- 1Department of Rheumatology and Clinical Immunology, Medical Center - University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany
- 2Center for Chronic Immunodeficiency (CCI), Medical Center - University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany
- 3Institute of Medical Bioinformatics and Systems Medicine, Medical Center - University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany
- 4Genomics Core Facility, European Molecular Biology Laboratory, Heidelberg, Germany
- 5Department of Oto-Rhino-Laryngology, Medical Center - University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany
- 6German Cancer Consortium (DKTK), Partner site Freiburg, a partnership between DKFZ and Medical Center - University of Freiburg, Freiburg, Germany
- 7Division of Genetics, Department of Biology, University of Erlangen, Erlangen, Germany
Background: The posttranslational modification of cellular macromolecules by glycosylation is considered to contribute to disease pathogenesis in autoimmune and inflammatory conditions. In a subgroup of patients with common variable immunodeficiency (CVID), the occurrence of such complications is associated with an expansion of naïve-like CD21low B cells during a chronic type 1 immune activation. The glycosylation pattern of B cells in CVID patients has not been addressed to date.
Objective: The objective of this study was to examine the surface glycome of B cells in patients with CVID and associated immune dysregulation.
Methods: We performed surface lectin staining on B cells from peripheral blood and tonsils, both ex vivo and after in vitro stimulation. Additionally, we examined the expression of glycosylation-related genes by RNAseq in naïve-like CD21low B cells ex vivo, as well as in naïve CD21pos B cells from healthy controls after in vitro stimulation.
Results: Unlike CD21pos B cells, naïve-like CD21low B cells from CVID patients and CD21low B cells from healthy controls exhibited a unique glycosylation pattern with high levels of α2,6 sialic acids and fucose. This hypersialylation and hyperfucosylation were particularly induced by activation with anti-IgM and interferon-γ (IFN-γ). Transcriptome analysis suggested that naïve-like CD21low B cells possess a comprehensively reorganised glycosylation machinery, with anti-IgM/IFN-γ having the potential to initiate these changes in vitro.
Conclusion: CD21low B cells are hypersialylated and hyperfucosylated. This may implicate altered lectin-ligand interactions on the cell surface potentially affecting the CD21low B-cell function. These glycome changes appear to be driven by the prominent type I immune response in complicated CVID patients. A better understanding of how altered glycosylation influences immune cell function could lead to new therapeutic strategies.
1 Introduction
Common variable immunodeficiency (CVID) is the most prevalent human antibody deficiency syndrome. In addition to recurrent infections, up to 60% of CVID patients present with noninfectious complications, such as autoimmunity, lymphoproliferation, and organ inflammation (CVIDc) (1). B-cell differentiation in these patients is typically characterised by a reduction in class-switched memory (cs mem) B cells and plasma cells. A subgroup of these patients further presents with an accumulation of naïve-like CD21low B cells, classified as CVID smB-21lo patients according to the EUROclass classification (2). CD21low B cells constitute a B-cell subset that is characterised by the common and uniquely high expression of the transcription factor T-box expressed in T cells (T-bet) (3), most prominently induced by interferon-γ (IFN-γ) and B-cell receptor (BCR) costimulation, the integrin CD11c, and a strongly divergent gene expression profile compared to conventional B-cell subsets (4, 5). CD21low B cells are infrequent in healthy individuals and expand not only in the peripheral blood of CVIDc patients but also in distinct autoimmune and infectious disease conditions such as systemic lupus erythematosus (SLE) (6), rheumatoid arthritis (RA) (7), Sjögren’s syndrome (8), chronic HIV infection (9), malaria (10) and others (11, 12). As the differentially expressed genes (DEGs) of CD21low B cells contained some enzymes involved in glycosylation, and, in addition, an altered glycome has been suggested to contribute to disease pathogenesis in autoimmune disorders (13–18), we set out to explore the glycome of naïve-like CD21low B cells of CVIDc patients in more detail. Indeed, our investigations uncovered, for the first time, comprehensive aberrations in the glycome of CD21low B cells in CVID smB-21lo patients and healthy individuals, which might, in part, be direct subjects to the transcriptional regulation of T-bet.
2 Materials and methods
2.1 Patient cohort and ethics statement
The current study included blood from 23 patients (14 women, nine mem; age: median 50 years ± 13 years) who fulfilled the diagnostic criteria for CVID (www.ESID.org). All patients were classified as CVID smB-21lo according to the EUROclass classification (2) and suffered from additional immune dysregulation, as listed in Supplementary Table S1. Control blood and tonsils were obtained from healthy donors (HD) without known immunodeficiency. All experiments were performed with ethical approval from local authorities (251/13 and 254/19) in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients, healthy individuals, or their parents. Tonsillectomies were performed for medical reasons.
2.2 Antibodies and lectins used in this study
The following antibodies and lectins were used: CD3 PE/Cy7 (UCHT1), CD4 BV421 (RPA-T4), sialyl-Lewis X (SLeX) AF488 (FH6), CD19 APC/Cy7 (HIB19), CD21 PE/Cy7 (Bu32), CD27 BV421 (M-T271), CD38 PerCP/Cy5.5 (HIT2), IgD BV785 (IA6-2), IgM BV605 (MHM-88) were obtained from BioLegend (San Diego, CA, USA). CD3 BUV395 (UCHT1), CD27 BV605 (L128), IgG AF700 (G18-145), and streptavidin APC (70312) for the detection of biotinylated lectins were obtained from BD Biosciences (Franklin Lakes, NJ, USA). CD19 PE/Cy7 (J3-119), and CD21 PE (BL13) were obtained from Beckman Coulter (Brea, CA, USA). IgD PE (polyclonal), IgD FITC (polyclonal), and IgM Cy5 (polyclonal) were obtained from SouthernBiotech (Birmingham, AL, USA). Sambucus nigra agglutinin (SNA) FITC (ZE0328), Lens culinaris agglutinin (LCA) FITC (ZD0104), Aleuria aurantia lectin (AAL) FITC (ZE0628), and Erythrina cristagalli lectin (ECL) biotinylated (ZB042) were obtained from Vector Laboratories (Newark, CA, USA). Lectin-binding properties and symbol nomenclature for graphical representations of glycans (19) are shown in Supplementary Figure S1. All glycans were created using the web software DrawGlycan (20).
2.3 Cell isolation
PBMCs were isolated from EDTA blood by Ficoll density gradient centrifugation following standard protocols. For in vitro cultures, naïve B cells were isolated using the naïve B-Cell Isolation Kit II (Miltenyi Biotec, Bergisch Gladbach, Rhineland, Germany) following the manufacturer’s instructions. To determine the purity of naïve B cells, cells were stained with IgD FITC, CD21 PE, CD38 PerCpCy5.5, CD3 PC7, IgM Cy5, CD19 APC-Cy7, CD4 BV421, and CD27 BV605. Purity was above 93%. Tonsil cells were obtained by mechanical dissociation. After isolation, cells were resuspended in IMDM (Life Technologies, Carlsbad, CA, USA) with 10% foetal calf serum (FCS; Biochrom, Cambridge, Cambridgeshire, United Kingdom).
2.4 Flow cytometric staining
Harvested cells were washed two to three times in phosphate-buffered saline (PBS; Lonza Basel, Switzerland) 1% bovine serum albumin (BSA; Sigma-Aldrich St. Louis and Burlington, MA, USA). Cell surface lectin staining was performed separately for each lectin (SNA FITC, LCA FITC, AAL FITC ECL biotinylated) for 15 min on ice. Cells were washed with PBS containing 1% BSA. For ECL, cells were subsequently stained with streptavidin APC for 15 min on ice. Antibody staining was then performed for 15 min at 4°C. PBMCs and tonsil cells were stained with IgD PE, CD38 PerCpCy5.5, CD21 PC7, CD19 APC-Cy7, and CD27 BV421 to determine the respective B-cell subsets. For flow cytometric analysis of naïve activated B cells, cells were stained with IgD PE and CD19 PC7, and DAPI was added to exclude dead cells prior to data acquisition. Data were acquired using an LSRFortessa (BD Biosciences) and analysed using FlowJo software (Treestar, Ashland, OR, USA).
2.5 In vitro culture assay
For B-cell activation, 50,000 isolated naïve B cells were stimulated with anti-IgM (5 µg/ml; SouthernBiotech, Birmingham, AL, USA), IFN-γ (50 ng/ml; BioLegend), CpG oligodeoxynucleotides (2.5 µg/ml, InvivoGen, San Diego, CA, USA), or cultivated without additional stimulus in IMDM supplemented with l-glutamine, l-glutathione, insulin, apo-transferrin (all Sigma-Aldrich), nonessential amino acids (Gibco, Waltham, MA, USA), and 10% FCS for 48 h as described before (5). Cells were harvested and processed for flow cytometry or RNAseq.
2.6 RNAseq
After stimulation, B cells were resuspended in RLT buffer (Qiagen, Shanghai, China) and frozen at − 80°C. RNA was isolated using the RNAeasy Micro Kit (Qiagen) according to the manufacturer’s instructions. Samples were processed and sequenced as described previously (5). Alignments and gene count tables were obtained for each sample using STAR (version 2.4.2a) on the GRCh38 genome and annotation (GRCh38.p7). Differential gene expression was analysed using the limma R package. An adjusted p-value (Benjamini and Hochberg) < 0.05 was considered significant. The original raw files are available in the Gene Expression Omnibus data repository of the National Center for Biotechnology Information (21) under the accession number GSE181739 for anti-IgM/IFN-γ in vitro-stimulated B cells or GSE148163 for naïve-like CD21low B cells ex vivo.
2.7 Statistics
Statistical analyses were performed using GraphPad Prism 10 (GraphPad Software Inc., San Diego, CA, USA). Normal distribution was determined using D’Agostino and Pearson or Shapiro–Wilk test. Data were analysed using repeated measures one-way ANOVA with Tukey’s multiple comparisons test for normally distributed results and Friedman’s test with Dunn’s multiple comparisons test for nonnormally distributed results, as indicated. p-values of less than 0.05 were considered significant: *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001. Error bars in all figures define the means ± standard deviation (SD).
3 Results
3.1 Cell surface glycome of CD21low B cells
To address the question of whether the glycome of CVID patients’ B cells, and especially of CD21low B cells, is altered, we performed lectin staining of B cells from CVID smB-21lo patients and compared these to their CD21pos B-cell counterparts in patients and HD. Glycan binding properties for SNA (22, 23), ECL (22, 24), AAL (22, 25), LCA (22, 26, 27), and anti-SLeX are shown in Supplementary Figure S1. The gating strategy for naïve, IgM memory (IgM mem), cs mem, and CD21low B cells is depicted in Figure 1A. Due to the low memory cell numbers in CVID patients, we restricted the analysis of CD21low B cells to the naïve B-cell compartment.
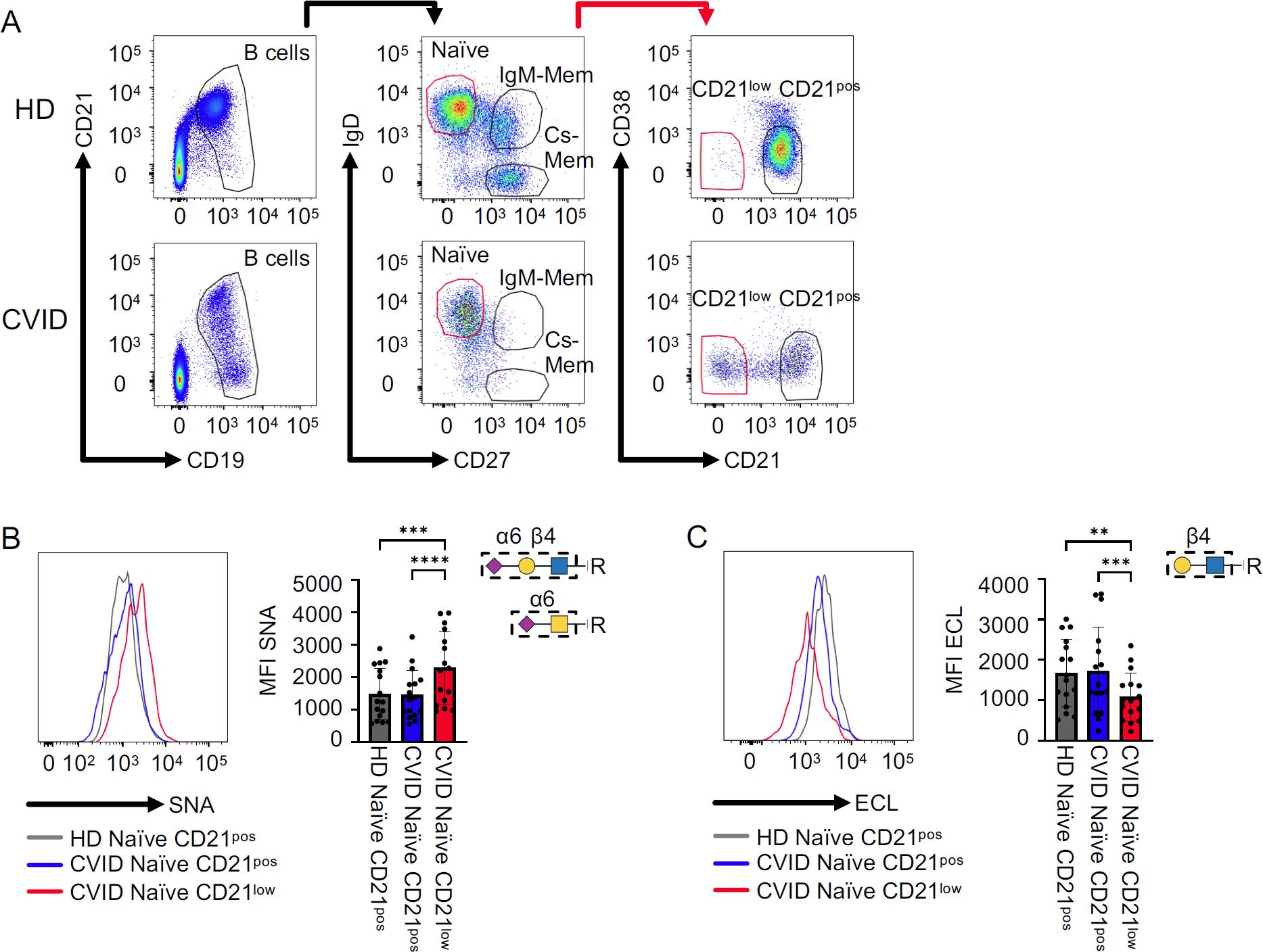
Figure 1. Sialylation on CVID patients’ B cells. (A) Naïve, IgM memory (IgM mem), and class-switched memory B cells (cs mem) were defined based on IgD and CD27 expression after gating on CD19posCD21pos-low B cells. Subpopulations were further differentiated into CD21pos and CD21low subsets. (B) Representative histogram for naïve B cells from one HD, naïve CD21pos, and naïve-like CD21low B cells from a CVID patient, with corresponding statistical analysis of mean fluorescence intensities (MFI) after SNA staining (n = 19). (C) Representative histogram for ECL staining as described in (A) (n = 16). Lectin-binding epitopes are illustrated. p-values as determined by one-way ANOVA with Tukey’s multiple comparisons test (B, C). **p < 0.01; ***p < 0.001; and ****p < 0.0001.
Lectin staining with α2,6 sialic acid-binding SNA (22, 23) revealed significantly increased levels of SNA ligands on the surface of naïve-like CD21low B cells from CVID patients compared to naïve CD21pos B cells from patients or HD. In contrast, patients’ naïve CD21pos B cells were comparable to those from HD (Figure 1B). ECL binds terminal β1,4-linked galactose (22, 24), representing an asialylated form of SNA ligands (Supplementary Figure S1). In line with increased SNA binding, ECL staining demonstrated a reduction of ligands on the surface of naïve-like CD21low B cells from patients compared to naïve CD21pos B cells from patients and HD. Naïve CD21pos B cells from patients and HD exposed ECL ligands to a similar extent (Figure 1C). Overall, these data reveal α2,6 hypersialylation of naïve-like CD21low B cells from CVID patients.
AAL binds to α1,3-, α1,4-, and α1,6-fucosylated glycans (22, 25). Lectin staining revealed an increased expression of AAL ligands on the surface of naïve-like CD21low B cells from CVID patients compared to naïve CD21pos B cells from patients or HD (Figure 2A). Similarly, lectin staining with LCA, which binds selectively to N-glycans with α1,6-linked core fucose (22, 26, 27), was increased on naïve-like CD21low B cells from CVID patients compared to naïve CD21pos B cells from patients and HD (Figure 2B). The mean fluorescence intensity (MFI) of both lectins was similar in naïve CD21pos B cells from patients and HD. Given the prominent role of SLeX, an α1,3 fucose-containing selectin ligand, on leukocyte trafficking, we analysed its expression on naïve B cells (28–33). Consistent with increased AAL staining, anti-SLeX staining demonstrated elevated levels of SLeX on patients’ naïve-like CD21low B cells compared to patients’ and HD’s naïve CD21pos B cells. Naïve CD21pos B cells from patients tended to have increased SLeX expression compared to naïve CD21pos B cells from HD, although this did not reach statistical significance (Figure 2C). The fucosylation pattern of naïve-like CD21low B cells supports increased α1,3-, α1,4-, and/or α1,6-linked fucose, increased core fucosylation of N-glycans, and increased SLeX epitopes relative to naïve CD21pos B cells.
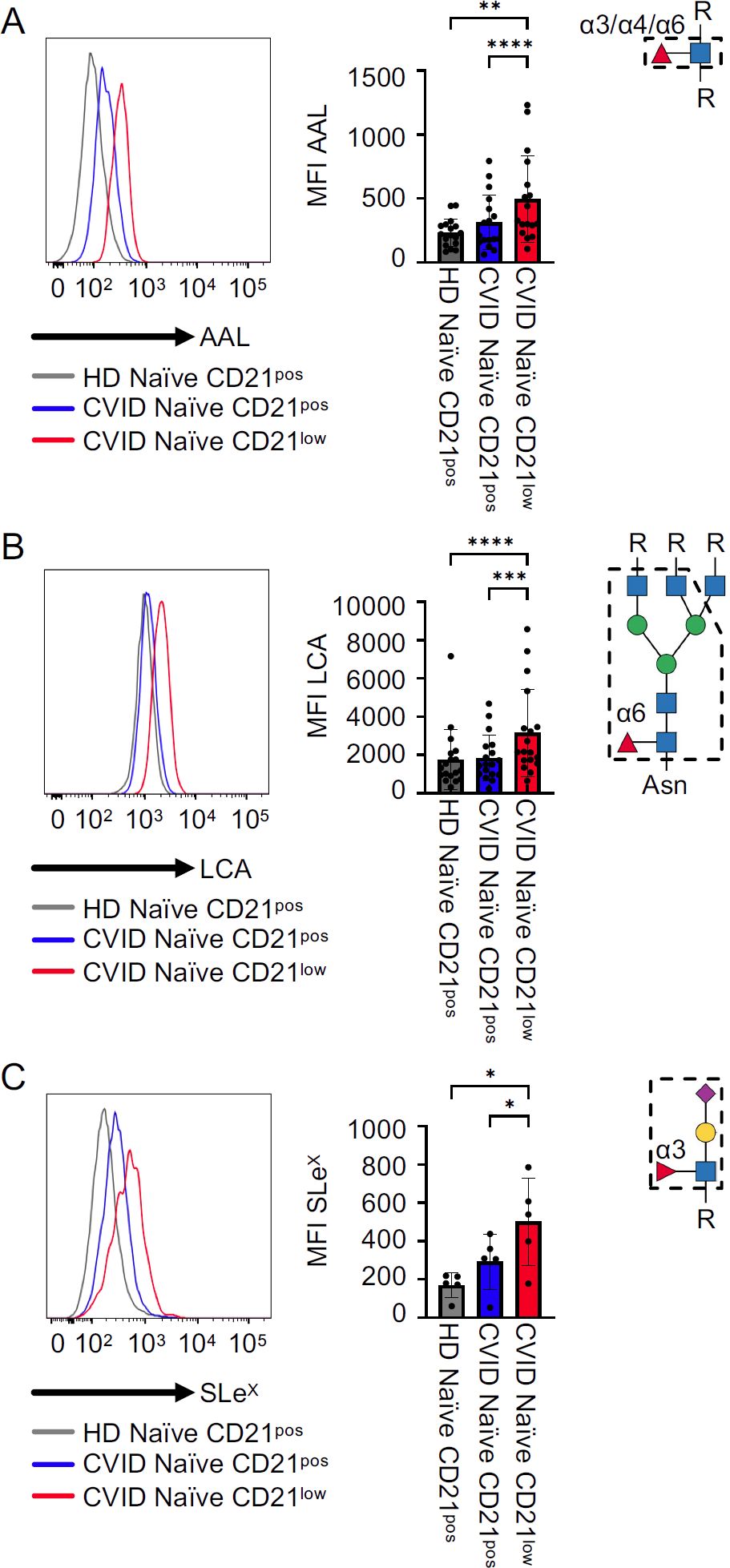
Figure 2. Fucosylation on CVID patients’ B cells. (A) Representative histogram for AAL in naïve CD21pos B cells from a HD and naïve CD21pos and naïve-like CD21low B cells from a CVID patient, with corresponding statistical analysis (n = 20). (B, C) LCA (n = 18) and anti-SLeX (n = 5) staining as described in (A). Lectin/antibody-binding epitopes are illustrated. p-values as determined by one-way ANOVA with Tukey’s multiple comparisons test (A, B) and Friedman’s test with Dunn’s multiple comparisons test (B). *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001.
We further tested whether external factors might influence the glycosylation of B cells in our patients. We did not observe any correlation between age or sex and the glycosylation of CD21low or CD21pos B cells in our patients (Supplementary Figures S2A, B). Genetic alterations (chromosome 22 aberrations, CTLA4 or KMT2D mutation) (Supplementary Figure S2C) and therapeutic treatments, such as low-dose steroids or sirolimus (Supplementary Figure S2D), did not affect the surface glycosylation pattern of naïve-like CD21low and CD21pos B cells. Lastly, we tested if noninfectious complications, which are common in CVID patients with an accumulation of the CD21low B-cell subset, were associated with glycome aberrations. We separated patients according to the presence of splenomegaly, granuloma formation, granulomatous lymphocytic interstitial lung disease (GLILD)/interstitial lung disease (ILD), lymphoproliferation (LP), autoimmune organ manifestation (AIO), autoimmune cytopenia (AIC), enteropathy, and hepatopathy. None of these clinical manifestations was specifically associated with significant changes in the surface glycome (Supplementary Figure S3). Overall, these results indicate: (A) that the glycosylation pattern of B cells in CVID is per se not strongly influenced by these factors and (B) that the alterations are characteristic for the naïve-like CD21low B-cell subset, in contrast to the CD21pos B-cell compartment.
3.2 Cell surface glycome of B-cell subsets from healthy controls
Next, to determine whether the altered glycome observed in CD21low B cells from CVID patients is unique to these cells, we compared the glycome of different B-cell subsets derived from peripheral blood and secondary lymphoid organs of HD. Due to the altered composition of CD21low B cells in HD, including classical or atypical memory B-cell subsets and only very low proportions of naïve-like CD21low B cells (34), the analysis in HD refers to “total” CD21low B cells, without differentiating CD27 and IgD expression. Also, in HD, increased SNA and reduced ECL staining corroborated hypersialylation of total CD21low B cells compared to CD21pos naïve, IgM mem, and cs mem B cells (Figures 3A, B), indicating that this glycosylation pattern is characteristic of the CD21low B-cell population and not limited to CVID. The degree of α2,6 surface sialylation was similar between the different blood-derived CD21pos B-cell subsets (Figure 3A) and, as previously published (16, 35), also between tonsillar B-cell populations (Supplementary Figures S4A, B). The level of terminal β1,4-linked galactose was significantly reduced in circulating cs mem B cells compared to naïve and IgM mem B cells, yet still increased compared to CD21low B cells (Figure 4B). This difference was not seen for the B-cell populations from secondary lymphoid tissue (Supplementary Figure S4C).
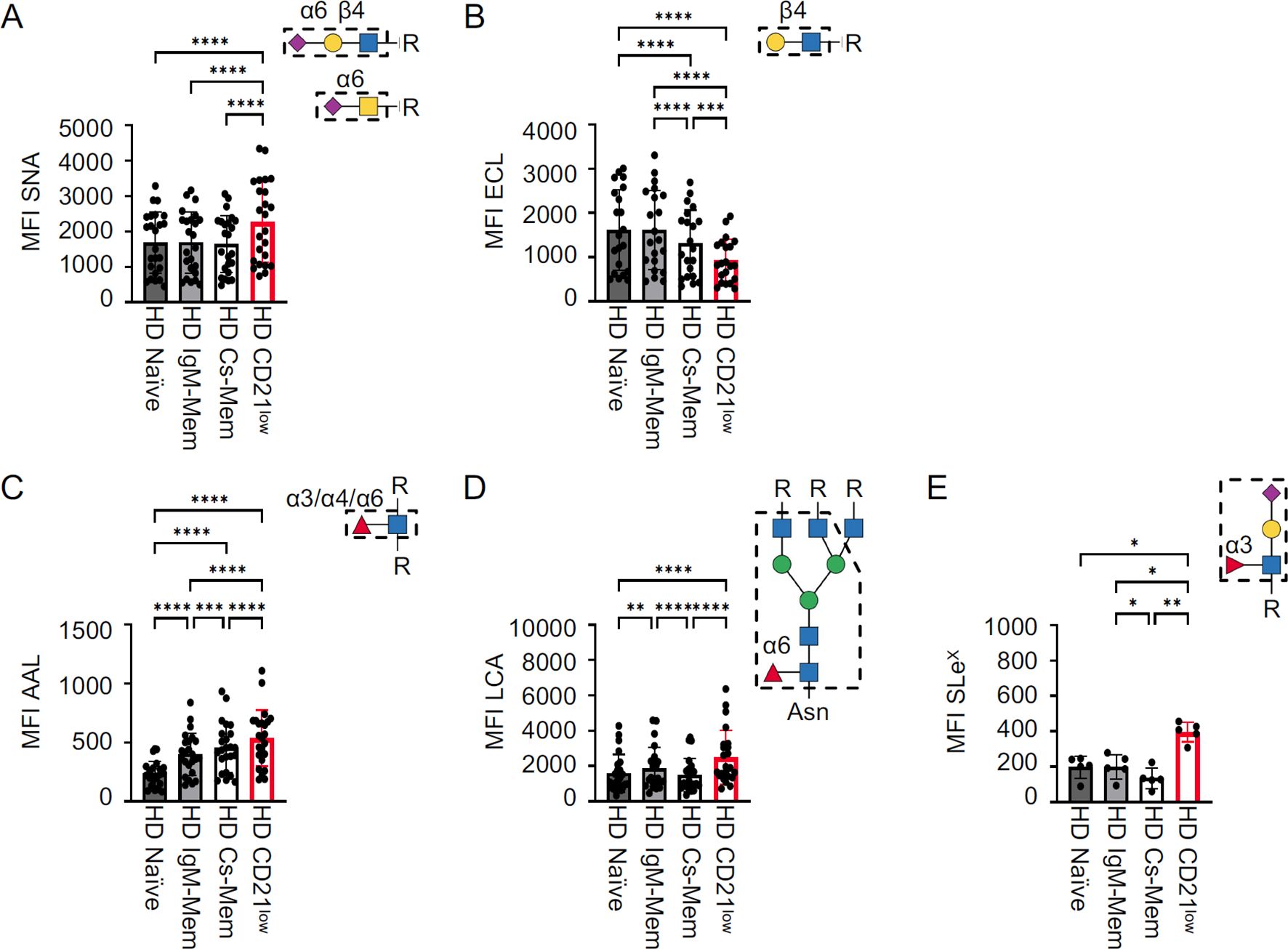
Figure 3. Cell surface glycome of HD B-cell subsets. Statistical analysis of the MFI for (A) SNA (n = 24), (B) ECL (n = 21), (C) AAL (n = 24), (D) LCA (n = 24), and (E) anti-SLeX (n = 5) for HD B-cell subpopulations from peripheral blood. CD21low B cells were gated from total B cells, while other subsets were determined from non-CD21low B cells. Lectin/antibody-binding epitopes are illustrated. p-values as determined by one-way ANOVA with Tukey’s multiple comparisons test (A–C, E) and Friedman’s test with Dunn’s multiple comparisons test (D). *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001.

Figure 4. Surface glycome of activated B cells. (A) Representative histogram showing the MFI of SNA in activated naïve B cells from a HD after 48 h of in vitro stimulation, as indicated, and statistical analysis of the ratios of the MFI of stimulated to unstimulated (n = 11). (B–D) Results after lectin staining with ECL (n = 5), AAL (n = 11), and LCA (n = 10) as described in (A). Lectin-binding epitopes are illustrated. p-values as determined by Friedman’s test with Dunn’s multiple comparisons test (A, D) and one-way ANOVA with Tukey’s multiple comparisons test (B, C). *p < 0.05; and **p < 0.01.
CD21low B cells from HD also revealed hyperfucosylation with increased expression of AAL, LCA, and anti-SLeX ligands (Figures 3C–E). Unlike the sialylation profile, fucosylation differed between the various CD21pos B-cell populations. We observed increasing levels of AAL ligands from the naïve to the cs mem B-cell compartment, still not reaching the level seen in CD21low B cells (Figure 3C), while LCA binding was slightly increased on IgM mem B cells compared to naïve and cs mem B cells (Figure 3D). Reduced expression of SLeX was observed in cs mem B cells (Figure 3E). Interestingly, in the secondary lymphoid tissue, the strongest AAL and LCA staining was detected in pre-germinal center cells and plasmablasts (Supplementary Figures S4D, E). Overall, HD CD21low B cells exhibited the same surface glycosylation pattern as naïve-like CD21low B cells from CVID patients, with the highest levels of α2,6 sialic acids of all investigated B-cell subsets, reduced levels of terminal β1,4 galactose, and increased levels of α1,3, α1,4, and/or α1,6 fucose, N-glycan core fucose, and SLeX. The direct comparison of total CD21low B cells in HD and CVID patient-derived naïve-like CD21low B cells did not reveal significant differences (Supplementary Figure S5). Minor changes were observed for AAL, which tended to be higher in HD, or LCA, which was slightly lower in HD (Supplementary Figure S5). These differences are most likely caused by the changes also observed in the CD21pos memory B-cell subsets compared to naïve, and thus reflect the different composition of the underlying CD21low B-cell subsets in HD and CVID (Figures 3C, D). Thus, the altered glycome of CD21low B cells is a general, yet unique and characteristic feature of this B-cell population.
3.3 Cell surface glycome after in vitro activation of naïve B cells
To investigate the impact of B-cell activation on the glycome, naïve CD21pos B cells from HD were cultured for 48 h in vitro with CpG, anti-IgM, anti-IgM/IFN-γ, or without stimulation. These conditions were chosen to distinguish the impact of BCR versus Toll-like receptor signals and, especially, the effect of the combined stimulation with anti-IgM/IFN-γ, which has been shown to provide an important signal for increased expression of the transcription factor T-bet and the differentiation of CD21low B cells (3, 5, 36).
All investigated stimuli increased the expression of SNA ligands on the B-cell surface after 48 h compared to unstimulated cells, but anti-IgM/IFN-γ stimulation induced significantly more α2,6-sialylated epitopes (Figure 4A). Unlike our observation of CD21low B cells ex vivo, B-cell activation also generally upregulated ECL ligands on the surface of activated cells, indicating a higher overall expression of terminal β1,4-linked galactose, irrespective of increased sialylation (Figure 4B). Analysis of fucose residues showed upregulation of AAL and LCA ligands upon each type of activation, reaching the highest levels after anti-IgM/IFN-γ stimulation (Figures 4C, D). In conclusion, all investigated conditions of B-cell activation led to the upregulation of SNA, ECL, AAL, and LCA ligands compared to unstimulated cells. A more potent role of anti-IgM/IFN-γ stimulation was visible for the upregulation of SNA, AAL, and LCA ligands, indicating a significantly stronger α2,6 hypersialylation, α1,3, α1,4, and/or α1,6 hyperfucosylation, and N-glycan core fucosylation under this condition.
3.4 Glycosylation-related genes in CD21low B cells and anti-IgM/IFN-γ-stimulated B cells
The transcription of glycosylation-related genes is a key factor in the cellular regulation of expressed glycans (37). Given the prominent role of anti-IgM/IFN-γ stimulation in the differentiation of the naïve-like CD21low B-cell phenotype (5), we compared DEGs from anti-IgM/IFN-γ-activated and unstimulated B cells from HD to DEGs of naïve-like CD21low B cells from CVID patients and naïve CD21pos B cells from HD ex vivo. A comparative analysis of selected genes involved in sialylation (Figure 5) and fucosylation (Figure 6) was performed. We focused on the expression of genes encoding for α2,6 sialyltransferases and α1,3, α1,4, or α1,6 fucosyltransferases, which are responsible for the generation of SNA, AAL, or LCA ligands, as well as genes contributing to the synthesis of the nucleotide sugars cytidine monophosphate (CMP)-sialic acid and guanosine diphosphate (GDP)-fucose as substrate for the respective glycosyltransferases.
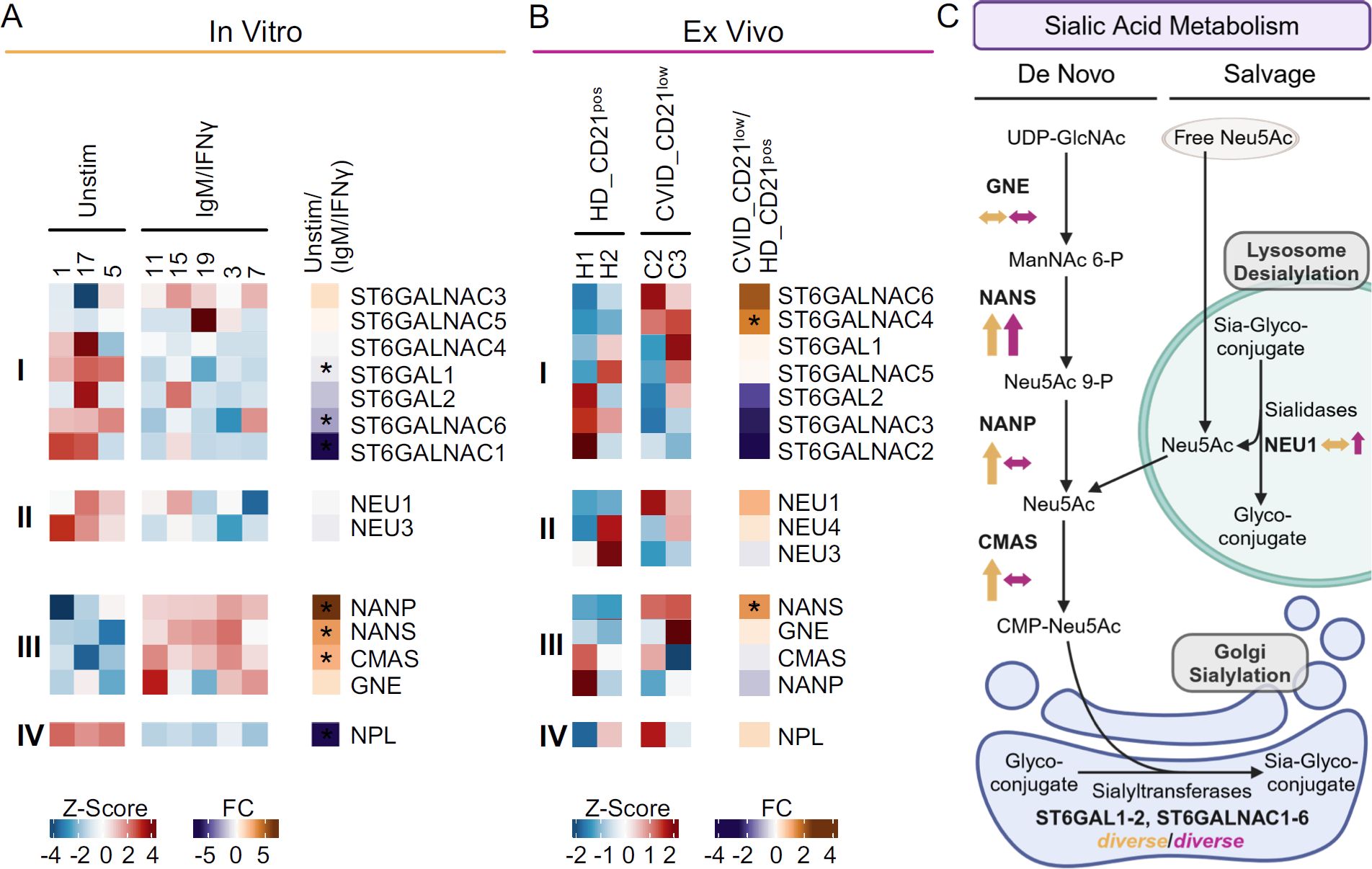
Figure 5. Sialylation-related genes in anti-IgM/IFN-γ-activated B cells and naïve-like CD21low B cells. (A) Heatmap of selected genes regulating sialylation, as determined by RNAseq of activated naïve B cells after in vitro stimulation with anti-IgM/IFN-γ compared to unstimulated cells. The expression of genes encoding for α2,6 sialyltransferases (i), sialidase (ii), enzymes involved in CMP-sialic acid de novo synthesis (iii), and sialic acid degradation (iv) is depicted. (B) Heatmap of sialylation-regulating genes identified by RNAseq of naïve-like CD21low B cells from CVID patients compared to naïve CD21pos B cells from HD ex vivo. (C) Schematic overview of gene expression involved in sialic acid metabolism after anti-IgM/IFN-γ in vitro stimulation (yellow) and in naïve-like CD21low B cells ex vivo (purple). Pathways include sialic acid salvage and CMP-Neu5Ac de novo synthesis as substrates for sialyltransferases (42, 43). Increased (↑), decreased (↓), or not clearly deviating (↔) gene expression is indicated. Large arrows indicate significant changes, while small ones indicate clear trends without reaching significance. The correct illustration of CMAS in the nucleus (77) was omitted for better visualisation. UDP, uridine diphosphate; GlcNAc, N-acetyl glucosamine; ManNAc, N-acetyl mannosamine; P, phosphate; Neu5Ac, N-acetyl neuraminic acid; CMP, cytidine monophosphate. (C) was created with BioRender.com. *p < 0.05.
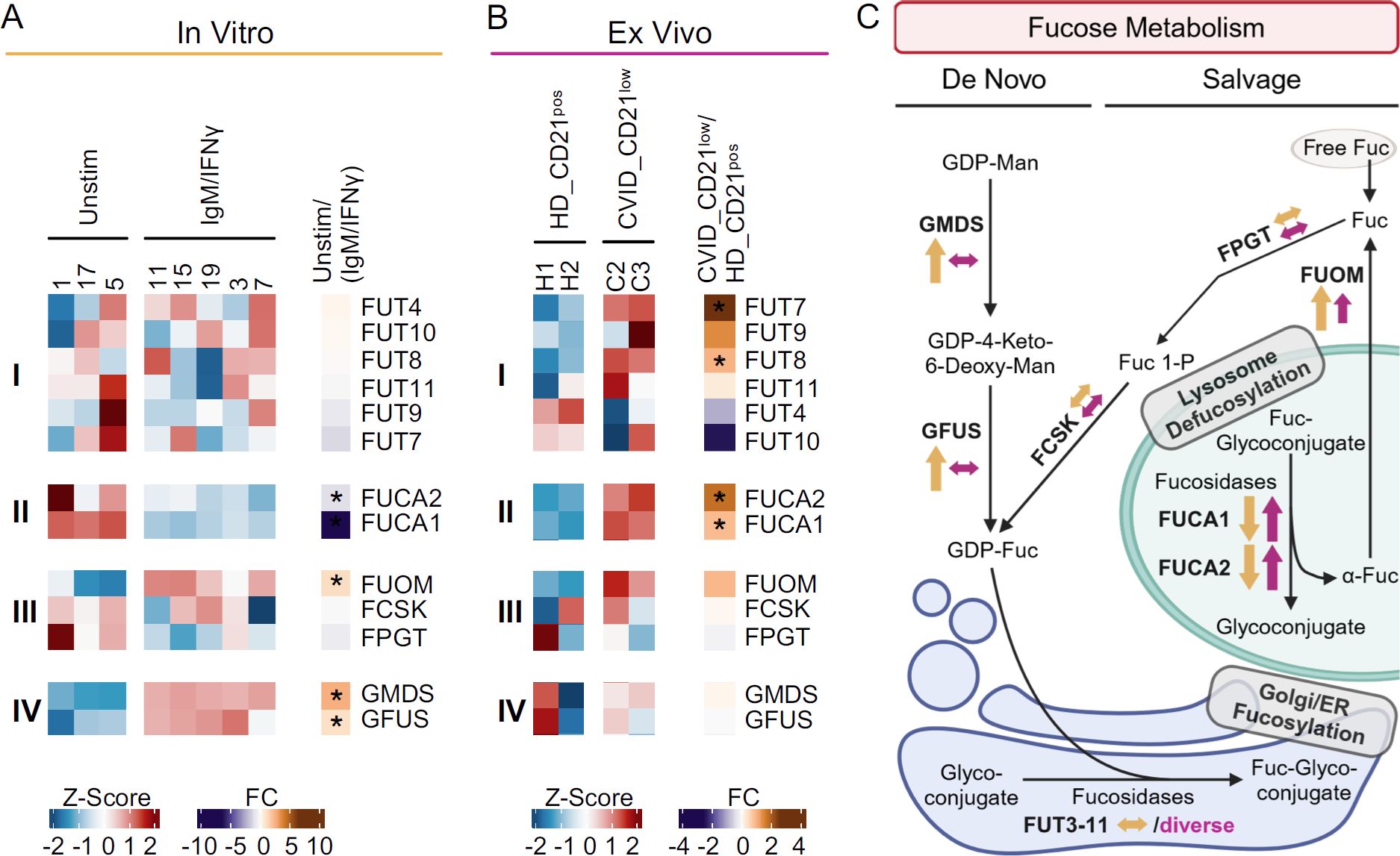
Figure 6. Fucosylation-related genes in anti-IgM/IFN-γ-activated B cells and naïve-like CD21low B cells. (A) Heatmap of selected genes of fucosylation in 48 h anti-IgM/IFN-γ-stimulated naïve B cells (IgM/IFN-γ) compared to unstimulated cells. The expression of genes encoding for α1,3, α1,4, α1,6 and fucosyltransferases (i), fucosidases (ii), enzymes of the GDP-fucose salvage (iii), and de novo synthesis (iv) is depicted. (B) Heatmap of genes in naïve-like CD21low B cells from CVID patients compared to naïve CD21pos B cells from HD, as described in (A). (C) Fucose salvage and GDP-fucose de novo synthesis as substrates for fucosyltransferases (48, 49). Increased (↑), decreased (↓), or not clearly deviating (↔) gene expression is indicated for anti-IgM/IFN-γ-stimulated B cells (yellow) and naïve-like CD21low B cells (purple). Large arrows indicate significance, while small ones a clear trend without reaching significance. The correct illustration of the extracellular localisation of FUCA2 (78) was omitted for better visualization. GDP, guanosine diphosphate; Man, mannose; Fuc, fucose; ER, endoplasmic reticulum. (C) was created with BioRender.com. *p < 0.05.
Anti-IgM/IFN-γ in vitro-stimulated B cells revealed diverse deviations in the expression of sialyltransferases that generate SNA epitopes by linking α2,6 sialic acid to galactose or N-acetyl galactosamine (38, 39). Only ST6GALNAC3 tended to be upregulated in the activated B cells, while ST6GALNAC4 and ST6GALNAC5 were not differentially expressed, ST6GAL2 tended to be downregulated, and ST6GAL1, ST6GALNAC1, and ST6GALNAC6 were significantly downregulated (Figure 5Ai). Genes for sialidases (also known as neuraminidases) that remove sialic acid from sialic acid-containing glycans (40, 41), such as NEU1 and NEU3, were not differentially expressed in vitro (Figure 5Aii). Genes regulating the de novo pathway of CMP-Neu5Ac synthesis (NANS, NANP, and CMAS but not GNE) were significantly upregulated in anti-IgM/IFN-γ-activated B cells (Figure 5Aiii) (42, 43). NPL, involved in the degradation of sialic acid, was significantly downregulated (Figure 5Aiv) (42, 44).
The analysis of differentially expressed genes in naïve-like CD21low B cells compared to naïve HD B cells ex vivo revealed a significant upregulation of ST6GALNAC4 and increased ST6GALNAC6 expression, though not reaching significance. The expression of other α2,6 sialyltransferases, such as ST6GAL1, ST6GAL2, ST6GALNAC2, ST6GALNAC3, and ST6GALNAC5 were unremarkable or tended to be downregulated (Figure 5Bi). The sialidase NEU1 tended to be upregulated in naïve-like CD21low B cells, while others, such as NEU3 and NEU4, were not differentially expressed (Figure 5Bii). In contrast to the in vitro- activated B cells, NANS was the only significantly upregulated gene related to the de novo synthesis of CMP-sialic acid in naïve-like CD21low B cells ex vivo (42, 43), while GNE, NANP, and CMAS were not differentially expressed (Figure 5Biii). NPL was not differentially expressed ex vivo (Figure 5Biv).
Overall, the results suggest an increased de novo synthesis of CMP-sialic acid in anti-IgM/IFN-γ-activated B cells and reduced sialic acid degradation potentially leading to increased substrate for sialyltransferases as the basis for α2,6 hypersialylation. In contrast, the α2,6 hypersialylation in naïve-like CD21low B cells may reflect a multifactorial basis involving the de novo and salvage synthesis of CMP-sialic acid and upregulation of individual sialyltransferases (Figure 5C).
None of the fucosyltransferases FUT3 to FUT11, linking fucose α1,3, α1,4, and/or α1,6 to N-acetyl glucosamine (45–47) and thus elicit AAL or LCA ligands, were differentially expressed in anti-IgM/IFN-γ-stimulated B cells (Figure 6Ai). In contrast, the FUCA1 and FUCA2 which remove fucose from fucosylated glycans, were significantly downregulated (Figure 6Aii) (40, 41). While activated B cells significantly upregulated FUOM, further genes of the fucose salvage pathway (FPGT, FCSK) were not differentially expressed (Figure 6Aiii) (48, 49). However, with GMDS and GFUS, all genes involved in the de novo synthesis of GDP-fucose were significantly upregulated in vitro (Figure 6Aiv) (48, 49).
Our analysis of naïve-like CD21low B cells ex vivo exhibited diverse expression of fucosyltransferases. FUT7, one of the α1,3 fucosyltransferases with the highest potency in creating the SLeX epitope (47, 50), and FUT8, the only known fucosyltransferase that can link fucose α1,6 to form N-glycan core fucose (45), were significantly upregulated (Figure 6Bi). The significant upregulation of both fucosidases, FUCA1 and FUCA2, in naïve-like CD21low B cells (Figure 6Bii) and the trend toward upregulation of FUOM suggests increased recycling of fucose in naïve-like CD21low B cells, in the presence of regular expression of the other genes involved in the fucose salvage pathway, FPGT and FCSK (Figure 6Biii). Genes of the de novo synthesis of GDP-fucose were not differentially expressed (Figure 6Biv).
As seen for sialylation, in vitro anti-IgM/IFN-γ-activated B cells upregulate genes involved in the de novo synthesis of GDP-fucose, potentially resulting in increased substrate for fucosyltransferases as basis for hyperfucosylation. In contrast, the basis in naïve-like CD21low B cells ex vivo seems to involve the upregulation of individual genes of fucosyltransferases and the fucose salvage pathway (Figure 6C).
4 Discussion
During lymphocyte development, the cellular glycome undergoes continuous changes that are believed to be relevant for differentiation, cell–cell interaction, and activation (51, 52). Although the understanding of the highly complex and dynamic glycome has advanced in recent years, the influence of specific glycan structures on individual cell types and differentiation phases, as well as their significance to immune system integrity and dysfunction, remains incomplete. Translation is further complicated by differences between humans and mice, despite conserved processes (53). In this study, we report the unique and characteristic glycosylation pattern of CD21low B cells.
Our data imply that naïve-like CD21low B cells from CVID patients and CD21low B cells from healthy controls exhibit a comprehensively restructured cell surface glycome, characterised by uniquely high levels of α2,6-linked sialic acid and fucose. The common upregulation of α2,6 sialyltransferases (ST6GALNAC4, ST6GALNAC6), along with NEU1 and NANS in naïve-like CD21low B cells from CVID patients, potentially increases substrate availability through enhanced sialic acid recycling and/or de novo synthesis of CMP-sialic acid (42, 43), suggesting a multifactorial basis for α2,6 hypersialylation in these cells. A multifactorial genesis likely underlies the hyperfucosylation observed in CD21low B cells, as the upregulation of FUT7, FUT8, FUCA1, FUCA2, and FUOM in naïve-like CD21low B cells from CVID patients suggests the relevance of fucosyltransferases and their substrate availability, potentially stemming from increased recycling of fucose (48, 49). While FUT8 encodes a fucosyltransferase uniquely responsible in linking fucose α1,6, thereby forming N-glycan core fucose (45), the fucosyltransferase encoded by FUT7 exhibits the highest potency in creating the α1,3 fucose-containing selectin ligand SLeX (50). The upregulation of these two fucosyltransferases in patients’ CD21low B cells may consequently contribute to the observed hyperfucosylation in these cells. Although the glycosidases NEU1, FUCA1, and FUCA2, which remove sialic acid and fucose from glycans (40, 41), are upregulated CD21low B cells, we observed increased α2,6 sialylation and α1,3, α1,4 and/or α1,6 fucosylation on CD21low B cells, suggesting a potentially ongoing reconstruction of the surface glycome. This hypothesis is strengthened by the increased activity of NEU1, which hydrolyses α2,3-linked sialic acid more rapidly than α2,6- or α2,8-linked sialic acid (40, 54), and by FUCA1 and FUCA2, which preferentially hydrolyse fucose α1,2-linked to galactose than fucose α1,3-, α1,4-, and α1,6-linked to N-acetyl glucosamine (41).
Interestingly, the CD21low B-cell-related CD11chigh B-cell population in SLE (55, 56) exhibits significant transcriptomic similarities in the expression of glycosylation-related genes, with marked upregulation of ST6GALNAC4, ST6GALNAC6, NEU1, NANS, FUT8, FUCA1, FUCA2, and FUOM (57). This further supports our hypothesis that hypersialylation and hyperfucosylation are general features of the CD21low B-cell population. In contrast, hypofucosylation has been reported in SLE-derived B cells without specific changes in the double-negative (CD27neg/IgDneg) atypical memory B-cell population (16). This finding clearly contrasts with the hyperfucosylation observed in naïve-like CD21low B cells in our study. Whether this represents a true difference between naïve-like CD21low B cells and double-negative (CD27neg/IgDneg) atypical memory B cells or a specific finding related to altered glycosylation in SLE requires further investigation.
Anti-IgM/IFN-γ stimulation provides a nonredundant signal during the differentiation of CD21low B cells and induces the initial changes toward the CD21low B-cell phenotype (5). Consistent with our hypothesis of increased sialylation and fucosylation on CD21low B cells ex vivo, the combined anti-IgM/IFN-γ stimulation was also the strongest inducer of α2,6 hypersialylation and α1,3, α1,4, and/or α1,6 hyperfucosylation in naïve B cells among all the investigated stimuli. Significant upregulation of NANS, NANP, CMAS, GMDS, and GFUS—the majority of genes involved in the de novo synthesis of CMP-sialic acid and GDP-fucose—suggests a central role for these pathways in vitro, potentially providing increased substrate for the respective sialyl- and fucosyltransferases (42, 43, 48, 49). Thus, anti-IgM/IFN-γ-driven B-cell activation most likely contributes to the altered glycome during the differentiation of CD21low B cells. Although presenting a similar surface glycosylation pattern, the mechanisms differ from those observed in naïve-like CD21low B cells ex vivo, suggesting potential differences in glycosylation during the differentiation and maintenance of these cells. Still, we cannot exclude the effects of limiting culture conditions on the availability of respective substrates.
The understanding of these aberrations in the glycome of normal human B cells remains ambiguous. In mice, increased sialylation limits the antigen-presenting potential (58–60). Sialic acids on antigen-presenting cells or the antigen itself can even induce B-cell tolerance, depending on interactions with Siglecs on the interacting cell, such as CD22 (Siglec 2) and/or Siglec G, the murine orthologue of Siglec 10 (61–64). Thus, in the context of increased frequency of autoreactive BCR specificities in the CD21low B-cell population (7, 8, 65, 66), hypersialylation may reflect an attempt to regulate an exacerbated immune response in autoimmune settings. On the other hand, reduced levels of terminal β1,4-linked galactose on CD21low B cells may actively contribute to immune dysregulation by limiting the secretion of highly expressed galectin 1 from naïve-like CD21low B cells, potentially favouring its intracellular retention (67). Since secreted galectin 1 counteracts Th1-driven immune responses (68, 69), reduced galectin 1 release by this B-cell subset may itself support a Th1 bias in its immediate neighbourhood, potentially contributing to a preferential type 1 immune response in different CD21low B-cell-associated disease conditions (3, 5, 70, 71). Moreover, upregulated SLeX on CD21low B cells may support their accumulation in inflamed tissues, such as the synovia of inflamed joints in rheumatoid arthritis, where the expression of SLeX, recognising endothelial selectins, is increased (28–33). Specific glycans mediate distinct functions. One of the most prominent examples is IgG: α2,6-linked sialylation of IgG conveys anti-inflammatory properties and reduces antibody-dependent cell-mediated cytotoxicity (ADCC) (72). Similarly, hyperfucosylation decreases ADCC (73). Notably, the secretion of IgG is low in antibody-deficient CVID patients. We cannot exclude the possibility that Ig substitution-mediated processes also play a role in the glycome changes observed in CD21low B cells from CVID patients, but given the strong similarities to the glycome of CD21low B cells in HD, this does not seem to be crucial. Unfortunately, the determination of the sialylation or fucosylation status of distinct surface proteins of CD21low B cells, such as CD45 or the BCR, known to be strongly influenced by glycosylation, was not feasible due to the limited number of primary B cells from immunodeficient patients.
It is noteworthy that increased sialylation, enhanced N-glycan core fucosylation, and elevated levels of SLeX on CD21low B cells are also seen in tumour cells, altering their cell interactions and modulating signal transduction (74). Tumour cells benefit from these aberrations, which foster their migration, promote their survival, and aid in immune evasion. These insights have opened new therapeutic strategies (75). The direct transfer of, for example, sialidase-conjugated antibodies mediating targeted desialylation of mammary carcinoma cells promoted the infiltration and activation of immune cells (76). A deeper understanding of the function and consequences of specific alterations in the glycome in human B cells is required before therapies targeting sialylation or fucosylation in CD21low B cells, or directly modulating glycan–lectin interactions, can be applied as a therapeutic approach in CVID.
In summary, naïve-like CD21low B cells from CVID patients and CD21low B cells from healthy controls exhibit a unique glycosylation pattern, expressing the highest levels of α2,6 sialic acid and high levels of α1,3-, α1,4-, and/or α1,6-linked fucose, including N-glycan core fucose and of SLeX. Unlike the more dynamic profile of fucosylation in different B-cell subpopulations, hypersialylation was unique for the CD21low B-cell phenotype. In vitro experiments indicate that these alterations are especially induced by the costimulation with anti-IgM/IFN-γ, which represents a key mechanism in the differentiation of the T-bet-expressing CD21low B-cell population. RNAseq data depict a fundamental reorganisation of the cellular glycosylation machinery during and after the differentiation of this unique B-cell subset. Our findings add a new aspect to the functional consequences of a chronic type I immune response in complicated CVID patients, which may become relevant if glycome-targeting strategies are incorporated into the therapeutic arsenal against autoimmune diseases.
Data availability statement
The datasets for anti-IgM/IFN-γ in vitro-stimulated B cells compared to unstimulated cells, and for naïve-like CD21low B cells ex vivo compared to HD CD21pos B cells, are available in the Gene Expression Omnibus data repository of the National Center for Biotechnology Information (21) under accession numbers GSE181739 and GSE148163, respectively.
Ethics statement
The studies involving humans were approved by Ethik-Kommission der Albert-Ludwigs-Universität Freiburg, 251/13 and 254/19. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants or their legal guardians.
Author contributions
PF: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. GA: Formal analysis, Investigation, Visualization, Writing – original draft. AM: Resources, Writing – original draft. SiG: Resources, Writing – original draft. IH: Investigation, Writing – original draft. SyG: Investigation, Writing – original draft. JL: Formal analysis, Investigation, Writing – original draft. VB: Supervision, Writing – original draft. TJ: Resources, Writing – original draft. MB: Formal analysis, Funding acquisition, Writing – original draft. LN: Writing – review & editing. RV: Writing – review & editing, Resources. KW: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. BK: Funding acquisition, Conceptualization, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the German Research Foundation TRR130 TP07 to KW. KW also receives support from the German Federal Ministry of Education and Research (BMBF) through a grant to the German Genetic Multi-Organ Auto-Immunity Network (GAIN), grant code 01GM2206A. BK receives support from the Deutsche Forschungsgemeinschaft (DFG; Project ID 545533246). Additionally, this work was supported by the DFG within the CRC1160 (Project ID 256073931-Z02 to MB), CRC/TRR167 (Project ID 259373024-Z01, MB), CRC1453 (Project ID 431984000-S1, MB), CRC1479 (Project ID: 441891347-S1 to MB), TRR 359 (Project ID 491676693-Z01, MB), and FOR 5476 UcarE (Project ID 493802833-P7, MB). Further support was provided by the German Federal Ministry of Education and Research (BMBF) within the Medical Informatics Funding Scheme PM4Onco-FKZ 01ZZ2322A (MB) and EkoEstMed-FKZ 01ZZ2015 (GA). We acknowledge the support of the Open Access Publication Fund of the University of Freiburg.
Acknowledgments
We thank the patients for their willingness to participate in this study, and the physicians and study nurses of the CCI for patient care, patient information, and support. Samples for this project were obtained from the CCI Biobank, which is a partner biobank/hub of the University Medical Center Freiburg and the Medical Faculty’s “Center for Biobanking - FREEZE”. We acknowledge the Lighthouse Core Facility unit for their excellent assistance with flow cytometry. Some figures were created with Biorender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1512279/full#supplementary-material
Glossary
AAL: Aleuria aurantia lectin
ADCC: antibody-dependent cell-mediated cytotoxicity
AIC: autoimmune cytopenia
AIO: autoimmune organ manifestation
APC: allophycocyanin
BCR: B-cell receptor
CMAS: cytidine monophosphate N-acetylneuraminic acid synthetase
Cs mem: class-switched memory
CMP: cytidine 5′-monophosphate
CVID: common variable immunodeficiency
DEGs: differentially expressed genes
ECL: Erythrina cristagalli lectin
FCSK: fucose kinase
FPGT: fucose-1-phosphate guanylyltransferase
FUCA1: alpha-L-fucosidase 1
FUCA2: alpha-L-fucosidase 2
FUT3-11: fucosyltransferase 3-11
FUOM: fucose mutarotase
GDP: guanosine 5′-diphosphate
GFUS: GDP-L-fucose synthase
GLILD: granulomatous lymphocytic interstitial lung disease
GMDS: GDP-mannose 4,6-dehydratase
GNE: glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
HD: healthy donor
IFN-γ: interferon γ
IgM mem: IgM memory
ILD: interstitial lung disease
IMDM: Iscove’s modified Dulbecco’s medium
LCA: Lens culinaris agglutinin
LP: lymphoproliferation
MFI: mean fluorescence intensity
NANP: N-acetylneuraminic acid phosphatase
NANS: N-acetylneuraminate synthase
NEU1: neuraminidase 1
NEU3: neuraminidase 3
NEU4: neuraminidase 4
NPL: N-acetylneuraminate pyruvate lyase
PBMCs: peripheral blood mononuclear cells
RA: rheumatoid arthritis
RNAseq: RNA-sequencing
SD: standard deviation
SLeX: sialyl-Lewis X
SNA: Sambucus nigra agglutinin
SLE: systemic lupus erythematosus
ST6GAL1-2: ST6 Beta-galactoside alpha-2,6-sialyltransferase 1-2
ST6GALNAC1-6: ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 1-6
T-bet: T-box expressed in T cells.
References
1. Chapel H, Lucas M, Lee M, Bjorkander J, Webster D, Grimbacher B, et al. Common variable immunodeficiency disorders: division into distinct clinical phenotypes. Blood. (2008) 112:277–86. doi: 10.1182/blood-2007-11-124545
2. Wehr C, Kivioja T, Schmitt C, Ferry B, Witte T, Eren E, et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood. (2008) 111:77–85. doi: 10.1182/blood-2007-06-091744
3. Unger S, Seidl M, van Schouwenburg P, Rakhmanov M, Bulashevska A, Frede N, et al. The TH1 phenotype of follicular helper T cells indicates an IFN-γ-associated immune dysregulation in patients with CD21low common variable immunodeficiency. J Allergy Clin Immunol. (2018) 141:730–40. doi: 10.1016/j.jaci.2017.04.041
4. Rakhmanov M, Keller B, Gutenberger S, Foerster C, Hoenig M, Driessen G, et al. Circulating CD21low B cells in common variable immunodeficiency resemble tissue homing, innate-like B cells. Proc Natl Acad Sci U.S.A. (2009) 106:13451–6. doi: 10.1073/pnas.0901984106
5. Keller B, Strohmeier V, Harder I, Unger S, Payne KJ, Andrieux G, et al. The expansion of human T-bethighCD21low B cells is T cell dependent. Sci Immunol. (2021) 6:eabh0891. doi: 10.1126/sciimmunol.abh0891
6. Wehr C, Eibel H, Masilamani M, Illges H, Schlesier M, Peter H-H, et al. A new CD21low B cell population in the peripheral blood of patients with SLE. Clin Immunol. (2004) 113:161–71. doi: 10.1016/j.clim.2004.05.010
7. Isnardi I, Ng Y-S, Menard L, Meyers G, Saadoun D, Srdanovic I, et al. Complement receptor 2/CD21- human naive B cells contain mostly autoreactive unresponsive clones. Blood. (2010) 115:5026–36. doi: 10.1182/blood-2009-09-243071
8. Saadoun D, Terrier B, Bannock J, Vazquez T, Massad C, Kang I, et al. Expansion of autoreactive unresponsive CD21-/low B cells in Sjögren’s syndrome-associated lymphoproliferation. Arthritis Rheumatol. (2013) 65:1085–96. doi: 10.1002/art.37828
9. Moir S, Malaspina A, Ogwaro KM, Donoghue ET, Hallahan CW, Ehler LA, et al. HIV-1 induces phenotypic and functional perturbations of B cells in chronically infected individuals. Proc Natl Acad Sci U.S.A. (2001) 98:10362–7. doi: 10.1073/pnas.181347898
10. Weiss GE, Crompton PD, Li S, Walsh LA, Moir S, Traore B, et al. Atypical memory B cells are greatly expanded in individuals living in a malaria-endemic area. J Immunol. (2009) 183:2176–82. doi: 10.4049/jimmunol.0901297
11. Kuzmina Z, Krenn K, Petkov V, Körmöczi U, Weigl R, Rottal A, et al. CD19(+)CD21(low) B cells and patients at risk for NIH-defined chronic graft-versus-host disease with bronchiolitis obliterans syndrome. Blood. (2013) 121:1886–95. doi: 10.1182/blood-2012-06-435008
12. Wang Z, Wang Z, Wang J, Diao Y, Qian X, Zhu N. T-bet-expressing B cells are positively associated with crohn’s disease activity and support th1 inflammation. DNA Cell Biol. (2016) 35:628–35. doi: 10.1089/dna.2016.3304
13. van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol. (2008) 9:593–601. doi: 10.1038/ni.f.203
14. Marth JD, Grewal PK. Mammalian glycosylation in immunity. Nat Rev Immunol. (2008) 8:874–87. doi: 10.1038/nri2417
15. Maverakis E, Kim K, Shimoda M, Gershwin ME, Patel F, Wilken R, et al. Glycans in the immune system and The Altered Glycan Theory of Autoimmunity: a critical review. J Autoimmun. (2015) 57:1–13. doi: 10.1016/j.jaut.2014.12.002
16. Morel M, Pochard P, Echchih W, Dueymes M, Bagacean C, Jousse-Joulin S, et al. Abnormal B cell glycosylation in autoimmunity: A new potential treatment strategy. Front Immunol. (2022) 13:975963. doi: 10.3389/fimmu.2022.975963
17. Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol. (2019) 15:346–66. doi: 10.1038/s41581-019-0129-4
18. Mahajan VS, Pillai S. Sialic acids and autoimmune disease. Immunol Rev. (2016) 269:145–61. doi: 10.1111/imr.2016.269.issue-1
19. Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, et al. Symbol nomenclature for graphical representations of glycans. Glycobiology. (2015) 25:1323–4. doi: 10.1093/glycob/cwv091
20. Cheng K, Zhou Y, Neelamegham S. DrawGlycan-SNFG: a robust tool to render glycans and glycopeptides with fragmentation information. Glycobiology. (2017) 27:200–5. doi: 10.1093/glycob/cww115
21. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. (2002) 30:207–10. doi: 10.1093/nar/30.1.207
22. Cummings RD, Darvill AG, Etzler ME, Hahn MG. Glycan-recognizing probes as tools. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al, editors. Essentials of Glycobiology, 3rd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press (2017). p. 48.
23. Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, Peumans WJ. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha2-6)Gal/GalNAc sequence. J Biol Chem. (1987) 262:1596–601. doi: 10.1016/S0021-9258(19)75677-4
24. Iglesias JL, Lis H, Sharon N. Purification and properties of a D-galactose/N-acetyl-D-galactosamine-specific lectin from Erythrina cristagalli. Eur J Biochem. (1982) 123:247–52. doi: 10.1111/j.1432-1033.1982.tb19760.x
25. Kochibe N, Furukawa K. Purification and properties of a novel fucose-specific hemagglutinin of Aleuria aurantia. Biochemistry. (1980) 19:2841–6. doi: 10.1021/bi00554a004
26. Gao C, Hanes MS, Byrd-Leotis LA, Wei M, Jia N, Kardish RJ, et al. Unique binding specificities of proteins toward isomeric asparagine-linked glycans. Cell Chem Biol. (2019) 26:535–547.e4. doi: 10.1016/j.chembiol.2019.01.002
27. Kornfeld K, Reitman ML, Kornfeld R. The carbohydrate-binding specificity of pea and lentil lectins. Fucose is an important determinant. J Biol Chem. (1981) 256:6633–40. doi: 10.1016/S0021-9258(19)69037-X
28. Angiari S. Selectin-mediated leukocyte trafficking during the development of autoimmune disease. Autoimmun Rev. (2015) 14:984–95. doi: 10.1016/j.autrev.2015.06.006
29. Barthel SR, Gavino JD, Descheny L, Dimitroff CJ. Targeting selectins and selectin ligands in inflammation and cancer. Expert Opin Ther Targets. (2007) 11:1473–91. doi: 10.1517/14728222.11.11.1473
30. Jin F, Wang F. The physiological and pathological roles and applications of sialyl Lewis x, a common carbohydrate ligand of the three selectins. Glycoconjugate J. (2020) 37:277–91. doi: 10.1007/s10719-020-09912-4
31. Kelly M, Hwang JM, Kubes P. Modulating leukocyte recruitment in inflammation. J Allergy Clin Immunol. (2007) 120:3–10. doi: 10.1016/j.jaci.2007.05.017
32. McEver RP. Selectins: initiators of leucocyte adhesion and signalling at the vascular wall. Cardiovasc Res. (2015) 107:331–9. doi: 10.1093/cvr/cvv154
33. Sperandio M, Gleissner CA, Ley K. Glycosylation in immune cell trafficking. Immunol Rev. (2009) 230:97–113. doi: 10.1111/j.1600-065X.2009.00795.x
34. Freudenhammer M, Voll RE, Binder SC, Keller B, Warnatz K. Naive- and memory-like CD21low B cell subsets share core phenotypic and signaling characteristics in systemic autoimmune disorders. J Immunol. (2020) 205:2016–25. doi: 10.4049/jimmunol.2000343
35. Macauley MS, Kawasaki N, Peng W, Wang S-H, He Y, Arlian BM, et al. Unmasking of CD22 co-receptor on germinal center B-cells occurs by alternative mechanisms in mouse and man. J Biol Chem. (2015) 290:30066–77. doi: 10.1074/jbc.M115.691337
36. Xu W, Zhang JJ. Stat1-dependent synergistic activation of T-bet for IgG2a production during early stage of B cell activation. J Immunol. (2005) 175:7419–24. doi: 10.4049/jimmunol.175.11.7419
37. Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. (2006) 126:855–67. doi: 10.1016/j.cell.2006.08.019
38. Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi M-A, Samyn-Petit B, Julien S, Delannoy P. The human sialyltransferase family. Biochimie. (2001) 83:727–37. doi: 10.1016/S0300-9084(01)01301-3
39. Takashima S, Tsuji S, Tsujimoto M. Characterization of the second type of human beta-galactoside alpha 2,6-sialyltransferase (ST6Gal II), which sialylates Galbeta 1,4GlcNAc structures on oligosaccharides preferentially. Genomic analysis of human sialyltransferase genes. J Biol Chem. (2002) 277:45719–28. doi: 10.1074/jbc.M206808200
40. Miyagi T, Yamaguchi K. Mammalian sialidases: physiological and pathological roles in cellular functions. Glycobiology. (2012) 22:880–96. doi: 10.1093/glycob/cws057
41. Johnson SW, Alhadeff JA. Mammalian α-L-fucosidases. Comp Biochem Physiol. (1991) 99B:479–88. doi: 10.1016/0305-0491(91)90327-A
42. Altheide TK, Hayakawa T, Mikkelsen TS, Diaz S, Varki N, Varki A. System-wide genomic and biochemical comparisons of sialic acid biology among primates and rodents: Evidence for two modes of rapid evolution. J Biol Chem. (2006) 281:25689–702. doi: 10.1074/jbc.M604221200
43. Bhide GP, Colley KJ. Sialylation of N-glycans: mechanism, cellular compartmentalization and function. Histochem Cell Biol. (2017) 147:149–74. doi: 10.1007/s00418-016-1520-x
44. Wen X-Y, Tarailo-Graovac M, Brand-Arzamendi K, Willems A, Rakic B, Huijben K, et al. Sialic acid catabolism by N-acetylneuraminate pyruvate lyase is essential for muscle function. JCI Insight. (2018) 3:e122373. doi: 10.1172/jci.insight.122373
45. Miyoshi E, Noda K, Yamaguchi Y, Inoue S, Ikeda Y, Wang W, et al. The α1-6-fucosyltransferase gene and its biological significance. Biochim Biophys Acta Gen Subj. (1999) 1473:9–20. doi: 10.1016/S0304-4165(99)00166-X
46. Mollicone R, Moore SEH, Bovin N, Garcia-Rosasco M, Candelier J-J, Martinez-Duncker I, et al. Activity, splice variants, conserved peptide motifs, and phylogeny of two new alpha1,3-fucosyltransferase families (FUT10 and FUT11). J Biol Chem. (2009) 284:4723–38. doi: 10.1074/jbc.M809312200
47. de Vries T, Knegtel RM, Holmes EH, Macher BA. Fucosyltransferases: structure/function studies. Glycobiology. (2001) 11:119R–28R. doi: 10.1093/glycob/11.10.119R
48. Becker DJ, Lowe JB. Fucose: biosynthesis and biological function in mammals. Glycobiology. (2003) 13:41R–53R. doi: 10.1093/glycob/cwg054
49. Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. (2008) 143:725–9. doi: 10.1093/jb/mvn011
50. Mondal N, Dykstra B, Lee J, Ashline DJ, Reinhold VN, Rossi DJ, et al. Distinct human α(1,3)-fucosyltransferases drive Lewis-X/sialyl Lewis-X assembly in human cells. J Biol Chem. (2018) 293:7300–14. doi: 10.1074/jbc.RA117.000775
51. Vicente MM, Leite-Gomes E, Pinho SS. Glycome dynamics in T and B cell development: basic immunological mechanisms and clinical applications. Trends Immunol. (2023) 44:585–97. doi: 10.1016/j.it.2023.06.004
52. Giovannone N, Antonopoulos A, Liang J, Geddes Sweeney J, Kudelka MR, King SL, et al. Human B cell differentiation is characterized by progressive remodeling of O-linked glycans. Front Immunol. (2018) 9:2857. doi: 10.3389/fimmu.2018.02857
53. North SJ, Chalabi S, Sutton-Smith M, Dell A, Haslam SM. Mouse and human glycomes. In: Cummings RD, Pierce JM, editors. Handbook of Glycomics, 1st. Academic Press/Elsevier, Amsterdam, Boston (2009). p. 263–327.
54. Miyagi T, Tsuiki S. Rat-liver lysosomal sialidase. Solubilization, substrate specificity and comparison with the cytosolic sialidase. Eur J Biochem. (1984) 141:75–81. doi: 10.1111/j.1432-1033.1984.tb08159.x
55. Gjertsson I, McGrath S, Grimstad K, Jonsson CA, Camponeschi A, Thorarinsdottir K, et al. A close-up on the expanding landscape of CD21-/low B cells in humans. Clin Exp Immunol. (2022) 210:217–29. doi: 10.1093/cei/uxac103
56. Rincon-Arevalo H, Wiedemann A, Stefanski A-L, Lettau M, Szelinski F, Fuchs S, et al. Deep phenotyping of CD11c+ B cells in systemic autoimmunity and controls. Front Immunol. (2021) 12:635615. doi: 10.3389/fimmu.2021.635615
57. Maul RW, Catalina MD, Kumar V, Bachali P, Grammer AC, Wang S, et al. Transcriptome and igH repertoire analyses show that CD11chi B cells are a distinct population with similarity to B cells arising in autoimmunity and infection. Front Immunol. (2021) 12:649458. doi: 10.3389/fimmu.2021.649458
58. Bagriaçik EÜ, Miller KS. Cell surface sialic acid and the regulation of immune cell interactions: the neuraminidase effect reconsidered. Glycobiology. (1999) 9:267–75. doi: 10.1093/glycob/9.3.267
59. Frohman M, Cowing C. Presentation of antigen by B cells: functional dependence on radiation dose, interleukins, cellular activation, and differential glycosylation. J Immunol. (1985) 134:2269–75. doi: 10.4049/jimmunol.134.4.2269
60. Silva M, Silva Z, Marques G, Ferro T, Gonçalves M, Monteiro M, et al. Sialic acid removal from dendritic cells improves antigen cross-presentation and boosts anti-tumor immune responses. Oncotarget. (2016) 7:41053–66. doi: 10.18632/oncotarget.v7i27
61. Duong BH, Tian H, Ota T, Completo G, Han S, Vela JL, et al. Decoration of T-independent antigen with ligands for CD22 and Siglec-G can suppress immunity and induce B cell tolerance in vivo. J Exp Med. (2010) 207:173–87. doi: 10.1084/jem.20091873
62. Macauley MS, Paulson JC. Siglecs induce tolerance to cell surface antigens by BIM-dependent deletion of the antigen-reactive B cells. J Immunol. (2014) 193:4312–21. doi: 10.4049/jimmunol.1401723
63. Macauley MS, Pfrengle F, Rademacher C, Nycholat CM, Gale AJ, von Drygalski A, et al. Antigenic liposomes displaying CD22 ligands induce antigen-specific B cell apoptosis. J Clin Invest. (2013) 123:3074–83. doi: 10.1172/JCI69187
64. Pfrengle F, Macauley MS, Kawasaki N, Paulson JC. Copresentation of antigen and ligands of Siglec-G induces B cell tolerance independent of CD22. J Immunol. (2013) 191:1724–31. doi: 10.4049/jimmunol.1300921
65. Tipton CM, Fucile CF, Darce J, Chida A, Ichikawa T, Gregoretti I, et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. (2015) 16:755–65. doi: 10.1038/ni.3175
66. Wang S, Wang J, Kumar V, Karnell JL, Naiman B, Gross PS, et al. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat Commun. (2018) 9:1758. doi: 10.1038/s41467-018-03750-7
67. Seelenmeyer C, Wegehingel S, Tews I, Künzler M, Aebi M, Nickel W. Cell surface counter receptors are essential components of the unconventional export machinery of galectin-1. J Cell Biol. (2005) 171:373–81. doi: 10.1083/jcb.200506026
68. Toscano MA, Bianco GA, Ilarregui JM, Croci DO, Correale J, Hernandez JD, et al. Differential glycosylation of TH1, TH2 and TH-17 effector cells selectively regulates susceptibility to cell death. Nat Immunol. (2007) 8:825–34. doi: 10.1038/ni1482
69. Motran CC, Molinder KM, Liu SD, Poirier F, Miceli MC. Galectin-1 functions as a Th2 cytokine that selectively induces Th1 apoptosis and promotes Th2 function. Eur J Immunol. (2008) 38:3015–27. doi: 10.1002/eji.200838295
70. Yang R, Avery DT, Jackson KJL, Ogishi M, Benhsaien I, Du L, et al. Human T-bet governs the generation of a distinct subset of CD11chighCD21low B cells. Sci Immunol. (2022) 7:eabq3277. doi: 10.1126/sciimmunol.abq3277
71. Park J, Munagala I, Xu H, Blankenship D, Maffucci P, Chaussabel D, et al. Interferon signature in the blood in inflammatory common variable immune deficiency. PloS One. (2013) 8:e74893. doi: 10.1371/journal.pone.0074893
72. Zhu W, Zhou Y, Guo L, Feng S. Biological function of sialic acid and sialylation in human health and disease. Cell Death Discovery. (2024) 10:415. doi: 10.1038/s41420-024-02180-3
73. Sun Y, Li X, Wang T, Li W. Core fucosylation regulates the function of pre-BCR, BCR and igG in humoral immunity. Front Immunol. (2022) 13:844427. doi: 10.3389/fimmu.2022.844427
74. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. (2015) 15:540–55. doi: 10.1038/nrc3982
75. Matsumoto Y, Ju T. Aberrant glycosylation as immune therapeutic targets for solid tumors. Cancers (Basel). (2023) 15. doi: 10.3390/cancers15143536
76. Gray MA, Stanczak MA, Mantuano NR, Xiao H, Pijnenborg JFA, Malaker SA, et al. Targeted glycan degradation potentiates the anticancer immune response in vivo. Nat Chem Biol. (2020) 16:1376–84. doi: 10.1038/s41589-020-0622-x
77. Kean EL, Münster-Kühnel AK, Gerardy-Schahn R. CMP-sialic acid synthetase of the nucleus. Biochim Biophys Acta Gen Subj. (2004) 1673:56–65. doi: 10.1016/j.bbagen.2004.04.006
Keywords: CD21low B cells, CVID, glycome, glycosylation, hypersialylation, hyperfucosylation, anti-IgM/IFNγ, T-bet
Citation: Felixberger PT, Andrieux G, Maul-Pavicic A, Goldacker S, Harder I, Gutenberger S, Landry JJM, Benes V, Jakob TF, Boerries M, Nitschke L, Voll RE, Warnatz K and Keller B (2025) CD21low B cells reveal a unique glycosylation pattern with hypersialylation and hyperfucosylation. Front. Immunol. 16:1512279. doi: 10.3389/fimmu.2025.1512279
Received: 16 October 2024; Accepted: 10 January 2025;
Published: 12 February 2025.
Edited by:
Aleš Janda, Ulm University Medical Center, GermanyReviewed by:
Catarina Gregório Martins, New University of Lisbon, PortugalChangsong Wang, The 989th Hospital of the PLA Joint Logistic Support Force, China
Copyright © 2025 Felixberger, Andrieux, Maul-Pavicic, Goldacker, Harder, Gutenberger, Landry, Benes, Jakob, Boerries, Nitschke, Voll, Warnatz and Keller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Klaus Warnatz, a2xhdXMud2FybmF0ekB1bmlrbGluaWstZnJlaWJ1cmcuZGU=
†These authors have contributed equally to this work and share last authorship
 Peter Tobias Felixberger
Peter Tobias Felixberger Geoffroy Andrieux
Geoffroy Andrieux Andrea Maul-Pavicic
Andrea Maul-Pavicic Sigune Goldacker
Sigune Goldacker Ina Harder1,2
Ina Harder1,2 Vladimir Benes
Vladimir Benes Till Fabian Jakob
Till Fabian Jakob Melanie Boerries
Melanie Boerries Lars Nitschke
Lars Nitschke Reinhard Edmund Voll
Reinhard Edmund Voll Klaus Warnatz
Klaus Warnatz Baerbel Keller
Baerbel Keller