- 1Center for Evolutionary and Theoretical Immunology, Department of Biology, University of New Mexico, Albuquerque, NM, United States
- 2Department of Anatomy, University of Otago, Dunedin, New Zealand
- 3College of Science and Engineering, Flinders University, Adelaide, SA, Australia
- 4South Australian Museum, Adelaide, SA, Australia
- 5School of Biological Sciences, The University of Adelaide, Adelaide, SA, Australia
Squamate reptiles are amongst the most successful terrestrial vertebrate lineages, with over 10,000 species across a broad range of ecosystems. Despite their success, squamates are also amongst the least studied lineages immunologically. Recently, a universal lack of γδ T cells in squamates due to deletions of the genes encoding the T cell receptor (TCR) γ and δ chains was discovered. Here, we begin to address how the loss of γδ T cells may have impacted the evolution of the squamate immune system. Using the skink Tiliqua rugosa, we found that squamates have not significantly increased the complexity of conventional T cell receptor beta (TCRβ or TRB) chain V regions compared to that of the nearest living squamate relative, the tuatara, Sphenodon punctatus or other amniotes. Our analyses include a putative new TCR locus. This novel locus contains V, D, and J gene segments that undergo V(D)J recombination, albeit with a limited number of gene segments in most squamate species. Based on conserved residues, the predicted protein chain would be expected to form a heterodimer with TCRα. This new TCR locus appears to be derived from an ancient duplication of the TRB locus and is homologous to the recently described T cell receptor epsilon (TRE). TRE is absent from the genomes of the tuatara and all Archosaurs examined and appears squamate specific.
Introduction
The term reptile describes a broad range of species across Sauropsida, the vertebrate clade that includes both the Archelosauria (birds, crocodilians, and turtles) and the Lepidosauria (1, 2). These two lineages diverged between 265-280 million years ago (MYA) (2, 3). The Lepidosauria contains two ancient lineages, the Rhynchocephalia with a single living species, the tuatara Sphenodon punctatus, and the Squamata, which are the lizards, snakes, and amphisbaenians (2, 4, 5). Squamata includes more than 10,000 species that occupy a broad range of environmental niches (4, 6–8).
Despite their evolutionary success, reptiles are arguably the least immunologically studied group of vertebrates (9–12). This is unfortunate as squamate reptiles provide many potential model species given their varying life-history traits including viviparity vs. oviparity, sexual reproduction vs. parthenogenesis, and adaptation to a wide range of ecosystems. Nonetheless, there remains comparatively few published squamate immune system studies (9–12).
With few exceptions, all jawed vertebrate immune systems have three distinct lineages of lymphocytes that are clonally unique due to somatic recombination of their antigen receptor genes (13). These receptors are the T cell receptors (TCR) expressed by αβ and γδ T cells and the immunoglobulins (Ig) expressed by B cells (13–15). Squamates lack γδ T cells due to major genomic deletions of the genes encoding the γδTCR chains (16). These deletions occurred after the Lepidosauria-Rhynchocephalia split approximately 260 MYA and appear to be squamate specific as S. punctatus has the genes encoding the TCR γ and δ chains (TRG and TRD) (5, 16, 17).
Here, we examine evidence for possible compensation of the loss of γδ T cells in squamates by investigating the complexity of the remaining TCR loci in a model species, the skink Tiliqua rugosa. This analysis includes investigating a potential new TCR chain, recently identified as TCRϵ (TRE), that appears to be squamate specific (18). We also provide evidence that this novel TCR is likely derived from a duplication of the TCRβ (TRB) locus.
Materials and methods
Animals
The T. rugosa spleen transcriptome data was generated from two animals, one from Western Australia and one from South Australia described previously in Morrissey et al. (16).
Genome annotation
The T. rugosa genome is being assembled and annotated as part of the Bioplatforms Australia - Australian Amphibian and Reptile Genomics Initiative (https://ausargenomics.com/). The animal used was the same individual, SAMAR71619 (South Australian Museum), used for one of the splenic transcriptomes. Briefly, Verkko (19) was used to generate a hybrid assembly of PacBio HiFi (https://data.bioplatforms.com/ausarg-pacbio-hifi/bpa-ausarg-pacbio-hifi-350719-da052873) and nanopore ultralong reads (https://data.bioplatforms.com/ausarg-ont-promethion/bpa-ausarg-ont-promethion-350780-pag18329), incorporating HiC reads (https://data.bioplatforms.com/ausarg-hi-c/bpa-ausarg-hi-c-350781-hcn7wdrxy) for extended phasing. The resulting pseudohaplotype assemblies and the unassigned contigs were scaffolded separately and together using YaHS (20). HiC contact maps were generated with PretextMap v0.1.90 (https://github.com/sanger-tol/PretextMap) and both haplotypes evaluated simultaneously for misjoins, haplotype switches and other assembly errors with PretextView v0.2.5 (https://github.com/sanger-tol/PretextView) as outlined in https://github.com/Nadolina/Rapid-curation-2.0. Each manually curated pseudohaplotype consists of 16 chromosome sized scaffolds and several unplaced contigs with an average haploid genome size of 1.69G.
Chromosomes containing TRB sequences were identified by BLASTn using putative variable (V) and constant (C) gene sequences from the transcriptome analyses (see below). The T. rugosa chromosome(s) containing TRB was chromosome 2 in both pseudohaplotypes. The S. punctatus genome assembly (ASM311381v1, GenBank accession number GCA_003113815.1) was also searched to identify scaffold(s) containing the TRB locus. GenBank TRB sequences from the chicken, Gallus gallus, were used to search the S. punctatus whole-genome assembly (accession number EF554755.1). The scaffold containing the S. punctatus TRB was scaffold QEPC01009940.1 (https://www.ncbi.nlm.nih.gov/).
Chromosomes containing TRE sequences in T. rugosa were identified by BLASTn using V and C gene sequences identified from the green anole (Anolis carolinensis), originally identified by Gambon-Deza (18) (accession number GCA_035594765.1, NC_085841.1). The T. rugosa chromosome(s) containing TRE was chromosome 1 in both pseudohaplotypes. The S. punctatus genome assembly (ASM311381v1, accession number GCA_003113815.1) was also searched to identify scaffold(s) containing either TRE or the TRE flanking genes (accession number QEPC01002436.1). Flanking genes were also identified in A. carolinensis (accession number GCA_035594765.1, NC_085841.1) and the American alligator (Alligator mississippiensis) (accession number GCA_030867095.1,NC_081825.1).
Transcriptome analysis
Previously published sequences were used to identify the TRB transcripts in T. rugosa (21). The TRBC region identified was used to identify transcripts in a previously published 454 transcriptome dataset (22). The outputs were analyzed for V regions and C regions based on conserved motifs. Identified partial sequences were then used to screen for full length sequences containing V or C regions. Sequences identified were then used to search the PacBio Isoseq transcriptomes with BLASTn in a local database, using the same process described above. Transmembrane regions were identified with DeepTMHMM-2.0 (23). The T. rugosa TRB sequences were previously deposited under GenBank accession numbers OL311598-OL311653 (https://www.ncbi.nlm.nih.gov/). The S. punctatus transcriptome assembly (GGNQ00000000.1) was searched using similar methods. GenBank accession numbers of all TRB sequences identified in the S. punctatus transcriptome are found in Supplementary Table 1.
To identify transcripts for TRE, the TREC region identified in the T. rugosa genome was used to analyze the same T. rugosa PacBio transcriptome (see above). Transcripts were then utilized to screen for sequences containing full-length V or C regions. Transmembrane regions were identified with DeepTMHMM-2.0 (23).
Annotation and characterization
Non-TCR gene models were predicted using GenSAS with references from non-mammalian vertebrates (24). BLAST was then used on all predicted coding sequences against the GenBank database. Genomic V, D, and J sequences were identified by recombination signal sequences (RSS) or comparison to available transcriptomic sequences (25). To identify or confirm RSS sequences, an RSS information content model (RIC) was used (26; https://www.itb.cnr.it/rss/index.html). NCBI’s BLASTp or tBLASTn algorithms were used to confirm V sequences and assess their similarity to TCR homologs from various species retrieved from GenBank (27). V gene nucleotide segments were then aligned with ClustalW (28). Gene segments were annotated following the international ImMunoGeneTics information system nomenclature (29). Gene segments were named according to their location from 5’ to 3’ end on the locus. V gene families were defined by sharing ≥80% nucleotide sequence identity based on ClustalW alignments (28).
Phylogenetic analysis
MEGAX was used to convert nucleotide sequences for both variable (V) genes and constant region genes (C) into amino acid residues which were then aligned with MUSCLE (30, 31). The aligned sequences were then used to construct phylogenetic trees using the neighbor-joining method (32). The trees were then visualized using iTOL (33).
Variable and constant gene sequences with accession numbers used in all phylogenetic analyses are found in Supplementary Tables 2–5. S. punctatus TRBV are found on scaffold QEPC01009940.1 (https://www.ncbi.nlm.nih.gov/). The Chinese alligator (Alligator sinensis) TRBV sequences were provided by Wang et al. (34). Opossum (Monodelphis domestica) TRB are also found in Parra et al. (35). Xenopus tropicalis and Ambystoma mexicanum TRBV sequences were provided by Jesus Martinez.
Percent nucleotide identity matrix
Germline nucleotide sequences were collected from both T. rugosa and S. punctatus (see above). Sequences were aligned via ClustalW (28). Gene segments were annotated following IMGT nomenclature. Families were defined by having ≥80% nucleotide identity in the ClustalW alignment (28). Analysis and visualization of the percent identity matrix generated by ClustalW was conducted using the R packages ggplot2 and reshape2 (28; RStudio 2024.4.2.764; 36–39).
Constant region analysis
TCR constant region sequences from Gallus gallus TRAC (MN646854.1), Gallus gallus TRBC (BAC67174), S. punctatus TRBC (GGNQ01096868.1), S. punctatus TRGC (GGNQ01074423.1), S. punctatus TRDC (GGNQ01087842.1), and T. rugosa TRAC (UYS90863.1), T. rugosa TRBC (UYS90848.1), and T. rugosa TREC were aligned via ClustalW (28). Sequences were then analyzed for transmembrane regions using DeepTMHMM-2.0 (23).
Results
Initially, we set out to characterize the TRB loci in the skink T. rugosa using the tuatara S. punctatus for comparative purposes. The T. rugosa TRB locus is located on chromosome 2 and is approximately 373kb in length (Figure 1A). The S. punctatus TRB locus is at least 718kb in length (Figure 1B). There is conserved synteny surrounding the TRB loci, which are flanked by DBHL (DBH-like monooxygenase protein 2) at the 5’ end and EPHB5-like (ephrin type-b receptor 5-like) at the 3’ end in both species (Figures 1, 2). This conserved synteny is maintained in several amniote species (Figure 2) (34, 35, 40–43).
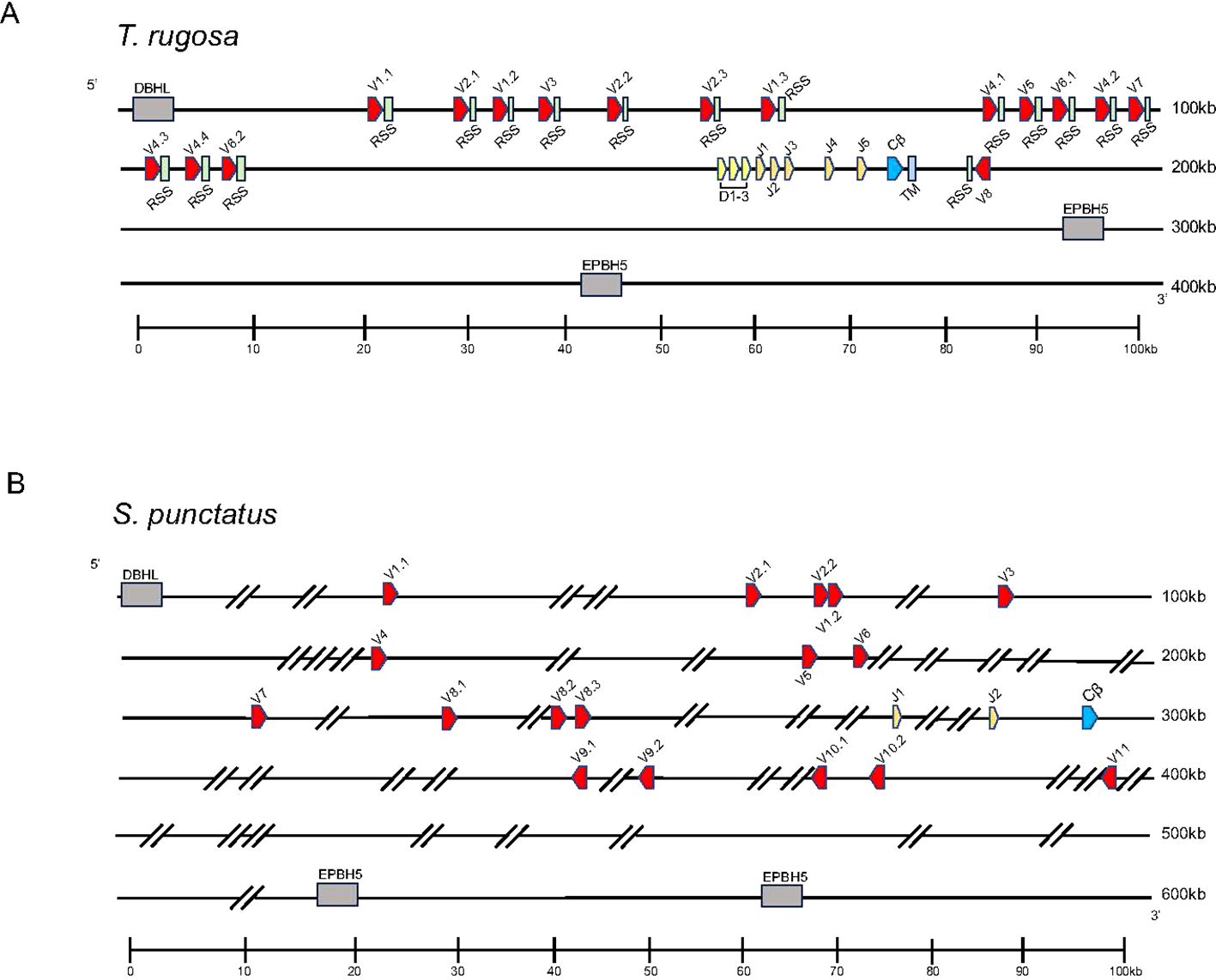
Figure 1. Map of the TRB loci. TRBV designated V (red), TRBD designated D (yellow), TRBJ designated J (orange), TRBC designated Cβ (blue), and the TRB transmembrane region designated TM (light blue) gene segments are numbered by their corresponding location in order across the locus. 23bp spacers are shown in light green near V segments. Other RSS are not shown for space. The flanking genes on the 5’ end of the locus was DBH-like monooxygenase protein 2 (DBHL). The 3’ flanking gene is ephrin type-b receptor 5-like (EPHB5-like). V segments are designated with a family member number followed by a period and a designated number according to their gene family. Transcriptional orientation is indicated by the direction of the arrow on each gene segment or exon. Arrows are not proportionate to the actual gene sizes. Gray boxes indicate flanking genes. Gaps in the genome are indicated by hatch marks. (A) Shows the skink TCR β locus in pseudohaplotype 2. The locus is ~373kb in length. Shown in the TRB locus on haplotype 2. (B) Shows the tuatara TCR β locus. The locus ~718kb in length.
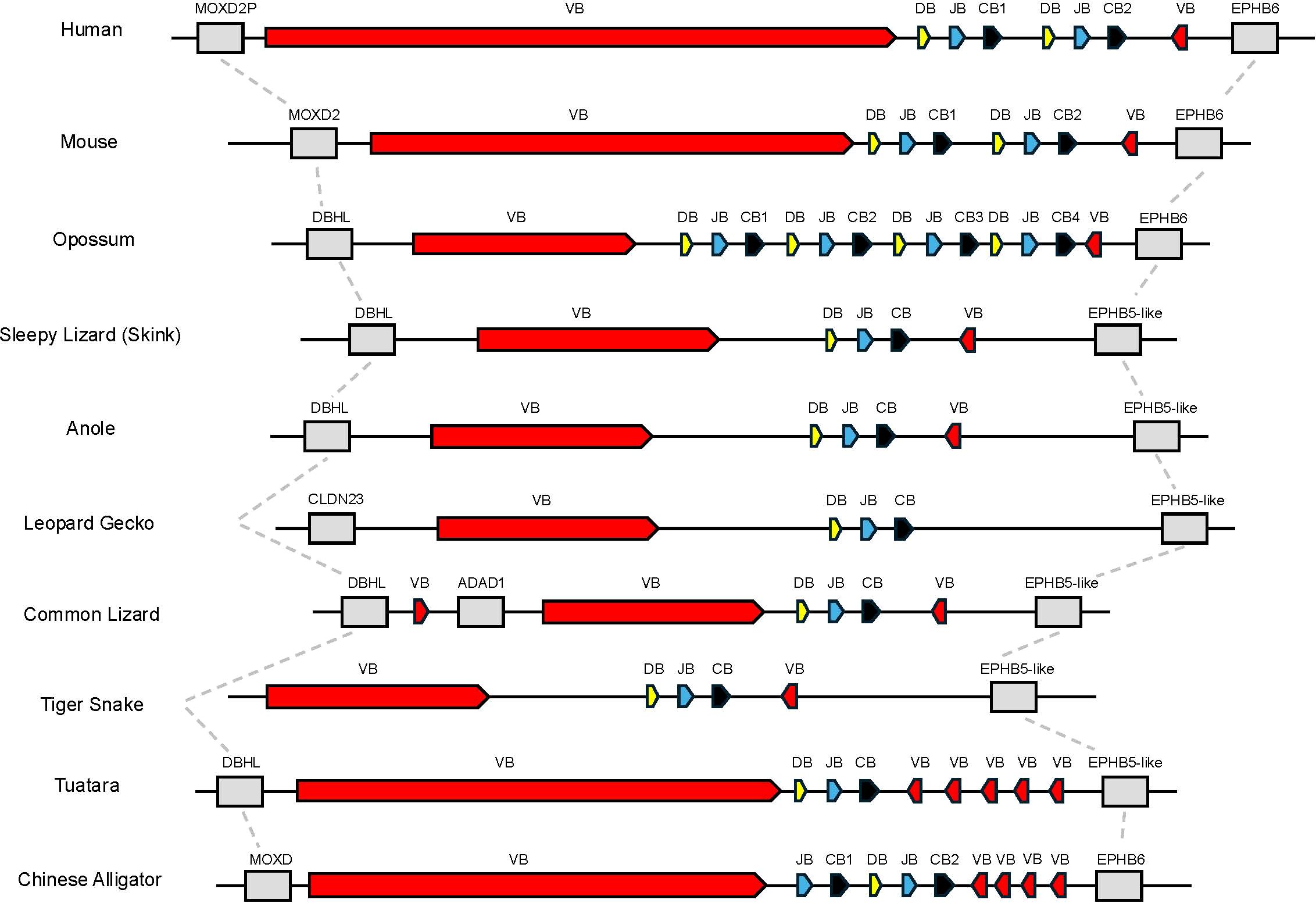
Figure 2. Comparison of the region containing TRB. Dashed lines connect genes with conserved synteny in this genomic region between species shown. Flanking genes are shown in grey. Monooxygenase, DBH-like 2 (MOXD2P/MOXD2/DBHL), ephrin type B receptor 6 or ephrin type b receptor 5-like (EPHB6 or EPHB5-like), claudin 23 (CLDN23), and adenosine deaminase containing protein 1 (ADAD1) are shown. Regions containing TRBV genes are labeled as VB shown in red, TRBD are labeled as DB shown in yellow, TRBJ are labeled as JB shown in blue, and TRBC are labeled as CB shown in black.
Both available T. rugosa TRB pseudohaplotypes were annotated and found to contain 15 and 16 TRBV gene segments in pseudohaplotypes 1 and 2, respectively. These gene segments could be classified into eight families based on nucleotide identity (Supplementary Figure 1A). All families were found in both pseudohaplotypes. Noteworthy was a single TRBV gene segment in an inverted reading frame relative to the rest of the locus on the 3’ side of TRBC (Figure 1A). As will be discussed later, inverted TRBV at the extreme 3’ end of the TRB locus is a feature shared with many other amniote species. Both T. rugosa pseudohaplotypes contained three TRBD, six TRBJ, and a single TRBC gene (Figure 1A). The T. rugosa TRBV sequences were flanked by a 23 base pair (bp) spacer and canonical CACAGTG heptamer (Figure 1A; 25, 44). The TRBD gene segments were flanked by a 12 bp spacer on the V proximal side and a 23 bp spacer on the C proximal side. Similarly, the TRBJ segments were flanked by a 12 bp spacer (not shown). In T. rugosa 100% of the RSS flanking the TRB V, D, and J segments were canonical (not shown). In other squamate species, the RSS appeared uniformly non-canonical e.g. CACAGCA (not shown). However, non-canonical RSS have been routinely shown to be functional (44). Across a wide variety of vertebrates, there is nucleotide conservation of TRBD genes (45). The most V proximal T. rugosa D segment, TRBD1, contains this conserved sequence (GGGACAGGGGGC) and is identical to TRBD sequences found in the A. carolinensis, the common lizard (Zootoca vivipara), and the mainland tiger snake (Notechis scutatus) (Supplementary Table 6).
Sphenodon punctatus has 17 TRBV gene segments which are classified into 11 gene families based on nucleotide identity (Figure 1B; Supplementary Figure 1B). Furthermore, five TRBV genes were inverted and found on the 3’ side of the single TRBC gene (Figure 1B). Two TRBJ gene segments were identified in S. punctatus, but no TRBD gene segments could be identified in the current genome, most likely due to gaps in the genome assembly (Figure 1B).
We compared the T. rugosa and S. punctatus TRBV sequences to TRBV of other vertebrate species in a phylogenetic analysis (Figure 3). TRBVs of both T. rugosa and S. punctatus were interspersed amongst the V genes of other vertebrates consistent with TRBV germline diversity being evolutionarily ancient (Figure 3). The exception is one clade that includes only mammalian TRBV (Figure 3). The 3’-inverted TRBV formed their own clade in the phylogenetic analysis despite low bootstrap values in multiple iterations of the tree including minimum evolution and maximum likelihood (Figure 3; Supplementary Figure 3A). This is consistent with a common ancestral inversion. We note that several amphibian TRBV from the axolotl Ambystoma mexicanum that are not inverted, also fall into this clade, whereas non-inverted TRBV from Xenopus tropicalis did not (Supplementary Figure 3).
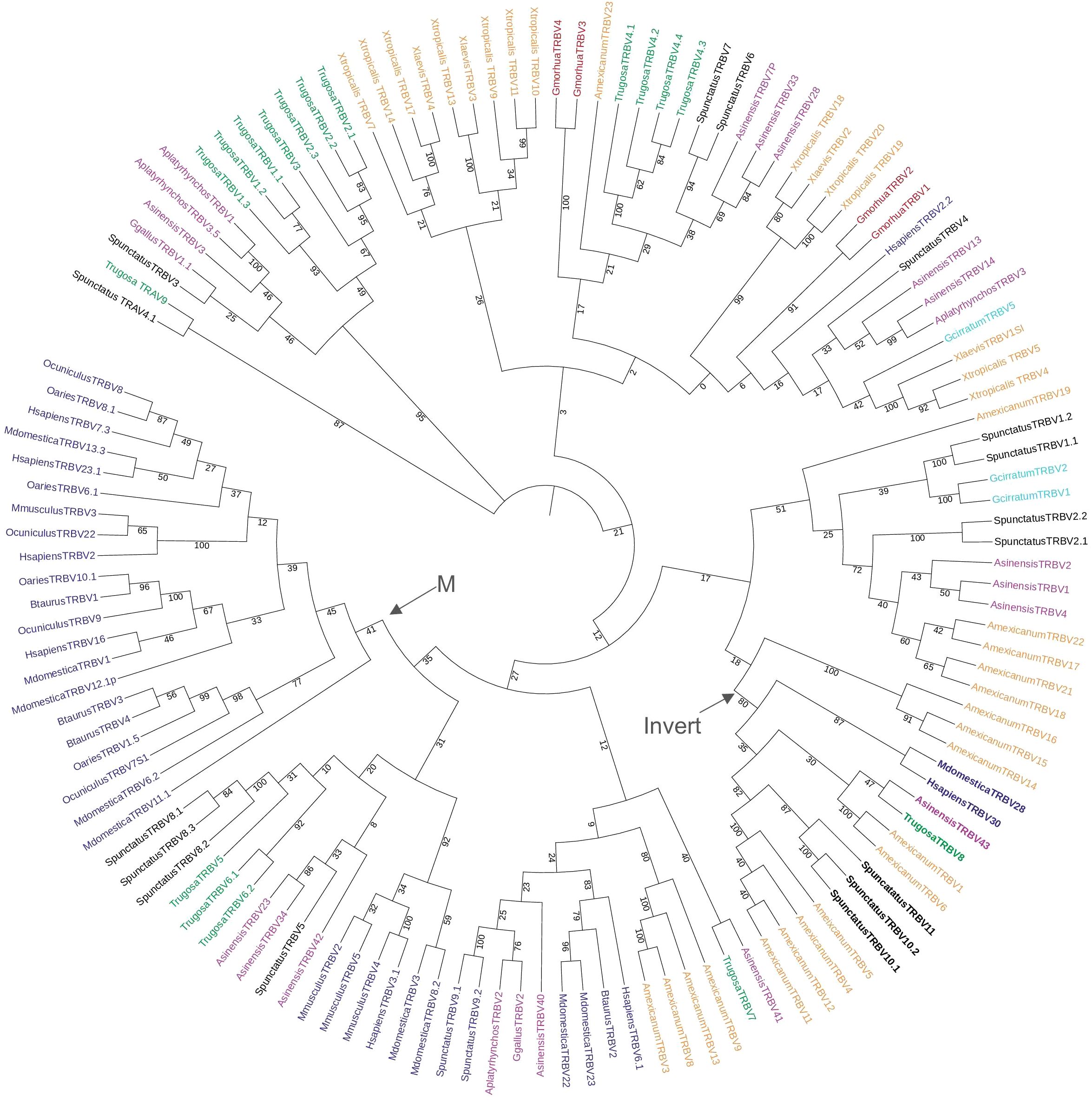
Figure 3. Neighbor-joining tree of vertebrate TRBV genes based on an amino acid alignment of TRBV sequences. Numbers on branches indicate bootstrap values on 1,000 replicates. Tree containing TRBV from 6 mammals, 1 squamate reptile, 1 Rhynchocephalian, 3 archelosaurs, 2 amphibians, 1 teleost fish, and 1 cartilaginous fish. The clade containing the inverted TRBVs is bolded and labeled with an arrow and “Invert”. Mammalian specific clade marked by arrow and “M.” TRBV families of squamates intersperse amongst the tree and 3’-inverted TRBVs consistently cluster together. Mammals included were short-tailed opossum (M. domestica), human (H. sapiens), mouse (M. musculus), cow (B. taurus), sheep (O. aries), and rabbit (O. cuniculus); the squamate is skink (T. rugosa); the Rhynchocephalian is tuatara (S. punctatus); the archelosaurs are Chinese alligator (A. sinensis), chicken (G. gallus), and duck (A. platyrhynchos); the amphibians are axolotl (A. mexicanum), African clawed frog (X. laevis), and Western clawed frog (X. tropicalis); the teleost is cod (G. morhua), and the cartilaginous fish is nurse shark (G. cirratum). Accession numbers of sequences used in the tree are found in Supplementary Table 2.
There were 38 TRB transcripts identified from the two T. rugosa spleen transcriptomes. Of those 38 sequences, 20 (52.6%) were complete enough at the 5’ end to show evidence of V(D)J recombination. Of those 20 transcripts, 16 (80%) were productively rearranged (Supplementary Figure 2A). The remaining transcripts contained out of frame V(D)J rearrangements that would not encode a functional TRB V domain.
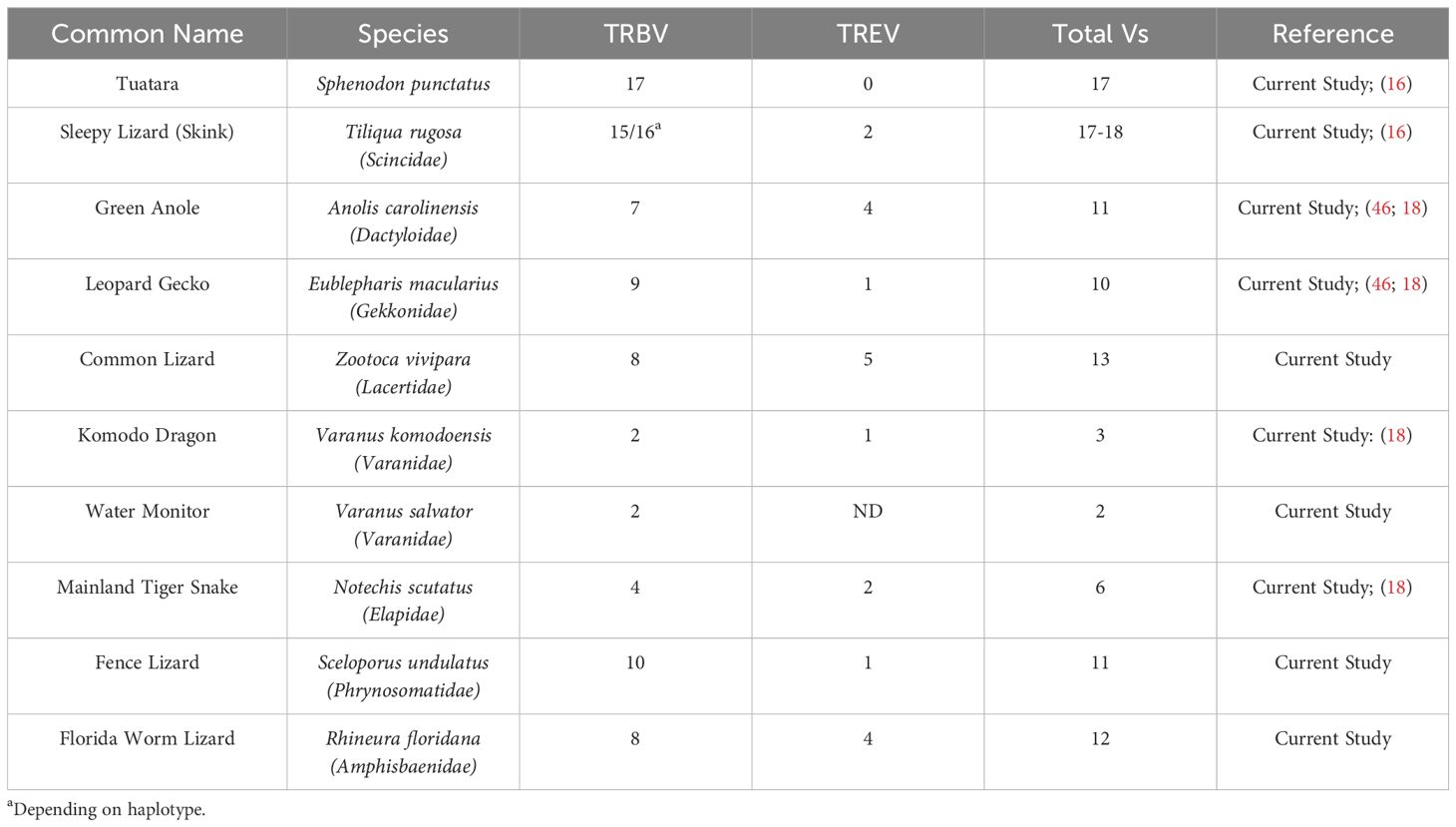
Analysis of the T. rugosa genome uncovered the presence of a third putative TCR locus similar to that recently described by Gambón-Deza, who designated it as TCR epsilon (TCRϵ or TRE) (18). The T. rugosa TRE locus is on chromosome 1 and is approximately 16kb in length from the most 5’ V to the 3’ C (Figure 4A). It contains 2 TREV, 1 TRED, 1 TREJ gene segments, and a single TREC in both pseudohaplotypes. As with the TRBV, the T. rugosa TREV sequences were both flanked by a 23 bp spacer and canonical heptamer (Figure 4; 25). The T. rugosa TRED gene segment was flanked by a 12 bp spacer on the V proximal side and a 23 bp spacer on the J proximal side. The T. rugosa TREJ segment was flanked by a 12 bp spacer (not shown). This pattern of spacers in the TRE locus is the same in several squamate species examined save for A. carolinensis (not shown). A. carolinensis had Vs flanked by both 12 bp spacers and 23 bp spacers and Js similarly flanked by both 12 and 23 bp spacers, demonstrating inversions that took place in the A. carolinensis TRE locus (not shown). We were unable to identify A. carolinensis TRED gene segments and therefore don’t know their RSS types (not shown, Supplementary Table 6). TRE was found in the genomes of Gekkonidae, Phrynosomatidae, Varanidae, Elapidae, Scincidae, Dactyloidae, Lacertidae, and Amphisbaenidae and was likely present in the last common ancestor of Squamates (Supplementary Table 6). In comparison to the genomes of other squamates, the T. rugosa TRE locus has among the lowest number of TREV gene segments (Table 1; Supplementary Table 6).
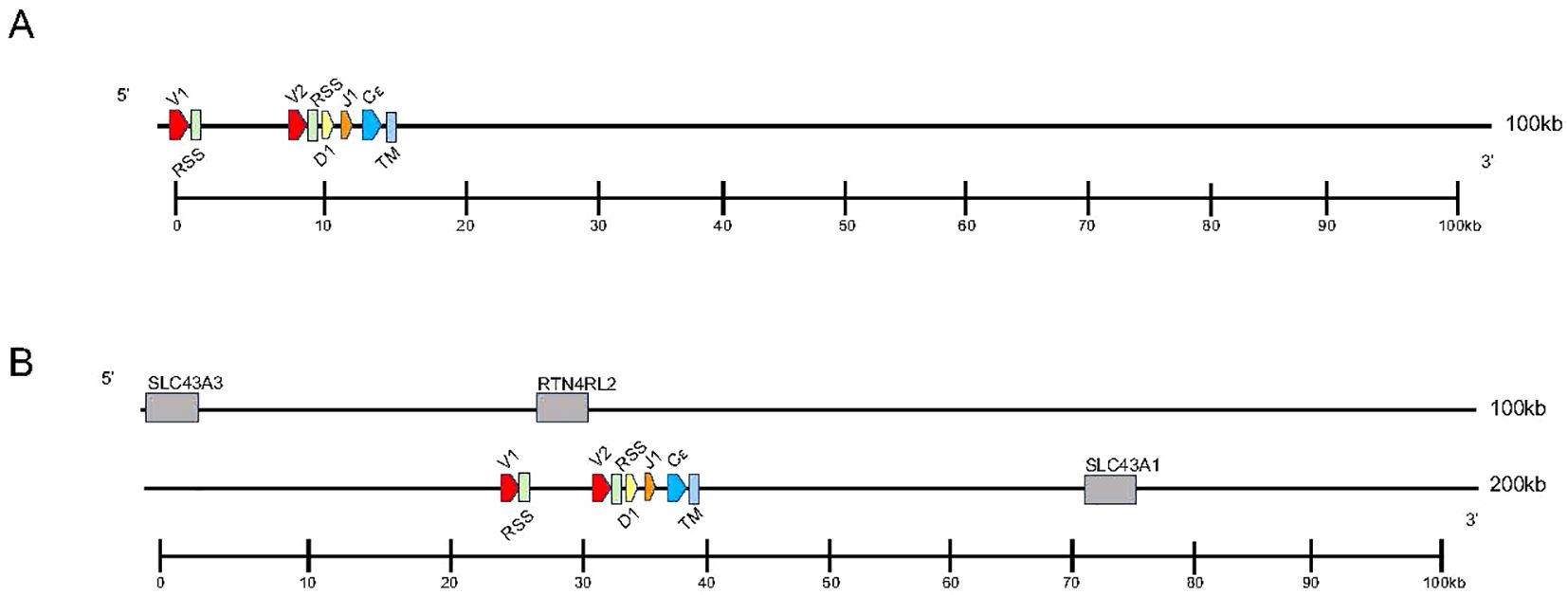
Figure 4. Map of the skink TRE locus. TREV (red), TREJ (orange), TRED (yellow), TREC (blue), and the TRE transmembrane region (light blue) gene segments are numbered by their corresponding location in order across the locus (nomenclature as seen in Figure 1). 23bp spacers are shown in light green near V segments. Other RSS are not shown for space. TREV segments are designated with a family member number. Transcriptional orientation is indicated by the direction of the arrow on each gene segment or exon. Arrows are not proportionate to the actual gene sizes. Gray boxes indicate flanking genes. (A) The skink TRE locus is ~16kb in length from V1 to TM region. (B) The flanking genes on the 5’ end of the locus are equilibrative nucleobase transporter 1 (SLC43A3) and reticulon-4 receptor-like 2 (RTN4RL2). The 3’ flanking gene is large neutral amino acids transporter small subunit (SLC43A1). The syntenic block containing TRE and the flanking genes is ~176kb.
There were 40 TRE sequences identified in two T. rugosa spleen transcriptomes. Twenty two of the 40 sequences (55%) were complete enough at the 5’ end to have evidence of being transcribed from a TRE locus that had undergone V(D)J recombination. Only three of the 22 (13.6%) were productively rearranged (Supplementary Figure 2B). Both TREV gene segments were found to be used in rearrangements (Supplementary Figure 2B). The majority of the transcripts contained out of frame V(D)J rearrangements that would not encode a functional TRE V domain.
To investigate the evolutionary origins of TRE, we searched for areas of synteny in the genomes of non-squamate reptiles, which lack TRE, compared with squamate TRE. In T. rugosa, TRE is flanked by RTN4RL2 (reticulon-4 receptor-like 2) and SLC43A3 (equilibrative nucleobase transporter 1) on the 5’ side and SLC43A1 (large neutral amino acids transporter small subunit 3) on the 3’ side (Figure 4B). This syntenic block was conserved in all reptiles examined (Figure 5). In T. rugosa, the flanking genes are 99 kb apart (Figure 5). In contrast, in the American alligator (Alligator mississippiensis), the distance between these genes is only 15kb (Figure 5). TRE could not be identified in the current S. punctatus genome, although absence due to gaps in the current assembly could not be ruled out (Figure 5). However, we had no difficulty identifying the S. punctatus TRA/D, TRB, and TRG loci (Figure 1B; 16). Moreover, we were unable to find TRE transcripts in an available S. punctatus blood transcriptome dataset, even though there was no difficulty identifying TRA, TRD, TRB, and TRG transcripts in this same dataset (Supplementary Table 1; 16).
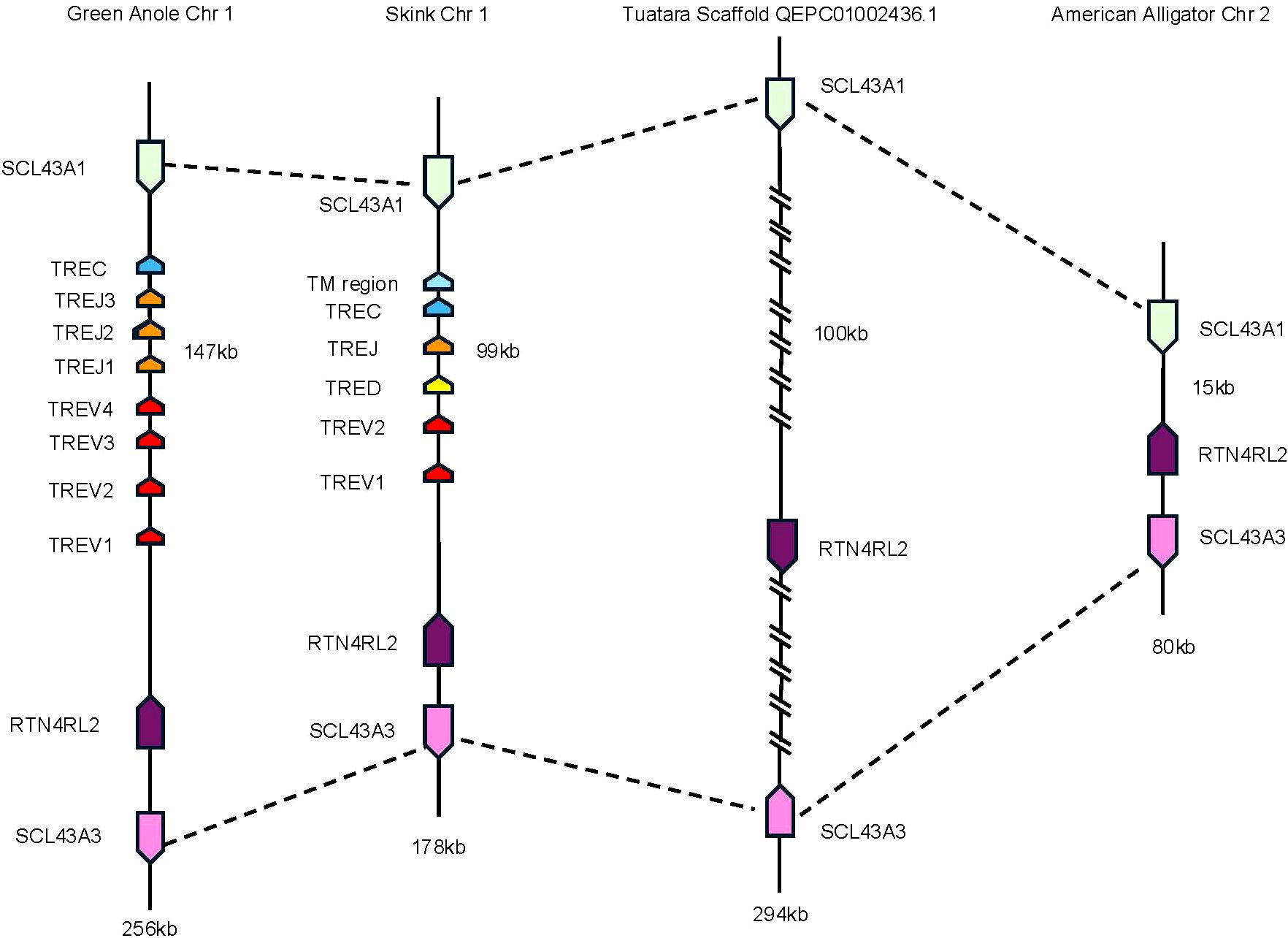
Figure 5. Comparison of the region containing TRE. Dashed lines connect genes with conserved synteny between the skink and tuatara. Genes flanking the TRE locus in the skink are equilibrative nucleobase transporter 1 (SLC43A3) (pink) and reticulon-4 receptor-like 2 (RTN4RL2) (dark purple) on the 5’ end and large neutral amino acids transporter small subunit (SLC43A1) (light green) on the 3’ end. The skink TRE locus is located between these genes (nomenclature as in Figures 1, 3). These same flanking genes were also identified in the tuatara, anole, and American alligator. Distance between RTN4RL2 and SCL43A1 shown next to each locus. The absence in the tuatara is either due to the absence of the locus or due to gaps in the scaffold. Gaps are indicated by hatch marks. Lengths of the complete loci are shown beneath each organism.
TREV genes were compared to other V genes found in immune receptors. There are five known TCR loci in amniotes, TRA, TRB, TRG, TRD, and TRM, and V genes from all five were included in the analysis (14, 35, 47; Figure 6A). Also included were V genes from the immunoglobulin heavy chain locus and both amniote light chain loci, kappa and lambda (Figure 6A). TREV consistently clustered with TRBV genes in multiple iterations of the tree including maximum likelihood and minimum evolution trees (Figure 6A; Supplementary Figure 3). Specifically, TREVs are the sister lineage to the 3’-inverted TRBV gene segments (Figure 6A; Supplementary Figure 3). We also compared the gene encoding the constant (C) domain of TRE to the C regions of the other five TCR, and it was most related to the C region genes encoding the TCRβ constant region (Figure 6B).
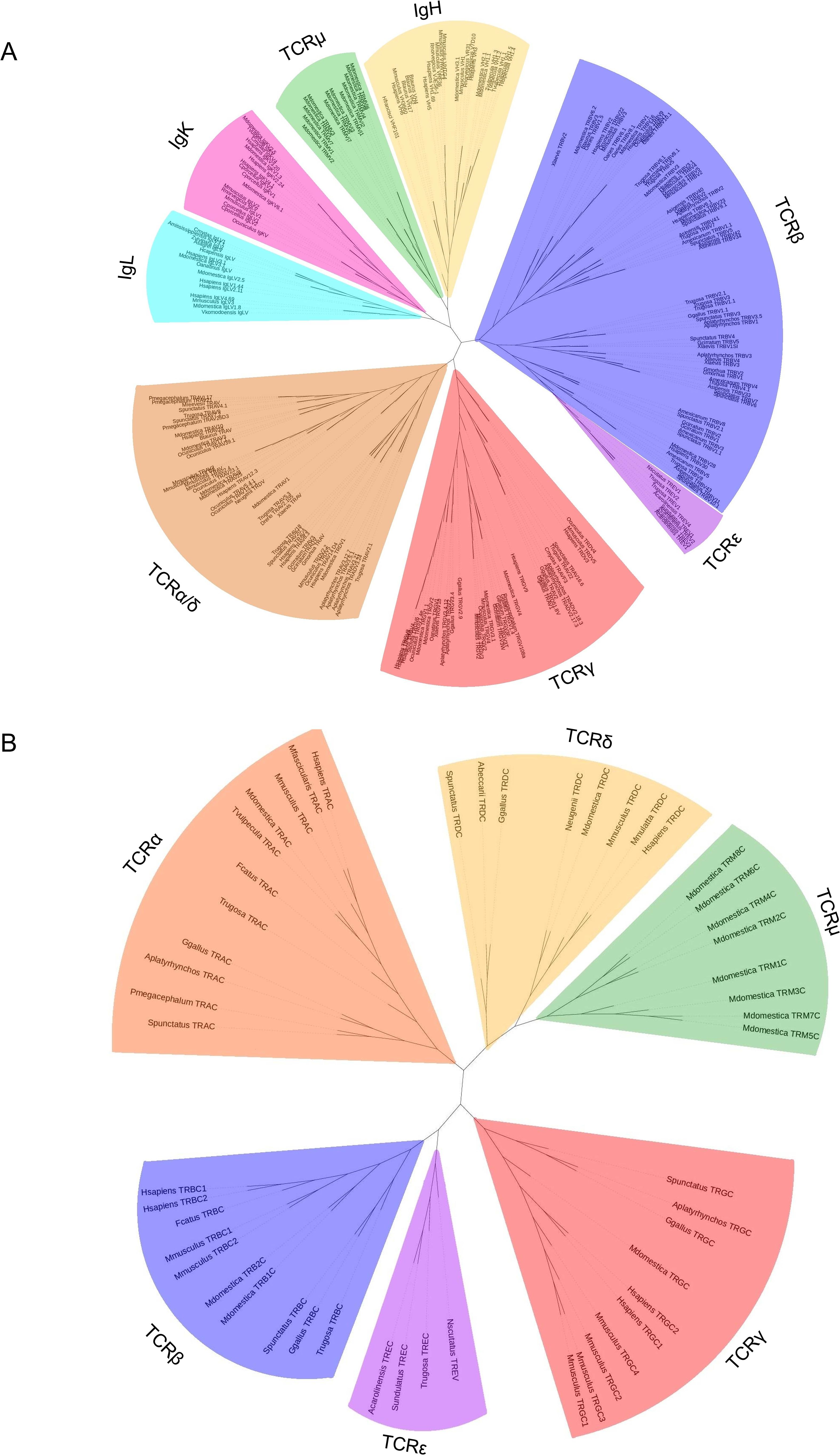
Figure 6. Neighbor-joining trees based on amino acid alignments of vertebrate variable (V) genes and constant regions (C). (A) V genes from both TCRs and Igs from several species. Vs from IgL are shown in light blue, IgK are shown in pink, IgH are shown in gold, TCRμ are shown in green, TCRβ are shown in light purple, TCRϵ are shown in dark purple, TCRγ are shown in red, and TCRα/δ are shown in red. (B) TCR constant regions from multiple species. Cs from TCRμ are shown in green, TCRγ are shown in red, TCRϵ are shown in dark purple, TCRβ are shown in light purple, TCRα are shown in orange, and TCRδ are shown in gold. Mammals included are humans (H. sapiens), crab eating macaque (M. fascicularis), sheep (O. aries), cow (B. taurus), pig (S. scrofa), rabbit (O. cuniculus), rat (R. norvegicus), cat (F. catus), short-tailed opossum (M. domestica), Tammar wallaby (N. eugenii), brushtail possum (T. vulpecula), and platypus (O. anatinus); squamates included are skink (T. rugosa), anole (A. carolinensis), mainland tiger snake (N. scutatus), and fence lizard (S. undulatus); the Rhynchocephalian is tuatara (S. punctatus); the archelosaurs are Chinese alligator (A. sinensis), Western bronze ground-dove (A. beccarii), chicken (G. gallus), duck (A. platyrhynchos), big headed turtle (P. megacephalum), Reeve’s turtle (M. reevesii), and green sea turtle (C. mydas), the amphibian is African clawed from (X. laevis), the teleost fish are cod (G. morhua), and zebrafish (D. rerio); and the cartilaginous fish are nurse shark (G. cirratum), and horned shark (H. francisci). Accession numbers of sequences used in 6A and 6B are found in Supplementary Tables 3 and 4 respectively.
Given TRE appears most related to TRB, we predicted the TCRϵ chain would likely pair with TCRα. For proper TCR heterodimer formation and interaction with the CD3 signaling complex, each TCR chain contains conserved arginine (Arg) and lysine (Lys) residues in the transmembrane region (47, 48). These conserved residues have an asymmetric pattern in the heterodimer, where one chain contains both Arg and Lys, while the other only Lys (Figure 7; Supplementary Figure 4) (47, 48). In a conventional αβTCR pair, the TCRα has Arg/Lys and the TCRβ has Lys only (Figure 7; Supplementary Figure 4). The same is true of squamate αβTCR (Figure 7; Supplementary Figure 4). The translated TCRϵ sequence has a conserved Lys at position 768, which is consistent with its ability to pair with TCRα and create a the TCR-CD3 complex (Figure 7; Supplementary Figure 4; 18).
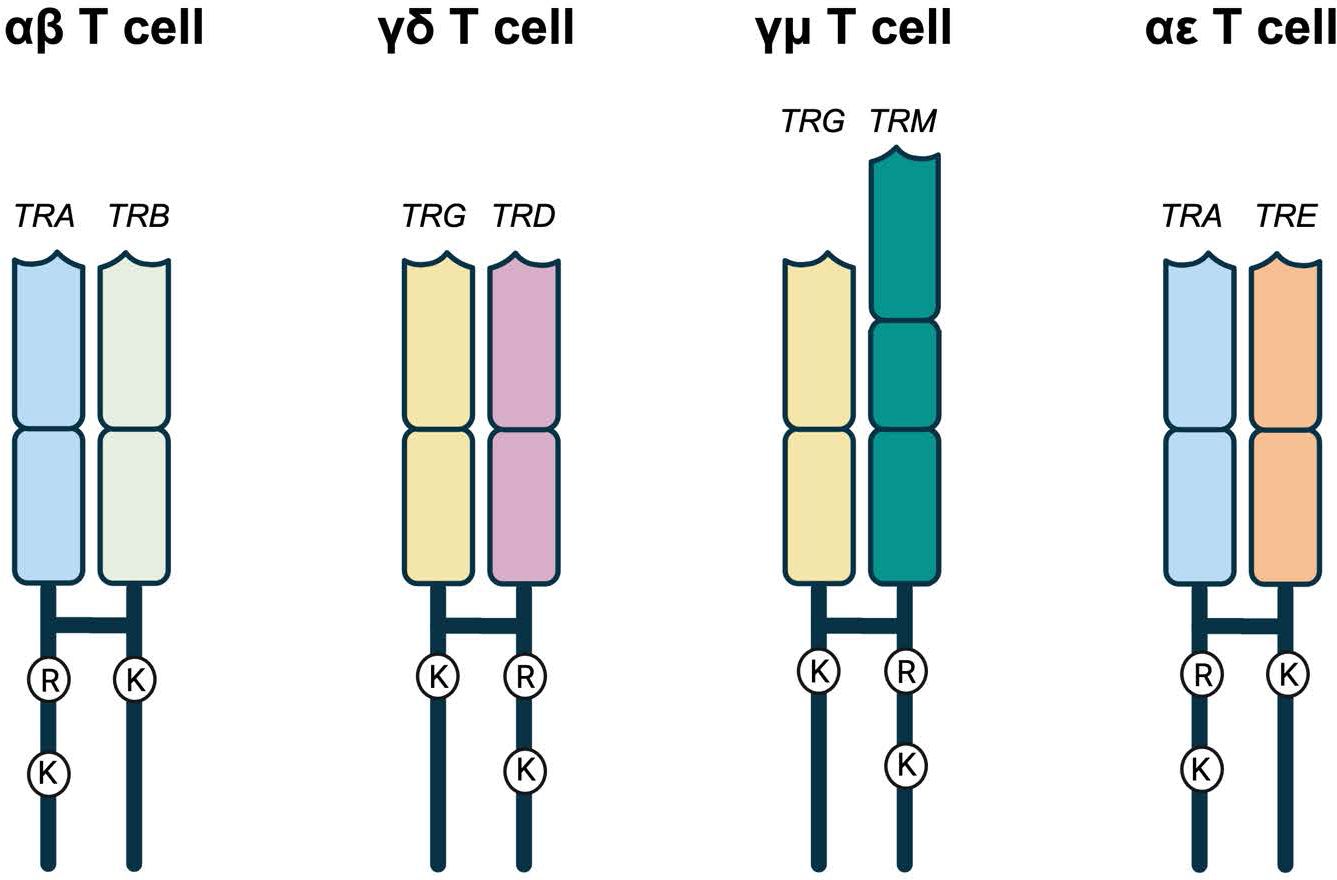
Figure 7. Representative TCRs with the amino acids in their transmembrane regions. αβ, γδ, and γμ represent the known amniote T cell receptors. In all three, there is an asymmetry of amino acids found in the transmembrane regions. One of the TCRs has an arginine (R) and a lysine (K) while the other has a single lysine (K). TRA, TRD, and TRM all have the R and K while TRB and TRG have the single K. In the potential squamate TCR, TRA still has the R and K while TRE has the single K that would allow it to potentially pair with TRA allowing for the possibility of αϵ T cells. Created in BioRender. Miller, R (2025). https://BioRender.com/v19g154.
Discussion
Squamate reptiles are amongst the most successful vertebrate lineages. More than 10,000 species occupy a broad range of ecosystems, from sea snakes to desert horned lizards. Despite their broad distribution and diversity, the squamates, and reptiles in general, remain amongst the least studied vertebrate lineages with respect to their immune systems, a shortcoming noted two decades ago (49). Indeed, most Sauropsid immunology has focused on a small number of species, mostly Archelosaurs, and has largely excluded the Lepidosaurs (9–12). What is known of reptile immune responses has primarily centered on innate immune responses with the conclusion that they may depend less on the adaptive response (11). Thankfully, the tools of genomics have increased the accessibility of many species to investigation, substantially enhancing the field of comparative biology, including comparative immunology.
The Australian skink species, T. rugosa, has several characteristics useful for a model squamate. They are abundant, widely distributed, and there is a 40 plus-year record of pathogen studies (50–54). Tiliqua rugosa is a host to multiple tick species that have been found to be vectors for blood pathogens such as rickettsia and apicomplexan protozoans (53–56). In the past, these tick species occupied distinct ecological zones (56). The tick boundary is known to shift between drier and wetter years, demonstrating how climate change might influence pathogen distribution (52, 56, 57).
We previously reported the lack of γδ T cells in squamates was due to deletions of the TRG and TRD loci needed to encode the TCRγ and TCRδ chains, respectively (16). Here, we investigate how the absence of the TCRγ and TCRδ chains may have influenced the remaining TCR genes. Our previous work showed little increase in the complexity of the TRA locus at the genomic level in the T. rugosa (16). Indeed, there is a relative decrease in complexity in the T. rugosa TRA locus, relative to S. punctatus which retains the TCRγ and TCRδ chains. Overall, there is comparatively low complexity in the available TRBV genes needed to assemble the exon encoding the TCRβ variable domain. Low numbers of TRBV genes appears to be the norm for Lepidosaurs (21, 46, 58). It is unlikely that an increase in the clonal diversity of αβ T cells, therefore, compensates for the loss of γδ T cells in squamates.
Surprising was the discovery that squamates have an additional locus that contains V, D and J segments like the genes encoding the conventional TCR and Ig. The T. rugosa locus is clearly homologous to a locus described recently by Gambón-Deza, who designated it as T cell receptor epsilon (TRE; 18). Analyses of the T. rugosa TRE gene segments are consistent with it being from a partial duplication of the TRB locus.
TRE was only found in the genomes of squamates, which lack γδ T cells, and not in non-squamate reptiles like S. punctatus, and A. mississippiensis (Figure 8) (16). This is consistent with the duplication giving rise to TRE occurring after the split between Rhynchocephalia and Squamata 250-280 MYA, and prior to the divergence of squamates more than 150 MYA (2, 5, 61). Analysis of the TREV genes revealed their relationship to a clade of TRBV that are in an inverted orientation and 3’ position in the TRB locus of most amniotes (34, 35, 40, 42, 43, 62). This inversion is also found in salmonids and some amphibians, consistent with it occurring earlier in vertebrate evolution (63; Jesus Martinez personal communication). From these observations emerges a model for the evolution of the TRB locus in amniotes and the origin of the TRE locus in squamates (Figure 9). Beginning with an ancestral TRB locus (Figure 9A) a family of TRBV translocated to an inverted location 3’ of the constant region genes (Figure 9B). Within the squamates, there was a translocation of a cluster of TRBV-D-J-C genes likely to another giving rise to TRE (Figure 9C). These duplications and translocations have resulted in the current conventional TRB locus in all amniotes and TRE in squamates (Figure 9D).
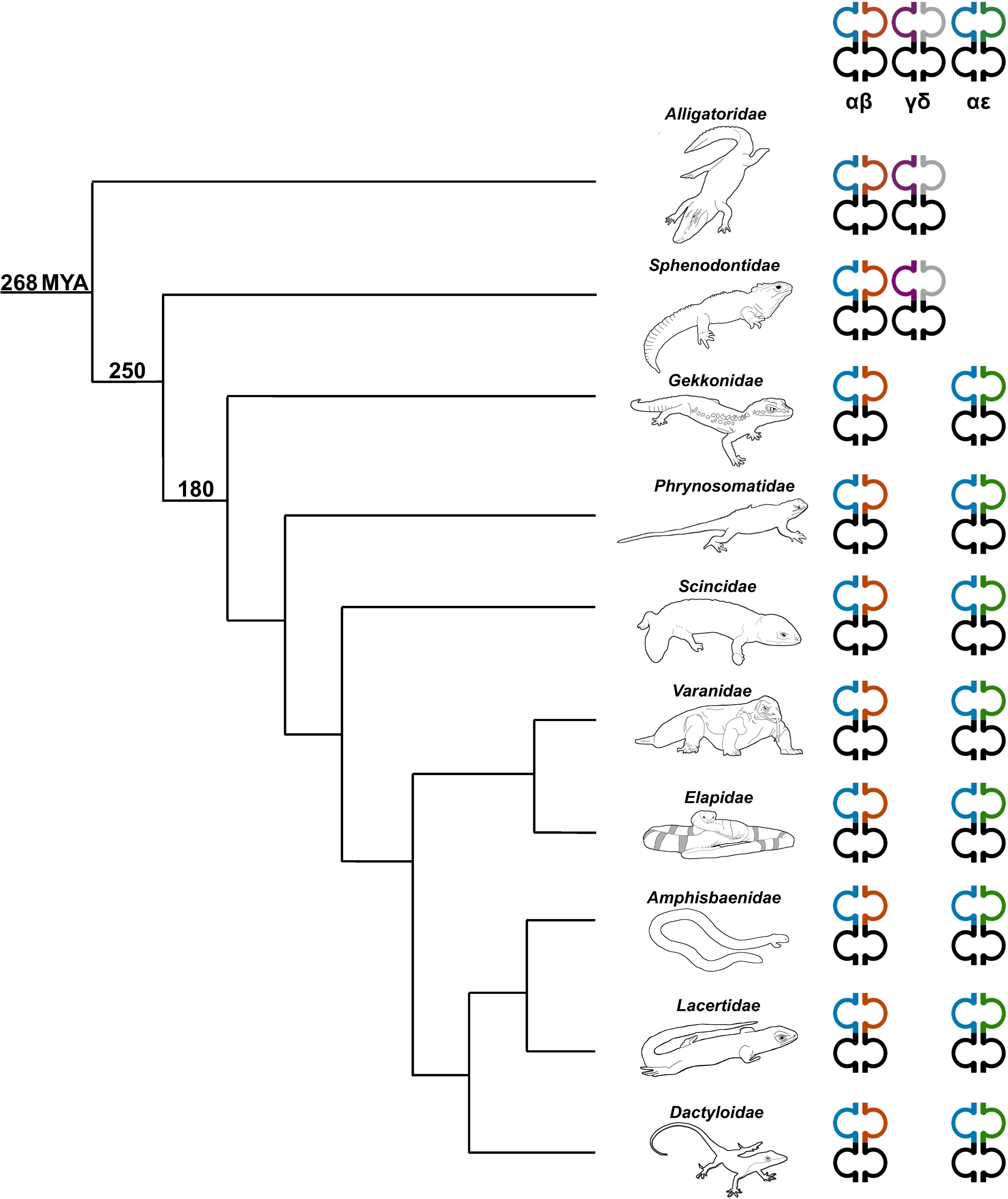
Figure 8. Phylogenetic relationship illustrating the diversity of TCR content in representative sauropsids. Representatives from several families were used including the Florida worm lizard (Amphisbaenidae), common lizard (Lacertidae), green anole (Dactyloidae), Komodo dragon (shown) and water monitor (not shown) (Varanidae), mainland tiger snake (Elapidae), skink (Scincidae), fence lizard (Phrynosomatidae), leopard gecko (Gekkonidae), tuatara (Sphenodontidae), and American alligator (Alligatoridae) (59). The number on each clade indicates approximate predicted divergence times in millions of years (MYA) (2, 5, 60). Heterodimer pairs are indicated at the top of each TCR chain type. TRA is shown in blue, TRB is shown in orange, TRG is shown in purple, TRD is shown in grey, and TRE is shown in green.
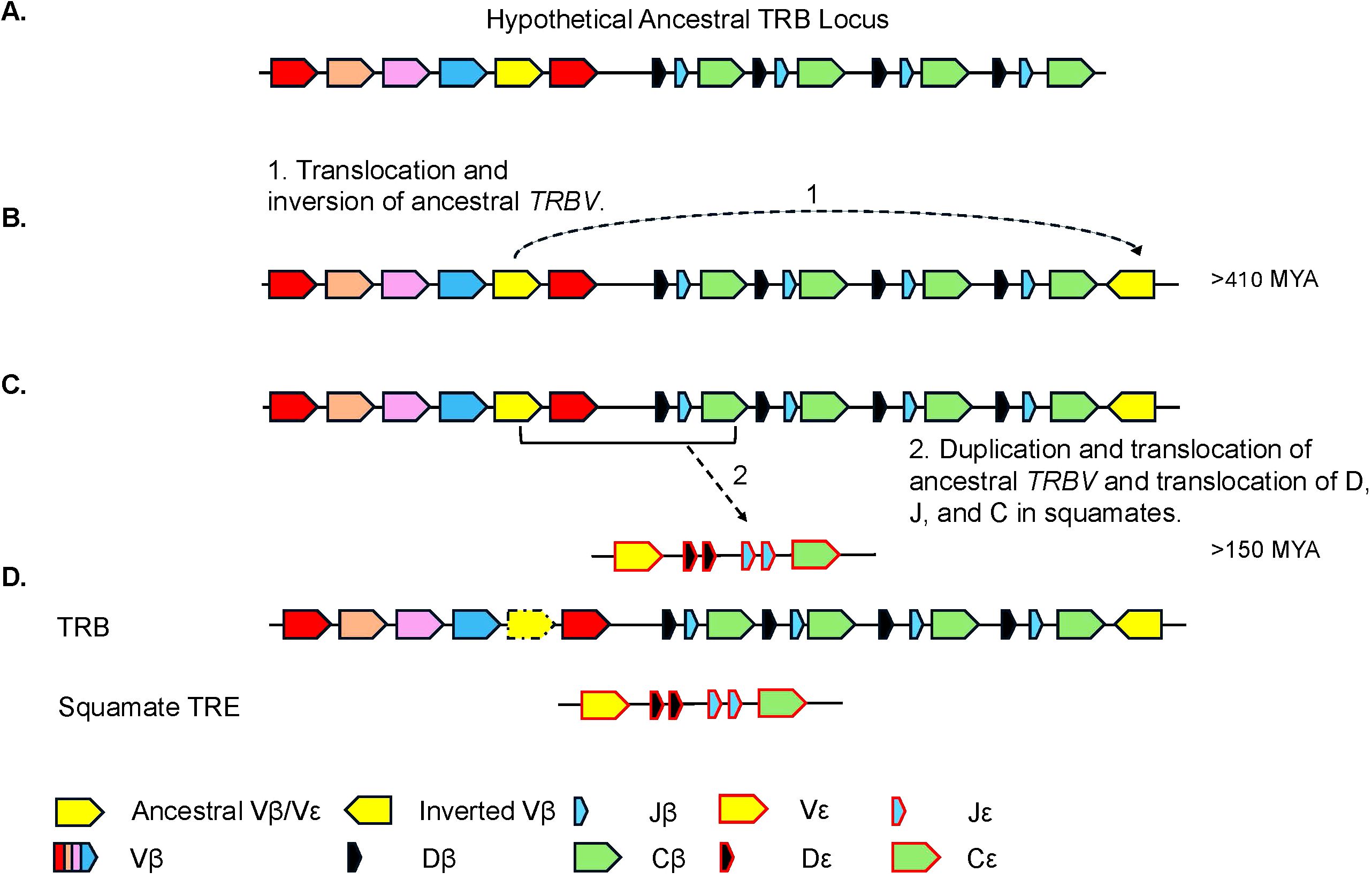
Figure 9. Proposed model for the evolution of the TRB and TRE loci. (A) Proposed model of the ancestral TRB locus. Highlighted in yellow is the TRBV gene segment(s) that is/are ancestral to the extant inverted TRBV and TREV gene segments. Other TRBV families are shown in additional colors. (B) Model hypothesizing (arrow 1) the duplication and inversion of the TRBV gene segment(s) within the TRB locus, currently found in several species. (C) The duplication and translocation of the TRB V-D-J-C (arrow 2) region to create the TRE locus. (D) Generic common TRB locus including the locus found in squamates and the squamate specific TRE locus. Presence of TRBV gene that gave rise to the inversion is species dependent and shown with dashed arrow.
Inverted V gene segment(s) are common to amniote TRB loci, are recombined in the αβ T cell repertoires, and detectable in transcriptomes. (34, 35, 40, 42, 43, 62–66; Supplementary Figure 2A). Indeed, inversions of genomic regions at the Ig and TCR loci are not uncommon throughout evolution (64, 65). Therefore, it is not clear if there is a fitness advantage to having these inverted V genes. The evidence that the TREV are most related to the inverted TRBV may simply reflect the plasticity of the locus that gave rise to the inversions in the first place locus that gave rise to the inversions in the first place.
In conventional T cells, the pairing of TCRα with TCRβ and TCRγ with TCRδ appears strictly enforced and remarkably conserved (14, 67, 68). However, there is precedence for T cell receptor gene duplications giving rise to novel TCR forms. To date, these novel TCR forms have involved specifically duplications of the TRD locus. In some birds, the TRD locus has been duplicated with the second locus using antibody heavy chain V gene segments in place of conventional TRVD (69, 70). Although it has not been physically demonstrated, it is likely the chains encoded by this second TRD locus also pair with the TCRγ chain. In mammals, duplications of the TRD locus gave rise to the genes encoding the T cell receptor μ chain (47, 71). The TCRμ chain has an unusual structure by having three extracellular immunoglobulin domains, however TCRμ has been shown to physically pair with TCRγ creating the γμTCR (72). γμ T cells are unique to mammals and only found in extant marsupials and monotremes (47, 73). The TRE locus would represent the first example of the evolution of a novel TCR due to duplications of the TRB locus which, like TRD undergoes recombination of V, D, and J gene segments. Marsupials and monotremes also have conventional γδ T cells, consistent with TRG pairing with either TRD or TRM. If TRE pairs with TRA, as predicted, this would demonstrate that, like TRG, TRA can pair with multiple partners, TRB or TRE in this case. This would be consistent with the TCR loci that undergo V to J recombination having greater promiscuity in their pairing possibilities.
As noted above, the TCR locus duplications found so far have involved either TRB or TRD and not TRA or TRG. The TRB and TRD loci are rearranged first in developing αβ and γδ T cells, respectively. Although much of early γδ T cell development remains a mystery, much is known about αβ T cell development, notably the role the TCRβ chain plays as a developmental checkpoint (74). Having a second TRB or TRB-like locus that encodes chains that pair with TCRα may provide additional options for successful αβ T cell development. This may be particularly important for species dependent on αβ T cells due to lacking γδ T cells. In addition, TCR chains encoded by combinations of V, D, and J gene segments, such as TRD and TRB, typically have increased diversity. Such increased diversity may again provide an evolutionary advantage to species lacking T cell subsets.
Most transcripts encoded by the TRE locus found in two T. rugosa spleen transcriptome databases were non-functional. Nonetheless they contained evidence of having been transcribed from genes assembled by somatic V(D)J recombination. There is also evidence of TRE being transcribed in other squamate reptiles including in a transcriptome of the many-banded krait, Bungarus multicinctus (18). Similarly, the majority of TRB transcripts (58%) were also non-functional. It was surprising to find such a large percentage (86.4%) of non-functional transcripts for TRE in a T. rugosa peripheral lymphoid organ. Though there were more functional transcripts for TRB than TRE it appears common for TCRs to have fewer functional transcripts in T. rugosa. Whether this is due to poor selection in the thymus, development occurring outside the thymus, or nonsense-mediated decay of TCR transcripts remains unknown (75). The high percentage of non-functional transcripts does not appear to be common to all recombined immune genes, however, as most of the Ig transcripts for both heavy and light chains are productively rearranged (not shown, unpublished data).
It is also possible that the spleen is not the primary site of mature αϵ T cells in squamates. Indeed, αϵ T cells may be found in locations that are associated with γδ T cells, such as the skin, gut, or other epithelial sites (76–78). It is known that the thymus of certain reptiles including squamates can develop seasonally, however, how this affects the development of T cells, when αβ T cells develop in squamates, and their relationship to potential αϵ T cells is unknown (9, 79). Further research into the location of αϵ T cells, the timing of their development, their function, and their ligands is necessary.
Conclusion
The lack of γδ T cells in squamates provides natural models with which to study evolutionary compensation to the wholesale loss of cell lineages in the adaptive immune system. Here, we confirm that the lack of γδ T cells has not resulted in increased genomic complexity of the genes encoding the potential αβTCR repertoire. Indeed, we have confirmed that αβTCR complexity is generally low in squamates compared to other amniote lineages. Noteworthy is duplication of the TRB locus giving rise to the TRE locus in squamates. TRE adds to the list of gene duplications giving rise to extra TCR loci not found in well-studied model species such as laboratory mice or humans. Whether αϵ T cells are compensating for the loss of γδ T cells in squamates is unknown. They do not appear to increase the potential overall diversity of T cells available to the host animal. The presence of functional or location differences between conventional αβ and the αϵ T cells remains to be determined.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by the institutional committees of the University of Otago and Flinders University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. KAM: Formal analysis, Investigation, Writing – review & editing. KJM: Formal analysis, Investigation, Methodology, Writing – review & editing. KZ: Resources, Writing – review & editing. NG: Data curation, Resources, Writing – review & editing. MG: Funding acquisition, Resources, Writing – review & editing. TB: Data curation, Formal analysis, Resources, Writing – review & editing. RM: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Science Foundation Award IOS-2103367, National Science Foundation Graduate Research Fellowship Award DGE-1939267, and Australian Research Council Discovery Grant DP200102880.
Acknowledgments
We thank Drs. Lijing Bu and Marjian Posavi for their assistance and bioinformatics support, Dr. Xifeng Wang, Institute of Zoology, Chinese Academy of Science, Beijing, China, for sharing the Alligator sinensis TRBV sequences, and Dr. Jesus Martinez Chronic Infectious Diseases and Cancer Division, Center for Research on Infectious Diseases, National Institute of Public Health, Cuernavaca, Morelos, Mexico for the Xenopus tropicalis and Ambystoma mexicanum TRBV sequences. We thank the UNM Center for Advanced Research Computing for providing the high-performance computing and large-scale storage resources used in this work. We would like to acknowledge the contribution of the Australian Amphibian and Reptile Genomics consortium in the generation of data used in this publication. The Australian amphibian and reptile genomics initiative is supported by funding from Bioplatforms Australia through the Australian Government’s National Collaborative Research Infrastructure Strategy (NCRIS), the Australian National University, the University of Canberra, the Australian Museum, Museums Victoria and the South Australian Museum.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1524471/full#supplementary-material
References
1. Nesbitt SJ. The early evolution of archosaurs: relationships and the origin of major clades. Bull Am Museum Natural History. (2011) 352:1–292. doi: 10.1206/352.1
2. Jones ME, Anderson CL, Hipsley CA, Müller J, Evans SE, Schoch RE. Integration of molecules and new fossils supports a Triassic origin for Lepidosauria (lizards, snakes, and tuatara). BMC Evol Biol. (2013) 13:208. doi: 10.1186/1471-2148-13-208
3. Hugall AF, Foster R, Lee MS. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Systematic Biol. (2007) 56:543–63. doi: 10.1080/10635150701477825
4. Pincheira-Donoso D, Bauer AM, Meiri S, Uetz P. Global taxonomic diversity of living reptiles. PloS One. (2013) 8:e59741. doi: 10.1371/journal.pone.0059741
5. Gemmell NJ, Rutherford K, Prost S, Tollis M, Winter D, Macey JR, et al. The tuatara genome reveals ancient features of amniote evolution. Nature. (2020) 584:403–9. doi: 10.1038/s41586-020-2561-9
6. Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature. (2021) 592:737–46. doi: 10.1038/s41586-021-03451-0
8. Lee MS, Palci A. Serpentes (Snakes). eLS. (2013) 2:111–7. doi: 10.1002/9780470015902.a0001543.pub2
9. Zimmerman LM, Vogel LA, Bowden RM. Understanding the vertebrate immune system: insights from the reptilian perspective. J Exp Biol. (2010) 213:661–71. doi: 10.1242/jeb.038315
10. Pettinello R, Dooley H. The immunoglobulins of cold-blooded vertebrates. Biomolecules. (2014) 4:1045–69. doi: 10.3390/biom4041045
11. Rios FM, Zimmerman LM. Immunology of reptiles. eLS. (2015), 1–7. doi: 10.1002/9780470015902.a0026260
12. Zimmerman LM. Adaptive immunity in reptiles: conventional components but unconventional strategies. Integr Comp Biol. (2022) 62:1572–83. doi: 10.1093/icb/icac022
13. Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. (2010) 11:47–59. doi: 10.1038/nrg2703
14. Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT, Litman GW. [amp]]alpha;, β, γ, and δ T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity. (1997) 6:1–11. doi: 10.1016/S1074-7613(00)80237-X
15. Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol. (2005) 35:936–45. doi: 10.1002/eji.200425760
16. Morrissey KA, Sampson JM, Rivera M, Lijing B, Hansen VL, Gemmell NJ, et al. Comparison of reptilian genomes reveals deletions associated with the natural loss of γδ T cells in squamates. J Immunol. (2022) 208:1960–7. doi: 10.4049/jimmunol.210115
17. Herrera-Flores JA, Stubbs TL, Benton MJ. Macroevolutionary patterns in Rhynchocephalia: is the tuatara (Sphenodon punctatus) a living fossil? Palaeontology. (2017) 60:319–28. doi: 10.5061/dryad.568jh
18. Gambon-Deza FG. A new T cell receptor in squamata reptiles. BioRxiv. (2023), 2023–09. doi: 10.1101/2023.09.04.556186
19. Rautiainen M, Nurk S, Walenz BP, Logsdon GA, Porubsky D, Rhie A, et al. Telomere-to-telomere assembly of diploid chromosomes with Verkko. Nat Biotechnol. (2023) 41:1474–82. doi: 10.1038/s41587-023-01662-6
20. Zhou C, McCarthy SA, Durbin R. YaHS: yet another Hi-C scaffolding tool. Bioinformatics. (2023) 39:btac808. doi: 10.1093/bioinformatics/btac808
21. Gambón-Deza F, Olivieri DN. Immunoglobulin and T cell receptor genes in Chinese crocodile lizard Shinisaurus crocodilurus. Mol Immunol. (2018) 101:160–6. doi: 10.1016/j.molimm.2018.06.263
22. Ansari TH, Bertozzi T, Miller RD, Gardner MG. MHC in a monogamous lizard–Characterization of class I MHC genes in the Australian skink Tiliqua rugosa. Dev Comp Immunol. (2015) 53:320–7. doi: 10.1016/j.dci.2015.07.012
23. Hallgren J, Tsirigos KD, Pedersen MD, Almagro Armenteros JJ, Marcatili P, Nielsen H, et al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv. (2022), 2022–04. doi: 10.1101/2022.04.08.487609
24. Humann JL, Lee T, Ficklin S, Main D. Structural and functional annotation of eukaryotic genomes with genSAS. Methods Mol Biol. (2019) 1962:29–51. doi: 10.1007/978-1-4939-9173-0_3
25. Bassing CH, Alt FW, Hughes MM, D'Auteuil M, Wehrly TD, Woodman BB, et al. Recombination signal sequences restrict chromosomal V (D) J recombination beyond the 12/23 rule. Nature. (2000) 405:583–6. doi: 10.1038/35014635
26. Cowell LG, Davila M, Ramsden D, Kelsoe G. Computational tools for understanding sequence variability in recombination signals. Immunol Rev. (2004) 200:57–69. doi: 10.1111/j.0105-2896.2004.00171.x
27. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
28. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. (1994) 22:4673–80. doi: 10.1093/nar/22.22.4673
29. IMGT. The international ImMunoGeneTics information system . Available online at: http://www.imgt.org (Accessed February 9, 2023).
30. Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. (2004) 5:1–19. doi: 10.1186/1471-2105-5-113
31. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis cross computing platforms. Mol Biol Evolution. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
32. Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci. (2004) 101:11030–5. doi: 10.1073/pnas.0404206101
33. Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. (2021) 49:W293–6. doi: 10.1093/nar/gkab301
34. Wang X, Wang P, Wang R, Wang C, Bai J, Ke C, et al. Analysis of TCRβ and TCRγ genes in Chinese alligator provides insights into the evolution of TCR genes in jawed vertebrates. Dev Comp Immunol. (2018) 85:31–43. doi: 10.1016/j.dci.2018.01.009
35. Parra ZE, Baker ML, Hathaway J, Lopez AM, Trujillo J, Sharp A, et al. Comparative genomic analysis and evolution of the T cell receptor loci in the opossum Monodelphis domestica. BMC Genomics. (2008) 9:1–19. doi: 10.1186/1471-2164-9-111
36. Posit team. RStudio: Integrated Development Environment for R. Boston, MA: Posit Software, PBC (2024). Available at: http://www.posit.co/ (Accessed August 6, 2024).
37. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing (2021). Available at: https://www.R-project.org/ (Accessed August 6, 2024).
38. Wickham H. Reshaping data with the reshape package. J Stat Software. (2007) 21:1–20. doi: 10.18637/jss.v021.i12
39. Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag (2016). Available at: https://ggplot2.tidyverse.org (Accessed August 6, 2024)978-3-319-24277-4.
40. Lefranc M-P, Lefranc G. The T cell receptor FactsBook. San Diego: Academic Press (2001). p. 398.
41. Connelley T, Aerts J, Law A, Morrison WI. Genomic analysis reveals extensive gene duplication within the bovine TRB locus. BMC Genomics. (2009) 10:1–20. doi: 10.1186/1471-2164-10-192
42. Antonacci R, Giannico F, Ciccarese S, Massari S. Genomic characteristics of the T cell receptor (TRB) locus in the rabbit (Oryctolagus cuniculus) revealed by comparative and phylogenetic analyses. Immunogenetics. (2014) 66:255–66. doi: 10.1007/s00251-013-0754-1
43. Giannico F, Massari S, Caputi Jambrenghi A, Soriano A, Pala A, Linguiti G, et al. The expansion of the TRB and TRG genes in domestic goats (Capra hircus) is characteristic of the ruminant species. BMC Genomics. (2020) 21:1–17. doi: 10.1186/s12864-020-07022-x
44. Ramsden DA, Baetz K, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. (1994) 22:1785–96. doi: 10.1093/nar/22.10.1785
45. Levinson M, Khass M, Burrows PD, Schroeder HW. Germline-enforced enrichment for charged amino acids in TCR beta chain (TCRβ) complementarity determining region 3 (CDR-B3) alters T cell development, repertoire content, and antigen recognition. Immunogenetics. (2023) 75:1–13. doi: 10.1007/s00251-023-01304-w
46. Olivieri DN, Mirete-Bachiller S, Gambón-Deza F. Insights into the evolution of IG genes in amphibians and reptiles. Dev Comp Immunol. (2021) 114:103868. doi: 10.1016/j.dci.2020.103868
47. Parra ZE, Baker ML, Schwarz RS, Deakin JE, Lindblad-Toh K, Miller RD. A unique T cell receptor discovered in marsupials. Proc Natl Acad Sci. (2007) 104:9776–81. doi: 10.1073/pnas.0609106104
48. Call ME, Wucherpfennig KW. The T cell receptor: critical role of the membrane environment in receptor assembly and function. Annu Rev Immunol. (2005) 23:101–25. doi: 10.1146/annurev.immunol.23.021704.115625
49. Warr GW, Chapman RW, Smith LC. Evolutionary immunobiology: new approaches, new paradigms. Dev Comp Immunol. (2003) 27:257–62. doi: 10.1016/s0145-305x(02)00070-8
50. Bull CM, Pamula Y, Schulze L. Parturition in the sleepy lizard, Tiliqua rugosa. J Herpetology. (1993) 27:489–92. doi: 10.2307/1564848
51. Bull CM, Cooper SJ, Baghurst BC. Social monogamy and extra-pair fertilization in an Australian lizard, Tiliqua rugosa. Behav Ecol Sociobiology. (1998) 44:63–72. doi: 10.1007/s002650050515
52. Godfrey SS, Gardner MG. Lizards, ticks and contributions to Australian parasitology: C. Michael Bull, (1947–2016). Int J Parasitology: Parasites Wildlife. (2017) 6:295–8. doi: 10.1016/j.ijppaw.2017.08.010
53. Norval G, Ross KE, Sharrad RD, Gardner MG. Taking stock: a review of the known parasites of the sleepy lizard, Tiliqua rugosa (Gray 1825), a common lizard endemic to Australia. Trans R Soc South Australia. (2019) 143:216–34. doi: 10.1080/03721426.2019.1595946
54. Norval G, Gardner MG. The natural history of the sleepy lizard, Tiliqua rugosa (Gray 1825)–Insight from chance observations and long-term research on a common Australian skink species. Austral Ecol. (2019) 45:410–7. doi: 10.1111/aec.12715
55. Bower DS, Brannelly LA, McDonald CA, Webb RJ, Greenspan SE, Vickers M, et al. A review of the role of parasites in the ecology of reptiles and amphibians. Austral Ecology. (2019) 44:433–48. doi: 10.1111/aec.12695
56. Barnden B, Slender AL, Sharrad RD, Gardner MG. Changes in parasite species distributions could be driven by host range expansions: the case of hybridisation between two Australian reptile ticks. Aust J Zoology. (2023) 70:153–63. doi: 10.1071/ZO23010
57. Bull CM, Burzacott D. Temporal and spatial dynamics of a parapatric boundary between two Australian reptile ticks. Mol Ecol. (2001) 10:639–48. doi: 10.1046/j.1365-294x.2001.01214.x
58. Olivieri DN, Von Haeften B, Sánchez-Espinel C, Faro J, Gambón-Deza F. Genomic V exons from whole genome shotgun data in reptiles. Immunogenetics. (2014) 66:479–92. doi: 10.1007/s00251-014-0784-3
59. Vidal N, Hedges SB. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. Comptes Rendus Biologies. (2009) 332:129–39. doi: 10.1016/j.crvi.2008.07.010
60. Ezcurra MD, Scheyer TM, Butler RJ. The origin and early evolution of Sauria: reassessing the Permian saurian fossil record and the timing of the crocodile-lizard divergence. PloS One. (2014) 9:e89165. doi: 10.1371/journal.pone.0089165
61. Mulcahy DG, Noonan BP, Moss T, Townsend TM, Reeder TW, Sites Jr, et al. Estimating divergence dates and evaluating dating methods using phylogenomic and mitochondrial data in squamate reptiles. Mol Phylogenet Evolution. (2012) 65:974–91. doi: 10.1016/j.ympev.2012.08.018
62. Lui F, Li J, Lin IYC, Yang X, Ma J, Chen Y, et al. The genome resequencing of TCR loci in gallus gallus revealed their distinct evolutionary features in avians. ImmunoHorizons. (2021) 4:33–46. doi: 10.4049/immunohorizons.1900095
63. Boudinot P, Novas S, Jouneau L, Mondot S, Lefranc MP, Grimholt U, et al. Evolution of T cell receptor beta loci in salmonids. Front Immunol. (2023) 14:823145. doi: 10.3389/fimmu.2022.823145
64. Zhang Y, Li X, Ba Z, Lou J, Gaertner KE, Zhu T, et al. Molecular basis for differential Igk versus Igh V (D) J joining mechanisms. Nature. (2024) 630:1–9. doi: 10.1038/s41586-024-07477-y
65. Korman AJ, Maruyama J, Raulet DH. Rearrangement by inversion of a T-cell receptor delta variable region gene located 3'of the delta constant region gene. Proc Natl Acad Sci. (1989) 86:267–71. doi: 10.1073/pnas.86.1.267
66. Malissen M, McCoy C, Blanc D, Trucy J, Devaux C, Schmitt-Verhulst, et al. Direct evidence for chromosomal inversion during T-cell receptor β-gene rearrangements. Nature. (1986) 319:28–33. doi: 10.1038/319028a0
67. Morath A, Schamel WW. [amp]]alpha;β and γδ T cell receptors: Similar but different. J leukocyte Biol. (2020) 107:1045–55. doi: 10.1002/JLB.2MR1219-233R
68. Attaf M, Legut M, Cole DK, Sewell AK. The T cell antigen receptor: the Swiss army knife of the immune system. Clin Exp Immunol. (2015) . 181:1–18. doi: 10.1111/cei.12622
69. Parra ZE, Lillie M, Miller RD. A model for the evolution of the mammalian t-cell receptor α/δ and μ loci based on evidence from the duckbill Platypus. Mol Biol Evol. (2012) 29:3205–14. doi: 10.1093/molbev/mss128
70. Liang C, Sun L, Zhu Y, Zhao A, Liu H, He K. Macroevolution of avian T cell receptor C segments using genomic data. Immunogenetics. (2023) . 75:531–41. doi: 10.1007/s00251-023-01322-8
71. Parra ZE, Mitchell K, Dalloul RA, Miller RD. A second TCRδ locus in Galliformes uses antibody-like V domains: insight into the evolution of TCRδ and TCRμ genes in tetrapods. J Immunol. (2012) 188:3912–9. doi: 10.4049/jimmunol.1103521
72. Morrissey KA, Wegrecki M, Praveena T, Hansen VL, Bu L, Sivaraman KK, et al. The molecular assembly of the marsupial γμ T cell receptor defines a third T cell lineage. Science. (2021) 371:1383–8. doi: 10.1126/science.abe7070
73. Wang X, Parra ZE. Miller RD Platypus TCRμ provides insight into the origins and evolution of a uniquely mammalian TCR locus. J Immunol. (2011) . 187:5246–54. doi: 10.4049/jimmunol.1101113
74. Dutta A, Zhao B, Love PE. New insights into TCR β-selection. Trends Immunol. (2021) 42:735–50. doi: 10.1016/j.it.2021.06.005
75. Delpy L, Sirac C, Magnoux E, Duchez S, Cogné M. RNA surveillance down-regulates expression of nonfunctional κ alleles and detects premature termination within the last κ exon. Proc Natl Acad Sci. (2004) 101:7375–80. doi: 10.1073/pnas.0305586101
76. Jameson J, Ugarte K, Chen N, Yachi P, Fuchs E, Boismenu R, et al. A role for skin γδ T cells in wound repair. Science. (2002) 296:747–9. doi: 10.1126/science.1069639
77. Hayday AC. [amp]]gamma;δ T cell update: adaptate orchestrators of immune surveillance. J Immunol. (2019) 203:311–20. doi: 10.4049/jimmunol.1800934
78. Binz C, Bubke A, Sandrock I, Prinz I. [amp]]alpha;β T cells replacing dermal and epidermal γδ T cells in Tcrd–/– mice express an MHC-independent TCR repertoire. Eur J Immunol. (2021) 51:2618–32. doi: 10.1002/eji.202149243
Keywords: squamate, T cells, comparative immunology, gene loss, gene duplication
Citation: Sampson JM, Morrissey KA, Mikolajova KJ, Zimmerly KM, Gemmell NJ, Gardner MG, Bertozzi T and Miller RD (2025) Squamate reptiles may have compensated for the lack of γδTCR with a duplication of the TRB locus. Front. Immunol. 15:1524471. doi: 10.3389/fimmu.2024.1524471
Received: 07 November 2024; Accepted: 16 December 2024;
Published: 09 January 2025.
Edited by:
Salvatrice Ciccarese, University of Bari Aldo Moro, ItalyReviewed by:
Pierre Boudinot, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceDavid Vermijlen, Université libre de Bruxelles, Belgium
Copyright © 2025 Sampson, Morrissey, Mikolajova, Zimmerly, Gemmell, Gardner, Bertozzi and Miller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert D. Miller, cmRtaWxsZXJAdW5tLmVkdQ==
 Jordan M. Sampson
Jordan M. Sampson Kimberly A. Morrissey
Kimberly A. Morrissey Kieran J. Mikolajova1
Kieran J. Mikolajova1 Neil J. Gemmell
Neil J. Gemmell Michael G. Gardner
Michael G. Gardner Terry Bertozzi
Terry Bertozzi Robert D. Miller
Robert D. Miller