
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 17 February 2025
Sec. Cancer Immunity and Immunotherapy
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1536569
This article is part of the Research Topic Exosomes in Renal Cell Cancer: A New Era in Diagnosis and Treatment View all articles
Aptamer-based immunotherapy can be a new hope for treating solid tumors with personalized and specific approaches toward cancer therapies. Aptamers are small synthetic single-stranded nucleic acids that may bring in a paradigm shift in treating solid tumors. These are highly selective drugs applied in cellular immunotherapy, cytokine modulation, and immune checkpoint suppression. This review provides an overview of the recent advances in aptamer-based technologies with specific key clinical trials involving AON-D21 and AM003. Aptamers are potently active in immune regulation and tumor targeting. However, aptamer stability and bioavailability are seriously compromised by the issues relating to renal clearance and rapid degradation through nucleases. The latter are reviewed here along with novel improvements, some of which involve chemical modifications that greatly enhance stability and prolong the circulation time; exemplary such modifications are PEGylation, cholesterol conjugation, and the synthesis of circular nucleic acids. The regulatory aspect is also crucial. For example, in addition to specific strategies to prevent drug-drug interactions (DDIs) in cancer remediation medications, this paper underscores the need of risk assessment, particularly because of immunogenicity and organ failure. The use of aptamers is expanded by the development of SOMAmers, X-aptamers, and bioinformatics. To make aptamer-based drugs a major part of cancer treatment, future research should concentrate more on resolving existing issues and expanding their beneficial uses.
Carcinogenesis encompasses not just genetic events but also heritable modifications in gene expression that occur without alterations to the DNA sequence, resulting in the transformation of normal cells into malignant ones. Cancer is characterized by these aberrant genetic and epigenetic alterations (1). Immune cells such as T and B lymphocytes, mast cells, natural killer cells, dendritic cells (DCs), polymorphonuclear cells, macrophages, and non-immune cells including endothelial and stromal cells coexist alongside tumor cells. They develop complex relationships with these cells, which are crucial in determining the progression and features of the tumor (2).
Cancer may have originated in the times when unicellular organisms developed into multi-cellular organisms. So far, the oldest evidence of cancer has been that in a fossil record of a shell-less stem turtle, Pappochelys rosinae, from 240 million years ago showing osteosarcoma. Until recently, tumor surgery, cytotoxic chemotherapy drugs, and radiation therapy remained the cornerstones in cancer treatment. Only from the beginning of the twenty-first century did cancer treatment with immunotherapy emerge. Inspired by the case of a soldier with a cancerous tumor regressing in an infection with Streptococcus bacteria, Coley proposed in the late 19th century that cancer could be treated through activated immune response (3).
William Coley, a bone surgeon in New York, invented “Coley’s Toxins” to treat osteosarcoma patients. His first patient, a young woman with hand osteosarcoma, died from metastatic illness after surgery. Coley’s experience inspired him to study hospital medical records for 90 sarcoma patients, focusing on one patient’s illness progression. His work is considered the “Father of Immunotherapy”. Coley discovered a case report of a sarcoma patient whose tumor shrunk after contracting erysipelas, a skin infection. He wondered if it was possible to cause erysipelas in cancer patients. German physician Friedrich Fehleisen isolated the bacteria responsible for erysipelas as Streptococcus in 1883. Coley tested his hypothesis and began administering injections of the bacteria to patients with sarcoma and finally settled on heat-killed Streptococcus pyogenes and Serratia marcescens. Coley, a pioneer in the study of cancer, injected over 1000 cancer patients and published over 150 articles. His toxins were later reevaluated in a controlled experiment, showing anticancer effects. Ehrlich believed that cancer incidence is rare, but abnormal cell production is frequent, suggesting a host defense system against cancer. Although experimental proof was unavailable, Coley’s work remains significant. Burnet and Thomas expanded on these concepts more than 50 years later, coining the term “immune surveillance” theory (3).
The idea of immunotherapy is a revolutionary approach that uses the body’s immune system to target and destroy cancer cells was first postulated by Paul Ehrlich in 1909 (4), offering a more targeted and long-lasting response compared to conventional treatments (3). This strategy combines cytokine therapies, cancer vaccines, adoptive cell therapy (CAR-T cells), and immune checkpoint inhibitors (5). As the field develops, its influence grows alongside established cancer treatments and new breakthroughs.
Multidisciplinary cancer treatment is being revolutionized by cancer immunotherapy, which is also opening up new therapeutic avenues. It continues to be included into multidisciplinary cancer care because, in certain cases, it can provide more patients long-lasting disease management (6). For many years, the foundations of cancer treatment have been radiation, chemotherapy, surgery, or a combination of these. Most patients with advanced solid tumors are not candidates for surgical removal, even if it may be curative in some circumstances (7). These treatments lack of specificity results in a high rate of recurrence and significant toxicity (8).
Additionally, the physical makeup of the tumor environment, including increased stiffness, high interstitial fluid pressure (IFP), and solid stress, poses significant challenges to delivering cancer drugs effectively—especially in dense tumors (9, 10). These forces compress blood vessels and block drugs from reaching the tumor core. High IFP limits drug movement, while solid stress from proteins like collagen further restricts diffusion (11). Combining treatments that address these physical barriers with standard therapies is showing promise in boosting drug delivery and improving outcomes for resistant tumors (12). Hence, innovative delivery methods should be explored to optimize therapeutic concentration in solid tumors and to overcome the delivery obstacles created by physical forces within the tumor microenvironment (2, 13).
Aptamer technology was developed in the 1990s, simultaneously, by two independent groups, Tuerk and Gold (14) and Ellington and Szostak (15). Aptamers known as “chemical antibodies” are a class of small single-stranded DNA or RNA oligonucleotides, with a three-dimensional structure enabling the target binding and specificity such as proteins, cells, or small molecules (16). Aptamer-based immunotherapy is a new and extremely promising method for immune regulation and cancer treatment. Aptamers have the ability to directly inhibit tumor cells, therefore serving as therapeutic agents. They may also be used in targeted drug delivery systems, which reduce toxicity to healthy tissues by directing medications precisely to tumor cells (17).
This article examines the innovative application of aptamers in cancer immunotherapy, with a focus on solid tumors. We describe synthesized single-stranded nucleic acids called aptamers and the ways in which bioinformatics techniques are advancing their development. The difficulties in using oligonucleotides to target solid tumors successfully are also covered, including problems with stability and intracellular delivery. In addition, we talked about FDA regulations for oligonucleotide-based products and the current clinical trials using the AON-D21 and AM003 aptamers.
Aptamer-based immunotherapy can be categorized into several key approaches, each leveraging the unique properties of aptamers to engage the immune system and target cancer cells making aptamers a promising tool in cancer immunotherapy. These categories include immune checkpoint inhibition, cytokine modulation, cellular immunotherapy and bispecific aptamer (Figure 1).
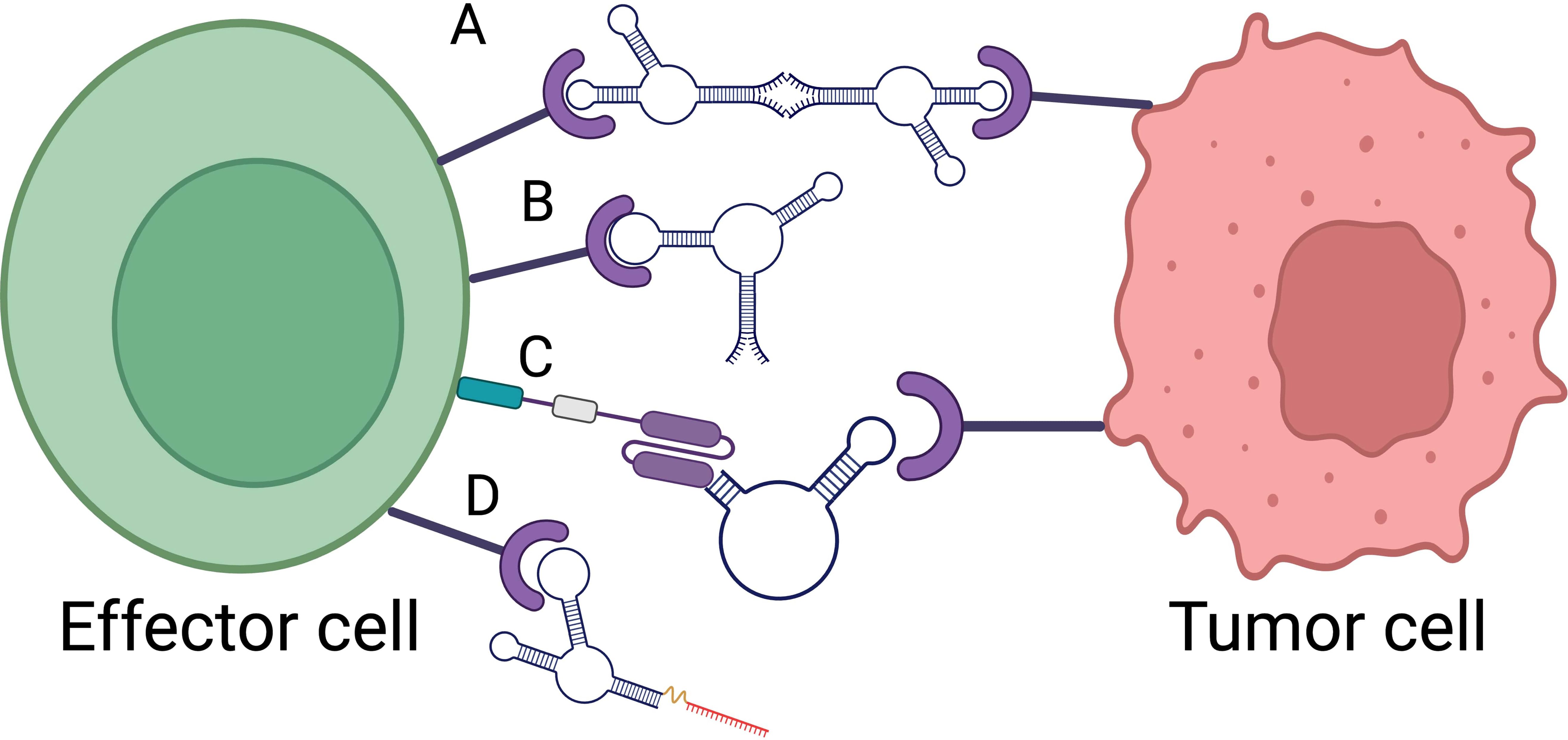
Figure 1. Aptamer based immunotherapy categories. (A) Bispecific aptamer. (B) Immune checkpoint inhibitor. (C) Car-ap. (D) Aptamer conjugated siRNA. Created with BioRender.com.
Immune checkpoint inhibitors block the proteins needed by cancer cells, such as CTLA-4, to avoid immune destruction. It aims to restore the immune system’s ability to detect and eliminate tumour cells. In this regard, a new DNA aptamer, CTLA-4 aptamer (aptCTLA-4), preferentially increased T cell proliferation while greatly inhibiting tumor development. AptCTLA-4 acts against the CTLA-4 protein, which is a key negative regulator of T cell activation. With a high affinity attachment to CTLA-4, a dissociation constant of 11.84 nM was reported. AptCTLA-4 blocks CTLA-4 from binding with its ligands, B7-1 (CD80) and B7-2 (CD86). This interaction is important because it inhibits T cell activation since CTLA-4 and CD28 compete for binding with B7 proteins, explained by Hossen in 2023 (18). Inhibition of CTLA-4 increases the lymphocyte proliferation rate, enhancing T cell activation and proliferation (19). Aptamers are less immunogenic than monoclonal antibodies, reducing the adverse immune-related events associated with them (20, 21).
A study compares Ipilimumab, an anti-CTLA-4 mAb with CTLA-4 aptamer, highlighting both advantages and disadvantages. AptCTLA-4 increases cytotoxicity T lymphocytes and tumor infiltrating cells, preventing tumor development in vivo while maintaining mice’s body weight. The half-life (22) of AptCTLA-4 is less than the antibodies. However, due to lack of clinical investigation, concerns about safety and effectiveness exist. Patients with these mAbs show long-lasting clinical improvements and sometimes extended survival (23, 24), but are linked to severe immune-related adverse effect including skin lesion, colitis, endocrinopathies (24) and systemic toxicity indicated by noticeable weight loss in mice (23).
The Programmed Death-1 inhibitory receptor and its ligand (PD-1/PD-L1) are essential for immunological suppression. Tasuku Honjo’s team at Kyoto University discovered a lymphoid cell surface protein called programmed cell death protein 1, which they believe triggers apoptosis. They identified PD-1 as an immunological receptor that inhibits or negatively regulated adaptive immune responses. In an artificial activation model, the interaction of PD-1/PD-L1 inhibited T-cell proliferation and cytokine production, indicating an internal inhibitory mechanism for autoreactive lymphocyte activation. Tumor cells that express PD-L1 on their surface hamper CD8+ T lymphocytes’ cytolytic effector activities. This also showed that this inhibition might be a potential way to preventing tumor cell escape (25). The FDA approved nivolumab, pembrolizumab, and atezolizumab in 2014 and 2016, respectively for treating metastatic melanoma, urothelial carcinoma, and non-small cell lung cancer, respectively, using humanized PD-1 mAbs (26).
Antibody can inhibit the interaction by reversing tumor immune evasion and produce strong antitumor reactions. However, dermatological toxicities, diarrhea, colitis, endocrine toxicities, hepatic toxicities, pneumonitis are general adverse events (26). However, a team has identified a DNA aptamer, MP7, that specifically binds to murine PD-1 receptor’s extracellular domain with high affinity. Functional assays demonstrated that MP7 effectively inhibits PD-L1 mediated suppression of IL2 secretion in primary T cells, thereby restoring T-cell function. To enhance the aptamer’s in vivo stability and circulation time, MP7 was conjugated with a 40kDa PEG moiety. This PEG-MP7 retained its ability to block the interaction. In vivo experiments using murine colon carcinoma model revealed the treatment with PEG-MP7 significantly suppressed tumor growth. Importantly, the study reported that the anti-PD1 aptamer did not induce off target effects, such as TLR9-triggered cytokine release (27).
Furthermore, the XA library constructed by Yang and associates consists of approximately 2×109 beads, each containing roughly 3×103 copies of a distinct chemically modified DNA strand. XA candidates are combinations of natural and modified nucleotides such as phosphorodithioate and several modified forms of dU. Such a broad scope of chemicals enhances interactions with targets to increase binding selectivity. The library was used to find two DNA aptamers, XA-PD1-78 and XA-PDL1-82, that bind with human PD-1 and PD-L1 proteins respectively. With cell lines of overexpression, the binding affinity and specificity were verified after aptamers with proper modification were produced. The binding intensities of these aptamers were equal to PD-1 and PD-L1 antibodies. These XAs are a synthetic substitute for antibodies not only for research but also for therapeutics (28).
Effective cytokine level manipulation during cancer therapies may be possible with aptamers. Targeting CD25, Axin-1 siRNAs complexed with the 4-1BB-binding oligonucleotide aptamer. CD8+ T cells that have been activated express the receptor 4-1BB. In activated CD8+ T cells, the 4-1BB aptamer-CD25 siRNA combination effectively reduced CD25 mRNA and protein levels in vitro. Treatment enhanced the antitumor response in mice models, both with a cellular vaccine and local radiation therapy (29) by promoting the development of long-lasting memory CD8+ T cells (30). Aptamers can deliver cytokines directly to the tumor site, enhancing immune cell recruitment and activation in the tumor microenvironment without causing systemic toxicity highlighting the importance of localized delivery in regulating aptamer-cytokine interactions (31). Downregulation and upregulation are a part of controlled modulation of cytokine levels. IL-2 upregulation at the site of 4-1BB expressing CD8+ T cells amplify immune activation while CD25 downregulation suppress immunosuppressive pathways (29), rebalancing the immune response toward anti-tumor activity.
Such fine tuning is critical in clinical applications to achieve the best therapeutic results minimizing the risk of cytokine release syndrome (CRS) and associated adverse effects (32). Since elevated blood levels are associated with increased toxicities, reducing plasma exposure while preserving efficacy has been the primary goal for the clinical development of cytokine treatments (33). Therefore, it is expected that successful therapeutic cytokine treatments need information of each patient’s unique immune profile and the ability to track changes related to the cytokine therapy (34). Dose titration and controlled-release formulations to regulate cytokine levels and minimize systemic exposure such as microsphere encapsulating cytokines (35).
Aptamers can be used to enhance the specificity of T-cell therapies. CAR-ap stands for Chimeric Antigen Receptor-specific binding Aptamer developed to overcome tumor immune escape mechanisms. Designed to specifically bind to CAR-positive cells, particularly those expressing CAR19-T. It allows for traceless enrichment of CAR-positive cells without the use of traditional methods that may leave markers. It also enables accurate in vivo monitoring of CAR-T cell expansion, providing insights into treatment efficacy. CAR-ap-enriched CAR19-T cells demonstrate comparable antitumor activity to those enriched using antibodies (36). Aptamer-guided cellular immunotherapies can be customized based on the tumor’s unique antigen profile, improving the efficacy and safety of treatment.
The development of a bi-specific aptamer (Bi-apt) platform for cancer therapeutics is formed by two monomeric aptamers, which can specifically bind to two kinds of targets simultaneously with high affinity (37). In the context of cancer therapeutics, both the bispecific aptamers can modulate the interaction between immune effector cells and tumor cells, and promote immune cell activation and tumor cell lysis via recruiting more lymphocytes around tumor cells (38). A highly stable CD16/PDL1 bi-specific aptamer was introduced by Zheng in 2022 (39) that may directly draw in CD16-positive natural killer (NK) cells to interact with tumor cells that express PD-L1, thereby mediating a strong antitumor immunity. CP-bi-apt also can be used as an immune checkpoint inhibitor to block up-regulated PD-L1, and thus, the function of NK cells can be restored, which promotes robust antitumor immune responses. By physically bringing immune cells in proximity to tumor cells, bispecific aptamers enhance the likelihood of immune cell-mediated killing of cancer cells.
Aptamers’ versatility allows for their integration into combination therapies, enhancing their therapeutic potential (40). These therapies can prevent tumor development and metastasis, reduce systemic toxicities (41), and lead to innovative cancer treatment strategies like aptamer-conjugated drug delivery systems, aptamer radiotherapy synergy, and aptamer-mediated immune cell recruitment with immunotherapy.
Aptamers can be used as targeting agents to deliver chemotherapeutic drugs, siRNAs, or nanocarriers directly to tumor cells, reducing off-target effects and systemic toxicity (42). A study found a novel HER2 aptamer (HB5) through SELEX and used it as a ligand to supply doxorubicin (Dox) to breast cancer cells in vitro. Apt-Dox, an aptamer-doxorubicin combination, decreased its toxic effects against HER2-negative breast cancer cells while maintaining Dox’s cytotoxicity against HER2-positive cells (43). Wang presented a novel approach for automated and modular production of next-generation ApDCs for targeted drug delivery applications using solid-phase synthesis techniques. This method allows multiple drug moieties to be conjugated onto a single aptamer, resulting in high drug-loading capacity and ease of use. Similar concept with different drug called mertansine (DM1) also been carried out by (44).
Radiation therapy (RT) aiding over 50% of cancer patients every year is one of the most popular therapies for solid tumors. It gives tumor tissue the highest possible radiation dosage (45). In one study, gold nanoparticles conjugated with the AS1411 aptamer was used to test the radiosensitization effect of 4MeV electron radiation on cancer cells. Clonogenic test and Au cell uptake data combined indicated that the aptamer has increased radiation-induced destruction of cells through Au absorption. Cancer cells become more sensitive to 4 MeV electron beams as a result only with AS1411/GNPs and not with GNP alone (46). Similar study conducted by Kardani to reduce mir-155 in breast cancer, they employed complex of Au nanoparticle AS1411 aptamer antagomir 155 (47).
Aptamers’ unique three-dimensional structure, achieved through intramolecular contacts, provides thermal stability and flexibility, essential for aptamer-target interaction (48). Aptamer were suitable for large scale preparation and easily modified. Notably, aptamers lack overt immunogenicity and exhibit favorable tissue permeability and safety in vivo (8). Aptamer made up of 12-30kda which is 15 times smaller than antibody (49).
Aptamers adopt a wide variety of structural motifs, such as stems, hairpins, bulges, pseudoknots, and G-quadruplexes (50). Nucleic acid, hydrogen bonds, hydrophobic interactions, and van der Waals interactions play crucial roles in stabilizing the aptamer’s three-dimensional structure and ensuring selective binding to targets (51). Aromatic stacking (π-π interactions) and electrostatic forces are also integral to the binding mechanism (21). Aptamers’ binding affinities toward target proteins can be tuned by modifying their structural makeup (52). There are favored amino acid-base hydrogen bonds. Arginine and lysine strongly favor guanine base while asparagine and glutamine prefer adenine. Arginine also makes a larger interaction with thymine and adenine (53). Hydrogen bonds facilitate the majority of aptamer-target binding (54). Phosphate groups are crucial for the creation of hydrogen bonds alongside nucleic acid bases. Side chains of a protein function as hydrogen bond contributors, while the phosphate groups in aptamers typically receive hydrogen bonds (55, 56). Aptamers, being composed of nucleic acids, have negatively charged phosphate backbones. This charge property enables them to interact with positively charged target proteins, facilitating electrostatic interactions (57). This electrostatic attraction often draws aptamers to positively charged protein regions, stabilizing the binding and contributing to the specificity and high affinity of the aptamer-protein interaction (58).
Aptamer-protein interactions may also be influenced by the hydrophobic interaction of aromatic rings with the aliphatic side chains of proteins (59). Furthermore, it has been verified that aromatic rings take part in π−π stacking, mostly because of the π orbitals’ overlap (60). The last and most significant is the van der Waals force, a crucial binding interaction, arises from the mutual attraction of dipoles and induced dipoles (61), with its strength significantly influenced by the surface dimensions (60). Aptamer-protein interactions are well known for their high sensitivity and specificity in detecting biomolecules, even with minimal changes in structure and content. This ability is attributed to aptamers’ conformational flexibility and their capacity to undergo structural changes upon binding to target molecules, allowing for precise and selective recognition (58).
Aptamers offer superior technology over antibodies. Unlike antibodies, which usually need to be stored at −80°C to avoid permanently denature and aggregate at higher temperatures (62), they can firstly renature after heat denaturation and remain stable at room temperature for years (63). Aptamers, smaller than antibodies, minimal toxicity, and scalability of production, overcome physiological barriers like blood-brain barriers, enhancing therapeutic treatments and gaining access to underdeveloped nations, making them useful for biodefense and accessing underdeveloped nations (64) The pros and cons of antibody and aptamer are tabulated in Table 1.
Aptamer was isolated usually using the conventional Systematic Evolution of Ligands by EXponential enrichment (SELEX) method, containing synthetic random nucleotide sequences length between 20-100 mers, and conserve regions at 5’ and 3’ oligonucleotides which serving as primers site for PCR amplification. The library typically contains 1015 – 1024 distinct molecules, providing a wide range of potential configurations, allowing researchers to select aptamers with the highest affinity for a ligand. SELEX generates aptamer-protein complexes by combining the library and target. The process involves removing non-specific sequences and eluding potential aptamers followed by PCR amplification. Double-stranded PCR amplicons are then separated for single stranded DNA preparation for continuation of SELEX round. SELEX undergoes multiple selection rounds to nature specific aptamer sequence and it normally took around 10–15 cycles shown in Figure 2 (65). Enrichment of SELEX cycle was observed and selected enriched cycle was then cloned and sequence in order to identify specific sequence candidates.
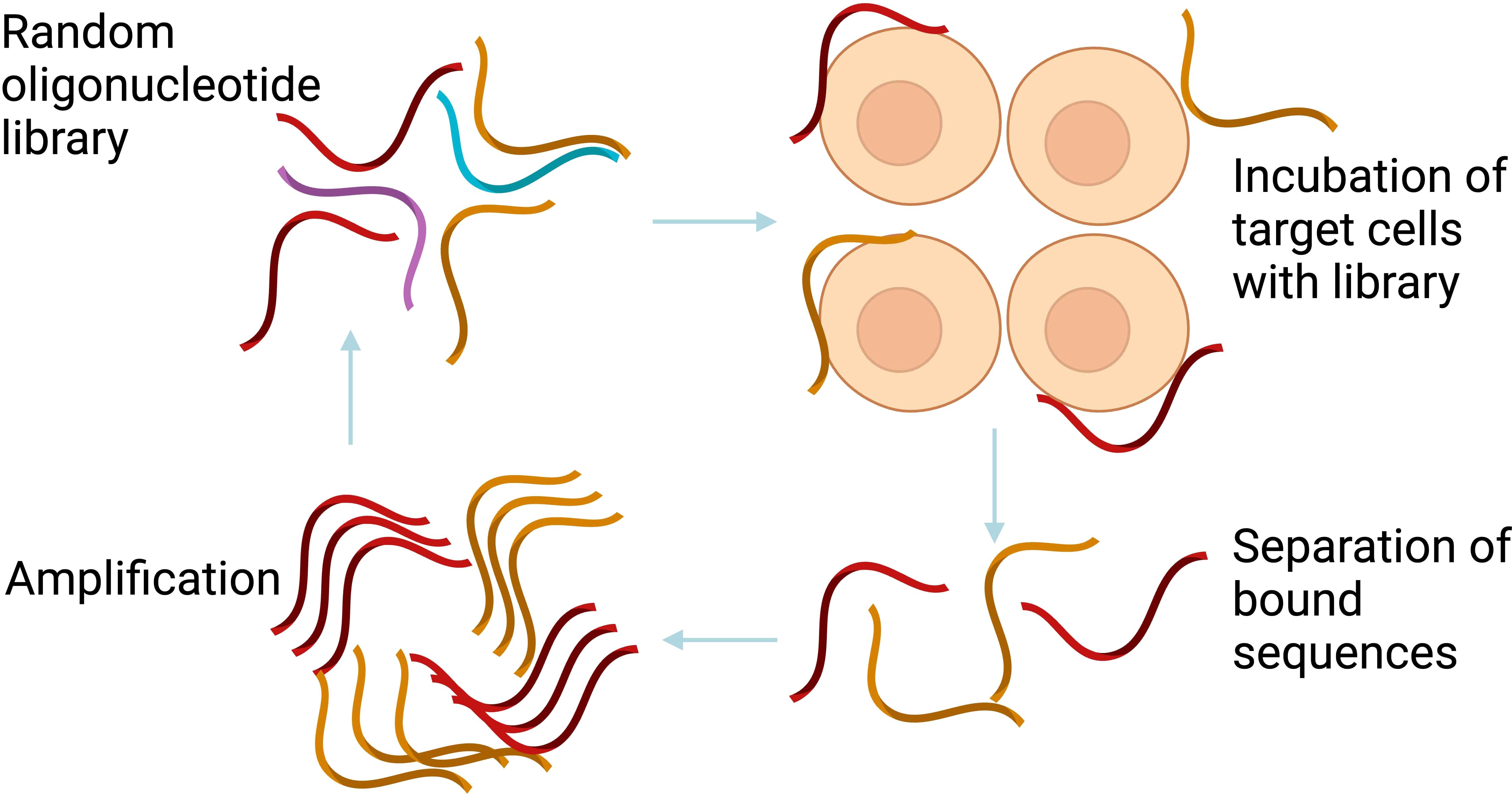
Figure 2. The conventional SELEX process. The conventional SELEX involves repeated rounds of selection and amplification to isolate high-affinity aptamers. Created with BioRender.com.
Aptamers for DNA and RNA are produced differently. The recovery step involves the use of conventional PCR for DNA aptamers, which are more durable and cost-effective. In contrast, RT-PCR is required for RNA aptamers, which display stronger intra-strand interactions and varied three-dimensional structures, enhancing affinity and specificity (66).
Conventional SELEX is a tedious and lengthy process, it needs very highly trained personnel. It has been reported to have a poor success rate to isolate very specific aptamer (67), leading to the development of in silico SELEX, a computational-based selection that can identify high affinity aptamer, shorten the aptamer selection process within an hour and provide additional information that can be retrieved such aptamer-protein binding site.
Chushak & Stone created the first in silico SELEX study by creating an extensive approach for selecting RNA aptamers (68). In order to choose aptamers with binding affinities to desired targets, the approach entails selecting RNA sequences based on their secondary structure, creating a library of 3D structures, and virtually screening the candidates shown in Figure 3. This technology can expedite the experimental procedures and ultimate aptamer selection by reducing the in vitro SELEX by 4–5 orders of magnitude (65).
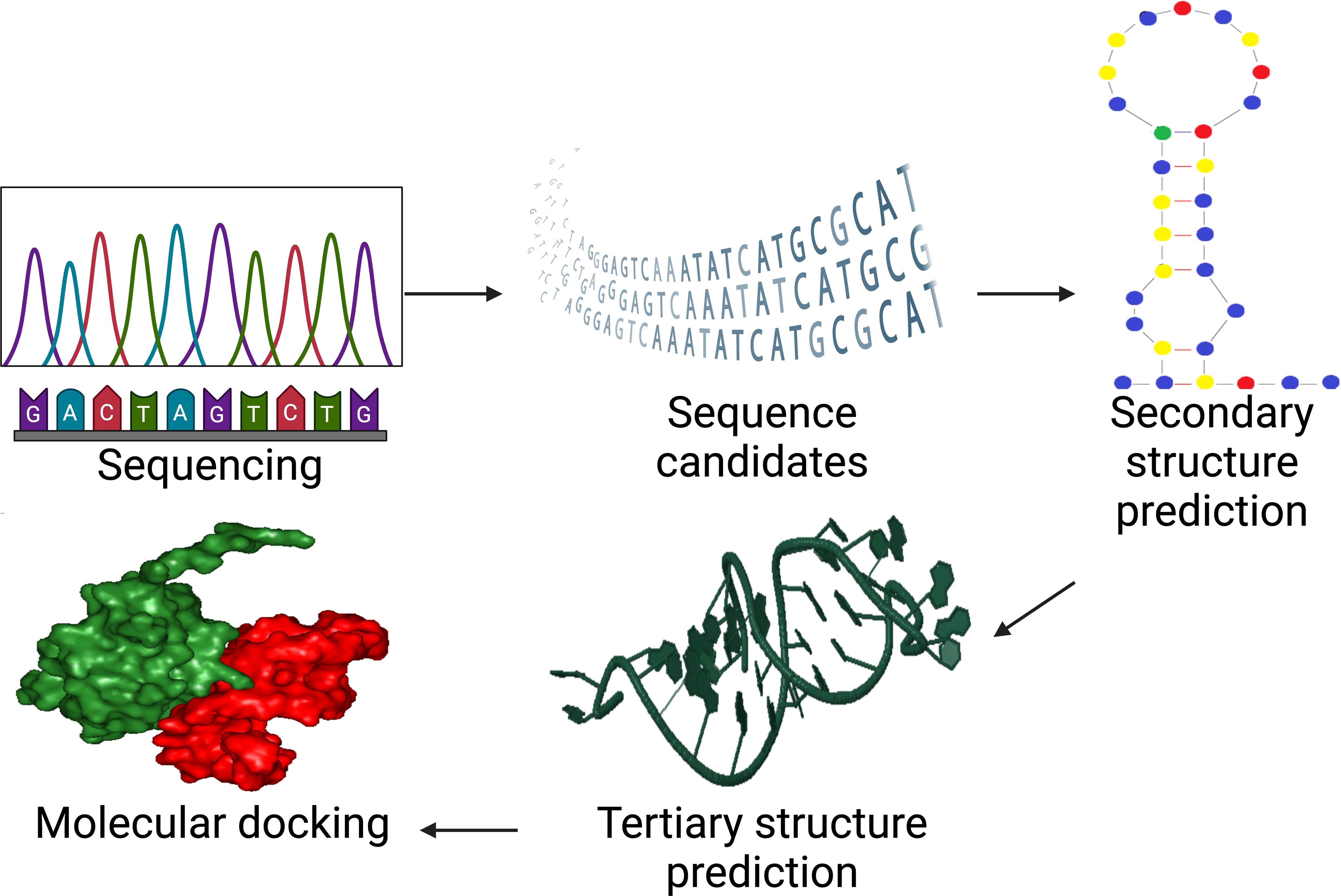
Figure 3. In silico SELEX process for aptamer selection whereas after experimental SELEX at amplification step, it incorporates sequencing, structure prediction, and molecular docking to accelerate aptamer design and optimization. Created with BioRender.com.
In addition, since in-silico SELEX doesn’t require the purchase of chemicals, lab supplies, or technical staff, it is less expensive than in vitro SELEX. These technologies improve library design by enhancing internal random sequence length, randomization values, chemical orientation, and primer annealing affinity in constant areas. The goal is to streamline the conventional SELEX process by reducing the need for physical experimentation (67).
Convenient and accurate aptamer creation methods have been researched, and computer-based approaches for selecting aptamers using aptamer structure prediction have been established (69). A new modern in silico method was developed to find high affinity aptamers by using 3D structural modeling and natural tRNA sequences. Bio-computation can optimize these aptamers, modeling and testing them through computational docking (70) by a compilation technology introduced by Biogenes Technologies called APTCAD shown in Figure 4. The selected RNA sequence from https://gtrnadb.ucsc.edu/ was then truncated in relation to sequence structure involving secondary structure prediction and tertiary structure optimization, followed by structural docking simulation between target molecule and aptamer candidates.

Figure 4. Computer interface for a software platform called APTCAD developed by Biogenes Technologies, where aptamers are designed and tested using computational tools. Natural tRNA sequences are selected from GtRNAdb, and in silico methods such as secondary and tertiary structure prediction, molecular docking, and dynamics simulations are employed to optimize aptamer candidates. Created with BioRender.com.
The candidate with the least energy is chosen, and molecular dynamics simulation is used to measure stability and binding energy. The binding affinity between the aptamer and target is determined, and chemical modification or point mutation can be used to increase it (Figure 4) (71). The secondary structure of an aptamer is crucial in binding to a specific target substance. The prediction of the tertiary structure is significantly influenced by its secondary structure (72). The Multiple Fold (mfold) web server is a widely used bioinformatics tool for identifying secondary structures of ssDNA (73), using thermodynamic approaches like Turner’s thermodynamics table and free-energy minimization algorithms to forecast hybridization and melting temperatures (74). This online service is accessible at http://unafold.rna.albany.edu (73).
Aptamer screening aids in predicting the tertiary structures of single-stranded oligonucleotide sequences, providing molecular insights into their dynamics and structure (67). An integrated pipeline was developed to predict DNA tertiary structure using RNA-based programs, addressing the lack of available computational tools for straight modelling (75, 76). RNA Composer is a widely utilized bioinformatics tool for transforming ssDNA secondary structures into RNA tertiary models (77) at http://rnacomposer.ibch.poznan.pl/Home. The method employs a motif template-based strategy to predict intricate structures such as pseudoknotted loops and multibranched loops (78).
The conversion of RNA tertiary structures into DNA tertiary structures can be achieved by altering bases, sugar backbones, and sugar residues (76) using Discovery Studio visualizer software version 3.5 (71). Once the tertiary structures of ssDNA have been identified, molecular docking must be applied to these structures (79). A computer technology called molecular docking uses the lowest ΔG binding energy to forecast the development of an aptamer-target complex (80).
Using atomistic simulations, molecular docking also helps determine the aptamer’s binding strength against the target. Apart from interactions, aptamers’ malleability, their flexibility or rigidity, impacts their binding to the target. Stiff aptamers bind to the target’s epitope without structural changes, similar to a lock and key concept. Flexible aptamers can alter their shape to fit the target’s epitope, enhancing their binding performance (67). While there are several reliable molecular docking simulation programs available, AutoDock Vina, Glide, and AutoDock Gold have been proven to be the most effective choices with the highest ratings, while SwissDock, PatchDock, and CB Dock are other free and user-friendly resources, as shown in Table 2.
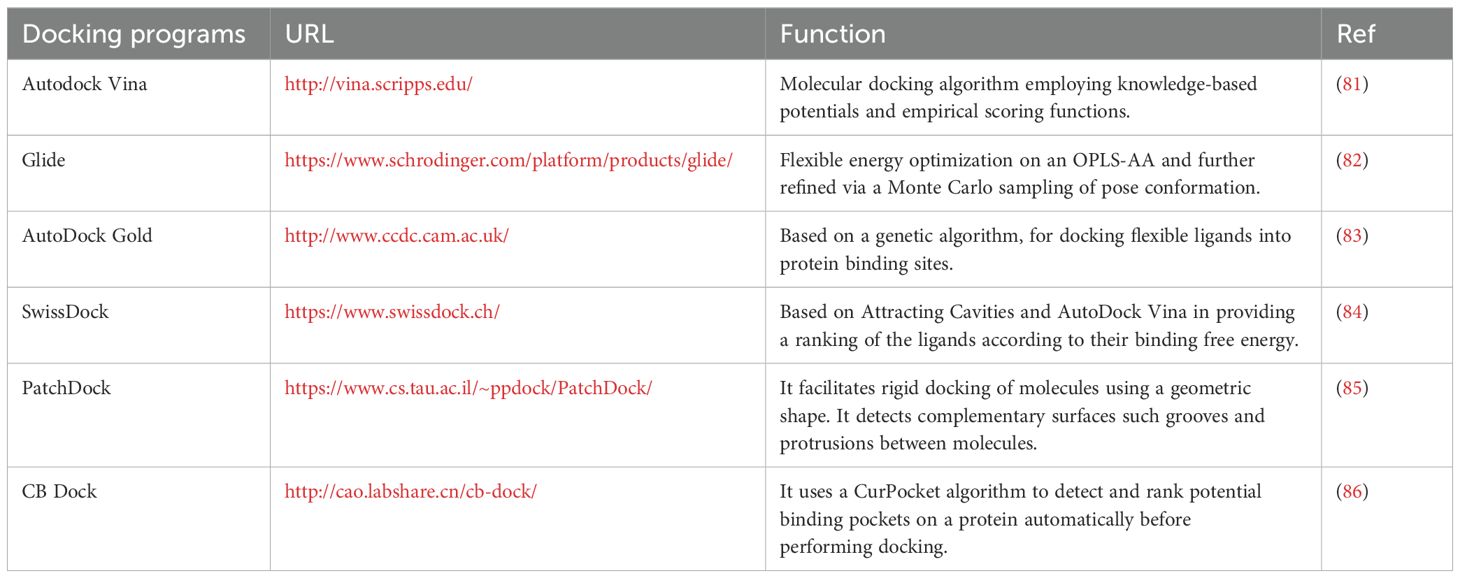
Table 2. Top options molecular docking software along with their URL address and a short description of their function.
Conventional SELEX is experimentally driven and allows for direct, real-world validation of aptamer-target interactions, but it is time-consuming and labor-intensive. In silico SELEX accelerates the discovery process through computational screening, making it faster and more efficient, though it requires experimental follow-up to confirm the predicted results. DNA aptamer development using APTCAD in silico methods is becoming more popular as a practical way to provide a greater understanding of aptamer-target interaction.
A growing number of aptamers have entered clinical trials and are being tested as drugs of treatment for solid tumor immunotherapy (Table 3).
AON-D21 is a continuation of NOX-D19 and NOX-D20 that were previously discussed. With a 7–8 hours plasma half-life, NOX-D20 demonstrated a much longer half-life in circulation. By delaying the onset of multiorgan failure and the advancement of sepsis, NOX-D20 increases survival (87).
AON-D21 is a PEG-modified L-aptamer (Spiegelmer) that binds to C5a and C5 in both humans and mice. Without interfering with C5 cleavage, it selectively blocks C5a from binding with its receptors. AON-D21, a plasma stable anti-C5a L-aptamer, in the in vivo model of A549M1 cell bone metastases. C5 or C5a was utilized as a negative control as the Spiegelmer of the reverse AON-D21 sequence, RevAON-D21, is unable to bind any of them. Every other day starting the day before the A549M1 cells were injected intracardiac, AON-D21 and revAON-D21 were administered intraperitoneally (10 mg/kg) in saline. The tumor volume was calculated and measured with the use of an electronic calculator. C5aR1 activation induces the production of osteoclastogenic factors and raises the risk of bone metastases from lung cancer (88).
A DNA pattern known as the CpG motif, which is abundant in unmethylated Cytosine-phosphate Guanine (CpG) nucleotides and common in bacterial DNA, triggers an immunological response in the host. The result of Aummune’s cutting-edge customized therapeutic platform, which finds functional aptamers capable of killing tumor target cells, is AM003, a Bispecific Personalized Aptamer with a T cell engager arm and a tumor-targeting arm. After hybridization, AM003’s two ssDNA aptameric arms are intended to generate a new immunostimulant CpG pattern. Intratumoral injection of AM003 significantly changed the tumor’s immunological environment by increasing the invasion of B and CD8+ CD4+ T cells. By enhancing T cell infiltration, stimulating the innate immune system, and facilitating subsequent efficient T cell activation through the compound’s T cell engager arm, AM003 tackles the problems associated with immunological resistance (89).
Although aptamers hold significant promise, most have failed to achieve the required safety and efficacy benchmarks in human clinical trials. A number of factors limit their use in therapeutics, including rapid degradation through nucleases, rapid clearance via renal filtration, and a lack of chemical diversity among others (90). Aptamers consist of nucleotides, and hence they might be degraded by nucleases under most in vivo or in vitro tissue culture conditions (91). These could cause strand breaks in either one or both strands of the nucleic acid, which will render it ineffective. Nucleases are enzymes capable of cleaving the phosphodiester bonds that join nucleotides in DNA and/or RNA. While the endonucleases can cut at different points along the polynucleotide strand, either single-strand or double-strand breaks, there exist exonucleases-which cleave nucleic acids from the ends, either 5′ or 3′ (92).
Because of their relatively small size, approximately 10-20 kDa or less than 5 nm in size, aptamers eliminated from the blood very fast after in vivo administration. By intravenous administration, they get cleared off the blood within approximately 10 minutes (93). Renal filtration can rapidly excrete small aptamers when they are introduced into the bloodstream, even if stabilizing backbone alterations are utilized (66). This reduces their therapeutic effectiveness since they do not remain in the body long enough to perform their intended function. These challenges make their development more complex and limit their wide application. Nevertheless, this property is not necessarily a drawback. It will rely on the intended application of the aptamer. On the other hand, due to their small dimensions, aptamers show improved tissue permeability and can reach their target more efficiently before being cleared by the kidneys (94).
Aptamers, unlike antibodies assembled into amino acids, are composed of four nucleotides: guanine, cytosine, adenosine, and thymine or uracil, compared with 20 amino acids in the case of antibodies (95, 96). Aptamers, as a result of their highly minimalist chemical structure devoid of even side chains and complex functional groups, have limited structural and chemical diversity. Their ability to effectively engage some targets would therefore be diminished (91, 97). The inventive structural development of aptamers has therefore been pushed forward into potentially more viable routes for aptamer-based therapeutics by the numerous problems that have been found in their development, such as those covered above.
From a clinical translation viewpoint, the most major challenge for aptamer medicines is their stability, particularly their sensitivity to fast nuclease breakdown and renal clearance. These obstacles significantly restrict their absorption and therapeutic effectiveness, necessitating chemical changes to optimize their pharmacokinetics. Chemical modifications are strategies that can improve stability and lengthen circulation duration, but they cannot resolve this issue adequately. Despite these advances, aptamers’ half-lives remain much shorter than those of monoclonal antibodies (mAbs) (62). Among the key issues that may answer the question of whether aptamers can remain intact and functional within biological systems, stability is the cornerstone challenge for their successful clinical translation. Stability is crucial for aptamer’s functionality in biological system (98). It therefore constitutes the primary obstacle to their effective clinical translation.
There is no documented evidence of tumor resistance to aptamer-based therapies in clinical settings till date. However, tumor resistance mechanism must be considered while developing any treatment approach including aptamers (99).
Improving the stability of unmodified aptamers is critical, especially in biological environments where nucleases can degrade them quickly. Here are several strategies to enhance the stability of unmodified aptamers. Modifying the 3’ and 5’ ends shields aptamers from exonucleases, while phosphorothioate linkages enhance protection against endonucleases (100).
Resistance mechanism in tumors such as multidrug resistance (MDR) also pose a challenge for aptamer-based therapies. One study demonstrated that aptamer d3 binding was revealed to block the MDR of the tumor cells and increase the accumulation of intracellular anticancer drug, which lead to a boost to the cell killing (101). Multifunctional aptamers, which target multiple pathways can further reduce the likelihood of resistance alongside with combination therapies reducing reliance on a single mechanism (102).
Adding modifications like inverted thymidine (inverted T) at the 3’ end is the most common modification (103). The presence of the inverted dT had a modest approximately threefold effect on stability according to Kratschmer’s study. The synthesis of aptamers with 3’ inverted thymidine modification requires the use of modified CPG, where the 5’ hydroxyl group of the first nucleoside is attached, and then the chain is elongated in the typical 3’→5’ direction (104). Incorporating a 3’ inverted dT residue can also prolong duration to several hours instead of an hour for DNA’s half-lives (105). Pegaptanib includes a 40 kDa polyethylene glycol (PEG) unit at the 5’ end to aid in renal clearance, along with a 3’-3’-linked deoxythymidine residue to enhance resistance to nuclease degradation (106). Despite these modifications, pegaptanib maintained an exceptionally high affinity for its VEGF165 target and demonstrated prolonged in vitro stability stated by (107).
Modifying the phosphodiester backbone changes the metabolic pathway, improving nuclease-resistant stability, as seen with phosphorothioate linkages (108). In this modification, one of the non-bridging oxygen atoms in the phosphodiester bond is substituted with a sulfur atom (109). In 1967, Eckstein (108) first introduced the phosphorothioate (PS) modification strategy to improve RNA’s resistance to ribonucleases (RNase). During the oxidation step, the standard iodine solution was replaced with phenylacetyl disulfide solution (5 g in 2 ml pyridine) to achieve PS modification. Since phenylacetyl disulfide lacks stereoselectivity, the resulting oligonucleotides are racemic mixtures (108). Based on research conducted by researcher Wu, the PS modified sequence WW-24 demonstrated outstanding selective anti-melanoma activity (A375 cells, ∼25 nM, 80%), targeting both hnRNP A1 and hnRNP A2/B1, exhibiting a stronger antitumor effect and prolonged accumulation time in vivo (110).
A circular nucleic acid (CAN) is defined by its closed-loop structure formed through covalent bonding between the two ends of a linear nucleic acid molecule (111). Most of the circular nucleic acids are prepared through the chemical or enzymatic ligation of linear oligonucleotides (112). Chemical ligation in this respect generally refers to the use of cyanogen bromide, BrCN, or 1-ethyl-3-(3′-dimethylaminopropyl) carbodiimide for connecting DNA-RNA hybrids. However, this technique also has its drawbacks of giving low ligation efficiency, and it may have some biosafety risks. Another drawback of the technique is that it generates 2′, 5′-phosphodiester bonds instead of the natural 3′, 5′-phosphodiester bonds. One of the typical protocols for the generation of cDNA involves the use of ligase for the circularization process (113). T4 DNA ligase is one among the commonly used ligases in this process that catalyzes the ligation of the 3′-hydroxy and 5′-phosphate ends of a ssDNA molecule with the help of a splint DNA strand (112).
Additionally, the enzyme CircLigase (114) or ssDNA ligase (115) can be employed to catalyze the joining of ssDNA that has complementary ends. Recently, a nuclease-resistant circular bivalent aptamer system was created, offering enhanced stability and improved tumor-targeting abilities in vivo (116, 117). This system, formed by ligating two linear aptamer sequences, demonstrated increased thermal and physical stability in biological environments while showing superior tumor cell targeting (118).
To address the quick renal filtration issue, two most common methods have been reported, such as attaching cholesterol and PEG (119). More importantly, PEGylation has already been established in extending the circulation life of various therapeutics, including proteins and small molecules, which in turn drives substantial progress in biologics development and drug-loaded nanoparticles (120). PEGylation was first conceptualized as a method to overcome the problem of immunogenicity of non-human derived proteins intended for human use, and the concept was first envisioned by Frank Davis in the late 1960s. PEG was chosen because it formed the hydrophilic component of a clinically utilized block copolymer (Pluronic, made up of PEG and polypropylene glycol). Davis theorized that attaching PEG to proteins or small molecules would make them less detectable by the immune system, thereby reducing immune responses while extending their circulation and functional lifespan (121).
The PEGylation can be attributed to the increase in hydrodynamic size, which leads to a reduction in renal filtration due to PEG conjugation (122). PEG with a molecular weight below 30 kDa is removed from the body via the kidneys, above 20 kDa is excreted through the feces (123). Macugen™ (pegaptanib sodium, Pfizer) is another significant PEGylated small molecule drug or aptamer. It is the first aptamer-based therapeutic approved for human use working as a selective antagonist of vascular endothelial growth factor (VEGF) (124). The FDA has approved Avacincaptad Pegol (Izervay) from Iveric Bio/Astellas for geographic atrophy due to age-related macular degeneration. It is the second RNA aptamer and complement-targeted drug to receive FDA approval for this condition (125).
Cholesterol conjugation at 5’ end improves aptamer pharmacokinetic properties by creating a cholesteryl-oligonucleotide (cholODN) complex with low density lipoprotein (LDL). This complex exhibits exceptional resistance to nuclease hydrolysis, extending half-life (117, 126). A modified RNA aptamer inhibited Hepatitis C virus replication in vitro. It was later derivatized with cholesterol to form chol-aptamer, which successfully inhibited HCV RNA replication in a cell-based system. Systemic administration was well-tolerated by mice and increased plasma retention time (127). By increasing aptamer hydrophobicity, cholesterol lowers plasma clearance rate and lengthens half-life. Aptamers combined with cholesterol have better biodistribution patterns and longer circulation half-lives, according to preclinical research (119).
Gold and colleagues developed SOMAmer, a method that mimics protein side chains by attaching amide groups to deoxyuridine triphosphate at the 5′ position aggrandizing the diversity of nucleic acids (128). In their respective investigations, researchers Li (129) and Ji (130) highlighted how this alteration improves binding affinities, provides hydrogen bonding partners, and avoids linkage bond rotations. SOMAmers incorporate modified nucleotide bases to make aptamers more effective at interacting with hard-to-reach sites that classical aptamers struggle to access. One goal is to enable aptamers to interact with hydrophobic regions, which can be crucial for the activity of toxins but are challenging for traditional nucleotides to target (131). SOMAmers interact with target molecules during the SELEX process through more hydrophobic and less polar interactions, involving fewer hydrogen bonds and charge–charge interactions than standard aptamers. They demonstrate slower dissociation rates (over 30 minutes) while avoiding covalent and permanent bonds that are often linked to higher dissociation constant (Kd) values (132, 133). An added benefit of these modifications is the significantly extended half-life of the aptamers in vivo (133). Even though SOMAmers has extensive purification and characterization compared to standard SELEX, they exhibit stable chemical structures, great nuclease resistance, and binding properties equivalent to antibodies. Personalized SOMAmer reagents for a specific protein target may be made faster and cheaper than antibodies (134).
Similarly, a new generation of hybrid aptamers, called X-aptamers, has been developed incorporating amino-acid-like side chains or drug-like ligands (X). AM Biotechnologies, Houston, USA, developed the X-aptamer selection kit, which was used to pick X-aptamers (135). These combine monothiophosphate-backbone-substituted aptamers for enhanced stability and protein binding with chemically modified uridine (referred to as X) that allows drug-like compounds or additional functionalities to be attached to the aptamers. The substitution of one of the oxygen atoms in the phosphate group with sulfur is the modification that makes the aptamer resistant to nuclease degradation (136). Utilizing this technology, selection of X-aptamers against two immune checkpoint proteins, PD-1 and PD-L1, have been developed (28). X-Aptamers were considered to be the next generation aptamers as X-aptamer showed therapeutic potential in acute pancreatitis (137). Generally, a bead-based X-aptamer library was used to find an X-aptamer that interacts to the target precisely (135). The PCR results were combined after 25 cycles and submitted to the manufacturer for Illumina next-generation sequencing. The company synthesized potential x-aptamers with a 5’ biotin tag, requiring 1-2 weeks for the entire procedure, which includes synthesis and sequencing (136).
Furthermore, selection of the best chemical modification approach, such as PEGylation, cholesterol conjugation, or phosphorothioate linkage for aptamers, depends on tumor type, patient characteristics, and therapeutic application has been discussed in general by (138). PEGylation is the best modification for aptamer stability and half-life circulation in solid tumors with poorly vascularized regions with dense extracellular matrix that leads to slower drug uptake, such as lung, colon, and breast cancer (139). Dosing may need adjustment due to individual differences such as liver and kidney function (140).
Cholesterol modified aptamers improve cellular uptake and enhanced interaction lipid bilayer hence its very beneficial for targeting lipid-rich tumors such as ovarian, prostate and glioblastoma (140–142). Patient-specific lipid profiles should thus be considered when formulating and delivering cholesterol-conjugated aptamer treatments in order to provide clear therapeutic recommendations (143).
PS linkages enhance aptamer stability and binding affinity, making them ideal for pancreatic cancer (144), brain tumors due to their ability to cross the blood brain barrier (140), also melanoma since phosphorotioated aptamer have special anti-melanoma effect (110). However, they can also increase immunogenicity and trigger an inflammatory cytokine response, requiring cautious treatment.
The term “drug development platform technology” describes the broad variety of tools, processes, and platforms set up with the purpose of further increasing the efficiency in development and manufacturing of pharmaceuticals. They are key drivers in accelerating the research of drugs, preclinical trials, clinical testing, and medicinal production (145). Regulatory frameworks are equally crucial in this development. Organizations such as the EMA (146) provide standardized regulations to ensure the safety, efficacy, and quality of drugs created using platform technologies, simplifying regulatory approval processes. The FDA introduced the most significant guideline in May 2024 (147), establishing criteria for the platform technology designation under Section 506K of the FD&C Act, aimed at improving drug development efficiency.
FDA recommends oligonucleotide therapeutic evaluations, including QTc interval prolongation assessment, immunogenicity risk, hepatic and renal impairment assessments, drug-drug interaction studies (148). The most important factors in assessing cardiac risk are the QT intervals, which are mediated by the flow of ions such as sodium, potassium, and calcium across receptors in the cell membrane (149). These may cause severe conditions including syncope, cardiac arrest, and sudden death especially when QTc is further extended. Normal QTc intervals normally are normally measured as < 450ms for men and < 460ms for women (150). A baseline ECG should be taken. It may be repeated once the drug reaches steady state. A baseline ECG should be taken and may be repeated once the drug reaches steady state (151).
Assessment of the risk of immunogenicity is a major component in the development of biotherapeutic drugs and is also one of the factors taken into consideration in the overall benefit-risk evaluation. This generally includes an assessment of the likelihood of inducing an immune response and any potential clinical consequences if such a response occurs (152). As the field advances with immunomodulatory multi-specific therapies, pharmacological or mechanism of action (MoA)-based immunogenic risk factors are becoming increasingly significant (153). However, the risk of an immune response can be lowered by the molecular weight of certain biotherapeutics (154), and high doses may promote long-term immune tolerance. Additionally, it is important to consider the patient’s immune status and prior exposure to relevant treatments (155).
A comprehensive immunogenicity risk evaluation includes a summary of the drug’s target, its intended use, and its structure. It considers sequence-related risks by identifying which sequences may bind to immune cells and evaluating the risks associated with the drug’s mechanism. Population-based risks are also assessed to determine if specific groups may have varying incidences of immune responses. Additionally, the evaluation addresses quality attribute-related risks, such as impurities, and includes factors like the expected half-life, dosing regimen, sampling plan, and a schedule for sharing immunogenicity data (155).
Healthy patients in the renal study needed to have a creatinine clearance of ≥90 mL/min as estimated by the Cockcroft-Gault equation. The hepatic study requires a creatinine clearance of at least 60 mL/min. The hepatic impairment trial was an open-label, phase 1 parallel-group, multicenter research designed to assess PK, safety, and tolerability in people with mild, moderate, and severe liver impairment to healthy participants. In other words, liver damage reduces intrinsic drug clearance due to decreased functional mass of liver cells and lower hepatic blood flow caused by liver fibrosis scarring (156).
In cases of severe hepatic impairment, the drug’s initial dose might need to be reduced and closely monitored. According to FDA guidelines, studies aimed at estimating the impact of renal function on drugs with a molecular weight below 69 kDa on their PK should be evaluated. Therapeutic drug monitoring (TDM) could be employed for dose adjustment (157). Myotoxicity, another potentially serious and severe side effect, may cause rhabdomyolysis, acute renal failure, disseminated intravascular coagulation, and death (158, 159).
Drug-drug interactions (DDIs) pose a serious global health threat because their adverse effects can be lethal (160), not just from the prescription drugs, but dietary and herbal supplements are also dangerous (161). One out of every 25 chances of supplements usage with prescription drugs leads to DDIs (162). While most DDIs are mild, some may be severe or life-threatening and may necessitate medical intervention. DDIs occurring with anticancer drugs may have serious consequences on both the treatment outcomes and toxicity. Evidence-based electronic surveillance for DDI is now crucial to increasing treatment efficacy while ensuring the safety of patients. In recent years, within the last ten years, several therapeutic drug managements (TDM) have been developed to manage DDIs (162). Physiological changes with age, food interactions, and other factors can alter drug absorption, distribution, metabolism, or excretion. New technologies can further enable the creation of more innovations in personalized medicine, therefore supporting the management of drug interactions (163).
A study on the first-in-human whole-body dynamic pharmacokinetics of aptamer incorporates a range of tests and evaluation to monitor and manage potential adverse effects. Preclinical toxicity evaluation is one of the crucial steps in the process where a single dose toxicity test was conducted to assess the aptamer’s safety. This involved monitoring dosage levels, clinical observation as well as changes in body weights, hematological markers, bone marrow micronucleus formation. Additional tests such as the mutagenic, carcinogenic and genotoxicity test were performed, including a reverse mutation assay and chromosomal aberration test in mammalian cells. In clinical studies, patient enrolment and clinical study design involve detailed recording of patient characteristic such as diagnosis of cancer or a medical history of cancer. Prior to treatment, a comprehensive physical examination is performed including the assessment of vital signs, routine blood tests, liver and kidney function, blood human chorionic gonadotropin (HCG) for females and electrocardiogram. These clinical assessments ensure that patients are suitable candidates for the treatment and provide baseline data for monitoring adverse effects during the study. In addition, whole-body dynamic PET imaging is employed for pharmacokinetics studies to observe biodistribution and the clearance patterns of the aptamer. This imaging provides real-time data on how the aptamer interacts with the body (164).
The advancements in aptamer-based immunotherapy for solid tumors underscore the potential for this technology to revolutionize cancer treatment. However, future study needed especially on stability since it still remains a significant barrier. Imidazole thymine is a novel discovery that has not yet been extensively studied or validated yet, but it mediated duplex stabilization (165, 166) compared to conventional chemical alteration. Expanding the strategy of aptamer integration with immune cells like Natural Killer (NK) to enhance efficacy for more personalized targeted cancer treatment is an attractive area for future study. The surface of NK cells was used to anchor aptamers for cell-specificity. This approach has yet to be explored (167).
Aptamer-based immunotherapy, in short, stands at the edge of revolutionizing cancer treatment, especially treatments against solid tumors. This is considered a very promising emerging field because the aptamers have special advantages in front of the traditional therapies, such as high specificity, lower immunogenicity, and ease of modification. Aptamers can act not only as therapeutic entities per se but also as vehicles and immune system modulators in the context of different treatment modalities that range from checkpoint inhibition to cytokine modulation, cellular immunotherapy, and bispecific aptamers.
It is indicated that aptamers may have a good future in solid tumor immunotherapy, according to trials such as AON-D21 and AM003. However, various limitations arise due to aptamer instability in vivo and their very short half-life, which results from rapid degradation by nucleases or renal clearance. Such are surmountable with particular chemical modifications including PEGylation, cholesterol conjugation, and circular nucleic acid, which enhance the stability of aptamers, improve pharmacokinetics, and prolong the circulation time.
According to the regulatory guidelines, it is also pinpointed that development should consider immunogenicity and impairment of organs, including renal and hepatic function, which may affect drug metabolism and safety. Drug-drug interactions remain important, especially in patients with cancers undergoing treatments; thus, thorough evaluations and personalized treatment approaches are called for.
Further advances in bioinformatics-driven aptamer design, in silico SELEX methods, and next-generation aptamer technologies, such as SOMAmers and X-aptamers, are foreseen to result in enhanced therapeutic efficacy and broader clinical use against cancer and other diseases. Remaining challenges regarding stability, delivery, and regulatory compliance will require research emphases in the future to fully exploit the potential for aptamer-based therapies in the clinic.
Personalized medicine and improved management of drug interactions will ensure optimum treatment for tumors, especially when new therapeutic combinations become available. Further studies in aptamer-based immunotherapy should be able to overcome the present obstacles and offer new effective treatments with less side effects to patients affected by solid tumors and beyond.
SM: Conceptualization, Writing – original draft, Writing – review & editing. GT: Conceptualization, Supervision, Writing – review & editing. YT: Funding acquisition, Resources, Writing – review & editing. KM: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The review was funded by Biogenes Technologies.
SM and YT were employed by Biogenes Technologies.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that this study received funding from Biogenes Technologies. The authors associated to this entity where responsible for the decision to publish and preparation of the manuscript.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Jin N, George TL, Otterson GA, Verschraegen C, Wen H, Carbone D, et al. Advances in epigenetic therapeutics with focus on solid tumors. Clin Epigenet. (2021) 13:1–27 . doi: 10.1186/s13148-021-01069-7
2. Giraldo NA, Sanchez-Salas R, Peske JD, Vano Y, Becht E, Petitprez F, et al. The clinical role of the TME in solid cancer. Br J Cancer. (2018) 120:45–53. doi: 10.1038/s41416-018-0327-z
3. Carlson RD, Flickinger JC, Snook AE. Talkin’ toxins: From Coley’s to Modern Cancer immunotherapy. Toxins. (2020) 12:241. doi: 10.3390/toxins12040241
4. DeLucia DC, Lee JK. Development of cancer immunotherapies. Cancer Treat Res. (2022) 183:1–48. doi: 10.1007/978-3-030-96376-7_1
5. Şimşek GÖ. Immunotherapy in the treatment of cancer: Today and Tomorrow. Curr Mol Biol Rep. (2024) 10:54–64. doi: 10.1007/s40610-024-00161-0
6. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. (2020) 17:807–21. doi: 10.1038/s41423-020-0488-6
7. Debela DT, Muzazu SG, Heraro KD, Ndalama MT, Mesele BW, Haile DC, et al. New approaches and procedures for cancer treatment: Current perspectives. SAGE Open Med. (2021) 9:1–10 . doi: 10.1177/20503121211034366
8. He S, Du Y, Tao H, Duan H. Advances in aptamer-mediated targeted delivery system for cancer treatment. Int J Biol Macromolecules. (2023) 238:124173. doi: 10.1016/j.ijbiomac.2023.124173
9. El-Tanani M, Rabbani SA, Babiker R, Rangraze I, Kapre S, Palakurthi SS, et al. Unraveling the tumor microenvironment: Insights into cancer metastasis and therapeutic strategies. Cancer Letters. (2024) 591:216894. doi: 10.1016/j.canlet.2024.216894
10. Kalli M, Poskus MD, Stylianopoulos T, Zervantonakis IK. Beyond matrix stiffness: Targeting force-induced cancer drug resistance. Trends Cancer. (2023) 9:937–54. doi: 10.1016/j.trecan.2023.07.006
11. Liu Y, Liang J, Zhang Y, Guo Q. Drug resistance and tumor immune microenvironment: An overview of current understandings (review). Int J Oncol. (2024) 65:1–20. doi: 10.3892/ijo.2024.5684
12. ter Linden E, Abels ER, van Solinge TS, Neefjes J, Broekman ML. Overcoming barriers in glioblastoma—advances in Drug Delivery Strategies. Cells. (2024) 13:998. doi: 10.3390/cells13120998
13. Guha P, Heatherton KR, O’Connell KP, Alexander IS, Katz SC. Assessing the future of solid tumor immunotherapy. Biomedicines. (2022) 10:655. doi: 10.3390/biomedicines10030655
14. Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. (1990) 249:505–10. doi: 10.1126/science.2200121
15. Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. (1990) 346:818–22. doi: 10.1038/346818a0
16. Kumar Kulabhusan P, Hussain B, Yüce M. Current perspectives on aptamers as diagnostic tools and therapeutic agents. Pharmaceutics. (2020) 12:646. doi: 10.3390/pharmaceutics12070646
17. Mahmoudian F, Ahmari A, Shabani S, Sadeghi B, Fahimirad S, Fattahi F. Aptamers as an approach to targeted cancer therapy. Cancer Cell Int. (2024) 24:1–22. doi: 10.1186/s12935-024-03295-4
18. Hossen MM, Ma Y, Yin Z, Xia Y, Du J, Huang JY, et al. Current understanding of CTLA-4: From mechanism to autoimmune diseases. Front Immunol. (2023) 14:1198365. doi: 10.3389/fimmu.2023.1198365
19. Huang B-T, Lai W-Y, Chang Y-C, Wang J-W, Yeh S-D, Lin EP-Y, et al. A CTLA-4 antagonizing DNA aptamer with antitumor effect. Mol Ther - Nucleic Acids. (2017) 8:520–8. doi: 10.1016/j.omtn.2017.08.006
20. Xiong H, Liu L, Liu X, Jiang H, Wang X. Aptamer-based immune drug systems (aptidcs) potentiating cancer immunotherapy. Chemistry. (2023) 5:1656–80. doi: 10.3390/chemistry5030114
21. Domsicova M, Korcekova J, Poturnayova A, Breier A. New insights into Aptamers: An alternative to antibodies in the detection of molecular biomarkers. Int J Mol Sci. (2024) 25:6833. doi: 10.3390/ijms25136833
22. Lipson EJ, Drake CG. Ipilimumab: An anti-CTLA-4 antibody for metastatic melanoma. Clin Cancer Res. (2011) 17:6958–62. doi: 10.1158/1078-0432.ccr-11-1595
23. Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. New Engl J Med. (2010) 363:711–23. doi: 10.1056/nejmoa1003466
24. Bertrand A, Kostine M, Barnetche T, Truchetet M-E, Schaeverbeke T. Immune related adverse events associated with anti-CTLA-4 antibodies: Systematic review and meta-analysis. BMC Med. (2015) 13:1–14. doi: 10.1186/s12916-015-0455-8
25. Prasad V, Kaestner V. Nivolumab and Pembrolizumab: Monoclonal antibodies against programmed cell death-1 (PD-1) that are interchangeable. Semin Oncol. (2017) 44:132–5. doi: 10.1053/j.seminoncol.2017.06.007
26. Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. (1992) 11:3887–95. doi: 10.1002/j.1460-2075.1992.tb05481.x
27. Prodeus A, Abdul-Wahid A, Fischer NW, Huang EH-B, Cydzik M, Gariépy J. Targeting the PD-1/PD-L1 immune evasion axis with DNA aptamers as a novel therapeutic strategy for the treatment of disseminated cancers. Mol Ther - Nucleic Acids. (2015) 4:1–10. doi: 10.1038/mtna.2015.11
28. Wang H, Lam CH, Li X, West DL, Yang X. Selection of PD1/PD-L1 X-aptamers. Biochimie. (2018) 145:125–30. doi: 10.1016/j.biochi.2017.09.006
29. Rajagopalan A, Berezhnoy A, Schrand B, Puplampu-Dove Y, Gilboa E. Aptamer-targeted attenuation of il-2 signaling in CD8 + T cells enhances antitumor immunity. Mol Ther. (2017) 25:54–61. doi: 10.1016/j.ymthe.2016.10.021
30. Raskov H, Orhan A, Christensen JP, Gögenur I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br J Cancer. (2020) 124:359–67. doi: 10.1038/s41416-020-01048-4
31. Hinrichs CS, Gattinoni L, Restifo NP. Programming CD8+ T cells for effective immunotherapy. Curr Opin Immunol. (2006) 18:363–70. doi: 10.1016/j.coi.2006.03.009
32. Li X, Shao M, Zeng X, Qian P, Huang H. Signaling pathways in the regulation of cytokine release syndrome in human diseases and intervention therapy. Signal Transduction Targeted Ther. (2021) 6:1–15. doi: 10.1038/s41392-021-00764-4
33. Hutmacher C, Neri D. Antibody-cytokine fusion proteins: Biopharmaceuticals with immunomodulatory properties for cancer therapy. Advanced Drug Delivery Rev. (2019) 141:67–91. doi: 10.1016/j.addr.2018.09.002
34. Pires IS, Hammond PT, Irvine DJ. Engineering strategies for immunomodulatory cytokine therapies: Challenges and clinical progress. Advanced Ther. (2021) 4:1–29. doi: 10.1002/adtp.202100035
35. Egilmez N, Kilinc M, Gu T, Conway T. Controlled-release particulate cytokine adjuvants for cancer therapy. Endocrine, Metab Immune Disorders-Drug Targets. (2007) 7:266–70. doi: 10.2174/187153007782794335
36. Zhou H, Abudureheman T, Zheng W, Yang L, Zhu J, Liang A, et al. Car-aptamers enable traceless enrichment and monitoring of car-positive cells and overcome tumor immune escape. Advanced Sci. (2023) 11:1–17. doi: 10.1002/advs.202305566
37. Miao Y, Gao Q, Mao M, Zhang C, Yang L, Yang Y, et al. Bispecific aptamer chimeras enable targeted protein degradation on cell membranes. Angewandte Chemie Int Edition. (2021) 60:11267–71. doi: 10.1002/anie.202102170
38. Thomas BJ, Porciani D, Burke DH. Cancer immunomodulation using Bispecific Aptamers. Mol Ther - Nucleic Acids. (2022) 27:894–915. doi: 10.1016/j.omtn.2022.01.008
39. Zheng A, Du Y, Wang Y, Zheng Y, Ning Z, Wu M, et al. CD16/PD-L1 bi-specific aptamer for cancer immunotherapy through recruiting NK cells and acting as immunocheckpoint blockade. Mol Ther - Nucleic Acids. (2022) 27:998–1009. doi: 10.1016/j.omtn.2022.01.010
40. Patil SJ, Thorat VM, Koparde AA, Bhosale RR, Bhinge SD, Chavan DD, et al. Theranostic applications of scaffolds in current biomedical research. Cureus. (2024) 16(10):1–18. doi: 10.7759/cureus.71694
41. Venkatesan S, Chanda K, Balamurali MM. Recent advancements of Aptamers in cancer therapy. ACS Omega. (2023) 8:32231–43. doi: 10.1021/acsomega.3c04345
42. Liu Z, Duan J-H, Song Y-M, Ma J, Wang F-D, Lu X, et al. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in vitro. J Trans Med. (2012) 10:1–10. doi: 10.1186/1479-5876-10-148
43. Wang R, Zhu G, Mei L, Xie Y, Ma H, Ye M, et al. Automated modular synthesis of aptamer–drug conjugates for targeted drug delivery. J Am Chem Society. (2014) 136:2731–4. doi: 10.1021/ja4117395
44. Jeong HY, Kim H, Lee M, Hong J, Lee JH, Kim J, et al. Development of HER2-specific aptamer-drug conjugate for breast cancer therapy. Int J Mol Sci. (2020) 21:9764. doi: 10.3390/ijms21249764
45. Yazdian-Robati R, Bayat P, Oroojalian F, Zargari M, Ramezani M, Taghdisi SM, et al. Therapeutic applications of AS1411 Aptamer, an update review. Int J Biol Macromolecules. (2020) 155:1420–31. doi: 10.1016/j.ijbiomac.2019.11.118
46. Mehrnia SS, Hashemi B, Mowla SJ, Nikkhah M, Arbabi A. Radiosensitization of breast cancer cells using AS1411 aptamer-conjugated gold nanoparticles. Radiat Oncol. (2021) 16:1–12. doi: 10.1186/s13014-021-01751-3
47. Kardani A, Yaghoobi H, Alibakhshi A, Khatami M. Inhibition of mir-155 in MCF-7 breast cancer cell line by gold nanoparticles functionalized with Antagomir and AS1411 Aptamer. J Cell Physiol. (2020) 235:6887–95. doi: 10.1002/jcp.29584
48. Xiao X, Li H, Zhao L, Zhang Y, Liu Z. Oligonucleotide Aptamers: Recent advances in their screening, molecular conformation and therapeutic applications. Biomedicine Pharmacotherapy. (2021) 143:112232. doi: 10.1016/j.biopha.2021.112232
49. Kim D-H, Seo J-M, Shin K-J, Yang S-G. Design and clinical developments of aptamer-drug conjugates for targeted cancer therapy. Biomaterials Res. (2021) 25:1–12. doi: 10.1186/s40824-021-00244-4
50. Daems E, Moro G, Campos R, De Wael K. Mapping the gaps in chemical analysis for the characterisation of aptamer-target interactions. TrAC Trends Analytical Chem. (2021) 142:116311. doi: 10.1016/j.trac.2021.116311
51. Amero P, Khatua S, Rodriguez-Aguayo C, Lopez-Berestein G. Aptamers: Novel therapeutics and potential role in neuro-oncology. Cancers. (2020) 12:2889. doi: 10.3390/cancers12102889
52. Li L, Xu S, Yan H, Li X, Yazd HS, Li X, et al. Nucleic acid aptamers for molecular diagnostics and therapeutics: Advances and perspectives. Angewandte Chemie Int Edition. (2020) 60:2221–31. doi: 10.1002/anie.202003563
53. Luscombe NM. Amino acid-base interactions: A three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. (2001) 29:2860–74. doi: 10.1093/nar/29.13.2860
54. Zon G. Recent advances in aptamer applications for analytical biochemistry. Analytical Biochem. (2022) 644:113894. doi: 10.1016/j.ab.2020.113894
55. Almazar CA, Mendoza MV, Rivera WL. In silico approaches for the identification of aptamer binding interactions to Leptospira spp. cell surface proteins. Trop Med Infect Disease. (2023) 8:125. doi: 10.3390/tropicalmed8020125
56. Mili M, Bachu V, Kuri PR, Singh NK, Goswami P. Improving synthesis and binding affinities of nucleic acid aptamers and their therapeutics and diagnostic applications. Biophys Chem. (2024) 309:107218. doi: 10.1016/j.bpc.2024.107218
57. Navaneeth AG, Karthikeyan S. A comprehensive investigation of the biophysical approach for aptamer functionalized nanoparticles in Cancer therapy: A review. RSC Pharmaceutics. (2024) 1:879–903. doi: 10.1039/d3pm00027c
58. Troisi R, Balasco N, Autiero I, Vitagliano L, Sica F. Structural insights into protein–aptamer recognitions emerged from experimental and computational studies. Int J Mol Sci. (2023) 24:16318. doi: 10.3390/ijms242216318
59. Ji C, Wei J, Zhang L, Hou X, Tan J, Yuan Q, et al. Aptamer–protein interactions: From regulation to biomolecular detection. Chem Rev. (2023) 123:12471–506. doi: 10.1021/acs.chemrev.3c00377
60. Krissanaprasit A, Key CM, Pontula S, LaBean TH. Self-assembling nucleic acid nanostructures functionalized with Aptamers. Chem Rev. (2021) 121:13797–868. doi: 10.1021/acs.chemrev.0c01332
61. Plach M, Schubert T. Biophysical characterization of aptamer-target interactions. Adv Biochem Engineering/Biotechnology. (2019) 174:1–15. doi: 10.1007/10_2019_103
62. Ma H, Ó’Fágáin C, O’Kennedy R. Antibody stability: A key to performance - analysis, influences and improvement. Biochimie. (2020) 177:213–25. doi: 10.1016/j.biochi.2020.08.019
63. Ni S, Zhuo Z, Pan Y, Yu Y, Li F, Liu J, et al. Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl Materials Interfaces. (2020) 13:9500–19. doi: 10.1021/acsami.0c05750
64. Sarafraz M, Nakhjavani M, Shigdar S, Christo FC, Rolfe B. Modelling of mass transport and distribution of Aptamer in blood-brain barrier for tumour therapy and cancer treatment. Eur J Pharmaceutics Biopharmaceutics. (2022) 173:121–31. doi: 10.1016/j.ejpb.2022.03.004
65. Chinchilla-Cárdenas DJ, Cruz-Méndez JS, Petano-Duque JM, García RO, Castro LR, Lobo-Castañón MJ, et al. Current developments of Selex Technologies and prospects in the Aptamer selection with clinical applications. J Genet Eng Biotechnol. (2024) 22:100400. doi: 10.1016/j.jgeb.2024.100400
66. Zhou J, Rossi J. Erratum: Aptamers as targeted therapeutics: Current potential and challenges. Nat Rev Drug Discovery. (2017) 16:440–0. doi: 10.1038/nrd.2017.86
67. Lee SJ, Cho J, Lee B-H, Hwang D, Park J-W. Design and prediction of Aptamers assisted by in silico methods. Biomedicines. (2023) 11:356. doi: 10.3390/biomedicines11020356
68. Chushak Y, Stone MO. In silico selection of RNA aptamers. Nucleic Acids Res. (2009) 37:1–9. doi: 10.1093/nar/gkp408
69. Buglak AA, Samokhvalov AV, Zherdev AV, Dzantiev BB. Methods and applications of in silico aptamer design and modeling. Int J Mol Sci. (2020) 21:8420. doi: 10.3390/ijms21228420
70. Soon S, Aina Nordin N. In silico predictions and optimization of aptamers against streptococcus agalactiae surface protein using computational docking. Materials Today: Proc. (2019) 16:2096–100. doi: 10.1016/j.matpr.2019.06.097
71. Navien TN, Thevendran R, Hamdani HY, Tang T-H, Citartan M. In silico molecular docking in DNA aptamer development. Biochimie. (2021) 180:54–67. doi: 10.1016/j.biochi.2020.10.005
72. Sullivan R, Adams MC, Naik RR, Milam VT. Analyzing secondary structure patterns in DNA aptamers identified via compels. Molecules. (2019) 24:1572. doi: 10.3390/molecules24081572
73. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. (2003) 31:3406–15. doi: 10.1093/nar/gkg595
74. Sato K, Akiyama M, Sakakibara Y. RNA secondary structure prediction using deep learning with thermodynamic integration. Nat Commun. (2021) 12:1–9. doi: 10.1038/s41467-021-21194-4
75. Sabri MZ, Hamid AA, Hitam SM, Rahim Mohd Z. The assessment of three dimensional modelling design for single strand DNA aptamers for computational chemistry application. Biophys Chem. (2020) 267:106492. doi: 10.1016/j.bpc.2020.106492
76. Jeddi I, Saiz L. Three-dimensional modeling of single stranded DNA hairpins for Aptamer-based biosensors. Sci Rep. (2017) 7:1–13. doi: 10.1038/s41598-017-01348-5
77. Wang Q-L, Cui H-F, Du J-F, Lv Q-Y, Song X. In silico post-SELEX screening and experimental characterizations for acquisition of high affinity DNA aptamers against carcinoembryonic antigen. RSC Advances. (2019) 9:6328–34. doi: 10.1039/c8ra10163a
78. Biesiada M, Purzycka KJ, Szachniuk M, Blazewicz J, Adamiak RW. Automated RNA 3D structure prediction with RNAComposer. Methods Mol Biol. (2016) 1490:199–215. doi: 10.1007/978-1-4939-6433-8_13
79. Sherman M, Contreras L. Computational approaches in design of nucleic acid-based therapeutics. Curr Opin Biotechnol. (2018) 53:232–9. doi: 10.1016/j.copbio.2017.12.001
80. Mathur N, Sai Chandragiri S, Sarita, Shandily S, Mukeshbhai Santoki K, Navinchandra Vadhavana N, et al. In silico docking: Protocols for computational exploration of Molecular Interactions. Unravelling Mol Docking - From Theory to Pract [Working Title]. (2024). doi: 10.5772/intechopen.1005527
81. Trott O, Olson AJ. Autodock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. (2009) 31:455–61. doi: 10.1002/jcc.21334
82. Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, et al. Glide: A new approach for rapid, accurate docking and scoring. 2. enrichment factors in database screening. J Medicinal Chem. (2004) 47:1750–9. doi: 10.1021/jm030644s
83. Agu PC, Afiukwa CA, Orji OU, Ezeh EM, Ofoke IH, Ogbu CO, et al. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci Rep. (2023) 13:1–18. doi: 10.1038/s41598-023-40160-2
84. Bugnon M, Röhrig UF, Goullieux M, Perez MAS, Daina A, Michielin O, et al. SwissDock 2024: Major enhancements for small-molecule docking with attracting cavities and autodock vina. Nucleic Acids Res. (2024) 52:1–9 . doi: 10.1093/nar/gkae300
85. Mashiach E, Schneidman-Duhovny D, Peri A, Shavit Y, Nussinov R, Wolfson HJ. An integrated suite of fast docking algorithms. Proteins: Structure Function Bioinf. (2010) 78:3197–204. doi: 10.1002/prot.22790
86. Liu Y, Grimm M, Dai W, Hou M, Xiao Z-X, Cao Y. CB-Dock: A web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacologica Sinica. (2019) 41:138–44. doi: 10.1038/s41401-019-0228-6
87. Hoehlig K, Maasch C, Shushakova N, Buchner K, Huber-Lang M, Purschke WG, et al. A novel c5a-neutralizing mirror-image (L-)aptamer prevents organ failure and improves survival in experimental sepsis. Mol Ther. (2013) 21:2236–46. doi: 10.1038/mt.2013.178
88. Ajona D, Zandueta C, Corrales L, Moreno H, Pajares MJ, Ortiz-Espinosa S, et al. Blockade of the complement c5a/C5AR1 axis impairs lung cancer bone metastasis by CXCL16-mediated effects. Am J Respir Crit Care Med. (2018) 197:1164–76. doi: 10.1164/rccm.201703-0660oc
89. Hanin N--, Lavi E, Pode Z, Dahan S, Buravenkov V, Carasso LS, et al. 1129 a novel CPG motif introduced into a bispecifc AM003 compound for the treatment of solid tumors. Regular Young Investigator Award Abstracts. (2022) 10:1129. doi: 10.1136/jitc-2022-sitc2022.1129
90. Odeh F, Nsairat H, Alshaer W, Ismail MA, Esawi E, Qaqish B, et al. Aptamers Chemistry: Chemical Modifications and conjugation strategies. Molecules. (2019) 25:3. doi: 10.3390/molecules25010003
91. Bukari B, Samarasinghe RM, Noibanchong J, Shigdar SL. Non-invasive delivery of therapeutics into the brain: The potential of aptamers for targeted delivery. Biomedicines. (2020) 8:120. doi: 10.3390/biomedicines8050120
92. Zhang H, Vandesompele J, Braeckmans K, De Smedt SC, Remaut K. Nucleic acid degradation as barrier to gene delivery: A guide to understand and overcome nuclease activity. Chem Soc Rev. (2024) 53:317–60. doi: 10.1039/d3cs00194f
93. Kelly L, Maier KE, Yan A, Levy M. A comparative analysis of cell surface targeting aptamers. Nat Commun. (2021) 12:1–14. doi: 10.1038/s41467-021-26463-w
94. Zhou G, Latchoumanin O, Hebbard L, Duan W, Liddle C, George J, et al. Aptamers as targeting ligands and therapeutic molecules for overcoming drug resistance in cancers. Advanced Drug Delivery Rev. (2018) 134:107–21. doi: 10.1016/j.addr.2018.04.005
95. Minchin S, Lodge J. Understanding biochemistry: Structure and function of Nucleic Acids. Essays Biochem. (2019) 63:433–56. doi: 10.1042/ebc20180038
96. Patrick GL. Nucleic acids: Structure and function. Introduction to Medicinal Chem. (2023) 7:73–85. doi: 10.1093/hesc/9780198866664.003.0007
97. Kohn EM, Konovalov K, Gomez CA, Hoover GN, Yik AK, Huang X, et al. Terminal alkyne-modified DNA aptamers with enhanced protein binding affinities. ACS Chem Biol. (2023) 18:1976–84. doi: 10.1021/acschembio.3c00183
98. Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discovery. (2010) 9:537–50. doi: 10.1038/nrd3141
99. Yoon S, Rossi JJ. Emerging cancer-specific therapeutic aptamers. Curr Opin Oncol. (2017) 29:366–74. doi: 10.1097/cco.0000000000000389
100. Ye Z, Chen H, Weinans H, van der Wal B, Rios JL. Novel aptamer strategies in combating bacterial infections: From diagnostics to therapeutics. Pharmaceutics. (2024) 16:1140. doi: 10.3390/pharmaceutics16091140
101. Wen X, Huang Z, Yang X, He X, Li L, Chen H, et al. Development of an aptamer capable of multidrug resistance reversal for tumor combination chemotherapy. Proc Natl Acad Sci. (2024) 121:1–11. doi: 10.1073/pnas.2321116121
102. Zhou G, Wilson G, Hebbard L, Duan W, Liddle C, George J, et al. Aptamers: A promising chemical antibody for cancer therapy. Oncotarget. (2016) 7:13446–63. doi: 10.18632/oncotarget.7178
103. Fàbrega C, Aviñó A, Eritja R. Chemical modifications in nucleic acids for therapeutic and diagnostic applications. Chem Rec. (2021) 22:1–10. doi: 10.1002/tcr.202100270
104. Niederlender S, Fontaine J-J, Karadjian G. Potential applications of aptamers in veterinary science. Veterinary Res. (2021) 52:1–20. doi: 10.1186/s13567-021-00948-4
105. Kratschmer C, Levy M. Effect of chemical modifications on aptamer stability in serum. Nucleic Acid Ther. (2017) 27:335–44. doi: 10.1089/nat.2017.0680
106. Sharda N, Khandelwal P, Zhang L, Caceres-Cortes J, Marathe P, Chimalakonda A. Pharmacokinetics of 40 KDA polyethylene glycol (PEG) in mice, rats, Cynomolgus monkeys and predicted pharmacokinetics in humans. Eur J Pharm Sci. (2021) 165:105928. doi: 10.1016/j.ejps.2021.105928
107. Lee J-H, Canny MD, De Erkenez A, Krilleke D, Ng Y-S, Shima DT, et al. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF 165. Proc Natl Acad Sci. (2005) 102:18902–7. doi: 10.1073/pnas.0509069102
108. Huang Z, Wang D, Zhang Q, Zhang Y, Peng R, Tan W. Leveraging aptamer-based DNA nanotechnology for bioanalysis and cancer therapeutics. Accounts Materials Res. (2024) 5:438–52. doi: 10.1021/accountsmr.3c00249
109. Hyjek-Składanowska M, Vickers TA, Napiórkowska A, Anderson BA, Tanowitz M, Crooke ST, et al. Origins of the increased affinity of phosphorothioate-modified therapeutic nucleic acids for proteins. J Am Chem Society. (2020) 142:7456–68. doi: 10.1021/jacs.9b13524
110. Wu J, Wang S, Li X, Zhang Q, Yang J, Ma Y, et al. Selective anti-melanoma effect of phosphothioated aptamer encapsulated by neutral cytidinyl/cationic lipids. Front Cell Dev Biol. (2021) 9:660233. doi: 10.3389/fcell.2021.660233
111. Li J, Mohammed-Elsabagh M, Paczkowski F, Li Y. Circular nucleic acids: Discovery, functions and applications. ChemBioChem. (2020) 21:1547–66. doi: 10.1002/cbic.202000003
112. Yan Y, Chang D, Xu Y, Chang Y, Zhang Q, Yuan Q, et al. Engineering a ligase binding DNA aptamer into a templating DNA scaffold to guide the selective synthesis of circular DNAzymes and DNA aptamers. J Am Chem Society. (2023) 145:2630–7. doi: 10.1021/jacs.2c12666
113. Chen X, Lu Y. Circular RNA: biosynthesis in vitro. Front Bioengineering Biotechnol. (2021) 9:787881. doi: 10.3389/fbioe.2021.787881
114. Li Q, Zhang S, Li W, Ge Z, Fan C, Gu H. Programming CircLigase catalysis for DNA rings and Topologies. Analytical Chem. (2020) 93:1801–10. doi: 10.1021/acs.analchem.0c04668
115. Ma Z, Chen H, Yang Y, Gao S, Yang J, Cui S, et al. Characterization of an ssdna ligase and its application in aptamer circularization. Analytical Biochem. (2024) 685:115409. doi: 10.1016/j.ab.2023.115409
116. Aljohani MM, Cialla-May D, Popp J, Chinnappan R, Al-Kattan K, Zourob M. Aptamers: Potential diagnostic and therapeutic agents for blood diseases. Molecules. (2022) 27:383. doi: 10.3390/molecules27020383
117. Kuai H, Zhao Z, Mo L, Liu H, Hu X, Fu T, et al. Circular bivalent aptamers enable in vivo stability and recognition. J Am Chem Society. (2017) 139:9128–31. doi: 10.1021/jacs.7b04547
118. Jiang Y, Pan X, Chang J, Niu W, Hou W, Kuai H, et al. Supramolecularly engineered circular bivalent aptamer for enhanced functional protein delivery. J Am Chem Society. (2018) 140:6780–4. doi: 10.1021/jacs.8b03442
119. Ma Y, Zhang Y, Chen Z, Tian Y, Zhang G. The modification strategies for enhancing the metabolic stabilities and pharmacokinetics of aptamer drug candidates. Pharm Sci. (2024). doi: 10.5772/intechopen.112756
120. Ibrahim M, Ramadan E, Elsadek NE, Emam SE, Shimizu T, Ando H, et al. Polyethylene Glycol (PEG): The nature, immunogenicity, and role in the hypersensitivity of pegylated products. J Controlled Release. (2022) 351:215–30. doi: 10.1016/j.jconrel.2022.09.031
121. Davis FF. The origin of pegnology. Advanced Drug Delivery Rev. (2002) 54:457–8. doi: 10.1016/s0169-409x(02)00021-2
122. Gao Y, Joshi M, Zhao Z, Mitragotri S. pegylated therapeutics in the clinic. Bioengineering Trans Med. (2023) 9:1–28. doi: 10.1002/btm2.10600
123. Hoang Thi TT, Pilkington EH, Nguyen DH, Lee JS, Park KD, Truong NP. The importance of poly(ethylene glycol) alternatives for overcoming peg immunogenicity in drug delivery and Bioconjugation. Polymers. (2020) 12:298. doi: 10.3390/polym12020298
124. Ng EW, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discovery. (2006) 5:123–32. doi: 10.1038/nrd1955
125. Mullard A. FDA approves second RNA aptamer. Nat Rev Drug Discovery. (2023) 22:774–4. doi: 10.1038/d41573-023-00148-z
126. Otte D-M, Choukeife M, Patwari T, Mayer G. Nucleic acid aptamers: From basic research to clinical applications. Handb Chem Biol Nucleic Acids. (2022), 1–25. doi: 10.1007/978-981-16-1313-5_25-1
127. Chandola C, Neerathilingam M. Aptamers for targeted delivery: Current challenges and future opportunities. Role Novel Drug Delivery Vehicles Nanobiomedicine. (2020). doi: 10.5772/intechopen.84217
128. Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for Biomarker Discovery. PloS One. (2010) 5:1–17. doi: 10.1371/journal.pone.0015004
129. Li Y, Tam WW, Yu Y, Zhuo Z, Xue Z, Tsang C, et al. The application of aptamer in biomarker discovery. biomark Res. (2023) 11:1–17. doi: 10.1186/s40364-023-00510-8
130. Ji D, Feng H, Liew SW, Kwok CK. Modified nucleic acid aptamers: Development, characterization, and biological applications. Trends Biotechnol. (2023) 41:1360–84. doi: 10.1016/j.tibtech.2023.05.005
131. Ascoët S, De Waard M. Diagnostic and therapeutic value of aptamers in envenomation cases. Int J Mol Sci. (2020) 21:3565. doi: 10.3390/ijms21103565
132. Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, et al. Nucleic acid ligands with protein-like side chains: Modified Aptamers and their use as diagnostic and therapeutic agents. Mol Ther - Nucleic Acids. (2014) 3:1–13. doi: 10.1038/mtna.2014.49
133. Chan KY, Kinghorn AB, Hollenstein M, Tanner JA. Chemical modifications for a next generation of Nucleic Acid Aptamers. ChemBioChem. (2022) 23:1–14. doi: 10.1002/cbic.202200006
134. Wu D, Katilius E, Olivas E, Dumont Milutinovic M, Walt DR. Incorporation of slow off-rate modified aptamers reagents in single molecule array assays for cytokine detection with ultrahigh sensitivity. Analytical Chem. (2016) 88:8385–9. doi: 10.1021/acs.analchem.6b02451
135. He W, Elizondo-Riojas M-A, Li X, Lokesh GL, Somasunderam A, Thiviyanathan V, et al. Correction to X-aptamers: A bead-based selection method for random incorporation of Druglike moieties onto next-generation aptamers for enhanced binding. Biochemistry. (2012) 51:9592–2. doi: 10.1021/bi301447a
136. Lokesh GL, Wang H, Lam CH, Thiviyanathan V, Ward N, Gorenstein DG, et al. X-aptamer selection and validation. Methods Mol Biol. (2017) 1632:151–74. doi: 10.1007/978-1-4939-7138-1_10
137. Liu W, Bi J, Ren Y, Chen H, Zhang J, Wang T, et al. Targeting extracellular CIRP with an X-aptamer shows therapeutic potential in acute pancreatitis. iScience. (2023) 26:107043. doi: 10.1016/j.isci.2023.107043
138. Ni S, Yao H, Wang L, Lu J, Jiang F, Lu A, et al. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int J Mol Sci. (2017) 18:1683. doi: 10.3390/ijms18081683
139. Kuszyk BS, Corl FM, Franano FN, Bluemke DA, Hofmann LV, Fortman BJ, et al. Tumor transport physiology. Am J Roentgenology. (2001) 177:747–53. doi: 10.2214/ajr.177.4.1770747
140. Healy JM, Lewis SD, Kurz M, Boomer RM, Thompson KM, Wilson C, et al. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm Res. (2004) 21:2234–46. doi: 10.1007/s11095-004-7676-4
141. Park JH, Jiang Y, Zhou J, Gong H, Mohapatra A, Heo J, et al. Genetically engineered cell membrane–coated nanoparticles for targeted delivery of dexamethasone to inflamed lungs. Sci Adv. (2021) 7:1–8. doi: 10.1126/sciadv.abf7820
142. Zhang R, Qin X, Kong F, Chen P, Pan G. Improving cellular uptake of therapeutic entities through interaction with components of cell membrane. Drug Delivery. (2019) 26:328–42. doi: 10.1080/10717544.2019.1582730
143. Jin H-R, Wang J, Wang Z-J, Xi M-J, Xia B-H, Deng K, et al. Lipid metabolic reprogramming in tumor microenvironment: From mechanisms to therapeutics. J Hematol Oncol. (2023) 16:1–33. doi: 10.1186/s13045-023-01498-2
144. Yang Q, Peng Y, Deng Z, Zhang D, Long C-Y, Zhang G-R, et al. Regulating the properties of XQ-2D for targeted delivery of therapeutic agents to pancreatic cancers. Natl Sci Rev. (2023) 10:1–7. doi: 10.1093/nsr/nwad113g
145. Vass P, Akdag DS, Broholm GE, Kjaer J, Humphreys AJ, Ehmann F. Enabling technologies driving drug research and development. Front Med. (2023) 10:1122405. doi: 10.3389/fmed.2023.1122405
146. Giannuzzi V, Bertolani A, Torretta S, Reggiardo G, Toich E, Bonifazi D, et al. Innovative Research Methodologies in the EU Regulatory Framework: An analysis of EMA qualification procedures from a pediatric perspective. Front Med. (2024) 11:1369547. doi: 10.3389/fmed.2024.1369547
147. FDA. Platform Technology Designation Program for Drug Development Guidance for Industry (2024). Available online at: https://www.fda.gov/media/178928/ (Accessed October 18, 2024).
148. Rogers H, Adeniyi O, Ramamoorthy A, Bailey S, Pacanowski M. Clinical Pharmacology Studies supporting oligonucleotide therapy development: An assessment of therapies approved and in development between 2012 and 2018. Clin Trans Science. (2021) 14:468–75. doi: 10.1111/cts.12945
149. Indraratna P, Tardo D, Delves M, Szirt R, Ng B. Measurement and management of QT interval prolongation for general physicians. J Gen Internal Med. (2019) 35:865–73. doi: 10.1007/s11606-019-05477-7
150. Berger FA, van der Sijs H, Becker ML, van Gelder T, van den Bemt PM. Development and validation of a tool to assess the risk of QT Drug-Drug Interactions in clinical practice. BMC Med Inf Decision Making. (2020) 20:1–12. doi: 10.1186/s12911-020-01181-3
151. Khatib R, Sabir FR, Omari C, Pepper C, Tayebjee MH. Managing drug-induced QT prolongation in clinical practice. Postgraduate Med J. (2020) 97:452–8. doi: 10.1136/postgradmedj-2020-138661
152. Kroenke MA, Milton MN, Kumar S, Bame E, White JT. Immunogenicity risk assessment for multi-specific therapeutics. AAPS J. (2021) 23:1–13. doi: 10.1208/s12248-021-00642-5
153. Deshaies RJ. Multispecific Drugs Herald a new era of biopharmaceutical innovation. Nature. (2020) 580:329–38. doi: 10.1038/s41586-020-2168-1
154. Wen Y, Jawa V. The impact of product and process related critical quality attributes on immunogenicity and adverse immunological effects of Biotherapeutics. J Pharm Sci. (2021) 110:1025–41. doi: 10.1016/j.xphs.2020.12.003
155. Jawa V, Maamary J, Swanson M, Zhang S, Montgomery D. Implementing a clinical immunogenicity strategy using preclinical risk assessment outputs. J Pharm Sci. (2022) 111:960–9. doi: 10.1016/j.xphs.2022.01.032
156. Zhao D, Long X, Fan H, Wang J. Effects of hepatic or renal impairment on the pharmacokinetics of immune checkpoint inhibitors. Am J Cancer Res. (2022) 12:4892–903.
157. Nafziger AN, Arscott KA, Cochrane K, Skobieranda F, Burt DA, Fossler MJ. The influence of renal or hepatic impairment on the pharmacokinetics, safety, and tolerability of Oliceridine. Clin Pharmacol Drug Dev. (2019) 9:639–50. doi: 10.1002/cpdd.750
158. Fletcher EP, Sahre M, Hon YY, Balakrishnan A, Zhou L, Sun Q, et al. Impact of organ impairment on the pharmacokinetics of therapeutic peptides and proteins. AAPS J. (2023) 25:1–11. doi: 10.1208/s12248-023-00819-0
159. Alhassani RY, Bagadood RM, Balubaid RN, Barno H, Alahmadi MO, Ayoub NA. Drug therapies affecting renal function: An overview. Cureus. (2021) 13(11):1–11. doi: 10.7759/cureus.19924
160. Peabody J, Acelajado MC, Robert T, Hild C, Schrecker J, Paculdo D, et al. Drug-Drug Interaction Assessment and identification in the primary care setting. J Clin Med Res. (2018) 10:806–14. doi: 10.14740/jocmr3557w
161. Axon DR, Vanova J, Edel C, Slack M. Dietary supplement use, knowledge, and perceptions among student pharmacists. Am J Pharm Education. (2017) 81:92. doi: 10.5688/ajpe81592
162. van Leeuwen RWF, le Comte M, Reyners AKL, van den Tweel A, van Vlijmen B, Kwee W, et al. Evidence- and consensus-based guidelines for drug-drug interactions with anticancer drugs; a practical and universal tool for Management. Semin Oncol. (2022) 49:119–29. doi: 10.1053/j.seminoncol.2022.03.002
163. Rakshith UR, Nair SP, Ranganath AH, Halagali P. Drug interactions: Mechanisms, assessment and management strategies. J Young Pharmacists. (2024) 16:447–55. doi: 10.5530/jyp.2024.16.58
164. Ding D, Zhao H, Wei D, Yang Q, Yang C, Wang R, et al. The first-in-human whole-body dynamic pharmacokinetics study of aptamer. Research. (2023) 6:1–12. doi: 10.34133/research.0126
165. Verdonck L, Buyst D, de Vries A-M, Gheerardijn V, Madder A, Martins JC. Tethered imidazole mediated duplex stabilization and its potential for aptamer stabilization. Nucleic Acids Res. (2018) 46:11671–86. doi: 10.1093/nar/gky1062
166. De Clercq E, Eckstein F, Merigan TC. Interferon induction increased through chemical modification of a synthetic polyribonucleotide. Science. (1969) 165:1137–9. doi: 10.1126/science.165.3898.1137
Keywords: aptamer, tumor, regulatory, chemical modification, clinical trials, immunotherapy
Citation: Mathavan S, Tam YJ, Mustaffa KMF and Tye GJ (2025) Aptamer based immunotherapy: a potential solid tumor therapeutic. Front. Immunol. 16:1536569. doi: 10.3389/fimmu.2025.1536569
Received: 29 November 2024; Accepted: 27 January 2025;
Published: 17 February 2025.
Edited by:
Stergios Boussios, Canterbury Christ Church University, United KingdomReviewed by:
Xiaohong Tan, Bowling Green State University, United StatesCopyright © 2025 Mathavan, Tam, Mustaffa and Tye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gee Jun Tye, Z2VlanVuQHVzbS5teQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.