- Department of Biochemistry, Faculty of Life Sciences, Aligarh Muslim University, Aligarh, India
Generating antibodies targeting native membrane proteins presents various challenges because these proteins are often embedded in the lipid bilayer, possess various extracellular and intracellular domains, and undergo post-translational modifications. These properties of MPs make it challenging to preserve their stable native conformations for immunization or antibody generation outside of the membranes. In addition, MPs are often hydrophobic due to their membrane-spanning regions, making them difficult to solubilize and purify in their native form. Therefore, employing purified MPs for immunogen preparation may result in denaturation or the loss of native structure, rendering them inadequate for producing antibodies recognizing native conformations. Despite these obstacles, various new approaches have emerged to address these problems. We outline recent advancements in designing and preparing immunogens to produce antibodies targeting MPs. Strategies outlined here are relevant for producing antibodies for research, diagnostics, and therapies and designing immunogens for vaccination purposes.
1 Introduction
The immune response is the defense mechanism of our body against substances it perceives as foreign or dangerous, generally marked as antigens (1). When the immune system detects an antigen, it attempts to attack and destroy the target molecule. The recognition molecules utilized by the immune system are either membrane-bound receptors or soluble proteins. This response has two key components, namely cellular immunity and humoral immunity. The former involves immune cells directly targeting and destroying non-self molecules, cancer cells, and whole pathogens, whereas the latter relies on B cells produced antibodies that bind to antigens, neutralizing them (2). B cells recognize solvent-exposed regions of an antigen, also called B cell epitope, that interact with both secreted and membrane-bound immunoglobulins (3). Based on their immunogenic potential B cell epitopes can be classified as immunodominant, immunogenic, and non-immunogenic (4). On the other hand, T cell epitopes are peptides derived from antigens, presented on the surface of antigen-presenting cells bound to MHC molecules, where the T cell receptor recognizes them. Most T cell epitopes are linear, whereas B cell epitopes can be either linear (10%) or conformational (90%). Therefore, preserving conformational epitopes while designing immunogens for raising antibodies is extremely important. Epitope identification in antigens is crucial for understanding disease mechanisms, immune monitoring, and designing epitope-based immunogens for both vaccines and developing antibodies for therapeutic, diagnostic, and research purposes (5). While several experimental approaches can identify B cell epitopes, including determining the three-dimensional (3D) structure of antigen-antibody complexes and screening peptide libraries for antibody binding, recently, B cell epitope prediction tools have also been developed (6).
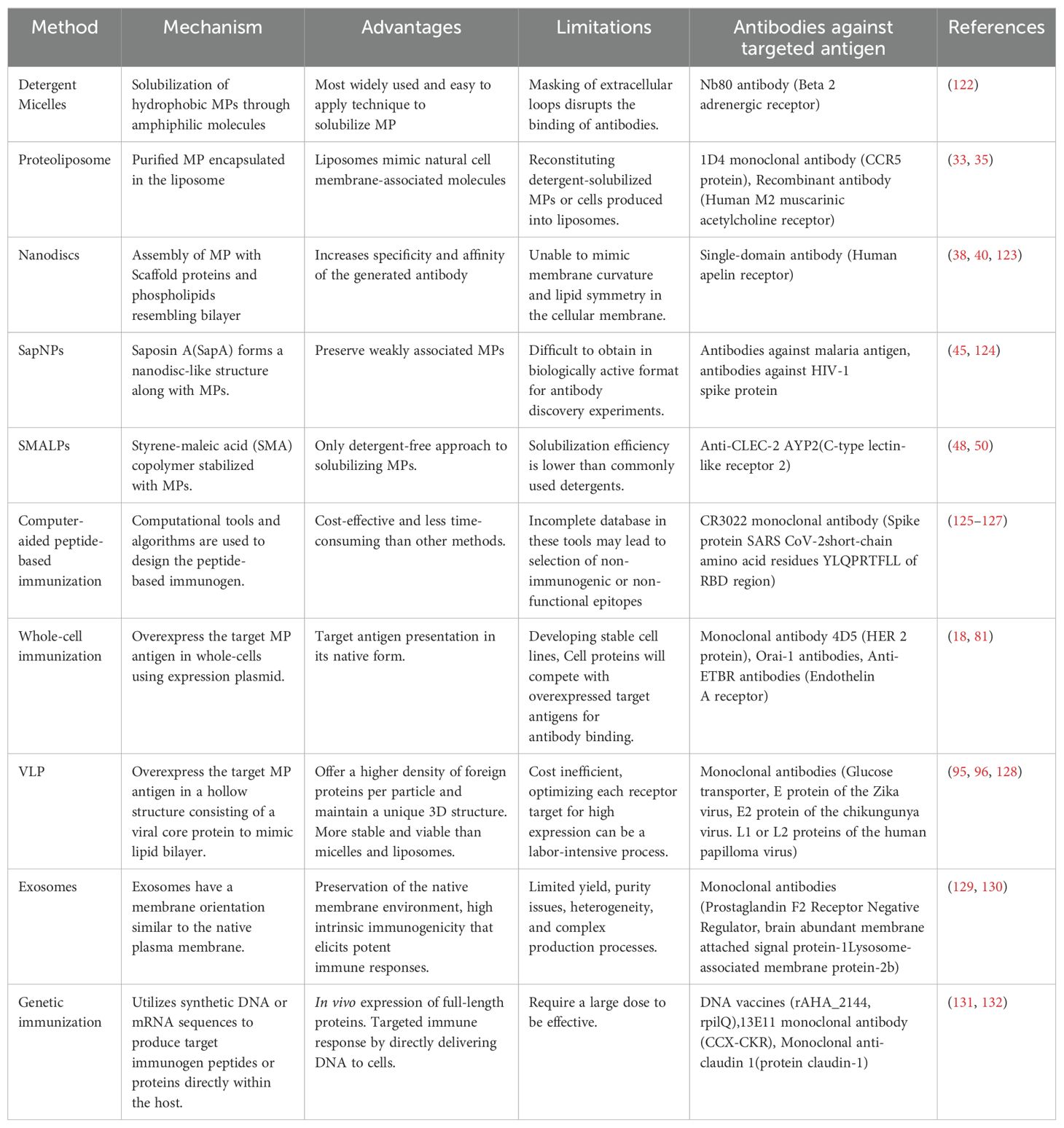
The ability of the immune system to produce specific and high-affinity antibodies against foreign antigens is harnessed for several purposes, including research and diagnostics, enabling the development of tools for detecting and analyzing biomolecules. This ability of immune system is also used in vaccination, where long-term disease prevention is achieved by stimulating the creation of memory cells and antibodies in response to exposure to a harmless form of an antigen (7, 8). Antibodies are indispensable for advancing our understanding of membrane protein's (MPs) structure, function, localization, transport, and interaction with various ligands (9). MPs account for over 20-30% of all cellular proteins encoded by the human genome and are essential for numerous cellular functions, including material transport, signal transduction, intercellular recognition, ligand-receptor binding, and cell adhesion, making them crucial therapeutic targets (10). MP can exist in various forms characterized by hydrophobic transmembrane spanning domain and including cell signaling receptors such as G protein-coupled receptors (GPCRs), ion channels, transporters, tight junction proteins, and signaling molecules, however, they remain underrepresented in Protein Data Bank, a worldwide repository for structural data (Figure 1) (9, 11–13). Advances in protein production and the secretion capacities of microbial hosts have positioned MPs at the forefront of therapeutic research, with MPs accounting for over 60% of current drug targets for various ailments (14). With the rapidly evolving nature of pathogens, as seen by SARS-CoV-2 during the pandemic, the efficient soluble production and study of MP domains involved in pathogenicity have become critical. The variations in the MPs that are target of the immune system against pathogens often possess characteristics that enable them to evade immune responses and antibody treatments, posing significant threats to public health. Since MP serves as a key entry point for critical molecules and pathogens, antibody production against MP immunogen has therapeutic advantages (15). MPs role in cellular interactions and pathogen recognition, when used as immunogens, can specifically elicit immune responses against pathogens. As a result, they hold significant potential for use in vaccine development and in generating antibodies for diagnostics and therapeutic applications. Despite numerous applications of antibodies in understanding MP biology, the structural intricacy of MPs makes it difficult to produce high-quality antibodies against them. It is challenging for MPs to retain their native conformation outside the membrane or their natural environment due to their hydrophobic regions that are naturally embedded in the lipid bilayer (16). Moreover, MPs only reveal a small fraction of their structure on the cell surface, which limits the accessibility of the epitopes available to generate antibodies (17). These restrictions make it more challenging to describe MPs and create efficient treatments targeting them thoroughly.
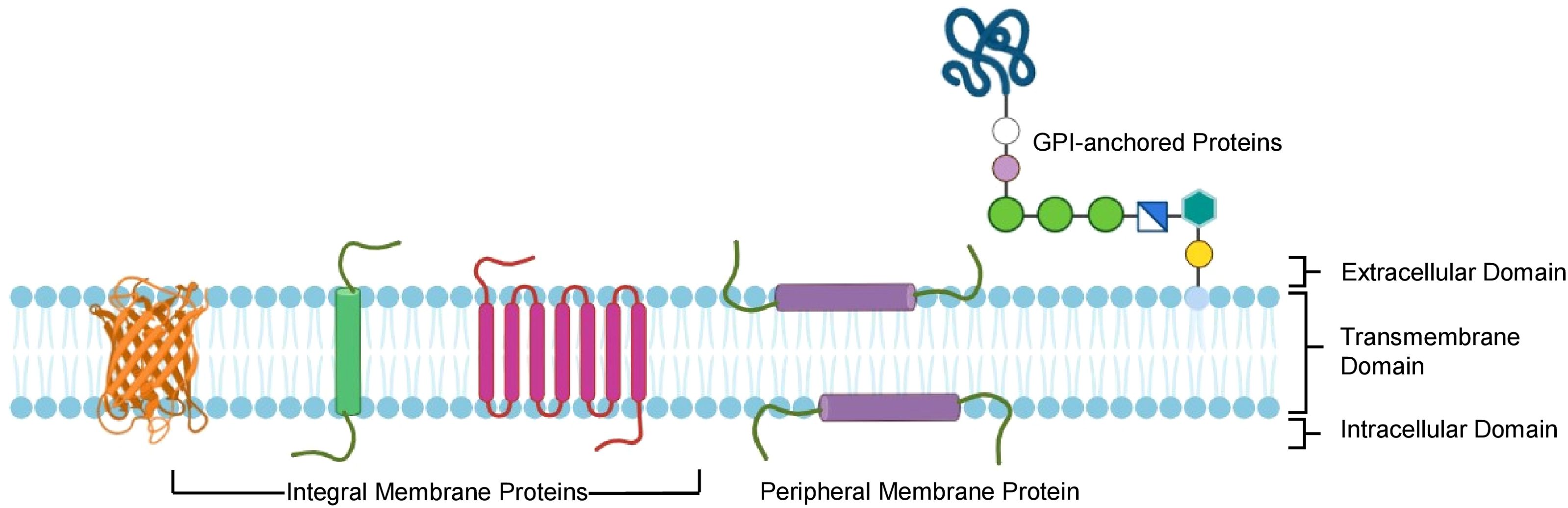
Figure 1. Different types of membrane proteins and their domains. Showing membrane protein structure and orientation within the lipid bilayer.
Traditional methods for generating MP antibodies, such as immunization with reconstituted MP or peptides, face several drawbacks, such as limited 3D presentation of MPs, their instability in purified forms, and the restricted solvent exposure of accessible epitopes. Conformational epitopes, essential for functional antibodies, are frequently missed in peptide-based immunization, making these antibodies unsuitable for applications like MP co-crystallization (18). Exposure of MPs to non-native environments can induce conformational changes, leading to low-resolution structures and potential misinterpretation of their structure and function when isolated as pure proteins. Purification of MP such as GPCR, N-methyl-D-aspartate (NMDA) receptors, and epidermal growth factor receptor (EGFR) frequently disrupts the lipid-protein interactions, membrane anchoring, and dimerization necessary to maintain their active conformations. As a result, these proteins lose their structural integrity, making them unsuitable for immunization (19, 20). For instance, Methanocaldococcusjannaschii S2P (MjS2P) protein crystallized in detergent showed multiple conformations, complicating the identification of its physiological structure (21). Similarly, γ-secretase reconstituted in an amphipol A8-35, a class of amphiphilic polymers that make it possible to keep MP soluble in detergent-free aqueous solution, yielded a high-resolution structure, whereas detergent use resulted in lower resolution (22). Another bottleneck for generating antibodies against MPs is the lack of effective screening methods that utilize MPs in their native environment.
Recent advancements in technology have improved the strategies for immunogen designing and preparation while enhancing the efficiency of antibody production and screening. Development of new strategies relying on membrane-based and nanoparticle-based technologies, genetic immunization, and flow cytometry-based transfection have shown promising results in MP immunization and antibody development. In addition, advancements in computational methods have significantly enhanced immunogen design by enabling the analysis of protein sequences, structural modeling, and the assessment of immunogenicity and antigenicity. Artificial Intelligence (AI) based structural prediction tools, such as AlphaFold2, have shown great potential to improve immunogen design processes by providing more accurate structural prediction of MPs (23).
This review outlines recent advancements in strategies for immunogen design for developing antibodies against MPs. Full-length MPs or their specific extra-cellular domains (ECDs) can be purified in their native form or reconstituted into desired membrane mimetics for use as immunogens. The commonly used membrane mimetics are categorized into two approaches: detergent- and nanoformulation-based strategies, which use purified proteins; and membrane-based strategies, which express the target Protein in its native lipid environment and avoid protein purification. The immunogen design and preparation strategies discussed here are crucial for producing antibodies for research, diagnostics, therapeutic applications, and designing immunogens for vaccination purposes.
2 Stabilizing the conformation of purified proteins
Soluble proteins have long been used as antigens to immunize animals to generate antibodies as they are easy to produce and stimulate both humoral and cell-mediated immunity. Many factors influence the use of soluble proteins as immunogens; the most crucial is their inability to retain their native conformation and function during the purification and preparation of immunogens, which is essential to produce antibodies that recognize the native proteins (24). The successful production of antibodies using any method will depend on the accessibility of epitopes on the target MP. However, using full MP as an immunogen is considered more suitable for producing antibodies than focusing on the ECD because full MP may present unique conformational epitopes that are not accessible or lose their native structure in truncated extracellular fragments. Moreover, ECD also result in low immunogenicity because of their conserved sequence (25). In most cases, MPs are purified using recombinant methods and extracted using detergents to solubilize the proteins in detergent micelles (26). To keep MP in its native environment, purified proteins are incorporated into liposomes to form proteoliposomes. Although detergents, micelles, and liposome-based methods are extremely helpful for solubilization and stabilization of MPs for immunogen generation, these approaches come with their own notable limitations. For instance, detergents and micelles often ablate critical protein-lipid interactions due to their inability to mimic the native lipid environment. Similarly, despite the closeness of liposome-based methods in mimicking natural lipid bilayers, they present challenges due to the aggregation tendency of the membrane, making it difficult to obtain proteoliposomes in a homogenous and stable state (27). Another approach to assemble and stabilize the membrane structure involves incorporating detergent-solubilized and purified MPs into nanoparticles such as nanodiscs, Saposin lipid nanoparticles (SapNPs), and Styrene-maleic acid-lipidparticles (SMALPs) into an artificial bilayer mimicking the native environment (28). In addition, nano-based platforms also facilitate antibody discovery, validation, and characterization through ELISA and surface plasmon resonance.
2.1 Detergent micelles
MP solubilization, an essential step for antigen generation, requires masking the hydrophobic surface of integral MPs (29). This can be achieved by micelles which are spherical structures generated by the self-assembly of amphipathic molecules such as surfactants or lipids in aqueous media. They have a hydrophobic core and a hydrophilic outer shell, allowing them to contain hydrophobic molecules within their core. Micelles are highly effective in solubilizing MPs with broad hydrophobic surfaces that are otherwise difficult to retain in solution. Immunization with micelle protein complex, sometimes integrated with adjuvant, elicits an effective immune response (Figure 2A). Advances in MP purification utilizing protein-detergent micelles have been fueled by the development of more efficient detergents, such as neopentyl glycols (30). But as the use of detergents has increased in recent years, the obstacles they pose during antibody generation are also being highlighted. The detergents with longer hydrophobic acyl chains are the most stabilizing, but they mask the ECD, which prevents immune response against the masked region. Moreover, after immunization, detergents may dissociate from MP, thus resulting in denaturation and possible conformational epitope loss. Introducing point mutations into the transmembrane helices of proteins is a strategy to overcome the normally low stability of complex MPs in detergent micelles (31). Employing specific point mutations can result in very stable MPs that continue to function in detergent micelles even with short acyl chains. A technique developed by Heptares therapeutics referred to as stabilized receptors or StaR has employed the thermo-stabilized β1-adrenergic receptor in a StaR boost configuration to isolate agonist antibodies and negative allosteric modulators (32).
Figure 2. Detergents and nanoformulations-based strategies for preparing membrane protein immunogens using purified proteins. Membrane protein solubilization and purification can be achieved with or without using detergent as an antigen. Mixing detergent-purified protein with lipids into different formats helps in immunization. (A) Detergents extract hydrophobic membrane proteins from the plasma membrane by forming micelles. (B) Liposomome-based reconstitution of membrane protein. (C) Wrapping the lipid bilayer with membrane scaffold protein to form nanodisc. (D) Peptidiscs are formed by wrapping with a lipid-bilayer by peptides such as 37-amino acid amphipathic ApoA1-mimetic peptide. (E) Saposin A protein is used as a Scaffold Protein to form SapNPs. (F) A Detergent-free method that relies on SMA co-polymer form SMALPs. MP, Membrane Protein; SapNP, Saposin lipid Nanoparticle; SMA, Styrene-Maleic Acid; SMALPs, Styrene-Maleic Acid Lipid Particles.
2.2 Proteoliposome
Reconstitution into liposomes is a common method for the functional characterization of MP. The lipid environment is essential for maintaining complex MPs’ correct conformation and function. Liposomes, which mimic natural cell membranes with their lipid bilayer structure, are commonly used as they can encapsulate and transport membrane-associated molecules (33). Typically, liposomes form through self-assembling pure lipids or lipid mixtures (34). MP from various pathogens are being assessed to be incorporated into liposomes as potential vaccines. When compared to detergent-extracted MP, proteoliposomes provide more native-like lipid configuration to complex MPs, enhancing immunogenicity. Liposomes can fuse with the plasma membrane to deliver the antigen directly into cells, allowing it to be processed via the endogenous pathway and thereby eliciting Cytotoxic T Lymphocytes responses (CTL). Overexpressing recombinant MPs in conventional cellular systems can be challenging due to insufficient membrane insertion, precipitation of newly synthesized proteins, or cytotoxic effects caused by significant disruptions to the host cell’s metabolism. Considerable time and effort are required to optimize cell culture conditions, stabilize and solubilize the target MP, purify it, and successfully reconstitute it into proteoliposomes. This system has been used to produce highly effective antibodies targeting the human M2 muscarinic acetylcholine receptor and CCR5 receptor (33, 35).
Another approach to preparing liposomes integrated with MP relies on cell-free translation system. When liposomes are added to a cell-free translation system, the synthesized MP integrates directly into the liposomal lipid membrane, eliminating the need for laborious steps like MP purification and proteoliposome reconstitution (Figure 2B) (36). A further modification of the cell-free system incorporates adjuvant-containing liposomes with monophosphoryl lipid A (MPLA), which adjusts the lipid composition to improve the reproducibility and stability of MPs. Additionally, this system produces large amounts of MP antigens, with MPLA acting as a toll-like receptor 4 agonist to enhance antibody production by stimulating B cells (37). However, preparing proteoliposomes for immunization presents significant challenges, including reconstituting detergent-solubilized MPs into liposomes with correct orientation or directly incorporating MPs produced in cells into the liposome structure. Despite these technical difficulties, proteoliposome immunization remains a valuable method for MP immunization.
2.3 Nanodiscs
Nanodiscs rely on scaffold proteins and phospholipids to assemble a bilayer that closely resembles the native environment of the MP (28). The nanodisc platform was originally conceptualized as a lipid bilayer stabilized by a hydrophobic belt formed by two copies of amphipathic membrane scaffold proteins (MSPs) (38). MSP self-assembles around the MP in the presence of a lipid, capturing the protein associated with the lipid in the process. SpyCatcher-SpyTag technology is one of the recent advances in MSP engineering, leading to a tenfold higher yield in protein extraction (39). As nanodiscs preserve the structural and functional integrity of the MPs, they provide an interesting approach to developing MP immunogens for antibody generation. In order to prepare a nanodisc-based MP immunogen, the detergent-solubilized MP is incorporated into nanodiscs, which are prepared by assembling lipids such as POPC (1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine) or POPE (1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphoethanolamine) and MSP Apolipoprotein A1 (ApoA1), or its derivatives such as MSP1D1, MSP1E3D1, MSP1D1ΔH5, and MSP2N2 (Figure 2C). Following validation, the resulting nanodisc-protein complex is formulated with an adjuvant to maintain stability and enhance the immune response. Nanodiscs incorporated with MPs have been successfully used to generate antibodies against the human apelin receptor (APLNR), immune checkpoint targets like PD-L1, ion channels like Kvv1.3, (39) and Influenza virus proteins matrix-2 and hemagglutinin, resulting in the increased specificity and affinity of the generated antibodies (38, 40).
2.4 Peptidiscs
Peptidisc, an alternative to nanodiscs, developed by Duong et al., is composed of short amphipathic peptides, such as the 37-amino acid ApoA1-mimetic peptide known as the nanodisc scaffold peptide (41). These peptides encircle and embed MPs within a lipid bilayer, mimicking the natural membrane environment (Figure 2D) (42). Peptidiscs can reconstitute MPs of varying sizes and topologies from prokaryotes and eukaryotes, offering a distinct advantage over nanodiscs (41). Peptidiscs are particularly advantageous because they provide a more stable and native-like setting for MPs, which are often unstable outside their natural environment. This stability makes them ideal for structural and functional studies, including X-ray crystallography, cryo-electron microscopy (cryo-EM), and drug screening. Despite the potential of nanodiscs for MP stabilization, they face challenges like limited size range, difficulty incorporating large proteins, and complicated preparation. Peptidiscs address these limitations of nanodiscs by using a flexible peptide scaffold that can accommodate even larger proteins with simplified preparation and improved stability.
2.5 Saposin lipid nanoparticles
Saposin-based nanoparticles (SapNPs), a new alternative tool for MP stabilization, are a class of nanoparticles derived from saposins (SapA), which are small lipid-binding proteins, forming nanodisc-like structures around MPs. In SapNPs, an MP integrated into a lipid bilayer is enclosed by multiple copies of SapA proteins, which preserves their structural and functional integrity (Figure 2E) (43). The flexibility of the SapA scaffold and its ability to adapt to the size of target MPs, accommodating the variable transmembrane regions within the SapNPs, make it an attractive alternative to nanodiscs (43). SapNPs solubilized MPs are used for both immunizations, enabling the immune system to produce specific antibodies against the delivered protein antigens and probes to sort antigen-specific B cells and for in vitro high throughput screening experiments (44). SapNPs incorporating malaria antigens have demonstrated the ability to elicit strong antibody responses. These nanoparticles help in better antigen stability and targeted delivery, enhancing immunogenicity against malaria. In cancer vaccine research, SapNPs have been used to deliver tumor-associated antigens. The adjuvanticity of SapA boosts the immune system’s ability to recognize and produce antibodies against these antigens, aiding in tumor rejection (45).
Based on the success of nanoparticle-based membrane solubilization methods, Salipro Biotech has pioneered the development of saposin-based membrane solubilization systems such as Direct MX for the stabilization of MPs in their native lipid environment (46). This method is advantageous because it relies on mild detergent, which preserves weakly associated MPs, helping maintain their native interactions. Like nanodiscs and SMALPs, the Salipro platform has been successfully applied to a wide range of targets, including GPCRs, ion channels, and other integral MPs, facilitating breakthroughs in drug discovery and antibody development.
2.6 Styrene-maleic acid lipid particles
SMALPs are similar to nanodiscs as they both utilize lipid-based systems to stabilize MPs in a native-like environment. However, unlike nanodiscs, which utilize MSPs to stabilize lipid bilayer, the bilayer in SMALPs is stabilized by an amphipathic styrene-maleic acid (SMA) copolymer. The copolymer inserts into the membrane and extracts a patch of the bilayer along with the embedded MP, creating a lipid particle around the protein (Figure 2F) (47). Unlike nanodisc, SMALPs offer a detergent-free approach to solubilizing MPs and are particularly advantageous for working with larger MPs (48). Similar to nanodiscs, SMALPs have been used to prepare immunogen for antibody production through the preservation of MPs in their natural state. The transmembrane influenza matrix-2 protein, for instance, was integrated into SMALPs, allowing the target domain to be isolated for antibody production. Using this method, a number of antibodies against the cytoplasmic domain of M2 were effectively generated and verified (49). SMALPs have also been utilized in research on human C-type lectin-like receptor 2 (CLEC-2), enabling activation for the production of antibodies (50). This illustrates the adaptability of SMALPs in producing antibodies against MP targets.
3 Computer-aided peptide-based immunization
Using peptides for immunization not only alters the need to produce full-length proteins but also directs antibodies to the particular epitopes of interest (51, 52). The peptide-based immunization strategy often depends on synthesizing peptides with short sequences corresponding to the extracellularly exposed portions of MPs or expressing the same regions as fusion proteins or soluble proteins (53). Therefore, peptide-based immunization eliminates the need to purify the entire MP. However, the linear shape of short peptides may lead to discovery of antibodies that do not recognize the native structure of proteins or conformational epitopes unless antibody screening is carried out using native proteins. This problem can be partially resolved by using cyclic or conformationally constrained peptides, which may help retain specific secondary structures by restricting the peptide flexibility, thereby resulting in antibodies recognizing conformational epitopes (54). PEPSCAN recently developed a process known as the chemical linkage of peptides onto scaffolds (CLIPS) that enables the synthesis of mono, di, or tricyclic limited peptides. This technique facilitates the production of conformation-specific antibodies and has been successfully applied to produce functional anti CXCR2 antibodies (55).
Peptide based immunization also allows raising antibodies against post-translational modifications (PTMs) of MPs. PTMs such as phosphorylation, glycosylation, and acetylation can be added to create synthetic peptides that mirror the protein’s native state, increasing the chance of developing high-affinity, functional antibodies against MPs (56). The conventional approach to immunogen designing involves large proteins or whole organisms that lead to an unnecessary antigenic load and induce the chances of allergy (57). This can be avoided by using peptide-based immunogen comprising short immunogenic peptide fragments, capable of evoking strong and targeted immune responses, thereby avoiding the chances of the allergenic response (58). However, the design of peptide-based immunogens for MPs is a challenging task due to the unique structural characteristics of MPs, particularly their hydrophobic regions and complex topologies (59).
Computational approaches have emerged as powerful tools to address these challenges, enabling the rational design of immunogens that can evoke a targeted immune response. Computational approaches for epitope prediction can facilitate the selection of both linear and conformational epitopes from trans MPs, paving the way for their development into peptides and soluble proteins suitable for use as antigens in antibody production help in identifying immunogenic regions within MPs (Figure 3) (60). Recent advancements in computational immunology, particularly in immuno-informatics and structural bioinformatics, have significantly enhanced our ability to identify promising epitopes on MPs (57, 61, 62).
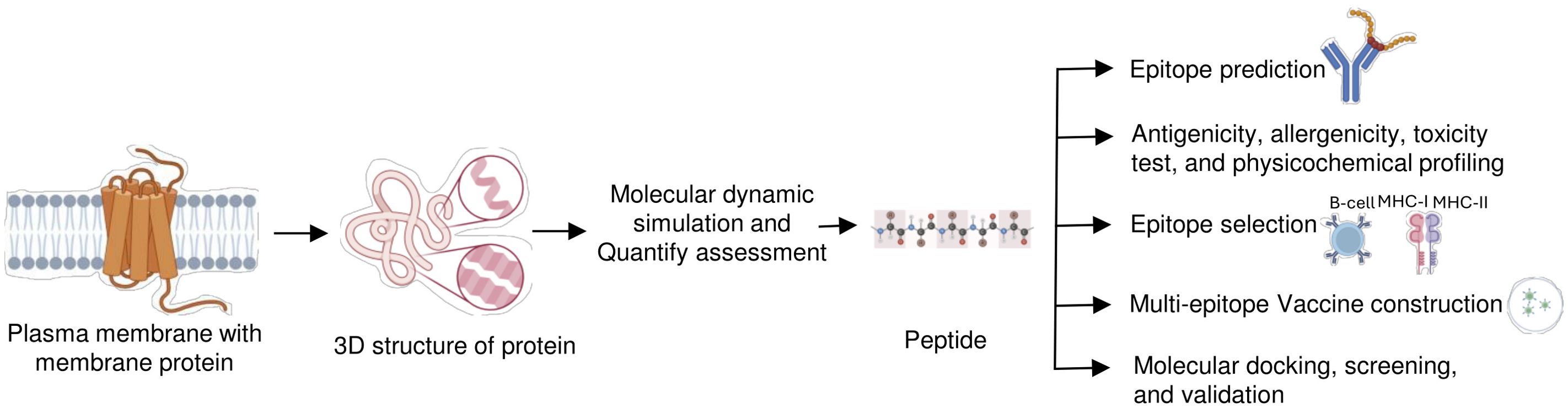
Figure 3. Computer-aided peptide-based immunization strategy. A computational analysis approach is used to develop the peptide-based immunogen that presents ideal features for antibody development. In this approach, first identification and selection of the membrane protein-specific region that will be immunogenic. Immunogenic peptide determination is via antibody interaction and multi-epitope construction, followed by bioinformatic tools and algorithms for testing antigenicity, and physicochemical profiling. Epitopes can then be optimized to best display their antigenic potential.
A critical strategy in the design of peptide-based immunogens is the identification of surface-exposed epitopes, which are required for producing antibodies capable of recognizing and neutralizing native MPs. Tools such as the VaxiJen v2.0 server are used to predict antigenicity by analyzing the physicochemical properties of protein sequences (63). In contrast, AllerTOP v2.0 server is used to distinguish the designed peptide, the immunogenic one, and the non-immunogenic one. These tools enable the screening of a large number of peptide candidates to identify peptides that are more likely to generate immune responses without causing adverse reactions (64, 65).
Traditional methods of epitope identification often rely on protein structures and need to account for the membrane context of proteins. A computational technique such as AlphaFold2 provides a massive transformational advantage over the existing methods (23, 66). AlphaFold2 utilizes the deep-learning algorithm that helps to determine the exact 3D structures of proteins, including the MPs, which are notoriously very challenging to study experimentally. AlphaFold also enhances the precision of predicting surface exposed epitopes in MPs, which is vital for eliciting a robust immune reaction with the accurate model. This capability greatly simplifies the immunogen design process by reducing reliance on experimental techniques and improving epitope prediction precision (67, 68). Although many computational techniques have been created to help in B-cell epitope prediction because experimental methods are expensive and time-consuming. At first, sequence-based techniques were the mainstay of these techniques. But with recent developments in protein structure prediction—like the innovation made by AlphaFold2—structure-based methods have also become strong substitutes to predict B cell epitope. GraphBepi is one such structure-based tool that leverages the advancements in protein structure prediction to improve the accuracy of B-cell epitope identification (69). Additionally, applications such as RoseTTAFold2 and ESMFold also help in modeling the structure of integral MP (70). The peptide-based immunogen formulations are further advanced through innovative methods, such as the design of multi-peptide epitopes derived from various regions of the target protein (71). Additionally, various tools available at the Immune Epitope Database can be employed to predict the immunogenicity scores of single or multi-epitope constructs, enabling the development of immunogens capable of eliciting robust and diverse immune responses (72, 73).
By predicting the most promising epitopes and ensuring they are non-toxic, non-allergenic, and well presented on MHC, computational methods help optimize the design of immunogens that can raise antibodies capable of recognizing native structure of MPs. For example, the peptide-based epitope “YLQPRTFLL” is located within the receptor binding domain (RBD) of SARS-CoV-2 spike glycoprotein, a trans membrane-associated protein crucial for viral entry through its RBD. The computational tools and algorithms predicted that this peptide possesses a high binding affinity to a prevalent HLA a, suggesting its potential to provoke a vigorous CTL (71, 74, 75). In addition, leveraging AlphaFold2 simplifies and accelerates the workflow by effectively forecasting the configuration of MP, which facilitates the logical choice of epitopes that are more likely to provoke robust and targeted immune responses (76, 77). The innovations in the computational design of immunogens and advanced structural prediction technologies such as AlphaFold2 signify a transformative change in developing vaccines and therapeutic antibodies targeting the MPs (66). The ability to efficiently design peptide-reduced risk of adverse immune responses offers promising prospects for the future of immunotherapy and vaccine development.
4 Membrane-based immunogen preparations
MPs either partially interact with or are fully embedded within the cellular membrane, characterized by their hydrophobic transmembrane domains, which cause them to aggregate, particularly when misfolded (78). Therefore, micelles, cell membranes mimicking lipid bilayer systems, or polymers are often necessary to solubilize and stabilize MPs. These methods enable more precise immune responses by maintaining the function and stability of MPs. The key membrane-based immunization strategies to produce anti-membrane antibodies are described below.
4.1 Whole-cell immunization
Whole-cell immunization effectively generates antibodies against MPs by expressing them in their native conformation on the cell surface to overcome the challenges of in vitro antigen preparation, making them suitable for a wide range of applications. Preparing pure, homogeneous, and conformationally stable MP antigens from plasma membranes is challenging. Hence, whole-cell antigen presentation preserves the MP in a native environment with correct and functional folding. In this approach, cells are engineered to express the MP of interest and are subsequently used to immunize animals (Figure 4A). Whole-cell immunization targets relevant functional epitopes of MPs that play essential physiological roles, helping the immune system to produce accurate antibodies. A key challenge with whole-cell immunization is the limited expression of MPs on the cell surface, which often necessitates strategies for their overexpression. Recent developments of flow cytometry-based transfection strategies such as MaxCyte flow electroporation efficiently express and isolate cell expression target proteins that are otherwise difficult to express (79). Such a strategy can achieve a transfection efficiency of up to 2 × 1011 cells without significant loss of viability to create stable cell lines (79). Such advanced strategies can successfully transfect and isolate cells even when the transfection efficiency is very low (80). Since most MP do not exist in abundance, overexpressing relevant epitopes in an expression system can increase the chances of immunogenicity.
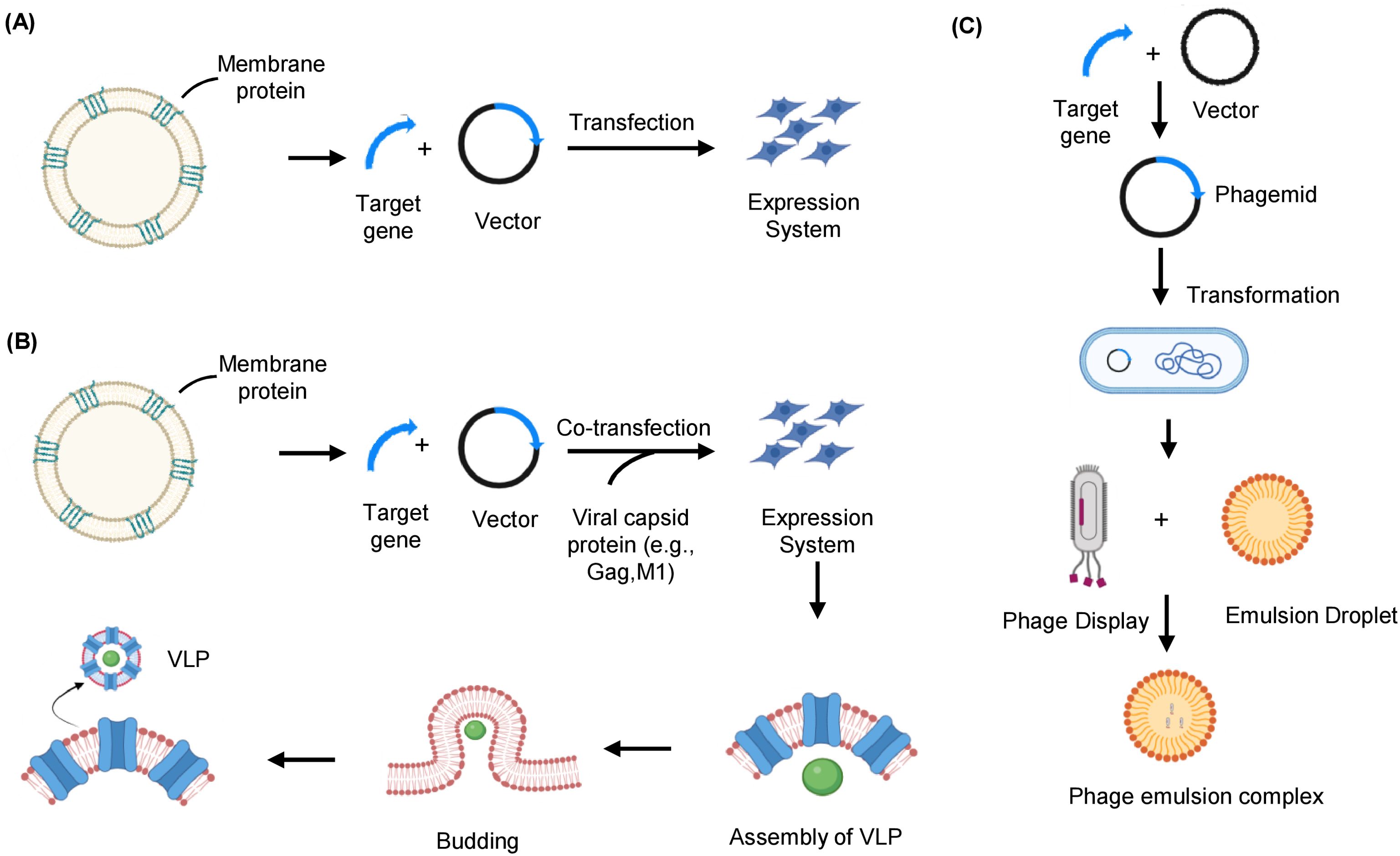
Figure 4. Native membrane-based strategies to express and prepare membrane protein immunogens. (A) Whole cells overexpressing the target membrane protein of interest can be used as immunogens. The whole cell’s entire proteome is injected into the host cell’s body, which considers it non-self and develops an immune response. (B) VLPs are produced by co-expressing the target antigen with viral capsid protein in the mammalian expression system. The capsid protein self-assembles at the plasma membrane, where the membrane protein of interest is overexpressed and buds off to form a VLP that serves as an immunogen for delivery into a host cell. (C) The target membrane protein gene segment is integrated with the phage genome, expressed on the phage coat. The phage displayed with the target membrane protein is encapsulated in an emulsion droplet to immunize the host. VLP, Virus-like particle.
Whole-cell immunization has successfully generated antibodies against various MPs, such as the HER2 protein in breast cancer and GPCRs. A monoclonal antibody 4D5, targeting the ECD of the HER2 protein, effectively inhibits the growth of HER2 overexpressing breast tumor cells and enhances their sensitivity to TNF-α (18). Immunizing with cells that express membrane peptides or proteins has successfully generated antibodies that modulate the function of ion channels, or GPCRs (51). For example, researchers generated antibodies against GPCR 5 by co-transfecting cells with the receptor peptide and receptor activity-modifying proteins, enabling the expression of the MP region. In contrast, antibodies targeting ion channels like Orai-1 and P2X7 have been developed for treating autoimmune and inflammatory diseases, respectively (81, 82). Bacterial ghost platforms, consisting of intact bacterial cell envelopes emptied of their contents, are also utilized to express or deliver various antigens, plasmid DNA encoding protein epitopes or present MP for immunization. The ghost vaccine method serves as an efficient carrier platform, addressing the potential poor immunogenicity of protein subunits and DNA-encoded antigens by effectively delivering them to antigen-presenting cells (83). Noncapsular bacterial endotoxin surrounding some bacteria can also be used to elicit an immune response. For example, to combat Klebsiella pneumoniae infections, a natural conjugate was developed by combining the bacterium’s O-antigen, a surface component that triggers an immune response, with bacterial outer MPs to strengthen the immune response (84). Although whole-cell immunization approach holds great potential, developing stable cell lines that express MPs at sufficient levels requires significant time and effort. However, once established, a single cell can be clonally expanded to consistently produce high MP levels, which researchers can use for antibody production and store indefinitely (85). Human Embryonic Kidney 293 (HEK-293) cells (86) and insect cells are commonly used to generate transient and stable expression systems to overexpress MPs. Whole-cell immunization using an engineered cell line is helpful for MPs that are challenging to isolate or purify, as it allows MPs to remain in their natural conformation on the cell surface (45).
4.2 Virus-like particle
In addition to other lipid bilayer-based membranous formats, besides proteoliposomes and nanodiscs, which resemble native biological membranes, including virus-like particles (VLPs) (87). As MPs are typically present in low quantities in their natural hosts, it is necessary to overexpress them in heterologous systems for sufficient production. VLPs are hollow structures consisting of a viral core protein encapsulated by portions of the cell membrane with MPs and receptors expressed in their native conformation. VLPs mimic the native structure of viruses but are non-infectious due to the absence of viral genetic material (88). These vesicular structures, typically 20 to 200 nm in diameter, are enriched with the MP of interest up to 100-fold and can be generated from a cell population that overexpresses the protein on its surface (89). The VLPs are produced by transfecting insect or mammalian cells with the retroviral core protein (gag) expression vectors. The capsid protein produced from the transfected vector spontaneously self-assembles to form VLPs. As gag protein naturally assembles at the plasma membrane and buds off from host cells that overproduce the target MP, VLPs displaying MP of interest (MP-VLPs) can be produced by co-expression of gag protein with the MP of interest (Figure 4B) (87, 90). A key step in successful VLP production is selecting the appropriate expression host system. The choice of an ideal system depends on various factors, such as the specific target, required quantity, and intended downstream applications.
There are several benefits to using VLPs for phage display (91). Compared to other carriers, VLPs offer a higher density of foreign proteins per particle and maintain a unique 3D structure, which is crucial for presenting conformational epitopes. As compared to whole cells, VLPs can provide a high concentration of target MP with fewer host cell MPs. VLPs can also be stored for prolonged periods at -80°C without significant degradation or loss of functionality when compared with other membrane-based formats (92). Due to the viral gag protein functioning both as an adjuvant and in VLP formation, the use of external adjuvants or toxins to enhance immunogenicity is not required. Preserving MPs in their natural phospholipid bilayer in a conformationally stable state is a significant advantage of this strategy, making VLPs suitable for both immunizations as well as antibody screening and validation (93). VLPs have been successfully used for both vaccination and antibody production for diagnostic purposes. For example, neuraminidase containing N1 VLP derived from the H1N1 influenza virus, when immunized in mice, induced virus-specific antibodies as well as reduced NA inhibition activity (94). The E protein of the Zika virus (specifically domain ED-II), the E2 protein of the chikungunya virus, and the L1 or L2 proteins of the human papillomavirus are utilized as antigens in VLP vaccines (95, 96).
4.3 Phage emulsion technique
The use of bacteriophages, viruses that infect bacteria, as direct immunogens are very well understood, but their use as a tool to generate antibodies indirectly acting as a carrier is also crucial. The phage display technique is based on genetically altering the phage DNA to allow the expression of a segment of peptide or protein on the phage surface (97). The DNA sequence of the protein of interest is inserted near the nucleotide sequence, which encodes for one of the phage coat proteins. Following phage infection, the inserted peptide or protein is displayed on the phage surface as a combined product of the genes generating the coat protein and the cloned protein (Figure 4C) (98).
The phage emulsion technique is a well-adapted version of the phage display technique for antibody generation. This method involves integrating a protein, peptide, or even synthetic epitope on the surface of bacteriophages and mixing them with an aqueous-organic phase system to form a stable emulsion. The bacteriophage emulsion complex is further used for immunization. The bacteriophage particles are effective carriers that closely resemble natural infections by displaying the associated antigens in a 3D conformation (99). The immune system perceives the phages as foreign particles and immediately starts an, immune response against the antigen they display. The emulsification procedure is crucial to create a stable environment for antigen and bacteriophage association. Thus, the antigen’s exposure to the immune system is prolonged, leading to a robust immune response and antibody generation (100). This method offers a strong foundation for creating new immunogens and vaccines and studying the relationships between specific antigens and the immune system (101). Phage micro-emulsion technology provides many essential benefits over existing phage-display techniques. One benefit is that each clone’s signal is enhanced because all phage copies are contained in a single droplet (102). However, there are still many challenges to the phage emulsion technique, such as difficulty in emulsion formation utilizing complex MPs as targets, high cost, etc. (103), which future research is expected to address. The filamentous phages are generally employed in the phage emulsion technique, but recently, various other bacteriophages are being modified, such as lytic phages, which can display more prominent antigens with their ability to carry larger foreign DNA fragments (104).
4.4 Exosomes
Exosomes are extracellular vesicles that carry a variety of cellular components, including DNA, RNA, MP, lipids, and both cytosolic and cell-surface proteins. Membranous nanovesicles, typically 50-100 nm in diameter, form within late endosomal compartments through the invagination of multivesicular bodies (MVB) membranes. The endosomal membranes involved in their formation are known as intraluminal vesicles (ILVs). Mature MVBs undergo exocytosis to release ILVs as exosomes into the extracellular environment after fusing with the plasma membrane (105, 106). As a result of this formation process, exosomes have a membrane orientation similar to the native plasma membrane, with ECDs facing outward and cytosolic proteins, along with small RNAs, enclosed within the lumen. After their secretion, exosomes protect their cargo and deliver it to the recipient cells by endocytosis or fusion without compromising the intrinsic function of the cargo (107). These exosomes involve cell-to-cell communication, immune response modulation, and intercellular signaling and are of high pathophysiological relevance in various diseases, including cancer (108, 109).
Exosomes offer several advantages as immunogens in anti-MP antibody production, including the lack of need for sequence modification, preservation of the native membrane environment, high intrinsic immunogenicity that elicits strong immune responses, and excellent stability for both short and long term storage. Additionally, exosomes do not require adjuvants and exhibit minimal cytotoxicity. Intracellular exosomes can be isolated and used to deliver proteins and RNA from other cells into ex vivo cell cultures. Exosomes can be purified from in vitro cultures of various cell types and can be loaded with a diverse range of biological molecules, including cytosolic proteins, membrane receptors, and nucleic acids (110).
MP bearing exosomes have successfully contributed to the development of numerous anti-GPCR antibodies (111, 112). Notably, exosomes secreted from dendritic cells, known as dexosomes, demonstrate exceptional immune capabilities in vivo (113). An extracellular vesicle-based technology enables the selective recruitment of viral membrane antigens containing WW domains onto the surface of WW domain–activated extracellular vesicles (WAEVs). By fusing viral MPs or peptides to WW domains, MP antigens can be efficiently displayed on the surface of WAEVs, eliciting robust antibody production that explicitly targets the corresponding viruses. This approach likely preserves the natural conformation of MPs, as specific antibodies, including neutralizing antibodies, recognize them (114). A newer approach in membrane engineering involves hybrid exosomes formed by merging exosome membranes with liposomes through the freeze-thaw method (115). The exosome liposome fusion technology is promising for loading therapeutic compounds into exosomes, making it easier for MPs to be incorporated into the exosomes. However, limitations of using exosomes for anti-MP antibody production include limited yield, purity issues, heterogeneity, complex production processes, and potential regulatory challenges.
4.5 Genetic immunization
The genetic immunization approach, involving DNA and mRNA immunization, relies on expression vectors or utilizes synthetic mRNA sequences to produce encoding the target immunogen peptides or proteins directly within the host. These expression vectors or mRNA are typically delivered to the host intradermally through various methods such as injection of naked DNA or mRNA using a simple needle or biolistic delivery of DNA-coated particles into dermal cells, transdermal patches, and electroporation-based and viral-based methods (116–118). Each of these studies used DNA of mRNA sequences encoding for soluble proteins or the soluble domains of membrane-anchored proteins (Figure 5). Genetic immunization overcomes the problem associated with peptide or protein-based immunization for antibody production against MPs because the gene-immunized host naturally produces, folds, and alters the MP antigen in its natural membrane context.
Figure 5. DNA-based immunization to generate membrane protein antibodies. DNA encoding membrane protein can be introduced directly into a host, allowing it to express the intact immunogens and generate antibodies against it.
One of the challenges of traditional protein-based immunization is the production of full-length MP immunogens in their native, membrane-associated form using recombinant protein techniques. The problem is even more significant, particularly for proteins like multispanning transmembrane proteins such as GPCRs and ion channels. DNA immunization overcomes these obstacles by enabling in vivo expression of full-length proteins, eliciting a targeted immune response against the MP by directly delivering DNA to cells, and inducing the formation of the desired proteins, effectively turning the cells into a source of antigen production to stimulate an immune response (118). DNA-based immunization for SARS-CoV-2 envelope and MPs in mice, provided protection against the disease (119). DNA expression vectors coated with gold particles injected into shaved and depilated skin cells of camelid using gene gun delivery methods expressed MP on transfected cells and led to the generation of nanobodies targeting MPs (120). RNA immunization reduces the risk of DNA integration into the host genome by directly delivering the target protein to the cytosol, improving the safety profile of nucleic acid, and enabling quicker immune response. Recently, a self-amplifying RNA-based immunogen containing the additional sequences of nsP1-4 proteins allowed their self-replication within the host cells. This helps in achieving a robust immune response with smaller doses compared to conventional RNA vaccines (121). Taken together, DNA and RNA-based immunization strategies, have demonstrated significant potential in inducing immune responses against MP.
5 Conclusion
Producing functional anti-MP antibodies requires the preparation and immunization of the protein of interest in its native form. In this review, we discussed various strategies for preparing and immunizing MP so that antibodies produced can recognize both linear as well as conformational epitopes of MPs, for therapeutic purposes (Table 1). The choice of antigen format for antibody discovery is dependent on compatible solubilization and purification methods for the target of choice. Selecting an appropriate method for immunogen production is challenging, as each approach comes with distinct advantages and limitations and the downstream antibody discovery platform used. The formats used range from simple soluble regions of the protein, such as soluble ECDs or peptides, to full-length MPs purified in detergent or lipidic environments, to complex membranous environments, such as VLPs, exosomes, and whole cells. Although generating specific antibodies from peptide-based immunizations eliminates the need to produce full-length transmembrane protein, the linear structure of peptides restricts the establishment of conformational epitopes and makes it more difficult to find effective antibodies. These problems can be partially tackled by complementing peptide-based immunogen design with computational modeling and prediction tools. A more recent advancement in AI-based structure prediction tools, such as AlphaFold2, RoseTTAFold2, and ESMFold, has great potential to predict the 3D structure of proteins directly from the amino acid sequence for more precise identification of immunogenic areas that can be used as antigens. Further advancements in AI-based tools make de novo protein design, epitope, and paratope identification, soluble analogous of MP, and native conformation analysis a realistic vision.
Biophysical techniques like X-ray crystallography, single-particle cryo-electron microscopy, and receptor-ligand binding assays are used to determine 3D structures. However, these techniques do not work in the native environment; therefore, the protein must be separated from the membrane and examined in vitro in a lipid or detergent environment. MP extracted through traditional methods such as detergents may destabilize MP as well as mask key extracellular loops of MPs and their dissociation from the membrane. Novel technologies like amphiols, nanodiscs, and SMALPS help overcome these limitations by replicating the composition of plasma membranes. While nanoparticle-based technologies have been widely used in MP structure elucidation (133), their role in the immunization of MPs remains limited. Despite their success in structural studies, the limited use of nanoparticle-based technologies for immunization is largely due to stability issues post-immunization. Nanoparticle-based formulations may also sometimes induce aggregation or alter the native conformation of MPs, affecting both immunogenicity and specificity. Additionally, maintaining the structural and functional characteristics of MPs throughout the MP reconstitution process is still very challenging. The variability incorporated during immunogen preparation can affect reproducibility in immunogen design. In order to minimize conformational changes, aggregation, and unpredictability, it is imperative to develop better membrane-mimetic systems that more precisely mimic the native lipid environment of MPs. Cryo-electron microscopy and other advanced characterization techniques are crucial for evaluating the structural consistency and integrity of MPs in different formulations. The development of innovative adjuvants tailored for MP immunization, as well as scalable and reproducible production techniques, will help ensure reliable antibody production against MPs.
Overexpressed MPs can be utilized for immunization either as whole cells expressing MPs or as MP-containing VLPs and exosomes produced by cells. Although VLPs evoke powerful immune responses and are more robust than whole-cell immunization, allowing fewer non-relevant targets on the surface, they can be expensive and time-consuming to produce. While exosomes can deliver MP in native form with a topology similar to the native plasma membrane, they face challenges such as heterogeneity, complex isolation processes, and limited scalability. Cell-free expression systems has emerged as powerful techniques for the expression of MP. Recent developments in detergent-free modified cellular lysates enable cell-free expression of target proteins, which can be coupled with direct reconstitution of newly synthesized proteins into membrane vesicles such as liposomes. This approach avoids common challenges such as poor membrane insertion, precipitation of newly synthesized proteins, or cytotoxic effects caused by excessive strain on the host cell’s metabolism. The chosen, optimized antigen formats can then be employed to drive antibody discovery through the appropriate use of strategies that leverage display technologies, B cell platforms, hybridoma, or a combination of these approaches to guarantee the isolation of a diverse panel of antibodies.
The effort required to create purified proteins or native membrane structures to generate antibodies can be bypassed by genetically immunizing the host organism. The SARS-CoV-2 pandemic demonstrated the effectiveness of RNA-based vaccines developed in a significantly shorter timeframe. However, challenges remain in ensuring that the expressed protein is properly processed, presented, and elicits a strong immune response. Future improvements in RNA delivery systems, such as more efficient lipid nanoparticles, and improvements in RNA synthesis and delivery technologies, are needed to make this approach for broader implementation. Further, the development of a well-optimized standard protocol to select the optimal antigen format and antibody generation platform for specific targets is also necessary. In addition, developing better strategies for screening membrane antibodies is another critical area in enhancing the production of specific antibodies. These advancements will pave the way for more efficient, accurate, and scalable antibody-generation processes, particularly for challenging targets like MPs.
Author contributions
AM: Visualization, Writing – original draft, Writing – review & editing. NF: Writing – original draft, Writing – review & editing. AF: Visualization, Writing – original draft, Writing – review & editing. MP: Writing – original draft, Writing – review & editing. MK: Writing – review & editing. AH: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We sincerely apologize to the numerous authors whose valuable studies we could not discuss due to space limitations. We would like to acknowledge the support and facilities provided by the Department of Biochemistry, Faculty of Life Sciences, AMU, Aligarh.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Marshall JS, Warrington R, Watson W, Kim HL. An introduction to immunology and immunopathology. Allergy Asthma Clin Immunol. (2018) 14:49. doi: 10.1186/s13223-018-0278-1
2. Honjo T, Reth M, Radbruch A, Alt F, Neuberger M. Molecular biology of B cells. ScienceDirect (2004). Available online at: https://www.sciencedirect.com/book/9780323958950/molecular-biology-of-b-cells.
3. Ahmad TA, Eweida AE, Sheweita SAJT. B-cell epitope mapping for the design of vaccines and effective diagnostics. Trials in Vaccinology (2016) 5:71–83. doi: 10.1016/j.trivac.2016.04.003
4. Biavasco R, De Giovanni M. The relative positioning of B and T cell epitopes drives immunodominance. Vaccines (Basel). (2022) 10. doi: 10.3390/vaccines10081227
5. Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Fundamentals and methods for T- and B-cell epitope prediction. J Immunol Res. (2017) 2017:2680160. doi: 10.1155/2017/2680160
6. Van Regenmortel MH. What is a B-cell epitope? Methods Mol Biol. (2009) 524:3–20. doi: 10.1007/978-1-59745-450-6_1
7. Salk JE. Studies in human subjects on active immunization against poliomyelitis. I. A preliminary report of experiments in progress. J Am Med Assoc. (1953) 151:1081–98. doi: 10.1001/jama.1953.13.1081
8. Burnett DL, Schofield P, Langley DB, Jackson J, Bourne K, Wilson E, et al. Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens. Proc Natl Acad Sci U.S.A. (2020) 117:22341–50. doi: 10.1073/pnas.2005102117
9. Douthwaite JA, Finch DK, Mustelin T, Wilkinson TC. Development of therapeutic antibodies to G protein-coupled receptors and ion channels: Opportunities, challenges and their therapeutic potential in respiratory diseases. Pharmacol Ther. (2017) 169:113–23. doi: 10.1016/j.pharmthera.2016.04.013
10. Yin H, Flynn AD. Drugging membrane protein interactions. Annu Rev BioMed Eng. (2016) 18:51–76. doi: 10.1146/annurev-bioeng-092115-025322
11. Li ZL, Buck M. Beyond history and on a roll: The list of the most well-studied human protein structures and overall trends in the protein data bank. Protein Sci. (2021) 30:745–60. doi: 10.1002/pro.v30.4
12. Jelokhani-Niaraki M. Membrane proteins: structure, function and motion. Int J Mol Sci. (2022) 24. doi: 10.3390/ijms24010468
13. Kubota Y, Shitara K. Zolbetuximab for Claudin18.2-positive gastric or gastroesophageal junction cancer. Ther Adv Med Oncol. (2024) 16:17588359231217967. doi: 10.1177/17588359231217967
14. Errey JC, Fiez-Vandal C. Production of membrane proteins in industry: The example of GPCRs. Protein Expr Purif. (2020) 169:105569. doi: 10.1016/j.pep.2020.105569
15. Stephens AD, Wilkinson T. Discovery of therapeutic antibodies targeting complex multi-spanning membrane proteins. BioDrugs. (2024) 38:769–94. doi: 10.1007/s40259-024-00682-1
16. Pandey A, Shin K, Patterson RE, Liu XQ, Rainey JK. Current strategies for protein production and purification enabling membrane protein structural biology. Biochem Cell Biol. (2016) 94:507–27. doi: 10.1139/bcb-2015-0143
17. McCusker EC, Bane SE, O'Malley MA, Robinson AS. Heterologous GPCR expression: a bottleneck to obtaining crystal structures. Biotechnol Prog. (2007) 23:540–7. doi: 10.1021/(ISSN)1520-6033
18. Hudziak RM, Lewis GD, Winget M, Fendly BM, Shepard HM, Ullrich A. p185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitizes human breast tumor cells to tumor necrosis factor. Mol Cell Biol. (1989) 9:1165–72. doi: 10.1128/mcb.9.3.1165-1172.1989
19. Hansen KB, Wollmuth LP, Bowie D, Furukawa H, Menniti FS, Sobolevsky AI, et al. Structure, function, and pharmacology of glutamate receptor ion channels. Pharmacol Rev. (2021) 73:298–487. doi: 10.1124/pharmrev.120.000131
20. Wagner A, Galicia-Andrés E, Teufl M, Gold L, Obinger C, Sykacek P, et al. Identification of activating mutations in the transmembrane and extracellular domains of EGFR. Biochemistry. (2022) 61:2049–62. doi: 10.1021/acs.biochem.2c00384
21. Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. (2007) 318:1608–12. doi: 10.1126/science.1150755
22. Lu P, Bai XC, Ma D, Xie T, Yan C, Sun L, et al. Three-dimensional structure of human γ-secretase. Nature. (2014) 512:166–70. doi: 10.1038/nature13567
23. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. (20217873) 596:583–9. doi: 10.1038/s41586-021-03819-2
24. Errasti-Murugarren E, Bartoccioni P, Palacín M. Membrane protein stabilization strategies for structural and functional studies. Membranes (Basel). (2021) 11. doi: 10.3390/membranes11020155
25. Gurdap CO, Wedemann L, Sych T, Sezgin E. Influence of the extracellular domain size on the dynamic behavior of membrane proteins. Biophys J. (2022) 121:3826–36. doi: 10.1016/j.bpj.2022.09.010
26. Krishnarjuna B, Ramamoorthy A. Detergent-free isolation of membrane proteins and strategies to study them in a near-native membrane environment. Biomolecules. (2022) 12. doi: 10.3390/biom12081076
27. Vitrac H, Mallampalli V, Bogdanov M, Dowhan W. The lipid-dependent structure and function of LacY can be recapitulated and analyzed in phospholipid-containing detergent micelles. Sci Rep. (2019) 9:11338. doi: 10.1038/s41598-019-47824-y
28. Henrich E, Peetz O, Hein C, Laguerre A, Hoffmann B, Hoffmann J, et al. Analyzing native membrane protein assembly in nanodiscs by combined non-covalent mass spectrometry and synthetic biology. Elife. (2017) 6. doi: 10.7554/eLife.20954
29. Anandan A, Vrielink A. Detergents in membrane protein purification and crystallisation. Adv Exp Med Biol. (2016) 922:13–28. doi: 10.1007/978-3-319-35072-1_2
30. Chae PS, Rasmussen SG, Rana RR, Gotfryd K, Chandra R, Goren MA, et al. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods. (2010) 7:1003–8. doi: 10.1038/nmeth.1526
31. Robertson N, Jazayeri A, Errey J, Baig A, Hurrell E, Zhukov A, et al. The properties of thermostabilised G protein-coupled receptors (StaRs) and their use in drug discovery. Neuropharmacology. (2011) 60:36–44. doi: 10.1016/j.neuropharm.2010.07.001
32. Hutchings CJ, Cseke G, Osborne G, Woolard J, Zhukov A, Koglin M, et al. Monoclonal anti-β1-adrenergic receptor antibodies activate G protein signaling in the absence of β-arrestin recruitment. MAbs. (2014) 6:246–61. doi: 10.4161/mabs.27226
33. Mirzabekov T, Kontos H, Farzan M, Marasco W, Sodroski J. Paramagnetic proteoliposomes containing a pure, native, and oriented seven-transmembrane segment protein, CCR5. Nat Biotechnol. (2000) 18:649–54. doi: 10.1038/76501
34. Jesorka A, Orwar O. Liposomes: technologies and analytical applications. Annu Rev Anal Chem (Palo Alto Calif). (2008) 1:801–32. doi: 10.1146/annurev.anchem.1.031207.112747
35. Suharni Y, Arakawa T, Hino T, Abe H, Nakada-Nakura Y, Sato Y, et al. Proteoliposome-based selection of a recombinant antibody fragment against the human M2 muscarinic acetylcholine receptor. Monoclon Antib Immunodiagn Immunother. (2014) 33:378–85. doi: 10.1089/mab.2014.0041
36. Zhou W, Takeda H. Production of immunizing antigen proteoliposome using cell-free protein synthesis system. Methods Mol Biol. (2018) 1868:49–67. doi: 10.1007/978-1-0716-3682-4_9
37. Hashimoto Y, Zhou W, Hamauchi K, Shirakura K, Doi T, Yagi K, et al. Engineered membrane protein antigens successfully induce antibodies against extracellular regions of claudin-5. Sci Rep. (2018) 8:8383. doi: 10.1038/s41598-018-26560-9
38. Dominik PK, Borowska MT, Dalmas O, Kim SS, Perozo E, Keenan RJ, et al. Conformational chaperones for structural studies of membrane proteins using antibody phage display with nanodiscs. Structure. (2016) 24:300–9. doi: 10.1016/j.str.2015.11.014
39. Zhang S, Ren Q, Novick SJ, Strutzenberg TS, Griffin PR, Bao H. One-step construction of circularized nanodiscs using SpyCatcher-SpyTag. Nat Commun. (2021) 12:5451. doi: 10.1038/s41467-021-25737-7
40. Ju MS, Ahn HM, Han SG, Ko S, Na JH, Jo M, et al. A human antibody against human endothelin receptor type A that exhibits antitumor potency. Exp Mol Med. (2021) 53:1437–48. doi: 10.1038/s12276-021-00678-9
41. Carlson ML, Young JW, Zhao Z, Fabre L, Jun D, Li J, et al. The Peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution. Elife. (2018) 7. doi: 10.7554/eLife.34085
42. Kariyazono H, Nadai R, Miyajima R, Takechi-Haraya Y, Baba T, Shigenaga A, et al. Formation of stable nanodiscs by bihelical apolipoprotein A-I mimetic peptide. J Pept Sci. (2016) 22:116–22. doi: 10.1002/psc.v22.2
43. Lyons JA, Bøggild A, Nissen P, Frauenfeld J. Saposin-lipoprotein scaffolds for structure determination of membrane transporters. Methods Enzymol. (2017) 594:85–99. doi: 10.1016/bs.mie.2017.06.035
44. Kanonenberg K, Smits SHJ, Schmitt L. Functional reconstitution of hlyB, a type I secretion ABC transporter, in saposin-A nanoparticles. Sci Rep. (2019) 9:8436. doi: 10.1038/s41598-019-44812-0
45. Reff ME, Carner K, Chambers KS, Chinn PC, Leonard JE, Raab R, et al. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood. (1994) 83:435–45. doi: 10.1182/blood.V83.2.435.435
46. Frauenfeld J, Löving R, Armache JP, Sonnen AF, Guettou F, Moberg P, et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods. (2016) 13:345–51. doi: 10.1038/nmeth.3801
47. Sahu ID, Dixit G, Reynolds WD, Kaplevatsky R, Harding BD, Jaycox CK, et al. Characterization of the human KCNQ1 voltage sensing domain (VSD) in lipodisq nanoparticles for electron paramagnetic resonance (EPR) spectroscopic studies of membrane proteins. J Phys Chem B. (2020) 124:2331–42. doi: 10.1021/acs.jpcb.9b11506
48. Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EA, et al. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc Natl Acad Sci U.S.A. (2014) 111:18607–12. doi: 10.1073/pnas.1416205112
49. Velappan N, Micheva-Viteva S, Adikari SH, Waldo GS, Lillo AM, Bradbury ARM. Selection and verification of antibodies against the cytoplasmic domain of M2 of influenza, a transmembrane protein. MAbs. (2020) 12:1843754. doi: 10.1080/19420862.2020.1843754
50. Clark JC, Martin EM, Morán LA, Di Y, Wang X, Zuidscherwoude M, et al. Divalent nanobodies to platelet CLEC-2 can serve as agonists or antagonists. Commun Biol. (2023) 6:376. doi: 10.1038/s42003-023-04766-6
51. Sasaki Y, Kosaka H, Usami K, Toki H, Kawai H, Shiraishi N, et al. Establishment of a novel monoclonal antibody against LGR5. Biochem Biophys Res Commun. (2010) 394:498–502. doi: 10.1016/j.bbrc.2010.02.166
52. Cox JH, Hussell S, Søndergaard H, Roepstorff K, Bui JV, Deer JR, et al. Antibody-mediated targeting of the Orai1 calcium channel inhibits T cell function. PloS One. (2013) 8:e82944. doi: 10.1371/journal.pone.0082944
53. Dodd RB, Wilkinson T, Schofield DJ. Therapeutic monoclonal antibodies to complex membrane protein targets: antigen generation and antibody discovery strategies. BioDrugs. (2018) 32:339–55. doi: 10.1007/s40259-018-0289-y
54. Zhang Y, Pool C, Sadler K, Yan HP, Edl J, Wang X, et al. Selection of active ScFv to G-protein-coupled receptor CCR5 using surface antigen-mimicking peptides. Biochemistry. (2004) 43:12575–84. doi: 10.1021/bi0492152
55. Richelle GJJ, Ori S, Hiemstra H, van Maarseveen JH, Timmerman P. General and facile route to isomerically pure tricyclic peptides based on templated tandem CLIPS/cuAAC cyclizations. Angew Chem Int Ed Engl. (2018) 57:501–5. doi: 10.1002/anie.201709127
56. van Els CA, Corbière V, Smits K, van-Gaans-van-den-Brink JA, Poelen MC, Mascart F, et al. Toward understanding the essence of post-translational modifications for the mycobacterium tuberculosis immunoproteome. Front Immunol. (2014) 5:361. doi: 10.3389/fimmu.2014.00361
57. Kar T, Narsaria U, Basak S, Deb D, Castiglione F, Mueller DM, et al. A candidate multi-epitope vaccine against SARS-CoV-2. Sci Rep. (2020) 10:10895. doi: 10.1038/s41598-020-67749-1
58. Gómara MJ, Riedemann S, Vega I, Ibarra H, Ercilla G, Haro I. Use of linear and multiple antigenic peptides in the immunodiagnosis of acute hepatitis A virus infection. J Immunol Methods. (2000) 234:23–34. doi: 10.1016/S0022-1759(99)00196-9
59. Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. (1999) 112:531–52. doi: 10.1385/1-59259-584-7:531
60. Rahman N, Ali F, Basharat Z, Shehroz M, Khan MK, Jeandet P, et al. Vaccine design from the ensemble of surface glycoprotein epitopes of SARS-coV-2: an immunoinformatics approach. Vaccines (Basel). (2020) 8. doi: 10.3390/vaccines8030423
61. Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. (2007) 450:663–9. doi: 10.1038/nature06384
62. Guarra F, Colombo G. Computational methods in immunology and vaccinology: design and development of antibodies and immunogens. J Chem Theory Comput. (2023) 19:5315–33. doi: 10.1021/acs.jctc.3c00513
63. Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. (2007) 8:4. doi: 10.1186/1471-2105-8-4
64. Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP v.2–a server for in silico prediction of allergens. J Mol Model. (2014) 20:2278. doi: 10.1007/s00894-014-2278-5
65. Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Raghava GP. Peptide toxicity prediction. Methods Mol Biol. (2015) 1268:143–57. doi: 10.1007/978-1-4939-2285-7_7
66. Omidi A, Møller MH, Malhis N, Bui JM, Gsponer J. AlphaFold-Multimer accurately captures interactions and dynamics of intrinsically disordered protein regions. Proc Natl Acad Sci U.S.A. (2024) 121:e2406407121. doi: 10.1073/pnas.2406407121
67. Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. (2022) 50:D439–d444. doi: 10.1093/nar/gkab1061
68. Varadi M, Bertoni D, Magana P, Paramval U, Pidruchna I, Radhakrishnan M, et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. (2024) 52:D368–d375. doi: 10.1093/nar/gkad1011
69. Zeng Y, Wei Z, Yuan Q, Chen S, Yu W, Lu Y, et al. Identifying B-cell epitopes using AlphaFold2 predicted structures and pretrained language model. Bioinformatics. (2023) 39. doi: 10.1093/bioinformatics/btad187
70. Nguyen PT, Harris BJ, Mateos DL, González AH, Murray AM, Yarov-Yarovoy V. Structural modeling of ion channels using AlphaFold2, RoseTTAFold2, and ESMFold. Channels (Austin). (2024) 18:2325032. doi: 10.1080/19336950.2024.2325032
71. Singh A, Thakur M, Sharma LK, Chandra K. Designing a multi-epitope peptide based vaccine against SARS-CoV-2. Sci Rep. (2020) 10:16219. doi: 10.1038/s41598-020-73371-y
72. Bibi S, Ullah I, Zhu B, Adnan M, Liaqat R, Kong WB, et al. In silico analysis of epitope-based vaccine candidate against tuberculosis using reverse vaccinology. Sci Rep. (2021) 11:1249. doi: 10.1038/s41598-020-80899-6
73. Sharma R, Rajput VS, Jamal S, Grover A, Grover S. Author Correction: An immunoinformatics approach to design a multi-epitope vaccine against Mycobacterium tuberculosis exploiting secreted exosome proteins. Sci Rep. (2021) 11:16844. doi: 10.1038/s41598-021-96314-7
74. Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. (2020) 9:382–5. doi: 10.1080/22221751.2020.1729069
75. Mitra D, Pandey J, Jain A, Swaroop S. In silico design of multi-epitope-based peptide vaccine against SARS-CoV-2 using its spike protein. J Biomol Struct Dyn. (2022) 40:5189–202. doi: 10.1080/07391102.2020.1869092
76. Bryant P, Pozzati G, Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun. (2022) 13:1265. doi: 10.1038/s41467-022-28865-w
77. Liu J, Guo Z, Wu T, Roy RS, Quadir F, Chen C, et al. Enhancing alphafold-multimer-based protein complex structure prediction with MULTICOM in CASP15. Commun Biol. (2023) 6:1140. doi: 10.1038/s42003-023-05525-3
78. Gonzalez-Garcia M, Fusco G, De Simone A. Membrane interactions and toxicity by misfolded protein oligomers. Front Cell Dev Biol. (2021) 9:642623. doi: 10.3389/fcell.2021.642623
79. Li LH, Shivakumar R, Feller S, Allen C, Weiss JM, Dzekunov S, et al. Highly efficient, large volume flow electroporation. Technol Cancer Res Treat. (2002) 1:341–50. doi: 10.1177/153303460200100504
80. Kim TK, Eberwine JH. Mammalian cell transfection: the present and the future. Anal Bioanal Chem. (2010) 397:3173–8. doi: 10.1007/s00216-010-3821-6
81. Lin FF, Elliott R, Colombero A, Gaida K, Kelley L, Moksa A, et al. Generation and characterization of fully human monoclonal antibodies against human Orai1 for autoimmune disease. J Pharmacol Exp Ther. (2013) 345:225–38. doi: 10.1124/jpet.112.202788
82. Buell G, Chessell IP, Michel AD, Collo G, Salazzo M, Herren S, et al. Blockade of human P2X7 receptor function with a monoclonal antibody. Blood. (1998) 92:3521–8. doi: 10.1182/blood.V92.10.3521
83. Batah AM, Ahmad TA. The development of ghost vaccines trials. Expert Rev Vaccines. (2020) 19:549–62. doi: 10.1080/14760584.2020.1777862
84. Ahmad TA, Haroun M, Hussein AA, El Ashry el SH, El-Sayed LH. Development of a new trend conjugate vaccine for the prevention of Klebsiella pneumoniae. Infect Dis Rep. (2012) 4:e33. doi: 10.4081/idr.2012.e33
85. Schiedner G, Hertel S, Bialek C, Kewes H, Waschütza G, Volpers C. Efficient and reproducible generation of high-expressing, stable human cell lines without need for antibiotic selection. BMC Biotechnol. (2008) 8:13. doi: 10.1186/1472-6750-8-13
86. Chaudhary S, Pak JE, Pedersen BP, Bang LJ, Zhang LB, Ngaw SM, et al. Efficient expression screening of human membrane proteins in transiently transfected Human Embryonic Kidney 293S cells. Methods. (2011) 55:273–80. doi: 10.1016/j.ymeth.2011.08.018
87. Willis S, Davidoff C, Schilling J, Wanless A, Doranz BJ, Rucker J. Virus-like particles as quantitative probes of membrane protein interactions. Biochemistry. (2008) 47:6988–90. doi: 10.1021/bi800540b
88. Jones JW, Greene TA, Grygon CA, Doranz BJ, Brown MP. Cell-free assay of G-protein-coupled receptors using fluorescence polarization. J Biomol Screen. (2008) 13:424–9. doi: 10.1177/1087057108318332
89. Mohsen MO, Gomes AC, Vogel M, Bachmann MF. Interaction of viral capsid-derived virus-like particles (VLPs) with the innate immune system. Vaccines (Basel). (2018) 6. doi: 10.3390/vaccines6030037
90. Endres MJ, Jaffer S, Haggarty B, Turner JD, Doranz BJ, O'Brien PJ, et al. Targeting of HIV- and SIV-infected cells by CD4-chemokine receptor pseudotypes. Science. (1997) 278:1462–4. doi: 10.1126/science.278.5342.1462
91. Huang R, Kiss MM, Batonick M, Weiner MP, Kay BK. Generating recombinant antibodies to membrane proteins through phage display. Antibodies (Basel). (2016) 5. doi: 10.3390/antib5020011
92. Caldeira JC, Peabody DS. Stability and assembly in vitro of bacteriophage PP7 virus-like particles. J Nanobiotechnology. (2007) 5:10. doi: 10.1186/1477-3155-5-10
93. Lynch A, Meyers AE, Williamson AL, Rybicki EP. Stability studies of HIV-1 Pr55gag virus-like particles made in insect cells after storage in various formulation media. Virol J. (2012) 9:210. doi: 10.1186/1743-422X-9-210
94. Kim KH, Lee YT, Park S, Jung YJ, Lee Y, Ko EJ, et al. Neuraminidase expressing virus-like particle vaccine provides effective cross protection against influenza virus. Virology. (2019) 535:179–88. doi: 10.1016/j.virol.2019.07.008
95. Boigard H, Alimova A, Martin GR, Katz A, Gottlieb P, Galarza JM. Zika virus-like particle (VLP) based vaccine. PloS Negl Trop Dis. (2017) 11:e0005608. doi: 10.1371/journal.pntd.0005608
96. Yadav R, Zhai L, Tumban E. Virus-like particle-based L2 vaccines against HPVs: where are we today? Viruses. (2019) 12. doi: 10.3390/v12010018
97. Sioud M. Phage display libraries: from binders to targeted drug delivery and human therapeutics. Mol Biotechnol. (2019) 61:286–303. doi: 10.1007/s12033-019-00156-8
98. Aghebati-Maleki L, Bakhshinejad B, Baradaran B, Motallebnezhad M, Aghebati-Maleki A, Nickho H, et al. Phage display as a promising approach for vaccine development. J BioMed Sci. (2016) 23:66. doi: 10.1186/s12929-016-0285-9
99. Prisco A, De Berardinis P. Filamentous bacteriophage fd as an antigen delivery system in vaccination. Int J Mol Sci. (2012) 13:5179–94. doi: 10.3390/ijms13045179
100. Puapermpoonsiri U, Spencer J, van der Walle CF. A freeze-dried formulation of bacteriophage encapsulated in biodegradable microspheres. Eur J Pharm Biopharm. (2009) 72:26–33. doi: 10.1016/j.ejpb.2008.12.001
101. Kiss MM, Babineau EG, Bonatsakis M, Buhr DL, Maksymiuk GM, Wang D, et al. Phage ESCape: an emulsion-based approach for the selection of recombinant phage display antibodies. J Immunol Methods. (2011) 367:17–26. doi: 10.1016/j.jim.2010.09.034
102. Wang J, Tan Y, Ling J, Zhang M, Li L, Liu W, et al. Highly paralleled emulsion droplets for efficient isolation, amplification, and screening of cancer biomarker binding phages. Lab Chip. (2021) 21:1175–84. doi: 10.1039/D0LC01146K
103. Malik DJ, Sokolov IJ, Vinner GK, Mancuso F, Cinquerrui S, Vladisavljevic GT, et al. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv Colloid Interface Sci. (2017) 249:100–33. doi: 10.1016/j.cis.2017.05.014
104. Jaroszewicz W, Morcinek-Orłowska J, Pierzynowska K, Gaffke L, Węgrzyn G. Phage display and other peptide display technologies. FEMS Microbiol Rev. (2022) 46. doi: 10.1093/femsre/fuab052
105. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. (2020) 367. doi: 10.1126/science.aau6977
106. Tenchov R, Sasso JM, Wang X, Liaw WS, Chen CA, Zhou QA. Exosomes─Nature's lipid nanoparticles, a rising star in drug delivery and diagnostics. ACS Nano. (2022) 16:17802–46. doi: 10.1021/acsnano.2c08774
107. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. (2014) 3. doi: 10.3402/jev.v3.24641
108. Salunkhe S, Dheeraj M, Chitkara BD, Mittal A. Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: Strategies and significance. J Control Release. (2020) 326:599–614. doi: 10.1016/j.jconrel.2020.07.042
109. Xu F, Luo S, Lu P, Cai C, Li W, Li C. Composition, functions, and applications of exosomal membrane proteins. Front Immunol. (2024) 15:1408415. doi: 10.3389/fimmu.2024.1408415
110. El Safadi D, Mokhtari A, Krejbich M, Lagrave A, Hirigoyen U, Lebeau G, et al. Exosome-mediated antigen delivery: unveiling novel strategies in viral infection control and vaccine design. Vaccines (Basel). (2024) 12. doi: 10.3390/vaccines12030280
111. Chaput N, Théry C. Exosomes: immune properties and potential clinical implementations. Semin Immunopathol. (2011) 33:419–40. doi: 10.1007/s00281-010-0233-9
112. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. (2019) 18:75. doi: 10.1186/s12943-019-0991-5
113. Pitt JM, André F, Amigorena S, Soria JC, Eggermont A, Kroemer G, et al. Dendritic cell-derived exosomes for cancer therapy. J Clin Invest. (2016) 126:1224–32. doi: 10.1172/JCI81137
114. Choi S, Yang Z, Wang Q, Qiao Z, Sun M, Wiggins J, et al. Displaying and delivering viral membrane antigens via WW domain-activated extracellular vesicles. Sci Adv. (2023) 9:eade2708. doi: 10.1126/sciadv.ade2708
115. Sato YT, Umezaki K, Sawada S, Mukai SA, Sasaki Y, Harada N, et al. Engineering hybrid exosomes by membrane fusion with liposomes. Sci Rep. (2016) 6:21933. doi: 10.1038/srep21933
116. Bergmann-Leitner ES, Leitner WW. Vaccination Using Gene-Gun Technology. Methods Mol Biol. (2015) 1325:289–302. doi: 10.1007/978-1-4939-2815-6_22
117. Peyrassol X, Laeremans T, Gouwy M, Lahura V, Debulpaep M, Van Damme J, et al. Development by Genetic Immunization of Monovalent Antibodies (Nanobodies) Behaving as Antagonists of the Human ChemR23 Receptor. J Immunol. (2016) 196(6):2893–901. doi: 10.4049/jimmunol.1500888
118. Hansen DT, Craciunescu FM, Fromme P, Johnston SA, Sykes KF. Generation of high-specificity antibodies against membrane proteins using DNA-gold micronanoplexes for gene gun immunization. Curr Protoc Protein Sci. (2018) 91:29.20.21–29.20.22. doi: 10.1002/0471140864.2018.91.issue-1
119. Chen J, Deng Y, Huang B, Han D, Wang W, Huang M, et al. DNA vaccines expressing the envelope and membrane proteins provide partial protection against SARS-coV-2 in mice. Front Immunol. (2022) 13:827605. doi: 10.3389/fimmu.2022.827605
120. Eden T, Menzel S, Wesolowski J, Bergmann P, Nissen M, Dubberke G, et al. A cDNA Immunization Strategy to Generate Nanobodies against Membrane Proteins in Native Conformation. Front Immunol. (2017) 8:1989. doi: 10.3389/fimmu.2017.01989
121. Bloom K, van den Berg F, Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. (2021) 28:117–29. doi: 10.1038/s41434-020-00204-y
122. Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, et al. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. (2011) 469:175–80. doi: 10.1038/nature09648
123. Ma Y, Ding Y, Song X, Ma X, Li X, Zhang N, et al. Structure-guided discovery of a single-domain antibody agonist against human apelin receptor. Sci Adv. (2020) 6:eaax7379. doi: 10.1126/sciadv.aax7379
124. Drulyte I, Gutgsell AR, Lloris-Garcerá P, Liss M, Geschwindner S, Radjainia M, et al. Direct cell extraction of membrane proteins for structure-function analysis. Sci Rep. (2023) 13:1420. doi: 10.1038/s41598-023-28455-w
125. Zhao J, Nussinov R, Wu WJ, Ma B. In silico methods in antibody design. Antibodies (Basel). (2018) 7. doi: 10.3390/antib7030022
126. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-coV-2. Cell Host Microbe. (2020) 27:671–680.e672. doi: 10.1016/j.chom.2020.03.002
127. Rijal P, Donnellan FR. A review of broadly protective monoclonal antibodies to treat Ebola virus disease. Curr Opin Virol. (2023) 61:101339. doi: 10.1016/j.coviro.2023.101339
128. Tucker DF, Sullivan JT, Mattia KA, Fisher CR, Barnes T, Mabila MN, et al. Isolation of state-dependent monoclonal antibodies against the 12-transmembrane domain glucose transporter 4 using virus-like particles. Proc Natl Acad Sci U.S.A. (2018) 115:E4990–e4999. doi: 10.1073/pnas.1716788115
129. Dooley K, McConnell RE, Xu K, Lewis ND, Haupt S, Youniss MR, et al. A versatile platform for generating engineered extracellular vesicles with defined therapeutic properties. Mol Ther. (2021) 29:1729–43. doi: 10.1016/j.ymthe.2021.01.020
130. Xu X, Liang Y, Li X, Ouyang K, Wang M, Cao T, et al. Exosome-mediated delivery of kartogenin for chondrogenesis of synovial fluid-derived mesenchymal stem cells and cartilage regeneration. Biomaterials. (2021) 269:120539. doi: 10.1016/j.biomaterials.2020.120539
131. Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterol 139(3). (2010) 953-964:964.e951–954. doi: 10.1053/j.gastro.2010.05.073
132. Takatsuka S, Sekiguchi A, Tokunaga M, Fujimoto A, Chiba J. Generation of a panel of monoclonal antibodies against atypical chemokine receptor CCX-CKR by DNA immunization. J Pharmacol Toxicol Methods. (2011) 63:250–7. doi: 10.1016/j.vascn.2010.12.003
Keywords: immunogen, membrane protein, antibody, lipid bilayer, native structure
Citation: Mahboob A, Fatma N, Faraz A, Pervez M, Khan MA and Husain A (2025) Advancements in the conservation of the conformational epitope of membrane protein immunogens. Front. Immunol. 16:1538871. doi: 10.3389/fimmu.2025.1538871
Received: 03 December 2024; Accepted: 03 February 2025;
Published: 28 February 2025.
Edited by:
Saad Tayyab, UCSI University, MalaysiaReviewed by:
Nayoon Jang, Seoul National University, Republic of KoreaAmit Verma, Jamia Millia Islamia, India
Copyright © 2025 Mahboob, Fatma, Faraz, Pervez, Khan and Husain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Afzal Husain, YWZ6YWwuYmNAYW11
 Aisha Mahboob
Aisha Mahboob Nishat Fatma
Nishat Fatma Ahmed Faraz
Ahmed Faraz Muntaha Pervez
Muntaha Pervez Mohammad Afeef Khan
Mohammad Afeef Khan Afzal Husain
Afzal Husain