
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 06 February 2025
Sec. Cancer Immunity and Immunotherapy
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1514780
This article is part of the Research Topic Exploring KLF4's Role in Immune Cell Function and Disease Progression View all articles
Krüppel-like factors (KLFs) regulate various biological processes such as cell proliferation, migration, invasion, and differentiation as gene transcription factors. Signaling pathways which mediated by KLF4 and KLF4 have a sophisticated role in tumors due to multiple factors, including the types or stage of tumors. KLF4 plays a promoter role in tumorigenesis and development, or tumor suppressor as a context-dependent anti- and pro-inflammatory factor. KLF4 over-expression increases CD8+T cell differentiation and enhances the antitumor immunity. This review aims to provide information about the relationship of KLF4 in immunity with tumors and to guide the future study.
Tumor-immune interactions are a critical driver of tumorigenesis and progression while also offering targets for preventive strategies or novel therapies. The tumor microenvironment (TME) and its unique immune milieu plays a dual role in both suppressing tumor growth and promoting tumor progression as multi-facted regulation is crucial for tumor immune evasion, TME formation, and the modulation of anti-tumor immune responses. However, the molecular networks connecting immunity and tumor progression are dense and interconnected, highlighting the necessity of identifying key “bridge” molecules that link immunity and tumor biology for the development of novel therapeutic strategies.
Interestingly, certain genes, such as TGF-β (1), Notch (2), Runx (3), E2f (4), CDKN1A (5), and Krüppel-like factor 4 (KLF4) (6), exhibit dual functionality in tumors. A single gene may serve as both a tumor suppressor and an oncogene in specific cancers; this paradox underscores the intricacy of tumor biology. Among dual-function transcription factors, KLF4, a prominent member of the Krüppel-like factor family, demonstrates this context-dependent dual role in tumor biology and immune system regulation. How exactly does KLF4 serve as a critical “bridge” connecting immunity and tumor biology by simultaneously regulating the tumor microenvironment and immune responses? These questions warrant deeper exploration to uncover KLF4’s potential as a pivotal molecular target in cancer immunotherapy.
KLF4 is a zinc finger-containing transcription factor belonging to the Specificity Protein (SP)/KLF family (7). The gene encoding KLF4 (Klf4) was first isolated by Shields et al. in 1996 (8). As a transcription factor, KLF4 plays diverse roles in the development and progression of multiple diseases across varying tissues and organs, as well as in multiple biological processes. In the context of cancer, KLF4 and its interacting molecular pathways exhibit complex, multifaceted roles depending on the type or stage of the tumor.
KLF4 is frequently linked to cancer stem cells (CSCs), which possess the ability to self-renew and differentiate into multiple tumor cell types. As CSCs are well-known contributors to cancer recurrence, metastasis, and resistance to chemotherapy tolerance, the central role of KLF4 in CSC regulation underscores its importance in tumor progression and therapy resistance (9, 10).
Despite significant advances in KLF4 regulatory mechanisms in tumorigenesis, research on KLF4’s role within the immune system and its relationship to cancer remains limited. This review aims to explore the biological and molecular mechanisms of KLF4 within the immune system, with a particular focus on its interactions with tumor immunity and its dual role as a switchable oncogene/suppressor. Additionally, this review will outline potential future research directions, emphasizing KLF4’s function in both normal immune cells and tumor immunity.
The structure of KLF4 is comprised of multiple functional domains that confer specific transcriptional regulatory functions. Protein databases list the canonical sequence of KLF4_HUMAN at 513 amino acids but, in contrast, most experimental functional studies utilize KLF4_MOUSE, whose canonical sequence has a standard length of 483 amino acids. With the rapid development of AI technology, particularly tools such as AlphaFold (11, 12), the protein structure of KLF4 has been predicted with high accuracy, laying the foundation for further investigation into the mechanisms of KLF4 in disease. The structure of KLF4 is shown in Figure 1. The Klf4 gene is primarily composed of five exons and four introns, encoding the KLF4 protein, which has three highly conserved C2H2-type zinc finger motifs at its carboxyl terminus. These zinc finger motifs are the defining characteristic of KLF4, enabling it to bind specifically to GC-rich regions in DNA, such as CACCC boxes, thereby regulating the transcription of downstream target genes (13). The normal function of KLF4 depends on the integrity of zinc finger domain. Crystallographic studies have revealed that the first zinc finger motif inhibits KLF4’s ability to promote self-renewal and block differentiation while the two C-terminal zinc finger motifs are essential for DNA binding specificity, facilitating the terminal differentiation of macrophages. Disruption of KLF4’s DNA-binding function, as indicated by crystallographic data, may impair macrophage differentiation and has been implicated in hematologic malignancies, such as monocytic leukemia (14).
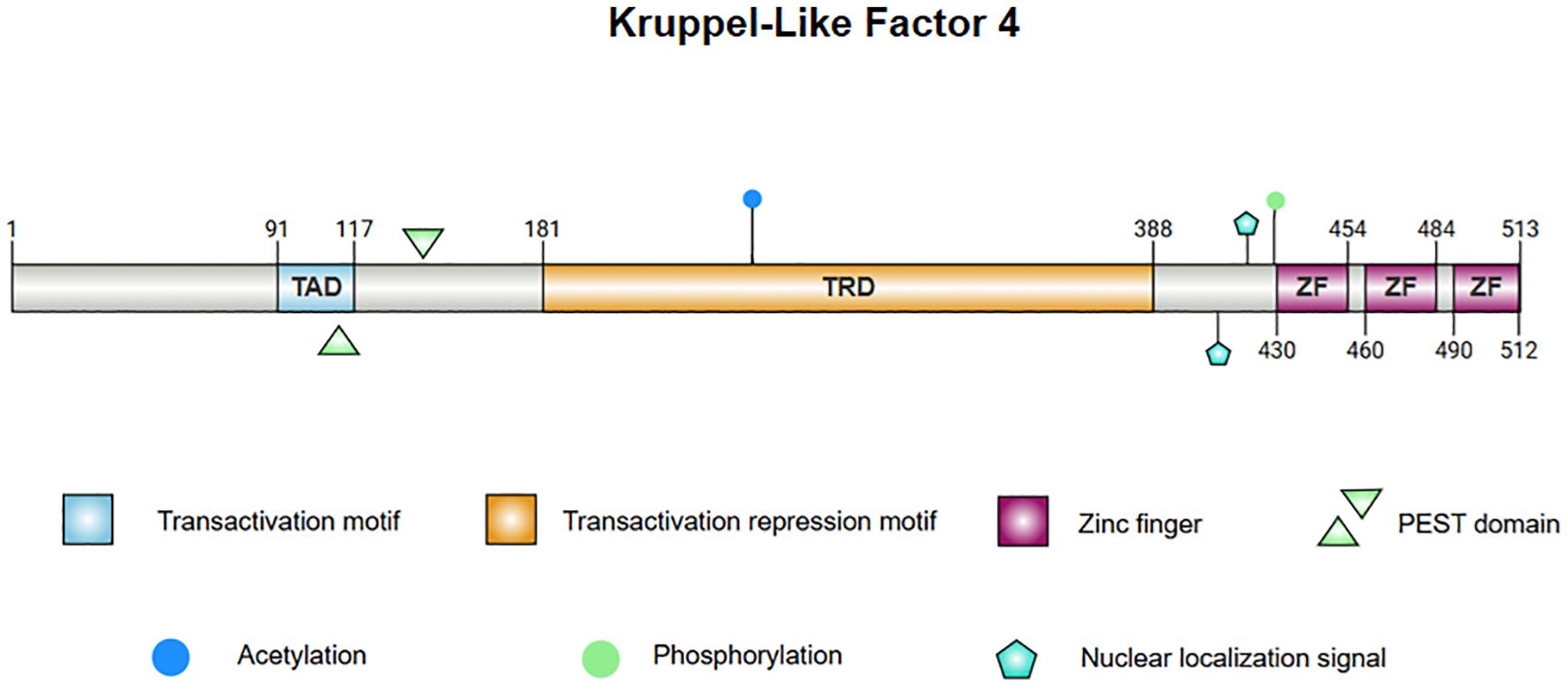
Figure 1. The protein structure of KLF4. 1. Linear diagram of the KLF4 protein, visualizing the protein sequence and functional regions. 2. The central image depicts the superimposed structure of KLF4 predicted by AlphaFold and the X-ray diffraction-derived protein model, demonstrating consistency in regions with high confidence. The left panel presents the AlphaFold-predicted structure of KLF4, with the Predicted Aligned Error (PAE) set to 300×300. The right panel illustrates the partial structure of KLF4 obtained from the PDB database via X-ray diffraction. Note: Internal confidence score (commonly referred to as pLDDT, Predicted Local Distance Difference Test): pLDDT > 90: Dark blue; 70 < pLDDT ≤ 90: Light blue; 50 < pLDDT ≤ 70: Yellow; pLDDT ≤ 50: Orange or red.
KLF4’s N-terminus contains both transcriptional activation and repression domains. A proline/serine-rich transactivation domain is located between amino acid residues 91 and 117, which promotes the expression of downstream genes. Adjacent to this is a repressive domain between residues 181 and 388, which collectively determine KLF4’s specificity in regulating transcriptional activity (15–17). KLF4 also contains two nuclear localization signals (NLS), one located near and the other upstream of the zinc finger motifs, which facilitate KLF4’s entry into the nucleus for transcriptional regulation (15). Additionally, KLF4 harbors a proline (P), glutamic acid (E), serine (S), and threonine (T) (PEST) region which is involved in protein degradation and regulates its own stability (18). In lymphomas and leukemias, mutations in this region may inhibit ubiquitination and degradation of KLF4 in tumors, contributing to malignant progression. Mutations in the PEST region may also be linked to early-stage pancreatic ductal adenocarcinoma (PDA) (19).
KLF4 contains several functional mutation sites, with certain key residues responsible for phosphorylation and acetylation modifications (8, 20, 21). These modifications affect KLF4’s stability and activity, and mutations at these sites alter its function in specific cancer contexts. For instance, the A472D mutation in KLF4 may play a critical role in the development of resistance to the anti-EGFR monoclonal antibody cetuximab in patients with metastatic colorectal cancer (mCRC) (22). Overall, this complex structure enables KLF4 to promote both stem cell differentiation and the maintenance of cellular homeostasis, while also playing a dual role in tumorigenesis.
To fully understand the role of KLF4 in tumor immunity, it is essential to first examine its functions in the normal immune system. As a critical transcription factor, KLF4 is involved in the differentiation, activation, and migration of immune cells, while also playing a key role in maintaining immune homeostasis. The relationship between KLF4 and the maturation and differentiation of immune cells during hematopoiesis is shown in Figure 2.
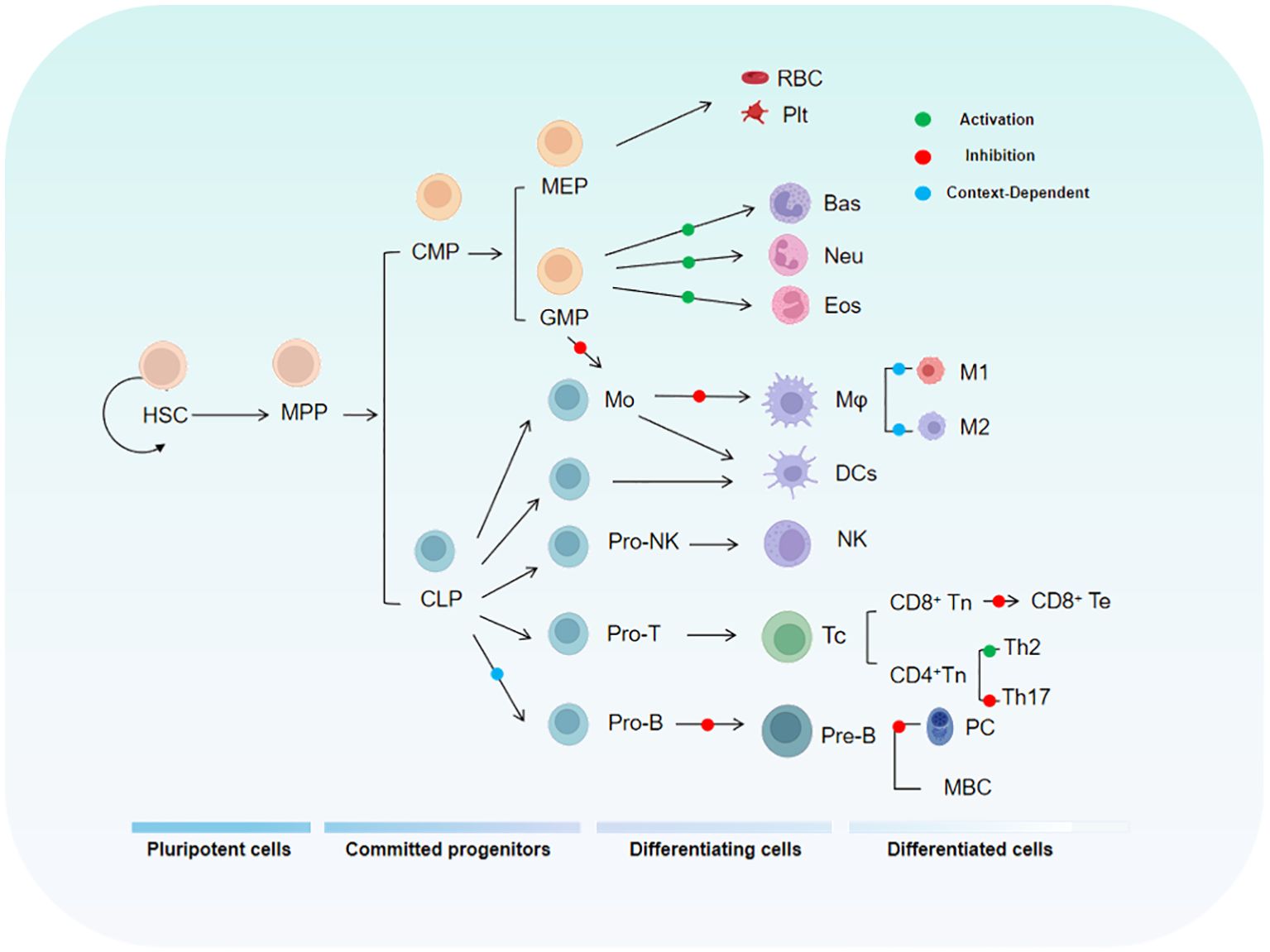
Figure 2. The diagram illustrates the relationship between KLF4 and the hematopoietic hierarchy. HSC, Hematopoietic Stem Cell; MPP, Multipotent Progenitor; CMP, Common Myeloid Progenitor; MEP, Megakaryocyte-Erythroid Progenitor; GMP, Granulocyte-Monocyte Progenitor.
During T cell development, multiple transcription factors regulate the expression of specific genes in series and stages, including hematopoiesis, thymic differentiation, and peripheral T cell specialization. KLF4 has been implicated in the differentiation, activation, quiescence, and homing of various T cell subpopulations (23). KLF4 is highly expressed in thymocytes and mature T cells but is rapidly downregulated following T cell activation (24) as this regulatory mechanism maintains a swift immune response while preventing autoimmunity. Studies have shown that specific deletion of the Klf4 gene in CD4+ T cells in the mouse thymus significantly inhibits the differentiation of Th17 cells and the expression of IL-17, demonstrating that KLF4 directly regulates Th17 differentiation and function via potential direct binding to the Il17a promoter (24). In vitro experiments by Yamada, Mamonkin, and colleagues showed that, under normal conditions, KLF4 suppressed the proliferation of naïve CD8+ T cells, potentially through the regulation of p21 expression in T cells (25). However, subsequent in vivo experiments revealed that the proliferation, maturation, and distribution of naïve CD8+ T cells were not significantly affected by Klf4 deletion. Conversely, under infection conditions, Klf4 deletion significantly increased the proliferation of naïve CD8+ T cells and the formation of memory T cells, indicating that the regulation of CD8+ naïve and memory T cell proliferation and differentiation by KLF4 varies by circumstance and that molecular sensors exist to switch it for such purposes (24, 26). KLF4’s Lys-48-linked ubiquitination is mediated by the E3 ubiquitin ligase Mule, which is crucial for T cell entry into S phase and proliferation. When Mule is absent, KLF4 accumulates, leading to E2F2 upregulation and activation of cell cycle inhibitors p21 and p27, thereby inhibiting T cell proliferation. Additionally, KLF4 expression is reduced in CD8+ T cells lacking KLF4, a condition that promotes excessive proliferation of CD8+ T cells (27). Tussiwand and colleagues demonstrated that KLF4 is an essential factor for inducing Th2 cell responses in classical dendritic cells (cDCs) expressing IRF4 (28).
Monocytes and macrophages are central for both innate and adaptive immunity and are closely associated with the pathogenesis of acute and chronic inflammatory diseases, as well as cancer (29). Recent studies have shown that KLF4 is a critical regulator of monocyte commitment, differentiation, and macrophage activation (30–32). KLF4 is highly expressed in monocytes and in vivo experiments have demonstrated that monocytic differentiation in KLF4-deficient wild-type mice is significantly impaired, with reductions in bone marrow monocytes, as well as decreases in both resident monocytes in the spleen and circulating monocytes. This may be related to the ability of intact KLF4 to transactivate the monocyte-specific CD14 promoter by binding to KLF common sites, thereby promoting monocyte differentiation. Reports indicate that KLF4 is expressed in a monocyte- and stage-specific manner during myelopoiesis. Forced expression of KLF4 out of sequence can induce a mature monocyte phenotype in promyelocytic HL-60 cells, characterized by significant upregulation of myeloid markers CD11b and CD14 and morphological changes in monocytes. In macrophages, Noti and colleagues found that KLF4 regulates the differentiation marker CD11d (33). KLF4 in macrophages is induced by pro-inflammatory stimuli such as IFN-γ, LPS, and TNF-α, while it is suppressed by the anti-inflammatory growth factor TGF-β1. Knockdown of KLF4 reduces the induction of inducible nitric oxide synthase (iNOS) by IFN-γ and/or LPS, while enhancing the response to TGF-β1 and Smad3 signaling. Mechanistically, KLF4 likely activates the iNOS promoter by binding to KLF4-binding sites and interacting with NF-κB family member p65. Additionally, KLF4 inhibits TGF-β1- and Smad3-mediated induction of plasminogen activator inhibitor-1 by competing with Smad3 for binding to the co-activator p300/CBP. In another study, acetylation of KLF4 at lysine residues 225 and 229, mediated by p300/CBP, was found to inhibit KLF4’s ability to activate downstream targets, suggesting that this specific acetylation modification limits KLF4’s transcriptional activation capacity (34). In the RAW264.7 macrophage cell line, KLF4 has been shown to positively regulate the expression of HMGB1, a late-stage inflammatory mediator. The HMGB1 promoter contains two potential KLF4-binding sites, and transfection of KLF4 expression plasmids or KLF4 antisense oligonucleotides into RAW264.7 macrophages increased the expression of HMGB1 in both the nucleus and cytoplasm, while KLF4 deficiency led to reduced HMGB1 expression (35). Finally, research by Alder et al. showed that in KLF4-deficient chimeric mice, circulating inflammatory monocytes (CD115+Gr1+) were almost undetectable (35). CD115+Gr1+ monocytes are capable of differentiating into macrophages and migrating to sites of inflammation, suggesting that the loss of KLF4 affects the generation and migration of pro-inflammatory monocytes. This highlights the critical role of KLF4 in monocyte development and mobilization.
Research indicates that KLF4 plays an important role in B cell development and proliferation of induced B cells as t expression level of Klf4 is low in pro-B cells but progressively increases in pre-B cells (36). KLF4 induces B cell proliferation by regulating cyclin D2 and, in Klf4-deficient mice, the number of pre-B cells in the bone marrow and mature B cells in the spleen is reduced (36). In the absence of Klf4, B cell DNA synthesis decreases while G1 phase cell numbers increase, suggesting that Klf4 deficiency impedes cellular proliferation. Other studies have shown that, in B cell-specific KLF4-deficient mice, the number of pre-B cells in the bone marrow and mature B cells in the spleen is moderately reduced, while bone marrow numbers of pro-B cells remain constant (37). These differing results highlight the functional variability of KLF4 in different environments, emphasizing its multifunctionality. In vitro experiments by Schoenhals and colleagues demonstrated that KLF4 can induce memory B cells to finally differentiate into long-lived quiescent plasma cells. Moreover, overexpression of Klf4 can promote the differentiation of plasma blasts into early plasma cells and long-lived plasma cells by inhibiting apoptosis and upregulating pathways involved in plasma cell differentiation (38).
Neutrophils are the most abundant circulatory leukocytes and are key to both innate and adaptive immune responses (39, 40). Granulocytes, such as neutrophils, and macrophages originate from a common myeloid progenitor (CMP) and granulocyte/macrophage progenitors in the bone marrow (41, 42), suggesting that KLF4 is likely involved in the maturation of immature granulocytes. Research has shown that the overexpression of KLF4 in CMPs promotes monocyte differentiation while almost completely inhibiting the generation of granulocytes, indicating that KLF4 promotes monocyte differentiation while simultaneously inhibiting granulocyte differentiation. In the bipotent promyelocytic cell line HL-60, forced overexpression of KLF4 blocks the granulocyte differentiation-promoting effect of all-trans retinoic acid (used to induce granulocyte differentiation) (32). This finding further supports KLF4’s role in inhibiting granulocyte differentiation. However, literature on the role of KLF4 in granulocytes is currently limited and further research is needed to elucidate the molecular mechanisms involved and to validate its impact on granulopoiesis in vivo.
KLF4 exhibits a context-dependent dual role in tumor biology, functioning as a tumor suppressor in some contexts while acting as an oncogene in others. This duality is largely dependent on specific tumor types (as detailed in Table 1), tumor grade, and stage, as well as its role in modulating tumor microenvironment (TME) responses and therapy-induced immunity. In most solid and hematologic malignancies, KLF4 acts as a tumor suppressor by inhibiting cell proliferation and promoting apoptosis. However, in certain cancers, KLF4 demonstrates oncogenic properties, likely associated with tumor stem cell differentiation. The complex functions of KLF4 in cancer arise from its regulation across multiple molecular levels. At the transcriptional level, KLF4 expression is regulated by epigenetic mechanisms, including promoter methylation, histone modifications, and non-coding RNAs (such as microRNAs and circRNAs), which collectively determine its expression patterns in different tumors (92). For instance, in cancers such as urothelial carcinoma, renal cell carcinoma, medulloblastoma, colorectal adenocarcinoma, and gastric cancer, KLF4 is often silenced due to promoter hypermethylation, which recruits DNA-binding proteins or histone deacetylases (HDACs), leading to chromatin compaction and transcriptional repression (93–97). In contrast, in head and neck squamous cell carcinoma (HNSCC), KLF4 exhibits oncogenic functions through interactions with super-enhancers or other chromatin activation mechanisms (98). At the post-transcriptional level, KLF4 mRNA stability and translation efficiency are primarily regulated by non-coding RNAs, while RNA methylation modifications also play a crucial role, influencing RNA localization, splicing, translation, and degradation, which in turn affect cancer progression (74, 99–101). At the post-translational level, KLF4 undergoes various modifications, such as phosphorylation, acetylation, ubiquitination, and SUMOylation, which impact its stability, subcellular localization, and interactions with co-activators or inhibitors (102, 103). These regulatory mechanisms are context-specific and will be discussed further in the context of various cancers in the following sections.
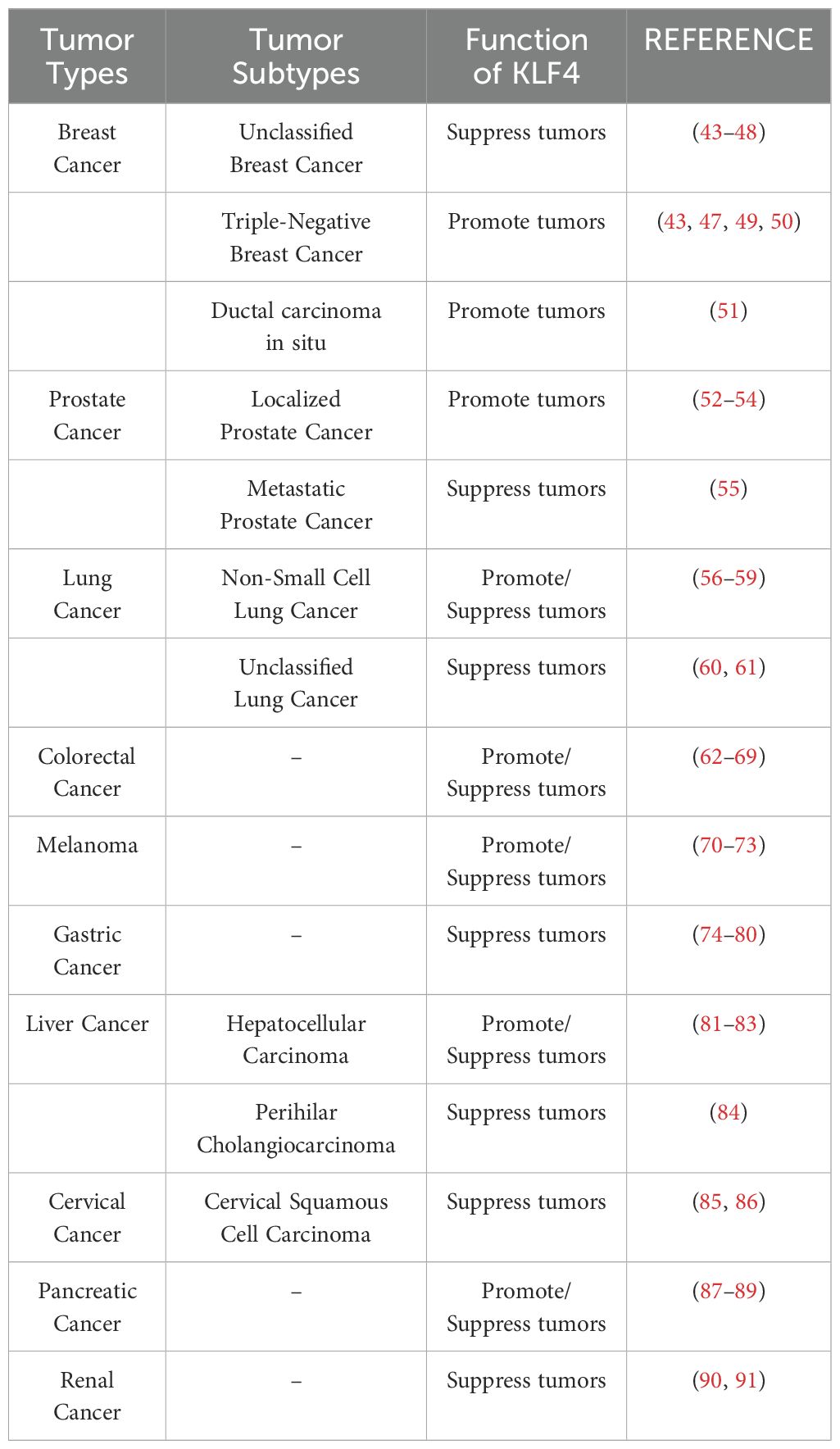
Table 1. The role of KLF4 in major cancers, a summary of the top ten cancers by incidence (based on the latest 2024 SEER database report).
KLF4 role switching in breast cancer may be associated with the stage and subtype of the disease as it has been implicated in promoting the progression of low-grade primary ductal carcinoma (104) while suppressing highly aggressive triple-negative breast cancer (TNBC). As a prognostic marker in TNBC (105), KLF4 inhibits the transcription of EGFR, thereby attenuating downstream signaling pathways (e.g., MAPK and PI3K/AKT pathways). This inhibition suppresses TNBC cell proliferation, migration, and invasion, while also increasing EGFR inhibitor erlotinib sensitivity (43).
The regulation of KLF4 involves multiple upstream factors and downstream targets, affecting its transcription, splicing, and protein degradation processes. Among upstream regulators, AR and DYRK2 promote KLF4 transcription (106), while NFI-C positively regulates its expression (107). Additionally, DDX3X is responsible for splicing the primary transcript of KLF4 (108). Non-coding RNAs such as miR-7 and miR-1233-3p negatively regulate KLF4 expression (44, 109, 110). At the protein level, ATXN3 and FBXO32 modulate KLF4 stability through protein degradation pathways (111, 112).
Regarding downstream targets, KLF4 positively regulates E-cadherin, LASS2, PFKP, and S100A14 (113–116), influencing therapy response and cellular proliferation in breast cancer. These regulatory mechanisms highlight the critical role of KLF4 in determining the biological behavior of breast cancer cells.
KLF4 may suppress the initiation and progression of hepatocellular carcinoma (HCC), with its expression levels closely linked to patient prognosis (117). Moreover, the expression of KLF4 in HCC tissues is generally lower than in adjacent non-tumor tissues. In HCC, KLF4 acts as a transcription factor, directly activating the expression of CD9, CD81, EpCAM, and CD133/Prominin-1 (81, 118) to suppress HCC cell proliferation through the EGFR and JNK signaling pathways. Additionally, KLF4 suppresses the growth and migration of human liver cancer cells by upregulating P-cadherin (CDH3) expression. This inhibitory effect is dependent on the regulation of the GSK-3β signaling pathway (82).
However, KLF4 also demonstrates oncogenic properties in HCC which are closely related to its cancer stem cell (CSC) maintenance capacity. MEOX2 activates KLF4 transcription, further enhancing the stemness traits of CSCs, thereby promoting liver cancer growth and recurrence (83). This contrasting regulatory effect suggests that the role of KLF4 in HCC is complex and may depend on the specific status of its upstream and downstream molecular networks.
KLF4 plays a crucial role in the development and progression of gastric cancer and its expression is influenced by multiple regulatory factors. In gastric cancer, KLF4 acts as a tumor suppressor. For instance, KLF4 upregulates SAMHD1, inhibiting the MAPK/p38 signaling pathway, thereby suppressing the progression of gastric cancer (75). Additionally, KLF4 exerts tumor-suppressive effects by inhibiting the transcription of CXCL8 (76). Among KLF4’s downstream target genes, iASPP, PODXL, and STK33 are negatively regulated by KLF4 (77, 119, 120), and these genes are closely associated with gastric cancer progression and prognosis.
The regulation of KLF4 is influenced by several upstream factors. CagA and TET1 inhibit KLF4 expression by methylating its promoter region (121). The transcription factor STAT5A further suppresses KLF4 transcription, promoting tumor progression, but KLF4 overexpression can counteract the oncogenic effects of STAT5A (74). Additionally, some studies suggest that KLF4 overexpression increases the sensitivity of gastric cancer cells to cisplatin and TRAIL, promoting apoptosis through DR4/DR5 upregulation, enhancing cell death induced by these treatments (78).
KLF4 is also regulated by various non-coding RNAs, since miR-32, miR-103, miR-135b-5p, and miR-155 negatively regulate KLF4 expression (79, 122–125), while LINC00673 also negatively modulates KLF4 expression (126). In contrast, the long, non-coding RNA SNHG5 positively regulates KLF4 expression and promotes gastric cancer development (122). These complex regulatory mechanisms highlight the multifaceted role of KLF4 in gastric cancer biology.
Although substantial research supports the tumor-suppressive role of KLF4, its function in colorectal cancer (CRC) remains controversial. In colon cancer, KLF4 reduces autophagy in cancer cells by inhibiting RAB26 expression while promoting apoptosis and enhancing response to 5-fluorouracil (5-FU) (127). The RNA-binding protein MEX3A sustains cancer cells in an undifferentiated and proliferative state, enhancing radiotherapy resistance by suppressing KLF4 expression and activating the WNT signaling pathway (62). Additionally, KLF4 regulates chemoresistance in CRC. In oxaliplatin-resistant CRC cells, reduced KLF4 expression weakens its transcriptional repression of PiHL, resulting in PiHL upregulation and activation of the EZH2/HMGA2/PI3K/Akt signaling pathway to promote oxaliplatin resistance (63). Conversely, KLF4 expression sensitizes cancer cells to cisplatin cytotoxicity, likely through the KLF4-HMGB1/hTERT signaling axis (128).
Interestingly, KLF4 exhibits opposing roles depending on regulation by TCFL5 isoforms. TCFL5-E8 reduces KLF4 mRNA levels, potentially suppressing its tumor-promoting effects while CHA upregulates KLF4, possibly facilitating tumor progression (64). The role of miRNAs in CRC has been extensively reviewed (129), with mechanisms primarily involving the regulation of tumor proliferation, metastasis, and angiogenesis through KLF4 targeting. For example, miR-25-3p not only targets KLF4 in CRC cells but also transfers through exosomes to endothelial cells, suppressing both KLF4 and KLF2 expression (130). Conversely, miR-7-5p inhibits cancer stem cell (CSC) traits and enhances radiosensitivity in CRC by regulating KLF4 (65).
KLF4 plays an important role in non-solid tumors (e.g., lymphoma and leukemia), with its functions again dependent on tumor type and co-regulatory mechanisms. In B-cell lymphoma and various leukemia lineages, KLF4 expression is markedly reduced, potentially due to promoter hypermethylation observed in several hematologic malignancies (131, 131). This epigenetic alteration, characterized by DNA methylation, shown in Figure 3, is associated with KLF4 downregulation and gene silencing as observed in pediatric patients with T-cell acute lymphoblastic leukemia (T-ALL), where the loss of KLF4 significantly promotes NOTCH1-driven T-ALL development (132). In contrast, KLF4 expression is strongly upregulated in multiple myeloma cell lines (133). Besides epigenetic regulation, factors such as apoptosis, proliferation control, and microenvironmental influences significantly impact KLF4’s role in hematologic malignancies. Low KLF4 expression, on the other hand, may play a critical role in resisting apoptosis and promoting uncontrolled proliferation. One important pro-apoptotic KLF4 mechanism involves BAK1 activation, which occurs independently of caspases. In pre-B cell leukemia models, KLF4 enhances apoptosis by suppressing anti-apoptotic proteins BCL2L1/BCL-xL (134). Additionally, in cHL, KLF4 is repressed by Notch1 and lacks activation by PU.1, further amplifying its suppression (31, 135, 136).
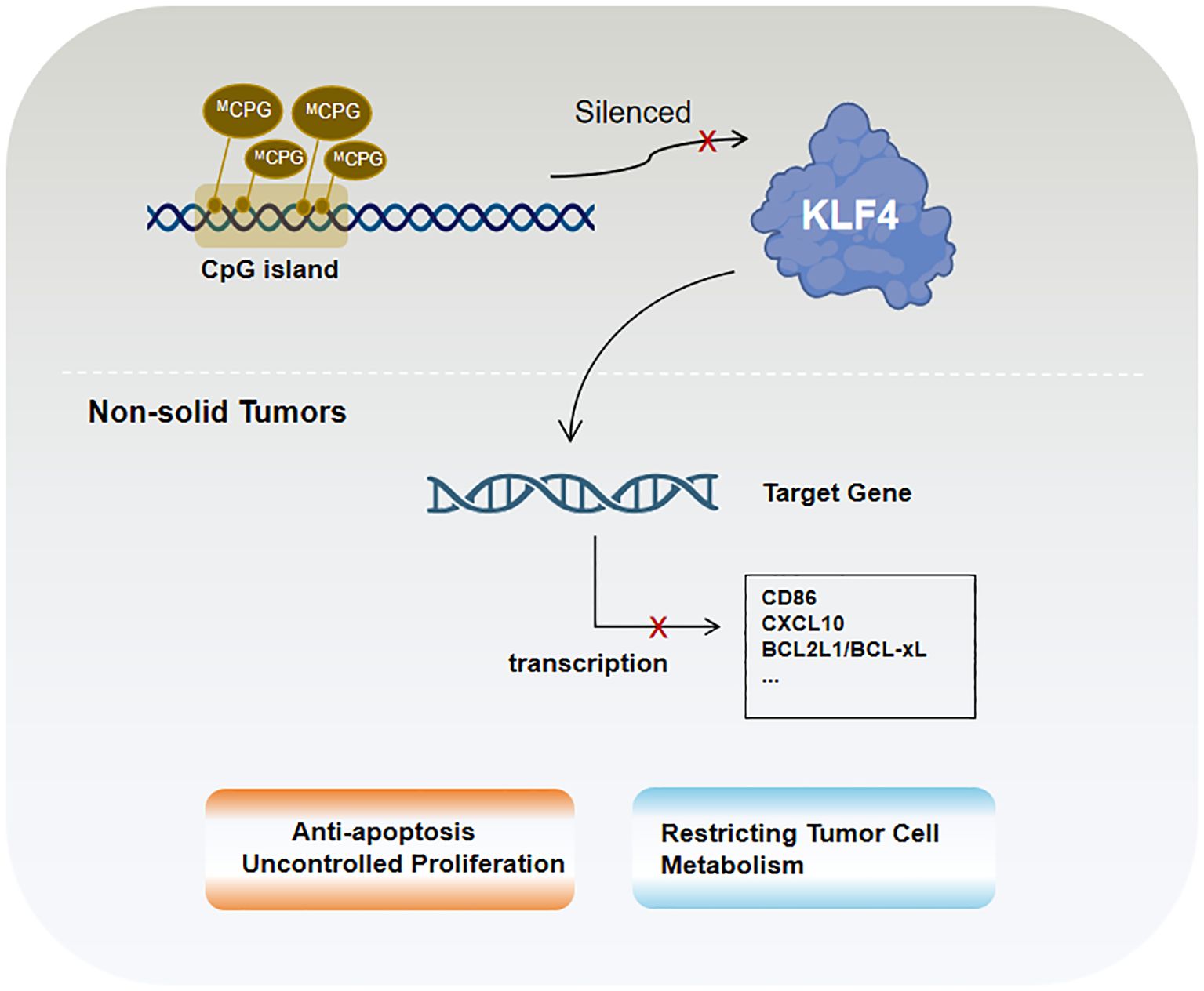
Figure 3. KLF4 in non-solid tumors is primarily regulated by epigenetic mechanisms, particularly CpG island methylation.
In B-cell acute lymphoblastic leukemia (B-ALL) mouse models, KLF4 inhibits ABL-induced pre-B cell transformation by inducing apoptosis and CDKN1A-mediated cell cycle arrest. Studies have shown that co-treatment with the tyrosine kinase inhibitor imatinib and KLF4 overexpression enhances apoptotic effects, suggesting their synergistic role in promoting tumor cell death. KLF4 also negatively regulates CD86 and CXCL10 (137), thereby reducing the recruitment of bystander immune cells, such as T lymphocytes, to tumor cells. Furthermore, KLF4 interacts with other critical factors in hematologic malignancies. In Burkitt lymphoma (BL), KLF4 acts as a tumor suppressor by reducing MYC expression and its downstream target EZH2 while activating tumor suppressors such as TXNIP (137). These effects collectively decrease reactive oxygen species (ROS)-induced damage and constrain tumor cell metabolism. In most cases of acute myeloid leukemia (AML), the homeobox transcription factor CDX2 promotes leukemogenesis by suppressing KLF4 (137). Moreover, in follicular lymphoma (FL), diffuse large B-cell lymphoma (DLBCL), and BL, low KLF4 expression is likely a prerequisite for high MSC/ABF-1 expression. MSC/ABF-1 inhibits E47/E12 transcriptional activity, silencing B-cell-specific gene expression programs and further enhancing tumor cell proliferation (133).
The interplay between tumors and the immune system is highly complex. On one hand, the immune system plays an anti-tumor role by recognizing and eliminating abnormal cells, including cancer cells. On the other hand, tumors can evade immune surveillance and attack by utilizing immune escape mechanisms and modulating immune checkpoints. The role of KLF4 in anti-tumor immunity is crucial and deserves special attention as studies have shown that, in hepatocellular carcinoma (HCC), KLF4 is significantly associated with CD8+ T cells, Th1 cells, dendritic cells (DCs), B cells, natural killer (NK) cells, macrophages, and their molecular markers (138). KLF4 plays an important role in regulating the differentiation and function of CD8+ T cells, influencing their ability to recognize and kill tumor cells. Additionally, KLF4 is closely related to macrophage polarization, regulating their polarization toward the pro-inflammatory M1 type or the anti-inflammatory M2 type and thereby affecting tumor immune escape and progression. KLF4 is also involved in regulating inflammatory responses, determining the immune status of the TME by balancing pro-inflammatory and anti-inflammatory factors. These actions reveal the complex role of KLF4 in tumor immunity, warranting further investigation.
CD8+ T cells play a critical role in tumor immunity. As cytotoxic T lymphocytes, they can directly recognize and kill tumor cells, thus exerting anti-tumor effects. CD8+ T cells play an essential defense role in immune surveillance of tumors (139) and in many types of cancer, CD8+ T cell infiltration correlates with improved patient survival. Given that KLF4 has been shown to significantly influence CD8+ T cell proliferation and differentiation in normal T cell biology, it raises the question of whether KLF4 similarly affects CD8+ T cell differentiation in the tumor microenvironment (TME), thereby impacting anti-tumor efficacy. Studies have indicated that in HCC patients with high KLF4 expression, the tumor immune microenvironment (TIME) is characterized by increased CD8+ T cell infiltration, suggesting a potential correlation between KLF4 expression and CD8+ T cell infiltration (138).
Immune-suppressive factors present in the TME, such as TGF-β, IL-10, and tumor-associated macrophages (TAMs), indirectly affect the anti-tumor capacity of CD8+ T cells. In prostate cancer, during T cell receptor activation, KLF4-deficient TAMs inhibit the expression of ELF4 and KLF4, releasing initial CD8+ T cells from quiescence and activating pro-inflammatory and pro-atherosclerotic pathways, leading to CD8+ T cell proliferation and enhanced immune response that suppresses prostate cancer growth (140). The removal of CD8+ T cells eliminates the inhibitory effects of KLF4 deficiency on prostate cancer growth. The specific molecular mechanism may involve mTOR, which inhibits KLF4 expression via downstream ERK and PI3K signaling. Additionally, research has shown that KLF4 suppresses the recruitment of CD8+ T cells to the TME via the Hedgehog signaling pathway, further contributing to immune suppression in the TME, likely by inhibiting the production of C-X-C Motif Chemokine Ligand 9 (CXCL9) and CXCL10 in TAMs (141).
CD8+ T cell exhaustion is a phenomenon observed during chronic inflammatory responses in persistent viral infections or cancer (142–144). Exhausted CD8+ T cells, due to prolonged exposure to antigens and inflammatory stimuli, gradually lose proliferation, cytokine secretion, and cytotoxic killing capacities. Exhaustion is regulated by the upregulation of several inhibitory receptors, such as PD-1, LAG-3, and TIM-3, which suppress T cell function (145–147). Tumor cells contribute to the inhibition and exhaustion of CD8+ T cells by upregulating immune checkpoint molecules, antigen loss, and altering the tumor microenvironment (148). KLF4 is closely associated with genes regulating immune checkpoints, particularly those involved in T cell exhaustion, as elevated levels of immune checkpoint molecules in the TME may increase KLF4 expression, resulting in diminished T cell function and promoting exhaustion. Additionally, some studies have indicated that KLF4 can activate and reactivate exhausted CD8+ T cells, promoting their differentiation into transient effector subpopulations and enhancing their anti-tumor activity (149). In melanoma, immunotherapy targeting the PD-1 receptor can upregulate KLF4 expression, thereby improving the survival rate of melanoma patients. The underlying mechanism may involve an increase in KLF4 expression on CD8 tumor-infiltrating lymphocytes (TILs) and an elevated number of transient effector CD8+ T cells expressing KLF4. In tumor-specific CD8+ T cells, high KLF4 expression upregulates AP-1 family factors, such as c-Jun, conferring transient effector characteristics upon CD8+ T cells (150–152). Research has demonstrated that c-Jun and BATF (basic leucine zipper ATF-like transcription factor), enhance CAR T cell effector function and even confers resistance to T cell exhaustion (152). This is consistent with KLF4’s role in upregulating c-Jun, underscoring KLF4 as critical for maintaining CD8+ effector function. In tumor-specific CD8+ T cells, KLF4 enhances effector function by regulating the expression of AP-1 family factors, such as c-Jun, thereby counteracting terminal exhaustion. The absence of KLF4 impairs CD8+ T cell differentiation and function, making them more susceptible to exhaustion and weakening the anti-tumor response.
In addition to CD8+ T cells, macrophage content is significantly higher in the tumor tissues of HCC patients with high KLF4 expression compared to low KLF4 expression (138). Tumor-associated macrophages (TAMs) can be classified into M1 and M2 types: M1 macrophages exhibit pro-inflammatory characteristics, activating immune responses and killing tumor cells, while M2 macrophages tend to be anti-inflammatory, promoting tumor growth and metastasis. Thus, macrophages play a dual role in tumor immunity, with their function depending on their polarization state (153, 154). KLF4 influences macrophage polarization toward either the M1 or M2 phenotype, acting as a key regulatory factor that not only induces M2 genetic programming but also maintains the balance by inhibiting M1 target gene expression. Additionally, KLF4 plays an essential role in maintaining the immune rhythm of macrophages; changes in its expression levels can therefore negatively affect immune function, leading to tumorigenesis (155).
Research has demonstrated KLF4 regulation of M2 polarization through its SUMOylation status, with specific protease SUMO-specific Protease 1 (SENP1) responsible for de-SUMOylating KLF4. This process drives macrophage polarization toward the M1 type, with the SENP1-KLF4 axis playing a key role in conjunction with the NF-κB signaling pathway. Moreover, KLF4 cooperates with Stat6 to induce M2 genetic programming and inhibits M1 target gene expression by sequestering co-activators necessary for NF-κB activation (156). In the Hedgehog signaling pathway, tumor cells secrete sonic hedgehog (SHH), a Hedgehog ligand that drives TAMs toward M2 polarization (156).
In diverse tumor types, KLF4 expression is closely associated with macrophage polarization. In liver cancer, miR-206 targets KLF4, suppressing its expression and promoting M1 macrophage polarization, thereby effectively preventing hepatocellular carcinoma. This mechanism may involve the KLF4/C-C Motif Chemokine Ligand 2 (CCL2)/C-C Motif Chemokine Receptor 2(CCR2) axis (157). In gastric cancer, exosomes derived from gastric cancer cells carry the long non-coding RNA (lncRNA) HCG18, which reduces the levels of miR-875-3p, thereby increasing KLF4 expression and promoting M2 macrophage polarization (158). In non-small cell lung cancer (NSCLC), KLF4 expression is upregulated and closely associated with macrophage infiltration and M2 polarization, while miR-34a-5p targets KLF4 and reverses macrophage polarization, regulating immune molecules such as IL-1β, thus showing therapeutic potential for NSCLC (58).
Furthermore, KLF4 plays an important role in inhibiting pro-inflammatory genes in M2 macrophages. Its absence weakens IL-4’s ability and suppresses M1 target genes. In immune-related thrombocytopenia (ITP), low-dose DAC promotes M2 macrophage polarization and inhibits inflammatory responses by enhancing KLF4’s binding to the PPARγ promoter (159). Overall, since KLF4 plays a crucial role in regulating macrophage polarization and function, understanding its mechanisms will contribute to the development of new tumor therapeutic strategies. The molecular mechanism by which KLF4 regulates macrophage polarization is shown in Figure 4.
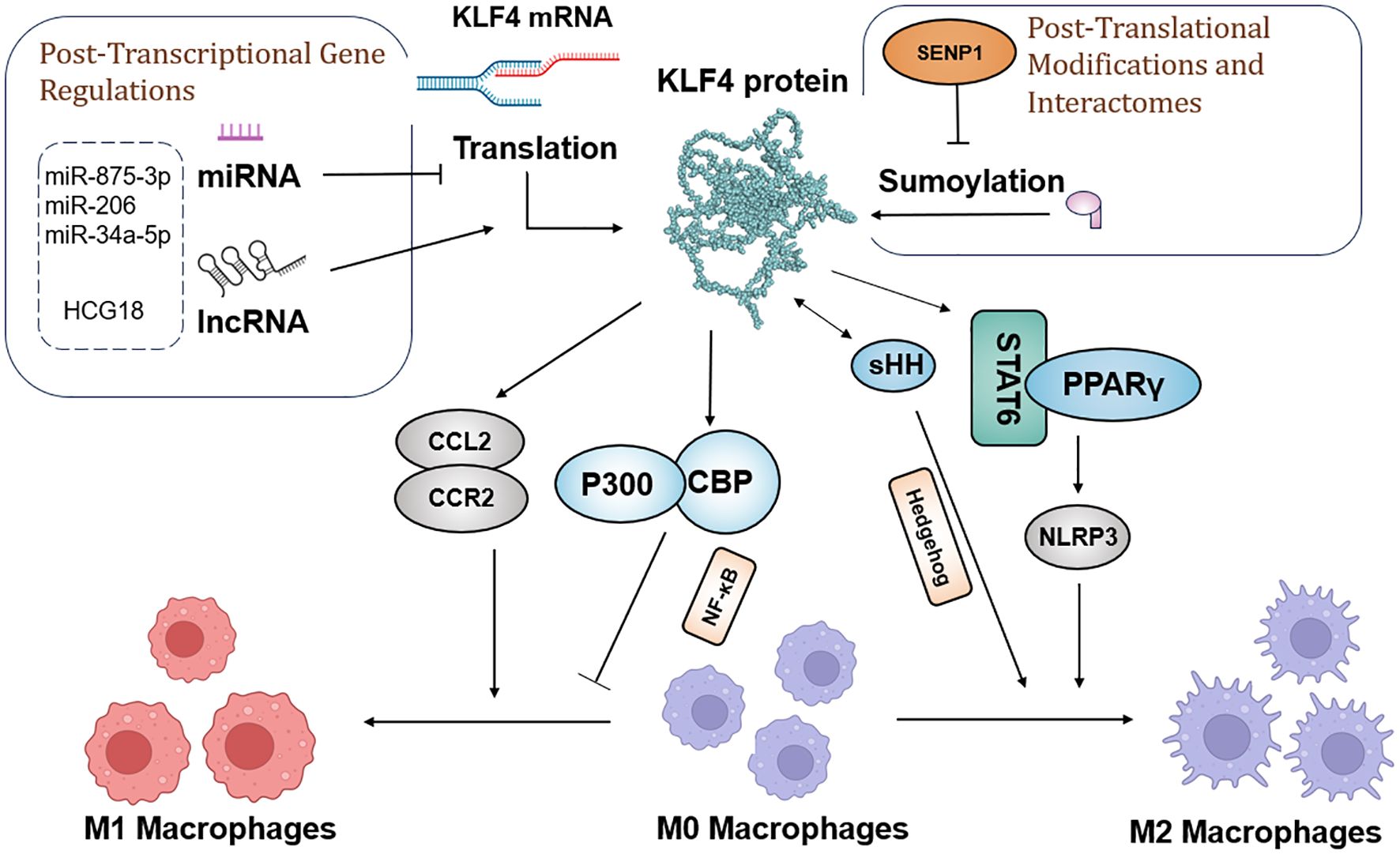
Figure 4. Schematic model of the molecular mechanisms by which KLF4 regulates macrophage polarization in the tumor environment, before and after transcription and translation.
Macrophage activation plays a central role in the onset and progression of chronic inflammation. Pro-inflammatory factors such as interferon-γ (IFN-γ), lipopolysaccharide (LPS), and tumor necrosis factor-α (TNF-α) promote macrophage activation, while anti-inflammatory factors like TGF-β suppress inflammation. This regulatory mechanism is crucial in both inflammatory diseases and cancer progression, with KLF4 playing a significant role in regulating macrophage pro-inflammatory responses, potentially acting as a key target in the TGF-β1/Smad3 signaling pathway (160). Studies have shown that KLF4 is essential for the expression of the macrophage activation marker iNOS, induced by IFN-γ or LPS, a process partially mediated by KLF4’s interaction with p65 (RelA), a member of the NF-κB family (30). Additionally, KLF4 is an IFN-γ target gene in cancer cell lines, with its function mediated by Stat1. Further research by Tetreault et al. demonstrated that KLF4 overexpression in mouse esophageal epithelial cells activates pro-inflammatory factors such as TNF-α, CXCL5, Granulocyte Colony-Stimulating Factor(G-CSF), and IL-1α, a process that also relies on NF-κB, ultimately leading to the development of esophageal squamous cell carcinoma (161). Hp infection can induce the expression of miR-135-5b by activating the pro-inflammatory NF-κB signaling pathway. miR-135-5b binds to the 3’ UTR of the Klf4 gene, leading to reduced Klf4 expression and promoting the development of gastric cancer (124).
Studies have shown that KLF4 activity can be modulated by exogenous drugs, indicating novel therapeutic approaches for cancer treatment. Several studies have reported that known compounds exert antitumor effects by acting on KLF4 in diverse types of cancer. For instance, in triple-negative breast cancer, the small molecule inhibitor WX2-43 blocks KLF4 methylation by inhibiting the Protein Arginine Methyltransferase 5 (PRMT5) -KLF4 interaction, showing significant antitumor efficacy (162). Kenpaullone inhibits KLF4 by suppressing CDK1/cyclin B and GSK-3β, reducing proliferation and migration of breast cancer cells in canine mammary cancer and inducing cancer cell death, although its effects in humans require further investigation (163). In colorectal cancer, sulforaphane induces KLF4 and enhances KLF4-p21 signaling, contributing to the inhibition of cancer cell growth and promoting differentiation (164). Regorafenib, a multi-kinase inhibitor, significantly suppresses tumor growth and angiogenesis in hepatocellular carcinoma (HCC), with effects similar to those of DC-101 (an anti-VEGFR2 antibody). However, Regorafenib is more effective in activating T cells and promoting M1 macrophage polarization. It also reverses M2 macrophage polarization by inhibiting p38 kinase phosphorylation and downstream Creb1/KLF4 activity (165). The small molecule compound APTO-253, which induces KLF4, was confirmed to have antitumor activity in a 2015 Phase I multicenter study (166). Recent research has also demonstrated that APTO-253 enhances immune responses in leukemia, rendering cancer cells more susceptible to NK cell-mediated termination. Increased KLF4 activity leads to elevated MICA expression on AML cells which, in turn, strengthens NK cell-mediated immune surveillance (167). KLF4 is also a downstream protein of Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) and its expression can enhance PPARγ levels. Pioglitazone, a PPARγ agonist, stabilizes KLF4 protein by activating the AKT pathway and reducing KLF4 ubiquitination, suggesting its potential as a KLF4-targeting drug (168, 169). Decitabine can enhance KLF4’s binding to the regulatory region of PPARγ, potentially increasing PPARγ transcriptional activity. Additionally, as a KLF4 methylation inhibitor, decitabine upregulates KLF4 expression by inhibiting methylation of the KLF4 promoter, possibly through suppression of the EMT pathway (170). Moreover, KLF4 can sensitize cancer cells to several established therapies, including cisplatin (124, 128, 171–173), cetuximab (22), mesalazine (174), and gefitinib (175), suggesting the potential for combinatory use with other anticancer drugs in future clinical trials. In summary, drugs targeting KLF4 are gradually being discovered for their potential to impact malignant tumors, but further mechanistic studies and clinical validation are necessary. Drugs targeting KLF4 and their mechanisms are listed in Table 2.
Previous reports indicate that KLF4 and its interacting molecules and pathways are involved in various types of cancer, leading to a reasonable hypothesis that KLF4 can serve as a potential target for cancer therapy. However, most experimental results have been based on simple assays performed at either the RNA or protein level. While KLF4 may exhibit high expression at the transcriptional level, its protein expression may not correspond due to various regulatory mechanisms, leading to the mistaken assumption that it has an inhibitory (or promotive) effect. Additionally, the lack of a ligand-binding domain renders the development of targeted drugs particularly challenging. Nevertheless, using KLF4 as a biomarker for tumor diagnosis and prognosis presents a promising option. However, given that KLF4 displays divergent, and at times even completely opposite, expression patterns in different stages and environments of tumors, its specific role in clinical and pathological grading and staging of cancer requires further exploration.
This article focuses on KLF4’s functions in various immune cells and its relationship with cancer and immunity. KLF4 exerts profound effects on multiple immune cells under physiological conditions and, in tumors, it primarily influences progression by regulating T cell subset differentiation and macrophage polarization states to regulate therapy resistance. In recent years, immunotherapy has made remarkable strides and has become a breakthrough in cancer treatment by activating or modulating the immune response to recognize and attack tumor cells. Notably, Adoptive Cell Transfer (ACT) and ICI therapies have made significant progress in both solid and hematologic malignancies by either directly modifying and expanding autologous T cells or relieving tumor-induced T cell suppression, thereby actively or passively modulating the immune response. This underscores the potential of leveraging immune cells, particularly T cells, to eliminate tumor cells. Moreover, the immunological roles of KLF4 in cancer help clarify the interaction between innate and adaptive immunity within the tumor microenvironment, providing new opportunities for developing strategies to further enhance antitumor immunity.
YJ: Writing – original draft. WX: Writing – original draft. BM: Writing – review & editing. YS: Writing – review & editing, Conceptualization, Supervision.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Siegel PM, Massagué J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. (2003) 3:807–21. doi: 10.1038/nrc1208
2. Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. (2003) 3:756–67. doi: 10.1038/nrc1186
3. Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. (2005) 5:376–87. doi: 10.1038/nrc1607
4. Dannenberg JH, te Riele HP. The retinoblastoma gene family in cell cycle regulation and suppression of tumorigenesis. Results Probl Cell Differ. (2006) 42:183–225. doi: 10.1007/400_002
5. Roninson IB. Oncogenic functions of tumour suppressor p21(Waf1/Cip1/Sdi1): association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett. (2002) 179:1–14. doi: 10.1016/s0304-3835(01)00847-3
6. Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. (2020) 5:8. doi: 10.1038/s41392-020-0110-5
7. Dang DT, Pevsner J, Yang VW. The biology of the mammalian Krüppel-like family of transcription factors. Int J Biochem Cell Biol. (2000) 32:1103–21. doi: 10.1016/s1357-2725(00)00059-5
8. Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem. (1996) 271:20009–17. doi: 10.1074/jbc.271.33.20009
9. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. (2001) 414:105–11. doi: 10.1038/35102167
10. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. (2017) 23:1124–34. doi: 10.1038/nm.4409
11. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. (2021) 596:583–9. doi: 10.1038/s41586-021-03819-2
12. Varadi M, Bertoni D, Magana P, Bordin N, Meyer A, Valkov E, et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. (2024) 52:D368–d375. doi: 10.1093/nar/gkad1011
13. Yu K, Zheng B, Han M, Wen JK. ATRA activates and PDGF-BB represses the SM22α promoter through KLF4 binding to, or dissociating from, its cis-DNA elements. Cardiovasc Res. (2011) 90:464–74. doi: 10.1093/cvr/cvr017
14. Schuetz A, Nana D, Rose C, Zocher G, Milanovic M, Koenig B, et al. The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell Mol Life Sci. (2011) 68:3121–31. doi: 10.1007/s00018-010-0618-x
15. Shields JM, Yang VW. Two potent nuclear localization signals in the gut-enriched Krüppel-like factor define a subfamily of closely related Krüppel proteins. J Biol Chem. (1997) 272:18504–7. doi: 10.1074/jbc.272.29.18504
16. Yet SF, McA’Nulty MM, Folta SC, Yen J, Yoshizumi M, Chin MT, et al. Humana EZF. Krüppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J Biol Chem. (1998) 273:1026–31. doi: 10.1074/jbc.273.2.1026
17. Geiman DE, Ton-That H, Johnson JM, Yang VW. Transactivation and growth suppression by the gut-enriched Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res. (2000) 28:1106–13. doi: 10.1093/nar/28.5.1106
18. Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. (2006) 27:23–31. doi: 10.1093/carcin/bgi243
19. Wei D, Wang L, Yan Y, Jia Y, Guo W, Wang Y, et al. KLF4 is essential for induction of cellular identity change and acinar-to-ductal reprogramming during early pancreatic carcinogenesis. Cancer Cell. (2016) 29:324–38. doi: 10.1016/j.ccell.2016.02.005
20. Wang X, Xie X, Ni JY, Li J, Zhang Y, Chen H, et al. USP11 promotes renal tubular cell pyroptosis and fibrosis in UUO mice via inhibiting KLF4 ubiquitin degradation. Acta Pharmacol Sin. (2024). doi: 10.1038/s41401-024-01363-z
21. Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. (2010) 3:ra3. doi: 10.1126/scisignal.2000475
22. Ye S, Hu X, Ni C, Li X, Wang J, Zhang Y, et al. KLF4 p.A472D mutation contributes to acquired resistance to cetuximab in colorectal cancer. Mol Cancer Ther. (2020) 19:956–65. doi: 10.1158/1535-7163.Mct-18-1385
23. Cao Z, Sun X, Icli B, Wara AK, Feinberg MW. Role of Kruppel-like factors in leukocyte development, function, and disease. Blood. (2010) 116:4404–14. doi: 10.1182/blood-2010-05-285353
24. An J, Golech S, Klaewsongkram J, Zhang Y, Subedi K, Huston GE, et al. Krüppel-like factor 4 (KLF4) directly regulates proliferation in thymocyte development and IL-17 expression during Th17 differentiation. FASEB J. (2011) 25:3634–45. doi: 10.1096/fj.11-186924
25. Yamada T, Park CS, Mamonkin M, Lacorazza HD. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Krüppel-like factors KLF4 and KLF2. Nat Immunol. (2009) 10:618–26. doi: 10.1038/ni.1730
26. Mamonkin M, Shen Y, Lee PH, Puppi M, Park CS, Lacorazza HD. Differential roles of KLF4 in the development and differentiation of CD8+ T cells. Immunol Lett. (2013) 156:94–101. doi: 10.1016/j.imlet.2013.09.008
27. Hao Z, Sheng Y, Duncan GS, Su Y, Li WY, Li Q, et al. K48-linked KLF4 ubiquitination by E3 ligase Mule controls T-cell proliferation and cell cycle progression. Nat Commun. (2017) 8:14003. doi: 10.1038/ncomms14003
28. Tussiwand R, Everts B, Grajales-Reyes GE, Kretzer NM, Iwata A, Bagaitkar J, et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. (2015) 42:916–28. doi: 10.1016/j.immuni.2015.04.017
29. Hao NB, Lü MH, Fan YH, Cao YL, Zhang ZR, Yang SM. Macrophages in tumor microenvironments and the progression of tumors. Clin Dev Immunol. (2012) 2012:948098. doi: 10.1155/2012/948098
30. Feinberg MW, Cao Z, Wara AK, Lebedeva MA, SenBanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. (2005) 280:38247–58. doi: 10.1074/jbc.M509378200
31. Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. (2007) 26:4138–48. doi: 10.1038/sj.emboj.7601824
32. Alder JK, Georgantas RW 3rd, Hildreth RL, Brinkman DC, Hargreaves DC, Hasan W, et al. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol. (2008) 180:5645–52. doi: 10.4049/jimmunol.180.8.5645
33. Noti JD, Johnson AK, Dillon JD. The leukocyte integrin gene CD11d is repressed by gut-enriched Kruppel-like factor 4 in myeloid cells. J Biol Chem. (2005) 280:3449–57. doi: 10.1074/jbc.M412627200
34. Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. (2007) 282:33994–4002. doi: 10.1074/jbc.M701847200
35. Liu J, Liu Y, Zhang H, Chen G, Wang K, Xiao X. KLF4 promotes the expression, translocation, and releas eof HMGB1 in RAW264.7 macrophages in response to LPS. Shock. (2008) 30:260–6. doi: 10.1097/shk.0b013e318162bef7
36. Klaewsongkram J, Yang Y, Golech S, Katz J, Kaestner KH, Weng NP. Krüppel-like factor 4 regulates B cell number and activation-induced B cell proliferation. J Immunol. (2007) 179:4679–84. doi: 10.4049/jimmunol.179.7.4679
37. Yusuf I, Kharas MG, Chen J, Peralta RQ, Wang Y, Rosen M, et al. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. (2008) 20:671–81. doi: 10.1093/intimm/dxn024
38. Schoenhals M, Jourdan M, Seckinger A, Rème T, Robert N, Caraux A, et al. Forced KLF4 expression increases the generation of mature plasma cells and uncovers a network linked with plasma cell stage. Cell Cycle. (2016) 15:1919–28. doi: 10.1080/15384101.2016.1191709
39. Hume DA. The mononuclear phagocyte system. Curr Opin Immunol. (2006) 18:49–53. doi: 10.1016/j.coi.2005.11.008
40. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. (2008) 8:958–69. doi: 10.1038/nri2448
41. Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. (2007) 26:726–40. doi: 10.1016/j.immuni.2007.06.004
42. Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. (2000) 404:193–7. doi: 10.1038/35004599
43. Roberts MS, Anstine LJ, Finke VS, Maharjan P, Brattain MG, Wang J, et al. KLF4 defines the efficacy of the epidermal growth factor receptor inhibitor, erlotinib, in triple-negative breast cancer cells by repressing the EGFR gene. Breast Cancer Res. (2020) 22:66. doi: 10.1186/s13058-020-01305-7
44. Lu M, Wu Y, Zeng B, Zhang Q, Wang X, Wang J, et al. CircEHMT1 inhibits metastatic potential of breast cancer cells by modulating miR-1233-3p/KLF4/MMP2 axis. Biochem Biophys Res Commun. (2020) 526:306–13. doi: 10.1016/j.bbrc.2020.03.084
45. Chen D, Yuan M, Ye Q, Zhou H, Sun W, Zhang J, et al. Cyanidin-3-O-glucoside inhibits epithelial-to-mesenchymal transition, and migration and invasion of breast cancer cells by upregulating KLF4. Food Nutr Res. (2020) 64. doi: 10.29219/fnr.v64.4240
46. Zhang L, Zhou Q, Qiu Q, Xu J, Zhang X, Wang X, et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer. (2019) 18:144. doi: 10.1186/s12943-019-1080-5
47. Zhou Y, Yang J, Chen C, Zhang W, Liu A, Zhao H, et al. Polyphyllin III-Induced Ferroptosis in MDA-MB-231 Triple-Negative Breast Cancer Cells can Be Protected Against by KLF4-Mediated Upregulation of xCT. Front Pharmacol. (2021) 12:670224. doi: 10.3389/fphar.2021.670224
48. Zhao Q, Shang Y, Lü J, Chen L, Wang P, Zhang H, et al. miR-29a-KLF4 signaling inhibits breast tumor initiation by regulating cancer stem cells. Int Immunopharmacol. (2024) 130:111797. doi: 10.1016/j.intimp.2024.111797
49. Wang R, Yu W, Zhu T, Li M, Chen Y, Liu H, et al. MED27 plays a tumor-promoting role in breast cancer progression by targeting KLF4. Cancer Sci. (2023) 114:2277–92. doi: 10.1111/cas.15757
50. Jung E, Lee YH, Ou S, Kim TY, Shin SY. EGR1 regulation of vasculogenic mimicry in the MDA-MB-231 triple-negative breast cancer cell line through the upregulation of KLF4 expression. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms241814375
51. Wei Y, Li H, Qu Q. miR-484 suppresses endocrine therapy-resistant cells by inhibiting KLF4-induced cancer stem cells in estrogen receptor-positive cancers. Breast Cancer. (2021) 28:175–86. doi: 10.1007/s12282-020-01152-6
52. Qu G, Zhang Y, Duan H, Li X, Wang J, Chen Z, et al. ARPC5 is transcriptionally activated by KLF4, and promotes cell migration and invasion in prostate cancer via up-regulating ADAM17: ARPC5 serves as an oncogene in prostate cancer. Apoptosis. (2023) 28:783–95. doi: 10.1007/s10495-023-01827-3
53. Jiang Z, Zhang Y, Chen X, Wu P, Chen D. Long non-coding RNA LINC00673 silencing inhibits proliferation and drug resistance of prostate cancer cells via decreasing KLF4 promoter methylation. J Cell Mol Med. (2020) 24:1878–92. doi: 10.1111/jcmm.14883
54. Feng F, Liu H, Chen A, Zhang X, Wang Y, Li J, et al. miR-148-3p and miR-152-3p synergistically regulate prostate cancer progression via repressing KLF4. J Cell Biochem. (2019) 120:17228–39. doi: 10.1002/jcb.28984
55. Tassone E, Bradaschia-Correa V, Xiong X, Chen S, Lin Z, Li W, et al. KLF4 as a rheostat of osteolysis and osteogenesis in prostate tumors in the bone. Oncogene. (2019) 38:5766–77. doi: 10.1038/s41388-019-0841-3
56. Ma X, Wang L, Shi G, Sun S. The deubiquitinase OTUD1 inhibits non-small cell lung cancer progression by deubiquitinating and stabilizing KLF4. Thorac Cancer. (2022) 13:761–70. doi: 10.1111/1759-7714.14320
57. Lu G, Yao Y, Zhang X, Cui D, Zhou J. Deguelin attenuates non-small-cell lung cancer cell metastasis by upregulating PTEN/KLF4/EMT signaling pathway. Dis Markers. (2022) 2022:4090346. doi: 10.1155/2022/4090346
58. Arora S, Singh P, Ahmad S, Kumar S, Sharma P, Verma R, et al. Comprehensive integrative analysis reveals the association of KLF4 with macrophage infiltration and polarization in lung cancer microenvironment. Cells. (2021) 10. doi: 10.3390/cells10082091
59. Tuo Z, Liang L, Zhou R. LINC00852 is associated with poor prognosis in non-small cell lung cancer patients and its inhibition suppresses cancer cell proliferation and chemoresistance via the hsa-miR-145-5p/KLF4 axis. J Gene Med. (2021) 23:e3384. doi: 10.1002/jgm.3384
60. Zhuan B, Lu Y, Chen Q, Wang X, Zhang H, Li J, et al. Overexpression of the long noncoding RNA TRHDE-AS1 inhibits the progression of lung cancer via the miRNA-103/KLF4 axis. J Cell Biochem. (2019) 120:17616–24. doi: 10.1002/jcb.29029
61. Yu T, Chen X, Zhang W, Li J, Gao F, Wang Y, et al. KLF4 regulates adult lung tumor-initiating cells and represses K-Ras-mediated lung cancer. Cell Death Differ. (2016) 23:207–15. doi: 10.1038/cdd.2015.85
62. Yang X, Li G, Tian Y, Zhou J, Wang C, Liu M, et al. Identifying the E2F3-MEX3A-KLF4 signaling axis that sustains cancer cells in undifferentiated and proliferative state. Theranostics. (2022) 12:6865–82. doi: 10.7150/thno.76619
63. Deng X, Kong F, Li S, Wang X, Zhou Y, Zhang H, et al. A KLF4/PiHL/EZH2/HMGA2 regulatory axis and its function in promoting oxaliplatin-resistance of colorectal cancer. Cell Death Dis. (2021) 12:485. doi: 10.1038/s41419-021-03753-1
64. Galán-Martínez J, Stamatakis K, Sánchez-Gómez I, Vázquez-Cuesta S, Gironés N, Fresno M. Isoform-specific effects of transcription factor TCFL5 on the pluripotency-related genes SOX2 and KLF4 in colorectal cancer development. Mol Oncol. (2022) 16:1876–90. doi: 10.1002/1878-0261.13085
65. Shang Y, Zhu Z, Zhang Y, Wang X, Li J, Chen H, et al. MiR-7-5p/KLF4 signaling inhibits stemness and radioresistance in colorectal cancer. Cell Death Discovery. (2023) 9:42. doi: 10.1038/s41420-023-01339-8
66. Yuan L, Meng Y, Xiang J. KLF4 induces colorectal cancer by promoting EMT via STAT3 activation. Dig Dis Sci. (2024) 69:2841–55. doi: 10.1007/s10620-024-08473-y
67. Babaei K, Khaksar R, Zeinali T, Mohammadi M, Azizi M, Rahmani A, et al. Epigenetic profiling of MUTYH, KLF6, WNT1 and KLF4 genes in carcinogenesis and tumorigenesis of colorectal cancer. Biomedicine (Taipei). (2019) 9:22. doi: 10.1051/bmdcn/2019090422
68. Yang VW, Liu Y, Kim J, Shroyer KR, Bialkowska AB. Increased genetic instability and accelerated progression of colitis-associated colorectal cancer through intestinal epithelium-specific deletion of klf4. Mol Cancer Res. (2019) 17:165–76. doi: 10.1158/1541-7786.Mcr-18-0399
69. Anuja K, Kar M, Chowdhury AR, Sen A, Ghosh S, Gupta P, et al. Role of telomeric RAP1 in radiation sensitivity modulation and its interaction with CSC marker KLF4 in colorectal cancer. Int J Radiat Biol. (2020) 96:790–802. doi: 10.1080/09553002.2020.1721609
70. Li X, Jiang Y, Wang Y, Zhang Q, Liu J, Wang H, et al. KLF4 suppresses anticancer effects of brusatol via transcriptional upregulating NCK2 expression in melanoma. Biochem Pharmacol. (2024) 223:116197. doi: 10.1016/j.bcp.2024.116197
71. Riverso M, Montagnani V, Stecca B. KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth. Oncogene. (2017) 36:3322–33. doi: 10.1038/onc.2016.481
72. Liu J, Cao F, Li X, Zhang Y, Wang T, Chen H, et al. ITIH5, a p53-responsive gene, inhibits the growth and metastasis of melanoma cells by downregulating the transcriptional activity of KLF4. Cell Death Dis. (2021) 12:438. doi: 10.1038/s41419-021-03707-7
73. Zhang D, Lin J, Chao Y, Wang Z, Li H, Liu X, et al. Regulation of the adaptation to ER stress by KLF4 facilitates melanoma cell metastasis via upregulating NUCB2 expression. J Exp Clin Cancer Res. (2018) 37:176. doi: 10.1186/s13046-018-0842-z
74. Zang Y, Tian Z, Wang D, Chen L, Zhao H, Zhang X, et al. METTL3-mediated N(6)-methyladenosine modification of STAT5A promotes gastric cancer progression by regulating KLF4. Oncogene. (2024) 43:2338–54. doi: 10.1038/s41388-024-03085-2
75. Chen Z, Jiang Z, Meng L, Wang X, Zhang H, Zhou Y, et al. SAMHD1, positively regulated by KLF4, suppresses the proliferation of gastric cancer cells through MAPK p38 signaling pathway. Cell Cycle. (2022) 21:2065–78. doi: 10.1080/15384101.2022.2085356
76. Liu Z, Wu X, Tian Y, Wang J, Zhang Q, Li M, et al. H. pylori infection induces CXCL8 expression and promotes gastric cancer progress through downregulating KLF4. Mol Carcinog. (2021) 60:524–37. doi: 10.1002/mc.23309
77. Zhang J, Zhu Z, Wu H, Chen L, Wang Y, Liu J, et al. PODXL, negatively regulated by KLF4, promotes the EMT and metastasis and serves as a novel prognostic indicator of gastric cancer. Gastric Cancer. (2019) 22:48–59. doi: 10.1007/s10120-018-0833-y
78. To HTN, Le QA, Bui HTT, Park JH, Kang D. Modulation of spheroid forming capacity and TRAIL sensitivity by KLF4 and nanog in gastric cancer cells. Curr Issues Mol Biol. (2022) 45:233–48. doi: 10.3390/cimb45010018
79. Chen Z, Gao Y, Gao S, Song D, Feng Y. MiR-135b-5p promotes viability, proliferation, migration and invasion of gastric cancer cells by targeting Krüppel-like factor 4 (KLF4). Arch Med Sci. (2020) 16:167–76. doi: 10.5114/aoms.2019.87761
80. Zhu M, Zhang N, He S. Transcription factor KLF4 modulates microRNA-106a that targets Smad7 in gastric cancer. Pathol Res Pract. (2019) 215:152467. doi: 10.1016/j.prp.2019.152467
81. Karagonlar ZF, Akbari S, Karabicici M, Ergin G, Becer E, Atabey N, et al. A Novel Function for KLF4 in Modulating the De-differentiation of EpCAM(-)/CD133(-) nonStem Cells into EpCAM(+)/CD133(+) Liver Cancer Stem Cells in HCC Cell Line HuH7. Cells. (2020) 9. doi: 10.3390/cells9051198
82. Li L, Yu S, Wu Q, Dou N, Li Y, Gao Y. KLF4-mediated CDH3 upregulation suppresses human hepatoma cell growth and migration via GSK-3β Signaling. Int J Biol Sci. (2019) 15:953–61. doi: 10.7150/ijbs.30857
83. Chen J, Li H, Zhang B, Wang X, Zhao Y, Liu M, et al. ABI2-mediated MEOX2/KLF4-NANOG axis promotes liver cancer stem cell and drives tumour recurrence. Liver Int. (2022) 42:2562–76. doi: 10.1111/liv.15412
84. Zhang X, Wang W, Lu C, Zhang H. KLF4 suppresses the proliferation of perihilar cholangiocarcinoma by negatively regulating GDF15 and phosphorylating AKT. Oncol Rep. (2023) 50. doi: 10.3892/or.2023.8659
85. Yucel Polat A, Ayva ES, Gurdal H, Ozdemir BH, Gur Dedeoglu B. MiR-25 and KLF4 relationship has early prognostic significance in the development of cervical cancer. Pathol Res Pract. (2021) 222:153435. doi: 10.1016/j.prp.2021.153435
86. Yang WT, Zheng PS. Promoter hypermethylation of KLF4 inactivates its tumor suppressor function in cervical carcinogenesis. PloS One. (2014) 9:e88827. doi: 10.1371/journal.pone.0088827
87. Liu D, Jin Y, Wu J, Zhu H, Ye D. MiR-135b-5p is an oncogene in pancreatic cancer to regulate GPRC5A expression by targeting transcription factor KLF4. Cell Death Discovery. (2022) 8:23. doi: 10.1038/s41420-022-00814-y
88. Ganguly K, Krishn SR, Rachagani S, Batra SK, Jain M, Patel A, et al. Secretory mucin 5AC promotes neoplastic progression by augmenting KLF4-mediated pancreatic cancer cell stemness. Cancer Res. (2021) 81:91–102. doi: 10.1158/0008-5472.Can-20-1293
89. Feng L, Wang J, Zhang J, Li Y, Chen X, Zhou H, et al. Comprehensive analysis of E3 ubiquitin ligases reveals ring finger protein 223 as a novel oncogene activated by KLF4 in pancreatic cancer. Front Cell Dev Biol. (2021) 9:738709. doi: 10.3389/fcell.2021.738709
90. Li H, Wang J, Xiao W, Zhao X, Liu Q, Zhang Y, et al. Epigenetic alterations of Krüppel-like factor 4 and its tumor suppressor function in renal cell carcinoma. Carcinogenesis. (2013) 34:2262–70. doi: 10.1093/carcin/bgt189
91. Chen W, Wang H, Lu Y, Zhang R, Sun T, Lin J, et al. GTSE1 promotes tumor growth and metastasis by attenuating of KLF4 expression in clear cell renal cell carcinoma. Lab Invest. (2022) 102:1011–22. doi: 10.1038/s41374-022-00797-5
92. Frazzi R. KLF4 is an epigenetically modulated, context-dependent tumor suppressor. Front Cell Dev Biol. (2024) 12:1392391. doi: 10.3389/fcell.2024.1392391
93. Choi BJ, Cho YG, Song JW, Kim CJ, Kim SY, Nam SW, et al. Altered expression of the KLF4 in colorectal cancers. Pathol Res Pract. (2006) 202:585–9. doi: 10.1016/j.prp.2006.05.001
94. Cho YG, Song JH, Kim CJ, Nam SW, Yoo NJ, Lee JY, et al. Genetic and epigenetic analysis of the KLF4 gene in gastric cancer. Apmis. (2007) 115:802–8. doi: 10.1111/j.1600-0463.2007.apm_643.x
95. Xu J, Lü B, Xu F, Wang X, Chen Y, Zhang Y, et al. Dynamic down-regulation of Krüppel-like factor 4 in colorectal adenoma-carcinoma sequence. J Cancer Res Clin Oncol. (2008) 134:891–8. doi: 10.1007/s00432-008-0353-y
96. Nakahara Y, Northcott PA, Li M, Kongkham PN, Smith C, Yan H, et al. Genetic and epigenetic inactivation of Kruppel-like factor 4 in medulloblastoma. Neoplasia. (2010) 12:20–7. doi: 10.1593/neo.91122
97. Li H, Wang J, Xiao W, Zhao X, Liu Q, Zhang Y, et al. Epigenetic inactivation of KLF4 is associated with urothelial cancer progression and early recurrence. J Urol. (2014) 191:493–501. doi: 10.1016/j.juro.2013.08.087
98. Tsompana M, Gluck C, Sethi I, Joshi I, Rodriguez R, Thakur J, et al. Reactivation of super-enhancers by KLF4 in human Head and Neck Squamous Cell Carcinoma. Oncogene. (2020) 39:262–77. doi: 10.1038/s41388-019-0990-4
99. Zhu X, Wang J, Zhang H, Li Y, Chen L, Zhao M, et al. Downregulated KLF4, induced by m6A modification, aggravates intestinal barrier dysfunction in inflammatory bowel disease. Cell Mol Life Sci. (2024) 81:470. doi: 10.1007/s00018-024-05514-7
100. Chen Y, Liu M, Lu M, Luo L, Han Z, Liu X. Exploring the impact of m(6)A modification on immune diseases: mechanisms and therapeutic implication. Front Immunol. (2024) 15:1387582. doi: 10.3389/fimmu.2024.1387582
101. Han X, Zhu Y, Ke J, Wang L, Chen Y, Zhang H, et al. Progression of m(6)A in the tumor microenvironment: hypoxia, immune and metabolic reprogramming. Cell Death Discovery. (2024) 10:331. doi: 10.1038/s41420-024-02092-2
102. Wang K, Zhou W, Cai Q, Cheng J, Cai R, Xing R. SUMOylation of KLF4 promotes IL-4 induced macrophage M2 polarization. Cell Cycle. (2017) 16:374–81. doi: 10.1080/15384101.2016.1269045
103. Jha K, Kumar A, Bhatnagar K, Sharma R, Singh V, Gupta P, et al. Modulation of Krüppel-like factors (KLFs) interaction with their binding partners in cancers through acetylation and phosphorylation. Biochim Biophys Acta Gene Regul Mech. (2024) 1867:195003. doi: 10.1016/j.bbagrm.2023.195003
104. Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. (2004) 10:2709–19. doi: 10.1158/1078-0432.ccr-03-0484
105. Nagata T, Shimada Y, Sekine S, Matsui K, Takahashi H, Ishibashi M, et al. KLF4 and NANOG are prognostic biomarkers for triple-negative breast cancer. Breast Cancer. (2017) 24:326–35. doi: 10.1007/s12282-016-0708-1
106. Mimoto R, Imawari Y, Hirooka S, Takeyama H, Yoshida K. Impairment of DYRK2 augments stem-like traits by promoting KLF4 expression in breast cancer. Oncogene. (2017) 36:1862–72. doi: 10.1038/onc.2016.349
107. Lee HK, Lee DS, Park JC. Nuclear factor I-C regulates E-cadherin via control of KLF4 in breast cancer. BMC Cancer. (2015) 15:113. doi: 10.1186/s12885-015-1118-z
108. Cannizzaro E, Bannister AJ, Han N, Alendar A, Kouzarides T. DDX3X RNA helicase affects breast cancer cell cycle progression by regulating expression of KLF4. FEBS Lett. (2018) 592:2308–22. doi: 10.1002/1873-3468.13106
109. Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. (2013) 73:1434–44. doi: 10.1158/0008-5472.Can-12-2037
110. Akalay I, Tan TZ, Kumar P, Janji B, Berchem G, Noman MZ, et al. Targeting WNT1-inducible signaling pathway protein 2 alters human breast cancer cell susceptibility to specific lysis through regulation of KLF-4 and miR-7 expression. Oncogene. (2015) 34:2261–71. doi: 10.1038/onc.2014.151
111. Zou H, Chen H, Zhou Z, Wan Y, Liu Z. ATXN3 promotes breast cancer metastasis by deubiquitinating KLF4. Cancer Lett. (2019) 467:19–28. doi: 10.1016/j.canlet.2019.09.012
112. Zhou H, Liu Y, Zhu R, Li Y, Wang J, Zhang X, et al. FBXO32 suppresses breast cancer tumorigenesis through targeting KLF4 to proteasomal degradation. Oncogene. (2017) 36:3312–21. doi: 10.1038/onc.2016.479
113. Tiwari N, Meyer-Schaller N, Arnold P, Antoniadis H, Pachkov M, van Nimwegen E, et al. Klf4 is a transcriptional regulator of genes critical for EMT, including Jnk1 (Mapk8). PloS One. (2013) 8:e57329. doi: 10.1371/journal.pone.0057329
114. Fan SH, Wang YY, Wu ZY, Zhang J, Li TT, Chang YX, et al. AGPAT9 suppresses cell growth, invasion and metastasis by counteracting acidic tumor microenvironment through KLF4/LASS2/V-ATPase signaling pathway in breast cancer. Oncotarget. (2015) 6:18406–17. doi: 10.18632/oncotarget.4074
115. He H, Li S, Chen H, Chen J, Yang Y, Liu X, et al. 12-O-tetradecanoylphorbol-13-acetate promotes breast cancer cell motility by increasing S100A14 level in a Kruppel-like transcription factor 4 (KLF4)-dependent manner. J Biol Chem. (2014) 289:9089–99. doi: 10.1074/jbc.M113.534271
116. Moon JS, Kim HE, Koh E, Park SH, Jin W. Krüppel-like factor 4 (KLF4) activates the transcription of the gene for the platelet isoform of phosphofructokinase (PFKP) in breast cancer. J Biol Chem. (2011) 286:23808–16. doi: 10.1074/jbc.M111.236737
117. Sun H, Peng Z, Tang H, Xie D, Jia Z, Ma Z, et al. Loss of KLF4 and consequential downregulation of Smad7 exacerbate oncogenic TGF-β signaling in and promote progression of hepatocellular carcinoma. Oncogene. (2017) 36:2957–68. doi: 10.1038/onc.2016.447
118. Li Y, Yu S, Li L, Zhang J, Wang L, Gao Y, et al. KLF4-mediated upregulation of CD9 and CD81 suppresses hepatocellular carcinoma development via JNK signaling. Cell Death Dis. (2020) 11:299. doi: 10.1038/s41419-020-2479-z
119. Kong F, Sun T, Kong X, Xie D, Li Z, Xie K. Krüppel-like factor 4 suppresses serine/threonine kinase 33 activation and metastasis of gastric cancer through reversing epithelial-mesenchymal transition. Clin Cancer Res. (2018) 24:2440–51. doi: 10.1158/1078-0432.Ccr-17-3346
120. Wang L, Li Y, Li L, Zhang H, Liu X, Chen J, et al. Role of Kruppel-like factor 4 in regulating inhibitor of apoptosis-stimulating protein of p53 in the progression of gastric cancer. Oncol Lett. (2018) 15:6865–72. doi: 10.3892/ol.2018.8203
121. Zhao R, Liu Z, Xu W, Wang X, Zhang Y, Zhou H, et al. Helicobacter pylori infection leads to KLF4 inactivation in gastric cancer through a TET1-mediated DNA methylation mechanism. Cancer Med. (2020) 9:2551–63. doi: 10.1002/cam4.2892
122. Zhao L, Han T, Li Y, Chen X, Wang J, Liu M, et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. (2017) 31:893–903. doi: 10.1096/fj.201600994R
123. Zheng J, Liu Y, Qiao Y, Zhang L, Lu S. miR-103 promotes proliferation and metastasis by targeting KLF4 in gastric cancer. Int J Mol Sci. (2017) 18. doi: 10.3390/ijms18050910
124. Shao L, Chen Z, Soutto M, Zhang J, Wang Y, Xu W, et al. Helicobacter pylori-induced miR-135b-5p promotes cisplatin resistance in gastric cancer. FASEB J. (2019) 33:264–74. doi: 10.1096/fj.201701456RR
125. Ou Y, Ren H, Zhao R, Liu W, Yan L, Zhang Q, et al. Helicobacter pylori CagA promotes the Malignant transformation of gastric mucosal epithelial cells through the dysregulation of the miR-155/KLF4 signaling pathway. Mol Carcinog. (2019) 58:1427–37. doi: 10.1002/mc.23025
126. Ba MC, Long H, Cui SZ, He JR, Jiang YZ, Zeng MS, et al. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget. (2017) 8:95542–53. doi: 10.18632/oncotarget.20980
127. Zheng Y, Wu J, Chen H, Li X, Wang Z, Zhang Y, et al. KLF4 targets RAB26 and decreases 5-FU resistance through inhibiting autophagy in colon cancer. Cancer Biol Ther. (2023) 24:2226353. doi: 10.1080/15384047.2023.2226353
128. Yadav SS, Kumar M, Varshney A, Yadava PK. KLF4 sensitizes the colon cancer cell HCT-15 to cisplatin by altering the expression of HMGB1 and hTERT. Life Sci. (2019) 220:169–76. doi: 10.1016/j.lfs.2019.02.005
129. Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Pol. (2012) 59:467–74. doi: 10.18388/abp.2012_2079
130. Zeng Z, Li Y, Pan Y, Wang J, Tang Y, Wang X, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. (2018) 9:5395. doi: 10.1038/s41467-018-07810-w
131. Bennett LB, Schnabel JL, Kelchen JM, Futscher BW, Baldwin DA, Licht JD, et al. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes Chromosomes Cancer. (2009) 48:828–41. doi: 10.1002/gcc.20687
132. Shen Y, Park CS, Suppipat K, Peng J, Sharma R, Balasubramanian S, et al. Inactivation of KLF4 promotes T-cell acute lymphoblastic leukemia and activates the MAP2K7 pathway. Leukemia. (2017) 31:1314–24. doi: 10.1038/leu.2016.339
133. Guan H, Xie L, Leithäuser F, Flory E, O’Connell T, Garraway LA, et al. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood. (2010) 116:1469–78. doi: 10.1182/blood-2009-12-256446
134. Kharas MG, Yusuf I, Scarfone VM, Yang W, Green MR, Halpern J, et al. KLF4 suppresses transformation of pre-B cells by ABL oncogenes. Blood. (2007) 109:747–55. doi: 10.1182/blood-2006-03-011106
135. Ushmorov A, Leithäuser F, Sakk O, Juldanny M, Bruderlein S, Schwab M, et al. Epigenetic processes play a major role in B-cell-specific gene silencing in classical Hodgkin lymphoma. Blood. (2006) 107:2493–500. doi: 10.1182/blood-2005-09-3765
136. Ushmorov A, Ritz O, Hummel M, Leithäuser F, Klapper W, Möller P, et al. Epigenetic silencing of the immunoglobulin heavy-chain gene in classical Hodgkin lymphoma-derived cell lines contributes to the loss of immunoglobulin expression. Blood. (2004) 104:3326–34. doi: 10.1182/blood-2003-04-1197
137. MaChado L, Jarrett R, Morgan S, Dunlop L, Bell A, Angus B, et al. Expression and function of T cell homing molecules in Hodgkin’s lymphoma. Cancer Immunol Immunother. (2009) 58:85–94. doi: 10.1007/s00262-008-0528-z
138. Chen D, Zhu Q, Li T, Wang Y, Liu X, Zhang H, et al. KLF4 loss in hepatocellular carcinoma: Improving prognostic prediction and correlating immune infiltrates. Front Genet. (2023) 14:1106952. doi: 10.3389/fgene.2023.1106952
139. Prieto J, Melero I, Sangro B. Immunological landscape and immunotherapy of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. (2015) 12:681–700. doi: 10.1038/nrgastro.2015.173
140. Barakat DJ, Suresh R, Barberi T, Pienta KJ, Simons BW, Friedman AD. Absence of myeloid Klf4 reduces prostate cancer growth with pro-atherosclerotic activation of tumor myeloid cells and infiltration of CD8 T cells. PloS One. (2018) 13:e0191188. doi: 10.1371/journal.pone.0191188
141. Petty AJ, Li A, Wang X, Dai R, Heyman B, Hsu D, et al. Hedgehog signaling promotes tumor-associated macrophage polarization to suppress intratumoral CD8+ T cell recruitment. J Clin Invest. (2019) 129:5151–62. doi: 10.1172/jci128644
142. Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. (1998) 188:2205–13. doi: 10.1084/jem.188.12.2205
143. Gallimore A, Glithero A, Godkin A, Tissot AC, Lorang CG, Feng L, et al. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J Exp Med. (1998) 187:1383–93. doi: 10.1084/jem.187.9.1383
144. Schietinger A, Philip M, Krisnawan VE, Chiu EY, Delrow JJ, Basom RS, et al. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity. (2016) 45:389–401. doi: 10.1016/j.immuni.2016.07.011
145. Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity. (2016) 45:415–27. doi: 10.1016/j.immuni.2016.07.021
146. Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. (2016) 537:417–21. doi: 10.1038/nature19330
147. He R, Hou S, Liu C, Zhang A, Bai Q, Han M, et al. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature. (2016) 537:412–28. doi: 10.1038/nature19317
148. Beltra JC, Manne S, Abdel-Hakeem MS, Kurachi M, Giles JR, Chen Z, et al. Developmental relationships of four exhausted CD8(+) T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity. (2020) 52:825–841.e8. doi: 10.1016/j.immuni.2020.04.014
149. Nah J, Seong RH. Krüppel-like factor 4 regulates the cytolytic effector function of exhausted CD8 T cells. Sci Adv. (2022) 8:eadc9346. doi: 10.1126/sciadv.adc9346
150. Macián F, García-Rodríguez C, Rao A. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. (2000) 19:4783–95. doi: 10.1093/emboj/19.17.4783
151. Jain J, McCaffrey PG, Miner Z, Wison CB, Rao A, Hogan PG, et al. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. (1993) 365:352–5. doi: 10.1038/365352a0
152. Kel A, Kel-Margoulis O, Babenko V, Wingender E. Recognition of NFATp/AP-1 composite elements within genes induced upon the activation of immune cells. J Mol Biol. (1999) 288:353–76. doi: 10.1006/jmbi.1999.2684
153. Medrek C, Pontén F, Jirström K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. (2012) 12:306. doi: 10.1186/1471-2407-12-306
154. Yamaguchi T, Fushida S, Yamamoto Y, Tsukada T, Kinoshita J, Oyama K, et al. Tumor-associated macrophages of the M2 phenotype contribute to progression in gastric cancer with peritoneal dissemination. Gastric Cancer. (2016) 19:1052–65. doi: 10.1007/s10120-015-0579-8
155. Blacher E, Tsai C, Litichevskiy L, Wright C, Willet K, Creighton C, et al. Aging disrupts circadian gene regulation and function in macrophages. Nat Immunol. (2022) 23:229–36. doi: 10.1038/s41590-021-01083-0
156. Wang K, Xiong J, Lu Y, Wang L, Tian T. SENP1-KLF4 signalling regulates LPS-induced macrophage M1 polarization. FEBS J. (2023) 290:209–24. doi: 10.1111/febs.16589
157. Liu N, Wang X, Steer CJ, Song G. MicroRNA-206 promotes the recruitment of CD8(+) T cells by driving M1 polarisation of Kupffer cells. Gut. (2022) 71:1642–55. doi: 10.1136/gutjnl-2021-324170
158. Xin L, Wu Y, Liu C, Zhang Y, Wang X, Li H, et al. Exosome-mediated transfer of lncRNA HCG18 promotes M2 macrophage polarization in gastric cancer. Mol Immunol. (2021) 140:196–205. doi: 10.1016/j.molimm.2021.10.011
159. Shao X, Xu P, Ji L, Wang J, Zhang H, Liu Y, et al. Low-dose decitabine promotes M2 macrophage polarization in patients with primary immune thrombocytopenia via enhancing KLF4 binding to PPARgamma promoter. Clin Transl Med. (2023) 13:e1344. doi: 10.1002/ctm2.1344
160. Leng Z, Li Y, Zhou G, Chen X, Wang Y, Zhao L, et al. Krüppel-like factor 4 regulates stemness and mesenchymal properties of colorectal cancer stem cells through the TGF-β1/Smad/snail pathway. J Cell Mol Med. (2020) 24:1866–77. doi: 10.1111/jcmm.14882
161. Tetreault MP, Wang ML, Yang Y, Travis J, Carter P, Kaestner KH, et al. Klf4 overexpression activates epithelial cytokines and inflammation-mediated esophageal squamous cell cancer in mice. Gastroenterology. (2010) 139:2124–2134.e9. doi: 10.1053/j.gastro.2010.08.048
162. Zhou Z, Feng Z, Hu D, Zhang W, Zhou J, Han B, et al. A novel small-molecule antagonizes PRMT5-mediated KLF4 methylation for targeted therapy. EBioMedicine. (2019) 44:98–111. doi: 10.1016/j.ebiom.2019.05.011
163. Tien YT, Chang MH, Chu PY, Lin CS, Liu CH, Liao AT. Downregulation of the KLF4 transcription factor inhibits the proliferation and migration of canine mammary tumor cells. Vet J. (2015) 205:244–53. doi: 10.1016/j.tvjl.2014.12.031
164. Traka MH, Chambers KF, Lund EK, Goodlad RA, Johnson IT, Mithen RF. Involvement of KLF4 in sulforaphane- and iberin-mediated induction of p21(waf1/cip1). Nutr Cancer. (2009) 61:137–45. doi: 10.1080/01635580802348641
165. Ou DL, Chen CW, Hsu CL, Lin L, Lee BS, Chang CY, et al. Regorafenib enhances antitumor immunity via inhibition of p38 kinase/Creb1/Klf4 axis in tumor-associated macrophages. J Immunother Cancer. (2021) 9. doi: 10.1136/jitc-2020-001657
166. Cercek A, Wheler J, Murray PE, Zhou S, Saltz L. Phase 1 study of APTO-253 HCl, an inducer of KLF4, in patients with advanced or metastatic solid tumors. Invest New Drugs. (2015) 33:1086–92. doi: 10.1007/s10637-015-0273-z
167. Alkhayer R, Ponath V, Frech M, Zhao X, Wang Y, Liu L, et al. KLF4-mediated upregulation of the NKG2D ligand MICA in acute myeloid leukemia: a novel therapeutic target identified by enChIP. Cell Commun Signal. (2023) 21:94. doi: 10.1186/s12964-023-01118-z
168. Song Y, Li X, Liu F, Zhu H, Shen Y. Isoalantolactone alleviates ovalbumin−induced asthmatic inflammation by reducing alternatively activated macrophage and STAT6/PPAR−γ/KLF4 signals. Mol Med Rep. (2021) 24. doi: 10.3892/mmr.2021.12340
169. Li S, Zhou Q, He H, Zhao Y, Liu Z. Peroxisome proliferator-activated receptor γ agonists induce cell cycle arrest through transcriptional regulation of Kruppel-like factor 4 (KLF4). J Biol Chem. (2013) 288:4076–84. doi: 10.1074/jbc.M111.317487
170. Xiao X, Tang W, Yuan Q, Peng L, Yu P. Epigenetic repression of Krüppel-like factor 4 through Dnmt1 contributes to EMT in renal fibrosis. Int J Mol Med. (2015) 35:1596–602. doi: 10.3892/ijmm.2015.2189
171. Liu M, Li X, Peng KZ, Wang H, Zhang Y, Chen J, et al. Subcellular localization of Klf4 in non-small cell lung cancer and its clinical significance. BioMed Pharmacother. (2018) 99:480–5. doi: 10.1016/j.biopha.2018.01.090
172. Jia Y, Zhang C, Zhou L, Xu H, Shi Y, Tong Z. Micheliolide overcomes KLF4-mediated cisplatin resistance in breast cancer cells by downregulating glutathione. Onco Targets Ther. (2015) 8:2319–27. doi: 10.2147/ott.S88661
173. Zhang L, Li X, Chao Y, Wang J, Liu Z, Huang X, et al. KLF4, a miR-32-5p targeted gene, promotes cisplatin-induced apoptosis by upregulating BIK expression in prostate cancer. Cell Commun Signal. (2018) 16:53. doi: 10.1186/s12964-018-0270-x
174. Parenti S, Montorsi L, Fantini S, Guidoboni M, Pini R, Russo A, et al. KLF4 mediates the effect of 5-ASA on the β-catenin pathway in colon cancer cells. Cancer Prev Res (Phila). (2018) 11:503–10. doi: 10.1158/1940-6207.Capr-17-0382
175. Feng W, Xie Q, Liu S, Wang Y, Zhang L, Wu J, et al. Krüppel-like factor 4 promotes c-Met amplification-mediated gefitinib resistance in non-small-cell lung cancer. Cancer Sci. (2018) 109:1775–86. doi: 10.1111/cas.13601
176. Choi H, Roh J. LH-induced Transcriptional Regulation of Klf4 Expression in Granulosa Cells Occurs via the cAMP/PKA Pathway and Requires a Putative Sp1 Binding Site. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21197385
Keywords: KLF4, tumor microenvironment (TME), immune regulation, oncogenesis, tumor immunology
Citation: Ju Y, Xiao W, Mathis BJ and Shi Y (2025) KLF4: a multifunctional nexus connecting tumor progression and immune regulation. Front. Immunol. 16:1514780. doi: 10.3389/fimmu.2025.1514780
Received: 21 October 2024; Accepted: 17 January 2025;
Published: 06 February 2025.
Edited by:
Yichyi Lai, Chung Shan Medical University, TaiwanReviewed by:
Raffaele Frazzi, Azienda Unità Sanitaria Locale - IRCCS di Reggio Emilia, ItalyCopyright © 2025 Ju, Xiao, Mathis and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Shi, ODI2MjUzMzhAcXEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.