- 1Doctoral Degree Program of Translational Medicine, National Yang Ming Chiao Tung University and Academia Sinica, Taipei, Taiwan
- 2Immunology Research Center, National Health Research Institute, Zhunan, Taiwan
- 3Institute of Clinical Medicine and Institute of Microbiology and Immunology, National Yang Ming Chiao Tung University, Taipei, Taiwan
- 4Department of Medical Research, Taipei Veterans General Hospital, Taipei, Taiwan
- 5Master Program in Clinical Genomics and Proteomics, School of Pharmacy, Taipei Medical University, Taipei, Taiwan
CASK, a MAGUK family scaffold protein, regulates gene expression as a transcription co-activator in neurons. However, the mechanism of CASK nucleus translocation and the regulatory function of CASK in myeloid cells remains unclear. Here, we investigated its role in H5N1-infected macrophages. We found that H5N1 triggers CASK nuclear translocation via PKR and SRC signaling. HCK, a SRC family kinase, enhances CASK phosphorylation at S395 via CDK5, facilitating CASK’s nuclear entry. Knocking out CASK in myeloid cells specifically reduces interferon-alpha (IFNA) production by hindering the nuclear export of Ifna mRNA, while leaving its mRNA levels unchanged. Myeloid-specific CASK knockout (KO) mice display exacerbated lung inflammation, which correlates with reduced IFNA levels during H5N1 infection. Interactome studies show that H5N1 triggers associations between CASK and CCT4, STIP1, and TNK1. These associations recruit IRF7, POLR2C, TAF15, HNRNPs, and CRM1, enabling the CASK complex to bind to the Ifna promoter, bind co-transcriptionally to Ifna mRNA, and facilitate CRM1-dependent Ifna mRNA export. This underscores CASK’s critical role in the antiviral response.
Introduction
Calcium/calmodulin-dependent serine protein kinase (CASK) is a scaffold protein belonging to the membrane-associated guanylate kinases (MAGUKs) family (1). The human CASK protein, composed of 926 amino acids, includes a calmodulin domain, two L27 domains, one PDZ domain, one SH3 domain, and a guanylate kinase domain. These domains are known to function as protein–protein interaction modules, making CASK a key scaffold protein for assembling large signaling complexes (1). Previous studies have shown that CASK is expressed at both the pre- and postsynaptic sides of excitatory synapses and is crucial for cerebrocortical development. Mutations in CASK have been linked to mental retardation, autism-spectrum disorders (2, 3), epilepsy (4), cerebellar hypoplasia (5, 6), and X-linked intellectual disability (7). CASK has been shown to interacts with Tbr-1, a T-box transcription factor that is involved in forebrain development (8). Although CASK is primarily located in the juxta-membrane region of neuronal cells, co-transfection of CASK and Tbr-1 results in nuclear translocation and binding to specific DNA sequences in a complex with Tbr-1 to induce the transcription of T-element-containing genes (9), suggesting that CASK acts as a co-activator for gene transcription. However, the mechanism by which CASK translocates from the membrane to the nucleus remains unknown.
In addition to neurons, CASK is also expressed in other cell lineages, including immune and non-immune cells. However, the functions of CASK in non-neuronal cells are not yet well understood. CASK is abundantly expressed in macrophages and is secreted from macrophages via exosomes. Interestingly, CASK is detectable in the serum of patients with focal and segmental glomerulosclerosis (FSGS), a condition that causes nephrotic syndrome with a risk of progressing to end-stage renal disease. Moreover, recombinant CASK alters the permeability of a monolayer of podocytes and increases the motility of podocytes (10), suggesting that CASK, either alone or in the context of exosomes, may act as an endogenous danger signal to trigger inflammatory reactions (10).
Macrophages play a pivotal role in immune responses, serving as a key component of the host defense against pathogens such as the H5N1 influenza A virus (IAV). The H5N1 influenza A virus (IAV) can establish productive infections in macrophages (11), activating intracellular viral RNA sensors such as RIG-I (12, 13). These sensors initiate signaling cascades that induce the transcription of type I interferons (14, 15) (including interferon-beta and interferon-alpha) and pro-inflammatory cytokines (16). This activation results in a cytokine storm (17, 18), a characteristic double-edged phenomenon of H5N1 infection. While the cytokine response helps control viral spread, it simultaneously contributes to immunopathology, highlighting the complex interplay between host defense mechanisms and viral pathogenesis.
Previous studies have shown that CASK can translocate into the nucleus and function as a transcription co-activator (9), regulating gene expression in neurons. We hypothesize that CASK’s ability to regulate gene expression may be comparable to other scaffold proteins, such as MAVS (19) and NEMO (20), which are known to orchestrate antiviral signaling by serving as platforms for assembling signaling complexes. This hypothesis is supported by the modular domains of CASK that enable diverse protein-protein interactions, positioning it as a potential regulator of gene expression in response to cellular stress or infection. In this study, we aim to investigate whether CASK is involved in the signal transduction pathway regulating the H5N1-induced cytokine storm in macrophage, and if so, to elucidate the underlying molecular mechanisms.
Confocal fluorescence microscopy reveals that H5N1 infection triggers CASK nuclear translocation. Kinase inhibitor screening identifies PKR and SRC signaling as key regulators of CASK’s nuclear entry. Overexpression of the SRC-family kinase HCK and an EGFP-CASK fusion protein in 293T cells, followed by IP-LC-MS/MS analysis, shows a 10-fold increase in S395 phosphorylation. Conversely, the S395A mutation diminishes the HCK-induced nuclear EGFP signal. In H5N1-infected cells, CASK knockout selectively reduces interferon-alpha (Ifna) production without affecting its mRNA levels. RNA-FISH demonstrates a marked increase in the nuclear-to-cytoplasmic ratio of Ifna mRNA in CASK-deficient macrophages, indicating CASK’s role in Ifna mRNA nuclear export. In vivo, myeloid-specific CASK knockout mice exhibit more severe lung inflammation, correlating with significantly decreased lung IFNA levels. Interactome studies (IP-LC-MS/MS and NCBI database analysis) reveal enhanced associations between CASK and CCT4, STIP1, and TNK1 under H5N1 infection. These primary interactions recruit secondary interactors such as IRF7, POLR2C, HNRNPs, and CRM1, facilitating CASK complex binding to the Ifna promoter, co-transcriptional mRNA binding, and more efficient CRM1-dependent mRNA export. In summary, our study elucidates the mechanisms governing CASK nuclear translocation and CASK-mediated IFNA expression in response to H5N1 infection. This work provides crucial insights into the intricate interplay between viral infection, cellular signaling, and the host immune response, highlighting CASK’s pivotal role in orchestrating antiviral defenses.
Results
Upregulation and nucleus translocation of CASK in H5N1-IAV-infected macrophage
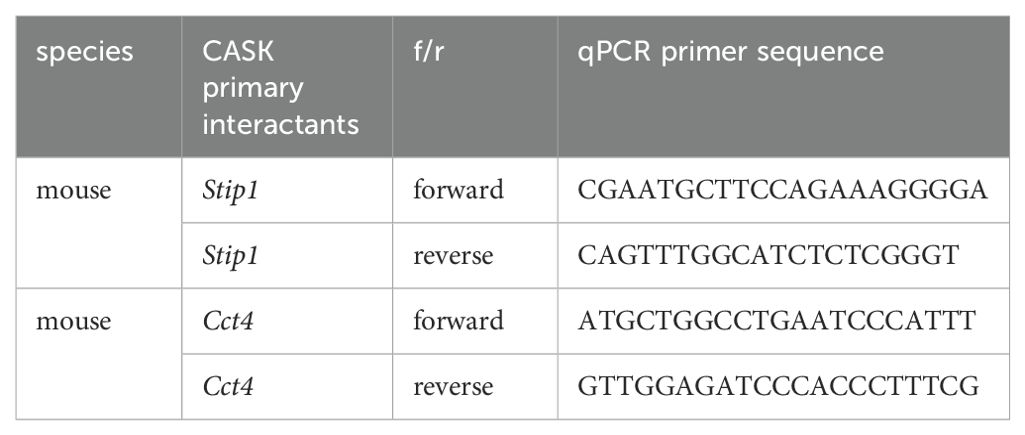
To investigate the roles of CASK in myeloid cells, we incubated mouse bone marrow-derived macrophages (BMDMs) with H5N1-IAV and examined CASK expression and subcellular localization. We observed that the overall expression of CASK increased in a time-dependent manner (Figure 1A). In contrast, nuclear CASK expression increased within the nucleus at 2 h and peaked at 4 h (Figure 1B). The cytosolic abundance of CASK is significantly higher than its nuclear counterpart, making direct side-by-side comparisons challenging. Therefore, we focused our analysis on the time course of CASK expression within the nuclear fraction. Evidence of macrophage infection by H5N1-IAV was confirmed by the expression of the non-structural protein NS1 starting from 4 h post-infection (Figure 1B). These observations indicate that H5N1-IAV infection upregulates CASK expression and triggers its translocation into the nucleus, starting at 2 h and peaking at 4 h post-infection. Immunofluorescence confocal microscopy analysis confirmed the upregulation and nuclear translocation of CASK (Figure 1C), with CASK intensity quantified in Figure 1D.
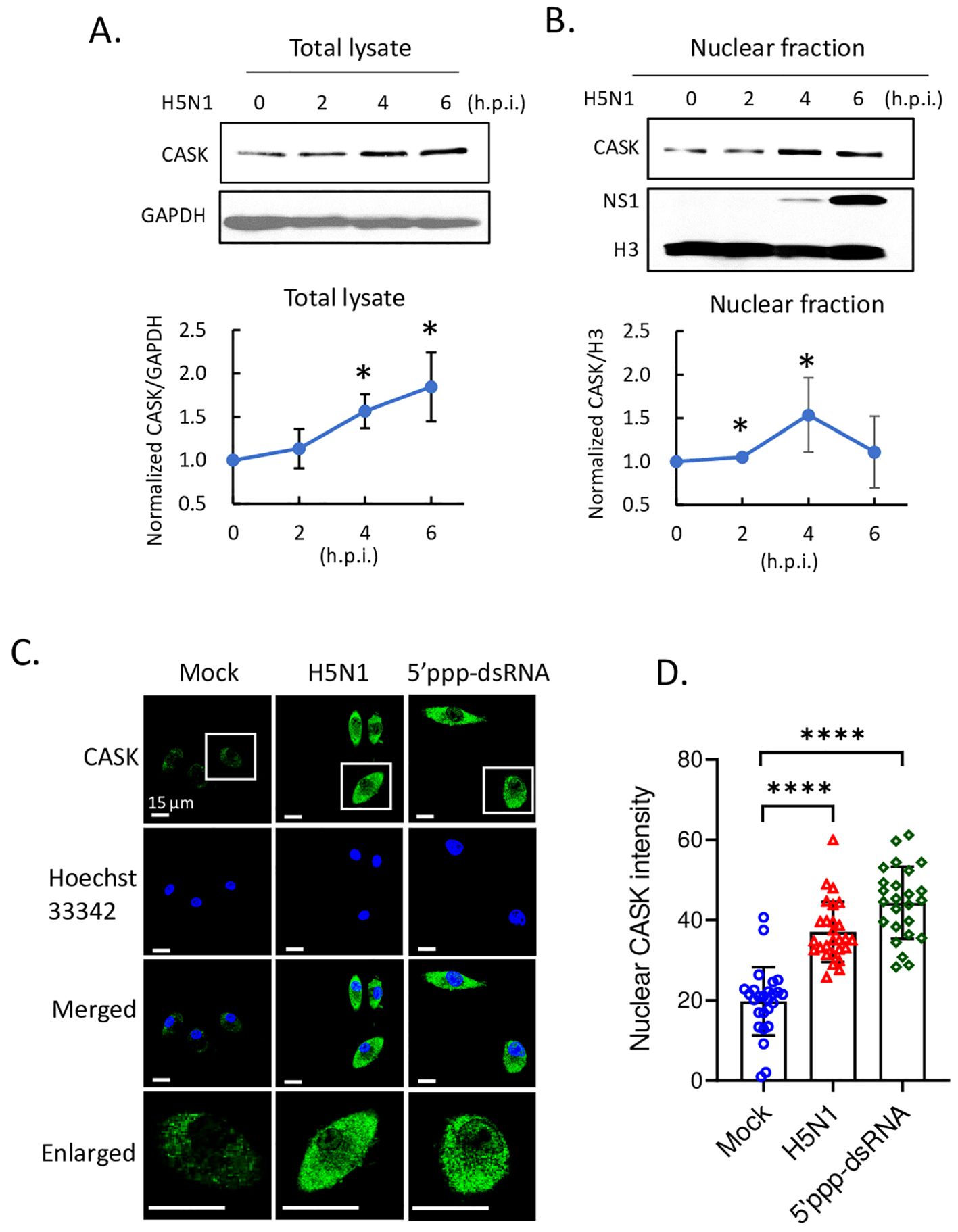
Figure 1. H5N1 infection and RIG-I ligand stimulation trigger CASK upregulation and nucleus entry. Mouse primary GM-MΦ infected with H5N1 MOI=1 for 1 hour and harvested at 0, 2, 4, 6 hours post infection (h.p.i.). (A) Total lysate analyzed by SDS-PAGE and western blot. (B) H5N1 infected primary GM-MΦ are fractionated into nuclear fraction and then analyzed by SDS-PAGE and western blot. H3, histone H3, nuclear marker. NS1, influenza a virus nonstructural protein 1. (A, B) Quantification data are from three independent experiments. (C) primary GM-MΦ are seeded onto coverslip in 24 well and infected with H5N1, MOI=1, or transfected with 5’ppp dsRNA 400 ng/ml, for 3 hours and then fixed with 4% paraformaldehyde (Electron Microscopy Sciences, 15710), stained with anti-CASK antibody, anti-mouse Alexa488 secondary antibody and Hoechst33342, and analyze nuclear CASK by confocal fluorescence microscope. (D) Nuclear CASK intensity is analyzed using LAS-X software. *p < 0.05, ****p < 0.0001 (Student’s t-test).
To elucidate how H5N1-IAV upregulates CASK expression and promotes nuclear translocation, BMDMs were incubated with various ligands capable of activating different nucleic acid receptors and sensors. Notably, transfected 5’ppp dsRNA (a ligand of RIG-I and PKR) and transfected poly (I:C) (a ligand of RIG-I/MDA-5) induced CASK upregulation and nuclear translocation similar to H5N1-IAV, whereas un-transfected poly (I:C) (a TLR3 ligand) and CL097 (a TLR-7/8 ligand) only increased CASK expression in the juxta-membrane region (Supplementary Figure S1).
CASK deficiency impaired the production of IFNA in response to H5N1-IAV by disrupting the nuclear export of IFN-α mRNA
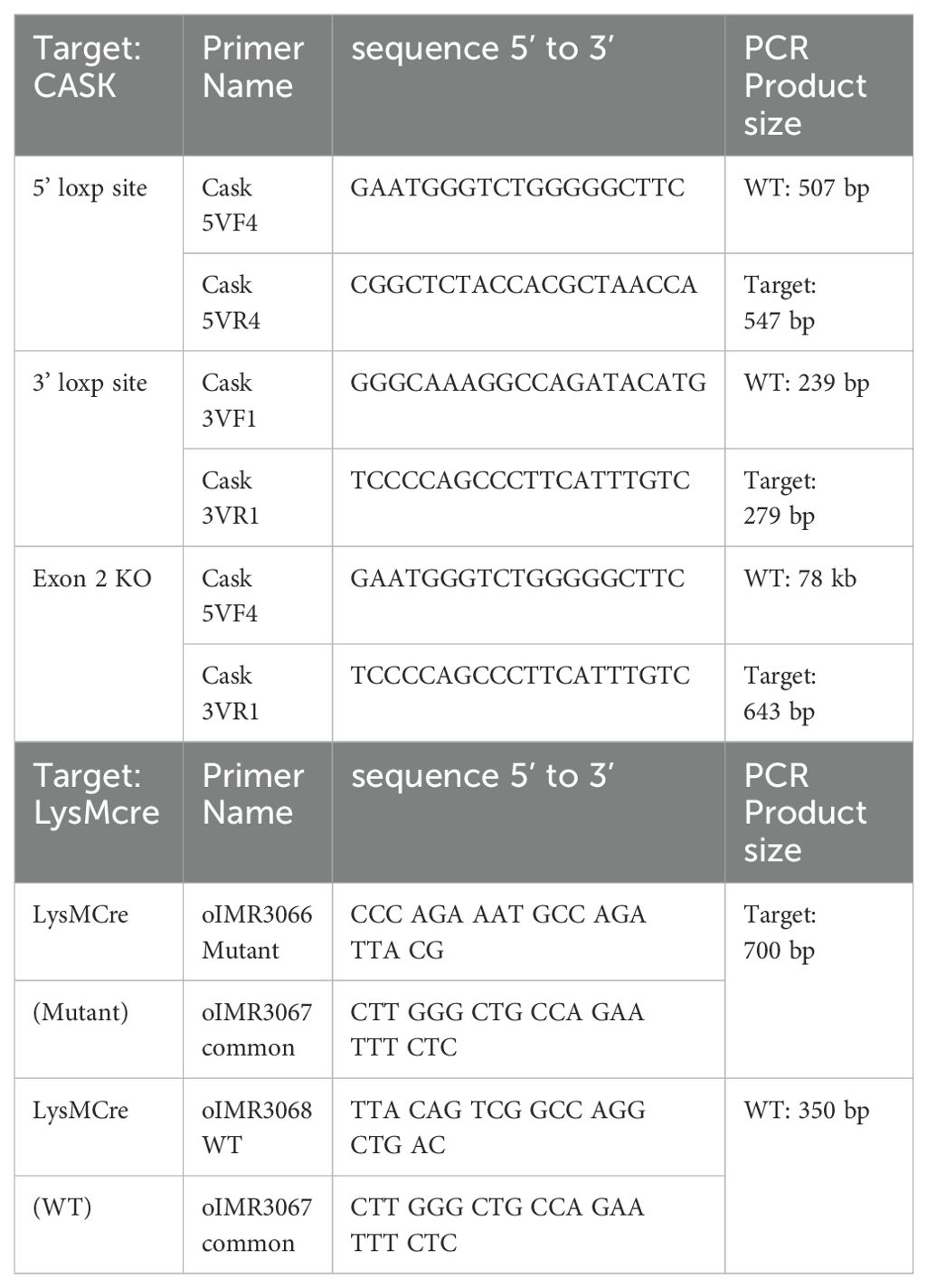
As systemic knockout of Cask is neonatal-lethal in mice (21), we generated mice with tissue-specific Cask knockout by introducing LoxP sites flanking Cask exon 2 on X chromosome (Supplementary Figure S2A). To achieve myeloid-specific knockout of Cask, we crossed heterozygous floxed females (Cask f/x) with LysM-Cre male (Supplementary Figure S2B). As a result, the male offspring (Cask f/y LysM-Cre+) carried the myeloid-specific Cre, leading to the deletion of CASK in myeloid cells such as macrophages and granulocytes (22). We first confirmed the deletion of exon 2 of Cask in genomic DNA by PCR (Supplementary Figure S2C, D) in macrophages from Cask f/y LysM-Cre+ mice. SDS-PAGE and western blot analysis further validated the absence of CASK protein in these CASK-deficient macrophages (Supplementary Figure S2E).
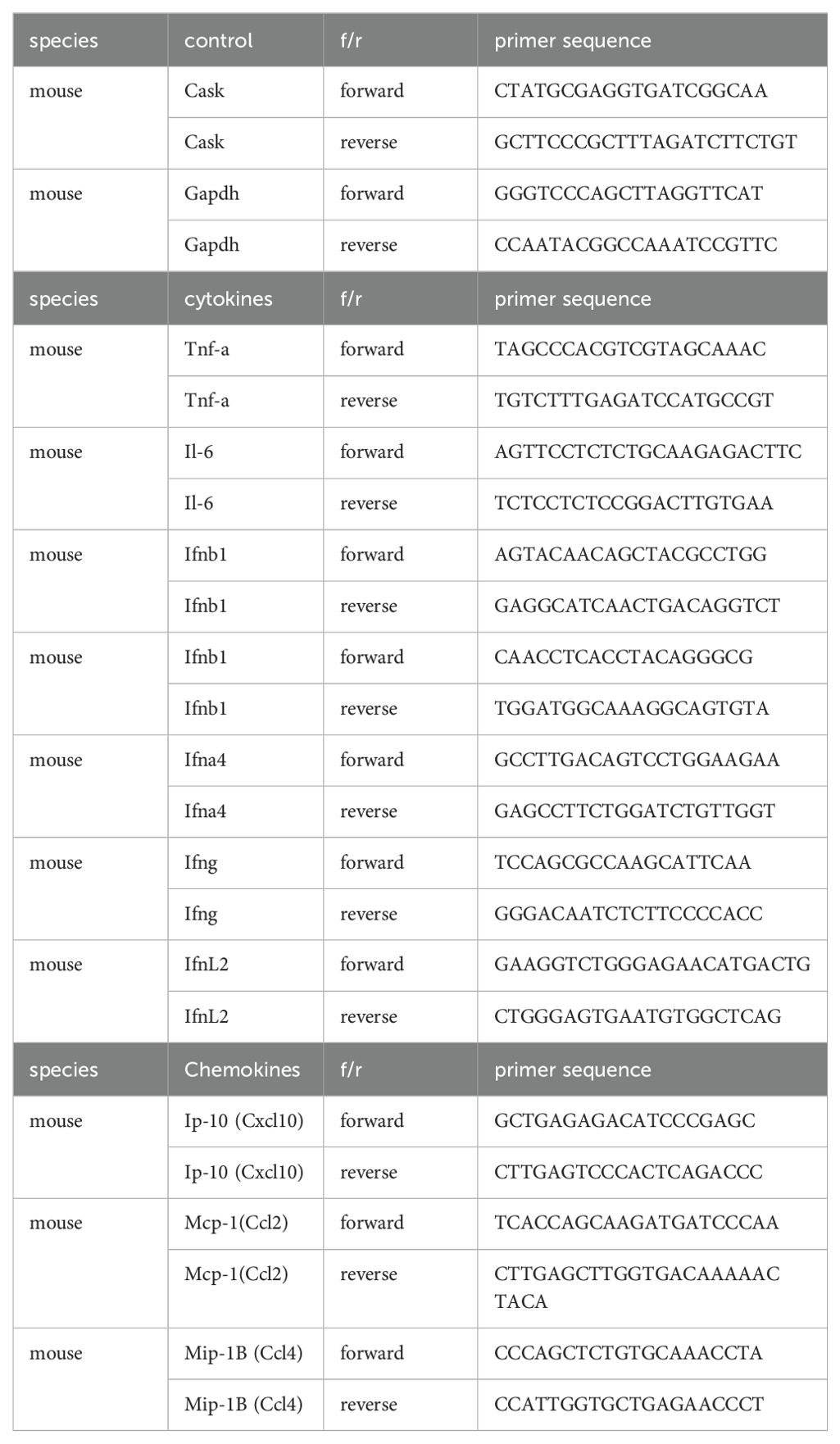
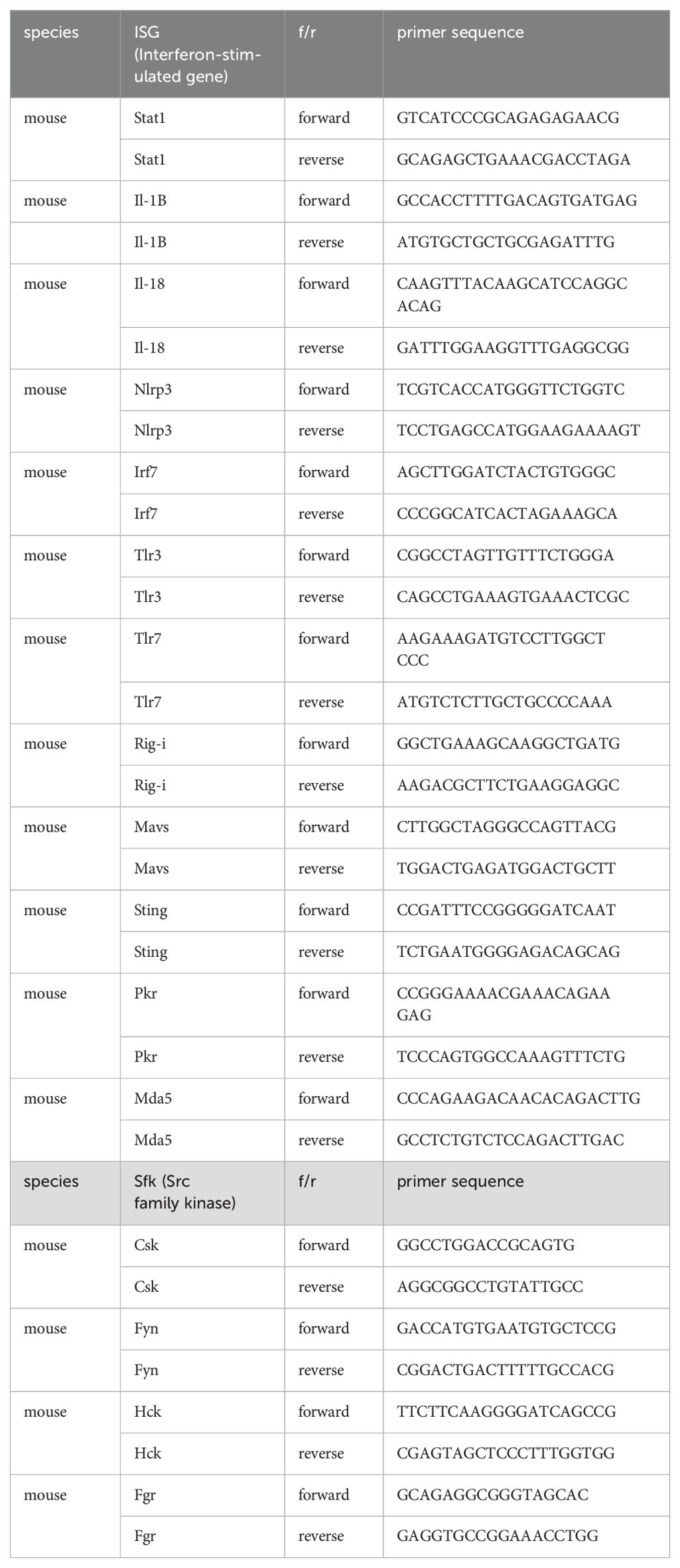
Upon H5N1-IAV infection, macrophages secrete interferon alpha (IFN-α) and pro-inflammatory cytokines (16, 23). To investigate the impact of CASK on cytokine production, we measured the levels of IFN-β, IFN-α, IFN-6, TNF-α, IL-6, IL-1β and IP-10 in the culture supernatant of macrophages at 12 h post-H5N1-IAV (MOI=1) infection. Surprisingly, we observed a 68% reduction in IFN-α in CASK-deficient GM-MΦ, while the levels of other 6 cytokines (IFNB, IFNG, TNFA, IL-6, IL1B, IP-10) remained unaffected (Figure 2A). Despite the 68% decrease in protein expression, the mRNA level of Ifna4 remained unchanged when comparing WT and Cask KO macrophages (Figure 2B). Consistent with the decreased IFN-α level at 12 hours post-infection, the mRNA levels of various interferon-stimulated genes (ISGs) were significantly reduced in Cask knockout (KO) macrophages. The extent of transcriptional induction of these ISGs, including Mda5, Rig-I, Irf7, Stat1, Cxcl10, and Ccl2, reflects the signal intensity of type I interferons. In contrast, the mRNA levels of other interferons (Ifnb1, Ifnl2 and Ifng), proinflammatory cytokines (Il1b, Il18), Pkr, Tlr7 and the inflammasome gene Nlrp3 remained unaffected when comparing WT and Cask KO macrophages (Supplementary Figure S3). In addition, CASK-deficiency does not affect H5N1 IAV viral gene expression (NP and M, Supplementary Figure S3L, M) or viral titer (Supplementary Figure S3N).
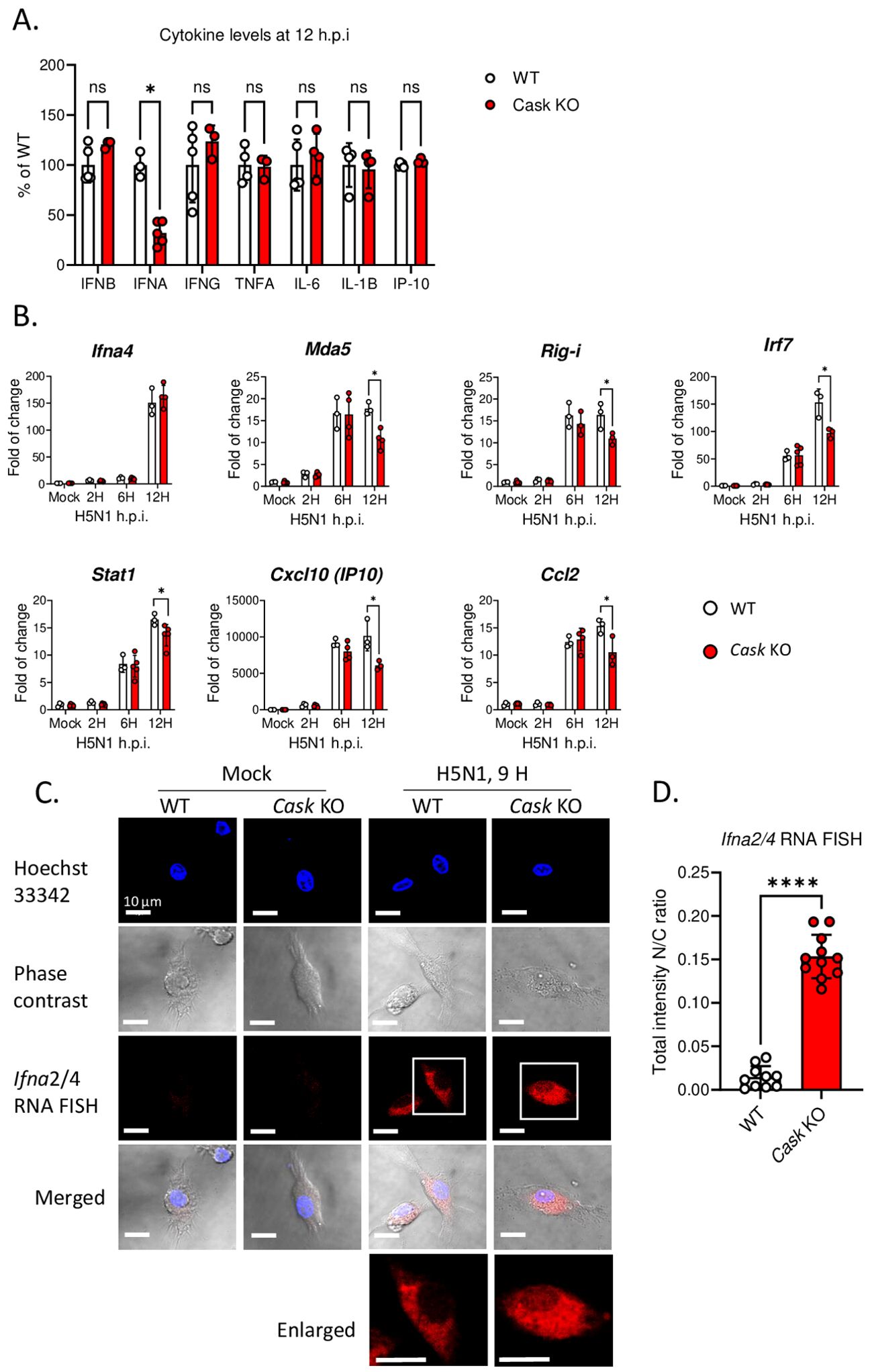
Figure 2. CASK deficiency selectively blunts H5N1-induced IFNA production through perturbation of Ifna mRNA export. (A) Primary GM-MΦ derived from CASK wild type mouse (WT=CaskwtLysMcre+) and myeloid-specific CASK deficient mouse (Cask KO=CaskfloxLysMcre+) were infected with H5N1, MOI=1, for 12 hours, and the culture supernatant were collected and subjected to ELISA analysis of IFNB, IFNA, IFNG, TNFA, IL6, IL-1B, and IP-10. (B) Primary GM-MΦ derived from WT and Cask KO were infected with H5N1, MOI=1, for 2, 6, 12 hours, RNA were extracted for RT-qPCR to evaluate mRNA levels of Ifna4 and interferon-stimulated genes: Mda-5, Rig-I, Irf7, Stat1, Ip-10 and Ccl2. (C) Primary GM-MΦ derived from WT and Cask KO were seeded onto coverslip and infected with H5N1, MOI=1, for 9 hours, cells were then fixed, stained with Hoechst 33342 and hybridize with fluorescent oligonucleotide probes specifically recognize Ifna2 and Ifna4 mRNA sequences. This RNA FISH (fluorescence in situ hybridization) samples were then analyzed by confocal fluorescence microscope. Nucleus and cytoplasm fluorescence intensity were analyzed by MetaMorph software. Total intensity N/C ratio= (fluorescence integrated intensity of nucleus)/(fluorescence integrated intensity of cytoplasm) of each cell were calculated and shown in (D). Scale bar=10 μm. *p < 0.05, ****p < 0.0001 (Student’s t-test).
To understand how CASK-deficiency reduces IFN-α secretion without affecting its transcription, we first ruled out the possibility of CASK involvement in the IFN-α secretory pathway. Our findings revealed no intracellular accumulation of IFN-α in CASK-deficient macrophages following IAV infection (Supplementary Figure S4). We then investigated whether the nuclear export of Ifna mRNA was impaired in CASK-deficient macrophages. Using fluorescence in situ hybridization (RNA-FISH), we observed that most Ifna mRNA was efficiently exported to the cytosol in wild type macrophage infected with H5N1. In contrast, in CASK-deficient macrophages, a higher percentage of Ifna2/4 mRNA remained in the nucleus (nucleus/cytosol ratio: 0.15) at 9 h post H5N1-IAV infection in the CASK-deficient macrophages (Figures 2C, D). This suggests that H5N1-IAV induces CASK translocation into the nucleus to facilitate the export of Ifna mRNA to the cytosol.
Regulation of CASK expression and nuclear translocation by PKR and Src kinases
We further investigated the signaling pathway responsible for the upregulation and nuclear translocation of CASK. Given the crucial role of PKR in antiviral responses (24) and the activation of Src family kinases (SFKs) by influenza virus (25), we examined whether PKR and Src family kinases contributed to the elevation and nuclear translocation of CASK following H5N1-IAV infection. Using confocal microscopy-based screening with various kinase inhibitors (unpublished data), we found that PKR inhibition most effectively suppressed CASK upregulation (Figure 3A). The Src inhibitor (PP2) successfully blocked CASK nuclear translocation. The quantification of nuclear CASK is shown in Figure 3B.
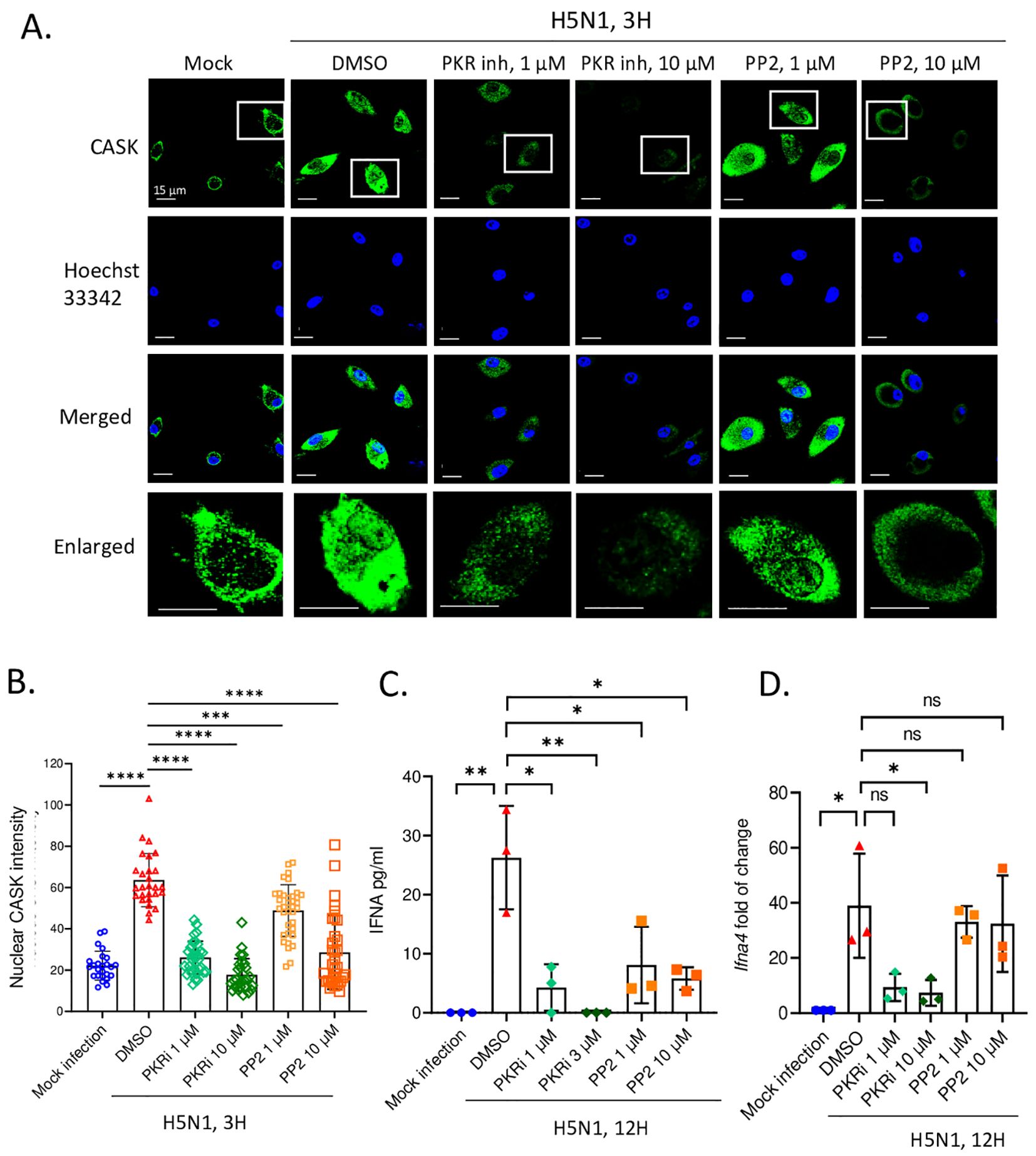
Figure 3. PKR and SRC inhibitors suppress H5N1-induced CASK nuclear translocation and repress IFNA production at transcriptional and post transcriptional levels, respectively. (A-D) Primary GM-MΦ are the attached cells derived from freshly isolated mouse bone marrow cells cultured for 7 days with 10 ng/mL mouse GM-CSF. H5N1 infection was performed in serum-free RPMI medium for 1 hour at 37°C, MOI=1. (A) Primary GM-MΦ are seeded onto coverslip in 24-well overnight and infected with H5N1, MOI=1, for 3 hours and then fixed with 4% paraformaldehyde (Electron Microscopy Sciences, 15710), stained with anti-CASK antibody, anti-mouse Alexa488 antibody and Hoechst 33342, then analyze nuclear translocation of CASK by confocal fluorescence microscope. Scale bar=15 μm. (A) Inhibitors are diluted to the indicated concentrations in RPMI 10% serum culture medium and incubate with H5N1 infected GM-Mf for 3 hours. (B) Quantification of nucleus intensity by LAS-X software to reflect the effects of inhibitors on CASK nucleus translocation. (C, D) H5N1-infected Primary GM-MΦ were treated with indicated kinase inhibitors for 12 hours and culture supernatants were analyzed by IFNA2/4 ELISA (C); cells were harvested with Trizol at 12-hours post-infection and total RNA were extracted. Ifna4 mRNA levels were analyzed by RT-qPCR (D). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student’s t-test).
Of particular interest is the observation that PKR inhibition not only suppressed the production of IFN-α but also its transcription (Figures 3C, D), while the Src inhibitor (PP2) selectively impeded IFN-α translation without affecting its transcription (Figures 3C, D). These data suggest that the Src kinase inhibitor specifically restrains CASK nuclear trans location and IFN-α secretion without interfering with transcription. Importantly, these findings demonstrate a strong positive correlation between the extent of CASK nuclear translocation and the abundance of secreted IFN-α, underscoring the critical role of CASK in modulating the antiviral response.
HCK contributes to CASK nucleus translocation during H5N1-IAV infection
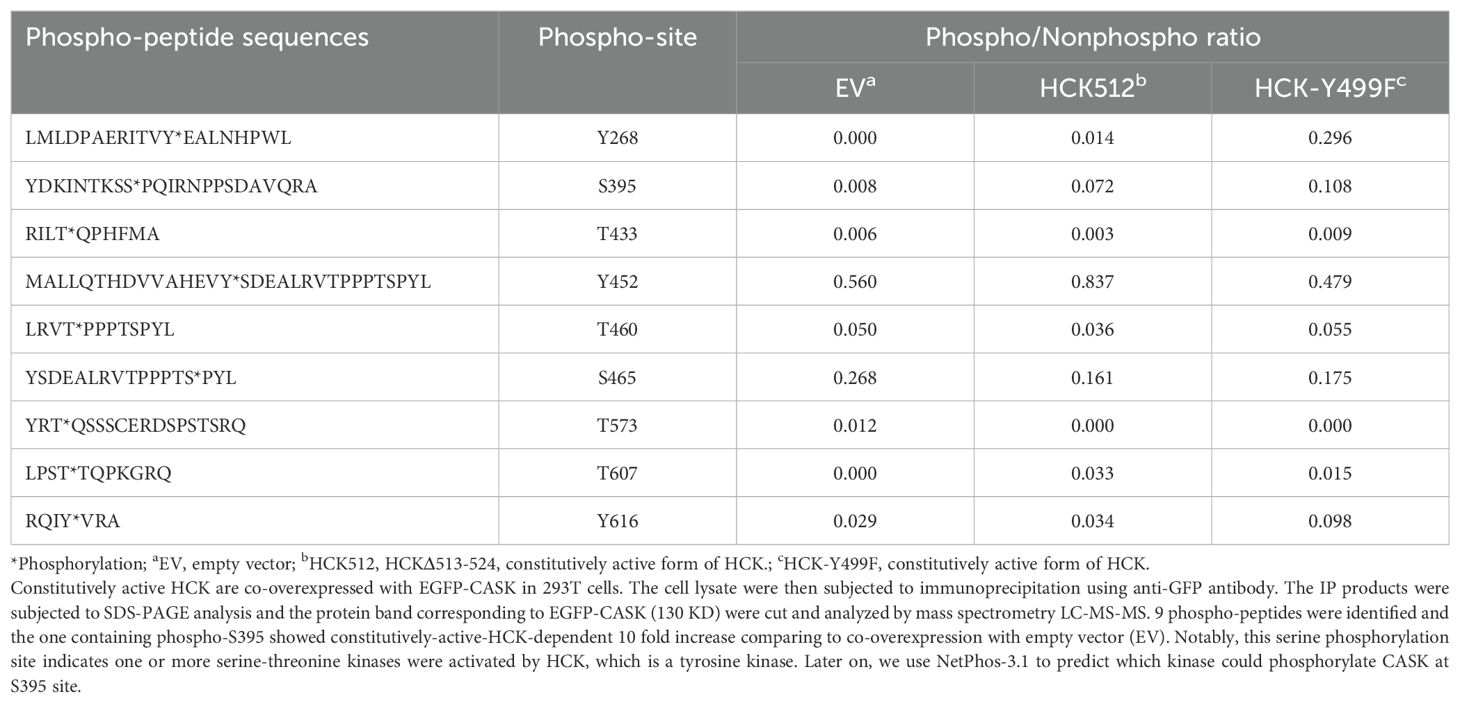
Since the SRC inhibitor PP2 is a general inhibitor for Src family kinases (SFKs), which includes nine members (Csk, Yes, Fyn, Fgr, Lck, Hck, Blk, Lyn, and Frk), we aimed to identify the specific member responsible for the nuclear translocation of CASK. First, we examined the expression of SFKs in macrophages (Csk, Fyn, Fgr, and Hck) following H5N1-IAV infection (Figure 4A). This observation suggests that Hck is the most abundant SFK and may contribute to virus-induced nuclear translocation of CASK. To test this, we generated constitutively active forms of HCK: HCKΔ513-524 (deletion mutant) and HCK Y499F mutant (Y→F mutation) (26), then co-transfected them with an EGFP-CASK construct into 293T cells. Immunoprecipitation with anti-GFP mAb, followed by trypsin digestion and LC-MS/MS analysis, revealed that compared to the control group, the phospho/nonphospho ratio at residue Serine 395 (S395) of CASK increased approximately 10-fold (0.008 to 0.072 by HCKΔ513-524, 0.008 to 0.108 by HCK Y499F), while the other 8 potential phosphorylation sites did not show significant alterations (Table 1).
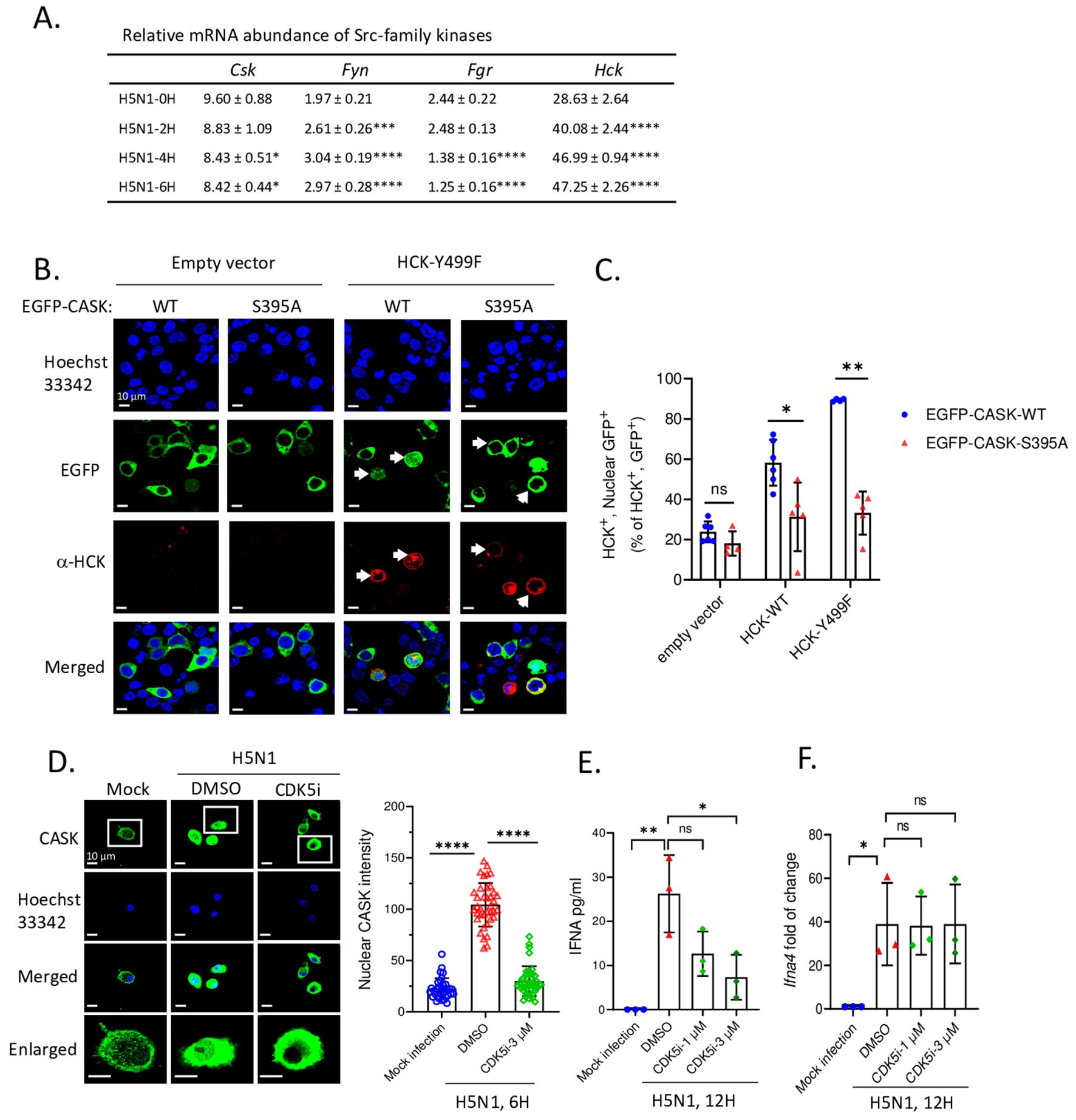
Figure 4. HCK regulates CASK S395 phosphorylation and nuclear translocation via CDK5 during H5N1 infection. (A) There are 4 out of 9 Src-family kinase members express in myeloid cells. Its abundance in H5N1-infected GM-MΦ are quantified by RT-qPCR. (B) 293T cells were seeded onto coverslip and transfected with EGFP-CASK (either wild-type or S395A) and co-overexpressed with either pCMV-empty vector, pCMV-HCK-WT or pCMV-HCK-Y499F. 24 hours after transfection, cells were fixed and stained with anti-HCK antibody, anti-mouse APC conjugated antibody and Hoechst 33342, then analyzed with fluorescence confocal microscope. HCK-expressing cells are counted as either nuclear GFP positive or negative from 15 fields for each treatment group and the statistical analysis is shown in (C). (D) Primary GM-MΦ were mock infected or infected with H5N1, MOI=1. Cells were seeded onto coverslip and treated with DMSO or CDK5 inhibitor 3 μM for 6 hours and then fixed, stained and analyzed by fluorescence confocal microscope. Nuclear CASK intensity from the images acquired in (D) is calculated by Las X software. (E, F) H5N1-infected GM-MΦ were treated with CDK5 inhibitor for 12 hours and culture supernatant were analyzed by IFNA2/4 ELISA (E); cells were lyzed with Trizol, total RNA were extracted and Ifna4 mRNA levels are analyzed by RT-qPCR (F). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student’s t-test).
To confirm the phosphorylation of S395 of CASK is required for nuclear translocation, we generated an EGFP-CASK S395A mutant (S→A mutation) to prevent CASK phosphorylation. In the absence of HCK Y499F, both the wild type (CASK S395) and mutant (CASK S395A) remained in the cytosol (left two columns, Figure 4B). In contrast, transfection of HCK Y499F (red color) induced the translocation of wild type CASK (green color) into the nucleus (3rd column from left, Figure 4B), an effect not observed with the CASK S395A mutant (4th column from left, Figure 4B). The percentage of nuclear GFP-CASK (60% and 90% in WT and 35% in S395A) resulting from the overexpression of wild type HCK and HCK Y499F is shown in Figure 4C. Thus, we concluded that activated HCK induces the phosphorylation of CASK at Ser395, facilitating its nuclear translocation.
Since HCK, a tyrosine kinase, is unlikely to directly phosphorylate S395 of CASK, we used the NetPhos-3.1 software (27) to predict the potential serine-threonine kinase that could phosphorylate CASK at S395. Our analysis revealed that cyclin-dependent kinase 5 (CDK5) (Table 2) is the most likely candidate kinase downstream to HCK. This finding aligns with previous reports showing that CDK5 phosphorylates CASK at S395 in neurons (28). To confirm the involvement of CDK5 in CASK nuclear translocation, we incubated macrophage with H5N1-IAV in the presence of a CDK5 inhibitor. While CASK upregulation remained unaffected, the H5N1-IAV-induced nuclear translocation of CASK (Nuclear CASK intensity average: 105) was suppressed by the CDK5 inhibitor (Nuclear CASK intensity average: 26) (Figure 4D). Thus, we concluded that H5N1-IAV infection activates HCK to phosphorylate CASK at residue S395 via CDK5, thereby inducing the nuclear translocation of CASK. Moreover, the CDK5 inhibitor also decreased H5N1-induced IFN-α production (from average 27 pg/ml in DMSO treatment to average 7 in CDK5 inhibitor 3 µM treatment) (Figure 4E) without suppressing Ifna mRNA levels (Figure 4F). These observations further confirm the crucial role of the HCK-CDK5 axis in regulating CASK nuclear translocation during H5N1-IAV infection.
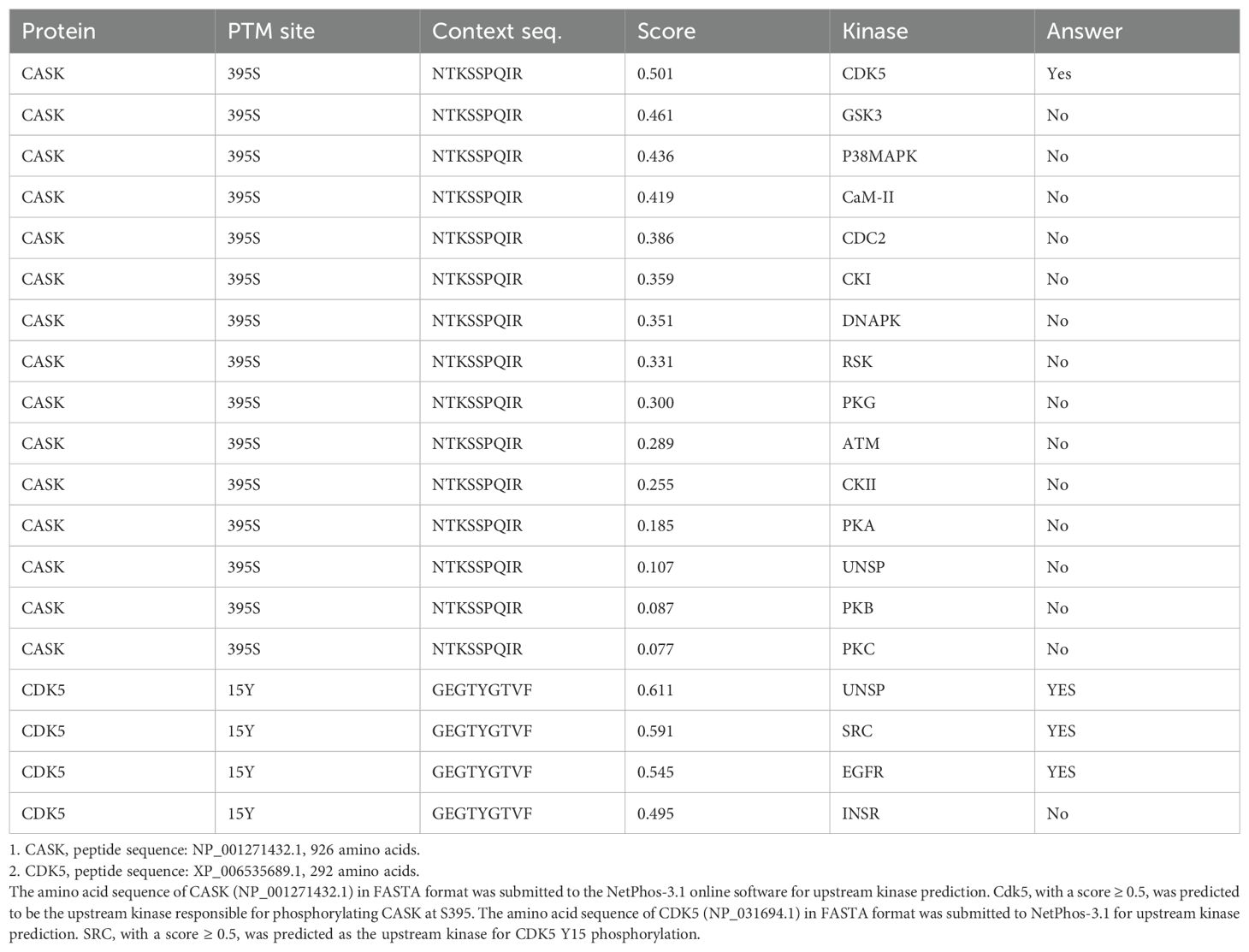
Table 2. Prediction of upstream kinases phosphorylating CASK at S395 and CDK5 at Y15 by NetPhos-3.1.
Myeloid-specific CASK knockout in mice reduce IFN-α production and exacerbate lung inflammation during H5N1-IAV LD50 challenge
To assess the impact of CASK deficiency on attenuating H5N1-IAV-induced IFN-α production in vivo, we intranasally challenged myeloid-specific Cask knockout mice with a median lethal dose (LD50) of H5N1-IAV and then examined lung IFN-α production and histopathology. We observed significantly higher peri-bronchial leukocyte infiltration, alveolar cellularity, and thickening in the lungs of CASK-deficient mice compared to wild-type mice at day 5 post-infection (Figures 5A, B). The extent of H5N1-IAV-induced lung immunopathology in CASK-deficient mice corresponds with the reduced IFN-α production in the lungs (Figure 5C). These findings suggest that CASK plays a crucial role in regulating IFN-α production and contributes to the host’s defense against viral infection.
Figure 5. Myeloid-specific CASK knockout in mice reduce IFNA production and exacerbate lung inflammation during H5N1 LD50 challenge. CaskwtLysMCre+ (WT) and CaskfloxLysMcre+(Cask KO) mice were infected with H5N1 by intranasal challenge of LD50 (750 pfu/mouse), and sacrificed at post-infection day 3 (C) or day 5 (A, B). (A) Haematoxylin and eosin staining of left lung cross section. Left lung histopathology shows peri-bronchial leukocyte infiltration in H5N1 infected groups. (B) The severity of lung inflammation in response to H5N1 infection at post-infection day 5 were quantified by inflammation score. The inflammation score evaluates two parts, peri-bronchial infiltration (0-4 points, 0, none; 1, mild, infiltrate ≤4 cells thick; 2, moderate, infiltrate 5-10 cells thick; 3, severe, infiltrate 20%-50% visualized lumens; 4, diffuse, infiltrate >50% visualized lumens), and alveolar involvement (0-3 points, 0, none; 1, mild, patchy increased cellularity/thickening; 2, moderate, 25%-50% visualized lung with increased cellularity/thickening; 3, severe, > 50% visualized lung with increased cellularity/thickening). Lung section scoring was performed at 4X magnification and divided one whole section into 15 fields. The inflammation scores of 15 fields were averaged to represent an overall score from one mouse (n=3-5). (C) Left lungs were collected at post infection day 3 and lung homogenates were analyzed by IFNA2/4 ELISA. *p < 0.05, ***p < 0.001 (Student’s t-test).
IP-LC-MS/MS and meta-analysis of NCBI database reveal CASK interactants involving in processing of mRNA and CRM1-mediated mRNP nuclear export
We then investigated how nuclear CASK regulates the export of Ifna mRNA to the cytosol. As CASK is a scaffold protein known to interact with many proteins via its PDZ, L27, SH3, and GK domain (1), we explored whether CASK interacts with nuclear proteins involved in the export of messenger ribonucleoprotein (mRNP) to the cytosol. To address this question, macrophages were incubated with H5N1-IAV for 3 h, followed by a 2-step ultracentrifugation process to harvest cytosolic and nuclear fractions. The nuclear fractions were then incubated with Dynabeads® coupled with anti-CASK antibody or an isotype control. Nuclear proteins associated with endogenous CASK were pulled down by the ani-CASK antibody-coupled Dynabeads® then subjected to in-solution trypsin digestion, and analyzed via liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS).
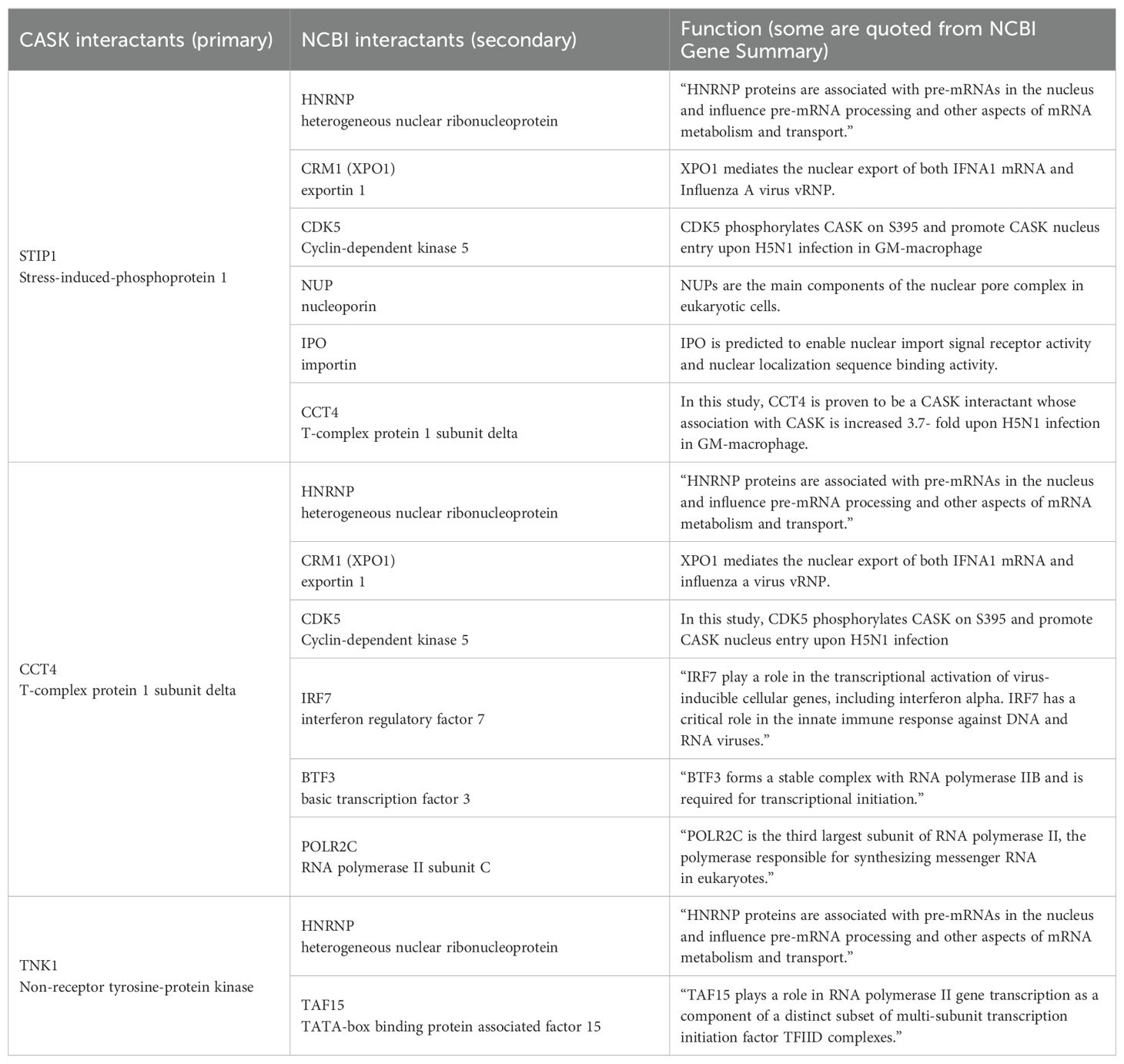
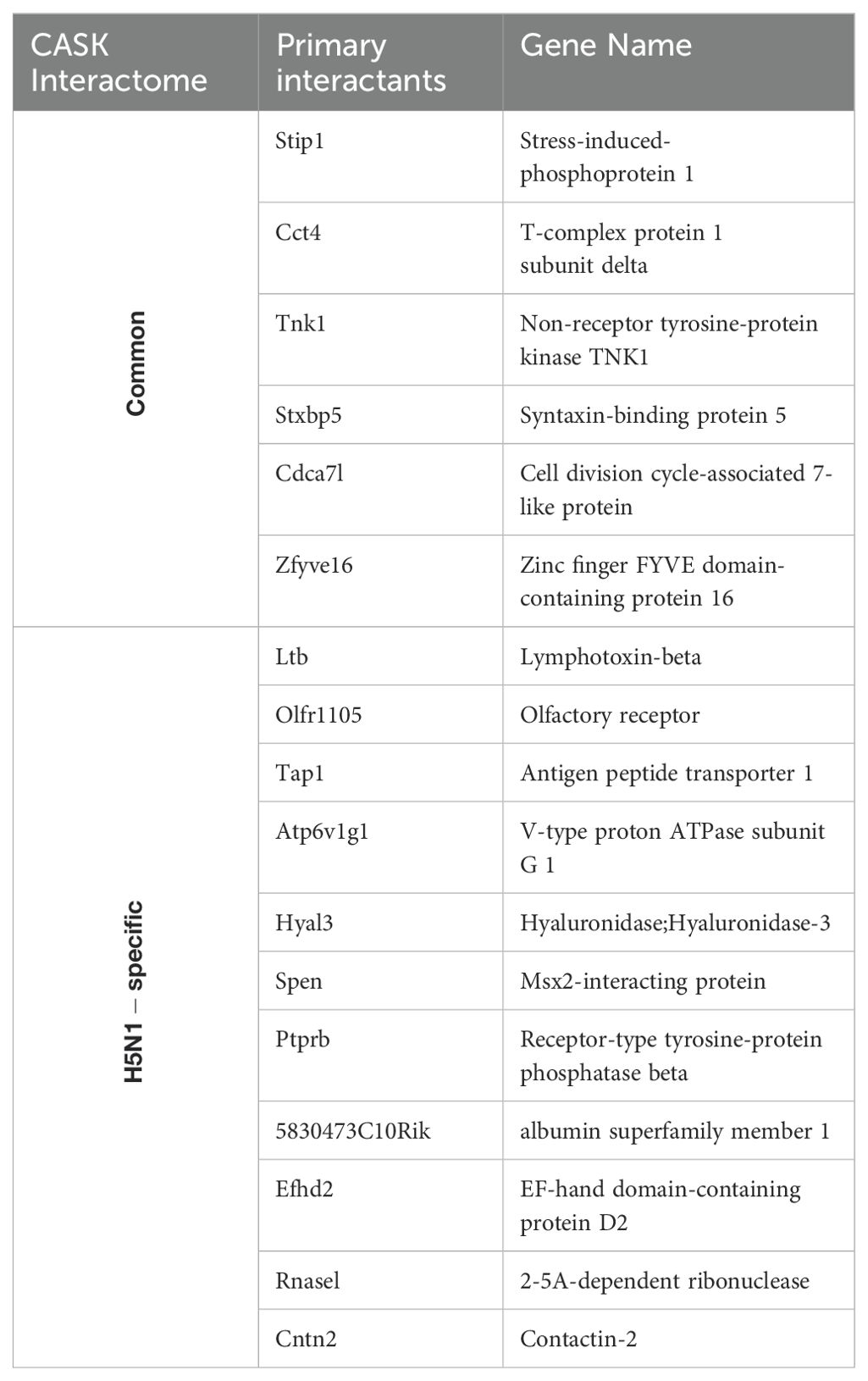
Following H5N1-IAV infection, we observed increased associations between CASK and STIP1 (5.57-fold), CCT4 (3.74-fold), and TNK1 (1.96-fold) (Figure 6A). Additionally, we identified potential associated proteins (secondary interactants) linked with STIP1, CCT4, and TNK1 (Figure 6A, Table 3) using data from the NCBI database. We also identified 11 nuclear proteins specifically associated with CASK post-H5N1-IAV infection (Table 4). The interactome study using CASK IP-LC-MS/MS (Figure 6A, Table 4) and meta-analysis of NCBI/Gene/interactions (Table 3) suggest that pre-existing nuclear CASK forms a complex with primary interactants (STIP1, CCT4, TNK1) in macrophages before stimulation. As CASK is phosphorylated at residue S395, these observations suggest that phosphorylated CASK (pCASK-S395) enters the nucleus and recruits more STIP1, CCT4, and TNK1 to form a complex. Consistent with the effect of CASK deficiency on IFN-α, siRNA-mediated knockdown of STIP1 and CCT4 significantly reduces IFN-α production by around 40% (Figure 6B).
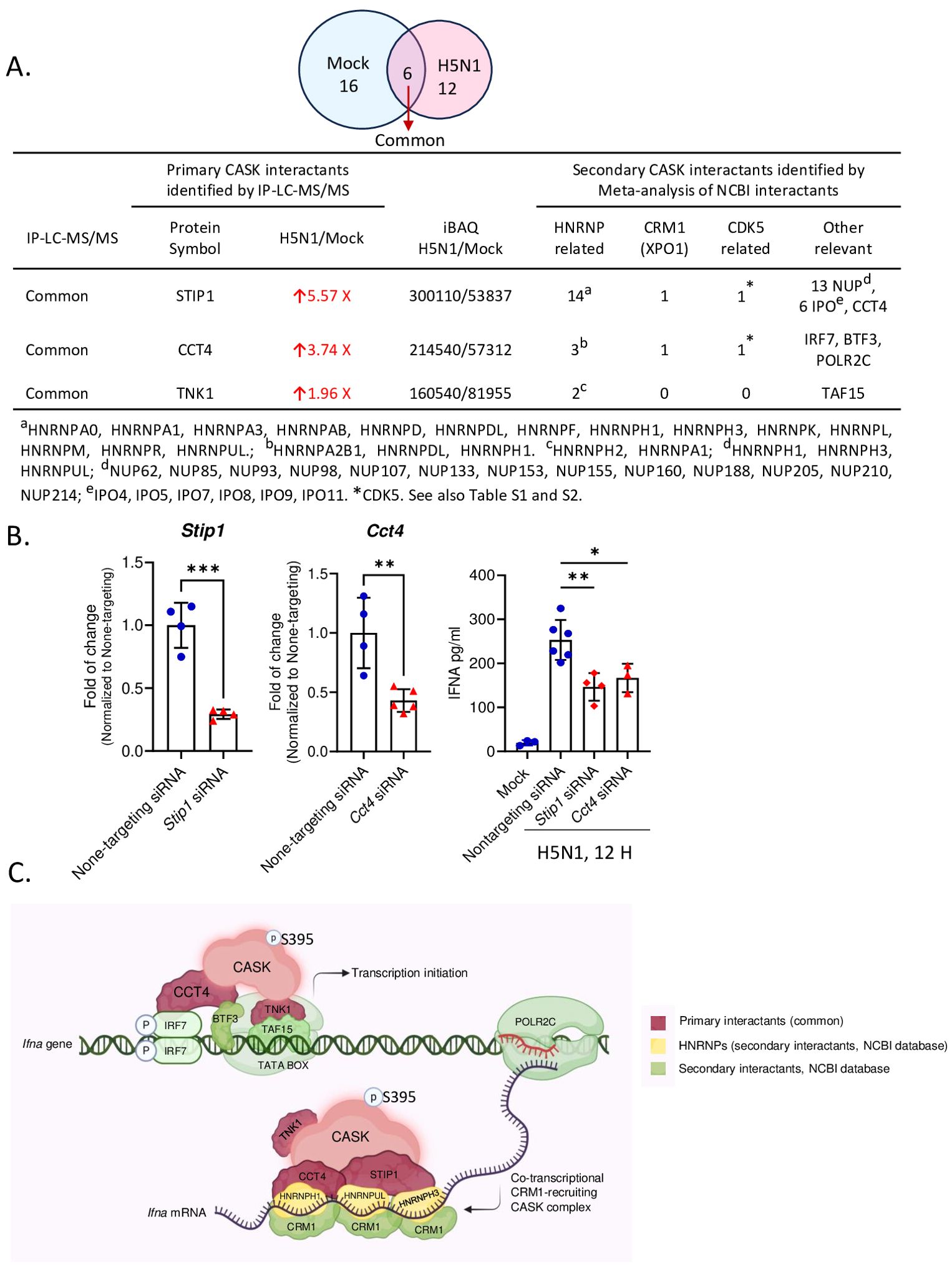
Figure 6. IP-LC-MS/MS analysis of H5N1-infected GM-MΦ and meta-analysis unveiled CASK-interacting proteins involving in mRNA processing and CRM1-mediated mRNA nuclear export. (A) Immunoprecipitation coupled LC-MS/MS analysis of nuclear fractions from H5N1-infected GM-MΦ (MOI=1, 6 hours post-infection) using anti-CASK antibody-conjugated Dynabeads identified CASK-interacting proteins upregulated upon infection. Subsequent NCBI interaction analysis prioritized CASK interactants involved in mRNA export (HNRNPs and CRM1), CASK regulating kinases (CDK5-related), and other relevant proteins, facilitating mechanistic modeling of CASK’s role in anti-viral responses. (B) Silencing of Cct4 and Stip1 suppressed H5N1-induced IFNA production in GM-MΦ. GM-MΦ were transfected with siRNAs targeting Cct4 and Stip1 for 48 hours prior to H5N1 infection (MOI=1) for 12 hours. Knockdown efficiencies were confirmed by RT-qPCR analysis of Cct4 and Stip1 mRNA levels in total RNA extracts. Culture supernatants were collected for measurement of IFNA protein levels by ELISA. (C) The proposed model elucidates how the nuclear CASK complex regulates IFNA mRNA nuclear export in GM-MΦ upon H5N1 infection. Following infection, IRF7 undergoes phosphorylation, translocate into the nucleus, and bind to IFNA gene promoter to initiate transcription. Concurrently, S395 phosphorylated CASK enters the nucleus and interacts with CCT4, STIP1, and TNK1. Through CCT4’s interaction with IRF7 and TNK1’s with TATA-box binding protein TAF15, the CASK complex is recruited to IFNA promoters. Subsequently, via CCT4’s interaction with POLR2C, the CASK complex tracks along with the transcribing IFNA mRNA. The CASK complex then, facilitated by CCT4 and STIP1, binds the nascent IFNA mRNA through RNA-binding HNRNPs and recruits the nuclear export factor CRM1 onto the transcript. This Ifna mRNA-coupled CASK complex could counteract the competition posed by influenza vRNP for CRM1-mediated nuclear export. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test).
Based on these findings, we propose a mechanism model explaining how the CASK complex facilitates the nuclear export of Ifna mRNA (Figure 6C). The model elucidates the process by which the nuclear CASK complex regulates Ifna mRNA nuclear export in GM-MΦ upon H5N1 infection. Following viral infection, IRF7 undergoes phosphorylation, translocates to the nucleus, and binds to the Ifna gene promoter, initiating transcription. Simultaneously, CASK, phosphorylated at S395, enters the nucleus and interacts with CCT4, STIP1, and TNK1. The CASK complex is recruited to Ifna promoters through CCT4’s interaction with IRF7 and TNK1’s association with the TATA-box binding protein TAF15. Subsequently, via CCT4’s interaction with POLR2C, the CASK complex tracks along with the transcribing Ifna mRNA. Facilitated by CCT4 and STIP1, the CASK complex binds to the nascent Ifna mRNA through RNA-binding HNRNPs and recruits the nuclear export factor CRM1 onto the transcript (29). This Ifna mRNA-coupled CASK complex potentially counteracts the competition posed by influenza viral ribonucleoprotein (vRNP) for CRM1-mediated nuclear export, thereby ensuring the efficient export of host antiviral transcripts. The whole picture of this study is summarized in Figure 7 to indicate that the phosphorylated CASK enters nucleus and forms a complex to facilitate CRM1-mediated IFNA mRNP export during H5N1 infection.
Figure 7. Phosphorylated CASK enters nucleus and forms a complex to facilitate CRM1-mediated Ifna mRNP export during H5N1 infection When the H5N1 virus infects macrophages, PKR senses the 5’ tri-phosphate group of the viral genomic RNA, activating a signaling pathway that induces the expression of IFN-α. PKR activity is crucial for the transcriptional activation of Ifna, as treatment with a PKR inhibitor suppresses both CASK and IFN-α expression. During H5N1 infection, the expression and activity of HCK are upregulated, positively correlating with CDK5-mediated phosphorylation of CASK at S395. The S395-phosphorylated CASK translocates to the nucleus, where it forms a complex with STIP1, CCT4, and TNK1, as confirmed by IP-LC-MS/MS analysis. This CASK complex is recruited to the Ifna promoters through CCT4’s interaction with IRF7. Additionally, the complex associates with nascent Ifna mRNA via secondary interactions with HNRNPs and facilitates CRM1 recruitment to the transcript, enabling CRM1-mediated nuclear export. This Ifna mRNA-coupled CASK complex may counteract the competitive binding of influenza vRNP to CRM1, thereby supporting efficient nuclear export of Ifna mRNA.
Discussion
CASK, a multifaceted scaffolding protein, is involved in various cellular processes such as synapse formation, neuronal development, and gene expression regulation. Mutations in the human CASK gene are associated with X-linked brain malformations and mental retardation (2). Despite its abundant expression in myeloid cells, CASK’s role in immune response regulation remains unexplored. Although primarily localized in the cytosol, co-transfection of CASK with Tbr-1, a crucial T-box transcription factor in forebrain development, triggers CASK’s translocation into the nucleus, where it binds to a specific DNA sequence called the T-element to regulate gene expression (9). However, the signals governing CASK’s movement from the membrane to the nucleus, as well as its role in regulating gene expression in immune cells upon activation, have not been investigated.
In this study, we found that H5N1-IAV infection activates PKR, leading to the upregulation of CASK expression and its subsequent translocation to the nucleus. This translocation of CASK is initiated by the activation of the Src family kinase HCK, which induces the phosphorylation of CASK at residue S395 via CDK5. Notably, mutation of CASK at residue S395 abolishes nuclear translocation triggered by HCK, indicating the crucial role of CASK phosphorylation in this process. Based on our data, we hypothesize that the nuclear translocation of CASK in response to H5N1 infection requires two distinct signals: activation of PKR and the HCK-CDK5 axis. PKR is activated by 5’ppp dsRNA, while, according to the literature, the upstream signal for HCK may involve TLR3. TLR3 activation directly induces the phosphorylation of SRC at Y416 (30), leading to its activation. Notably, HCK is a member of the SRC-family kinases (SFK). This two-signal hypothesis explains why TLR3 activation alone is insufficient to drive the nuclear translocation of CASK.
PKR is known to phosphorylate eIF2α, which inhibits the recycling of eIF2β, thereby blocking translation initiation and exerting a global translational inhibition effect. However, studies have shown that under PKR activation, a subset of mRNAs with complex secondary structures and multiple upstream short open reading frames (uORFs) in their 5’ UTR can escape this inhibition. This is because the translation of these mRNAs is regulated at the elongation stage rather than initiation (31). Notably, the mouse Cask mRNA transcript is 8,376 bp long, with a 490 bp 5’ UTR and a 5,106 bp 3’ UTR. Similarly, the human CASK mRNA transcript is 8,749 bp long, featuring a 566 bp 5’ UTR and a 5,487 bp 3’ UTR. These extended 5’ and 3’ UTRs likely play a critical role in regulating the translation of Cask mRNA, enabling CASK expression under PKR activation.
Moreover, CASK regulates the transport of Ifna mRNA from the nucleus to the cytosol, with a significant decrease in IFN-α secretion in CASK-deficient macrophages during H5N1 infection. Furthermore, the myeloid-specific knockout of CASK impedes IFN-α secretion and exacerbates pulmonary inflammation following H5N1-IAV infection. Although CASK knockout does not affect viral replication or titer, it significantly impairs the nuclear export of Ifna mRNA, leading to a reduction in interferon-alpha (IFN-α) secretion. This reduction disrupts the activation of interferon-stimulated genes (ISGs), which are essential for controlling inflammation and orchestrating antiviral defense. As a result, the absence of CASK creates a cytokine signaling imbalance, allowing unchecked pro-inflammatory responses to dominate. This dysregulation contributes to severe inflammatory phenotypes, including heightened lung inflammation in CASK-deficient mice. Furthermore, the findings suggest that without CASK, competition for CRM1-mediated nuclear export machinery favors viral mRNA, further undermining the host’s ability to mount an effective immune response. These observations highlight the significant role of CASK in host defense against H5N1 IAV infection through the regulation of IFN-α production and reveal a novel mechanism of CASK-mediated regulation of gene expression via facilitating mRNA transportation from the nucleus to the cytosol.
It is interesting to note that CDK5 phosphorylate residue S395 located in the L27 domain of CASK protein. The L27 domain, protein-protein interaction module, can form heteromeric complexes that integrate multiple scaffold proteins into supramolecular assemblies required for the establishment and maintenance of cell polarity (32). The L27 domain can occur singly or in duplication in association with other domains such as PDZ, SH3, the guanylate kinase domain, or the serine/threonine protein kinase domain, as seen in the CASK protein. Interestingly, SAP97, a membrane-associated guanylyl kinase (MAGUK) protein crucial for trafficking various proteins essential to cellular functions, undergoes phosphorylation by CaMKII kinase at S39 located within the N-terminal L27 domain. This phosphorylation event modulates the trafficking of SAP97 and its associated proteins (33). Consequently, phosphorylation of the L27 domain appears to play a pivotal role in regulating protein assembly among members of MAGUK family.
In the nervous system, CDK5 is involved in neuron migration, neurite outgrowth and support, and synaptogenesis. Furthermore, CDK5 phosphorylates the L-type voltage-dependent Ca++ channel (L-VDCC), inhibiting the exocytosis of insulin and thereby regulating glucose-stimulated insulin secretion (34). Given CASK’s involvement in brain development and insulin vesicle secretion within pancreatic beta-cells, it would be interesting to ask whether nuclear translocation of CASK is involved in CDK5-mediated neuron migration, synaptogenesis, and insulin secretion. In addition to CDK5, CASK is phosphorylated by protein kinase A (PKA) at residue T724 in the guanylate kinase domain to increase its interaction with Tbr1 (35). Furthermore, it has been reported that CDK5 phosphorylates residue S51, S395, and T846 of CASK in neuronal cells, resulting in CASK distribution to membrane to promote synaptogenesis (28). In this study, we found that H5N1-IAV causes CASK phosphorylation at S395 via the HCK-CDK5 axis, resulting in the nuclear translocation of CASK in macrophages. Interestingly, we did not detect tyrosine phosphorylation of CASK after H5N1 infection. Instead, we found that CASK is phosphorylated at S395, and a CDK5 inhibitor is able to suppress CASK’s nuclear translocation. This observation suggests that HCK-mediated CASK translocation is regulated through CDK5 activation, thus it is interesting to investigate how HCK regulates CDK5 activation in macrophages in the future.
While HCK plays a crucial role in regulating IAV-induced nuclear translocation, the mechanism by which IAV infection activates HCK remains unclear. Currently, there is no conclusive evidence supporting the activation of HCK via PKR- or RIG-I-mediated pathways. However, it has been demonstrated that TLR ligands can activate HCK through a MyD88-dependent mechanism (36). Since IAV is known to activate TLR3 (37), which signals via MyD88 and TRIF, it is plausible that IAV-induced HCK activation occurs through the TLR3-MyD88-HCK pathway. Additionally, the PKR inhibitor is as effective as the CDK inhibitor in suppressing CASK nuclear translocation, raising the intriguing possibility that PKR may directly phosphorylate CASK and interact with the HCK-CDK5 axis. Furthermore, whether the PKR and SRC can activate CASK independently need to be explored in the future.
CASK selectively regulates IFN-alpha but not IFN-beta, suggesting an evolutionary advantage in having these two antiviral type I IFNs governed by distinct regulatory mechanisms. This diversity potentially confers greater benefits in overcoming viral attacks. While there is only one IFN-beta gene, mice possess 14 Ifna isotypes and humans have 13. Influenza A virus NS1 protein interacts with NFX1, the mRNA export receptor for bulk mRNA transport, thereby blocking the nuclear export of poly-A tailed mRNAs (38), including IFN-beta. In contrast, Ifna mRNA nuclear export is CRM1-mediated (39). The high nucleotide sequence conservation in the coding regions containing cis-active structures (40) suggests that all Ifna isotypes likely utilize a CRM1-dependent export pathway. In the context of influenza A virus infection, competition for CRM-1 mediated mRNA export machinery between viral ribonucleoprotein (vRNP, viral genome) (41) and Ifna mRNA is intense. Our findings elucidate a CASK-dependent host strategy that counteracts this competition, favoring the nuclear export of Ifna mRNA.
In addition to H5N1 influenza A virus, many other RNA and DNA viruses exploit the host’s CRM1-mediated nuclear export pathway for viral mRNA export. These include Human Immunodeficiency Virus 1, Human T-cell leukemia virus type 1, Rous Sarcoma Virus, Prototype foamy virus, and Human papillomavirus (42). Whether CASK has evolved to counteract these viruses’ competition for CRM1-mediated nuclear export and promotes the expression of host interferon-α (IFNA) remains to be investigated.
The model presented in Figure 7 is based on interactome data obtained through co-immunoprecipitation followed by proteomic analysis. In addition to host proteins, we identified the interaction between the IAV nucleoprotein (NP) with CASK. Previous studies have demonstrated that IAV NP directly interacts with CRM1 (41), suggesting that CASK may associate with IAV NP either via CRM1 or through direct interaction. Further investigation, including independent validation of these proteomics findings through IP experiments, is needed to determine whether CASK can directly interact with viral proteins, primary interactants and secondary interactants. Moreover, the use of a phosphorylation-null CASK mutant (CASK S395A) could clarify whether CASK phosphorylation contributes to IFN-α expression.
Our findings indicate that CASK’s role in Ifna mRNA export may extend beyond H5N1 infection, offering a framework to understand similar mechanisms in other viral contexts. These results fill a critical gap in understanding the intracellular processes governing Ifna mRNA export in macrophages and its broader implications for immune responses. The described mechanism—CASK-mediated nuclear translocation and CRM1-dependent mRNA export—could be leveraged to address competition between host and viral mRNAs for nuclear export machinery. Future research should investigate CASK’s involvement in regulating immune responses across various viral infections to elucidate its therapeutic potential further.
This study uncovers a novel role for CASK as a key regulator of Ifna mRNA export during H5N1 infection. By serving as a hub for protein-protein interactions, CASK facilitates the efficient nuclear export of antiviral transcripts. This function is analogous to that of other scaffold proteins, such as MAVS and NEMO, which coordinate antiviral signaling. Notably, our findings suggest that CASK’s regulatory functions might also apply to other RNA viruses requiring robust interferon responses for viral control. Supporting this hypothesis, our data (not shown) indicate that dengue virus, another single-stranded RNA virus, similarly induces the nuclear translocation of CASK in primary macrophages. These insights not only clarify CASK’s mechanism of action but also underscore its potential as a therapeutic target to mitigate hyperinflammatory responses during severe viral infections.
In addition to Ifna mRNA export, CASK may also influence Ifna expression at the translational level. This is supported by the observation that cytoplasmic Ifna mRNA is more abundant than nuclear Ifna mRNA, yet the differences between WT and CASK KO mice remain relatively subtle (Figure 2). Considering CASK’s function as a scaffold protein, investigating its cytoplasmic interactome following IAV infection would be a valuable approach to better understand its role in the cytoplasm.
Methods
Reagents and antibodies
Culture media and supplements were purchased from Invitrogen GIBCO. Chemical reagents were from Sigma. Granulocyte–macrophage-colony-stimulating factor (GM-CSF) was from R&D systems. PKR inhibitor was purchased from Santa Cruz (sc-204200) and SFK inhibitor (PP2) was purchased from sigma (529573). 5’pppRNA (tlrl-3prna), 5’ppp dsRNA control (tlrl-3prnac), CL097 (tlrl-c97) and polyI:C (tlrl-pic) were purchased from InvivoGen. Antibodies used in western blot, immunostaining and immunoprecipitation are listed as follows (Table M1):
Culture of mouse primary GM-Mϕ
Male mice aged 8-10 weeks were euthanized, and bone marrow cells were harvested from their femurs and tibias. The isolated bone marrow cells were then washed with 1× RBC lysis buffer to eliminate red blood cells. Subsequently, the nucleated cells were precipitated by centrifugation at 430 g for 5 minutes at room temperature. After discarding the supernatant, the cell pellet was resuspended in 5 ml of RPMI culture medium. This medium consisted of RPMI-1640 (Hyclone, GE Healthcare Life Science) supplemented with 10% FBS (Sigma-Aldrich), 1× penicillin-streptomycin (Gibco), 50 mM 2-mercaptoethanol (Sigma-Aldrich), and 10 ng/mL mouse GM-CSF (R&D systems). A total of 5×106 cells were then seeded into a 9 cm non-coated, sterile petri dish. The culture was maintained by replenishing with fresh medium every 2-3 days for a total of 7 days prior to experimental use.
H5N1 infection of primary GM-Mϕ
Throughout this study, an MOI of 1 was employed for H5N1 infection of GM-MΦ. For RNA isolation and ELISA experiments, GM-MΦ were seeded in 12-well culture plates at a density of 1×106 cells per well. Each well was infected with 0.5 ml of virus suspension (2×106 pfu/mL H5N1) and incubated at 37°C for 1 hour. Subsequently, the virus-containing medium was replaced with 1 ml of fresh culture medium per well. For Western blot experiments, 2×107 GM-MΦ were infected with 4 ml of virus suspension (5×106 pfu/mL H5N1) in 50 ml conical tubes and incubated at 37°C for 1 hour. Following incubation, the virus-containing medium was removed, and the cells were transferred to 9-cm Petri dishes containing 10 ml of RPMI culture medium per dish. H5N1 infected GM-MΦ were then incubated at 37°C for indicated hours post infection.
Stimulation of RIG-I, PKR and MDA5 through transfection of the PRR ligand 5’pppRNA and poly(I:C) in GM-Mϕ
GM-MΦ (1×105) were seeded overnight on acid-etched coverslips (High Precision Microscope Cover Glasses, 12 mm, No. 1.5H; Paul Marienfeld GmbH & Co. KG, 0117520) placed in 24-well plates with 0.5 mL of medium per well, prior to experimentation. Transfection of 5’pppRNA and poly(I:C) was performed using Lipofectamine 2000 (Invitrogen, 11668027). For the transfection mixture: Combine 50 μL of OPTI-MEM (Gibco, 31985070) with 2 μL of 0.1 μg/μL ligand stock in a 1.5 mL Eppendorf tube labeled “A.” In a separate 1.5 mL Eppendorf tube labeled “B,” mix 50 μL of OPTI-MEM with 2 μL of Lipofectamine 2000. Gently mix the contents of tubes A and B and incubate for 20 minutes at room temperature. The transfection mixture was then added to the GM-MΦ cultures in the 24-well plates containing 400 μL of culture medium per well. Cells were incubated at 37°C in a 5% CO2 incubator for 3 or 6 hours, after which they were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, 15710).
Fluorescent confocal microscopy
GM-MΦ or 293T cells (1×105) were seeded overnight on acid-etched coverslips (High precision microscope cover glasses 12 mm, No. 1.5H (Paul Marienfeild GmbH & co. KG, 0117520)) in 24-well plates, with 0.5 mL medium per well, prior to experimentation. Following treatment, cells were washed and fixed with 300 μl of 4% paraformaldehyde (Electron Microscopy Sciences, 15710) in PBS for 20 minutes, then washed twice with 300 μl of 1× PBS. Cells were permeabilized with 0.5% Triton X-100 in 1× PBS for 10 minutes, followed by two PBS washes. Blocking was performed using 10% BSA in PBS for 1 hour at room temperature, followed by two additional PBS washes. Primary antibody incubation was conducted overnight at 4°C using antibodies diluted 1:50 in 3% BSA/PBS. For CASK staining, anti-CASK antibody (Sigma, SAB5200069) was used, while anti-HCK antibody (Santa Cruz, 101428) was employed for HCK staining. After two PBS washes, cells were incubated with secondary antibodies (diluted 1:200) for 1 hour. Goat anti-mouse Alexa-488 (Jackson ImmunoResearch Labs, 115-545-146) was used for CASK detection, and donkey anti-mouse-Alexa 647 (Jackson ImmunoResearch Labs, 715-605-150) for HCK detection. Following secondary antibody staining, cells were washed twice with 1× PBS and incubated with 0.1 μg/mL Hoechst 33342 in 1× PBS for 30 minutes at room temperature. After two final PBS washes, slides were prepared for imaging. Image acquisition was performed using a Leica TCS SP8 X-FALCON confocal microscope. To determine optimal fluorescence gain settings, secondary-antibody-only controls were employed, which should exhibit minimal signal under the selected gain values.
Influenza A virus H5N1 propagation and titer determination by plaque assay
H5N1 in this study is derived from recombinant PR8 strain expressing HA and NA from VN1203 (VN HA, NA) and was generated as described previously (43). The multi-basic HA cleavage site within VN HA, NA strain is replaced by that of a low-pathogenicity avian influenza virus (44). The H5N1 virus is cultured and expanded using MDCK cells. Virus titer is determined using MDCK cells for plaque assay (45).
Cytokine measurement by ELISA and multiplex immunoassay
The concentrations of mouse IFN-α in media from macrophages were measured by ELISA according to the manufacturer’s instructions (Invitrogen, BMS6027).
The level of mouse IL-1β, IFN-β, IL-6, TNF-α, IP-10 and IFN-γ in GM-Mϕ culture supernatant was measured by Multi-Plex Immunoassay (MPI) performed by Inflammation Core Facility (Institute of Biomedical Sciences, Academia Sinica, Taiwan). Antibody conjugated magnetic beads were incubated with cytokine containing samples, washed and incubated with biotinylated antibody and subsequently Streptavidin-Phycoerythrin (PE). The fluorescence levels of the beads were measured by Bio-Plex® 200 system (Bio-Rad, USA) and the concentration of the cytokines were calculated with standard. All assays were protected from light and performed at room temperature.
Extraction of total cell lysate for western blot and ELISA
Prepare a cell lysis solution containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1X Phosphatase Inhibitor Cocktail (Roche, 04906837001), and 1X Protease Inhibitor Cocktail (Roche, 04693132001). Resuspend the cell pellet in the lysis solution and incubate on a rotary tube mixer at 4°C for 1 hour. Centrifuge the lysed cells at 16,000 g for 10 minutes at 4°C. Collect the supernatant (total cell lysate) into a clean, labeled Eppendorf tube. Dilute the total cell lysate 10X or 20X, and measure the protein concentration using the DC Protein Assay Kit (Bio-Rad, 5000111). For western blot, adjust the samples to equal protein concentrations and mix with 6X sample buffer. Load 25 μl of the samples and separate them by SDS-PAGE using a constant voltage of 50–100V. For IFNA ELISA, the total cell lysate was subjected to buffer exchange using Pierce Protein Concentrators PES, 10K MWCO (Thermo Scientific, 88513) spin columns. The proteins were then resuspended in 120 μl of 1X PBS for the following ELISA experiments.
Extraction of nuclear proteins for western blot
Nuclear and cytoplasmic fractions were separated using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo, 78833). To enhance protein preservation, phosphatase inhibitor cocktail (Roche, 04906837001) and protease inhibitor cocktail (Roche, 04693132001) were added to the extraction reagents, with one-quarter tablet of each per 1 mL of CERI reagent (Thermo, 78833) and 0.5 mL of NER reagent (Thermo, 78833).
For each treatment, 2×107 cells were utilized. Cells were washed with 1× PBS and transferred to a 1.5 mL microcentrifuge tube, then pelleted by centrifugation at 500 g for 3 minutes. The supernatant was carefully removed, leaving the cell pellet as dry as possible. The cell pellet was resuspended in 200 μL of ice-cold CERI and incubated on ice for 10 minutes. Subsequently, 11 μL of ice-cold CERII (Thermo, 78833) was added, and the mixture was homogenized by pipetting. After 1 minute of incubation on ice, the tube was centrifuged at 16,000 g for 5 minutes. The supernatant (cytoplasmic extract) was transferred to a clean microcentrifuge tube. To minimize cytosolic contamination, the insoluble pellet was washed with 1 mL of 1× PBS, and the supernatant was carefully removed, leaving the pellet as dry as possible.
The insoluble pellet containing nuclei was then suspended in 100 μL of ice-cold NER. The suspension was vortexed at the highest setting for 15 seconds every 10 minutes, for a total of 5 cycles. Following this, the tube was centrifuged at 16,000 g for 10 minutes. The resulting supernatant (nuclear extract) was transferred to a clean microcentrifuge tube. Protein concentrations in each fraction were determined using the DC™ Protein Assay Kit.
RNA isolation, reverse transcription and qPCR
Monolayer cells in 12-well (1×106 cells) are lysed with 0.4 ml Trizol®Reagent (Invitrogen, 15596-026), homogenizing the lysate by vortex for 1 minute. Using TriRNA Pure Kit (Geneaid, TRP200) for RNA extraction and following the manufacture’s instruction. RNA concentrations were measured using NanoDrop Microvolume Spectrophotometers.
Reverse transcription was performed using RevertAid™ First Strand cDNA Synthesis Kit (Thermo, K1622). 1 µg of RNA was dissolve in RNase-free water in a final volume of 23 µl followed by adding 2 µl of 100 µM oligo(dT) primer and 1 µl of RiboLock™ Ribonuclease Inhibitor (20U/µl) (Thermo Scientific, EO0381), then incubate at 65°C for 5 minutes. Placing the tube on ice and add the following components: 5X reaction buffer 8 µl, 10 mM dNTP mix 4 µl, and Reverse transcriptase 2 µl. Mix gently then incubate the mixture at 42°C for 60 minutes. Terminating the reaction by heating at 72°C for 10 minutes, and store cDNA samples in -20°C.
40 µl cDNA samples are diluted to 200 µl for the following real-time PCR reaction. Using 2 µl diluted cDNA, 5 µl 2× Luminars Color HiGreen qPCR Master Mix (Thermo Scientific, K0393), 2.4 µl d2H2O, 0.6 µl 10 µM (forward primer + reverse primer) for each reaction in 384 well. The reaction condition is as follows: hot start 95°C, 5 minutes, denaturing 95°C, 10 seconds, annealing 60°C, 1 minute, elongation 72°C, 10 seconds, 40 cycles from denaturing to elongation. Primers for qPCR are listed as follows (Tables M2 and M3):
Myeloid-specific CASK knockout mice: generation, breeding strategy and genotyping
Generation of CASK flox mice by CRISPR-mediated gene editing
Caskflox mice were generated by CRISPR-mediated pronuclei genome editing. Lox P sites are designed to be flanking CASK exon 2 following a stepwise procedure. Lox P site was first inserted into 5 prime side of CASK exon 2 in pronuclei and followed by in vitro fertilization and zygote microinjection into surrogate mother mice. The littermates are screened for 5 prime lox P site by genotyping. Pronuclei from the oocytes of female carriers are used for 3 prime lox P site insertion and in vitro fertilization. The littermates are screened for 5 prime lox P site and 3 prime lox P site. Double positive male carriers are founders of CASKflox strain.
Caskflox mice are crossed with C57BL/6JNarl mice (provided by National Laboratory Animal Center (NLAC), NARLabs, Taiwan) for 3 generations to dilute and minimize potential CRISPR-mediated off-target effects. N4 Caskflox mice are then crossed with LysM-Cre mice to generate myeloid-specific Cask knockout offsprings. Male offsprings with genotype Caskflox LysMCre+ are myeloid-specific Cask KO and Caskwt LysMCre+ are littermate controls.
For genotyping, mouse genomic DNA is obtained from 3 mm tail-tip cut overnight digested using 200 µl Tail Digestion Buffer (0.1M Tris-HCl, 5mM EDTA, 0.5% SDS, 0.2M NaCl) with 0.1 mg/ml Proteinase K (Thermo Scientific, EO0491). After overnight digestion, add 200 µl isopropanol to precipitate gDNA. Centrifuge at 16000 g for 10 minutes at room temperature. Remove and discard supernatant. Air dry the pellet then dissolve gDNA in 60 µl autoclaved d2H2O. gDNA is then used for genotyping PCR. The PCR condition is as follows: Initial denaturation 95°C 1 minute, denaturation 95°C 20 seconds, annealing 58°C, 20 seconds, elongation 72°C, 1 minute, final extension 72°C 7 minutes, 50 cycles from denaturation to elongation. Each mouse was genotyped for 5 prime loxP site, 3 prime loxP site, LysMCre mutant and LysMcre wildtype. Genotyping primers are listed as follows (Tables M4):
C57BL/6J mice were purchased from the National Laboratory Animal Center (NLAC, Taiwan). All mice were bred and maintained in specific pathogen-free conditions at Academia Sinica SPF animal facility (AS core). Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at AS core (protocol ID 19-02-1288). In this study, we used 8–12-week-old male.
Interferon alpha mRNA subcellular localization measured by RNA FISH and fluorescence confocal microscopy
The probe sequences are designed using online software (Stellaris Probe Designer version 4.2) (46). Specific probes, 24 probes each, for Ifna2 and Ifna4 are combined to represent Interferon alpha mRNA, and ordered as Stellaris RNA FISH probes, custom assay with Quasar 570 dye. Probes are reconstituted to 12.5 µM using TE buffer (10 mM Tris-HCl, 1mM EDTA, pH8.0). Probe sequences are listed as follows (Tables M5):
Hybridization mixture contains 125 nM probe diluted using Stellaris RNA FISH Hyrbidizatiob Buffer (Biosearch, SMF-HB1-10). Mouse primary GM macrophage were seeded onto coverslip at the density of 6X104/well in 24-well, and subjected to H5N1 infection for indicated time points before fixation using paraformaldehyde (4% paraformaldehyde (Electron Microscopy Sciences, 15710)) in 1XPBS (RNase free) for 15 minutes. The fixed cells are then washed with 1XPBS (RNase free) and immersed in 70% ethanol 1XPBS (RNase free) to make plasma membrane permeable. Use 0.5 ml wash buffer A (20% Wash Buffer A stock, SMF-WA1-60, 10% deionized formamide in ultrapure nuclease-free water) to immerse the coverslips (High precision microscope cover glasses 12 mm, No. 1.5H (Paul Marienfeild GmbH & co. KG, 0117520)) for 5 minutes before hybridization. For hybridization step, 100 µl of hybridization mixture were used to immerse the coverslip and incubate at 37°C for 4 hours in dark. Followed by wash step 1, use 0.5 ml wash buffer A (20% wash buffer A, SMF-WA1-60, 10% deionized formamide in ultrapure nuclease-free water) to immerse the coverslips at 37°C for 30 minutes in dark. Aspire the wash buffer and stain the fixed cells with DAPI using 0.5 ml DAPI nuclear stain (0.5 ng/ml DAPI in Wash buffer A) at 37°C for 30 minutes in dark. Aspire DAPI nuclear stain and immerse the coverslip with wash buffer B (20% Wash Buffer B stock, SMF-WB1-20) at room temperature for 5 minutes in dark. Dry the coverslips by dripping off wash buffer B and mount the coverslip onto microscope slide using anti-fade mounting solution. Seal the coverslip with nail polish and allow to dry before storing at -20°C. Use confocal microscope to capture 570 nm emission light and record interferon alpha mRNA subcellular localization. RNA FISH confocal images were analyzed for nucleus/cytoplasmic ratio using MetaMorph software.
CASK EGFP fusion protein and constitutively active HCK constructs
CASK coding sequence were constructed into pEGFP-N vector (Addgene, 6085-1) in EcoRI site to make N-terminal EGFP fusion CASK. Constitutively active HCK(Y499F) were constructed into pCDNA4-V5/His-A vector (Addgene, V86120) in EcoRI site by site-directed mutagenesis.
Co-overexpression of CASK-EGFP fusion protein and HCKCA in 293T
15 µg of CASK EGFP fusion protein vector and 15 µg of constitutively active HCK Y499F (HCKCA) were mixed in 500 µl Opti-MEM (Gibco, 31985070) with 30 µl of Lipofectamine 2000 (Invitrogen, 11668027) to form complex for transfection of each 10-cm culture dish (1× 107) 293T cells for 24 hours.
Identification of HCK-induced CASK phosphorylation site by IP-LC-MS/MS
10 × 107 293T cells were transfected with CASK-EGFP and HCKCA for 24 hours to yield 10 mg of protein for immunoprecipitation. Lysate was precleared by 40 µl of 50% Protein A beads (rProtein A Sepharose Fast Flow (GE Healthcare, 17-1279-02)) in final 1 ml wash buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.05% Triton-X100, 1X Phosphatase inhibitor cocktail (Roche, 04906837001), 1X Protease inhibitor cocktail (Roche, 04693132001)), and on rotary tube mixer at 4°C for 1 hour.
To pull down CASK-EGFP fusion protein, we use anti-GFP protein A beads (rProtein A Sepharose Fast Flow (GE Healthcare, 17-1279-02)). Preparation of Anti-GFP Protein A Beads: Mix 40 µl of 50% protein A with 5 µl anti-GFP (Abcam, ab290) in final 1 ml binding buffer. The binding buffer contains 50 mM Tris-HCl, 150 mM NaCl, 0.1% Triton-X100, 1 pill of phosphatase inhibitor cocktail (Roche, 04906837001) and 1 pill of protease inhibitor cocktail (Roche, 04693132001). Incubate the mixture on a rotary tube mixer at 4°C for 2 hours. Wash with 1 ml wash buffer for 3 times by microcentrifuge at 1600g for 1 minute.
Precleared lysate (10 mg) and anti-GFP Sepharose beads were incubated in a final volume of 1 mL at 4°C for 2 hours on a rotary tube mixer. The beads were washed three times with 1 mL wash buffer, once with 1 mL wash buffer containing 0.1% SDS, and once with 1 mL PBS. 50 μL of 2× SDS sample buffer was added to the post-IP protein A Sepharose beads, which were then boiled at 95°C for 10 minutes before SDS-PAGE. SDS-PAGE was performed using a 10% SDS-acrylamide gel, and the gel was stained with Rapid stain (786-31, G-BIOSCIENCES) to visualize the CASK-EGFP band. The gel band corresponding to the CASK-EGFP molecular weight (130 kDa) was excised. In-gel digestion of CASK-EGFP was performed using trypsin (Thermo Scientific, 90059) and chymotrypsin (Thermo Scientific, 90056). The resulting peptides were extracted and subjected to LC-MS/MS for screening of CASK phosphorylation modifications. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD057889 and 10.6019/PXD057889 for “CASK phosphorylation pattern in the presence of constitutively active HCK”.
Prediction of phosphorylation sites and its corresponding upstream kinases by NetPhos-3.1
We submit full length CASK 926 amino acids sequence (NP_001271432.1) and CDK5 292 amino acids sequence (NP_031694.1) in FASTA format, respectively, for NetPhos-3.1 (DTU Health Tech) prediction. Our IP-LC-MS/MS study shows that CASK S935 is upregulated upon co-transfection with constitutively active HCK. We then use NetPhos3.1 to predicts which kinase among the 17 kinases (ATM, CKI, CKII, CaM-II, DNAPK, EGFR, GSK3, INSR, PKA, PKB, PKC, PKG, RSK, SRC, cdc2, cdk5 and p38MAPK.) can phosphorylate CASK at S395. CDK5 has a score above 0.5 and hence the positive prediction. We then ask which tyrosine kinase is upstream to CDK5. NetPhos3.1 shows that SRC has a score above 0.5 (0.591), a positive prediction kinase on CDK Y15 site.
Intranasal challenge of H5N1 LD50 (750 pfu) to CASKfloxLysMCre and CASKwtLysMCre mice, lung lysate preparation for IFNA ELISA and lung histopathology
8-12-week old CaskfloxLysMCre+ and CaskwtLysMCre+ mice were anesthetized by intraperitoneal. injection of 150 mg/kg ketamine (Imalgene 1000, Boehringer Ingelheim) and 30 mg/kg Xylazine (Rompun, Dechra). Followed by intranasal inoculation with H5N1 at LD50 (750 pfu in 20 ul serum-free DMEM/mouse) through left nostril. Mice body weights were recorded daily and mice were sacrificed by isoflurane anesthesia followed by cervical dislocation on days 3 and 5 post-infection. The left lungs were collected and homogenized in 1 ml of 1 X PBS supplemented with protease inhibitors (Roche, REF04693132001) and phosphatase inhibitor (Roche, REF04693132001) using MagNA Lyser (Roche) and MagNA Lyser Green Beads (Roche) at 6500 speed, 40 seconds twice with a 10 minutes interval on ice. Lung homogenates were then centrifuged at 160000g for 10 minutes, 4°C, and supernatant were collected and store at -80°C overnight, and were used in IFNA ELISA (Mouse IFN-α (Invitrogen, BMS6027) right next day to prevent loss of IFNA in the homogenates.
For histopathology analysis, the left lungs were fixed in 10% formalin for at least 3 days and then sent to Taiwan Mouse Clinic for paraffin embedding, tissue sectioning and H&E staining. Lung tissue section H&E slides were scanned into “.svs” files by the Leica Aperio GT450. We use the histological scoring system for inflammation in the lungs of mice published by H. K. Bayes et al. (47). Briefly, there are two parts of the scoring system, part one is peribronchial infiltration scoring 0-4, the other part is alveolar involvement scoring 0-3. For peribronchial infiltration, 0=none, 1=mild (infiltrate ≤ 4 cells thick), 2=moderate (infiltrate 5-10 cells thick), 3=severe (infiltrate 25-50% visualized lumens), 4=diffuse (infiltrate > 50% visualized lumens). For alveolar involvement, 0=none, 1=mild (patchy increased cellularity/thickening), 2=moderate (25-50% visualized lung with increased cellularity/thickening), 3=severe (>50% visualized lung with increased cellularity/thickening). We use 4X magnification for histological scoring of left lung cross section. Each cross section is dividing into 15 fields for scoring and then averaged to represent the final score.
Identification of CASK interactome in resting and activated GM-macrophage by IP-LC-MS/MS
Dynabeads Antibody Coupling Kit (Thermo Fisher, 14311D) was used to covalently couple anti-CASK polyclonal antibody to magnetic beads for immunoprecipitation. Control antibody is Mouse IgG1 (Sigma, M7894), CASK polyclonal antibody (Abnova, PAB27198). Covalent conjugation protocol follows the instruction of Dynabeads Antibody Coupling Kit. Briefly, 10 mg of dynabeads were conjugated with 100 µg of antibody. Activated conjugated beads were used for immunoprecipitation assay. Equal volume (bring volume to 1ml by adding 1% Triton X100 lysis buffer) of nuclear extract from the same treatment were incubated with anti-IgG1 dynabeads (2mg) or anti-CASK dynabeads (2mg). The extract and dynabeads mixture were kept in resuspending using a rotary mixer in 4°C overnight (around 20 hours). The dynabeads were then washed 3 times, 2-minute wash each time, once with 1 ml 0.05% Triton X100 (50 mM Tris-HCl pH7.4, 150 mM NaCl, 0.05% Triton X100, phosphatase inhibitor (Roche, 04906837001) 1 pill, protease inhibitor cocktail (Roche, 04639132001) 1 pill in final 4ml), twice with wash buffer (50 mM Tris-HCl pH7.4, 150 mM NaCl, phosphatase inhibitor (Roche, 04906837001) 1 pill, protease inhibitor cocktail (Roche, 04639132001) 1 pill in final 4ml). Put eppendorf upside down to air dry all remaining liquids before elution. For elution, use 33 µl 0.1 M Glycine pH=3 to immerse the dynabeads and incubate at 55°C for 10 mins. Collect 30 µl of supernatant from each sample and bring the pH to neutral by adding 1.5 µl 1 M ammonium bicarbonate. Store the neutralized sample at -80°C before subjected to LC-MS-MS experiment. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD058016 and 10.6019/PXD058016 for “Identification of CASK nuclear interactome in resting and H5N1-activated GM-macrophage by IP-LC-MS/MS”.
LC-MS/MS method
Samples were analyzed by LC-ESI-MS using an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an EASY-nLC 1200 system and EASY-spray source (both from Thermo, San Jose, CA, US). The digestion solution (5 μL) was injected at a flow rate of 1 μL/min onto an EASY column (C18, 0.075 mm × 150 mm, ID 3 μm; Thermo Scientific). Chromatographic separation was performed using 0.1% formic acid in water as mobile phase A and 0.1% formic acid in 80% acetonitrile as mobile phase B, with a flow rate of 300 nL/min. The gradient employed ranged from 5% buffer B at 2 min to 40% buffer B at 50 min.
Full-scan MS conditions were as follows: mass range m/z 375-1500 (AGC target 5E5) with lock mass, resolution 60,000 at m/z 200, and maximum injection time of 50 ms. MS/MS was run in top speed mode with 3 s cycles, using CID for protein identification or HCD and EThcD for phosphopeptide analysis. Dynamic exclusion duration was set to 60 s with a 10 ppm tolerance around the selected precursor and its isotopes. The electrospray voltage was maintained at 1.8 kV, and the capillary temperature was set at 275°C.
siRNA knockdown of CASK primary interactant Stip1 and Cct4
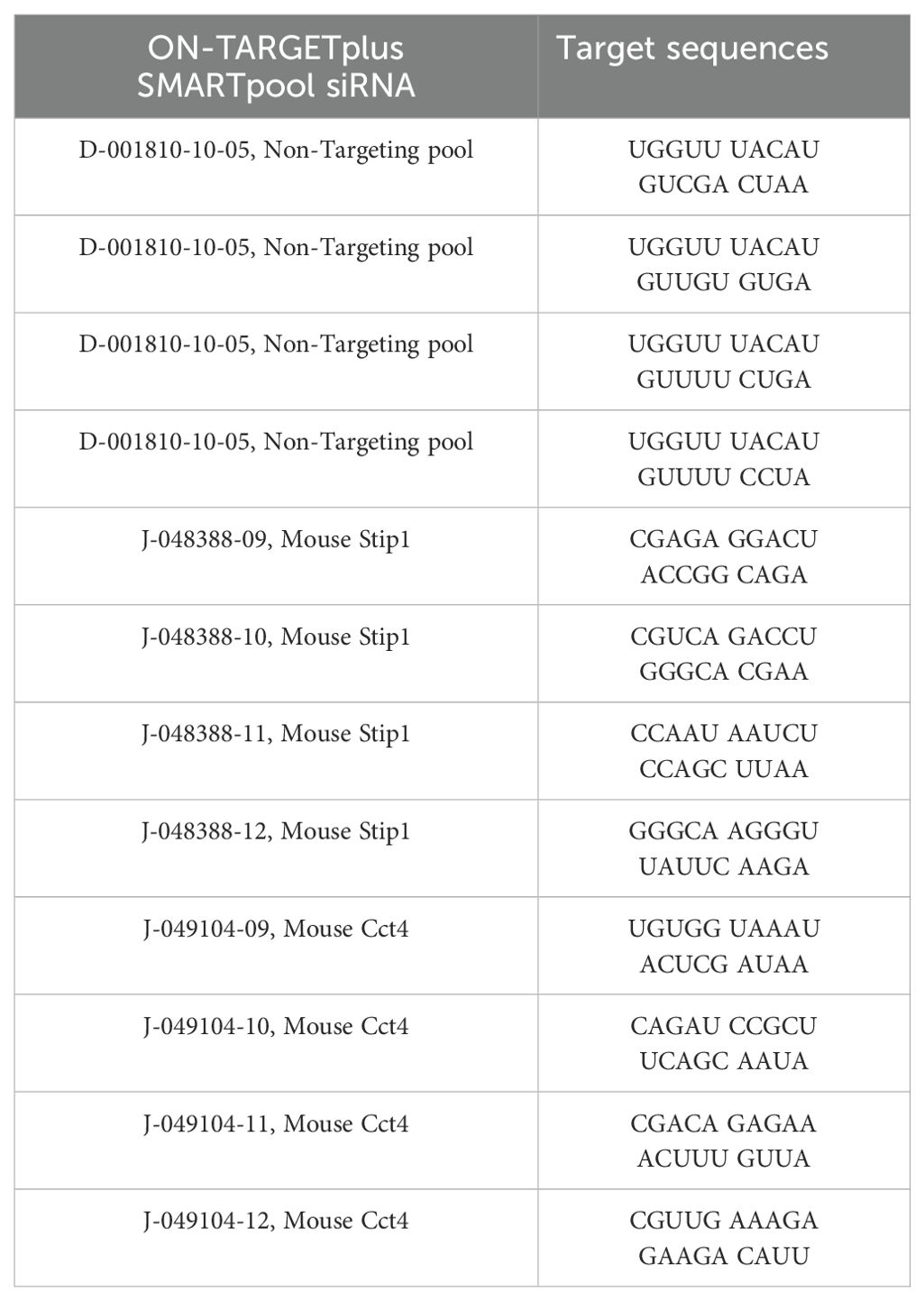
GM-MΦ 7x105 cells (in 12-well per well) were transfected with (25 µM) siRNAs targeting Cct4 and Stip1 for 48 hours prior to H5N1 infection (MOI=1) for 12 hours. Transfection is performed using HiPerFect transfection reagent (Cat. No. 301704, QIAGEN) 12 µL + 5 µL 5µM SMARTpool siRNA in 200 µL Opti-MEM (Cat. No. 31985070, ThermoFisher Scientific) incubate for 10 minutes, and then add drop by drop into 12-well (200 µL RPMI culture medium) to reach final volume of 400 µL. Incubate at 37°C for 4 hours, then add 600 µL RPMI culture medium to reach final volume of 1 mL. Stip1 siRNA (L-048388-01-0005, ON-TARGETplus SMARTpool, Dharmacon) and Cct4 siRNA (L-049104-01-0005, ON-TARGETplus SMARTpool, Dharmacon) sequences are listed as follows (Tables M6):
Knockdown efficiencies were confirmed by RT-qPCR analysis of Cct4 and Stip1 mRNA levels in total RNA extracts. qPCR primer sequences for Stip1 and Cct4 are listed as follows (Tables M7):
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism software (version 9.0; GraphPad Software Inc., San Diego, CA, USA). Data are presented as mean ± standard deviation (SD). Statistical significance between two datasets was determined using either an unpaired, two-tailed Student’s t-test for parametric data or a Mann-Whitney test for nonparametric data. For multiple group comparisons, parametric data were analyzed using one-way ANOVA followed by the Bonferroni post hoc test, while nonparametric data were analyzed using the Kruskal–Wallis test followed by Dunn’s post hoc test. Across all experiments, significance levels were denoted as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by Academia Sinica Institutional Animal Care & Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing, Visualization. PS: Formal analysis, Methodology, Supervision, Visualization, Investigation, Writing – review & editing. SH: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work is supported by NSTC 113-2320-B-400-026 and National Health Research Institutes IM-112-PP-01; IM-112-SP-01; IM-113-PP-01; IM-113-SP-01; IM-113-SP-10; and IM-113-SP-13.
Acknowledgments
We thank the “Transgenic Mouse Model Core Facility of the National Core Facility for Biopharmaceuticals, National Science and Technology Council, Taiwan” and the “Gene Knockout Mouse Core Laboratory of National Taiwan University Center of Genomic and Precision Medicine” for their technical services. We are grateful to the GRC Mass Spectrometry Facility of Academia Sinica for their assistance with MS analysis. We also thank the Taiwan Mouse Clinic, Academia Sinica, and Taiwan Animal Consortium for their technical support in lung tissue paraffin embedding, sectioning, and H&E staining. We are grateful to National Laboratory Animal Center (NLAC), NARLabs, Taiwan, for technical support in pathology analysis. Additionally, we acknowledge the Academia Sinica Inflammation Core Facility, IBMS, for their technical support. The core facility is funded by the Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-113-A9). Lastly, we thank Yao-An Yu for her technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. During the preparation of this work the authors used Claude and ChatGPT in order to correct grammar. After using this tool/service, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1513713/full#supplementary-material
Supplementary Table 1 | Source data-confocal images.
Supplementary Table 2 | Nuclear CASK interactome raw data.
Supplementary Table 3 | Source data.
References
1. Zhu J, Shang Y, Zhang M. Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat Rev Neurosci. (2016) 17:209–23. doi: 10.1038/nrn.2016.18
2. Hsueh YP. Calcium/calmodulin-dependent serine protein kinase and mental retardation. Ann Neurol. (2009) 66:438–43. doi: 10.1002/ana.21755
3. Huang TN, Hsueh YP. Calcium/calmodulin-dependent serine protein kinase (CASK), a protein implicated in mental retardation and autism-spectrum disorders, interacts with T-Brain-1 (TBR1) to control extinction of associative memory in male mice. J Psychiatry Neurosci. (2017) 42:37–47. doi: 10.1503/jpn.150359
4. Giacomini T, Nuovo S, Zanni G, Mancardi MM, Cusmai R, Pepi C, et al. CASK related disorder: Epilepsy and developmental outcome. Eur J Paediatr Neurol. (2021) 31:61–9. doi: 10.1016/j.ejpn.2021.02.006
5. Wu S, Jiang C, Li J, Zhang G, Shen Y, Wang J. A novel missense variant in the CASK gene causes intellectual developmental disorder and microcephaly with pontine and cerebellar hypoplasia. BMC Med Genomics. (2022) 15:127. doi: 10.1186/s12920-022-01275-z
6. Patel PA, Hegert JV, Cristian I, Kerr A, LaConte LEW, Fox MA, et al. Complete loss of the X-linked gene CASK causes severe cerebellar degeneration. J Med Genet. (2022) 59(11):1044–57. doi: 10.1136/jmedgenet-2021-108115
7. Dubbs H, Ortiz-Gonzalez X, Marsh ED. Pathogenic variants in CASK: Expanding the genotype-phenotype correlations. Am J Med Genet A. (2022) 188:2617–26. doi: 10.1002/ajmg.a.v188.9
8. Butz S, Okamoto M, Sudhof TC. A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell. (1998) 94:773–82. doi: 10.1016/S0092-8674(00)81736-5
9. Samuels BA, Hsueh YP, Shu T, Liang H, Tseng HC, Hong CJ, et al. Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature. (2000) 404:298–302. doi: 10.1038/35005118
10. Zhang X, Herr F, Vernochet A, Lorenzo HK, Beaudreuil S, Durrbach A. CASK, the soluble glomerular permeability factor, is secreted by macrophages in patients with recurrent focal and segmental glomerulo-sclerosis. Front Immunol. (2020) 11:875. doi: 10.3389/fimmu.2020.00875
11. Westenius V, Makela SM, Julkunen I, Osterlund P. Highly pathogenic H5N1 influenza A virus spreads efficiently in human primary monocyte-derived macrophages and dendritic cells. Front Immunol. (2018) 9:1664. doi: 10.3389/fimmu.2018.01664
12. Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. (2020) 20:537–51. doi: 10.1038/s41577-020-0288-3
13. Lee MK, Kim HE, Park EB, Lee J, Kim KH, Lim K, et al. Structural features of influenza A virus panhandle RNA enabling the activation of RIG-I independently of 5’-triphosphate. Nucleic Acids Res. (2016) 44:8407–16. doi: 10.1093/nar/gkw525
14. Munoz-Moreno R, Martinez-Romero C, Garcia-Sastre A. Induction and evasion of type-I interferon responses during influenza A virus infection. Cold Spring Harb Perspect Med. (2021) 11(10):a038414. doi: 10.1101/cshperspect.a038414
15. McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol. (2015) 15:87–103. doi: 10.1038/nri3787
16. Cheung CY, Poon LL, Lau AS, Luk W, Lau YL, Shortridge FK, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. (2002) 360:1831–7. doi: 10.1016/S0140-6736(02)11772-7
17. Guo XJ, Thomas PG. New fronts emerge in the influenza cytokine storm. Semin Immunopathol. (2017) 39:541–50. doi: 10.1007/s00281-017-0636-y
18. Fajgenbaum DC, June CH. Cytokine storm. N Engl J Med. (2020) 383:2255–73. doi: 10.1056/NEJMra2026131
19. Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. (2005) 122:669–82. doi: 10.1016/j.cell.2005.08.012
20. Belgnaoui SM, Paz S, Samuel S, Goulet ML, Sun Q, Kikkert M, et al. Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell Host Microbe. (2012) 12:211–22. doi: 10.1016/j.chom.2012.06.009
21. Atasoy D, Schoch S, Ho A, Nadasy KA, Liu X, Zhang W, et al. Deletion of CASK in mice is lethal and impairs synaptic function. Proc Natl Acad Sci USA. (2007) 104:2525–30. doi: 10.1073/pnas.0611003104
22. Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. (1999) 8:265–77. doi: 10.1023/A:1008942828960
23. Teng O, Chen ST, Hsu YL, Sia SF, Cole S, Valkenburg SA, et al. CLEC5A-mediated enhancement of the inflammatory response in myeloid cells contributes to influenza virus pathogenicity in vivo. J Virol. (2017) 91(1):e01813-16. doi: 10.1128/JVI.01813-16
24. Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, et al. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. (2000) 13:129–41. doi: 10.1016/S1074-7613(00)00014-5
25. Bavagnoli L, Dundon WG, Garbelli A, Zecchin B, Milan A, Parakkal G, et al. The PDZ-ligand and Src-homology type 3 domains of epidemic avian influenza virus NS1 protein modulate human Src kinase activity during viral infection. PloS One. (2011) 6:e27789. doi: 10.1371/journal.pone.0027789
26. Byeon SE, Yi YS, Oh J, Yoo BC, Hong S, Cho JY, et al. The role of Src kinase in macrophage-mediated inflammatory responses. Mediators Inflammation. (2012), 512926. doi: 10.1155/2012/512926
27. Li F, Li C, Marquez-Lago TT, Leier A, Akutsu T, Purcell AW, et al. Quokka: a comprehensive tool for rapid and accurate prediction of kinase family-specific phosphorylation sites in the human proteome. Bioinformatics. (2018) 34:4223–31. doi: 10.1093/bioinformatics/bty522
28. Samuels BA, Hsueh YP, Shu T, Liang H, Tseng HC, Hong CJ, et al. Cdk5 promotes synaptogenesis by regulating the subcellular distribution of the MAGUK family member CASK. Neuron. (2007) 56:823–37. doi: 10.1016/j.neuron.2007.09.035
29. Pinol-Roma S. HnRNP proteins and the nuclear export of mRNA. Semin Cell Dev Biol. (1997) 8:57–63. doi: 10.1006/scdb.1996.0122
30. Yamashita M, Chattopadhyay S, Fensterl V, Zhang Y, G. C. Sen GC. A TRIF-independent branch of TLR3 signaling. J Immunol. (2012) 188:2825–33. doi: 10.4049/jimmunol.1103220
31. Chesnokova E, Bal N, Kolosov P. Kinases of eIF2a Switch Translation of mRNA Subset during Neuronal Plasticity. Int J Mol Sci. (2017) 18(10):2213. doi: 10.3390/ijms18102213
32. Doerks T, Bork P, Kamberov E, Makarova O, Muecke S, Margolis B. L27, a novel heterodimerization domain in receptor targeting proteins Lin-2 and Lin-7. Trends Biochem Sci. (2000) 25:317–8. doi: 10.1016/S0968-0004(00)01599-1
33. Mauceri D, Cattabeni F, Di Luca M, Gardoni F. Calcium/calmodulin-dependent protein kinase II phosphorylation drives synapse-associated protein 97 into spines. J Biol Chem. (2004) 279:23813–21. doi: 10.1074/jbc.M402796200
34. Wei FY, Nagashima K, Ohshima T, Saheki Y, Lu YF, Matsushita M, et al. Cdk5-dependent regulation of glucose-stimulated insulin secretion. Nat Med. (2005) 11:1104–8. doi: 10.1038/nm1299
35. Huang TN, Chang HP, Hsueh YP. CASK phosphorylation by PKA regulates the protein-protein interactions of CASK and expression of the NMDAR2b gene. J Neurochem. (2010) 112:1562–73. doi: 10.1111/j.1471-4159.2010.06569.x
36. Lantermans HC, Minderman M, Kuil A, Kersten MJ, Pals ST, Spaargaren M. Identification of the SRC-family tyrosine kinase HCK as a therapeutic target in mantle cell lymphoma. Leukemia. (2021) 35:881–6. doi: 10.1038/s41375-020-0934-6
37. An W, Lakhina S, Leong J, Rawat K, Husain M. Host innate antiviral response to influenza A virus infection: from viral sensing to antagonism and escape. Pathogens. (2024) 13(7):561. doi: 10.3390/pathogens13070561
38. Zhang K, Xie Y, Munoz-Moreno R, Wang J, Zhang L, Esparza M, et al. Structural basis for influenza virus NS1 protein block of mRNA nuclear export. Nat Microbiol. (2019) 4:1671–9. doi: 10.1038/s41564-019-0482-x
39. Kimura T, Hashimoto I, Nagase T, Fujisawa J. CRM1-dependent, but not ARE-mediated, nuclear export of IFN-alpha1 mRNA. J Cell Sci. (2004) 117:2259–70. doi: 10.1242/jcs.01076
40. Kimura T, Hashimoto I, Nishizawa M, Ito S, Yamada H. Novel cis-active structures in the coding region mediate CRM1-dependent nuclear export of IFN-alpha 1 mRNA. Med Mol Morphol. (2010) 43:145–57. doi: 10.1007/s00795-010-0492-5
41. Eisfeld AJ, Neumann G, Kawaoka Y. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol. (2015) 13:28–41. doi: 10.1038/nrmicro3367
42. Guo J, Zhu Y, Ma X, Shang G, Liu B, Zhang K. Virus infection and mRNA nuclear export. Int J Mol Sci. (2023) 24(16):12593. doi: 10.3390/ijms241612593
43. Hoffmann E, Neuman G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA. (2000) 97:6108–13. doi: 10.1073/pnas.100133697
44. Li S, Liu C, Klimov A, Subbarao K, Perdue ML, Mo D. Recombinant influenza A virus vaccines for the pathogenic human A/Hong Kong/97 (H5N1) viruses. J Infect Dis. (1999) 179:1132–8. doi: 10.1086/jid.1999.179.issue-5
45. Huprikar J, Rabinowitz S. A simplified plaque assay for influenza viruses in Madin-Darby kidney (MDCK) cells. J Virol Methods. (1980) 1:117–20. doi: 10.1016/0166-0934(80)90020-8
46. Mor A, White A, Zhang K, Thompson M, Esparza L, Munoz-Moreno R, et al. Influenza virus mRNA trafficking through host nuclear speckles. Nat Microbiol. (2016) 1:16069. doi: 10.1038/nmicrobiol.2016.69
Keywords: CASK, H5N1, influenza A virus, macrophage, nuclear entry, interferon-alpha, CRM1, mRNA export
Citation: Huang J-Y, Sung P-S and Hsieh S-L (2025) Regulation of interferon alpha production by the MAGUK-family protein CASK under H5N1 infection. Front. Immunol. 15:1513713. doi: 10.3389/fimmu.2024.1513713
Received: 18 October 2024; Accepted: 16 December 2024;
Published: 09 January 2025.
Edited by:
Emilia Lecuona, Northwestern University, United StatesCopyright © 2025 Huang, Sung and Hsieh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shie-Liang Hsieh, c2xoc2llaEBuaHJpLmVkdS50dw==
 Jing-Ying Huang
Jing-Ying Huang Pei-Shan Sung
Pei-Shan Sung Shie-Liang Hsieh
Shie-Liang Hsieh