
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 16 April 2025
Sec. Cancer Immunity and Immunotherapy
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1548292
This article is part of the Research TopicCell Models and Preclinical Validation of Immune-mediating Biological TherapiesView all articles
Antibody-dependent cellular cytotoxicity (ADCC) is a critical mechanism by which therapeutic antibodies leverage the immune system to target and eliminate cancer cells. The key agents of ADCC are natural killer (NK) cells, specifically targeting antibody-covered cancer cells through the CD16 receptor. While other immune cells and Fc receptors can contribute and enhance ADCC, NK cells and the CD16 receptor are crucial for the efficacy of cancer therapies such as trastuzumab, cetuximab and rituximab. Co-culture assays are essential for understanding the mechanisms of these therapies, overcoming resistance and optimizing novel therapeutic antibodies. This review highlights the importance of measuring ADCC to assess the efficacy of therapeutic antibodies. Here we also present the various in vitro models and assay methodologies available for studying ADCC, comparing the strengths and limitations of approaches like using PBMCs to better reflect real-life conditions or NK cell lines for standardization. It also covers different readouts for ADCC, either focusing on effector cells activation, including reporter and degranulation assays or in the target cell killing, including different molecule release assays, flow cytometry and immunofluorescence techniques. Selecting the best model for studying ADCC is crucial for the translational significance of therapeutic antibody research.
Therapeutic antibodies have revolutionized the field of cancer therapy. These agents can engage the immune system to attack the cancer cells through antibody-dependent cellular cytotoxicity (ADCC). These antibodies are designed to recognize cancer-specific epitopes or targets overexpressed by the cancer cells. Many of these antibodies require Fc-FcγR interactions for their antitumor activity (1). Upon binding to cancer cells via the fab region, the Fc domain of these antibodies engages FcγR on immune cells. This interaction triggers the activation of signaling pathways in immune cells that result in release of cytotoxic vesicles containing perforin and granzymes to induce cell death in the targeted cancer cells. Using this mechanism, we can direct the immune system specifically towards the cancer.
One such therapy is trastuzumab, a monoclonal antibody targeting the extracellular domain of HER2. Trastuzumab is the first-line treatment for HER2-postive (HER2+) breast cancer and has been leading the ADCC rock band for over 20 years (2–4). With trastuzumab leading the way, over 40 other antibody-based therapies (5) have been approved in the clinic (6, 7). Many of these antibody-based targeting therapies such as cetuximab exert their function by inhibiting specific signaling pathways (8) and some of them have been further improved by attaching a cytotoxic payload that is internalized into the cancer cell upon antibody binding. It is worth noting that these antibody-drug conjugates such as trastuzumab-emtansine (T-DM1) and trastuzumab-deruxtecan (T-DXd) retain the ability of trastuzumab of triggering ADCC, combining this immune-mediated mechanism with the delivery of potent cytotoxic agents for enhanced antitumor efficacy (9, 10).
Not all therapeutic antibodies target tumor cells. Immune checkpoint inhibitors, for instance, modulate the immune system to enhance anti-tumor responses. The role of ADCC in these therapies remains unclear. In some cases, antibodies are designed to deliberately eliminate ADCC to prevent adverse effects (11). However, in others such as anti-CTLA-4 antibodies, which block T-cell inhibition during antigen presentation, ADCC may play a beneficial role. While eliminating T cells is not the intended outcome, studies suggest that ADCC-mediated depletion of intratumoral regulatory T cells can enhance tumor control (12–14). This principle may also be applied to PD-1/PD-L1 inhibitors. In particular, PD-L1 blocking antibodies could improve efficacy by promoting ADCC against tumor cells (15, 16). Further research is needed to clarify the contribution of ADCC to the mechanisms of these antibodies.
Though the efficacy of these therapies depends largely on the interaction with immune cells, many in vitro studies studying trastuzumab efficacy and resistance rely on assays measuring proliferation or cell viability, often disregarding the immune component. This can result in discoveries in vitro with less translational potential (17). While proliferation assays are easier, in-depth studies of ADCC using suitable in vitro models provide more relevant information about the efficacy of antibody-based therapeutics revealing which factors influence the success or failure of this rock band (18–28).
Natural killer (NK) cells are the main agent responsible for killing cancer cells through antibody-dependent cellular cytotoxicity. NK cells are generally characterized as CD3 negative CD56 positive cells (29) and comprise 5 to 20% of the peripheral blood mononuclear cells (PBMCs) (30). Two subsets of NK cells have been described which express different cell surface molecules and are functionally distinct. 95% of the NK cells in the blood are CD56dimCD16+. This subset is highly cytotoxic and due to its expression of CD16 it is capable of antibody recognition and is the primary mediator of ADCC. The other CD56brightCD16- subset is mainly present in the lymph nodes and, while capable of cytokine production, only exerts low cytotoxicity (31).
Patient studies have provided evidence for the role of NK cells in therapeutic antibody efficacy. In HER2+ breast cancer patients, infiltration of NK cells is increased in response to neoadjuvant trastuzumab treatment, suggesting their involvement in trastuzumab action (32). In addition, studies isolating NK cells from patients show that those with higher levels of NK cells have a longer progression free survival (PFS) (33), while the patients with no in vitro ADCC results did not respond to treatment (34). This correlation is not only confined to breast cancer, as NK cell activity is also associated with relapse-free survival of colorectal cancer patients treated with cetuximab (35).
In vitro assays have been employed to separately study the contribution of the different immune cell subsets to ADCC. Kute et al., demonstrated in vitro that NK cells are twice more cytotoxic against HER2+ breast cancer cells than monocytes (36). While the cell killing mediated by NK cells is almost exclusively reliant on trastuzumab, the monocyte-mediated killing is predominantly antibody independent. Regarding macrophages, addition of trastuzumab increases cancer cell killing by these immune cells in vitro through antibody-dependent phagocytosis (37). In accordance with Kute et al, less than 50% of the cell killing mediated by the macrophages is trastuzumab dependent. In the case of B and T lymphocytes, they also show low levels of cytotoxicity, though partially mediated by NK contamination (36). Several studies have identified a specific subset of γδ T cells capable of ADCC. Infusions of this subset in mice models resulted in an increase in trastuzumab efficiency (38). Although this T cell subset offers an opportunity for immune therapy, it is unlikely to be a main contributor to trastuzumab efficacy (39).
Further evidence of the significance of NK cells in ADCC is provided by studies depleting specific immune cell subsets from the PBMC population before co-culturing with target cells. While the depletion of monocytes does not significantly affect cytotoxicity of the PBMCs (40), the depletion of NK cells reduces or eliminates antibody-mediated cellular cytotoxicity in vitro (40–42) and in mouse models (43, 44).
ADCC performing immune cells are activated through the signaling of Fc gamma receptors (FcγR). These receptors specifically bind to the Fc domain of the antibodies coating the target cells (45) and are crucial for linking antibody-mediated immune responses to cellular effector functions such as ADCC, phagocytosis and immune regulation. The main Fc receptors that bind IgG antibodies are FcγRI (CD64), FcγRIIa (CD32A), FcγRIIb (CD32B), FcγRIIc (CD32C), FcγRIIIa (CD16A) and FcγRIIIb (CD16B). Primarily they are activating receptors, only FcγRIIb inhibits activation of immune cells to prevent excessive inflammation (46).
FcγRI binds monomeric IgG molecules and is expressed on phagocytotic cells. Out of all Fc receptors the FcγRI has the highest affinity, but its constitutive saturation attenuates its function. The FcγRIIIb receptor is expressed in high amounts by neutrophils. However, this receptor lacks internal signaling motifs and acts as decoy receptor. FcγRIIc can enhance ADCC but is functionally inactive in 80% of humans due to a premature stop codon. The FcγRIIIa and FcγRIIa activating receptors along with the inhibitory FcγRIIb receptor, participate in antibody-dependent cellular phagocytosis (ADCP), antibody-dependent cellular cytotoxicity (ADCC), as well as in the induction of cytokines and chemokines. FcγRIIa and FcγRIIIa are both present in monocytes, macrophages and dendritic cells. In these subsets of immune cells antibody-mediated activation is regulated by the balance between activating and inhibitory FcγRs. In contrast, NK cells predominantly express the activating receptor FcγRIIIa and are not inhibited by FcγRIIb (46).
More details about these receptors such as their structure, domains, genetic variability, affinity and functions have been previously extensively reviewed (47–49).
The strength of ADCC is influenced by several factors, including IgG subclass, glycosylation and antibody polymorphisms (45), but also by polymorphisms in the Fc receptors recognizing the antibody. Polymorphisms in these Fc receptors generate variants that bind to the Fc region of antibodies with different affinities thereby mediating sensitivity to therapy. HER2+ breast cancer patients with FcγRIIIa-158 V/V genotype exhibited a higher objective response rate (ORR) and a longer PFS in response to trastuzumab than patients with FcγRIIIa-158 F/V or F/F variants (50). The same relationship between FcγRIIIa polymorphisms and response to therapeutic antibodies is observed in non-Hodgkin lymphoma patients treated with rituximab (51, 52) and efficacy of cetuximab in metastatic colorectal cancer patients (53, 54).
Furthermore, PBMCs harboring FcγRIIIa and FcγRIIa high-affinity variants were also significantly associated with higher trastuzumab-mediated ADCC in vitro, which was even higher in PBMCs with the combination of both variants (50). However, in other studies only blocking of FCγRIIIa, but not FCγRIIa, greatly reduced ADCC of PBMCs (42), indicating a predominant role for FCγRIIIa. This receptor is also the only one capable of independently triggering cytotoxicity. Its activation can be enhanced or inhibited by costimulatory or inhibitory receptors, but their signaling is not required (55).
Co-culture assays have been instrumental in identifying mechanisms of resistance and developing strategies to overcome them. Several studies have combined CRISPR screens with high-throughput ADCC assays to uncover novel mechanisms. On the immune side, these studies have identified cytokines and NK cell subsets that promote resistance in melanoma (18). On the tumor side, they revealed that loss and overexpression of certain proteins confers resistance to daratumumab for multiple myeloma (19). Hypothesis-driven studies have further identified the expression of the membrane receptor CD64 as a mediator of resistance to anti-CD123 for AML using in vitro ADCC assays. This discovery led to the development of a novel anti-CD123 antibody with added NKp46-engaging molecules to restore sensitivity. Consistent with the in vitro data, this novel antibody performed well in both mouse models and in primates and is now in the preclinical stage (20). Another example of identification of a resistance mechanism resulting in novel drug development is the role of ICAM-1 downregulation in conferring resistance to trastuzumab. This finding prompted the development of CAR-NK cells to reduce the need for ICAM-1 or other co-stimulation (21). Additionally, another study tested combination of existing antibody drugs and small molecule kinase inhibitors to overcome resistance (22).
In developing novel therapeutic antibodies and drug combinations, in vitro ADCC assays are essential at the preclinical evaluation and characterization stage. For antibody selection, in vitro ADCC assays were used to test multiple antibodies against DDK1 peptide-HLA-A2 complex for efficacy and the specificity, selecting the best one for further testing in mice (23). Comparing novel antibodies, bemarituzumab directed against FGFR2b, demonstrated higher ADCC than existing antibodies and is specific for FGFR2b (24). In another study, in vitro ADCC assays demonstrated that inhibiting endocytosis effectively induces ADCC for the antibodies cetuximab, trastuzumab and avelumab (25). Furthermore, in vitro ADCC studies were used to investigate the mechanism of amivantamab, a bispecific antibody targeting EGFR and MET, which is now approved in several countries for treatment NSCLC patients (26). These assays also tested the effect of AFM24, a bispecific antibody targeting EGFR and CD16, on different immune cells and its effectiveness against various tumor cell subsets to identify which patients can benefit (27). Similarly, the efficacy of zenocutuzumab against different breast cancer subsets was characterized, leading to a phase I/II clinical trial for NRG1-fusion cancers (28).
Natural killer cells activate ADCC and eradicate the cancer cells through the signaling of FcγRIIIa (CD16). Co-culture assays using both cancer cells and immune cells are used to study this ADCC in vitro. While cancer cells are often the primary focus as the target cells, some considerations should be taken into account regarding the choice of effector cells. PBMCs are the most frequently used immune cells for ADCC assays, as they can be isolated from donors and are not modified to adapt to laboratory culture. However, the use of PBMCs presents several challenges. First, donor blood needs to be available and the PBMCs need to be isolated using a lengthy procedure. More importantly, studies show a high batch variability depending on the donor’s health, age and unknown factors resulting in differences in number or cytotoxic potential of the NK subset (36). Part of this can be circumvented by isolating the NK cells or even the CD16-positive NK cells from the blood, but this requires more time and laborious procedures and results in a loss of cells in the process. Even separating donors based on their CD16 variants does not eliminate the high variability (56).
For those without access to donor blood or those who wish to standardize their experiments or do high-throughput cytotoxicity screens, a cell line-based system offers many advantages. A cytotoxically active NK cell line is flexible, infinitely scalable and has a high reproducibility. Many studies have used NK cell lines for measuring ADCC (56–63). The difficulty of working with cell lines lies in that few NK cell lines have been established, and most lines lose expression of the CD16 receptor in culture, impairing their ability to mediate antibody-dependent cytotoxicity (64). To bypass this limitation, stable overexpression of CD16 restores antibody recognition and antibody-dependent cellular cytotoxicity of the NK cell lines. These models have been successfully established in NK-92 (59, 60), KHYG-1 (58, 61, 62) and NKL (56) NK cell lines. With the adequate cellular model selected, attention turns to the technical readouts of the antibody-dependent cellular cytotoxicity.
In measuring ADCC, the focus can be placed on either the immune cells or the target cells. Initially, recognition of the Fc domain of the targeting antibody by CD16 leads to activation of the immune cells. This activation triggers changes in receptor expression on the immune cell surface and the release of the toxic granules which results in killing of the cancer cells. A wide array of assays is currently available to analyze the different aspects of effector activation and the final target killing (Figure 1, Table 1).
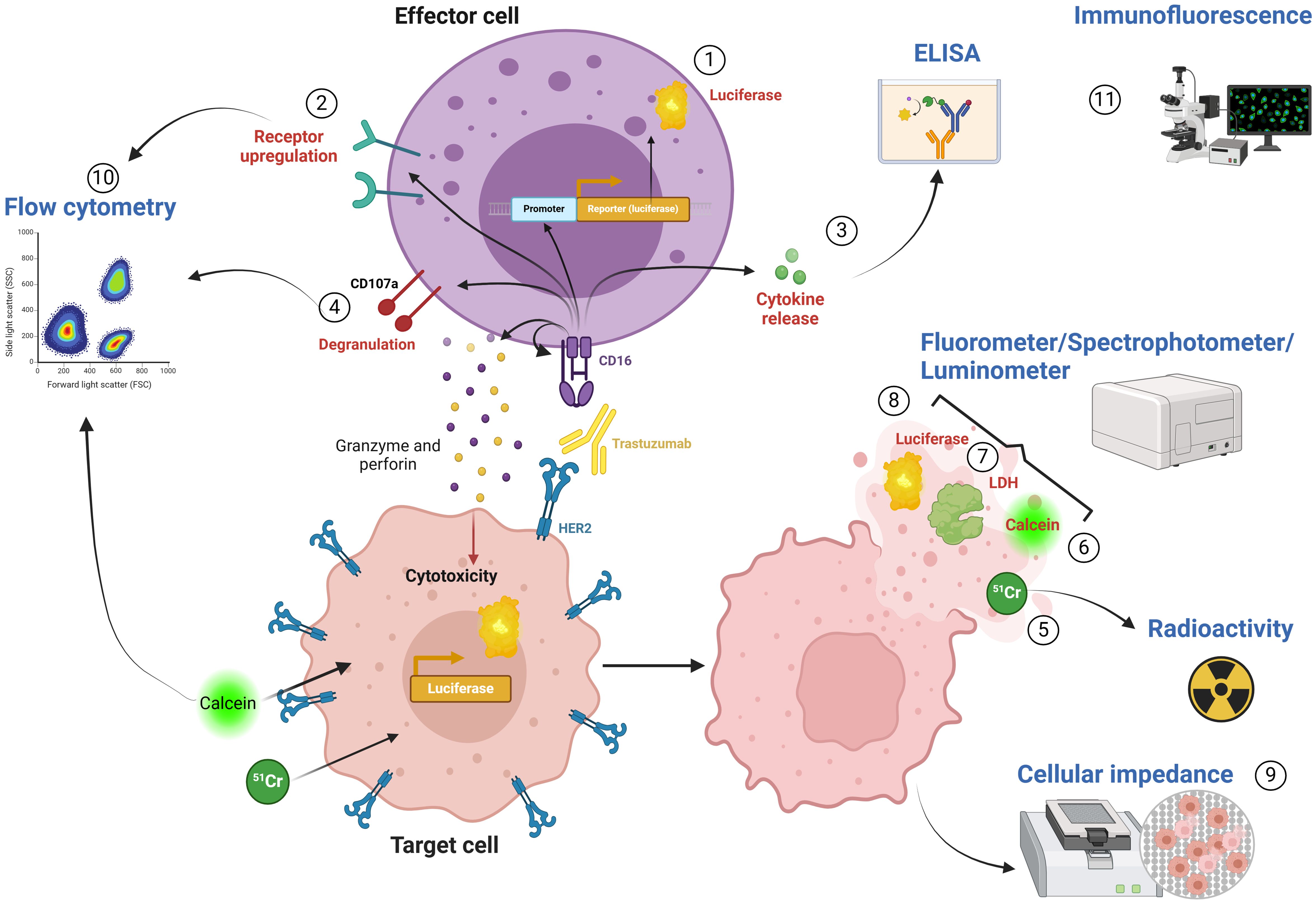
Figure 1. Technical readouts of ADCC. 1) Luciferase reporter assay: Luciferase expression upon CD16 activation in FcγRIIIA/NFAT-RE/luc2 engineered Jurkat T cell reporter cell line. 2) Receptor upregulation: NK cell activation can be assessed by measuring the expression of activating and inhibitory receptors on the cell surface. 3) Cytokine release: Cytokine secretion can be measured in the supernatant using ELISA assays. 4) Degranulation assay: Mobilization of receptor CD107a can be used as an indicator of granzyme- and perforin-containing vesicle release. 5) 51Cr-release assay: Radioactivity of 51Cr released into the medium can be quantified using a gamma-counter. 6) Calcein-AM assay: Released calcein can be measured via fluorometer or fluorescence retained in live cells can be measured by flow cytometry. 7) LDH release assay: LDH released by lysed target cells can be assessed by spectrophotometer. 8) Bioluminescence assay: Cytotoxicity can be measured as a decrease in bioluminescence signal upon cell death using a luminometer. 9) Cellular impedance: Cellular impedance measures the loss of impedance of the electron flow after cell lysis. 10) Flow cytometry: Flow cytometry allows several measurements including cell viability, receptor upregulation (2) and degranulation (4). 11) Immunofluorescence: Immunofluorescence allows for the visualization of cell killing by staining cells with viability or cell death stains.
Recognition of a cancer cell by CD16 via an antibody triggers downstream signaling through the NFAT family of transcription factors. This signaling cascade forms the basis for several readouts used to measure NK cell activation.
One method for measuring immune cell activation is through luciferase reporter assay. Coupling an NFAT response element to the luciferase gene results in strong luciferase expression upon CD16 activation (63). The FcγRIIIA/NFAT-RE/luc2 engineered Jurkat T cell reporter cell line is not capable of cell lysis but shows similar intracellular activation of NFAT as both primary and engineered NK cells. This cell line provides an easy and precise readout of immune cell activation. However, it is limited in physiological representation of the NK cell. Jurkat cells express a different set of receptors and though CD16 does not require co-receptors these can enhance activation. Furthermore, killing of the cancer cells is not measured by this type of assay. It can be applied for testing antibody batches, screening novel anti-CD16 monoclonal antibodies (65) and screening of engineered mAbs potency for ADCC (66).
NK cell activation also results in changes in expression of receptors on the cell membrane and release of cytokines. These changes can be used as a readout of activation without the need for an engineered reporter system. Secretion of cytokines, such as IFNγ can be measured in the supernatant using a simple ELISA assay or multiplex immunoassays such as the ProcartaPlex Luminex system (20, 21, 67). Cytokine release assays are a straightforward readout of activation of NK cells and can easily be combined with other assays using the co-culture supernatant to measure target cell lysis such as the lactate dehydrogenase (LDH) assay. Production of cytokines can also be stained intracellularly, or NK cell activation can be measured through assessing the expression of activating and inhibitory receptors on the cell surface by flow cytometry (68, 69). Typical markers expressed by target-activated NK cells are activation markers such as CD69, degranulation markers such as CD107a, key effector cytokines such as TNFα and IFNγ, and chemokines such as MIP-1α and MIP-1β. These markers are expressed in an activation-dependent manner and indicate both the type of immune response and strength of activation (20, 21).
One receptor expressed on activated NK cells that can be used as a more functional marker is CD107a. Mobilization of CD107a to the cell surface indicates the release of granzyme- and perforin-containing vesicles and is therefore more closely linked to cytotoxic killing of the cancer cells. This assay can be used to distinguish between cytotoxic effector cells and those exhibiting only cytokine production or other effector functions (69–72).
The recognition of an antibody by NK cells triggers the clustering of receptors and the formation of an immune synapse, resulting in a strong binding between the two cells. This binding can be measured using flow cytometry by staining of both the target and immune cell and selecting events positive for both cell types (21). To better understand the formation of the immune synapse and the processes leading to NK activation the type and quantity of proteins in the synapse can be assessed. Examples include detection of F-actin at the synapse using confocal microscopy to evaluate NK cell spreading in response to target stiffness (73), and detection of ICAM-1 and CD16 clustering using TIRF microscopy (21).
Activation and degranulation of NK cells may or may not lead to killing of the cancer cells, depending on the strength of the immune response and the sensitivity of the cancer cells. To assess cancer cell lysis, various readouts are available.
The classical way of measuring cytotoxicity is using the Chromium-51 release assay (74, 75). Target cells are pre-labeled with Na51CrO, which is incorporated into intracellular proteins, and co-cultured with effector cells to allow ADCC to take place. Lysis of the cancer cells releases the 51Cr into the medium, which is harvested and quantified using a gamma counter (76). While this method is valued for its simplicity and high sensitivity, 51Cr has a short half-life and can be spontaneously released by the target cells, affecting data accuracy. The main downside of this technique is the use of radioactive materials. These require special training, handling and disposal and have risks of harming both the individual and the environment. Due to these limitations, the Calcein-AM assay was proposed as a similar but safer alternative (77).
In the Calcein-AM assay, instead of radioactive chromium, the target cell is labeled with fluorescent calcein which can be measured rapidly and with high sensitivity in the supernatant from lysed cells. In addition, it also allows for the assessment of the cytotoxicity by measuring the fluorescence retained in live cells after quenching the signal released by lysed cells. The main advantage of this technique is that cells and supernatant can be recovered and reused for further assays (i.e. ELISA) which is not possible when using radioactive labeling. However, compared to 51Cr, calcein has a higher spontaneous leakage, although it is better than other fluorescent molecules (61, 69, 77, 78).
LDH release assay is based on a similar principle of release of intracellular contents during cell lysis. However, this assay measures the release of the cytoplasmic enzyme lactate dehydrogenase and does not use an exogenous dye. Unlike Calcein-AM or 51Cr, this method does not require pre-labelling of the target cells, thereby reducing the preparation time and cell loss, ideal for patient samples. As well as with the Calcein-AM assay, only part of the supernatant is taken for LDH measurement, and the remainder can be used for other experiments such as a cytokine release assay (60, 62, 79, 80).The principal disadvantage of this technique is spontaneous release of LDH into the supernatant. Although almost negligible for the majority of the target cells, the spontaneous LDH release can be high for the NK cells (62). This results in an increase of the background signal that must be subtracted to calculate the percentage of cytotoxicity. Thereby, LDH assay requires different background controls and is highly dependent on proper normalization of the data.
A cost-effective alternative for measuring cytotoxicity is the luciferase-based bioluminescence assay, which evaluates the luciferase activity within healthy cells compared to luciferase release into the supernatant upon cell lysis. Bioluminescence is produced when luciferase interacts with its substrate in the presence of ATP but ceases upon cell death. Thereby, cytotoxicity is quantified as a decrease in bioluminescence. Key advantages of this assay include high sensitivity (76), no interference from effector cells and no pre-labelling of the target cells. Therefore, this assay is well-suited for studies using unique target cell lines. Luciferase expressing target cells can also be co-cultured with other cell types (i.e. stromal cells) without confounding the results (81). Furthermore, this assay allows for time-course measurements from the same sample. The primary limitation is the need to generate stable luciferase-expressing cell lines. However, once a stable target cell line is established, these cells or surroundings can be modified to easily assess the effect of changes in the tumor cells, environment or effector cells on cytotoxicity (75, 82–84).
Cellular impedance assay provides a label-free, real-time approach to monitor cell killing. It measures the electron flow impeded by cells attached to the interface. Cell lysis results in a loss of impedance, which can be continuously monitored. It does however require the target cells to adhere and reach the exponential growth phase before effector cells can be added. However, growing the target cells past 100% confluence will result in cell death and therefore the time of starting the co-culture needs to be well monitored (85). While it is most suited for adherent target cells (36, 59, 86), it can be adjusted to include suspension target cells by tethering the cells to interface (87). A major drawback of this technique is that it requires expensive specialized equipment. This, however, allows for highly sensitive label-free real-time assay.
Flow cytometry is a highly sensitive and versatile technique for analyzing single cells, capable of assessing subpopulations of both target and effector cells and simultaneously measuring cell death and receptor expression. However, flow cytometry is relatively expensive, technically demanding and challenging to standardize. The analysis is highly dependent on the staining efficiency and accurate gating of subpopulations. Additionally, distinguishing between NK cells and target cells typically requires labelling at least one subset or preferably both, to avoid cross-contamination. Incomplete staining of the effector cells can significantly affect the results, particularly at high effector:target ratios. Similarly, loss of fluorescent reporter expression in even a small subset of effector cells can hinder accurate separation.
Target cells are typically pre-labelled with a live-cell dye, while a cell death marker is used to assess cytotoxicity (70, 78, 88–93). Alternatively, target cells can be engineered to stably express a fluorescent protein, and cytotoxicity can be measured as the absence of these cells after co-culture. Flow cytometry measurement of cytotoxicity can be combined with commercials assays, like Pantoxilux (OncoImmunin®), identifying the enzymes responsible for cell killing through a fluorogenic substrate whose cleavage by granzyme B and caspases leads to fluorescence in dying cells (94). This assay has demonstrated that trastuzumab-mediated ADCC specific killing depends on granzyme B and caspase activity in HER2+ breast cancer cells (36).
Although flow cytometry offers a higher sensitivity than many other assays, a lower cytotoxicity is often observed (61, 91) due to loss of dead target cells in labelling and washing steps. This is especially pronounced in the case of adherent cells where the additional trypsinization step can induce further cell loss. Under optimal conditions, flow cytometry is highly sensitive, reliable for low target cell numbers (61), and cell death can be analyzed alongside markers expressed by target and immune cells. This, for instance, facilitates the identification of relationships between NK cell phenotype and the subsequent killing potential (93).
While methods like ELISA, luciferase and flow cytometry give precise quantitative data, immunofluorescence allows for visualization of cell killing. By staining cells with a viability dye, real-time quantification of cell death can be performed using a widefield incubator microscope like the Incucyte® or a confocal microscope for more detail (70, 83). Immunofluorescence, however, is a laborious technique, difficult to standardize and requires extensive optimization of staining and analysis parameters to distinguish between variation and biological alterations. It is preferred for immobile adherent cells but has also been performed with suspension cells (83). Immunofluorescence allows for gaining insight into the underlying mechanism of cytotoxicity by quantifying effector cell recruitment, interaction time and effector activity (95).
Most of the current ADCC approaches are performed in 2D allowing tumor and NK cells to passively interact or are performed in semi 3D by centrifuging tumor and NK cells to form 3D aggregates. However, for physiological representation tumor cells should be grown in 3D spheroids (95–97), which makes quantifying cytotoxicity complex, time-consuming and low throughput. Although establishing spheroids before co-culture creates more variability, automated injection of spheroids greatly reduces variability and improves standardization. Furthermore, precise localization of the spheroids reduces imaging time, which can be a major difficulty in high-throughput immunofluorescence-based assays (98).
To accurately recapitulate the in vivo mechanism, more stromal cells and even fluidics need to be incorporated into the ADCC model. Ayuso et al. developed a microfluidic device consisting of a hydrogel containing spheroids and lumens, which can be covered with endothelial cells to create artificial blood vessels. NK cells and therapeutic antibodies can be introduced in these vessels. The readouts of this model include time-lapse microscopy to assess antibody dynamics and NK cell dynamics, qPCR to assess receptor and chemokine expression, and microscopy to assess 3D cytotoxicity (99). Chernyavska et al. used a similar model with multiple channels that can incorporate target cells in 2D or 3D, along with antibodies and NK cells. While microscopy is the primary readout of cytotoxicity, cytokines can be measured in the medium, and cells can be extracted for gene expression analysis. This model can also be adjusted to incorporate endothelial cells to mimic blood vessels (100). With further development these models can be adapted to include additional stromal cells within the spheroids and hydrogel, thereby increasing their complexity (101).
Cancer-immune co-culture experiments are crucial when studying therapeutic antibodies capable of engaging the immune system. By closely replicating the in vivo condition, these models enhance our understanding of the mode of action, optimize drug discovery and help identify new biomarkers of therapy resistance. Especially as the focus in drug discovery shifts from antigen targeting to altering antibody interactions with the Fc receptors, co-culture systems are indispensable for testing both novel and modified antibodies, evaluating their ability to activate immune cells and induce cytotoxicity (45). Furthermore, these assays can be used to detect unwanted immunogenicity that could lead to reduced efficacy or allergic reactions, which is especially critical for emerging antibody-derived molecules such as nanobodies (87, 102). Using in vitro assays to assess desired and adverse reactions to novel drugs allows researchers to efficiently pre-screen antibodies, reducing reliance on animal models while saving time, cost and resources in drug discovery and optimization.
The choice of experimental model depends on the study goals. For initial screening, NK cell lines offer higher reproducibility and easier manipulation. For validation experiments, PBMCs are preferred, providing a closer representation of the in vivo immune responses and enabling the matching of patient target and effector cells. Multiple readouts are available focusing either on NK activation or target lysis. NK activation can be assessed through methods such as reporter, signaling or degranulation assays. However, while these methods are suitable for screening antibodies for CD16 activation, they do not capture the NK cytotoxic function of killing the cancer cell. Other factors around or within the target cells play a role in whether NK cell activation results in ADCC, which is especially relevant when studying resistance to therapy. For more translational relevance, readouts focusing on cancer cell killing, such as intracellular molecule release assays, bioluminescence, flow cytometry and impedance assays, are indispensable. Advanced techniques like immunofluorescence, and 3D tumor spheroid systems aim to further improve physiological representation.
Each method offers specific advantages, such as real-time monitoring or label-free detection, but also facing challenges like background noise or technical complexity. Despite the difficulties, use of co-culture systems is essential for refining drug discovery and tailoring therapies to overcome resistance effectively.
RV: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. UA: Visualization, Writing – original draft, Writing – review & editing. AR: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported in part by grants to AR-S: Marie Skłodowska-Curie Actions Individual Fellowship (MSCA-IF-894191); Ikerbasque Foundation; Victoria’s Secret Global Fund for Women’s Cancers Career Development Award, in partnership with Pelotonia & AACR (PC # 1159511); Basque Department of Industry, Tourism, and Trade (Elkartek); the Ministerio de Ciencia e Innovación (MICINN; PID2023-150421OB-I00 and Ramón y Cajal Program); CRIS Emerging Leader Grant from the Fundación CRIS Contra el Cáncer and the VI Proyecto FERO – GHD awarded by the FERO Foundation.
Graphical images were created using BioRender.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Osorio JC, Smith P, Knorr DA, Ravetch JV. The antitumor activities of anti-CD47 antibodies require Fc-FcγR interactions. Cancer Cell. (2023) 41:2051–2065.e6. doi: 10.1016/J.CCELL.2023.10.007
2. Ruiz-saenz A, Moasser MM. Targeting HER2 by combination therapies Ana. J Clin Oncol. (2018) 36:808–11. doi: 10.1200/JCO.2017.77.1899
3. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2. New Engl J Med. (2001) 344(11):783–92. doi: 10.1056/nejm200103153441101
4. Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN. The HER-2 receptor and breast cancer: ten years of targeted anti–HER-2 therapy and personalized medicine. Oncologist. (2009) 14:320–68. doi: 10.1634/THEONCOLOGIST.2008-0230
5. Goydel RS, Rader C. Antibody-based cancer therapy. Oncogene. (2021) 40:3655. doi: 10.1038/S41388-021-01811-8
6. Smith MR. Rituximab (monoclonal anti-CD20 antibody): mechanisms of action and resistance. Oncogene. (2003) 22:7359–68. doi: 10.1038/sj.onc.1206939
7. Graham J, Muhsin M, Kirkpatrick P. Cetuximab. Nat Rev Drug Discovery. (2004) 3:549–50. doi: 10.1038/NRD1445
8. Bokemeyer C, Van Cutsem E, Rougier P, Ciardiello F, Heeger S, Schlichting M, et al. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer. (2012) 48:1466–75. doi: 10.1016/J.EJCA.2012.02.057
9. Junttila TT, Li G, Parsons K, Phillips GL, Sliwkowski MX. Trastuzumab-DM1 (T-DM1) retains all the mechanisms of action of trastuzumab and efficiently inhibits growth of lapatinib insensitive breast cancer. Breast Cancer Res Treat. (2011) 128:347–56. doi: 10.1007/S10549-010-1090-X/FIGURES/4
10. Ogitani Y, Aida T, Hagihara K, Yamaguchi J, Ishii C, Harada N, et al. DS-8201a, A novel HER2-targeting ADC with a novel DNA topoisomerase I inhibitor, demonstrates a promising antitumor efficacy with differentiation from T-DM1. Clin Cancer Res. (2016) 22:5097–108. doi: 10.1158/1078-0432.CCR-15-2822
11. Hamilton G, Rath B. Avelumab: combining immune checkpoint inhibition and antibody-dependent cytotoxicity. Expert Opin Biol Ther. (2017) 17:515–23. doi: 10.1080/14712598.2017.1294156
12. Gan X, Shan Q, Li H, Janssens R, Shen Y, He Y, et al. An anti-CTLA-4 heavy chain-only antibody with enhanced Treg depletion shows excellent preclinical efficacy and safety profile. Proc Natl Acad Sci U.S.A. (2022) 119(32). doi: 10.1073/PNAS.2200879119
13. Vargas FA, Furness AJS, Litchfield K, Joshi K, Rosenthal R, Ghorani E, et al. Fc effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell. (2018) 33:649–663.e4. doi: 10.1016/J.CCELL.2018.02.010
14. Selby MJ, Engelhardt JJ, Quigley M, Henning KA, Chen T, Srinivasan M, et al. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol Res. (2013) 1:32–42. doi: 10.1158/2326-6066.CIR-13-0013
15. Boyerinas B, Jochems C, Fantini M, Heery CR, Gulley JL, Tsang KY, et al. Antibody-dependent cellular cytotoxicity activity of a novel anti-PD-L1 antibody avelumab (MSB0010718C) on human tumor cells. Cancer Immunol Res. (2015) 3:1148–57. doi: 10.1158/2326-6066.CIR-15-0059
16. Park JE, Kim SE, Keam B, Park HR, Kim S, Kim M, et al. Anti-tumor effects of NK cells and anti-PD-L1 antibody with antibody-dependent cellular cytotoxicity in PD-L1-positive cancer cell lines. J Immunother Cancer. (2020) 8(2). doi: 10.1136/JITC-2020-000873
17. Moasser MM. Inactivating amplified HER2: challenges, dilemmas, and future directions. Cancer Res. (2022) 82(16):2811–20. doi: 10.1158/0008-5472.CAN-22-1121
18. Hofman T, Ng SW, Garcés-Lázaro I, Heigwer F, Boutros M, Cerwenka A. IFNγ mediates the resistance of tumor cells to distinct NK cell subsets. J Immunother Cancer. (2024) 12:e009410. doi: 10.1136/JITC-2024-009410
19. Liu J, Xing L, Li J, Wen K, Liu N, Liu Y, et al. Epigenetic regulation of CD38/CD48 by KDM6A mediates NK cell response in multiple myeloma. Nat Commun. (2024) 15:1367. doi: 10.1038/S41467-024-45561-Z
20. Gauthier L, Virone-Oddos A, Beninga J, Rossi B, Nicolazzi C, Amara C, et al. Control of acute myeloid leukemia by a trifunctional NKp46-CD16a-NK cell engager targeting CD123. Nat Biotechnol. (2023) 41:1296. doi: 10.1038/S41587-022-01626-2
21. Eitler J, Rackwitz W, Wotschel N, Gudipati V, Murali Shankar N, Sidorenkova A, et al. CAR-mediated targeting of NK cells overcomes tumor immune escape caused by ICAM-1 downregulation. J Immunother Cancer. (2024) 12(2). doi: 10.1136/JITC-2023-008155
22. Muraro E, Montico B, Lum B, Colizzi F, Giurato G, Salvati A, et al. Antibody dependent cellular cytotoxicity-inducing anti-EGFR antibodies as effective therapeutic option for cutaneous melanoma resistant to BRAF inhibitors. Front Immunol. (2024) 15:1336566. doi: 10.3389/FIMMU.2024.1336566
23. Qian J, Wang Q, Xiao L, Xiong W, Xian M, Su P, et al. Development of therapeutic monoclonal antibodies against DKK1 peptide-HLA-A2 complex to treat human cancers. J Immunother Cancer. (2024) 12:e008145. doi: 10.1136/JITC-2023-008145
24. Xiang H, Chan AG, Ahene A, Bellovin DI, Deng R, Hsu AW, et al. Preclinical characterization of bemarituzumab, an anti-FGFR2b antibody for the treatment of cancer. MAbs. (2021) 13:1981202. doi: 10.1080/19420862.2021.1981202
25. Chew HY, De Lima PO, Gonzalez Cruz JL, Banushi B, Echejoh G, Hu L, et al. Endocytosis inhibition in humans to improve responses to ADCC-mediating antibodies. Cell. (2020) 180:895–914.e27. doi: 10.1016/J.CELL.2020.02.019
26. Cho BC, Simi A, Sabari J, Vijayaraghavan S, Moores S, Spira A. Amivantamab, an epidermal growth factor receptor (EGFR) and mesenchymal-epithelial transition factor (MET) bispecific antibody, designed to enable multiple mechanisms of action and broad clinical applications. Clin Lung Cancer. (2023) 24:89–97. doi: 10.1016/J.CLLC.2022.11.004
27. Wingert S, Reusch U, Knackmuss S, Kluge M, Damrat M, Pahl J, et al. Preclinical evaluation of AFM24, a novel CD16A-specific innate immune cell engager targeting EGFR-positive tumors. MAbs. (2021) 13:1950264. doi: 10.1080/19420862.2021.1950264
28. Schram AM, Odintsov I, Espinosa-Cotton M, Khodos I, Sisso WJ, Mattar MS, et al. Zenocutuzumab, a HER2xHER3 bispecific antibody, is effective therapy for tumors driven by NRG1 gene rearrangements. Cancer Discovery. (2022) 12:1233. doi: 10.1158/2159-8290.CD-21-1119
29. Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene. (2008) 27:5932–43. doi: 10.1038/onc.2008.267
30. Becker PSA, Suck G, Nowakowska P, Ullrich E, Seifried E, Bader P, et al. Selection and expansion of natural killer cells for NK cell-based immunotherapy. Cancer Immunol Immunother. (2016) 65:477–84. doi: 10.1007/S00262-016-1792-Y
31. Milush JM, Long BR, Snyder-Cappione JE, Cappione AJ, York VA, Ndhlovu LC, et al. Functionally distinct subsets of human NK cells and monocyte/DC-like cells identified by coexpression of CD56, CD7, and CD4. Blood. (2009) 114:4823. doi: 10.1182/BLOOD-2009-04-216374
32. Arnould L, Gelly M, Penault-Llorca F, Benoit L, Bonnetain F, Migeon C, et al. Trastuzumab-based treatment of HER2-positive breast cancer: an antibody-dependent cellular cytotoxicity mechanism? Br J Cancer. (2006) 94:259. doi: 10.1038/SJ.BJC.6602930
33. Beano A, Signorino E, Evangelista A, Brusa D, Mistrangelo M, Polimeni MA, et al. Correlation between NK function and response to trastuzumab in metastatic breast cancer patients. J Transl Med. (2008) 6:25. doi: 10.1186/1479-5876-6-25
34. Varchetta S, Gibelli N, Oliviero B, Nardini E, Gennari R, Gatti G, et al. Elements related to heterogeneity of antibody-dependent cell cytotoxicity in patients under trastuzumab therapy for primary operable breast cancer overexpressing Her2. Cancer Res. (2007) 67:11991–9. doi: 10.1158/0008-5472.CAN-07-2068
35. Rocca YS, Roberti MP, Juliá EP, Pampena MB, Bruno L, Rivero S, et al. Phenotypic and functional dysregulated blood NK cells in colorectal cancer patients can be activated by cetuximab plus IL-2 or IL-15. Front Immunol. (2016) 7:413. doi: 10.3389/FIMMU.2016.00413
36. Kute T, Stehle JR, Ornelles D, Walker N, Delbono O, Vaughn JP. Understanding key assay parameters that affect measurements of trastuzumab-mediated ADCC against Her2 positive breast cancer cells. Oncoimmunology. (2012) 1:810. doi: 10.4161/ONCI.20447
37. Shi Y, Fan X, Deng H, Brezski RJ, Rycyzyn M, Jordan RE, et al. Trastuzumab triggers phagocytic killing of high HER2 cancer cells in vitro and in vivo by interaction with fcγ Receptors on macrophages. J Immunol. (2015) 194:4379–86. doi: 10.4049/JIMMUNOL.1402891
38. Capietto A-H, Martinet L, Fournié J-J. Stimulated γδ T cells increase the in vivo efficacy of trastuzumab in HER-2+ Breast cancer. J Immunol. (2011) 187:1031–8. doi: 10.4049/JIMMUNOL.1100681
39. Pizzamiglio S, Ciniselli CM, Triulzi T, Gargiuli C, De Cecco L, De Azambuja E, et al. Integrated molecular and immune phenotype of HER2-positive breast cancer and response to neoadjuvant therapy: A NeoALTTO exploratory analysis. Clin Cancer Res. (2021) 27:6307–13. doi: 10.1158/1078-0432.CCR-21-1600
40. Abdullah N, Greenman J, Pimenidou A, Topping KP, Monson JRT. The role of monocytes and natural killer cells in mediating antibody- dependent lysis of colorectal tumour cells. Cancer Immunol Immunother. (1999) 48:517–24. doi: 10.1007/S002620050600/METRICS
41. Xiao H, Woods EC, Vukojicic P, Bertozzi CR. Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc Natl Acad Sci U.S.A. (2016) 113:10304–9. doi: 10.1073/pnas.1608069113
42. Richards JO, Karki S, Lazar GA, Chen H, Dang W, Desjarlais JR. Optimization of antibody binding to FcγRIIa enhances macrophage phagocytosis of tumor cells. Mol Cancer Ther. (2008) 7:2517–27. doi: 10.1158/1535-7163.MCT-08-0201
43. Arora J, Ayyappan S, Yin C, Smith BJ, Lemke-Miltner CD, Wang Z, et al. T-cell help in the tumor microenvironment enhances rituximab-mediated NK-cell ADCC. Blood. (2024) 143:1816–24. doi: 10.1182/BLOOD.2023023370
44. Pham TT, Chenoweth A, Patel N, Banu A, Osborn G, Blower PJ, et al. In vivo PET imaging of 89Zr-labeled natural killer cells and the modulating effects of a therapeutic antibody. J Nucl Med. (2024) 65:1035. doi: 10.2967/JNUMED.124.267876
45. de Taeye SW, Bentlage AEH, Mebius MM, Meesters JI, Lissenberg-Thunnissen S, Falck D, et al. FcγR binding and ADCC activity of human IgG allotypes. Front Immunol. (2020) 11:740/FULL. doi: 10.3389/FIMMU.2020.00740/FULL
46. Musolino A, Gradishar WJ, Rugo HS, Nordstrom JL, Rock EP, Arnaldez F, et al. Role of Fcγ receptors in HER2-targeted breast cancer therapy. J Immunother Cancer. (2022) 10:e003171. doi: 10.1136/JITC-2021-003171
47. Frampton S, Smith R, Ferson L, Gibson J, Hollox EJ, Cragg MS, et al. Fc gamma receptors: Their evolution, genomic architecture, genetic variation, and impact on human disease. Immunol Rev. (2024) 328:65. doi: 10.1111/IMR.13401
48. Galvez-Cancino F, Simpson AP, Costoya C, Matos I, Qian D, Peggs KS, et al. Fcγ receptors and immunomodulatory antibodies in cancer. Nat Rev Cancer. (2024) 24:51–71. doi: 10.1038/S41568-023-00637-8
49. Gogesch P, Dudek S, van Zandbergen G, Waibler Z, Anzaghe M. The role of Fc receptors on the effectiveness of therapeutic monoclonal antibodies. Int J Mol Sci. (2021) 22:8947. doi: 10.3390/IJMS22168947
50. Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G, et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol. (2008) 26:1789–96. doi: 10.1200/JCO.2007.14.8957
51. Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcγRIIIa gene. Blood. (2002) 99:754–8. doi: 10.1182/BLOOD.V99.3.754
52. Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol. (2003) 21:3940–7. doi: 10.1200/JCO.2003.05.013
53. Rodríguez J, Zarate R, Bandres E, Boni V, Hernández A, Sola JJ, et al. Fc gamma receptor polymorphisms as predictive markers of Cetuximab efficacy in epidermal growth factor receptor downstream-mutated metastatic colorectal cancer. Eur J Cancer. (2012) 48:1774–80. doi: 10.1016/J.EJCA.2012.01.007
54. Zhang W, Gordon M, Schultheis AM, Yang D, Nagashima F, Azuma M, et al. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J Clin Oncol. (2007) 25:3712–8. doi: 10.1200/JCO.2006.08.8021
55. Bryceson YT, Ljunggren HG, Long EO. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. (2009) 114:2657–66. doi: 10.1182/BLOOD-2009-01-201632
56. Chung S, Lin YL, Reed C, Ng C, Cheng ZJ, Malavasi F, et al. Characterization of in vitro antibody-dependent cell-mediated cytotoxicity activity of therapeutic antibodies — Impact of effector cells. J Immunol Methods. (2014) 407:63–75. doi: 10.1016/J.JIM.2014.03.021
57. Drexler HG, Matsuo Y. Malignant hematopoietic cell lines: in vitro models for the study of natural killer cell leukemia–lymphoma. Leukemia. (2000) 14:777–82. doi: 10.1038/sj.leu.2401778
58. Mishima Y, Terui Y, Mishima Y, Kuniyoshi R, Matsusaka S, Mikuniya M, et al. High reproducible ADCC analysis revealed a competitive relation between ADCC and CDC and differences between FcγRllla polymorphism. Int Immunol. (2012) 24:477–83. doi: 10.1093/INTIMM/DXS048
59. Cheng ZF, Li HK, Yang HP, Lee CY, Tang SW, Lin YL, et al. A novel endogenous CD16-Expressing Natural Killer Cell for cancer immunotherapy. Biochem Biophys Rep. (2021) 26:100935. doi: 10.1016/J.BBREP.2021.100935
60. Schnueriger A, Grau R, Sondermann P, Schreitmueller T, Marti S, Zocher M. Development of a quantitative, cell-line based assay to measure ADCC activity mediated by therapeutic antibodies. Mol Immunol. (2011) 48:1512–7. doi: 10.1016/J.MOLIMM.2011.04.010
61. Tanaka M, Ishige A, Yaguchi M, Matsumoto T, Shirouzu M, Yokoyama S, et al. Development of a simple new flow cytometric antibody-dependent cellular cytotoxicity (ADCC) assay with excellent sensitivity. J Immunol Methods. (2019) 464:74–86. doi: 10.1016/J.JIM.2018.10.014
62. Vincken R, Ruiz-Saenz A. A co-culture model system to quantify antibody-dependent cellular cytotoxicity in human breast cancer cells using an engineered natural killer cell line. STAR Protoc. (2023) 4(2). doi: 10.1016/j.xpro.2023.102224
63. Hsieh YT, Aggarwal P, Cirelli D, Gu L, Surowy T, Mozier NM. Characterization of FcγRIIIA effector cells used in in vitro ADCC bioassay: Comparison of primary NK cells with engineered NK-92 and Jurkat T cells. J Immunol Methods. (2017) 441:56–66. doi: 10.1016/j.jim.2016.12.002
64. Gunesch JT, Angelo LS, Mahapatra S, Deering RP, Kowalko JE, Sleiman P, et al. Genome-wide analyses and functional profiling of human NK cell lines. Mol Immunol. (2019) 115:64–75. doi: 10.1016/J.MOLIMM.2018.07.015
65. Van Faassen H, Jo DH, Ryan S, Lowden MJ, Raphael S, Mackenzie CR, et al. Incorporation of a novel CD16-specific single-domain antibody into multispecific natural killer cell engagers with potent ADCC. Mol Pharm. (2021) 18:2375–84. doi: 10.1021/ACS.MOLPHARMACEUT.1C00208
66. Kurogochi M, Mori M, Osumi K, Tojino M, Sugawara SI, Takashima S, et al. Glycoengineered monoclonal antibodies with homogeneous glycan (M3, G0, G2, and A2) using a chemoenzymatic approach have different affinities for FcγRIIIa and variable antibody-dependent cellular cytotoxicity activities. PloS One. (2015) 10:e0132848. doi: 10.1371/JOURNAL.PONE.0132848
67. Shete A, Suryawanshi P, Chavan C, Kulkarni A, Godbole S, Ghate M, et al. Development of IFN-γ secretory ELISPOT based assay for screening of ADCC responses. J Immunol Methods. (2017) 441:49–55. doi: 10.1016/J.JIM.2016.12.001
68. Bryceson YT, Fauriat C, Nunes JM, Wood SM, Björkström NK, Long EO, et al. Functional analysis of human NK cells by flow cytometry. Methods Mol Biol. (2010) 612:335. doi: 10.1007/978-1-60761-362-6_23
69. Felgueres MJ, Esteso G, García-Jiménez ÁF, Dopazo A, Aguiló N, Mestre-Durán C, et al. BCG priming followed by a novel interleukin combination activates Natural Killer cells to selectively proliferate and become anti-tumour long-lived effectors. Sci Rep. (2024) 14:13133. doi: 10.1038/S41598-024-62968-2
70. Pesini C, Hidalgo S, Arias MA, Santiago L, Calvo C, Ocariz-Díez M, et al. PD-1 is expressed in cytotoxic granules of NK cells and rapidly mobilized to the cell membrane following recognition of tumor cells. Oncoimmunology. (2022) 11(1). doi: 10.1080/2162402X.2022.2096359
71. Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. (2004) 294:15–22. doi: 10.1016/J.JIM.2004.08.008
72. Schönberg K, Hejazi M, Uhrberg M. Protocol for the clonal analysis of NK cell effector functions by multi-parameter flow cytometry. Methods Mol Biol. (2012) 903:381–92. doi: 10.1007/978-1-61779-937-2_26
73. Friedman D, Simmonds P, Hale A, Bere L, Hodson NW, White MRH, et al. Natural killer cell immune synapse formation and cytotoxicity are controlled by tension of the target interface. J Cell Sci. (2021) 134:jcs258570. doi: 10.1242/JCS.258570
74. Brunner KT, Mauel J, Cerottini J-C, Chapuis B. Quantitative assay of the lytic action of immune lymphoid cells of 51Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology. (1968) 14:181.
75. Hartl FA, Beck-Garcìa E, Woessner NM, Flachsmann LJ, Cárdenas RMHV, Brandl SM, et al. Noncanonical binding of Lck to CD3ϵ promotes TCR signaling and CAR function. Nat Immunol. (2020) 21:902–13. doi: 10.1038/s41590-020-0732-3
76. Karimi MA, Lee E, Bachmann MH, Salicioni AM, Behrens EM, Kambayashi T, et al. Measuring cytotoxicity by bioluminescence imaging outperforms the standard chromium-51 release assay. PloS One. (2014) 9(2). doi: 10.1371/JOURNAL.PONE.0089357
77. Cholujová D, Jakubíková J, Kubeš M, Arendacká B, Sapák M, Ihnatko R, et al. Comparative study of four fluorescent probes for evaluation of natural killer cell cytotoxicity assays. Immunobiology. (2008) 213:629–40. doi: 10.1016/J.IMBIO.2008.02.006
78. Gillissen MA, Yasuda E, de Jong G, Levie SE, Go D, Spits H, et al. The modified FACS calcein AM retention assay: A high throughput flow cytometer based method to measure cytotoxicity. J Immunol Methods. (2016) 434:16–23. doi: 10.1016/J.JIM.2016.04.002
79. Decker T, Lohmann-Matthes ML. A quick and simple method for the quantitation of lactate dehydrogenase release in measurements of cellular cytotoxicity and tumor necrosis factor (TNF) activity. J Immunol Methods. (1988) 115:61–9. doi: 10.1016/0022-1759(88)90310-9
80. Iida S, Misaka H, Inoue M, Shibata M, Nakano R, Yamane-Ohnuki N, et al. Nonfucosylated Therapeutic IgG1 Antibody Can Evade the Inhibitory Effect of Serum Immunoglobulin G on Antibody-Dependent Cellular Cytotoxicity through its High Binding to FcγRIIIa. Clin Cancer Res. (2006) 12:2879–87. doi: 10.1158/1078-0432.CCR-05-2619
81. McMillin DW, Delmore J, Weisberg E, Negri JM, Geer DC, Klippel S, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anti-cancer drug activity. Nat Med. (2010) 16:483. doi: 10.1038/NM.2112
82. Matta H, Gopalakrishnan R, Choi S, Prakash R, Natarajan V, Prins R, et al. Development and characterization of a novel luciferase based cytotoxicity assay. Sci Rep. (2018) 8(1):199. doi: 10.1038/S41598-017-18606-1
83. Meléndez AV, Velasco Cárdenas RMH, Lagies S, Strietz J, Siukstaite L, Thomas OS, et al. Novel lectin-based chimeric antigen receptors target Gb3-positive tumour cells. Cell Mol Life Sci. (2022) 79:1–25. doi: 10.1007/s00018-022-04524-7
84. Schäfer H, Schäfer A, Kiderlen AF, Masihi KN, Burger R. A highly sensitive cytotoxicity assay based on the release of reporter enzymes, from stably transfected cell lines. J Immunol Methods. (1997) 204:89–98. doi: 10.1016/S0022-1759(97)00040-9
85. Kanemaru H, Mizukami Y, Kaneko A, Kajihara I, Fukushima S. A protocol for quantifying lymphocyte-mediated cytotoxicity using an impedance-based real-time cell analyzer. STAR Protoc. (2022) 3:101128. doi: 10.1016/J.XPRO.2022.101128
86. Zhu J, Wang X, Xu X, Abassi YA. Dynamic and label-free monitoring of natural killer cell cytotoxic activity using electronic cell sensor arrays. J Immunol Methods. (2006) 309:25–33. doi: 10.1016/J.JIM.2005.10.018
87. Cerignoli F, Abassi YA, Lamarche BJ, Guenther G, Ana DS, Guimet D, et al. In vitro immunotherapy potency assays using real-time cell analysis. PloS One. (2018) 13:e0193498. doi: 10.1371/JOURNAL.PONE.0193498
88. Jedema I, van der Werff NM, Barge RMY, Willemze R, Falkenburg JHF. New CFSE-based assay to determine susceptibility to lysis by cytotoxic T cells of leukemic precursor cells within a heterogeneous target cell population. Blood. (2004) 103:2677–82. doi: 10.1182/BLOOD-2003-06-2070
89. Duensing TD, Watson SR. Assessment of antibody-dependent cellular cytotoxicity by flow cytometry. Cold Spring Harb Protoc. (2018) 2018:138–41. doi: 10.1101/pdb.prot093815
90. Gómez-Román VR, Florese RH, Patterson LJ, Peng B, Venzon D, Aldrich K, et al. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. J Immunol Methods. (2006) 308:53–67. doi: 10.1016/J.JIM.2005.09.018
91. Yamashita M, Kitano S, Aikawa H, Kuchiba A, Hayashi M, Yamamoto N, et al. A novel method for evaluating antibody-dependent cell-mediated cytotoxicity by flowcytometry using cryopreserved human peripheral blood mononuclear cells. Sci Rep. (2016) 6:1–10. doi: 10.1038/srep19772
92. Richard J, Veillette M, Batraville LA, Coutu M, Chapleau JP, Bonsignori M, et al. Flow cytometry-based assay to study HIV-1 gp120 specific antibody-dependent cellular cytotoxicity responses. J Virol Methods. (2014) 208:107–14. doi: 10.1016/J.JVIROMET.2014.08.003
93. Bennett-Boehm MMC, Mahr AR, Hartwell ST, Regan AK, Weber IS, Blackmon A, et al. Development and implementation of natural killer cell simultaneous ADCC and direct killing assay. Heliyon. (2023) 9:e22991. doi: 10.1016/J.HELIYON.2023.E22991
94. Packard BZ, Telford WG, Komoriya A, Henkart PA. Granzyme B activity in target cells detects attack by cytotoxic lymphocytes. J Immunol. (2007) 179:3812–20. doi: 10.4049/JIMMUNOL.179.6.3812
95. Dekkers JF, Alieva M, Cleven A, Keramati F, Wezenaar AKL, van Vliet EJ, et al. Uncovering the mode of action of engineered T cells in patient cancer organoids. Nat Biotechnol. (2022) 41:60–9. doi: 10.1038/s41587-022-01397-w
96. Gopal S, Kwon SJ, Ku B, Lee DW, Kim J, Dordick JS. 3D tumor spheroid microarray for high-throughput, high-content natural killer cell-mediated cytotoxicity. Commun Biol. (2021) 4:1–14. doi: 10.1038/s42003-021-02417-2
97. Booij TH, Price LS, Danen EHJ. 3D cell-based assays for drug screens: challenges in imaging, image analysis, and high-content analysis. Slas Discovery. (2019) 24:615. doi: 10.1177/2472555219830087
98. Liao CY, Engelberts P, Ioan-Facsinay A, Klip JE, Schmidt T, Ruijtenbeek R, et al. CD3-engaging bispecific antibodies trigger a paracrine regulated wave of T-cell recruitment for effective tumor killing. Commun Biol. (2024) 7(1):983. doi: 10.1038/S42003-024-06682-9
99. Ayuso JM, Truttschel R, Gong MM, Humayun M, Virumbrales-Munoz M, Vitek R, et al. Evaluating natural killer cell cytotoxicity against solid tumors using a microfluidic model. Oncoimmunology. (2019) 8(3). doi: 10.1080/2162402X.2018.1553477
100. Chernyavska M, Hermans CKJC, Chan C, Baumann N, Rösner T, Leusen JHW, et al. Evaluation of immunotherapies improving macrophage anti-tumor response using a microfluidic model. Organs-on-a-Chip. (2022) 4:100019. doi: 10.1016/J.OOC.2022.100019
101. Gjorevski N, Avignon B, Gérard R, Cabon L, Roth AB, Bscheider M, et al. Neutrophilic infiltration in organ-on-a-chip model of tissue inflammation. Lab Chip. (2020) 20:3365–74. doi: 10.1039/D0LC00417K
Keywords: antibody-dependent cellular cytotoxicity, cancer immune co-cultures, natural killer cells, ADCC readouts, therapeutic antibodies, immunotherapy
Citation: Vincken R, Armendáriz-Martínez U and Ruiz-Sáenz A (2025) ADCC: the rock band led by therapeutic antibodies, tumor and immune cells. Front. Immunol. 16:1548292. doi: 10.3389/fimmu.2025.1548292
Received: 19 December 2024; Accepted: 27 March 2025;
Published: 16 April 2025.
Edited by:
Alja Zottel, University of Ljubljana, SloveniaReviewed by:
Weifeng Xu, Merck (United States), United StatesCopyright © 2025 Vincken, Armendáriz-Martínez and Ruiz-Sáenz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Ruiz-Sáenz, YXJ1aXpAY2ljYmlvZ3VuZS5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.