- Department of Dermatology, Jacobs School of Medicine and Biomedical Sciences, University at Buffalo, Buffalo, NY, United States
Autoimmune diseases (AID) are defined by immune dysregulation characterized by specific humoral and/or cell mediated responses directed against the body’s own tissues. Cytokines in particular play a pivotal role in the pathogenesis of AID, with proinflammatory cytokines contributing to the initiation and propagation of autoimmune inflammation, whereas anti-inflammatory cytokines facilitate regression of inflammation and recovery from acute phases of the disease. Parallel work by our group evaluating a comprehensive set of pro- and anti-inflammatory serum cytokines in Pemphigus vulgaris (PV) as well as Alopecia areata (AA) uncovered a similar pattern of inheritance specific immune dysregulation in these two distinct autoimmune skin diseases. In AA, we found healthy control subjects who are blood related to AA patients exhibit the same cytokine dysregulation in Th1 and Th17 pathways as do patients with AA. In PV, patients as well as individuals who are healthy but yet carry certain PV-associated HLA alleles (termed here as HLA-matched controls) share a similar, but not fully overlapping pattern of cytokine expression that is distinct from control subjects who do not type for these HLA alleles. Specifically, PV patients as well as HLA-matched controls demonstrate immunological activation of several pro-inflammatory-, Th17-, Th2-pathway associated cytokines, and the chemokine IL-8. Thus, in both AA and PV, we reveal cytokine dysregulations that are linked to genetic background. The presence of disease promoting pathways in not only patients, but also genetically related, but healthy control individuals further evokes the novel hypothesis that there may be co-existing disease counteracting immune protective mechanisms at play in thwarting the threat of disease in genetically predisposed individuals who, despite harboring disease associated immune imbalances, remain healthy. Our data underscore the known tendency of AID to cluster in families and support the notion of the shared genetic/common cause hypothesis across multiple AID.
1 Introduction
Pemphigus vulgaris (PV) is a life-threatening chronic autoimmune blistering condition affecting both the skin and mucous membranes, caused by autoantibodies targeting desmosomal proteins, which results in acantholysis in the suprabasilar epidermis, ultimately leading to blistering of the skin and/or mucosa. In addition to this autoantibody(autoAb)-mediated epidermal damage, it is well established there is co-existing systemic and local inflammation driven by cytokine expression (1). Cytokine secretion, in particular, results from T cell activation resulting in increased tissue inflammation. T cells also promote the survival and differentiation of B cells, thus allowing for continued autoantibody production (2).
In PV, there is a strong genetic association with two specific HLA haplotypes - HLA-DR4 (DRB1*0402) and DR6 (DQB1*0503); approximately 80% of North American patients express one, or both of these alleles (3). It is important to note that the presence of these PV-associated susceptibility alleles is not exclusive to patients, as the vast majority of individuals expressing these alleles remain disease free. Thus, HLA genes are not in themselves sufficient to cause disease; additional genetic and environmental factors (most of which are yet to be identified) are required (4).
Alopecia areata (AA) is also autoimmune in nature, characterized by nonscarring hair loss. Clinical manifestation of disease ranges from small-well circumscribed patches of hair loss limited to the scalp to diffuse hair loss over the entire body (5, 6). The exact etiopathogenesis of AA remains unclear, but again, both genetic and environmental factors have been implicated (6–9).
AA has a prevalence of 1.7% in the general population (10). However, familial aggregation data in AA patients has demonstrated an estimated lifetime risk of AA in siblings to be 7.1%, 7.8% in parents of AA patients, and 5.7% in offspring of AA patients (11). Twin studies have found a concordance rate of 42-55% in monozygotic twins and 0-10% in dizygotic twins (12, 13). Certain HLA alleles have also been linked to AA pathogenesis, however, the association is not nearly as strong as in PV (6, 14).
The high concordance rates in identical twins in AA (12, 13) and the strong HLA-associations seen in PV (15), along with the known tendency of AID to cluster in families (16), provide strong support for a clear genetic undergirding operative in each of these conditions. We have previously reported on the concept of familial inheritance in AA (17–19) and in PV (20–22), and here directly compare the impact of genetic background on cytokine patterns relevant to these two diseases. For this comparison, we particularly draw on data from two previous studies by our group on AA (18) and PV (23) to gain insight into patterns of cytokine dysregulation that are shared in patients as well as disease free but at-risk individuals (genetically susceptible healthy subjects) and distinct from healthy individuals not carrying any known genetic risk elements.
A multitude of studies have looked specifically at cytokine dysregulation in PV and in AA. PV had generally been considered a T helper (Th)2 disease, while AA was seen as predominantly Th1 mediated (24, 25). Newer studies have also implicated activation of the Th17 cytokine profile in both diseases (1, 23, 24, 26–28). However, none of the previous studies stratified their control populations based on their status of blood relation (in the case of AA) or HLA haplotype (in the case of PV). Applying this stratification to the control groups we studied allowed us to isolate the impact of genetic background on key immune mechanisms relevant to disease pathogenesis.
2 Methods
This work provides a new interpretation of data based on two separate but parallel studies examining cytokine dysregulation in AA and PV, Van Acker et al. (18) and Schwartz et al. (23). In short, for AA, we analyzed a number of Th1 and Th17 related cytokines, including interleukin (IL)-1β, IL-6, IL-10, IL-17A, IL-21, IL-22, IL-23, interferon (IFN)γ and tumor necrosis factor (TNF)α by ELISA in 64 patients with a diagnosis of AA, 16 unaffected relatives of AA patients, and 16 unaffected non-relative control subjects. At the time of the initial study, our subgroups were classified according to the National Alopecia Areata Registry (29) as Alopecia Areata persistent (n=17), Alopecia Areata transitory (n=15), Alopecia Universalis (n=16), and Alopecia Totalis (n=16). None of the AA patients were receiving systemic immunomodulating treatments at the time of their blood draw. Statistical analysis was conducted using Kruskall-Wallis non-parametric comparison method for analyzing population means. For PV, we analyzed 20 cytokines across multiple T helper cell and pro-inflammatory pathways including IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-9, IL-10, IL-12, IL-13, IL-15, IL-17, IL-21, IL-22, IL-23, TNFα, IFNγ, MCP-1, and Eotaxin by multiplexed bead array assays in 116 PV patients, 15 healthy control subjects that carry PV-associated HLA-alleles and 14 healthy controls that did not carry PV-associated HLA-alleles. Our cohort of PV patients was further broken down by disease activity (active disease (n=75) and remission (n=55)), disease phenotype (mucosal (n=48), mucocutaneous (n=48), cutaneous (n=3) and undetermined (n=4)), and therapy status (off therapy (n=40), minimal therapy (n=34) and more than minimal therapy (n=56)). Statistical analysis was conducted using heteroscedastic T-tests. For detailed methodologies, as well as additional demographic details, please see Van Acker et al. (18) and Schwartz et al. (23).
3 Results
3.1 Healthy relatives of AA patients and healthy controls carrying PV-associated HLA alleles exhibit cytokine dysregulation similar to their respective patient population
In AA, including patients as well as healthy relatives and healthy non-relative control subjects, we found the Th1-associated cytokines IFNγ and TNFα and the Th17-associated cytokines IL-17A and IL-23 to be significantly upregulated in all AA patients in comparison to unaffected non-relatives (unrelated controls). Figure 1A shows a representative plot of these findings for TNFα. A heatmap representation of our z-score transformed data with supervised clustering shows that first degree relatives exhibit a marked increase in serum cytokine concentration similar to AA patients but distinct from controls not related to AA patients across multiple cytokines examined (Figure 1B). These data indicate that there is an inheritance-specific dysregulation within both the Th1- and Th17 cytokine pathways. No statistically significant differences were found when comparing subtypes of disease to one another. No significant differences were found for any of the other cytokines we measured.

Figure 1. (A) Representative box and whisker plot of TNFα levels among Alopecia areata patients, genetically susceptible healthy controls (first degree relatives of Alopecia areata patients) and not genetically susceptible healthy controls (individuals with no family history of any autoimmune disease). (B) Heatmap of the distribution of four cytokines that were found to be significantly elevated in Alopecia areata patients and genetically susceptible healthy controls (first degree relatives of Alopecia areata patients) when compared to not genetically susceptible healthy controls (individuals with no family history of any autoimmune disease). Heatmap of z-score transformed data was created with Morpheus software. https://software.broadinstitute.org/morpheus.
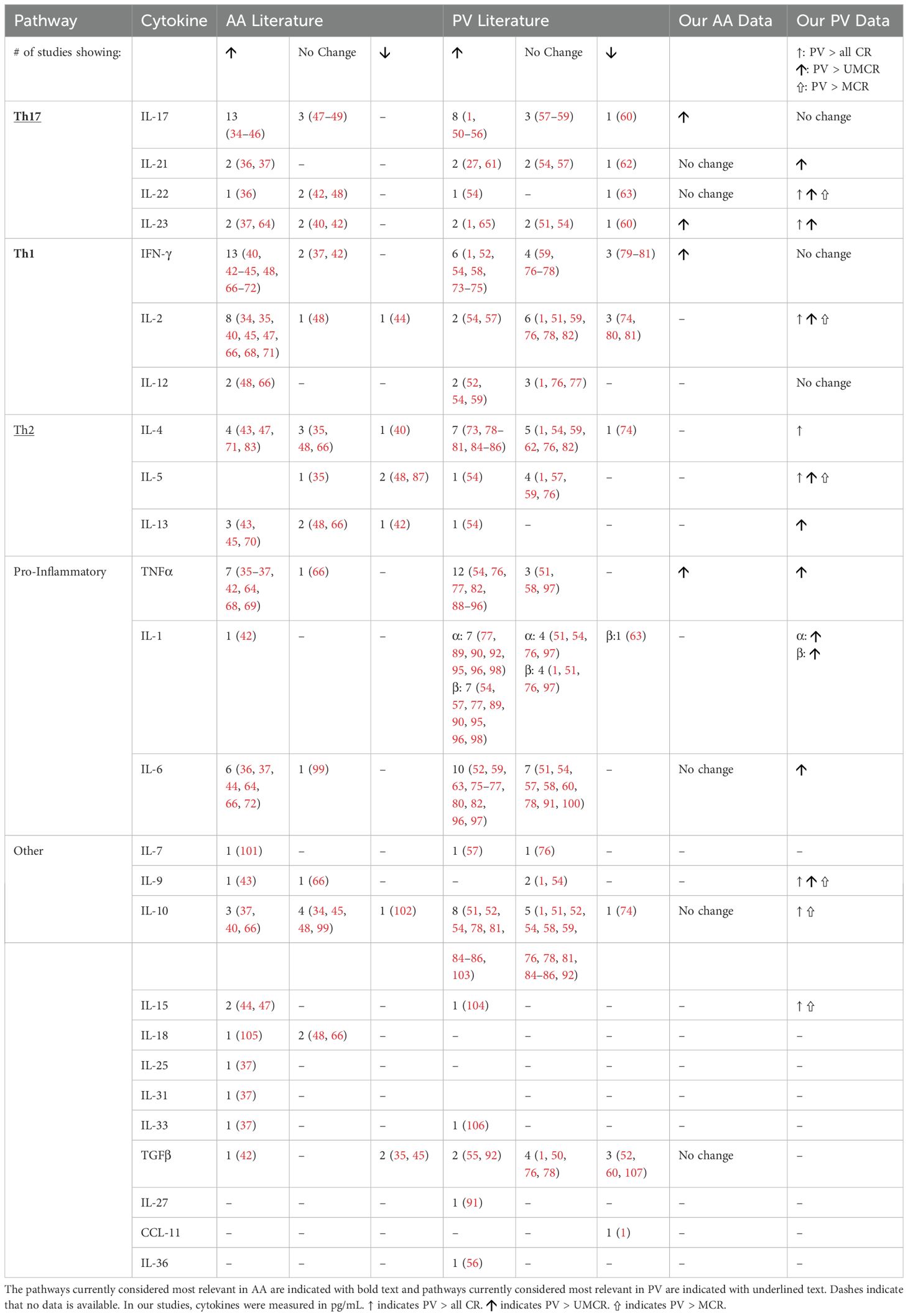
For PV, we uncovered a pattern mirroring what we observed in AA-genetically susceptible individuals, in this case linked to influence of HLA-association on cytokine expression. For a distinct set of cytokines (Th17: IL-21, IL-23; Th1: TNFα; Pro-Inflammatory: IL-1α, IL-1β, IL-6; Th2: IL-13; and chemokine: IL-8), we found that HLA-matched controls exhibited elevations in cytokines levels in a number of cases similar to patients. Figure 2A shows a representative plot of these findings for TNFα. Figure 2B displays these findings across multiple cytokines in heatmap format. While the mean levels of cytokines in HLA-matched controls were not quite as elevated as patients, we found no statistically relevant difference when comparing these two groups. However, healthy controls unmatched for HLA-susceptibility alleles had significantly lower cytokine levels when compared to patients. Thus, similar to our findings in AA, our data suggest an inheritance-specific dysregulation within numerous cytokine pathways (pro-inflammatory, Th2 and Th17) in healthy controls carrying genetic susceptibility for PV.
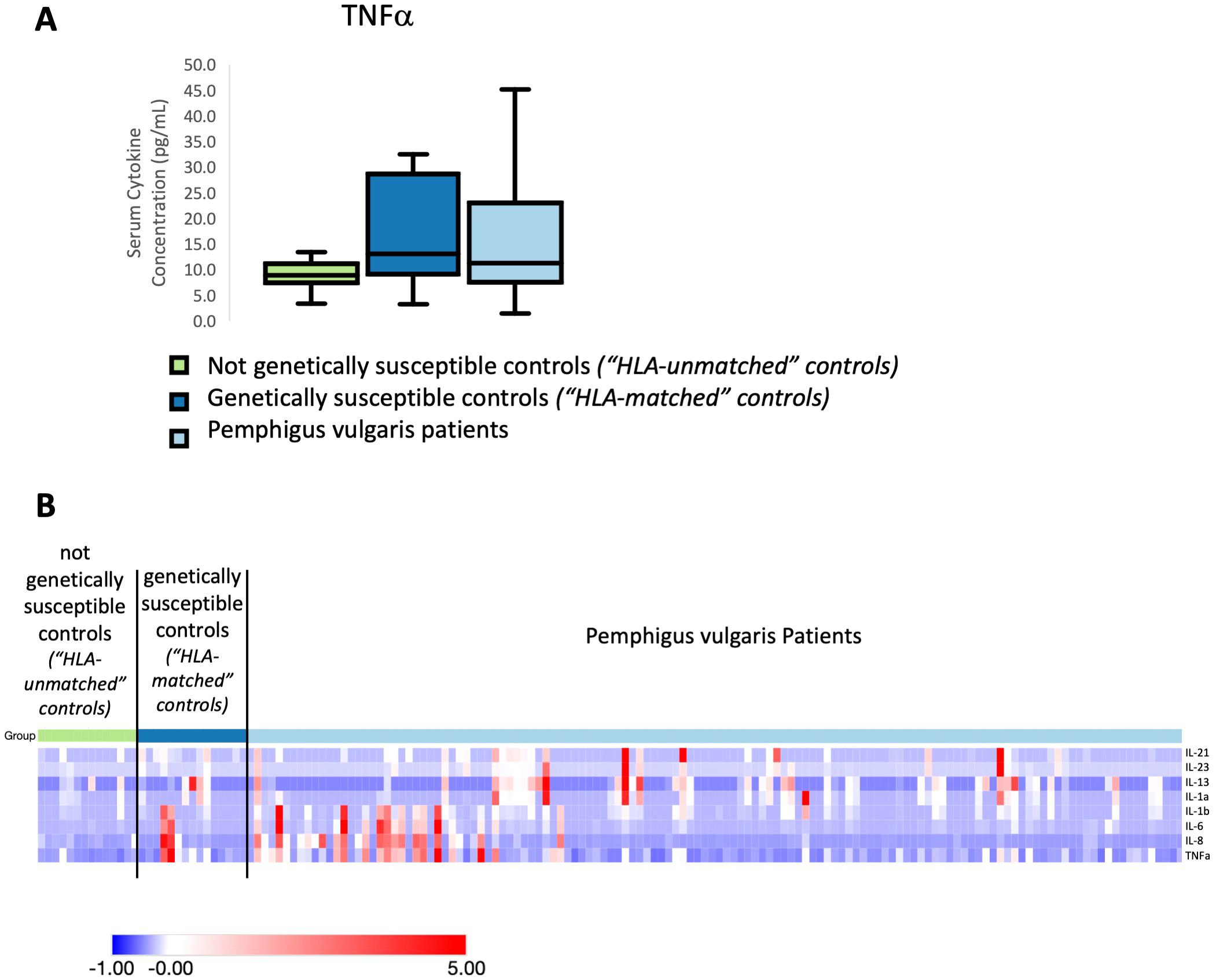
Figure 2. (A) Representative box and whisker plot of TNFα levels among PV patients, genetically susceptible healthy controls (“HLA-matched”) and not genetically susceptible (“HLA-unmatched”). (B) Heatmap of the distribution of cytokines that were found to be significantly elevated in Pemphigus vulgaris patients and genetically susceptible healthy controls (express Pemphigus vulgaris-associated HLA alleles) when compared to not genetically susceptible healthy controls (do not express Pemphigus vulgaris-associated HLA alleles). Heatmap of z-score transformed data was created with Morpheus software. https://software.broadinstitute.org/morpheus.
3.2 Inheritance linked cytokine patterns in PV and AA invoke novel concepts regarding autoimmune risk
We see similar patterns of genetically-based cytokine dysregulation in both AA and PV with at-risk individuals exhibiting cytokine concentrations in line with that seen in patients. Moreover, and notably, patients and at-risk individuals in both PV and AA share a dysregulation of IL-23 and TNFα across both diseases, supporting the notion of shared or common autoimmune pathomechanisms.
However, in addition to these shared cytokine changes in PV and AA, we also find dysregulation in other pathways and cytokines in PV - in particular, certain pro-inflammatory cytokines (IL-1α), chemokines (IL-8), and the Th2 pathway (IL-13) - that had not been analyzed in our AA studies. Of note, we found IL-2, IL-5, IL-22, and IL-9 to be significantly higher in PV patients compared to both HLA-matched controls and HLA-unmatched controls (Figure 3), reflecting changes unlinked to HLA type and suggesting cytokine activation is not strictly HLA driven, implicating the existence of additional genetic (and/or environmental) factors operative in patients.
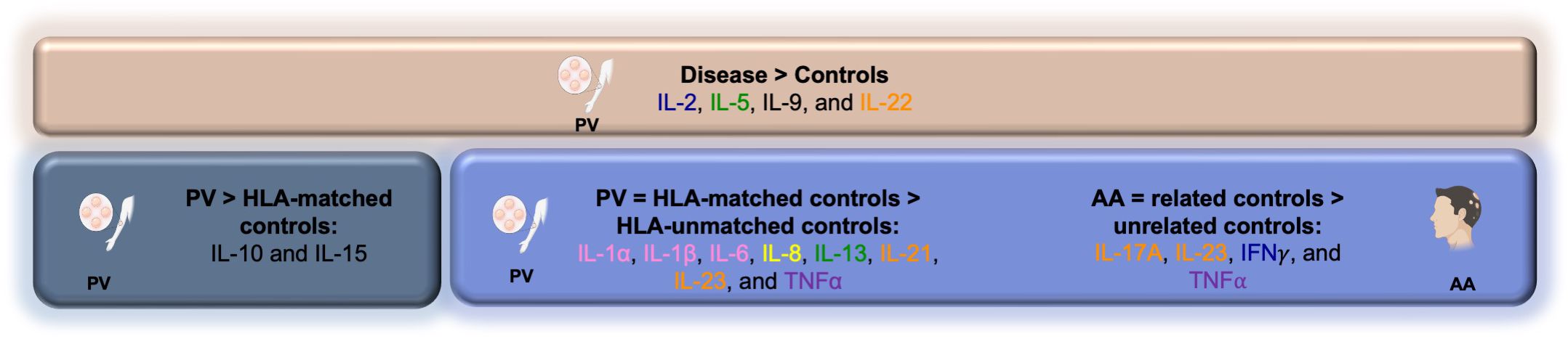
Figure 3. Summary schematic outlining cytokine dysregulation among PV and AA patients and control groups.
Finally, we further postulated that due to the presence of AA/PV-associated cytokine profiles in genetically at-risk but healthy individuals, there may be mechanisms that actively counteract this (partial) autoimmune activation that operate to keep them disease free. In support of this hypothesis, we found certain cytokines such as IL-10 and IL-15 to be downregulated in HLA-matched healthy controls when compared to PV patients (Figure 3), yet they are not downregulated in non-HLA matched healthy controls compared to PV patients. These data suggest that a downregulation of certain cytokines may serve as one component of a necessary neutralizing force aimed at mitigating the existing partial (auto)immune activation seen in genetically susceptible, but healthy controls.
4 Discussion
The pathogenesis of both PV and AA is certainly multifactorial, influenced by a combination of patient genetics as well as environmental factors that ultimately lead to immune dysregulation. Highlighting cytokines as key effectors of the immune cascade, a multitude of studies in both conditions have attempted to identify the extent of dysregulation of numerous cytokines with potential relevance to disease pathogenesis (summarized in Table 1). A majority of studies indicate that AA is Th1- and Th17-driven disease (28), whereas PV appears to be driven by dysregulations in the Th2 and Th17 pathways (25). In accordance with previous studies, we also found find strong evidence for the activation of these pathways in PV and AA patients (Table 1).
Remarkably, in AA we report an inheritance-specific dysregulation of Th1 and Th17 pathway cytokines with an elevation in all AA clinical subtypes as well as first degree relatives in comparison to unrelated control subjects. Equally striking, we find healthy controls matched for the known PV-susceptibility alleles DRB1*0402 and/or DQB1*0503 (“HLA-matched”) exhibit an elevation of pro-inflammatory cytokines that is similar to that seen in PV patients when compared to HLA-unmatched controls (healthy controls who do not express the PV-associated susceptibility alleles) for Th2, Th17 and generally proinflammatory pathways. These data invoke a novel paradigm of disease susceptibility whereby unknown genetic elements in the case of AA or HLA molecules linked to PV susceptibility in the case of PV predispose individuals to a limited activation of inflammatory cytokines regardless of disease manifestation.
Thus, perplexingly, it appears that some genetically at-risk individuals, on the basis of their HLA status (PV) and/or other shared genetic background (AA), harbor disease-promoting inflammatory pathways, yet for reasons that are unclear, remain disease free. In support of this assumption, our group has previously found that in PV, HLA-matched controls share an upregulation of certain autoantibodies with patients when compared to HLA-unmatched controls (20). Furthermore, we found total antioxidant capacity to be similarly diminished in HLA-matched control and PV patients, but not in HLA unmatched controls (30). Thus, our work emphasizes that individuals who may have genetic susceptibility to these autoimmune skin diseases may be skewed towards activation of certain inflammatory pathways. On the other hand, it may be the case that HLA and/or other genetically driven differences in themselves are insufficient to produce the full scope of autoimmune dysregulation required to cross the threshold necessary to induce disease-specific phenotype. This idea is supported by the presence of cytokine dysregulations unique to PV patients alone, separate from HLA-matched control subjects (i.e. for IL-2, IL-5, IL-22, and IL-9). The latter findings indicate that cytokine dysregulation is not entirely HLA driven. Ultimately, disease manifestation is most likely to rely on the presence of additional genetic and/or environmental factors.
Genome-wide association studies (GWAS) may prove useful in identification of these additional genetic factors which contribute to disease. In PV, GWAS are limited, notably due to the low prevalence of the disease, however one study by Sarig et al. identified an association between the pro-apoptotic molecule ST18 and PV in a group of 100 Jewish PV patients and 400-age and population-matched controls (31). Little was reported on immune pathways within this work. In AA, however, GWAS have identified several immune-related pathways. In particular, literature has identified genes responsible for activation of IL-21 and IL-2, as well as the IL-2/IL-21, IL-2RA and HLA class II loci all to be implicated in AA (14, 32). This matches the Th1 and Th17 cytokine dysregulation we find in our work. However, GWAS have also found polymorphisms in the promotor region of IL-13 to be associated with AA (33).
Perhaps most intriguingly, a disease relevant, and at least partial, autoimmune activation in individuals that share genetic risk elements evokes a further novel hypothesis that there may be additional mechanisms at play which prevent these at-risk individuals exhibiting disease-like inflammation from progressing to frank clinical disease. It may be the case, for example, that specific counter-regulatory immune mechanisms offset the disease-driving cytokine dysregulation. One component of the necessary neutralizing force against the existing partial autoimmune activation seen in PV HLA-matched controls may be the downregulation of other specific cytokines, such as we observed for IL-10 and IL-15. Previous work in our lab supports this concept within both PV and AA. In gene expression studies in PV, our group has previously identified a specific ‘protection signature’ in HLA-matched controls, indicating that these individuals down- or up-regulate a specific set of genes that is otherwise similarly expressed in PV patients and healthy individuals not matched for PV-associated HLA alleles (HLA-unmatched controls) (22). Interestingly, a downregulation of the IL-13RA1 gene is part of this signature, supporting the importance of cytokine dysregulation in disease development or prevention thereof. Echoing this theme, previous gene microarray work in our lab uncovered specific “inheritance,” “disease,” and “severity” transcriptional signatures within AA patients, healthy relatives (related controls), and healthy non-relatives (unrelated controls) (19).
Figure 4 summarizes our findings regarding these novel concepts regarding disease risk, and the immune requirements and limitations linked to disease development. Our data allow for a more complete and deeper understanding of how cytokine pathways are altered in PV and AA pathogenesis and outline a potential role for HLA haplotype/familial relation in disease.
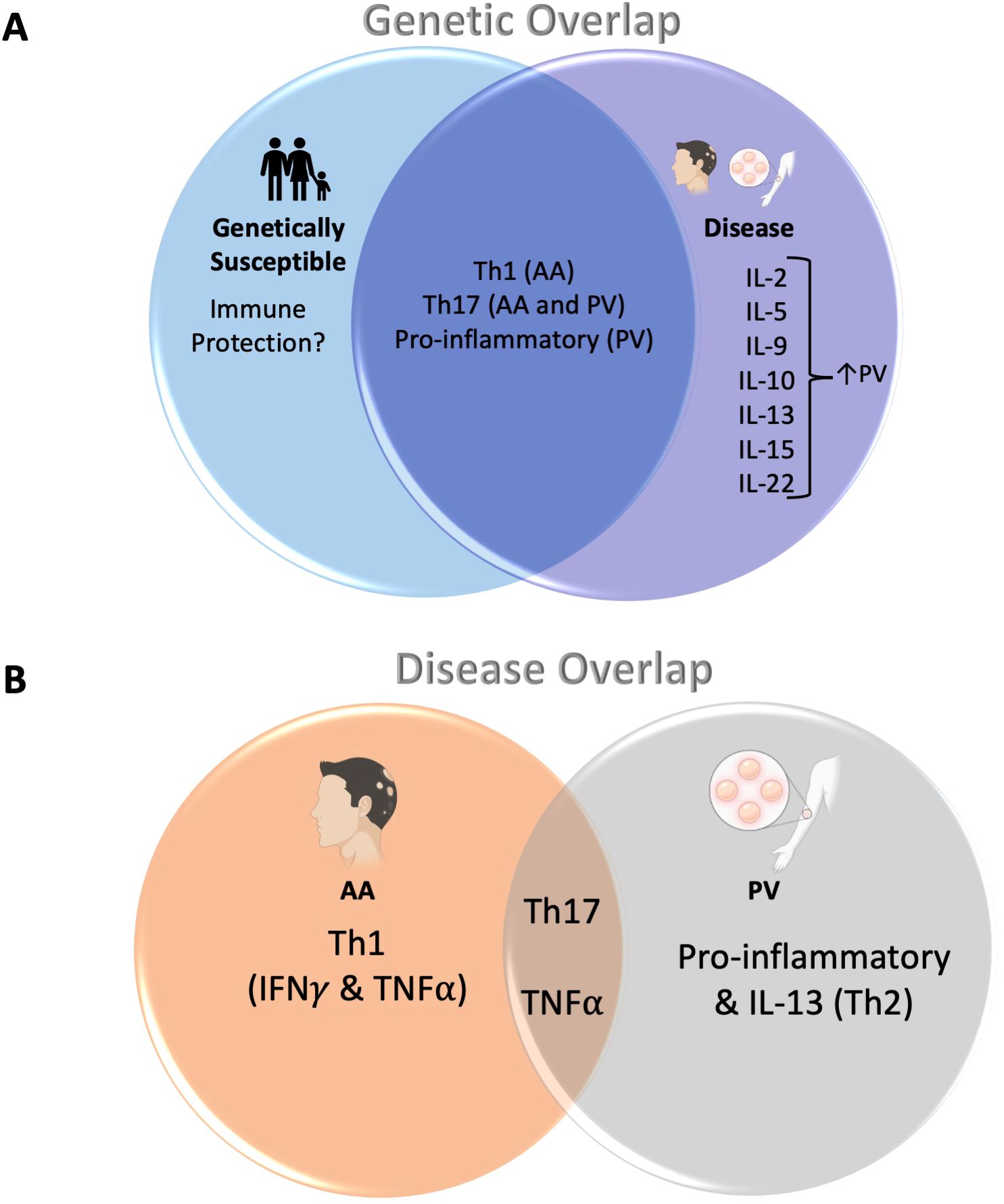
Figure 4. (A) Cytokine dysregulation shared across related controls/HLA-matched controls and patients with AA/PV: The overlapping portion of the Venn diagram shows cytokine pathways that are shared between both patients with AA and PV, as well as their genetically susceptible counterparts (related controls or HLA-matched controls, respectively). The portion to the right show cytokines that were found to be upregulated in disease alone (note: outside of IL-10 and IL-22, these cytokines were not analyzed for AA patients). (B) Cytokine dysregulation shared across both AA and PV: Both AA and PV share a dysregulation in Th17-pathway associated cytokines as well as TNFα (overlapping area of the Venn diagram). In addition, AA shows dysregulation in Th1-pathway cytokines IFNγ and TNFα (left). PV, on the other hand, shows an additional dysregulation in other pro-inflammatory cytokines (IL-1α, IL-1β, IL-6, IL-8) as well as the Th2 pathway cytokine IL-13 (right).
In summary, our data uncover a genetic underpinning to Th1- and Th17-related cytokine dysregulation in AA and to Th2-, Th17- and pro-inflammatory cytokine dysregulation in PV that are not restricted to patients. Our work supports the novel hypothesis that healthy individuals related to AA patients as well as individuals genetically susceptible to PV exhibit disease-associated immune disturbances that in themselves fall below the threshold required for disease development, and/or possess counter-regulatory mechanisms that prevent progression to active disease.
Though the work presented here focuses solely on peripheral cytokine dysregulation in AA and PV, target tissues of these diseases such as hair follicles in AA and keratinocytes in PV may harbor their own genetic variations and respond to an altered cytokine milieu differently. Our studies do not shed light on the effect of peripheral cytokine dysregulation on these target tissues. More work is thus required to identify how and if the peripheral cytokine dysregulation we find ultimately affects target tissues, and whether the cytokine dysregulation in genetically susceptible but healthy controls would extend to target tissues. Such work would need to focus on both lesional and non-lesional skin in patients, whereas genetically susceptible individuals could provide only non-lesional skin samples for comparison.
Nevertheless, knowledge of the extent and limitations of genetic contribution to disease provides new details regarding the mechanistic road map underlying the development of AA and PV, and shines a spotlight on the murky and previously underrecognized and understudied question of why certain individuals develop disease while others do not. Moreover, our data add to the knowledge base of how and why particular autoimmune diseases tend to cluster within families in support of the common gene/cause hypothesis. This work provides a stepping stone for further investigation to the precise mechanisms that produce disease, and ultimately those that may prevent it as well. Future work will be necessary to pinpoint potential disease-counteracting immune mechanisms in genetically predisposed individuals that could inform an entirely new approach to disease management and therapeutics.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the institutional review board (IRB) of Weill-Cornell Medical College (IRB 0998-398), Michigan State University (IRB 05-1034), and the University at Buffalo (IRB 456887). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
RS: Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. KS: Data curation, Methodology, Writing – review & editing. AS: Conceptualization, Investigation, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Timoteo RP, da Silva MV, Miguel CB, Silva DA, Catarino JD, Rodrigues Junior V, et al. Th1/Th17-related cytokines and chemokines and their implications in the pathogenesis of pemphigus vulgaris. Mediators Inflammation. (2017) 2017:7151285. doi: 10.1155/2017/7151285
2. Fang H, Li Q, Wang G. The role of T cells in pemphigus vulgaris and bullous pemphigoid. Autoimmun Rev. (2020) 19:102661. doi: 10.1016/j.autrev.2020.102661
3. Baker J, Seiffert-Sinha K, Sinha AA. Patient genetics shape the autoimmune response in the blistering skin disease pemphigus vulgaris. Front Immunol. (2022) 13:1064073. doi: 10.3389/fimmu.2022.1064073
4. Sinha AA. The genetics of pemphigus. Dermatol Clin. (2011) 29:381–91, vii. doi: 10.1016/j.det.2011.03.020
5. Sterkens A, Lambert J, Bervoets A. Alopecia areata: a review on diagnosis, immunological etiopathogenesis and treatment options. Clin Exp Med. (2021) 21:215–30. doi: 10.1007/s10238-020-00673-w
6. Zhou C, Li X, Wang C, Zhang J. Alopecia areata: an update on etiopathogenesis, diagnosis, and management. Clin Rev Allergy Immunol. (2021) 61:403–23. doi: 10.1007/s12016-021-08883-0
7. Alexis AF, Dudda-Subramanya R, Sinha AA. Alopecia areata: autoimmune basis of hair loss. Eur J Dermatol. (2004) 14:364–70.
8. Dudda-Subramanya R, Alexis AF, Siu K, Sinha AA. Alopecia areata: genetic complexity underlies clinical heterogeneity. Eur J Dermatol. (2007) 17:367–74. doi: 10.1684/ejd.2007.0231
9. Rajabi F, Drake LA, Senna MM, Rezaei N. Alopecia areata: a review of disease pathogenesis. Br J Dermatol. (2018) 179:1033–48. doi: 10.1111/bjd.16808
10. Safavi KH, Muller SA, Suman VJ, Moshell AN, Melton LJ 3rd. Incidence of alopecia areata in Olmsted County, Minnesota, 1975 through 1989. Mayo Clin Proc. (1995) 70:628–33. doi: 10.4065/70.7.628
11. Blaumeiser B, van der Goot I, Fimmers R, Hanneken S, Ritzmann S, Seymons K, et al. Familial aggregation of alopecia areata. J Am Acad Dermatol. (2006) 54:627–32. doi: 10.1016/j.jaad.2005.12.007
12. Rodriguez TA, Fernandes KE, Dresser KL, Duvic M, National Alopecia Areata R. Concordance rate of alopecia areata in identical twins supports both genetic and environmental factors. J Am Acad Dermatol. (2010) 62:525–7. doi: 10.1016/j.jaad.2009.02.006
13. Jackow C, Puffer N, Hordinsky M, Nelson J, Tarrand J, Duvic M. Alopecia areata and cytomegalovirus infection in twins: genes versus environment? J Am Acad Dermatol. (1998) 38:418–25. doi: 10.1016/s0190-9622(98)70499-2
14. Betz RC, Petukhova L, Ripke S, Huang H, Menelaou A, Redler S, et al. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat Commun. (2015) 6:5966. doi: 10.1038/ncomms6966
15. Lee E, Lendas KA, Chow S, Pirani Y, Gordon D, Dionisio R, et al. Disease relevant HLA class II alleles isolated by genotypic, haplotypic, and sequence analysis in North American Caucasians with pemphigus vulgaris. Hum Immunol. (2006) 67:125–39. doi: 10.1016/j.humimm.2005.09.003
16. Cardenas-Roldan J, Rojas-Villarraga A, Anaya JM. How do autoimmune diseases cluster in families? A systematic review and meta-analysis. BMC Med. (2013) 11:73. doi: 10.1186/1741-7015-11-73
17. Bhanusali DG, Sachdev A, Olson MA, Gerlach JA, Sinha AA. PTPN22 profile indicates a novel risk group in Alopecia areata. Hum Immunol. (2014) 75:81–7. doi: 10.1016/j.humimm.2013.09.003
18. Van Acker MM, Schwartz RR, Andrews K, Seiffert-Sinha K, Sinha AA. Inheritance-specific dysregulation of Th1- and Th17-associated cytokines in alopecia areata. Biomolecules. (2023) 13:1285. doi: 10.3390/biom13091285
19. Coda AB, Qafalijaj Hysa V, Seiffert-Sinha K, Sinha AA. Peripheral blood gene expression in alopecia areata reveals molecular pathways distinguishing heritability, disease and severity. Genes Immun. (2010) 11:531–41. doi: 10.1038/gene.2010.32
20. Sajda T, Hazelton J, Patel M, Seiffert-Sinha K, Steinman L, Robinson W, et al. Multiplexed autoantigen microarrays identify HLA as a key driver of anti-desmoglein and -non-desmoglein reactivities in pemphigus. Proc Natl Acad Sci U.S.A. (2016) 113:1859–64. doi: 10.1073/pnas.1525448113
21. Bhanusali DG, Sachdev A, Rahmanian A, Gerlach JA, Tong JC, Seiffert-Sinha K, et al. HLA-E*0103X is associated with susceptibility to Pemphigus vulgaris. Exp Dermatol. (2013) 22:108–12. doi: 10.1111/exd.2013.22.issue-2
22. Dey-Rao R, Seiffert-Sinha K, Sinha AA. Genome-wide expression analysis suggests unique disease-promoting and disease-preventing signatures in Pemphigus vulgaris. Genes Immun. (2013) 14:487–99. doi: 10.1038/gene.2013.44
23. Schwartz RR, Seiffert-Sinha K, Sinha AA. Cytokine profiling reveals HLA-linked Th2 and Th17 driven immune activation in pemphigus vulgaris patients and genetically susceptible healthy controls. Front Immunol. (2024) 15. doi: 10.3389/fimmu.2024.1500231
24. Giordano CN, Sinha AA. Cytokine pathways and interactions in alopecia areata. Eur J Dermatol. (2013) 23:308–18. doi: 10.1684/ejd.2013.2042
25. Giordano CN, Sinha AA. Cytokine networks in Pemphigus vulgaris: An integrated viewpoint. Autoimmunity. (2012) 45:427–39. doi: 10.3109/08916934.2012.697593
26. Holstein J, Solimani F, Baum C, Meier K, Pollmann R, Didona D, et al. Immunophenotyping in pemphigus reveals a T(H)17/T(FH)17 cell-dominated immune response promoting desmoglein1/3-specific autoantibody production. J Allergy Clin Immunol. (2021) 147:2358–69. doi: 10.1016/j.jaci.2020.11.008
27. Morsy H, Zedan H, Kassim M, Mwafey IM, Negm D. Serum IL-21 level in patients with pemphigus: Before and after treatment. Dermatol Ther. (2020) 33:e14482. doi: 10.1111/dth.14482
28. Waskiel-Burnat A, Osinska M, Salinska A, Blicharz L, Goldust M, Olszewska M, et al. The role of serum Th1, Th2, and Th17 cytokines in patients with alopecia areata: clinical implications. Cells. (2021) 10:3397. doi: 10.3390/cells10123397
29. Duvic M, Christiano AM, Hordinsky MK, Norris DA, Price VH, Amos CI. The national alopecia areata registry-update. J Investig Dermatol Symp Proc. (2013) 16:S53. doi: 10.1038/jidsymp.2013.20
30. Shah AA, Dey-Rao R, Seiffert-Sinha K, Sinha AA. Increased oxidative stress in pemphigus vulgaris is related to disease activity and HLA-association. Autoimmunity. (2016) 49:248–57. doi: 10.3109/08916934.2016.1145675
31. Sarig O, Bercovici S, Zoller L, Goldberg I, Indelman M, Nahum S, et al. Population-specific association between a polymorphic variant in ST18, encoding a pro-apoptotic molecule, and pemphigus vulgaris. J Invest Dermatol. (2012) 132:1798–805. doi: 10.1038/jid.2012.46
32. Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. (2010) 466:113–7. doi: 10.1038/nature09114
33. Jagielska D, Redler S, Brockschmidt FF, Herold C, Pasternack SM, Garcia Bartels N, et al. Follow-up study of the first genome-wide association scan in alopecia areata: IL13 and KIAA0350 as susceptibility loci supported with genome-wide significance. J Invest Dermatol. (2012) 132:2192–7. doi: 10.1038/jid.2012.129
34. Aljabali MA, Kuts L. Serum levels of IL-2 and IL-17A are related to clinical type and severity of alopecia areata. Wiad Lek. (2022) 75:263–7. doi: 10.36740/WiadLek
35. Alzolibani AA, Rasheed Z, Bin Saif G, Al-Dhubaibi MS, Al Robaee AA. Altered expression of intracellular Toll-like receptors in peripheral blood mononuclear cells from patients with alopecia areata. BBA Clin. (2016) 5:134–42. doi: 10.1016/j.bbacli.2016.03.006
36. Atwa MA, Youssef N, Bayoumy NM. T-helper 17 cytokines (interleukins 17, 21, 22, and 6, and tumor necrosis factor-alpha) in patients with alopecia areata: association with clinical type and severity. Int J Dermatol. (2016) 55:666–72. doi: 10.1111/ijd.2016.55.issue-6
37. Bain K, McDonald E, Moffat F, Tutino M, Castelino M, Barton A, et al. Alopecia areata is characterized by dysregulation in systemic type 17 and type 2 cytokines, which may contribute to disease-associated psychological morbidity. Br J Dermatol. (2020) 182:130–7. doi: 10.1111/bjd.18008
38. El-Morsy EH, Eid AA, Ghoneim H, Al-Tameemi KA. Serum level of interleukin-17A in patients with alopecia areata and its relationship to age. Int J Dermatol. (2016) 55:869–74. doi: 10.1111/ijd.12994
39. Elela MA, Gawdat HI, Hegazy RA, Fawzy MM, Abdel Hay RM, Saadi D, et al. B cell activating factor and T-helper 17 cells: possible synergistic culprits in the pathogenesis of Alopecia Areata. Arch Dermatol Res. (2016) 308:115–21. doi: 10.1007/s00403-016-1617-z
40. Gautam RK, Singh Y, Gupta A, Arora P, Khurana A, Chitkara A. The profile of cytokines (IL-2, IFN-gamma, IL-4, IL-10, IL-17A, and IL-23) in active alopecia areata. J Cosmet Dermatol. (2020) 19:234–40. doi: 10.1111/jocd.12970
41. Hatif ST, Hussein TA, Muhieldden AAR. EVALUATION OF IL17-A SERUM LEVELS IN IRAQI PATIENTS WITH ALOPECIA AREATA AND ALOPECIA UNIVERSALIS. Biochem Cell Arch. (2020) 20:6419–25.
42. Loh SH, Moon HN, Lew BL, Sim WY. Role of T helper 17 cells and T regulatory cells in alopecia areata: comparison of lesion and serum cytokine between controls and patients. J Eur Acad Dermatol Venereology. (2018) 32:1028–33. doi: 10.1111/jdv.2018.32.issue-6
43. Manimaran RP, Ramassamy S, Rajappa M, Chandrashekar L. Therapeutic outcome of diphencyprone and its correlation with serum cytokine profile in alopecia areata. J Dermatol Treat. (2022) 33:324–8. doi: 10.1080/09546634.2020.1752887
44. Tabara K, Kozłowska M, Jędrowiak A, Bienias W, Kaszuba A. Serum concentrations of selected proinflammatory cytokines in children with alopecia areata. Adv Dermatol Allergology/Postępy Dermatologii i Alergologii. (2019) 36:63–9. doi: 10.5114/ada.2019.82826
45. Tembhre M, Sharma V. T-helper and regulatory T-cell cytokines in the peripheral blood of patients with active alopecia areata. Br J Dermatol. (2013) 169:543–8. doi: 10.1111/bjd.2013.169.issue-3
46. Tomaszewska KA, Kozlowska M, Kaszuba A, Lesiak A, Narbutt J, Zalewska-Janowska AM. Increased serum levels of interleukin-17 in patients with alopecia areata and non-segmental vitiligo. Postepy Dermatol Alergol. (2022) 39:195–9. doi: 10.5114/ada.2022.113612
47. Askin O, Yucesoy SN, Coskun E, Engin B, Serdaroglu S. Evaluation of the level of serum Interleukins (IL-2, IL-4, IL-15 andIL-17) and its relationship with disease severity in patients with alopecia areata. Bras Dermatol. (2021) 96:551–7. doi: 10.1016/j.abd.2021.03.006
48. Gong Y, Zhao Y, Zhang X, Qi S, Li S, Ye Y, et al. Serum level of IL-4 predicts response to topical immunotherapy with diphenylcyclopropenone in alopecia areata. Exp Dermatol. (2020) 29:231–8. doi: 10.1111/exd.13758
49. Morsy H, Maher R, Negm D. Correlation between serum IL-17A level and SALT score in patients with alopecia areata before and after NB-UVB therapy. J Cosmet Dermatol. (2018) 17:533–7. doi: 10.1111/jocd.2018.17.issue-3
50. Joshi N, Minz RW, Anand S, Parmar NV, Kanwar AJ. Vitamin D deficiency and lower TGF-beta/IL-17 ratio in a North Indian cohort of pemphigus vulgaris. BMC Res Notes. (2014) 7:536. doi: 10.1186/1756-0500-7-536
51. Kowalski EH, Kneibner D, Kridin K, Amber KT. Serum and blister fluid levels of cytokines and chemokines in pemphigus and bullous pemphigoid. Autoimmun Rev. (2019) 18:526–34. doi: 10.1016/j.autrev.2019.03.009
52. Singh PK, Das S, Rai G, Ansari MA, Dar SA, Singh T, et al. A snapshot of T cell subset cytokines in pemphigus vulgaris: A cross-sectional study. Cureus. (2022) 14:e29890. doi: 10.7759/cureus.29890
53. Arakawa M, Dainichi T, Yasumoto S, Hashimoto T. Lesional Th17 cells in pemphigus vulgaris and pemphigus foliaceus. J Dermatol Sci. (2009) 53:228–31. doi: 10.1016/j.jdermsci.2008.09.008
54. You S, Ouyang J, Wu Q, Zhang Y, Gao J, Luo X, et al. Comparison of serum cytokines and chemokines levels and clinical significance in patients with pemphigus vulgaris-A retrospective study. Exp Dermatol. (2024) 33:e15173. doi: 10.1111/exd.15173
55. Asothai R, Anand V, Das D, Antil PS, Khandpur S, Sharma VK, et al. Distinctive Treg associated CCR4-CCL22 expression profile with altered frequency of Th17/Treg cell in the immunopathogenesis of Pemphigus Vulgaris. Immunobiology. (2015) 220:1129–35. doi: 10.1016/j.imbio.2015.06.008
56. Zebrowska A, Wozniacka A, Juczynska K, Ociepa K, Waszczykowska E, Szymczak I, et al. Correlation between IL36alpha and IL17 and activity of the disease in selected autoimmune blistering diseases. Mediators Inflammation. (2017) 2017:8980534. doi: 10.1155/2017/8980534
57. Huang S, Mao J, Zhou L, Xiong X, Deng Y. The imbalance of gut microbiota and its correlation with plasma inflammatory cytokines in pemphigus vulgaris patients. Scand J Immunol. (2019) 90:e12799. doi: 10.1111/sji.2019.90.issue-3
58. Lee SH, Hong WJ, Kim SC. Analysis of serum cytokine profile in pemphigus. Ann Dermatol. (2017) 29:438–45. doi: 10.5021/ad.2017.29.4.438
59. Masjedi M, Esmaeil N, Saffaei A, Abtahi-Naeini B, Pourazizi M, Haghjooy Javanmard S, et al. Cytokine indexes in pemphigus vulgaris: perception of its immunpathogenesis and hopes for non-steroidal treatment. Iran J Pharm Res. (2017) 16:1223–9.
60. Gholibeigian Z, Izad M, Daneshpazhooh M, Mortazavi H, Salehi Z, Behruzifar S, et al. Decreased serum levels of interleukin-17, interleukin-23, TGF-beta in pemphigus vulgaris patients, and their association with disease phase. Dermatol Ther. (2020) 33:e14071. doi: 10.1111/dth.14071
61. Akman Karakas A, Ergun E, Sayin Ekinci N, Toylu A, Uzun S, Alpsoy E. The role of T follicular helper cells in clinical remission and relapse in patients with pemphigus treated with rituximab. Acta Dermatovenerol Croat. (2023) 31:72–9.
62. Shahbazian P, Izad M, Daneshpazhooh M, Mortazavi H, Salehi Z, Behruzifar S, et al. Decreased serum levels of interleukin-4 and interleukin-21 in new pemphigus vulgaris patients, but not chronic patients with inactive disease compared to healthy controls. Dermatol Pract Concept. (2021) 11:e2021035. doi: 10.5826/dpc.1102a35
63. Mortazavi H, Esmaili N, Khezri S, Khamesipour A, Vasheghani Farahani I, Daneshpazhooh M, et al. The effect of conventional immunosuppressive therapy on cytokine serum levels in pemphigus vulgaris patients. Iran J Allergy Asthma Immunol. (2014) 13:174–83.
64. Bilgic O, Sivrikaya A, Unlu A, Altinyazar HC. Serum cytokine and chemokine profiles in patients with alopecia areata. J Dermatolog Treat. (2016) 27:260–3. doi: 10.3109/09546634.2015.1093591
65. Pouralibaba F, Babaloo Z, Pakdel F, Abdollahian T, Pourzare S. Elevated levels of interleukin-23 in sera of patients with pemphigus vulgaris. Iran J Immunol. (2012) 9:261–5. doi: IJIv9i4A7
66. Barahmani N, Lopez A, Babu D, Hernandez M, Donley S, Duvic M. Serum T helper 1 cytokine levels are greater in patients with alopecia areata regardless of severity or atopy. Clin Exp Dermatol. (2010) 35:409–16. doi: 10.1111/j.1365-2230.2009.03523.x
67. Arca E, Muşabak U, Akar A, Erbil AH, Taştan HB. Interferon-gamma in alopecia areata. Eur J Dermatol. (2004) 14:33–6.
68. Kasumagic-Halilovic E, Prohic A, Cavaljuga S. Tumor necrosis factor-alpha in patients with alopecia areata. Indian J Dermatol. (2011) 56:494–6. doi: 10.4103/0019-5154.87124
69. Omar SI, Hamza AM, Eldabah N, Habiba DA. IFN-α and TNF-α serum levels and their association with disease severity in Egyptian children and adults with alopecia areata. Int J Dermatol. (2021) 60:1397–404. doi: 10.1111/ijd.v60.11
70. Song T, Pavel AB, Wen H-C, Malik K, Estrada Y, Gonzalez J, et al. An integrated model of alopecia areata biomarkers highlights both TH1 and TH2 upregulation. J Allergy Clin Immunol. (2018) 142:1631–1634.e13. doi: 10.1016/j.jaci.2018.06.029
71. Teraki Y, Imanishi K, Shiohara T. Cytokines in alopecia areata: contrasting cytokine profiles in localized form and extensive form (alopecia universalis). Acta Dermato-Venereologica. (1996) 76:421–3. doi: 10.2340/0001555576421423
72. Tomaszewska K, Kozłowska M, Kaszuba A, Lesiak A, Narbutt J, Zalewska-Janowska A. Increased serum levels of IFN-γ, IL-1β, and IL-6 in patients with alopecia areata and nonsegmental vitiligo. Oxid Med Cell Longevity. (2020) 2020:5693572. doi: 10.1155/2020/5693572
73. Das D, Anand V, Khandpur S, Sharma VK, Sharma A. T helper type 1 polarizing gammadelta T cells and Scavenger receptors contribute to the pathogenesis of Pemphigus vulgaris. Immunology. (2018) 153:97–104. doi: 10.1111/imm.2018.153.issue-1
74. Hertl M. Humoral and cellular autoimmunity in autoimmune bullous skin disorders. Int Arch Allergy Immunol. (2000) 122:91–100. doi: 10.1159/000024364
75. Stern JN, Keskin DB, Barteneva N, Zuniga J, Yunis EJ, Ahmed AR. Possible role of natural killer cells in pemphigus vulgaris - preliminary observations. Clin Exp Immunol. (2008) 152:472–81. doi: 10.1111/j.1365-2249.2008.03638.x
76. D’Auria L, Bonifati C, Mussi A, D’Agosto G, De Simone C, Giacalone B, et al. Cytokines in the sera of patients with pemphigus vulgaris: interleukin-6 and tumour necrosis factor-alpha levels are significantly increased as compared to healthy subjects and correlate with disease activity. Eur Cytokine Netw. (1997) 8:383–7.
77. Keskin DB, Stern JNH, Fridkis-Hareli M, Razzaque Ahmed A. Cytokine profiles in pemphigus vulgaris patients treated with intravenous immunoglobulins as compared to conventional immunosuppressive therapy. Cytokine. (2008) 41:315–21. doi: 10.1016/j.cyto.2007.12.007
78. Takahashi H, Amagai M, Tanikawa A, Suzuki S, Ikeda Y, Nishikawa T, et al. T helper type 2-biased natural killer cell phenotype in patients with pemphigus vulgaris. J Invest Dermatol. (2007) 127:324–30. doi: 10.1038/sj.jid.5700527
79. El-Darouti MA, Hegazy RA, Abdel Hay RM, El Hawary MS, Tawdy AM, Fawzy MM, et al. Study of T helper 1 and T helper 2 responses in pemphigus vulgaris patients receiving interferon alpha 2a injections in addition to a standard protocol therapy: a randomized controlled trial. Arch Dermatol Res. (2015) 307:299–307. doi: 10.1007/s00403-014-1522-2
80. Lin MS, Swartz SJ, Lopez A, Ding X, Fernandez-Vina MA, Stastny P, et al. Development and characterization of desmoglein-3 specific T cells from patients with pemphigus vulgaris. J Clin Invest. (1997) 99:31–40. doi: 10.1172/JCI119130
81. Satyam A, Khandpur S, Sharma VK, Sharma A. Involvement of T(H)1/T(H)2 cytokines in the pathogenesis of autoimmune skin disease-Pemphigus vulgaris. Immunol Invest. (2009) 38:498–509. doi: 10.1080/08820130902943097
82. Eid HE-G. Basiouny, Cytokine profiles in the sera of Egyptian patients with oral pemphigus vulgaris. Braz J Oral Sci. (2011) 10:83–7.
83. Attia EA, El Shennawy D, Sefin A. Serum interleukin-4 and total immunoglobulin E in nonatopic alopecia areata patients and HLA-DRB1 typing. Dermatol Res Pract. (2010) 2010:503587. doi: 10.1155/2010/503587
84. Rizzo C, Fotino M, Zhang Y, Chow S, Spizuoco A, Sinha AA. Direct characterization of human T cells in pemphigus vulgaris reveals elevated autoantigen-specific Th2 activity in association with active disease. Clin Exp Dermatol. (2005) 30:535–40. doi: 10.1111/j.1365-2230.2005.01836.x
85. Wucherpfennig KW, Yu B, Bhol K, Monos DS, Argyris E, Karr RW, et al. Structural basis for major histocompatibility complex (MHC)-linked susceptibility to autoimmunity: charged residues of a single MHC binding pocket confer selective presentation of self-peptides in pemphigus vulgaris. Proc Natl Acad Sci U.S.A. (1995) 92:11935–9. doi: 10.1073/pnas.92.25.11935
86. Yokoyama T, Amagai M. Immune dysregulation of pemphigus in humans and mice. J Dermatol. (2010) 37:205–13. doi: 10.1111/j.1346-8138.2009.00797.x
87. Gong Y, Luo L, Li L, He X, Lu W, Sha X, et al. Diphenylcyclopropenone plays an effective therapeutic role by up-regulating the TSLP/OX40L/IL-13 pathway in severe alopecia areata. Exp Dermatol. (2021) 30:278–83. doi: 10.1111/exd.14254
88. Alecu M, Alecu S, Coman G, Gălăţescu E, Ursaciuc C. ICAM-1, ELAM-1, TNF-alpha and IL-6 in serum and blister liquid of pemphigus vulgaris patients. Roumanian Arch Microbiol Immunol. (1999) 58:121–30.
89. Feliciani C, Toto P, Amerio P, Pour SM, Coscione G, Shivji G, et al. In vitro and in vivo expression of interleukin-1alpha and tumor necrosis factor-alpha mRNA in pemphigus vulgaris: interleukin-1alpha and tumor necrosis factor-alpha are involved in acantholysis. J Invest Dermatol. (2000) 114:71–7. doi: 10.1046/j.1523-1747.2000.00835.x
90. Feliciani C, Toto P, Wang B, Sauder DN, Amerio P, Tulli A. Urokinase plasminogen activator mRNA is induced by IL-1alpha and TNF-alpha in in vitro acantholysis. Exp Dermatol. (2003) 12:466–71. doi: 10.1034/j.1600-0625.2002.120415.x
91. Hennerici T, Pollmann R, Schmidt T, Seipelt M, Tackenberg B, Mobs C, et al. Increased frequency of T follicular helper cells and elevated interleukin-27 plasma levels in patients with pemphigus. PloS One. (2016) 11:e0148919. doi: 10.1371/journal.pone.0148919
92. Khozeimeh F, Savabi O, Esnaashari M. Evaluation of interleukin-1alpha, interleukin-10, tumor necrosis factor-alpha and transforming growth factor-beta in the serum of patients with pemphigus vulgaris. J Contemp Dent Pract. (2014) 15:746–9. doi: 10.5005/jp-journals-10024-1610
93. Lopez-Robles E, Avalos-Diaz E, Vega-Memije E, Hojyo-Tomoka T, Villalobos R, Fraire S, et al. TNFalpha and IL-6 are mediators in the blistering process of pemphigus. Int J Dermatol. (2001) 40:185–8. doi: 10.1046/j.1365-4362.2001.01083.x
94. Ragab N, Abdallah M, El-Gohary E, Elewa R. Stress and serum TNF-alpha levels may predict disease outcome in patients with pemphigus: a preliminary study. Cutis. (2011) 87:189–94.
95. Feliciani C, Toto P, Amerio P. In vitro C3 mRNA expression in Pemphigus vulgaris: complement activation is increased by IL-1alpha and TNF-alpha. J Cutan Med Surg. (1999) 3:140–4. doi: 10.1177/120347549900300306
96. Ludwig RJ, Schmidt E. Cytokines in autoimmune bullous skin diseases. Epiphenomena or contribution to pathogenesis? G Ital Dermatol Venereol. (2009) 144:339–49.
97. Narbutt J, Lukamowicz J, Bogaczewicz J, Sysa-Jedrzejowska A, Torzecka JD, Lesiak A. Serum concentration of interleukin-6 is increased both in active and remission stages of pemphigus vulgaris. Mediators Inflammation. (2008) 2008:875394. doi: 10.1155/2008/875394
98. Bhol KC, Desai A, Kumari S, Colon JE, Ahmed AR. Pemphigus vulgaris: the role of IL-1 and IL-1 receptor antagonist in pathogenesis and effects of intravenous immunoglobulin on their production. Clin Immunol. (2001) 100:172–80. doi: 10.1006/clim.2001.5061
99. Ataseven A, Saral Y, Godekmerdan A. Serum cytokine levels and anxiety and depression rates in patients with alopecia areata. Eurasian J Med. (2011) 43:99. doi: 10.5152/eajm.2011.22
100. Metwally D, Fawzy M, ElKalioby M, Hegazy R, Hay RA, Abd-Elreheem H, et al. Assessment of the quality of life, prevalence of depression, and the level of interleukin 6 in patients with pemphigus vulgaris. Acta Dermatovenerol Croat. (2020) 28:57–62.
101. Dai Z, Wang EHC, Petukhova L, Chang Y, Lee EY, Christiano AM. Blockade of IL-7 signaling suppresses inflammatory responses and reverses alopecia areata in C3H/HeJ mice. Sci Adv. (2021) 7:eabd1866. doi: 10.1126/sciadv.abd1866
102. Ma X, Chen S, Jin W, Gao Y. Th1/Th2 PB balance and CD200 expression of patients with active severe alopecia areata. Exp Ther Med. (2017) 13:2883–7. doi: 10.3892/etm.2017.4312
103. Bhol KC, Rojas AI, Khan IU, Ahmed AR. Presence of interleukin 10 in the serum and blister fluid of patients with pemphigus vulgaris and pemphigoid. Cytokine. (2000) 12:1076–83. doi: 10.1006/cyto.1999.0642
104. Kheirodin M, Tehranchinia Z, Ketabi Y, Tavakolpour S, Dadkhahfar S, Faghankhani M, et al. Elevated serum levels of interleukin-15 in pemphigus vulgaris patients: a potential therapeutic target. Dermatol Pract Concept. (2022) 12:e2022118. doi: 10.5826/dpc.1203a118
105. Lee D, Hong SK, Park SW, Hur DY, Shon JH, Shin JG, et al. Serum levels of IL-18 and sIL-2R in patients with alopecia areata receiving combined therapy with oral cyclosporine and steroids. Exp Dermatol. (2010) 19:145–7. doi: 10.1111/j.1600-0625.2009.00937.x
106. Bakr RM, Sayed DS, Abd-Elkader AS, Kamel AA, Badran AY. Does interleukin-33 level correlate with the activity of Pemphigus vulgaris?: A case-control study. Dermatol Ther. (2021) 34:e14605. doi: 10.1111/dth.14605
Keywords: pemphigus, alopecia, cytokine, HLA, Th17, Th2, autoimmunity
Citation: Schwartz RR, Seiffert-Sinha K and Sinha AA (2025) Healthy individuals genetically at-risk for the development of Pemphigus vulgaris or Alopecia areata share disease-like cytokine dysregulation. Front. Immunol. 15:1500284. doi: 10.3389/fimmu.2024.1500284
Received: 23 September 2024; Accepted: 13 December 2024;
Published: 13 January 2025.
Edited by:
Ming Yu Lien, China Medical University Hospital, TaiwanReviewed by:
Xuming Mao, University of Pennsylvania, United StatesYan Lu, The First Affiliated Hospital of Nanjing Medical University, China
Copyright © 2025 Schwartz, Seiffert-Sinha and Sinha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Animesh A. Sinha, YWFzaW5oYUBidWZmYWxvLmVkdQ==
 Rebekah R. Schwartz
Rebekah R. Schwartz Kristina Seiffert-Sinha
Kristina Seiffert-Sinha Animesh A. Sinha
Animesh A. Sinha