
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Immunol. , 05 February 2025
Sec. Cancer Immunity and Immunotherapy
Volume 16 - 2025 | https://doi.org/10.3389/fimmu.2025.1492296
This article is part of the Research Topic Early Detection, Cancer Interception, Et Al: Translating the Multifaceted Use of Liquid Biopsy to the Management of Early Disease View all 5 articles
 Daria Kravchuk1
Daria Kravchuk1 Alexandra Lebedeva2,3*
Alexandra Lebedeva2,3* Olesya Kuznetsova2,4
Olesya Kuznetsova2,4 Alexandra Kavun2
Alexandra Kavun2 Anastasiia Taraskina2
Anastasiia Taraskina2 Ekaterina Belova2,3,5
Ekaterina Belova2,3,5 Tatiana Grigoreva2,3
Tatiana Grigoreva2,3 Egor Veselovsky2
Egor Veselovsky2 Vladislav Mileyko2,3
Vladislav Mileyko2,3 Vladislav Nikulin4
Vladislav Nikulin4 Lidia Nekrasova6
Lidia Nekrasova6 Alexey Tryakin4
Alexey Tryakin4 Mikhail Fedyanin1,4,7
Mikhail Fedyanin1,4,7 Maxim Ivanov2,3
Maxim Ivanov2,3Microsatellite instability (MSI) is a widely studied molecular signature, which is associated with long-term benefit in patients treated with immune checkpoint inhibitor therapy. This approach has been proven to be effective in the treatment of patients with MSI-positive colorectal cancer (CRC). Analysis of serial liquid biopsy samples allows to detect changes in the tumor in response to therapy. Typically, somatic mutations are used for tracing the dynamics of the tumor, and the assessment of DNA signatures such as MSI is not currently used for these purposes. Here, we describe a case of a MSI-positive CRC, who received nivolumab monotherapy. Sequential sampling of the patient’s plasma demonstrated an increase in MSI burden (bMSI), which was found to correlate with the increase of driver mutation burden one month after starting nivolumab, and hyperprogressive disease. Thus, analysis of bMSI in liquid biopsy via NGS may be a promising method for timely assessment of the treatment effectiveness received by patients with MSI-positive CRC.
Microsatellite instability (MSI) is a phenomenon of hypermutation occurring at microsatellite loci, which are tracts of short repetitive DNA motifs. MSI comes as the shortening of the mononucleotide tracts due to failure to correct DNA polymerase slippage mistakes due to defects in the mismatch repair system (dMMR) (1, 2). MSI/dMMR occurs among various tumor types (3), and is associated with improved outcomes of patients treated with immune checkpoint inhibitors (ICI) (4–6). The effectiveness of ICI has been well demonstrated for patients with MSI-positive colorectal cancer (CRC) in numerous clinical studies (7–12). Standard diagnostic methods such as immunohistochemistry (IHC) or polymerase chain reaction (PCR) are routinely used in clinical practice for the assessment of MSI/dMMR and provide a binary assessment of MSI/dMMR (positive or negative) (13). Next generation sequencing (NGS) is an emerging method for assessing MSI (13–15). The advantages of assessing MSI status using NGS include the ability to study a larger number of short tandem repeats (STRs), as well as to simultaneously analyze the mutational profile and mutational burden (TMB) of the tumor (13). Much like routine methods, standard NGS pipelines classify tumors as stable or unstable based on the percentage of unstable STRs (14). Recent studies have demonstrated excellent sensitivity of NGS-based assays for MSI analysis in liquid biopsies (16). However, current binary classification of STRs only allows to analyze the percentage of unstable STRs. This approach does not allow for a quantitative assessment of MSI burden as the percentage of unstable DNA fragments in the sample (17). Similarly to the analysis of variant allele frequencies of mutations, the quantitative assessment of MSI might reflect the clonality of the tumor, as well as can be utilized to measure tumor dynamics when liquid biopsy is used. The use of liquid biopsy makes it possible to detect early changes in response to therapy that cannot be detected by standard imaging methods such as computer tomography (CT) (18–22). Typically, somatic mutations are used for tracing the dynamics of the tumor. However, the assessment of DNA signatures such as MSI is not currently used for these purposes. Here, we report a clinical case of a colorectal cancer patient with MSI confirmed by PCR and NGS. The patient was treated with ICI, eventually demonstrating hyperprogression. An algorithm for qualitative measurement of MSI burden was used for tracking its changes in serial on-treatment plasma samples (bMSI). Here, for the first time, we demonstrate that the dynamics of bMSI correlate with driver mutational burden and disease progression.
A 40-year-old male was diagnosed with sigmoid cancer in November 2021. He underwent right-sided hemicolectomy on 23 November 2021. Histologically, colon adenocarcinoma pT3N1bM0 was then confirmed. CT scan in December 2021 revealed no signs of continued tumor growth and a lesion in upper lobe (S4) of the right lung up to 18 mm with no enlargement in size compared to previous CT scans. The patient then underwent 6 cycles of adjuvant chemotherapy with XELOX and 3 cycles of capecitabine monotherapy from December 2021 to July 2022. In January 2022, in between treatment cycles, the patient underwent PET/CT scan, which did not confirm a lesion in the lung as metastatic due to low 18FDG uptake. In April 2022, no KRAS/NRAS/BRAF alterations were found via PCR, and the IHC for HER2 was negative. However, following the results of a 5-loci PCR, the tumor was found to be MSI-positive. In line with the patient’s decision, no subsequent germline testing to rule out Lynch syndrome was performed. Following the completion of adjuvant chemotherapy, the patient underwent PET/CT in August 2022, which revealed the enlargement of size and contrast accumulation in S4 of the right lung (up to 5 cm) and the appearance of a new lesion, measuring up to 3 cm in the right hepatic lobe (S8). As it was not clear whether an inflamed retention cyst or metastatic lesion was observed in the lung, a bronchoscopy with brush-biopsy was performed. Consequent cytology did not reveal any atypical cells, and the lesion was determined a cyst. The patient was offered to undergo a liver biopsy to confirm the progression of the disease, but he declined and decided to remain under observation.
The next follow-up visit was in February 2023. The patient underwent CT that revealed the same lesions: 55x36 mm in S4 of the right lung and 34x27 mm in the S8 of the liver. Due to the enlarged size of the lesions, the patient was offered to undergo systemic therapy first. As the primary tumor was MSI-positive, monotherapy with nivolumab was recommended. The patient was then enrolled in an observational clinical trial BLOOMSI (NCT06414304). As part of the trial, the pre-treatment blood plasma sample was collected on 27.03.2023. On March 28th 2023, the patient started nivolumab. During the time on treatment, two serial plasma samples were collected, 17 and 31 days after therapy initiation (on 13.04.2023 and 27.04.2023, respectively). On May 2nd 2023, while receiving the 3rd cycle, the patient complained of limb weakness and mild joint pain. Following the recommendation of a general practitioner, he started to take еtoricoxib with short term pain relief. The patient received a total of 4 cycles of nivolumab, and the symptoms were increasing. After the discontinuation of nivolumab, a post-treatment blood plasma sample was obtained (on 11.05.2023, 45 days after the start of treatment). In the beginning of June, patient underwent radiography of the hip joints and CT of the brain, but no abnormalities were found. Further CT scan of the lumbosacral region of the spine revealed metastases of the vertebrae in Th11-S5 and pathological fractures of the L2, L4 vertebrae bodies. Metastases were also found in pelvic bones leading to their destruction, with the spread of the pathological process to soft tissue at the level of the iliac crests and lumbar lymph nodes. Furthermore, adrenal glands and liver were affected by metastatic lesions. Decompression laminectomy at the level of L4 vertebrae with tumor removal with microsurgical reconstruction of the L4 root nerve with posterior stabilization was performed on July 7th. Subsequent histology confirmed the colorectal origin of the metastatic lesion. Following the surgery, the patient did not receive any systemic treatment. In August 2023, he received treatment for lumbosacral abscess after laminectomy and died a month later (Figures 1, 2).
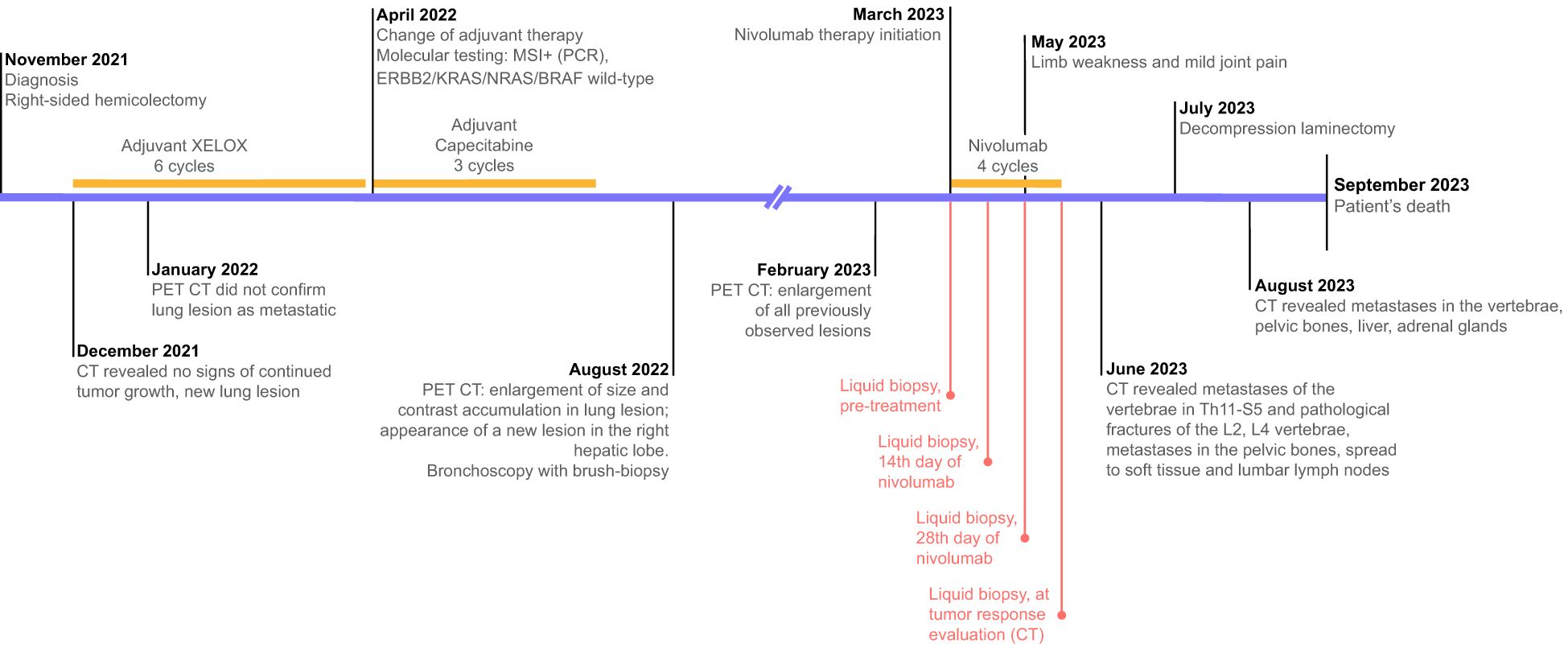
Figure 1. Treatment timeline and diagnostic procedures. Text in red reflects the timeline of the collection of serial liquid biopsy samples.
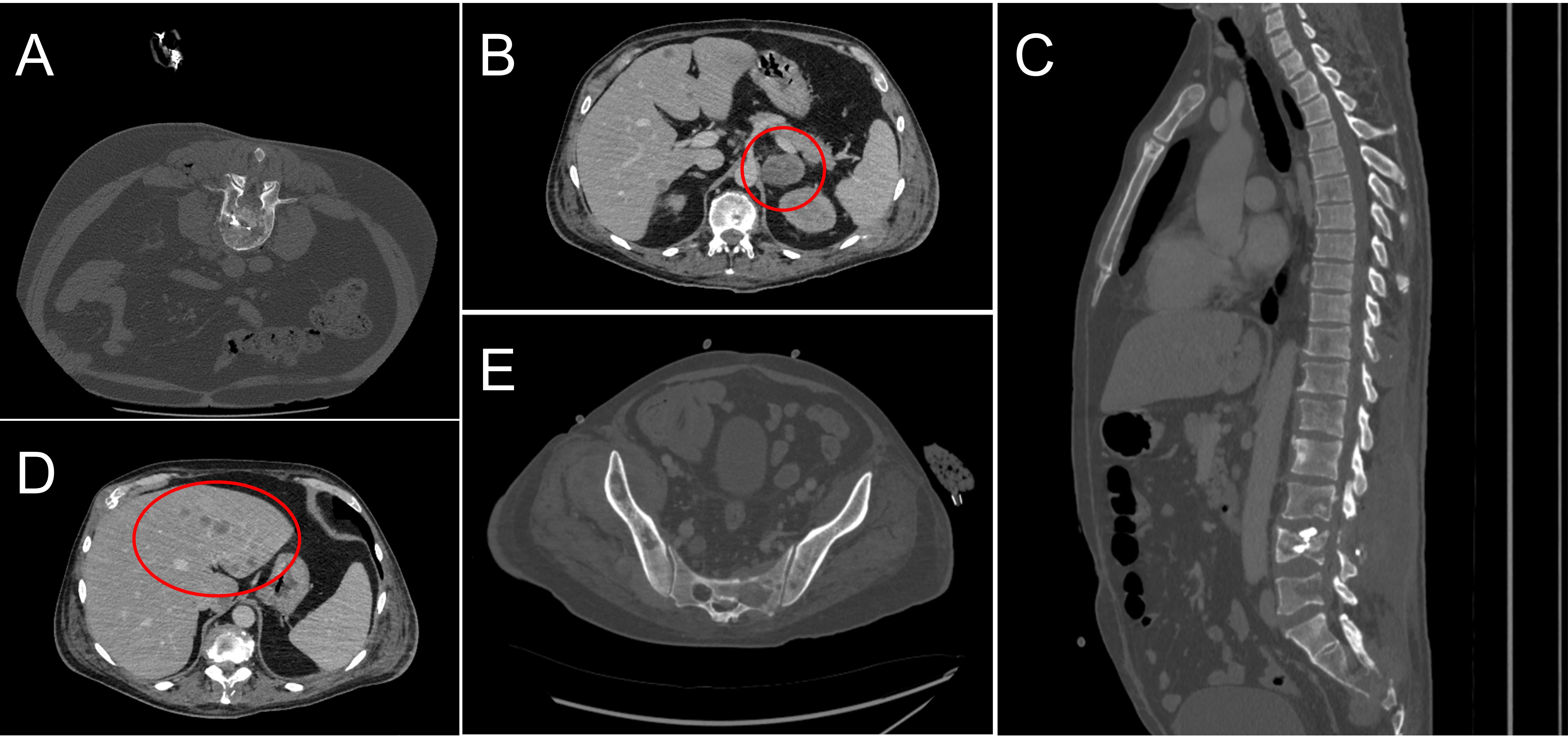
Figure 2. Computed tomography scans of the patient’s metastatic sites in the course of the treatment. (A) Metastasis and pathologic fracture in L4 revealed in June 2023; (B) Metastatic lesions in the adrenal glands, August 2023; (C) Metastatic lesions in the Th11-S5 vertebrae, June 2023; (D) Metastatic lesions in the liver, August 2023; (E) Destruction of pelvic bones, August 2023.
Consistent with current treatment guidelines, MSI was evaluated via standard 5-loci PCR panel, uncovering MSI positivity in the patient’s primary tumor sample. Based on the results of PCR, the patient was included in the local observational clinical trial evaluating the dynamics of MSI burden in serial samples from patients receiving ICI. As part of the trial, confirmatory MSI/dMMR testing was performed using a 4-antibody IHC (loss of two proteins was observed) and NGS via Solo test Atlas Pro panel covering 34 commonly altered cancer-related genes and 30 MSI mononucleotide tandem repeats. NGS was performed on primary tumor and serial plasma (liquid biopsy, LB) samples received prior to the start of ICI therapy, on the 17th, 31st days on therapy, as well as after the discontinuation of ICI.
NGS revealed point mutations in KRAS, PIK3CA, PTEN and TP53 (the latter was only observed in the FFPE sample) in the pre-treatment FFPE and LB samples (Figure 3). Of note, initial PCR testing for common CRC alterations in colorectal cancer found no KRAS mutations, whereas NGS testing of a tumor sample found a rare hotspot KRAS p.Lys117Asn mutation (23) with a VAF of 22%. MSIsensor2 (14) revealed a slight decrease unstable STRs by 3% at the 14th day after therapy initiation and gradual increase by 7% and 12% in the subsequent liquid biopsy samples during ICI treatment (73%, 71%, 76% and 85% for baseline, 14th day, 28th day and 1st control LB, respectively). KRAS, PIK3CA and PTEN driver mutations were identified in LB samples with almost the same variant allele frequency (13.9, 15.6 and 17.3% respectively) (Table 1), similarly to the molecular profile seen in primary FFPE sample (22.1%, 18.2% and 18.7%, respectively) alluding to its homogeneous representation across tumor clones in primary tumor. Across serial LB samples driver mutations demonstrated positive dynamics at 14th day on therapy (decrease by 41%, 35% and 8% for KRAS, PIK3CA and PTEN mutations, respectively), and negative dynamics afterwards, although PTEN mutation demonstrated dissimilar dynamics contrasting to ones of PIK3CA and KRAS hinting at the positive selection of PTEN-mutated clone during the treatment (Figure 3B).
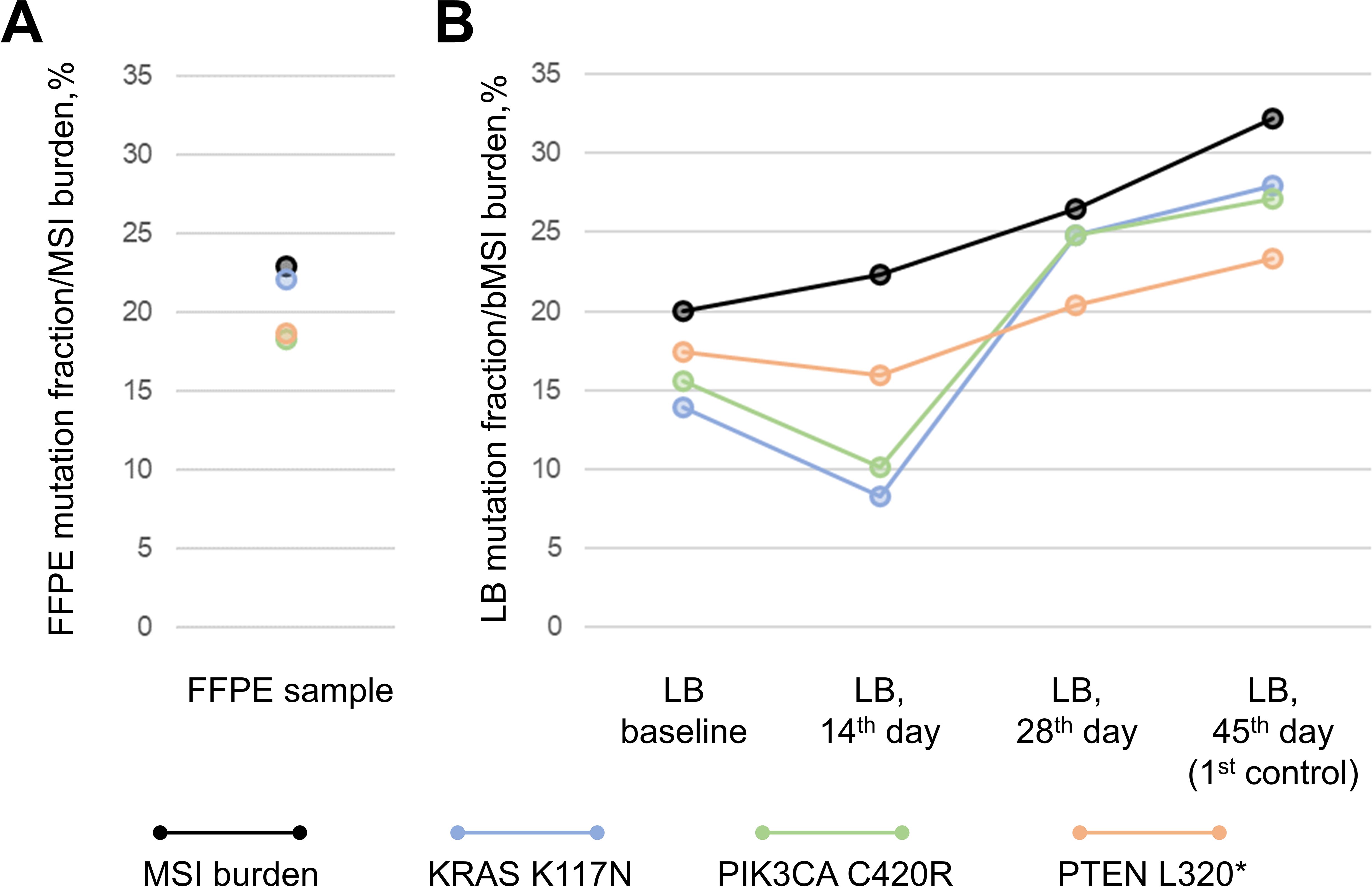
Figure 3. Molecular profile of FFPE sample (A) and dynamics of driver mutations and DNA signatures during the course of immunotherapy as traced via serial liquid biopsy samples (LB) (B). *premature stop-codon.
To assess MSI quantitatively in FFPE sample, i.e. to estimate the percentage of tumor cells exhibiting MSI, we calculated the ratio of total amount of sequencing reads in NGS data supporting DNA fragments with altered STR sequences (i.e. demonstrating shortage of reference STR length by 4b.p. and more) to the total amount of sequencing reads supporting any STR sequences. The calculated percentage was close to the VAF of KRAS, PIK3CA and PTEN mutations (Figure 3A), suggesting that quantitatively estimated MSI burden was correlated with tumor clonality. Similarly to the FFPE sample, MSI burden was in line with VAFs of driver mutations in pre-treatment LB sample. Further tracing of bMSI burden across serial LB samples demonstrated a gradual increase without significant drop-outs seen for KRAS and PIK3CA mutations hinting at co-evolution of tumor clones exhibiting PTEN mutation and MSI.
MSI/dMMR is a known biomarker of ICI response across tumor types, including colorectal cancer (26, 27). Gold standard methods for MSI/dMMR detection include PCR and IHC, however in recent years NGS has been recognized as a potentially decisive tool for selecting candidates for ICI (13). Although tumor samples are considered preferential for MSI/dMMR testing, various issues, including insufficient tumor purity and overall suboptimal tissue quality might result in ambiguous results (28). Large-scale studies suggest that liquid biopsy might perform as well as tumor samples for identifying MSI-positive patients who are candidates for ICI (20, 29, 30).
A high concordance was observed between baseline tissue and plasma mutations (Table 1). Evaluation of MSI dynamics in liquid biopsies has been reported to correlate with response to immunotherapy or lack thereof (31, 32). Additionally, serial ctDNA analysis has been shown to be prognostic of tumor dynamics, with an increase of ctDNA indicating progressive disease (33–36). In our patient, while an increase in MSI burden was immediate and persistent, the increase of driver and passenger mutational burden was delayed in both pre- and on-treatment serial samples. Moreover, bMSI increase preceded the first clinical signs of disease progression, whereas an increase in mutational burden coincided with disease progression (Figure 3). Since no CT imaging was done in the course of nivolumab treatment, we could not compare the results of imaging studies to LB analysis, however, the results of the latter are consistent with the patient’s symptoms, which were later confirmed to have originated from progressive disease. Previous studies have suggested that analysis of LB has the potential to outperform standard radiographic imaging for predicting treatment outcomes (37), however no studies have reported the utility of MSI burden as a screening tool for monitoring tumor dynamics in the course of ICI. While these findings are interesting, they require further validation in prospective clinical trials to validate the role of bMSI for tumor response monitoring in MSI-positive patients.
It is worth mentioning that initial tumor testing with PCR, in contrast with NGS, did not reveal a KRAS p.Lys117Asn hotspot mutation (23). Precise determination of KRAS mutational status is crucial for tailoring therapy for patients with advanced MSS/pMMR colorectal cancer (38). Although seen at lower rates than mutations affecting codons 12, 13 and 61, the KRAS codon 117 mutation is considered a ‘standard’ KRAS mutation, indicating that all standard testing methodologies used in the clinic should be able to detect this mutation. For instance, in KEYNOTE-177, no benefit from pembrolizumab monotherapy was observed for patients with MSI and RAS mutations (11). However, as seen in the CheckMate 8HW trial, the presence of RAS mutations does not interfere with the efficacy of anti-PD-1 and anti-CTLA-4 combination therapy for patients with MSI-positive CRC (39), suggesting that dual ICI might have been preferential for this patient. Results of these studies suggest that oncogenic KRAS mutations have a potential to influence the patient’s outcome following nivolumab monotherapy. However, this was not observed in our case, as we saw significant degression of KRAS-mutated clones at day 14 after therapy initiation in contrast to tumor clones possessing PTEN-mutation and MSI.
The term hyperprogression has been a topic of debate among the scientific community, however since it indicates the state of rapid progression of disease in the course of immunotherapy, it seems applicable for our patient’s case (40). Hyperprogression in response to immunotherapy is often linked to MDM2 or MDM4 amplification, however, targeted NGS sequencing performed for our patient did not allow for MDM2/4 analysis, since these genes were not included in the panel design (41). Furthermore, the small panel size did not allow for the evaluation of tumor mutational burden (TMB). TMB is an important predictive biomarker of ICI benefit, reflecting a number of mutations occurring in tumor cells. Tumors with high TMB tend to be more immunogenic and more responsive to ICI therapy (42). Even in the presence of MSI, patients with low TMB demonstrate poor outcomes when treated with ICI (43). However, given the relatively low amount of passenger mutations observed in our patient’s samples, ultra-high TMB could be potentially ruled out, as in samples with ultra-high TMB harbor passenger mutations in at least one of the genes analyzed by our panel. For instance, 60% and 76% of patients included in the MSK MetTropism study with TMB>20 Mut/Mb and >40 Mut/Mb, respectively, had at least one passenger mutation in the analyzed genes (44). Additionally, some authors have linked EGFR amplifications to the state of hyperprogression in response to immunotherapy (40), however, our patient was EGFR amplification-negative. However, since the information on MDM2/4/EGFR amplifications was unavailable due to the panel design, the question of whether the patient’s hyperprogressive disease was associated with molecular correlates. Although high TMB could be potentially ruled out in this case, precise TMB estimation could not be performed, which could be considered a significant limitation of the study, since TMB is an important biomarker of ICI benefit. Moreover, the activation of T-helpers has gained attention as another mechanism of ICI resistance (45). Finally, 30-40% of patients treated with ICI fail to respond to therapy (46).
Overall, our patient’s case highlights the potential of ctDNA MSI burden as a tool for monitoring tumor response and clonal evolution in the course of ICI in patients with MSI-positive colorectal cancer. Further study of the clinical application of MSI burden by liquid biopsy could potentially allow timely assessment of treatment effectiveness in patients before visible signs of progression over time among patients with colorectal and other MSI-positive solid tumors.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving humans were approved by ethics committee at the Blokhin National Medical Research Center of Oncology Research Center. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
DK: Investigation, Resources, Writing – original draft, Writing – review & editing. AL: Conceptualization, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. OK: Formal analysis, Writing – original draft, Writing – review & editing. AK: Visualization, Writing – original draft, Writing – review & editing. ATa: Formal analysis, Writing – original draft, Writing – review & editing. EB: Visualization, Writing – original draft, Writing – review & editing. TG: Investigation, Writing – original draft, Writing – review & editing. EV: Formal analysis, Writing – original draft, Writing – review & editing. VM: Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing. VN: Resources, Writing – original draft, Writing – review & editing. LN: Resources, Writing – original draft, Writing – review & editing. ATr: Project administration, Supervision, Writing – original draft, Writing – review & editing. MF: Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. MI: Conceptualization, Formal analysis, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The research was supported by the Russian Science Foundation (Project No. 22-75-10154; https://rscf.ru/project/22-75-10154/).
AL, OK, AK, ATa, EB, TG, VM, and MI are currently employed by OncoAtlas LLC. EV ended employment in the past 24 months.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Schlötterer C. Genome evolution: Are microsatellites really simple sequences? Curr Biol. (1998) 8:R132–4. doi: 10.1016/S0960-9822(98)70989-3
2. Shia J. Evolving approach and clinical significance of detecting DNA mismatch repair deficiency in colorectal carcinoma. Semin Diagn Pathol. (2015) 32:352–61. doi: 10.1053/j.semdp.2015.02.018
3. Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. (2017) 1–15. doi: 10.1200/PO.17.00073
4. Maio M, Ascierto PA, Manzyuk L, Motola-Kuba D, Penel N, Cassier PA, et al. Pembrolizumab in microsatellite instability high or mismatch repair deficient cancers: updated analysis from the phase II KEYNOTE-158 study. Ann Oncol. (2022) 33:929–38. doi: 10.1016/j.annonc.2022.05.519
5. Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair–deficient cancer: results from the phase II KEYNOTE-158 study. JCO. (2020) 38:1–10. doi: 10.1200/JCO.19.02105
6. Geoerger B, Kang HJ, Yalon-Oren M, Marshall LV, Vezina C, Pappo A, et al. Pembrolizumab in paediatric patients with advanced melanoma or a PD-L1-positive, advanced, relapsed, or refractory solid tumour or lymphoma (KEYNOTE-051): interim analysis of an open-label, single-arm, phase 1–2 trial. Lancet Oncol. (2020) 21:121–33. doi: 10.1016/S1470-2045(19)30671-0
7. Le DT, Diaz LA Jr, Kim TW, Van Cutsem E, Geva R, Jäger D, et al. Pembrolizumab for previously treated, microsatellite instability–high/mismatch repair–deficient advanced colorectal cancer: final analysis of KEYNOTE-164. Eur J Cancer. (2023) 186:185–95. doi: 10.1016/j.ejca.2023.02.016
8. Overman MJ, Lonardi S, Wong KYM. et al: durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair–deficient/microsatellite instability–high metastatic colorectal cancer. JCO. (2018) 36:773–9. doi: 10.1200/JCO.2017.76.9901
9. Segal NH, Wainberg ZA, Overman MJ. et al: Safety and clinical activity of durvalumab monotherapy in patients with microsatellite instability–high (MSI-H) tumors. JCO. (2019) 37:670–0. doi: 10.1200/JCO.2019.37.4_suppl.670
10. Oh CR, Kim JE, Hong YS, Kim SY, Ahn JB, Baek JY, et al. Phase II study of durvalumab monotherapy in patients with previously treated microsatellite instability-high/mismatch repair-deficient or POLE-mutated metastatic or unresectable colorectal cancer. Intl J Cancer. (2022) 150:2038–45. doi: 10.1002/ijc.v150.12
11. Diaz LA Jr, Shiu K-K, Kim T-W, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. (2022) 23:659–70. doi: 10.1016/S1470-2045(22)00197-8
12. Lenz H-J, Van Cutsem E, Luisa Limon M, Wong KYM, Hendlisz A, Aglietta M, et al. First-Line nivolumab plus low-Dose ipilimumab for microsatellite instability-High/Mismatch repair-Deficient metastatic colorectal cancer: the phase II checkMate 142 study. JCO. (2022) 40:161–70. doi: 10.1200/JCO.21.01015
13. Luchini C, Bibeau F, Ligtenberg MJL, Singh N, Nottegar A, Bosse T, et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol. (2019) 30:1232–43. doi: 10.1093/annonc/mdz116
14. Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. (2013) 30:1015–6. doi: 10.1093/bioinformatics/btt755
15. Haraldsdottir S. Microsatellite instability testing using next-generation sequencing data and therapy implications. JCO Precis Oncol. (2017) 1:1–4. doi: 10.1200/PO.17.00189
16. Tieng FYF, Abu N, Lee L-H, Ab Mutalib NS. Microsatellite instability in colorectal cancer liquid biopsy—Current updates on its potential in non-invasive detection, prognosis and as a predictive marker. Diagnostics. (2021) 11:544. doi: 10.3390/diagnostics11030544
17. Yu F, Leong KW, Makrigiorgos A, Adalsteinsson VA, Ladas I, Ng K, et al. NGS-based identification and tracing of microsatellite instability from minute amounts DNA using inter-Alu-PCR. Nucleic Acids Res. (2020) 49:e24–4. doi: 10.1093/nar/gkaa1175
18. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. (2012) 486:532–6. doi: 10.1038/nature11156
19. Siena S, Sartore-Bianchi A, Garcia-Carbonero R, Karthaus M, Smith D, Tabernero J, et al. Dynamic molecular analysis and clinical correlates of tumor evolution within a phase II trial of panitumumab-based therapy in metastatic colorectal cancer. Ann Oncol. (2018) 29:119–26. doi: 10.1093/annonc/mdx504
20. Kasi PM, Klempner SJ, Starr JS, Shergill A, Bucheit LA, Weipert C, et al. Clinical utility of microsatellite instability (MSI-H) identified on liquid biopsy in advanced gastrointestinal cancers (aGI). JCO. (2022) 40:56–6. doi: 10.1200/JCO.2022.40.4_suppl.056
21. Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer. JAMA Oncol. (2016) 2:1310. doi: 10.1001/jamaoncol.2016.1279
22. Fribbens C, O’Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, et al. Plasma ESR1 mutations and the treatment of estrogen receptor–positive advanced breast cancer. JCO. (2016) 34:2961–8. doi: 10.1200/JCO.2016.67.3061
23. Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. (2015) 34:155–63. doi: 10.1038/nbt.3391
24. Li L, Hu Y, Xu Y, Tang S. Mathematical modeling the order of driver gene mutations in colorectal cancer. PloS Comput Biol. (2023) 19:e1011225. doi: 10.1371/journal.pcbi.1011225
25. Chao JY, Chang H-C, Jiang J-K, Yang CY, Chen FH, Lai YL, et al. Using bioinformatics approaches to investigate driver genes and identify BCL7A as a prognostic gene in colorectal cancer. Comput Struct Biotechnol J. (2021) 19:3922–9. doi: 10.1016/j.csbj.2021.06.044
26. André T, Shiu K-K, Kim TW, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab in microsatellite-instability–high advanced colorectal cancer. N Engl J Med. (2020) 383:2207–18. doi: 10.1056/NEJMoa2017699
27. O’Malley DM, Bariani GM, Cassier PA, Marabelle A, Hansen AR, De Jesus Acosta A, et al. Pembrolizumab in patients with microsatellite instability–high advanced endometrial cancer: results from the KEYNOTE-158 study. JCO. (2022) 40:752–61. doi: 10.1200/JCO.21.01874
28. Yakushina V, Kavun A, Veselovsky E, Grigoreva T, Belova E, Lebedeva A, et al. Microsatellite instability detection: the current standards, limitations, and misinterpretations. JCO Precis Oncol. (2023) 7. doi: 10.1200/PO.23.00010
29. Vidula N, Lipman A, Kato S, Weipert C, Hesler K, Azzi G, et al. Detection of microsatellite instability high (MSI-H) status by targeted plasma-based genotyping in metastatic breast cancer. NPJ Breast Cancer. (2022) 8:117. doi: 10.1038/s41523-022-00490-2
30. Chakrabarti S, Bucheit L, Starr JS, Innis-Shelton R, Shergill A, Dada H, et al. Detection of microsatellite instability-high (MSI-H) by liquid biopsy predicts robust and durable response to immunotherapy in patients with pancreatic cancer. J Immunother Cancer. (2022) 10::e004485. doi: 10.1136/jitc-2021-004485
31. Casas-Arozamena C, Cortegoso A, Piñeiro-Perez R, Abalo A, Arias E, Sampayo V, et al. Improving the management of endometrial cancer patients through the use of liquid biopsy analyses: A case report. IJMS. (2022) 23:8539. doi: 10.3390/ijms23158539
32. Kasi PM, Budde G, Krainock M, Circulating tumor DNA. (ctDNA) serial analysis during progression on PD-1 blockade and later CTLA-4 rescue in patients with mismatch repair deficient metastatic colorectal cancer. J Immunother Cancer. (2022) 10:e003312. doi: 10.1136/jitc-2021-003312
33. Gray ES, Rizos H, Reid AL, Boyd SC, Pereira MR, Lo J, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. (2015) 6:42008–18. doi: 10.18632/oncotarget.v6i39
34. Xi L, Pham TH-T, Payabyab EC, Sherry RM, Rosenberg SA, Raffeld M. Circulating tumor DNA as an early indicator of response to T-cell transfer immunotherapy in metastatic melanoma. Clin Cancer Res. (2016) 22:5480–6. doi: 10.1158/1078-0432.CCR-16-0613
35. Chang-Hao-Tsao S, Weiss J, Hudson C, Christophi C, Cebon J, Behren A, et al. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci Rep. (2015) 5:11198. doi: 10.1038/srep11198
36. Fitzgerald S, Blenkiron C, Stephens R, Mathy JA, Somers-Edgar T, Rolfe G, et al. Dynamic ctDNA mutational complexity in patients with melanoma receiving immunotherapy. Mol Diagn Ther. (2023) 27:537–50. doi: 10.1007/s40291-023-00651-4
37. Assaf ZJF, Zou W, Fine AD, Socinski MA, Young A, Lipson D, et al. A longitudinal circulating tumor DNA-based model associated with survival in metastatic non-small-cell lung cancer. Nat Med. (2023) 29:859–68. doi: 10.1038/s41591-023-02226-6
38. NCCN guidelines version 1.2024 NCCN clinical practice guidelines in oncology (NCCN guidelines®): colon cancer . Available online at: https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf (Accessed 11 Marth 2024).
39. Andre T, Elez E, Van Cutsem E, Jensen LH, Bennouna J, Mendez G, et al. Nivolumab (NIVO) plus ipilimumab (IPI) vs chemotherapy (chemo) as first-line (1L) treatment for microsatellite instability-high/mismatch repair-deficient (MSI-H/dMMR) metastatic colorectal cancer (mCRC): First results of the CheckMate 8HW study. JCO. (2024) 42:LBA768–8. doi: 10.1200/JCO.2024.42.3_suppl.LBA768
40. Adashek JJ, Subbiah IM, Matos I, Garralda E, Menta AK, Ganeshan DM, et al. Hyperprogression and immunotherapy: fact, fiction, or alternative fact? Trends Cancer. (2020) 6:181–91. doi: 10.1016/j.trecan.2020.01.005
41. Sehgal K. Hyperprogression in patients with cancer receiving immune checkpoint inhibitors. JAMA Netw Open. (2021) 4:e211839. doi: 10.1001/jamanetworkopen.2021.1839
42. Li Y, Ma Y, Wu Z, Zeng F, Song B, Zhang Y, et al. Tumor mutational burden predicting the efficacy of immune checkpoint inhibitors in colorectal cancer: A systematic review and meta-analysis. Front Immunol. (2021) 12. doi: 10.3389/fimmu.2021.751407
43. Manca P, Corti F, Intini R, Mazzoli G, Miceli R, Germani MM, et al. Tumour mutational burden as a biomarker in patients with mismatch repair deficient/microsatellite instability-high metastatic colorectal cancer treated with immune checkpoint inhibitors. Eur J Cancer. (2023) 187:15–24. doi: 10.1016/j.ejca.2023.03.029
44. Nguyen B, Fong C, Luthra A, Smith SA, DiNatale RG, Nandakumar S, et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell. (2022) 185:563–575.e11. doi: 10.1016/j.cell.2022.01.003
45. Zhu M, Ganesan C, Flanagan S, Yoon HH. Life-threatening “hyper-progression” on immunotherapy revealed as pseudo-progression in DNA mismatch repair deficiency: a case report. J Gastrointestinal Oncol. (2023) 14:435–41. doi: 10.21037/jgo-22-709
Keywords: colorectal cancer, liquid biopsy, hyperprogression, next-generation sequencing, dMMR
Citation: Kravchuk D, Lebedeva A, Kuznetsova O, Kavun A, Taraskina A, Belova E, Grigoreva T, Veselovsky E, Mileyko V, Nikulin V, Nekrasova L, Tryakin A, Fedyanin M and Ivanov M (2025) Dynamics of blood microsatellite instability (bMSI) burden predicts outcome of a patient treated with immune checkpoint inhibitors: a case report of hyperprogressive disease. Front. Immunol. 16:1492296. doi: 10.3389/fimmu.2025.1492296
Received: 06 September 2024; Accepted: 16 January 2025;
Published: 05 February 2025.
Edited by:
Ralf Weiskirchen, RWTH Aachen University, GermanyReviewed by:
Elena Lastraioli, University of Florence, ItalyCopyright © 2025 Kravchuk, Lebedeva, Kuznetsova, Kavun, Taraskina, Belova, Grigoreva, Veselovsky, Mileyko, Nikulin, Nekrasova, Tryakin, Fedyanin and Ivanov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandra Lebedeva, bGViZWRldmFfYV9hXzFAc3RhZmYuc2VjaGVub3YucnU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.