- 1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, School of Marine Sciences, Ningbo University, Ningbo, China
- 2Laboratory of Biochemistry and Molecular Biology, School of Marine Sciences, Ningbo University, Ningbo, China
- 3Key Laboratory of Aquacultural Biotechnology of Ministry of Education, Ningbo University, Ningbo, China
Neutrophil extracellular traps (NETs) are crucial for the immune defense of many organisms, serving as a potent mechanism for neutrophils to capture and eliminate extracellular pathogens. While NETosis and its antimicrobial mechanisms have been well studied in mammals, research on NETs formation in teleost fish remains limited. In this study, we used the large yellow croaker (Larimichthys crocea) as the study model to investigate NETosis and its role in pathogen defense. Our results showed that infection with Pseudomonas plecoglossicida could induce NETosis. To further explore the underlying mechanism, we performed transcriptome analysis and western blotting, which revealed that P. plecoglossicida triggers NETosis through activation of the autophagy pathway. Inhibition of autophagy significantly reduced NET production, highlighting its critical role in this process. Furthermore, our studies demonstrated that NETs exert a bacteriostatic effect, significantly suppressing the growth of P. plecoglossicida. Taken together, our findings reveal that autophagy regulates NETosis in large yellow croaker and underscore the essential role of NETs in bacterial defense, providing new insights into immune responses in teleost fish.
Introduction
Neutrophils, which are polymorphonuclear and granular leukocytes, constitute 50%-70% of all circulating leukocytes in humans and play a critical role in the innate immune system (1). In mammals, there cells are primarily generated in the bone marrow and subsequently released into the bloodstream, where they can rapidly migrate to infection or inflammation sites via the leukocyte adhesion cascade (2, 3). Upon reaching the affected area, neutrophils initiate powerful antimicrobial responses as the first line of defense against pathogens (4–6). These responses include degranulation (7), the generation of reactive oxygen species (ROS) (8–10), the initiation of phagocytosis (11), and the formation of the neutrophil extracellular traps (NETs) (12, 13). Ultimately, these processes, often in collaboration with macrophages, contribute to the effective elimination of pathogens (14).
NETs are fibrous structures composed of chromatin that is condensed and decorated with histones, antimicrobial proteins, and enzymes derived from neutrophil granules (15, 16). First described by Brinkmann et al. in 2004 (17), NETs have become a focal point in the study of neutrophil immune functions. The formation of NETs, or NETosis, can occur in two main forms: suicidal, where neutrophils undergo programmed cell death during NETs release; and vital, where neutrophils remain active while releasing NETs. Both forms of NETs can capture and kill a wide variety of pathogens, including bacteria, viruses, fungi, and parasites (18–20). In mammals, NETs formation can be triggered by various of exogenous stimuli, such as phorbol 12-myristate 13-acetate (PMA), lipopolysaccharides (LPS), as well as fungi, parasites, bacteria, and viruses (21, 22). Moreover, regulatory mechanisms controlling NETosis have also been identified. For example, activation of autophagy (via inhibition of the mTOR pathway or activation of cGAS-STING signaling pathway) promotes the formation of NETs (23, 24), whereas inhibition of autophagy (use PtdIns3K inhibitor) reduces the release of NETs (25), suggesting that autophagy is essential for NETosis. Research on NETs extends beyond mammals to teleost fish species, including the fathead minnow (Pimephales promelas) (26), zebrafish (Danio rerio) (27), common carp (Cyprinus carpio) (28), turbot (Scophthalmus maximus) (29, 30), tongue sole (Cynoglossus semilaevis) (31), and rainbow trout (Oncorhynchus mykiss) (32) among other teleost fish. In turbot, NETs can be induced in vitro by Edwardsiella piscicida (E. piscicida) and have been shown to trap and kill bacteria (29). Similarly, in tongue sole, NETosis can be triggered by Pseudomonas fluorescens (P. fluorescens) and Vibrio harveyi (V. harveyi), with the resulting NETs exhibiting bactericidal activity (30). Despite these findings, the mechanisms underlying NETosis in teleost fish remain less well understood compared to mammals, particularly with respect to the role of autophagy in regulating this process. Given the evolutionarily conservation of autophagy, it is crucial to understand whether and how autophagy signaling regulates NETosis in teleost fish models.
The large yellow croaker (Larimichthys crocea) is a commercially important species in China’s marine aquaculture industry. However, bacterial infections, especially visceral white spot disease (VDPD) caused by Pseudomonas plecoglossicida (P. plecoglossicida), pose significant challenges to the industry’s growth (33, 34). P. plecoglossicida is a pathogen not only of the large yellow croaker but also of other farmed fish species, such as Epinephelus coioides and Plecoglossus altivelis, and is equally pathogenic (35, 36), where it induces inflammation and cell death (37). In this study, we used the large yellow croaker as a model to investigate the regulatory mechanisms of NETosis. We observed that P. plecoglossicida infection induced both autophagy and NETosis in neutrophils. Notably, the autophagy pathway was activated in neutrophils upon infection, and inhibition of autophagy was found to significantly reduce the formation of NETs. Furthermore, we confirmed the antibacterial properties of the NETs produced in response to P. plecoglossicida. In summary, our findings provide new insights into the regulatory role of autophagy in NETosis of teleost fish. This study contributes to a better understanding of immune responses in aquaculture and highlights the potential of targeting autophagy pathways to enhance fish immune defenses against bacterial infections.
Materials and methods
Experimental animals
Healthy large yellow croaker (Larimichthys crocea) (~60g) were sourced from a croaker farming base in Xiangshan harbor (Zhejiang, China) and then maintained in a recirculating aquatic system at Ningbo University for a period exceeding 2 weeks prior to conducting the experiment. The fish were kept at 22°C and fed daily with commercial diet. Feeding was terminated 48 h prior to the sampling experiment. The entire animal husbandry practices and experimental methodologies were approved by Institutional Animal Care and Use Committee of Ningbo University (permit number 12841).
Neutrophil isolation
To gain neutrophils, we first sampled the large yellow croaker head kidney, and the head kidney was treated with DMEM (supplemented with 5% FBS) and mechanically disaggregated on a 100-μm nylon cell strainers (BD Biosciences, Franklin Lakes, USA). Then, the obtained cell suspensions were placed into 61% (1.080g/mL) to 72% (1.094g/mL) percoll (GE Healthcare, Chalfont St. Giles, UK) discontinuous density gradient and centrifuged at 400 g for 30 min at 4°C. The neutrophils at the interface were collected and washed with DMEM (supplemented with 5% FBS). Subsequently, the purity of the collected neutrophils was verified by flow cytometry SSC and FSC, and the category of granulocytes was verified by using Giemsa staining according to the instructions, and the high purity neutrophils obtained were pending further analysis.
RNA isolation and quantitative real-time PCR analysis
Total RNA from neutrophils was extracted with TRIzol Reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s protocol. Briefly, the purity and concentration of the extracted RNA was carried out by spectrophotometry (NanoPhotometer NP80 Touch), and the integrity of the RNA was determined by agarose gel electrophoresis. The quantified RNA samples were then used for cDNA synthesis (Invitrogen, Carlsbad, CA, USA). Thereafter, qPCR was performed on a 7,500 qPCR system (Applied Biosystems) with the cDNA using the EvaGreen 2 × qPCR Master mix (Yeasen, Shanghai, China). All samples were performed following conditions: 95°C for 5 min, followed by 38 cycles at 95°C for 10 s and at 60°C for 30 s. A dissociation protocol was carried out after thermos cycling to confirm only one amplicon of each gene. β-actin was used as the housekeeping gene for the normalization of target gene expression. The relative expression level of all target genes was determined using the Pfaffl method (38). The primer sequences used for qPCR are listed in Supplementary Table S3.
Sequencing and analyses
The neutrophil samples of the control group and the infected group were sent to Wuhan Benagen Technologies Co., Ltd. (Wuhan, China). Briefly, the RNA was extracted using the Total RNA Kit I, and the concentration and integrity of RNA were checked by NanoDrop One spectrophotometer (NanoDrop Technologies, Wilmington, DE) and Agilent 2100 Bioanalyzer, respectively. Thereafter, the RNA was used for stranded RNA sequencing library preparation using a MGIEasy RNA Library Prep Kit for BGI® following the manufacturer’s instruction. PCR products underwent enrichment, quantification procedures, and ultimately underwent sequencing using the DNBSEQ. The clean reads were aligned to the genome of the large yellow croaker. Use gffcompare program to compare merged transcripts with known transcripts of the genome, discovering new transcripts and new genes to complement existing annotations. Differential expression analysis of two groups was performed using the DESeq2. The FDR ≤ 0.05 and Fold Change ≥ 2 was set as the threshold for significantly differential expression. For further analysis of the DEGs, we carried out a KEGG enrichment using kofam_scan software to identify the major pathways that were significantly enriched after P. plecoglossicida treatment.
SDS-PAGE and western blot
The neutrophil intracellular proteins were resolved on 15% SDS-PAGE gel (Bio-Rad, Hercules, USA) under reducing conditions, and the gels were transferred onto a 0.22 μm PVDF membrane and blocked in 0.05% PBST with 5% skim milk. Then, the membrane was incubated with anti-p62/SQSTM1, anti-LC3B polyclonal rabbit antibody (Servicebio, Wuhan, China), or anti-β-actin monoclonal rabbit antibody (ABclonal Technology, Wuhan, China) for 1 h on a shaker. After washing four times with 0.05% PBST (5 min/time), goat anti-rabbit IgG was added and incubated for 45 min at room temperature. After washing four times with 0.05% PBST (5 min/time), the reaction bands were visualized with the ECL Substrate (Bio-Rad, Hercules, USA) and then scanned by Tanon-5200 Chemiluminescent Imaging System (Tanon Science & Technology). The densitometry of target band was analyzed with ImageJ software.
Immunofluorescence microscopy
The microscopic observation of NETs was consistent with that previously reported (29). Briefly, P. plecoglossicida was cultured in TSB medium to an OD600 of 0.8 at 26°C. Neutrophils (~2 × 105) were inoculated onto glass slides that had been treated with 0.001% polylysine (Sigma, St. Louis, MO, USA) and placed in a 24-well cell culture plates. Cells were left to rest for 2 h and then treated with P. plecoglossicida (~ 106 CFU) at 26°C for 2 and 4 h. Control cells were left untreated. Control cells were left untreated. Under a fluorescence microscope, SYTOX Green (Beyotime, Shanghai, China) was added to the cells, and after incubation for 5 min, the cells were washed three times with PBS. Cells were then fixed with 4% paraformaldehyde (Sigma-Aldrich, St. Louis, USA) for 25 min and stained with 4’,6-diamidino-2-phenylindole (DAPI) (1 mg/ml; Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. Scanning electron microscopy (SEM) was performed as reported (39).
Autophagy assay
To detect the percentage of autophagy of neutrophils after P. plecoglossicida treatment. According to the manufacturer’s instructions with sight modifications, briefly, the large yellow croaker neutrophils (3 × 106 cells/well in a volume of 200 μL) were collected after treatment with P. plecoglossicida (1 × 106 CFU) for 1, 2, and 4 h or pre-treatment with autophagy inhibitor 3-methyladenine (3-MA) (GlpBio, Wuhan, China) for 1 h and then with P. plecoglossicida (1 × 106 CFU) for 1, 2, and 4 h. Control cells were untreated. MDC (Beyotime, Shanghai, China) were added to the neutrophils and incubated for 20 min at 26°C and analyzed using a fluorescence microplate reader (Molecular Devices, Sunnyvale, USA) at 360 nm excitation and 512 nm emission and expressed as relative fluorescence units (RFU).
Proliferation of P. plecoglossicida captured by NETs
To investigate the effect of NETs on the proliferation of P. plecoglossicida, we examined the survival of bacteria captured by NETs. Briefly, P. plecoglossicida was cultured in TSB medium to an OD600 of 0.8 at 26°C. Bacteria were collected by centrifugation at 14,000 g and washed three times with PBS and resuspended. Neutrophils (~2 × 105 cells/well in a volume of 200 μL) were seeded in a 96-well cell culture plate and stimulated with PMA (1 μg/mL) at 26°C for 2 h. The cells were centrifuged at 400 g for 5 min, and 150 μL of the supernatant was discarded. 50 μL of DMEM medium with or without 100 U/ml DNase I was then added to the cells. P. plecoglossicida (~104 CFU) was then added to the culture plates. centrifugation at 800 g for 10 min brought the bacteria into close contact with the neutrophils. The plates were then incubated at 26°C for 1, 3, and 6 h. At the end of the incubation, the contents of each well (bacteria + neutrophils and NETs) were collected and serially diluted, and one part of the diluted contents was plated on TSA agar plates to observe subsequent colony formation, and the other part was seeded into TSB medium for incubation and the OD values were determined spectrophotometrically at 2, 4, 6, 8, 10, 12, 14, and 16 h after incubation, respectively.
Quantification of NETs
To quantify NETs, we used the method of Parker et al. (40). Briefly, neutrophils (3 × 106 cells/well in a volume of 200 μL) were suspended in HBSS and inoculated in black 96-well plates. Cells were treated with P. plecoglossicida (1 × 106 CFU) for 1, 2, and 4 h or pre-treated with 3-MA for 1 h followed by P. plecoglossicida (1 × 106 CFU) for 1, 2 and 4 h. Control cells were untreated. After treatment, SYTOX Green was added to the cells and incubated for 5 min. Fluorescence was then quantified using a fluorescence microplate reader at 488 nm excitation and 536 nm emission and expressed as RFU.
Statistical analysis
Statistical evaluations were performed using GraphPad Prism 9. Data comparisons between groups were determined by Student’s t test. Data were expressed as the mean ± SEM, p < 0.05 was considered statistically significant.
Results
Large yellow croaker NETosis induced by P. plecoglossicida
To investigate the impact of P. plecoglossicida on neutrophils in large yellow croaker, we isolated head kidney granulocytes using a percoll discontinuous density gradient (55/61%) separation method. Flow cytometry analysis revealed that the high purity cell population (88.6%) we obtained closely matched the granulocyte distribution within the head kidney leukocytes, indicating that the isolated cells were predominantly granulocytes (Figure 1A). To confirm the identity of these cells as neutrophils, we conducted Giemsa staining, which showed that the cytoplasm of these cells was predominantly stained mauve (neutrophils), rather than light red (eosinophils) or purple (basophils), further supporting that the isolated granulocytes were primarily neutrophils (Figure 1B). Subsequently, we co-incubated P. plecoglossicida with neutrophils for 2 and 4 h, respectively, and observed the formation of NETs using immunofluorescence microscopy (Figure 1C; Supplementary Figure S1). This confirmed our hypothesis that P. plecoglossicida induces NETosis in neutrophils.
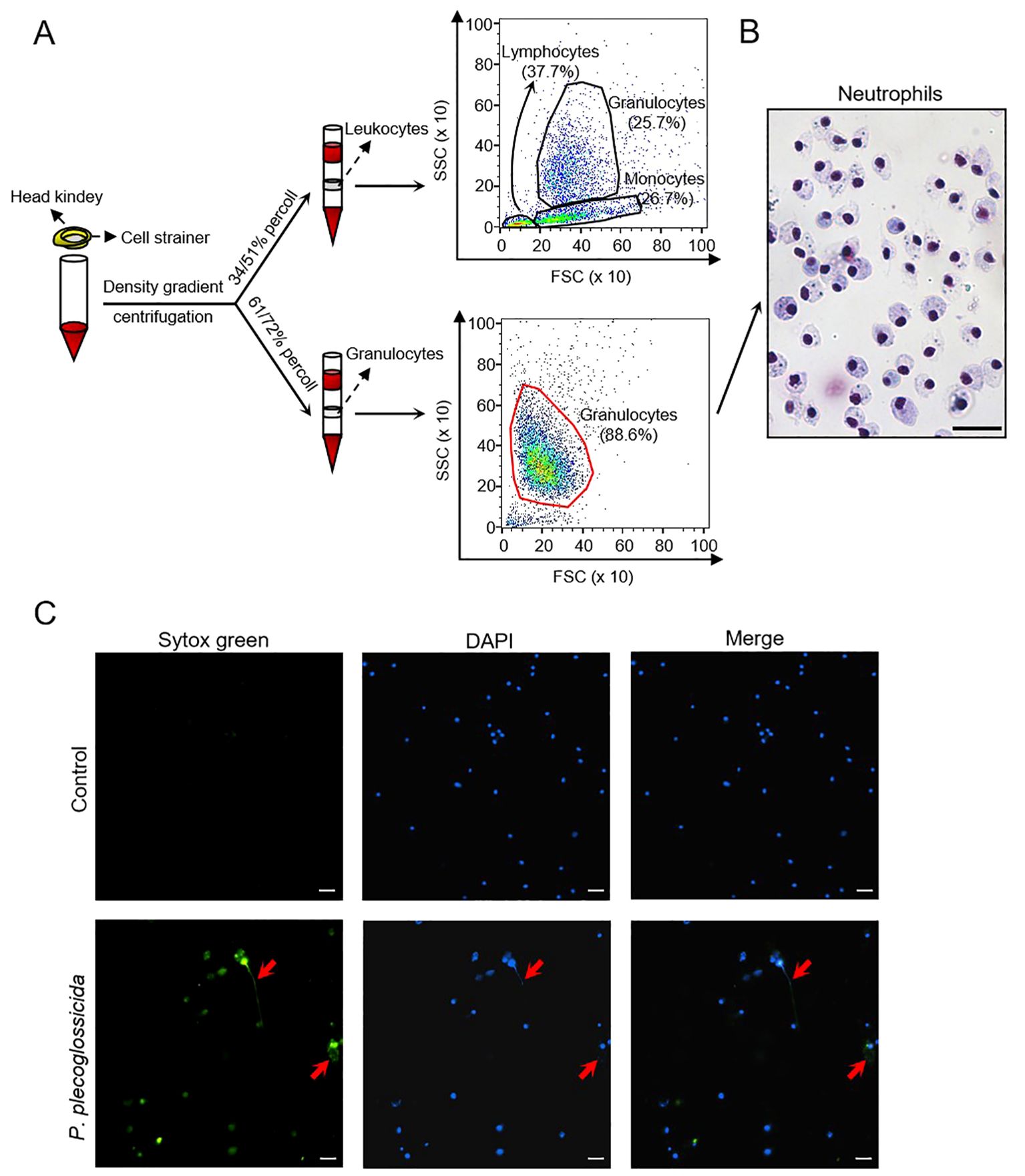
Figure 1. Isolation and identification of large yellow croaker neutrophils and the effect of P. plecoglossicida on the NETs formation. (A) Isolation of leucocytes and neutrophils from the head kidney and validation by flow cytometry. (B) Giemsa staining of neutrophils. Scale bar, 20 μm. (C) Neutrophils in the control and P. plecoglossicida groups (P. plecoglossicida incubated with neutrophils for 2 h) were stained with SYTOX Green and DAPI and analyzed by immunofluorescence microscopy. Arrows indicate NETs, Scale bar, 20 μm.
Analysis of transcriptomic changes in large yellow croaker neutrophils after in vitro P. plecoglossicida treatment
To elucidate the mechanisms underlying the induction of NETosis by P. plecoglossicida, we constructed RNA-Seq libraries from six samples, comprising three controls (LiC group) and three infected with P. plecoglossicida (LiE group). These libraries were sequenced using the Illumina platform. Following quality filtering, we obtained a total of 257,097,340 clean reads (Supplementary Table S1). Subsequently, these clean reads were mapped back to the Larimichthys crocea genome, with each sample achieving a mapping rate exceeding 90% (Supplementary Table S2).
Through statistical analysis, 1,577 differential expressed genes (DEGs) were identified in the LiE/LiC group (618 up-regulated and 959 down-regulated). The expression pattern of DEGs was presented in the volcano plots (Figure 2A). To further investigate the changes in DEGs after P. plecoglossicida treatment of neutrophils, we performed Gene Ontoloy (GO) enrichment (biological process) and KEGG pathway enrichment analysis. As illustrated in Supplementary Figure S2, metabolic processes were notably enriched, suggesting that cellular vital activities and nutrient metabolism were significantly impacted by P. plecoglossicida infection. In KEGG pathway enrichment analysis, numerous essential signaling pathways were successfully identified. The results indicated enrichment in pathways integral to neutrophil life activities, including autophagy (closely associated with the formation of NETs), ubiquitin mediated proteolysis (a critical protein degradation mechanism within the cell that plays a pivotal role in cell biological processes), mitophagy, ribosome biogenesis in eukaryotes, and the cell cycle (relevant to the regulation of cell proliferation post-bacterial infection) (Figure 2B). Despite the significant enrichment of autophagy signaling pathways, some pathways related to NETs formation were not enriched, such as regulation of actin cytoskeleton and mTOR signaling pathways, which have been well-demonstrated in mammals (24, 41–43). Consequently, we examined these pathways in the transcriptome data and found that regulation of actin cytoskeleton is promoted following P. plecoglossicida infection, whereas the mTOR pathway is suppressed (Figure 2C). To validate the RNA-Seq expression levels, we detected the expression levels of the corresponding genes in Figure 2C using qRT-PCR. The relative mRNA expression levels determined by RNA-Seq and qRT-PCR were significantly correlated (Figures 2C–G), demonstrating that RNA-Seq and qRT-PCR possess comparable sensitivity and accuracy for assessing gene expression in vitro.
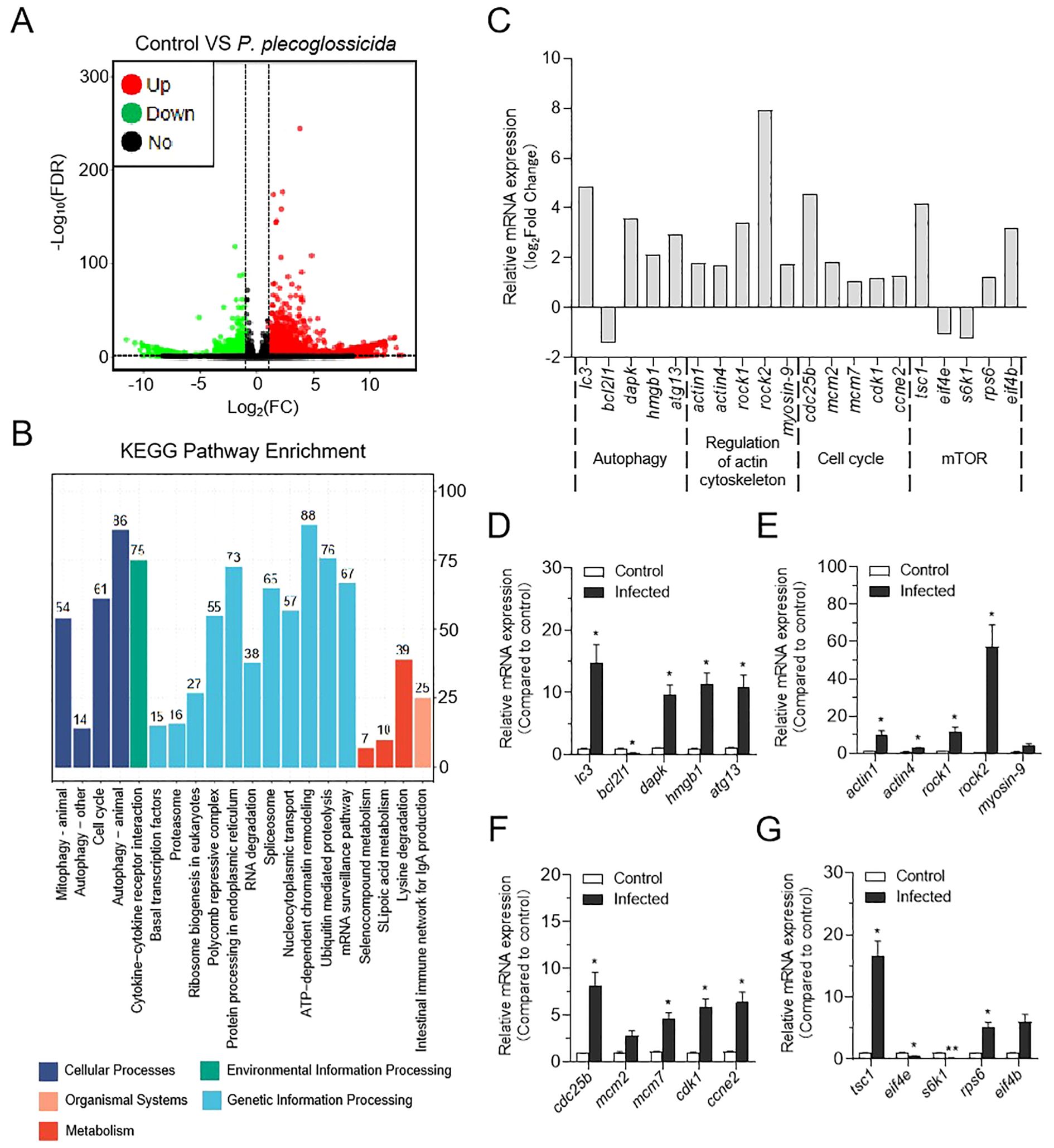
Figure 2. Effect of P. plecoglossicida infection on transcriptome changes in neutrophils of the large yellow croaker. (A) Volcano plot showing the distribution of DEGs in the P. plecoglossicida infection group compared to the control group. (B) Counts of DEGs in each KEGG pathway. Different colors mean different classes. (C) The expression of major genes in the autophagy, regulatory actin cytoskeleton, cell cycle, and mTOR pathways in RNA sequencing. (D–G) qPCR validation of DEGs in the pathways of autophagy, regulation of the actin cytoskeleton, cell cycle, and mTOR (n = 3 per group). Statistical differences were performed by Student’s t-test. Data in (D–G) are representative of at least three independent experiments (Mean ± SEM). *P < 0.05, **P < 0.01.
P. Plecoglossicida induces autophagy in large yellow croaker neutrophils
In mammals, autophagy has been implicated in the induction of NETosis. To investigate whether a similar process occurs in teleost fish, we examined the transcriptome of neutrophils infected with P. plecoglossicida. Our analysis revealed that the autophagy signaling pathway was activated upon infection (Figure 3A), and several key genes within this pathway were significantly up-regulated (Figure 3B). These transcriptomic findings led us to hypothesize that P. plecoglossicida infection could trigger autophagy in neutrophils. To substantiate this speculation, we conducted further analyses at the protein and cellular levels. Western blot results demonstrated that the expression of p62/SQSTM1 protein was markedly reduced, and the ratio of LC3-II/LC3-I was significantly elevated in neutrophils from the infected group compared to the control group (Figures 3C, D). Additionally, fluorescence microplate reader results also confirmed that neutrophils exhibited more pronounced autophagy following infection (Figure 3E). Collectively, these results substantiate the induction of autophagy in neutrophil by P. plecoglossicida.
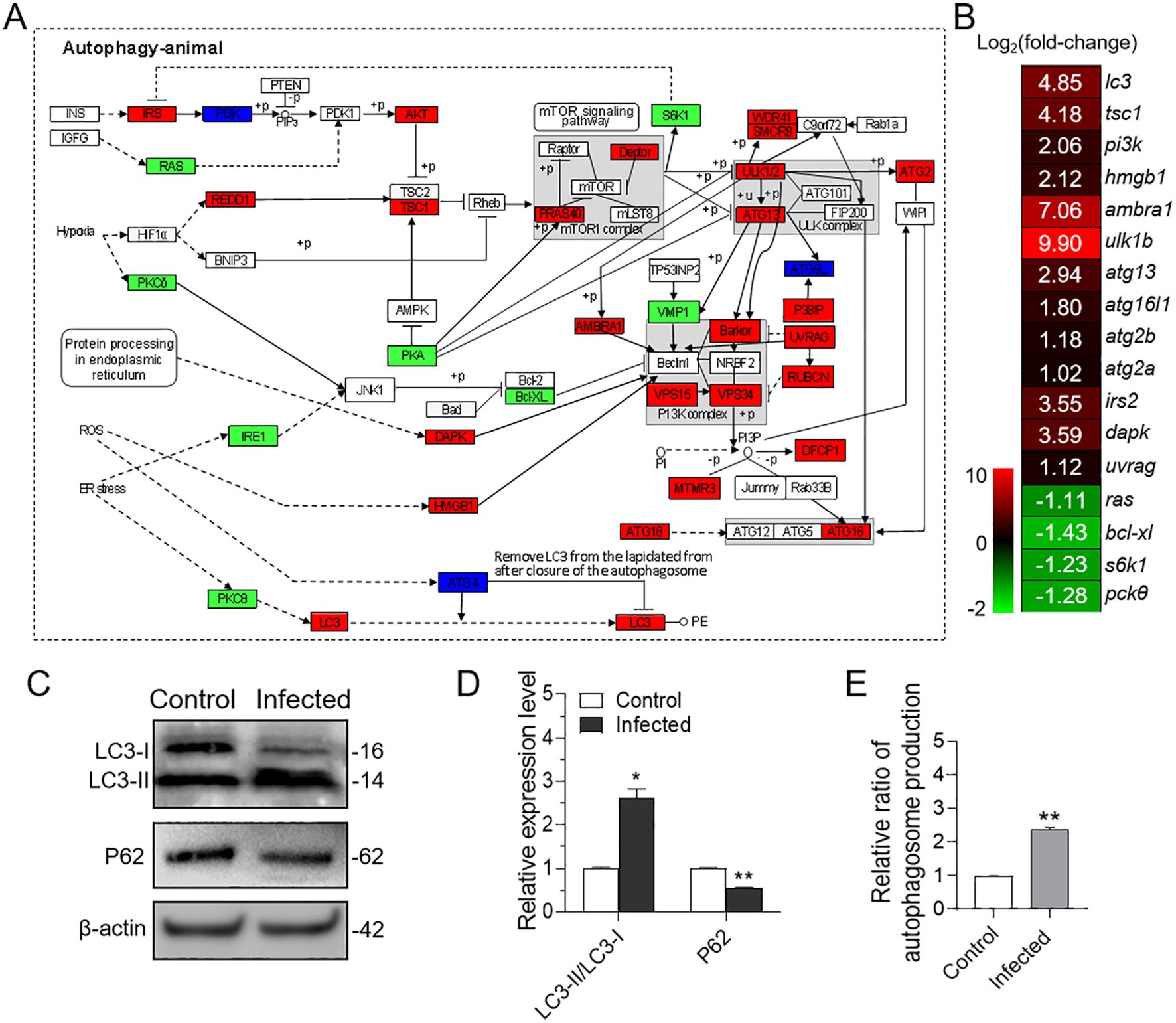
Figure 3. Effect of in vitro infection of neutrophils with P. plecoglossicida on autophagy. (A) Both of red, green, and deep blue shading boxes represent molecules of the autophagy pathway identified in head kidney neutrophils of large yellow croaker. And the red boxes indicate the up-regulated DEGs in this pathway, the green boxes indicate the down-regulated DEGs in this pathway, and the deep blue boxes indicate both up-regulated and down-regulated DEGs in this pathway. (B) Differential expression genes involved in the autophagy pathway were analyzed after P. plecoglossicida infection. The color gradient represents highly up-regulated (red) to highly down-regulated (green) genes. (C) Immunoblot analysis of LC3 and p62/SQSTM1 protein levels in neutrophils infected with P. plecoglossicida for 2 h. (D) The relative proportions of LC3-II/LC3-I and p62/SQSTM1 in the infected compared with the control group were evaluated by densitometric analysis of immunoblots from (C) (n = 3 per group). (E) Neutrophils were infected with or without P. plecoglossicida for 2 h, and autophagy fluorescence intensity was detected (n = 3 per group). Statistical differences were performed by Student’s t-test. Data in (D, E) are representative of at least three independent experiments (Mean ± SEM). *P < 0.05, **P < 0.01.
Autophagy regulates NETosis in large yellow croaker
To explore the role of autophagy in regulating NETosis, we established a model of autophagy inhibition using 3-MA. Fluorescence microplate reader analysis revealed that neutrophils pretreated with 3-MA exhibited a significant decrease in autophagy levels following infection with P. plecoglossicida at 1, 2, and 4 h post-infection, as compared to the infected group (Figure 4A). Subsequently, we further validated the impact of autophagy on NETosis through similar fluorescence microplate reader assays. The results indicated that the formation of NETs was notably reduced in neutrophils pretreated with 3-MA and then infected, at both 2 and 4 h post-infection, relative to the infected group. Meanwhile, the infected group demonstrated a significant increase in NETs formation compared to the control group (Figure 4B). Collectively, these findings underscore the critical role of autophagy in the NETosis process, wherein activation of autophagy enhances NETosis, whereas inhibition of autophagy dampens it.
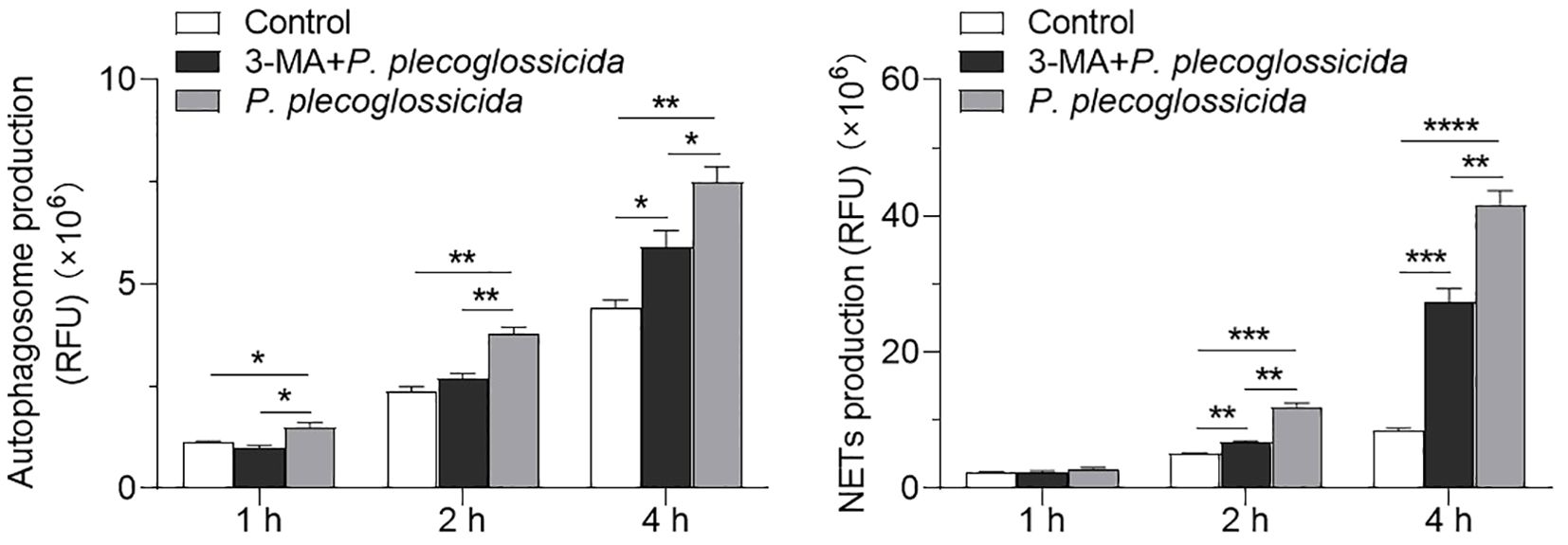
Figure 4. Effect of the inhibition of autophagy on the NETs formation in neutrophils. (A) Large yellow croaker neutrophils were treated with or without 3-MA and then infected with P. plecoglossicida for different times to detect autophagy production. The control group was untreated (n = 3 per group). (B) Large yellow croaker neutrophils were treated with or without 3-MA and then infected with P. plecoglossicida for different times to detect NETs production. The control group was untreated (n = 3 per group). Statistical differences were performed by Student’s t-test. Data in (A, B) are representative of at least three independent experiments (Mean ± SEM). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
NETs possess the capacity to inhibit the proliferation of P. plecoglossicida
In order to investigate the impact of NETs on bacterial growth, we conducted growth experiments. The results showed that compared with P. plecoglossicida infection group (NETs-positive group), the P. plecoglossicida + DNase I group (NETs-negative group) exhibited a greater number of colony formation on TSA agar plates following incubation with neutrophils (Figures 5A–D). Additionally, the NETs-negative group demonstrated a more rapid growth rate of P. plecoglossicida in TSB medium compared to the NETs-positive group (Figures 5E, F). These findings confirm that neutrophil NETs are capable of capturing and suppressing the growth of P. plecoglossicida, emphasizing their significant role in the resistance to bacterial invasion.
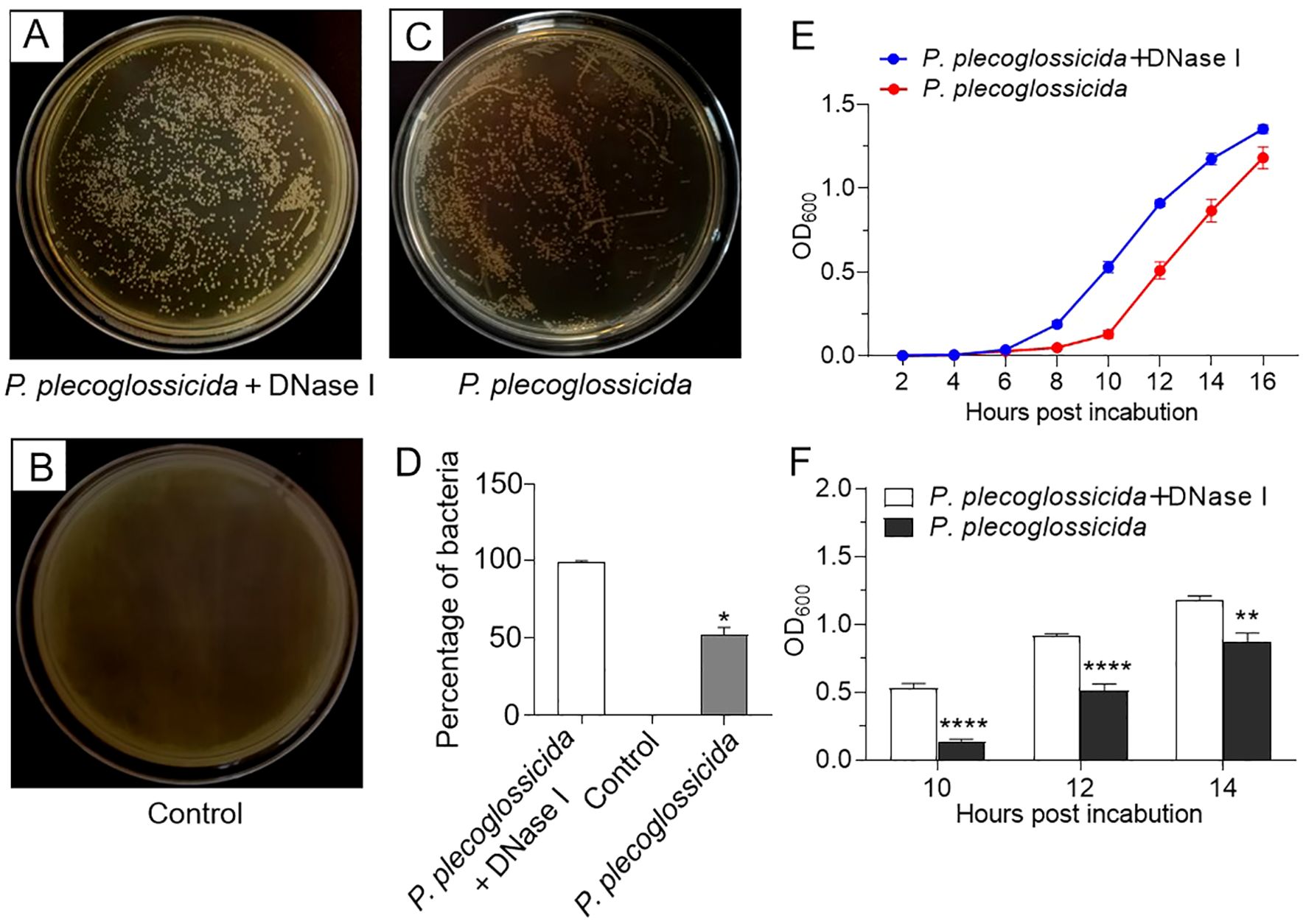
Figure 5. Effects of neutrophil NETs on survival and growth of P. plecoglossicida. (A–C) Large yellow croaker neutrophils were pretreated with PMA for 2h and then infected with P. plecoglossicida before being treated with or without DNase I for 6 h, and the collected mixtures (bacteria + neutrophils and NETs) were inoculated into the TSA agar plates to observe colony formation. The control group was untreated. (D) Graph showing the number of colonies in (A–C) (n = 3 per group). (E) Large yellow croaker neutrophils were infected with P. plecoglossicida and then treated with or without DNase I for 6 h, and the collected mixtures (bacteria + neutrophils and NETs) were inoculated into the TSB medium to observe the OD value of the P. plecoglossicida growth curve (n = 3 per group). (F) Statistical plots of OD values at 10, 12, and 14 h in F (n = 3 per group). Statistical differences were performed by Student’s t-test. Data in (D–F) are representative of at least three independent experiments (Mean ± SEM). *P < 0.05, **P < 0.01, ****P < 0.0001.
Discussion
Neutrophils are the first line of defense in innate immunity, rapidly mobilizing to sites of infection or tissue damage. Historically, their bactericidal activity have been attributed primarily to phagocytosis, where microorganisms are engulfed and subsequently destroyed within phagosomes by various proteases. Additionally, neutrophils release antimicrobial proteins from their granules, which can act both intracellularly on phagosomes and extracellularly to eliminate pathogens (44). However, the paradigm shifted in 2004 when Brinkmann et al. revealed that a novel mechanism: neutrophils can extrude DNA to form NETs, which have the ability to capture and kill bacteria (17). This discovery transformed our understanding of neutrophil function and sparked widespread interest in the study of NETs, particularly within mammalian models. In contrast, research on NETs in fish has lagged behind, and relatively few studies have explored the regulatory mechanisms underlying NETs formation. To date, only a handful of reports have confirmed the regulatory roles of ROS, nitric oxide (NO), myeloperoxidase (MPO), and pyroptotic signaling in NETosis (30, 31). Therefore, to expand our understanding of NETs formation in fish, we investigated the mechanism of P. plecoglossicida-induced NETosis in large yellow croaker.
Previous studies has shown that several aquaculture-relevant bacteria, including E. piscicida, P. fluorescens, and V, harveyi, can induce NETosis (30, 31). Building upon this foundation, our current study focused on the effect of P. plecoglossicida on large yellow croaker neutrophils. Upon bacterial stimulation, we observed that these neutrophils released filamentous, net-like structures, consistent with the formation of NETs. To further investigate this phenomenon, we used double staining with SYTOX Green (a DNA dye that easily passes through damaged cytoplasmic membranes but not through the plasma membranes of living cells) and DAPI (a DNA dye that penetrates intact cell membranes to stain the nucleus). The staining results showed that both the neutrophils and the extracellular structures were labeled, suggesting that the NETs formation in these cells was associated with progressive cell death. This observation is consistent with the phenomenon of suicidal NETosis in mammals, where the formation of NETs is often linked to cell death (45).
To further explore the potential molecular mechanisms underlying P. plecoglossicida-induced NETosis in large yellow croaker, we analyzed the effects of P. plecoglossicida on neutrophils at the transcriptome level. When the organism is infected by pathogens, a range of cellular life activities are affected, such as cell survival, death, and metabolism of substances (proteins and nucleic acids). Based on the results of the transcriptome KEGG signaling pathway and GO enrichment (biological processes) analyses, our results also revealed that a significant number of pathways associated with autophagy, cell cycle, ubiquitin mediated proteolysis, and ribosome biogenesis in eukaryotes, and metabolism-related processes were highly enriched in neutrophils. Previous research has established that autophagy plays a role in various neutrophil functions, such as differentiation, phagocytosis, cytokine production, degranulation, cell death, and NETosis. Inhibition of autophagy has been shown to prevent the formation of NETs in neutrophils (46, 47). Notably, our transcriptome and qPCR data indicated that the autophagy signaling pathway was markedly activated in neutrophils following infection with P. plecoglossicida, with numerous autophagy-related genes significant up-regulated. This suggests that, akin to mammals, autophagy may regulate NETosis in large yellow croaker. Additionally, studies in mammals have demonstrated that the mTOR signaling pathway and regulation of actin cytoskeleton are closely associated with the formation of NETs, and their inhibition leads to impaired NETosis (24, 41–43). Although our analysis did not show enrichment for these pathways, examination of their respective genes revealed that both the mTOR signaling pathway and actin cytoskeleton regulation were suppressed at the overall level post-infection. This finding suggests that while these pathways may not be directly enriched in our transcriptome data, they could still play a role in the regulation of NETosis in large yellow croaker, potentially in conjunction with autophagy.
Autophagy is an intracellular degradation process that transports cytoplasmic material to lysosomes or autophagic vesicles for degradation. This process is crucial in a wide range of physiological and pathological conditions across species from yeast to mammals. To measure autophagic activity, we employed immunoblotting to assess key markers of autophagy, including LC3 and p62/SQSTM1 (48). In mammals, LC3-I is conjugated to phosphatidylethanolamine (PE) to form LC3-II during autophagy, while p62/SQSTM1 levels decrease upon autophagy induction (49, 50). Western blot analysis revealed that P. plecoglossicida infection significantly increased the LC3-II/LC3-I ratio in neutrophils, while p62/SQSTM1 levels were notably reduced, confirming the induction of autophagy. Subsequently, to further investigate the relationship between autophagy and NETs formation, we treated neutrophils with 3-MA, a non-selective inhibitor of autophagy that blocks phosphatidylinositol 3-kinase (PI3K) activity (51, 52). In mammals, 3-MA inhibits autophagy and attenuates the release of NETs in neutrophils during Gram-negative sepsis (53). In our study, with large yellow croaker, we observed that treating neutrophils with 3-MA prior to P. plecoglossicida infection led to a significant decrease in both autophagy and NETs production compared to the group directly infected with P. plecoglossicida. This inhibition of autophagy corresponded with reduced NETs production. Intriguingly, we found that autophagy preceded NETs formation, suggesting a temporal relationship between autophagy and NETosis.
NETs have been shown to capture and eliminate various pathogens, though their effectiveness can vary depending on the pathogen and environmental conditions. For instance, mammalian NETs can trap Streptococcus pneumoniae and Streptococcus aureus without killing them (54, 55), whereas they can directly eradicate Streptococcus flexneri and Candida albicans (17, 56). A similar pattern is observed in fish, where turbot NETs can eliminate Escherichia coli but not P. fluorescens (29), and tongue sole NETs can immobilize and disrupt the replication of V. harveyi and P. fluorescens without directly killing them. Notably, NETs did not significantly impede the proliferation of Edwardsiella tarda (E. tarda), indicating that E. tarda may possess a mechanism to counteract the antimicrobial effects of NETs (31). In the case of large yellow croaker, we observed that NETs captured P. plecoglossicida and slowed its growth. After a 6-hour incubation with neutrophils, both bacterial counts and OD measurements revealed that the growth of P. plecoglossicida was significantly slower compared to the NETs-negative group (P. plecoglossicida + DNase I group). Interestingly, neutrophils incubated with P. plecoglossicida for 1 or 3 h did not show a significant difference compared to the NETs-negative group (data not shown). These results collectively indicate that while NETs may limit bacterial growth, they do not necessarily result in complete bacterial eradication.
In conclusion, our study provides compelling evidence that autophagy signaling pathways are pivotal in regulating the formation of NETs in large yellow croaker neutrophils during infection with P. plecoglossicida (Figure 6). Our findings suggest that autophagy is essential for the process of NETosis and that NETs capture and suppressed the proliferation of P. plecoglossicida. These results highlight the evolutionary conservation of the autophagy signaling pathway in regulating NETs formation and offer valuable insights into the immune responses of teleost fish, with potential implications for enhancing disease resistance in aquaculture.
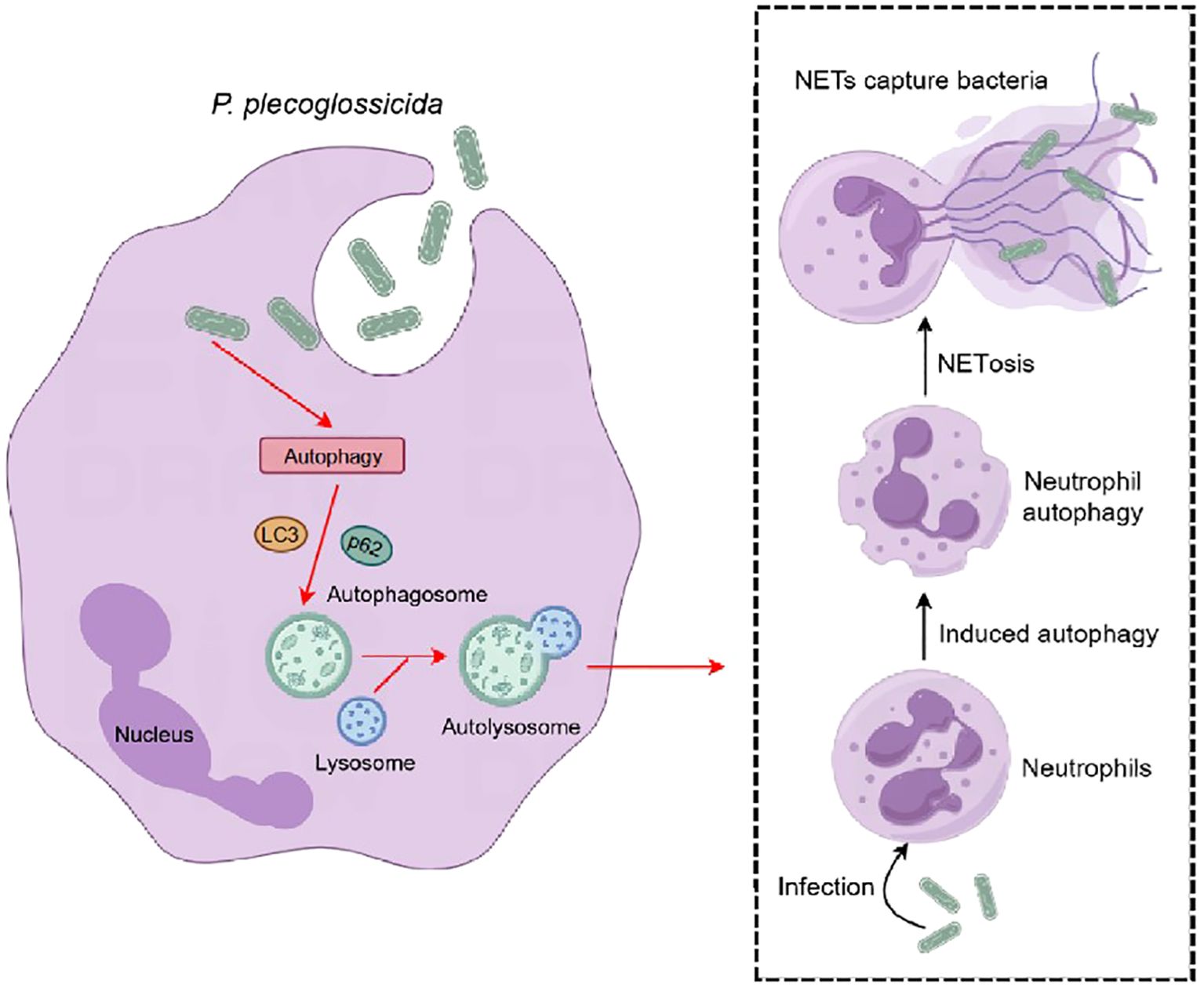
Figure 6. Pattern diagram of P. plecoglossicida induced NETosis in the large yellow croaker. After infection with P. plecoglossicida, the autophagy pathway is activated in large yellow croaker neutrophils, which in turn causes autophagy in neutrophils and induces NETosis, and the formation of NETs can capture the pathogenic bacterium P. plecoglossicida to prevent its invasion of the organism.
Data availability statement
The data presented in the study are deposited in the NCBI Bioproject repository, accession number PRJNA1197011.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of Ningbo University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
J-FC: Investigation, Methodology, Visualization, Writing – original draft. JC: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32473194), and the One Health Interdisciplinary Research Project of Ningbo University (no. HZ202201).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1521080/full#supplementary-material
Supplementary Figure 1 | Effect of P. plecoglossicida on the formation of neutrophil NETs. Neutrophils in P. plecoglossicida groups (P. plecoglossicida incubated with neutrophils for 4 h) were stained with DAPI and analyzed by immunofluorescence microscopy. Arrows indicate NETs, Scale bar, 20 μm.
Supplementary Figure 2 | Effect of P. plecoglossicida infection on GO Enrichment (Biological Process) in neutrophils of the large yellow croaker. The top 20 GO Enrichment (Biological Process) terms for DEGs.
References
1. Mayadas TN, Cullere X, Lowell CA. The multifaceted functions of neutrophils. Annu Rev Pathol. (2014) 9:181–218. doi: 10.1146/annurev-pathol-020712-164023
2. Chavakis E, Choi EY, Chavakis T. Novel aspects in the regulation of the leukocyte adhesion cascade. Thromb Haemost. (2009) 102:191–7. doi: 10.1160/TH08-12-0844
3. Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. (2007) 7:678–89. doi: 10.1038/nri2156
4. Borregaard N. Neutrophils, from marrow to microbes. Immunity. (2010) 33:657–70. doi: 10.1016/j.immuni.2010.11.011
5. Gazendam RP, van de Geer A, Roos D, van den Berg TK, Kuijpers TW. How neutrophils kill fungi. Immunol Rev. (2016) 273:299–311. doi: 10.1111/imr.12454
6. Nauseef WM. How human neutrophils kill and degrade microbes: an integrated view. Immunol Rev. (2007) 219:88–102. doi: 10.1111/j.1600-065X.2007.00550.x
7. Bedouhène S, Dang PM, Hurtado-Nedelec M, El-Benna J. Neutrophil degranulation of azurophil and specific granules. Methods Mol Biol. (2020) 2087:215–22. doi: 10.1007/978-1-0716-0154-9_16
8. Rieger AM, Konowalchuk JD, Grayfer L, Katzenback BA, Havixbeck JJ, Kiemele MD, et al. Fish and mammalian phagocytes differentially regulate pro-inflammatory and homeostatic responses in vivo. PloS One. (2012) 7:e47070. doi: 10.1371/journal.pone.0047070
9. Katzenback BA, Belosevic M. Isolation and functional characterization of neutrophil-like cells, from goldfish (Carassius auratus L.) kidney. Dev Comp Immunol. (2009) 33:601–11. doi: 10.1016/j.dci.2008.10.011
10. Wilhelm Filho D. Reactive oxygen species, antioxidants and fish mitochondria. Front Biosci. (2007) 12:1229–37. doi: 10.2741/2141
11. Rosales C, Uribe-Querol E. Phagocytosis: a fundamental process in immunity. BioMed Res Int. (2017) 2017:9042851. doi: 10.1155/2017/9042851
12. Deniset JF, Kubes P. Recent advances in understanding neutrophils. F1000Res. (2016) 5:2912. doi: 10.12688/f1000research.9691.1
13. Brinkmann V. Neutrophil extracellular traps in the second decade. J Innate Immun. (2018) 10:414–21. doi: 10.1159/000489829
14. Greenlee-Wacker MC. Clearance of apoptotic neutrophils and resolution of inflammation. Immunol Rev. (2016) 273:357–70. doi: 10.1111/imr.12453
15. Yipp BG, Petri B, Salina D, Jenne CN, Scott BN, Zbytnuik LD, et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking. vivo Nat Med. (2012) 18:1386–93. doi: 10.1038/nm.2847
16. Neeli I, Radic M. Knotting the NETs: analyzing histone modifications in neutrophil extracellular traps. Arthritis Res Ther. (2012) 14:115. doi: 10.1186/ar3773
17. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. (2004) 303:1532–5. doi: 10.1126/science.1092385
18. Burgener SS, Schroder K. Neutrophil extracellular traps in host defense. Cold Spring Harb Perspect Biol. (2020) 12:a037028. doi: 10.1101/cshperspect.a037028
19. Castanheira FVS, Kubes P. Neutrophils and NETs in modulating acute and chronic inflammation. Blood. (2019) 133:2178–85. doi: 10.1182/blood-2018-11-844530
20. Twaddell SH, Baines KJ, Grainge C, Gibson PG. The emerging role of neutrophil extracellular traps in respiratory disease. Chest. (2019) 156:774–82. doi: 10.1016/j.chest.2019.06.012
21. von Köckritz-Blickwede M, Nizet V. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J Mol Med (Berl). (2009) 87:775–83. doi: 10.1007/s00109-009-0481-0
22. Kenny EF, Herzig A, Krüger R, Muth A, Mondal S, Thompson PR, et al. Diverse stimuli engage different neutrophil extracellular trap pathways. Elife. (2017) 6:e24437. doi: 10.7554/eLife.24437
23. Skendros P, Mitsios A, Chrysanthopoulou A, Mastellos DC, Metallidis S, Rafailidis P, et al. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J Clin Invest. (2020) 130:6151–7. doi: 10.1172/JCI141374
24. Pan J, Fei CJ, Hu Y, Wu XY, Nie L, Chen J. Current understanding of the cGAS-STING signaling pathway: Structure, regulatory mechanisms, and related diseases. Zool Res. (2023) 44:183–218. doi: 10.24272/j.issn.2095-8137.2022.464
25. Itakura A, McCarty OJ. Pivotal role for the mTOR pathway in the formation of neutrophil extracellular traps via regulation of autophagy. Am J Physiol Cell Physiol. (2013) 305:C348–354. doi: 10.1152/ajpcell.00108.2013
26. Palić D, Ostojić J, Andreasen CB, Roth JA. Fish cast NETs: neutrophil extracellular traps are released from fish neutrophils. Dev Comp Immunol. (2007) 31:805–16. doi: 10.1016/j.dci.2006.11.010
27. Palić D, Andreasen CB, Ostojić J, Tell RM, Roth JA. Zebrafish (Danio rerio) whole kidney assays to measure neutrophil extracellular trap release and degranulation of primary granules. J Immunol Methods. (2007) 319:87–97. doi: 10.1016/j.jim.2006.11.003
28. Brogden G, von Köckritz-Blickwede M, Adamek M, Reuner F, Jung-Schroers V, Naim HY, et al. [amp]]beta;-Glucan protects neutrophil extracellular traps against degradation by Aeromonas hydrophila in carp (Cyprinus carpio). Fish Shellfish Immunol. (2012) 33:1060–4. doi: 10.1016/j.fsi.2012.08.009
29. Chi H, Sun L. Neutrophils of Scophthalmus maximus produce extracellular traps that capture bacteria and inhibit bacterial infection. Dev Comp Immunol. (2016) 56:7–12. doi: 10.1016/j.dci.2015.11.005
30. Zhao J, Chen W, Zhang Y, Liu Q, Yang D, Wang Z. Bacterial infection induces pyroptotic signaling-mediated neutrophil extracellular traps (NETs) formation in turbot (Scophthalmus maximus). Fish Shellfish Immunol. (2022) 127:982–90. doi: 10.1016/j.fsi.2022.07.026
31. Zhao ML, Chi H, Sun L. Neutrophil extracellular traps of Cynoglossus semilaevis: production characteristics and antibacterial effect. Front Immunol. (2017) 8:290. doi: 10.3389/fimmu.2017.00290
32. Van AP, Álvarez de Haro N, Bron JE, Desbois AP. Chromatin extracellular trap release in rainbow trout, Oncorhynchus mykiss (Walbaum, 1792). Fish Shellfish Immunol. (2020) 99:227–38. doi: 10.1016/j.fsi.2020.01.040
33. Li C, Wang S, Ren Q, He T, Chen X. An outbreak of visceral white nodules disease caused by Pseudomonas plecoglossicida at a water temperature of 12°C in cultured large yellow croaker (Larimichthys crocea) in China. J Fish Dis. (2020) 43:1353–61. doi: 10.1111/jfd.13206
34. Tang Y, Xin G, Zhao LM, Huang LX, Qin YX, Su YQ, et al. Novel insights into host-pathogen interactions of large yellow croakers (Larimichthys crocea) and pathogenic bacterium Pseudomonas plecoglossicida using time-resolved dual RNA-seq of infected spleens. Zool Res. (2020) 41:314–27. doi: 10.24272/j.issn.2095-8137.2020.035
35. Kobayashi T, Imai M, Ishitaka Y, Kawaguchi Y. Histopathological studies of bacterial haemorrhagic ascites of ayu, Plecoglossus altivelis (Temminck & Schlegel). J Fish Dis. (2004) 27:451–7. doi: 10.1111/j.1365-2761.2004.00563.x
36. Yuan B, Zhao LM, Zhuang ZX, Wang XR, Fu Q, Huang HB, et al. Transcriptomic and metabolomic insights into the role of the flgK gene in the pathogenicity of Pseudomonas plecoglossicida to orange-spotted grouper (Epinephelus coioides). Zool Res. (2022) 43:952–65. doi: 10.24272/j.issn.2095-8137.2022.216
37. Dai T, Zhao Z, Zhu T, Fei C, Nie L, Chen J. The anti-inflammatory role of zDHHC23 through the promotion of macrophage M2 polarization and macrophage necroptosis in large yellow croaker (Larimichthys crocea). Front Immunol. (2024) 15:1401626. doi: 10.3389/fimmu.2024.1401626
38. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. (2001) 29:e45. doi: 10.1093/nar/29.9.e45
39. Ng TH, Chang SH, Wu MH, Wang HC. Shrimp hemocytes release extracellular traps that kill bacteria. Dev Comp Immunol. (2013) 41:644–51. doi: 10.1016/j.dci.2013.06.014
40. Parker H, Dragunow M, Hampton MB, Kettle AJ, Winterbourn CC. Requirements for NADPH oxidase and myeloperoxidase in neutrophil extracellular trap formation differ depending on the stimulus. J Leukoc Biol. (2012) 92:841–9. doi: 10.1189/jlb.1211601
41. Liu ML. Functional actin cytoskeleton is required in early stage of NETosis induction. Proc Natl Acad Sci U.S.A. (2020) 117:22653–4. doi: 10.1073/pnas.2013790117
42. Mannherz HG, Budde H, Jarkas M, Hassoun R, Malek-Chudzik N, Mazur AJ, et al. Reorganization of the actin cytoskeleton during the formation of neutrophil extracellular traps (NETs). Eur J Cell Biol. (2024) 103:151407. doi: 10.1016/j.ejcb.2024.151407
43. Sprenkeler EGG, Tool ATJ, Henriet SSV, van Bruggen R, Kuijpers TW. Formation of neutrophil extracellular traps requires actin cytoskeleton rearrangements. Blood. (2022) 139:3166–80. doi: 10.1182/blood.2021013565
44. Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. (2013) 13:159–75. doi: 10.1038/nri3399
45. Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. (2007) 176:231–41. doi: 10.1083/jcb.200606027
46. Huang Z, Zhang H, Fu X, Han L, Zhang H, Zhang L, et al. Autophagy-driven neutrophil extracellular traps: The dawn of sepsis. Pathol Res Pract. (2022) 234:153896. doi: 10.1016/j.prp.2022.153896
47. Remijsen Q, Vanden Berghe T, Wirawan E, Asselbergh B, Parthoens E, De Rycke R, et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. (2011) 21:290–304. doi: 10.1038/cr.2010.150
48. Jiang P, Mizushima N. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods. (2015) 75:13–8. doi: 10.1016/j.ymeth.2014.11.021
49. Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. (2001) 276:1701–6. doi: 10.1074/jbc.C000752200
50. Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. (2002) 277:13739–44. doi: 10.1074/jbc.M200385200
51. Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. (2010) 285:10850–61. doi: 10.1074/jbc.M109.080796
52. Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discovery. (2017) 16:487–511. doi: 10.1038/nrd.2017.22
53. Kambas K, Mitroulis I, Apostolidou E, Girod A, Chrysanthopoulou A, Pneumatikos I, et al. Autophagy mediates the delivery of thrombogenic tissue factor to neutrophil extracellular traps in human sepsis. PloS One. (2012) 7:e45427. doi: 10.1371/journal.pone.0045427
54. Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol. (2006) 16:401–7. doi: 10.1016/j.cub.2006.01.056
55. Menegazzi R, Decleva E, Dri P. Killing by neutrophil extracellular traps: fact or folklore? Blood. (2012) 119:1214–6. doi: 10.1182/blood-2011-07-364604
Keywords: Pseudomonas plecoglossicida, Larimichthys crocea, neutrophil extracellular traps, autophagy, antibacterial
Citation: Cao J-F and Chen J (2024) Pseudomonas plecoglossicida infection induces neutrophil autophagy-driven NETosis in large yellow croaker Larimichthys crocea. Front. Immunol. 15:1521080. doi: 10.3389/fimmu.2024.1521080
Received: 01 November 2024; Accepted: 22 November 2024;
Published: 23 December 2024.
Edited by:
Junji Zhu, Cleveland Clinic, United StatesReviewed by:
Qingpi Yan, Jimei University, ChinaZuberwasim Sayyad, Lerner Research Institute, United States
Copyright © 2024 Cao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiong Chen, Y2hlbmppb25nQG5idS5lZHUuY24=
 Jia-Feng Cao
Jia-Feng Cao Jiong Chen
Jiong Chen