burger MM. Cell adhesion and histocompatibility in sponges. . Microsc Res Tech. (1999) 44:204–18. doi: 10.1002/(SICI)1097-0029(19990215)44:4<204::AID-JEMT2>3.0.CO;2-I
104. Ereskovsky A, Borisenko IE, Bolshakov FV, Lavrov AI. Whole-body regeneration in sponges: diversity, fine mechanisms, and future prospects. Genes (Basel). (2021) 12(4). doi: 10.3390/genes12040506
105. Guerardel Y, Czeszak X, Sumanovski LT, Karamanos Y, Popescu O, Strecker G. Molecular fingerprinting of carbohydrate structure phenotypes of three porifera proteoglycan-like glyconectins. J Biol Chem. (2004) 279:15591–603. doi: 10.1074/jbc.M308928200
Keywords: IgSF, arthropods, mollusks, nervous system, immune system
Citation: Li H, Zhang Y, Zhu Y, Zhao Q, Xu J, Li X, Zhao L, Li H, Liu M, Qian Y, Zhang X and Chen K (2025) Functional insights into immunoglobulin superfamily proteins in invertebrate neurobiology and immunity. Front. Immunol. 16:1552151. doi: 10.3389/fimmu.2025.1552151
Received: 27 December 2024; Accepted: 13 March 2025;
Published: 02 April 2025.
Edited by:
Humberto Lanz-Mendoza, National Institute of Public Health, MexicoReviewed by:
Xiaobo Zhang, Zhejiang University, ChinaCopyright © 2025 Li, Zhang, Zhu, Zhao, Xu, Li, Zhao, Li, Liu, Qian, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Li, aG9uZ3l1ODg5MjZAempzcnUuZWR1LmNu; Keda Chen, Y2hlbmtkQHpqc3J1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.