
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 09 April 2025
Sec. Autoimmune and Autoinflammatory Disorders : Autoimmune Disorders
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1551552
This article is part of the Research Topic Exploring the Immune-Metabolic Network in Diabetes View all articles
Introduction: Genome-wide association study identified C1QTNF6 as a candidate gene for type 1 diabetes (T1D) in Caucasians. We aimed to investigate if rs229541 in C1QTNF6 conferred susceptibility to T1D in Chinese, independent of DR-DQ genotypes and if this gene polymorphism affected the clinical profiles of T1D.
Methods: In this case–control study, genotypes of C1QTNF6 rs229541 were obtained from 1278 patients with T1D and 1282 nondiabetic controls using MassARRAY.
Results: Genotypic (P = 0.0210) and allelic (P = 0.0084) frequencies were significantly different between the T1D group and the control group. When the model was adjusted for DR-DQ genotypes, G allele carriers were observed less often in the T1D group (P = 0.0423, OR 0.82, 95% CI 0.68-0.99) than in the control group, and the G allele was associated with reduced T1D risk(P = 0.0167, OR 0.83, 95% CI 0.71-0.97). T1D patients who were homozygous for the G allele showed a higher positive rate of ZnT8A than carriers of the A allele (P = 0.0171, OR 1.88, 95% CI 1.12-3.16). By detection of fasting C-peptide, G allele carriers exhibited a lower frequency of beta-cell failure compared to those with A/A genotype (P = 0.0058, OR 0.70, 95% CI 0.54-0.90). C1QTNF6 was not found to be correlated with GADA, IA-2A or age at T1D diagnosis.
Discussion: The polymorphism in C1QTNF6 was independently associated with T1D risk in Chinese and broadly modified clinical features of the disease. This loci might be utilized to construct genetic risk model in combination with the well-known DR-DQ region for future screening of genetically T1D prone individuals among Chinese.
Type 1 diabetes (T1D) is generally considered to be an endocrine disorder initiated by an autoimmune attack targeting pancreatic beta-cells, and susceptibility to T1D can be attributed to both genetic and environmental elements (1). In addition to the predominant genetic determinants of T1D risk located within the HLA region (1), over 70 non-HLA loci that confer modest susceptibility to T1D individually have been identified in several genome-wide association studies and meta-analyses over the past 14 years (2–4). Furthermore, these findings have already been used into clinical practice: a genetic risk score constructed by a cluster of non-HLA SNPs in combination with DR-DQ information has shown efficacy in discriminating between diabetes subtypes and new-born screening for T1D-susceptible individuals (5).
However, the abovementioned associations with T1D are mostly reported in Caucasians and cannot be directly applied to other ethnic groups. For instance, the mutant T allele of the well-established PTPN22 has rarely been observed in Asian populations (6), whereas the DR9 haplotype is popular in East Asian patients and rare among Caucasians (7). Hence, the notable heterogeneity of T1D across diverse populations indicates the need to delineate the genetics of this disease in non-Caucasian populations, as association studies of gene polymorphisms are rare in these groups. For Chinese patients, only a limited number of genes have been identified as candidates for T1D, including PTPN22 (6), CTLA4 (8), ERBB3 (9) and STAT4 (10). Thus, the vast majority of non-HLA genes that indicate T1D risk among Caucasians remain unverified in Chinese Han individuals.
Gene polymorphism rs229541, an intron of complement 1q tumour necrosis factor-related protein 6 (C1QTNF6, also known as CTRP6), was identified as a T1D-related locus in a prior genome-wide association study among Caucasians (2). The protein encoded by C1QTNF6 has proven to be a pattern recognition molecule that binds to a wide variety of endogenous or microorganic ligands, followed by recruitment of collectin-11. It subsequently initiates the complement cascade and decomposition of C4 and is thus involved in the immune response (11). Overlapping risk variants within C1QTNF6 have been found among various autoimmune disorders, evidenced by rs229541 in rheumatoid arthritis (12), rs229527 (in tight linkage disequilibrium with rs229541) in generalized vitiligo (13) and rs229527 in Graves’ disease (14), indicating a common molecular pathway underlying these diseases. To the best of our knowledge, the role of C1QTNF6 polymorphisms in the etiopathogenesis of T1D among non-Caucasian populations has not been validated thus far.
Therefore, the current research was carried out to determine 1. whether C1QTNF6 rs229541 conferred susceptibility to T1D of Chinese origin; 2. whether the effect stemmed from the polymorphism itself or was merely a reflection of the uneven distribution of DR-DQ genotypes among participants; and 3. whether this variant modulated clinical profiles of T1D, including autoantibody status, age at diagnosis and beta-cell function.
A total of 1280 T1D patients were identified in the Department of Metabolism and Endocrinology of the Second Xiangya Hospital. The inclusion criteria were as follows: 1. diagnosis of diabetes based on the criteria put forth by the WHO in 1999; 2. acute disease onset and dependency on insulin administration after diagnosis; and 3. positivity of at least one of the three islet autoantibodies, i.e., glutamic acid decarboxylase antibody (GADA), protein tyrosine phosphatase antibody (IA-2A), and zinc transporter 8 antibody (ZnT8A). A total of 1293 control subjects with normal glucose tolerance but without a family history of diabetes were recruited from the Department of Physical Examination in the Second Xiangya Hospital. All the participants declared themselves to be of Chinese Han ethnicity. This research was approved by the Ethics Committee of the Second Xiangya Hospital and was conducted in line with the Declaration of Helsinki guidelines. All the participants or guardians of children with T1D gave their written informed consent and were completely aware of the study procedures.
Both fasting and postprandial C-peptide levels were detected with a chemiluminescence approach from the Adiva Centaur System kit. Approximately 70% of T1D subjects were tested within 1 year of disease onset, while the remaining 30% were tested at least 1 year after disease onset. Radioligand assays were performed in duplicate to detect autoantibodies against islets, including GADA, IA-2A and ZnT8A. All T1D patients underwent these assays at diagnosis, and positivity was confirmed by repetition to ensure that case subjects negative for all autoantibodies and those with equivocal results were not enrolled. MassARRAY (Sequenom, the US) was applied to genotype C1QTNF6 rs229541 followed by quality control, in which 2 samples from T1D patients and 11 samples from control subjects failed and were excluded from statistical analysis, to ensure the accuracy and reliability of the data analysis. The overall detection rate for this SNP was 99.49%. The methods of HLA genotyping were described in a previous publication by our group (15), and DRB1-DQA1-DQB1 haplotypes were constructed using PHASE version 2.0 (16). HLA haplotypes other than the susceptible DR3, DR4 and DR9 were integrated and defined as DRX.
Data were all computed in SPSS version 22.0 (IBM, USA). Continuous variables with normal distributions were expressed as the mean ± standard deviation and compared via Student’s t test, whereas nonnormally distributed variables were expressed as the median and interquartile range and compared via the Mann–Whitney U test. Categorical variables were expressed as n (%) and compared using the chi-squared test. The chi-squared test was also performed to directly compare genotypic and allelic frequencies with the Hardy-Weinberg equilibrium. The following analyses were all conducted using stepwise logistic regression, which included DRB1-DQA1-DQB1 genotypes (DRX/DRX as the reference) as covariates to adjust for HLA information. Under the assumption of a dominant model, G/A and G/G genotypes were encoded as 1 and A/A genotype as 0. In the recessive model, G/A and A/A genotypes were set as 0 and G/G as 1. For the additive model, which calculated the effect for each copy of the minor allele G, the A/A genotype was defined as 0, G/A as 1 and G/G as 2. When investigating the clinical relevance of gene polymorphism, positivity of islet autoantibodies, early disease onset and worse beta-cell function were encoded as 1, and negativity of islet autoantibodies, late disease onset and better beta-cell function were encoded as 0. P <0.05 was considered statistically significant.
Demographic and clinical characteristics for 1278 T1D subjects and 1282 nondiabetic controls included in the further analysis are summarized in Table 1. Briefly, there were no significant differences in sex between the two groups (P = 0.9196), and T1D subjects were younger (P < 0.0001). As expected, T1D patients were remarkably leaner (P < 0.0001) and had higher levels of fasting plasma glucose (P < 0.0001) and 2-hour postprandial plasma glucose (P < 0.0001) than controls. The median age at onset among the T1D group was 21.0 (12.0-34.0) years, and 86.5% of the patients manifested diabetic ketoacidosis when the disease occurred. The patients showed an average HbA1c level of 10.5 ± 3.7%, a median fasting C-peptide level of 82.6 (29.5-167.0) pmol/L and a median postprandial C-peptide level of 163.6 (54.9-340.5) pmol/L. The disease duration was documented when C-peptide levels were determined. GADA, IA-2A and ZnT8A were positive in 90.2%, 49.5% and 34.5% of the T1D patients, respectively.
Genotype information of both T1D subjects (P = 0.1176) and control subjects (P = 0.8129) did not show departure from the Hardy-Weinberg equilibrium. The genotypic and allelic frequencies of C1QTNF6 rs229541 as well as the analyses on the dominant model, recessive model and additive model are all shown in Table 2. Frequencies of A/A, G/A and G/G genotypes differed between the T1D group and the control group (P = 0.0210). In the dominant model, which was adjusted for DR-DQ genotypes, the T1D group exhibited a lower proportion of G allele carriers (P = 0.0423, OR 0.82, 95% CI 0.68-0.99). However, this protective effect was not observed for the G/G genotype in the recessive model, which was also adjusted for DR-DQ genotypes (P = 0.0587). The minor allele G was less often seen in cases than in controls (26.4% vs. 29.7%, P = 0.0084). In the additive model, which calculated the per copy effect of an allele, the protection by minor allele G (P = 0.0093) remained statistically significant after adjusting for DR-DQ genotypes (P = 0.0167, OR 0.83, 95% CI 0.71-0.97).
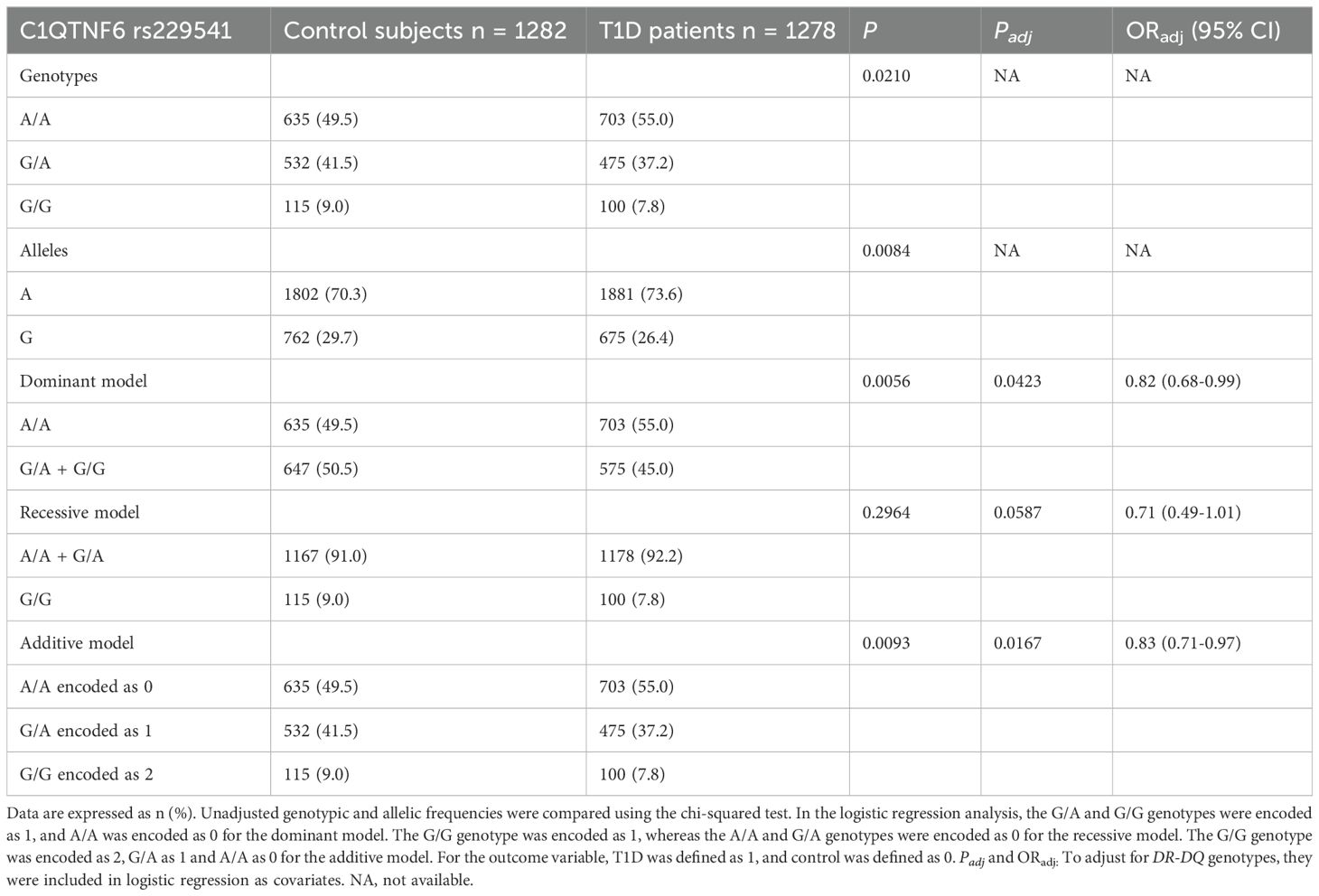
Table 2. Frequency distributions of C1QTNF6 rs229541 genotypes and alleles between Chinese patients with T1D and healthy controls.
We next analysed the autoantibody status (Table 3), age at onset (Table 4) and C-peptide levels (Table 5) of T1D patients with data broken down by genotypes of C1QTNF6 rs229541 and adjustment for DR-DQ genotypes. No correlation was observed for GADA or IA-2A in any of the 3 genetic models tested (all adjusted P values > 0.05). However, in the recessive model, patients with the G/G genotype showed a significantly higher positive rate of ZnT8A than carriers of the A allele. (47.0% vs. 33.5%, P = 0.0171, OR 1.88, 95% CI 1.12-3.16). There was no significant association between ZnT8A and C1QTNF6 rs229541 in the dominant model or in the additive model (both P values > 0.05).
When T1D participants were separated into those with childhood onset (age at onset < 15 years) and those without childhood onset (age at onset ≥ 15 years), genotypic information of C1QTNF6 rs229541 did not alter the risk for early disease onset under all genetic models tested (all P values > 0.05), as shown in Table 4. Furthermore, when the cut-off point for age at diagnosis was set to 18 years – which is considered the beginning of adulthood among Chinese individuals – there was still no significant association between the C1QTNF6 polymorphism and susceptibility to early disease manifestation under in dominant model, recessive model or additive model (all P values > 0.05).
A fasting C-peptide level < 50 pmol/L or a postprandial C-peptide level < 100 pmol/L was defined as worse beta-cell function, and those with fasting C-peptide ≥ 50 pmol/L or postprandial C-peptide ≥ 100 pmol/L were considered to have better beta-cell function. In the additive model for the analysis of fasting C-peptide, a protective effect was revealed for the G allele (P = 0.0296, OR 0.80, 95% CI 0.65-0.98), as shown in Table 5. Moreover, in the dominant model, G allele carriers exhibited a dramatically lower frequency of worse beta-cell function than those with the A/A genotype (31.9% vs. 38.7%, P = 0.0058, OR 0.70, 95% CI 0.54-0.90). Nonetheless, such a protective effect on beta-cell function was not observed for C1QTNF6 rs229541 in the analysis of postprandial C-peptide, irrespective of the genetic models assumed (all P values > 0.05).
To the best of our knowledge, the first genome-wide association study conducted among Chinese individuals identified BTN3A1, GATA3, and SUOX as susceptibility loci for T1D, in addition to HLA region, PTPN22, CTLA4, ERBB3 and STAT4 (17). The handful of genomic locations confirmed thus far among Chinese patients indicate that the genetic architecture of T1D has yet to be fully elucidated, and several obstacles have hampered the attempt to identify additional determinants of T1D risk. First, given the greatest contribution to T1D by HLA signals, we cannot conclude that certain non-HLA alleles are independently linked to the disease unless we adjust for DR-DQ markers. The second problem to be resolved when dealing with genes showing modest effect size is that a sufficient number of samples is necessary to maximize statistical power. Third, considering that T1D can appear at any age and that most new-onset T1D cases in China occur among adults (18), studies that examine only childhood-onset T1D patients can hardly provide a comprehensive interpretation of the genetic background for Chinese individuals. Fourth, as China is a melting pot of various races and because some ethnic minorities in China are genetically closer to Caucasians than to Han individuals (19), the results can be misleading if heterogenous populations are enrolled. The current study enrolled a considerably large sample of genetically homogeneous T1D subjects with Han ethnicity, thus covering all age groups. Additionally, the models examined herein were adjusted for DR-DQ genotypes to determine the true effect of C1QTNF6 rs229541 in Chinese individuals.
As shown in Table 2, the genotypes and alleles of rs229541 were heterogeneously distributed between patients and control subjects, with a significant decrease in G allele frequency observed among patients. The additive model has become the most important model for calculating the per copy effect of each allele. In this model adjusted for DR-DQ genotypes, the A allele conferred susceptibility to T1D with an OR of 1.20 (OR 0.83 for G allele), in accordance with a previous genome-wide association study (2) in Caucasians (OR 1.15-1.16 for A allele). These findings suggest that C1QTNF6 rs229541 plays a minor but indispensable role in both ethnic groups. A higher percentage of G allele carriers (dominant model) was observed in controls than in T1D patients, indicating that one copy of minor allele G was sufficient to reduce T1D risk. However, such protection was not confirmed in carriers of the G/G genotype compared to those with the A allele (recessive model), likely due to the risk of the A/A genotype counterbalanced by protection of the G/A genotype when combined and the relatively low counts and frequencies of minor allele G homozygotes in this population.
Full-blown T1D is usually preceded by seroconversion to the presence of autoantibodies against pancreatic islets (1). Both HLA (20) and non-HLA genes (21) exert their function on altering the specificity of the antibody initiating autoimmune process, i.e., GADA or IAA as the first autoantibody. Three islet autoantibodies were routinely detected in our department prior to the diagnosis of classic T1D. Although the data presented here did not support the correlation of C1QTNF6 rs229541 with GADA or IA-2A, it was intriguing that patients with the susceptible A allele exhibited a lower positive rate of ZnT8A than those homozygous for the G allele. Such a seemingly paradoxical phenomenon could also be seen in the negative association of HLA-A*24 (a well-established HLA Class I allele susceptible to T1D) with IA-2A (22) and ZnT8A (23). E.M. Balke described this inverse relationship as “a delayed antigen spreading of humoral autoimmunity” in carriers of certain risk alleles (24). We next categorized the age at diagnosis into either early or late onset to determine whether the allelic variation in C1QTNF6 could partially explain the prevalence of late T1D onset in Chinese patients. Regardless of whether the cut-off age was 15 years or 18 years and regardless of the genetic models used, genotypic frequencies of rs229541 did not significantly differ between the two groups with distinct onset ages. Both FCP and PCP were measured in this research to evaluate the beta-cell function of patients with T1D, and a lower frequency of FCP < 50 pmol/L was observed for patients with the protective G allele in comparison with those carrying the A/A genotype. Accumulating evidence gathered from multiple populations supports the effect of polymorphisms in non-HLA genes, such as PTPN22, FCRL3 (25) and vitamin D receptor gene (26), on modulating beta-cell function in T1D subjects. Nonetheless, one major limitation of the current research was that only cross-sectional data of islet autoantibodies and C-peptide were available here. Therefore, we were not able to investigate whether the C1QTNF6 polymorphism alone or in combination with other genetic variants could predict the appearance of autoantibodies, the progression to clinical T1D or the fast decline in beta-cell function in Chinese patients.
Fortunately, studies of Caucasian populations managed to build prospective cohorts that carefully followed up genetically susceptible children from birth to the initiation of islet autoimmunity and ultimately to T1D diagnosis; both encouraging and negative results have been reported. For instance, C1QTNF6 rs229541 has been shown to predict the risk of islet autoimmunity instead of progression to T1D in DAISY children (27), whereas the methylation of this gene was not correlated with T1D development in another study conducted by the same group (28). The MIDIA study (29) detected enteroviral RNA in stool samples from Norwegian children, genotyped both T1D susceptibility SNPs and those within innate immune genes and found that the protective genotype of C1QTNF6 rs229541 was related to a lower enterovirus frequency, thus emphasizing the interplay between genetics and environmental factors when scrutinizing the etiology of T1D.
As C1QTNF6 plays an active role in a wide variety of endocrine and inflammatory pathways, C1QTNF6-related pathophysiological processes or diseases include but are by no means limited to T1D. Polycystic ovary syndrome (PCOS) is a common reproductive endocrine disorder, the pathogenesis of which features low-grade chronic inflammation. S. Yan and his colleagues found that the expression of C1QTNF6 was elevated in serum from both PCOS patients and PCOS mice compared to the corresponding controls. They further demonstrated that the levels of proinflammatory factors, including CRP, IL6 and TNFα, were increased in C1QTNF6-overexpressing PCOS mice and that this protein regulated the inflammatory response through the AKT/NF-κB signaling pathway (30). J. Wang revealed that miR-29b-3p modulated particulate matter-induced inflammatory responses via the C1QTNF6/AMPK pathway (31), which provided new insights into the molecular mechanisms behind chronic inflammation in respiratory diseases. A study by the Institute of Medical Science from the University of Tokyo showed a higher expression of C1QTNF6 in mouse models with rheumatoid arthritis. C1QTNF6 (-/-) mice were extremely prone to induced arthritis because of augmented complement activation. C1QTNF6 specifically inhibited the alternative pathway in the complement system by competing against factor B for C3 (H2O) binding. Intra-articular injection of arthritis-induced mice with recombinant human C1QTNF6 cured the disease (32), which suggested that this protein is a novel target for the treatment of inflammatory diseases. Researchers from Israel proposed C1QTNF6 as a link between immunity and metabolism, as this protein responds to a rapid change in the calorie intake of mice, triggering a “physiological inflammatory response” that suppresses the expansion of adipose tissue (33). For the development of malignancies, upregulation of C1QTNF6 was observed for stage I lung adenocarcinoma in contrast to the adjacent noncancer tissues. Knockdown of C1QTNF6 markedly dampened cell proliferation, migration and invasion, which was negatively regulated by miR-29a-3p (34). A similar role of C1QTNF6 was also recognized in oral squamous cell carcinoma, as C1QTNF6 knockdown led to cell cycle arrest at phase G2/M and enhancement of cancer cell apoptosis (35). Overall, the publications described above indicate that C1QTNF6 is actively engaged in a broad spectrum of pathophysiological events, including immunity directed towards either self-tissues or external stimuli, lipid metabolism and homeostasis as well as oncogenesis across multiple disciplines.
Additionally, the studies mentioned further suggest that the polymorphism rs229541 in C1QTNF6 is associated with immune responses in T1D. C1QTNF6’s involvement in the complement system may also affect the clearance of apoptotic cells and the resolution of inflammation. By regulating complement activation, C1QTNF6 could prevent excessive tissue damage and autoimmunity, potentially limiting the progression of β-cell destruction in T1D patients. The G allele of rs229541 may alter immune responses by affecting the function of antigen-presenting cells or regulatory T cells, both of which are essential for maintaining immune tolerance (24, 30). Our study found that the G allele of rs229541 in C1QTNF6 is positively correlated with ZnT8A positivity, which may be related to the “delayed antigen spreading of humoral autoimmunity” mentioned earlier. C1QTNF6 may impact the development of islet autoantibodies by regulating B cell activation and plasma cell differentiation (36). The immune-modulatory role of C1QTNF6 has been recognized in other autoimmune diseases, such as rheumatoid arthritis, where its expression and function in immune cells contribute to either promoting or suppressing autoimmunity, depending on the genetic context (32). Moreover, C1QTNF6 may regulate the inflammatory process through key immune signaling pathways such as NF-κB and AKT, playing a crucial role in the modulation of cytokine levels and immune cell activation (37). By influencing these pathways, C1QTNF6 could help shape the immune microenvironment within the pancreatic islets, potentially altering the initiation and progression of autoimmune responses. These findings suggest that C1QTNF6 could have a broader impact on the regulation of immune responses in T1D, making it an important candidate for further investigation in the context of genetic susceptibility and immune tolerance.
Taken together, the data presented herein reveal the role of the C1QTNF6 polymorphism rs229541 in the pathogenesis of T1D of Chinese origin, and it also modulates biochemical profiles, including islet autoantibodies and C-peptide levels, independent of DR-DQ genotypes. However, it is important to interpret these results with caution, as several limitations should be considered. First, the sample population was limited to Han Chinese individuals, which may affect the generalizability of the results to other ethnic groups. Further studies involving diverse populations and larger sample sizes are needed to confirm these findings. Second, environmental factors such as viral infections, diet, and other exposures were not accounted for, despite their known influence on T1D development. Future studies should incorporate these factors to better understand the genetic-environmental interactions in T1D. Lastly, as mentioned earlier, the cross-sectional design limits our ability to infer causality. Longitudinal studies are necessary to explore the temporal relationship between genetic variations and disease onset. Future research should examine whether this gene polymorphism aids in predicting the clinical course of T1D from genetic susceptibility and seroconversion to clinical manifestation in non-Caucasian populations.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by the Ethics Committee of the Second Xiangya Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
JL: Data curation, Formal Analysis, Writing – original draft, Writing – review & editing. YX: Formal Analysis, Methodology, Writing – review & editing. ZX: Methodology, Resources, Writing – review & editing. XL: Methodology, Resources, Writing – review & editing. GH: Software, Visualization, Writing – review & editing. JH: Project administration, Supervision, Writing – review & editing. ZZ: Funding acquisition, Project administration, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (81820108007, 82200933), the Fundamental Research Funds for the Central Universities of Central South University (2021zzts0381), the Natural Science Foundation of Hunan Province (2021JC0003), and the Science and Technology Innovation Program of Hunan Province (2024RC3054).
The authors thank all the doctors, nurses, case and control individuals for their devotion to this investigation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1551552/full#supplementary-material
Supplementary Figure 1 | Distribution of C-peptide levels in Chinese T1D patients stratified by C1QTNF6 rs229541 genotypes. This boxplot presents the distribution of C-peptide levels (pmol/L) among Chinese T1D patients, comparing individuals carrying the G/A and G/G genotypes (combined) with those harboring the A/A genotype. Group differences were evaluated using the Mann-Whitney U test. Statistical significance is denoted as *P < 0.05, **P < 0.01, and ***P < 0.001.
1. Ilonen J, Lempainen J, Veijola R. The heterogeneous pathogenesis of type 1 diabetes mellitus. Nat Rev Endocrinol. (2019) 15:635–50. doi: 10.1038/s41574-019-0254-y
2. Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. (2008) 40:1399–401. doi: 10.1038/ng.249
3. Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. (2009) 41:703–7. doi: 10.1038/ng.381
4. Onengut-Gumuscu S, Chen WM, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. (2015) 47:381–6. doi: 10.1038/ng.3245
5. Sharp SA, Rich SS, Wood AR, Jones SE, Beaumont RN, Harrison JW, et al. Development and standardization of an improved type 1 diabetes genetic risk score for use in newborn screening and incident diagnosis. Diabetes Care. (2019) 42:200–7. doi: 10.2337/dc18-1785
6. Pei Z, Chen X, Sun C, Du H, Wei H, Song W, et al. A novel single nucleotide polymorphism in the protein tyrosine phosphatase N22 gene (PTPN22) is associated with Type 1 diabetes in a Chinese population. Diabetes Med. (2014) 31:219–26. doi: 10.1111/dme.2014.31.issue-2
7. Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. (2008) 57:1084–92. doi: 10.2337/db07-1331
8. Jin P, Xiang B, Huang G, Zhou Z. The association of cytotoxic T-lymphocyte antigen-4 + 49A/G and CT60 polymorphisms with type 1 diabetes and latent autoimmune diabetes in Chinese adults. J endocrinological Invest. (2015) 38:149–54. doi: 10.1007/s40618-014-0162-x
9. Sun C, Wei H, Chen X, Zhao Z, Du H, Song W, et al. ERBB3-rs2292239 as primary type 1 diabetes association locus among non-HLA genes in Chinese. Meta gene. (2016) 9:120–3. doi: 10.1016/j.mgene.2016.05.003
10. Bi C, Li B, Cheng Z, Hu Y, Fang Z, Zhai A. Association study of STAT4 polymorphisms and type 1 diabetes in Northeastern Chinese Han population. Tissue Antigens. (2013) 81:137–40. doi: 10.1111/tan.2013.81.issue-3
11. Kirketerp-Møller N, Bayarri-Olmos R, Krogfelt KA, Garred P. C1q/TNF-related protein 6 is a pattern recognition molecule that recruits collectin-11 from the complement system to ligands. J Immunol. (2020) 204:1598–606. doi: 10.4049/jimmunol.1901316
12. Eyre S, Hinks A, Bowes J, Flynn E, Martin P, Wilson AG, et al. Overlapping genetic susceptibility variants between three autoimmune disorders: rheumatoid arthritis, type 1 diabetes and coeliac disease. Arthritis Res Ther. (2010) 12:R175. doi: 10.1186/ar3139
13. Jin Y, Birlea SA, Fain PR, Gowan K, Riccardi SL, Holland PJ, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med. (2010) 362:1686–97. doi: 10.1056/NEJMoa0908547
14. Zhao SX, Xue LQ, Liu W, Gu ZH, Pan CM, Yang SY, et al. Robust evidence for five new Graves’ disease risk loci from a staged genome-wide association analysis. Hum Mol Genet. (2013) 22:3347–62. doi: 10.1093/hmg/ddt183
15. Luo S, Lin J, Xie Z, Xiang Y, Zheng P, Huang G, et al. HLA genetic discrepancy between latent autoimmune diabetes in adults and type 1 diabetes: LADA China study no. 6. J Clin Endocrinol Metab. (2016) 101:1693–700. doi: 10.1210/jc.2015-3771
16. Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. (2003) 73:1162–9. doi: 10.1086/379378
17. Zhu M, Xu K, Chen Y, Gu Y, Zhang M, Luo F, et al. Identification of novel T1D risk loci and their association with age and islet function at diagnosis in autoantibody-positive T1D individuals: based on a two-stage genome-wide association study. Diabetes Care. (2019) 42:1414–21. doi: 10.2337/dc18-2023
18. Weng J, Zhou Z, Guo L, Zhu D, Ji L, Luo X, et al. Incidence of type 1 diabetes in China, 2010-13: population based study. BMJ. (2018) 360:j5295. doi: 10.1136/bmj.j5295
19. Liu S, Huang S, Chen F, Zhao L, Yuan Y, Francis SS, et al. Genomic analyses from non-invasive prenatal testing reveal genetic associations, patterns of viral infections, and Chinese population history. Cell. (2018) 175:347–59.e14. doi: 10.1016/j.cell.2018.08.016
20. Ilonen J, Hammais A, Laine AP, Lempainen J, Vaarala O, Veijola R, et al. Patterns of β-cell autoantibody appearance and genetic associations during the first years of life. Diabetes. (2013) 62:3636–40. doi: 10.2337/db13-0300
21. Lempainen J, Laine AP, Hammais A, Toppari J, Simell O, Veijola R, et al. Non-HLA gene effects on the disease process of type 1 diabetes: From HLA susceptibility to overt disease. J Autoimmun. (2015) 61:45–53. doi: 10.1016/j.jaut.2015.05.005
22. Howson JM, Stevens H, Smyth DJ, Walker NM, Chandler KA, Bingley PJ, et al. Evidence that HLA class I and II associations with type 1 diabetes, autoantibodies to GAD and autoantibodies to IA-2, are distinct. Diabetes. (2011) 60:2635–44. doi: 10.2337/db11-0131
23. Howson JM, Krause S, Stevens H, Smyth DJ, Wenzlau JM, Bonifacio E, et al. Genetic association of zinc transporter 8 (ZnT8) autoantibodies in type 1 diabetes cases. Diabetologia. (2012) 55:1978–84. doi: 10.1007/s00125-012-2540-2
24. Balke EM, Balti EV, van der Auwera B, Weets I, Costa O, Demeester S, et al. Accelerated progression to type 1 diabetes in the presence of HLA-A*24 and -B*18 is restricted to multiple islet autoantibody-positive individuals with distinct HLA-DQ and autoantibody risk profiles. Diabetes Care. (2018) 41:1076–83. doi: 10.2337/dc17-2462
25. Pawłowicz M, Filipów R, Krzykowski G, Stanisławska-SaChadyn A, Morzuch L, Kulczycka J, et al. Coincidence of PTPN22 c.1858CC and FCRL3 -169CC genotypes as a biomarker of preserved residual β-cell function in children with type 1 diabetes. Pediatr Diabetes. (2017) 18:696–705. doi: 10.1111/pedi.12429
26. Habibian N, Amoli MM, Abbasi F, Rabbani A, Alipour A, Sayarifard F, et al. Role of vitamin D and vitamin D receptor gene polymorphisms on residual beta cell function in children with type 1 diabetes mellitus. Pharmacol reports: PR. (2019) 71:282–8. doi: 10.1016/j.pharep.2018.12.012
27. Frederiksen BN, Steck AK, Kroehl M, Lamb MM, Wong R, Rewers M, et al. Evidence of stage- and age-related heterogeneity of non-HLA SNPs and risk of islet autoimmunity and type 1 diabetes: the diabetes autoimmunity study in the young. Clin Dev Immunol. (2013) 2013:417657. doi: 10.1155/2013/417657
28. Carry PM, Vanderlinden LA, Johnson RK, Dong F, Steck AK, Frohnert BI, et al. DNA methylation near the INS gene is associated with INS genetic variation (rs689) and type 1 diabetes in the Diabetes Autoimmunity Study in the Young. Pediatr Diabetes. (2020) 21:597–605. doi: 10.1111/pedi.12995
29. Witsø E, Cinek O, Tapia G, Brorsson CA, Stene LC, Gjessing HK, et al. Genetic determinants of enterovirus infections: polymorphisms in type 1 diabetes and innate immune genes in the MIDIA study. Viral Immunol. (2015) 28:556–63. doi: 10.1089/vim.2015.0067
30. Yan S, Ding J, Zhang Y, Wang J, Zhang S, Yin T, et al. C1QTNF6 participates in the pathogenesis of PCOS by affecting the inflammatory response of granulosa cells‡. Biol reproduction. (2021) 105:427–38. doi: 10.1093/biolre/ioab094
31. Wang J, Zhu M, Ye L, Chen C, She J, Song Y. MiR-29b-3p promotes particulate matter-induced inflammatory responses by regulating the C1QTNF6/AMPK pathway. Aging. (2020) 12:1141–58. doi: 10.18632/aging.102672
32. Murayama MA, Kakuta S, Inoue A, Umeda N, Yonezawa T, Maruhashi T, et al. CTRP6 is an endogenous complement regulator that can effectively treat induced arthritis. Nat Commun. (2015) 6:8483. doi: 10.1038/ncomms9483
33. Lahav R, Haim Y, Bhandarkar NS, Levin L, Chalifa-Caspi V, Sarver D, et al. CTRP6 rapidly responds to acute nutritional changes, regulating adipose tissue expansion and inflammation in mice. Am J Physiol Endocrinol Metab. (2021) 321:E702–e13. doi: 10.1152/ajpendo.00299.2021
34. Lin G, Lin L, Lin H, Xu Y, Chen W, Liu Y, et al. C1QTNF6 regulated by miR-29a-3p promotes proliferation and migration in stage I lung adenocarcinoma. BMC pulmonary Med. (2022) 22:285. doi: 10.1186/s12890-022-02055-2
35. Song X, Li L, Shi L, Liu X, Qu X, Wei F, et al. C1QTNF6 promotes oral squamous cell carcinoma by enhancing proliferation and inhibiting apoptosis. Cancer Cell Int. (2021) 21:666. doi: 10.1186/s12935-021-02377-x
36. Qu HX, Cui L, Meng XY, Wang ZJ, Cui YX, Yu YP, et al. C1QTNF6 is overexpressed in gastric carcinoma and contributes to the proliferation and migration of gastric carcinoma cells. Int J Mol Med. (2019) 43:621–9. doi: 10.3892/ijmm.2018.3978
Keywords: type 1 diabetes, Genetics, C1QTNF6, Autoimmunity, Beta-cell function
Citation: Liu J, Xia Y, Xie Z, Li X, Huang G, Hu J and Zhou Z (2025) Association of C1QTNF6 gene polymorphism with risk and clinical features of type 1 diabetes in Chinese: implications for ZnT8A and beta-cell function. Front. Immunol. 16:1551552. doi: 10.3389/fimmu.2025.1551552
Received: 07 January 2025; Accepted: 21 March 2025;
Published: 09 April 2025.
Edited by:
Beatrice Dufrusine, University of Teramo, ItalyReviewed by:
Kuanfeng Xu, Nanjing Medical University, ChinaCopyright © 2025 Liu, Xia, Xie, Li, Huang, Hu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiguang Zhou, emhvdXpoaWd1YW5nQGNzdS5lZHUuY24=; Jingyi Hu, aHVqaW5neWkwMDY2QGNzdS5lZHUuY24=
†These authors have contributed equally to this work and share last authorship
‡ORCID: Zhiguang Zhou, orcid.org/0000-0002-0374-1838
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.