- 1First Clinical Medical College, Guangzhou University of Chinese Medicine, Guangzhou, China
- 2Department of Oncology, Chongqing Hospital, The First Affiliated Hospital of Guangzhou University of Chinese Medicine, Chongqing, China
- 3Department of Oncology, The First Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, China
- 4Guangdong Clinical Research Academy of Chinese Medicine, Guangzhou, China
- 5Lingnan Medical Research Center, Guangzhou University of Chinese Medicine, Guangzhou, China
- 6Department of Oncology, Shenshan Hospital, The First Affiliated Hospital of Guangzhou University of Chinese Medicine, Shanwei, China
Background: Recently there has been an increasing number of studies have explored apoptosis mechanisms in lung cancer (LC). However, no researchers have conducted a bibliometric analysis of the most cited articles in this field.
Objective: To examine the top 100 most influential and cited publications on apoptosis in non-small cell lung cancer (NSCLC) from 2004 to 2023, summarizing research trends and key focus areas.
Methods: This study utilized the Web of Science Core Database (WOSCC) to research NSCLC apoptosis from 2004 to 2023, using keyword selection and manual screening for article searches. Bibliometrix package of R software 4.3.1 was used to generate distribution statistics for the top ten institutions, journals and authors. Citespace6.2. R6 was used to create the visualization maps for keyword co-occurrence and clustering. VOSviewer1.6.19 was used to conduct cluster analysis of publishing countries (regions), with data exported to SCImago Graphica for geographic visualization and cooperation analysis. VOSviewer1.6.19 was used to produced co-citation maps of institutions, journals, authors, and references.
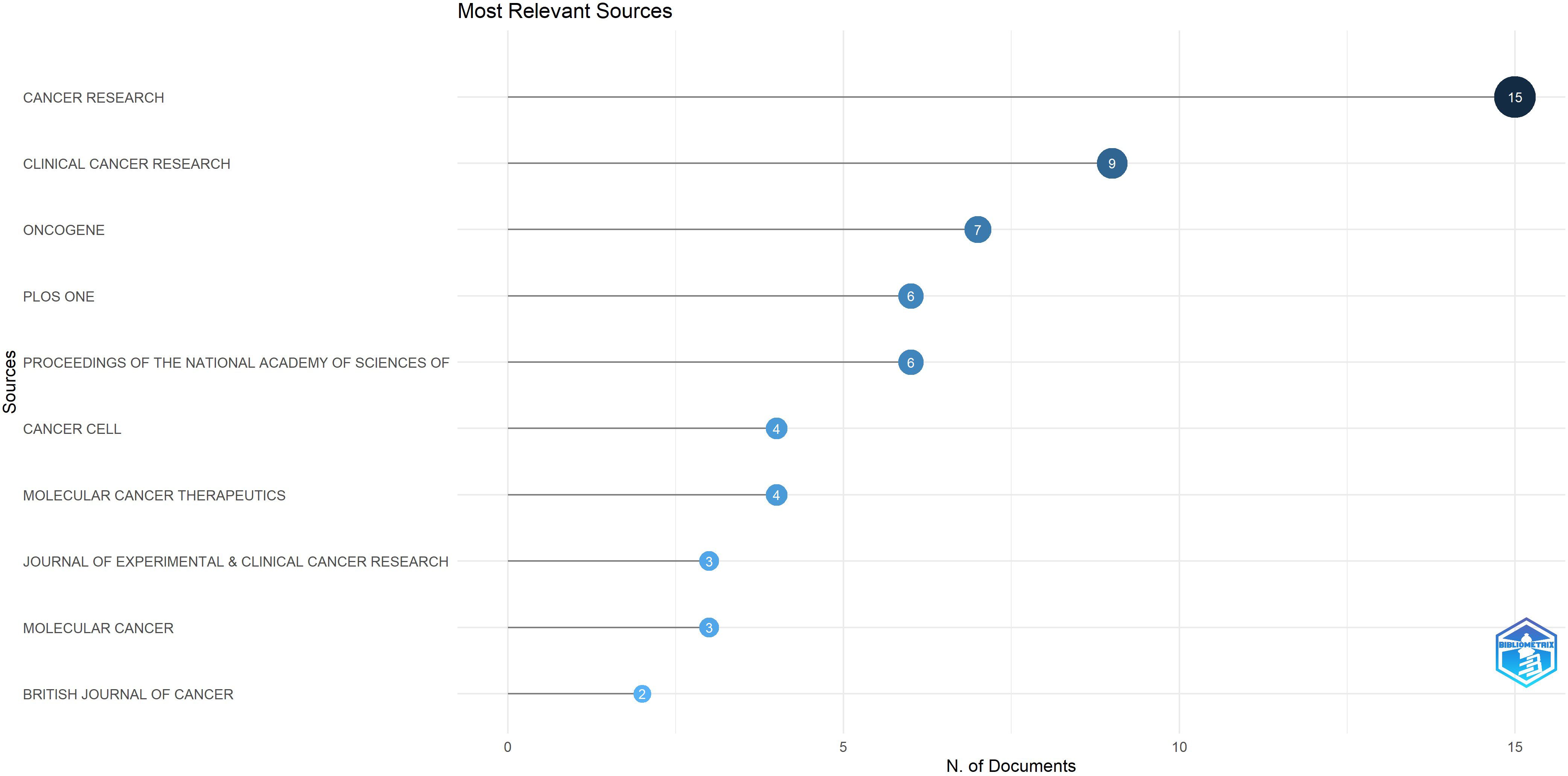
Results: From 2004 to 2023, 13316 articles were retrieved, and the top 100 most cited were chosen. These were authored by 934 individuals from 269 institutions across 18 countries and appeared in 45 journals. Citations ranged from 150 to 1,389, with a median of 209.5. The most influential articles appeared in 2005 and 2007 (n=13). The leading countries (regions), institutions, journals and authors were identified as the United States (n=60), Harvard University (n=64), CANCER RESEARCH (n=15), SUN M and YANG JS (n=6). The top five keywords were “expression”, “activation”, “apoptosis”, “pathway” and “gefitinib”. This study indicates that enhancing apoptosis through circular RNA regulation and targeting the Nrf2 signaling pathway could become a key research focus in recent years.
Conclusion: Apoptosis has been the subject of extensive research over many years, particularly in relation to its role in the pathogenesis, diagnosis, and treatment of NSCLC. This study aims to identify highly influential articles and forecast emerging research trends, thereby offering insights into novel therapeutic targets and strategies to overcome drug resistance. The findings are intended to serve as a valuable reference for scholars engaged in this field of study.
1 Introduction
Lung cancer (LC) is one of the most common cancers in the world and is the foremost cause of cancer-related mortality, characterized by high incidence and mortality rates (1). Non-small cell lung cancer (NSCLC) represents the predominant subtype of LC, comprising 80%-85% of cases (2). The occurrence of apoptosis plays an important role in the development of cancer, so it is particularly important to study the mechanism of apoptosis in NSCLC for therapeutic advancements. The mechanisms of apoptosis encompass endogenous pathways, which are activated by factors such as DNA damage, oxidative stress and a variety of other stress conditions, as well as extrinsic pathways, which are initiated by cell membrane proteins associated with death receptors (3, 4). Alterations in upstream regulatory factors within the two pathways may lead to the imbalance between cell proliferation and apoptosis (5).
Bibliometrics uses mathematical and statistical techniques to quantitatively and qualitatively assess publications within a given research domain, thereby facilitating the monitoring of research trends and serving as a diagnostic tool for the field (6, 7). Bibliometrics serves as a tool for assessing the current state of research, identifying research hotspots, and predicting future research trajectories (8). Citespace, a scientific visualization software package developed by Dr. Chaomei Chen from Drexel University, USA. It operates within a Java runtime environment and offers direct visual effects (9). VOSviewer is a software application designed for the construction and visualization of bibliometric maps. It also provides a viewer, which provides a detailed check on the scientific nature of cartography (10). The software is equipped with robust capabilities for conducting co-occurrence and co-citation analyses (11). Additionally, its data can be exported to SCImago Graphica for enhanced geographic visualization and collaboration analysis, thereby improving the aesthetic quality of the visual representations. Bibliometrix package of R software 4.3.1 serves as a comprehensive language and environment for performing statistical analyses and generating graphical representations of data (12). As a versatile analytical tool developed in R, Bibliometrix is particularly well-suited for the scientific mapping of bibliometric data (13).
The number of citations of an article serves as a significant metric for assessing its scholarly impact, with a high number of citations often indicating its recognition and validation within the academic community. Despite this, an analysis of the most influential articles in the domain of NSCLC apoptosis research has not yet been conducted. In this study, we selected the top 100 most frequently cited articles and analyzed their quality and characteristics in order to provide references for researchers.
2 Data and methods
2.1 Data source
Related publications on NSCLC apoptosis research were retrieved from the Web of Science Core Database (WOSCC) on January 1, 2024, with a search formula of TS=(Carcinoma, Non-Small-Cell Lung OR Carcinoma, Non Small Cell Lung OR Carcinomas, Non-Small-Cell Lung OR Lung Carcinoma, Non-Small-Cell OR Lung Carcinomas, Non-Small-Cell OR Non-Small-Cell Lung Carcinomas OR Non-Small-Cell Lung Carcinoma OR Non Small Cell Lung Carcinoma OR Carcinoma, Non-Small Cell Lung OR Non-Small Cell Lung Carcinoma OR Non-Small Cell Lung Cancer OR Non small Cell Lung Cancer OR NSCLC) AND TS=(Apoptosis OR Classic Apoptosis OR Apoptosis, Classic OR Classic Apoptoses OR Programmed Cell Death, Type I OR Classical Apoptosis OR Apoptosis, Classical OR Apoptosis, Intrinsic Pathway OR Apoptoses, Intrinsic Pathway OR Intrinsic Pathway Apoptoses OR Intrinsic Pathway Apoptosis OR Apoptosis, Extrinsic Pathway OR Apoptoses, Extrinsic Pathway OR Extrinsic Pathway Apoptoses OR Extrinsic Pathway Apoptosis OR Caspase-Dependent Apoptosis OR Apoptosis, Caspase-Dependent OR Caspase Dependent Apoptosis OR Programmed Cell Death OR Cell Death, Programmed OR cell apoptosis OR cells apoptosis). The time span is 2004-2023, the article type is “article”, and the language of the article is “English”. The article was excluded if its title and abstract indicated inconsistency with the study’s topic. Subsequently, the 100 most-cited articles were selected for inclusion (see Figure 1). Complete records and references were exported and saved in plain text format for analysis using bibliometrics tools.
2.2 Visual analysis methods
The bibliometric visualization analysis was conducted using the Bibliometrix package of R software 4.3.1, Citespace6.2.R6, VOSviewer1.6.19 and SCImago Graphica 1.0.46. Citespace6.2.R6 was used to de-duplicate the exported plain text format, after which there were still 100 articles. The free statistics website (https://bibliometric.com/) was utilized to analyze publication time. Bibliometrix package of R software 4.3.1 was used to generate distribution statistics for the top ten institutions, journals, and authors. Additionally, Citespace version 6.2.R6 was used to create a visualization map for keyword co-occurrence and clustering, which served to elucidate the temporal distribution of research hotspots and to forecast future research trends. VOSviewer1.6.19 was used to conduct a cluster analysis of the publishing country (region). The resulting data were subsequently exported to SCImago Graphica for geographic visualization and collaboration analysis. Additionally, VOSviewer was used to construct co-citation maps of institutions, journals, authors, and references. These visualizations effectively highlight the articles that have significantly contributed to the advancement and development of research within this discipline.
3 Results
3.1 General data analysis
On January 1, 2024, a total of 13,881 articles were retrieved from WOSCC using the specified search formula. Following the screening process outlined in Figure 1, we identified the top 100 most frequently cited publications (14–113). Subsequently, we extracted detailed information on these publications, including their titles, journals, first authors, publication years, total citations and average annual citations (refer to the Supplementary Materials for further details). Among these selected articles, the number of citations ranged from 150 to 1,389, with a median of 209.5 and an average of 259.5 citations per year. In a seminal 2004 publication in the journal SCIENCE, SORDELLA R. et al. (2004) (14) published an article that has garnered the highest number of citations (1,389 times). Their research elucidates the selective activation of Akt and the signal transducer and activator of transcription (STAT) signaling pathways by mutated epidermal growth factor receptors (EGFRs). Furthermore, the study demonstrates that gefitinib effectively inhibits these pathways, thereby inducing apoptosis.
3.2 Analysis of publication time
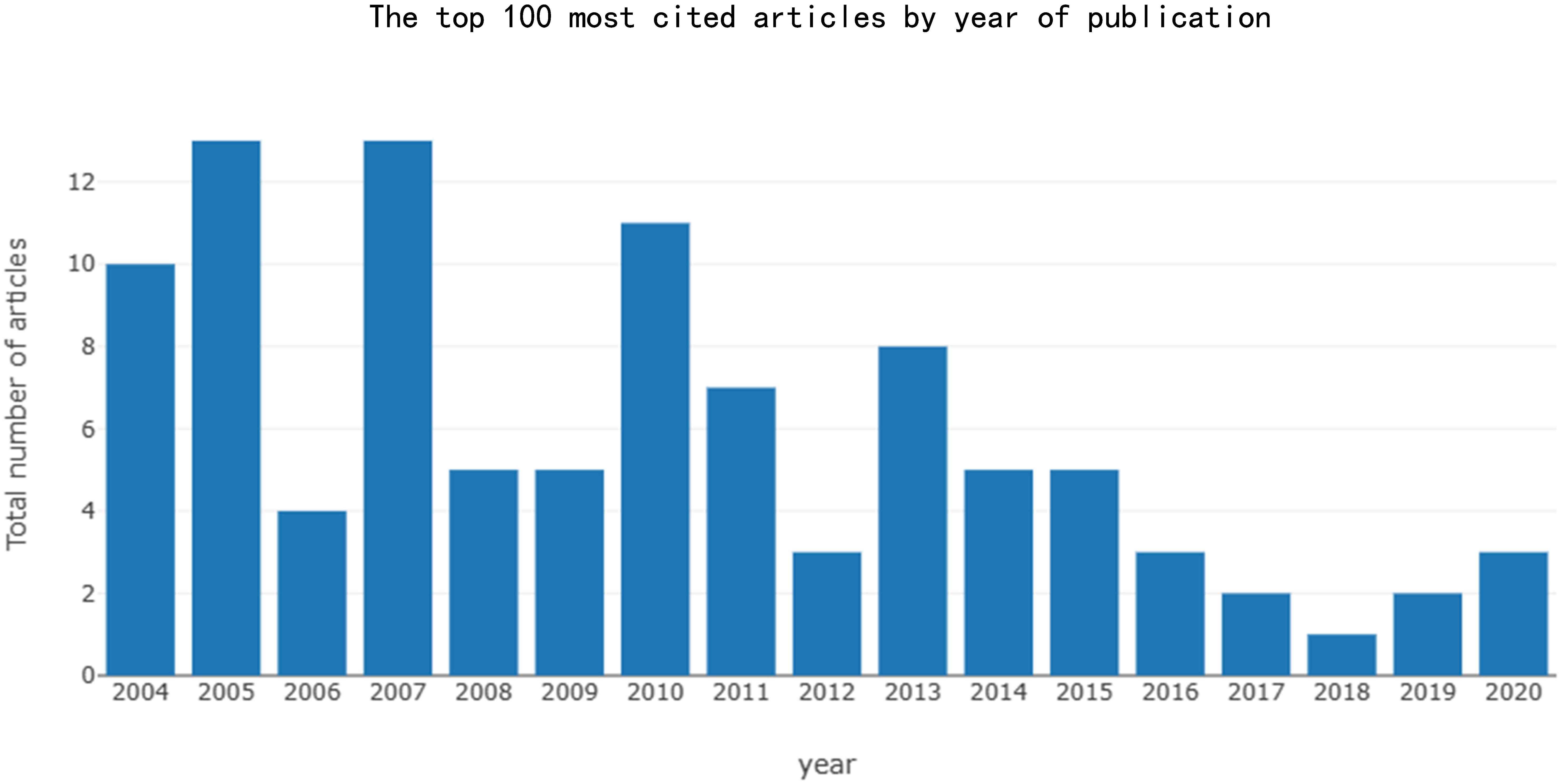
Among the top 100 most-cited articles, the publication period spans from 2004 to 2020. There is a overall trend of decreasing article quantity over time, with an average annual growth rate in the number of publications of -7.25%. The most influential articles were produced in 2005 and 2007 (n=13) (see Figure 2). The earliest article was published by BRÖER LE. et al. (2004) (83) in CANCER RESEARCH journal in 2004, elucidated the pivotal role of the lysosomal protease cathepsin B in facilitating cell death. This study provided compelling evidence for a novel cathepsin B-dependent cell death pathway, which is induced by microtubule stabilizers in NSCLC cells.
3.3 Analysis of countries (regions)
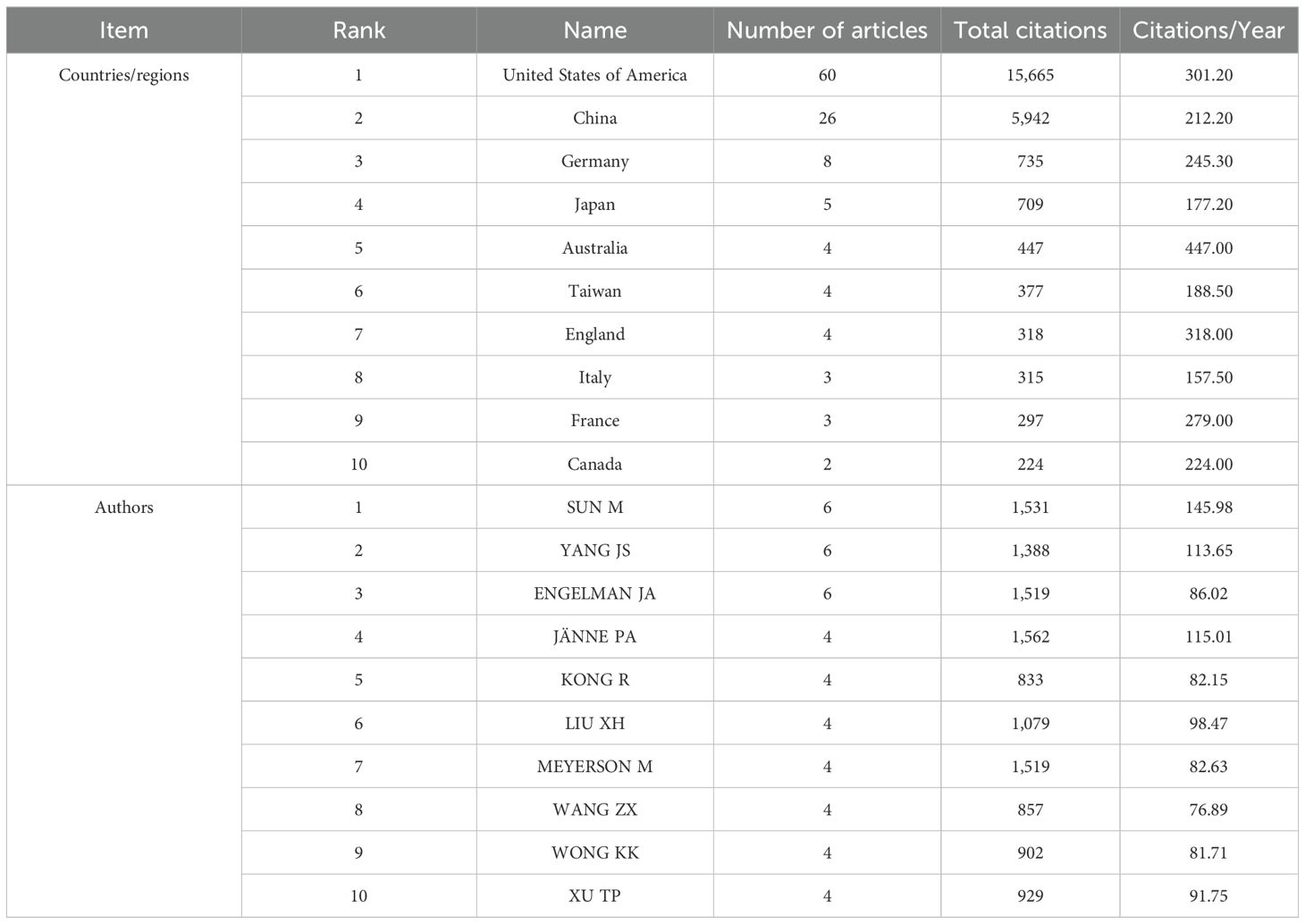
Based on the data presented in Table 1, the top 100 most-cited articles originate from 18 countries (regions). The United States leads with the highest number of publications is from the United States (n=60), followed by China (n=26) and Germany (n=8). Notably, only the United States and China have published more than 20 articles among all the countries (region). The world map (Figure 3) shows the distribution of published countries (regions) is mainly in North America, East Asia and Western Europe. In contrast, Africa and South America have no top 100 most-cited articles in this field. This suggests a significant imbalance in the geographical distribution of influential research articles. Enhancing international collaboration is essential to advancing the development of this research domain. The United States not only leads in the volume of academic publications but also demonstrates the highest level of international collaboration. Furthermore, Germany, China and Italy also exhibit significant cooperation with other countries (regions) (see Figure 4).
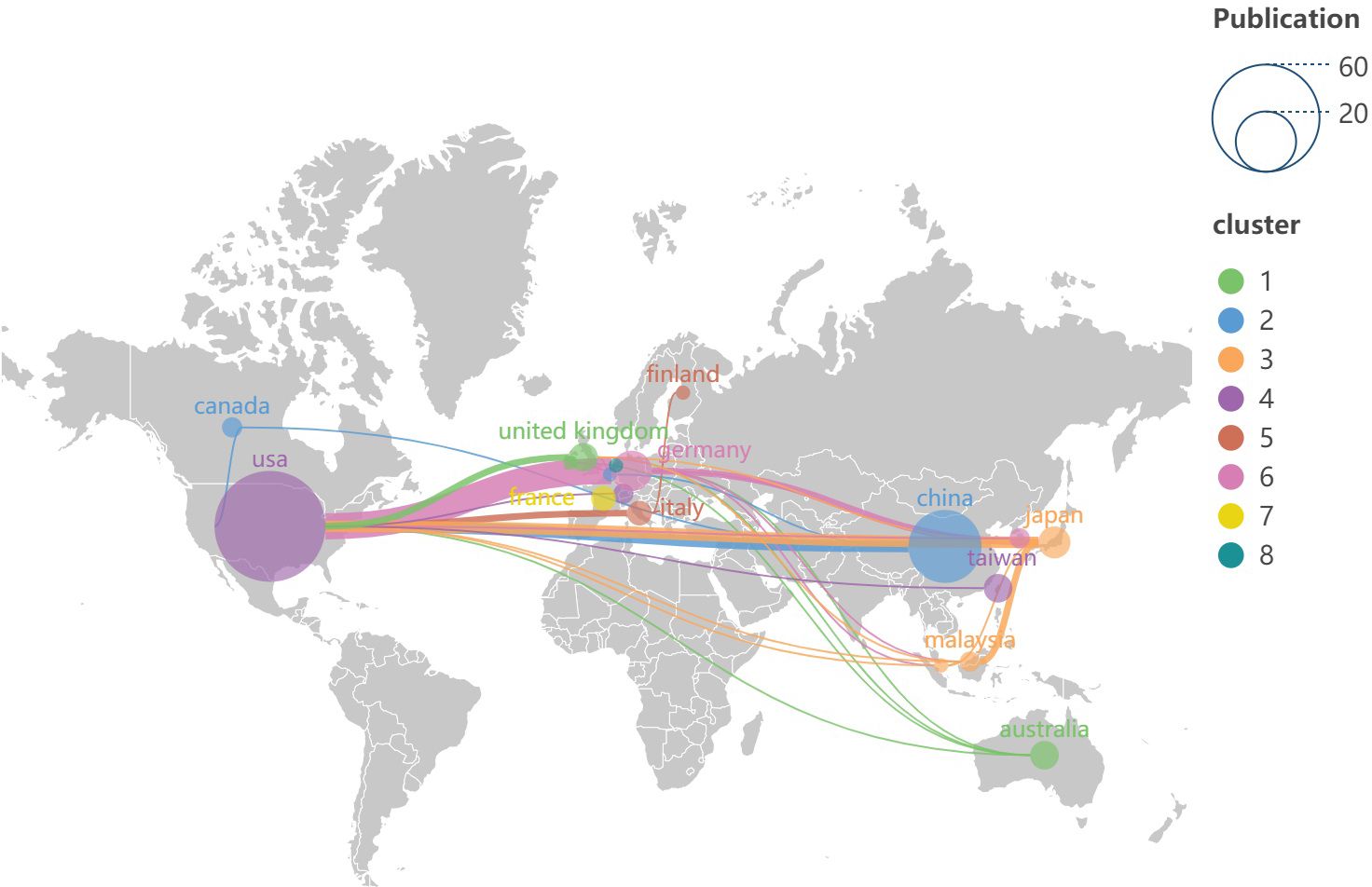
Figure 3. Distribution of countries and regions published on the world map. Each node symbolizes a country (region), with the size of the node indicating the volume of published articles originating from that country (region). The lines connecting nodes illustrate the degree of collaborative activity between the countries (regions). The color of each node corresponds to the cluster classification shown on the right side of the figure.
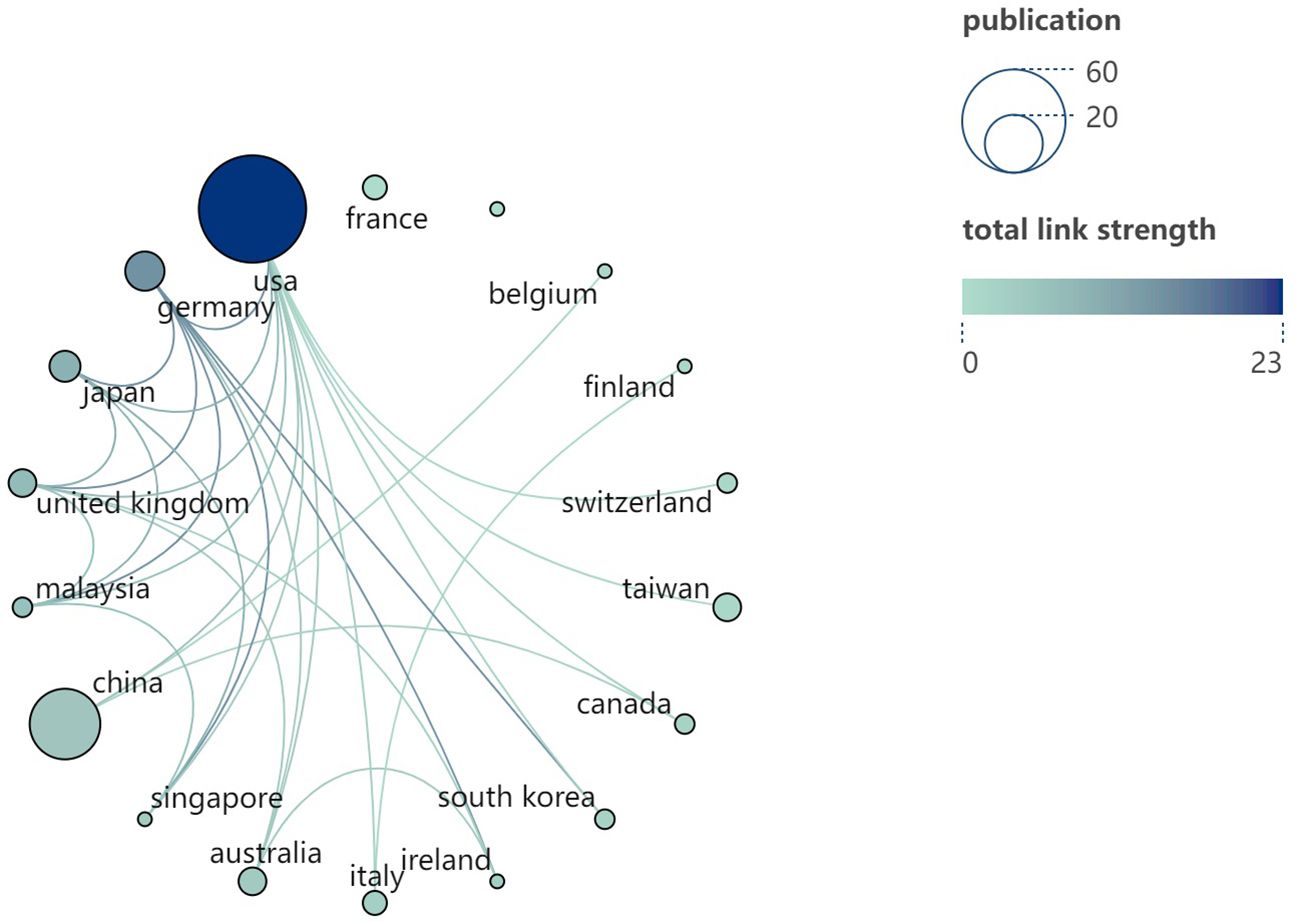
Figure 4. Cooperation relationship diagram between countries (regions). Each node symbolizes a country (region). The size of each node is indicative of the number of articles published within that country (region). The color of the node, as well as the connecting lines, signifies the aggregate number of interactions between the node in question and other nodes. Additionally, the color of the node correlates with the total link strength, as depicted on the right side of the visual representation.
3.4 Analysis of institutions
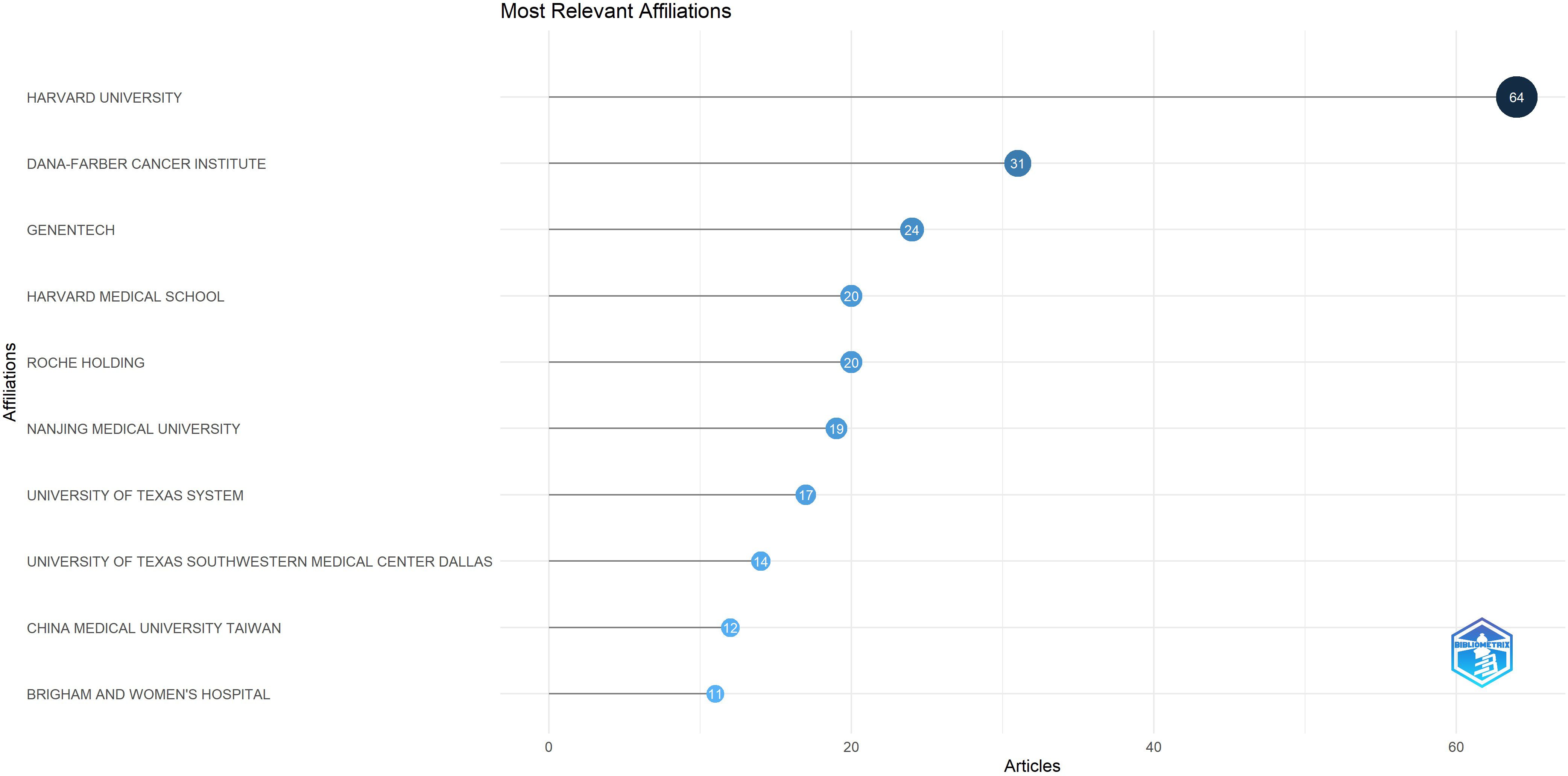
A total of 269 institutions contributed to the publication of the high-impact article within this field. Among these, 5 institutions published 15 or more articles (n≥15). The leading three institutions are Harvard University (n=64), Dana-Farber Cancer Institute (n=31) and Genentech (n=24). As illustrated in Figure 5, which presents a statistical chart of the top 10 institutions, the United States holds a significant standing in this domain. Apart from a select few leading global institutions, there is minimal variation in the number of publications produced by most institutions. Figure 6 illustrates a deficiency in collaboration among international institutions. Predominantly, cooperation occurs internally within individual countries, with the United States’ domestic institutions lead in fostering close partnerships in this domain.

Figure 6. Co-authoring relationship diagram between publishing institutions. Each node symbolizes an institution, with the size of the node corresponding to the volume of articles published by that institution. The connecting lines illustrate co-authorship relationships between institutions, with the line thickness indicating the strength of these collaborative efforts. Additionally, the color of each node signifies the cluster to which it belongs, with each cluster being represented by a distinct color.
3.5 Analysis of journals
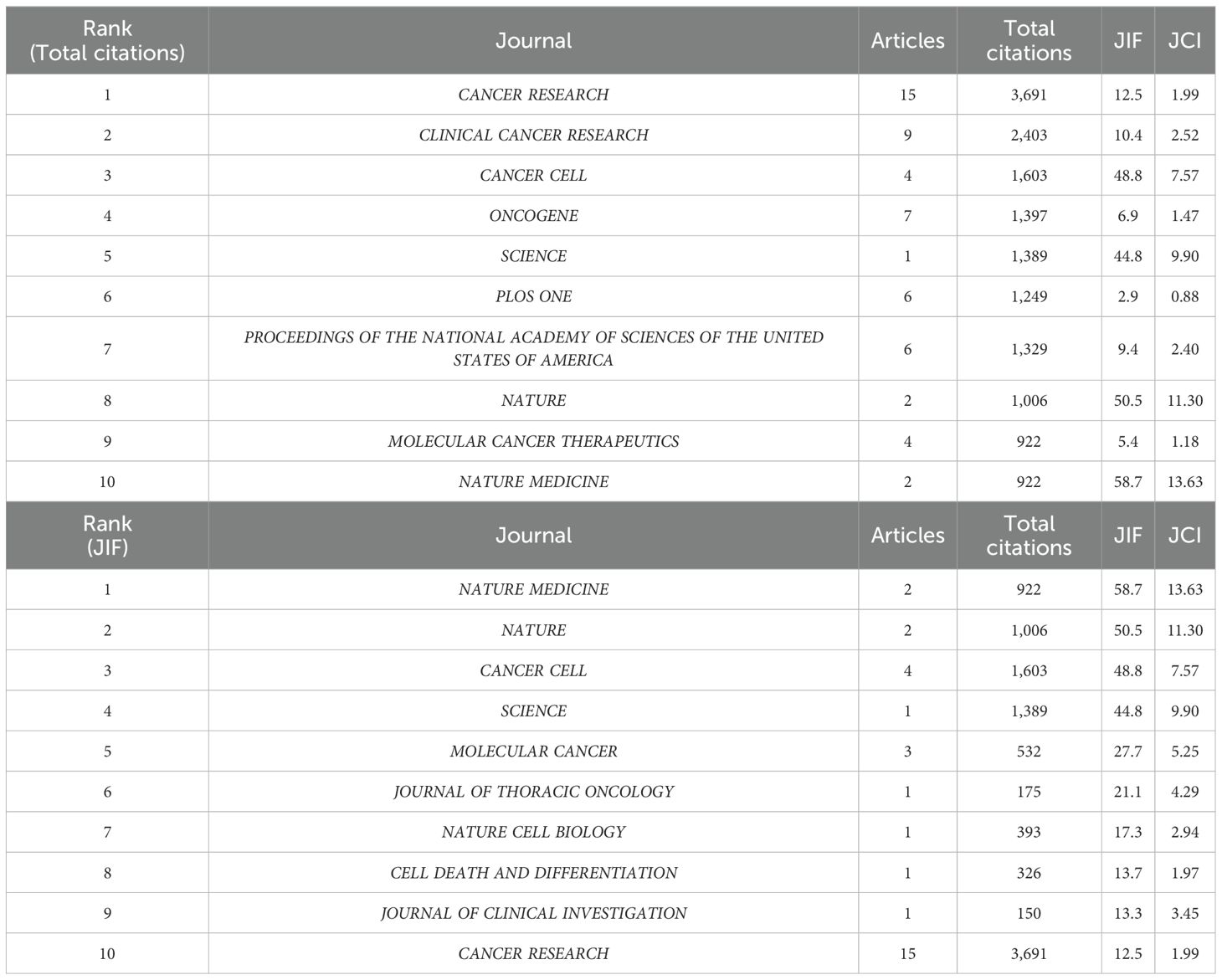
The top 100 highly cited articles retrieved in this study were published across 45 distinct journals, with Impact Factors (IF) ranging from 1.6 to 58.7 as of 2023. Notably, 80% of these journals are classified within the first quartile (Q1) according to the 2023 Journal Citation Reports (JCR) (see Supplementary Material). Among these, CANCER RESEARCH holds the highest position, contributing 15 articles (see Figure 7) and also leading in total citations, with 3,691 citations. However, CANCER RESEARCH IF 12.5 in 2023 was not the highest among the 45 journals analyzed. NATURE MEDICINE, (n = 2; IF(2023) was 58.7), demonstrated a significant advantage in terms of IF. All journals ranked within the top 10 for publication volume achieved a Q1 classification in the 2023 JCR rankings. The total citations and journal IF of 45 journals were sorted (see Supplementary Materials for details), and the top 10 journals were selected respectively (see Table 2). Among the top ten journals ranked by total citations, 8 have accrued more than 1,000 citations each. Notably, despite having published only a single article, the journal SCIENCE ranks fifth in total citations, underscoring its significant influence. All of the top ten journals possess an IF exceeding 10, indicating their status as leading publications within their respective fields. This suggests that the majority of highly influential articles in this study were published in high-impact journals (refer to Supplementary Material).
3.6 Analysis of authors
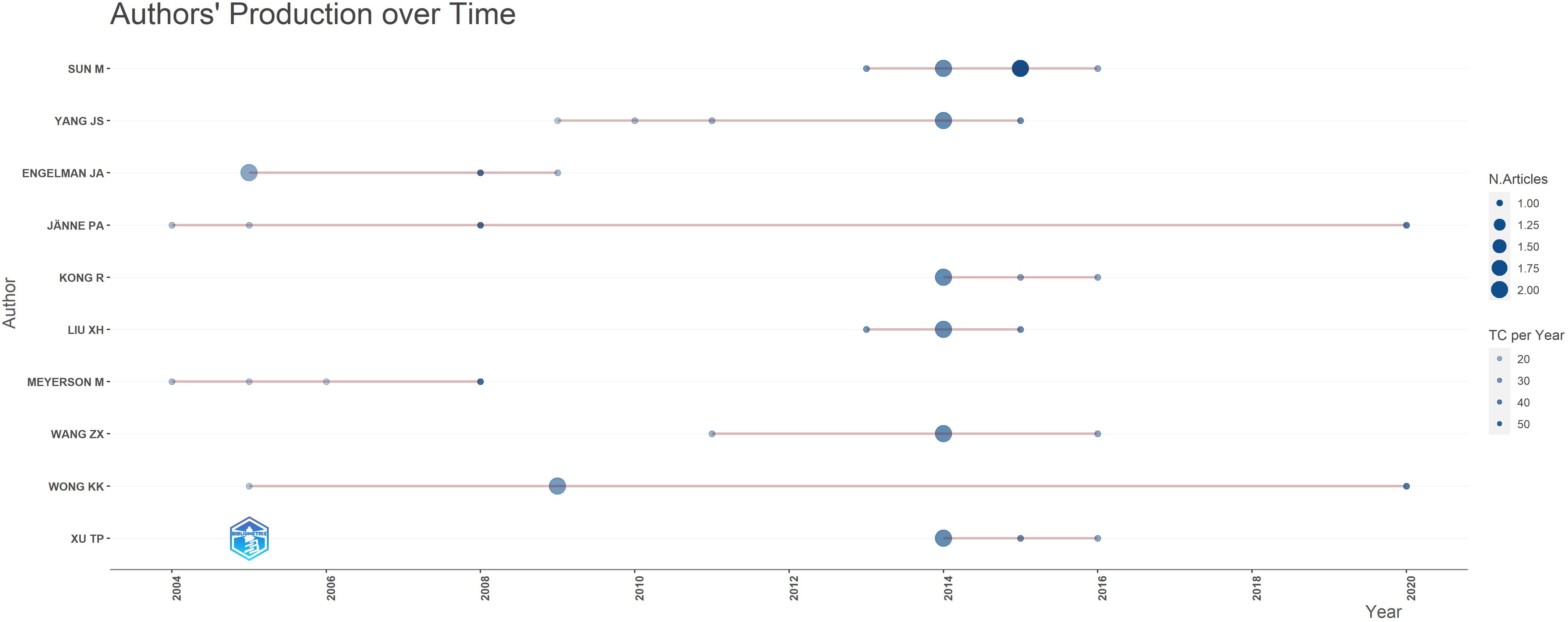
A total of 934 authors participated in the production of high-impact studies within the field. Notably, SUN M and YANG JS emerged as the most prolific authors, each having published 6 articles. However, the works authored by SUN M have garnered a higher total number of citations, as well as a greater average number of citations per year, compared to those authored by YANG JS (refer to Table 1). Based on the publication time distribution diagram for the top 10 authors (Figure 8), there was a notable concentration of articles published in 2014, with several authors belonging to the same research team. For instance, researchers SUN M, YANG JS, XU TP, LIU XH, KONG R and WANG ZX, affiliated with Nanjing Medical University, have contributed significantly to high-impact studies investigating the pivotal role of non-coding RNA (ncRNA) in the proliferation and apoptosis of NSCLC (26, 30, 39, 49, 64, 66, 79). ENGELMAN JA, WONG KK and MEYERSON M, affiliated with Harvard Medical School, collaborate on research focused on developing effective strategies for the treatment of EGFR-mutated LC (42, 44, 52, 59, 65, 67). As illustrated in Figure 9, the collaboration diagram among authors does not constitute a cohesive network. Instead, author collaboration predominantly occurs within individual research teams. Notably, several key research teams have been identified, including BRONSON RT, THOMALE J, LIFSHITS E, ENGELMAN JA and others. The researchers continue to novel strategies to address EGFR mutations in NSCLC. Their findings indicate that targeting heat shock protein 90, along with the simultaneous inhibition of the PI3K-mTOR and MEK signaling pathways may constitute promising therapeutic approaches for these mutations (44, 59). Furthermore, they have identified distinct characteristics of cisplatin-resistant subpopulations within NSCLC cells (72, 102). These scholars, primarily affiliated with prestigious academic institutions, participate in international collaborations to enhance knowledge sharing and collective expertise.

Figure 9. Co-authorship relationship diagram between authors. Each node symbolizes an author, with the size of the node indicating the number of articles published by that author. The connecting lines denote co-authorship relationships, where the thickness of the lines reflects the strength of these collaborations. The color of each node corresponds to the timeline of publication, as depicted in the graph located at the bottom right.
3.7 Analysis of keywords
There are a total of 539 keywords (including keywords that indicate the relevant meaning) in these 100 articles and there are 7 keywords that appear more than 10 times. Keyword co-occurrence analysis serves as an effective tool for identifying prominent topics within this research domain. Figure 10 shows the visual network diagram of keyword co-occurrence, in which the keyword “expression” appears most frequently, followed by “activation”, “apoptosis”, “pathway” and “gefitinib”.
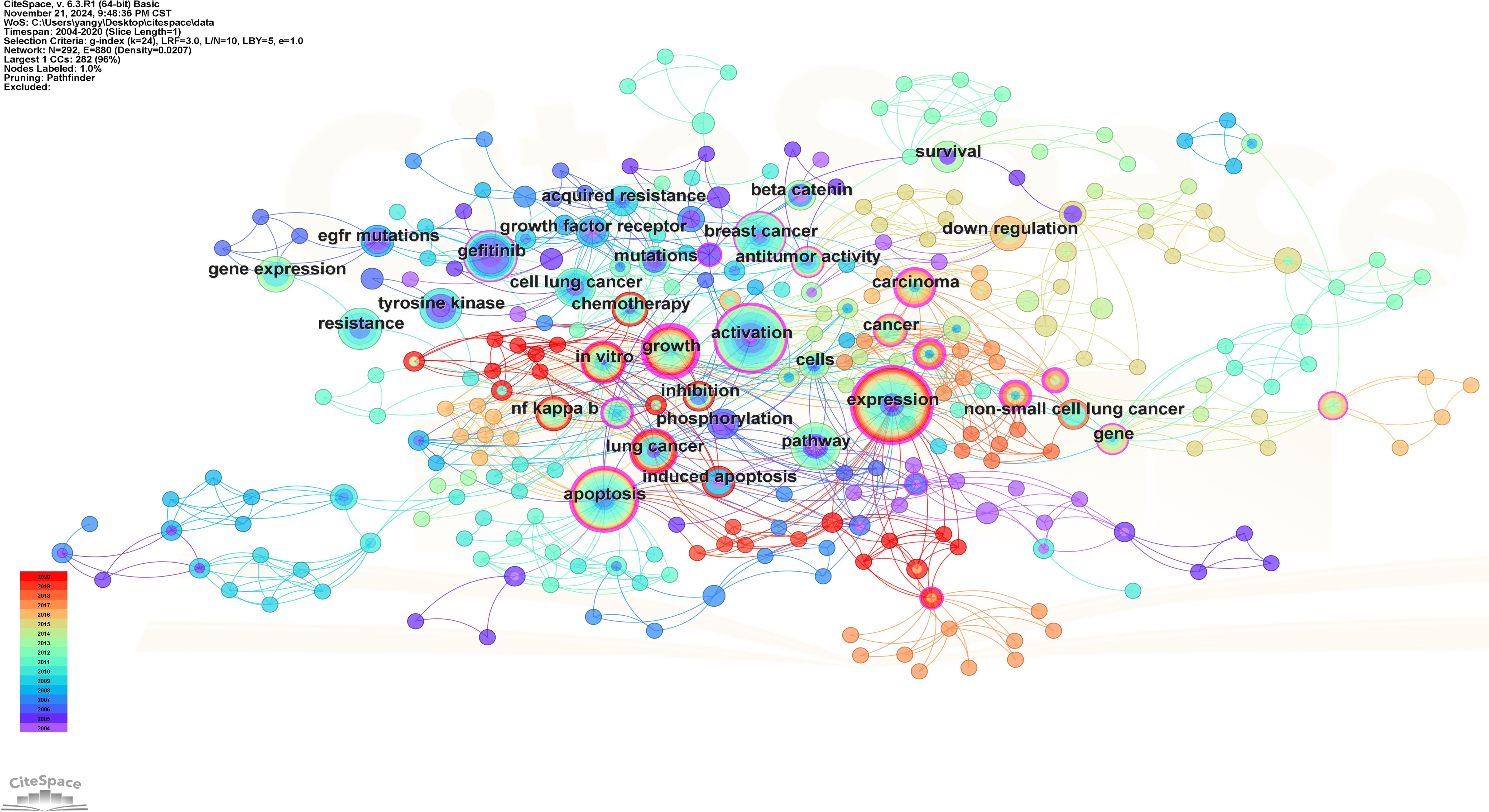
Figure 10. Keywords co-occurrence map. Each node symbolizes a specific keyword, with the size of each node reflecting the frequency of the keyword’s occurrence; a higher frequency results in a larger node. The connecting lines between nodes signify the co-occurrence of keywords. Additionally, the color of each node corresponds to the temporal diagram presented on the left.
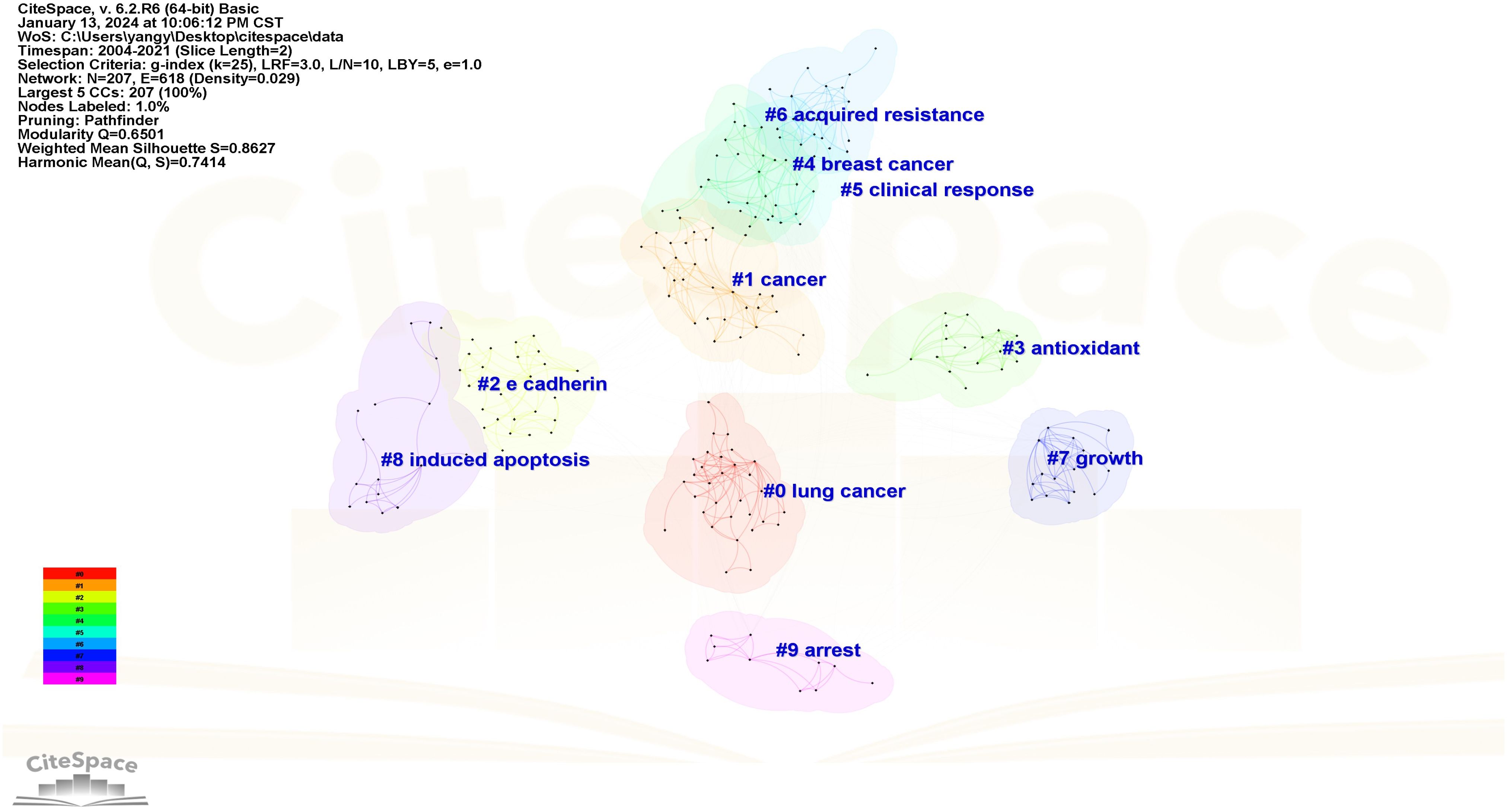
The Log-likelihood Rate (LLR) algorithm in Citespace is used for cluster analysis. The analysis results delineated the keywords into 10 distinct clusters, each represented by a unique color. The clustering modularity value (Q) is 0.6501 and the average clustering contour value (S) is 0.862 7. These metrics suggest that the cluster structure is both significant and reasonable (Figure 11). The smaller the cluster label, the more keywords contained in the cluster. So the cluster with the most keywords is clusters #0, which focuses on the study of apoptosis of mitochondrial pathway. Clusters #1, #5, and #6 explore EGFR tyrosine kinase inhibitors (TKI) to inhibit signal transduction and accelerate apoptosis. Clusters #3 focuses on the relationship between antioxidant system and apoptosis. Clusters #2, #4 and #7 focus on the pathways and bioactive substances that promote cell proliferation and anti-apoptosis. Clusters #8 and #9 examine the link between tumor suppressor genes and proteins encoded by tumor suppressor genes and cell apoptosis (see Supplementary Material). The keyword clustering time plot (Figure 12) shows that “mesenchymal stem cells”, “let-7-miRNA”, “IL-1β” and “cellular senescence” have been prominent keywords recently dicating current research trends and potential future hotspots in this field.
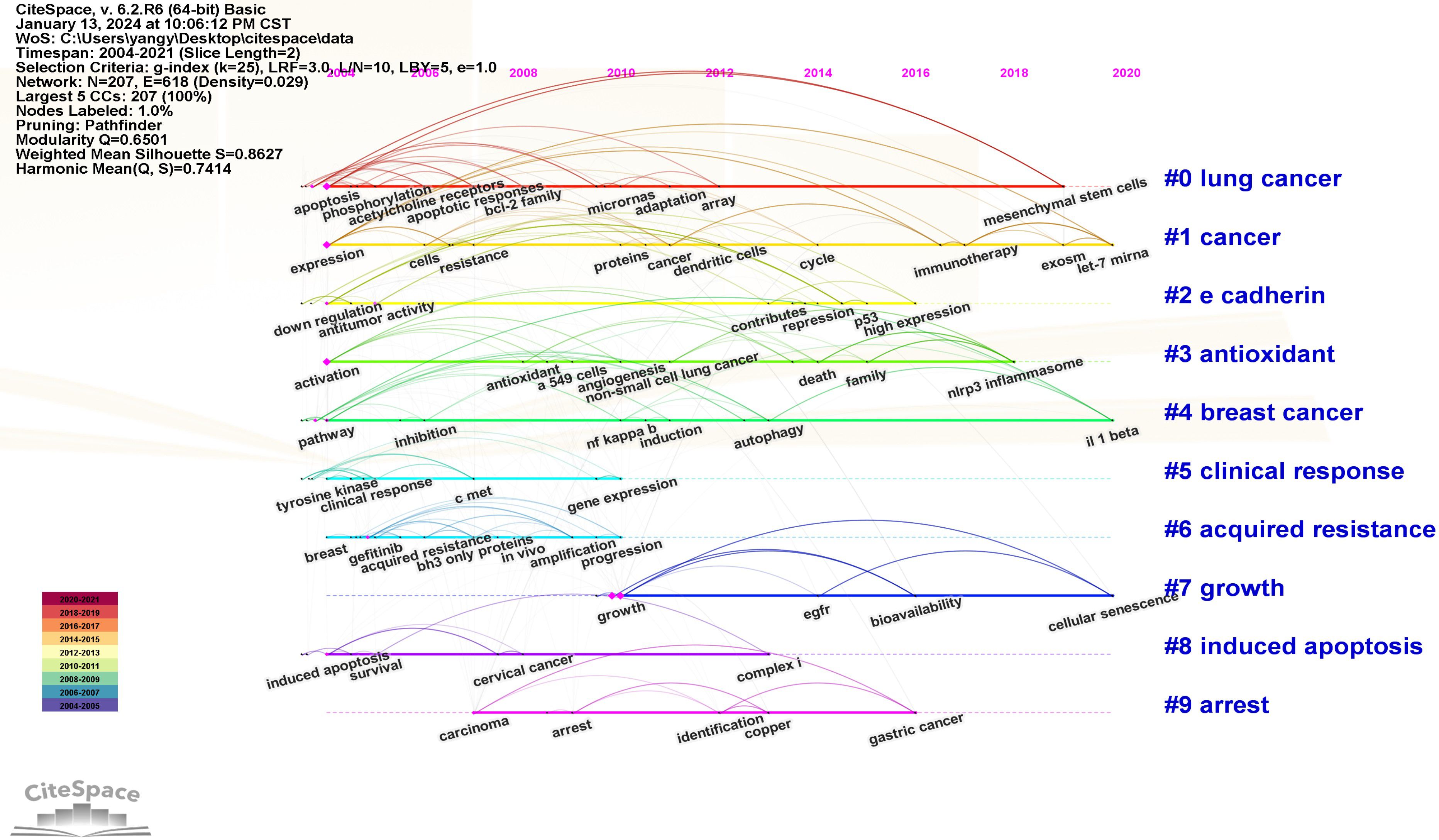
Figure 12. Keywords clustering timeline diagram. The placement of a node along the horizontal axis indicates the initial occurrence of the corresponding keyword. A node positioned further to the right on the horizontal axis signifies a later appearance of the keyword.
3.8 Analysis of references
A total of 3,855 references were referenced in this study, with 15 cited over 5 times by these 100 highly cited articles. The top 10 references in the number of co-citations are listed in Table 3. These were analyzed and visualized using VOSviewer, revealing a tightly connected network that underpins and advances the research field (see Supplementary Material). The first cited reference is the EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy written by PAEZ JG. et al(2004) (114), which has been cited 21 times in these 100 articles, with a total of 7,773 citations. This study reported that EGFR mutations can predict the sensitivity of LC patients to gefitinib. Secondly, Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib, written by LYNCH TJ. et al. (2004) (115), was cited 18 times in the 100 articles, with a total of 9,113 citations. This study sequenced the EGFR gene in 16 LC patients treated with gefitinib, finding gene mutations in 8 of the 9 patients who responded well, but none in the 7 who did not. This is the first evidence linking EGFR mutations to drug efficacy. These two influential articles initiated precise targeted therapy for LC. Later studies (116, 117) revealed that some tumors progressing after EGFR-TKI treatment had mutations at exon 20, site 790 of the EGFR gene, leading to gefitinib resistance. The ISEL study (118) found that gefitinib did not significantly improve overall survival in the general or adenocarcinoma populations, though non-smokers and Asian patients may benefit. However, at the same time, the large phase III study BR21 (119) demonstrated that erlotinib extends survival and alleviates symptoms in previously treated NSCLC patients compared to placebo, which further confirmed the status of EGFR-TKI. A phase 3 open-label study (IPASS) (120) found gefitinib to be more effective than carboplatin plus paclitaxel for initially treating lung adenocarcinoma in East Asian non-smokers or former mild smokers, suggesting that patients with EGFR mutations benefit most from gefitinib as a first-line treatment. It can be seen that in a series of multi-center clinical studies carried out from 2003 to 2005 showed expanding clinical benefits and increased significance of gefitinib for patients with EGFR mutation, making it a major research focus.
3.9 Analysis of research hotspots
Given that the articles included in this study do not encompass articles from 2021 onwards, in order to understand the research hotspots in this field in recent years, the top 10 most-cited articles published between 2021 and March 8, 2024 were retrieved. Using the same retrieval method described previously (see Supplementary Materials for specific articles). Subsequently, the authors’ keywords were compiled, and synonymous keywords were consolidated to quantify their frequency of occurrence. Considering the top ranking “non-small cell lung cancer” and “apoptosis” as thematic keywords in this study, these terms were excluded from further analysis. Among the remaining keywords, “circular RNA”, “Nrf2”, “autophagy” and “ferroptosis” emerged as the most frequently occurring, each appearing twice. This keyword analysis suggests that recent research in this field may focus on promoting apoptosis in NSCLC by regulating circular RNA (circRNA) and targeting the nuclear factor erythroid 2-related factor 2 (Nrf2) signaling pathway.
4 Discussion
This study seeks to identify the top 100 most-cited articles pertaining to the apoptosis of NSCLC over past two decades and to analyze their bibliometric characteristics. The volume of research on NSCLC apoptosis has shown a consistent upward trend in recent years, reflecting the growing scholarly interest in this area. To investigate the current state of research and emerging focal points within this domain, this study compiles and examines the most frequently cited articles from the past twenty years, thereby offering a valuable reference for future research endeavors.
LC ranks as the second most prevalent malignancy globally, with approximately 1,796,144 fatalities reported in 2020. The overall 5-year survival rate for LC patients ranges from 10% to 20% following diagnosis (1). However, for those diagnosed at stage IV, the 5-year survival rate diminishes to a mere 5%. Notably, the majority of patients are diagnosed at an advanced stage of the disease (121). The therapeutic approaches for NSCLC typically encompass surgical intervention, chemotherapy, radiotherapy, molecular targeted therapy, immunotherapy, among others (122). However, the emergence of personalized treatment strategies for NSCLC has notably enhanced patient prognosis, particularly through the advancements in molecular targeted therapy and immunotherapy (123). Recent studies indicate that the mortality rate of NSCLC has declined more significantly than its incidence rate in recent years, coinciding with the approval of targeted therapies (124). Currently, numerous targets and targetable pathways have been identified, including EGFR, ALK, RET, MET, PI3K/AKT/mTOR, RAS-MAPK, and NTRK/ROS1, among others (125, 126).
In this study, while the annual number of studies on apoptosis in NSCLC has consistently increased, the number of highly cited articles exhibits a fluctuating pattern and has demonstrated a negative growth trend over the past two decades. Upon reviewing the titles and abstracts of the 100 highly cited articles included in this analysis, they can be categorized into two main categories that articles are nearly equivalent in quantity. The first area of focus pertains to the mechanisms of apoptosis induced by pharmacological agents and compounds, as well as the mechanisms underlying drug resistance. Specifically, 17 articles address targeted therapies, primarily those involving EGFR-TKIs; 16 articles explore the effects of chemical compounds, including nicotine and curcumin; and 6 articles examine the role of chemotherapeutic agents. The other category pertains to the relationship between the expression of genes, proteins and other biomarkers and the process of apoptosis. Specifically, 17 articles focus on the involvement of related proteins in mediating apoptosis, 11 articles examine the direct or indirect role of microRNAs in regulating multiple apoptotic pathways, and 7 articles explore the regulation of apoptosis by long non-coding RNAs (lncRNA). The United States and China are identified as the first and second leading countries (regions) in terms of published articles (Table 1). The United States holds the distinction of having the highest number of published articles, with a total of 60. Between 2004 and 2007, the United States contributed to over 80% of published articles. However, post-2008, there was a noticeable decline in the proportion of articles originating from the United States. The proportion of published articles originating from China has shown a continuous increase since 2013. Since that time, the annual number of highly cited articles from China has surpassed those from other countries. This trend underscores the significant contributions made by both the United States and China in this field. Regarding publishing institutions, the United States maintains a dominant position, with seven of the top 10 most productive institutions being based there, highlighting the critical role of the United States in this area of research.
The 100 articles analyzed were published in 45 distinct journals. Although none of these articles appeared in the top four medical journals, over 60% were published in journals with a high IF and were published in journals classified as JCR Q1, indicating that the field of NSCLC apoptosis research is well-established and has garnered significant attention from the academic community. Notably, some studies have been featured in prestigious scientific journals such as NATURE and SCIENCE. Additionally, publications have appeared in leading cell biology journals, including NATURE MEDICINE, CANCER CELL, NATURE CELL BIOLOGY, CELL DEATH AND DIFFERENTIATION. In this study, it was observed that the majority of articles with a higher number of citations were published in journals possessing a higher IF. However, journals with a greater number of co-citations did not necessarily exhibit a higher IF. Among the 45 journals analyzed, the lowest IF recorded was 1.6, demonstrating that even journals with a lower IF can produce highly cited articles. Furthermore, previous research has indicated that there is no statistically significant correlation between high citations counts and a journal’s IF (127). This finding underscores the importance of prioritizing the quality of individual papers alongside the journal IF.
Highly cited articles frequently represent the research focal points within a specific field, with keywords serving as significant indicators (128). Among the 100 articles analyzed, the most frequently occurring keyword is “expression”, followed by “activation”, “apoptosis”, “pathway and “gefitinib”. In the research articles examined, apoptosis is primarily induced through the regulation of the expression of microRNAs, tumor suppressors, EGFR, cell cycle proteins, and anti-apoptotic proteins, among others. Simultaneously, the study examined the activation of signaling pathways, transcription factors and various proteases involved in the regulation of apoptosis. The primary focus was on the activation of diverse signaling pathways and their impact on apoptosis. For instance, the activation of pathways such as PI3K/AKT, Wnt/β-catenin, and NF-κB by drugs or compounds plays a crucial role in modulating apoptosis in NSCLC. The findings indicate that the apoptosis pathway and the mechanisms by which drugs induce apoptosis remain among the most prominent topics of interest in the field.
Research priorities are subject to change over time. Consequently, a further examination and analysis of the top 10 most highly cited articles post-2021 revealed that the predominant keywords include “circular RNA”, “Nrf2”, “autophagy” and “ferroptosis”. This suggests that the regulation of circRNA and the targeting of the Nrf2 signaling pathway to induce apoptosis in NSCLC may constitute a significant focus of contemporary research. The dysregulated expression and control of circRNA are recognized as playing a crucial regulatory role in NSCLC (129). Two of the studies included in the analysis have demonstrated that silencing tumor-derived exosomal circRNA_102481 can enhance apoptosis in EGFR-TKIs resistant NSCLC, while the loss of circRNA vacuolar membrane protein 1 similarly promotes apoptosis of cisplatin-resistant NSCLC (129, 130). Additionally, one of the articles reported that kaempferol effectively induces down-regulation of Nrf2 mRNA and disrupts Nrf2 downstream signaling in a redox-independent manner, thereby facilitating apoptosis in NSCLC (131). Four articles have concurrently examined ferroptosis and apoptosis, as well as the interplay between autophagy and apoptosis, thereby analyzing the relationship and interactions between the processes. This research has significantly enhanced the understanding of cell death mechanisms (132–135).
The principal mechanisms of cancer cell death encompass apoptosis, pyroptosis, autophagy and necrosis, with apoptosis being particularly significant (136). Apoptosis is characterized as a gene-regulated, programmed cell death process that maintains cellular homeostasis without eliciting an inflammatory response. It can be initiated via an extrinsic pathway, triggered by cell surface death receptors, or an intrinsic pathway, primarily activated by mitochondrial damage. Both pathways predominantly involve the mediation of effector caspases (137–139). The canonical mechanism of apoptosis involves the activation of the caspase pathway. Initiator caspases, including caspase-2, -8, and -9, play a critical role in triggering the apoptotic cascade by activating downstream effector caspases, such as caspase-3, -6, and -7, which ultimately execute the apoptotic process (140–142). The exogenous apoptotic pathway is activated by various death receptor stimuli, including Fas, tumor necrosis factor (TNF) receptors, and the TNF-related apoptosis-inducing ligand receptor (TRAILR), among others. These membrane receptors, which participate in the apoptotic process, are members of the TNF receptor superfamily. Within this family, TNF and Fas serve as the primary ligands that activate these receptors, thereby initiating the TNF signaling pathway (143, 144). Endogenous apoptotic pathways are activated by factors such as DNA damage, energy deprivation, and hypoxia, leading to the dephosphorylation and cleavage of pro-apoptotic proteins, their translocation to the mitochondria, and the subsequent induction of apoptosis (143). Additionally, the endoplasmic reticulum (ER) plays a critical role in apoptosis. Dysfunction in protein folding within the ER results in the accumulation of unfolded or misfolded proteins in its lumen, thereby inducing ER stress and contributing to the apoptotic process (145).
Numerous signaling pathways are implicated in the regulation of apoptosis in NSCLC, including but not limited to STAT3, Wnt/β-catenin, NF-κB, PI3K/AKT, mTOR, MAPK/Slug, ROS, p53, and Nrf2. These pathways can modulate apoptosis through both endogenous and exogenous mechanisms (146). Additionally, a range of pro-apoptotic regulatory factors are involved this process, including cytochrome C, Smac/DIABLO, Omi/HtrA2, AIF, and endonuclease G, among others (147). Numerous genes and proteins play crucial roles in the mechanism of apoptosis. For instance, TP53 is a significant tumor suppressor gene that can arrest the cell cycle and promote apoptosis. P53, a widely studied pro-apoptotic transcription factor, is a key participant in the cell cycle regulation. Apoptosis signal induced by cancer cells is p53-dependent (148, 149). For instance, the BCL-2 protein family plays a crucial role in the regulation of apoptosis and is categorized into three distinct subgroups: anti-apoptotic proteins (including BCL-2, BCL-xL, BCL-w, MCL-1, and BFL-1), multi-BH domain pro-apoptotic proteins (such as BAK, BAX, and BOK), and BH3-domain only pro-apoptotic proteins (such as BIM, BAD, BID, NOXA, BIK, HRK, BMF and PUMA) (150, 151). Venetoclax, the sole globally approved Bcl-2 selective inhibitor, has received approval for the treatment of relapsed or refractory chronic lymphocytic leukemia and acute myeloid leukemia in elderly patients. However, its clinical indications for other diseases remain under investigation (152).
Consequently, targeting the apoptotic pathway represents a potent strategy for the treatment of NSCLC. Anticancer agents exert their effects by inducing apoptosis and arresting the cell cycle (153). These agents facilitate apoptosis by inhibiting the expression of anti-apoptotic proteins, upregulating pro-apoptotic proteins, decreasing mitochondrial membrane potential, and promoting the release of cytochrome C (154, 155). Mutations in EGFR, human epidermal growth factor receptor-2 (HER-2), or Kirsten rat sarcoma virus oncogene homolog (KRAS) result in increased tumor cell proliferation and diminished apoptosis (156). A critical mechanism by which EGFR-TKIs exert therapeutic effects in EGFR-mutated NSCLC is through the induction of apoptosis. This process is mediated by the regulation of BCL-2 family protein expression, with the pro-apoptotic protein BIM plays a pivotal role (157, 158). Nevertheless, drug resistance in patients with NSCLC patients with NSCLC remains a significant challenge. Drug resistance can be categorized into primary and acquired types. Primary drug resistance is present in a minor subset of cancer cells that have not previously undergone treatment, whereas acquired drug resistance frequently proves difficult to circumvent (159). The mechanisms underlying drug resistance in LC are predominantly associated with gene mutations, gene deletions, gene amplifications, pharmacokinetics factors, targeting of oncogenes, and drug-induced apoptosis (160, 161). Inhibition of apoptosis represents a principal factor contributing to multidrug resistance in tumors, with the PI3K/AKT signaling pathway playing a crucial role in the regulation of this resistance in cancer (162). Residual cancer cells that persist following drug treatment can act as reservoirs for acquired drug resistance. Therefore, augmenting initial drug-induced apoptosis and minimizing residual lesions is a promising strategy to mitigate drug resistance (65). Currently, research on EGFR-TKI has progressed to the fourth generation, with compounds such as EAI045, OBX02-011, LS-106 and CH7233163 under investigation. These fourth-generation drugs are anticipated to address multiple mechanisms of drug resistance, including those associated with third-generation inhibitors, which are predominantly exemplified by resistance to osimertinib (163). Inhibition of apoptosis has implications for tumor drug resistance. In the context of hematological malignancies, the use of BCL-2 family inhibitors in combination therapies has been shown to effectively enhance apoptosis and diminish drug resistance (164, 165). A Phase I clinical trial investigating the combination of osimertinib and ABT-263 (Navitoclax) for the treatment of LC patients with EGFR mutations, following the failure of EGFR-TKI therapy, utilized a dosage regimen of 80 mg/day of osimertinib and 150 mg/day of Navitoclax. The treatment was both efficacious and well-tolerated by the patient cohort (164). A study has demonstrated that the upregulation of BCL2L1 expression constitutes the primary alteration observed in cells that develop resistance to EGFR-TKI (166).
LncRNAs constitute a significant focus within the literature reviewed in this study. These molecules are intricately associated with the pathogenesis of NSCLC and are critical in modulating drug resistance mechanisms. Evidence indicates that lncRNAs contribute to the development of cisplatin resistance in LC, potentially influencing resistance to taxanes and acquired resistance to EGFR-TKIs (167). The lncRNA/PTEN axis is pivotal in modulating the sensitivity of LC cells to chemotherapeutic agents. Research indicates that miRNA-221 contributes to the development of cisplatin resistance in LC by down-regulating PTEN (168). The suppression of LINC01296 has been shown to significantly enhance the sensitivity of NSCLC to paclitaxel (169). LncRNAs play a crucial role in the emergence and progression of resistance to EGFR-TKIs in NSCLC by modulating various molecular pathways. Certain lncRNAs enhance the expression of oncogenes, thereby facilitating tumor cell proliferation, survival, and resistance to EGFR-TKI. Additionally, some lncRNAs suppress the expression of tumor suppressor genes, induce the epithelial-mesenchymal transition (EMT) process, and ultimately contribute to EGFR-TKI resistance. This resistance is partially mediated through the activation of the Rho-associated protein kinase (ROCK) and mesenchymal-epithelial transition factor (MET) signaling pathways. Certain lncRNAs suppress the expression of tumor suppressor genes and induce the EMT process, ultimately contributing to resistance against EGFR-TKIs. This resistance is partially mediated through the activation of the ROCK and MET signaling pathways (170). CircRNAs represent a novel category of endogenous ncRNAs characterized by a covalently closed-loop structure. Currently, substantial evidence suggests that circRNAs are capable of modulating gene expression across various levels. These regulatory functions include influencing the transcription of their parental genes, affecting the splicing processes of their linear counterparts, serving as microRNA sponges, modulating the activity of RNA-binding proteins, and facilitating the encoding of peptides (171). Aberrant circRNAs play a significant role in various developmental processes of malignant tumors, such as tumorigenesis, growth, metastasis, apoptosis, angiogenesis, and resistance to chemotherapy (172). CircRNAs have been identified as crucial elements within the intricate regulatory network of the KRAS pathway across different cancer types. Investigating the involvement of circRNAs in the KRAS pathway could enhance strategies for cancer detection and treatment (173). Currently, numerous studies have identified that certain circRNAs are down-regulated in tumors, potentially contributing to the inhibition of tumor chemotherapy resistance. Conversely, their up-regulation may enhance chemotherapy resistance (174). These circRNAs can modulate the resistance of various tumors to conventional chemotherapy or immunotherapy, either positively or negatively (172). Consequently, circRNAs hold promise as potential diagnostic, prognostic, and therapeutic targets for cancer. Furthermore, they offer a promising avenue for research aimed at addressing the challenge of drug resistance in LC. Nrf2 is a pivotal transcription factor that plays an essential role in the oxidative stress response. It possesses the capacity to modulate apoptosis in human cancer cells, exhibiting a dualistic function. The interaction between Nrf2 and apoptosis processes can significantly influence cancer progression (175). The accumulation of Nrf2 within cancer cells, along with its cytoprotective effects, can be attributed to somatic mutations in Nrf2, Keap1, or CUL3. Additionally, the down-regulation of Keap1, as well as the interactions of p62/Sqstm1 and p21 with Keap1, can impact Nrf2 levels (176). Mutations within the Keap1/Nrf2 signaling pathway have been implicated in the development of resistance to cancer chemotherapy, targeted therapies, and radiotherapy (177). Yoon Jin Lee. et al. (2020) (178) demonstrated that Nrf2 can enhance the survival rate of cancer cells by inhibiting apoptosis. The overexpression of Nrf2 may be associated with tumorigenesis and tumor progression. In cancer cells, the overexpression of NRF2, resulting from constitutive activation or hyperactivation, facilitates their survival in environments rich in ROS, which is closely associated with enhanced proliferation and increased resistance to chemotherapy (179). Currently, research on NRF2 activators is extensive, whereas investigations into NRF2 inhibitors remain limited (180). NRF2 represents a promising therapeutic target or a potential avenue for targeted therapy, given the frequent occurrence of molecular pathway disruptions or loss-of-function mutations in tumors.
5 Conclusion
According to the search, no bibliometric study on the same topic as this study was found. This study utilized the WOSCC, recognized as a relatively authoritative citation database (181). Through an analysis of the 100 most highly cited articles, the study identified the most influential countries (regions), institutions, journals and authors in the field of NSCLC apoptosis. Using bibliometric methods, the research summarized the current status and key areas of interest within this field and projected future research trends based on the temporal distribution of keywords.
Apoptosis-based therapeutic strategies have undergone extensive development over the years. Currently, significant advancements have been achieved in the formulation of novel pharmacological agents and in addressing the challenge of existing drug resistance. Despite the absence of approved anti-apoptotic drugs specifically for NSCLC, these therapies hold considerable potential for future applications. The resistance to anti-tumor drugs remains a formidable challenge within oncology, with the suppression of apoptosis identified as a principal factor contributing to the multidrug resistance observed in tumors. LncRNAs, circRNAs and Nrf2 represent promising therapeutic targets and strategies for addressing cancer drug resistance. The advancement of drugs targeting these molecules holds significant clinical value. Nonetheless, the development of Nrf2 inhibitors aimed at overcoming tumor multidrug resistance remains in its nascent stages, with limited research conducted on these inhibitors to date. NcRNAs have been demonstrated to play a critical role in the development of drug resistance across various therapeutic modalities, including chemotherapy, targeted therapy, immunotherapy, and others, with particular emphasis on lncRNAs and circRNAs. Modulating dysregulated ncRNAs presents a promising therapeutic strategy to counteract drug resistance. Continued investigation into their regulatory mechanisms in anti-tumor drug resistance is essential for advancing the treatment of NSCLC. Consequently, it is anticipated that this area will become a focal point of future research endeavors. Despite these advancements, significant challenges remain in the development of novel pharmacological agents and in overcoming resistance to existing treatments. Consequently, additional research is unequivocally warranted to advance future developments. It is anticipated that such efforts will yield significant breakthroughs in the diagnosis, treatment, and mitigation of drug resistance in cancer.
6 Limitations
Nevertheless, this study is subject to certain limitations: (1) The analysis was conducted using the WOSCC, yet numerous other databases, such as PubMed and Scopus, exist. The citation counts of articles vary across these databases (182), potentially affecting the study’s analytical outcomes based on the database selection. (2) Articles of significant influence may not have accrued sufficient citations to be included in the top 100 due to their recent publication, potentially omitting emerging research trends from the study. (3) The citation count of an article can be influenced by various factors, including the prestige of the publishing journal and the incidence of self-citation, among others. Consequently, while citation count serves as a significant metric for assessing an article’s impact, it should not be considered the sole indicator. (4) Furthermore, this study exclusively included English-language articles from the WOSCC, which may result in the exclusion of potentially influential articles published in other languages.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
LM: Conceptualization, Data curation, Software, Writing – original draft. JZ: Data curation, Writing – original draft. ZD: Writing – original draft, Data curation. PL: Visualization, Writing – original draft. JG: Supervision, Validation, Writing – review & editing. ZL: Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Traditional Chinese Medicine Inheritance and Innovation Center Research Project (Project No. 2023QN12), the Guangdong Provincial Medical Research Fund Project (Project No. B2019148), and the Guangzhou Science and Technology Plan Project (Project No. 201707010299).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1512349/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020:GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Chen P, Liu Y, Wen Y, Zhou C. Non-small cell lung cancer in China. Cancer Commun. (2022) 42:937–70. doi: 10.1002/cac2.12359
3. Favaloro B, Allocati N, Graziano V, Di Ilio C, De Laurenzi V. Role of apoptosis in disease. Aging (Albany NY). (2012) 4:330–49. doi: 10.18632/aging.100459
4. Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. (2020) 17:395–417. doi: 10.1038/s41571-020-0341-y
5. Chang W, Lin E, Tsai M, Huang M, Kuo P. Isolinderalactone inhibits proliferation of A549 human non−small cell lung cancer cells by arresting the cell cycle at the G0/G1 phase and inducing a Fas receptor and soluble Fas ligand-mediated apoptotic pathway. Mol Med Rep. (2014) 9:1653–9. doi: 10.3892/mmr.2014.2015
6. Durieux V, Gevenois PA. Bibliometric indicators: quality measurements of scientific publication. Radiology. (2010) 255:342–51. doi: 10.1148/radiol.09090626
7. Liao H, Mi X, Xu Z. A survey of decision-making methods with probabilistic linguistic information: bibliometrics, preliminaries, methodologies, applications and future directions. Fuzzy Optim Decis Making. (2020) 19:81–134. doi: 10.1007/s10700-019-09309-5
8. Yu Y, Li Y, Zhang Z, Gu Z, Zhong H, Zha Q, et al. A bibliometric analysis using VOSviewer of publications on COVID-19. Ann Trans Med. (2020) 8:816. doi: 10.21037/atm-20-4235
9. Song J, Zhang H, Dong W. A review of emerging trends in global PPP research: analysis and visualization. Scientometrics. (2016) 107:1111–47. doi: 10.1007/s11192-016-1918-1
10. Van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. (2010) 84:523–38. doi: 10.1007/s11192-009-0146-3
11. Liao H, Tang M, Luo L, Li C, Chiclana F, Zeng X-J. A bibliometric analysis and visualization of medical big data research. Sustainability. (2018) 10:166. doi: 10.3390/su10010166
12. Wang S, Zhou H, Zheng L, Zhu W, Zhu L, Feng D, et al. Global trends in research of macrophages associated with acute lung injury over past 10 years: A bibliometric analysis. Front Immunol. (2021) 12:669539. doi: 10.3389/fimmu.2021.669539
13. Aria M, Cuccurullo C. bibliometrix: An R-tool for comprehensive science mapping analysis. J Informetrics. (2017) 11:959–75. doi: 10.1016/j.joi.2017.08.007
14. Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. (2004) 305:1163–7. doi: 10.1126/science.1101637
15. Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. (2007) 17:1298–307. doi: 10.1016/j.cub.2007.06.068
16. Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. (2008) 14:4275–83. doi: 10.1158/1078-0432.CCR-08-0168
17. Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. (2011) 471:110–4. doi: 10.1038/nature09779
18. Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J, et al. Autocrine TNFα signaling renders human cancer cells susceptible to smac-mimetic-induced apoptosis. Cancer Cell. (2007) 12:445–56. doi: 10.1016/j.ccr.2007.08.029
19. Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of k-Ras-Dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. (2010) 18:282–93. doi: 10.1016/j.ccr.2010.08.013
20. Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, et al. Death-receptor o-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. (2007) 13:1070–7. doi: 10.1038/nm1627
21. Ng KP, Hillmer AM, Chuah CTH, Juan WC, Ko TK, Teo ASM, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. (2012) 18:521–8. doi: 10.1038/nm.2713
22. Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. (2013) 23:143–58. doi: 10.1016/j.ccr.2012.12.008
23. Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PloS Med. (2007) 4:1669–80. doi: 10.1371/journal.pmed.0040315
24. Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of slug. Nat Cell Biol. (2009) 11:694–U22. doi: 10.1038/ncb1875
25. Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. (2007) 446(7137):815–9. doi: 10.1038/nature05697
26. Amann J, Kalyankrishna S, Massion PP, Ohm JE, Girard L, Shigematsu H, et al. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res. (2005) 65:226–35.
27. Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. (2013) 13:461. doi: 10.1186/1471-2407-13-461
28. Leung ELH, Fiscus RR, Tung JW, Tin VPC, Cheng LC, Sihoe ADL, et al. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PloS One. (2010) 5:e14062. doi: 10.1371/journal.pone.0014062
29. Leverson JD, Zhang H, Chen J, Tahir SK, Phillips DC, Xue J, et al. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax). Cell Death Disease. (2015) 6:e1590. doi: 10.1038/cddis.2014.561
30. Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. (2015) 14:268–77. doi: 10.1158/1535-7163.MCT-14-0492
31. Puisségur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death And Differentiation. (2011) 18:465–78. doi: 10.1038/cdd.2010.119
32. Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, et al. miR-15a and miR-16 are implicated in cell cycle regulation in a rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. (2009) 69:5553–9. doi: 10.1158/0008-5472.CAN-08-4277
33. Tracy S, Mukohara T, Hansen M, Meyerson M, Johnson BE, Jänne PA, et al. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. (2004) 64:7241–4. doi: 10.1158/0008-5472.CAN-04-1905
34. Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. (2008) 68:7975–84. doi: 10.1158/0008-5472.CAN-08-1401
35. Bey EA, Bentle MS, Reinicke KE, Dong Y, Yang CR, Girard L, et al. An NQO1-and PARP-1-mediated cell death pathway induced in non-small-cell lung cancer cells by β-lapachone. Proc Of Natl Acad Of Sci Of United States Of America. (2007) 104:11832–7. doi: 10.1073/pnas.0702176104
36. Morgillo F, Woo JK, Kim ES, Hong WK, Lee HY. Heterodimerization of insulin-like growth factor receptor/epidermal growth factor receptor and induction of survivin expression counteract the antitumor action of erlotinib. Cancer Res. (2006) 66:10100–11. doi: 10.1158/0008-5472.CAN-06-1684
37. Cragg MS, Kuroda J, Puthalakath H, Huang DCS, Strasser A. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PloS Med. (2007) 4:1681–90. doi: 10.1371/journal.pmed.0040316
38. Johnson FM, Saigal B, Talpaz M, Donato NJ. Dasatinib (BMS-354825) tyrosine kinase inhibitor suppresses invasion and induces cell cycle arrest and apoptosis of head and neck squamous cell carcinoma and non-small cell lung cancer cells. Clin Cancer Res. (2005) 11:6924–32. doi: 10.1158/1078-0432.CCR-05-0757
39. Wang R, Wang ZX, Yang JS, Pan X, De W, Chen LB. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14). Oncogene. (2011) 30:2644–58. doi: 10.1038/onc.2010.642
40. Gills J, Lo Piccolo J, Tsurutani J, Shoemaker RH, Best CJM, Abu-Asab MS, et al. Nelfinavir, a lead HIV protease inhibitor, is a broad-spectrum, anticancer agent that induces endoplasmic reticulum stress, autophagy, and apoptosis in vitro and in vivo. Clin Cancer Res. (2007) 13:5183–94. doi: 10.1158/1078-0432.CCR-07-0161
41. Lutterbach B, Zeng QW, Davis LJ, Hatch H, Hang GZ, Kohl NE, et al. Lung cancer cell lines harboring MET gene amplification are dependent on met for growth and survival. Cancer Res. (2007) 67:2081–8. doi: 10.1158/0008-5472.CAN-06-3495
42. Mukohara T, Engelman JA, Hanna NH, Yeap BY, Kobayashi S, Lindeman N, et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. JNCI-Journal Of Natl Cancer Institute. (2005) 97:1185–94. doi: 10.1093/jnci/dji238
43. Pukac L, Kanakaraj P, Humphreys R, Alderson R, Bloom M, Sung C, et al. HGS-ETR1, a fully human TRAIL-receptor 1 monoclonal antibody, induces cell death in multiple tumour types in vitro and in vivo. Br J Of Cancer. (2005) 92:1430–41. doi: 10.1038/sj.bjc.6602487
44. Faber AC, Li DA, Song Y, Liang MC, Yeap BY, Bronson RT, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Of Natl Acad Of Sci Of United States Of America. (2009) 106:19503–8. doi: 10.1073/pnas.0905056106
45. Dasgupta P, Kinkade R, Joshi B, DeCook C, Haura E, Chellappan S, et al. Proc Of Natl Acad Of Sci Of United States Of America. (2006) 103:6332–7. doi: 10.1073/pnas.0509313103
46. Zhu XL, Wang XY, Wei SZ, Chen Y, Chen Y, Fan XB, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. (2017) 284:2170–82. doi: 10.1111/febs.14132
47. Stabile LP, Lyker JS, Gubish CT, Zhang WP, Grandis JR, Siegfried JM. Combined targeting of the estrogen receptor and the epidermal growth factor receptor in non-small cell lung cancer shows enhanced antiproliferative effects. Cancer Res. (2005) 65:1459–70. doi: 10.1158/0008-5472.CAN-04-1872
48. Tsurutani J, Castillo SS, Brognard J, Granville CA, Zhang CY, Gills JJ, et al. Tobacco components stimulate akt-dependent proliferation and NFκB-dependent survival in lung cancer cells. Carcinogenesis. (2005) 26:1182–95. doi: 10.1093/carcin/bgi072
49. Shi XF, Sun M, Liu HB, Yao YW, Kong R, Chen FF, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinogenesis. (2015) 54:E1–E12. doi: 10.1002/mc.22120
50. Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. (2010) 70:1793–803. doi: 10.1158/0008-5472.CAN-09-3112
51. Liu XG, Yue P, Zhou ZM, Khuri FR, Sun SY. Death receptor regulation and celecoxib-induced apoptosis in human lung cancer cells. Jnci-Journal Of Natl Cancer Institute. (2004) 96:1769–80. doi: 10.1093/jnci/djh322
52. Kobayashi S, Ji HB, Yuza Y, Meyerson M, Wong KK, Tenen DG, et al. An alternative inhibitor overcomes resistance caused by a mutation of the epidermal growth factor receptor. Cancer Res. (2005) 65:7096–101. doi: 10.1158/0008-5472.CAN-05-1346
53. Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. (2008) 27:3845–55. doi: 10.1038/onc.2008.6
54. Chen G, Umelo IA, Lv SS, Teugels E, Fostier K, Kronenberger P, et al. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PloS One. (2013) 8:e60317. doi: 10.1371/journal.pone.0060317
55. Wu SH, Hang LW, Yang JS, Chen HY, Lin HY, Chiang JH, et al. Curcumin induces apoptosis in human non-small cell lung cancer NCI-H460 cells through ER stress and caspase cascade- and mitochondria-dependent pathways. Anticancer Res. (2010) 30:2125–33.
56. Storozhuk Y, Hopmans SN, Sanli T, Barron C, Tsiani E, Cutz JC, et al. Metformin inhibits growth and enhances radiation response of non-small cell lung cancer (NSCLC) through ATM and AMPK. Br J Of Cancer. (2013) 108:2021–32. doi: 10.1038/bjc.2013.187
57. You L, He B, Xu ZD, Uematsu K, Mazieres J, Mikami I, et al. Inhibition of wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene. (2004) 23:6170–4. doi: 10.1038/sj.onc.1207844
58. Turkson J, Kim JS, Zhang SM, Yuan J, Huang M, Glenn M, et al. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther. (2004) 3:261–9.
59. Shimamura T, Lowell AM, Engelman JA, Shapiro GI. Epidermal growth factor receptors harboring kinase domain mutations associate with the heat shock protein 90 chaperone and are destabilized following exposure to geldanamycins. Cancer Res. (2005) 65:6401–8. doi: 10.1158/0008-5472.CAN-05-0933
60. Sos ML, Fischer S, Ullrich R, Peifer M, Heuckmann JM, Koker M, et al. Identifying genotype-dependent efficacy of single and combined PI3K-and MAPK-pathway inhibition in cancer. Proc Of Natl Acad Of Sci Of United States Of America. (2009) 106:18351–6. doi: 10.1073/pnas.0907325106
61. He B, You L, Uematsu K, Xu ZD, Lee AY, Matsangou M, et al. A monoclonal antibody against wnt-1 induces apoptosis in human cancer cells. Neoplasia. (2004) 6:7–14. doi: 10.1016/S1476-5586(04)80048-4
62. Morgillo F, Kim WY, Kim ES, Ciardiello F, Hong WK, Lee HY. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells totreatment with gefitinib. Clin Cancer Res. (2007) 13:2795–803. doi: 10.1158/1078-0432.CCR-06-2077
63. Wang G, Li X, Huang F, Zhao J, Ding H, Cunningham C, et al. Antitumor effect of β-elemene in non-small-cell lung cancer cells is mediated via induction of cell cycle arrest and apoptotic cell death. Cell And Mol Life Sci. (2005) 62:881–93. doi: 10.1007/s00018-005-5017-3
64. Wan L, Sun M, Liu GJ, Wei CC, Zhang EB, Kong R, et al. Long noncoding RNA PVT1 promotes non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther. (2016) 15:1082–94. doi: 10.1158/1535-7163.MCT-15-0707
65. Kurppa KJ, Liu Y, To C, Zhang TH, Fan MY, Vajdi A, et al. Treatment-induced tumor dormancy through YAP-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell. (2020) 37:104. doi: 10.1016/j.ccell.2019.12.006
66. Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. (2014) 5:e1298. doi: 10.1038/cddis.2014.256
67. Alvarez JV, Greulich H, Sellers WR, Meyerson M, Frank DA. Signal transducer and activator of transcription 3 is required for the oncogenic effects of non-small-cell lung cancer-associated mutations of the epidermal growth factor receptor. Cancer Res. (2006) 66:3162–8. doi: 10.1158/0008-5472.CAN-05-3757
68. Haura EB, Zheng Z, Song LX, Cantor A, Bepler G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin Cancer Res. (2005) 11:8288–94. doi: 10.1158/1078-0432.CCR-05-0827
69. Chen LJ, Nan A, Zhang N, Jia YY, Li X, Ling YH, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. (2019) 18:13. doi: 10.1186/s12943-019-0943-0
70. Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu L, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. (2012) 31:2794–804. doi: 10.1038/onc.2011.449
71. Gan PP, Pasquier E, Kavallaris M. Class III β-tubulin mediates sensitivity to chemotherapeutic drugs in non-small cell lung cancer. Cancer Res. (2007) 67:9356–63. doi: 10.1158/0008-5472.CAN-07-0509
72. Barr MP, Gray SG, Hoffmann AC, Hilger RA, Thomale J, O’Flaherty JD, et al. Generation and characterisation of cisplatin-resistant non-small cell lung cancer cell lines displaying a stem-like signature. PloS One. (2013) 8:e54193. doi: 10.1371/journal.pone.0054193
73. Xin MG, Deng XM. Nicotine inactivation of the proapoptotic function of bax through phosphorylation. J Of Biol Chem. (2005) 280:10781–9. doi: 10.1074/jbc.M500084200
74. Hsu YL, Kuo PL, Lin CC. Proliferative inhibition, cell-cycle dysregulation, and induction of apoptosis by ursolic acid in human non-small cell lung cancer A549 cells. Life Sci. (2004) 75:2303–16. doi: 10.1016/j.lfs.2004.04.027
75. Gao JW, Qiu XY, Xi GM, Liu HB, Zhang F, Lv TF, et al. Downregulation of GSDMD attenuates tumor proliferation via the intrinsic mitochondrial apoptotic pathway and inhibition of EGFR/Akt signaling and predicts a good prognosis in non-small cell lung cancer. Oncol Rep. (2018) 40:1971–84. doi: 10.3892/or.2018.6634
76. Song LX, Coppola D, Livingston S, Cress D, Haura EB. Mcl-1 regulates survival and sensitivity to diverse apoptotic stimuli in human non-small cell lung cancer cells. Cancer Biol Ther. (2005). doi: 10.4161/cbt.4.3.1496
77. Li TH, Ling YH, Goldman ID, Perez-Soler R. Schedule-dependent cytotoxic synergism of pemetrexed and erlotinib in human non-small cell lung cancer cells. Clin Cancer Res. (2007) 13:3413–22. doi: 10.1158/1078-0432.CCR-06-2923
78. Teng JF, Mei QB, Zhou XG, Tang Y, Xiong R, Qiu WQ, et al. Polyphyllin VI induces caspase-1-mediated pyroptosis via the induction of ROS/NF-κB/NLRP3/GSDMD signal axis in non-small cell lung cancer. Cancers. (2020) 12:193. doi: 10.3390/cancers12010193
79. Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. (2014) 13:68. doi: 10.1186/1476-4598-13-68
80. Li MQ, Song WT, Tang ZH, Lv SX, Lin L, Sun H, et al. Nanoscaled poly(L-glutamic acid)/doxorubicin-amphiphile complex as pH-responsive drug delivery system for effective treatment of nonsmall cell lung cancer. ACS Appl Materials Interfaces. (2013) 5:1781–92. doi: 10.1021/am303073u
81. Hong WJ, Xue M, Jiang J, Zhang YJ, Gao XW. Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Of Exp Clin Cancer Res. (2020) 39:149. doi: 10.1186/s13046-020-01648-1
82. Aqil F, Kausar H, Agrawal AK, Jeyabalan J, Kyakulaga AH, Munagala R, et al. Exosomal formulation enhances therapeutic response of celastrol against lung cancer. Exp And Mol Pathology. (2016) 101:12–21. doi: 10.1016/j.yexmp.2016.05.013
83. Broüker LE, Huisman C, Span SW, Rodriguez JA, Kruyt FAE, Giaccone G. Cathepsin b mediates caspase-independent cell death induced by microtubule stabilizing agents in non-small cell lung cancer cells. Cancer Res. (2004) 64:27–30. doi: 10.1158/0008-5472.CAN-03-3060
84. Ji BC, Hsu WH, Chung JG. Gallic acid induces apoptosis via caspase-3 and mitochondrion-dependent pathways in vitro and suppresses lung xenograft tumor growth in vivo. J Of Agric And Food Chem. (2009) 57:7596–604. doi: 10.1021/jf901308p
85. Gong XM, Li XF, Jiang T, Xie HK, Zhu ZF, Zhou F, et al. Combined radiotherapy and anti-PD-L1 antibody synergistically enhances antitumor effect in non-small cell lung cancer. J Of Thorac Oncol. (2017) 12:1085–97. doi: 10.1016/j.jtho.2017.04.014
86. Su H, Bodenstein C, Dumont RA, Seimbille Y, Dubinett S, Phelps ME, et al. Monitoring tumor glucose utilization by positron emission tomography for the prediction of treatment response to epidermal growth factor receptor kinase inhibitors. Clin Cancer Res. (2006) 12:5659–67. doi: 10.1158/1078-0432.CCR-06-0368
87. Sos ML, Rode HB, Heynck S, Peifer M, Fischer F, Klüter S, et al. Chemogenomic profiling provides insights into the limited activity of irreversible EGFR inhibitors in tumor cells expressing the T790M EGFR resistance mutation. Cancer Res. (2010) 70:868–74. doi: 10.1158/0008-5472.CAN-09-3106
88. Sos ML, Rode HB, Heynck S, Peifer M, Fischer F, Klüter S, et al. Chemogenomic profiling provides insights into the limited activity of irreversible EGFR inhibitors in tumor cells expressing the T790M EGFR resistance mutation. Cancer Res. (2010) 70:868–74. doi: 10.1158/0008-5472.CAN-09-3106
89. Tajeddine N, Galluzzi L, Kepp O, Hangen E, Morselli E, Senovilla L, et al. Hierarchical involvement of bak, VDAC1 and bax in cisplatin-induced cell death. Oncogene. (2008) 27:4221–32. doi: 10.1038/onc.2008.63
90. Xiong SD, Zheng YJ, Jiang P, Liu RH, Liu XM, Chu YW, et al. MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int J Of Biol Sci. (2011) 7:805–14. doi: 10.7150/ijbs.7.805
91. Ye X, Li Y, Stawicki S, Couto S, Eastham-Anderson J, Kallop D, et al. An anti-axl monoclonal antibody attenuates xenograft tumor growth and enhances the effect of multiple anticancer therapies. Oncogene. (2010) 29:5254–64. doi: 10.1038/onc.2010.268
92. Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ, et al. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol Therapy-Nucleic Acids. (2016) 5:e385. doi: 10.1038/mtna.2016.94
93. Wang S, Han XH, Wang JF, Yao JR, Shi YK. Antitumor effects of a novel chromosome region maintenance 1 (CRM1) inhibitor on non-small cell lung cancer cells in vitro and in mouse tumor xenografts. PloS One. (2014) 9:e89848. doi: 10.1371/journal.pone.0089848
94. Wei Z, Yuan S, Chen YZ, Yu SY, Hao JG, Luo JQ, et al. Enhanced antitumor efficacy by paclitaxel-loaded pluronic P123/F127 mixed micelles against non-small cell lung cancer based on passive tumor targeting and modulation of drug resistance. Eur J Of Pharmaceutics And Biopharmaceutics. (2010) 75:341–53. doi: 10.1016/j.ejpb.2010.04.017
95. Droemann D, Albrecht D, Gerdes J, Ulmer AJ, Branscheid D, Vollmer E, et al. Human lung cancer cells express functionally active toll-like receptor 9. Respir Res. (2005) 6:1. doi: 10.1186/1465-9921-6-1
96. Shishodia S, Koul D, Aggarwal BB. Cyclooxygenase (COX)-2 inhibitor celecoxib abrogates TNF-induced NF-κB activation through inhibition of activation of IκBα kinase and akt in human non-small cell lung carcinoma: Correlation with suppression of COX-2 synthesis. J Of Immunol. (2005) 173:2011–22. doi: 10.4049/jimmunol.173.3.2011
97. Ren WH, Hou JF, Yang CG, Wang HJ, Wu ST, Wu YB, et al. Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21-5p delivery. J Of Exp Clin Cancer Res. (2019) 38:62. doi: 10.1186/s13046-019-1027-0
98. Ye MX, Zhang J, Zhang J, Miao Q, Yao LB, Zhang J. Curcumin promotes apoptosis by activating the p53-miR-192-5p/215-XIAP pathway in non-small cell lung cancer. Cancer Letters. (2015) 357:196–205. doi: 10.1016/j.canlet.2014.11.028
99. David O, LeBeau H, Brody AR, Friedman M, Jett J. Phospho-akt overexpression in non-small cell lung cancer confers significant stage-independent survival disadvantage. Chest. (2004) 125:152S–S. doi: 10.1378/chest.125.5_suppl.152S-a
100. Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S, Navab R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and bcl-2 in lung squamous carcinoma, gejiu city, china. PloS One. (2014) 9:e103698. doi: 10.1371/journal.pone.0103698
101. Ong CC, Jubb AM, Haverty PM, Zhou W, Tran V, Truong T, et al. Targeting p21-activated kinase 1 (PAK1) to induce apoptosis of tumor cells. Proc Of Natl Acad Of Sci Of United States Of America. (2011) 108:7177–82. doi: 10.1073/pnas.1103350108
102. Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. (2010) 24:837–52. doi: 10.1101/gad.1897010
103. Stathopoulos GT, Sherrill TP, Cheng DS, Scoggins RM, Han W, Polosukhin VV, et al. Epithelial NF-κB activation promotes urethane-induced lung carcinogenesis. Proc Of Natl Acad Of Sci Of United States Of America. (2007) 104:18514–9. doi: 10.1073/pnas.0705316104
104. Miyamoto S, Inoue H, Nakamura T, Yamada M, Sakamoto C, Urata Y, et al. Coxsackievirus B3 is an oncolytic virus with immunostimulatory properties that is active against lung adenocarcinoma. Cancer Res. (2012) 72:2609–21. doi: 10.1158/0008-5472.CAN-11-3185
105. Hayami S, Yoshimatsu M, Veerakumarasivam A, Unoki M, Iwai Y, Tsunoda T, et al. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol Cancer. (2010) 9:59. doi: 10.1186/1476-4598-9-59
106. Yang Y, Li H, Hou SC, Hu B, Liu J, Wang J, et al. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PloS One. (2013) 8:e65309. doi: 10.1371/journal.pone.0065309
107. Song LB, Xiong HP, Li J, Liao WT, Wang L, Wu JH, et al. Sphingosine kinase-1 enhances resistance to apoptosis through activation of PI3K/Akt/NF-κB pathway in human non-small cell lung cancer. Clin Cancer Res. (2011) 17:1839–49. doi: 10.1158/1078-0432.CCR-10-0720
108. Toulany M, Kehlbach R, Florczak U, Rodman HP, Wang SM, Chen JY, et al. Targeting of AKT1 enhances radiation toxicity of human tumor cells by inhibiting DNA-PKcs-dependent DNA double-strand break repair. Mol Cancer Ther. (2008) 7:1772–81. doi: 10.1158/1535-7163.MCT-07-2200
109. Singh A, Weddings M, Wakabayashi N, Bunz F, Biswal S. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxidants Redox Signaling. (2010) 13:1627–37. doi: 10.1089/ars.2010.3219
110. Kang J, Kim E, Kim W, Seong KM, Youn H, Kim JW, et al. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of notch-1 expression in non-small cell lung cancer cell lines. J Of Biol Chem. (2013) 288:27343–57. doi: 10.1074/jbc
111. Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, et al. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. (2015) 34:3240–50. doi: 10.1038/onc.2014.267
112. Glasauer A, Seine LA, Diebold LP, Mazar AP, Chandel NS. Targeting SOD1 reduces experimental non-small-cell lung cancer. J Clin Invest. (2014) 124:117–28. doi: 10.1172/JCI71714
113. Zachar Z, Marecek J, Mature C, Gupta S, Stuart SD, Howell K, et al. Non-redox-active lipoate derivates disrupt cancer cell mitochondrial metabolism and are potent anticancer agents in vivo. J Mol Medicine-Jmm. (2011) 89:1137–48. doi: 10.1007/s00109-011-0785-8
114. Paez JG, Jaünne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. (2004) 304:1497–500. doi: 10.1126/science.1099314
115. Lynch TJ, Bwll DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. (2004) 350:2129–39. doi: 10.1056/NEJMOA040938
116. Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. (2005) 352:786–92. doi: 10.1056/NEJMoa044238
117. Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarci-nomas to getinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PloS Med. (2005) 2:225–35. doi: 10.1371/journal.pmed.0020073
118. Thatcher N, Chang A, Parikh P, Pereira JR, Ciuleanu T, von Pawel J, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer). Lancet. (2005) 366:1527–37. doi: 10.1016/S0140-6736(05)67625-8
119. Shepherd FA, Rodrigues JP, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. (2005) 353:123–32. doi: 10.1056/NEJMoa050753
120. Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. (2009) 361:947–57. doi: 10.1056/NEJMoa0810699
121. Xu L, Huang X, Lou Y, Xie W, Zhao H. Regulation of apoptosis, autophagy and ferroptosis by non−coding RNAs in metastatic non−small cell lung cancer. Exp Ther Med. (2022) 23:352. doi: 10.3892/etm.2022.11279
122. Alexander M, Kim SY, Cheng HY. Update 2020: management of non-small cell lung cancer. Lung. (2020) 198:897–907. doi: 10.1007/s00408-020-00407-5
123. Wang MN, Herbst RS, Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med. (2021) 27:1345–56. doi: 10.1038/s41591-021-01450-2
124. Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin K, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med. (2020) 383:640–9. doi: 10.1056/NEJMoa1916623
125. Majeed U, Manochakian R, Zhao Y, Lou Y. Targeted therapy in advanced non-small cell lung cancer: current advances and future trends. J Hematol Oncol. (2021) 14:108. doi: 10.1186/s13045-021-01121-2
126. Yuan M, Huang LL, Chen JH, Wu J, Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Sig Transduct Target Ther. (2019) 4:61. doi: 10.1038/s41392-019-0099-9
127. Ahmad P, Arshad AI, Della Bella E, Khurshid Z, Stoddart M. Systemic manifestations of the periodontal disease: A bibliometric review. Molecules. (2020) 25:4508. doi: 10.3390/molecules25194508
128. Duan SL, Qi L, Li MH, Liu LF, Wang Y, Guan X. The top 100 most-cited papers in pheochromocytomas and paragangliomas: A bibliometric study. Front Oncol. (2022) 12:993921. doi: 10.3389/fonc.2022.993921
129. Yang B, Teng F, Chang L, Wang J, Liu D, Cui Y, et al. Tumor-derived exosomal circRNA_102481 contributes to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small cell lung cancer. Aging (Albany NY). (2021) 13:m13264–13286. doi: 10.18632/aging.203011
130. Xie HY, Yao J, Wang YX, Ni B. Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Delivery. (2022) 29:1257–71. doi: 10.1080/10717544.2022.2057617
131. Fouzder C, Mukhuty A, Kundu R. Kaempferol inhibits Nrf2 signalling pathway via downregulation of Nrf2 mRNA and induces apoptosis in NSCLC cells. Arch Biochem Biophys. (2020) 697:108700. doi: 10.1016/j.abb.2020.108700
132. Zhang QT, Yi HM, Yao H, Lu L, He G, Wu M, et al. Artemisinin derivatives inhibit non-small cell lung cancer cells through induction of ROS-dependent apoptosis/ferroptosis. J Cance. (2021) 12:4075–85. doi: 10.7150/jca.57054
133. Wu HX, Liu AW. Long non-coding RNA NEAT1 regulates ferroptosis sensitivity in non-small-cell lung cancer. J Int Med Res. (2021) 49:300060521996183. doi: 10.1177/0300060521996183
134. Xie CF, Zhou X, Liang CH, Li XT, Ge MM, Chen Y, et al. Apatinib triggers autophagic and apoptotic cell death via VEGFR2/STAT3/PD-L1 and ROS/Nrf2/p62 signaling in lung cancer. J Exp Clin Cancer Res. (2021) 40:266. doi: 10.1186/s13046-021-02069-4
135. Guo HJ, Ding H, Tang X, Liang M, Li S, Zhang J, et al. Quercetin induces pro-apoptotic autophagy via SIRT1/AMPK signaling pathway in human lung cancer cell lines A549 and H1299 in vitro. Thorac Cancer. (2021) 12:1415–22. doi: 10.1111/1759-7714.13925
136. Jiang MX, Qi L, Li LS, Li YJ. The caspase-3/GSDME signal pathway as a switch between apoptosis and pyroptosis in cancer. Cell Death Discovery. (2020) 6:112. doi: 10.1038/s41420-020-00349-0
137. Rogers C, Fogers T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. (2017) 8:14128. doi: 10.1038/ncomms14128
138. Boice A, Bouchier-Hayes L. Targeting apoptotic caspases in cancer. Biochim Biophys Acta-Molecular Cell Res. (2020) 1867:118688. doi: 10.1016/j.bbamcr.2020.118688
139. Van Opdenbosch N, Lamkanfi M. Caspases in cell death, inflammation, and disease. Immunity. (2019) 50:1352–64. doi: 10.1016/j.immuni.2019.05.020
140. Wang Y, Li S, Weng L, Du H, Wang J, Xu X. LASS2 overexpression enhances early apoptosis of lung cancer cells through the caspase−dependent pathway. Oncol Rep. (2022) 48:220. doi: 10.3892/or.2022.8435
141. Mahib MR, Hosojima S, Kushiyama H, Kinoshita T, Shiroishi T, Suda T, et al. Caspase-7 mediates caspase-1-induced apoptosis independently of Bid. Microbiol Immunol. (2020) 64:143–52. doi: 10.1111/1348-0421.12756
142. Mccomb S, Chan PK, Guinot A, Hartmannsdottir H, Jenni S, Dobay MP, et al. Efficient apoptosis requires feedback amplification of upstream apoptotic signals by effector caspase-3 or -7. Sci Adv. (2019) 5:eaau9433. doi: 10.1126/sciadv.aau9433
143. Liu GB, Pei F, Yang FQ, Li L, Amin AD, Liu S, et al. Role of autophagy and apoptosis in non-small-cell lung cancer. Int J Mol Sci. (2017) 18:367. doi: 10.3390/ijms18020367
144. Xu XB, Lai YY, Hua ZC. Apoptosis and apoptotic body: disease message and therapeutic target potentials. Biosci Rep. (2019) 39:BSR20180992. doi: 10.1042/BSR20180992
145. Shi PF, Zhang ZH, Xu J, Zhang L, Cui H. Endoplasmic reticulum stress−induced cell death as a potential mechanism for targeted therapy in glioblastoma. Int J Oncol. (2021) 59:60. doi: 10.3892/ijo.2021.5240
146. Luo YQ, Lu JY, Yu P, Sun JY, Hu YF, Meng XL, et al. Targeting lncRNAs in programmed cell death as a therapeutic strategy for non-small cell lung cancer. Cell Death Discovery. (2022) 8:159. doi: 10.1038/s41420-022-00982-x
147. Lalier L, Vallette F, Manon S. Bcl-2 family members and the mitochondrial import machineries: the roads to death. Biomolecules. (2022) 12:162. doi: 10.3390/biom12020162
148. Schofield JH, Schafer ZT. Regulators mount up: the metabolic roles of apoptotic proteins. Front Cell Death. (2023) 2:1223926. doi: 10.3389/fceld.2023.1223926
149. Sánchez-Rivera FJ, Ryan J, Soto-Feliciano YM, Beytagh MC, Xuan L, Feldser DM, et al. Mitochondrial apoptotic priming is a key determinant of cell fate upon p53 restoration. Natl Acad Sci. (2021) 118:23:e2019740118. doi: 10.1073/pnas.2019740118
150. Shen Q, Li J, Mai JH, Zhang Z, Fisher A, Wu XY, et al. Sensitizing non-small cell lung cancer to BCL-xL-targeted apoptosis. Cell Death Dis. (2018) 9:986. doi: 10.1038/s41419-018-1040-9
151. Zhang CAL, Wu LJ, Liu XC, Gao JM, Liu S, Wu J, et al. Discovery of novel PTP1B inhibitors derived from the BH3 domain of proapoptotic bcl-2 proteins with antidiabetic potency. ACS Med Chem Lett. (2021) 12:1017–23. doi: 10.1021/acsmedchemlett.1c00174
152. Kayagaki N, Webster JD, Newton K. Control of cell death in health and disease. Annu Rev Pathol Mech Dis. (2024) 19:157–80. doi: 10.1146/annurev-pathmechdis-051022-014433
153. Kim HS, Oh HN, Kwak AW, Kim E, Lee MH, Seo JH, et al. Deoxypodophyllotoxin inhibits cell growth and induces apoptosis by blocking EGFR and MET in gefitinib-resistant non-small cell lung cancer. J Microbiol Biotechnol. (2021) 31:559–69. doi: 10.4014/jmb.2101.01029
154. Ke WW, Zhao XX, Lu ZM. Foeniculum vulgare seed extract induces apoptosis in lung cancer cells partly through the down-regulation of Bcl-2. Biomedicine Pharmacotherapy. (2021) 135:111213. doi: 10.1016/j.biopha.2020.111213
155. Arunmanee W, Ecoy GAU, Khine HEE, Duangkaew M, Prompetchara E, Chanvorachote P, et al. Colicin N mediates apoptosis and suppresses integrin-modulated survival in human lung cancer cells. Molecules. (2020) 25:816. doi: 10.3390/molecules25040816
156. Xu Y, Su Z, Li J, Wang Q, Meng G, Zhang Y, et al. Role of RNA−binding protein 5 in the diagnosis and chemotherapeutic response of lung cancer. Oncol Lett. (2019) 17:2013–9. doi: 10.3892/ol.2018.9818
157. Faber AC, Corcoran RB, Eei H, Sequist LV, Waltman BA, Chung E, et al. BIM expression in treatment-naïve cancers predicts responsiveness to kinase inhibitors. Cancer Discovery. (2011) 1:352–65. doi: 10.1158/2159-8290.CD-11-0106
158. Nishihara S, Yamaoka T, Ishikawa F, Higuchi K, Hasebe Y, Manabe R, et al. Mechanisms of EGFR-TKI-induced apoptosis and strategies targeting apoptosis in EGFR-mutated non-small cell lung cancer. Genes. (2022) 13:2183. doi: 10.3390/genes13122183
159. Bain NT, Wang Y, Arulananda S. Minimal residual disease in EGFR-mutant non-small-cell lung cancer. Front Oncol. (2022) 12:1002714. doi: 10.3389/fonc.2022.1002714
160. Ma X, Liang AL, Liu YJ. Research progress on the relationship between lung cancer drug-resistance and microRNAs. J Cancer. (2020) 10:6865–75. doi: 10.7150/jca.31952
161. Zang H, Peng J, Wang W, Fan S. Roles of microRNAs in the resistance to platinum based chemotherapy in the non-small cell lung cancer. J Cancer. (2017) 8:3856–61. doi: 10.7150/jca.21267
162. Liu R, Chen YW, Liu GZ, Li CX, Song YR, Cao ZW, et al. PI3K/AKT pathway as a key link modulates the multidrug resistance of cancers. Cell Death Dis. (2020) 11:797. doi: 10.1038/s41419-020-02998-6
163. Cheng Z, Cui H, Wang Y, Yang J, Lin C, Shi X. et al: The advance of the third-generation EGFR-TKI in the treatment of non-small cell lung cancer. Oncol Rep. (2024) 51:16. doi: 10.3892/or.2023.8675
164. Bertino EM, Gentzler RD, Clifford S, Kolesar J, Muzikansky A, Haura EB, et al. Phase IB study of osimertinib in combination with navitoclax in EGFR-mutant NSCLC following resistance to initial EGFR therapy (ETCTN 9903). Clin Cancer Res. (2021) 27:1604–11. doi: 10.1158/1078-0432.CCR-20-4084
165. Ahn IE, Davids MS. Therapeutic targeting of apoptosis in chronic lymphocytic leukemia. Semin Hematol. (2024) 61:109–18. doi: 10.1053/j.seminhematol.2024.01.015
166. Izumi M, Fujii M, Kobayashi IS, Ho V, Kashima Y, Udagawa H, et al. Integrative single-cell RNA-seq and spatial transcriptomics analyses reveal diverse apoptosis-related gene expression profiles in EGFR-mutated lung cancer. Cell Death Dis. (2024) 15:580. doi: 10.1038/s41419-024-06940-y
167. Wang M, Fu Y, Zhong C, Gacche RN, Wu P. Long non-coding RNA and Evolving drug resistance in lung cancer. Heliyon. (2023) 9:e22591. doi: 10.1016/j.heliyon.2023.e22591
168. Shelash SI, Shabeeb IA, Ahmad I, Saleem HM, Bansal P, Kumar A, et al. lncRNAs’p potential roles in the pathogenesis of cancer via interacting with signaling pathways; special focus on lncRNA-mediated signaling dysregulation in lung cancer. Med Oncol. (2024) 41:310. doi: 10.1007/s12032-024-02536-w
169. Li YL, Zhang H, Guo J, Li WQ, Wang XY, Zhang CY, et al. Downregulation of LINC01296 suppresses non-small-cell lung cancer via targeting miR-143-3p/ATG2B. Acta Biochim Et Biophys Sin. (2021) 53:1681–90. doi: 10.1093/abbs/gmab149
170. Ge X, Shen ZC, Yin YX. Comprehensive review of LncRNA-mediated therapeutic resistance in non-small cell lung cancer. Cancer Cell Int. (2024) 24:369. doi: 10.1186/s12935-024-03549-1
171. Cheng J, Meng JL, Zhu L, Peng Y. Exosomal noncoding RNAs in Glioma: biological functions and potential clinical applications. Mol Cancer. (2020) 19:66. doi: 10.1186/s12943-020-01189-3
172. Chao FM, Zhang Y, Lv L, Wei YQ, Dou XY, et al. Extracellular Vesicles Derived circSH3PXD2A Inhibits Chemoresistance of Small Cell Lung Cancer by miR-375-3p/YAP1. Int J Nanomedicine. (2023) 18:2989–3006. doi: 10.2147/IJN.S407116
173. Hussain MS, Moglad E, Afzal M, Bansal P, Kaur H, Deorari M, et al. Circular RNAs in the KRAS pathway: emerging players in cancer progression. Pathol Res Pract. (2024) 256:155259. doi: 10.1016/j.prp.2024.155259
174. Li Q, Zhang YH, Jin PK, Chen YP, Zhang CC, Geng XC, et al. New insights into the potential of exosomal circular RNAs in mediating cancer chemotherapy resistance and their clinical applications. BioMed Pharmacother. (2024) 177:117027. doi: 10.1016/j.biopha.2024.117027
175. Tian Y, Tang LX, Wang X, Ji YQ, Tu YY. Nrf2 in human cancers: biological significance and therapeutic potential. Am J Cancer Res. (2024) 14:3935–61. doi: 10.62347/LZVO6743
176. Dindi UMR, Al-Ghamdi S, Alrudian NA, Dayel SB, Abuderman AA, Saad Alqahtani M, et al. Ameliorative inhibition of sirtuin 6 by imidazole derivative triggers oxidative stress-mediated apoptosis associated with Nrf2/Keap1 signaling in non-small cell lung cancer cell lines. Front Pharmacol. (2024) 14:1335305. doi: 10.3389/fphar.2023.1335305
177. Aboulkassim T, Tian XH, Liu Q, Qiu DH, Hancock M, Wu JH, et al. A NRF2 inhibitor selectively sensitizes KEAP1 mutant tumor cells to cisplatin and gefitinib by restoring NRF2-inhibitory function of KEAP1 mutants. Cell Rep. (2003) 42:113104. doi: 10.1016/j.celrep.2023.113104
178. Lee YJ, Kim WI, Bae JH, Cho MK, Lee SH, Nam HS, et al. Overexpression of Nrf2 promotes colon cancer progression via ERK and AKT signaling pathways. Ann Surg Treat Res. (2020) 98:159–67. doi: 10.4174/astr.2020.98.4.159
179. Wadowski P, Juszczak M, Wozniak K. NRF2 modulators of plant origin and their ability to overcome multidrug resistance in cancers. Int J Mol Sci. (2024) 25:11500. doi: 10.3390/ijms252111500
180. Lv BB, Xing SS, Wang ZQ, Chen Y, Wang QJ, Bian YY, et al. NRF2 inhibitors: Recent progress, future design and therapeutic potential. Eur J Of Medicinal Chem. (2024) 279:116822. doi: 10.1016/j.ejmech.2024.116822
181. Kuang XY, Zhong ZH, Liang W, Huang SZ, Luo RJ, Luo H, et al. Bibliometric analysis of 100 top cited articles of heart failure-associated diseases in combination with machine learning. Front Cardiovasc Med. (2023) 10:1158509. doi: 10.3389/fcvm.2023.1158509
Keywords: non-small cell lung cancer, apoptosis, cited articles, bibliometrics, visual analysis
Citation: Ma L, Zhang J, Dai Z, Liao P, Guan J and Luo Z (2025) Top 100 most-cited articles on apoptosis of non-small cell lung cancer over the past two decades: a bibliometrics analysis. Front. Immunol. 15:1512349. doi: 10.3389/fimmu.2024.1512349
Received: 16 October 2024; Accepted: 09 December 2024;
Published: 13 January 2025.
Edited by:
Huanling Lai, Guangzhou National Laboratory, ChinaReviewed by:
Cong Xu, Nanjing Medical University, ChinaXiaoqing Fan, Columbia University, United States
Jun Cai, Jiangxi Science and Technology Normal University, China
Copyright © 2025 Ma, Zhang, Dai, Liao, Guan and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jieshan Guan, ZG9jdG9yZ2pzQDE2My5jb20=; Zhijie Luo, bHVvemhpamllNTM0NkBnenVjbS5lZHUuY24=
 Leshi Ma
Leshi Ma Jing Zhang1
Jing Zhang1 Zi Dai
Zi Dai Zhijie Luo
Zhijie Luo