- Department of Nephrology, Rheumatology, Endocrinology and Metabolism, Faculty of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, Okayama, Japan
Poly-ADP-ribosylation (PARylation) is a post-translational modification in which ADP-ribose is added to substrate proteins. PARylation is mediated by a superfamily of ADP-ribosyl transferases known as PARPs and influences a wide range of cellular functions, including genome integrity maintenance, and the regulation of proliferation and differentiation. We and others have recently reported that PARylation of SH3 domain-binding protein 2 (3BP2) plays a role in bone metabolism, immune system regulation, and cytokine production. Additionally, PARylation has recently gained attention as a target for cancer treatment. In this review, we provide an overview of PARylation, its involvement in several signaling pathways related to cancer immunity, and the potential of combination therapies with PARP inhibitors and immune checkpoint inhibitors.
Introduction
Poly-ADP-ribosylation (PARylation) is a post-translational modification in which ADP-ribose is added to substrate proteins. PARylation is mediated by a superfamily of ADP-ribosyl transferases known as PARPs and has a wide range of effects on cellular functions, including proliferation and differentiation. Additionally, PARylation-mediated post-transcriptional modifications have recently gained attention as targets for cancer treatment. In this review, we provide an overview of PARylation and its involvement in several signaling pathways related to cancer immunity. Lastly, we will discuss the relationship between PARylation and immune checkpoint inhibitors.
ADP-ribosylation
ADP-ribosylation is a reversible post-translational modification that is required for regulation of molecular interactions (1, 2). During ADP-ribosylation, nicotinamide adenine dinucleotide (NAD+) is consumed as a donor and split into ADP-ribose and nicotinamide (NAM), resulting in addition of ADP-ribose to a substrate (2, 3). The addition of a single ADP-ribose is referred to as mono-ADP ribosylation (MARylation). In contrast, the reaction that adds two or more ADP-ribose units or creates a branched structure is called poly-ADP-ribosylation (PARylation) (4, 5). PARylation primarily affects proteins, although previous reports have shown that it can also modify nucleic acids (6, 7).
ADP-ribosylation is mediated by the ADP-ribosyl transferase superfamily (ARTs), which comprises 23 families including diphtheria toxin-like ARTs (ARTDs) and cholera toxin-like ARTs (ARTCs). ARTDs consist of 17 members, referred to as PARPs (PARP1-PARP16), with PARP5A and PARP5B also called tankyrase 1 (TNKS1) and tankyrase 2 (TNKS2), respectively (2, 4, 5, 8, 9). Among the ARTD members, only PARP1, PARP2, TNKS1, and TNKS2 exhibit PARylation activity, and they are referred to as poly-ARTs, despite the name PARP historically being derived from poly ADP-ribose polymerase. In contrast, the other members (PARP3, PARP4, PARP6-12, PARP14-16) possess MARylation activity and are referred to as mono-ARTs. PARP13 is thought to be inactive due to a defect in its NAD+ binding residues (5, 10).
ADP-ribosylation by “writers” such as PARPs is recognized by “reader” proteins that contain specific modules or motifs, including macro domains, PAR binding zinc finger (PBZ) domains, WWE domains, and the PAR-binding motif (9, 11, 12). Ubiquitin E3 ligase ring finger protein 146 (RNF146) is a reader protein in which the WWE domain detects PARylation by binding to the iso-ADP-ribose moiety (13, 14). ADP-ribosylation is quickly terminated by the removal of ADP-ribose by “eraser” proteins such as poly(ADP-ribose) glycohydrolase (PARG), MacroD1, MacroD2, terminal ADP-ribose protein glycohydrolase 1 (TARG1), and the ADP-ribose hydrolase (ARH) members ARH1 and ARH3 (15–17).
PARylation regulates a wide range of molecular functions, including transcription, RNA regulation, mitosis, telomere length maintenance, cell-cycle regulation, cellular differentiation, DNA damage response, protein degradation, ubiquitination, metabolism, and innate and adaptive immunity, among many others (3, 15, 18–25) (Table 1).
PARylation modulates DNA damage response and transcription
DNA is constantly exposed to endogenous and exogenous damage, requiring frequent restoration to maintain genome integrity (87). PARP1, an abundant nuclear protein, plays a crucial role in the early phase of DNA damage response (DDR). When single-strand breaks (SSBs) are detected by PARP1 or PARP2, PARylation occurs on PARP1 (self-PARylation) or DDR-associated proteins (25, 46). PAR recruits the scaffold protein XRCC1 and its partner proteins, facilitating the repair of SSBs (46). Both PARylation on PARP1 and histones induces chromatin decompaction, thereby promoting transcription (26, 27). In response to double-strand breaks (DSBs), homologous recombination (HR) and non-homologous end-joining (NHEJ) are the primary mechanisms for repair. During HR, activated PARP1 recruits the MRE11-RAD50-NBS1 complex to the sites of damage (25). PARP1 inhibits the classical pathway of NHEJ by binding to DSBs in direct competition with Ku70/80 proteins and promotes alternative NHEJ (Alt-EJ) by recruiting MRN and CtIP (25, 48). Since the accumulation of DNA damage contributes to the pathophysiology of tumorigenesis, neurodegeneration, and premature aging, DDR is a critical mechanism for preventing these diseases (47).
Nuclear stress, such as heat shock, activates PARP1, leading to the PARylation of poly(A) polymerase (PAP). This modification prevents PAP from binding to target mRNA and inhibits subsequent 3’ mRNA processing, resulting in decreased mRNA synthesis (35). Additionally, splicing is regulated by PARP1-mediated PARylation of heterogeneous nuclear ribonucleoproteins (hnRNPs) hrp38 and squid in Drosophila (32).
Role of PARylation in mitosis and telomere length maintenance
In cell division, mitotic spindle formation is a crucial mechanism for the segregation of chromosomes into two daughter cells (88). Spindle orientation is determined by Gαi-LGN-NuMA complex, which regulates the extent of microtubule-pulling forces (89). It has been reported that NuMA localizes tankyrase1 to spindle poles and that tankyrase1 PARylates NuMA at the onset of mitosis (39, 40). Miki, another protein associated with mitosis, has also been shown to undergo PARylation by tankyrase1 during late G2 and prophase. This modification translocates Miki to mitotic centrosomes from the Golgi apparatus, anchoring CG-NAP, which serves as a scaffold for the γ-tubulin ring complex. Tankyrase1 knockdown impairs spindle formation and causes mitotic defects in prometaphase, such as preanaphase arrest, chromosome scattering, and pseudometaphase, highlighting the importance of PARylation in normal cell division (39–41).
Telomeres, nucleoprotein structures located at the ends of chromosomes, play a key role in maintaining genome integrity (90, 91). Chromosome duplication presents intrinsic challenges, including the inability of DNA polymerases to fully replicate the ends of chromosomes and the misrecognition of chromosome ends as DSBs, leading to improper repair. Telomeres address these issues and prevent genome instability by protecting chromosome ends (90, 91).
Telomeres are associated with a six-subunit protein complex called Shelterin, which consists of TRF1, TRF2, Rap1, TIN2, TPP1, and POT1 (92). TRF1 negatively regulates telomerase activity by limiting its accessibility for DNA (93, 94). Tankyrase-mediated PARylation of TRF1 inhibits the binding between TRF1 and DNA, allowing telomerase to extend telomeres (44, 95). TIN2 forms a ternary complex with TRF1 and tankyrase, repressing PARylation on TRF1 (96). Since telomeres harbor PARP1 activation sites, PARP1 is considered a potential inhibitor of telomere activity (92). TRF2 and TIN2 have been reported to protect telomeres from PARP1 independently (97).
PARylation regulates the ubiquitin-proteasome system
The ubiquitin-proteasome system (UPS) is a pivotal mechanism that controls the stability of intracellular proteins, modulating processes such as the cell cycle, apoptosis, transcription, and protein quality control (98, 99). Three classes of enzymes, ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3), facilitate the addition of ubiquitin chains to substrate proteins, which are subsequently recognized and degraded by the proteasome into peptide chains (99). Since numerous proteins, including oncoproteins and tumor suppressor proteins, are regulated by the UPS, abnormal ubiquitination has been reported to contribute to the development of various cancers (98, 100).
PI31 is an evolutionarily conserved protein that was initially identified as a suppressor of proteasome (101); however, subsequent studies have also suggested that PI31 activates the 20S core protease (102, 103). PI31 undergoes tankyrase-mediated PARylation, which decreases its affinity for 20S proteasome α-subunits, thereby reducing the inhibitory effect of PI31 (60). This modification enhances the binding and sequestration of dp27 and dS5b from 19S regulatory particles, promoting 26S proteasome assembly (60). In summary, tankyrase modulates proteasome activity through PARylation of PI31.
The well-known role of tankyrase is PARylation-mediated proteasomal degradation. Tankyrase PARylates substrates such as AXIN, PTEN, TRF1, RNF146, 3BP2, BLZF1, and CASC3 (104, 105). The E3-ubiquitin ligase RNF146 recognizes these PARylation modifications and ubiquitinates the substrate proteins, leading to their proteasomal degradation (104, 105). The significance of PARylation-mediated proteasomal degradation in several signaling pathways will be discussed in later sections.
PARylation as a modulator of innate immunity
It has been reported that PARP inhibitors (PARPis) reduced the transcription and release of lipopolysaccharide (LPS)-induced inflammation mediators, including TNF-α, IL-1, IL-6, and nitrite (NO2-), in murine bone marrow-derived macrophages (106). Additionally, macrophages derived from PARP1-deficient mice showed defective nuclear factor kappa B (NK-κB) activation and decreased production of TNF-α and IFN-γ in response to LPS (61). These PARP1-deficient mice were protected from death due to septic shock, highlighting the importance of PARP1 as an inflammatory mediator (61). In a zymosan-induced peritonitis model, a previous study showed that inhibition of poly (ADP-ribosyl) synthetase (PARS) suppressed neutrophil recruitment to sites of inflammation through postcapillary venules, providing protection against organ injury (69). PARP1 forms a complex with the subunits of NF-κB (p50 and p65 (RelA)), a key regulator of transcription involved in immune response and inflammation (107). The interaction between PARP1 and p65 was shown to be essential for NF-κB-dependent transcription of the iNOS and P-selectin promoters in B and T cells (107). In the inflammatory state induced by LPS, extracellular signal-regulated kinases 1/2 (ERK1/2) directly activate PARP1 through phosphorylation at serine 372 or threonine 373 (62), inducing PARylation-mediated activation of p65 and subsequent transcriptions of proinflammatory genes (63). The non-receptor tyrosine kinase c-Abl also activates PARP1 via phosphorylation at tyrosine 829, resulting in PARylation of p65 (64). Notably, extracellular PAR is recognized by Toll-like receptor 2 (TLR2) or TLR4 on macrophages, triggering cytokine release in humans and mice, suggesting that PAR may function as a damage-associated molecular pattern (DAMP) (108).
Tankyrase regulates bone metabolism and the immune system via PARylation-mediated proteasomal degradation of 3BP2
SH3 domain-binding protein-2 (3BP2) was initially identified as a protein binding to the Src homology 3 (SH3) domain of Abl (109, 110). Subsequent studies revealed that 3BP2 functions as an adaptor protein, forming a signaling complex with SYK (111), SRC (112, 113), and VAV (114), thereby regulating intracellular signaling pathways. Gain-of-function missense mutations in the SH3BP2 gene have been identified as the cause of cherubism, which is an autosomal dominant disorder characterized by facial swelling owing to severe craniofacial bone destruction and subsequent fibrous tissue masses (115, 116). Cherubism model mice with a mutation in the Sh3bp2 gene exhibit hyperactivity of macrophages and osteoclasts, leading to systemic inflammation and bone loss (116, 117).
Prof. Robert Rottapel’s lab at the University of Toronto, Canada has provided mechanistic insights by demonstrating that the gain-of-function mutations protect 3BP2 from tankyrase-mediated PARylation and proteasomal degradation (118, 119) (Figure 1). The missense mutation uncouples 3BP2 from the proteasomal degradation, leading to its accumulation in cells and hyperactivation of its substrates SYK, SRC, and VAV. Consequently, the loss of tankyrase-mediated degradation of 3BP2 underlies the pathophysiology of cherubism (118).
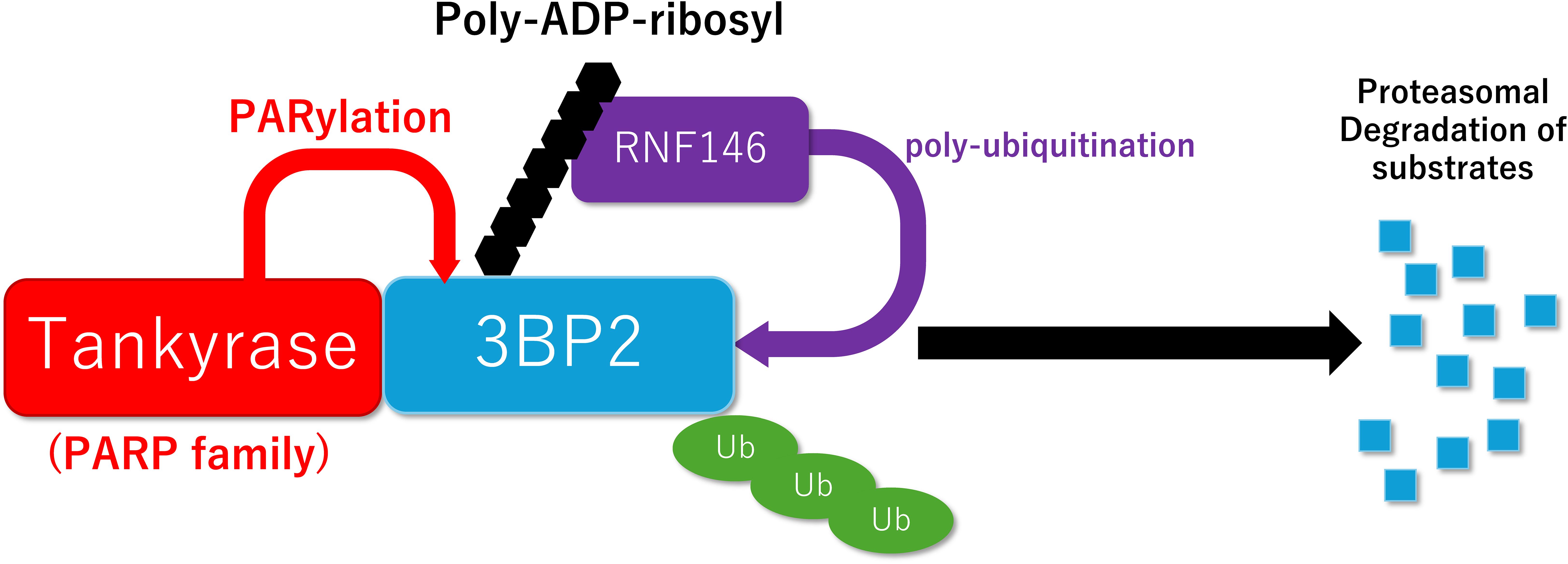
Figure 1. Schematic model of PARylation-mediated protein degradation. Tankyrase promotes poly-ADP-ribosylation (PARylation) of 3BP2, which creates a recognition site for RNF146, leading to ubiquitylation and subsequent proteasomal degradation of 3BP2.
Furthermore, they and we have discovered that 3BP2-induced activation of ABL and SRC is required for both RUNX2-mediated osteoblastogenesis and NFATc1-mediated osteoclastogenesis, respectively (112, 120). We have shown that conditional knockout of Rnf146 leads to stabilization of AXIN in osteoblasts and osteoclasts, resulting in phenotypes resembling osteoporosis and cleidocranial dysplasia (CCD), respectively (121, 122). Tankyrase regulates the TLR signaling pathways via PARylation-mediated degradation of 3BP2, and dysregulation of 3BP2 leads to autoinflammatory phenotypes, including severe inflammatory bowel disease (123, 124). 3BP2 is also required for proliferation and activation of T cells (125) and B cells (126) as well as for optimal neutrophil chemoattractant responses and host defense (127). Altogether, PARylation-mediated degradation of 3BP2 is a crucial mechanism for regulation of bone metabolism and the immune system (128–131).
Wnt/β-catenin signaling pathway
Wnt/β-catenin signaling is involved in embryonic development and cell homeostasis (132–134). The Wnt signaling pathway is primarily divided into three pathways: the canonical, β-catenin-dependent pathway and the non-canonical Wnt/Ca2+ (calcium) and Wnt/PCP (planar cell polarity) pathways (135). AXIN negatively regulates the canonical pathway by acting as a scaffold protein that forms the destruction complex (DC), which includes the tumor suppressor protein APC and the two serine-threonine kinases CK1α/δ and GSK3α/β (134).
Dysregulation of the Wnt/β-catenin pathway is associated with various types of cancer, including colorectal cancer (136), hepatocellular carcinoma (137), cholangiocarcinoma (138), lung cancer (139), hematological malignancies (140), and melanoma (141). As a result, the Wnt/β-catenin pathway has emerged as a potential target for cancer therapy (134, 142, 143), although such therapies are not yet in practical use.
Huang et al. reported that the tankyrase inhibitor XAV939 stabilizes AXIN and inhibits the Wnt/β-catenin pathway. They revealed that AXIN binds to tankyrase in the tankyrase-binding domain (TBD) and undergoes PARylation and ubiquitination (56). In addition to inducing proteolysis, tankyrase promotes accumulation of AXIN in the stimulatory signalosome and enhances the interaction between AXIN and the Wnt co-receptor LRP6 in response to Wnt stimulation (144). Tankyrase can also promote Wnt/β-catenin signaling in a manner independent of its PARP catalytic activity. Crystal structure analysis of tankyrase revealed that tankyrase polymerizes on its sterile alpha motif (SAM) domains, which are required for both tankyrase-dependent Wnt signaling and intact PARylation activity (145–147). Several in vitro studies have shown that XAV939 inhibits the Wnt/β-catenin signaling pathway and suppresses the proliferation of various types of cancer (56, 148–150). Additionally, multiple reports suggest that upregulation of the Wnt/β-catenin pathway contributes to resistance to a PARPi (151, 152), and combined therapies targeting both PARP and the Wnt/β-catenin pathway have demonstrated a synergistic effect (151–153).
PI3K-AKT signaling pathway
The phosphatidylinositol 3-kinase (PI3K)-AKT signaling pathway promotes cell survival, growth, differentiation, proliferation and glucose homeostasis in response to various stimuli (154). When membrane receptors, including receptor tyrosine kinases (RTKs) and G-protein-coupled receptors (GPCRs), receive extracellular signals, class I PI3K catalyzes the phosphorylation of phosphatidylinositol-bisphosphate (PIP2) to generate phosphatidylinositol-3,4,5-triphosphate (PIP3), which activates AKT and various types of AKT-dependent downstream signaling. Conversely, PTEN negatively regulates this pathway by dephosphorylating PIP3, converting it back to PIP2 (155). Upregulation of PI3K and downregulation of PTEN are recognized as tumorigenic (156, 157). In fact, a meta-analysis of cancer genome sequencing studies revealed that PIK3CA (which encodes one of the class1 PIK3 isoforms) and PTEN were the second and third most frequently mutated genes in human cancers (158).
PTEN has been reported to be regulated by PARylation-mediated degradation (Figure 2). Double knockdown of tankyrase 1/2 stabilized PTEN and downregulated AKT signaling, leading to the suppression of colon carcinoma proliferation (58). Additionally, investigation of human colon carcinoma samples revealed that tankyrase was upregulated and negatively correlated with PTEN expression (58). PARP1 also indirectly inhibits PTEN expression through PARylation-mediated degradation of its master regulator, Snail. Inhibition of PARP1 prevents doxorubicin-induced PTEN suppression, suggesting that combined therapy with a PARP1 inhibitor and cytotoxic drugs could be a promising treatment strategy (159, 160). Moreover, a recent study showed that AKT activation induces nuclear localization of glutamyl-prolyl-tRNA synthetase (EPRS1), which binds to PARP1 and activates PARylation, thereby contributing to breast cancer cell survival (161). Some preclinical studies have shown that PI3K inhibition enhances sensitivity to PARPis by suppressing HR repair (162, 163). In a phase I trial evaluating the efficacy of the PARPi olaparib combined with the AKT inhibitor capivasertib for advanced solid tumors, the combination therapy was well tolerated (164). Regarding efficacy, 44.6% of patients (25 out of 56 patients) had clinical benefits, including complete remission (CR), partial remission (PR), or stable disease (SD) lasting more than 4 months (164).
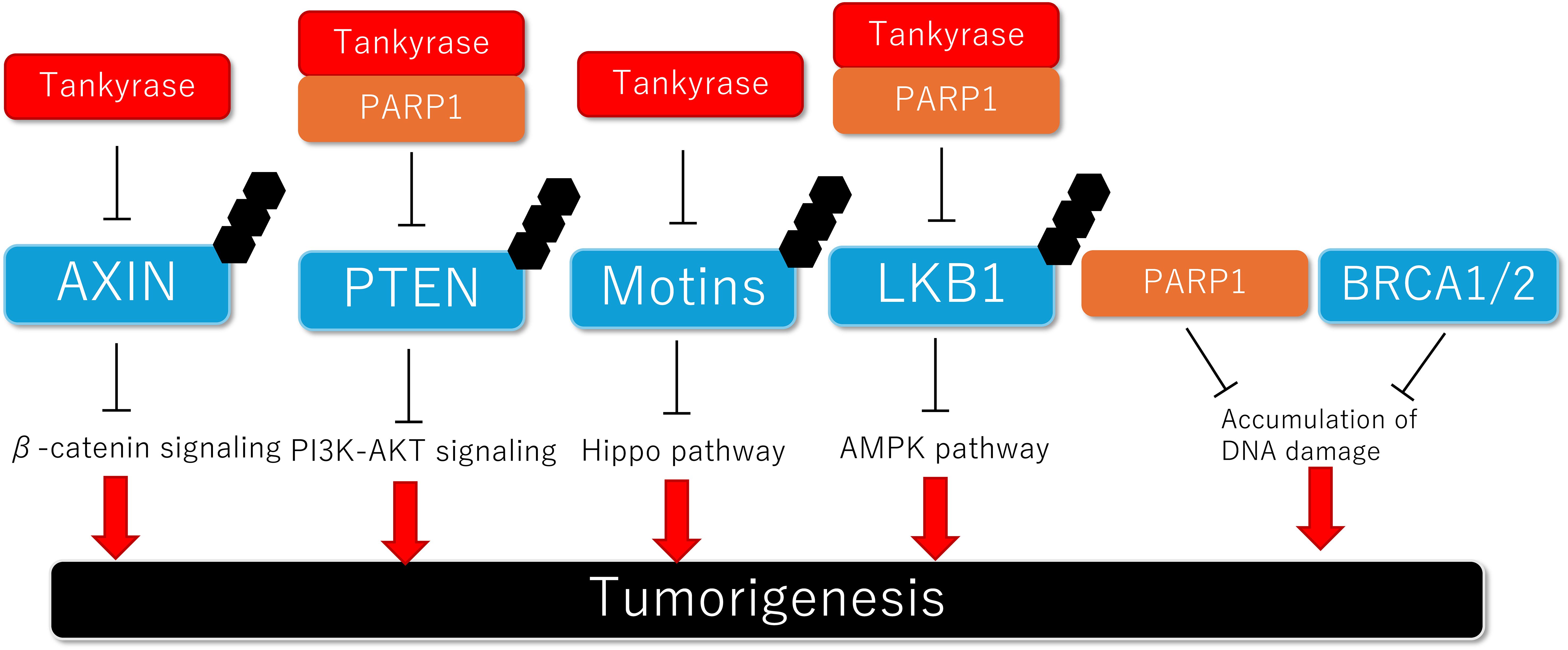
Figure 2. Schematic model of the association between PARylation-mediated protein degradation and tumorigenesis.
Hippo pathway
The Hippo pathway is a highly conserved growth control system that regulates cell, tissue or organ growth. This system can be activated by a broad range of extracellular stimuli, including changes in tight junctions and adherence junctions, energy stress, heat shock, osmotic stress, glycogen accumulation and mechanical forces. Downstream, Yes-associated protein (YAP) and its paralog, transcriptional co-activator PDZ-binding motif (TAZ), are phosphorylated and retained in the cytoplasm, leading to repression of the pro-growth transcriptional activity (165). Dysregulation of the Hippo pathway is thought to promote tumorigenesis, although it is not a direct trigger for cancer development (166–168).
Motin family proteins (Motins), including Angiomotin (AMOT), Angiomotin like 1 (AMOTL1) and Angiomotin like 2 (AMOTL2), are known to negatively regulate YAP by retaining it in the cytosol (169–172). Wang et al. reported that tankyrase interacts with all of the Motin family proteins (AMOT, AMOTL1, and AMOTL2) and PARylates them, leading to RNF146-mediated ubiquitination and subsequent proteasomal degradation (Figure 2) (57). Besides, RNF166 has been found to recognize tankyrase-mediated PARylation on Motins (AMOT and AMOTL2). RNF166 interacted with AMOT more strongly than did RNF146, leading to K48-linked polyubiquitination and degradation of AMOT. Overexpression of RNF166 resulted in elevated YAP activity and colorectal cancer progression (173). Several in vivo studies have shown that tankyrase inhibitors stabilize Motins, thereby suppressing the oncogenic function or drug resistance mediated by YAP (57, 174, 175). In contrast, tankyrase has been shown to maintain the Crumbs complex, which regulates tight junctions and resistance to epithelial-to-mesenchymal transitions (EMT), by modulating the expression of Motins. That study suggested that tankyrase inhibition could induce cancer progression (176).
LKB1/AMPK pathway
LKB1 was originally identified as a tumor suppressor gene located on human chromosome 19p13 and it is responsible for Peutz-Jeghers syndrome, an autosomal dominant inherited disorder characterized by hamartomatous polyps and mucocutaneous pigmentation (177, 178) and also an increased risk for various malignancies including gastrointestinal, gynecological, colorectal, pancreatic, and lung cancers (179–181). LKB1 encodes serine/threonine kinase LKB1, which forms a complex with STRADα and MO25 and phosphorylates AMP-activated protein kinase (AMPK) (182). LKB1 suppresses tumorigenesis through activation of AMPK (Figure 2), maintenance of cell polarity, and regulation of the cell cycle (183). Approximately 30% of human non-small cell lung cancers and 20% of cervical cancers harbor mutations in LKB1 (184–186).
It was reported that tankyrases repress LKB1 activity through PARylation at Glu130/138 and promote K63-linked ubiquitination by RNF146, thereby blocking the formation of the LKB1/STRAD/MO25 complex (59). Additionally, both in vitro and in vivo studies showed that inhibition of tankyrase repressed tumorigenesis by activating LKB1 and AMPK (59), providing further evidence for the potential of tankyrase inhibitors as anti-cancer drugs.
In a study on LKB1-mutant lung cancer, Long et al. found that LKB1 mutation caused deficiencies in the DNA damage repair process and hyperactivation of PARP1, leading to the PARylation of STAT1 (187). This modification inactivated STAT1 and resulted in downregulation of the interferon-gamma (IFNγ) response. Furthermore, the PARP1 inhibitor olaparib restored STAT1 phosphorylation and the IFNγ response.
BRCA1/2 mutation and PARP inhibitors
Breast cancer has the highest prevalence and mortality rate among malignancies in women worldwide (188). It is classified immunohistochemically based on positivity of estrogen receptor (ER), progesterone receptor (PR), and HER2 (189). The absence of these markers defines triple-negative breast cancer (TNBC), an aggressive subtype (189, 190). Approximately 5% of breast cancer patients have germline pathogenic variants in cancer disposition genes, with BRCA1 and BRCA2 being the major ones (191, 192). Germline mutations in BRCA1/2 are also prevalent in ovarian, prostate and pancreatic cancers (193, 194).
In the DDR, BRCA1 and BRCA2 are crucial proteins for homologous recombination repair (HR), a process by which DNA is synthesized using a homologous DNA molecule as a template. Germline mutations in BRCA1/2 lead to accumulation of DNA damage and subsequent tumorigenesis. Tumor cells with BRCA1/2 mutation rely on alternative repair pathways, such as PARP1-mediated repairing (Figure 2). Therefore, PARPis exhibit anti-cancer effects through a mechanism known as synthetic lethality (25, 48).
In 2014, the U.S. Food and Drug Administration (FDA) and European Medicines Agency (EMA) approved the first PARPi, olaparib, as maintenance therapy for platinum-sensitive advanced ovarian cancer with germline mutations in BRCA1/2. Currently, four different PARPis (olaparib, talazoparib, rucaparib, and niraparib) are available for treatment of ovarian, breast, pancreatic, and prostate cancers (195–199).
Bromodomain-containing protein 7 (BRD7) is a tumor suppressor protein that regulates cell cycle progression and transcriptional regulation (200). It has been reported that PARP1 regulates BRD7 expression through PARylation-mediated ubiquitination, enhancing the survival of cancer cells. PARP1 inhibition not only suppresses cell proliferation but also sensitizes cancer cells to DNA-damaging chemotherapy, suggesting the potential for combined therapies using PARPis and chemotherapeutic drugs (201).
Immune checkpoint inhibitors
T cell responses to antigen recognition by the T cell receptor (TCR) are regulated by a balance between co-stimulatory and inhibitory signals, also known as immune checkpoints (202). Well-studied immune checkpoints include programmed death-1 or its ligand (PD-1/PD-L1) as well as cytotoxic T-lymphocyte-associated antigen 4 (CTLA4). Immune checkpoint inhibitors (ICIs) enhance immune responses against malignancies by blocking these pathways (202).
PD-1 is expressed on activated CD4+ or CD8+ T cells, monocytes, natural killer T cells, B cells, and dendritic cells (203). PD-1 ligation dampens TCR signaling, cytokine release, and cell viability, while co-stimulation with CD28 can reverse these effects (203). PD-L1 is expressed by various cell types, including immune cells and tumor cells, in response to IFN-γ produced by activated T cells (204). Many human cancers, including breast, urothelial, ovarian, and pancreatic cancers, express tumor-associated PD-L1 (204). Binding of PD-L1 to PD-1 induces effector T cell exhaustion and immune evasion by tumor cells, leading to malignancy progression (203–205). Another immune checkpoint, CTLA4, is expressed on T cells and is essential for the function of regulatory T cells (206). CTLA4 and CD28 are homologous glycoproteins of the immunoglobulin superfamily (207) and share identical ligands, CD80 and CD86 (202). CTLA4 has a higher affinity than that of CD28 for these ligands, resulting in the inhibition of T cell activation (202). Since cancer cells exploit these checkpoints to evade host immune surveillance, ICIs act by blocking these pathways and activating the anti-tumor immunity of T cells (208). The U.S. FDA has approved three categories of ICIs: anti-CTLA4 inhibitor (ipilimumab), anti-PD-1 inhibitors (nivolumab, pembrolizumab, and cemiplimab), and anti-PD-L1 inhibitors (atezolizumab, durvalumab, and avelumab). Therapies with these ICIs are used for a wide range of malignancies including melanoma, breast cancer, non-small lung cancer, renal cell carcinoma, urothelial carcinoma, gastric cancer, colorectal cancer, and many others (209–211).
PARP inhibitors/tankyrase inhibitors and immune checkpoint inhibitors show a synergistic effect
Despite the durable response rate of an ICI, many patients experience primary or acquired resistance (212), highlighting the need for new strategies, such as multidrug therapies. Several preclinical studies have suggested a synergistic effect between DDR inhibition and ICIs. PARP inhibition leads to the upregulation of PD-L1 and suppression of anticancer immunity, while PD-L1 blockade re-sensitizes PARPi-treated cancer cells to T cell cytotoxicity (213, 214). Mechanistically, inhibitors of DDR components, such as PARP or checkpoint kinase 1 (CHK1), increase cytosolic damaged DNA and activate the STING/TBK1/IRF3 innate immune pathway. This activation results in the upregulation of PD-L1, IFN-β, IFN-γ, and CCL5, which drives CD8+ T cell infiltration into tumors (215–217). Additionally, a PARPi has been reported to enhance PD-L1 expression by preventing PARP1-mediated dephosphorylation on STAT3 (218).
Tankyrase inhibition may also enhance the efficacy of ICIs. Spranger et al. classified metastatic human cutaneous melanoma samples into two groups based on T cell signatures: non-T-cell inflamed group and T-cell-inflamed group (219). Gene expression analysis revealed that the non-T-cell-inflamed group exhibited upregulation of Wnt/β-catenin signaling compared to that in the T-cell-inflamed group (219). This evidence suggests that activated Wnt/β-catenin signaling may inhibit antitumor T cell response. As mentioned above, tankyrase can activate Wnt/β-catenin signaling through PARylation-mediated degradation of AXIN, indicating that tankyrase inhibition could serve as a potential therapy to enhance the anticancer effect of T cells in combination with ICIs. In mouse models, tankyrase inhibitors (G007-LK and OM-153) have been shown to suppress Wnt/β-catenin signaling, leading to activation of T cell-mediated antitumor responses induced by PD-1 inhibition and suppression of melanoma growth (220, 221).
Clinical trials on combined therapies
There has been an increasing number of clinical trials to evaluate the efficacy of combination therapies involving PARPis and ICIs. In breast cancer, the results of combination therapy have been inconsistent (222–224). A previous consensus paper recommended adjuvant treatment combining pembrolizumab with a PARP inhibitor for a limited population in patients with TNBC harboring BRCA1/2 mutations (197). A randomized phase Ib/II trial was carried out to compere two combination therapies for pancreatic cancers: niraparib with nivolumab and niraparib with ipilimumab (225). The rate of progression-free survival (PFS) at 6 months was higher in the latter group (20.6% vs 59.6%), although adverse events were more frequently observed in the latter group (225). Combination therapies for pancreatic cancer are currently being examined in multiple studies (226). Although PARPis are also crucial drugs for ovarian cancer, prolonged usage often leads to the development of PARPi resistance (227). The use of ICIs is an attractive strategy for overcoming the problem of PARPi resistance. A phase I/II study in which the combination of niraparib and pembrolizumab was evaluated showed promising tolerability and antitumor activity (228). The efficacy and safety of combination therapies involving PARPi and ICIs are currently being investigated in several ongoing studies (227). In contrast, combination therapies appear to be less effective in prostate cancer. In a phase Ib/II trial, patients with metastatic castration-resistant prostate cancer were treated with pembrolizumab plus olaparib (229). The median radiographic PFS (rPFS) and overall survival (OS) rates were 4.5 months and 14 months, respectively (229). In a subsequent phase III trial, this combination was compared to abiraterone or enzalutamide in patients pretreated with androgen receptor signaling inhibitors and docetaxel (230). That study was discontinued for futility due to a lack of improvement in rPFS and OS in the interim analysis, despite a higher objective response rate (17% vs. 0%) (230). The development of this combination therapy has been halted, and there are no ongoing phase III trials for prostate cancer (231).
Conclusion
We have discussed the associations between PARylation and several signaling pathways involved in cancer generation and progression. Since PARylation regulates a wide range of cellular functions, it represents an attractive target for cancer therapy. Further research is needed to develop appropriate treatment strategies, including combination therapies.
Author contributions
KM: Writing – original draft, Writing – review & editing. YM: Writing – original draft, Writing – review & editing. JW: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Luscher B, Butepage M, Eckei L, Krieg S, Verheugd P, Shilton BH. ADP-ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem Rev. (2018) 118:1092–136. doi: 10.1021/acs.chemrev.7b00122
2. Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. (2012) 13:411–24. doi: 10.1038/nrm3376
3. Suskiewicz MJ, Prokhorova E, Rack JGM, Ahel I. ADP-ribosylation from molecular mechanisms to therapeutic implications. Cell. (2023) 186:4475–95. doi: 10.1016/j.cell.2023.08.030
4. Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. (2010) 35:208–19. doi: 10.1016/j.tibs.2009.12.003
5. Luscher B, Ahel I, Altmeyer M, Ashworth A, Bai P, Chang P, et al. ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. (2022) 289:7399–410. doi: 10.1111/febs.16142
6. Musheev MU, Schomacher L, Basu A, Han D, Krebs L, Scholz C, et al. Mammalian N1-adenosine PARylation is a reversible DNA modification. Nat Commun. (2022) 13:6138. doi: 10.1038/s41467-022-33731-w
7. Schuller M, Raggiaschi R, Mikolcevic P, Rack JGM, Ariza A, Zhang Y, et al. Molecular basis for the reversible ADP-ribosylation of guanosine bases. Mol Cell. (2023) 83:2303–15 e6. doi: 10.1016/j.molcel.2023.06.013
8. Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. (2006) 7:517–28. doi: 10.1038/nrm1963
9. Hottiger MO. SnapShot: ADP-ribosylation signaling. Mol Cell. (2015) 58:1134– e1. doi: 10.1016/j.molcel.2015.06.001
10. Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, et al. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. (2014) 5(1):4426. doi: 10.1038/ncomms5426
11. Karlberg T, Langelier MF, Pascal JM, Schuler H. Structural biology of the writers, readers, and erasers in mono- and poly(ADP-ribose) mediated signaling. Mol Aspects Med. (2013) 34:1088–108. doi: 10.1016/j.mam.2013.02.002
12. Teloni F, Altmeyer M. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. (2016) 44:993–1006. doi: 10.1093/nar/gkv1383
13. Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, et al. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. (2012) 26:235–40. doi: 10.1101/gad.182618.111
14. DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, et al. Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature. (2015) 517:223–6. doi: 10.1038/nature13826
15. Gupte R, Liu Z, Kraus WL. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. (2017) 31:101–26. doi: 10.1101/gad.291518.116
16. O’Sullivan J, Tedim Ferreira M, Gagne JP, Sharma AK, Hendzel MJ, Masson JY, et al. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat Commun. (2019) 10:1182. doi: 10.1038/s41467-019-08859-x
17. Rack JGM, Palazzo L, Ahel I. (ADP-ribosyl)hydrolases: structure, function, and biology. Genes Dev. (2020) 34:263–84. doi: 10.1101/gad.334631.119
18. Bock FJ, Chang P. New directions in poly(ADP-ribose) polymerase biology. FEBS J. (2016) 283:4017–31. doi: 10.1111/febs.13737
19. Palazzo L, Mikoc A, Ahel I. ADP-ribosylation: new facets of an ancient modification. FEBS J. (2017) 284:2932–46. doi: 10.1111/febs.14078
20. Kim DS, Challa S, Jones A, Kraus WL. PARPs and ADP-ribosylation in RNA biology: from RNA expression and processing to protein translation and proteostasis. Genes Dev. (2020) 34:302–20. doi: 10.1101/gad.334433.119
21. Azarm K, Smith S. Nuclear PARPs and genome integrity. Genes Dev. (2020) 34:285–301. doi: 10.1101/gad.334730.119
22. Eisemann T, Pascal JM. Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity. Cell Mol Life Sci. (2020) 77:19–33. doi: 10.1007/s00018-019-03366-0
23. Szanto M, Bai P. The role of ADP-ribose metabolism in metabolic regulation, adipose tissue differentiation, and metabolism. Genes Dev. (2020) 34:321–40. doi: 10.1101/gad.334284.119
24. Zimmerlin L, Zambidis ET. Pleiotropic roles of tankyrase/PARP proteins in the establishment and maintenance of human naive pluripotency. Exp Cell Res. (2020) 390:111935. doi: 10.1016/j.yexcr.2020.111935
25. Huang D, Kraus WL. The expanding universe of PARP1-mediated molecular and therapeutic mechanisms. Mol Cell. (2022) 82:2315–34. doi: 10.1016/j.molcel.2022.02.021
26. Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. (2008) 20:294–302. doi: 10.1016/j.ceb.2008.03.006
27. Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. (2010) 39:8–24. doi: 10.1016/j.molcel.2010.06.017
28. Martinez-Zamudio R, Ha HC. Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Mol Cell Biol. (2012) 32:2490–502. doi: 10.1128/MCB.06667-11
29. Lu Y, Fu W, Xing W, Wu H, Zhang C, Xu D. Transcriptional regulation mechanism of PARP1 and its application in disease treatment. Epigenet Chromatin. (2024) 17:26. doi: 10.1186/s13072-024-00550-w
30. Rajawat J, Shukla N, Mishra DP. Therapeutic targeting of poly(ADP-ribose) polymerase-1 (PARP1) in cancer: current developments, therapeutic strategies, and future opportunities. Med Res Rev. (2017) 37:1461–91. doi: 10.1002/med.21442
31. Gagne JP, Hunter JM, Labrecque B, Chabot B, Poirier GG. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem J. (2003) 371:331–40. doi: 10.1042/BJ20021675
32. Ji Y, Tulin AV. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res. (2009) 37:3501–13. doi: 10.1093/nar/gkp218
33. Malanga M, Czubaty A, Girstun A, Staron K, Althaus FR. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J Biol Chem. (2008) 283:19991–8. doi: 10.1074/jbc.M709495200
34. Isabelle M, Moreel X, Gagne JP, Rouleau M, Ethier C, Gagne P, et al. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. (2010) 8:22. doi: 10.1186/1477-5956-8-22
35. Di Giammartino DC, Shi Y, Manley JL. PARP1 represses PAP and inhibits polyadenylation during heat shock. Mol Cell. (2013) 49:7–17. doi: 10.1016/j.molcel.2012.11.005
36. Ji Y, Tulin AV. Poly(ADP-ribose) controls DE-cadherin-dependent stem cell maintenance and oocyte localization. Nat Commun. (2012) 3:760. doi: 10.1038/ncomms1759
37. Rodriguez MI, Gonzalez-Flores A, Dantzer F, Collard J, de Herreros AG, Oliver FJ. Poly(ADP-ribose)-dependent regulation of Snail1 protein stability. Oncogene. (2011) 30:4365–72. doi: 10.1038/onc.2011.153
38. Ji Y, Tulin AV. Post-transcriptional regulation by poly(ADP-ribosyl)ation of the RNA-binding proteins. Int J Mol Sci. (2013) 14:16168–83. doi: 10.3390/ijms140816168
39. Chang W, Dynek JN, Smith S. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem J. (2005) 391:177–84. doi: 10.1042/BJ20050885
40. Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. (2005) 7:1133–9. doi: 10.1038/ncb1322
41. Ozaki Y, Matsui H, Asou H, Nagamachi A, Aki D, Honda H, et al. Poly-ADP ribosylation of Miki by tankyrase-1 promotes centrosome maturation. Mol Cell. (2012) 47:694–706. doi: 10.1016/j.molcel.2012.06.033
42. Kim MK, Dudognon C, Smith S. Tankyrase 1 regulates centrosome function by controlling CPAP stability. EMBO Rep. (2012) 13:724–32. doi: 10.1038/embor.2012.86
43. Muoio D, Laspata N, Fouquerel E. Functions of ADP-ribose transferases in the maintenance of telomere integrity. Cell Mol Life Sci. (2022) 79:215. doi: 10.1007/s00018-022-04235-z
44. Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr Biol. (2000) 10:1299–302. doi: 10.1016/s0960-9822(00)00752-1
45. Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. EMBO J. (2007) 26:4867–78. doi: 10.1038/sj.emboj.7601903
46. Caldecott KW. DNA single-strand break repair. Exp Cell Res. (2014) 329:2–8. doi: 10.1016/j.yexcr.2014.08.027
47. Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. (2010) 40:179–204. doi: 10.1016/j.molcel.2010.09.019
48. Groelly FJ, Fawkes M, Dagg RA, Blackford AN, Tarsounas M. Targeting DNA damage response pathways in cancer. Nat Rev Cancer. (2023) 23:78–94. doi: 10.1038/s41568-022-00535-5
49. Saenz L, Lozano JJ, Valdor R, Baroja-Mazo A, Ramirez P, Parrilla P, et al. Transcriptional regulation by poly(ADP-ribose) polymerase-1 during T cell activation. BMC Genomics. (2008) 9:171. doi: 10.1186/1471-2164-9-171
50. Nasta F, Laudisi F, Sambucci M, Rosado MM, Pioli C. Increased Foxp3+ regulatory T cells in poly(ADP-Ribose) polymerase-1 deficiency. J Immunol. (2010) 184:3470–7. doi: 10.4049/jimmunol.0901568
51. Zhang P, Nakatsukasa H, Tu E, Kasagi S, Cui K, Ishikawa M, et al. PARP-1 regulates expression of TGF-beta receptors in T cells. Blood. (2013) 122:2224–32. doi: 10.1182/blood-2013-05-503250
52. Luo X, Nie J, Wang S, Chen Z, Chen W, Li D, et al. Poly(ADP-ribosyl)ation of FOXP3 protein mediated by PARP-1 protein regulates the function of regulatory T cells. J Biol Chem. (2015) 290:28675–82. doi: 10.1074/jbc.M115.661611
53. Navarro J, Gozalbo-Lopez B, Mendez AC, Dantzer F, Schreiber V, Martinez C, et al. PARP-1/PARP-2 double deficiency in mouse T cells results in faulty immune responses and T lymphomas. Sci Rep. (2017) 7:41962. doi: 10.1038/srep41962
54. Galindo-Campos MA, Bedora-Faure M, Farres J, Lescale C, Moreno-Lama L, Martinez C, et al. Coordinated signals from the DNA repair enzymes PARP-1 and PARP-2 promotes B-cell development and function. Cell Death Differ. (2019) 26:2667–81. doi: 10.1038/s41418-019-0326-5
55. Aldinucci A, Gerlini G, Fossati S, Cipriani G, Ballerini C, Biagioli T, et al. A key role for poly(ADP-ribose) polymerase-1 activity during human dendritic cell maturation. J Immunol. (2007) 179:305–12. doi: 10.4049/jimmunol.179.1.305
56. Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. (2009) 461:614–20. doi: 10.1038/nature08356
57. Wang W, Li N, Li X, Tran MK, Han X, Chen J. Tankyrase inhibitors target YAP by stabilizing angiomotin family proteins. Cell Rep. (2015) 13:524–32. doi: 10.1016/j.celrep.2015.09.014
58. Li N, Zhang Y, Han X, Liang K, Wang J, Feng L, et al. Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev. (2015) 29:157–70. doi: 10.1101/gad.251785.114
59. Li N, Wang Y, Neri S, Zhen Y, Fong LWR, Qiao Y, et al. Tankyrase disrupts metabolic homeostasis and promotes tumorigenesis by inhibiting LKB1-AMPK signalling. Nat Commun. (2019) 10:4363. doi: 10.1038/s41467-019-12377-1
60. Cho-Park PF, Steller H. Proteasome regulation by ADP-ribosylation. Cell. (2013) 153:614–27. doi: 10.1016/j.cell.2013.03.040
61. Oliver FJ, Menissier-de-Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, et al. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. (1999) 18:4446–54. doi: 10.1093/emboj/18.16.4446
62. Kauppinen TM, Chan WY, Suh SW, Wiggins AK, Huang EJ, Swanson RA. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc Natl Acad Sci U.S.A. (2006) 103:7136–41. doi: 10.1073/pnas.0508606103
63. Liu L, Ke Y, Jiang X, He F, Pan L, Xu L, et al. Lipopolysaccharide activates ERK-PARP-1-RelA pathway and promotes nuclear factor-kappaB transcription in murine macrophages. Hum Immunol. (2012) 73:439–47. doi: 10.1016/j.humimm.2012.02.002
64. Bohio AA, Sattout A, Wang R, Wang K, Sah RK, Guo X, et al. c-abl-mediated tyrosine phosphorylation of PARP1 is crucial for expression of proinflammatory genes. J Immunol. (2019) 203:1521–31. doi: 10.4049/jimmunol.1801616
65. Ditsworth D, Zong WX, Thompson CB. Activation of poly(ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J Biol Chem. (2007) 282:17845–54. doi: 10.1074/jbc.M701465200
66. Yang Z, Li L, Chen L, Yuan W, Dong L, Zhang Y, et al. PARP-1 mediates LPS-induced HMGB1 release by macrophages through regulation of HMGB1 acetylation. J Immunol. (2014) 193:6114–23. doi: 10.4049/jimmunol.1400359
67. Walko TD 3rd, Di Caro V, Piganelli J, Billiar TR, Clark RS, Aneja RK. Poly(ADP-ribose) polymerase 1-sirtuin 1 functional interplay regulates LPS-mediated high mobility group box 1 secretion. Mol Med. (2015) 20:612–24. doi: 10.2119/molmed.2014.00156
68. Kim C, Wang XD, Yu Y. PARP1 inhibitors trigger innate immunity via PARP1 trapping-induced DNA damage response. Elife. (2020) 9:e60637. doi: 10.7554/eLife.60637
69. Szabo C, Lim LH, Cuzzocrea S, Getting SJ, Zingarelli B, Flower RJ, et al. Inhibition of poly (ADP-ribose) synthetase attenuates neutrophil recruitment and exerts antiinflammatory effects. J Exp Med. (1997) 186:1041–9. doi: 10.1084/jem.186.7.1041
70. Cameron AM, Castoldi A, Sanin DE, Flachsmann LJ, Field CS, Puleston DJ, et al. Inflammatory macrophage dependence on NAD(+) salvage is a consequence of reactive oxygen species-mediated DNA damage. Nat Immunol. (2019) 20:420–32. doi: 10.1038/s41590-019-0336-y
71. Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. (2011) 13:461–8. doi: 10.1016/j.cmet.2011.03.004
72. Bai P, Nagy L, Fodor T, Liaudet L, Pacher P. Poly(ADP-ribose) polymerases as modulators of mitochondrial activity. Trends Endocrinol Metab. (2015) 26:75–83. doi: 10.1016/j.tem.2014.11.003
73. Ying W, Chen Y, Alano CC, Swanson RA. Tricarboxylic acid cycle substrates prevent PARP-mediated death of neurons and astrocytes. J Cereb Blood Flow Metab. (2002) 22:774–9. doi: 10.1097/00004647-200207000-00002
74. Ye DZ, Tai MH, Linning KD, Szabo C, Olson LK. MafA expression and insulin promoter activity are induced by nicotinamide and related compounds in INS-1 pancreatic beta-cells. Diabetes. (2006) 55:742–50. doi: 10.2337/diabetes.55.03.06.db05-0653
75. Andrabi SA, Umanah GK, Chang C, Stevens DA, Karuppagounder SS, Gagne JP, et al. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci U.S.A. (2014) 111:10209–14. doi: 10.1073/pnas.1405158111
76. Fouquerel E, Goellner EM, Yu Z, Gagne JP, Barbi de Moura M, Feinstein T, et al. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. (2014) 8:1819–31. doi: 10.1016/j.celrep.2014.08.036
77. Hopp AK, Gruter P, Hottiger MO. Regulation of glucose metabolism by NAD(+) and ADP-ribosylation. Cells. (2019) 8(8):890. doi: 10.3390/cells8080890
78. Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, et al. Poly(ADP-ribose) polymerase-2 [corrected] controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma [corrected] heterodimer. J Biol Chem. (2007) 282:37738–46. doi: 10.1074/jbc.M701021200
79. Bai P, Canto C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. (2012) 16:290–5. doi: 10.1016/j.cmet.2012.06.016
80. Szanto M, Brunyanszki A, Marton J, Vamosi G, Nagy L, Fodor T, et al. Deletion of PARP-2 induces hepatic cholesterol accumulation and decrease in HDL levels. Biochim Biophys Acta. (2014) 1842:594–602. doi: 10.1016/j.bbadis.2013.12.006
81. Marton J, Peter M, Balogh G, Bodi B, Vida A, Szanto M, et al. Poly(ADP-ribose) polymerase-2 is a lipid-modulated modulator of muscular lipid homeostasis. Biochim Biophys Acta Mol Cell Biol Lipids. (2018) 1863:1399–412. doi: 10.1016/j.bbalip.2018.07.013
82. Erener S, Mirsaidi A, Hesse M, Tiaden AN, Ellingsgaard H, Kostadinova R, et al. ARTD1 deletion causes increased hepatic lipid accumulation in mice fed a high-fat diet and impairs adipocyte function and differentiation. FASEB J. (2012) 26:2631–8. doi: 10.1096/fj.11-200212
83. Szanto M, Gupte R, Kraus WL, Pacher P, Bai P. PARPs in lipid metabolism and related diseases. Prog Lipid Res. (2021) 84:101117. doi: 10.1016/j.plipres.2021.101117
84. Yeh TY, Sbodio JI, Tsun ZY, Luo B, Chi NW. Insulin-stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase. Biochem J. (2007) 402:279–90. doi: 10.1042/BJ20060793
85. Yeh TY, Beiswenger KK, Li P, Bolin KE, Lee RM, Tsao TS, et al. Hypermetabolism, hyperphagia, and reduced adiposity in tankyrase-deficient mice. Diabetes. (2009) 58:2476–85. doi: 10.2337/db08-1781
86. Su Z, Deshpande V, James DE, Stockli J. Tankyrase modulates insulin sensitivity in skeletal muscle cells by regulating the stability of GLUT4 vesicle proteins. J Biol Chem. (2018) 293:8578–87. doi: 10.1074/jbc.RA117.001058
87. Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. (2009) 461:1071–8. doi: 10.1038/nature08467
88. Prosser SL, Pelletier L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol. (2017) 18:187–201. doi: 10.1038/nrm.2016.162
89. Lechler T, Mapelli M. Spindle positioning and its impact on vertebrate tissue architecture and cell fate. Nat Rev Mol Cell Biol. (2021) 22:691–708. doi: 10.1038/s41580-021-00384-4
90. Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, et al. A human telomeric protein. Science. (1995) 270:1663–7. doi: 10.1126/science.270.5242.1663
92. de Lange T. Shelterin-mediated telomere protection. Annu Rev Genet. (2018) 52:223–47. doi: 10.1146/annurev-genet-032918-021921
93. van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. (1997) 385:740–3. doi: 10.1038/385740a0
94. Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, et al. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol Cell Biol. (2002) 22:3474–87. doi: 10.1128/MCB.22.10.3474-3487.2002
95. Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. (2008) 90:83–92. doi: 10.1016/j.biochi.2007.07.012
96. Ye JZ, de Lange T. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat Genet. (2004) 36:618–23. doi: 10.1038/ng1360
97. Schmutz I, Timashev L, Xie W, Patel DJ, de Lange T. TRF2 binds branched DNA to safeguard telomere integrity. Nat Struct Mol Biol. (2017) 24:734–42. doi: 10.1038/nsmb.3451
98. Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. (2002) 82:373–428. doi: 10.1152/physrev.00027.2001
99. Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. (2009) 78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607
100. Liu F, Chen J, Li K, Li H, Zhu Y, Zhai Y, et al. Ubiquitination and deubiquitination in cancer: from mechanisms to novel therapeutic approaches. Mol Cancer. (2024) 23:148. doi: 10.1186/s12943-024-02046-3
101. Chu-Ping M, Slaughter CA, DeMartino GN. Purification and characterization of a protein inhibitor of the 20S proteasome (macropain). Biochim Biophys Acta. (1992) 1119:303–11. doi: 10.1016/0167-4838(92)90218-3
102. Bader M, Benjamin S, Wapinski OL, Smith DM, Goldberg AL, Steller H. A conserved F box regulatory complex controls proteasome activity in Drosophila. Cell. (2011) 145:371–82. doi: 10.1016/j.cell.2011.03.021
103. Yang BJ, Han XX, Yin LL, Xing MQ, Xu ZH, Xue HW. Arabidopsis PROTEASOME REGULATOR1 is required for auxin-mediated suppression of proteasome activity and regulates auxin signalling. Nat Commun. (2016) 7:11388. doi: 10.1038/ncomms11388
104. Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. (2011) 13:623–9. doi: 10.1038/ncb2222
105. Zamudio-Martinez E, Herrera-Campos AB, Munoz A, Rodriguez-Vargas JM, Oliver FJ. Tankyrases as modulators of pro-tumoral functions: molecular insights and therapeutic opportunities. J Exp Clin Cancer Res. (2021) 40:144. doi: 10.1186/s13046-021-01950-6
106. Hauschildt S, Scheipers P, Bessler W, Schwarz K, Ullmer A, Flad HD, et al. Role of ADP-ribosylation in activated monocytes/macrophages. Adv Exp Med Biol. (1997) 419:249–52. doi: 10.1007/978-1-4419-8632-0_31
107. Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J Biol Chem. (2001) 276:45588–97. doi: 10.1074/jbc.M106528200
108. Krukenberg KA, Kim S, Tan ES, Maliga Z, Mitchison TJ. Extracellular poly(ADP-ribose) is a pro-inflammatory signal for macrophages. Chem Biol. (2015) 22:446–52. doi: 10.1016/j.chembiol.2015.03.007
109. Cicchetti P, Mayer B, Thiel G, Baltimore D. Identification of a protein that binds to the SH3 region of abl and is similar to bcr and GAP-rho. Science. (1992) 257:803–6. doi: 10.1126/science.1379745
110. Ren R, Mayer B, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. (1993) 259:1157–61. doi: 10.1126/science.8438166
111. Deckert M, Tartare-Deckert S, Hernandez J, Rottapel R, Altman A. Adaptor function for the Syk kinases-interacting protein 3BP2 in IL-2 gene activation. Immunity. (1998) 9:595–605. doi: 10.1016/s1074-7613(00)80657-3
112. Levaot N, Simoncic PD, Dimitriou ID, Scotter A, La Rose J, Ng AH, et al. 3BP2-deficient mice are osteoporotic with impaired osteoblast and osteoclast functions. J Clin Invest. (2011) 121:3244–57. doi: 10.1172/JCI45843
113. GuezGuez A, Prod’homme V, Mouska X, Baudot A, Blin-Wakkach C, Rottapel R, et al. 3BP2 adapter protein is required for receptor activator of NFkappaB ligand (RANKL)-induced osteoclast differentiation of RAW264.7 cells. J Biol Chem. (2010) 285:20952–63. doi: 10.1074/jbc.M109.091124
114. Foucault I, Le Bras S, Charvet C, Moon C, Altman A, Deckert M. The adaptor protein 3BP2 associates with VAV guanine nucleotide exchange factors to regulate NFAT activation by the B-cell antigen receptor. Blood. (2005) 105:1106–13. doi: 10.1182/blood-2003-08-2965
115. Ueki Y, Tiziani V, Santanna C, Fukai N, Maulik C, Garfinkle J, et al. Mutations in the gene encoding c-Abl-binding protein SH3BP2 cause cherubism. Nat Genet. (2001) 28:125–6. doi: 10.1038/88832
116. Ueki Y, Lin CY, Senoo M, Ebihara T, Agata N, Onji M, et al. Increased myeloid cell responses to M-CSF and RANKL cause bone loss and inflammation in SH3BP2 “cherubism” mice. Cell. (2007) 128:71–83. doi: 10.1016/j.cell.2006.10.047
117. Kittaka M, Yoshimoto T, Schlosser C, Rottapel R, Kajiya M, Kurihara H, et al. Alveolar bone protection by targeting the SH3BP2-SYK axis in osteoclasts. J Bone Miner Res. (2020) 35:382–95. doi: 10.1002/jbmr.3882
118. Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, et al. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. (2011) 147:1324–39. doi: 10.1016/j.cell.2011.10.045
119. Guettler S, LaRose J, Petsalaki E, Gish G, Scotter A, Pawson T, et al. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. (2011) 147:1340–54. doi: 10.1016/j.cell.2011.10.046
120. Matsumoto Y, La Rose J, Kent OA, Wagner MJ, Narimatsu M, Levy AD, et al. Reciprocal stabilization of ABL and TAZ regulates osteoblastogenesis through transcription factor RUNX2. J Clin Invest. (2016) 126:4482–96. doi: 10.1172/JCI87802
121. Matsumoto Y, La Rose J, Lim M, Adissu HA, Law N, Mao X, et al. Ubiquitin ligase RNF146 coordinates bone dynamics and energy metabolism. J Clin Invest. (2017) 127:2612–25. doi: 10.1172/JCI92233
122. Matsumoto Y, Larose J, Kent OA, Lim M, Changoor A, Zhang L, et al. RANKL coordinates multiple osteoclastogenic pathways by regulating expression of ubiquitin ligase RNF146. J Clin Invest. (2017) 127:1303–15. doi: 10.1172/JCI90527
123. Prod’Homme V, Boyer L, Dubois N, Mallavialle A, Munro P, Mouska X, et al. Cherubism allele heterozygosity amplifies microbe-induced inflammatory responses in murine macrophages. J Clin Invest. (2015) 125:1396–400. doi: 10.1172/JCI71081
124. Matsumoto Y, Dimitriou ID, La Rose J, Lim M, Camilleri S, Law N, et al. Tankyrase represses autoinflammation through the attenuation of TLR2 signaling. J Clin Invest. (2022) 132(7):e140869. doi: 10.1172/JCI140869
125. Dimitriou ID, Lee K, Akpan I, Lind EF, Barr VA, Ohashi PS, et al. Timed regulation of 3BP2 induction is critical for sustaining CD8(+) T cell expansion and differentiation. Cell Rep. (2018) 24:1123–35. doi: 10.1016/j.celrep.2018.06.075
126. Chen G, Dimitriou ID, La Rose J, Ilangumaran S, Yeh WC, Doody G, et al. The 3BP2 adapter protein is required for optimal B-cell activation and thymus-independent type 2 humoral response. Mol Cell Biol. (2007) 27:3109–22. doi: 10.1128/MCB.01014-06
127. Chen G, Dimitriou I, Milne L, Lang KS, Lang PA, Fine N, et al. The 3BP2 adapter protein is required for chemoattractant-mediated neutrophil activation. J Immunol. (2012) 189:2138–50. doi: 10.4049/jimmunol.1103184
128. Matsumoto Y, Rottapel R. PARsylation-mediated ubiquitylation: lessons from rare hereditary disease Cherubism. Trends Mol Med. (2023) 29:390–405. doi: 10.1016/j.molmed.2023.02.001
129. Asano Y, Matsumoto Y, Wada J, Rottapel R. E3-ubiquitin ligases and recent progress in osteoimmunology. Front Immunol. (2023) 14:1120710. doi: 10.3389/fimmu.2023.1120710
130. Asano Y, Matsumoto Y, He F, Katsuyama T, Katsuyama E, Tsuji S, et al. Pharmacologic inhibition of PARP5, but not that of PARP1 or 2, promotes cytokine production and osteoclastogenesis through different pathways. Clin Exp Rheumatol. (2023) 41:1735–45. doi: 10.55563/clinexprheumatol/qf55h8
131. Matsumoto Y, Rottapel R. Bone dynamics and inflammation: Lessons from rarediseases. Immunol Med (Review). (2020) 43(2):61–4. doi: 10.1080/25785826.2020.1720104
132. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. (2006) 127:469–80. doi: 10.1016/j.cell.2006.10.018
133. Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. (2012) 149:1192–205. doi: 10.1016/j.cell.2012.05.012
134. Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. (2017) 169:985–99. doi: 10.1016/j.cell.2017.05.016
135. Zhang X, Yu X. Crosstalk between Wnt/beta-catenin signaling pathway and DNA damage response in cancer: a new direction for overcoming therapy resistance. Front Pharmacol. (2023) 14:1230822. doi: 10.3389/fphar.2023.1230822
136. Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan C, et al. The lncRNA NEAT1 activates Wnt/beta-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. (2018) 11:113. doi: 10.1186/s13045-018-0656-7
137. Cao MQ, You AB, Zhu XD, Zhang W, Zhang YY, Zhang SZ, et al. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J Hematol Oncol. (2018) 11:12. doi: 10.1186/s13045-018-0555-y
138. He S, Tang S. WNT/β-catenin signaling in the development of liver cancers. Biomed Pharmacother. (2020) 132:110851. doi: 10.1016/j.biopha.2020.110851
139. Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. (2013) 13:11–26. doi: 10.1038/nrc3419
140. Ge X, Wang X. Role of Wnt canonical pathway in hematological Malignancies. J Hematol Oncol. (2010) 3:33. doi: 10.1186/1756-8722-3-33
141. Gajos-Michniewicz A, Czyz M. WNT signaling in melanoma. Int J Mol Sci. (2020) 21(14):4852. doi: 10.3390/ijms21144852
142. Pai SG, Carneiro BA, Mota JM, Costa R, Leite CA, Barroso-Sousa R, et al. Wnt/beta-catenin pathway: modulating anticancer immune response. J Hematol Oncol. (2017) 10:101. doi: 10.1186/s13045-017-0471-6
143. Zhang Y, Wang X. Targeting the Wnt/beta-catenin signaling pathway in cancer. J Hematol Oncol. (2020) 13:165. doi: 10.1186/s13045-020-00990-3
144. Yang E, Tacchelly-Benites O, Wang Z, Randall MP, Tian A, Benchabane H, et al. Wnt pathway activation by ADP-ribosylation. Nat Commun. (2016) 7:11430. doi: 10.1038/ncomms11430
145. Mariotti L, Templeton CM, Ranes M, Paracuellos P, Cronin N, Beuron F, et al. Tankyrase requires SAM domain-dependent polymerization to support wnt-beta-catenin signaling. Mol Cell. (2016) 63:498–513. doi: 10.1016/j.molcel.2016.06.019
146. Riccio AA, McCauley M, Langelier MF, Pascal JM. Tankyrase sterile alpha motif domain polymerization is required for its role in wnt signaling. Structure. (2016) 24:1573–81. doi: 10.1016/j.str.2016.06.022
147. Pillay N, Mariotti L, Zaleska M, Inian O, Jessop M, Hibbs S, et al. Structural basis of tankyrase activation by polymerization. Nature. (2022) 612:162–9. doi: 10.1038/s41586-022-05449-8
148. Pan F, Shen F, Yang L, Zhang L, Guo W, Tian J. Inhibitory effects of XAV939 on the proliferation of small-cell lung cancer H446 cells and Wnt/beta-catenin signaling pathway. vitro Oncol Lett. (2018) 16:1953–8. doi: 10.3892/ol.2018.8790
149. Li C, Zheng X, Han Y, Lv Y, Lan F, Zhao J. XAV939 inhibits the proliferation and migration of lung adenocarcinoma A549 cells through the WNT pathway. Oncol Lett. (2018) 15(6):8973–82. doi: 10.3892/ol.2018.8491
150. Stakheev D, Taborska P, Strizova Z, Podrazil M, Bartunkova J, Smrz D. The WNT/beta-catenin signaling inhibitor XAV939 enhances the elimination of LNCaP and PC-3 prostate cancer cells by prostate cancer patient lymphocytes. vitro Sci Rep. (2019) 9:4761. doi: 10.1038/s41598-019-41182-5
151. Fukumoto T, Zhu H, Nacarelli T, Karakashev S, Fatkhutdinov N, Wu S, et al. N(6)-methylation of adenosine of FZD10 mRNA contributes to PARP inhibitor resistance. Cancer Res. (2019) 79:2812–20. doi: 10.1158/0008-5472.CAN-18-3592
152. Yamamoto TM, McMellen A, Watson ZL, Aguilera J, Ferguson R, Nurmemmedov E, et al. Activation of Wnt signaling promotes olaparib resistant ovarian cancer. Mol Carcinog. (2019) 58:1770–82. doi: 10.1002/mc.23064
153. Kaur A, Lim JYS, Sepramaniam S, Patnaik S, Harmston N, Lee MA, et al. WNT inhibition creates a BRCA-like state in Wnt-addicted cancer. EMBO Mol Med. (2021) 13:e13349. doi: 10.15252/emmm.202013349
154. Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. (2006) 7:606–19. doi: 10.1038/nrg1879
155. Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol: Mech Dis. (2009) 4:127–50. doi: 10.1146/annurev.pathol.4.110807.092311
156. Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. (2018) 19:547–62. doi: 10.1038/s41580-018-0015-0
157. Álvarez-Garcia V, Tawil Y, Wise HM, Leslie NR. Mechanisms of PTEN loss in cancer: It’s all about diversity. Semin Cancer Biol. (2019) 59:66–79. doi: 10.1016/j.semcancer.2019.02.001
158. Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. (2014) 505:495–501. doi: 10.1038/nature12912
159. Lin Y, Kang T, Zhou BP. Doxorubicin enhances Snail/LSD1-mediated PTEN suppression in a PARP1-dependent manner. Cell Cycle. (2014) 13:1708–16. doi: 10.4161/cc.28619
160. Mariano G, Ricciardi MR, Trisciuoglio D, Zampieri M, Ciccarone F, Guastafierro T, et al. PARP inhibitor ABT-888 affects response of MDA-MB-231 cells to doxorubicin treatment, targeting Snail expression. Oncotarget. (2015) 6:15008–21. doi: 10.18632/oncotarget.3634
161. Zin I, China A, Khan K, Nag JK, Vasu K, Deshpande GM, et al. AKT-dependent nuclear localization of EPRS1 activates PARP1 in breast cancer cells. Proc Natl Acad Sci U.S.A. (2024) 121:e2303642121. doi: 10.1073/pnas.2303642121
162. Ibrahim YH, Garcia-Garcia C, Serra V, He L, Torres-Lockhart K, Prat A, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discovery. (2012) 2:1036–47. doi: 10.1158/2159-8290.CD-11-0348
163. Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmana J, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discovery. (2012) 2:1048–63. doi: 10.1158/2159-8290.CD-11-0336
164. Yap TA, Kristeleit R, Michalarea V, Pettitt SJ, Lim JSJ, Carreira S, et al. Phase I trial of the PARP inhibitor olaparib and AKT inhibitor capivasertib in patients with BRCA1/2- and non-BRCA1/2-mutant cancers. Cancer Discovery. (2020) 10:1528–43. doi: 10.1158/2159-8290.CD-20-0163
165. Franklin JM, Wu Z, Guan K-L. Insights into recent findings and clinical application of YAP and TAZ in cancer. Nat Rev Cancer. (2023) 23:512–25. doi: 10.1038/s41568-023-00579-1
166. Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. (2014) 158:185–97. doi: 10.1016/j.cell.2014.06.003
167. Wang Y, Xu X, Maglic D, Dill MT, Mojumdar K, Ng PK, et al. Comprehensive molecular characterization of the hippo signaling pathway in cancer. Cell Rep. (2018) 25:1304–17 e5. doi: 10.1016/j.celrep.2018.10.001
168. Luo J, Deng L, Zou H, Guo Y, Tong T, Huang M, et al. New insights into the ambivalent role of YAP/TAZ in human cancers. J Exp Clin Cancer Res. (2023) 42:130. doi: 10.1186/s13046-023-02704-2
169. Wang W, Huang J, Chen J. Angiomotin-like proteins associate with and negatively regulate YAP1. J Biol Chem. (2011) 286:4364–70. doi: 10.1074/jbc.C110.205401
170. Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. (2011) 286:7018–26. doi: 10.1074/jbc.C110.212621
171. Moleirinho S, Guerrant W, Kissil JL. The Angiomotins–from discovery to function. FEBS Lett. (2014) 588:2693–703. doi: 10.1016/j.febslet.2014.02.006
172. Amirifar P, Kissil J. The role of Motin family proteins in tumorigenesis-an update. Oncogene. (2023) 42:1265–71. doi: 10.1038/s41388-023-02677-8
173. Li Y, Zhang X, Liu N, Liu R, Zhang W, Chen L, et al. RNF166 promotes colorectal cancer progression by recognizing and destabilizing poly-ADP-ribosylated angiomotins. Cell Death Dis. (2024) 15:211. doi: 10.1038/s41419-024-06595-9
174. Troilo A, Benson EK, Esposito D, Garibsingh RA, Reddy EP, Mungamuri SK, et al. Angiomotin stabilization by tankyrase inhibitors antagonizes constitutive TEAD-dependent transcription and proliferation of human tumor cells with Hippo pathway core component mutations. Oncotarget. (2016) 7:28765–82. doi: 10.18632/oncotarget.9117
175. Wang H, Lu B, Castillo J, Zhang Y, Yang Z, McAllister G, et al. Tankyrase inhibitor sensitizes lung cancer cells to endothelial growth factor receptor (EGFR) inhibition via stabilizing angiomotins and inhibiting YAP signaling. J Biol Chem. (2016) 291:15256–66. doi: 10.1074/jbc.M116.722967
176. Campbell CI, Samavarchi-Tehrani P, Barrios-Rodiles M, Datti A, Gingras AC, Wrana JL. The RNF146 and tankyrase pathway maintains the junctional Crumbs complex through regulation of angiomotin. J Cell Sci. (2016) 129:3396–411. doi: 10.1242/jcs.188417
177. Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. (1998) 18:38–43. doi: 10.1038/ng0198-38
178. Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. (1998) 391:184–7. doi: 10.1038/34432
179. Giardiello FM, Welsh SB, Hamilton SR, Offerhaus GJ, Gittelsohn AM, Booker SV, et al. Increased risk of cancer in the Peutz-Jeghers syndrome. N Engl J Med. (1987) 316:1511–4. doi: 10.1056/NEJM198706113162404
180. van Lier MG, Mathus-Vliegen EM, Wagner A, van Leerdam ME, Kuipers EJ. High cumulative risk of intussusception in patients with Peutz-Jeghers syndrome: time to update surveillance guidelines? Am J Gastroenterol. (2011) 106:940–5. doi: 10.1038/ajg.2010.473
181. van Lier MG, Westerman AM, Wagner A, Looman CW, Wilson JH, de Rooij FW, et al. High cancer risk and increased mortality in patients with Peutz-Jeghers syndrome. Gut. (2011) 60:141–7. doi: 10.1136/gut.2010.223750
182. Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, et al. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. (2003) 13:2004–8. doi: 10.1016/j.cub.2003.10.031
183. Li N, Huang D, Lu N, Luo L. Role of the LKB1/AMPK pathway in tumor invasion and metastasis of cancer cells (Review). Oncol Rep. (2015) 34:2821–6. doi: 10.3892/or.2015.4288
184. Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. (2002) 62:3659–62.
185. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. (2008) 455:1069–75. doi: 10.1038/nature07423
186. Wingo SN, Gallardo TD, Akbay EA, Liang MC, Contreras CM, Boren T, et al. Somatic LKB1 mutations promote cervical cancer progression. PloS One. (2009) 4:e5137. doi: 10.1371/journal.pone.0005137
187. Long LL, Ma SC, Guo ZQ, Zhang YP, Fan Z, Liu LJ, et al. PARP inhibition induces synthetic lethality and adaptive immunity in LKB1-mutant lung cancer. Cancer Res. (2023) 83:568–81. doi: 10.1158/0008-5472.CAN-22-1740
188. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. (2020) 70:7–30. doi: 10.3322/caac.21590
189. Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discovery. (2019) 9:176–98. doi: 10.1158/2159-8290.CD-18-1177
190. Howlader N, Altekruse SF, Li CI, Chen VW, Clarke CA, Ries LA, et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst. (2014) 106. doi: 10.1093/jnci/dju055
191. Lang GT, Shi JX, Hu X, Zhang CH, Shan L, Song CG, et al. The spectrum of BRCA mutations and characteristics of BRCA-associated breast cancers in China: Screening of 2,991 patients and 1,043 controls by next-generation sequencing. Int J Cancer. (2017) 141:129–42. doi: 10.1002/ijc.30692
192. Hu C, Hart SN, Gnanaolivu R, Huang H, Lee KY, Na J, et al. A population-based study of genes previously implicated in breast cancer. New Engl J Med. (2021) 384:440–51. doi: 10.1056/NEJMoa2005936
193. Riaz N, Blecua P, Lim RS, Shen R, Higginson DS, Weinhold N, et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun. (2017) 8:857. doi: 10.1038/s41467-017-00921-w
194. Cheng HH, Shevach JW, Castro E, Couch FJ, Domchek SM, Eeles RA, et al. BRCA1, BRCA2, and associated cancer risks and management for male patients. JAMA Oncol. (2024) 10(9):1272–81. doi: 10.1001/jamaoncol.2024.2185
195. O’Malley DM, Krivak TC, Kabil N, Munley J, Moore KN. PARP inhibitors in ovarian cancer: A review. Target Oncol. (2023) 18:471–503. doi: 10.1007/s11523-023-00970-w
196. Morganti S, Marra A, De Angelis C, Toss A, Licata L, Giugliano F, et al. PARP inhibitors for breast cancer treatment: A review. JAMA Oncol. (2024) 10:658–70. doi: 10.1001/jamaoncol.2023.7322
197. Zambelli A, Cortesi L, Gaudio M, Arpino G, Bianchini G, Caruso F, et al. Parp-inhibitors in the therapeutic landscape of breast cancer patients with BRCA1 and BRCA2 pathogenic germline variants: An Italian consensus paper and critical review. Cancer Treat Rev. (2024) 130:102815. doi: 10.1016/j.ctrv.2024.102815
198. Desai D, Khandwala P, Parsi M, Potdar R. PARP inhibitors: shifting the paradigm in the treatment of pancreatic cancer. Med Oncol. (2021) 38:61. doi: 10.1007/s12032-021-01507-9
199. Cai M, Song XL, Li XA, Chen M, Guo J, Yang DH, et al. Current therapy and drug resistance in metastatic castration-resistant prostate cancer. Drug Resist Update. (2023) 68:100962. doi: 10.1016/j.drup.2023.100962
200. Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. (2012) 586:2692–704. doi: 10.1016/j.febslet.2012.04.045
201. Hu K, Wu W, Li Y, Lin L, Chen D, Yan H, et al. Poly(ADP-ribosyl)ation of BRD7 by PARP1 confers resistance to DNA-damaging chemotherapeutic agents. EMBO Rep. (2019) 20(5):e46166. doi: 10.15252/embr.201846166
202. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. (2012) 12:252–64. doi: 10.1038/nrc3239
203. Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. (2008) 26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331
204. Wang X, Teng F, Kong L, Yu J. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther. (2016) 9:5023–39. doi: 10.2147/OTT.S105862
205. Ichikawa M, Chen L. Role of B7-H1 and B7-H4 molecules in down-regulating effector phase of T-cell immunity: novel cancer escaping mechanisms. Front Biosci. (2005) 10:2856–60. doi: 10.2741/1742
206. Oyewole-Said D, Konduri V, Vazquez-Perez J, Weldon SA, Levitt JM, Decker WK. Beyond T-cells: functional characterization of CTLA-4 expression in immune and non-immune cell types. Front Immunol. (2020) 11:608024. doi: 10.3389/fimmu.2020.608024
207. Brunet JF, Denizot F, Luciani MF, Roux-Dosseto M, Suzan M, Mattei MG, et al. A new member of the immunoglobulin superfamily–CTLA-4. Nature. (1987) 328:267–70. doi: 10.1038/328267a0
208. Sharma P, Allison JP. Dissecting the mechanisms of immune checkpoint therapy. Nat Rev Immunol. (2020) 20:75–6. doi: 10.1038/s41577-020-0275-8
209. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. (2018) 359:1350–5. doi: 10.1126/science.aar4060
210. Shiravand Y, Khodadadi F, Kashani SMA, Hosseini-Fard SR, Hosseini S, Sadeghirad H, et al. Immune checkpoint inhibitors in cancer therapy. Curr Oncol. (2022) 29:3044–60. doi: 10.3390/curroncol29050247
211. Sharma P, Siddiqui BA, Anandhan S, Yadav SS, Subudhi SK, Gao J, et al. The next decade of immune checkpoint therapy. Cancer Discovery. (2021) 11:838–57. doi: 10.1158/2159-8290.CD-20-1680
212. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. (2017) 168:707–23. doi: 10.1016/j.cell.2017.01.017
213. Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, et al. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res. (2017) 23:3711–20. doi: 10.1158/1078-0432.CCR-16-3215
214. Sato H, Niimi A, Yasuhara T, Permata TBM, Hagiwara Y, Isono M, et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun. (2017) 8:1751. doi: 10.1038/s41467-017-01883-9
215. Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J Natl Cancer Inst. (2017) 109(1):djw199. doi: 10.1093/jnci/djw199
216. Chabanon RM, Muirhead G, Krastev DB, Adam J, Morel D, Garrido M, et al. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J Clin Invest. (2019) 129:1211–28. doi: 10.1172/JCI123319
217. Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discovery. (2019) 9:646–61. doi: 10.1158/2159-8290.CD-18-1020
218. Ding L, Chen X, Xu X, Qian Y, Liang G, Yao F, et al. PARP1 suppresses the transcription of PD-L1 by poly(ADP-ribosyl)ating STAT3. Cancer Immunol Res. (2019) 7:136–49. doi: 10.1158/2326-6066.CIR-18-0071
219. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. (2015) 523:231–5. doi: 10.1038/nature14404
220. Waaler J, Mygland L, Tveita A, Strand MF, Solberg NT, Olsen PA, et al. Tankyrase inhibition sensitizes melanoma to PD-1 immune checkpoint blockade in syngeneic mouse models. Commun Biol. (2020) 3:196. doi: 10.1038/s42003-020-0916-2
221. Brinch SA, Amundsen-Isaksen E, Espada S, Hammarstrom C, Aizenshtadt A, Olsen PA, et al. The tankyrase inhibitor OM-153 demonstrates antitumor efficacy and a therapeutic window in mouse models. Cancer Res Commun. (2022) 2:233–45. doi: 10.1158/2767-9764.CRC-22-0027
222. Domchek SM, Postel-Vinay S, Im SA, Park YH, Delord JP, Italiano A, et al. Olaparib and durvalumab in patients with germline BRCA-mutated metastatic breast cancer (MEDIOLA): an open-label, multicentre, phase 1/2, basket study. Lancet Oncol. (2020) 21:1155–64. doi: 10.1016/S1470-2045(20)30324-7
223. Fanucci KA, Pilat MJ, Shyr D, Shyr Y, Boerner SA, Durecki D, et al. Abstract CT145: Olaparib +/- atezolizumab in patients with BRCA-mutated (BRCAmt) locally advanced unresectable or metastatic (advanced) breast cancer: an open-label, multicenter, randomized phase II trial. Cancer Res. (2023) 83:CT145–CT. doi: 10.1158/1538-7445.Am2023-ct145
224. Rugo H, Robson M, Im S-A, Dalenc F, Ruiz EY, Im Y-H, et al. Abstract GS01-05: pembrolizumab + Olaparib vs pembrolizumab + Chemotherapy after induction with pembrolizumab + Chemotherapy for locally recurrent inoperable or metastatic TNBC: randomized open-label phase 2 KEYLYNK-009 study. Cancer Res. (2024) 84:GS01–5-GS-5. doi: 10.1158/1538-7445.Sabcs23-gs01-05
225. Reiss KA, Mick R, Teitelbaum U, O’Hara M, Schneider C, Massa R, et al. Niraparib plus nivolumab or niraparib plus ipilimumab in patients with platinum-sensitive advanced pancreatic cancer: a randomised, phase 1b/2 trial. Lancet Oncol. (2022) 23:1009–20. doi: 10.1016/S1470-2045(22)00369-2
226. Buckley CW, O’Reilly EM. Next-generation therapies for pancreatic cancer. Expert Rev Gastroenterol Hepatol. (2024) 18:55–72. doi: 10.1080/17474124.2024.2322648
227. Bi R, Chen L, Huang M, Qiao Z, Li Z, Fan G, et al. Emerging strategies to overcome PARP inhibitors’ resistance in ovarian cancer. Biochim Biophys Acta Rev Cancer. (2024) 1879:189221. doi: 10.1016/j.bbcan.2024.189221
228. Konstantinopoulos PA, Waggoner S, Vidal GA, Mita M, Moroney JW, Holloway R, et al. Single-arm phases 1 and 2 trial of niraparib in combination with pembrolizumab in patients with recurrent platinum-resistant ovarian carcinoma. JAMA Oncol. (2019) 5:1141–9. doi: 10.1001/jamaoncol.2019.1048
229. Yu EY, Piulats JM, Gravis G, Fong PCC, Todenhofer T, Laguerre B, et al. Pembrolizumab plus olaparib in patients with metastatic castration-resistant prostate cancer: long-term results from the phase 1b/2 KEYNOTE-365 cohort A study. Eur Urol. (2023) 83:15–26. doi: 10.1016/j.eururo.2022.08.005
230. Yu EY, Park SH, Goh JCH, Shin SJ, Mehra N, McDermott R, et al. 1362MO Pembrolizumab + olaparib vs abiraterone (abi) or enzalutamide (enza) for patients (pts) with previously treated metastatic castration-resistant prostate cancer (mCRPC): Randomized open-label phase III KEYLYNK-010 study. Ann Oncol. (2022) 33:S1163–S4. doi: 10.1016/j.annonc.2022.07.1494
Keywords: PARylation, cancer, post-transcriptional regulation, ubiquitylation, immune system
Citation: Matsumoto K, Matsumoto Y and Wada J (2025) PARylation-mediated post-transcriptional modifications in cancer immunity and immunotherapy. Front. Immunol. 16:1537615. doi: 10.3389/fimmu.2025.1537615
Received: 01 December 2024; Accepted: 17 February 2025;
Published: 11 March 2025.
Edited by:
Duanwu Zhang, Fudan University, ChinaReviewed by:
Yuan Tian, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, ChinaCopyright © 2025 Matsumoto, Matsumoto and Wada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoshinori Matsumoto, eW1hdHN1bW90b0Bva2F5YW1hLXUuYWMuanA=
 Kazuya Matsumoto
Kazuya Matsumoto Yoshinori Matsumoto
Yoshinori Matsumoto Jun Wada
Jun Wada