- 1School of Marine Sciences, State Key Laboratory for Biocontrol/Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Guangdong Provincial Key Laboratory of Marine Resources and Coastal Engineering & Guangdong Provincial Observation and Research Station for Marine Ranching of the Lingdingyang Bay, Sun Yat-sen University, Guangzhou, China
- 2School of Life Sciences, Guangdong Province Key Laboratory for Aquatic Economic Animals, Sun Yat-sen University, Guangzhou, China
Stimulator of interferon genes (STING) is a key connector protein in interferon (IFN) signaling, crucial for IFN induction during the activation of antiviral innate immunity. In mammals, ring finger protein 5 (RNF5) functions as an E3 ubiquitin ligase, mediating STING regulation through K150 ubiquitylation to prevent excessive IFN production. However, the mechanisms underlying RNF5’s regulation of STING in teleost fish remain unknown. This study investigated the regulatory role of the mandarin fish (Siniperca chuatsi) RNF5 (scRNF5) in the STING-mediated antiviral immune response and identified the specific regulatory sites on scSTING. Furthermore, an examination of scRNF5 expression patterns in virus-infected cells revealed its responsiveness to mandarin fish ranavirus (MRV) infection. The ectopic expression of scRNF5 suppressed scSTING-mediated IFN signaling and facilitated MRV replication. Co-immunoprecipitation experiments indicated an interaction between scRNF5 and scSTING. The further experiments demonstrated that scRNF5 exerted its inhibitory effect by promoting the degradation of scSTING, which was observed to be blocked by MG132 treatment. Ubiquitination assays with various scSTING mutants showed that scRNF5 catalyzed the ubiquitination of scSTING at K135 and K155 residues. Furthermore, we provided evidence that scRNF5 significantly attenuated scSTING-dependent antiviral immunity by targeting negative regulators within the scSTING signaling cascade. This study underscored that RNF5 negatively regulated the STING-mediated IFN signaling pathway in mandarin fish, attenuated STING’s antiviral activity, and facilitated STING degradation via the ubiquitin-proteasome pathway at two novel lysine sites (K135 and K155). Our work offered valuable insights into the regulatory mechanisms of STING-mediated signaling in teleost fish, paving the way for further research.
1 Introduction
Stimulator of interferon genes (STING), also known as TMEM173, MITA, ERIS, MPYS or NET23, is a transmembrane adaptor protein critical for the innate immune response to pathogenic cytoplasmic DNA (1, 2). Upon recognition of pathogenic DNA, signals converge on STING. The activated STING is then translocated to perinuclear endosomes along with TANK-binding kinase 1 (TBK1) (3). TBK1 phosphorylates and activates interferon regulatory factors (IRFs) and nuclear factor kappa B (NF-κB), thus initiating the production of type I interferon (IFN-I) and other immune response genes (1, 4, 5). STING activity is tightly regulated through various post-translational modifications, including phosphorylation and ubiquitination in mammals (6). Among these regulatory mechanisms, ubiquitination of STING has emerged as a key process, exerting both positive and negative effects on its antiviral signaling (7).
Ring finger protein 5 (RNF5), also referred to as G16 or RMA1, is an E3 ubiquitin ligase localized to the endoplasmic reticulum (8). Early research indicates that RNF5, identified as a novel regulator of cell movement, suppresses cell motility by promoting paxillin ubiquitination (9). Subsequent studies have demonstrated the RNF5’s involvement in muscular dystrophies, myogenesis, and the regulation of various malignancies (9–11). Furthermore, RNF5 facilitates K48-linked ubiquitination and degradation of STING and mitochondrial antiviral signaling protein (MAVS), thereby suppressing host antiviral responses (12, 13). RNF5 inhibits STING/IRF3 signaling, limiting the antiviral response to type I IFN in herpes simplex virus keratitis (14). The V protein of Newcastle disease virus interacts with MAVS, promoting K48-linked ubiquitination and RNF5-mediated degradation (15). In zebrafish, the NLRX1 isoform specifically recruits the E3 ubiquitin ligase RNF5, facilitating K48-linked ubiquitination and leading to proteasome-dependent degradation of STING, ultimately downregulating the IFN antiviral response (16). Beyond degrading STING and MAVS, RNF5 also directly inhibits the function of other virus-associated proteins, potentially modulating the viral life cycle. For instance, RNF5 targets viral antiviral signaling proteins for degradation, thereby hindering viral proliferation and spread (17). Moreover, studies have shown that RNF5 competitively inhibits viral proteins, impeding the replication of specific viruses, further supporting its role in the antiviral process (18). STING homologs have been identified in various teleost fish species, such as grass carp (Ctenopharyngodon idella), grouper (Epinephelus coioides), black carp (Mylopharyngodon piceus) and mandarin fish (Siniperca chuatsi), playing a crucial role in the antiviral signaling pathway (19–25). In black carp, RNF5 mediates STING and MAVS ubiquitination, thereby negatively regulating the antiviral innate immune response (26, 27). However, the precise mechanism by which RNF5 regulates STING activity via ubiquitination and the specific site of STING ubiquitination, remains unknown in teleost fish.
Mandarin fish (S. chuatsi) is a highly valuable aquaculture species in China’s aquaculture market (28). Intensive aquaculture practices, driven by the human demand for animal protein, have led to high-density farming of mandarin fish. However, these practices frequently result in disease outbreaks, with the mandarin fish ranavirus (MRV), a double-stranded DNA virus belonging to the Ranavirus genus of Iridoviridae family, posing a significant threat (29). Therefore, elucidating the innate immune regulatory mechanisms in fish will advance research on antiviral immunity and therapeutics.
In this study, the regulatory role of the S. chuatsi RNF5 (scRNF5) in the STING-mediated antiviral immune response and pinpointed its specific regulatory sites were investigated. Our findings will offer valuable insights for the further elucidating the regulatory mechanisms underlying STING-mediated signaling in teleost fish.
2 Materials and methods
2.1 Animal, cells, and virus
Mandarin fish (S. chuatsi), averaging 50 grams in weight, were sourced from a farm in Guangdong province, China. Prior to the experiment, the fish were acclimated for two weeks in a laboratory-controlled, recirculating freshwater system at 27°C. All animal procedures were conducted in accordance with Guangdong Province’s animal experimentation regulations and approved by the ethics committee of Sun Yat-sen University. The mandarin fish fry (MFF-1) cell line was cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Grand Island, USA), supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, USA), and maintained at 27°C in a humidified atmosphere with 5% CO2 (30). Fathead minnow (FHM) cells (ATCC CCL-42) were maintained in M199 medium supplemented with 10% FBS (Gibco, Grand Island, USA) at 27°C (31). The mandarin ranavirus (MRV) strain NH-1609 (Accession number: MG941005) was isolated from moribund hybrid mandarin fish in Naihai, Guangdong, China, and is preserved in our laboratory (31).
2.2 Antibodies and reagents
Mouse monoclonal anti-c-Myc antibody and mouse monoclonal anti-Flag antibody were obtained from Sigma–Aldrich (St. Louis, USA). The anti-Ha tag antibody was purchased from Cell Signaling Technology (CST, Danvers, USA). Anti-GAPDH was obtained from Abways (Shanghai, China), and rabbit polyclonal anti-mrvORF097L antibody was produced in our laboratory (32). Secondary antibodies included horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (H+L) and HRP-conjugated goat anti-rabbit IgG (H+L), both supplied by Promega (Madison, USA). The proteasome inhibitor MG132, cycloheximide (CHX), 3-Methyladenine (3-MA), and chloroquine (CQ) were all purchased from MedChemExpress (New Jersey, USA). Lodoacetamide (IAA) was obtained from CST (Danvers, USA).
2.3 Protein analysis
For protein domain analysis, SMART (Simple Modular Architecture Research Tool, a web resource for the identification and annotation of protein domains and the analysis of protein domain architectures was used to compartmentalize the protein domains of scRNF5 in Genomic Mode (33). For evolutionary tree, protein sequences were retrieved from GenBank (34) and RefSeq (35) databases and aligned by ClustalW in MEGA v11.0.13 (a software contains a large collection of methods and tools of computational molecular evolution) (36). Then the evolutionary tree was constructed in MEGA using the Neighbor-Joining method with 1,000 bootstrap replications, Jones-Taylor-Thornton (JTT) model, and complete deletion. The evolutionary tree was visualized using iTOL (an online tool for the display, annotation and management of phylogenetic and other trees) (37), incorporating cartoon elements from the Generic Diagramming Platform (a comprehensive database of high-quality biomedical graphics) (38). For protein three-dimensional (3–D) structure predictions, protein sequences were obtained from RefSeq databases, and Alphafold Server (a web server can predict the joint structure of complexes including proteins, nucleic acids, small molecules, ions and modified residues) (39) was employed to predict the protein structures. For protein amino acid sequence alignment, protein sequences were obtained from GenBank and RefSeq databases and DNAMAN v9.0 (a one-for-all software package for molecular biology applications) was used to align with Dynamic Alignment and Default Parameters.
2.4 Plasmid construction and cell transfection
Total RNAs were extracted, and cDNAs were synthesized following previously described methods (25). scRNF5 and scSTING were constructed via polymerase chain reaction (PCR), cloned, and then inserted into pCMV-Myc-N and pCMV-Flag-N vectors (Takara, Japan). This process generated the plasmids: pCMV-Myc-scRNF5, pCMV-Flag-scSTING, pCMV-Flag-scSTING (1–140), pCMV-Flag-scSTING(141-417), pCMV-Flag-scSTING(1-180), and pCMV-Flag-scSTING(181-417) plasmids Various STING constructs, including Ha-scSTING(1-180)KR, Flag-scSTINGCKR, Flag-scSTINGCKRK20, Flag-scSTINGCKRK95, Flag-scSTINGCKRK117, Flag-scSTINGCKRK122, Flag-scSTINGCKRK135 and Flag-scSTINGCKRK155, were synthesized by Beijing Tsingke Biotechnology (China). The primers used for gene cloning, which contained restriction enzyme sites, are listed in Supplementary Material 1. The plasmids were transfected into FHM cells using Fugene HD (Promega, USA), and into MFF-1 cells using Transfect EZ 3000 Plus (eLGbio, China), adhering to the manufacturers’ standard protocols.
2.5 Tissue expression profiles of scRNF5
To examine the tissue distribution of scRNF5 in healthy fish, total RNAs were extracted from various tissues including the gills, fins, spleen, intestine, brain, head kidney, hind kidney, middle kidney, blood, fat, heart, liver, and muscle. Extraction was performed using the SV Total RNA Isolation Kit (Promega, USA) following the manufacturer’s instructions. The expression levels of scRNF5 in these tissues were then quantified by quantitative reverse-transcription PCR (qRT-PCR) using the primers listed in Supplementary Material 1.
2.6 Dual-luciferase reporter assays
MFF-1 cells were cultured in 24-well plates and co-transfected with pRL-TK and IFN-β-Luc plasmids (Promega, USA), and the indicated plasmids or empty vector. After 36 h, cells were harvested, lysed in 200 µL of Passive Lysis Buffer (Promega, USA), and subjected to the Dual-Luciferase Reporter Gene Assay Kit (Promega, USA) following the manufacturer’s instructions. Luciferase activities were measured using Glomax instrument (Promega, USA). Experiments were performed in triplicate, and data represented the mean of at least three independent experiments. All experiments were conducted in at least three independent trials with three technical replicates.
2.7 RNA extraction and qRT-PCR analysis
Total RNA was extracted from cells or tissues using the SV Total RNA Isolation Kit (Promega, USA) and reverse transcribed to synthesize first-strand cDNA using the Evo M-MLV qPCR RT Kit (AG, China), following manufacturers’ protocols. qRT-PCR reactions were performed with SYBR premix ExTaq (Takara, Japan) on a LightCycler 480 instrument (Roche Diagnostics, Switzerland), as previously described (40). Expression levels of immune-related genes and viral genes were normalized to scβ-actin gene. Primer sets for the scRNF5, scSTING, scβ-actin, mrvMCP, mrvICP18, and mrvDNA polymerase (mrvDNA-Poly) genes are described in Supplementary Material 1. Data were analyzed using Q-gene statistics add-in and unpaired sample t-test. Statistical significance was set at p<0.05, with high significance at p<0.01. All data were expressed as mean ± standard deviation (SD).
2.8 Co-immunoprecipitation and western blot analysis
For Co-IP, FHM cells were seeded in 25 cm² dishes, cultured overnight, and co-transfected with 4 μg of vector. After 24 h, cells were lysed using IP lysis buffer (Beyotime, China) with protease inhibitor cocktail III (Merck Millipore, USA), and whole-cell lysates were centrifuged at 12,000 ×g for 10 minutes at 4°C. Supernatants were collected for immunoprecipitation (IP) using anti-Flag or anti-Myc beads (Alpa Life Bio, China), as previously described [36]. Beads were added to the supernatant, incubated for 4 h at 4°C, washed, and proteins were collected by centrifugation and heated to 100°C for 10 minutes for WB analysis. Protein bands were detected using the High-sig ECL western blotting substrate (Tanon, China) and visualized with the Amersham Image Quant 800 system (Cytiva, USA).
2.9 Viral titer determination
MFF-1 cells were seeded into 96-well plates, cultured overnight to exceeding 80% confluence, and infected with serial 10-fold dilutions of MRV-containing samples, with eight wells per dilution. Eight wells containing 100 μL of virus-free culture medium served as negative controls, and plates were incubated at 27°C for seven days, as previously described (41, 42). The number of positive and negative wells was recorded to calculate the TCID50 using the Spearman-Karber method, as previously described (41, 42).
3 Results
3.1 Sequence and phylogenetic analysis of scRNF5
To investigate the role of RNF5 in mandarin fish, we cloned the full-length cDNA of scRNF5 gene and constructed expression plasmids. Sequence analysis showed that the scRNF5 coding sequence (NCBI accession number: XP_044026571.1) consists of 648 nucleotides. It encodes a 216 amino acids (aas) protein with two transmembrane domains (TM) and a ring-finger domain (RING), weighting 23.2 kDa (Figure 1A). 3–D structure predictions showed that the spatial structure of scRNF5 and the spatial arrangement of its domains were similar to its mammalian ortholog (Figure 1B). Phylogenetic analysis clustered scRNF5 with RNF5 proteins from other fish, distinguishing it from the other RNF5 family members (Figure 1C; Supplementary Material 2).
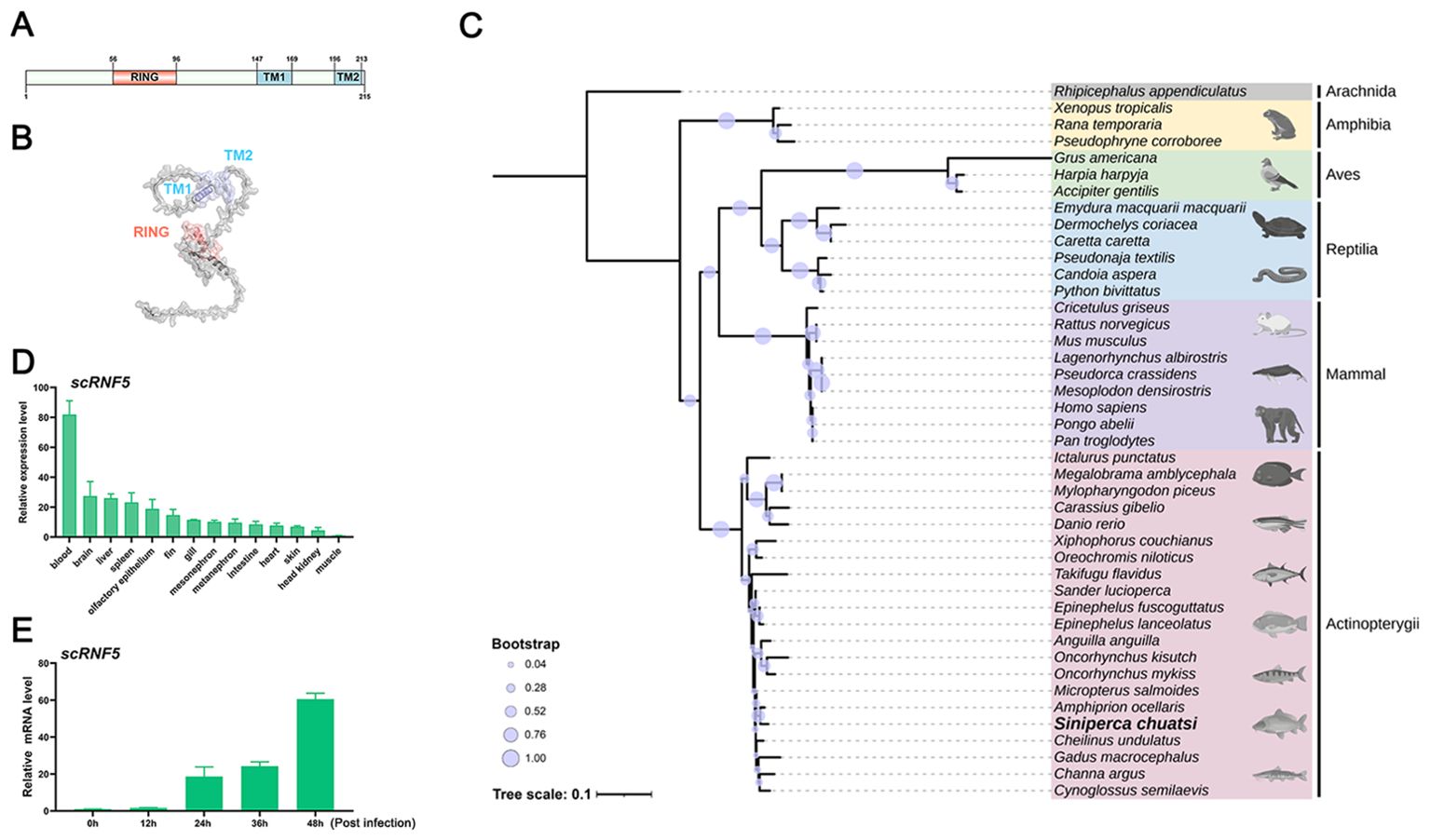
Figure 1. Molecular characteristics of scRNF5. (A, B) Domain and 3–D structure prediction of scRNF5. (C) A phylogenetic tree was generated using the MEGA v11.0.13 program, based on RNF5 sequences from various vertebrate species (GenBank accession numbers are provided in the Supplementary Material 2). (D) Tissue distributions of scRNF5 expressions in different tissues of mandarin fish. Samples were collected from the blood, brain, liver, spleen, fin, gill, intestine, heart, skin, head kidney, and muscle of three healthy mandarin fish, and the expression levels of the scRNF5 were detected by qRT-PCR. (E) Expression of scRNF5 in response to MRV infection. scRNF5 mRNA levels in MFF-1 cells were measured by qRT-PCR and normalized to the reference β-actin gene after infection with MRV at 0.1 MOI.
3.2 Expression patterns of scRNF5 in healthy fish and virus-infected cells
To assess the tissue-specific expression of RNF5 in mandarin fish, we used qRT-PCR to analyze transcript levels of scRNF5 across various tissues, including blood, brain, liver, spleen, fin, gill, intestine, heart, skin, head kidney, and muscle. As shown in Figure 1D, scRNF5 transcripts were detected in all sampled tissues from healthy individuals, with notably higher expression in blood, approximately 81-fold higher than in muscle, highlighting its tissue-specific expression. In MRV-infected cells, scRNF5 expression exhibited a time-dependent increase, with up-regulation levels reaching 18-, 24-, and 60-fold at 24, 36, and 48 h post-infection, respectively (Figure 1E), suggested that scRNF5 responded to MRV infection.
3.3 scRNF5 facilitated MRV replication
MFF-1 cells expressing scRNF5 or a control vector (pCMV-myc) were infected with MRV, and subsequent cells were then harvested for qRT-PCR, WB, and TCID50 analysis. The results showed that overexpression of scRNF5 (Figure 2A) reduced the expression level of scIFN-h gene (Figure 2B); but the expression levels of the and MRV viral genes (mrvMCP, mrvICP18, mrvDNA-Poly) were significantly higher in cells expressing scRNF5 compared to the control group (Figures 2C–E). Furthermore, both the levels of viral titers (Figure 2F) and viral protein mrvORF097L (Figure 2G) were markedly elevated in the scRNF5 group compared to the control. These results indicated that scRNF5 facilitated MRV replication.
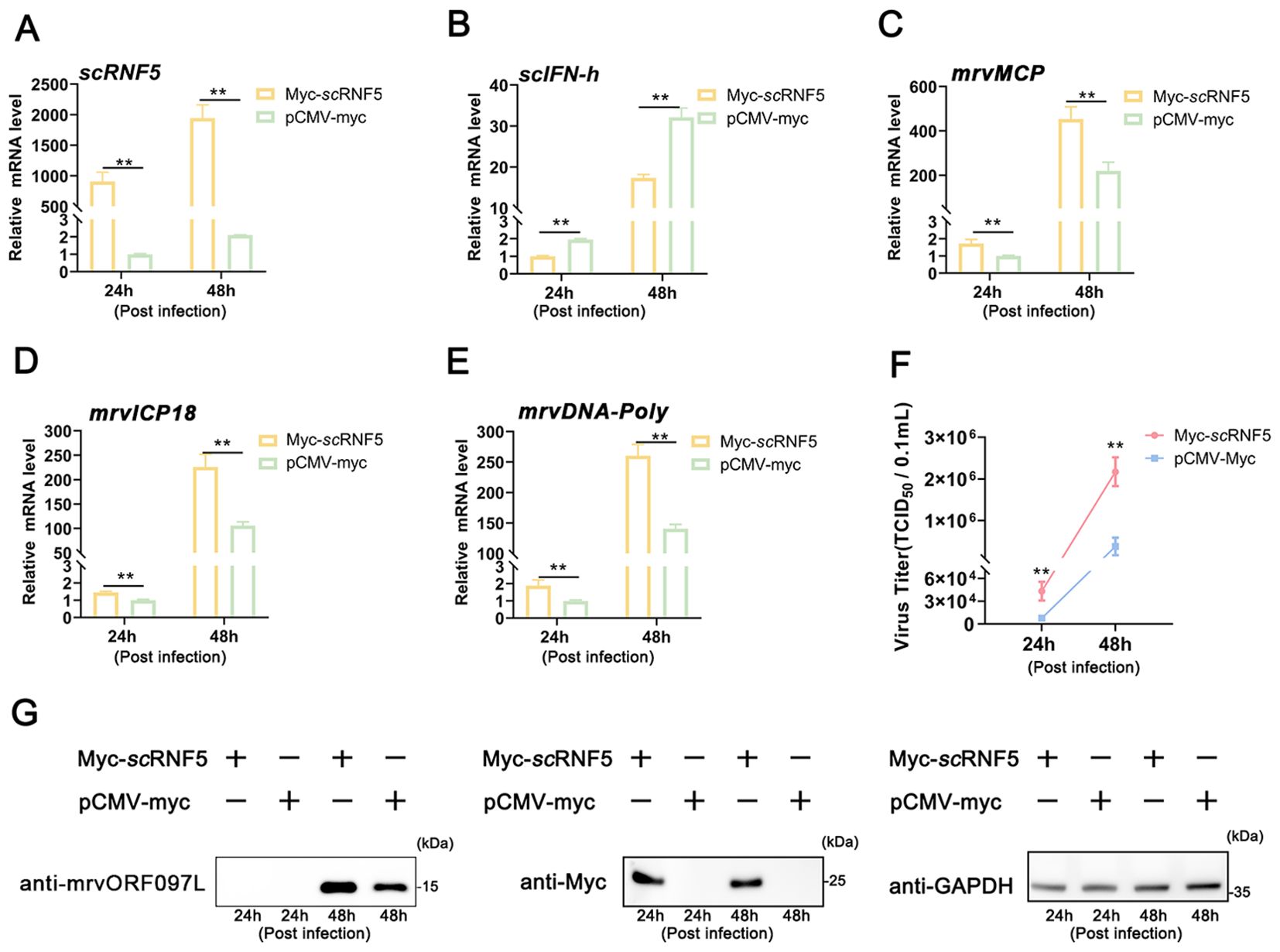
Figure 2. Overexpression of scRNF5 enhances MRV infection. Cells were transfected with scRNF5-myc or pCMV-myc for 24 h, then infected with MRV, and harvested at the indicated time points for qRT-PCR analysis. (A) Expression levels of scRNF5 in MRV-infected cells at indicated times. (B) Expression levels of scIFN-h genes in MRV-infected cells at various time points. (C–E) Expression levels of mrvMCP, mrvICP18, mrvDNA-Poly genes in MRV-infected cells at indicated times. For qRT-PCR analysis, the baseline was established as the group exhibiting the lowest relative expression among all groups, with this value set to 1. All groups represented different time points following viral infection. (F) Virus infection was measured on a 96-well cell culture plate via the finite dilution method. (G) Protein levels of mrvORF097L were detected via WB analysis. Vertical bars represent ± SD (n = 3). Statistical significance is indicated by asterisks, with ** referring to p < 0.01.
3.4 scRNF5 suppressed scSTING-mediated IFN signaling
In mammals, STING serves as a key adaptor in innate immune responses triggered by pathogenic DNA or RNA, but its antiviral activity is attenuated by RNF5 (12). Previously, we found that overexpressing scSTING enhances IFN promoter activity in MFF-1 cells (25). To further explored the role of scRNF5 in the IFN signaling pathway, MFF-1 cells were co-transfected with plasmids expressing scRNF5 and/or scSTING, followed by a luciferase reporter assay. The results showed that scSTING, but not scRNF5, significantly activated the IFN-β promoter. Notably, scRNF5 inhibited scSTING-mediated IFN-β-luc activity in a dose-dependent manner when co-expressed with scSTING (Figures 3A, B). Furthermore, co-transfection experiments revealed that the expression levels of IFN-I signaling related genes (scIFN-h, scMx, scISG15 genes; Figures 3C–E) and pro-inflammatory factor (scTNF-α gene; Figure 3F) in cells co-expressing scSTING and scRNF5 were significantly lower than those in cells co-expressing scSTING and control vector pCMV-myc (p<0.01). These results suggested that scRNF5 might suppress the scSTING-mediated IFN signaling pathway.
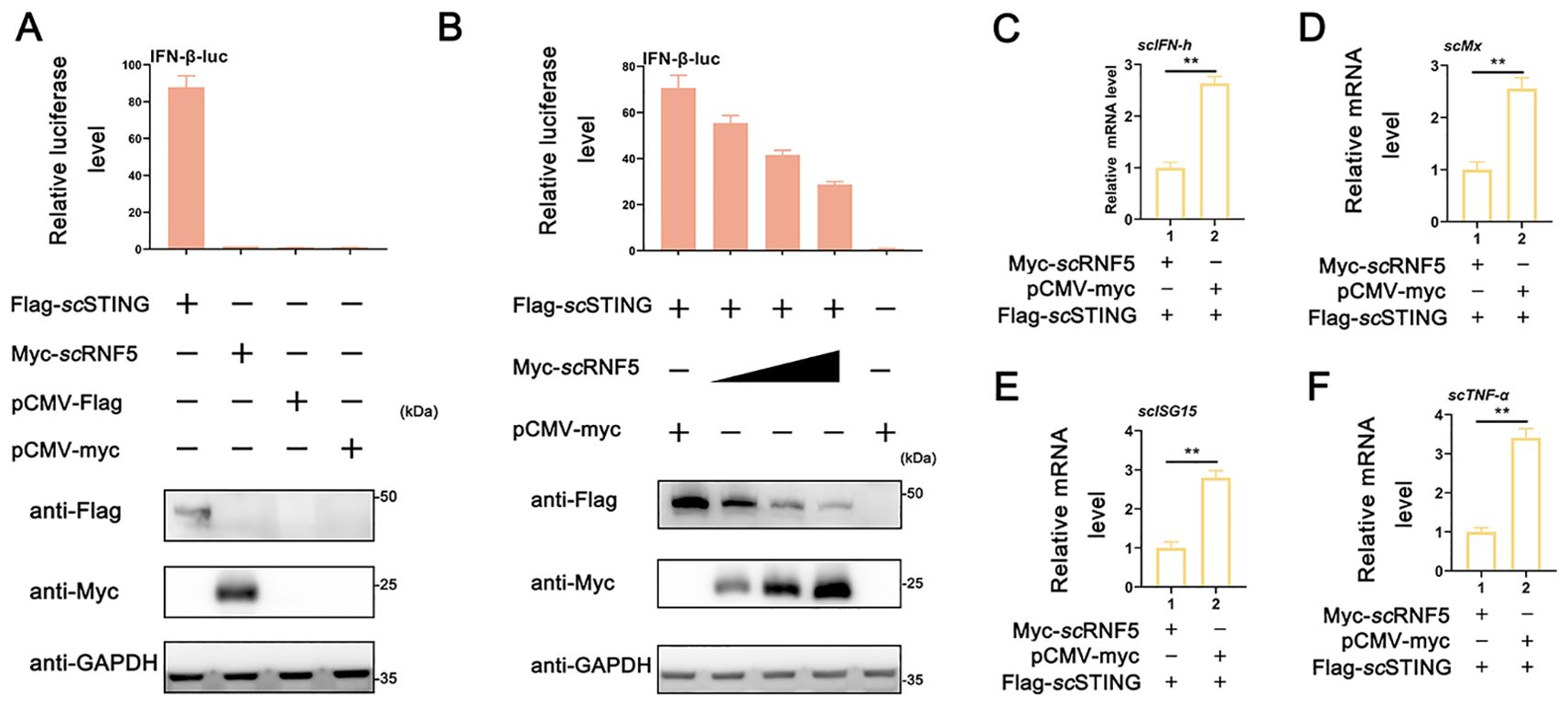
Figure 3. Suppression of scSTING-triggered IFN signaling by scRNF5. (A) Relative luciferase activities of IFN-β-luc were measured relative to controls after cells were co-transfected with scSTING-flag and scRNF5-myc. Cells transfected with pCMV-myc alone served as the negative control. (B) MFF-1 cells were co-transfected with the reporter plasmids pRL-TK and IFN-β-luc, as well as the indicated plasmids scRNF5 and scSTING in 24-well plates. A reporter assay was performed 36 h post-transfection. (C–F) Cells were collected 24 h post-transfection after co-transfection scRNF5 or vector alone alongside scSTING, and levels of expression levels of scIFN-h (C), scMx (D), scISG15 (E) and scTNF-α (F) were assessed. Data shown are representative of three independent experiments (n=3). Statistical analysis revealed significant differences (**p < 0.01).
3.5 scRNF5 targeted scSTING for degradation via ubiquitination pathway
To further explored the mechanism of how scRNF5 regulated the scSTING IFN signaling pathway, we conducted a Co-IP assay to analyze the interaction between scSTING and scRNF5. Figure 4A shows a distinct band representing scRNF5 in the Flag-scSTING immunoprecipitated protein complex. Likewise, Figure 4B depicts a distinct band for scSTING in the Myc-scRNF5 immunoprecipitated protein complex. These findings confirmed the interaction between scRNF5 and scSTING. Upon co-transfection of scRNF5 with scSTING, WB analysis showed a significant reduction in scSTING protein levels in the RNF5-expressing group (Figure 4C). To further investigate this effect, we employed a gradient of scRNF5 expression and observed a clear inverse relationship; increasing scRNF5 levels correlated with a progressive decrease in scSTING protein levels (Figure 4D). Next, we explored the mechanism by which scRNF5 mediated scSTING degradation. Co-transfected of scRNF5 and scSTING, followed by treatment with various drugs including MG132 (specific proteasome inhibitor), 3-MA (autophagy Inhibitor), CQ, and NH4Cl (lysosomal inhibitors), revealed that MG132 effectively blocked scRNF5-mediated scSTING degradation (Figure 4E). Dose-dependent experiments with MG132 further confirmed that it inhibited scRNF5-mediated scSTING degradation in a dose-dependent manner (Figure 4F). Based on these results, we hypothesized that scRNF5 mediated scSTING degradation through ubiquitination. Ubiquitination experiments on scSTING in the presence of scRNF5 revealed a significant increase in scSTING ubiquitination levels compared to control samples (Figure 4G). These observations suggested that scRNF5 targeted scSTING for degradation via ubiquitination pathway.
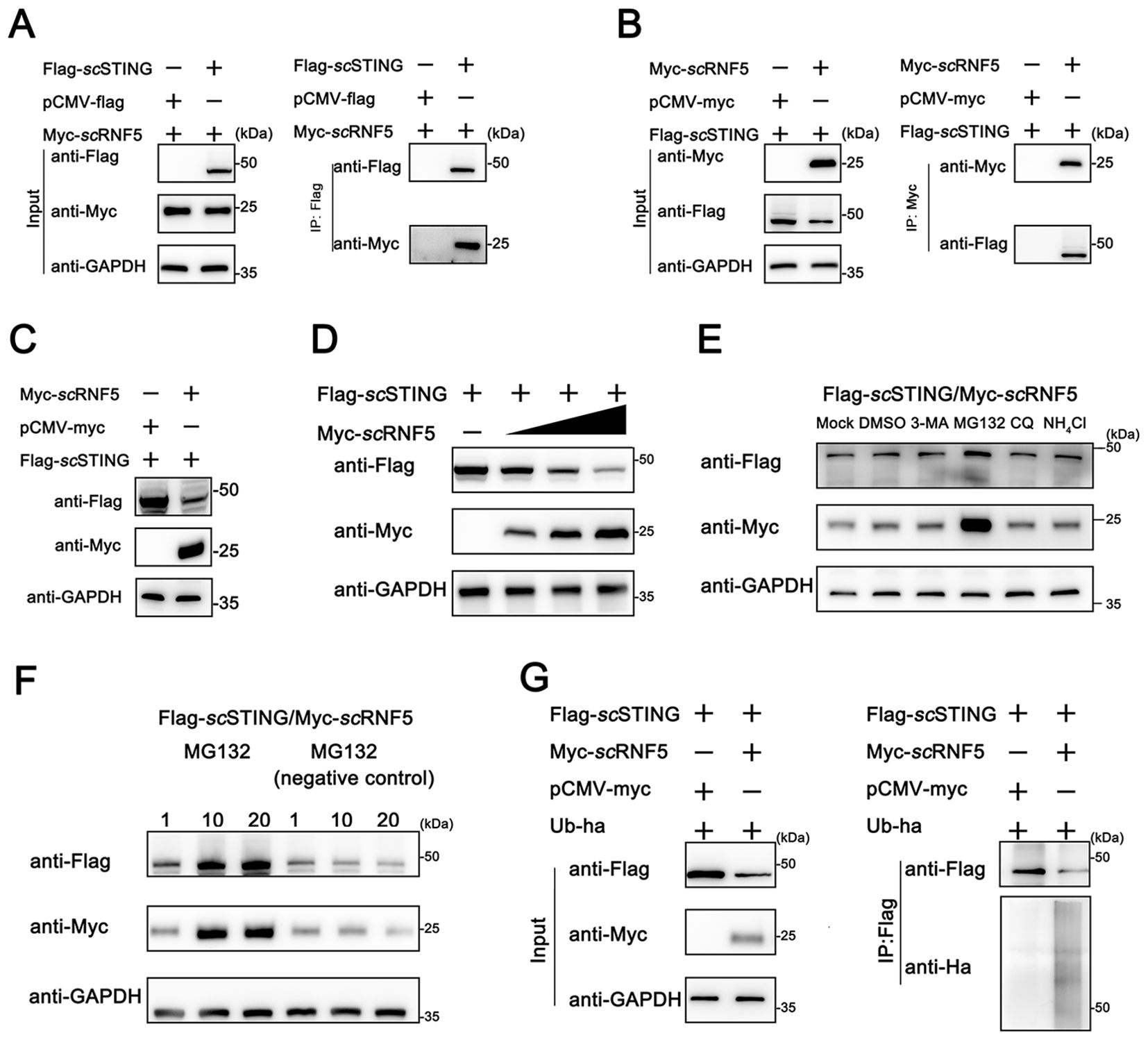
Figure 4. Ubiquitination-mediated degradation of scSTING by scRNF5. (A, B) FHM cells were transfected with pCMV-Myc-scRNF5 or empty vector and pCMV-Flag-scSTING plasmids. Co-IP and WB analyses were performed for detection. IB: immunoblotting; IP: immunoprecipitation. (C) Western blotting analyses were used for detection. FHM cells were transfected with pCMV-Myc-scRNF5 or empty vector and pCMV-Flag-scSTING plasmids. (D) scRNF5 overexpression leads to dose-dependent STING degradation. FHM cells were seeded into 6-well plates, incubated overnight, and co-transfected with 2 μg Flag-scSTING and Myc-scRNF5 (0.5, 1, and 2 μg, respectively), After 24 h, lysates were immunoblotted with anti-Flag, anti-Myc, and anti-GAPDH antibodies. (E) The effects of MG132, 3-MA, CQ and NH4Cl on scRNF5-induced scSTING degradation were assessed. FHM cells in 6-well plates were transfected with Flag-scSTING (2 μg) and Myc-scRNF5 (2 μg). At 24 h post-transfection (hpt), cells were treated with DMSO or the respective inhibitors (MG132: 20 mM, 3-MA: 5 mM, CQ: 10 mM, NH4Cl: 20 mM). After 12 h, lysates were immunoblotted using anti-Flag, anti-Myc, and anti-GAPDH antibodies. (F) FHM cells were transfected and treated as in (E), but with MG132 concentrations of 1, 10, and 20 mM, and a negative control (MG-132). Lysates were immunoblotted as before. (G) Ubiquitination of scSTING was assessed in FHM cells co-expressing Flag-scSTING, ha-ubiquitin, and either Myc-scRNF5 (lane 2) or empty vector (lane 1). Flag-scSTING was immunoprecipitated with anti-Flag, and poly-ubiquitin chains were detected with anti-Ha.
3.6 scRNF5 ubiquitination site on scSTING located in the first 180 aas
To identify the specific ubiquitination site of scSTING by scRNF5, we constructed various truncation mutants of scSTING (Figure 5A) and conducted degradation experiments. Initial results showed that scRNF5 degraded both scSTING (1–140) and scSTING(141-417) (Figure 5B). However, further segmentation analysis showed that scRNF5 only degraded scSTING(1-180), but not scSTING (181-417) (Figure 5C), implying the ubiquitination site is located within the first 180 aas of scSTING. Moreover, ubiquitination experiments on scSTING(1-180) and scSTING(181-417) confirmed significant ubiquitination of scSTING(1-180) by scRNF5 (Figure 5D), further indicating that the ubiquitination occurred within the 1 to 180 aas region.
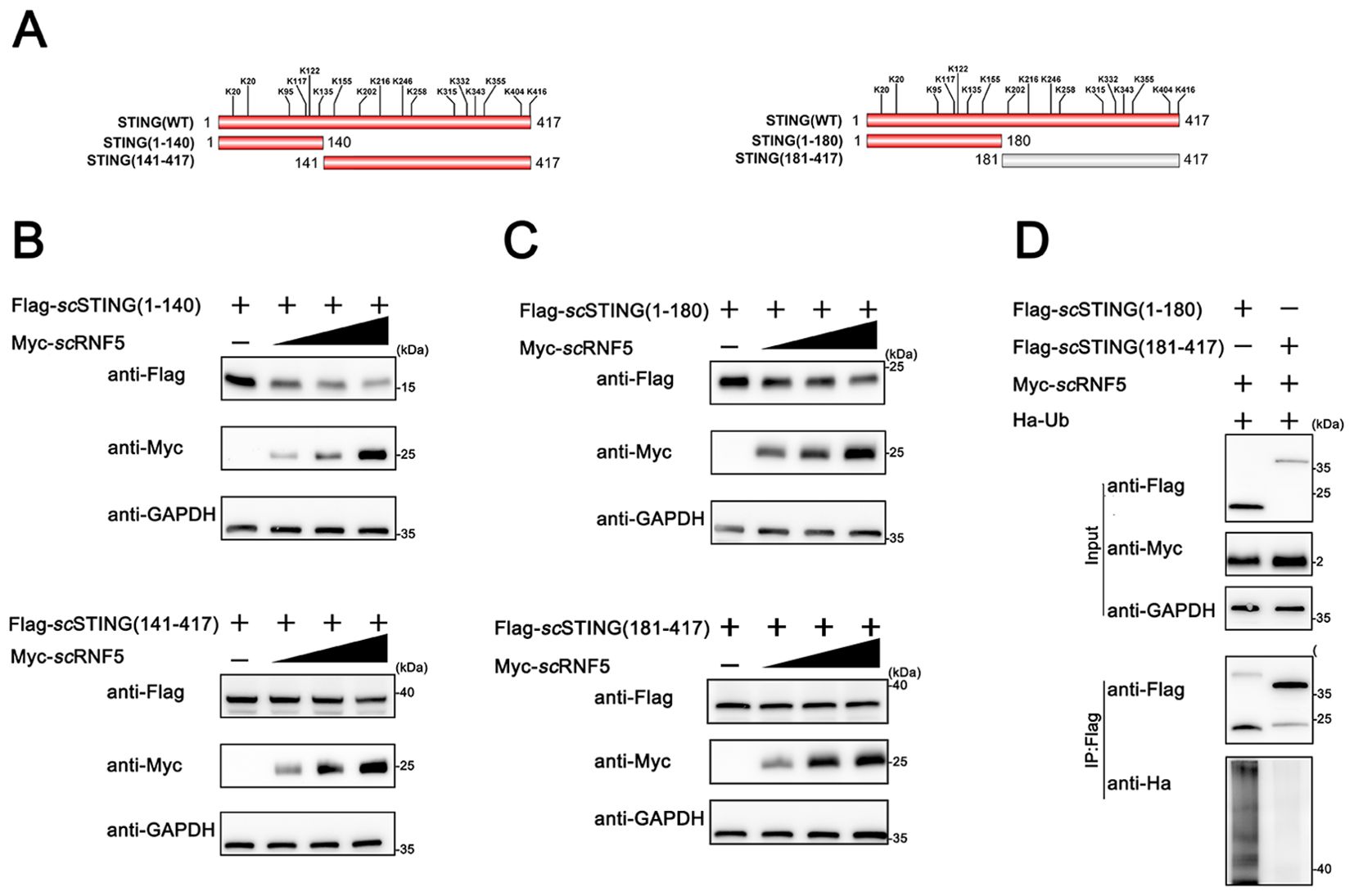
Figure 5. scRNF5 ubiquitinated scSTING within its N-terminal 180 aas. (A) Lysine (K) residues positions in truncated STING variants. (B) FHM cells were seeded overnight in 6-well plates and co-transfected with 2 μg of either Flag-scSTING(1-140) or Flag-scSTING(141-417), along with Myc-scRNF5 at concentrations of 0.5, 1, and 2 μg. After 24 h, lysates were immunoblotted using anti-Flag, anti-Myc, and anti-GAPDH antibodies. (C) Similarly, FHM cells were seeded and co-transfected as in (B), but with Flag-scSTING(1-180) or Flag-scSTING(181-417). Lysates were processed for immunoblotting as before. (D) Ubiquitination of scSTING fragments was assessed. FHM cells were co-expressed with Myc-scRNF5, Ha-ubiquitin, and either Flag-scSTING(1-180) or Flag-scSTING(181-417). Flag-scSTING was immunoprecipitated using anti-Flag, and poly-ubiquitin chains were detected with anti-Ha.
3.7 scRNF5 catalyzed the ubiquitination of scSTING at two lysine residues K135 and K155
Ubiquitination primarily targets lysine residues in various proteins, although other aas, like cysteine, can also undergo this post-translational modification (25). Mammalian RNF5 suppresses K48-linked ubiquitination at the K150 site, promoting proteasomal degradation (12). To investigated RNF5-mediated ubiquitination of STING involves amino acid residues in mandarin fish, we substituted all C and K residues in scSTING with R, generating a variant named scSTINGCKR. Moreover, we constructed a variant where all K residues in scSTING(1-180) were replaced with R, named scSTING(1-180)KR. Degradation experiments observed that scRNF5 could no longer degrade scSTINGCKR (Figure 6A) and scSTING(1-180)KR (Figure 6B), suggesting scRNF5-mediated ubiquitination of scSTING specifically targeted lysine residues. As shown in Figure 6C, the 1-180 aa region of scSTING contains six lysine residues. Next, we generated six mutants, each restoring one of the original K residue in the scSTINGCKR, and designed scSTINGCKR20K, scSTINGCKR95K, scSTINGCKR117K, scSTINGCKR122K, scSTINGCKR135K, and scSTINGCKR155K, respectively. We then performed co-transfection with the mutants and scRNF5 was to identify the specific K residue targeted for degradation. The results showed that scRNF5 had no degradation effect on scSTINGCKR20K, scSTINGCKR95K, scSTINGCKR117K, scSTINGCKR122K (Figures 6D–G). However, scRNF5 regained its ability to degrade scSTINGCKR when K mutations were restored at positions K135 and K155 (Figures 6H–I). Those results suggested that scRNF5 catalyzed the ubiquitination of scSTING specifically at lysine residues K135 and K155.
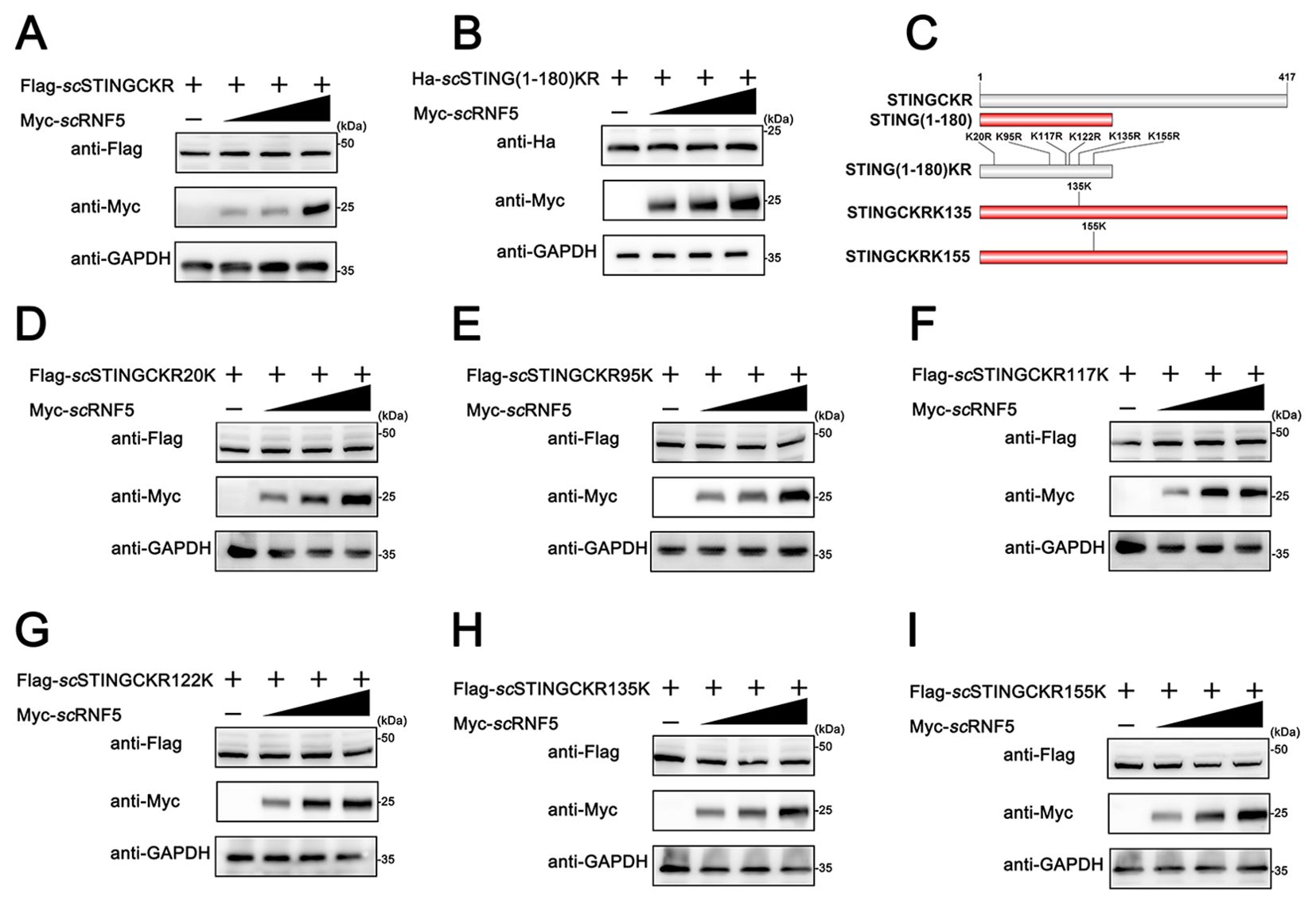
Figure 6. scRNF5 targeted scSTING for ubiquitination at K135 and K155. A (A, B) FHM cells were co-transfected with 2 μg of Flag-scSTINGCKR/Ha-scSTING(1-180)KR and Myc-scRNF5 (0.5, 1 and 2 μg) for 24 h. Lysates were then IB with anti-Ha, anti-Myc, and anti-GAPDH Abs. (C) Schematic of scSTING mutants used in this study. (D–I) FHM cells were co-transfected with 2 μg of various Flag-tagged scSTING mutants (Flag-scSTINGKR20K, Flag-scSTINGKR95K, Flag-scSTINGKR117K, Flag-scSTINGKR122K, Flag-scSTINGKR135K or Flag-scSTINGKR155K) and Myc-scRNF5 (0.5, 1 and 2 μg) for 24 h. Lysates were then subjected to immunoblotted with anti-Flag, anti-Myc, and anti-GAPDH Abs.
Furthermore, to compare the differences in scRNF5 targeting of scSTING lysine residues between mammals and other vertebrates, we performed homologous sequence alignment of STING proteins from mammals, birds, reptiles, amphibians and fish. Interestingly, STING from most fish species contains two lysine residues at positions K135 and K155, whereas STING from other vertebrates (mammals, birds, reptiles, and amphibians) does not contain this two lysine (Figure 7), suggesting potential differences in the negative regulation of STING in teleost fish compared to other vertebrates.
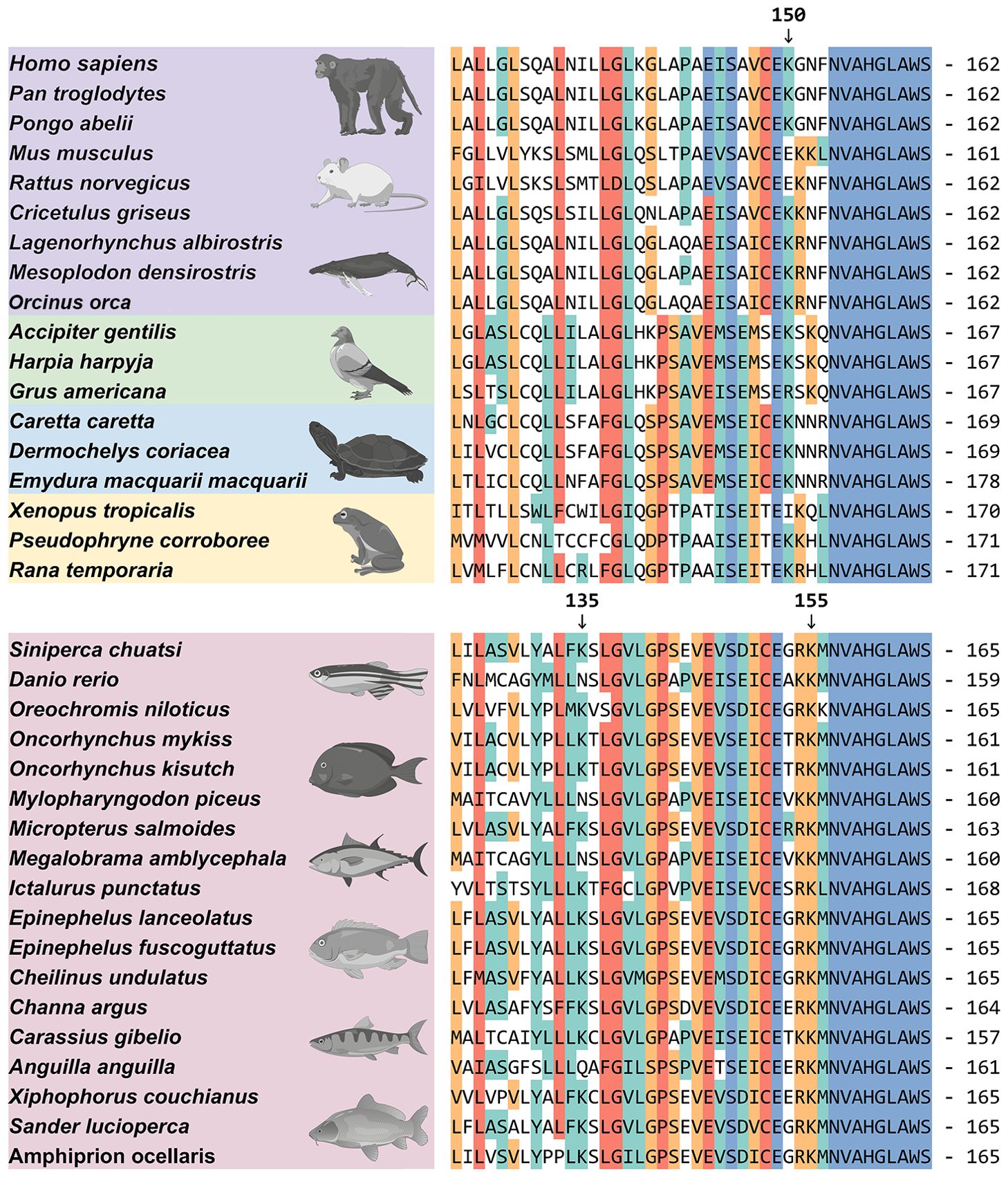
Figure 7. Protein sequence alignment of STING. The protein amino acid alignment reveals the conserved aa sequences and lysine site distributions of STING across different species. Accession numbers for these sequences were provided in the Supplementary Material 2. Arrows indicate lysine at the 150th position in hsSTING and lysine at the 135th and 155th positions in scSTING.
3.8 Negative regulation of scSTING-mediated antiviral activity by scRNF5
To investigate whether the antiviral ability of scSTING was also regulated by scRNF5, MFF-1 cells co-expressing scRNF5 and scSTING were infected with MRV. Viral titer was then assayed from collected supernatants, while cells were harvested for qRT-PCR and WB analysis. The results showed that cells co-expressing scSTING and scRNF5 (Figures 8A, B) had increased the expression levels of the scIFN-h gene (Figure 8C) and MRV viral genes (mrvMCP, mrvICP18, mrvDNA-Poly) (Figures 8D–F). In addition, these cells exhibited higher viral titers (TCID50) and more abundant viral proteins (mrvORF097L) compared to control cells transfected with scSTING alone (Figures 8G, H). These results suggested that the significant negative regulation of scRNF5 on scSTING-mediated antiviral innate immunity.
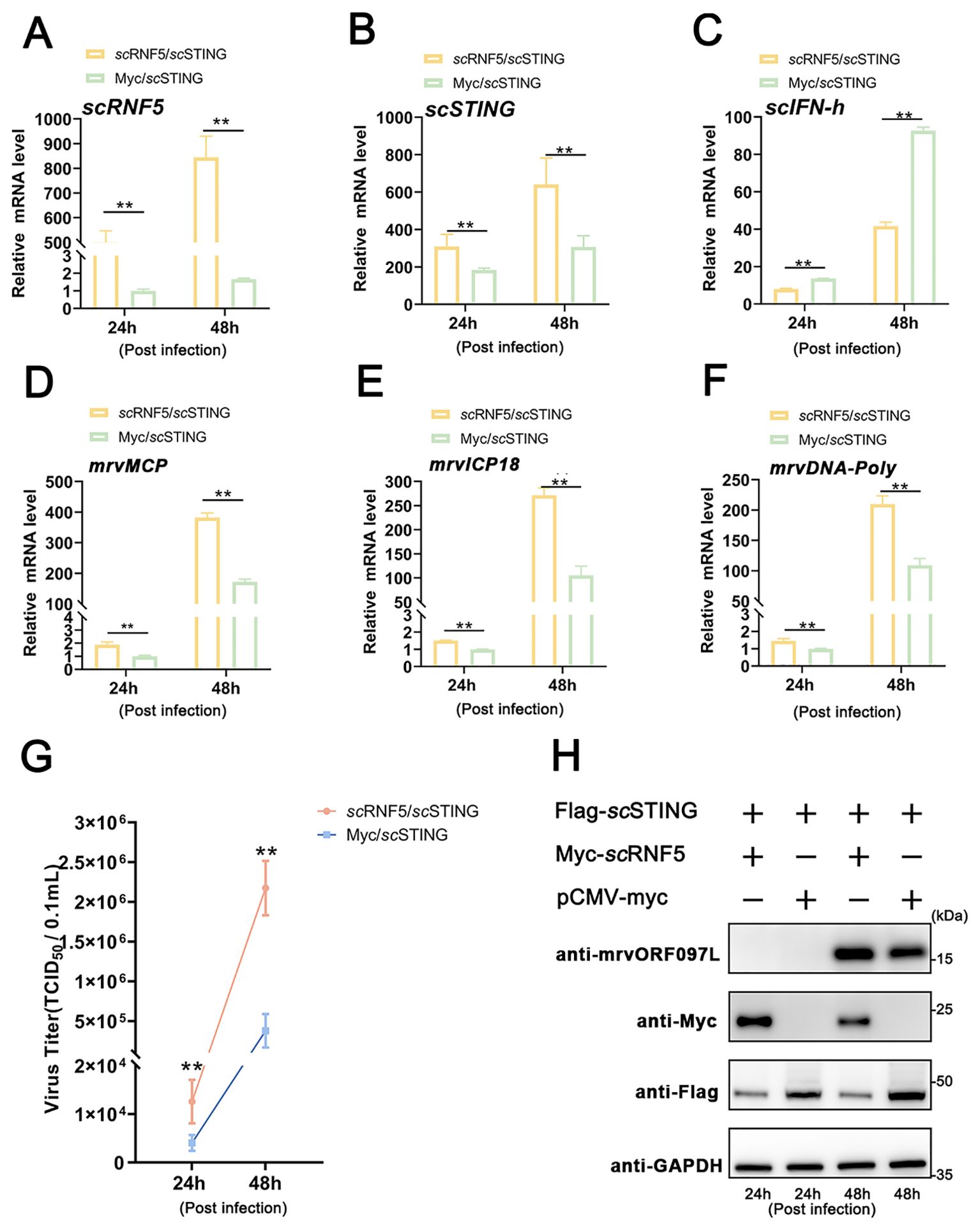
Figure 8. scRNF5 inhibited scSTING-mediated antiviral activity. (A) MFF-1 cells were infected with MRV and harvested for RT-qPCR, WB, and virus titers analysis independently at the indicated time points. Expression levels of scRNF5 (A), scSTING (B), scIFN-h (C), mrvMCP (D), mrvICP18 (E), mrvDNA-Poly (F) in MRV-infected cells at indicated times. For qRT-PCR analysis, the baseline was established as the group exhibiting the lowest relative expression among all groups, with this value set to 1. All groups represented different time points following viral infection. (G) Virus titers were measured on a 96-well cell culture plate using the finite dilution method. (H) WB analysis was performed to detect levels of viral protein mrvORF097L. The left two lanes indicate samples taken at 24 h post-infection, while the right two lanes indicate samples taken at 48 h post-infection. Vertical bars represent ± SD (n = 3). Statistical significance is indicated by asterisks, with ** referring to p < 0.01.
4 Discussion
IFNs are crucial antiviral agents in innate immunity, but their unregulated production can harm the organism, potentially leading to severe outcomes. Therefore, maintaining balanced IFN production is essential for organism homeostasis (43). Upon activation, STING undergoes post-translational modifications, such as phosphorylation and ubiquitination (44). TRIM56 induce K63-linked ubiquitination at residue K150 of STING, thereby activating the STING/IFN signaling pathway (45). Autocrine motility factor receptor facilitates STING activation through K27-linked ubiquitination at multiple sites (46). Conversely, human ubiquitin specific peptidase 13 suppresses K27-linked ubiquitination of STING, reducing its interaction with TBK1 (47). Our study showed that RNF5 regulated STING by facilitating its ubiquitination and destabilization, thereby negatively modulating the activity of STING in mandarin fish.
One key ubiquitination and regulation site on mammalian STING is K150. RNF5 suppresses K48-linked ubiquitination at the K150 site, promoting proteasomal degradation (12). RNF26 enhances STING stability by facilitating K11-linked polyubiquitination at K150, while inhibiting RNF5-induced STING ubiquitination (48). The deubiquitinase cylindromatosis protein removes K48-linked ubiquitination at K150 (49). Our data showed that scRNF5-mediated ubiquitination of scSTING at two lysine residue leads to proteasome degradation. scRNF5 regulated two ubiquitination sites on scSTING: K155 (corresponding to mammalian STING K150) and an extra site at K135. Ubiquitination at either of these sites is sufficient to trigger degradation, suggesting potential differences in the negative regulation of STING between teleost fish and other vertebrates.
Furthermore, E3 ligase plays a pivotal role in substrate determination during ubiquitination and have been extensively studied. Recent research has shown that in black carp, RNF5 regulates STING degradation through the K48-linked ubiquitin-proteasome pathway (27). Black carp RNF5 also interacts with MAVS, negatively regulating the MAVS/IFN signaling pathway (26). Blood plays a vital role in the immune system, particularly in mounting immune responses. The tissue distribution and expression of RNF5 in mandarin fish during MRV infection reveal a high level of expression in the blood, suggesting RNF5 might contribute to the immune response in mandarin fish. Furthermore, in terms of virus promotion, our data showed that scRNF5 overexpression in MFF-1 cells significantly suppressed IFN/ISG transcription, augmented MRV-encoded proteins, and increased virus titers, attenuating STING-mediated IFN expression and antiviral activity.
Our findings suggest several promising strategies for preventing and controlling MRV disease in mandarin fish. One approach involves incorporating scRNF5 inhibitors into mandarin fish feed, which may modulate immune responses and reduce MRV infection; nevertheless, additional research is needed to confirm its efficacy and safety. Another strategy entails identifying the K135/155 mutant scSTING in wild mandarin fish, providing a valuable genetic resource for breeding disease-resistant strains. Once these mutants are identified, they can be integrated into selective breeding programs to enhance the disease resistance of farmed fish. Furthermore, introduction of R135/155 scSTING into mandarin fish via transgenic techniques offers an innovative approach to bolster their antiviral defenses. Although this strategy is promising, it also poses challenges in technology, regulation, and public acceptance. Overall, these strategies have the potential to substantially improve both the disease resistance and production efficiency of mandarin fish aquaculture.
In summary, our study highlighted that RNF5 negatively regulated the STING-mediated IFN signaling pathway in mandarin fish, attenuated STING’s antiviral capacity, and facilitated STING degradation via the ubiquitin-proteasome pathway at K135 and K155 resides (Figure 9). This study offers valuable insights into the regulatory mechanisms of STING-mediated signaling in teleost fish, presenting novel prospects for disease-resistant breeding and drug target identification, thereby paving the way for further elucidation.
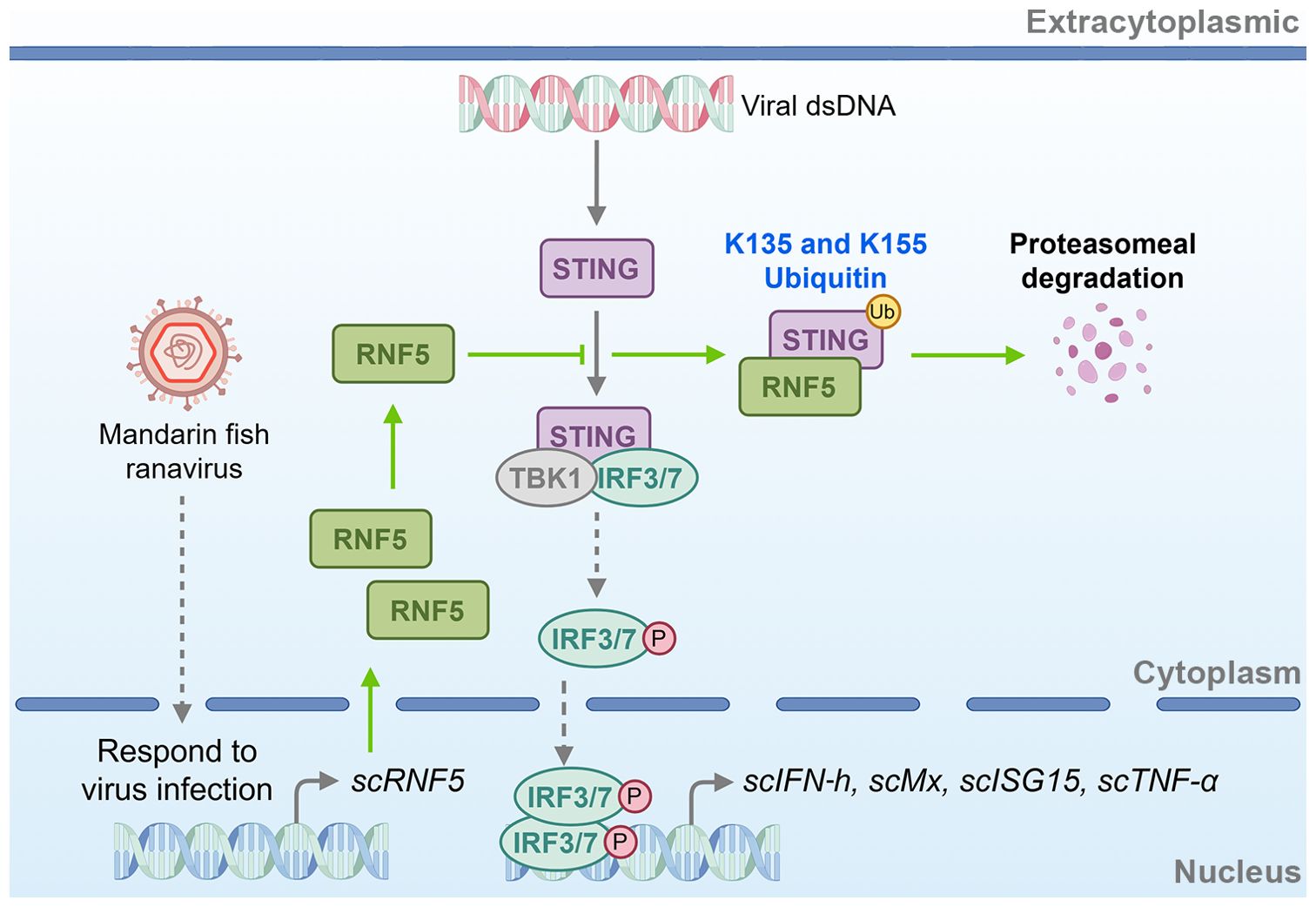
Figure 9. Schematic of RNF5-mediated negative regulation of STING signaling in mandarin fish. scRNF5 interacts with scSTING, ubiquitinating lysine residues at positions 135 and 155. This ubiquitination marks scSTING for proteasomal degradation, preventing IFN antiviral interferon response. By promoting scSTING degradation, scRNF5 suppresses the host’s antiviral immune response, thereby facilitating viral replication.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Laboratory Animals of Sun Yat-sen University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XQ: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. CL: Data curation, Formal analysis, Investigation, Writing – original draft. ML: Formal analysis, Visualization, Writing – original draft. ZQ: Data curation, Investigation, Writing – original draft. YY: Investigation, Resources, Writing – original draft. SW: Investigation, Resources, Writing – original draft. JH: Funding acquisition, Project administration, Resources, Writing – original draft. CG: Conceptualization, Formal analysis, Funding acquisition, Project administration, Visualization, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (No. 2022YFE0203900), the China Agriculture Research System (No. CARS-46), the Guangdong Key Research and Development Program (Nos. 2022B1111030001), the Guangdong Province Basic and Applied Basic Research Fund Project (No. 2023A1515110395), and the Science and Technology Plan Project of Guangdong Province (No. 2024B1212040007).
Acknowledgments
Authors would also thank www.figdraw.com for cartoon components. The authors would like to acknowledge all study participants and individuals who contributed to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1525376/full#supplementary-material
Supplementary Material 1 | Primers used in this study.
Supplementary Material 2 | Accession numbers of RNF and STING family members sequences in the NCBI.
References
1. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. (2008) 455:674–78. doi: 10.1038/nature07317
2. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. (2008) 29:538–50. doi: 10.1016/j.immuni.2008.09.003
3. Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. (2009) 461:788–92. doi: 10.1038/nature08476
4. Phelan T, Little MA, Brady G. Targeting of the cGAS-STING system by DNA viruses. Biochem Pharmacol. (2020) 174:113831. doi: 10.1016/j.bcp.2020.113831
5. Sun W, Li Y, Chen L, Chen H, You F, Zhou X, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U.S.A. (2009) 106:8653–58. doi: 10.1073/pnas.0900850106
6. Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol. (2020) 21:501–21. doi: 10.1038/s41580-020-0244-x
7. Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. (2009) 458:430–37. doi: 10.1038/nature07959
8. Adir O, Bening-Abu-Shach U, Arbib S, Henis-Korenblit S, Broday L. Inactivation of the Caenorhabditis elegans RNF-5 E3 ligase promotes IRE-1-independent ER functions. Autophagy. (2021) 17:2401–14. doi: 10.1080/15548627.2020.1827778
9. Didier C, Broday L, Bhoumik A, Israeli S, Takahashi S, Nakayama K, et al. RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol Cell Biol. (2003) 23:5331–45. doi: 10.1128/MCB.23.15.5331-5345.2003
10. Delaunay A, Bromberg KD, Hayashi Y, Mirabella M, Burch D, Kirkwood B, et al. The ER-bound RING finger protein 5 (RNF5/RMA1) causes degenerative myopathy in transgenic mice and is deregulated in inclusion body myositis. PloS One. (2008) 3:e1609. doi: 10.1371/journal.pone.0001609
11. Bromberg KD, Kluger HM, Delaunay A, Abbas S, DiVito KA, Krajewski S, et al. Increased expression of the E3 ubiquitin ligase RNF5 is associated with decreased survival in breast cancer. Cancer Res. (2007) 67:8172–79. doi: 10.1158/0008-5472.CAN-07-0045
12. Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, et al. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. (2009) 30:397–407. doi: 10.1016/j.immuni.2009.01.008
13. Zhong B, Zhang Y, Tan B, Liu TT, Wang YY, Shu HB. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J Immunol. (2010) 184:6249–55. doi: 10.4049/jimmunol.0903748
14. Liu Z, Xia L. E3 ligase RNF5 inhibits type I interferon response in herpes simplex virus eratitis through the STING/IRF3 signaling pathway. Front Microbiol. (2022) 13:944101. doi: 10.3389/fmicb.2022.944101
15. Sun Y, Zheng H, Yu S, Ding Y, Wu W, Mao X, et al. Newcastle disease virus V protein degrades mitochondrial antiviral signaling protein to inhibit host type I interferon production via E3 ubiquitin ligase RNF5. J Virol. (2019) 93:e00322-19. doi: 10.1128/JVI.00322-19
16. Zhao X, An L, Gong X, Dan C, Qu Z, Sun H, et al. A zebrafish NLRX1 isoform downregulates fish IFN responses by targeting the adaptor STING. J Virol. (2024) 98:e180123. doi: 10.1128/jvi.01801-23
17. Li Z, Hao P, Zhao Z, Gao W, Huan C, Li L, et al. The E3 ligase RNF5 restricts SARS-CoV-2 replication by targeting its envelope protein for degradation. Signal Transduct Target Ther. (2023) 8:53. doi: 10.1038/s41392-023-01335-5
18. Ge J, Zhang L. RNF5: inhibiting antiviral immunity and shaping virus life cycle. Front Immunol. (2023) 14:1324516. doi: 10.3389/fimmu.2023.1324516
19. Fan Y, Sun Y-B, Zhang T-K, Liu T-K, et al. Fish MITA serves as a mediator for distinct fish IFN gene activation dependent on IRF3 or IRF7. J Immunol (Baltimore Md.: 1950). (2011) 187:2531–39. doi: 10.4049/jimmunol.1101547
20. Stéphane B, Emilie M, Annie L, Julie B, Michel B, Mossman KL. Both STING and MAVS fish orthologs contribute to the induction of interferon mediated by RIG-I. PloS One. (2012) 7:e47737. doi: 10.1371/journal.pone.0047737
21. Feng X, Yang C, Zhang Y, Peng L, Chen X, Rao Y, et al. Identification, characterization and immunological response analysis of stimulator of interferon gene (STING) from grass carp Ctenopharyngodon idella. Dev Comp Immunol. (2014) 45:163–76. doi: 10.1016/j.dci.2014.03.001
22. Huang Y, Ouyang Z, Wang W, Yu Y, Li P, Zhou S, et al. Antiviral role of grouper STING against iridovirus infection. Fish Shellfish Immunol. (2015) 47:157–67. doi: 10.1016/j.fsi.2015.09.014
23. Su JG, Nie GX, Cao XL, Chen JJ. Identification and expression analysis of the sting gene, a sensor of viral DNA, in common carp (Cyprinus carpio). J Fish Biol. (2016) 88:1949–64. doi: 10.1111/jfb.12960
24. Lu L, Wang X, Wu S, Song X, Zou Z, Xie X, et al. Black carp STING functions importantly in innate immune defense against RNA virus. Fish Shellfish Immunol. (2017) 70:13–24. doi: 10.1016/j.fsi.2017.08.037
25. Qin XW, He J, Yu Y, Liu C, Luo ZY, Li ZM, et al. The roles of mandarin fish STING in innate immune defense against Infectious spleen and kidney necrosis virus infections. Fish Shellfish Immunol. (2020) 100:80–9. doi: 10.1016/j.fsi.2020.02.062
26. Yan J, Qiao G, Wang E, Peng Y, Yu J, Wu H, et al. Negatively regulation of MAVS-mediated antiviral innate immune response by E3 ligase RNF5 in black carp. Fish Shellfish Immunol. (2023) 134:108583. doi: 10.1016/j.fsi.2023.108583
27. Yan J, Qiao G, Yin Y, Wang E, Xiao J, Peng Y, et al. Black carp RNF5 inhibits STING/IFN signaling through promoting K48-linked ubiquitination and degradation of STING. Dev Comp Immunol. (2023) 145:104712. doi: 10.1016/j.dci.2023.104712
28. Yu Y, He J, Liu W, Li Z, Weng S, He J, et al. Molecular characterization and functional analysis of hypoxia-responsive factor prolyl hydroxylase domain 2 in mandarin fish (Siniperca chuatsi). Animals (Basel). (2023) 13:1556. doi: 10.3390/ani13091556
29. Dong C, Wang Z, Weng S, He J. Occurrence of a lethal ranavirus in hybrid mandarin (Siniperca scherzeri × Siniperca chuatsi) in Guangdong, South China. Vet Microbiol. (2017) 203:28–33. doi: 10.1016/j.vetmic.2017.02.006
30. Dong C, Weng S, Shi X, Xu X, Shi N, He J. Development of a mandarin fish Siniperca chuatsi fry cell line suitable for the study of infectious spleen and kidney necrosis virus (ISKNV). Virus Res. (2008) 135:273–81. doi: 10.1016/j.virusres.2008.04.004
31. Li ZM, Qin XW, Zhang Q, He J, Liang MC, Li CR, et al. Mandarin fish von Hippel-Lindau protein regulates the NF-kappaB signaling pathway via interaction with IkappaB to promote fish ranavirus replication. Zool Res. (2024) 45:990–1000. doi: 10.24272/j.issn.2095-8137.2023.392
32. Pan W, Liang M, You Y, Li Z, Weng S, He J, et al. Viral genomic methylation and the interspecies evolutionary relationships of ranavirus. PloS Pathog. (2024) 20(11):e1012736. doi: 10.1371/journal.ppat.1012736
33. Letunic I, Khedkar S, Bork P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. (2021) 49:D458–60. doi: 10.1093/nar/gkaa937
34. Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, et al. GenBank. Nucleic Acids Res. (2013) 41:D36–42. doi: 10.1093/nar/gks1195
35. O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. (2016) 44:D733–45. doi: 10.1093/nar/gkv1189
36. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–27. doi: 10.1093/molbev/msab120
37. Letunic I, Bork P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. (2024) 52(W1):W78–82. doi: 10.1093/nar/gkae268
38. Jiang S, Li H, Zhang L, Mu W, Zhang Y, Chen T, et al. Generic Diagramming Platform (GDP): a comprehensive database of high-quality biomedical graphics. Nucleic Acids Res. (2024) gkae973. doi: 10.1093/nar/gkae973
39. Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. (2024) 630:493–500. doi: 10.1038/s41586-024-07487-w
40. Qin XW, Luo ZY, Pan WQ, He J, Li ZM, Yu Y, et al. The interaction of mandarin fish DDX41 with STING evokes type I interferon responses inhibiting ranavirus replication. Viruses. (2022) 15:58. doi: 10.3390/v15010058
41. Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Epidemiol. (1938) 27(3):493–7. doi: 10.1093/oxfordjournals.aje.a118408
42. Zeng R, Pan W, Lin Y, He J, Luo Z, Li Z, et al. Development of a gene-deleted live attenuated candidate vaccine against fish virus (ISKNV) with low pathogenicity and high protection. Iscience. (2021) 24:102750. doi: 10.1016/j.isci.2021.102750
43. Jefferies CA. Regulating IRFs in IFN driven disease. Front Immunol. (2019) 10:325. doi: 10.3389/fimmu.2019.00325
44. Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. (2012) 5:ra20. doi: 10.1126/scisignal.2002521
45. Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. (2010) 33:765–76. doi: 10.1016/j.immuni.2010.10.013
46. Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, et al. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. (2014) 41:919–33. doi: 10.1016/j.immuni.2014.11.011
47. Sun H, Zhang Q, Jing YY, Zhang M, Wang HY, Cai Z, et al. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat Commun. (2017) 8:15534. doi: 10.1038/ncomms15534
48. Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, et al. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PloS Pathog. (2014) 10:e1004358. doi: 10.1371/journal.ppat.1004358
Keywords: innate immunity, interferons, RNF5, STING, ubiquitination
Citation: Qin X, Li C, Liang M, Qian Z, You Y, Weng S, He J and Guo C (2024) Ring finger protein 5 mediates STING degradation through ubiquitinating K135 and K155 in a teleost fish. Front. Immunol. 15:1525376. doi: 10.3389/fimmu.2024.1525376
Received: 09 November 2024; Accepted: 25 November 2024;
Published: 11 December 2024.
Edited by:
Zhiwei Liao, The Ohio State University, United StatesReviewed by:
Jianfei Lu, Ningbo University, ChinaQuanyuan Wan, Houston Methodist Research Institute, United States
Copyright © 2024 Qin, Li, Liang, Qian, You, Weng, He and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changjun Guo, Z2NoYW5nakBtYWlsLnN5c3UuZWR1LmNu
†ORCID: Changjun Guo, orcid.org/0000-0003-3223-6176
 Xiaowei Qin1
Xiaowei Qin1 Zhen Qian
Zhen Qian Changjun Guo
Changjun Guo