- Department of Gynecology, Beijing Obstetrics and Gynecology Hospital, Capital Medical University, Beijing Maternal and Child Health Care Hospital, Beijing, China
Background: Early diagnosis and treatment of endometriosis (EM) remain challenging because of the lack of knowledge about EM development. While oxidative stress (OS) has been associated with EM, the link is unclear. We explored OS-related genes (OSRGs) and their role in EM pathogenesis.
Material and methods: We combined two ectopic endometrium (EC) and eutopic endometrium (EU) datasets (GSE11691 and GSE25628) into a dataset for analysis. Bioinformatic analyses were used to identify differentially expressed genes (DEGs), OS-related genes (OSRGs), enriched pathways, competitive endogenous RNA network, and immune cell infiltration. Finally, real time-quantitative polymerase chain reaction (RT-qPCR) and Western blot (WB) were used to validate the expression of key OSRGs in clinical patient samples.
Results: Bioinformatic analysis identified 459 DEGs between EC and EU samples, including 67 OSRGs. A ceRNA network was established, encompassing 28 DE-OSRGs, 32 miRNAs, and 53 lncRNAs. Four key OSRGs (CYP17A1, NR3C1, ENO2, and NGF) were selected from protein-protein interaction network analysis. The RT-qPCR and WB analysis showed that these genes’ abnormal changes in RNA and protein levels were consistent with data in public databases. Weighted gene co-expression network analysis identified three immune-related OSRGs (CYP17A1, NR3C1, and NGF) and 20 lncRNAs that may regulate NR3C1 through 10 miRNAs.
Conclusion: The key OSRGs may function via multilayered networks in EM. We provide insights into EM and underscore the potential significance of OSRGs and the immune environment for diagnostic and prognosis evaluation.
1 Introduction
Endometriosis (EM) is a chronic disease characterized by the development of lesions, starting as speckled spots on the ovary and progressing into deep infiltrating cysts filled with brown fluid. These cysts, along with endometrioid tissue, are located outside the uterine cavity (1, 2). Ectopic lesions predominantly consist of interstitial cells and glands sensitive to hormonal fluctuations; these lesions are commonly found in the pelvic region, particularly affecting the ovaries, pelvic peritoneum, uterosacral ligament, and fallopian tubes (3). Individuals with EM commonly experience a range of symptoms, including pelvic pain, irregular menstrual bleeding, dysmenorrhea, difficulty in sexual intercourse, abdominal and pelvic pain during non-menstrual periods, dysuria, and gastrointestinal disturbances such as constipation and diarrhea. Approximately 70% of EM patients experience chronic pelvic pain, and 30%–50% encounter infertility (4). The increased surgical intervention decreased quality of life, and high prevalence of assisted reproductive technology caused by EM have led to a significant increase in social costs (5). While medical treatment can alleviate symptoms in up to 50–80% of cases, approximately 20% of patients still have symptoms (6–8). More effective treatments are lacking, mainly because the mechanism underlying the pathogenesis of EM remains unclear. Consequently, identifying novel biomarkers is imperative to deepen understanding of etiology and molecular underpinnings of EM and to identify novel targets for its clinical diagnosis and therapeutic intervention.
Ectopic endometrium (EC) undergoes three stages of “adhesion, invasion, and angiogenesis” to ultimately lead to the occurrence of EM (9). The related cytokines, extracellular matrix degradation, and angiogenesis processes are increased by oxidative stress (OS), leading to the occurrence of EM (10). OS is defined as a stress state in which the balance between oxidation and antioxidant systems is disrupted. A large number of oxygen-free radicals accumulate that exceeds the body’s ability to clear oxygen free radicals, leading to inflammatory infiltration of neutrophils and the production of pain-inducing factors such as macrophages, enhanced pain-related protease activity, promoted lipid peroxidation, and pelvic pain (11). Excessive oxygen free radicals can extensively oxidize and damage DNA, lipids, or proteins in cells, directly disrupting the cellular structure and physiological functions. They also serve as secondary messengers, indirectly leading to the occurrence and development of various diseases, such as EM, by activating related factors and signaling pathways (12, 13). Excessive reactive oxygen species (ROS) activation of the NF-κB signaling pathway upregulates expression of adhesion factor-1 (ICAM-1), alters the morphology of peritoneal epithelial cells, provides adhesion sites for ectopic endometrium, and upregulates interleukin-8 (IL-8) and tumor necrosis factor-β (TGF-β) inflammatory factors (9, 14, 15). Inflammatory factors and highly expressed superoxide dismutase (SOD2), which counteracts excessive oxygen-free radical stress, can mediate the Ras/Raf/MEK/ERK signaling pathway to stimulate extracellular regulatory protein kinase (ERK1/2) phosphorylation and upregulate levels of MMP-2 and MMP-9, accelerating extracellular matrix degradation and enhancing the proliferation, invasion, and migration of EM ectopic endometrial cells (16, 17). ROS also activates the MAPK/ERK signaling pathway to regulate c-Fos and c-Jun factors, inhibit cell apoptosis, and promote ectopic endometrial cell proliferation in EM (18).
While the role of OS in the pathogenesis of EM has been investigated, the molecular mechanism underlying EM pathogenesis is still limited and requires further research. Here, we performed a comparative transcriptome analysis of ectopic endometrium (EC) and eutopic endometrium (EU) samples to identify OS-related genes (OSRGs) and immune cells that are critical in EM development. Our studies results provide new insights into the pathogenesis of EM and underscore the potential significance of OSRGs and the immune environment for diagnostic values and prognosis evaluation in EM.
2 Materials and methods
2.1 Integrated bioinformatics analysis
2.1.1 Data sources
We downloaded four EM-related transcriptome datasets (GSE11691, GSE25628, GSE105764 and GSE105765) from GEO database (19–22). The nine EU samples and nine EC samples in the GSE11691 dataset and the seven EC samples and nine EU samples in the GSE25628 dataset were used as the combined dataset for the main analysis (20). The “ComBat” algorithm was applied to reduce batch effects caused by non-biotechnological biases in the expression profile of composite datasets. Principal component analysis (PCA) was performed on data before and after batch calibration. The GSE105764 and GSE105765 datasets were used to construct the competitive endogenous RNA (ceRNA) network, with each dataset containing eight EC samples and eight EU samples (22). The 1,150 OSRGs were derived from the GeneCards database (https://www.genecards.org/) with the keyword “oxidative stress related gene” by setting the criterion as “relevance score ≥ 7” (23).
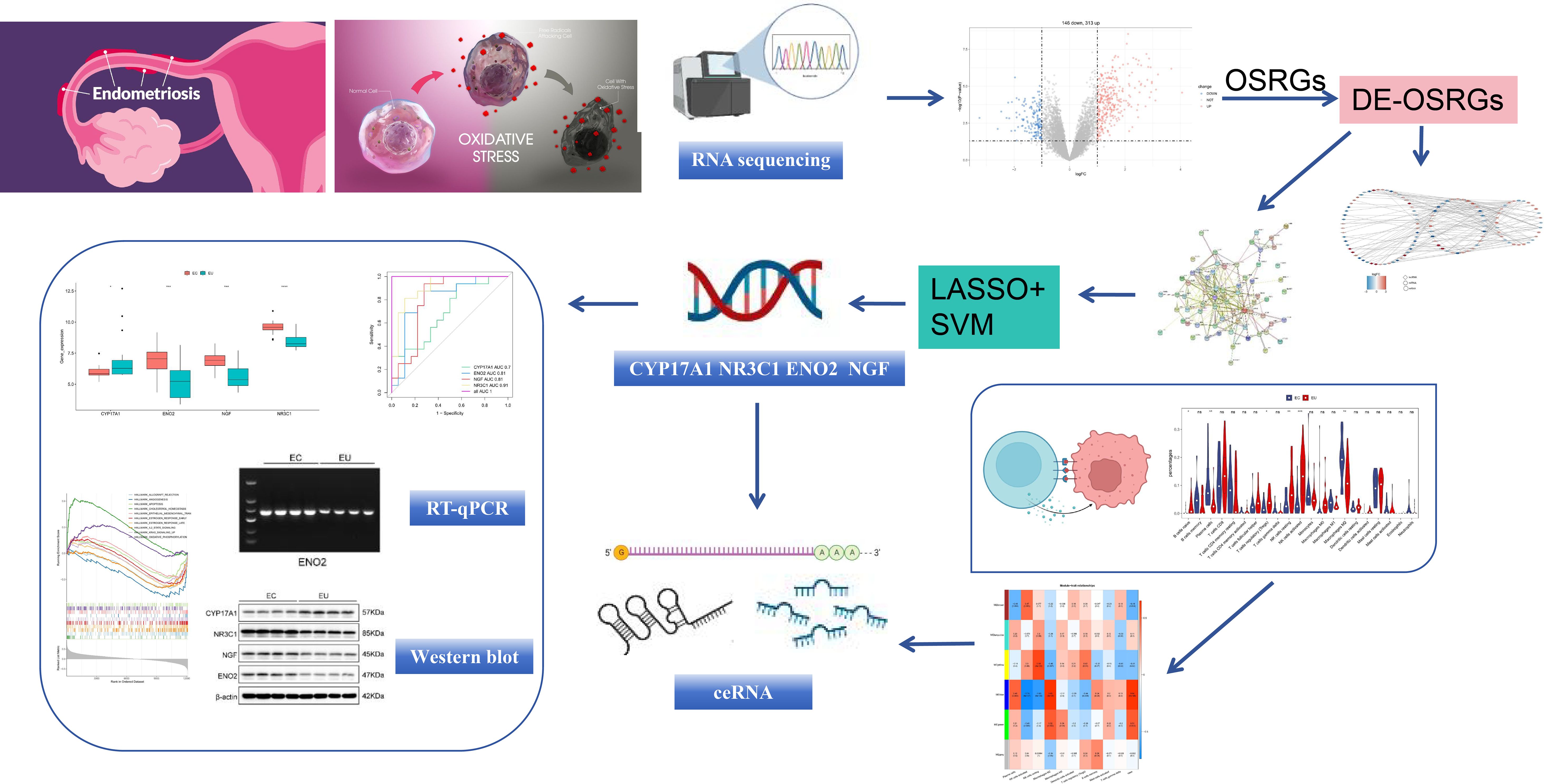
The overall study design is shown in Figure 1.
2.1.2 Identification of differentially expressed OSRGs
Differentially expressed genes (DEGs) from EC and EU samples in the combined dataset were determined using ‘limma’ package (v 3.46.0) (24, 25). P < 0.05 and |Log2FC|>1 were applied as the cut-off criteria. DE-OSRGs were acquired by overlapping DEGs and OSRGs by the ‘ggplot2’ package (v 3.3.2) (26).
2.1.3 Construction of the ceRNA regulation network
To investigate the molecular regulatory mechanisms of EM, differentially expressed miRNAs (DE-miRNAs) in the GSE105765 dataset were screened out via DESeq2 package (v 1.30.1) with P < 0.05 and |Log2FC|>1 (27). We also used the DESeq2 package to screen out differentially expressed lncRNAs (DE-lncRNAs) in the GSE105764 dataset with P < 0.05 and |Log2FC|>1. The starbase database (https://rnasysu.com/encori/) was used to predict the miRNAs that interact with DE-OSRGs. We determined the intersection of the miRNA predictions across DE-miRNAs and the starbase database. The lncRNAs corresponding to miRNAs were predicted in the starBase database. We then identified DE-lncRNAs that overlapped with the predicted results from starBase database to obtain the final lncRNAs.
2.1.4 Functional enrichment analysis of DE-OSRGs and PPI network
To explore potential biological functions and signaling pathways associated with DE-OSRGs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DE-OSRGs were completed via ‘clusterProfiler’ package (v 4.0.2). P <0.05 and count ≥ 1 were set as the threshold value (28). To explore the interactions between proteins coded by the detected DE-OSRGs, we used the search tool for the retrieval of interacting genes (STRING) database (https://string-db.org/), along with Cytoscape software (3.8.2) (29). This approach allowed us to examine the relationships among DE-OSRGs and assemble a protein-protein interaction (PPI) network. We selected DE-OSRGs with connectivity >5 for subsequent analysis, and the top 20 DE-OSRGs were shown.
2.1.5 Development of machine learning diagnostic models
For further exploration of key OSRGs, based on DE-OSRGs with connectivity >5 in PPI network, we performed least absolute selection and shrinkage operator (LASSO) Cox analysis via ‘glmnet’ package (v 4.1-4) and support vector machine recursive feature elimination (SVM-RFE) algorithm via ‘care’ package (v 6.0-92). The key OSRGs were obtained from the overlap of the two machine learning results (30, 31). To assess the diagnostic performance of key OSRGs, receiver operating characteristic (ROC) curves were plotted to calculate the area under the curve (AUC) for the combined dataset using the ‘pROC’ package (v 1.17.0.1) (32). The expression levels of key OSRGs were shown in the combined dataset.
2.1.6 Gene set enrichment analysis
To explore the biological roles and pathways linked to key OSRGs, GSEA of each key OSRG was executed by the ClusterProfiler package based on the correlation between key OSRGs and other genes. The threshold for enrichment significance was adj. P < 0.05, and the reference gene set was the KEGG gene set (33).
2.1.7 Immune infiltration analysis
To evaluate the immune cell composition within the immune landscape of EM patients, the CIBERSORT algorithm was applied to estimate the percentages of 22 immune cell types in the combined dataset. Violin plots were created to display the variations in immune cell infiltration between EC and EU samples (34, 35).
2.1.8 Weighted gene co-expression network analysis
To further identify immune-related module genes, WGCNA (version 1.70.3) was used (36). A cluster analysis was conducted to determine whether the exclusion of outlier samples was necessary to ensure accuracy of subsequent analyses. A soft threshold (β) was selected to construct the co-expression network. Genes were compared for similarity according to their proximity, and a phylogenetic tree of the genes was created. The dynamic tree-cutting algorithm was used to partition the modules, with a minimum requirement of 300 genes per module. Gene modules with a stronger correlation with differential immune cells were used as the immune-related module for further analysis (37).
2.1.9 Analysis of the ceRNA regulation network of key immune-related OSRGs
Key immune-related OSRGs were identified by intersecting the obtained immune-related module genes with key OSRGs. A ceRNA network was then constructed via key immune-related OSRGs.
2.2 Clinical samples
This study included 30 patients diagnosed with EM from Beijing Obstetrics and Gynecology Hospital that met the following inclusion criteria: body mass index of 19–25 kg/m2, no operative contraindication, non-vegetarian patients, and age between 20 and 45 years. The exclusion criteria were diabetes and other endocrine diseases and cardiopulmonary, liver, and serious gastrointestinal diseases. EC and EU tissues were collected from the same patient. All tissue samples were taken from tissues discarded during surgery. Samples were obtained after approval from our hospital’s ethics review committee. The samples were then snap-frozen in liquid nitrogen and stored for further analysis.
2.3 cDNA synthesis and reverse transcription quantitative polymerase chain reaction
RNA isolation and reverse transcription quantitative polymerase chain reaction (RT-qPCR) were performed as described previously (38). PCR reactions were conducted using 0.4 µM of forward and reverse primers, 1 µL of Light Cycler DNA Master SYBR Green I (10× concentrate, Roche), and 3 mM of MgCl2. To ensure full denaturation of cDNA prior to amplification, an initial step of denaturation was carried out at 95°C for 1 min and 15 sec to activate FastStart DNA polymerase. The reaction continued with desaturation at 95°C for 15 sec, annealing at 62°C for 10 sec, and extension for 40 cycles at 72°C for 25 seconds to amplify target genes and the endogenous control gene. We analyzed the amplification product and performed a melting curve analysis to check the integrity of the amplification after the last cycle. Data analysis was performed with LightCycler Relative Quantification Software, employing the second derivative max method for calculation. The primers used for RT-PCR analysis are as follows: CYP17A1, sense 5′-TTCAGCCGCACACCAACTAT-3′ and anti-sense 5′-GGACAG GGGCTGTG AGTTAC-3′; ENO2, sense 5′-TCGCTTTGCCGGACATAACT-3′ and anti-sense 5′-GACACAT CGTTCCCCCAAGT-3′; NR3C1, sense 5′-AGGAATAGAAACAGAAAGAGGTTGA -3′ and anti-sense 5′-ACTGGGGCTTGACAAAACCA-3′; and NGF, sense 5′-GCGCAGCGAGTT TTGGC-3′ and anti-sense 5′-GGATGGGATGATGACCGCTT-3′. The relative quantitation values were calculated using the ΔΔCt method. By subtracting the Ct value of the reference genes GAPDH/actin from the Ct value of the gene under investigation, the ΔCt value was derived. This was done to transform the output of PCR into risk scores. The ΔCt values of all samples were then used to calculate the average ΔCt value. Next, the difference between the ΔCt value and its average value (ΔΔCt) was obtained. Finally, -ΔΔCt values were used to calculate the relative expression level of the target gene through two exponentiations. The relative expression level is an alternative to the risk-scoring formula. After qRT-PCR, we verified the mRNA specificity of the key OSRGs with agarose gel electrophoresis. All experiments were performed at least three times.
2.4 Western blot analysis
Protein lysate preparation and western blotting analysis were performed as previously described (38). Protein extraction was performed using the ProteoExtract Native Membrane Protein Extraction Kit (M-PEK; Calbiochem, Darmstadt, Germany). The Quick Start Bradford protein assay (Thermo Fisher Scientific, Waltham, MA, USA) was used to determine protein concentration. We conducted SDS-PAGE with approximately 20 µg of protein sample loaded onto 7% triethyl acetate gels at 120 V for 2 h. Samples were not heated before electrophoresis to maintain the integrity of the target protein. Proteins were transferred to an Immobilization NC transfer membrane under 300 mA for 90 min. The membrane was blocked in 5% skim milk powder containing 0.1% Tween 20 (TBST) Tris buffer saline for 2 h and incubated overnight at 4°C with the rabbit monoclonal anti-Cytochrome P450 17A1/CYP17A1 antibody (ab134910; Abcam, Cambridge, UK), rabbit monoclonal anti-ENO2 antibody (ab133309; Abcam, Cambridge, UK), rabbit monoclonal anti-NGF antibody (ab52987; Abcam, Cambridge, UK), and rabbit monoclonal anti-NR3C1 antibody (ab3671; Abcam, Cambridge, UK) (1:1000 in TBST). The membrane was washed three times with TBST for 10 min each. Next, the membrane was incubated with the secondary antibody (1:5000 in TBST) conjugated with mouse anti-rabbit IgG horseradish peroxidase (HRP) at room temperature (RT) for 45 min. After washing the membrane three times for 10 minutes in TBST, the signal was recorded by digital imaging using ChemiDocTM XRS+and Image Lab™ Software (BIO-RAD, Hercules, CA, USA). β-actin was used as an internal control. We used ImageJ for density measurement and quantitative analysis (value=absorbance of the target band/β-actin absorbance). All experiments were performed at least three times.
2.5 Statistical analysis
All analyses were executed in R language (v 4.2.3). Differences between groups were analyzed by the Wilcoxon test. P < 0.05 was considered statistically significant.
3 Results
3.1 Identification of differentially expressed OSRGs from the combined dataset of EC and EU samples
To identify the differentially expressed OSRGs involved in EM, we first integrated two sample datasets (GSE11691 and GSE25628). The Datasets GSE11691 and GSE25628 underwent background adjustment and quantile normalization prior to their merger. Subsequently, we eliminated batch effects, and gene expression profiles were generated from 34 samples, encompassing 18 EU and 16 EC across 12,403 genes. Visual assessments using box plots and PCA confirmed the effective mitigation of batch effects (Figures 2A–D).

Figure 2. Visualization of the data processing for GSE11691 and GSE25628, highlighting the effects of background adjustment and quantile normalization. (A) The principal component analysis (PCA) results before correction. (B) The box plots before normalization. (C) The PCA results after correction. (D) The box plots after normalization.
Subsequent data analysis identified 459 DEGs, including 313 up-regulated and 146 down-regulated DEGs, from the combined EC and EU sample dataset. The volcano plot and heatmap showed the distribution of DEGs. (Figures 3A, B). By overlapping DEGs with OSRGs, we identified 67 differentially expressed OSRGs (DE-OSRGs) (Figure 3C).

Figure 3. Identification of differentially expressed genes (DEGs) between ectopic endometrium (EC) and eutopic endometrium (EU) samples. (A) Volcano plot representing the distribution of DEGs. (B) Heat map detailing the expression patterns of the DEGs. (C) Venn diagram showing the intersection of 67 DE-OSRGs between DEGs and OSRGs.
3.2 Construction of a ceRNA network of DE-OSRGs
We identified 286 DE-miRNAs (147 up-regulated and 139 down-regulated) from the GSE105765 dataset (Figure 4A). In addition, we identified 184 DE-lncRNA (98 up-regulated and 86 down-regulated) from the GSE105764 dataset (Figure 4B). A ceRNA network was constructed by overlapping DE-miRNA and DE-lncRNAs with the predicted results from the starBase database. Specifically, the ceRNA network analysis revealed that 28 DE-OSRGs could interact with 32 miRNAs, which could, in turn, interact with 53 lncRNAs (Figure 4C).

Figure 4. Construction of the competitive endogenous RNA (ceRNA) regulation network. (A) Volcano plot representing the distribution of differentially expressed miRNAs (DE-miRNAs). (B) Volcano plot representing the distribution of differentially expressed lncRNAs (DE-lncRNA). (C) ceRNA network encompassing mRNAs, miRNAs, and lncRNAs.
3.3 Function enrichment analysis and PPI network of DE-OSRGs
Following the previous analysis, DE-OSRGs were incorporated into the GO and KEGG analyses. DEGs were primarily associated with “cellular divalent inorganic cation homeostasis,” “leukocyte cell-cell adhesion,” “leukocyte migration,” “skeletal muscle organ development,” and “skeletal muscle tissue development” in GO entries (Figures 5A, B) and were mainly enriched in “African trypanosomiasis,” “apelin signaling pathway,” “cell adhesion molecules,” “fluid shear stress and atherosclerosis,” “malaria” and other KEGG pathways (Figures 5C, D). These analyses revealed that DE-OSRGs were associated with multiple biological functions and implicated disease-related pathways. To characterize the interaction of DE-OSRGs at the protein level, we established a PPI network containing 63 nodes and 220 edges; for instance, CYP17A1 was associated with FOXL2, CYP17A1, AKR1C2, and NR3C1 (Figure 5E). Figure 5F shows the top 20 genes with the highest connectivity.

Figure 5. Function enrichment analysis and protein-protein interaction (PPI) network. (A, B) Gene Ontology (GO) of DE-OSRGs. (C, D) Kyoto Encyclopedia of Genes and Genomes (KEGG) of DE-OSRGs. (E) PPI network of DE-OSRGs. (F) The top 20 genes with the highest connectivity.
3.4 Identification of key OSRGs
By least absolute selection and shrinkage operator (LASSO) algorithm, we identified seven genes (ACTA2, CYP17A1, ENO2, NGF, NR3C1, SELP, and SPP1) among the 31 DE-OSRGs with a connectivity score greater than 5 (Lambdamin = 0.066) (Figures 6A, B). Using the support vector machine recursive feature elimination (SVM-RFE) algorithm, five genes were screened out: CYP17A1, NR3C1, ENO2, NGF, and CCL2 (Figures 6, D). Finally, four key OSRGs, including CYP17A1, NR3C1, ENO2, and NGF were identified (Figure 6E). ROC curves indicated that the AUC of key OSRGs was greater than 0.7 in the combined dataset, demonstrating decent diagnostic performance of these key OSRGs for EM (Figure 6F). We then compared the expression levels of four key OSRGs in the combined dataset. NR3C1, ENO2, and NGF showed upregulated expression, while CYP17A1 showed down-regulated expression in EC compared with EU samples (P < 0.05) (Figure 6G).

Figure 6. The development of the machine learning diagnostic models and key OSRGs. (A, B) Among the 31 DE-OSRGs, ACTA2, CYP17A1, ENO2, NGF, NR3C1, SELP, and SPP1 were identified with a connectivity score greater than five by least absolute selection and shrinkage operator (LASSO) algorithm. (C, D) CYP17A1, NR3C1, ENO2, NGF, and CCL2 were pinpointed via the Support vector machine recursive feature elimination (SVM-RFE) algorithm. (E) CYP17A1, NR3C1, ENO2, and NGF were obtained by overlapping both machine learning results as key OSRGs. (F) Receiver operating characteristic (ROC) curve of the four key OSRGs. (G) Expression levels of four key OSRGs were compared in the combined dataset. *P < 0.05, ***P < 0.001, ****P < 0.0001.
3.5 Comparison and validation of four OSRGs at RNA and protein levels
To validate and compare the RNA levels between EC (n=15) and EU samples (n=15), PCR (Figure 7A) and RT-qPCR analysis (Figure 7B) were performed. The RNA expression of NR3C1, NGF, and ENO2 in the EC samples was upregulated compared with EU samples in the PCR results (Figure 7A). RT-qPCR analysis demonstrated that the RNA expression of NR3C1, NGF, and ENO2 in the EC samples was significantly higher than in the EU samples. Moreover, the RNA expression of CYP17A1 was downregulated in EC samples compared with EU samples in both PCR and RT-qPCR results.

Figure 7. The mRNA and protein levels of key OSRGs. (A)The agarose gel electrophoresis of PCR amplification products of four key OSRGs in the indicated samples. GAPDH was used as an endogenous control. (B) RT-qPCR of four key OSRGs. (C) Western blot of four key OSRGs. (D) Quantitative analysis of OSRG proteins from panel (C). β-actin was used as a loading control for western blot analysis. Data are presented as mean ± SD (n = 4). **P < 0.01.
To validate and compare the protein levels between EC and EU samples, we performed western bot analysis of the same EC (n=15) and UC samples (n=15) used for RNA analysis. As shown in Figures 7C, D, NR3C1, NGF, and ENO2 protein levels were significantly upregulated, while CYP17A1 protein levels were significantly downregulated in EC samples compared with EU samples, indicating that the abnormal changes of NR3C1, NGF, ENO2, and CYP17A1 proteins in EC samples may occur through transcriptional regulation. Notably, our results were consistent with the microarray data (GSE11691 and GSE25628).
3.6 Identification of key OSRG-related signaling pathways
To further determine the OSRG-related functions, GSEA was performed to characterize the potential pathways impacted by the key OSRGs. CYP17A1, NR3C1, ENO2, and NGF were co-enriched in epithelial-mesenchymal transition; CYP17A1, NR3C1, andENO2 were co-enriched in KRAS signaling; NR3C1, ENO2, and NGF were co-enriched in E2F targets, G2M checkpoint, MYC targets v1, and myogenesis pathways. These analyses revealed the importance of these OSRG-related pathways in the pathogenesis of EM (Figures 8A–D).

Figure 8. Gene set enrichment analysis (GSEA) results of key OSRG-related signaling pathways. (A) The CYP17A1-related signaling pathways. (B) The NR3C1-related signaling pathways. (C) The ENO2-related signaling pathways. (D) The NGF-related signaling pathways.
3.7 Difference in the composition of immune cells and immune-related OSRGs between EC and EU samples
Based on the combined dataset, we performed an immune infiltration analysis between EC and EU samples to assess the composition of immune cells in the immune microenvironment of EM patients. Figure 9A illustrates the proportion of 22 types of immune cells in each sample within the combined dataset. The proportion of six types of immune cells exhibited significant differences between EC and EU samples (P < 0.05) (Figure 9B). The proportion of plasma cells and M2 macrophages was significantly higher in EC samples (P < 0.05). Conversely, naïve B cells, regulatory T cells (Tregs), resting natural killer (NK) cells, and activated NK cells were significantly lower in EC samples compared with EU samples (P < 0.05). These results indicated that these immune cells might influence the development of EM.

Figure 9. Immune infiltration analysis from EC and EU samples. (A) The distribution of 22 distinct immune cell types across each specimen. (B) The proportion of six types of immune cells exhibited significant differences between EC and EU samples. *P < 0.05, **P < 0.01, ***P < 0.001, ns: P > 0.05.
To determine if OSRGs were involved in the immune microenvironment of EM pathogenesis, WGCNA was first constructed to identify immune-related module genes. Cluster analysis indicated the absence of outlier samples (Figure 10A). The optimal soft threshold (β) was finally chosen as 11 (R2 close to 0.85 and mean connectivity close to 0) (Figure 10B). By constructing a co-expression network and setting the minimum number of genes per gene modules to 300, six modules were obtained (Figures 10C, D). The MEblue module (case correlation = 0.64 and P < 0.0001) was taken as a key module with 2,284 genes according to scoring correlation (Figures 10E, F). After overlapping key OSRGs with immune-related module genes, three key immune-related OSRGs, including CYP17A1, NR3C1, and NGF, were identified (Figure 10G). Moreover, the ceRNA network showed that 20 lncRNAs regulated NR3C1 through 10 miRNAs, with a total 40 predicted interactions, such as NR3C1-hsa-miR-30a-5p-GATA3AS1, NR3C1-hsa-miR-196b-5p-LOXL1-AS1, and NR3C1-hsa-miR-182-5p-LINC01018 (Figure 10H).

Figure 10. Identification of immune-related module genes via weighted gene co-expression network analysis (WGCNA). (A) The results of cluster analysis indicated the absence of outlier samples. (B) The optimal soft threshold (β) was finally chosen as 11. (C, D) By constructing a co-expression network and setting the minimum number of genes per gene modules to 300, six modules were obtained. (E, F) According to the scoring correlation, the MEblue module was taken as a key module with 2,284 genes. (G) CYP17A1, NR3C1, and NGF were acquired by overlapping key OSRGs with immune-related module genes. (H) The ceRNA regulation network of NR3C1.
4 Discussion
EM is a progressive disease associated with severe pain, abdominal bloating, nausea, bowel movements, and infertility. Early diagnosis and treatment of EM are very important, but the diagnosis of EM is often delayed (39–42), which affects the efficient treatment of EM. The occurrence and development of EM have been shown to be associated with OS and immune filtration (43). However, detailed information on the link between OS and EM is limited. Here, we performed a comparative transcriptome analysis between EC and EU samples with extensive bioinformatic analysis and validation of key DE-OSRGs in laboratory study settings. We demonstrated for the first time that several key OSRGs, including NR3C1, NGF, ENO2, and CYP17A1, are critical in EM pathogenesis. These featured OSRGs may function via multilayered intracellular communications and networks that contribute to EM. Notably, the ROC curve revealed that these four OSRGs displayed commendable diagnostic performance.
CYP17A1, which is located on chromosome 10q24.3 with 8 exons and 7 introns, encodes a membrane-bound bifunctional monooxygenase that belongs to the CYP450 enzyme superfamily and plays a key role in the androgen and estrogen metabolism pathway (44, 45). CYP17A1 is a key enzyme in steroid hormone synthesis that synthesizes hormones such as progesterone, mineralocorticoids, glucocorticoids, androgens, and estrogen; these hormones can affect the activity and function of immune cells, thereby affecting the inflammatory response. Polymorphisms of the CYP17A1 gene might affect the metabolism of estrogen and contribute to the onset of EM (46, 47). Estrogen plays a crucial role in the occurrence and development of EM, and abnormal expression of CYP17A1 may lead to elevated estrogen levels, thereby promoting the growth of ectopic endometrial cells (48, 49). Therefore, CYP17A1 can serve as a potential therapeutic target to regulate the progression of EM by modulating its activity. ENO2 is an intracellular enzyme involved in glycolysis that catalyzes the dehydration of 2-phospho-D-glycerol esters to phosphoenolpyruvate. This conversion process can promote the conversion of glucose to pyruvate, a precursor that promotes the production of NADH molecules and high-energy ATP (50, 51). Many enzymes involved in the glycolytic pathway play important roles in biological processes, tumor progression, and disease occurrence (52–54). Research has shown that ENO2 is a reliable biomarker for diagnosing cancer with great potential for use in various tumors such as melanoma and seminoma (55). The mutation of ENO2 significantly reduces the phosphorylation level of PKM2, while PAK5mediated PKM2 phosphorylation can increase its enzyme activity, promote glucose metabolism, and support the survival and proliferation of EM cells in hypoxic environments (56, 57). Therefore, mutations in ENO2 may indirectly affect the metabolic status of EM by affecting the phosphorylation of PKM2. NR3C1 is a glucocorticoid receptor gene that participates in glucocorticoid signal transduction and plays a crucial role in immune and inflammatory responses. Corticosteroids are important regulatory factors in immune and inflammatory responses that can inhibit inflammatory responses and regulate the activity and function of immune cells. Mutations in the NR3C1 gene can lead to glucocorticoid resistance, affecting immune and inflammatory responses (58). Abnormalities in this function may lead to increased expression of inflammatory factors such as TNF-α, which can induce angiogenesis, promote endometrial cell proliferation and intercellular adhesion, and drive the formation and development of ectopic lesions (59, 60). Anaf et al. found that in the most symptomatic deep glandular nodules of EM, significant NGF expression and invasion of the perineum and neuro-endometrium were observed (61). Research has found that the nerve fiber density and NGF expression level are much higher in deep infiltrating EM than in superficial peritoneal EM (62). High expression of NGF can promote the regeneration and proliferation of nerve fibers, thereby increasing the sensitivity of nociceptors and exacerbating pain (63). These studies indicate that NGF is closely related to EM and its associated pelvic pain (64, 65). In summary, CYP17A1, ENO2, NR3C1, and NGF were associated with EM from different perspectives, such as hormone metabolism, glycolysis metabolism, inflammatory response, and neural correlation, which provided important evidence for analyzing the pathogenesis and exploring new management methods of EM.
We conducted validation studies and found that the mRNA and protein levels of NR3C1, NGF, and ENO2 were significantly elevated in EC compared with EU, whereas the levels of CYP17A1 was the opposite. This was consistent with the patterns observed in public databases used for our study. Based on these findings, we hypothesized that ENO2 could facilitate the conversion of glucose to pyruvate, thereby supplying the necessary energy for the adhesion and proliferation of ectopic endometrial cells, leading to the emergence and progression of EM. NR3C1 may lead to glucocorticoid resistance, impacting the body’s immunity and triggering inflammatory responses, which could precipitate EM. Moreover, CYP17A’s influence on estrogen metabolism might contribute to the progression of EM, while NGF could exacerbate pelvic pain in EM through OS. No studies on the association of ENO2 and NR3C1 with EM have been published. Our investigation is the first to link these genes with EM, thereby introducing a novel perspective for future research. Additional studies are required to confirm the involvement of these signature genes in the etiology of EM.
Enrichment analysis shows that CYP17A1, NR3C1, ENO2, and NGF are co-enriched in the epithelial-mesenchymal transition pathway (EMT), a key mechanism in morphogenesis and organogenesis (66). Numerous studies have shown that EMT is closely related to wound healing, fibrosis, tissue regeneration, inflammation, and cancer metastasis (67–69). EMT is intrinsically tied to EM development and formation (70). During this process, the epithelial cells of the endometrium are transformed into stromal cells through EMT, and these cells may then spread to the outside of the uterus through the blood or lymphatic system and form endometriotic lesions in other areas (71). Moreover, EMT appears to contribute to the invasion and metastasis of EM (72). CYP17A1, in addition to the EMT pathway, NR3C1 and ENO2 are co-enriched in the KRAS signaling pathway. EM is characterized by the extraneous growth of endometrial cells, which, during ectopic expansion, may distress surrounding tissues and organs, resulting in pain and infertility, among other symptoms. Studies have indicated that abnormal activation of the KRAS signaling pathway may play a crucial role in this process (72–74). KRAS activation accelerates endometrial cell proliferation and growth, expediting the formation and progression of ectopic lesions (75). KRAS activation may enhance cell mobility, facilitating the spread of endometrial cells beyond the uterus (76). Hence, investigating the role of the KRAS signaling pathway in EM could yield new therapeutic targets and strategies. Finally, NR3C1, ENO2, and NGF are co-enriched in G2M checkpoint, E2F targets, MYC targets V1, and myogenesis pathways. Research has found that G2M checkpoints play a vital role in the pathogenesis of EM. When cells are subjected to certain stress stimuli, such as DNA damage or abnormal cell division, G2M checkpoints are activated to prevent damaged cells from entering the division cycle (77). In EM, damaged endometrial cells may avoid excessive proliferation and carcinogenesis by activating G2M checkpoints (78). There is no evidence to suggest a direct relationship between EM with E2F targets, MYC targets V1, and myogenesis pathways. Any new research or discoveries in these areas would warrant further exploration.
Abnormal immune regulation is closely related to the pathogenesis of EM, manifested by weakened immune surveillance function and cytotoxic effects of immune killing cells, which cannot effectively clear EC (79, 80). In our investigation, we used CIBERSORT to assess the immune cell infiltration present in EC and EU specimens. The results indicated the involvement of various immune cell subsets in the onset and development of EM. We observed an increased presence of plasma cells and M2 macrophages, alongside a reduced infiltration of immature B cells, Tregs, resting NK cells, and activated NK cells. These fluctuations are potentially implicated in the emergence and progression of EM. As EM is an immune-related condition, the invasive and adhesive properties of endometrial cells are amplified because of the quantity and functionality of diverse immune cells within the peritoneal milieu. This includes plasma cells, M2 macrophages, NK cells, macrophages, dendritic cells, mast cells, and T cells. These cells facilitate angiogenesis and support the formation and persistence of ectopic growths. In EM, NGF may affect the immune response by regulating the differentiation and survival of T and B cells; CYP17A1 can regulate estrogen levels to promote the proliferation of Tregs, thereby inhibiting the TGF-β signaling pathway or reducing the number of Tregs, which may help enhance the immune system’s ability to clear ectopic lesions (81–83). They also aid in the implantation of EC cells, which may be transported back to the pelvic and peritoneal cavities with the menstrual blood, thereby circumventing immune surveillance (84). Endometrioid tissue may spread beyond its endometrial position during the menstrual cycle and implant in other locations; the most common is the formation of lesions by implanting pelvic structures (85). This damage attracts natural killer cells, macrophages, and cytotoxic T cells, and the immune response seems insufficient to clear them (86). The secretion of chemokines and cytokines in the peritoneal cavity can be increased by activating the inflammatory response, creating a microenvironment that promotes local angiogenesis and disrupts endometrial apoptosis, contributing to the development of EC tissue (87). This evidence combined with our current research findings underscores that the infiltration of various immune cells is pivotal in the context of EM and, as such, should be the focal point of future investigations.
The ceRNA network we developed showed that 20 lncRNAs contributed to the regulation of NR3C1 via 10 distinct miRNAs, forming a total of 40 interaction pairs. NR3C1 modulates the activity of steroid hormone receptors and plays a consequential role in EM, exhibiting a close association with the risk and severity of EM symptoms (88). miR-200a and miR-200b belong to the miR-200 family, which are the upstream targets of NR3C1. The miR-200 family regulates the transcription factor network involved in EMT, which is considered a key event in the progression of EM (65). Studies have shown that miR-200a and miR-200b have significant links to the development of EM, regulating the proliferation, apoptosis, and invasion of ectopic endometrial cells (89–93). Consistent with our results, miRNAs of the miR-200 family were downregulated in ectopic endometrium compared with the in-situ endometrium, and lower levels of the miR-200 family were believed to participate in the pathogenesis of EM by stimulating cell movement and inducing EMT processes (94–97). The recently identified LncRNA-MSC-AS1 has been associated with the genesis and progression of diverse cancers, including its role in promoting osteogenic differentiation in bone marrow–derived mesenchymal stem cells (98). LncRNA MSC-AS1 may also worsen the progression of nasopharyngeal carcinoma through upregulation of NR4A2 via sequestration of miR-524-5p (99). A study by Hu and colleagues indicated that MSC-AS1 modulates the proliferation and migration of kidney renal clear cell carcinoma by acting on the miR3924/WNT5A/β-catenin pathways (91). Additionally, a study by Chenglei and colleagues revealed a significant link between lncRNA MSC-AS1 and EM (100).
Bioinformatics results indicated that LncRNA MSC-AS1/NR3C1 was increased while miR-200b-8p was reduced. Based on the ceRNA mechanism, we speculate that lncRNA MSC-AS1 enhances the level of NR3C1 by absorbing miR-200b-8p, thereby exacerbating OS during the progression of EM. However, more studies and large-scale randomized controlled clinical trials are required to validate this vital OS-related axis in EM. Multiple reports have suggested a strong correlation between OS and the pathophysiology of EM (13, 101). The apoptotic endometrial tissue, macrophages, and red blood cells produced by retrograde menstruation promote the production of ROS (102). The increase in ROS levels induces a general inflammatory response, which is beneficial for the adhesion and growth ofendometrial cells detached from the peritoneal cavity (13). Research on the potential use of OS markers as diagnostic tests for EM is still in its early stages (103–105).
It is worth noting that there was no differential expression of eosinophils between EC and EA samples in our study. Previous studies have shown a close relationship between eosinophils and oxidative stress, and an increase in eosinophils is often associated with allergic reactions, autoimmune diseases, or certain tumors. However, the research on eosinophils in EM has not been reported (106). Our study suggests that eosinophils may not be directly associated with EM. In addition, there is currently no direct evidence about the role of CYP17A1, NR3C1, ENO2, and NGF genes, and their related miRNAs in regulating eosinophils. Although this study showed a decline in inactive B cells, regulatory B cells, and NK cells in EC, there is no direct evidence to suggest that the reduction of those cells would lead to an increase in eosinophils. The changes in these immune cells may reflect alterations in the immune microenvironment of EM patients rather than simply caused by changes in the number of specific immune cells. Although eosinophils play an important role in immune responses, the molecular mechanism underlying aberrant eosinophils contributing to EM is unclear, and future research is needed to further explore the specific mechanisms of eosinophils in EM.
Our research has several limitations. Firstly, the diagnostic performance of machine learning models has not been externally validated, which limits their widespread application in a wider population. Secondly, due to the small sample size, the generalizability of research results may be affected. Therefore, larger-scale clinical and basic research is needed to confirm the featured OSRGs we identified. In addition, although the CIBERSORT algorithm performs relatively low in evaluating bias compared to other algorithms, it still has inherent limitations in overestimating or underestimating cell subsets. Therefore, future research can optimize model performance by adjusting algorithm parameters and attempting to use more complex algorithms, such as deep learning, to further improve prediction accuracy. In order to enhance the reliability and application breadth of the model, more clinical data should be introduced for validation in the future. Moreover, increasing the sample size, especially including patients with EM from different types and stages, can improve the representativeness and application value of research results. Furthermore, expanded validation of miRNA, lncRNA, and other DEGs at RNA or protein can provide more solid experimental support for our findings. These efforts will provide a more comprehensive theoretical basis and practical guidance for the precise diagnosis and treatment of EM in the future.
In conclusion, we identified several featured OSRGs, including CYP17A1, ENO2, NGF, and NR3C1, critical in EM pathogenesis. Several types of immune cells, including plasma cells, M2 macrophages, Tregs, resting NK cells, naive B cells and activated NK cells are closely related to the pathogenesis of EM. The functional enrichment analysis results of this study also showed that the screened immune-related OS genes were mainly enriched in OS and immune-related pathways. Therefore, based on these findings, early diagnosis of EM can improve accuracy and timeliness by detecting the expression levels of these biomarkers. The development of drugs targeting these OSRGs was expected to alleviate EM symptoms by reducing oxidative stress, providing new targets for treatment. In addition, the infiltration pattern of immune cells can serve as a biomarker for evaluating the immune microenvironment, helping to determine the severity and progression of diseases. Finally, combining hormone therapy, immune regulation, and antioxidant therapy to develop a multi-target combination therapy strategy will help provide a more comprehensive intervention plan, thereby improving the clinical prognosis of patients. Overall, this study not only provides new insights into the pathogenesis, but also offers potential directions for clinical diagnosis and treatment of EM.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by ethics review committee of Beijing Obstetrics and Gynecology Hospital, Capital Medical University and Beijing Maternal and Child Health Care Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
CX: Data curation, Writing – original draft. CL: Software, Writing – review & editing. NL: Formal analysis, Writing – review & editing. WK: Conceptualization, Data curation, Writing – review & editing. YL: Methodology, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Beijing Municipal Excellent Talents Foundation (No.2018000021469G282).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1515490/full#supplementary-material
Abbreviations
OS, oxidative stress; EM, endometriosis; DEG, differentially expressed gene; EC, ectopic endometrium; EU, eutopic endometrium; OSRG, oxidative stress related gene; DE-OSRG, differentially expressed oxidative stress related gene; ceRNA, competitive endogenous RNA; SVM-RFE, recursive feature elimination; LASSO, selection operator; ROC, receiver operating characteristic; AUC, area under curve; WGCNA, weighted gene co-expression network analysis; ROS, reactive oxygen species.
References
1. Rogers PA, Adamson GD, Al-Jefout M, Becker CM, D’Hooghe TM, Dunselman GA, et al. Research priorities for endometriosis. Reprod Sci. (2017) 24:202–26. doi: 10.1177/1933719116654991
2. Johnson NP, Hummelshoj L, Adamson GD, Keckstein J, Taylor HS, Abrao MS, et al. World Endometriosis Society consensus on the classification of endometriosis. Hum Reprod. (2017) 32:315–24. doi: 10.1093/humrep/dew293
3. Burney RO, Giudice LC. Pathogenesis and pathophysiology of endometriosis. Fertil Steril. (2012) 98:511–9. doi: 10.1016/j.fertnstert.2012.06.029
4. Agarwal SK, Chapron C, Giudice LC, Laufer MR, Leyland N, Missmer SA, et al. Clinical diagnosis of endometriosis: a call to action. Am J Obstet Gynecol. (2019) 220:354.e1–354.e12. doi: 10.1016/j.ajog.2018.12.039
5. Vitagliano A, Noventa M, Quaranta M, Gizzo S. Statins as targeted “Magical pills” for the conservative treatment of endometriosis: may potential adverse effects on female fertility represent the “Dark side of the same coin”? A systematic review of literature. Reprod Sci. (2016) 23:415–28. doi: 10.1177/1933719115584446
6. Horne AW, Missmer SA. Pathophysiology, diagnosis, and management of endometriosis. BMJ. (2022) 379:e070750. doi: 10.1136/bmj-2022-070750
7. Buggio L, Barbara G, Facchin F, Frattaruolo MP, Aimi G, Berlanda N. Self-management and psychological-sexological interventions in patients with endometriosis: strategies, outcomes, and integration into clinical care. Int J Womens Health. (2017) 9:281–93. doi: 10.2147/IJWH.S119724
8. Vargas E, García-Moreno E, Aghajanova L, Salumets A, Horcajadas JA, Esteban FJ, et al. The mid-secretory endometrial transcriptomic landscape in endometriosis: a meta-analysis. Hum Reprod Open. (2022) 2022:hoac016. doi: 10.1093/hropen/hoac016
9. Da Broi MG, Navarro PA. Oxidative stress and oocyte quality: ethiopathogenic mechanisms of minimal/mild endometriosis-related infertility. Cell Tissue Res. (2016) 364:1–7. doi: 10.1007/s00441-015-2339-9
10. Nanda AKT, Banerjee P, Dutta M, Wangdi T, Sharma P, et al. Cytokines, angiogenesis, and extracellular matrix degradation are augmented by oxidative stress in endometriosis. Ann Lab Med. (2020) 40:390–7. doi: 10.3343/alm.2020.40.5.390
11. Clower L, Fleshman T, Geldenhuys WJ, Santanam N. Targeting oxidative stress involved in endometriosis and its pain. Biomolecules. (2022) 12:1055. doi: 10.3390/biom12081055
12. Asghari S, Valizadeh A, Aghebati-Maleki L, Nouri M, Yousefi M. Endometriosis: Perspective, lights, and shadows of etiology. BioMed Pharmacother. (2018) 106:163–74. doi: 10.1016/j.biopha.2018.06.109
13. Santulli P, SandrineFiorese M, LouisLemarechal H, Anne-ElodieBatteux F, DidierChapron C. Protein oxidative stress markers in peritoneal fluids of women with deep infiltrating endometriosis are increased. Hum Reproduction. (2015) 30(1):49-60. doi: 10.1093/humrep/deu290
14. Kyama CM, Overbergh L, Mihalyi A, Meuleman C, Mwenda JM, Mathieu C, et al. Endometrial and peritoneal expression of aromatase, cytokines, and adhesion factors in women with endometriosis. Fertil Steril. (2008) 89:301–10. doi: 10.1016/j.fertnstert.2007.02.057
15. Samimi M, Pourhanifeh MH, Mehdizadehkashi A, Eftekhar T, Asemi Z. The role of inflammation, oxidative stress, angiogenesis, and apoptosis in the pathophysiology of endometriosis: Basic science and new insights based on gene expression. J Of Cell Physiol. (2019) 234(11):19384–92. doi: 10.1002/jcp.v234.11
16. Barbe AM, Berbets AM, Davydenko IS, Koval HD, Yuzko VO, Yuzko OM. Expression and significance of matrix metalloproteinase-2 and matrix metalloproteinas-9 in endometriosis. J Med Life. (2020) 13:314–20. doi: 10.25122/jml-2020-0117
17. Chen C, Zhou Y, Hu C, Wang Y, Yan Z, Li Z, et al. Mitochondria and oxidative stress in ovarian endometriosis. Free Radic Biol Med. (2019) 136:22–34. doi: 10.1016/j.freeradbiomed.2019.03.027
18. Prefumo F, Rossi AC. Endometriosis, endometrioma, and ART results: Current understanding and recommended practices. Best Pract Res Clin Obstet Gynaecol. (2018) 51:34–40. doi: 10.1016/j.bpobgyn.2018.01.019
19. Tomimatsu T, Mimura K, Matsuzaki S, Endo M, Kumasawa K, Kimura T. Preeclampsia: maternal systemic vascular disorder caused by generalized endothelial dysfunction due to placental antiangiogenic factors. Int J Mol Sci. (2019) 20(17):4246. doi: 10.3390/ijms20174246
20. Hull ML, Escareno CR, Godsland JM, Doig JR, Johnson CM, Phillips SC, et al. Endometrial-peritoneal interactions during endometriotic lesion establishment. Am J Pathol. (2008) 173:700–15. doi: 10.2353/ajpath.2008.071128
21. Crispi S, Piccolo MT, D’Avino A, Donizetti A, Viceconte R, Spyrou M, et al. Transcriptional profiling of endometriosis tissues identifies genes related to organogenesis defects. J Cell Physiol. (2013) 228:1927–34. doi: 10.1002/jcp.v228.9
22. Zhao L, Gu C, Ye M, Zhang Z, Li L, Fan W, et al. Integration analysis of microRNA and mRNA paired expression profiling identifies deregulated microRNA-transcription factor-gene regulatory networks in ovarian endometriosis. Reprod Biol Endocrinol. (2018) 16:4. doi: 10.1186/s12958-017-0319-5
23. Wu Z, Wang L, Wen Z, Yao J. Integrated analysis identifies oxidative stress genes associated with progression and prognosis in gastric cancer. Sci Rep. (2021) 11:3292. doi: 10.1038/s41598-021-82976-w
24. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
25. Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat. (2016) 10:946–63. doi: 10.1214/16-AOAS920
26. Gustavsson EK, Zhang D, Reynolds RH, Garcia-Ruiz S, Ryten M. ggtranscript: an R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics. (2022) 38:3844–6. doi: 10.1093/bioinformatics/btac409
27. Wu M, Shang X, Sun Y, Wu J, Liu G. Integrated analysis of lymphocyte infiltration-associated lncRNA for ovarian cancer via TCGA, GTEx and GEO datasets. PeerJ. (2020) 8:e8961. doi: 10.7717/peerj.8961
28. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
29. Liu P, Xu H, Shi Y, Deng L, Chen X. Potential molecular mechanisms of plantain in the treatment of gout and hyperuricemia based on network pharmacology. Evid Based Complement Alternat Med. (2020) 2020:3023127. doi: 10.1155/2020/3023127
30. Li Y, Lu F, Yin Y. Applying logistic LASSO regression for the diagnosis of atypical Crohn’s disease. Sci Rep. (2022) 12:11340. doi: 10.1038/s41598-022-15609-5
31. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. (2013) 14:7. doi: 10.1186/1471-2105-14-7
32. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. (2011) 12:77. doi: 10.1186/1471-2105-12-77
33. Xu H, Pan Y. A prognostic fibroblast-related risk signature in colorectal cancer. Aging (Albany NY). (2021) 13:24251–70. doi: 10.18632/aging.203677
34. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. (2018) 1711:243–59. doi: 10.1007/978-1-4939-7493-1_12
35. Yang Z, Wei X, Pan Y, Xu J, Si Y, Min Z, et al. A new risk factor indicator for papillary thyroid cancer based on immune infiltration. Cell Death Dis. (2021) 12:51. doi: 10.1038/s41419-020-03294-z
36. Yang L, Ma TJ, Zhang YB, Wang H, An RH. Construction and Analysis of lncRNA-miRNA-mRNA ceRNA Network Identify an Eight-Gene Signature as a Potential Prognostic Factor in Kidney Renal Papillary Cell Carcinoma (KIRP). Altern Ther Health Med. (2022) 28:42–51.
37. Li A, He J, Zhang Z, Jiang S, Gao Y, Pan Y, et al. Integrated bioinformatics analysis reveals marker genes and potential therapeutic targets for pulmonary arterial hypertension. Genes (Basel). (2021) 12:1339. doi: 10.3390/genes12091339
38. Cheng-Mao X, Yan L, Li L, Hua J, Xiao-Ju W, Jie-Wen Z. Placental ABCA1 expression is increased in spontaneous preterm deliveries compared with iatrogenic preterm deliveries and term deliveries. BioMed Res Int. (2017) 2017:8248094. doi: 10.1155/2017/8248094
39. Zhu T, Zhang X. Research progress on the role of epithelial-mesenchymal transition in pathogenesis of endometriosis. Zhejiang Da Xue Xue Bao Yi Xue Ban. (2016) 45(4):439–45. doi: 10.3785/j.issn.1008-9292.2016.07.17
40. Ballard K, Lowton K, Wright J. What’s the delay? A qualitative study of women’s experiences of reaching a diagnosis of endometriosis. Fertility Sterility. (2006) 86:1296–301. doi: 10.1016/j.fertnstert.2006.04.054
41. Hudelist G, Fritzer N, Thomas A, Niehues C, Oppelt P, Haas D, et al. Diagnostic delay for endometriosis in Austria and Germany: causes and possible consequences. Hum Reprod. (2012) 27(12):3412–6. doi: 10.1093/humrep/des316
42. Nnoaham KE, Hummelshoj L, Webster P, d’Hooghe T, de Cicco Nardone F, de Cicco Nardone C, et al. Impact of endometriosis on quality of life and work productivity: a multicenter study across ten countries. Fertil Steril. (2011) 96:366–73.e8. doi: 10.1016/j.fertnstert.2011.05.090
43. Ito F, Yamada Y, Shigemitsu A, Akinishi M, Kaniwa H, Miyake R, et al. Role of oxidative stress in epigenetic modification in endometriosis. Reprod Sci. (2017) 24:1493–502. doi: 10.1177/1933719117704909
44. Picado-Leonard J, Miller WL. Cloning and sequence of the human gene for P450c17 (steroid 17 alpha-hydroxylase/17,20 lyase): similarity with the gene for P450c21. DNA. (1987) 6:439–48. doi: 10.1089/dna.1987.6.439
45. Jahromi MS, Tehrani FR, Noroozzadeh M, Zarkesh M, Ghasemi A, Zadeh-Vakili A. Elevated expression of steroidogenesis pathway genes; CYP17, GATA6 and StAR in prenatally androgenized rats. Gene. (2016) 593:167–71. doi: 10.1016/j.gene.2016.07.067
46. Nguyen PT, Conley AJ, Sneyd J, Lee RS, Soboleva TK, Shorten PR. The role of enzyme compartmentalization on the regulation of steroid synthesis. J Theor Biol. (2013) 332:52–64. doi: 10.1016/j.jtbi.2013.04.021
47. Al-Rubae’i SH, Naji TS, Turki KM. Common variation of the CYP17 gene in Iraqi women with endometriosis disease. Genom Data. (2017) 11:55–9. doi: 10.1016/j.gdata.2016.11.019
48. Lin Z, Li F, Zhang Y, Tan X, Luo P, Liu H. Analysis of astaxanthin molecular targets based on network pharmacological strategies. J Food Biochem. (2021) 45:e13717. doi: 10.1111/jfbc.13717
49. Zhao S, Xu Z. Lipoxin A4 inhibits the development of endometriosis in a mouse model by suppressing local estradiol synthesis. Prostaglandins Other Lipid Mediat. (2021) 153:106521. doi: 10.1016/j.prostaglandins.2020.106521
50. Liu CC, Wang H, Wang WD, Wang L, Liu WJ, Wang JH, et al. ENO2 promotes cell proliferation, glycolysis, and glucocorticoid-resistance in acute lymphoblastic leukemia. Cell Physiol Biochem. (2018) 46:1525–35. doi: 10.1159/000489196
51. Vizin T, Kos J. Gamma-enolase: a well-known tumour marker, with a less-known role in cancer. Radiol Oncol. (2015) 49:217–26. doi: 10.1515/raon-2015-0035
52. Pancholi V. Multifunctional alpha-enolase: its role in diseases. Cell Mol Life Sci. (2001) 58:902–20. doi: 10.1007/PL00000910
53. Díaz-Ramos A, Roig-Borrellas A, García-Melero A, López-Alemany R. [amp]]alpha;-Enolase, a multifunctional protein: its role on pathophysiological situations. J BioMed Biotechnol. (2012) 2012:156795. doi: 10.1155/2012/156795
54. Isgrò MA, Bottoni P, Scatena R. Neuron-specific enolase as a biomarker: biochemical and clinical aspects. Adv Exp Med Biol. (2015) 867:125–43. doi: 10.1007/978-94-017-7215-0_9
55. Yu Q, Tong C, Luo M, Xue X, Mei Q, Ma L, et al. Regulation of SESAME-mediated H3T11 phosphorylation by glycolytic enzymes and metabolites. PloS One. (2017) 12:e0175576. doi: 10.1371/journal.pone.0175576
56. Lu J, Wang X, Shi X, Jiang J, Liu L, Liu L, et al. PAK5-mediated PKM2 phosphorylation is critical for anaerobic glycolysis in endometriosis. Front Med. (2024) 18:1054–67. doi: 10.1007/s11684-024-1069-3
57. Liu L, Wu J, Qing L, Li J, Yang H, Ji A, et al. DNA methylation analysis of the NR3C1 gene in patients with schizophrenia. J Mol Neurosci. (2020) 70:1177–85. doi: 10.1007/s12031-020-01525-8
58. Panek M, Pietras T, Fabijan A, Zioło J, Wieteska Ł, Małachowska B, et al. The NR3C1 glucocorticoid receptor gene polymorphisms may modulate the TGF-beta mRNA expression in asthma patients. Inflammation. (2015) 38:1479–92. doi: 10.1007/s10753-015-0123-3
59. Liu M, Li Y, Yuan Y, Jiang M, Yin P, Yang D. Peri-implantation treatment with TNF-α inhibitor for endometriosis and/or adenomyosis women undergoing frozen-thawed embryo transfer: A retrospective cohort study. J Reprod Immunol. (2025) 167:104415. doi: 10.1016/j.jri.2024.104415
60. Anaf V, Simon P, El Nakadi I, Fayt I, Simonart T, Buxant F, et al. Hyperalgesia, nerve infiltration and nerve growth factor expression in deep adenomyotic nodules, peritoneal and ovarian endometriosis. Hum Reprod. (2002) 17:1895–900. doi: 10.1093/humrep/17.7.1895
61. Wang G, Tokushige N, Markham R, Fraser IS. Rich innervation of deep infiltrating endometriosis. Hum Reprod. (2009) 24:827–34. doi: 10.1093/humrep/den464
62. Liu Z, Li M, Zhang L, Shi X, Liao T, Jie L, et al. NGF signaling exacerbates KOA peripheral hyperalgesia via the increased TRPV1-labeled synovial sensory innervation in KOA rats. Pain Res Manag. (2024) 2024:1552594. doi: 10.1155/2024/1552594
63. Meijer OC, Koorneef LL, Kroon J. Glucocorticoid receptor modulators. Ann Endocrinol (Paris). (2018) 79:107–11. doi: 10.1016/j.ando.2018.03.004
64. Ishmael FT, Fang X, Houser KR, Pearce K, Abdelmohsen K, Zhan M, et al. The human glucocorticoid receptor as an RNA-binding protein: global analysis of glucocorticoid receptor-associated transcripts and identification of a target RNA motif. J Immunol. (2011) 186:1189–98. doi: 10.4049/jimmunol.1001794
65. Cho H, Park OH, Park J, Ryu I, Kim J, Ko J, et al. Glucocorticoid receptor interacts with PNRC2 in a ligand-dependent manner to recruit UPF1 for rapid mRNA degradation. Proc Natl Acad ences. (2015) 112:1540–9. doi: 10.1073/pnas.1409612112
66. Pei D, Shu X, Gassama-Diagne A, Thiery JP. Mesenchymal-epithelial transition in development and reprogramming. Nat Cell Biol. (2019) 21:44–53. doi: 10.1038/s41556-018-0195-z
67. Jolly MK, Ware KE, Gilja S, Somarelli JA, Levine H. EMT and MET: necessary or permissive for metastasis. Mol Oncol. (2017) 11:755–69. doi: 10.1002/mol2.2017.11.issue-7
68. Das V, Bhattacharya S, Chikkaputtaiah C, Hazra S, Pal M. The basics of epithelial-mesenchymal transition (EMT): A study from a structure, dynamics, and functional perspective. J Cell Physiol. (2019) 234:14535–55. doi: 10.1002/jcp.v234.9
69. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. (2019) 20:69–84. doi: 10.1038/s41580-018-0080-4
70. Grund EM, Kagan D, Tran CA, Zeitvogel A, Starzinski-Powitz A, Nataraja S, et al. Tumor necrosis factor-alpha regulates inflammatory and mesenchymal responses via mitogen-activated protein kinase kinase, p38, and nuclear factor kappaB in human endometriotic epithelial cells. Mol Pharmacol. (2008) 73:1394–404. doi: 10.1124/mol.107.042176
71. Matsuzaki S, Darcha C. Epithelial to mesenchymal transition-like and mesenchymal to epithelial transition-like processes might be involved in the pathogenesis of pelvic endometriosis. Hum Reprod. (2012) 27:712–21. doi: 10.1093/humrep/der442
72. Bilyk O, Coatham M, Jewer M, Postovit LM. Epithelial-to-mesenchymal transition in the female reproductive tract: from normal functioning to disease pathology. Front Oncol. (2017) 7:145. doi: 10.3389/fonc.2017.00145
73. Guo SW. Cancer-associated mutations in endometriosis: shedding light on the pathogenesis and pathophysiology. Hum Reprod Update. (2020) 26:423–49. doi: 10.1093/humupd/dmz047
74. Lac V, Verhoef L, Aguirre-Hernandez R, Nazeran TM, Tessier-Cloutier B, Praetorius T, et al. Iatrogenic endometriosis harbors somatic cancer-driver mutations. Hum Reprod. (2019) 34:69–78. doi: 10.1093/humrep/dey332
75. Yachida N, Yoshihara K, Suda K, Nakaoka H, Ueda H, Sugino K, et al. Biological significance of KRAS mutant allele expression in ovarian endometriosis. Cancer Sci. (2021) 112:2020–32. doi: 10.1111/cas.v112.5
76. Praetorius TH, Leonova A, Lac V, Senz J, Tessier-Cloutier B, Nazeran TM, et al. Molecular analysis suggests oligoclonality and metastasis of endometriosis lesions across anatomically defined subtypes. Fertil Steril. (2022) 118:524–34. doi: 10.1016/j.fertnstert.2022.05.030
77. Johnson DG, Walker CL. Cyclins and cell cycle checkpoints. Annu Rev Pharmacol Toxicol. (1999) 39:295–312. doi: 10.1146/annurev.pharmtox.39.1.295
78. Lee J, Banu SK, Rodriguez R, Starzinski-Powitz A, Arosh JA. Selective blockade of prostaglandin E2 receptors EP2 and EP4 signaling inhibits proliferation of human endometriotic epithelial cells and stromal cells through distinct cell cycle arrest. Fertil Steril. (2010) 93:2498–506. doi: 10.1016/j.fertnstert.2010.01.038
79. Unger MS, Marschallinger J, Kaindl J, Klein B, Johnson M, Khundakar AA, et al. Doublecortin expression in CD8+ T-cells and microglia at sites of amyloid-β plaques: A potential role in shaping plaque pathology. Alzheimers Dement. (2018) 14:1022–37. doi: 10.1016/j.jalz.2018.02.017
80. Yang HL, Zhou WJ, Gu CJ, Meng YH, Shao J, Li DJ, et al. Pleiotropic roles of melatonin in endometriosis, recurrent spontaneous abortion, and polycystic ovary syndrome. Am J Reprod Immunol. (2018) 80:e12839. doi: 10.1111/aji.2018.80.issue-1
81. Terracina S, Ferraguti G, Tarani L, Fanfarillo F, Tirassa P, Ralli M, et al. Nerve growth factor and autoimmune diseases. Curr Issues Mol Biol. (2023) 45:8950–73. doi: 10.3390/cimb45110562
82. Deng D, Wu Y, Wu K, Zeng N, Li W. Dihydroberberine alleviates Th17/Treg imbalance in premature ovarian insufficiency mice via inhibiting Rheb/mTOR signaling. Mol Med. (2024) 30:194. doi: 10.1186/s10020-024-00971-z
83. Li J, Liu L, Fan R. The PKM2/HIF-1α Axis is Involved in the Pathogenesis of Endometriosis via TGF-β1 under Endometrial Polyps. Front Biosci (Landmark Ed). (2024) 29:417. doi: 10.31083/j.fbl2912417
84. Capobianco A, Cottone L, Monno A, Manfredi AA, Rovere-Querini P. The peritoneum: healing, immunity, and diseases. J Pathol. (2017) 243:137–47. doi: 10.1002/path.2017.243.issue-2
85. Rzewuska AM, Żybowska M, Sajkiewicz I, Spiechowicz I, Żak K, Abramiuk M, et al. Gonadotropin-releasing hormone antagonists-A new hope in endometriosis treatment. J Clin Med. (2023) 12(3):1008. doi: 10.3390/jcm12031008
86. Chen S, Liu Y, Zhong Z, Wei C, Liu Y, Zhu X. Peritoneal immune microenvironment of endometriosis: Role and therapeutic perspectives. Front Immunol. (2023) 14:1134663. doi: 10.3389/fimmu.2023.1134663
87. Reusche N, Beineke A, Urhausen C, Beyerbach M, Schmicke M, Kramer S, et al. Proliferative and apoptotic changes in the healthy canine endometrium and in cystic endometrial hyperplasia. Theriogenology. (2018) 114:14–24. doi: 10.1016/j.theriogenology.2018.03.018
88. Peng X, Xia Y, Xie J, Liu H, Fan L, Yu C, et al. Mechanism of Thunberg Fritillaria in treating endometriosis based on network pharmacology and the effect of Peiminine on the MEK/ERK pathway. Am J Transl Res. (2022) 14:6196–209.
89. Rekker K, Saare M, Roost AM, Kaart T, Sõritsa D, Karro H, et al. Circulating miR-200-family micro-RNAs have altered plasma levels in patients with endometriosis and vary with blood collection time. Fertil Steril. (2015) 104:938–46.e2. doi: 10.1016/j.fertnstert.2015.06.029
90. Wang D, Luo Y, Wang G, Yang Q. CircATRNL1 promotes epithelial-mesenchymal transition in endometriosis by upregulating Yes-associated protein 1 in vitro. Cell Death Dis. (2020) 11:594. doi: 10.1038/s41419-020-02784-4
91. Hu W, Xie Q, Xu Y, Tang X, Zhao H. Integrated bioinformatics analysis reveals function and regulatory network of miR-200b-3p in endometriosis. BioMed Res Int. (2020) 2020:3962953. doi: 10.1155/2020/3962953
92. de Oliveira RZ, de Oliveira Buono F, Cressoni A, Penariol L, Padovan CC, Tozetti PA, et al. Overexpression of miR-200b-3p in menstrual blood-derived mesenchymal stem cells from endometriosis women. Reprod Sci. (2022) 29:734–42. doi: 10.1007/s43032-022-00860-y
93. Saare M, Rekker K, Laisk-Podar T, Sõritsa D, Roost AM, Simm J, et al. High-throughput sequencing approach uncovers the miRNome of peritoneal endometriotic lesions and adjacent healthy tissues. PloS One. (2014) 9:e112630. doi: 10.1371/journal.pone.0112630
94. Ohlsson Teague EM, van der Hoek KH, van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, et al. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. (2009) 23:265–75. doi: 10.1210/me.2008-0387
95. Filigheddu N, Gregnanin I, Porporato PE, Surico D, Perego B, Galli L, et al. Differential expression of microRNAs between eutopic and ectopic endometrium in ovarian endometriosis. J BioMed Biotechnol. (2010) 2010:369549. doi: 10.1155/2010/369549
96. Hawkins SM, Creighton CJ, Han DY, Zariff A, Anderson ML, Gunaratne PH, et al. Functional microRNA involved in endometriosis. Mol Endocrinol. (2011) 25:821–32. doi: 10.1210/me.2010-0371
97. Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. (2010) 16:142–65. doi: 10.1093/humupd/dmp034
98. Zhang N, Hu X, He S, Ding W, Wang F, Zhao Y, et al. LncRNA MSC-AS1 promotes osteogenic differentiation and alleviates osteoporosis through sponging microRNA-140-5p to upregulate BMP2. Biochem And Biophys Res Commun. (2019) 519:790–6. doi: 10.1016/j.bbrc.2019.09.058
99. Yao H, Yang L, Tian L, Guo Y, Li Y. LncRNA MSC-AS1 aggravates nasopharyngeal carcinoma progression by targeting miR-524-5p/nuclear receptor subfamily 4 group A member 2 (NR4A2). Cancer Cell Int. (2020) 20:138. doi: 10.1186/s12935-020-01202-1
100. Hu Z, Li L, Cheng P, Liu Q, Zheng X, Peng F, et al. lncRNA MSC-AS1 activates Wnt/β-catenin signaling pathway to modulate cell proliferation and migration in kidney renal clear cell carcinoma via miR-3924/WNT5A. J Cell Biochem. (2020) 121:4085–93. doi: 10.1002/jcb.v121.10
101. Gu C, Meng Y, Meng Q, Fan W, Ye M, Zhang Q, et al. Exploring the potential key incRNAs with endometriosis by construction of a ceRNA network. Int J Gen Med. (2021) 14:4161–70. doi: 10.2147/IJGM.S321648
102. González-Foruria I, Santulli P, Chouzenoux S, Carmona F, Chapron C, Batteux F. Dysregulation of the ADAM17/Notch signalling pathways in endometriosis: from oxidative stress to fibrosis. Mol Hum Reprod. (2017) 23:488–99. doi: 10.1093/molehr/gax028
103. Ngô C, Chéreau C, Nicco C, Weill B, Chapron C, Batteux F. Reactive oxygen species controls endometriosis progression. Am J Pathol. (2009) 175:225–34. doi: 10.2353/ajpath.2009.080804
104. Cheng SC, Liou CJ, Wu SJ, Lin CF, Huang TH, Huang WC. Neochlorogenic acid ameliorates allergic airway inflammation by suppressing type 2 immunity and upregulating HO-1 expression. Int Immunopharmacol. (2025) 146:113867. doi: 10.1016/j.intimp.2024.113867
105. Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. (2005) 30:142–50. doi: 10.1016/j.tibs.2005.01.005
Keywords: endometriosis, ectopic endometrium, eutopic endometrium, oxidative stress, machine learning, immune cells, lncRNA
Citation: Xie C, Lu C, Lv N, Kong W and Liu Y (2025) Identification and analysis of oxidative stress-related genes in endometriosis. Front. Immunol. 16:1515490. doi: 10.3389/fimmu.2025.1515490
Received: 23 October 2024; Accepted: 18 February 2025;
Published: 07 March 2025.
Edited by:
Chuanwen Fan, Linköping University, SwedenReviewed by:
Hao Wang, Shenzhen University General Hospital, ChinaPing Li, Chinese Academy of Sciences CAS), China
Copyright © 2025 Xie, Lu, Lv, Kong and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Liu, eW91bmdsZXVAY2NtdS5lZHUuY24=
 Chengmao Xie
Chengmao Xie Chang Lu
Chang Lu Yong Liu
Yong Liu