
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 07 March 2025
Sec. Microbial Immunology
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1508584
This article is part of the Research TopicNontuberculous Mycobacterial Pulmonary Disease: Immunopathogenesis and Immunological Risk FactorsView all 6 articles
Introduction: Mycobacterium abscessus is a pathogen recently associated with patients with chronic lung conditions such as bronchiectasis and cystic fibrosis. M. abscessus is an environmental bacterium but recent evidence suggests that the pathogen is also transmitted from host-to-host. Because M. abscessus is known to form biofilms on the respiratory mucosa the release of bacteria from the biofilm becomes an important aspect on the transmission of the infection.
Methods: A biofilm releasing system was established. A transposon library of M. abscessus was then screened to identify genes associated with the release from biofilms.
Results: Several enzymes and genes of unidentified function were linked with the ability to detach from the biofilm. It was also shown that detached bacteria were increased capable of establish a new biofilm, attach to epithelial cells, and infect macrophages. To determine the surface molecules linked with the ability to infect new hosts, a surface proteomic was performed, showing that detaching bacteria express many proteins do not present in biofilm bacteria.
Discussion: Detached M. abscessus, one of the possible infectious phenotypes, contains specific proteins and lipids in the surface that facilitate the infection of new hosts. In addition, we identified many small proteins that have the likelihood to be associated with the release of the biofilm bacteria.
Pulmonary infections caused by Mycobacterium abscessus have been increasingly identified in individuals with chronic lung pathologies, such as emphysema, bronchiectasis, and cystic fibrosis (1–3). All three subspecies of M. abscessus, i.e., M. abscessus subsp. abscessus, M. abscessus subsp. Bolletii, and M. abscessus subsp. masssiliense, are resistant to the majority of available antibiotics, making the treatment of patients quite challenging (3, 4).
There is evidence that M. abscessus has evolved recently in order to adapt to the airway environment of humans harboring the risk factors (5, 6). Not only is the infection becoming more common, but additional evidence of genome evolution has also emerged (7, 8). A few clones of the pathogen have been isolated from infections in the respiratory tracts of patients, which suggests rapid evolution allowing for adaptation to the environment.
One of the strategies by which environmental bacteria adapt to mammalian host infections is the inherent ability to form a biofilm (BF). Studies and clinical experience have shown that M. abscessus uses that resource to initiate and maintain a niche or niches on the airway mucosal surface in patients with chronic lung pathologies (9, 10). To achieve this, the bacterium rapidly needs to establish and develop the biofilm structure before being challenged by the host immune cells or anti-bacterial products secreted by the host cells. We have previously described how M. abscessus responds to the environmental conditions of the airways with the efficient formation of biofilm (9). It is apparent that the bacterium uses the magnesium concentration in the mucus of cystic fibrosis patients to quickly adhere to the respiratory mucosa and develop a robust biofilm (9–11). It is also puzzling that the pathogen uses the host DNA existing in the airways as a carbon source, which apparently also has a role in the transition from the planktonic to the biofilm phenotype (12).
Considering all the accumulated pieces of information and adding the recent evidence that M. abscessus can be also transmitted among cystic fibrosis patients, in many cases, within the shared environment of a clinic waiting room or other facilities (7), we hypothesized the bacteria being released from airway biofilms would most likely represent an infectious phenotype of the pathogen. In fact, the ability to detach from biofilm can be linked with additional site seeding in the airways which would have implications in the extension of the disease. Understanding the pathogenic mechanisms associated with the infection will allow for the development of prevention measures or potentially the elimination of the disease in the at-risk population.
To address the question, we established an in vitro set of assays to identify detached organisms and the genes associated with the ability. Our findings identified many genetic components pointing to an association with genes and encoded proteins linked to surface structures and the transport mechanisms that exist to export these proteins to the bacterial surface. Further studies determined that some of these proteins participate in the attachment to bronchial epithelial cells.
Mycobacterium abscessus subsp. abscessus strain 19977 was obtained from the American Type Culture Collection (ATCC). M. abscessus subsp. abscessus strains 00103 and 01715 (obtained from patients with cystic fibrosis) were a gift from Jerry Nick and Charles Daley from the National Jewish Health Hospital. All three strains were of the in vitro smooth phenotype. Unless otherwise noted, bacteria for assays were grown and maintained on Middlebrook 7H10 agar supplemented with 10% oleic acid, albumin, dextrose, and catalase (OADC; Hardy Diagnostics), referred to as 7H10. For liquid cultures, bacteria were grown in 7H9 broth supplemented with Tween-80 and OADC, referred to as 7H9. All the cultures were grown at 37°C for 3 to 5 days. The bacteria used in the assays were of the planktonic phenotype initially. Every assay was controlled by microscopic observation so that the inoculum completely dispersed.
Human monocyte THP-1 (TIB-202) and HEp-2 (CCL-23) pharyngeal epithelial cells were obtained from the ATCC and grown in RMPI-1640 (RPMI) and DMEM sterile media containing L-glutamine, 25 mM HEPES (Corning Life Sciences), and 10% fetal bovine serum (FBS, Gemini Bio-products, Sacramento, CA). Both cell types were obtained from the ATCC and maintained at 37°C with 5% CO2.
M. abscessus biofilms were formed by generating a turbid suspension of the bacteria in HBSS. Turbidity was measured via O.D. using an EPOCH spectrophotometer (Biotex) at 595nm to obtain a concentration of 3 x 108 CFU/mL. SCFM media were prepared as described previously (9, 13) and utilized as a biofilm formation condition in tandem with HBSS biofilm formation. For the biofilms formed in SCFM, bacteria were diluted to obtain 1 x 106 CFU/200 µL and biofilms formed in HBSS were diluted to obtain 1 x 107 CFU/200 µL. Secondary biofilms were established by collecting the supernatant from primary biofilms during the replicative stage (day 2) or non-replicative stage (day 5) of biofilm formation and then placed in fresh SCFM or HBSS. SCFM and HBSS were replaced with fresh media every 24 h so that the supernatants mainly contained released bacteria during biofilm formation. Secondary biofilms were established for 7 and 14 days. The biomass of the primary and secondary biofilms was quantified using the crystal violet assay (14) and absorbance was measured at 550 nm. CFU quantitation for the biofilms was obtained by mechanically disrupting the biofilms via pipetting, followed by serial dilutions and plating on 7H10 agar. Planktonic bacteria at 1 x 105, 1 x 106, and 1 x 107 CFU/mL served as biofilm controls.
The M. abscessus transposon library was generated as described previously (15). MycomarT7 (mmT7) is a temperature-dependent transposon-containing phagemid and was kindly provided by Eric Rubin (Harvard T.H. Chan School of Public Health, Boston, MA). MmT7 was propagated and titers were generated using M. smegmatis strain mc2 155 as described previously (15). M. abscessus was grown in 7H9 broth supplemented with 10% OADC and 0.1% Tween-80 at 37°C in a shaking incubator before transduction. Bacteria were then pelleted and washed with MP buffer (150 mM NaCl, 50 mM Tris-HCl, 10 mM Mg2SO4, 2 mM CaCl2) twice. Washed bacteria were resuspended in MP and buffered and infected with mmT7 at an MOI of 2. The transduction commenced at 37°C for 4 hours with intermittent mixing. Aliquots were plated on 7H10 containing 400 µg/mL kanamycin to obtain individual transposon mutants. A selection of mutants was screened for the presence of mmT7 by amplifying the kanamycin resistance gene with PCR.
The SCFM and HBSS biofilm conditions were screened to identify mutants unable to release bacteria to form secondary biofilms. For the SCFM biofilms, individual mmt7 mutants were selected and cultured in 7H9 broth containing 400 µg/mL kanamycin for 4 days at 37°C in a shaking incubator using a 96-deep-well plate format. After 4 days, bacteria were pelleted and resuspended in HBSS. Fresh SCFM was aliquoted into a flat bottom 96-well plate, and 50 µL of each mutant suspension was added to the plate. Biofilms were formed for 7 days in the dark at 25°C with supernatant replacement after 2 days and then the resulting supernatant was carefully removed to form a secondary biofilm. These secondary biofilms were allowed to form for another 7 days and then the O.D. of the supernatant was measured along with the biomass of the biofilm using the crystal violet assay. Mutants were selected based on diminished secondary biofilm biomass and low O.D. in the corresponding supernatant compared to the wild-type.
For HBSS biofilms, individual mmt7 mutant colonies were added to a 48-well plate containing 7H9 broth media and allowed to grow for 3-4 days. Once mutants had grown to 3 x 108 CFU/mL, 100 µL of the bacteria was placed in 900 µL of HBSS in a fresh 48-well plate to form biofilm for 7 days in the dark at 37°C with supernatant replacement after 2 days. After 7 days, the supernatant was carefully removed without disturbing the transposon mutant biofilm and placed in another fresh well containing HBSS. The supernatant of each of these mutants was allowed to form biofilm for another 7 days, and the O.D. at 595 nm was measured, along with a visual inspection for opaqueness. Wells that matched the O.D. of HBSS alone or had a low O.D. were considered deficient in release or secondary biofilm formation. These mutants were selected for Sanger sequencing using an ABI 3730 capillary sequence machine within the Center for Quantitative Life Sciences (CQLS) at Oregon State University.
Transposon mutants were reconfirmed in duplicate before sequencing as previously described (9). Briefly, mutants selected from the biofilm detachment screen were sequenced utilizing a previously reported LM-PCR assay with some changes (16). Cells were lysed using 1-mm-diameter glass beads in diH2O via mechanical homogenization. Lysates were centrifuged for 1 minute at 21,000 x g to pellet cell debris. DNA was purified from collected supernatant using a DNA clean and concentrate kit (Zymo Research) following the manufacturer’s protocol. Furthermore, 150 ng of purified DNA was utilized for single digestion with SalI (Thermo fast-digest enzyme) for 30 minutes at 37°C. LM-PCR adapter oligos were generated for SalI (Salgd+Salpt, see Table 1) by adding 45 mM of each oligonucleotide to 1x Taq DNase buffer plus MgCl2 and ligating by decreasing the temperature from 80°C to 4°C over an hour. After digestion, DNA for each mutant was ligated with the adapter oligos using T4 DNase ligase. The ligated product was used as the template in the LM-PCR reaction. The LM-PCR reaction utilized Gold 360 MasterMix (Applied Biosystems) and was performed using 97°C for 7 min, 40 cycles of 97°C for 30 s, 58°C for 1 min, and 72°C for 1 min 45 s, and then a final step of 72°C for 10 min. PCR products were visualized using gel electrophoresis and ethidium bromide. The bands of interest were excised and purified using a gel extraction kit (Thermo) and sequencing at the Center for Quantitative Life Sciences (CQLS) at Oregon State University. Sequencing results were blasted in the NCBI against M. abscessus to identify disrupted genes.
The MAB_4706c gene with native promoters was complemented by the use of the integrative pMV306 plasmid as previously described (17) with some changes. First, an apramycin-resistance gene was cloned into pMV306 to generate the pMV306-Apr plasmid. Genes were cloned into pMV306-Apr using primers designed for these specific regions. This product was then transformed into electrocompetent DH10β E. coli and grown on LB containing 400 µg/mL kanamycin and apramycin. Positive colonies were used for colony PCR to confirm apramycin resistance and gene of interest ligation. These constructs were extracted using a Qiagen Mini Prep kit as per the manufacturer’s protocol. Electrocompetent M. abscessus mutant cells were prepared by washing plate-grown bacteria four times via centrifugation at 2000 x g at 4°C for 10 minutes in an ice-cold 10% glycerol and 0.1% Tween-80 solution. Cells were stored at -80°C in 10% glycerol until use. Plasmids were electroporated into M. abscessus competent cells using a 0.2cm cuvette (BioRad) at 2500 V, 1000Ω, and 25µF. Bacteria were recovered in 7H9 for 2 to 3 hours and plated onto 7H10 containing 400 µg/mL of kanamycin and apramycin and positive colonies were screened via PCR as described above.
Biofilms were established for 7 days in either HBSS or SCFM as described above. THP-1 monocytes were stimulated with 100 ng/mL of IFN-γ for 24 h prior to biofilm contact. After biofilm maturation, supernatants were removed and replaced with stimulated THP-1 monocytes (1x105 cells/well) in RPMI media. The biofilms were mechanically lysed at days 1, 2, and 3 post THP-1 addition to determine macrophage activity against these biofilms as quantified by CFUs. Activated THP-1 cells were also added during the replicative stage and non-replicative stage of biofilm formation by replacing supernatants with RMPI media followed by CFU determination at days 1 and 3 post contact with monocytes via serial dilution.
THP-1 cells were seeded at 3 x 105 cells/500 µL of RPMI supplemented with 10% FBS containing 50 ng/mL phorbol 12-myristate 13-acetate (PMA) and incubated at 37°C. After 24 hours, PMA was removed and replaced with fresh complete media and incubated overnight at 37°C. Biofilms were formed as previously described in either HBSS or SCFM. At day 7, the biofilms were disrupted to release bacteria and then THP-1 macrophages were infected with primary biofilm or secondary biofilm bacteria for 1 hour, including a 10-min synchronization step at 150 x g. To kill extracellular bacteria, 200 µg/mL of amikacin was added to the monolayers for 2 hours. Cells were lysed post antibiotic treatment, at 24 hours, and 72 hours post infection. Lysates were serial diluted and plated on 7H10 agar for intracellular CFU quantification.
HEp-2 cells were seeded with DMEM media containing 10% FBS and allowed to achieve 90% confluency. Cells were then overlayed with ice-cold culture medium containing 106 CFU/mL bacteria to prevent internalization of bacteria by the epithelial cells. Wild-type M. abscessus 19977, secondary biofilm mutants, and a complemented strain were utilized for infection. The M. abscessus strains were allowed to bind to epithelial cells for 30 min at 4°C. Monolayers were then washed with ice-cold HBSS twice and lysed with 0.01% TritonX-100 and serially diluted on 7H10 agar for CFU determination.
To isolate surface proteins from bacteria, we utilized biotinylation followed by streptavidin-bound magnetic bead purification. We compared planktonic bacteria, bacteria with the biofilm phenotype in HBSS (7-day biofilm), bacteria that formed secondary biofilms in HBSS, and bacteria that formed secondary biofilms in SCFM. First, bacteria cells were washed twice with HBSS. After the 2nd wash, bacteria pellets were resuspended in 1 mg Sulfo-NHS-LC-biotin (Thermo Scientific) reconstituted in 1mL HBSS and then incubated for 30 min at 4°C with gentle rotation. Leftover biotin reagent was quenched with 10 mM glycine in HBSS for 10 min at room temperature under gentle rotation. Bacteria cells were washed twice with 1mL HBSS. After washing, the pellets were resuspended with GLIB buffer (10 mM EDTA, 2 mM EGTA, 0.1% tween-20, 6 M guanidinium HCl in PBS, pH 7.2) and then transferred to a 2 mL bead-beating tube with <0.1 mm glass beads.
Bacteria samples were lysed using an Omni Bead Ruptor (Omni Intl.) set to speed 4 with three cycles of 30 s between each cycle samples and were placed on ice to prevent degradation. To clear, the lysate samples were centrifuged for 1 min at room temperature at 21,000 x g. The supernatant was collected and passed through a 0.2 µm syringe filter to remove DNA and any large debris from the protein extract. The supernatant was transferred to a 2 mL protein lo-bind (Eppendorf) tube. Streptavidin Dynabeads (Thermo Scientific) were added to the extract and incubated for 1 hour in a rotary shaker at room temperature. The protein mixture was transferred into a µMacs magnetic column (Miltenyi Biotec). The beads were washed twice with GLIB buffer with an incubation step in between for 5 mins on a rotary shaker. After second GLIB wash, the beads were washed twice with PBS (0.05% tween-20 in PBS, pH 7.2) in a new clean tube with an incubation of 30 min on a rotary shaker at room temperature. Beads containing surface proteins were eluted in resuspension buffer (1% SDS, 10 mM EDTA in H2O) and then incubated at 65°C for 10 min to denature proteins from the beads for mass spectrometry.
To reduce the disulfide bonds of the proteins, the samples were incubated at 56°C for 1 hour with 5 mM dithiothreitol (ThermoFisher). The samples were then incubated with 10 mM iodoacetamide (MilliPore Sigma) for 1 hour at room temperature in the dark in order to carbamidomethylate the cysteine residues. The samples were digested overnight at 37°C using Trypsin Gold (Mass Spectrometry Grade, Promega). After digestion, the samples were spun down at 12,000 x g for 30 s to collect the condensate, and the digestion was stopped by the addition of 0.5% (v/v) trifluoroacetic acid. The samples were centrifuged at 12,000 x g for 10 minutes and then transferred to LC vials.
A Waters nanoAcquityTM UPLC system (Waters, Milford, MA) was coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Peptides were loaded onto a trap 2G nanoAcquity UPLC Trap Column (180um, 50mm, 5um) at a flow rate of 5 μL/min for 5 min. The results were obtained on a commercially available Acquity UPLC Peptide BEH C18 column (100um, 100mm, 1.7um). The column temperature was maintained at 37°C using the integrated column heater. Solvent A was 0.1% formic acid in LC-MS grade water and solvent B was 0.1% formic acid in LC-MS grade acetonitrile. The separation was performed at a flow rate of 0.5 μL/min, and using linear gradients of 3% to 10% B for 10 min, 10% to 30% B for 10 min, 30% to 70% B for 5 min, 70% to 95% B for 3 min, 95% to 3% B for 4 min, and 95% to 3% B for 3 min. Total method length was 35 min. The outlet of the column was connected to a Thermo Nanospray Flex ion source and +2300V were applied to the needle.
MS1 spectra were acquired at a resolution of 120,000 (at m/z 200) in the Orbitrap using a maximum IT of 50 ms and an automatic gain control (AGC) target value of 2E5. For MS2 spectra, up to 10 peptide precursors were isolated for fragmentation (isolation width of 1.6 Th, maximum IT of 10 ms, and AGC value of 2e4). Precursors were fragmented by HCD using 30% normalized collision energy (NCE) and analyzed in the Orbitrap at a resolution of 30,000. The dynamic exclusion duration of fragmented precursor ions was set to 30 s. Raw files were processed in Thermo Proteome Discoverer 2.3. Precursor ion mass tolerance was set to 5 ppm, while fragment ion mass tolerance was 0.02 Da. The SequestHT search engine was used to search against the Swissprot human and M. abscessus protein database. Only b and y ions were considered for peptide spectrum matching. MS1 precursor quantification was used for label-free quantitation of the peptides. Protein abundances were calculated as the sum of the abundances of unique peptides detected.
Statistical analyses were performed using GraphPad Prism 9 software. The comparisons between the treatment groups were analyzed using either t-tests or analysis of variance (ANOVA) with multiple comparisons where appropriate. Specific statistical tests are named in the figure legends in which they were used. A P-value of < 0.05 was considered significant.
A key aspect of the biofilm formation cycle is the bacteria’s ability to detach from the biofilm and establish a secondary or satellite biofilm. Growing evidence has been mounting of patient-to-patient transmission in hosts that have cystic fibrosis, but more information is needed to ascertain this infectious phenotype of M. abscessus. Previous work in our lab showed that M. abscessus can form robust biofilms in synthetic cystic fibrosis media (SCFM), and specifically that the colony forming units (CFUs) of the biofilm do not increase after day 4, while the matrix biomass continues to increase over time (9). Based on this, the capability of bacteria released from replicative stage biofilms and non-replicative stage biofilms to re-form biofilms was investigated. Supernatants of bacteria forming biofilms were carefully removed from wells on day 2 (replicative stage) and day 5 (non-replicative stage) and utilized to form biofilms. These secondary biofilms were developed in either a 1:1 ratio of supernatant to fresh media or unaltered transferred supernatant (Figure 1). Secondary biofilms were established for 7 and 14 days in Hank’s balanced salt solution (HBSS) or SCFM. The biomass was determined at both time points, and matched primary biofilms of wild-type bacteria were included under SCFM and HBSS formation conditions. Released bacteria from both the replicative stage (Figures 1A, B) and non-replicative stage biofilms (Figures 1C, D) were able to form secondary biofilms, especially when introduced to fresh media. The biomass of the SCFM biofilms formed with non-replicative stage bacteria matched the biomass of wild-type bacteria when added into fresh SCFM at both time points (Figures 1C, D). The biomass of the secondary biofilms transferred without fresh media remained significantly lower than the control groups in all conditions. There did not appear to be differences in the biomass of secondary biofilms formed by released bacteria in HBSS compared to wild-type bacteria except at day 14 for the replicative stage supernatants and day 7 for the non-replicative stage supernatants compared to the 107 bacteria control (Figures 1B, C, respectively).
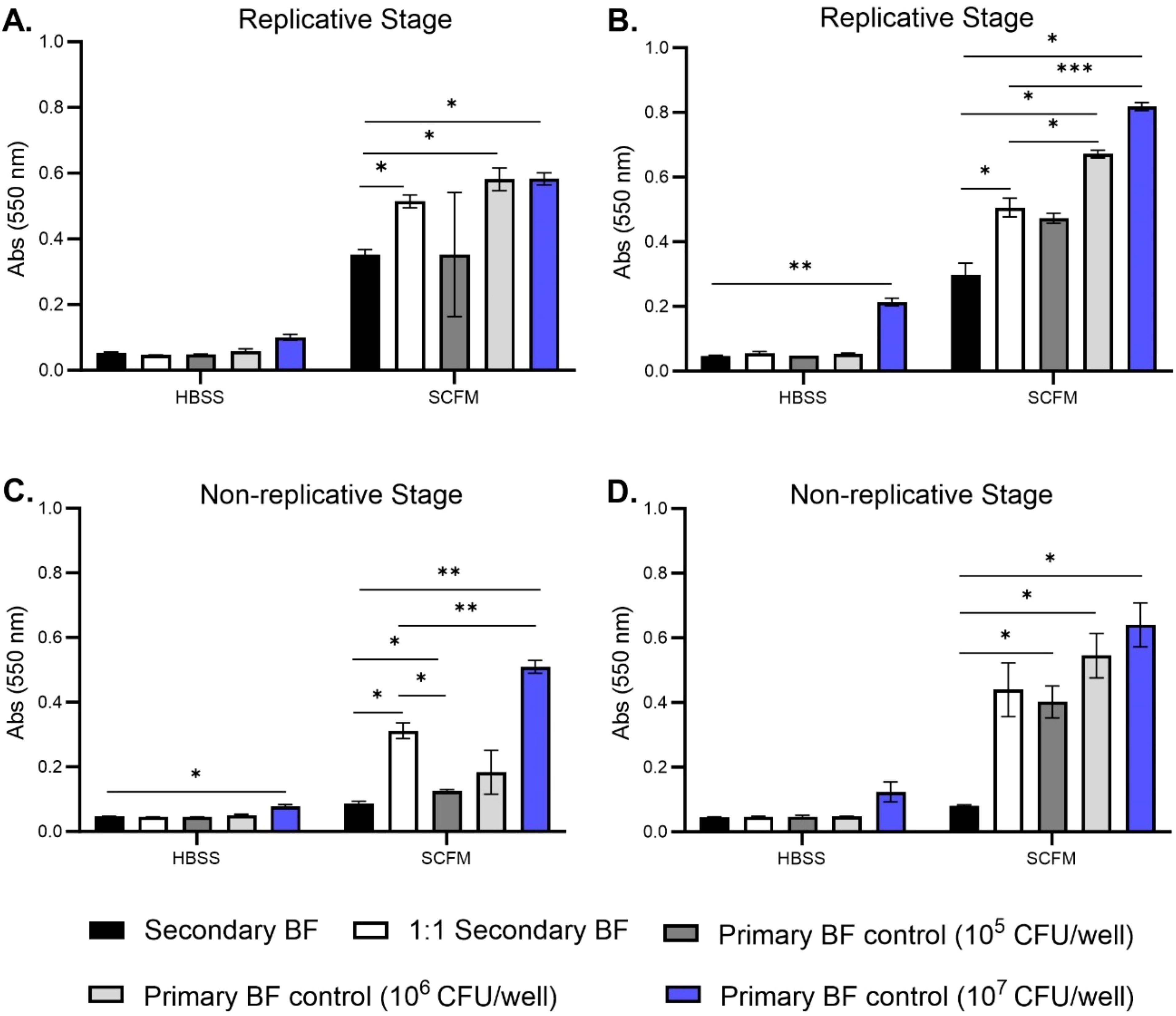
Figure 1. M. abscessus bacteria released from primary biofilms are capable of establishing secondary biofilms. M. abscessus bacteria released from replicative stage biofilms and non-replicative stage biofilms were utilized to reform secondary biofilms in both HBSS and SCFM conditions. Biomass was measured for replicative stage secondary biofilms at days 7 (A) and 14 (B). Biomass was measured for non-replicative stage secondary biofilms at days 7 (C) and 14 (D) via crystal violet assay. BF, biofilm. The error bars represent the SEM of three biological replicates. Statistical analysis was conducted using Brown–Forsythe and Welch ANOVA with a Dunnett’s T3 multiple comparison test; *** indicates p < 0.0001, ** indicates p < 0.001, * indicates p < 0.05.
Next, assays were performed to determine whether the replicative stage biofilm-released bacteria had any advantage in establishing secondary biofilms more quickly than the wild-type bacteria in SCFM conditions. For CFU determination, both the disrupted biofilms and released biofilms into the supernatant were enumerated. Similarly, the replicative stage released bacteria were placed in either a 1:1 ratio of supernatant to fresh SCFM media or unaltered transferred supernatant and the biomass and CFUs were measured at days 2, 5, and 7 during biofilm formation (Figure 2). As seen previously, all the conditions established more robust biomass by day 5, with day 7 staining showing the highest biomass [Figure 2A (9)]. Unaltered supernatants were significantly increased by day 2; however, by days 5 and 7, the secondary biofilm biomass was less than the primary wild-type biofilms (Figure 2A). The secondary biofilms, when given fresh media, had similar biomass accumulation to the wild-type. The CFU/well was obtained for the bacteria in the biofilm or detached phenotypes (Figure 2B). Bacteria replication (CFU/mL) in the secondary biofilm conditions increased on day 2 but plateaued by day 5 whereas the supernatant phenotypes dropped on day 2 but then increased by day 5, suggesting the bacteria were switching to biofilm-formation phenotypes between days 0 and 2 but increased biomass formation at later time points. Taken together, M. abscessus is able to release bacteria from biofilms, and these bacteria can form secondary biofilms similar to those formed by the wild-type.
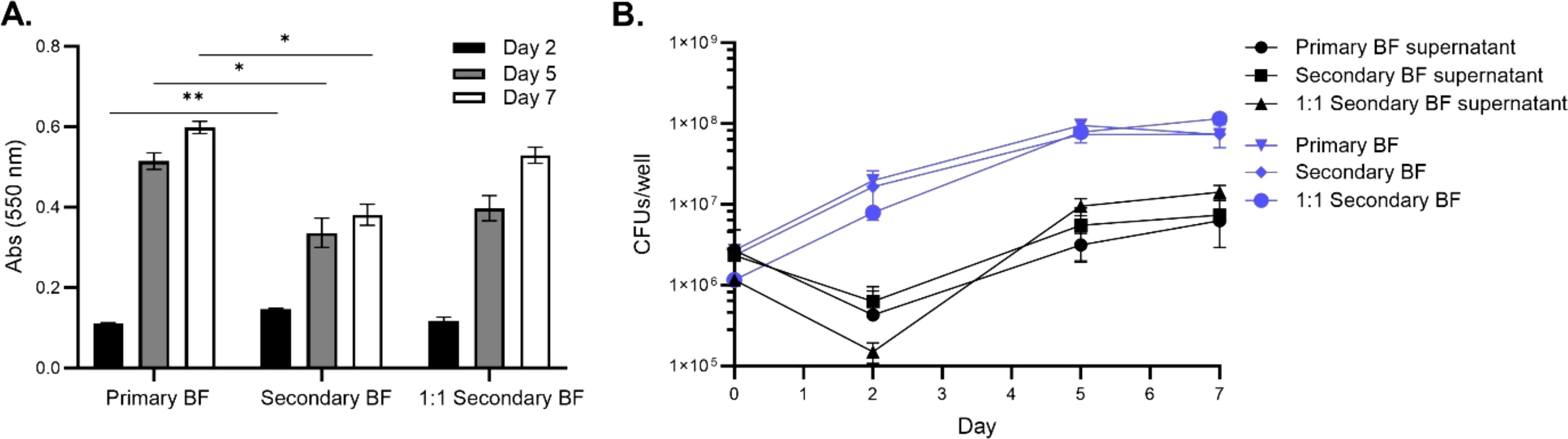
Figure 2. Secondary biofilms were established similarly to wild-type M. abscessus primary biofilms in SCFM. The replicative stage supernatants of SCFM primary biofilms were used to form secondary biofilms. The biomass was determined for the primary biofilm, unaltered supernatant forming a secondary biofilm, and a 1:1 ratio of supernatant with fresh SCFM forming a secondary biofilm (A). The CFU/well was determined for both the biofilm and released bacteria in the supernatant (B). For both assays, the timepoints were measured at days 2, 5, and 7 of the formation process. BF, biofilm. The error bars represent the SEM of two biological replicates. Statistical analysis was conducted using Brown–Forsythe and Welch ANOVA with a Dunnett’s T3 multiple comparison test; ** indicates p < 0.001, * indicates p < 0.05.
To verify whether the results obtained were only observed with the M. abscessus 19977 strain or were also seen with other strains isolated from patients with cystic fibrosis, the ability of the strain M. abscessus 00103 and M. abscessus 01715 were evaluated in comparison with the strain M. abscessus 19977 regarding the efficiency to form a new biofilm once it detaches from an existing biofilm. The results shown in Table 2 suggest that all three strains behaved similarly and that detaching from a biofilm is a more efficient manner to establish a new biofilm than when bacteria were obtained from an agar plate.
To better understand the mechanisms underpinning the release of M. abscessus from primary biofilms, an mmt7 transposon library was screened for mutant clones that do not release from primary biofilms in both HBSS and SCFM conditions. The mutants in this system were defined as being unable to establish a secondary biofilm. A supernatant containing released bacteria was utilized to form another biofilm, and each clone’s secondary biofilm was compared to that of the wild-type. In SCFM conditions, eight mutants were identified via ligation-mediated PCR (LM-PCR) and sequencing identified transposon insertion (Table 3). The transposon was inserted directly into two genes encoding hypothetical proteins [MAB_0274c (GXWXG domain), MAB_1222], although probable neighboring genes were affected if in an operon. Three membrane proteins [MAB_4706c (membrane-associated oxidoreductase complex, DoxX), MAB_2480 (Transmembrane), MAB_2301 (MmpL lipoprotein), MAB_0525c (LpqG lipoprotein)]. A chaperonin [MAB_0650 (GroEL)] and an enzyme [MAB_1277 (glycosyltransferase, modifies lipopolysaccharide]. In HBSS conditions, 15 mutants were selected and sequenced (Table 4). The transposon inserted into two tRNA’s (MAB_0275 and MAB_t5030c), along with two lipoproteins [MAB_3785c (LppF), MAB_0307c] and a translation elongation factor [MAB_1310 (BipA)]. Two genes encoded oxidoreductases [MAB_2438 (molybdopterin) and MAB_4628c]. One gene was a monooxygenase enzyme (MAB_4050c). A chaperonin [MAB_0650 (GroEL)] and a probable carbon starvation protein [MAB_1260c (CstA)] were also found. Two genes had domains suggesting kinase activity [MAB_3538 (DAGKc-domain) and MAB_1507 (hypothetical protein)]. An alkyl mercury lyase (MAB_4789c), a membrane protein [MAB_1170 (transports anions)], and transcriptional regulator (MAB_2089) were found. Further characterization is needed to understand the role these genes play in bacterial release from biofilms.
M. abscessus lung infections are associated with biofilms, however, the infection likely originates with bacteria released from a biofilm in an environmental setting or from another patient. The bacteria encounter the lung epithelium and must be able to invade or establish a niche to form biofilms (12). The ability of M. abscessus to bind to respiratory epithelial cells was assayed by utilizing the wild-type M. abscessus 19977 and secondary biofilm mutants from the previous section (Figure 3). Unfortunately, no secondary biofilm mutants from SCFM demonstrated a significant reduction in binding epithelial capacity compared to the wild-type (Figure 3A). Six HBSS secondary biofilm mutants had significantly inhibited binding compared to wild-type M. abscessus 19977 (Figure 3B). Three of these binding deficient mutants are hypothetical proteins [MAB_1170 (membrane transporter for anions), MAB_4789c (Alky mercury lyase in an operon with TetR gene for regulating MmpL transporters), and MAB_1507 (conserved hypothetical protein with kinase activity)]. The observation of kinases and more transporter regulation are also key factors in the binding to epithelial cells and not just for the release from biofilms. Other mutants may encode enzymes or signals that are involved in the interaction with epithelial cells. Next, released bacteria from both the SCFM and HBSS biofilms were collected 7 days after formation and utilized to infect epithelial cells. The bacteria released from the biofilms, whether formed in HBSS or SCFM, significantly increased the invasion of epithelial cells compared to planktonic M. abscessus after 1 hour of infection (Figure 4). This finding further supports the hypothesis that the released bacteria phenotype from established biofilms is an adaptation suited not only for reforming biofilms but also establishing an infection in the lungs.
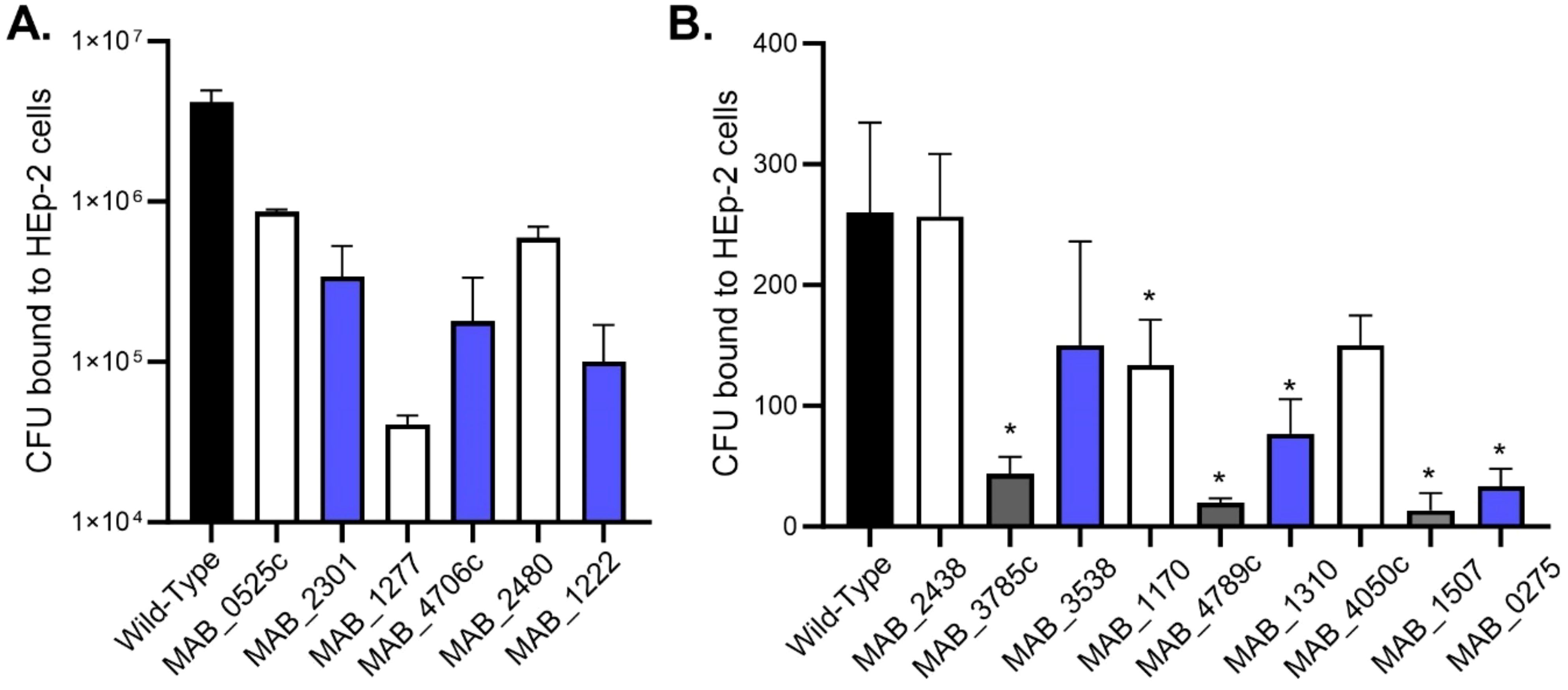
Figure 3. M. abscessus secondary biofilm mutants were deficient in binding the host epithelium. Wild-type and secondary biofilm deficient mutants identified in SCFM (A) or HBSS (B) media were incubated with HEp-2 epithelial cells at 4°C for 30 min. CFUs were recovered from epithelial cells. The error bars represent the SEM of 3 experiments. Statistical analysis was conducted using Brown–Forsythe and Welch ANOVA with a Dunnett’s T3 multiple comparison test. All mutants were compared to the wild-type; * indicates p < 0.05.
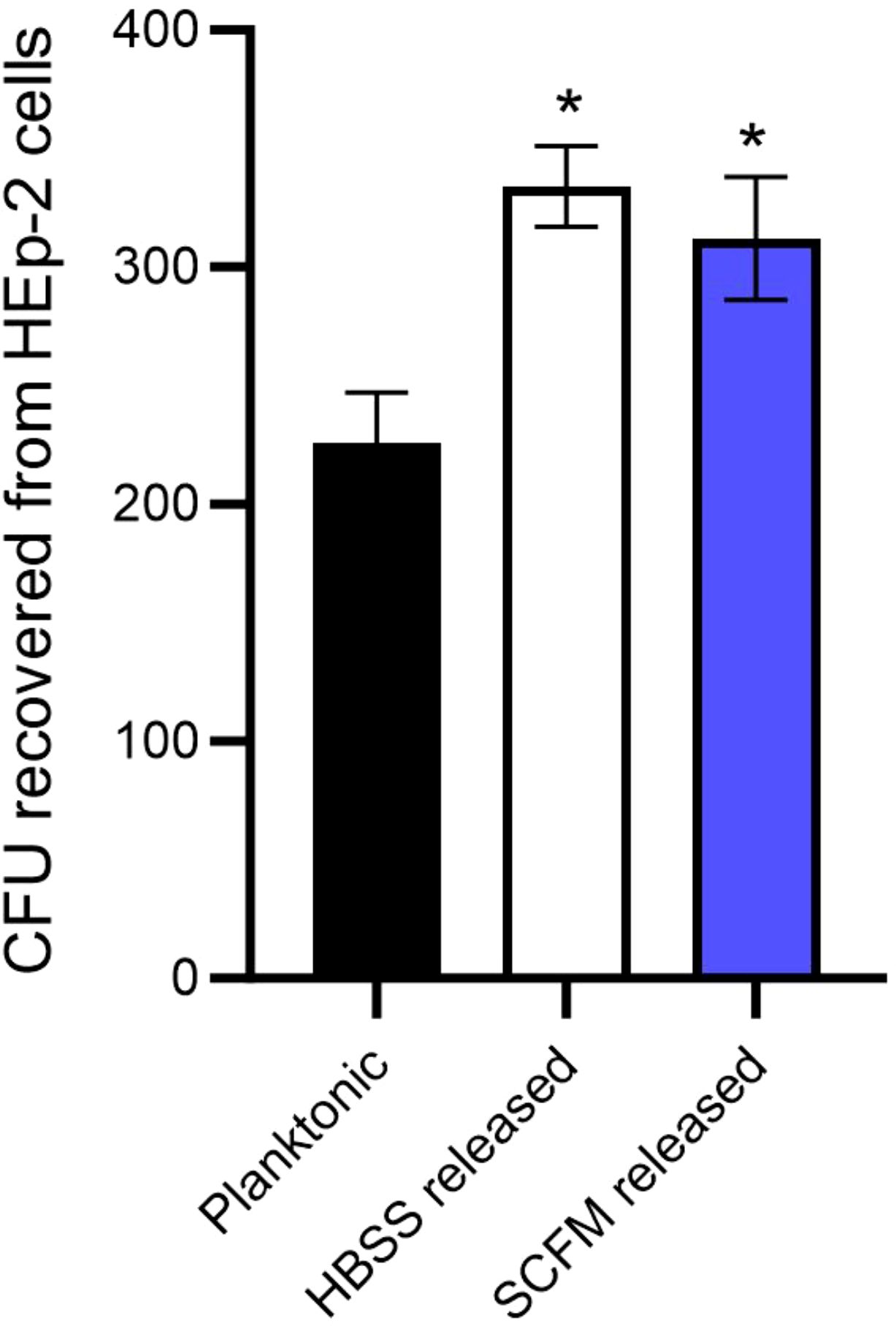
Figure 4. M. abscessus bacteria released from biofilms are more efficient at invading the epithelial cells. HEp-2 cells were infected with either planktonic M. abscessus or bacteria released from biofilms after 7 days in either HBSS or SCFM. The error bars represent SEM from three experiments. Statistical analysis was conducted using the Mann–Whitney t-test; * indicates p < 0.05.
MAB_4706c was cloned into pMV306 containing an apramycin-resistance gene and transformed into electrocompetent mutant MAB_4706c cells. Complemented MAB_4706c (ΔMAB_4706c) was tested against wild-type and mutant cells in its ability to form satellite biofilms and bind to epithelial cells (Figure 5). Secondary biofilms were formed in SCFM with wild-type, mutant 4706c, and ΔMAB_4706c, and absorbance readings were taken after 7 days (Figure 5A). ΔMAB_4706c had a similar optical density (O.D.) to wild-type M. abscessus 19977, while mutant 4706c biofilms had significantly decreased absorbance readings, suggesting complementation restores the bacteria’s ability to disperse from primary biofilms. The epithelial binding assay was repeated using the wild-type, mutant, and complemented bacteria (Figure 5B). ΔMAB_4706c had a similar binding capability to wild-type M. abscessus 19977, also restoring this phenotype. Taken together, the genes in the MAB_4706c operon play a major role in M. abscessus biofilm release and secondary attachment to the respiratory epithelium. MAB_4706c and MAB_4702c are both hypothetical proteins, while 4705c, 4704c, and 4703c are all membrane proteins in the MmpL family.
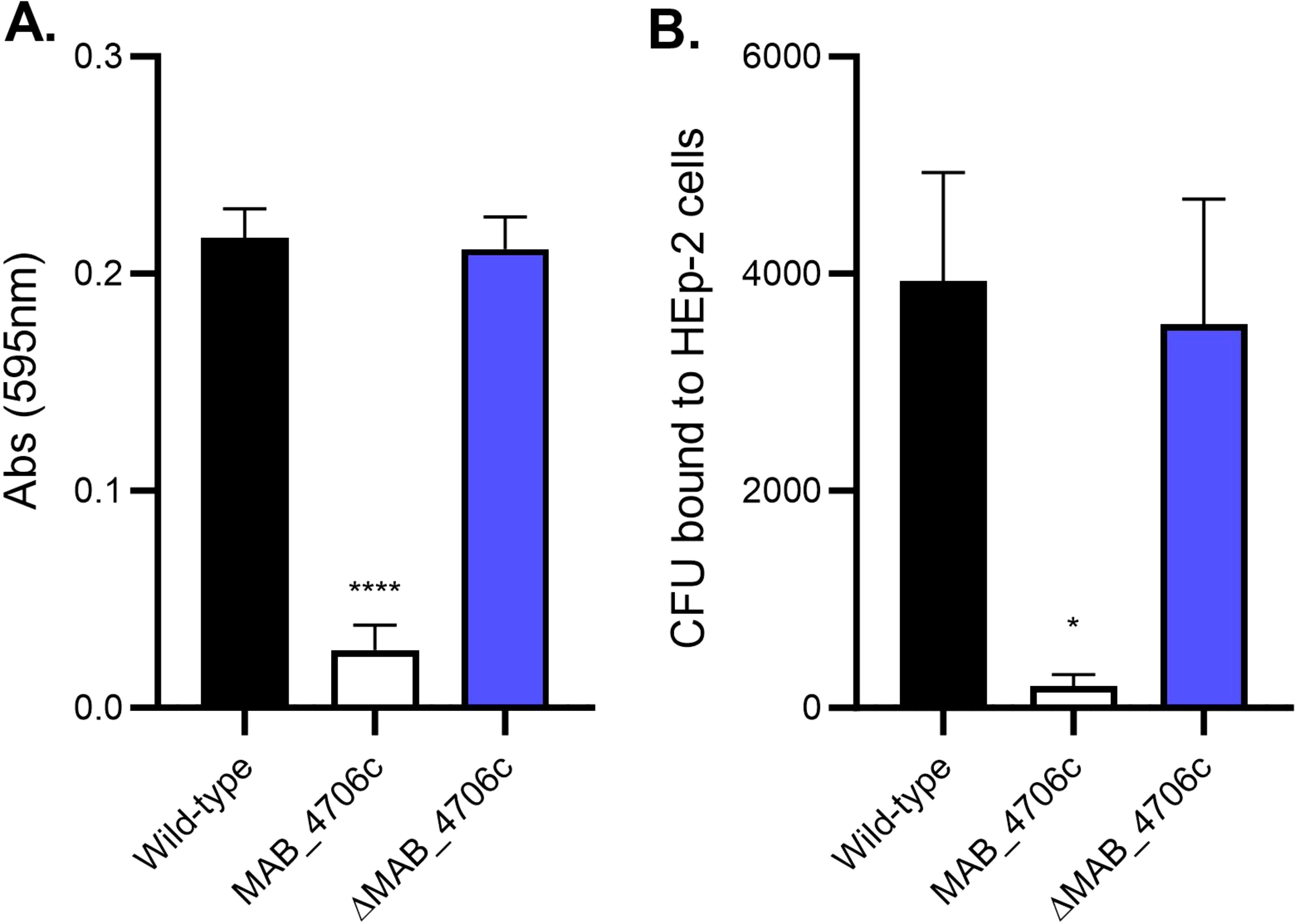
Figure 5. Complementation of MAB_4706c restores the wild-type phenotype. The ability of the bacteria to form secondary biofilms and bind to epithelial cells was determined. Wild-type M. abscessus 19977, mutant 4706c, and complemented MAB_4706c (ΔMAB_4706c) were utilized to form secondary biofilms (A) and assayed for epithelial binding (B). Data are representative of three biological replicates. Statistical analysis was conducted using Brown–Forsythe and Welch ANOVA with a Dunnett’s T3 multiple comparison test; **** indicates p < 0.0001, * indicates p < 0.05.
Macrophages are the first immune cells that bacteria encounter in the lung environment, whether they are circulating airway macrophages or tissue macrophages after penetrating the epithelial layer. Mycobacteria are obligate intracellular pathogens and can disseminate the infection by infecting secondary macrophages in a transitionary manner. The ability of the bacteria collected from primary biofilms and those of released bacteria to form secondary biofilms means they are able to invade and replicate in macrophages (Figure 6). Disrupted primary biofilm bacteria were more efficient at invading macrophage monolayers compared to planktonic bacteria (Figure 6A). Both released bacteria and disrupted biofilm phenotypes were able to grow by day 3 within the macrophages (Figures 6B, D). Bacteria released from secondary biofilms had a similar uptake to plate bacteria but may not necessarily confer an advantage (Figure 6C). Overall, M. abscessus released from biofilms or in the biofilm phenotype were able to invade and grow intracellularly, regardless of whether the biofilms were made in SCFM or HBSS.
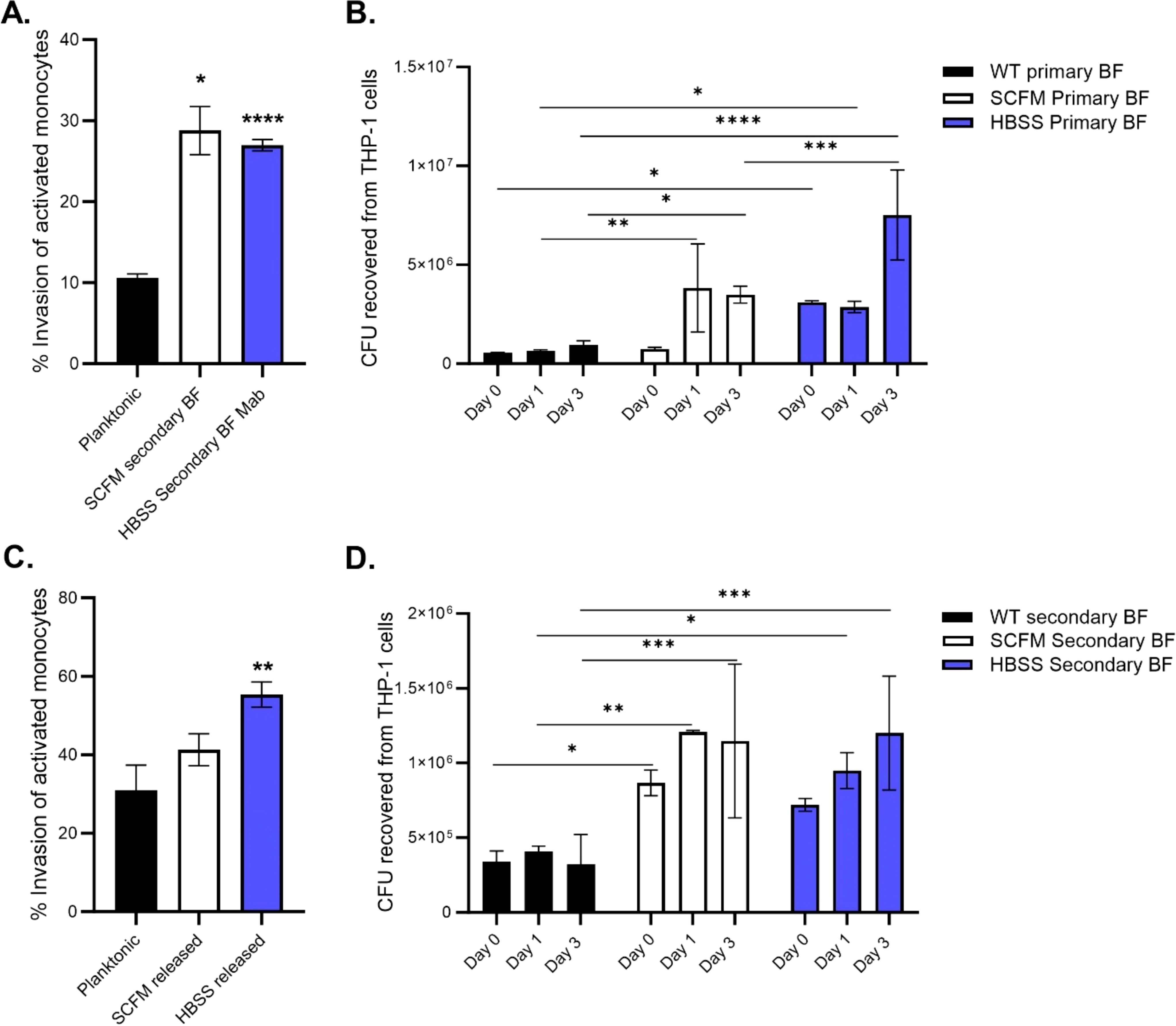
Figure 6. Macrophages ingest M. abscessus biofilm phenotypes more efficiently than planktonic bacteria. Bacteria from primary biofilms (A, B) or bacteria from secondary biofilms (C, D) were used to infect THP-1 macrophages. The invasion percentage (A, C) was determined after a 1-hour infection and surviving intracellular CFUs were obtained at days 0,1, and 3 (B, D) post-infection. Data are representative of two biological replicates. Statistical analysis was conducted using Brown–Forsythe and Welch ANOVA with a Dunnett’s T3 multiple comparison test (A, C) and 2-way ANOVA with Tukey’s multiple comparison test (B, D); **** indicates p < 0.0001, *** indicates p < 0.001, ** indicates p < 0.01, * indicates p < 0.05.
Next, IFN-γ-stimulated monocytes were added to mature biofilms in both SCFM and HBSS to determine whether the cells could eliminate biofilm CFUs (Figure 7). Monocytes did not decrease any of the biofilm CFUs at all time points: 24, 48, and 72 hours post addition (Figure 7A). Planktonic bacteria were significantly reduced by monocytes compared to the bacteria protected by biofilms. THP-1 monocytes were also added during the biofilm formation process in replicative stage biofilm (day 2) and non-replicative stage biofilm (day 5) (Figures 7B, C, respectively). The addition of phagocytic cells to biofilms did not contribute to a decrease in biofilm CFUs during the replication stage. However, during the stationary phase, biofilms established in HBSS were significantly reduced by day 3 compared to the HBSS or SCFM biofilm control. As before, the planktonic bacteria control had a significant reduction in CFUs compared to its biofilm counterparts. Taken together, bacteria released from biofilms or in the biofilm phenotype are able to combat macrophage killing but only confer an advantage if formed in SCFM and during the non-replicative stage of formation.
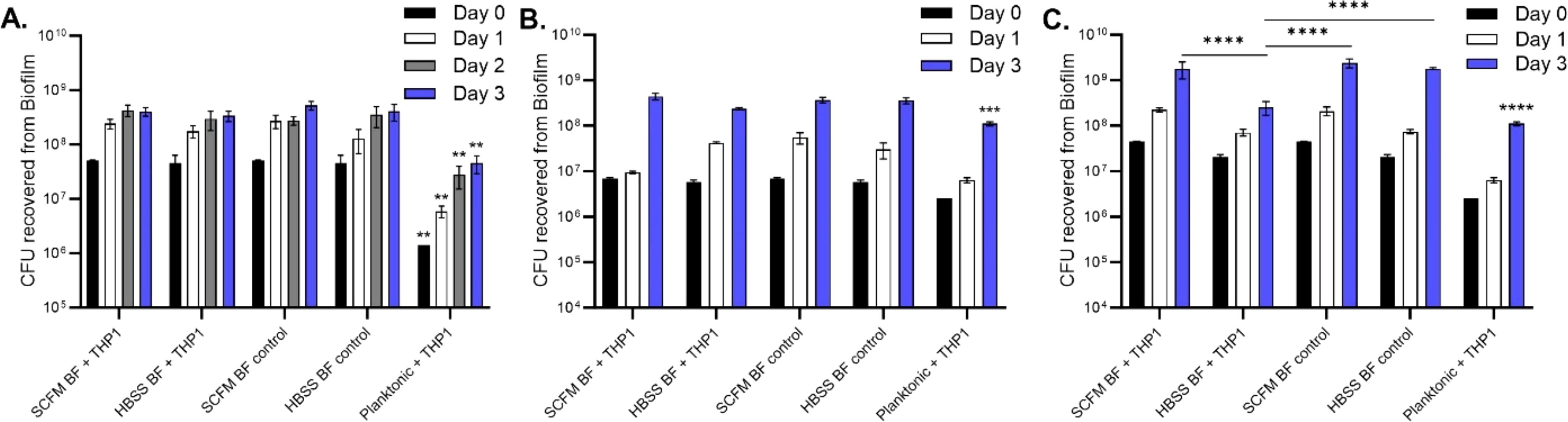
Figure 7. Monocytes were unable to kill M. abscessus biofilms. IFN-γ-stimulated monocytes were added to mature (7 day) biofilms formed in either SCFM or HBSS (A). Monocytes were also added to replicative stage biofilms (B) and non-replicative stage biofilms (C). The biofilms were disrupted and CFUs were determined at days 1, 2, or 3 post cell addition for biofilm survival. Planktonic bacteria served as a macrophage killing control. Data are representative of three biological replicates. Statistical analysis was conducted using 2-way ANOVA with Tukey’s multiple comparison test. **** indicates p < 0.0001, *** indicates p < 0.001, ** indicates p < 0.01.
The release of bacteria from biofilms may express a set of different proteins on the surface that then make them more capable of seeding and infecting different sites or hosts. We compared planktonic bacteria grown in 7H10 agar, bacteria with the biofilm phenotype (7 days biofilm), bacteria released from biofilm when in Middlebrook 7H9 broth, and bacteria released from biofilm in SCFM. The results, shown in a Venn diagram, demonstrated that bacteria released from biofilm expressed significantly more proteins than bacteria grown in plates (Figure 8). The bacteria found in biofilms accounted for 295 of the 380 proteins detected and are summarized in Tables 5–15. Planktonic bacteria accounted for 85 of the 380 proteins and are summarized in Tables 5, 6, 8, 15 and all the listed proteins overlapped with the other treatment groups examined. The bacteria released from biofilms expressed more proteins on their surface, suggesting a remarkable change in phenotype. It is notable that many of the proteins were different lipoproteins, of a small size, and uncharacterized, and there were many types of enzymes. We compared the transposon mutants that were found deficient in secondary biofilm formation to the surface proteins detected. Three mutants, MAB_0525c (all treatments), MAB_2301 [planktonic, secondary BF (7H9), secondary BF (SCFM), and primary BF (7H9)], and MAB_3538 [secondary BF (7H9) and secondary BF (SCFM)] were found in the proteomic lists. Interestingly, the latter mutant, MAB_3538, with kinase activity, was only found in the released biofilm groupings.
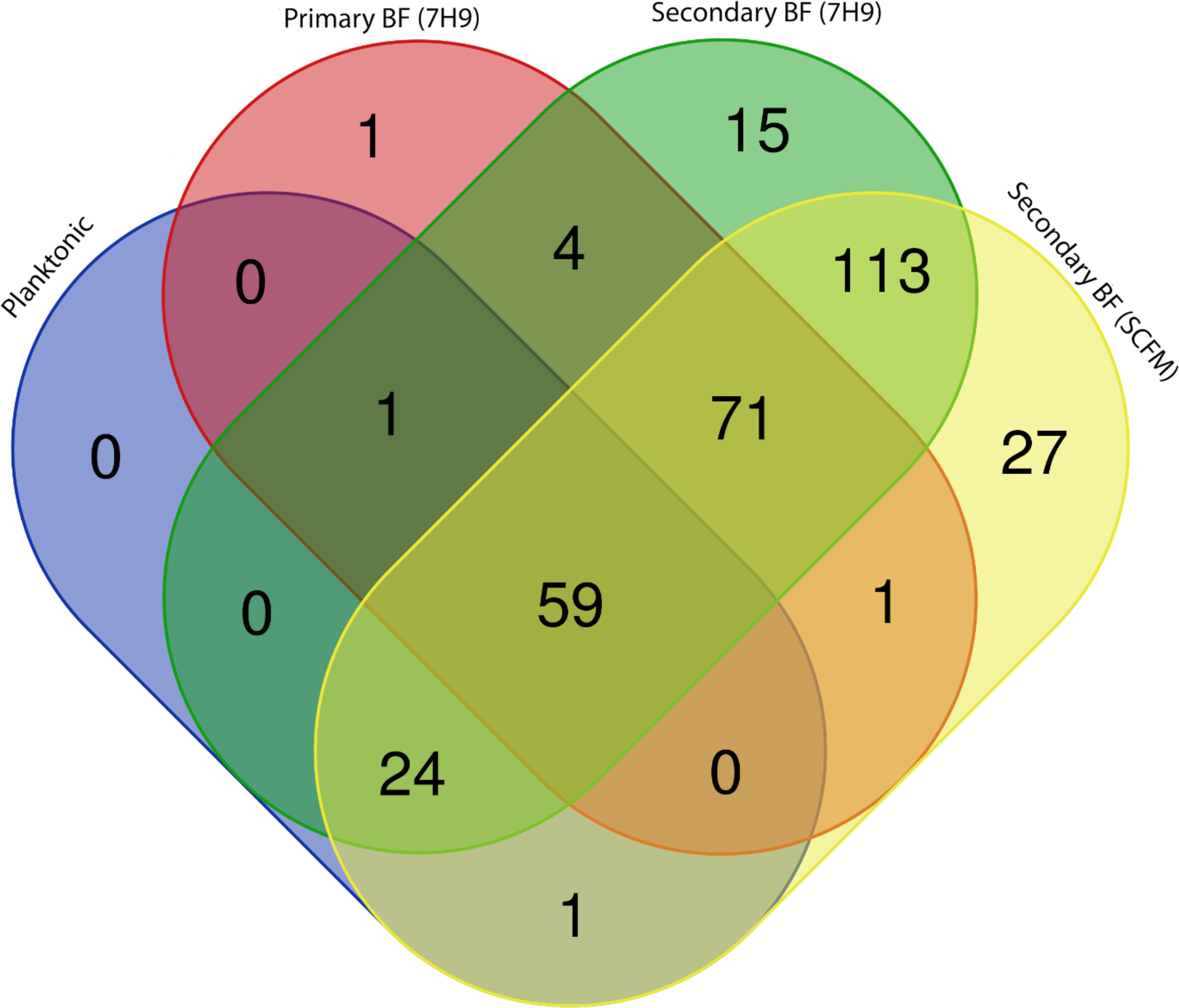
Figure 8. A Venn diagram depicting proteins determined through proteomic analysis. Each treatment group’s list of proteins was compared to each other to resolve overlaps. Graphical representation of Tables 1, 5–14. https://bioinformatics.psb.ugent.be/webtools/Venn/.

Table 5. Surface proteins unique to planktonic bacteria, primary biofilm (BF) (7H9), and secondary BF (7H9).
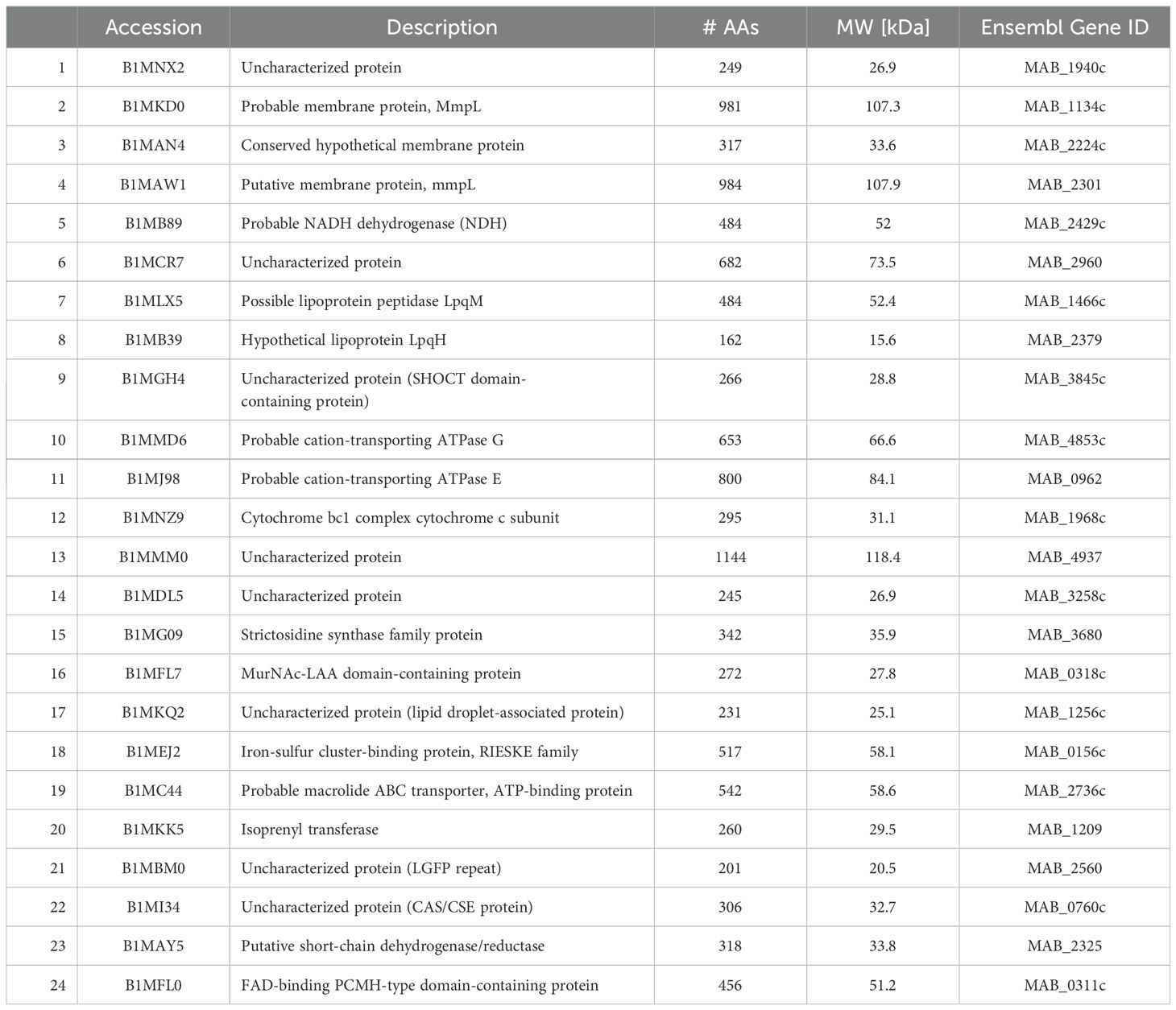
Table 6. Surface proteins identified in planktonic bacteria, secondary biofilm (BF) (7H9), and secondary BF (SCFM).
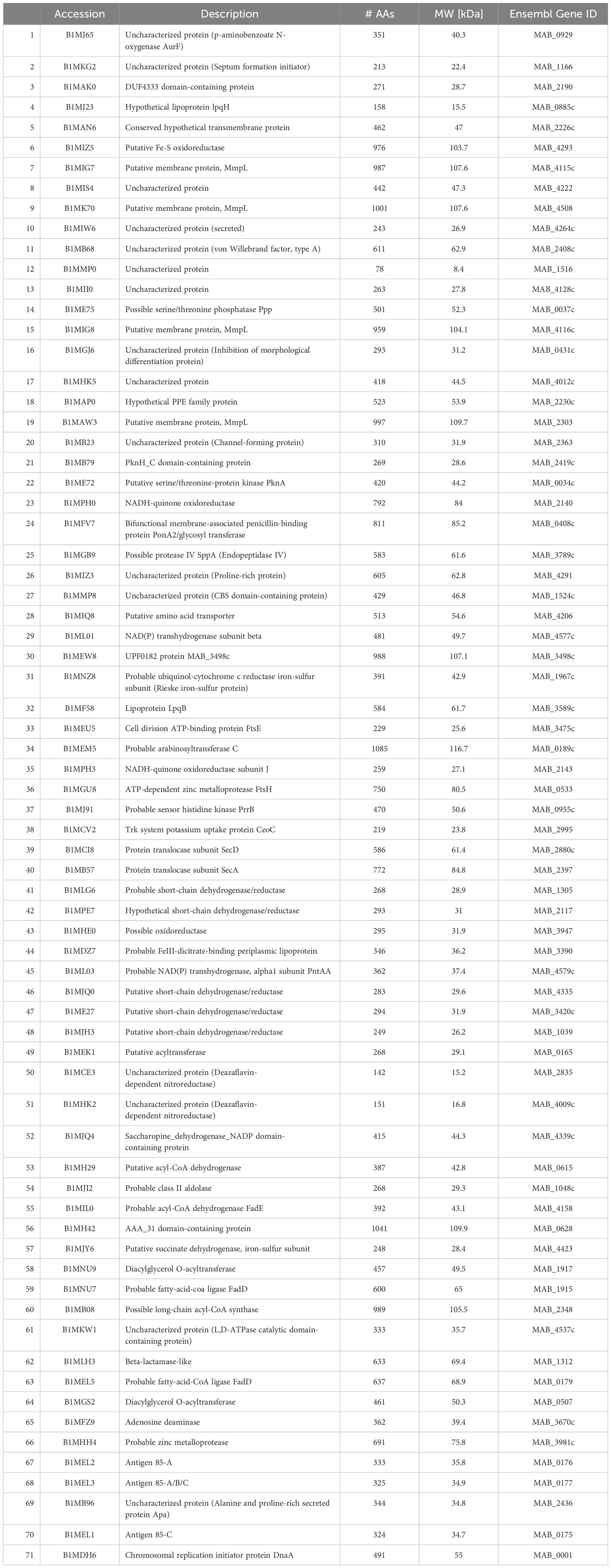
Table 7. Surface proteins unique to primary biofilm (BF) (7H9), secondary BF (7H9), and secondary BF (SCFM).
The intrinsic resistance of M. abscessus to available antimicrobials creates a major challenge for the treatment of the disease in high-risk populations (18, 19). Therefore, improving our understanding of the pathogenesis of infection in these populations is needed to develop alternative ways to prevent or treat the infectious condition.
M. abscessus, like other environmental microorganisms, has the ability to form biofilms on surfaces, which has been shown as a strategy used by the pathogen to establish a niche on the airway mucosa (12, 20). Past observations in both animal models and humans have demonstrated that M. abscessus biofilm is part of the pathogenesis of lung infections (4, 21). Recent studies have determined that M. abscessus cultured in SCFM, which mimics the mucus environment in the airways of patients with cystic fibrosis, forms a biofilm that differs from biofilms formed under water or in buffer conditions (22, 23), which indicates that in the presence of sputum contents in the airways M. abscessus differentiates and acquires a new phenotype. In addition, the biofilm’s extracellular matrix is formed by eDNA and glycol-phospholipids (21), which, in the case of Mycobacterium avium, has been shown to protect the bacteria against the action of phagocytic cells (24).
In this work, we demonstrate that bacteria in M. abscessus biofilm at some point are able to release from the initial biofilm mass and probably seed in an adjacent site in the airway. This information is important since in our model it was observed that bacteria in biofilms formed under SCFM sputum conditions, once detached, can establish another biofilm with significantly greater efficiency than bacteria in biofilm developed under phosphate buffer conditions. In fact, previous studies in the laboratory have determined that M. abscessus is capable of utilizing the magnesium concentration in the cystic fibrosis medium environment to quickly establish a niche and develop a robust biofilm (9). Furthermore, bacteria released from biofilms in the airways can be expelled in the sputum, which increases the chance of transmission by aerosols in the airway of a second individual. In fact, recent observations by diverse groups have suggested that non-tuberculous mycobacteria can be transmitted from an individual to a recipient, most likely in the environment of clinics that assist patients belonging to at-risk groups (6, 7, 25). The hypotheses and the observations in studies on necropsy in animals or lung transplantation in humans suggest that bacteria released from biofilms may be transported upward or downward depending on the flow of the air either leaving or entering the airways. A limitation of our study is that despite the hypothesis and the previous observations, we do not provide visual proof in this study. A previous study demonstrated the possibility that environmental mycobacteria can be transmitted from person to person, although the prolonged time between infection and disease makes the epidemiological connection very difficult. Using Caenorhabditis elegans as a model, M. avium infection could be transmitted directly from one host to another without the environmental step that was previously thought to be required (26, 27). A clear consequence of these findings is that an infection can be transmitted from host to host without the environmental step, which makes the bacterial phenotype released from the biofilm one of the potential phenotypes that transmit the infection.
Bacteria released from biofilm in cystic fibrosis medium were shown to be able to establish a secondary biofilm at the same rate as the primary biofilm (Figure 2). The SCFM biomass was more robust than the biofilm formed by bacteria released from HBSS-established biofilm, showing that bacteria in the presence of a cystic fibrosis environment can still efficiently seed in another location in the airways. In addition, detached M. abscessus invades epithelial cells more efficiently than planktonic bacteria, indicating that the process of binding to establish a primary biofilm and then the subsequent release and formation of a secondary biofilm is associated with bacterial structural modifications, most likely on the bacterial surface. The number of different proteins expressed by the bacterial surface released from biofilms in comparison with bacteria attached to the biofilm is striking. When comparing proteins uniquely expressed by the released bacteria that were not expressed in the other conditions, several uncharacterized proteins, an Mg++ transport, and a few lipoproteins and polyketide synthases were identified. Matching the mutant MAB_3538, which has kinase activity, in the detached biofilm groupings is an interesting finding. A definitive analysis of the function of these proteins is warranted.
Although the changes on the bacterial surface described in this study and in recent publications (22, 23) are responsible for the different interactions between the pathogen and host cells, an increased uptake by the phagocyte could be a plausible outcome. Our results also demonstrated that the ingested bacteria are not killed by macrophages and grow inside the phagocytic cells at a higher rate than planktonic bacteria. The question of why bacteria uptake is increased with better survival outcomes is pertinent here. Since significant changes occur during intracellular survival, one may hypothesize that the presence of adherent molecules on the bacterial surface is advantageous for disease progression. More research needs to be conducted to understand the effects of the identified genes during infections.
The ability of several different bacterial species to detach from biofilms seems to be dictated by a quorum-sensing mechanism (28). Quorum sensing is well-described for Gram-negative bacteria, mostly Pseudomonas aeruginosa, Vibrio sp. and Gram-positive bacteria such as Bacillus cereus and Staphylococcus aureus (28). There are three known quorum-sensing types. Type I utilizes N-acyl homoserine lactone, while the other two (types II and III) are regulated by other autoinducers, many of them peptides (28–30). Autoinducers accumulate in the environment as the bacteria population increases and stimulate the expression of membrane transporters and activation of histidine kinases inside the bacterium. P. aeruginosa has several interconnected quorum-sensing circuits that collectively regulate hundreds of genes (31). Pseudomonas has three major circuits that regulate approximately 10% of the bacterial genes (32). Two circuits respond to N-acyl homoserine lactone signals and a third one, the Pseudomonas quinolone signal system, uses the quinolone signal to interact with RHI receptors which also recognize homoserine lactone signals. These signals are detected by receptors present in the cytoplasm or in the membrane. Gram-positive bacteria, in contrast, use peptides as signaling molecules. They usually bind to membrane histidine kinase receptors that autophosphorylate, although, in some cases, the peptides are transported in the cytoplasm where they interact with the transcription factors (28). Mycobacterial proteins do not share any homology with known quorum-sensing-linked proteins of different bacteria. However, several of the small proteins identified in our work as being expressed under biofilm in SCFM and detachment phenotypes have signal peptides and are potentially exported in the biofilm setting. Other researchers are currently addressing this hypothesis regarding the function of surface proteins and their potential participation as signal proteins (13, 28, 33–35).
One should consider that detachment from biofilms may depend on the type and the status of the patient’s immune system, which can interfere with the dissemination of the pathogen in their lungs. It is also important to take into account the pathogen’s phenotype which can be influenced by the lung environment (36).
In summary, this work shows that M. abscessus can release from biofilms on the surface of mucosal epithelial cells and these released bacteria are very efficient in developing a new biofilm to bind to mucosal epithelial cells and infect macrophages. Based on the findings that M. abscessus can infect patients directly, the released bacteria can be considered an infectious phenotype. The proteomic sequences of the released bacterial surface proteins identified potential candidates involved in the detachment from biofilm and these need to be studied further.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
BK: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft. AL: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. LB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The reported work was funded by NIH grant AI152258, and San Francisco Microbiology Foundation grant # 102.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Floto RA, Haworth CS. The growing threat of nontuberculous mycobacteria in CF. J Cyst Fibros. (2015) 14:1–2. doi: 10.1016/j.jcf.2014.12.002
2. Martiniano SL, Nick JA, Daley CL. Nontuberculous mycobacterial infections in cystic fibrosis. Clin Chest Med. (2022) 43:697–716. doi: 10.1016/j.ccm.2022.06.010
3. Johansen MD, Herrmann J-L, Kremer L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. 7. Nat Rev Microbiol. (2020) 18:392–407. doi: 10.1038/s41579-020-0331-1
4. Ryan K, Byrd TF. Mycobacterium abscessus: shapeshifter of the mycobacterial world. Front Microbiol. (2018) 9:2642. doi: 10.3389/fmicb.2018.02642
5. Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Andrejak C, et al. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Eur Respir J. (2020) 56:2000535. doi: 10.1183/13993003.00535-2020
6. Bryant JM, Brown KP, Burbaud S, Everall I, Belardinelli JM, Rodriguez-Rincon D, et al. Stepwise pathogenic evolution of Mycobacterium abscessus. Science. (2021) 372:eabb8699. doi: 10.1126/science.abb8699
7. Ruis C, Bryant JM, Bell SC, Thomson R, Davidson RM, Hasan NA, et al. Dissemination of Mycobacterium abscessus via global transmission networks. Nat Microbiol. (2021) 6:1279–88. doi: 10.1038/s41564-021-00963-3
8. Miranda-CasoLuengo AA, Staunton PM, Dinan AM, Lohan AJ, Loftus BJ. Functional characterization of the Mycobacterium abscessus genome coupled with condition specific transcriptomics reveals conserved molecular strategies for host adaptation and persistence. BMC Genomics. (2016) 17:553. doi: 10.1186/s12864-016-2868-y
9. Keefe BF, Bermudez LE. Environment in the lung of cystic fibrosis patients stimulates the expression of biofilm phenotype in Mycobacterium abscessus. J Med Microbiol. (2022) 71:001467. doi: 10.1099/jmm.0.001467
10. Fennelly KP, Ojano-Dirain C, Yang Q, Liu L, Lu L, Progulske-Fox A, et al. Biofilm formation by mycobacterium abscessus in a lung cavity. Am J Respir Crit Care Med. (2016) 193:692–3. doi: 10.1164/rccm.201508-1586IM
11. Belardinelli JM, Li W, Avanzi C, Angala SK, Lian E, Wiersma CJ, et al. Unique features of mycobacterium abscessus biofilms formed in synthetic cystic fibrosis medium. Front Microbiol. (2021) 12:743126. doi: 10.3389/fmicb.2021.743126
12. Rose SJ, Babrak LM, Bermudez LE. Mycobacterium avium possesses extracellular DNA that contributes to biofilm formation, structural integrity, and tolerance to antibiotics. PloS One. (2015) 10:e0128772. doi: 10.1371/journal.pone.0128772
13. Palmer KL, Mashburn LM, Singh PK, Whiteley M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol. (2005) 187:5267–77. doi: 10.1128/JB.187.15.5267-5277.2005
14. Rose SJ, Bermudez LE. Identification of bicarbonate as a trigger and genes involved with extracellular DNA export in mycobacterial biofilms. mBio. (2016) 7:e01597–16. doi: 10.1128/mBio.01597-16
15. Leestemaker-Palmer AL, Bermudez LE. Mycobacterium abscessus infection results in decrease of oxidative metabolism of lung airways cells and relaxation of the epithelial mucosal tight junctions. Tuberculosis (Edinb). (2023) 138:102303. doi: 10.1016/j.tube.2023.102303
16. Lewis MS, Danelishvili L, Rose SJ, Bermudez LE. MAV_4644 Interaction with the Host Cathepsin Z Protects Mycobacterium avium subsp. hominissuis from Rapid Macrophage Killing. Microorganisms. (2019) 7:144. doi: 10.3390/microorganisms7050144
17. Danelishvili L, Rojony R, Carson KL, Palmer AL, Rose SJ, Bermudez LE. Mycobacterium avium subsp. hominissuis effector MAVA5_06970 promotes rapid apoptosis in secondary-infected macrophages during cell-to-cell spread. Virulence. (2018) 9:1287–300. doi: 10.1080/21505594.2018.1504559
18. Griffith DE, Girard WM, Wallace RJ. Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am Rev Respir Dis. (1993) 147:1271–8. doi: 10.1164/ajrccm/147.5.1271
19. Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother. (2012) 67:810–8. doi: 10.1093/jac/dkr578
20. Esteban J, García-Coca M. Mycobacterium biofilms. Front Microbiol. (2017) 8:2651. doi: 10.3389/fmicb.2017.02651
21. Blanchard JD, Elias V, Cipolla D, Gonda I, Bermudez LE. Effective Treatment of Mycobacterium avium subsp. hominissuis and Mycobacterium abscessus Species Infections in Macrophages, Biofilm, and Mice by Using Liposomal Ciprofloxacin. Antimicrob Agents Chemother. (2018) 62:e00440–18.
22. Wiersma CJ, Belardinelli JM, Avanzi C, Angala SK, Everall I, Angala B, et al. Cell Surface Remodeling of Mycobacterium abscessus under Cystic Fibrosis Airway Growth Conditions. ACS Infect Dis. (2020) 6:2143–54. doi: 10.1021/acsinfecdis.0c00214
23. Leestemaker-Palmer A, Ong T, Bermudez LE. Exposure of Mycobacteriodes abscessus to mucin affects bacterial phenotype. Sci Rep. (2024) 15:393.
24. Rose SJ, Bermudez LE. Mycobacterium avium biofilm attenuates mononuclear phagocyte function by triggering hyperstimulation and apoptosis during early infection. Infect Immun. (2014) 82:405–12. doi: 10.1128/IAI.00820-13
25. Kreutzfeldt KM, McAdam PR, Claxton P, Holmes A, Seagar AL, Laurenson IF, et al. Molecular longitudinal tracking of Mycobacterium abscessus spp. during chronic infection of the human lung. PloS One. (2013) 8:e63237.
26. Everman JL, Ziaie NR, Bechler J, Bermudez LE. Establishing Caenorhabditis elegans as a model for Mycobacterium avium subspecies hominissuis infection and intestinal colonization. Biol Open. (2015) 4:1330–5. doi: 10.1242/bio.012260
27. Bermudez LE, Rose SJ, Everman JL, Ziaie NR. Establishment of a Host-to-Host Transmission Model for Mycobacterium avium subsp. hominissuis Using Caenorhabditis elegans and Identification of Colonization-Associated Genes. Front Cell Infect Microbiol. (2018) 8:123. doi: 10.3389/fcimb.2018.00123
28. Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. (2012) 2:a012427. doi: 10.1101/cshperspect.a012427
29. Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PloS Pathog. (2008) 4:e1000052. doi: 10.1371/journal.ppat.1000052
30. Borlee BR, Geske GD, Blackwell HE, Handelsman J. Identification of synthetic inducers and inhibitors of the quorum-sensing regulator LasR in Pseudomonas aeruginosa by high-throughput screening. Appl Environ Microbiol. (2010) 76:8255–8. doi: 10.1128/AEM.00499-10
31. Lazar V. Quorum sensing in biofilms -How to destroy the bacterial citadels or their cohesion/power? Anaerobe. (2011) 117:280–5.
32. Mattman ME, Blackwell HE. Small molecules that modulate quorum sensing and control virulence in Pseudomonas aeruginosa. J Org Chem. (2010) 75:6737–6746. doi: 10.1021/jo101237e
33. Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. (1995) 92:12055–9. doi: 10.1073/pnas.92.26.12055
34. Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. (1994) 77:207–16. doi: 10.1016/0092-8674(94)90313-1
35. Lilley BN, Bassler BL. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol. (2000) 36:940–54. doi: 10.1046/j.1365-2958.2000.01913.x
Keywords: mycobacterium abscessus, biofilm, detachment, infectious phenotype, host bacterial surface, macrophages, uptake
Citation: Keefe B, Leestemaker-Palmer A and Bermudez LE (2025) The ability to detach from biofilms in the lung airways prior to transmission to another host is associated with the infectious phenotype of Mycobacterium abscessus. Front. Immunol. 16:1508584. doi: 10.3389/fimmu.2025.1508584
Received: 09 October 2024; Accepted: 17 January 2025;
Published: 07 March 2025.
Edited by:
Veronica Schmitz, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Juan José Valdez Alarcón, Universidad Michoacana de San Nicolás de Hidalgo, MexicoCopyright © 2025 Keefe, Leestemaker-Palmer and Bermudez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luiz E. Bermudez, bHVpei5iZXJtdWRlekBvcmdvbnN0YXRlLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.