- 1Department of Otorhinolaryngology, The Affiliated Fengcheng Hospital of Yichun University, Fengcheng, Jiangxi, China
- 2Department of Urology, The First People's Hospital of Xiushui, Jiujiang, Jiangxi, China
- 3The First Clinical College, Gannan Medical University, Ganzhou, Jiangxi, China
Urological tumors are an important disease affecting global human health, and their pathogenesis and treatment have been the focus of medical research. With the in - depth study of microbiomics, the role of the microbiome in urological tumors has gradually attracted attention. However, the current research on tumor - associated microorganisms mostly focuses on one type or one site, and currently, there is a lack of attention to the microbiome in the immunity and immunotherapy of urological tumors. Therefore, in this paper, we systematically review the distribution characteristics of the microbiome (including microorganisms in the gut, urine, and tumor tissues) in urologic tumors, the relationship with disease prognosis, and the potential mechanisms of microbial roles in immunotherapy. In particular, we focus on the molecular mechanisms by which the microbiome at different sites influences tumor immunity through multiple “messengers” and pathways. We aim to further deepen the understanding of microbiome mechanisms in urologic tumors, and also point out the direction for the future development of immunotherapy for urologic tumors.
1 Introduction
Urologic tumors commonly include prostate cancer, bladder cancer, and kidney cancer. They not only pose a great threat to the life and health of patients but also put a heavy burden on the global healthcare system. Epidemiological data surveys show that in 2020, the global incidence of prostate cancer was 1,414,259, that of bladder cancer was 573,278, and that of renal cancer was 431,288, resulting in 375,304, 212,536, and 179,368 deaths respectively (1, 2). Compared with the previous year, the incidence rates of the three diseases have increased significantly (3). In addition, all three diseases showed an increasing incidence with age, which was especially evident above 50 years of age. In addition, they also exhibit significant gender differences (3, 4). These figures present a dismal situation. Moreover, with the trend of population aging and economic development, the incidence of these diseases will continue to increase, and the resulting burden will also increase (5). The occurrence of urologic tumors is associated with a variety of factors, including but not limited to genetics, environment, and lifestyle habits. To date, the complex interactions of these factors have led to a pathogenesis of urological tumors that has not been fully elucidated. As medical research progresses, our understanding of these tumors has improved. For example, prostate cancer may be associated with obesity (6), fitness (7), diabetes mellitus (8), dietary patterns (9), and vitamin E supplements (10); bladder cancer may be associated with smoking (11, 12), parasitic infections, and chronic inflammation (13); and renal cancer may be closely related to obesity (14), fruit or vegetable intake (15, 16), smoking (17), hypertension (17–19), and other factors. Treatment methods for urological tumors mainly include surgery, radiotherapy, and chemotherapy. Due to the complexity and diversity of urological tumors, the therapeutic efficacy often varies from person to person. In addition, the high recurrence rate of urological tumors is also a major problem in current treatment. Therefore, finding more effective treatment methods to improve the survival rate and quality of life of patients has become an important topic in the current research field of urological tumors. With the rapid development of biomedical technology, our understanding of urological tumors has penetrated to the molecular level. This enables us to diagnose the disease more accurately, assess the prognosis and develop personalized treatment plans. At the same time, the development and clinical application of novel drugs have also brought new hope for the treatment of urological tumors. Despite the achievements, there is still a long way to go in the prevention and treatment of urological tumors.
The microbiome, a term referring to the sum total of all microorganisms living on and within the human body, encompasses not only bacteria but also a wide range of microbial species, such as fungi and viruses (20). There is a delicate and complex balance between these microscopic lifeforms and the human body, and they play a vital role in several ways (21). For maintaining human health, the microbiome is indispensable. It prevents the invasion and colonization of foreign pathogens by occupying ecological niches and producing antimicrobial substances, etc., thus protecting the host from infection (22). In addition, the microbiome aids in nutrient absorption and utilization in the human body. For example, gut microbes can break down indigestible food components like cellulose and produce beneficial substances such as short - chain fatty acids (SCFA), which provide the body with energy and promote the growth and repair of intestinal cells (23). Besides the above functions, the microbiome has a significant impact on the regulation of the immune system. It participates in the development and operation of the human immune system by interacting with immune cells, inducing immune tolerance or activating immune responses. This immunomodulatory effect helps maintain the homeostasis of the immune system and mobilizes immune resources against pathogens during disease onset (24). When the balance of the microbiome is disturbed, this harmonious symbiosis may turn into a pathogenic factor. Microbiome imbalances may contribute to the development and progression of various diseases, including but not limited to intestinal inflammation, metabolic diseases, and autoimmune diseases. The development of these diseases is often closely related to factors such as reduced microbiome diversity, over proliferation or absence of specific microbial species (23, 25–27). However, with the continuous development of detection techniques, tissues or organs previously considered sterile, such as the kidney and prostate, and urine have also been shown to contain small amounts of microbial communities (28, 29). Additionally, tumor tissues have been shown to be present in the tumor microbiota, which is considered part of the tumor microenvironment and plays an important role in tumor development (30). Overall, the microbiome, as an important component of the internal and external environment of the human body, plays an irreplaceable role in maintaining human health, regulating the immune system, and promoting nutrient absorption. An in - depth study of the interactions between the microbiome and the human body will not only help us better understand the mechanisms of the microbiome’s role in health and disease, but also provide new ideas and methods for disease prevention and treatment.
2 Microbiome in urinary tumors
In the human body, the gut flora was one of the first microbiomes to draw our attention. As early as reported in1944, the discovery of the gut microbiota (GM) confirmed that the microorganisms in the gut are an integral part of the human body and are closely related to human health (31). With the development of 16S rRNA sequencing, an increasing number of diseases, especially tumor diseases, have been found to be closely related to gut flora. In addition, advances in detection technology have enabled us to study microorganisms outside the gut, such as those in urine and tissues. Similar to the gut flora, these microorganisms located at different sites may be involved in tumorigenesis and development in various ways and may also serve as potential molecular markers for predicting tumor formation. Here, we review the microbiota at various sites associated with urological tumors and describe the pathways by which they are involved in tumor development. A preliminary summary of the role of these microorganisms in urinary tumors is presented.
2.1 Prostate cancer
The development of prostate cancer is influenced by various factors, including lifestyle changes, endocrinology, and inflammation. However, as researchers began to consider microorganisms, they realized that alterations in microbial levels were also strongly associated with prostate cancer development. A study by Boursi et al. confirmed a slight increase in the risk of prostate cancer after repeated exposure to antibiotics such as penicillin, quinolones, sulfonamides, and tetracyclines. They concluded that the prolonged use of these antibiotics disrupts the normal microbial structure of the gut, ultimately leading to an increased risk of prostate cancer (32). On the contrary, another study (33) showed that Astragaloside IV co - peptides derived from scorpion venom (PESV) treatment can inhibit prostate cancer tumor growth by restoring the GM and microbial metabolic homeostasis through the AGE - RAGE pathway. Similarly, an in vivo study (34) confirmed that fecal microbiota from healthy control mice significantly reduced TNF - α levels and inhibited tumor growth when transplanted into CRPC mice. This indicates that gut microbial homeostasis is important for prostate cancer development. Subsequently, two Mendelian randomization studies (35, 36) evaluated the association between several gut flora and prostate cancer, and the two studies yielded consistent results; specifically, the level of Alphaproteobacteria in the gut was negatively associated with the risk of prostate cancer. The above - mentioned studies initially identified gut microorganisms at the phylum level associated with prostate cancer development. Furthermore, a meta - analysis (37), which included 7 research papers with a total of 250 prostate cancer patients and 192 controls, further assessed the gut flora closely associated with prostate cancer at the bacterial phylum level. Their results showed that in terms of gut flora abundance, among prostate cancer patients, the abundances of Proteobacteria, Bacteroidia, Clostridia, Bacteroidales, Clostridiales, Prevotellaceae, Lachnospiraceae, Prevotella, Escherichia, Shigella, Faecalibacterium, and Bacteroides were higher. Conversely, in the control group, the abundances of Actinobacteria, Bacteroidetes, Firmicutes, Selenomonadales, Veillonella, and Megasphaera were higher. In addition, a study (38) that compared the gut flora of patients in the high - risk, low - risk, and control groups by 16S rRNA sequencing found that the levels of several specific gut microbes, namely Rikenellaceae, Alistipes, and Lachnospira, were significantly increased in the high - risk group. This suggests that the GM is not only closely associated with tumorigenesis but may also further influence tumor progression, such as invasion and metastasis.
However, instead of regarding the microbiota as an independent risk factor, we need to focus on its role in the host, specifically, what processes in the host it influences to contribute to tumorigenesis and progression. As previously mentioned, prostate cancer is associated with various factors, including poor lifestyle habits, endocrine metabolism, and inflammation, and we will continue to explore how the gut flora is involved in tumor progression through its participation in these factors. We know that poor dietary habits, such as a high - fat diet, are a major risk factor for prostate cancer, and that long - term intake of high - fat foods, especially those rich in saturated and trans fats, can lead to elevated androgen levels in the body. In addition, a study (39, 40) confirms that a high - fat diet also promotes prostate cancer progression by affecting the abundance of specific gut microbial species as well as the levels of genes involved in lipid metabolism. Altered endocrine levels are another major factor in prostate cancer development, and one study (41) found that through a functional analysis of the differential gut flora associated with prostate cancer, the majority of the GM were associated with steroid biosynthesis (5 - alpha reductase) metabolism. In addition, GM have been shown to activate local MAPK and PI3K pathways via their - derived short - chain fatty acids, which in turn affect IGF1 signaling and ultimately prostate cancer growth (41). Finally, the influence of the GM on host immunoinflammation is also a key factor in promoting tumor progression. Transplantation of feces from CRPC patients into normal control mice revealed an increase in SCFA-producing microorganisms, including Ruminococcus, Alistipes, and Phascolarctobacterium, and a corresponding increase in their intestinal levels of SCFAs. These SCFAs, on the one hand, enhanced PCa cell migration and invasion by inducing TLR3 - triggered autophagy to further activate NF - κB and MAPK signaling, and on the other hand, reprogrammed the tumor microenvironment through enhanced autophagy by releasing higher levels of the chemokine CCL20, which recruited more macrophage infiltration and simultaneously polarized them to the M2 type (42). Similarly, another study demonstrated that Cutibacterium acnes in the gut induces the expression of macrophage - immunity genes and can influence the levels of regulatory T cells within the tumor microenvironment (43). In addition, the levels of several pro - and anti - cancer molecules in the host are closely linked to the gut flora. Elevated levels of phenylacetylglutamine (PAGln) greatly increase the likelihood of developing high - risk prostate cancers, and the presence of microorganisms in the gut that catabolize the amino acid phenylalanine to produce PAGln is considered a significant risk for invasive prostate cancers (44, 45).
As the notion that urine is sterile has been refuted, researchers have begun to study the urinary microbiome and have found that, in addition to gut microbiome differences, there are also differences in the microbial composition of urine between prostate cancer patients and healthy individuals. A study by Hurst et al. (46) confirms that urinary microbes are associated with a higher risk of prostate cancer and identifies five prognostic species through bacterial testing of urinary sediment associated with poor prognosis. A systematic review of the urologic microbiome and prostate disease (47) also indicated that specific genera/species such as Streptococcus, Bacteroidetes, Faecalibacterium, Lactobacilli, and Acinetobacter were found in higher abundance in the urine of prostate cancer patients. In addition, Alanee et al. (48) collected urine from patients with transrectal prostate biopsies for a before - and - after comparison and found that when the prostate biopsy was negative, the level of Lactobacillus in the urine was high, whereas when the biopsy was positive, the level of Lactobacillus in the urine was reduced and replaced by a higher level of Prevotella bacteria. In addition to showing significant differences in the urinary microbiome compared to healthy controls, there were also differences in the urinary microbiomes of patients with different - grade tumors (49), implying that, like the gut flora, urinary microbes may be further involved in tumor progression. A study (50) confirmed that prostate tissue contains a large number of bacteria, mainly located in the gland, and that there is a pathophysiological correlation between its microbial composition and prostate cancer development. This finding provides an interesting perspective for prostate cancer research and treatment. Subsequently, several genomic studies (51, 52) comparing the macrogenomic differences between prostate tumors and adjacent benign tissues have also confirmed the non - sterile environment in the prostate gland and some differences in the microbial composition between prostate cancer tissues and benign tissues, such as a significantly higher abundance of Shewanella in tumor tissues and reduced species diversity in Staphylococcus saprophyticus and Vibrio parahaemolyticus. Similarly, the microbiota in the tumor tissue was strongly associated with the level of tumor progression or prognosis. It was found that high - grade prostate cancer tissues were enriched with more Cutibacterium, Pelomonas, and Corynebacterium genera compared to low - grade prostate cancer tissues. Furthermore, Kim (53, 54) compared the microbiota of biochemically recurrent and recurrence - free prostate cancer tissues and found that the abundance of Lactobacillus was relatively high in the recurrence - free group, further emphasizing the role of Lactobacillus in prostate defense.
2.2 Bladder cancer
Similar to prostate cancer, bladder carcinogenesis is closely related to the GM, despite not being part of the gastrointestinal tissue. A case - control study in China comparing the differences in GM between bladder cancer patients and normal controls found that the abundances of Clostridium cluster XI and Prevotella were reduced in bladder cancer patients (55). The effect of GM on bladder cancer can be achieved, on the one hand, by modulating the body’s inflammatory immune response. As He et al. found, normalization of the GM using radicicol thiols reduces the inflammatory and immune response, thereby preventing chemically - induced bladder cancer (56). On the other hand, GM metabolites are also important means by which the GM influences tumors in distal tissues. For example, the human GM can influence the toxic metabolism of nitrosamines, and its depletion significantly reduces the progression and severity of nitrosamine - induced bladder cancer (57). Similarly, metabolites of the GM can influence retinoic acid metabolism and thus the preventive effect of microbial A on bladder cancer via various pathways (58).
As a microbiota that can directly contact the bladder, in contrast to the GM, the urine microbiome has been extensively studied in relation to bladder cancer development. Several systematic reviews of the literature (59–61) have summarized the changes in the urine microbiome of bladder cancer patients, and we have also compiled them in Table 1. As demonstrated in the table, although there are large discrepancies among the results of these studies, probably due to differences in the characteristics of the study subjects and technical reasons for urine sampling, a more consistent trend can be summarized. For example, the abundances of Fusobacterium and Actinobacillus were higher in the urine of bladder cancer patients, while, in contrast, the abundances of Lactobacillus and Weyronella were higher in the urine of the control group. In addition to showing deviations from normal in the bladder cancer patients’ urine microbiome, more interestingly, it has been found that the urine microbiome composition of some patients with non - invasive bladder cancer slowly changes over time after transurethral resection and was even found to regress to previously normal levels in some recurrence - free tumor groups (53, 73). Similarly, the urinary microbiome was also strongly associated with patients’ postoperative survival and recurrence (74). In addition, there was also a difference in the abundance of the urine microbiome among patients with different tumor grades (64). This implies, on the one hand, that the urine microbiome plays an important role in bladder cancer and, on the other hand, suggests its potential as a prognostic marker for bladder cancer treatment. However, despite this, studies related to the urine microbiome have faced major challenges. One of the biggest problems is the collection and contamination of urine samples. Hourigan et al. compared mid - stream urine samples with those obtained by cystoscopy and found that in male patients, there was a significant difference between the urine microbiomes identified by the two urine collection methods (75). Similarly, there have been reports on the microbiota of bladder cancer tissues. Pederzoli ‘s study (62) showed a higher abundance of Burkholderia in tumor tissues compared to non - tumor tissues. Another study (72) that compared 22 cancerous tissues and 12 normal tissues adjacent to the cancer found lower relative abundances of Lactobacillus, Prevotella_9, and Ruminococcaceae, while Cupriavidus, Brucellaceae, Acinetobacter, Anoxybacillus, Escherichia, Shigella, Geobacillus, Pelomonas, Ralstonia, and Sphingomonas had higher relative abundances in cancer tissues. In addition, it has been shown (76) that microbiota detection in samples from cancerous tissues is highly reproducible and independent of the given bladder tissue collection site when compared to the urinary microbiome. In addition, microbial genera such as Akkermansia, Bacteroides, Clostridium sensu stricto, Enterobacter and Klebsiella are more representative in bladder cancer identification in tissues compared to the urinary microbiome.
2.3 Renal cancer
Unsurprisingly, the microbiota has also been reported to be associated with the development of renal malignancies. A Mendelian randomization study (77) involving 248,356 benign tumors and 238,678 renal malignancies genetically controlled for intestinal microbiota found that Actinobacteria, Intestinimonas and Veillonella were protective against renal carcinoma, whereas higher levels of intestinal Enterobacteriaceae increased the risk of kidney cancer. Another similar study (78) also showed that certain microbial genera in the gut, such as Alphaproteobacteria, Bacilli, Coprococcus2, Intestinimonas, Lachnoclostridium, Lactococcus, Ruminococcus torques and Eubacterium brachy, have been associated with renal cell carcinoma. Similarly, a study (79) evaluated the differential expression of microbiota in renal cancer tissues. A comparison of 16S rRNA sequencing of tumors and adjacent normal tissues of 24 patients with renal cell carcinoma revealed that some genera, such as Nitriliruptor, Deinococcus, Actinomyces, Gordonia, Pseudoclavibacter, Microlunatus, Amycolatopsis, Weissella, Brevundimonas and Phyllobacterium, were abundant in renal cancer tissues; however, other genera, such as Geothrix, Bifidobacterium, Paenisporosarcina, Alloiococcus, Caloramator, Allobaculum and Pseudoclavibacter, were also found to be abundant in renal cancer tissues, while Caloramator, Allobaculum, Rhodoplanes, Carludovica, Novosphingobium, Dechloromonas, Klebsiella, Coxiella and Pseudomonas were predominantly in normal tissues. Also, Heidler et al. (80) compared microbiome expression differences in benign and malignant kidney tumor tissues and found that the microbiomes of Aeromonas salmonicida, Pseudoalteromonas haloplanktis, Parageobacillus toebii, Trachelomonas volvocinopsis, Mycoplasma mycoides and Halomicrobium mukohataei are particularly common in cancer tissues.
It can be seen that microbiota colonizing the gut, urine, and tissues are closely associated with the occurrence, development, and treatment of urinary tumors. This indicates the important role of the gut microbiota in urinary tumors. In recent years, a great deal of research has been conducted on the mechanisms between microbiota and urinary tumors, and the involvement of microbiota in the regulation of tumor therapy has been continuously explored. However, summaries regarding the microbiome and immunotherapy of urinary tumors are scarce. Several recent studies have found that antibiotic intervention can influence tumor immunotherapy in certain tumors, which is thought to be associated with gut flora dysbiosis. However, these studies are only at a preliminary stage, and as summarized earlier, a considerable number of microbiota species located at different sites are strongly associated with urological tumors and are involved in tumor development through various pathways. Therefore, it is necessary to comprehensively compile these elements.
3 Role of microbiome in urinary tumor immunity
In the last two decades, modulation of the tumor immune response has emerged as a reliable option for tumor therapy. Similarly, as gene sequencing technology continues to evolve, there is growing evidence that the composition of the microbiota (both intestinal and extraintestinal) confers susceptibility to certain tumor diseases, as well as influencing cancer outcomes, and it has been demonstrated that this correlates with modulation of the immune response (81, 82). A large body of data (83–88) suggests that testing the microbiota of oncology patients can be used as a marker to predict a patient’s response to chemotherapy or immunotherapy, as well as to predict treatment-related toxicity. This means that shaping the microbiota is expected to be a new target for cancer treatment efficacy and toxicity. This exciting result has attracted the interest of many researchers, who have begun to employ a variety of strategies to shape the microbiota to enhance anticancer efficacy. Several preliminary trials (85, 87–89) have shown that fecal micro transplantation (FMT), in combination with immunotherapy, induces differential expression of T-cell and NK-cell related pathways to control tumors and improve the immune response; similarly, the use of probiotics (Lactobacillus and Bifidobacterium), whose substrates are selectively utilized by the host microorganisms to confer health benefits, can also promote immune modulation, among other things, to alleviate dysbiosis and enhance anticancer efficacy and immunity and immune checkpoint inhibitor (ICI) therapeutic effects (84, 90, 91). In addition, with the development of gene technology, some engineered microbial products have attracted the attention of a large number of researchers due to their unique advantages. To date, several research reports have confirmed that a variety of engineered bacterial strains reliably provide antitumor benefits in many different settings (92–94). However, despite the rapid development of the concept of the gut microbiota as a precision medicine tool over the past decade, the number of published studies on practical interventions to modify the gut microbiota is relatively limited and lacks specificity The promise of the gut microbiome as part of individualized treatment strategies (95). Further elucidation of the microbiota and tumor immunity mechanisms will help drive its further development.
As is known, immunotherapy comprises active immunotherapy and passive immunotherapy. In urologic tumors, the former mainly involves bladder BCG infusion, while the latter mainly includes monoclonal antibodies, such as some immunocheckpoint inhibitor drugs like PD - 1/PD - L1 and CTLA - 4 antibodies. Significant efficacy has been achieved in many malignant tumors, including urologic tumors (96–98). It is worth noting that there is growing evidence that these groups of organisms, to which we have previously paid little attention, play an important role in the use of these drugs for the treatment of urologic tumors. Knorr et al. classified patients with high - risk non - muscle - invasive bladder cancer as BCG non - responsive and BCG - responsive (defined as no evidence of tumor recurrence within 2 years of BCG treatment) and found that the Lactobacillus was significantly enriched in BCG - responders (99). The use of tyrosinase inhibitors and antibiotics prior to the use of nivolumab for advanced renal cell carcinoma significantly affects the treatment efficacy, which is thought to be closely related to the fact that tyrosinase inhibitor and antibiotic use affects the microbiological composition of patients (84). Similarly, another study has confirmed significant changes in the fecal microbial composition during the treatment of patients with metastatic renal cell carcinoma with PD - 1. Furthermore, this study found that patients with higher microbiota diversity tended to have a better response to treatment. All of this suggests that the composition of some specific microorganisms is closely related to tumor immunotherapy. However, this is insufficient to address the challenges of tumor immunotherapy in the clinic. Further exploration of various types of microbiota in tumor immunity and the molecular mechanisms of immunotherapy is the key to solving this problem.
A number of “checkpoint” mechanisms exist in the human immune system for regulating the strength of the immune response. Among them, cytotoxic T - lymphocyte - associated antigen 4 (CTLA - 4), PD - 1, and PD - L1 are the most intensively researched immune checkpoints. PD - L1 is a molecule expressed on the tumor cell membrane surface, while its receptor, PD - 1, is expressed on the T - cell surface. The binding of PD - L1 to PD - 1 negatively inhibits signaling initiation and ultimately inhibits T - cell activation, proliferation, and cytokine secretion (e.g., IFN - γ, IL - 2, etc.) (100). Since 2015, ICIs have been successively approved by the U.S. Food and Drug Administration (FDA) for the treatment of renal and uroepithelial cancers (101, 102). In addition, although many studies (103–105) have shown that ICIs exhibit limited therapeutic activity in prostate cancer, there is also evidence indicating some therapeutic promise of ICIs in prostate cancer (106). As we mentioned earlier, the balance of the microbiota plays an important role in host physiological functions, especially in shaping the body’s normal and abnormal immune responses, which in turn affects tumor promotion or suppression. This suggests that the role of the microbiota in antitumor immunosurveillance cannot be underestimated and could be a potential biomarker of response to ICIs. As hypothesized, the microbiota we focused on proved to be a key factor that may be partially responsible for the lack of response to treatment with ICIs. Evidence from preclinical mouse models suggests that Alistipes gavage restores the response of TNF-dependent tumors to immunotherapy, whereas Lactobacillus inhibit the response (107). Similarly, Sivan et al. (108) found that mice carrying different microbiota presented different results in terms of anti-tumor immunity and melanoma growth. Primarily, the abundance of Bifidobacterium was shown to correlate with T-cell responses, and anti-PD - L1 treatment combined with transoral feeding of these bacteria nearly eliminated tumor growth by increasing dendritic cell maturation and anti-tumor CD8+ T-cell activity. Evidence from multiple clinical patients has led to similar conclusions. In treated patients with metastatic melanoma, a variety of bacteria (Faecalibacterium prauznitzii, Holdemania filiformis and Dorea formicigenerans) were associated with anti-CTLA - 4 plus anti-PD - 1 clinical efficacy (85, 109). In patients with non-small cell lung cancer (NSCLC) or uroepithelial carcinoma treated with anti-PD - 1, baseline microbiota analyses showed that patients with clinical benefit had higher microbiota richness. A higher distribution of the Firmicutes, as well as Akkermansia and Alistipes, was found in responding patients (88). However, some patients with certain tumors exhibit primary resistance to ICIs. Moreover, even in some tumors where ICIs are generally beneficial, patients show large variations in treatment outcomes (96, 97, 110). Therefore, we need to further clarify the reasons for this and the composition of microorganisms associated with urologic tumors to provide a good starting point.
The Bacillus Calmette - Guérin (BCG) vaccine is an important means for preventing tumor recurrence in non - muscle - invasive bladder cancer (NMIBC). Additionally, BCG can also delay tumor progression and reduce the risk of tumor infiltration into the muscularis propria and even distant metastasis, and the therapeutic efficacy of BCG is more pronounced in high - risk NMIBC patients (111, 112). A recent study (113) evaluated the accuracy of the NMIBC scoring model in predicting disease progression in patients with high-risk stage T1 NMIBC treated with BCG. The study demonstrated that more than 70% of patients did not progress to muscle-invasive disease after intravesical BCG infusion following re-TURB, confirming the benefit of BCG therapy in these patients. Although the therapeutic mechanism of BCG is still unclear, available evidence suggests that innate and adaptive immunity play key roles in BCG treatment, which produces pathogen-associated molecular patterns that can be recognized by host pattern recognition receptors (e.g., TLR2, TLR4, and TLR9, etc.), which induces MyD88 signaling to regulate pro-inflammatory cytokine production, and MyD88 deficiency leads to reduced response to BCG treatment in mice (114). In the urine and bladder wall of BCG-treated patients, the number and activity of several innate immune cells, including macrophages (115), polymorphonuclear cells (116), dendritic cells (117), and natural killer cells (118), were altered, and removal of these cells abrogated the efficacy of BCG treatment in the mouse model. In terms of adaptive immunity, T cells are central to the treatment of bladder cancer with BCG, and their presence can be detected in the urine and bladder mucosa of patients treated with BCG for several months (119). In addition, BCG shifts the urinary cytokine milieu from Th2 to Th1 (120, 121), and changes in the levels of Th1-associated cytokines (e.g., IL-12 and interferon γ, etc.) are strongly correlated with the clinical response. The mechanisms by which BCG is used in the treatment of bladder cancer are multifaceted and interrelated. During the interaction with uroepithelial cells, BCG adheres to them, and the likely mechanism is that Mycobacterial fibronectin adhesion protein binds to host fibronectin, which in turn attaches to uroepithelial cells (122). Bladder cancer cells can uptake BCG, and after uptake they secrete a variety of immune-activating effectors, which may recruit or activate immune cells, although there is no definitive in vivo evidence that this uptake process is necessary for the efficacy of BCG (123). At the same time, BCG has a direct toxic effect on bladder cancer cells, which has been demonstrated in vitro in experiments with high ratios of BCG to cancer cells, but the in vivo situation is not clear (124, 125). Encouraging data show that BCG has been effective for more than 40 years, significantly reducing long-term tumor recurrence, risk of progression and mortality. A Southwest Oncology Group randomized study (126) showed a 5-year survival rate of 78% in the no BCG maintenance group compared to 83% in the maintenance group, suggesting that BCG immunotherapy is beneficial for patients with carcinoma in situ, as well as for selected patients with Ta and T1 bladder cancers. Similarly, Herr (127) evaluated 86 patients and found that the mortality rate decreased from 32% to 14% with BCG. A subsequent follow-up report (128) noted that patients who adhered to BCG therapy experienced a reduction in cancer mortality from 37% to 12% within three years. Despite the low number of serious side effects of BCG, some common adverse reactions such as urinary frequency, urgency, nocturia, bladder pain, low-grade fever, chills, and hematuria have been reported and most of the symptoms subside within 48 hours of BCG instillation (129). In addition, some rare adverse events have been reported, including Reiters syndrome (130), parotid infection (131), arteriovenous dermatocutaneous fistulae (132), psoas major muscle abscess, rupture of the iliac artery (133), and Poncet’s disease (134). Moreover, there are still some limitations of current BCG therapy. For example, the following adverse side effects have been reported during treatment (135). Secondly, in some immunocompromised individuals, such as the elderly, the therapeutic efficacy is greatly reduced (136), and tumor recurrence has been reported in about 30% of patients within the first 3 years of BCG treatment (137). Similarly, the microbiome has become a key target for addressing these issues, especially in urine, as it can come into close contact with BCG and tissues.
3.1 Role of gut microbiota in immunity to urologic tumors
When considering the influence of gut flora on tumor immunomodulation, especially in non - gastrointestinal tumors, the first question to consider is by what means the gut flora transmits regulatory information. The next step is to explore the tumor - immune - related pathways in which the gut flora is involved. A study on the microbial mechanisms of pancreatic ductal carcinoma (138) detected microorganisms within the tumor, and it was hypothesized that they entered and elicited a cancer response via retrograde migration from the duodenum to the pancreatic duct. This suggests that intestinal microorganisms can induce carcinogenesis through direct invasive effects on tissues. However, this explanation is not applicable to urinary tract tumors because, under normal conditions, microorganisms from the gastrointestinal tract are virtually incapable of directly invading the urinary tract. Some studies (139, 140) have summarized the pathophysiological links between the gut microbiota, intestinal permeability, and the genitourinary tract, providing a referable basis for our speculations, in addition to directly inducing carcinogenesis, host immune factors and immune cells are considered effective “messengers” for intestinal microorganisms. They can spread throughout the body by driving immune recognition in the intestinal epithelium or mesenteric lymph nodes and inducing immunomodulatory factors to enter the circulation. Subsequently, circulating immune factors and immune cells can transmit signals from the intestinal microbes to the tumor - immune microenvironment or tumor - draining lymph nodes (141). Finally, microbial metabolites are also among the factors that enable the gut flora to influence a wide range of pathophysiological processes in the host. They can also enter the blood circulation and be transported throughout the body, thereby not only regulating host metabolism (142) but also influencing the activity and function of various immune cells such as macrophages and B cells (143). Therefore, we began exploring the mechanism of immunomodulation of urological tumors by GM from these “messengers”.
Wang et al. (144) examined the efficacy of PD - 1 immunotherapy in MB49 tumor - bearing mice by performing FMT with Parabacteroides distasonis. It was found that the introduction of Parabacteroides distasonis enhanced the efficacy of PD - 1 immunotherapy, as evidenced by delayed tumor growth and increased densities of CD4+ T and CD8+ T cells in the tumor. In addition, transcriptome analysis further revealed that Parabacteroides distasonis supplementation by gavage further enhanced the levels of multiple anti - tumor immune pathways, including natural killer cell - mediated cytotoxicity, T - cell receptor signaling pathway, B - cell receptor signaling pathway, and chemokine signaling pathway. This study provides preliminary evidence that the GM modulates the immune response to urologic tumors through immunokines and immune cells acting as “messengers”. Another study (88) analyzed the immunological changes induced in mesenteric lymph nodes (mLN), tumor - draining lymph nodes (dLN), and tumor tissues after oral gavage of Akkermansia muciniphila and Enterococcus hirae. It was found that central memory (TCM) CD4+ T cells expressing the chemokine receptor CXCR3 were enriched in the mLN 48 h after injection. Subsequently, these CD4+ T cells were observed in the dLNs and tissues after being killed, respectively. This implies that the GM achieves its immune - killing effect on tumors by inducing immune cell differentiation in the mesenteric lymph nodes and reaching the tumor site through the circulation. It is worth noting that this study also found that specific gut microbes were strongly associated with tumor immunotherapeutic efficacy by modulating specific peripheral T - cell subset responses (e.g., Th1 and Tc1 cell responses against Akkermansia muciniphila and Tc1 responses against Enterococcus hirae) in the context of PD - 1 blockade therapy. This result once again suggests that there are “key members” of the GM that regulate the immune response to urologic tumors through specific signaling molecules.
We next focus on the role of another “messenger” of the gut flora in urologic tumor immunity. Liu ‘s in vivo and ex vivo studies (42) have amply demonstrated that GM - derived SCFA, as mediators linking the GM to prostate cancer, are closely associated with prostate cancer progression. On the one hand, their in vivo study found that colonizing the intestinal tract of mice with fecal suspensions from CRPC patients resulted in an increase in the number of SCFA - producing flora and a corresponding increase in the levels of SCFAs such as intestinal acetate and butyrate. On the other hand, in vitro experiments showed that SCFAs induced Toll - like receptor 3 (TLR3) activation and further induced autophagy activation in prostate cancer cells. Autophagy activation was further accompanied by NF - κB and MAPK signaling, ultimately regulating the invasion and immune - inflammatory response of prostate cancer cells. In addition, it was found that the increased level of autophagy resulted in the release of more chemokine CCL20 from prostate cancer cells, which could recruit more macrophages for infiltration and induce their polarization to M2 type for the regulation of the tumor immune microenvironment. Similarly, another study on SCFA and prostate carcinogenesis (145) noted that high - fat diet and antibiotic application affect SCFAs by interfering with the GM, and SCFAs can stimulate the level of insulin - like growth factor - 1 (IGF1) to promote the proliferation of prostate cancer cells. However, this study focused only on the proliferative effects of IGF1 on prostate cells. Given the current evidence that IGF1 interacts with M2 - like macrophages and autophagy (M2 - like tumour - associated macrophage - secreted IGF promotes thyroid cancer stemness and metastasis by activating the PI3K/AKT/mTOR pathway), it is reasonable to speculate that the IGF1 pathway regulated by SCFAs may also have a potential impact on prostate cancer tumor immunity. However, further experiments are required to directly confirm this. In addition, inosine, another metabolite of GM - derived SCFAs, has also been shown to be closely related to the tumor immune response. Roje ‘s study (57) found that serum from mice treated with anti - CTLA - 4 and colonized with Bifidobacterium pseudolongum inhibited tumor growth and induced strong anti - tumor immunity in the tumors and spleens of mice. Further metabolomics of the serum samples revealed that the purine metabolite inosine was the only metabolite that was significantly more abundant in the sera of Bifidobacterium pseudolongum monocolonized mice. It is worth noting that this study (57) also found that the effect of inosine on T cells requires sufficient co - stimulation (possibly through IL - 12 or IFN - γ production) to achieve efficient anti - tumor immunity. Specifically, in the presence of IFN - γ, inosine strongly promotes TH1 differentiation, whereas in the absence of IFN - γ, inosine exhibits an inhibitory effect on TH1 differentiation. This implies that the activation of GM and exogenous immune factors need to be considered to achieve better tumor immunotherapy. Additionally, this study found that in addition to mediating T - cell effects, inosine may also directly affect tumor cell survival by altering susceptibility to T - cell - mediated killing through direct action on tumor cells (Figure 1).
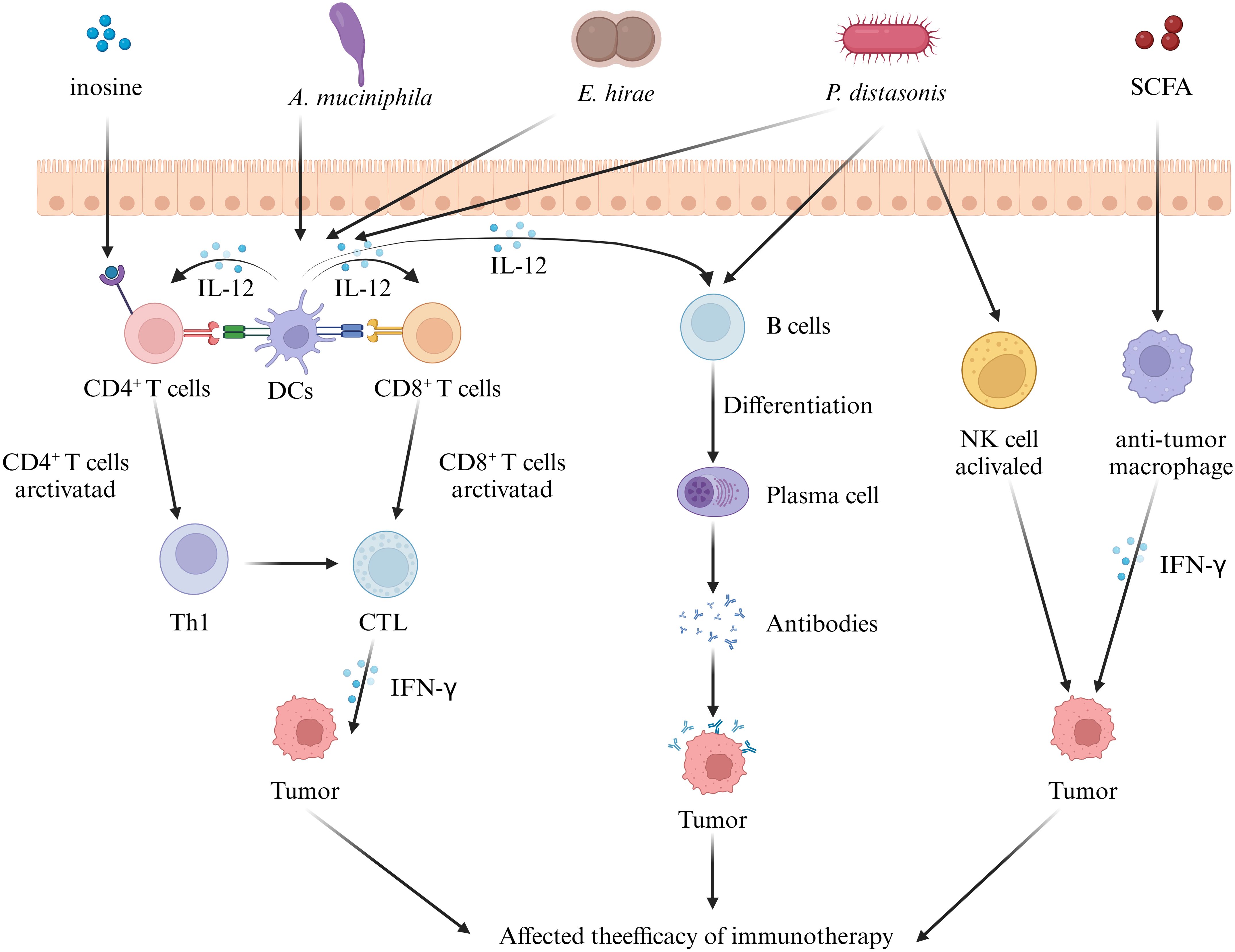
Figure 1. Effect of gut microbiota on tumor immunity in urologic tumors. Some intestinal microorganisms induce the differentiation of immune cells in mesenteric lymph nodes. These immune cells and cytokines reach urological tumor tissues via the circulation and influence tumor immunity by regulating the T - cell, B - cell, and NK - cell signaling pathways. In addition, the metabolites of the gut microbiota can also act as messengers, reach the tumor tissues, and regulate the immune response, ultimately influencing tumor immunotherapy. (IL, interleukin; SCFA, short chain fatty acid; DC, dendritic cell; CTL, cytotoxic t lymphocyte; IFN-γ, interferon-γ).
3.2 Urinary microbiome in tumor immunity
Unlike the GM, microorganisms in urine are in direct contact with urinary tract tissues. Consequently, the role of the urinary microbiome in tumor immunomodulation centers on the microbial action pathways. Evidence exists that some urinary microorganisms, such as Fusobacterium, Sphingobacterium and Enterococcus, may induce bladder cancer through schistosomiasis (146). In addition, a study by Shrestha et al. (49) found that some pro - inflammatory microorganisms associated with the urinary tract were enriched in the urine of prostate cancer patients. They suggested that these microorganisms may enter the prostate through the biotubes, causing chronic inflammation and atrophy. In addition, a specific type of prostate inflammation, granulomatous prostatitis (GP), has been shown to be closely related to urinary microorganisms (147). Specifically, damage to the epithelium and leakage of prostate secretions leads to reflux of bacterial products in the interstitium, which in turn triggers a strong foreign body inflammatory response in the prostate (148). In addition, BCG-induced GP has been reported in up to 75% of patients treated with BCG, due to reflux of BCG-contaminated urine from the bladder into the prostate (148). Although currently there is no evidence to support the notion that chronic inflammation of the prostate will eventually lead to cancer, it is also regarded as a suspected risk factor for prostate cancer development. We know that prolonged inflammation of mucosal epithelium significantly increases the likelihood of epithelial carcinogenesis, including the urinary tract epithelium. Repeated urinary tract infections can lead to chronic inflammation of the urinary mucosa, resulting in the release of inflammatory factors and reactive oxygen species, which can lead to carcinogenesis. Urinary tract infections have been shown to be closely linked to genitourinary tract microorganisms as well as intestinal microorganisms. Antibiotic therapy may disrupt the ecological balance of urinary microorganisms, exacerbating inflammation and leading to recurrence (149, 150). In addition, ecological imbalances in the gut flora, such as a decrease in SCFAs-producing bacteria, can disrupt the integrity of the intestinal barrier, triggering the “leaky gut syndrome,” which results in the displacement of bacteria and microbial-derived products (MDPs). Upon entering the circulatory system, these substances are able to activate innate immune cells, especially macrophages, causing them to produce pro-inflammatory cytokines, triggering localized inflammation and oxidative stress, creating a chronic inflammatory state (151). At the same time, persistent antigenic stimulation might cause functional depletion of innate immune cells and weaken local immune defenses, thus promoting recurrent urinary tract infections (152, 153). For example, some studies have found that female patients with recurrent urinary tract infections have reduced gut microbial abundance, with a significant reduction in flora associated with propionic acid and butyric acid production, and are associated with low levels of inflammation and specific immune states (154). In contrast, another hypothesis posits that the urinary microbiome may invade extracellular mechanisms that promote or inhibit bladder cancer development (155). We know that the extracellular mechanisms of tumors play a non - negligible role in tumor cell proliferation, invasion, and tumor immunity (156–159). Consequently, we first focus on the effect of urinary microbes on the extracellular matrix of urological tumors (primarily bladder cancer in this case). Under normal conditions, the mucosal barrier on the bladder surface restricts the direct contact between microorganisms in the urine and the extracellular matrix (160). However, when there is persistent bacterial translocation or tissue invasion, for example, when pathogenic bacteria invade and break the mucosal barrier, microorganisms in the urine can come into contact with the bladder tissue or even directly enter the extracellular matrix.
Specifically, microorganisms directly “invade” tissues by releasing a range of enzymes acting as extracellular virulence factors, which disrupt the host’s physical barriers and immune defense mechanisms (161). For example, some bacteria produce proteases that can disrupt the host’s cytokine regulation by directly acting on cytokines or degrading cytokine receptors (162, 163). These signaling factors include IL - 6 receptors, FAS ligands, and TNF and its receptors (164, 165). In addition, some bacteria release elastases that cleave not only cytokines and receptors but also factors and receptors of the complement system (166). For example, elastases from Pseudomonas aeruginosa can modulate the host immune response by inhibiting lymphocyte proliferation through IL - 2 inhibition and IFN - γ inactivation. Urinary microbes have also been shown to potentially influence tumor immunity by acting on T cells. A study on the urinary microbiota for prognostic prediction in non - muscle - invasive bladder cancer (74) demonstrated significant differences in the urinary microbiota between the recurrent and non - recurrent groups. The recurrent group exhibited an enrichment of Pseudomonas, Staphylococcus, Corynebacterium and Acinetobacter, and the tumor stroma showed increased infiltration of FoxP3+ regulatory T cells (FoxP3+ Treg), which has been shown to promote immune escape and tumor growth through various mechanisms (167–170). Further correlation analyses showed that the degree of FoxP3+ Treg infiltration was correlated with the urinary microbiology index, albeit not statistically significant, which was mainly attributed to the small number of samples tested. In addition, we further summarize the immune pathways related to the role of urinary microbes in the immunotherapy of urological tumors. An investigative study on urinary microbiology and bladder cancer (70) found that differences in the urinary microbiome were demonstrated not only in patients with non - neoplastic tumors and those with and without muscular infiltration but also in patients who responded and did not respond to BCG therapy. For example, Serratia and Brochothrix were present in patients who responded to BCG, and Negativicoccus, Escherichia, Shigella and Pseudomonas were significantly increased in patients who responded to BCG. Mechanistically, BCG has been shown to modulate tumor immunity by binding to fibronectin, thereby inducing CD8+ T and natural killer cells (171). In contrast, some uropathogens, such as Lactobacillus casei, that also bind fibronectin may influence the BCG therapeutic efficacy by enhancing fibronectin stimulation or competing with BCG for fibronectin binding. Another more direct evidence comes from a study investigating the urogenital flora of PD - L1 - positive and negative subjects (172). This study showed higher species richness in the PD - L1 - positive group, which increased with increasing PD - L1 positivity intensity. Meanwhile, the PD - L1 - positive group also showed an enrichment of some bacterial genera (e.g., Leptotrichia, Roseomonas and Propionibacterium) and a decrease in others (e.g., Prevotella and Massilia). The implication is that these bacteria not only play a key role in tumor immune escape and metastasis but may also serve as potential cofactors to enhance tumor immunotherapy (Figure 2).
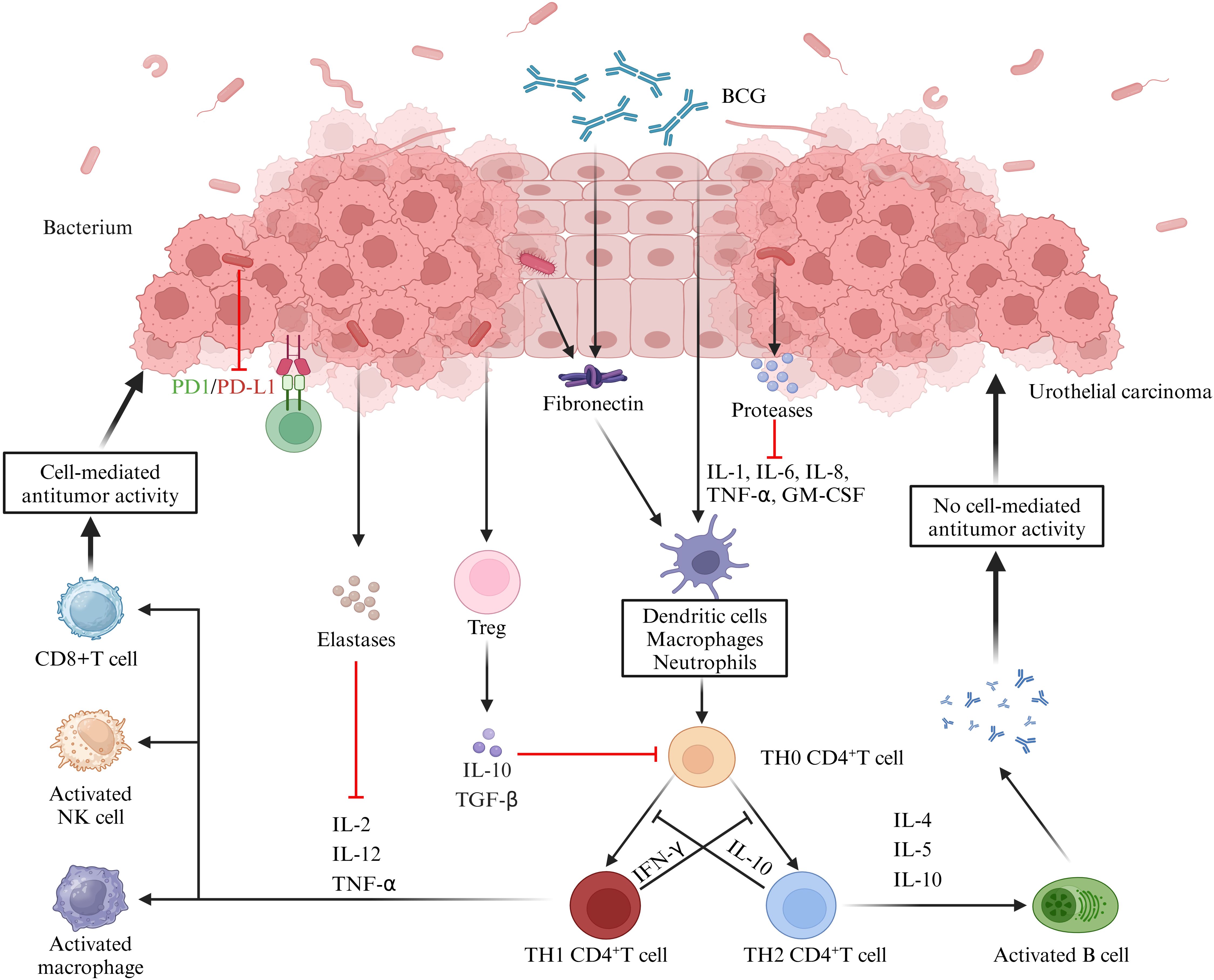
Figure 2. Role of the urine microbiome in the immunotherapy of uroepithelial carcinoma. BCG attaches to the uroepithelium and induces the release of cytokines and chemokines, thereby exerting anti - tumor immunity through the activation of cellular or humoral immunity. Microorganisms in the urine can interfere with these responses. Firstly, bacteria produce proteases or elastases that can directly act on cytokines or degrade cytokine receptors, thereby disrupting the regulation of immune factors. Secondly, some microorganisms can inhibit cell - mediated tumor killing by promoting the infiltration of Treg cells. Additionally, the cross - linking of microorganisms to collagen can further contribute to the therapeutic efficacy of BCG, while other microorganisms can directly inhibit PD - L1 expression, which is important for the immunotherapeutic effect of bladder cancer. (IL, interleukin; BCG, Bacillus Calmette-Guérin; TNF-α, tumor necrosis factor-α; GM-CSF, granulocyte-macrophage colony-stimulating factor; TGF-β, transforming growth factor-β; IFN-γ, interferon-γ).
3.3 Tissue/tumor microbiome
Research on tissue microbiota is still in its early stages, and much confusion persists. Firstly, the concept of tissue microbiome is being questioned. Although it has been shown that a variety of microbial DNAs can be detected in prostate cancer tissues (50, 173, 174), there are no direct studies reporting the identification of bacteria through visualization or localization in the corresponding tissue samples. In addition, since the presence of microorganisms is often considered pathogenic, even if microorganisms are present in the tissues, are these microorganisms, like those in our intestines, in a mutually beneficial symbiotic relationship with the host rather than having a pathogenic infectious role? Therefore, in the absence of clear direct evidence, we believe it is inappropriate to use the concept of tissue microbiome to summarize its relationship with immunity to urological tumors. Therefore, we directly focused our attention on intra - tumor microbes and concentrated on the relevant immunomodulatory role of tumor microbes in urologic tumors. It has been demonstrated (175, 176) that certain microorganisms may be present in the tumor itself as part of the tumor microenvironment, and they may be of circulating origin or from normal adjacent tissues. They are not randomly present in the tumor tissue; instead, they are specifically present and highly organized within tumor cells, epithelial cells, and immune cells (175, 177). In addition, it should be noted that although current evidence cannot indicate whether these microbes in tumors play a causal role in cancer development or serve as potential biomarkers for tumor presence (178, 179), they have been noted to be strongly associated with the tumor immune response and immunotherapy efficacy (176). However, this finding has been confirmed in urologic tumors, which are of interest to us. A study by Davidsson et al. (43) found higher Tregs infiltration in Cutibacterium acnes - positive prostate cancer tissues. Additionally, their in vitro findings showed that Cutibacterium acnes stimulation significantly increased the expression of PD - L1 and the chemokines CCL17 and CCL18, confirming the important role of microorganisms in tissues in the tumor immune microenvironment.
4 Conclusion and prospect
The microbiome in the human body, an integral part, is closely related to the development of urological tumors. Specific microorganism species may promote tumor cell proliferation and invasion by metabolizing carcinogens, inducing chronic inflammation, or interfering with cell signaling. Differences in microbiota characteristics are manifested not only between tumor and non - tumor patients but also among patients at different tumor stages. Studying these differentially expressed microorganisms is of great significance as they may serve as potential markers for tumorigenesis and progression and as important targets for tumor therapy. In addition, we note that microorganisms from various sites have an important impact on tumor immunity and tumor therapy in urologic tumors. They can enhance the immune system’s killing effect on tumor cells through their metabolites, the production of some enzymes, or their direct action by activating immune cells, inducing tumor cell apoptosis, or changing the tumor microenvironment. These roles of the microbiome and tumor immunity not only well explain the individual differences in the effectiveness of tumor immunotherapy but also, based on these findings, are expected to enable us to develop more efficient and safer microbial immunotherapy regimens, which will bring better therapeutic effects to urological tumor patients. It is worth noting that, as Roje ‘s study (57) pointed out, the effect of inosine, a gut microbe metabolite, on T cells requires sufficient co - stimulation to achieve potent anti - tumor immunity. Thus, in future studies, we cannot simply consider microbes as a single factor, and more interactions need to be further explored. In addition, microorganisms at different sites do not act independently and complementarily, and there are also interactions among them. In future studies, we need to further deepen our understanding of the role of the microbiome in urologic tumorigenesis and development. This includes further elucidating the interactions between microbes and tumor cells and the specific mechanisms by which microbes influence the tumor microenvironment and immune responses. By exploring these mechanisms, we expect to provide new ideas and approaches for treating urologic tumors.
Some significant achievements have been made in microbial modulation approaches for tumor treatment. For example, several studies in pre - clinical trials have shown that microbiota - centered interventions significantly improve the therapeutic outcomes of ICIs (180, 181). In the future, further developing microbial - based tumor vaccines and immunotherapy protocols is also an important research direction. We can utilize known microbial strains with antitumor activity or modify microbes through genetic engineering methods to more effectively stimulate the body’s antitumor immune response. In addition, with the rapid development of big data technologies in software engineering, we can use these technologies to optimize microbiome regulation strategies. For example, by constructing a microbiome database of urological tumor patients and performing algorithmic analysis, we can identify microbial markers closely related to tumorigenesis and development. This not only aids in predicting patient prognosis but also provides a basis for developing individualized treatment plans. Meanwhile, with the help of these technologies, we can also monitor real - time changes in the patient’s microbiome and adjust the treatment plan promptly to achieve the goal of precision medicine. In response to the previously mentioned limitations of microbial detection in tumor tissues, a series of in-tumor microbial detection technological tools have emerged in recent years to overcome these challenges. In addition to the commonly used 16S rRNA gene sequencing, whole metagenome-based shotgun sequencing (WMS) can determine the gene expression status of microorganisms in tumor tissues. By understanding the transcriptional activity of microorganisms, their functional status in the tissue can be inferred (182, 183). In addition, some high-resolution visualization techniques, such as fluorescence in situ hybridization (FISH) and immunofluorescence and immunohistochemistry, can be used to directly label microorganisms at the cellular or tissue level, so as to more accurately observe the distribution and localization of microorganisms in tumor tissues (184). It is even possible to construct images of microbial distribution in cancerous tissues in three dimensions, which can be used to explore the interrelationships between microbes and tumor cells as well as the surrounding stromal cells, and to determine whether the microbes are at a possible symbiotic interface or pathogenic invasion site (185). We firmly believe that with the development of technology, tissue microbes will also play a key role in tumor prediction and intervention, just like gut and urine microbes. Future research on the microbiome in urological tumors will focus on exploring the mechanisms of action, developing novel therapeutic regimens, strengthening multidisciplinary cooperation, and optimizing therapeutic strategies with advanced technologies. Through these efforts, we hope to provide more effective treatments for urologic tumor patients and improve their quality of life.
Author contributions
JZ: Formal analysis, Investigation, Writing – original draft. BX: Conceptualization, Funding acquisition, Methodology, Writing – original draft. PL: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. TC: Data curation, Validation, Writing – review & editing. HD: Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. HG: Data curation, Methodology, Investigation, Software, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Jiangxi Provincial Health Commission Research Project (No. SKJP220227287).
Acknowledgments
All figures are created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: A global overview and recent trends. Eur Urol. (2017) 71(1):96–108. doi: 10.1016/j.eururo.2016.06.010
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
3. Zi H, He S-H, Leng X-Y, Xu X-F, Huang Q, Weng H, et al. Global, regional, and national burden of kidney, bladder, and prostate cancers and their attributable risk factors, 1990-2019. Mil Med Res. (2021) 8:60. doi: 10.1186/s40779-021-00354-z
4. Deng T, Cai L, Chen Z, Guo J, Huang X, Ke CX, et al. Analysis of the burden of prostate cancer in China in 1990 and 2017. Yixue Xinzhi Zazhi. (2020) 30(4):252–9. doi: 10.12173/j.issn.1004-5511.2020.04.01
5. Culp MB, Soerjomataram I, Efstathiou JA, Bray F, Jemal A. Recent global patterns in prostate cancer incidence and mortality rates. Eur Urol. (2020) 77:38–52. doi: 10.1016/j.eururo.2019.08.005
6. Allott EH, Masko EM, Freedland SJ. Obesity and prostate cancer: weighing the evidence. Eur Urol. (2013) 63:800–9. doi: 10.1016/j.eururo.2012.11.013
7. Reiter-Brennan C, Dzaye O, Al-Mallah MH, Dardari Z, Brawner CA, Lamerato LE, et al. Fitness and prostate cancer screening, incidence, and mortality: Results from the Henry Ford Exercise Testing (FIT) Project. Cancer. (2021) 127:1864–70. doi: 10.1002/cncr.v127.11
8. Cai H, Xu Z, Xu T, Yu B, Zou Q. Diabetes mellitus is associated with elevated risk of mortality amongst patients with prostate cancer: a meta-analysis of 11 cohort studies. Diabetes Metab Res Rev. (2015) 31:336–43. doi: 10.1002/dmrr.v31.4
9. Peisch SF, Van Blarigan EL, Chan JM, Stampfer MJ, Kenfield SA. Prostate cancer progression and mortality: a review of diet and lifestyle factors. World J Urol. (2017) 35:867–74. doi: 10.1007/s00345-016-1914-3
10. Klein EA, Thompson IM, Tangen CM, Crowley JJ, Lucia MS, Goodman PJ, et al. Vitamin E and the risk of prostate cancer: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). Jama. (2011) 306:1549–56. doi: 10.1001/jama.2011.1437
11. Freedman ND, Silverman DT, Hollenbeck AR, Schatzkin A, Abnet CC. Association between smoking and risk of bladder cancer among men and women. Jama. (2011) 306:737–45. doi: 10.1001/jama.2011.1142
12. Wilhelm-Benartzi CS, Christensen BC, Koestler DC, Houseman EA, Schned AR, Karagas MR, et al. Association of secondhand smoke exposures with DNA methylation in bladder carcinomas. Cancer Causes Control. (2011) 22:1205–13. doi: 10.1007/s10552-011-9788-6
13. Ishida K, Hsieh MH. Understanding urogenital schistosomiasis-related bladder cancer: an update. Front Med (Lausanne). (2018) 5:223. doi: 10.3389/fmed.2018.00223
14. Bergström A, Hsieh CC, Lindblad P, Lu CM, Cook NR, Wolk A. Obesity and renal cell cancer–a quantitative review. Br J Cancer. (2001) 85:984–90. doi: 10.1054/bjoc.2001.2040
15. Liu B, Mao Q, Wang X, Zhou F, Luo J, Wang C, et al. Cruciferous vegetables consumption and risk of renal cell carcinoma: a meta-analysis. Nutr Cancer. (2013) 65:668–76. doi: 10.1080/01635581.2013.795980
16. Zhao J, Zhao L. Cruciferous vegetables intake is associated with lower risk of renal cell carcinoma: evidence from a meta-analysis of observational studies. PloS One. (2013) 8:e75732. doi: 10.1371/journal.pone.0075732
17. Macleod LC, Hotaling JM, Wright JL, Davenport MT, Gore JL, Harper J, et al. Risk factors for renal cell carcinoma in the VITAL study. J Urol. (2013) 190:1657–61. doi: 10.1016/j.juro.2013.04.130
18. Chien C-C, Han M-M, Chiu Y-H, Wang J-J, Chu C-C, Hung C-Y, et al. Epidemiology of cancer in end-stage renal disease dialysis patients: a national cohort study in Taiwan. J Cancer. (2017) 8(1):9–18. doi: 10.7150/jca.16550
19. Mazzucotelli V, Piselli P, Verdirosi D, Cimaglia C, Cancarini G, Serraino D, et al. De novo cancer in patients on dialysis and after renal transplantation: north-western Italy, 1997-2012. J Nephrol. (2017) 30:851–7. doi: 10.1007/s40620-017-0385-y
20. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. (2007) 449:804–10. doi: 10.1038/nature06244
21. Shi N, Li N, Duan X, Niu H. Interaction between the gut microbiome and mucosal immune system. Mil Med Res. (2017) 4:14. doi: 10.1186/s40779-017-0122-9
22. Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. (2019) 16:35–56. doi: 10.1038/s41575-018-0061-2
23. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. (2021) 19:55–71. doi: 10.1038/s41579-020-0433-9
24. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. (2016) 535:65–74. doi: 10.1038/nature18847
25. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Sci (New York N.Y.). (2012) 336(6080):489–93. doi: 10.1126/science.1219328
26. Marchesi JR, Adams DH, Fava F, Hermes GDA, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. (2016) 65:330–9. doi: 10.1136/gutjnl-2015-309990
27. de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. (2022) 71:1020–32. doi: 10.1136/gutjnl-2021-326789
28. Azevedo MM, Pina-Vaz C, Baltazar F. Microbes and cancer: friends or faux? Int J Mol Sci. (2020) 21(9):3115. doi: 10.3390/ijms21093115
29. Wong-Rolle A, Wei HK, Zhao C, Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell. (2021) 12:426–35. doi: 10.1007/s13238-020-00813-8
30. Gagliani N, Hu B, Huber S, Elinav E, Flavell RA. The fire within: microbes inflame tumors. Cell. (2014) 157:776–83. doi: 10.1016/j.cell.2014.03.006
31. Hungate RE. Studies on cellulose fermentation: I. The culture and physiology of an anaerobic cellulose-digesting bacterium. J Bacteriol. (1944) 48:499–513. doi: 10.1128/jb.48.5.499-513.1944
32. Boursi B, Mamtani R, Haynes K, Yang YX. Recurrent antibiotic exposure may promote cancer formation–Another step in understanding the role of the human microbiota? Eur J Cancer. (2015) 51:2655–64. doi: 10.1016/j.ejca.2015.08.015
33. You X, Qiu J, Li Q, Zhang Q, Sheng W, Cao Y, et al. Astragaloside IV-PESV inhibits prostate cancer tumor growth by restoring gut microbiota and microbial metabolic homeostasis via the AGE-RAGE pathway. BMC Cancer. (2024) 24:472. doi: 10.1186/s12885-024-12167-z
34. Lin G, Zhang F, Weng X, Hong Z, Ye D, Wang G. Role of gut microbiota in the pathogenesis of castration-resistant prostate cancer: a comprehensive study using sequencing and animal models. Oncogene. (2024) 43:2373–88. doi: 10.1038/s41388-024-03073-6
35. Liu X, Dong Q. Associations between gut microbiota and three prostate diseases: a bidirectional two-sample Mendelian randomization study. Sci Rep. (2024) 14:4019. doi: 10.1038/s41598-024-54293-5
36. Wei Z, Yang B, Tang T, Xiao Z, Ye F, Li X, et al. Gut microbiota and risk of five common cancers: A univariable and multivariable Mendelian randomization study. Cancer Med. (2023) 12:10393–405. doi: 10.1002/cam4.v12.9
37. Huang H, Liu Y, Wen Z, Chen C, Wang C, Li H, et al. Gut microbiota in patients with prostate cancer: a systematic review and meta-analysis. BMC Cancer. (2024) 24:261. doi: 10.1186/s12885-024-12018-x
38. Matsushita M, Fujita K, Motooka D, Hatano K, Fukae S, Kawamura N, et al. The gut microbiota associated with high-Gleason prostate cancer. Cancer Sci. (2021) 112:3125–35. doi: 10.1111/cas.v112.8
39. Lachance G, Robitaille K, Laaraj J, Gevariya N, Varin TV, Feldiorean A, et al. The gut microbiome-prostate cancer crosstalk is modulated by dietary polyunsaturated long-chain fatty acids. Nat Commun. (2024) 15:3431. doi: 10.1038/s41467-024-45332-w
40. Sato H, Narita S, Ishida M, Takahashi Y, Mingguo H, Kashima S, et al. Specific gut microbial environment in lard diet-induced prostate cancer development and progression. Int J Mol Sci. (2022) 23(4):2214. doi: 10.3390/ijms23042214
41. Gut microbiota differs significantly between men with and without prostate cancer. Cancer. (2023) 129:169–70. doi: 10.1002/cncr.v129.2
42. Liu Y, Zhou Q, Ye F, Yang C, Jiang H. Gut microbiota-derived short-chain fatty acids promote prostate cancer progression via inducing cancer cell autophagy and M2 macrophage polarization. Neoplasia. (2023) 43:100928. doi: 10.1016/j.neo.2023.100928
43. Davidsson S, Carlsson J, Greenberg L, Wijkander J, Söderquist B, Erlandsson A. Cutibacterium acnes induces the expression of immunosuppressive genes in macrophages and is associated with an increase of regulatory T-cells in prostate cancer. Microbiol Spectr. (2021) 9:e0149721. doi: 10.1128/spectrum.01497-21
44. O’Rourke K. Gut microbiome linked to aggressive prostate cancer. Cancer. (2022) 128:938. doi: 10.1002/cncr.v128.5
45. Reichard CA, Naelitz BD, Wang Z, Jia X, Li J, Stampfer MJ, et al. Gut microbiome-dependent metabolic pathways and risk of lethal prostate cancer: prospective analysis of a PLCO cancer screening trial cohort. Cancer Epidemiol Biomarkers Prev. (2022) 31:192–9. doi: 10.1158/1055-9965.EPI-21-0766
46. Hurst R, Meader E, Gihawi A, Rallapalli G, Clark J, Kay GL, et al. Microbiomes of urine and the prostate are linked to human prostate cancer risk groups. Eur Urol Oncol. (2022) 5:412–9. doi: 10.1016/j.euo.2022.03.006
47. Mjaess G, Karam A, Roumeguère T, Diamand R, Aoun F, McVary K, et al. Urinary microbiota and prostatic diseases: the key for the lock? A systematic review. Prostate Cancer Prostatic Dis. (2023) 26:451–60. doi: 10.1038/s41391-022-00602-w
48. Alanee S, El-Zawahry A, Dynda D, McVary K, Karr M, Braundmeier-Fleming A. Prospective examination of the changes in the urinary microbiome induced by transrectal biopsy of the prostate using 16S rRNA gene analysis. Prostate Cancer Prostatic Dis. (2019) 22:446–52. doi: 10.1038/s41391-018-0120-3
49. Shrestha E, White JR, Yu SH, Kulac I, Ertunc O, De Marzo AM, et al. Profiling the urinary microbiome in men with positive versus negative biopsies for prostate cancer. J Urol. (2018) 199:161–71. doi: 10.1016/j.juro.2017.08.001
50. Cavarretta I, Ferrarese R, Cazzaniga W, Saita D, Lucianò R, Ceresola ER, et al. The microbiome of the prostate tumor microenvironment. Eur Urol. (2017) 72:625–31. doi: 10.1016/j.eururo.2017.03.029
51. Salachan PV, Rasmussen M, Fredsøe J, Ulhøi B, Borre M, Sørensen KD. Microbiota of the prostate tumor environment investigated by whole-transcriptome profiling. Genome Med. (2022) 14:9. doi: 10.1186/s13073-022-01011-3
52. Feng Y, Ramnarine VR, Bell R, Volik S, Davicioni E, Hayes VM, et al. Metagenomic and metatranscriptomic analysis of human prostate microbiota from patients with prostate cancer. BMC Genomics. (2019) 20:146. doi: 10.1186/s12864-019-5457-z
53. Kim JH, Seo H, Kim S, Rahim MA, Jo S, Barman I, et al. Different prostatic tissue microbiomes between high- and low-grade prostate cancer pathogenesis. Int J Mol Sci. (2024) 25(16):8943. doi: 10.3390/ijms25168943
54. Kim JH, Seo H, Kim S, Ul-Haq A, Rahim MA, Jo S, et al. Biochemical recurrence in prostate cancer is associated with the composition of lactobacillus: microbiome analysis of prostatic tissue. Int J Mol Sci. (2023) 24(13):10423. doi: 10.3390/ijms241310423
55. He C, Li B, Huang L, Teng C, Bao Y, Ren M, et al. Gut microbial composition changes in bladder cancer patients: A case-control study in Harbin, China. Asia Pac J Clin Nutr. (2020) 29:395–403. doi: 10.6133/apjcn.202007_29(2).0022
56. He C, Huang L, Lei P, Liu X, Li B, Shan Y. Sulforaphane normalizes intestinal flora and enhances gut barrier in mice with BBN-induced bladder cancer. Mol Nutr Food Res. (2018) 62:e1800427. doi: 10.1002/mnfr.201800427
57. Roje B, Zhang B, Mastrorilli E, Kovačić A, Sušak L, Ljubenkov I, et al. Gut microbiota carcinogen metabolism causes distal tissue tumours. Nature. (2024) 632:1137–44. doi: 10.1038/s41586-024-07754-w
58. Luo P, Zheng L, Zou J, Chen T, Zou J, Li W, et al. Insights into vitamin A in bladder cancer, lack of attention to gut microbiota? Front In Immunol. (2023) 14:1252616. doi: 10.3389/fimmu.2023.1252616
59. Karam A, Mjaess G, Albisinni S, El Daccache Y, Farah M, Daou S, et al. Uncovering the role of urinary microbiota in urological tumors: a systematic review of literature. World J Urol. (2022) 40:951–64. doi: 10.1007/s00345-021-03924-x
60. Yacouba A, Tidjani Alou M, Lagier JC, Dubourg G, Raoult D. Urinary microbiota and bladder cancer: A systematic review and a focus on uropathogens. Semin Cancer Biol. (2022) 86:875–84. doi: 10.1016/j.semcancer.2021.12.010
61. Bukavina L, Isali I, Ginwala R, Sindhani M, Calaway A, Magee D, et al. Global meta-analysis of urine microbiome: colonization of polycyclic aromatic hydrocarbon-degrading bacteria among bladder cancer patients. Eur Urol Oncol. (2023) 6:190–203. doi: 10.1016/j.euo.2023.02.004
62. Pederzoli F, Ferrarese R, Amato V, Locatelli I, Alchera E, Lucianò R, et al. Sex-specific alterations in the urinary and tissue microbiome in therapy-naïve urothelial bladder cancer patients. Eur Urol Oncol. (2020) 3:784–8. doi: 10.1016/j.euo.2020.04.002
63. Oresta B, Braga D, Lazzeri M, Frego N, Saita A, Faccani C, et al. The microbiome of catheter collected urine in males with bladder cancer according to disease stage. J Urol. (2021) 205:86–93. doi: 10.1097/JU.0000000000001336
64. Wu P, Zhang G, Zhao J, Chen J, Chen Y, Huang W, et al. Profiling the urinary microbiota in male patients with bladder cancer in China. Front Cell Infect Microbiol. (2018) 8:167. doi: 10.3389/fcimb.2018.00167
65. Zeng J, Zhang G, Chen C, Li K, Wen Y, Zhao J, et al. Alterations in urobiome in patients with bladder cancer and implications for clinical outcome: A single-institution study. Front Cell Infect Microbiol. (2020) 10:555508. doi: 10.3389/fcimb.2020.555508
66. Bučević Popović V, Šitum M, Chow CT, Chan LS, Roje B, Terzić J. The urinary microbiome associated with bladder cancer. Sci Rep. (2018) 8:12157. doi: 10.1038/s41598-018-29054-w
67. Mai G, Chen L, Li R, Liu Q, Zhang H, Ma Y. Common core bacterial biomarkers of bladder cancer based on multiple datasets. BioMed Res Int. (2019) 2019:4824909. doi: 10.1155/2019/4824909
68. Yang JW, Wan S, Li KP, Chen SY, Yang L. Gut and urinary microbiota: the causes and potential treatment measures of renal cell carcinoma. Front Immunol. (2023) 14:1188520. doi: 10.3389/fimmu.2023.1188520
69. Hrbáček J, Tláskal V, Čermák P, Hanáček V, Zachoval R. Bladder cancer is associated with decreased urinary microbiota diversity and alterations in microbial community composition. Urol Oncol. (2023) 41:107.e15–107.e22. doi: 10.1016/j.urolonc.2022.09.018
70. Hussein AA, Elsayed AS, Durrani M, Jing Z, Iqbal U, Gomez EC, et al. Investigating the association between the urinary microbiome and bladder cancer: An exploratory study. Urol Oncol. (2021) 39:370.e9–370.e19. doi: 10.1016/j.urolonc.2020.12.011
71. Chipollini J, Wright JR, Nwanosike H, Kepler CY, Batai K, Lee BR, et al. Characterization of urinary microbiome in patients with bladder cancer: Results from a single-institution, feasibility study. Urol Oncol. (2020) 38:615–21. doi: 10.1016/j.urolonc.2020.04.014
72. Liu F, Liu A, Lu X, Zhang Z, Xue Y, Xu J, et al. Dysbiosis signatures of the microbial profile in tissue from bladder cancer. Cancer Med. (2019) 8:6904–14. doi: 10.1002/cam4.v8.16
73. Huang PY, Yang YC, Wang CI, Hsiao PW, Chiang HI, Chen TW. Increase in akkermansiaceae in gut microbiota of prostate cancer-bearing mice. Int J Mol Sci. (2021) 22(17):9626. doi: 10.3390/ijms22179626
74. Qiu Y, Gao Y, Chen C, Xie M, Huang P, Sun Q, et al. Deciphering the influence of urinary microbiota on FoxP3+ regulatory T cell infiltration and prognosis in Chinese patients with non-muscle-invasive bladder cancer. Hum Cell. (2022) 35:511–21. doi: 10.1007/s13577-021-00659-0
75. Hourigan SK, Zhu W, Wong SWW, Clemency NC, Provenzano M, Vilboux T, et al. Studying the urine microbiome in superficial bladder cancer: samples obtained by midstream voiding versus cystoscopy. BMC Urol. (2020) 20:5. doi: 10.1186/s12894-020-0576-z
76. Mansour B, Monyók Á, Makra N, Gajdács M, Vadnay I, Ligeti B, et al. Bladder cancer-related microbiota: examining differences in urine and tissue samples. Sci Rep. (2020) 10:11042. doi: 10.1038/s41598-020-67443-2
77. Huang Z, Ye B, Li W, Shi X. A commentary on ‘Causal effects of gut microbiota on renal tumor: a Mendelian randomization study’. Int J Surg. (2024) 110:5266–7. doi: 10.1097/JS9.0000000000001567
78. Yin Z, Liu B, Feng S, He Y, Tang C, Chen P, et al. A large genetic causal analysis of the gut microbiota and urological cancers: A bidirectional mendelian randomization study. Nutrients. (2023) 15(18):4086. doi: 10.3390/nu15184086
79. Wang J, Li X, Wu X, Wang Z, Zhang C, Cao G, et al. Uncovering the microbiota in renal cell carcinoma tissue using 16S rRNA gene sequencing. J Cancer Res Clin Oncol. (2021) 147:481–91. doi: 10.1007/s00432-020-03462-w
80. Heidler S, Lusuardi L, Madersbacher S, Freibauer C. The microbiome in benign renal tissue and in renal cell carcinoma. Urol Int. (2020) 104:247–52. doi: 10.1159/000504029
81. Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin. (2017) 67:326–44. doi: 10.3322/caac.21398
82. Pham F, Moinard-Butot F, Coutzac C, Chaput N. Cancer and immunotherapy: a role for microbiota composition. Eur J Cancer (Oxford England: 1990). (2021) 155:145–54. doi: 10.1016/j.ejca.2021.06.051
83. Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. (2019) 30:2012. doi: 10.1093/annonc/mdz224
84. Derosa L, Routy B, Fidelle M, Iebba V, Alla L, Pasolli E, et al. Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur Urol. (2020) 78:195–206. doi: 10.1016/j.eururo.2020.04.044
85. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Sci (New York N.Y.). (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
86. Hakozaki T, Richard C, Elkrief A, Hosomi Y, Benlaïfaoui M, Mimpen I, et al. The gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non-small cell lung cancer. Cancer Immunol Res. (2020) 8:1243–50. doi: 10.1158/2326-6066.CIR-20-0196
87. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Sci (New York N.Y.). (2018) 359:104–8. doi: 10.1126/science.aao3290
88. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Sci (New York N.Y.). (2018) 359:91–7. doi: 10.1126/science.aan3706
89. Shaikh FY, Gills JJ, Mohammad F, White JR, Stevens CM, Ding H, et al. Murine fecal microbiota transfer models selectively colonize human microbes and reveal transcriptional programs associated with response to neoadjuvant checkpoint inhibitors. Cancer Immunol Immunother. (2022) 71:2405–20. doi: 10.1007/s00262-022-03169-6
90. Kaźmierczak-Siedlecka K, Roviello G, Catalano M, Polom K. Gut Microbiota Modulation in the Context of Immune-Related Aspects of Lactobacillus spp. and Bifidobacterium spp. in Gastrointestinal Cancers. Nutrients. (2021) 13(8):2674. doi: 10.3390/nu13082674
91. Jacouton E, Chain F, Sokol H, Langella P, Bermúdez-Humarán LG. Probiotic strain lactobacillus casei BL23 prevents colitis-associated colorectal cancer. Front In Immunol. (2017) 8:1553. doi: 10.3389/fimmu.2017.01553
92. Liang K, Liu Q, Li P, Luo H, Wang H, Kong Q. Genetically engineered Salmonella Typhimurium: Recent advances in cancer therapy. Cancer Lett. (2019) 448:168–81. doi: 10.1016/j.canlet.2019.01.037
93. Leventhal DS, Sokolovska A, Li N, Plescia C, Kolodziej SA, Gallant CW, et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat Commun. (2020) 11:2739. doi: 10.1038/s41467-020-16602-0
94. Chowdhury S, Castro S, Coker C, Hinchliffe TE, Arpaia N, Danino T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med. (2019) 25:1057–63. doi: 10.1038/s41591-019-0498-z
95. Schupack DA, Mars RAT, Voelker DH, Abeykoon JP, Kashyap PC. The promise of the gut microbiome as part of individualized treatment strategies. Nat Rev Gastroenterol Hepatol. (2022) 19(1):7–25. doi: 10.1038/s41575-021-00499-1
96. Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. (2015) 373:1627–39. doi: 10.1056/NEJMoa1507643
97. Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. (2015) 373:1803–13. doi: 10.1056/NEJMoa1510665
98. Weber JS, D’Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. (2015) 16:375–84. doi: 10.1016/S1470-2045(15)70076-8
99. Knorr J, Lone Z, Werneburg G, Adler A, Agudelo J, Suryavanshi M, et al. An exploratory study investigating the impact of the bladder tumor microbiome on Bacillus Calmette Guerin (BCG) response in non-muscle invasive bladder cancer. Urol Oncol. (2024) 42:291.e1–291.e11. doi: 10.1016/j.urolonc.2024.04.011
100. Dermani FK, Samadi P, Rahmani G, Kohlan AK, Najafi R. PD-1/PD-L1 immune checkpoint: Potential target for cancer therapy. J Cell Physiol. (2019) 234:1313–25. doi: 10.1002/jcp.v234.2
101. Powles T, Staehler M, Ljungberg B, Bensalah K, Canfield SE, Dabestani S, et al. Updated EAU guidelines for clear cell renal cancer patients who fail VEGF targeted therapy. Eur Urol. (2016) 69:4–6. doi: 10.1016/j.eururo.2015.10.017
102. Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. (2016) 387:1909–20. doi: 10.1016/S0140-6736(16)00561-4
103. Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, et al. Phase I study of single-agent anti–programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. (2010) 28:3167–75. doi: 10.1200/JCO.2009.26.7609
104. Kwon ED, Drake CG, Scher HI, Fizazi K, Bossi A, van den Eertwegh AJM, et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. (2014) 15:700–12. doi: 10.1016/S1470-2045(14)70189-5
105. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. (2012) 366:2443–54. doi: 10.1056/NEJMoa1200690
106. Bracarda S, Altavilla A, Hamzaj A, Sisani M, Marrocolo F, Del Buono S, et al. Immunologic checkpoints blockade in renal cell, prostate, and urothelial Malignancies. Semin Oncol. (2015) 42:495–505. doi: 10.1053/j.seminoncol.2015.02.004
107. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Sci (New York N.Y.). (2013) 342:967–70. doi: 10.1126/science.1240527
108. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Sci (New York N.Y.). (2015) 350:1084–9. doi: 10.1126/science.aac4255
109. Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia (New York N.Y.). (2017) 19:848–55. doi: 10.1016/j.neo.2017.08.004
110. Gide TN, Wilmott JS, Scolyer RA, Long GV. Primary and acquired resistance to immune checkpoint inhibitors in metastatic melanoma. Clin Cancer Research: an Off J Am Assoc For Cancer Res. (2018) 24:1260–70. doi: 10.1158/1078-0432.CCR-17-2267
111. Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmström P-U, Choi W, et al. Bladder cancer. Lancet. (2016) 388:2796–810. doi: 10.1016/S0140-6736(16)30512-8
112. Lenis AT, Lec PM, Chamie K, Mshs MD. Bladder cancer: A review. JAMA. (2020) 324:1980–91. doi: 10.1001/jama.2020.17598
113. Contieri R, Hurle R, Paciotti M, Casale P, Saita A, Porpiglia F, et al. Accuracy of the European Association of Urology (EAU) NMIBC 2021 scoring model in predicting progression in a large cohort of HG T1 NMIBC patients treated with BCG. Minerva Urol Nephrol. (2023) 75:180–7. doi: 10.23736/S2724-6051.22.04953-9
114. Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front In Immunol. (2014) 5:461. doi: 10.3389/fimmu.2014.00461
115. Prescott S, James K, Hargreave TB, Chisholm GD, Smyth JF. Intravesical Evans strain BCG therapy: quantitative immunohistochemical analysis of the immune response within the bladder wall. J Urol. (1992) 147:1636–42. doi: 10.1016/S0022-5347(17)37668-1
116. Suttmann H, Riemensberger J, Bentien G, Schmaltz D, Stöckle M, Jocham D, et al. Neutrophil granulocytes are required for effective Bacillus Calmette-Guérin immunotherapy of bladder cancer and orchestrate local immune responses. Cancer Res. (2006) 66:8250–7. doi: 10.1158/0008-5472.CAN-06-1416
117. Beatty JD, Islam S, North ME, Knight SC, Ogden CW. Urine dendritic cells: a noninvasive probe for immune activity in bladder cancer? BJU Int. (2004) 94:1377–83. doi: 10.1111/j.1464-410X.2004.05176.x
118. Brandau S, Riemensberger J, Jacobsen M, Kemp D, Zhao W, Zhao X, et al. NK cells are essential for effective BCG immunotherapy. Int J Cancer. (2001) 92:697–702. doi: 10.1002/1097-0215(20010601)92:5<697::AID-IJC1245>3.0.CO;2-Z
119. Boccafoschi C, Montefiore F, Pavesi M, Pastormerlo M, Annoscia S, Lozzi C, et al. Immunophenotypic characterization of the bladder mucosa infiltrating lymphocytes after intravesical BCG treatment for superficial bladder carcinoma. Eur Urol. (1992) 21:304–8. doi: 10.1159/000474862
120. McAveney KM, Gomella LG, Lattime EC. Induction of TH1- and TH2-associated cytokine mRNA in mouse bladder following intravesical growth of the murine bladder tumor MB49 and BCG immunotherapy. Cancer Immunol Immunother. (1994) 39:401–6. doi: 10.1007/BF01534428
121. Luo Y, Chen X, O’Donnell MA. Role of Th1 and Th2 cytokines in BCG-induced IFN-gamma production: cytokine promotion and simulation of BCG effect. Cytokine. (2003) 21:17–26. doi: 10.1016/S1043-4666(02)00490-8
122. Zhao W, Schorey JS, Bong-Mastek M, Ritchey J, Brown EJ, Ratliff TL. Role of a bacillus Calmette-Guérin fibronectin attachment protein in BCG-induced antitumor activity. Int J Cancer. (2000) 86:83–8. doi: 10.1002/(SICI)1097-0215(20000401)86:1<83::AID-IJC13>3.0.CO;2-R
123. Becich MJ, Carroll S, Ratliff TL. Internalization of bacille Calmette-Guerin by bladder tumor cells. J Urol. (1991) 145:1316–24. doi: 10.1016/S0022-5347(17)38622-6
124. Chen F, Zhang G, Iwamoto Y, See WA. BCG directly induces cell cycle arrest in human transitional carcinoma cell lines as a consequence of integrin cross-linking. BMC Urol. (2005) 5:8. doi: 10.1186/1471-2490-5-8
125. Pook S-H, Rahmat JNB, Esuvaranathan K, Mahendran R. Internalization of Mycobacterium bovis, Bacillus Calmette Guerin, by bladder cancer cells is cytotoxic. Oncol Rep. (2007) 18:1315–20. doi: 10.3892/or.18.5.1315
126. Lamm DL, Blumenstein BA, Crissman JD, Montie JE, Gottesman JE, Lowe BA, et al. Maintenance bacillus Calmette-Guerin immunotherapy for recurrent TA, T1 and carcinoma in situ transitional cell carcinoma of the bladder: a randomized Southwest Oncology Group Study. J Urol. (2000) 163:1124–9. doi: 10.1016/S0022-5347(05)67707-5
127. Herr HW, Laudone VP, Badalament RA, Oettgen HF, Sogani PC, Freedman BD, et al. Bacillus Calmette-Guérin therapy alters the progression of superficial bladder cancer. J Clin Oncol. (1988) 6:1450–5. doi: 10.1200/JCO.1988.6.9.1450
128. Herr HW. Transurethral resection and intravesical therapy of superficial bladder tumors. Urologic Clinics North America. (1991) 18:525–8. doi: 10.1016/S0094-0143(21)00346-3
129. Lamm DL. Efficacy and safety of bacille Calmette-Guérin immunotherapy in superficial bladder cancer. Clin Infect Dis. (2000) 31 Suppl 3:S86–90. doi: 10.1086/314064
130. Ng KL, Chua CB. Reiter’s syndrome postintravesical Bacillus Calmette-Guérin instillations. Asian J Surg. (2017) 40:163–5. doi: 10.1016/j.asjsur.2014.01.016
131. Friedlander E, Martínez Pascual P, Montilla de Mora P, Scola Yurrita B. Bilateral parotid glands infection caused by Calmette-Guerin Bacillus after intravesical therapy for recurrent bladder cancer: a case report. Braz J Otorhinolaryngol. (2017) 83:726–9. doi: 10.1016/j.bjorl.2015.10.017
132. Torres-Blanco Á, Gómez-Palonés F, Edo-Fleta G. Arteriocutaneous fistula associated with bilateral femoral pseudoaneurysms caused by bacillus calmette-guérin. Apropos of a case and review of literature. Ann Vasc Surg. (2017) 39:291.e1–6. doi: 10.1016/j.avsg.2016.07.094
133. Leeman M, Burgers P, Brehm V, van Brussel JP. Psoas abscess after bacille Calmette-Guérin instillations causing iliac artery contained rupture. J Vasc Surg. (2017) 66:1236–8. doi: 10.1016/j.jvs.2017.02.038
134. Sampaio PCM, Lira YG, Ribeiro HYU, de Paula Moreira F, Gadelha MSM, da Cruz SFS. Poncet’s disease after the intravesical instillation of Bacillus Calmette-Guérin (BCG): a case report. BMC Res Notes. (2017) 10:416. doi: 10.1186/s13104-017-2606-9
135. Kustrimovic N, Bilato G, Mortara L, Baci D. The urinary microbiome in health and disease: relevance for bladder cancer. Int J Mol Sci. (2024) 25(3):1732. doi: 10.3390/ijms25031732
136. Oddens JR, Sylvester RJ, Brausi MA, Kirkels WJ, van de Beek C, van Andel G, et al. The effect of age on the efficacy of maintenance bacillus Calmette-Guérin relative to maintenance epirubicin in patients with stage Ta T1 urothelial bladder cancer: results from EORTC genito-urinary group study 30911. Eur Urol. (2014) 66:694–701. doi: 10.1016/j.eururo.2014.05.033
137. Pirzada MT, Ghauri R, Ahmed MJ, Shah MF, Nasir IUI, Siddiqui J, et al. Outcomes of BCG induction in high-risk non-muscle-invasive bladder cancer patients (NMIBC): A retrospective cohort study. Cureus. (2017) 9:e957. doi: 10.7759/cureus.957
138. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Sci (New York N.Y.). (2017) 357(6356):1156–60. doi: 10.1126/science.aah5043
139. Priadko K, Romano L, Olivieri S, Romeo M, Barone B, Sciorio C, et al. Intestinal microbiota, intestinal permeability and the urogenital tract: is there a pathophysiological link? J Physiol Pharmacol. (2022) 73(5). doi: 10.26402/jpp.2022.5.01
140. Romano L, Napolitano L, Crocetto F, Sciorio C, Sio MD, Miranda A, et al. Prostate and gut: Any relationship? A narrative review on the available evidence and putative mechanisms. Prostate. (2024) 84:513–24. doi: 10.1002/pros.24675
141. Morton AM, Sefik E, Upadhyay R, Weissleder R, Benoist C, Mathis D. Endoscopic photoconversion reveals unexpectedly broad leukocyte trafficking to and from the gut. Proc Natl Acad Sci U.S.A. (2014) 111:6696–701. doi: 10.1073/pnas.1405634111
142. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. (2013) 54:2325–40. doi: 10.1194/jlr.R036012
143. White CA, Pone EJ, Lam T, Tat C, Hayama KL, Li G, et al. Histone deacetylase inhibitors upregulate B cell microRNAs that silence AID and Blimp-1 expression for epigenetic modulation of antibody and autoantibody responses. J Immunol. (2014) 193:5933–50. doi: 10.4049/jimmunol.1401702
144. Wang B, Qiu Y, Xie M, Huang P, Yu Y, Sun Q, et al. Gut microbiota Parabacteroides distasonis enchances the efficacy of immunotherapy for bladder cancer by activating anti-tumor immune responses. BMC Microbiol. (2024) 24:237. doi: 10.1186/s12866-024-03372-8
145. Matsushita M, Fujita K, Hayashi T, Kayama H, Motooka D, Hase H, et al. Gut microbiota-derived short-chain fatty acids promote prostate cancer growth via IGF1 signaling. Cancer Res. (2021) 81:4014–26. doi: 10.1158/0008-5472.CAN-20-4090
146. Adebayo AS, Suryavanshi MV, Bhute S, Agunloye AM, Isokpehi RD, Anumudu CI, et al. The microbiome in urogenital schistosomiasis and induced bladder pathologies. PloS Negl Trop Dis. (2017) 11:e0005826. doi: 10.1371/journal.pntd.0005826
147. Crocetto F, Barone B, De Luca L, Creta M. Granulomatous prostatitis: a challenging differential diagnosis to take into consideration. Future Oncol. (2020) 16:805–6. doi: 10.2217/fon-2020-0185
148. Shukla P, Gulwani HV, Kaur S. Granulomatous prostatitis: clinical and histomorphologic survey of the disease in a tertiary care hospital. Prostate Int. (2017) 5:29–34. doi: 10.1016/j.prnil.2017.01.003
149. Rath T, Dieterich W, Kätscher-Murad C, Neurath MF, Zopf Y. Cross-sectional imaging of intestinal barrier dysfunction by confocal laser endomicroscopy can identify patients with food allergy in vivo with high sensitivity. Sci Rep. (2021) 11:12777. doi: 10.1038/s41598-021-92262-4
150. Paniagua-Contreras GL, Monroy-Pérez E, Díaz-Velásquez CE, Uribe-García A, Labastida A, Peñaloza-Figueroa F, et al. Whole-genome sequence analysis of multidrug-resistant uropathogenic strains of Escherichia coli from Mexico. Infect Drug Resist. (2019) 12:2363–77. doi: 10.2147/IDR.S203661
151. Ratajczak W, Rył A, Mizerski A, Walczakiewicz K, Sipak O, Laszczyńska M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim Pol. (2019) 66(1):1–12. doi: 10.18388/abp.2018_2648
152. Martel J, Chang S-H, Ko Y-F, Hwang T-L, Young JD, Ojcius DM. Gut barrier disruption and chronic disease. Trends Endocrinol Metab. (2022) 33:247–65. doi: 10.1016/j.tem.2022.01.002
153. Krneta T, Gillgrass A, Poznanski S, Chew M, Lee AJ, Kolb M, et al. M2-polarized and tumor-associated macrophages alter NK cell phenotype and function in a contact-dependent manner. J Leukoc Biol. (2017) 101:285–95. doi: 10.1189/jlb.3A1215-552R
154. Worby CJ, Schreiber HL, Straub TJ, van Dijk LR, Bronson RA, Olson BS, et al. Longitudinal multi-omics analyses link gut microbiome dysbiosis with recurrent urinary tract infections in women. Nat Microbiol. (2022) 7:630–9. doi: 10.1038/s41564-022-01107-x
155. Alfano M, Canducci F, Nebuloni M, Clementi M, Montorsi F, Salonia A. The interplay of extracellular matrix and microbiome in urothelial bladder cancer. Nat Rev Urol. (2016) 13:77–90. doi: 10.1038/nrurol.2015.292
156. Berndt A, Richter P, Kosmehl H, Franz M. Tenascin-C and carcinoma cell invasion in oral and urinary bladder cancer. Cell Adh Migr. (2015) 9:105–11. doi: 10.1080/19336918.2015.1005463
157. Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of Malignant tumour progression. Nat Rev Cancer. (2005) 5:744–9. doi: 10.1038/nrc1694
158. Iozzo RV. Tumor stroma as a regulator of neoplastic behavior. Agonistic and antagonistic elements embedded in the same connective tissue. Lab Invest. (1995) 73:157–60.
159. Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. (2012) 196:395–406. doi: 10.1083/jcb.201102147
160. Lilly JD, Parsons CL. Bladder surface glycosaminoglycans is a human epithelial permeability barrier. Surg Gynecol Obstet. (1990) 171:493–6.
161. Vollmer P, Walev I, Rose-John S, Bhakdi S. Novel pathogenic mechanism of microbial metalloproteinases: liberation of membrane-anchored molecules in biologically active form exemplified by studies with the human interleukin-6 receptor. Infect Immun. (1996) 64:3646–51. doi: 10.1128/iai.64.9.3646-3651.1996
162. Theander TG, Kharazmi A, Pedersen BK, Christensen LD, Tvede N, Poulsen LK, et al. Inhibition of human lymphocyte proliferation and cleavage of interleukin-2 by Pseudomonas aeruginosa proteases. Infect Immun. (1988) 56:1673–7. doi: 10.1128/iai.56.7.1673-1677.1988
163. Wilson M, Seymour R, Henderson B. Bacterial perturbation of cytokine networks. Infect Immun. (1998) 66:2401–9. doi: 10.1128/IAI.66.6.2401-2409.1998
164. Kayagaki N, Kawasaki A, Ebata T, Ohmoto H, Ikeda S, Inoue S, et al. Metalloproteinase-mediated release of human Fas ligand. J Exp Med. (1995) 182:1777–83. doi: 10.1084/jem.182.6.1777
165. Müllberg J, Durie FH, Otten-Evans C, Alderson MR, Rose-John S, Cosman D, et al. A metalloprotease inhibitor blocks shedding of the IL-6 receptor and the p60 TNF receptor. J Immunol. (1995) 155:5198–205. doi: 10.4049/jimmunol.155.11.5198
166. Wysocki AB, Bhalla-Regev SK, Tierno PM, Stevens-Riley M, Wiygul R-C. Proteolytic activity by multiple bacterial species isolated from chronic venous leg ulcers degrades matrix substrates. Biol Res Nurs. (2013) 15:407–15. doi: 10.1177/1099800412464683
167. Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. (2007) 204:1257–65. doi: 10.1084/jem.20062512
168. Saleh R, Elkord E. FoxP3+ T regulatory cells in cancer: Prognostic biomarkers and therapeutic targets. Cancer Lett. (2020) 490:174–85. doi: 10.1016/j.canlet.2020.07.022
169. Sawant DV, Yano H, Chikina M, Zhang Q, Liao M, Liu C, et al. Adaptive plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes tumor T cell exhaustion. Nat Immunol. (2019) 20:724–35. doi: 10.1038/s41590-019-0346-9
170. Sitkovsky M, Lukashev D, Deaglio S, Dwyer K, Robson SC, Ohta A. Adenosine A2A receptor antagonists: blockade of adenosinergic effects and T regulatory cells. Br J Pharmacol. (2008) 153 Suppl 1:S457–64. doi: 10.1038/bjp.2008.23
171. Zlotta AR, Drowart A, Van Vooren JP, de Cock M, Pirson M, Palfliet K, et al. Evolution and clinical significance of the T cell proliferative and cytokine response directed against the fibronectin binding antigen 85 complex of bacillus Calmette-Guerin during intravesical treatment of superficial bladder cancer. J Urol. (1997) 157:492–8. doi: 10.1016/S0022-5347(01)65185-1
172. Chen C, Huang Z, Huang P, Li K, Zeng J, Wen Y, et al. Urogenital microbiota: potentially important determinant of PD-L1 expression in male patients with non-muscle invasive bladder cancer. BMC Microbiol. (2022) 22:7. doi: 10.1186/s12866-021-02407-8
173. Yow MA, Tabrizi SN, Severi G, Bolton DM, Pedersen J, Giles GG, et al. Characterisation of microbial communities within aggressive prostate cancer tissues. Infect Agent Cancer. (2017) 12:4. doi: 10.1186/s13027-016-0112-7
174. Sfanos KS, Sauvageot J, Fedor HL, Dick JD, De Marzo AM, Isaacs WB. A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate. (2008) 68:306–20. doi: 10.1002/pros.20680
175. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Sci (New York N.Y.). (2020) 368:973–80. doi: 10.1126/science.aay9189
176. Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, Asraf O, Martino C, Nejman D, et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. (2022) 185(20):3789–806.e17. doi: 10.1016/j.cell.2022.09.005
177. Galeano Niño JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. (2022) 611:810–7. doi: 10.1038/s41586-022-05435-0
178. Cummins J, Tangney M. Bacteria and tumours: causative agents or opportunistic inhabitants? Infect Agent Cancer. (2013) 8:11. doi: 10.1186/1750-9378-8-11
179. Pope JL, Tomkovich S, Yang Y, Jobin C. Microbiota as a mediator of cancer progression and therapy. Transl Res. (2017) 179:139–54. doi: 10.1016/j.trsl.2016.07.021
180. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Sci (New York N.Y.). (2021) 371:602–9. doi: 10.1126/science.abb5920
181. Dizman N, Meza L, Bergerot P, Alcantara M, Dorff T, Lyou Y, et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: a randomized phase 1 trial. Nat Med. (2022) 28:704–12. doi: 10.1038/s41591-022-01694-6
182. Sharpton TJ. An introduction to the analysis of shotgun metagenomic data. Front Plant Sci. (2014) 5:209. doi: 10.3389/fpls.2014.00209
183. Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet. (2005) 6:805–14. doi: 10.1038/nrg1709
184. Zaki S, Blau DM, Hughes JM, Nolte KB, Lynfield R, Carr W, et al. CDC Grand Rounds: discovering new diseases via enhanced partnership between public health and pathology experts. MMWR Morb Mortal Wkly Rep. (2014) 63:121–6.
Keywords: microbiome, urologic neoplasms, tumor immunity, immunotherapy, inflammation
Citation: Zou J, Xu B, Gao H, Luo P, Chen T and Duan H (2024) Microbiome in urologic neoplasms: focusing on tumor immunity. Front. Immunol. 15:1507355. doi: 10.3389/fimmu.2024.1507355
Received: 07 October 2024; Accepted: 18 November 2024;
Published: 05 December 2024.
Edited by:
Sheila Spada, Hospital Physiotherapy Institutes (IRCCS), ItalyReviewed by:
Biagio Barone, ASL Napoli 1 Centro, ItalyLorenzo Romano, University of Campania Luigi Vanvitelli, Italy
Copyright © 2024 Zou, Xu, Gao, Luo, Chen and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huanglin Duan, MTU3Nzk4MzExNzlAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Jun Zou
Jun Zou Baisheng Xu2†
Baisheng Xu2† Peiyue Luo
Peiyue Luo Tao Chen
Tao Chen Huanglin Duan
Huanglin Duan