- 1Xi’an People’s Hospital (Xi’an Fourth Hospital), Affiliated People’s Hospital of Northwest University, Xi’an, China
- 2Technical University of Munich (TUM) School of Medicine and Health, Munich, Germany
- 3Department of General, Visceral, and Transplant Surgery, Ludwig-Maximilians University, Munich, Germany
CD8+ T cells are crucial cytotoxic components of the tumor immune system. In chronic inflammation, they become low-responsive, a state known as T cell exhaustion (TEX). The aim of immune checkpoint blockade is to counteract TEX, yet its dynamics in breast cancer remain poorly understood. This review defines CD8+ TEX and outlines its features and underlying mechanisms. It also discusses the primary mechanisms of CD8+ TEX in breast cancer, covering inhibitory receptors, immunosuppressive cells, cytokines, transcriptomic and epigenetic alterations, metabolic reprogramming, and exosome pathways, offering insights into potential immunotherapy strategies for breast cancer.
1 Introduction
Breast cancer (BC) is a prevalent cancer in women worldwide, with its mechanisms not yet fully understood (1). Immune cells play a crucial role from immunosurveillance in normal breast tissue to the progression of BC, including both primary and metastatic stages. The tumor microenvironment in BC shows an increase in immune cells like CD4+ and CD8+ granzyme B+ cytotoxic T cells, B cells, macrophages, and dendritic cells (DCs) (2). In estrogen receptor (ER)-positive tumors, neutrophils and natural killer (NK) cells are most positively associated, while resting memory T cells and CD8+ T cells are negatively associated. In contrast, ER-negative BC shows strong positive correlations with T regulatory and CD8+ cells, with similar negative correlations as ER-positive cases (3). Notably, even early-stage BC patients display exhaustion in tumor-associated CD8+ T cells (4), which are essential for eliminating pathogens and tumors (5). Chronic antigen exposure and inflammation in cancer can lead to CD8+ T cell exhaustion or altered differentiation (6), with exhausted T cells showing tumor reactivity and proliferation signatures (7). A higher CD8+ T cell exhaustion (TEX) score is linked to poorer disease-free survival in BC (8), highlighting the importance of CD8+ effector T cell activation versus exhaustion in BC progression and patient outcomes. Additionally, TEX can predict immunotherapy responses in ER-positive BC, underscoring its potential in treatment strategies (9). Recent studies have extensively explored the mechanisms of CD8+ TEX in BC. This article summarizes our current understanding of these mechanisms.
2 Definition and characteristics of CD8+ T cell exhaustion
Tex was first observed in murine models of lymphocytic choriomeningitis virus (LCMV) infection (10). It represents T cell dysfunction with a progressive loss of effector functions during chronic infections and cancer (11). Tex cells exhibit elevated inhibitory receptors (PD-1, CTLA-4, TIM-3, TIGIT, LAG-3), reduced antitumor cytokines (IFN-γ, IL-2, TNF) (6, 11, 12), increased tumor-promoting chemokines, altered transcription factors (TCF1, T-bet, TOX), metabolic issues, and decreased proliferation and survival (6, 11).
In BC, immune cell distribution varies by tumor subtype and between stromal and parenchymal regions (2). CD8+ Tex are present in certain ER+ and triple-negative breast cancers (TNBC), creating a unique tumor microenvironment (TME) with higher IFN-γ activity. Increased CD8+ Tex infiltration is linked to poorer overall and relapse-free survival in premenopausal ER+ BC patients (13). Conversely, TNBC exhibits a more immunosuppressive environment compared to HER2+ or luminal types, with more regulatory T cells (Tregs), exhausted CD8+ T cells, and plasma cells (14).
2.1 Mechanism of CD8+ T cell exhaustion
Persistent antigen stimulation is essential for inducing T cell exhaustion (15, 16). Additional key signals include pro-inflammatory cytokines (e.g., IFN-α/β, IL-6, IL-27), suppressive cytokines (e.g., IL-10, TGF-β), regulatory leukocytes (e.g., Tregs, immunoregulatory APCs), and TME factors like hypoxia, nutrient deprivation, and altered pH (17–19). Together with chronic TCR engagement, these signals cause sustained upregulation of inhibitory receptors, transcription factor changes, metabolic shifts, and unique transcriptional programs (20–22). This leads to diminished effector functions, altered homeostasis compared to memory T cells, and cell death from overstimulation, resulting in inadequate tumor control (23, 24). Exhausted T cells are regulated by transcriptional and epigenetic mechanisms, including key transcription factors such as NFAT (25), NR4A (26), TOX (27), PTPN2 (28), TCF-1 (29), and Eomes (30). In the BC tumor microenvironment, TEX significantly impacts immune escape and therapeutic response, with immune regulation central to TEX through inhibitory receptors (e.g., PD-1), cytokines (e.g., IL-10), and immunoregulatory cells like TAMs.
3 Inhibitory receptors
IRs include PD-1, CTLA-4, TIGIT, TIM-3, LAG-3, which are hallmarks of TEX. Exhausted T cells often express high levels of IRs, which act as immunomodulators, limiting immunopathology and promoting tolerance to self-antigens. In BC, the continued expression of inhibitory receptors is critical for TEX.
3.1 Programmed death protein 1
PD-1, an inhibitory transmembrane protein on T, B, and NK cells, blocks key T cell pathways (PI3K-AKT-mTOR, RAS-MEK-ERK) when binding PD-L1, leading to T cell exhaustion and conversion to Treg cells (31, 32). Immune checkpoint inhibitors targeting PD-1/PD-L1 are being explored to restore T cell function in BC treatments. In PD-1-high CD8+ T cells, miR-149-3p reduces apoptosis and downregulates exhaustion markers by targeting PD-1, TIM-3, BTLA, and Foxp1 mRNAs (33). Additionally, miR-424-5p targets PD-L1 and the PTEN/PI3K/AKT/mTOR pathway, potentially reversing T cell exhaustion (34, 35). BC model DCs overexpress PD-L1 and Gal-9, inducing TEX, but miRNA-5119 mimic-engineered DCs restored function, reduced PD-L1, and boosted immune responses (36). In BC, miRNA-138-5p lowers PD-L1, promoting apoptosis through Caspase-9/3 activation and cell cycle arrest, and affects cell motility and T-cell cytokines by interacting with MMP2, MMP9, vimentin, and E-cadherin (37).
3.2 Cytotoxic T lymphocyte-associated protein 4
CTLA-4 (CD152) is a transmembrane protein expressed on regulatory T cells (Tregs), CD4+, and CD8+ T cells, containing two cytoplasmic domains with tyrosine-based signaling motifs for signal transduction (38). Its extracellular domain, similar to CD28, competitively binds B7-1/2 (CD80/86) on antigen-presenting cells (APCs), modulating T cell function and preventing overactive immune responses (39). Additionally, CTLA-4 is present in the cytoplasm and on the surface of BC cells (40). Elevated serum CTLA-4 in BC patients versus healthy individuals highlights its role in BC pathogenesis and progression (41). Co-culturing CTLA-4+ BC cells with human dendritic cells (DCs) inhibits extracellular signal-regulated kinase and activating transcription factor 3 (ATF3), suppressing DC function and maturation (42). These impaired DCs further inhibit the proliferation of allogeneic CD4+/CD8+ T cells, Th1 differentiation, and cytotoxic T lymphocyte (CTL) function.
3.3 Lymphocyte-activation gene 3
Lymphocyte-activation gene 3 (LAG-3 or CD223) is an inhibitory immune checkpoint molecule found on the surface of various lymphocytes (43, 44), including CD4+ and CD8+ T cells, natural killer (NK) cells, NKT cells, and regulatory T (Treg) cells (45, 46). It shapes the tumor immune environment by inducing T cell exhaustion and limiting proliferation (47). LAG-3 binds ligands like fibrinogen-like protein 1 (FGL1) to suppress T cell activation and is often co-expressed with other checkpoints such as PD-1 or PD-L1 in BC (48–50). Additionally, Liu et al. found that LAG-3 may synergize with multiple immune checkpoints in the BC-induced immune response, including PD-L1, TIGIT, CTLA-4, ICOS, and IDO1 (51).
3.4 T-cell Ig and ITIM domain
TIGIT, a member of the immunoglobulin superfamily, is expressed on T lymphocytes and NK cells and is primarily involved in immune responses and inflammatory activities (52). Co-expression of TIGIT with other inhibitory receptors on exhausted CD8+ T-cell subsets has been observed in tumors (53). In early-stage breast cancer, high co-expression of TIGIT and other immune checkpoint receptors—including PD-1, CTLA-4, LAG-3, and TIM-3—on tumor-infiltrating lymphocytes correlates with increased disease aggressiveness (54). Elevated levels of TIGIT, TIM-3, and LAG-3 are found in locally advanced breast cancer patients with poor prognostic factors following neoadjuvant chemotherapy (55). TIGIT plays a role in immune and inflammatory responses similar to PD-1 in breast cancer (56), suggesting that TIGIT and PD-1 may synergistically promote the development of severely dysfunctional T cells, as reported in other cancers (57, 58).
3.5 T-cell immunoglobulin and mucin-domain containing-3
TIM-3 is a checkpoint receptor expressed on immune cells like dendritic cells, macrophages, and T cells (59–61), mediating immunosuppression through ligands such as phosphatidylserine, CEACAM-1, and galectin-9 (62, 63). In activated T cells, TIM-3 signaling induces exhaustion by decreasing proliferation, reducing effector cytokine production, and promoting apoptosis of cytotoxic T cells (64). In BC, TIM-3 is overexpressed, enhancing tumor cell proliferation, migration, invasion, and inhibiting apoptosis (65–67). High TIM-3 levels correlate with advanced clinical stages, lymph node metastasis, increased Ki67 expression, and poorer 5-year survival rates (65, 66). It promotes tumorigenesis by activating the NF-κB/STAT3 pathway and altering gene expression—upregulating CCND1, C-Myc, MMP1, TWIST, VEGF, and downregulating E-cadherin (68, 69). Additionally, TIM-3 affects tight junction dynamics by downregulating ZO-2, ZO-1, and occludin, which may enhance tumor invasion and migration. Polymorphisms in the TIM-3 gene are also linked to BC susceptibility, progression, and prognosis (70, 71).
Galectin-9 (Gal-9) is an immune checkpoint protein that promotes TEX and modulates the tumor microenvironment (72, 73). As a ligand for TIM-3, Gal-9 induces apoptosis in T cells and suppresses the cytotoxic function of antigen-specific CTLs (74). The TIM-3/Gal-9 pathway is involved in immune evasion by BC cells, which show higher levels of Gal-9 and TIM-3 compared to healthy mammary tissues, with both proteins co-localizing in tumor cells. Upregulation of LPHN2, along with expression of LPHN3 and FLRT3, is also detected in breast tumor cells. Activation of this pathway leads to Gal-9 translocation to the tumor cell surface, protecting them from CTL-induced apoptosis. In patients with TNBC, high Gal-9 expression correlates with positive PD-L1 expression on tumor cells (75).
3.6 V-set domain containing T cell activation inhibitor 1
B7-H4 is an inhibitory member of the B7 family and functions as an onco-fetal immune tolerance checkpoint (76, 77). Genetic anomalies in B7-H4 are associated with immune activation and fetal rejection in syngeneic pregnancy models. Similarly, in BC, B7-H4 is linked to tumor progression and correlates with exhaustion of CD8+ T cells. Hormonal assays have demonstrated that progesterone induces B7-H4 expression in both placental and BC cells (78).
In summary, exhaustion in CD8+ T cells is marked by the concurrent upregulation of multiple immune checkpoints (ICs), with the diversity and quantity of these ICs directly influencing the extent of T cell dysfunction (79–82). Despite the well-documented impacts of individual ICs on T cell functionality, investigations into the additive influence of these ICs on T cell exhaustion in BC remain scant. Nevertheless, compelling data supports the notion that dual IC inhibition, particularly incorporating PD-1, significantly surpasses single IC interventions in boosting tumor-associated CD8+ T cell efficacy, both in vitro and in vivo (83). Besides, nanodrug releasing anti-Galectin-9 antibody can exert local blockade of PD-1/Galectin-9/TIM-3 interaction to enhance effector T cells in BC via reversing the exhaustion (84).
4 Immunosuppressive cells and cytokines
In addition to cell surface IRs, immunoregulatory cells and cytokines in the TME—including myeloid-derived suppressor cells (MDSCs), TAMs, and Tregs—contribute to TEX. These immunosuppressive cells hinder immune rejection of malignant cells via cytokine secretion, promoting tumor progression and posing challenges to immunotherapy.
CXCR2+ MDSCs inversely correlate with infiltrating CD4+ and CD8+ T cells. These MDSCs facilitate BC progression and metastasis to lungs and lymph nodes, drive EMT via IL-6, and increase levels of immunosuppressive proteins (PD-1, PD-L1, LAG3, CTLA4, TIM-3) on T cells, contributing to their partial exhaustion mediated by IFN-γ (85).
TAMs are predominant immune cells in BC and can polarize into pro-inflammatory M1 or immunosuppressive M2 phenotypes (86–88). Progranulin (PGRN) promotes CD8+ TEX by inducing TAM polarization to M2 macrophages, which suppress T cell proliferation and activation via ICAM-1 interactions (89–91). Elevated CHI3L2 levels in TAMs correlate with poor prognosis in BC. Y-box binding protein 1 (YBX1) is positively associated with M2 macrophage infiltration and TEX markers IDO1 and CTLA4 in luminal BC (92).
IL1R2 in macrophages and BC cells contributes to an immunosuppressive TME. IL1β from TAMs activates IL1R2, increasing PD-L1 levels by promoting YY1 degradation. Inhibiting IL1R2 reduces macrophage recruitment, alters TAM polarization, decreases breast tumor-initiating cell self-renewal, and reduces CD8+ TEX (93). Sustained high levels of IL-2 in BC induce CD8+ TEX by persistently activating STAT5, which increases tryptophan hydroxylase 1 expression. This enzyme converts tryptophan to 5-hydroxytryptophan (5-HTP), activating the aryl hydrocarbon receptor (AhR) and leading to upregulation of inhibitory receptors and decreased cytokine production, impairing T cell function (94). Low IL-10 expression or loss of IL-10R-STAT3 signaling is linked to increased CD8+ TEX and reduced survival in BC patients (95). Conversely, in murine BC models, Heparanase (HPSE)-induced IL-10 upregulation promotes M2 macrophage polarization and TEX (96). Malignant BC cells transfer active TGF-β type II receptor (TβRII) via tumor-derived extracellular vesicles to CD8+ T cells, inducing SMAD3 activation and TEX (97). USP8 stabilizes TβRII by deubiquitination, enhancing its expression in plasma membranes and TEVs. USP8 promotes TGF-β/SMAD-induced EMT, invasion, metastasis, and facilitates TβRII+ circulating EVs to induce TEX and chemoimmunotherapy resistance (92). RAI14 levels correlate positively with M2 macrophage marker CD163 and TEX marker PD-1, indicating its association with M2 macrophage infiltration and TNBC (98).
5 Transcriptomics and epigenetic regulation
Transcriptomic and epigenetic mechanisms play crucial roles in TEX within BC. Epigenetic programming is central to regulating various T cell subsets. In TNBC patients, dysfunctional PD-1+ CD8+ T cells are enriched with EOMES and nLSD1p. LSD1 promotes TEX by controlling the nuclear localization of EOMES through bivalent post-translational modification at lysine 641, directly affecting T cells (99).
TWIST1, a transcription factor that binds to the PD-L1 promoter and significantly accelerates the exhaustion and death of cytotoxic CD8+ T cells in BC (100). Additionally, TWIST1 induces EMT in BC cells via the ITGB1-FAK/ILK signaling pathways and related downstream networks (101). The expression of transcription factor IRF8 facilitates TAM acquisition, antigen presentation of cancer cells, and induction of CTL exhaustion in BC (102). Thymocyte selection-associated high-mobility group box (TOX), involved in TEX during chronic infection and cancer, is associated with better prognosis in BC when highly expressed, as confirmed by survival analysis (103). Bromodomain and extra-terminal (BET) proteins are important in regulating the PD-1/PD-L1 pathway in BC. They mediate interferon-γ (IFN-γ) secretion from activated T cells and IFN-γ signaling in TNBC to induce PD-L1 expression (104).
METTL3 enhances PD-L1 expression in BC cells post-transcriptionally through an m6A-IGF2BP3-dependent mechanism, promoting mRNA stability and affecting the efficacy of tumor immunotherapy (105). Glycosylphosphatidylinositol (GPI) anchor biosynthesis, a common post-translational modification, is elevated in BC patients with severe TEX. Exhausted CD8+ T cells exhibit higher levels of GPI anchor biosynthesis than normal CD8+ T cells (106).
A comprehensive evaluation of mitochondrial DNA methylation (MTDM) in BC indicates that patients with high MTDM have increased proliferation rates and elevated CD8+ TEX, potentially related to the secretion of growth differentiation factor 15 (GDF15) by malignant breast epithelial cells in a high MTDM state (107).
6 Metabolic reprogramming
Metabolic reprogramming plays a pivotal role in TEX in BC. Metformin reduces tumor hypoxia and PD-L1 expression, enhances T cell function, and boosts the efficacy of immune checkpoint inhibitors, positioning it as a potential adjunct therapy for refractory tumors like TNBC. At high concentrations, metformin suppresses mTOR signaling and decreases transcription factors (T-bet, Eomes, STAT3), potentially limiting T cell proliferation and cytokine secretion. Notably, metformin reduces PD-1 expression in Jurkat cells and improves the PD-1/CD69 ratio in primary T cells, suggesting a potential to restore T cell activation and counter exhaustion (108).
In mouse BC models, a high-fat diet shifts PD-1- CD8+ non-exhausted T cells to PD-1+ CD8+ exhausted T cells, fostering tumor progression (109). Hypoxia-driven metabolic reprogramming also exacerbates T cell dysfunction and exhaustion. Hypoxia induces HIF1α-dependent epigenetic changes that suppress effector gene expression in T and NK cells, leading to immune dysfunction. Moreover, HIF1α-independent mechanisms promote TEX by upregulating co-inhibitory molecules like TIM-3 or via metabolic stress. Studies show that hypoxia-induced TEX in TNBC models confers resistance to anti-PD1 therapy (110). Additionally, hypoxic stress generates tRNA-derived fragments (tRFs) that bind and displace the 3’ UTR of the oncogenic RNA-binding protein YBX1, inhibiting oncogene stability and suppressing BC progression (111).
7 Exosome pathway
Exosomes play a crucial role in intercellular communication, with cancer cell-derived exosomes promoting tumor progression by modulating the tumor microenvironment. Tumor cells secrete PD-L1 via exosomes (exoPD-L1), which binds to PD-1 on CD8+ T cells at metastatic sites, inhibiting T cell activation and proliferation, leading to their functional inactivation and immunosuppression (112–115). ICAM1, a glycoprotein involved in immune functions and cancer progression, is secreted in exosomes and regulates the immunosuppressive microenvironment (116). Its interaction with LFA-1 is vital for exoPD-L1 binding to PD-1 (117). Knockdown of ICAM1 in BC cells reduces exosome-induced inhibition of CD8+ T cells, suggesting its role in TNBC bone metastasis (116). BC-derived exosomes also suppress CD8+ T cell glycolysis via an AKT-mTOR-dependent mechanism, contributing to immune escape (118). In migrating cancer cells, PD-L1 accumulates at the trailing edge, forming migrasomes that can be internalized by adjacent cells, increasing PD-L1 expression and immune suppression, while also releasing chemokines to promote cell migration within the tumor microenvironment (119). The role of migrasomes in CD8+ TEX remains understudied.
In summary, TEX is characterized by high expression of IRs, influenced by immune cells and cytokines, with potential mechanisms including transcriptomic, epigenetic, metabolic, and exosome-related pathways (Figure 1).
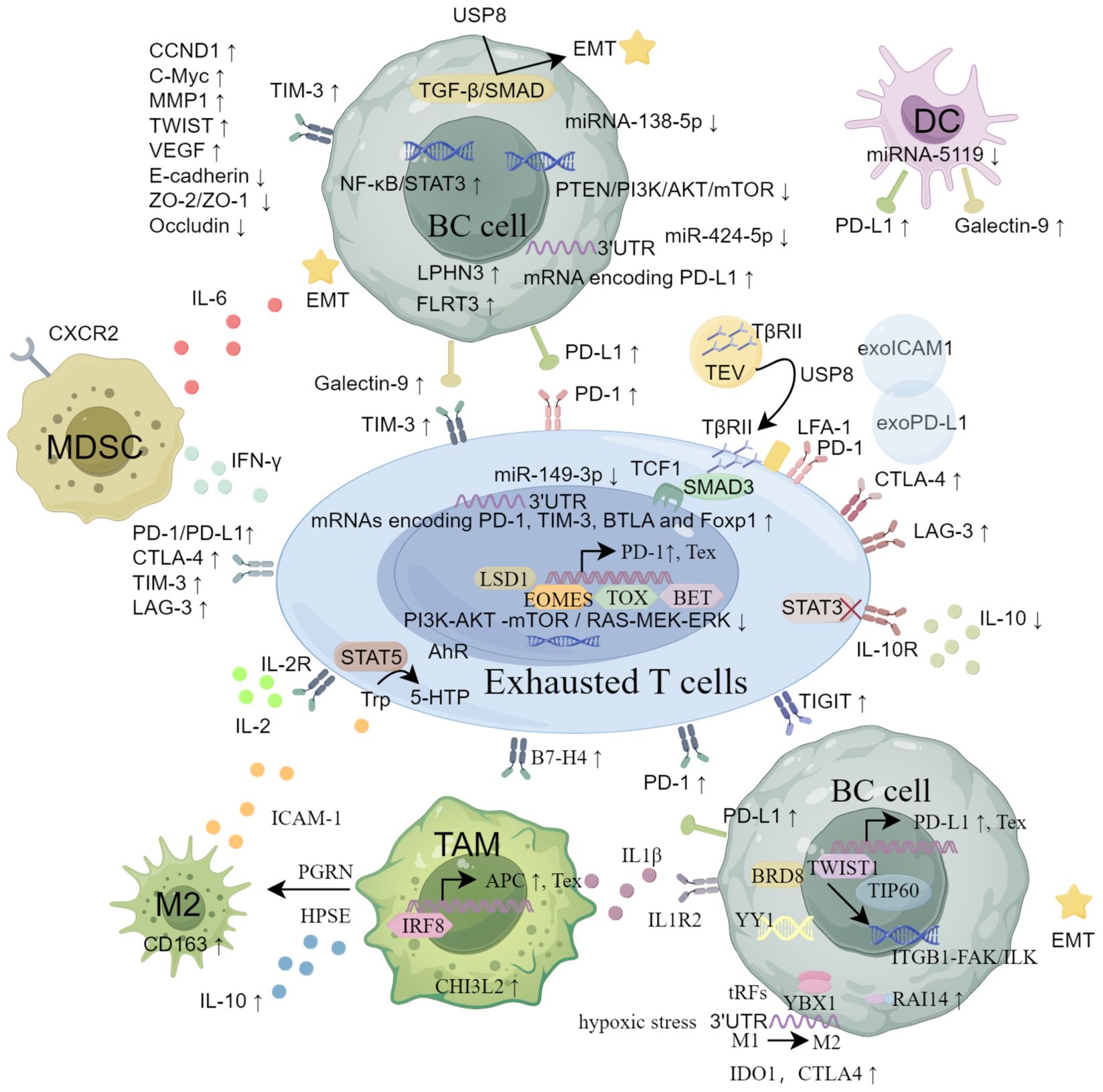
Figure 1. The main mechanisms of CD8+ Tex in breast cancer from the aspects of inhibitory receptors, immunosuppressive cells and cytokines, transcriptomics and epigenetic regulation, metabolic reprogramming and exosome pathway.
8 Discussion
CD8+ TEX is critical in immune evasion and progression of BC, involving upregulation of inhibitory receptors (PD-1, CTLA-4, LAG-3, TIM-3), immunosuppressive cells and cytokines, transcriptional and epigenetic alterations, metabolic reprogramming, and exosome pathways. These interactions within the tumor microenvironment diminish T cell function, facilitating tumor growth and metastasis
Understanding these mechanisms is essential for developing effective immunotherapies. While targeting inhibitory receptors with immune checkpoint inhibitors shows promise, their limited efficacy highlights the need for comprehensive approaches. Future research should elucidate molecular pathways of TEX in BC, including specific transcription factors, epigenetic modifications, and metabolic processes. Exploring combination therapies that address multiple aspects of T cell exhaustion may enhance efficacy. Personalizing immunotherapies based on each BC subtype’s unique immune landscape could further optimize patient outcomes. Investigating the role of exosomes and their impact on CD8+ T cell function may reveal new therapeutic targets. Integrating these insights can lead to novel interventions to prevent or reverse CD8+ TEX, ultimately improving survival and quality of life for BC patients.
Author contributions
HX: Funding acquisition, Writing – original draft, Writing – review & editing. XX: Writing – original draft. TL: Writing – original draft. HL: Writing – original draft. ZX: Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by grants from Xi’an People’s Hospital (Xi’an Fourth Hospital) Scientific Research Incubation Fund (No. LH-11).
Acknowledgments
The figure in this study was supported by FigDraw plate.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Landskron G, de la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. (2014) 2014:149185. doi: 10.1155/2014/149185
2. Goff SL, Danforth DN. The role of immune cells in breast tissue and immunotherapy for the treatment of breast cancer. Clin Breast Cancer. (2021) 21:e63–73. doi: 10.1016/j.clbc.2020.06.011
3. Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C. Patterns of immune infiltration in breast cancer and their clinical implications: A gene-expression-based retrospective study. PloS Med. (2016) 13:e1002194. doi: 10.1371/journal.pmed.1002194
4. Poschke I, De Boniface J, Mao Y, Kiessling R. Tumor-induced changes in the phenotype of blood-derived and tumor-associated T cells of early stage breast cancer patients. Int J Cancer. (2012) 131:1611–20. doi: 10.1002/ijc.v131.7
5. Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. (2012) 12:749–61. doi: 10.1038/nri3307
6. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. (2015) 15:486–99. doi: 10.1038/nri3862
7. Tietscher S, Wagner J, Anzeneder T, Langwieder C, Rees M, Sobottka B, et al. A comprehensive single-cell map of T cell exhaustion-associated immune environments in human breast cancer. Nat Commun. (2023) 14:98. doi: 10.1038/s41467-022-35238-w
8. Bense RD, Sotiriou C, Piccart-Gebhart MJ, Haanen JB, van Vugt MA, de Vries EG, et al. Relevance of tumor-infiltrating immune cell composition and functionality for disease outcome in breast cancer. J Natl Cancer Inst. (2017) 109:djw192. doi: 10.1093/jnci/djw192
9. Terranova-Barberio M, Pawlowska N, Dhawan M, Moasser M, Chien AJ, Melisko ME, et al. Exhausted T cell signature predicts immunotherapy response in ER-positive breast cancer. Nat Commun. (2020) 11:3584. doi: 10.1038/s41467-020-17414-y
10. Gallimore A, Glithero A, Godkin A, Tissot AC, Plückthun A, Elliott T, et al. Induction and exhaustion of lymphocytic choriomeningitis virus–specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I–peptide complexes. J Exp Med. (1998) 187:1383–93. doi: 10.1084/jem.187.9.1383
12. Blank CU, Haining WN, Held W, Hogan PG, Kallies A, Lugli E, et al. Defining ‘T cell exhaustion’. Nat Rev Immunol. (2019) 19:665–74. doi: 10.1038/s41577-019-0221-9
13. Egelston CA, Guo W, Tan J, Avalos C, Simons DL, Lim MH, et al. Tumor-infiltrating exhausted CD8+ T cells dictate reduced survival in premenopausal estrogen receptor-positive breast cancer. JCI Insight. (2022) 7:e153963. doi: 10.1172/jci.insight.153963
14. Ding S, Qiao N, Zhu Q, Tong Y, Wang S, Chen X, et al. Single-cell atlas reveals a distinct immune profile fostered by T cell-B cell crosstalk in triple negative breast cancer. Cancer Commun (Lond). (2023) 43:661–84. doi: 10.1002/cac2.12429
15. Bucks CM, Norton JA, Boesteanu AC, Mueller YM, Katsikis PD. Chronic antigen stimulation alone is sufficient to drive CD8+ T cell exhaustion. J Immunol. (2009) 182:6697–708. doi: 10.4049/jimmunol.0800997
16. Gavil NV, Scott MC, Weyu E, Smith OC, O’Flanagan SD, Wijeyesinghe S, et al. Chronic antigen in solid tumors drives a distinct program of T cell residence. Sci Immunol. (2023) 8:eadd5976. doi: 10.1126/sciimmunol.add5976
17. Shi H, Chen S, Chi H. Immunometabolism of CD8+ T cell differentiation in cancer. Trends Cancer. (2024) 10:610–26. doi: 10.1016/j.trecan.2024.03.010
18. Zhao J, Wang Z, Tian Y, Ning J, Ye H. T cell exhaustion and senescence for ovarian cancer immunotherapy. Semin Cancer Biol. (2024) 104-105:1–15. doi: 10.1016/j.semcancer.2024.07.001
19. Srirat T, Hayakawa T, Mise-Omata S, Nakagawara K, Ando M, Shichino S, et al. NR4a1/2 deletion promotes accumulation of TCF1+ stem-like precursors of exhausted CD8+ T cells in the tumor microenvironment. Cell Rep. (2024) 43:113898. doi: 10.1016/j.celrep.2024.113898
20. Zhang H, Hu Y, Shao M, Teng X, Jiang P, Wang X, et al. Dasatinib enhances anti-leukemia efficacy of chimeric antigen receptor T cells by inhibiting cell differentiation and exhaustion. J Hematol Oncol. (2021) 14:113. doi: 10.1186/s13045-021-01117-y
21. Maurice NJ, Berner J, Taber AK, Zehn D, Prlic M. Inflammatory signals are sufficient to elicit TOX expression in mouse and human CD8+ T cells. JCI Insight. (2021) 6:e150744. doi: 10.1172/jci.insight.150744
22. Hu Y, Kim JH, He K, Wan Q, Kim J, Flach M, et al. Scramblase TMEM16F terminates T cell receptor signaling to restrict T cell exhaustion. J Exp Med. (2016) 213:2759–72. doi: 10.1084/jem.20160612
23. Wang AZ, Mashimo BL, Schaettler MO, Sherpa ND, Leavitt LA, Livingstone AJ, et al. Glioblastoma-infiltrating CD8+ T cells are predominantly a clonally expanded GZMK+ effector population. Cancer Discovery. (2024) 14:1106–31. doi: 10.1158/2159-8290.CD-23-0913
24. Lan X, Mi T, Alli S, Guy C, Djekidel MN, Liu X, et al. Antitumor progenitor exhausted CD8+ T cells are sustained by TCR engagement. Nat Immunol. (2024) 25:1046–58. doi: 10.1038/s41590-024-01843-8
25. Martinez GJ, Pereira RM, Äijö T, Kim EY, Marangoni F, Pipkin ME, et al. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity. (2015) 42:265–78. doi: 10.1016/j.immuni.2015.01.006
26. Liu X, Wang Y, Lu H, Li J, Yan X, Xiao M, et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature. (2019) 567:525–9. doi: 10.1038/s41586-019-0979-8
27. Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. (2019) 571:211–8. doi: 10.1038/s41586-019-1325-x
28. LaFleur MW, Nguyen TH, Coxe MA, Miller BC, Yates KB, Gillis JE, et al. PTPN2 regulates the generation of exhausted CD8+ T cell subpopulations and restrains tumor immunity. Nat Immunol. (2019) 20:1335–47. doi: 10.1038/s41590-019-0480-4
29. Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. (2019) 51:840–855.e845. doi: 10.1016/j.immuni.2019.09.013
30. Lino CN, Barros-Martins J, Oberdörfer L, Walzer T, Prinz I. Eomes expression reports the progressive differentiation of IFN-γ-producing Th1-like γδ T cells. Eur J Immunol. (2017) 47:970–81. doi: 10.1002/eji.201646753
31. Swoboda A, Nanda R. Immune checkpoint blockade for breast cancer. Cancer Treat Res. (2018) 173:155–65. doi: 10.1007/978-3-319-70197-4
32. Sheppard K-A, Fitz LJ, Lee JM, Benander C, George JA, Wooters J, et al. PD-1 inhibits T-cell receptor induced phosphorylation of the ZAP70/CD3ζ signalosome and downstream signaling to PKCθ. FEBS Lett. (2004) 574:37–41. doi: 10.1016/j.febslet.2004.07.083
33. Zhang M, Gao D, Shi Y, Wang Y, Joshi R, Yu Q, et al. miR-149-3p reverses CD8(+) T-cell exhaustion by reducing inhibitory receptors and promoting cytokine secretion in breast cancer cells. Open Biol. (2019) 9:190061. doi: 10.1098/rsob.190061
34. Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, et al. miR-424 (322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. (2016) 7:11406. doi: 10.1038/ncomms11406
36. Zhang M, Shi Y, Zhang Y, Wang Y, Alotaibi F, Qiu L, et al. miRNA-5119 regulates immune checkpoints in dendritic cells to enhance breast cancer immunotherapy. Cancer Immunol Immunother. (2020) 69:951–67. doi: 10.1007/s00262-020-02507-w
37. Rasoolnezhad M, Safaralizadeh R, Hosseinpourfeizi MA, Banan-Khojasteh SM, Baradaran B. MiRNA-138-5p: A strong tumor suppressor targeting PD-L-1 inhibits proliferation and motility of breast cancer cells and induces apoptosis. Eur J Pharmacol. (2021) 896:173933. doi: 10.1016/j.ejphar.2021.173933
38. Zhang H, Mi J, Xin Q, Cao W, Song C, Zhang N, et al. Recent research and clinical progress of CTLA-4-based immunotherapy for breast cancer. Front Oncol. (2023) 13:1256360. doi: 10.3389/fonc.2023.1256360
39. Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. (2015) 161:205–14. doi: 10.1016/j.cell.2015.03.030
40. Contardi E, Palmisano GL, Tazzari PL, Martelli AM, Fala F, Fabbi M, et al. CTLA-4 is constitutively expressed on tumor cells and can trigger apoptosis upon ligand interaction. Int J Cancer. (2005) 117:538–50. doi: 10.1002/(ISSN)1097-0215
41. Erfani N, Razmkhah M, Ghaderi A. Circulating soluble CTLA4 (sCTLA4) is elevated in patients with breast cancer. Cancer Invest. (2010) 28:828–32. doi: 10.3109/07357901003630934
42. Chen X, Shao Q, Hao S, Zhao Z, Wang Y, Guo X, et al. CTLA-4 positive breast cancer cells suppress dendritic cells maturation and function. Oncotarget. (2017) 8:13703. doi: 10.18632/oncotarget.14626
43. Andrews LP, Marciscano AE, Drake CG, Vignali DA. LAG 3 (CD 223) as a cancer immunotherapy target. Immunol Rev. (2017) 276:80–96. doi: 10.1111/imr.2017.276.issue-1
44. Solinas C, Migliori E, De Silva P, Willard-Gallo KJC. LAG3: the biological processes that motivate targeting this immune checkpoint molecule in human cancer. Cancers. (2019) 11:1213. doi: 10.3390/cancers11081213
45. Ruffo E, Wu RC, Bruno TC, Workman CJ, Vignali DA. Lymphocyte-activation gene 3 (LAG3): The next immune checkpoint receptor. Semin Immunol. (2019) 42:101305. doi: 10.1016/j.smim.2019.101305
46. Camisaschi C, Casati C, Rini F, Perego M, De Filippo A, Triebel F, et al. LAG-3 expression defines a subset of CD4+ CD25highFoxp3+ regulatory T cells that are expanded at tumor sites. J Immunol. (2010) 184:6545–51. doi: 10.4049/jimmunol.0903879
47. Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discovery. (2018) 8:1069–86. doi: 10.1158/2159-8290.CD-18-0367
48. Wu S, Shi X, Wang J, Wang X, Liu Y, Luo Y, et al. Triple-negative breast cancer: intact mismatch repair and partial co-expression of PD-L1 and LAG-3. Front Immunol. (2021) 12:561793. doi: 10.3389/fimmu.2021.561793
49. Stovgaard ES, Kümler I, List-Jensen K, Roslind A, Christensen IJ, Høgdall E, et al. Prognostic and clinicopathologic associations of LAG-3 expression in triple-negative breast cancer. Appl Immunohistochem Mol Morphol. (2022) 30:62–71. doi: 10.1097/PAI.0000000000000954
50. Du H, Yi Z, Wang L, Li Z, Niu B, Ren G. The co-expression characteristics of LAG3 and PD-1 on the T cells of patients with breast cancer reveal a new therapeutic strategy. Int Immunopharmacol. (2020) 78:106113. doi: 10.1016/j.intimp.2019.106113
51. Liu Q, Qi Y, Zhai J, Kong X, Wang X, Wang Z, et al. Molecular and clinical characterization of LAG3 in breast cancer through 2994 samples. Front Immunol. (2021) 12:599207. doi: 10.3389/fimmu.2021.599207
52. Harjunpää H, Guillerey CJC, Immunology E. TIGIT as an emerging immune checkpoint. Clin Exp Immunol. (2020) 200:108–19. doi: 10.1111/cei.13407
53. Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8+ T cell effector function. Cancer Cell. (2014) 26:923–37. doi: 10.1016/j.ccell.2014.10.018
54. Zhang Q, Gao C, Shao J, Wang Z. TIGIT-related transcriptome profile and its association with tumor immune microenvironment in breast cancer. Biosci Rep. (2021) 41:BSR20204340. doi: 10.1042/BSR20204340
55. Abbasov A, Aktas Cetin E, Cabioglu N, Mollavelioglu B, Onder S, Emiroglu S, et al. Differential expression of Novel Immune Checkpoint receptors on Tumor infiltrating lymphocytes in patients with locally advanced breast Cancer after Neoadjuvant Chemotherapy. Neoplasma. (2021) 68:1079–90. doi: 10.4149/neo_2021_210127N141
56. Liu Q, Cheng R, Kong X, Wang Z, Fang Y, Wang J. Molecular and clinical characterization of PD-1 in breast cancer using large-scale transcriptome data. Front Immunol. (2020) 11:558757. doi: 10.3389/fimmu.2020.558757
57. Chauvin J-M, Pagliano O, Fourcade J, Sun Z, Wang H, Sander C, et al. TIGIT and PD-1 impair tumor antigen–specific CD8+ T cells in melanoma patients. J Clin Invest. (2015) 125:2046–58. doi: 10.1172/JCI80445
58. Yang Z-Z, Kim HJ, Wu H, Jalali S, Tang X, Krull JE, et al. TIGIT expression is associated with T-cell suppression and exhaustion and predicts clinical outcome and anti–PD-1 response in follicular lymphoma. Clin Cancer Res. (2020) 26:5217–31. doi: 10.1158/1078-0432.CCR-20-0558
59. Yan W, Liu X, Ma H, Zhang H, Song X, Gao L, et al. Tim-3 fosters HCC development by enhancing TGF-β-mediated alternative activation of macrophages. Gut. (2015) 64:1593–604. doi: 10.1136/gutjnl-2014-307671
60. de Mingo Pulido Á, Gardner A, Hiebler S, Soliman H, Rugo HS, Krummel MF, et al. TIM-3 regulates CD103+ dendritic cell function and response to chemotherapy in breast cancer. Cancer Cell. (2018) 33:60–74.e66. doi: 10.1016/j.ccell.2017.11.019
61. Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T, et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature. (2002) 415:536–41. doi: 10.1038/415536a
62. Sabatos-Peyton CA, Nevin J, Brock A, Venable JD, Tan DJ, Kassam N, et al. Blockade of Tim-3 binding to phosphatidylserine and CEACAM1 is a shared feature of anti-Tim-3 antibodies that have functional efficacy. Oncoimmunology (2018) 7:. doi: 10.1080/2162402X.2017.1385690
63. Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. (2005) 6:1245–52. doi: 10.1038/ni1271
64. Das M, Zhu C, Kuchroo VK. Tim-3 and its role in regulating anti-tumor immunity. Immunol Rev. (2017) 276:97–111. doi: 10.1111/imr.2017.276.issue-1
65. Cheng S, Han F, Xu Y, Qu T, Ju Y. Pathology E: Expression of Tim-3 in breast cancer tissue promotes tumor progression. Int J Clin Exp Pathol. (2018) 11:1157.
66. Zhang H, Xiang R, Wu B, Li J, Luo GJM. T-cell immunoglobulin mucin−3 expression in invasive ductal breast carcinoma: Clinicopathological correlations and association with tumor infiltration by cytotoxic lymphocytes. Mol Clin Oncol. (2017) 7:557–63. doi: 10.3892/mco.2017.1360
67. Sasidharan Nair V, El Salhat H, Taha RZ, John A, Ali BR, Elkord E. DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin Epigenet. (2018) 10:1–12. doi: 10.1186/s13148-018-0512-1
68. Cheng S, Han F, Xu Y, Qu T, Ju Y. Erratum: Expression of Tim-3 in breast cancer tissue promotes tumor progression. Int J Clin Exp Pathol. (2021) 14:855.
69. Cong Y, Cui Y, Zhu S, Cao J, Zou H, Martin TA, et al. Tim-3 promotes cell aggressiveness and paclitaxel resistance through NF-κB/STAT3 signalling pathway in breast cancer cells. Chin J Cancer Res. (2020) 32:564–79. doi: 10.21147/j.issn.1000-9604.2020.05.02
70. Wang Z, Liu X, Wang X, Chong T, Lin S, Wang M, et al. Polymorphisms in TIM-3 and breast cancer susceptibility in Chinese women: A case-control study. Oncotarget. (2016) 7:43703–12. doi: 10.18632/oncotarget.v7i28
71. Cheng S, Ju Y, Han F, Wang Y, Xu Y, Qu T, et al. T cell immunoglobulin- and mucin-domain-containing molecule 3 gene polymorphisms and susceptibility to invasive breast cancer. Ann Clin Lab Sci. (2017) 47:668–75. doi: 10.1007/s11033-012-1862-y
72. Sehrawat S, Reddy PB, Rajasagi N, Suryawanshi A, Hirashima M, Rouse BT. Galectin-9/TIM-3 interaction regulates virus-specific primary and memory CD8+ T cell response. PloS Pathog. (2010) 6:e1000882. doi: 10.1371/journal.ppat.1000882
73. Merani S, Chen W, Elahi S. The bitter side of sweet: the role of galectin-9 in immunopathogenesis of viral infections. Rev Med Virol. (2015) 25:175–86. doi: 10.1002/rmv.1832
74. Elahi S, Dinges WL, Lejarcegui N, Laing KJ, Collier AC, Koelle DM, et al. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat Med. (2011) 17:989–95. doi: 10.1038/nm.2422
75. Ju MH, Byun KD, Park EH, Lee JH, Han SH. Association of galectin 9 expression with immune cell infiltration, programmed cell death ligand-1 expression, and patient’s clinical outcome in triple-negative breast cancer. Biomedicines. (2021) 9:1383. doi: 10.3390/biomedicines9101383
76. Zang X, Loke PN, Kim J, Murphy K, Waitz R, Allison JP. B7x: a widely expressed B7 family member that inhibits T cell activation. Proc Natl Acad Sci USA. (2003) 100:10388–92. doi: 10.1073/pnas.1434299100
77. Prasad DV, Richards S, Mai XM, Dong CJI. B7S1, a novel B7 family member that negatively regulates T cell activation. Immunity. (2003) 18:863–73. doi: 10.1016/S1074-7613(03)00147-X
78. Yu J, Yan Y, Li S, Xu Y, Parolia A, Rizvi S, et al. Progestogen-driven B7-H4 contributes to onco-fetal immune tolerance. Cell. (2024) 187:4713–4732.e4719. doi: 10.1016/j.cell.2024.06.012
79. Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. (2009) 10:29–37. doi: 10.1038/ni.1679
80. Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. (2007) 27:670–84. doi: 10.1016/j.immuni.2007.09.006
81. Fourcade J, Sun Z, Benallaoua M, Guillaume P, Luescher IF, Sander C, et al. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J Exp Med. (2010) 207:2175–86. doi: 10.1084/jem.20100637
82. Fourcade J, Sun Z, Pagliano O, Guillaume P, Luescher IF, Sander C, et al. CD8(+) T cells specific for tumor antigens can be rendered dysfunctional by the tumor microenvironment through upregulation of the inhibitory receptors BTLA and PD-1. Cancer Res. (2012) 72:887–96. doi: 10.1158/0008-5472.CAN-11-2637
83. Zarour HM. Reversing T-cell dysfunction and exhaustion in cancer. Clin Cancer Res. (2016) 22:1856–64. doi: 10.1158/1078-0432.CCR-15-1849
84. Huang T, Zhang Q, Yi J, Wang R, Zhang Z, Luo P, et al. PEG-sheddable nanodrug remodels tumor microenvironment to promote effector T cell infiltration and revise their exhaustion for breast cancer immunotherapy. Small. (2023) 19:e2301749. doi: 10.1002/smll.202301749
85. Zhu H, Gu Y, Xue Y, Yuan M, Cao X, Liu Q. CXCR2(+) MDSCs promote breast cancer progression by inducing EMT and activated T cell exhaustion. Oncotarget. (2017) 8:114554–67. doi: 10.18632/oncotarget.23020
86. Zhao X, Qu J, Sun Y, Wang J, Liu X, Wang F, et al. Prognostic significance of tumor-associated macrophages in breast cancer: a meta-analysis of the literature. Oncotarget. (2017) 8:30576. doi: 10.18632/oncotarget.15736
87. Noy R, Pollard JWJI. Tumor-associated macrophages: from mechanisms to therapy. Immunity. (2014) 41:49–61. doi: 10.1016/j.immuni.2014.06.010
88. Joyce JA, Fearon DTJS. T cell exclusion, immune privilege, and the tumor microenvironment. Science. (2015) 348:74–80. doi: 10.1126/science.aaa6204
89. Pusztai C, Yusenko MV, Banyai D, Szanto A, Kovacs G. M2 macrophage marker chitinase 3-like 2 (CHI3L2) associates with progression of conventional renal cell carcinoma. Anticancer Res. (2019) 39:6939–43. doi: 10.21873/anticanres.13915
90. Liu T, Larionova I, Litviakov N, Riabov V, Zavyalova M, Tsyganov M, et al. Tumor-associated macrophages in human breast cancer produce new monocyte attracting and pro-angiogenic factor YKL-39 indicative for increased metastasis after neoadjuvant chemotherapy. Oncoimmunolgy. (2018) 7:e1436922. doi: 10.1080/2162402X.2018.1436922
91. Litviakov N, Tsyganov M, Larionova I, Ibragimova M, Deryusheva I, Kazantseva P, et al. Expression of M2 macrophage markers YKL-39 and CCL18 in breast cancer is associated with the effect of neoadjuvant chemotherapy. Cancer Chemother Pharmacol. (2018) 82:99–109. doi: 10.1007/s00280-018-3594-8
92. Xie F, Zhou X, Li H, Su P, Liu S, Li R, et al. USP8 promotes cancer progression and extracellular vesicle-mediated CD8+ T cell exhaustion by deubiquitinating the TGF-β receptor TβRII. EMBO J. (2022) 41:e108791. doi: 10.15252/embj.2021108791
93. Xia J, Zhang L, Peng X, Tu J, Li S, He X, et al. IL1R2 blockade alleviates immunosuppression and potentiates anti-PD-1 efficacy in triple-negative breast cancer. Cancer Res. (2024) 84:2282–96. doi: 10.1158/0008-5472.CAN-23-3429
94. Liu Y, Zhou N, Zhou L, Wang J, Zhou Y, Zhang T, et al. IL-2 regulates tumor-reactive CD8(+) T cell exhaustion by activating the aryl hydrocarbon receptor. Nat Immunol. (2021) 22:358–69. doi: 10.1038/s41590-020-00850-9
95. Hanna BS, Llaó-Cid L, Iskar M, Roessner PM, Klett LC, Wong JKL, et al. Interleukin-10 receptor signaling promotes the maintenance of a PD-1(int) TCF-1(+) CD8(+) T cell population that sustains anti-tumor immunity. Immunity. (2021) 54:2825–2841.e2810. doi: 10.1016/j.immuni.2021.11.004
96. Yang WJ, Shi L, Wang XM, Yang GW. Heparanase is a novel biomarker for immune infiltration and prognosis in breast cancer. Aging (Albany NY). (2021) 13:20836–52. doi: 10.18632/aging.203489
97. Xie F, Zhou X, Su P, Li H, Tu Y, Du J, et al. Breast cancer cell-derived extracellular vesicles promote CD8(+) T cell exhaustion via TGF-β type II receptor signaling. Nat Commun. (2022) 13:4461. doi: 10.1038/s41467-022-31250-2
98. Cui R, Zhao T, Bai C, Ji N, Hua J, Ren L, et al. High expression of RAI14 in triple-negative breast cancer participates in immune recruitment and implies poor prognosis through bioinformatics analyses. Front Pharmacol. (2022) 13:809454. doi: 10.3389/fphar.2022.809454
99. Tu WJ, McCuaig RD, Tan AHY, Hardy K, Seddiki N, Ali S, et al. Targeting nuclear LSD1 to reprogram cancer cells and reinvigorate exhausted T cells via a novel LSD1-EOMES switch. Front Immunol. (2020) 11:1228. doi: 10.3389/fimmu.2020.01228
100. Yu X, Xu J. TWIST1 drives cytotoxic CD8+ T-cell exhaustion through transcriptional activation of CD274 (PD-L1) expression in breast cancer cells. Cancers (Basel). (2024) 16:1973. doi: 10.3390/cancers16111973
101. Yang J, Hou Y, Zhou M, Wen S, Zhou J, Xu L, et al. Twist induces epithelial-mesenchymal transition and cell motility in breast cancer via ITGB1-FAK/ILK signaling axis and its associated downstream network. Int J Biochem Cell Biol. (2016) 71:62–71. doi: 10.1016/j.biocel.2015.12.004
102. Nixon BG, Kuo F, Ji L, Liu M, Capistrano K, Do M, et al. Tumor-associated macrophages expressing the transcription factor IRF8 promote T cell exhaustion in cancer. Immunity. (2022) 55:2044–2058.e2045. doi: 10.1016/j.immuni.2022.10.002
103. Arora M, Kumari S, Singh J, Chopra A, Chauhan SS. Expression pattern, regulation, and clinical significance of TOX in breast cancer. Cancer Immunol Immunother. (2021) 70:349–63. doi: 10.1007/s00262-020-02689-3
104. Andrieu GP, Shafran JS, Smith CL, Belkina AC, Casey AN, Jafari N, et al. BET protein targeting suppresses the PD-1/PD-L1 pathway in triple-negative breast cancer and elicits anti-tumor immune response. Cancer Lett. (2019) 465:45–58. doi: 10.1016/j.canlet.2019.08.013
105. Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. (2022) 21:60. doi: 10.1186/s12943-021-01447-y
106. Wu H, Wu Z, Li H, Wang Z, Chen Y, Bao J, et al. Glycosylphosphatidylinositol anchor biosynthesis pathway-based biomarker identification with machine learning for prognosis and T cell exhaustion status prediction in breast cancer. Front Immunol. (2024) 15:1392940. doi: 10.3389/fimmu.2024.1392940
107. Ma Y, Du J, Chen M, Gao N, Wang S, Mi Z, et al. Mitochondrial DNA methylation is a predictor of immunotherapy response and prognosis in breast cancer: scRNA-seq and bulk-seq data insights. Front Immunol. (2023) 14:1219652. doi: 10.3389/fimmu.2023.1219652
108. Repas J, Peternel L, Sourij H, Pavlin M. Low glucose availability potentiates the effects of metformin on model T cell activation and exhaustion markers. vitro. Front Endocrinol (Lausanne). (2023) 14:1216193. doi: 10.3389/fendo.2023.1216193
109. Kado T, Nawaz A, Takikawa A, Usui I, Tobe K. Linkage of CD8(+) T cell exhaustion with high-fat diet-induced tumourigenesis. Sci Rep. (2019) 9:12284. doi: 10.1038/s41598-019-48678-0
110. Ma S, Zhao Y, Lee WC, Ong LT, Lee PL, Jiang Z, et al. Hypoxia induces HIF1α-dependent epigenetic vulnerability in triple negative breast cancer to confer immune effector dysfunction and resistance to anti-PD-1 immunotherapy. Nat Commun. (2022) 13:4118. doi: 10.1038/s41467-022-31764-9
111. Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SFJC. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. (2015) 161:790–802. doi: 10.1016/j.cell.2015.02.053
112. Li C, Qiu S, Jin K, Zheng X, Zhou X, Jin D, et al. Tumor-derived microparticles promote the progression of triple-negative breast cancer via PD-L1-associated immune suppression. Cancer Lett. (2021) 523:43–56. doi: 10.1016/j.canlet.2021.09.039
113. Niu M, Liu Y, Yi M, Jiao D, Wu K. Biological characteristics and clinical significance of soluble PD-1/PD-L1 and exosomal PD-L1 in cancer. Front Immunol. (2022) 13:827921. doi: 10.3389/fimmu.2022.827921
114. Xie F, Xu M, Lu J, Mao L, Wang S. The role of exosomal PD-L1 in tumor progression and immunotherapy. Mol Cancer. (2019) 18:1–10. doi: 10.1186/s12943-019-1074-3
115. Yang Y, Li C-W, Chan L-C, Wei Y, Hsu J-M, Xia W, et al. Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res. (2018) 28:862–4. doi: 10.1038/s41422-018-0060-4
116. Chen M, Fu Z, Wu C. Tumor-derived exosomal ICAM1 promotes bone metastasis of triple-negative breast cancer by inducing CD8+ T cell exhaustion. Int J Biochem Cell Biol. (2024) 175:106637. doi: 10.1016/j.biocel.2024.106637
117. Zhang W, Zhong W, Wang B, Yang J, Yang J, Yu Z, et al. ICAM-1-mediated adhesion is a prerequisite for exosome-induced T cell suppression. Dev Cell. (2022) 57:329–343.e327. doi: 10.1016/j.devcel.2022.01.002
118. Choudhury A, Chatterjee S, Dalui S, Ghosh P, Daptary AH, Mollah GK, et al. Breast cancer cell derived exosomes reduces glycolysis of activated CD8 + T cells in a AKT-mTOR dependent manner. Cell Biol Int. (2024). doi: 10.1002/cbin.12241
Keywords: CD8, T cell exhaustion, breast cancer, tumor microenvironment, immunotherapy
Citation: Xie H, Xi X, Lei T, Liu H and Xia Z (2024) CD8+ T cell exhaustion in the tumor microenvironment of breast cancer. Front. Immunol. 15:1507283. doi: 10.3389/fimmu.2024.1507283
Received: 07 October 2024; Accepted: 20 November 2024;
Published: 09 December 2024.
Edited by:
Raquel Alarcon Rodriguez, University of Almeria, SpainReviewed by:
Yudong Cao, Central South University, ChinaCopyright © 2024 Xie, Xi, Lei, Liu and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanghang Xie, eGllaGFuZ2hhbmcwOTE1QDEyNi5jb20=
 Hanghang Xie
Hanghang Xie Xiaowei Xi
Xiaowei Xi Ting Lei1
Ting Lei1 Zhijia Xia
Zhijia Xia