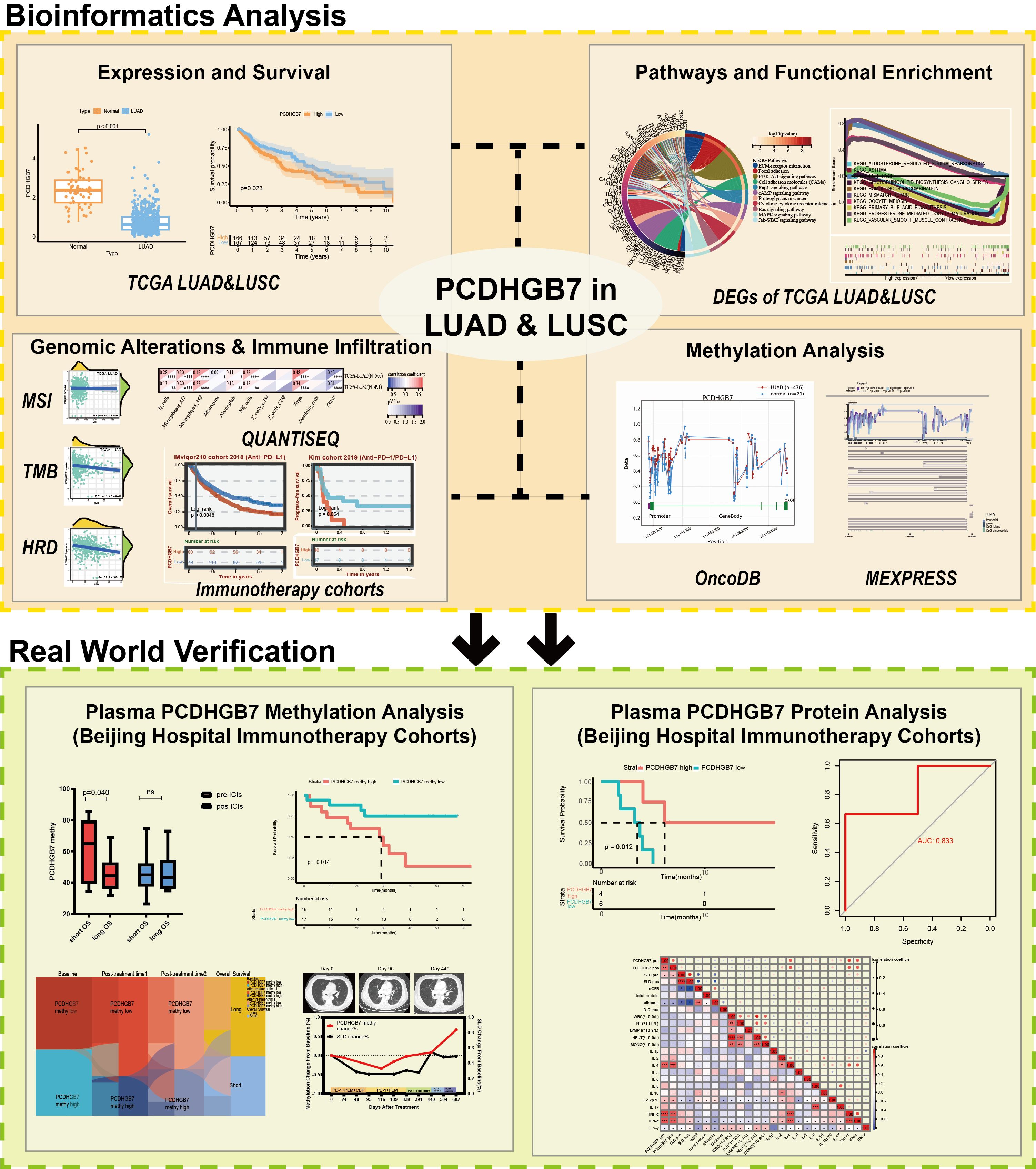
- 1Department of Oncology, Beijing Hospital, National Center of Gerontology, Institute of Geriatric Medicine, Chinese Academy of Medical Sciences, Beijing, China
- 2Beijing Hospital, National Center of Gerontology, Institute of Geriatric Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
Backgrounds: Protocadherin gamma subfamily B, 7 (PCDHGB7), a member of the protocadherin family, plays critical roles in neuronal connections and has been implicated in female reproductive system cancers. Its function in lung cancer has not been elucidated.
Methods: We comprehensively investigated PCDHGB7 expression, prognosis, biological function, methylation patterns, and it’s relationship with immune infiltration and immunotherapy response through public datasets (HPA, TCGA, GEO, OncoDB and MEXPRESS). Two lung cancer immunotherapy cohorts from our clinical center were enrolled to detect the relationship between methylation and protein levels of PCDHGB7 in plasma and immunotherapy outcomes.
Results: PCDHGB7 expression was downregulated in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) and associated with tumor prognosis. PCDHGB7 demonstrated a positive correlation with inhibitory immune cells and a negative correlation with tumor mutational burden (TMB) and homologous recombination deficiency (HRD). The methylation level of PCDHGB7 was upregulated in tumor tissue and negatively correlated with PCDHGB7 mRNA level. In immunotherapy cohort studies, patients with higher PCDHGB7 tissue expression showed worse prognosis. Patients with PCDHGB7 hypermethylation in baseline plasma had shorter progression-free survival (PFS) and overall survival (OS), while those with early reduction of PCDHGB7 methylation had the best prognosis. Plasma PCDHGB7 protein levels could predict responses to immune checkpoint inhibitors and function as a prognostic marker for PFS.
Conclusion: PCDHGB7 expression and methylation are prognostic and immunological biomarkers in non-small cell lung cancer. Plasma PCDHGB7 methylation and protein levels can be used as novel biomarkers for predicting the efficacy of immunotherapy in lung cancer.
1 Introduction
Lung cancer continues to be the leading cause of death in all demographics and a significant factor in the global decline in health-related quality of life (1). Immunotherapy has become a powerful and vital treatment approach with the introduction of immune checkpoint inhibitors (ICIs), such as anti-programmed death 1 (PD-1), anti-programmed death-ligand 1 (PD-L1), and anti-cytotoxic T lymphocyte-associated protein 4 (CTLA-4). Nonetheless, low response rates and drug resistance remain the main reasons, which hinder the efficacy of immunotherapy (2). Thus, tumor mutational burden (TMB), PD-L1, and other biomarkers have been used as indicators for predicting the effectiveness of immunotherapy (3, 4), which also played important roles in regulating the tumor microenvironment (TME) (5). However, identifying novel biomarkers for optimizing the therapeutic impact of immunotherapy is still in an urgent need since the existing indicators, such as PD-L1 and TMB, which rely on tumor tissue biopsy, still cannot meet the clinical needs due to the lack of invasive examination, inconsistent detection standards, and inability to detect dynamically.
Protocadherin gamma subfamily B, 7 (PCDHGB7) is a member of the protocadherins (PCDH) family, representing the largest group within the cadherin superfamily (6). The protein structure of PCDHGB7 comprises multiple β sheets and it is mainly localized to the vesicles and plasma membrane, with an additional presence detected in the nucleoplasm and cytosol (Supplementary Figures S1A, B). According to the Human Protein Atlas (HPA) database, PCDHGB7 exhibited high RNA expression in lung and cerebral cortex, as well as ovaries, endometrium, cervix and other female reproductive systems (Supplementary Figure S1C). At the protein level, PCDHGB7 was extensively expressed across multiple organs, with particularly high expression observed in the cervix, ovaries, and cerebral cortex. Low protein expression was also noted in the bronchial and pulmonary tissues (Supplementary Figure S1D). According to previous research, PCDHGB7 is involved in synaptic mobility, self- and mutual recognition, and the formation of the nervous system network (7–9). Of note, several studies have found that PCDHGB7 contributed to cancer development and functioned as a tumor suppressor gene. It was reported that protocadherins suppressed the development of tumors via multiple biological interventions, such as promoting cell cycle arrest, inducing apoptosis, and regulating the Wnt (Wingless and INT-1) pathway (10, 11). However, the role and function of PCDHGB7 in cancers have not been fully elucidated.
DNA methylation abnormalities have been shown to precede pathological changes (12, 13). Previous studies based on TCGA and GEO databases (n=7114) have demonstrated that PCDHGB7 was hypermethylated in all cancer types. Clinical cancer samples (n=727) detection revealed that PCDHGB7 hypermethylation could be a diagnostic biomarker for cancer detection in a variety of cancers (14). The level of PCDHGB7 methylation has been then proven to be an early and reliable diagnosis indicator of cervical cancer and has shown good diagnostic efficacy in the diagnosis of malignant body fluids (14–16). PCDHGB7 has been found to upregulated in lung cancer, while the specific function of PCDHGB7 remains to be further clarified.
In this study, we investigated the role of PCDHGB7 in lung cancers via integration of multiple bioinformatics methods. PCDHGB7-associated gene expression, biological function, genomic alterations, immune infiltration, and methylations were extensively studied in our work. The PCDHGB7 methylation and expression in plasma was also detected in lung cancer patients collected in our clinical center in order to reveal the association between PCDHGB7 and immunotherapy response. In summary, our research aim to contribute to a better understanding of the role of PCDHGB7 in lung cancer, and expose the potential of PCDHGB7 to be a novel biomarker for lung cancer.
2 Materials and methods
2.1 TCGA data acquisition and preprocessing
We downloaded the standardized pan-cancer dataset of The Cancer Genome Atlas (TCGA) repository (https://www.cancer.gov/ccg/research/genome-sequencing/tcga) from, the UCSC (https://xenabrowser.net/) database (accessed on May 17, 2024). Gene expression data and clinical parameters were sorted, merged, and normalized by PERL scripts and related R packages (R version: 4.3.3).
2.2 Expression analysis and prognostic analysis of PCDHGTB7
The architectural conformation and subcellular localization (Rh30, SK-MEL-30 and U2OS cell lines) of PCDHGTB7 were inferred from data accessible in the Human Protein Altas (HPA) repository (https://www.proteinatlas.org/). Using R package “survival” (version 4.3.3) to analyze the association between PCDHGB7 expression and prognosis (Data with survival time or follow-up time less than 30 days were excluded). Meanwhile, we use the online tool KMplot(https://kmplot.com/analysis/) to invoke the Gene Expression Omnibus (GEO) database (GSE19188, GES157011 and GSE102287) to draw Kaplan–Meier survival plots(accessed on May 17, 2024). The median values served as the cutoff points for high and low levels of PCDHGB7 expression in all analyses.
2.3 Immune infiltration and immunotherapy cohorts analysis of PCDHGB7
The examination of the associations between PCDHGB7 expression and the tumor mutation burden (TMB), microsatellite instability (MSI) as well as homologous recombination deficiency (HRD) from TCGA cohorts, was conducted through the Sanger Box platform. Pearson’s rank correlation test was executed, yielding both the partial correlation (cor) and corresponding p-value.
The ESTIMATE algorithm of LUAD and LUSC was used to analyze the disparity in the stromal score and the immune score with the package “estimate” (Version R4.2.1). Explorations into the connections between PCDHGB7 expression and tumor-infiltrating immune cells (TIICs), such as B cells, CD4+T memory cells, CD8+T cells, NK cells, monocytes, macrophages, among others was achieved through quanTIseq algorithm using package “quanTIseq” (Version R4.2.1).The immunotherapy cohorts of LUAD and LUSC were sourced from the BEST database (https://rookieutopia.com/appdirect/BEST/) (17).
2.4 Tumor stemness analysis of PCDHGB7
We obtained the methylation-based stemness score (DNAss) and epigenetically regulated DNA methylation-based stemness score (EREG-METHss) based on one-class logistic regression machine learning algorithm (OCLR) of LUAD and LUSC (18). Afterwards, we used the Pearson function in R studio to calculate the correlation between PCDHGB7 expression and tumor stemness indicators.
2.5 Methylation evaluation of PCDHGB7
OncoDB (19) (https://oncodb.org/) and MEXPRESS (20) (https://mexpress.be/) databases were used in our work to assess the PCDHGB7 DNA methylation levels.
2.6 Pathway and functional enrichment analyses
We retrieved RNA-Seq data of TCGA-LUAD and TCGA-LUSC and then stratified PCDHGB7 expression into high and low expression cohorts and conducted both Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) enrichment analyses on the differentially expressed genes between these two cohorts (LUAD: |logFC| > 1.5, p < 0.01, FDR < 0.01, LUSC: |logFC| > 1.0, p < 0.01, FDR < 0.01). KEGG and GO analysis were performed by the enrichGO function and enrichKEGG function in the “clusterProfiler” package of R studio, respectively. Gene set enrichment analysis (GSEA) was performed using GSEA software (version 4.1.0).
2.7 Detection of PCDHGB7 methylation in plasma
Patients from October 2018 to November 2022 with pathologically or immunohistochemically proven unresectable locally advanced or advanced non-small-cell lung cancer (ECOG score 0–2) in the Oncology Department of Beijing hospital were enrolled. All patients received monotherapy or a combination regimen containing PD-1inhibitors. RECIST 1.1 was used to evaluate the efficacy of solid tumors, and survival analysis was performed based on follow-up data. Peripheral blood plasma was collected before and at the early stage of treatment (after 2–4 courses of treatment), and for some patients, plasma collection occurred at several time points throughout treatment. All patients provided written informed consent. This study was approved by the ethics committee of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences, and Peking Union Medical College (Beijing, China; approved no. NCC2021C-527).
For sample preprocessing, 8–10 mL of whole blood was collected in a 10-mL purple-capped EDTA anticoagulant tube. The first centrifugation was performed within 4 h at 4°C, with the blood centrifuged at 3000 g for 10 min, and the upper layer of plasma was carefully transferred to a new 5-mL EP tube. Subsequently, a second centrifugation was performed at 4°C and 16, 000 g for 10 min, and the upper layer of plasma was carefully transferred again to a new 1.5-mL EP tube.
For DNA extraction and methylation analysis, ctDNA from plasma samples (400μL) was extracted using the Tiangen Enhanced Magnetic Beads Large Volume Free DNA Extraction Kit (DP720-02). Starting with 100 ng of DNA, bisulfite conversion (Zymo, D5006) was performed, followed by two rounds of nested PCR before pyrosequencing analysis to assess PCDHGB7 CpG methylation. DNA extraction and methylation analyses were performed by Shanghai Epiprobe Biotechnology Co., Ltd. Each sample requires 1mL of plasma for testing.
Some patients have already undergone targeted capture sequencing of 1021 genes in previous studies at our center, and PyClone was used to infer the molecular tumor burden index (mTBI) (21).
2.8 Detection of plasma PCDHGB7 protein levels
This was an observative real-world study performed in patients with locally advanced or advanced non-small-cell lung cancer (NSCLC) of Beijing Hospital between May 2020 and June 2023 who progressed after a list-one-line systematic therapy. Patients aged 65 or older with an ECOG score of 0–3 assessed by one tumor specialist were eligible for inclusion. Those who tested positive for driver genes were subsequently evaluated by two oncologists to determine their candidacy for immunotherapy following standard treatment protocols. All patients received PD-1 inhibitors (200 mg IV day 1, every 3 weeks) in combination with metronomic oral vinorelbine (30 mg, 3 times per week). After six cycles of combined therapy, patients who did not experience progression or for whom treatment was paused due to intolerable adverse reactions were switched to monotherapy with PD-1 inhibitors for maintenance treatment. Peripheral plasma was collected from all patients at the beginning and early stages of treatment (1–3 cycles after treatment). All patients provided written informed consent. The present study was approved by the ethics committee of Beijing Hospital (Beijing, China; approved no. 2023BJYYEC-196-01).
Plasma samples (50 μL) were analyzed using the SOMAscan Assay Kit for human serum, which measures the expression of 11000 types of human proteins using highly selective single-stranded modified Slow Off-rate Modified DNA Aptamers (SOMAmer) according to the manufacturer’s standard protocol (SomaLogic; Boulder, CO).
2.9 Statistical analysis
Data processing, statistical analysis, and visualization were performed comprehensively using the R 4.3.3 software package and GraphPad Prism 10.0 software. For datasets exhibiting a normal distribution, the unpaired Student’s t-test was applied. The associations between categorical variables were analyzed using Pearson’s chi-square test. Pearson’s correlation coefficients were used to evaluate the associations between two continuous variables. Considering the potential impact of skewed data, Spearman’s correlation analysis was also performed to ensure a comprehensive examination of the relationship. The prognostic value was evaluated by Kaplan–Meier analysis. A significance level of P < 0.05 (two-tailed) was considered statistically significant, and the false discovery rate (FDR) was controlled by the Benjamini–Hochberg procedure.
3 Results
3.1 PCDHGB7’s expression and survival research in lung adenocarcinoma and lung squamous cell carcinoma
The study’s procedural overview is depicted in Figure 1. Given the high expression of PCDHGB7 within pulmonary tissue, we aimed to elucidate the role of PCDHGB7 in lung cancer. PCDHGB7 showed significantly low expression in both LUAD (p <0.0001) and LUSC (p <0.0001) (Figure 2A). Next, we used R script to examine the prognostic relevance of PCDHGB7 expression in lung cancer. Figure 2B showed that low PCDHGB7 mRNA expression showed a trend of poor overall survival (OS) in patients with LUAD (p=0.062), while low PCDHGB7 mRNA expression was significantly associated with better OS in patients with LUSC(p=0.023) (Figure 2C). Due to the contradictory results of prognostic analysis, we further used the KMplot online resource to extract data from the Gene Expression Omnibus (GEO) database. The results of the GSE19188, GES157011 and GSE102287 datasets indicated that patients with high PCDHGB7 expression exhibited poor OS compared with those with low expression levels (Supplementary Figure S2). These results suggest that high PCDHGB7 expression could lead to a poor prognosis in patients with lung cancer.
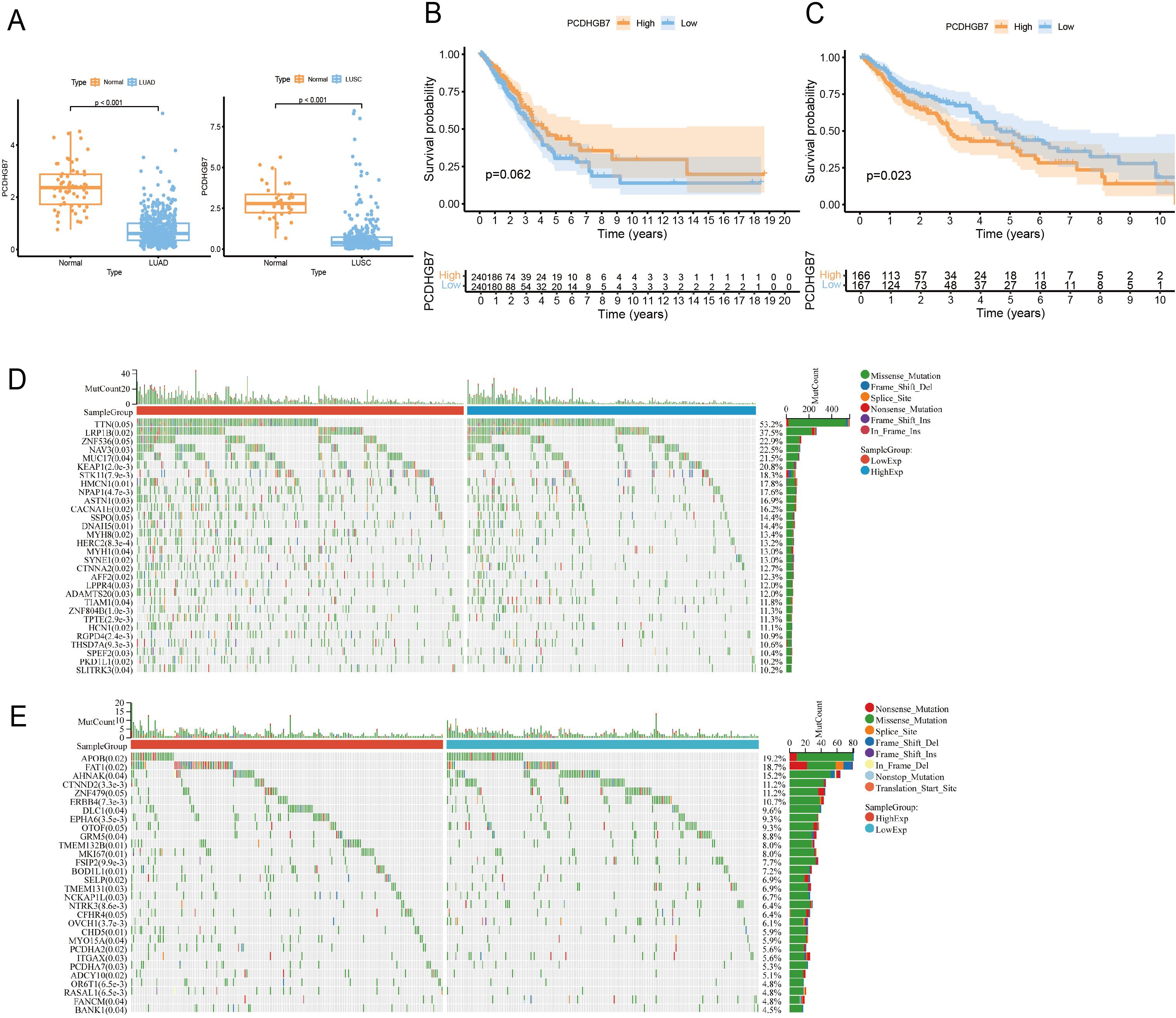
Figure 2. PCDHGB7’s expression, prognostic and mutations analysis in lung cancer. (A) PCDHGB7 expression in LUAD (normal n = 59, tumor n = 541. p <0.0001) and LUSC (normal n = 34, tumor n = 360, p <0.0001). (B) Kaplan-Meier survival analysis of PCDHGB7 expression in TCGA-LUAD(p=0.062). (C) Kaplan-Meier survival analysis of PCDHGB7 expression in TCGA-LUSC(p=0.023). Mutation landscape of LUAD (D) and LUSC (E).
3.2 PCDHGB7-associated mutation landscape in lung cancer
As gene mutations play an important role in lung cancer development, we next characterized the genomic mutation profile correlated with PCDHGB7 expression in lung cancer. PCDHGB7 expression in LUAD was associated with gene mutations such as LRP1B, STK11, and KEAP1. Of note, these genes mutations have been shown to have predictive effects on immune therapy for lung cancer in previous studies (Figure 2D) (4, 22). Meanwhile, patients with high expression of PCDHGB7 in LUSC were more likely to develop FAT1 mutations, which would contribute to tumor development (Figure 2E) (23).
3.3 PCDHGB7-related pathways and functional enrichment analysis in LUAD and LUSC
To further explore the biological function of PCDHGB7, we retrieved and downloaded the proteins closely interacting with PCDHGB7 from the STRING database and visualized the protein interaction network using Cytoscape. In addition to other members of the PCDH family, PCDHGB7 is closely related to proteins such as Protocadherin Fat 1(FAT1), mutL homolog 1(MLH1), and fragile histidine triad diadenosine triphosphatase (FHIT), which have been shown to be involved in DNA mismatch repair and related to tumor stemness (Figure 3A) (23, 24). Collectively, these data suggested that high PCDHGB7 expression may promote a hybrid EMT state due to dysfunction of FAT1 protein, tumor stemness, and metastasis.
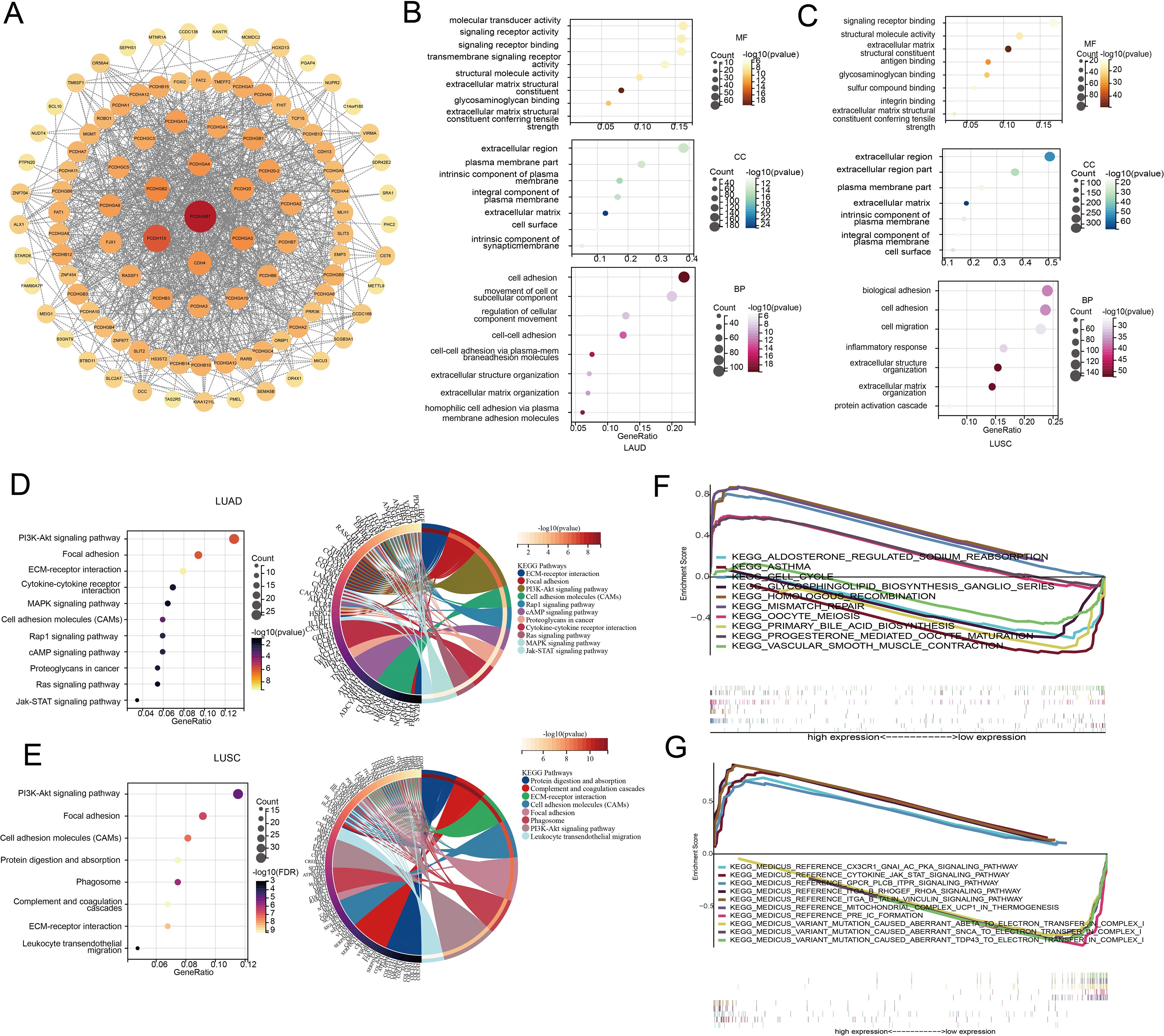
Figure 3. Pathway and functional enrichment analyses of LUAD and LUSC. (A) Protein interaction network of PCDHGB7 (constructed and visualized using Cytoscape software). (B) GO of LUAD. (C) GO of LUSC. (D) KEGG of LUAD. (E) KEGG of LUSC. (F) GSEA of LUAD. (G) GSEA of LUSC.
To investigate the function of PCDHGB7 in lung cancer with greater precision, we conducted GO and KEGG pathway enrichment analyses based on the differentially expressed genes between the PCDHGB7-high and PCDHGB7-low groups. The results of the GO analysis indicated that PCDHGB7 primarily contributes to the cellular component by being a part of the cell membrane, the intrinsic component of the plasma membrane, and the extracellular matrix. In terms of molecular function, PCDHGB7 is mainly involved in signal receptor binding and activity, as well as maintaining the structural integrity of the extracellular matrix. Regarding biological processes, PCDHGB7 predominantly participates in cell adhesion, homophilic cell-cell adhesion, and the organization of the extracellular matrix. (Figures 3B, C). KEGG analysis showed that PCDHGB7 was associated with typical tumor proliferation pathways including phosphatidylinositol 3’-kinase (PI3K)-Akt signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway, Janus kinase/signal transducers, and activators of transcription (JAK-stat) signaling pathway. PCDHGB7 also participated in leukocyte transactional migration, cytokine receptor interaction and other immune response processes (Figures 3D, E). GSEA revealed that the high expression of PCDHGB7 was associated with the upregulation of homologous recombination, mismatch repair, and cell cycle pathways in LUAD (Figure 3F), as well as the upregulation of the JAK-stat pathway and CX3CR1-GNA-AC-PKA signaling pathway in LUSCs (Figure 3G). The findings mentioned earlier indicated that PCDHGB7 is crucial for cell communication, tissue structure, and potentially for processes such as cell migration and differentiation. Additionally, PCDHGB7 plays a significant role in DNA homologous recombination and mismatch repair within lung cancer. It also contributes to pathways that are central to tumor proliferation. The involvement of PCDHGB7 in these critical biological processes suggests that it could be a pivotal factor in both the development and progression of lung cancer.
3.4 Genomic alterations of PCDHGB7 in LUAD and LUSC
The association between PCDHGB7 and biomarkers related to genomic alterations such MSI, TMB, and HRD was then assessed. There was no statistically significant connection between PCDHGB7 expression and MSI in either LUAD (r = -0.0064, p = 0.88) or LUSC (r = 0.026, p = 0.56) (Figure 4A). In the TMB analysis, PCDHGB7 expression showed a significant negative correlation with TMB in LUAD (r = -0.14, p = 0.0021), but no significant correlation was seen in LUSC (r = -0.029, p = 0.53) (Figure 4B). In contrast, PCDHGB7 expression showed a significant correlation with HRD, both in LUAD (r = -0.21, p<0.001) and LUSC (r = -0.18, p<0.001) (Figure 4C).
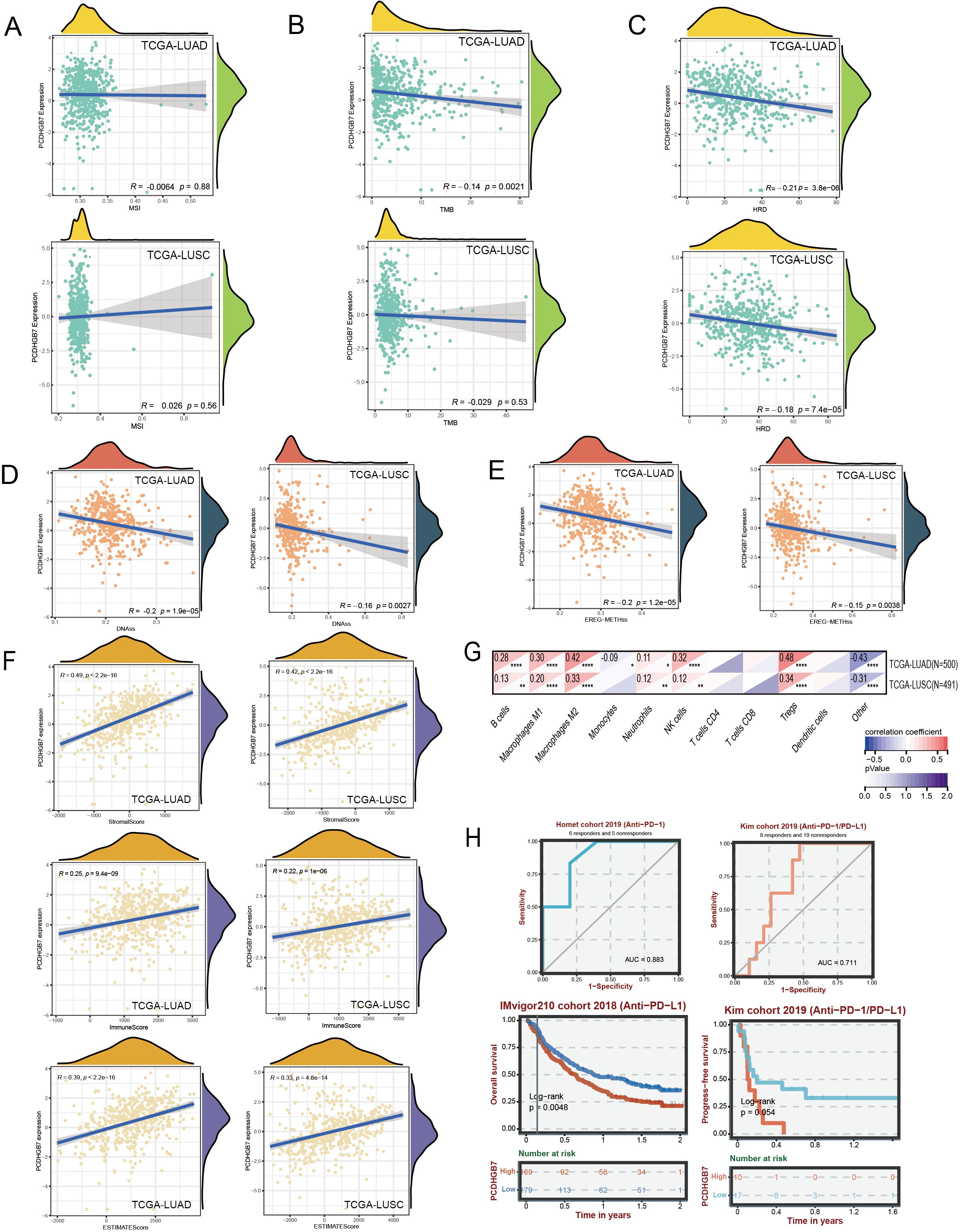
Figure 4. Genomic alterations and immune assessment of PCDHGB7 in lung cancer. (A) Correlation with microsatellite. (B) Correlation with tumor mutation burden. (C) Correlation withHRD. Correlation with homologous recombination repair defects: (D) DNAss score. (E) EREG-METHss score. (F) ESTIMATE scores of LUAD and LUSC. (G) Correlation of PCDHGB7 expression levels with immune cell infiltration based on quanTIseq (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (H) ROC curves and Kaplan-Meier survival analysis in the immunotherapy cohort analysis.
Cancer progression is mediated by the gradual loss of a differentiated phenotype and the acquisition of progenitor and stem cell-like features. Here, we used two types of one-class logistic regression machine learning algorithms to assess the degree of oncogenic dedifferentiation based on previously published results (18). The DNAss score (Figure 4D) and EREG-METHss score (Figure 4E) demonstrated that PCDHGB7 expression was negatively correlated with tumor stemness LUAD (r = -0.2, p<0.001) and LUSC (r = -0.16, p=0.0027). Empirical evidence from prior research has indicated that the stemness characteristics of neoplasms were correlated with tumor grade, processes of invasion and metastasis, as well as the reduction in PD-L1 expression has been observed (18). Consequently, these data suggest that PCDHGB7play a significant role in the pathobiological progression of tumor development.
3.5 Immune infiltration and real-world cohort immune response analysis of PCDHGB7
The TME fundamentally affects the progression of cancer, which is a complex of tumor cells, stromal elements, and immune components, the TME orchestrates intricate and dynamic interactions (25). The ESTIMATE algorithm has emerged as a robust computational tool for quantifying the infiltration of stromal and immune cells into tumor by calculating immune scores and stromal scores. The immune scores showed that PCDHGB7 expression was slightly positive correlated with immune infiltration in LUAD and LUSC (Figure 4F). Then we used mRNA-based immune infiltration prediction algorithm quanTIseq to further explore immune cell infiltration. The analysis of LUAD and LUSC both showed a significant positive correlation between PCDHGB7 and M2 macrophages (LUAD: r=0.42, P < 0.0001; LUSC: r=0.33, P < 0.0001) as well as Treg cells (LUAD: r=0.48, P < 0.0001; LUSC: r=0.34, P < 0.0001) compared to other immune cell subgroups (Figure 4G).
Analysis of mutation landscape and immune infiltration suggested that PCDHGB7 expression is related to the efficacy of lung cancer immunotherapy. To further decipher the effect of PCDHGB7 in real-world setting in patients with lung cancer undergoing immunotherapy, the BEST database was employed, and the data from the IMvigor210 cohort and Kim cohort showed that PCDHGB7 expression functioned as an effective predictor for immunotherapy response (Figure 4H AUC curves). Patients with high expression of PCDHGB7 exhibited significantly poor responses to immunotherapy (Figure 4H survival curves).
3.6 Methylation analysis of PCDHGB7 in LUAD and LUSC
Dysregulated DNA methylation profiles are associated with altered gene expression in tumors (26). To investigate the relationship between aberrant PCDHGB7 expression patterns and methylation, we used the OncoDB databases to explore abnormal PCDHGB7 methylation patterns in normal and tumor tissues. The data revealed that the methylation levels of multiple sites in PCDHGB7 gene were higher in tumor tissues than in normal tissues, especially in the potential promoter region (141417677 to 141419677) in both LUAD (Figure 5A) and LUSC (Figure 5B). Survival analysis revealed no significant correlation between the degree of methylation and OS in either LUAD (Figure 5C) or LUSC (Figure 5D). Additionally, we found that decreased PCDHGB7 expression may be linked to increased methylation in LUAD (Figure 5E) and LUSC (Figure 5F) when we used MEXPRESS to investigate the relationship between PCDHGB7 expression and CpG islands in lung cancer. The correlation coefficient and p-value of the methylation and expression correlation analysis of each DNA site are in Supplementary Figures S3 and S4. These findings implied that hypermethylation of the PCDHGB7 could contribute to the downregulated of PCDHGB7 expression in lung cancer.
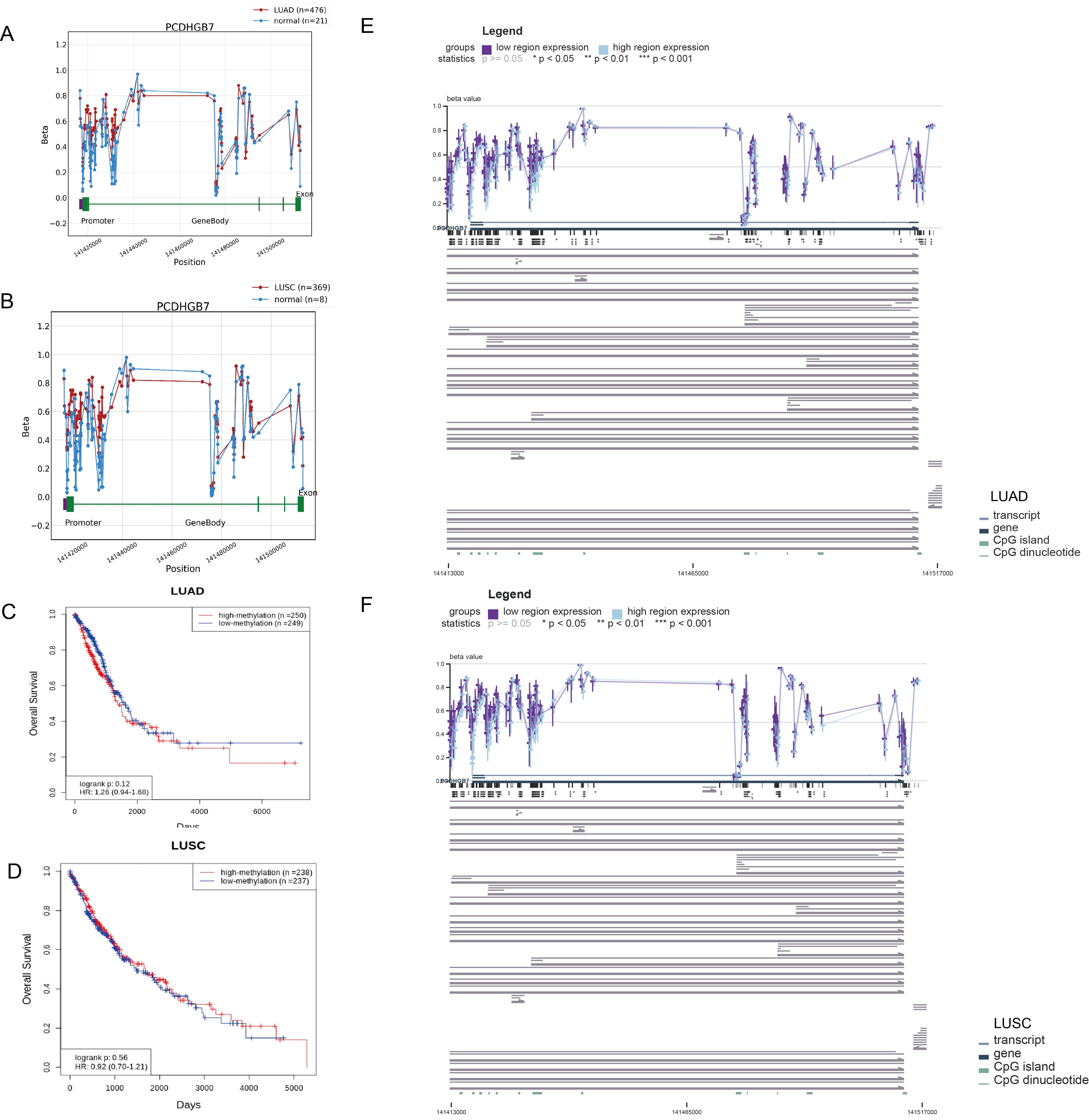
Figure 5. Methylation analysis of PCDHGB7 in LUAD and LUSC. (A) PCDHGB7 methylation comparison with normal tissues and survival analysis in LUAD. (B) PCDHGB7 methylation comparison with normal tissues and survival analysis in LUSC. Kaplan-Meier survival analysis of PCDHGB7 methylation in LUAD (C) and LUSC (D). Visualization of the methylation sites within the DNA sequence associated with gene expression was performed using MEXPRESS in LUAD (E) and LUSC (F). The expression of PCDHGB7 is represented by the blue line. Pearson’s correlation coefficients and p-values for methylation sites and are provided on the right side. *p < 0.01, **p < 0.01, ***p < 0.001.
3.7 PCDHGB7 methylation level in plasma is closely associated with immunotherapy efficacy
It has been reported that the level of PCDHGB7 methylation level in malignant body fluids serves as a promising biomarker for cancer, particularly the cervical cancer (14–16). However, it remains uncertain whether the PCDHGB7 methylation level in peripheral blood from patients with lung cancer could serve as a novel biomarker of immunotherapy efficacy. Therefore, we collected blood samples from patients with lung cancer before and after immunotherapy treatment for investigation. From October 2018 to November 2022, 32 patients were included in this study, with a median follow-up of 43.6 (30.9–62.4) months. The baseline characteristics of the enrolled patients are presented in Supplementary Table S1. The median age across is 65.5 years, with a slight male predominance (26 males, 82%). The pathological types are evenly distributed, with squamous cell carcinoma present in 13 patients (40.6%) and non-squamous cell carcinoma in 19 patients (59.4%). In terms of therapy, 20 patients (62.5%) received ICIs plus chemotherapy, 2 patients (6.3%) received ICIs plus chemotherapy plus anti-angiogenic therapy, 2 patients (6.3%) received immune plus anti-angiogenic therapy, and 9 patients (28.1%) received immune monotherapy. First-line therapy was administered to 24 patients (75%), and second-line and posterior line therapy to 8 patients (25%). Patients were divided into high- and low-methylation PCDHGB7 groups based on the baseline value of plasma PCDHGB7 methylation. Interestingly, patients with liver metastasis or a high baseline tumor load were more likely to have high plasma PCDHGB7 methylation.
Next, we explored PCDHGB7 methylation changes in peripheral blood at baseline and early post-treatment. PCDHGB7 methylation showed a downward trend after treatment, which was more significant in patients receiving first-line immunotherapy patients (total patients: 56.2 vs. 50.9; first line: 54.0 vs. 46.8, p = 0.034, Figures 6A, B). Besides, there was a significant correlation between the sum of longest diameters (SLD) of target lesions and PCDHGB7 methylation, both at baseline (r = 0.47, p = 0.0069) and after treatment (r = 0.54, p = 0.0015) (Figures 6D, E). Patients treated with first-line immunotherapy were divided into the long OS group (> 25 months) and the short OS group (≤ 25 months) based on the OS time. The short OS group had a higher degree of methylation at baseline (60.0 vs. 45.5, p = 0.040, Figure 6C).
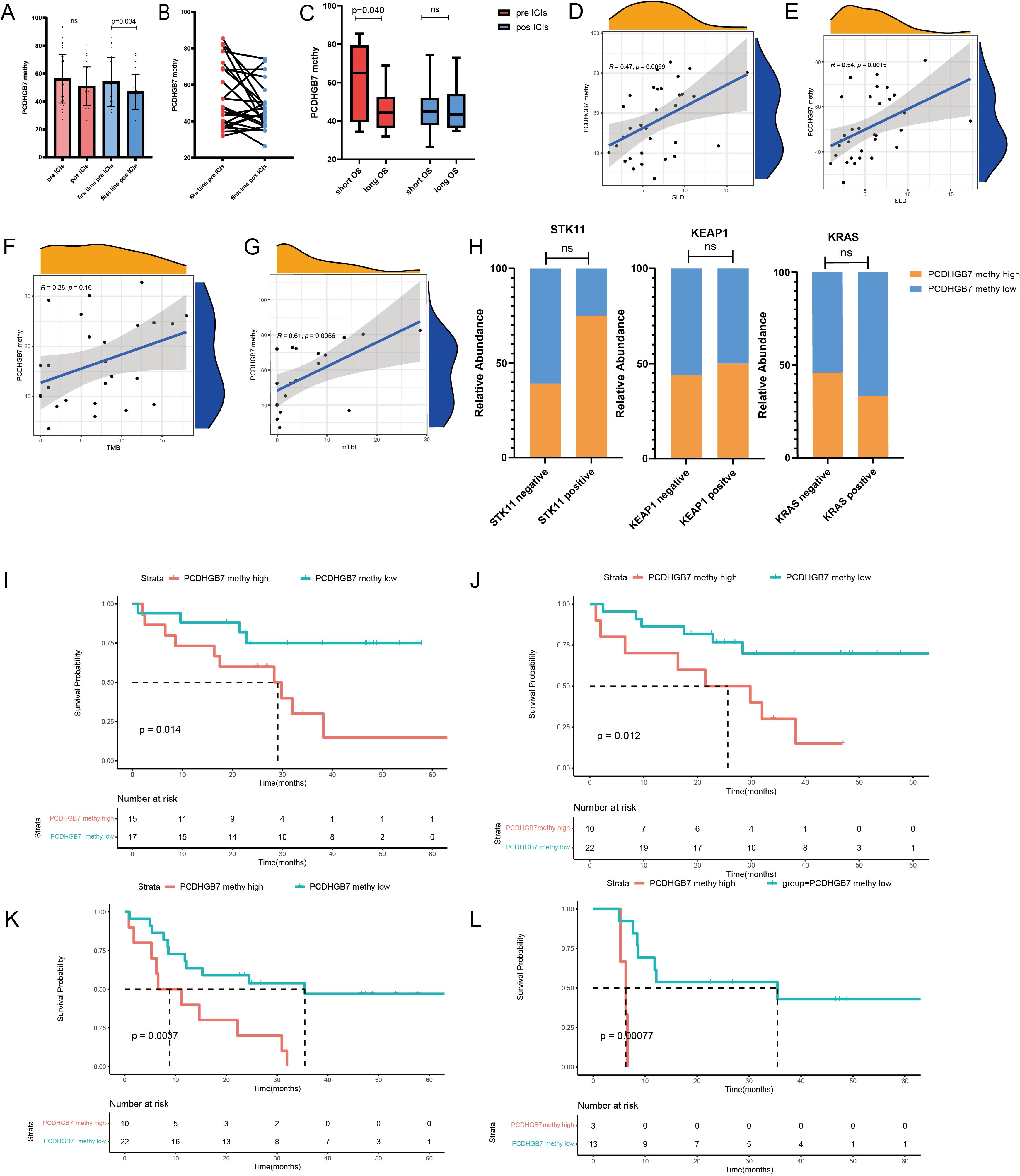
Figure 6. Plasma PCDHGB7 methylation analysis of immunotherapy in lung cancer. (A, B) Baseline and post-treatment PCDHGB7 methylation. (C) PCDHGB7 methylation before and after treatment was grouped according to efficacy. Association between PCDHGB7 methylation with treatment baseline SLD (D) and post-treatment SLD (E). Association between PCDHGB7 methylation and TMB (F) and mTBI (G). (H) Analysis of PCDHGB7 methylation and the STK11 KEAP1 KRAS mutation. Overall survival analysis of baseline PCDHGB7 methylation (I) and post-treatment PCDHGB7 methylation (J). Progression-free survival analysis of post-treatment PCDHGB7 methylation (K). Progression-free survival analysis of post-treatment PCDHGB7 methylation in patients with SD (L). ns, not significant.
The association between plasma PCDHGB7 methylation and gene mutations was equally notable, in that a statistically non-significant positive correlation (r = 0.28, p = 0.16) was identified between the tumor tissue TMB and plasma PCDHGB7 methylation (Figure 6F). In addition, there was a strong positive correlation between PCDHGB7 methylation and the plasma mTBI (r = 0.61, p = 0.0058) (Figure 6G). We also investigated the relationship between plasma PCDHGB7 methylation and immune-predictive marker mutations based on the results of TCGA. The results revealed that patients with mutations of STK11 (68.7 vs. 53.0, p = 0.103; χ2 = 1.776, p = 0.294) and KEAP1 (61.2 vs. 54.9, p = 0.636; χ2 = 0.27, p = 1.000) tended to have higher PCDHGB7 methylation, whereas patients with mutations of KRAS (55.3 vs. 55.3, p = 0.999; χ2 = 0.169, p = 0.869) correlated with lower PCDHGB7 methylation (Figure 6H).
Survival analysis showed that plasma PCDHGB7 methylation before treatment served as a predictive marker of immunotherapy efficacy, with patients with higher baseline plasma hypermethylation having a shorter OS (high vs. low 28.5 vs. 46.9 months, p = 0.014; Figure 6I). Patients with higher PCDHGB7 methylation levels after treatment had a shorter OS (high vs. low (23.7 vs. 52.2 months, p = 0.012, Figure 6J) and PFS (high vs. low (13.2 vs. 39.0 months, p = 0.0037, Figure 6K), particularly those with an initial tumor evaluation of stable disease after treatment (high vs. low (6.0 vs. 37.2 months, p < 0.001) (Figure 6L).
3.8 Dynamic change pattern of plasma PCDHGB7 methylation
Twenty-one patients who received PD-1 inhibitors combined with metronomic oral vinorelbine were enrolled in our study. Peripheral blood testing for PCDHGB7 methylation was conducted, and patients were divided into three groups: treatment-naive, 1–2 stages after treatment (post-treatment time point 1), and 3–4 stages after treatment (post-treatment time point 2). A Sankey diagram was used to show changes in methylation levels and OS in each patient at these three time points. The results indicated that patients with consistently high methylation throughout the study period and after treatment had shorter OS, whereas those with consistently low methylation had longer OS (Figure 7A). We created heatmaps of PCDHGB7 methylation levels at three time points (Figure 7B), and the methylation patterns of patients were divided into three categories based on the clustering results. The line chart shows the methylation changes in three modes: type A (high-down: the baseline methylation value of treatment was relatively high and continuously decreased with immunotherapy), type B (low-stable: both the baseline and post-treatment methylation levels remained low), and type C (up-down: the initial methylation level was low but increased and then decreased during the treatment process) (Figure 7C). PFS and OS analyses of these three metal patterns showed that patients with type A pattern had significantly better prognosis than those with type B and type C patterns, suggesting that dynamic monitoring of plasma methylation can effectively predict the efficacy of immunotherapy (Figures 7D, E) (type A, NA, 95% CI [24.50–NA]; type B, 11.17 months, 95% CI [6.57–NA]; type C, 6.93 months, 95% CI [5.27–NA] [p = 0.0044]; and OS, type A, NA, 95% CI [30.97–NA]; type B, 17.5 months, 95% CI [8.6–NA]; type C, 22.1 months, 95% CI [9.63–NA]; p = 0.014).
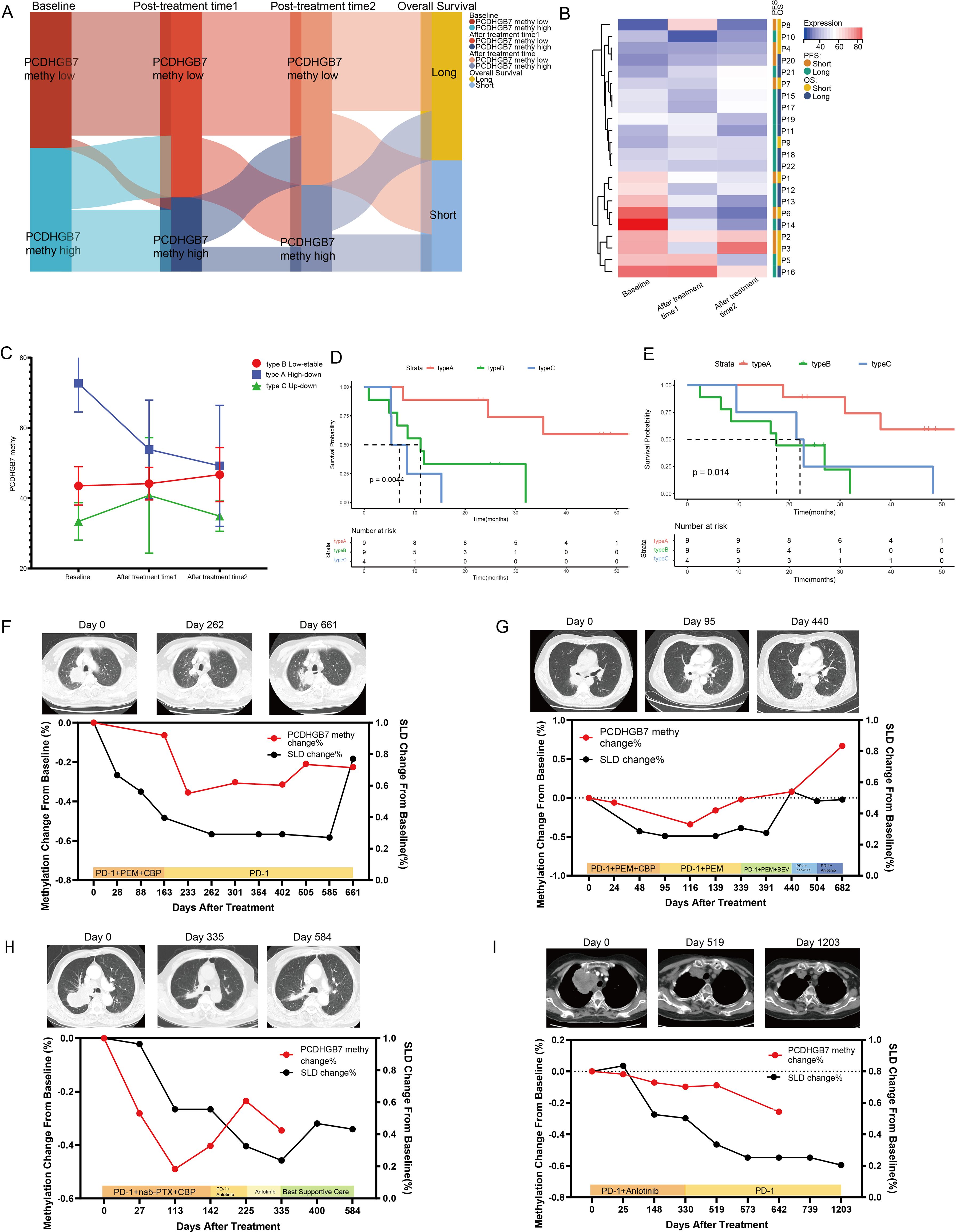
Figure 7. Dynamic changes in plasma PCDHGB7 methylation. (A) Sankey diagram depicting PCDHGB7 methylation at three time points and overall survival outcomes. (B) Heatmaps of PCDHGB7 methylation at three time points. (C) Line chart of the three PCDHGB7 methylation patterns. PFS (D) and OS (E) analyses of three methylation patterns. (F–I) Multipoint dynamic PCDHGB7 methylation and SLD changes.
Finally, we performed dynamic, all-encompassing multi-timepoint surveillance of a subset of patients who continued to receive immunotherapy. Analysis was performed on changes in SLDs on computed tomography (CT) scans. The patients depicted in Figures 7F–H initially received a combination of immunotherapy and chemotherapy, while their front-line treatment subsequently transitioned to a changed chemotherapy regimen or immunotherapy plus anti-angiogenic therapy following disease progression. In these three patients, a tendency toward declining to the lowest point and then increasing was observed for both PCDHGB7 methylation and SLDs. Notably, peripheral blood PCDHGB7 methylation events preceded the radiological recurrence of tumors on CT images by approximately 101–175 days, suggesting a potential epigenetic prelude to clinical relapse. The patient shown in Figure 7I received immunotherapy in addition to anti-angiogenic medication, and prolonged disease control was achieved for approximately 3 years, as evidenced by a steady decrease in plasma PCDHGB7 methylation levels.
3.9 Plasma PCDHGB7 protein level is a prognostic biomarker for lung cancer immunotherapy
We further analyzed plasma PCDHGB7 levels in a prospective clinical cohort of elderly patients with non-small cell lung cancer. Patients were treated with a PD-1 inhibitor combined with metagenomic oral vinorelbine as a posterior line therapy. The baseline clinical characteristics of the ten patients are presented in Supplementary Table S2. There are a total of 10 patients with a majority being male (70%) and a median age of 68 years. The majority of patients have an ECOG performance status of 0 (60%). Almost all patients are in TNM Stage IV (90%), with the pathological type split between non-squamous cell carcinoma (60%) and squamous cell carcinoma (40%). Metastases to the brain are present in 10% of the patients.
A significant increase in the plasma PCDHGB7 protein level was observed after treatment (810.0 vs. 863, p = 0.002, Figures 8A, B). Patients were further divided into two groups based on PFS. Patients in the long PFS group (≥ 5 months) had a higher degree of PCDHGB7 at baseline (708 vs. 1074, p = 0.034, Figure 8C). We also analyzed the relationship between PCDHGB7 protein expression and PFS at baseline and after treatment. The results showed that patients with higher levels of plasma PCDHGB7 protein had a longer PFS at baseline (PCDHGB7 high: 6.13 months, 95% CI [4.03–NA], PCDHGB7 low: 3.48 months, 95% CI [1.87–NA]) and after treatment (PCDHGB7 high: 5.57 months, 95% CI [4.03–NA], PCDHGB7 low: 3.48 months, 95% CI [1.67–NA]) (Figures 8D, E). According to the ROC curve analysis, the plasma protein level was a promising indicator of the efficacy of immunotherapy (Figures 8F, G).
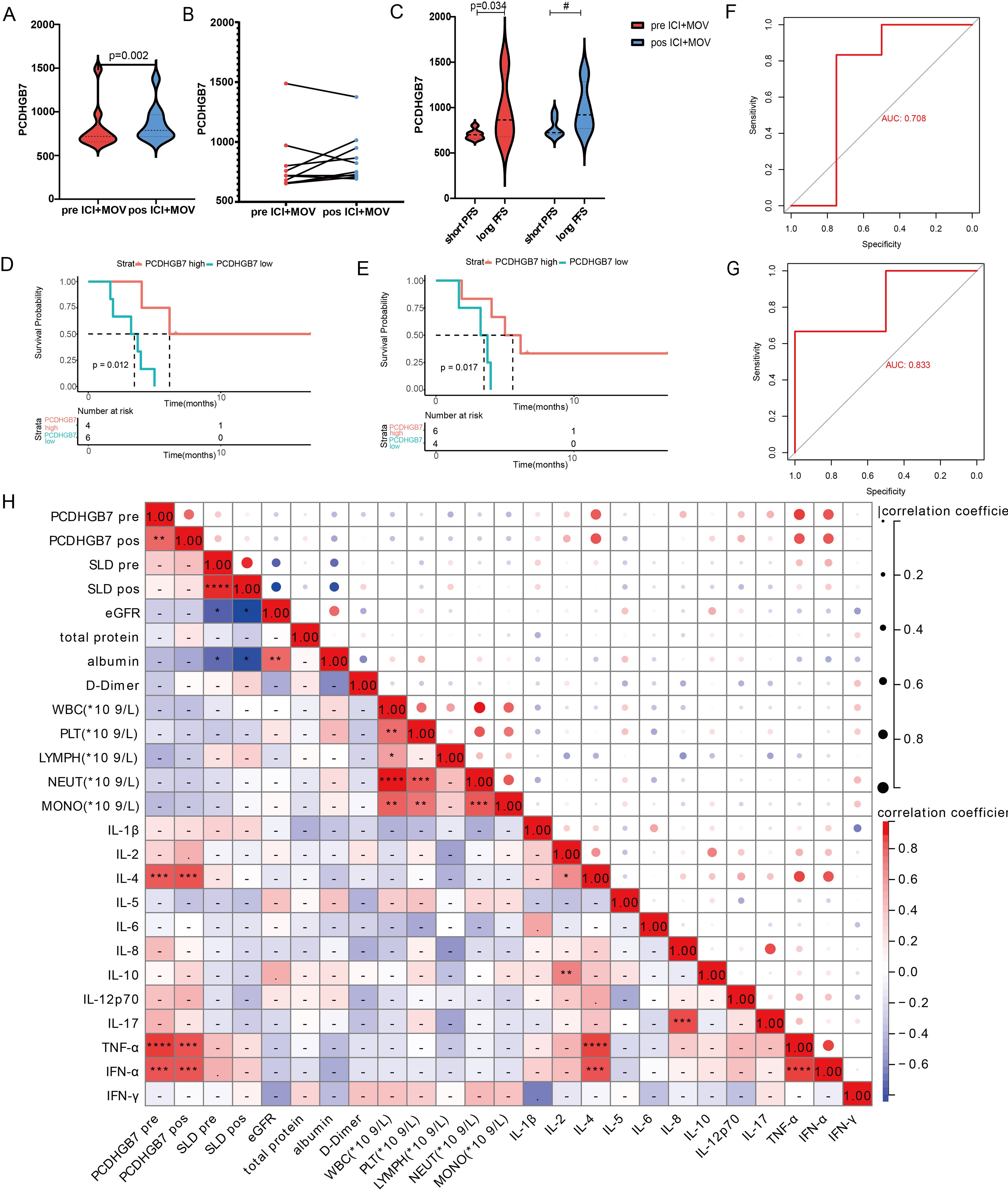
Figure 8. Analysis of baseline and post-treatment plasma PCDHGB7 protein levels. (A, B) Baseline and post-treatment plasma PCDHGB7 protein level. (C) Plasma PCDHGB7 before and after treatment was grouped according to efficacy. PFS analysis of baseline plasma PCDHGB7 (D) and post-treatment plasma PCDHGB7 (E). ROC curve analysis of baseline plasma PCDHGB7 (F) and post-treatment plasma PCDHGB7 (G). (H) Correlation matrix between plasma PCDHGB7 and various clinical indicators (*p< 0.05, **p< 0.01, ***p < 0.001, ****p < 0.0001). ns, not significant.
Further analysis was performed to examine the correlation between plasma PCDHGB7 protein levels and peripheral blood immune cells and cytokines, as well as various clinical parameters. Remarkably, PCDHGB7 protein levels exhibited significant positive associations with interleukin-4 (IL-4, p < 0.0001), tumor necrosis factor-alpha (TNF-α, p < 0.0001), and interferon-alpha (IFN-α, p < 0.001) (Figure 8H).
4 Discussion
PCDHs are a group of transmembrane proteins belonging to the cadherin superfamily and have been proven to play major tumor suppressor functions by inhibiting the proliferation and metastasis of cancer cells (6, 10). Unlike other members of the PCDHs family, the association between PCDHGB7 and cancer has not been fully understood. Few reports have been made to claim PCDHGB7 could be a novel marker for breast cancer, non-Hodgkin’s lymphoma, and hepatocellular carcinoma (27–29). Recently, PCDHGB7 methylation was identified as a diagnosis indicator for cervical cancer, endometrial cancer, and malignant body fluids which attracted more oncologists’ attention since its huge translational potential (14–16, 30). According to our comprehensive analysis, PCDHGB7 expression was downregulated in LUAD and LUSC and associated with tumor prognosis. PCDHGB7 was found to demonstrate a positive correlation with inhibitory immune cells and a negative correlation with TMB and HRD. The data suggested that PCDHGB7 may play an important role in tumor initiation and development by affecting the immune response and DNA damage in tumor microenvironment.
Similar to other investigations, we also found high expression of PCDHGB7 in lung cancer (14), we further deciphered the role of PCDHGB7 in lung cancer that has not yet been fully explored yet. Low gene expression is thought to have been caused by methylation of the DNA promoter region (31). Utilizing the OncoDB and MEXPRESS databases, we examined the methylation status of the PCDHGB7 gene in lung cancer and discovered a negative correlation between the methylation level and PCDHGB7 expression at most gene sites, particularly the promoter region (2 KB upstream of the gene). According to earlier research using public database analysis, over 80% of breast tumors display aberrant methylation of the PCDHGB7 gene. Meanwhile, the detection of PCDHGB7 mRNA expression in breast tissue and breast cancer cell lines e indicated that PCDHGB7 expression was downregulated in breast cancer tissue due to hypermethylation of the promoter region (32). Nevertheless, further immunohistochemical and lung cancer cell line detection data are required to ascertain the connection between PCDHGB7 promoter methylation and expression.
Although multiple data from TCGA LUSC and GEO databases indicate that high expression of PCDHGB7 is associated with poor prognosis, there is a potential contradiction in the prognostic role of PCDHGB7 in lung squamous cell carcinoma and adenocarcinoma, which requires further validation with larger sample sizes in the future. We believe that these potential differences may be due to different biological behaviors and molecular pathways related to LUAD and LUSC. Especially with the development of single-cell technology, studies have identified significant differences in the immune microenvironmental signals between LUAD and LUSC (33). In this study, PCHDGB7 showed different mutation landscapes in lung adenocarcinoma and lung squamous cell carcinoma, and the analysis of immune infiltrating cells was not the same, which may lead to its different prognostic value in the two types of lung cancer.
DNA mismatch repair (MMR) is an important DNA repair pathway that plays a critical role in DNA replication fidelity, mutation avoidance, and genome stability. A hypermutated phenotype in the genome caused by MMR deficiency ultimately results in MSI (34). Specifically, MMR-deficient cancers tend to be more sensitive to immune checkpoint blockade (35). Meanwhile, research continues on the HRD score as a biomarker of response to various therapies in non-small cell lung cancer. In our study, HRD analysis revealed PCDHGB7 expression showed a negative correlation with HRD in LUAD. According to the protein interaction network, PCDHGB7 shares a close relationship with proteins such as FAT1, MLH1, and the FHIT. Patients with LUSC who have low expression of PCDHGB7 are more likely to develop FAT1 mutations, while GSEA analysis revealed that high expression of PCDHGB was linked to the activation of homologous recombination, or mismatch repair, in LUAD (Figure 4F). These comprehensive investigations suggest that PCDHGB7 might play a significant role in lung cancer DNA mismatch repair. Doissy et al. demonstrated an exceptional duration of response of more than 20 months in a patient with LUSC who received platinum-based therapy and had a high HRD score (36). Although the specific mechanism by which PCDHGB7 participates in DNA MMR requires further exploration, the clinical significance of this phenomenon will grow with the extensive investigation into DNA damage repair inhibitors, such as poly-ADP ribose polymerase inhibitors, in patients with NSCLC (37).
Since the revolution of the new treatment paradigm of immunotherapy for lung cancer, the urgent need for novel predictive markers of the efficacy of immunotherapy has dramatically increased (38). The data from the IMvigor210 cohort and Kim cohort showed that patients with high expression of PCDHGB7 in tumor tissue exhibited significantly poor responses to immunotherapy, which may be due to its negative correlation with TMB, as well as its significant correlation with M2 macrophages and Treg cells. TMB is a widely used positive predictive indicator for the efficacy of immunotherapy in clinical practice (39). Numerous studies have shown that M2 macrophages play an important role in mediating tumor invasion and metastasis, immune suppression, and treatment resistance (40, 41). Infiltrating regulatory T cells (Tregs) are also important contributors to immunosuppressive and key targets for cancer immunotherapy (42). Meanwhile, we investigated the relationship between plasma PCDHGB7 methylation and immunotherapy response in patients with NSCLC who received immunotherapy at our center. Patients with baseline plasma hypermethylation had a shorter PFS and OS, while their PCDHGB7 methylation levels decreased after treatment. A specific clinical reason for the predictive diagnostic function of PCDHGB7 methylation can be found in its association with ctDNA, mTBI, and tumor burden. Furthermore, using methylation detection at baseline and two treatment time periods, we attempted to derive the PCDHGB7 methylation alteration pattern. Similar to the monitoring mode for the continuous clearance of ctDNA, the results indicated that patients with early reduction in DNA methylation had the best prognosis (43, 44). Combined with the fact that the methylation results may reveal the disease progression of patients in advance of imaging, PCDHGB7 methylation monitoring shows better stability and health economics effects than ctDNA and exosome detection; moreover, because of its noninvasive multi-point dynamic detection, prospective studies with larger samples can be conducted in the future to verify the above results.
Finally, our data showed that patients with higher plasma PCDHGB7 levels had better outcomes of immune checkpoint inhibitor treatment, and both baseline and post-treatment protein levels had good diagnostic efficacy for PFS. Notably, the PCDHGB7 protein is associated with two common inflammatory stimuli, IL-4 and TNF-α. IL-4 plays a critical role in the humoral immune response, antibody generation, and T and B cell development (45), while recent studies have suggested that TNF-α may participate in the killing effect of immunotherapy against tumors (46). Furthermore, our ensemble pathway enrichment analysis indicated that PCDHGB7 may affect the efficacy of lung cancer immunotherapy by regulating the interaction between cytokines and their paired receptors.
In summary, our research findings indicate that increased methylation of PCHDGB7 in lung adenocarcinoma and lung squamous cell carcinoma tissues leads to decreased expression compared to normal tissues. In lung cancer tissues, high expression of PCDHGB7 is associated with multiple pathways, such as homologous recombination, DNA mismatch repair, and JAK-stat pathway, while significantly increasing the infiltration of immune system negatively regulated cells. Therefore, high expression of PCDHGB7 may lead to disorders in DNA damage repair and anti-tumor immune regulation, resulting in poor responsiveness of lung cancer patients to immunotherapy. We further hypothesize that the methylation status of PCDHGB7 in ctDNA can reflect the overall epigenetic state of the tumor, and this state has a more accurate predictive value for the efficacy of immunotherapy. The dynamic changes in methylation and expression levels of plasma PCDHGB7 can serve as biomarkers for immunotherapy of lung cancer.
While our study provides preliminary insights into the potential of PCDHGB7 as a predictive biomarker for immunotherapy, the limitations underscore the need for further research. Our ability to explore the predictive role of PCDHGB7 in immunotherapy was constrained by the availability of tumor tissue for PCDHGB7 detection. This constraint prevented us from elucidating potential contradictions in the predictive role of PCDHGB7 between tumor tissue and peripheral blood. Secondly, the insufficient sample size of the single clinical center limits the generalizability of the results drawn in this study. In addition to the clinical limitations, there is a pressing need for further cellular and animal studies to explore the biological functions of PCDHGB7 in lung cancer. Addressing these limitations through larger, multicenter studies and deeper biological investigations will be instrumental in advancing our understanding of PCHDGB7’s role in immunotherapy and its potential as a clinical tool for personalized treatment strategies.
5 Conclusion
Our data illustrated that PCDHGB7 expression and methylation are prognostic and immunological biomarkers in non-small cell lung cancer. Plasma PCDHGB7 methylation and protein levels can be used as novel biomarkers for predicting the efficacy of immunotherapy against lung cancer.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Some datasets presented in this article are not readily available due to the ethical considerations and intellectual property regulations that govern the sharing of clinical data and plasma assay data. Requests to access these datasets should be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by committee of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences, and Peking Union Medical College committee of Beijing Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
YY: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. XN: Writing – review & editing. JG: Methodology, Writing – review & editing. YT: Writing – review & editing. LH: Writing – review & editing. XW: Writing – review & editing. PZ: Writing – review & editing. JM: Data curation, Writing – review & editing. LL: Funding acquisition, Project administration, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National High Level Hospital Clinical Research Funding (BJ-2023-073) and Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (CIFMS)(2021-I2M-1-012).
Acknowledgments
We are grateful to the contributors of data to public databases used in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1516628/full#supplementary-material
References
1. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. (2024) 74:12–49. doi: 10.3322/caac.21820
2. Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. (2021) 16:223–49. doi: 10.1146/annurev-pathol-042020-042741
3. Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. (2019) 30:44–56. doi: 10.1093/annonc/mdy495
4. Probing mutant kras, stk11, keap1 in nsclc. Cancer Discovery. (2022) 12:2226. doi: 10.1158/2159-8290.Cd-nb2022-0054
5. Petitprez F, Meylan M, de Reyniès A, Sautès-Fridman C, Fridman WH. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front Immunol. (2020) 11:784. doi: 10.3389/fimmu.2020.00784
6. Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. (2002) 14:557–62. doi: 10.1016/s0955-0674(02)00365-4
7. Junghans D, Haas IG, Kemler R. Mammalian cadherins and protocadherins: about cell death, synapses and processing. Curr Opin Cell Biol. (2005) 17:446–52. doi: 10.1016/j.ceb.2005.08.008
8. Morishita H, Yagi T. Protocadherin family: diversity, structure, and function. Curr Opin Cell Biol. (2007) 19:584–92. doi: 10.1016/j.ceb.2007.09.006
9. Sano K, Tanihara H, Heimark RL, Obata S, Davidson M, St John T, et al. Protocadherins: A large family of cadherin-related molecules in central nervous system. EMBO J. (1993) 12:2249–56. doi: 10.1002/j.1460-2075.1993.tb05878.x
10. Mah KM, Weiner JA. Regulation of wnt signaling by protocadherins. Semin Cell Dev Biol. (2017) 69:158–71. doi: 10.1016/j.semcdb.2017.07.043
11. Shan M, Su Y, Kang W, Gao R, Li X, Zhang G. Aberrant expression and functions of protocadherins in human Malignant tumors. Tumour Biol. (2016) 37:12969–81. doi: 10.1007/s13277-016-5169-9
12. Sinjab A, Han G, Wang L, Kadara H. Field carcinogenesis in cancer evolution: what the cell is going on? Cancer Res. (2020) 80:4888–91. doi: 10.1158/0008-5472.Can-20-1956
13. Ushijima T, Clark SJ, Tan P. Mapping genomic and epigenomic evolution in cancer ecosystems. Science. (2021) 373:1474–9. doi: 10.1126/science.abh1645
14. Dong S, Lu Q, Xu P, Chen L, Duan X, Mao Z, et al. Hypermethylated pcdhgb7 as a universal cancer only marker and its application in early cervical cancer screening. Clin Transl Med. (2021) 11:e457. doi: 10.1002/ctm2.457
15. Mao Z, Dong S, Yan Y, Wang C, Li W, Wang L, et al. Diagnosis of Malignant body fluids via cancer-universal methylation in cell-free DNA. JCI Insight. (2024) 9:e175482. doi: 10.1172/jci.insight.175482
16. Cao D, Yang Z, Dong S, Li Y, Mao Z, Lu Q, et al. Pcdhgb7 hypermethylation-based cervical cancer methylation (Cerme) detection for the triage of high-risk human papillomavirus-positive women: A prospective cohort study. BMC Med. (2024) 22:55. doi: 10.1186/s12916-024-03267-5
17. Liu Z, Liu L, Weng S, Xu H, Xing Z, Ren Y, et al. Best: A web application for comprehensive biomarker exploration on large-scale data in solid tumors. J Big Data. (2023) 10:12. doi: 10.1186/s40537-023-00844-y
18. Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. (2018) 173:338–54.e15. doi: 10.1016/j.cell.2018.03.034
19. Tang G, Cho M, Wang X. Oncodb: an interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. (2022) 50:D1334–d9. doi: 10.1093/nar/gkab970
20. Koch A, De Meyer T, Jeschke J, Van Criekinge W. Mexpress: visualizing expression, DNA methylation and clinical tcga data. BMC Genomics. (2015) 16:636. doi: 10.1186/s12864-015-1847-z
21. Yang F, Tang M, Cui L, Bai J, Yu J, Gao J, et al. Prognostic and predictive impact of molecular tumor burden index in non-small cell lung cancer patients. Thorac Cancer. (2023) 14:3097–107. doi: 10.1111/1759-7714.15098
22. Yu J, Fan Z, Zhou Z, Zhang P, Bai J, Li X, et al. Tp53 and lrp1b co-wild predicts improved survival for patients with lusc receiving anti-pd-L1 immunotherapy. Cancers (Basel). (2022) 14:3382. doi: 10.3390/cancers14143382
23. Pastushenko I, Mauri F, Song Y, de Cock F, Meeusen B, Swedlund B, et al. Fat1 deletion promotes hybrid emt state, tumour stemness and metastasis. Nature. (2021) 589:448–55. doi: 10.1038/s41586-020-03046-1
24. Niu Z, Jiang D, Shen J, Liu W, Tan X, Cao G. Potential role of the fragile histidine triad in cancer evo-dev. Cancers (Basel). (2023) 15:1144. doi: 10.3390/cancers15041144
25. Xiao Y, Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther. (2021) 221:107753. doi: 10.1016/j.pharmthera.2020.107753
26. Torres CM, Biran A, Burney MJ, Patel H, Henser-Brownhill T, Cohen AS, et al. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science. (2016) 353:aaf1644. doi: 10.1126/science.aaf1644
27. Wang X, Jia J, Gu X, Zhao WW, Chen C, Wu W, et al. Screening of breast cancer methylation biomarkers based on the tcga database. Int J Gen Med. (2021) 14:9833–9. doi: 10.2147/ijgm.S322857
28. Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, Al-Kuhlani M, et al. Discovery of novel epigenetic markers in non-Hodgkin’s lymphoma. Carcinogenesis. (2007) 28:60–70. doi: 10.1093/carcin/bgl092
29. Chang YS, Chou YP, Chung CC, Lee YT, Yen JC, Jeng LB, et al. Molecular classification of hepatocellular carcinoma using wnt-hippo signaling pathway-related genes. Cancers (Basel). (2022) 14:4580. doi: 10.3390/cancers14194580
30. Yuan J, Mao Z, Lu Q, Xu P, Wang C, Xu X, et al. Hypermethylated pcdhgb7 as a biomarker for early detection of endometrial cancer in endometrial brush samples and cervical scrapings. Front Mol Biosci. (2021) 8:774215. doi: 10.3389/fmolb.2021.774215
31. Koch A, Joosten SC, Feng Z, de Ruijter TC, Draht MX, Melotte V, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. (2018) 15:459–66. doi: 10.1038/s41571-018-0004-4
32. Hou S, Shan M, Gao C, Feng X, Yang Y, Zhang R, et al. Pcdhgb7 increases chemosensitivity to carboplatin by inhibiting hspa9 via inducing apoptosis in breast cancer. Dis Markers. (2019) 2019:6131548. doi: 10.1155/2019/6131548
33. Wang C, Yu Q, Song T, Wang Z, Song L, Yang Y, et al. The heterogeneous immune landscape between lung adenocarcinoma and squamous carcinoma revealed by single-cell rna sequencing. Signal Transduction Targeted Ther. (2022) 7:289. doi: 10.1038/s41392-022-01130-8
34. Baretti M, Le DT. DNA mismatch repair in cancer. Pharmacol Ther. (2018) 189:45–62. doi: 10.1016/j.pharmthera.2018.04.004
35. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to pd-1 blockade. Science. (2017) 357:409–13. doi: 10.1126/science.aan6733
36. Diossy M, Sztupinszki Z, Borcsok J, Krzystanek M, Tisza V, Spisak S, et al. A subset of lung cancer cases shows robust signs of homologous recombination deficiency associated genomic mutational signatures. NPJ Precis Oncol. (2021) 5:55. doi: 10.1038/s41698-021-00199-8
37. Khaddour K, Felipe Fernandez M, Khabibov M, Garifullin A, Dressler D, Topchu I, et al. The prognostic and therapeutic potential of DNA damage repair pathway alterations and homologous recombination deficiency in lung cancer. Cancers (Basel). (2022) 14:4580. doi: 10.3390/cancers14215305
38. Yang Y, Liu H, Chen Y, Xiao N, Zheng Z, Liu H, et al. Liquid biopsy on the horizon in immunotherapy of non-small cell lung cancer: current status, challenges, and perspectives. Cell Death Dis. (2023) 14:230. doi: 10.1038/s41419-023-05757-5
39. Mino-Kenudson M, Schalper K, Cooper W, Dacic S, Hirsch FR, Jain D, et al. Predictive biomarkers for immunotherapy in lung cancer: perspective from the international association for the study of lung cancer pathology committee. J Thorac Oncol: Off Publ Int Assoc Study Lung Cancer. (2022) 17:1335–54. doi: 10.1016/j.jtho.2022.09.109
40. Wang S, Wang J, Chen Z, Luo J, Guo W, Sun L, et al. Targeting M2-like tumor-associated macrophages is a potential therapeutic approach to overcome antitumor drug resistance. NPJ Precis Oncol. (2024) 8:31. doi: 10.1038/s41698-024-00522-z
41. Toledo B, Zhu Chen L, Paniagua-Sancho M, Marchal JA, Perán M, Giovannetti E. Deciphering the performance of macrophages in tumour microenvironment: A call for precision immunotherapy. J Hematol Oncol. (2024) 17:44. doi: 10.1186/s13045-024-01559-0
42. Kumagai S, Itahashi K, Nishikawa H. Regulatory T cell-mediated immunosuppression orchestrated by cancer: towards an immuno-genomic paradigm for precision medicine. Nat Rev Clin Oncol. (2024) 21:337–53. doi: 10.1038/s41571-024-00870-6
43. Li W, Liu JB, Hou LK, Yu F, Zhang J, Wu W, et al. Liquid biopsy in lung cancer: significance in diagnostics, prediction, and treatment monitoring. Mol Cancer. (2022) 21:25. doi: 10.1186/s12943-022-01505-z
44. Sinoquet L, Jacot W, Quantin X, Alix-Panabières C. Liquid biopsy and immuno-oncology for advanced nonsmall cell lung cancer. Clin Chem. (2023) 69:23–40. doi: 10.1093/clinchem/hvac166
45. McCormick SM, Heller NM. Commentary: il-4 and il-13 receptors and signaling. Cytokine. (2015) 75:38–50. doi: 10.1016/j.cyto.2015.05.023
Keywords: PCDHGB7, methylation, lung cancer, immunotherapy, biomarkers
Citation: Yuan Y, Nie X, Gao J, Tian Y, He L, Wang X, Zhang P, Ma J and Li L (2025) Deregulated methylation and expression of PCDHGB7 in patients with non-small cell lung cancer: a novel prognostic and immunological biomarker. Front. Immunol. 16:1516628. doi: 10.3389/fimmu.2025.1516628
Received: 24 October 2024; Accepted: 13 January 2025;
Published: 30 January 2025.
Edited by:
Guan-Jun Yang, Ningbo University, ChinaCopyright © 2025 Yuan, Nie, Gao, Tian, He, Wang, Zhang, Ma and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Li, bGlsaW5fNTFAaG90bWFpbC5jb20=
 Yue Yuan
Yue Yuan Xin Nie
Xin Nie Jiayi Gao1,2
Jiayi Gao1,2 Liuer He
Liuer He Lin Li
Lin Li