- 1Translational Medical Innovation Center, Zhangjiagang TCM Hospital Affiliated to Nanjing University of Chinese Medicine, Zhangjiagang, Jiangsu, China
- 2Department of Endocrinology, Zhangjiagang TCM Hospital Affiliated to Nanjing University of Chinese Medicine, Zhangjiagang, Jiangsu, China
- 3Department of Reproduction, Zhangjiagang TCM Hospital Affiliated to Nanjing University of Chinese Medicine, Jiangsu, Zhangjiagang, China
- 4Department of Surgery, Thomas E. Starzl Transplantation Institute, University of Pittsburgh School of Medicine, Pittsburgh, PA, United States
Achieving sustained activity and tolerance in of allogeneic grafts after post-transplantation remains a substantial challenge. The response of the immune system to “non-self” MHC-antigenic peptides initiates a crucial phase, wherein blocking positive co-stimulatory signals becomes imperative to ensure graft survival and tolerance. MicroRNAs (miRNAs) inhibit mRNA translation or promote mRNA degradation by complementary binding of mRNA seed sequences, which ultimately affects protein synthesis. These miRNAs exhibit substantial promise as diagnostic, prognostic, and therapeutic candidates for within the realm of solid organ transplantations. Current research has highlighted three members of the T cell immunoglobulin and mucin domain (TIM) family as a novel therapeutic avenue in transplantation medicine and alloimmunization. The interplay between miRNAs and TIM proteins has been extensively explored in viral infections, inflammatory responses, and post-transplantation ischemia-reperfusion injuries. This review aims to elucidate the distinct roles of miRNAs and TIM in transplantation immunity and delineate their interdependent relationships in terms of targeted regulation. Specifically, this investigation sought seeks to uncover the potential of miRNA interaction with TIM, aiming to induce immune tolerance and bolster allograft survival after transplantation. This innovative strategy holds substantial promise in for the future of transplantation science and practice.
1 Introduction
Allogeneic transplantation is the primary treatment for patients with end-stage diseases and severe trauma. Imbalances in the activation and suppression of the immune system, systemic dysfunction of the transplanted organ, and infections all contribute to the failure of allogeneic transplantations (1–3). In many cases, autologous transplantation is not feasible due to physiological restrictions (4, 5). Consequently, allogeneic transplantation remains the only viable solution in such scenarios. However, graft rejection remains a major obstacle leading to graft loss (6).
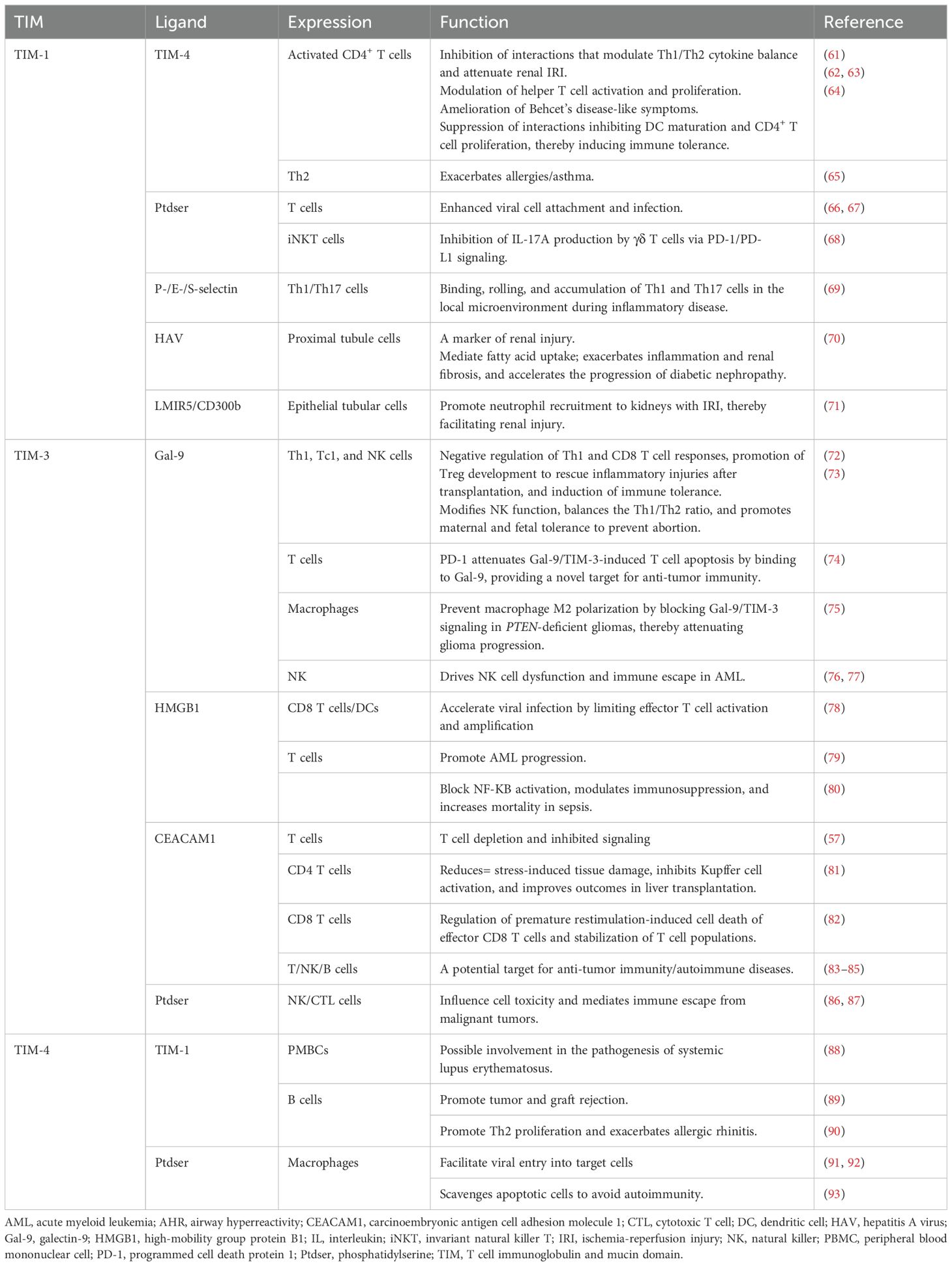
The T cell immunoglobulin and mucin domain (TIM) gene family comprises a series of genes encoding type 1 glycoprotein-like structural domains expressed on cell membranes that crucially regulate immune responses (7). Members of the TIM gene family, such as TIM-1, TIM-3, and TIM-4, exhibit structural characteristics that are conserved in both mice and humans (8). Initially identified as a susceptibility gene for asthma and allergy, TIM-1 is preferentially expressed on Th2 cells and linked to atopic and autoimmune diseases (9). TIM-3 is expressed on innate and adaptive immune cells, including mast cells, dendritic cells (DCs), macrophages, and Th1 and Tc1 cells, and acts as an inhibitory receptor that promotes Th1 apoptosis and reduces the production of inflammatory factors (10–12). TIM-4 is solely expressed on the surface of antigen-presenting cells (APCs), facilitating phagocytosis of apoptotic cells and modulating T cell responses (7, 13). Ongoing research underscores the extensive role of TIM proteins in immune tolerance and transplant rejection (14–16).
MicroRNAs (miRNAs), single-stranded RNAs approximately 22 nucleotides long, selectively and specifically regulate post-transcriptional gene expression (17). Recently, miRNAs have demonstrated specific and impactful biological effects, serving to establish immune tolerance following solid organ transplantation (18). Thus, miRNAs exhibit potential as diagnostic, predictive, and therapeutic markers for allograft rejection (19).
Both miRNAs and TIM proteins have wide applications in immune tolerance induction and transplantation (20). The interaction between miRNAs and TIM proteins in cancer therapy has been extensively studied (21, 22) (Table 1). However, their effects on allograft rejection models remain unclear. Thus, this review aims to discuss recent advancements in understanding the TIM–miRNA network and explore its potential applications in solid organ transplantation and immune tolerance.
2 TIM gene family
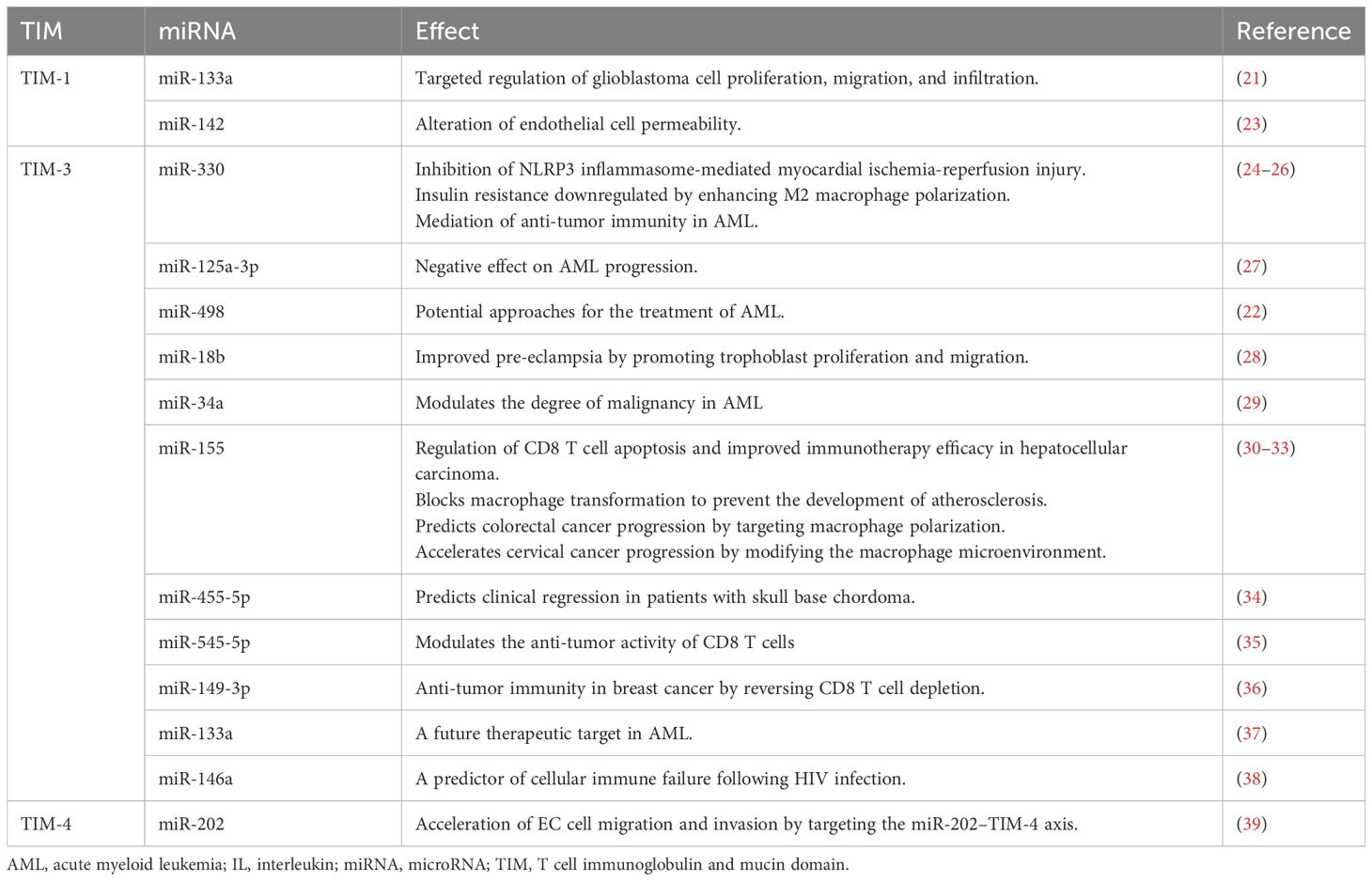
The TIM genes are located on mouse chromosome 11B1.1 and human chromosome 5q33.5, which are regions associated with various atopic/autoimmune diseases such as asthma and allergies (40). The TIM family comprises eight murine members (four coding genes, TIM-1–TIM-4, and four noncoding genes, TIM-5–TIM-8) and three human members (TIM-1, TIM-3, and TIM-4) (41). TIM proteins share a similar structure, encompassing an immunoglobulin domain, mucin-like domain, transmembrane region, and cytoplasmic domain containing tyrosine-phosphorylated motifs (except for TIM-4) (12) (Figure 1). Based on gene sequence similarity, murine TIM-2 shares structural and functional similarities with murine TIM-1, and is considered a direct homolog of human TIM-1 (42).
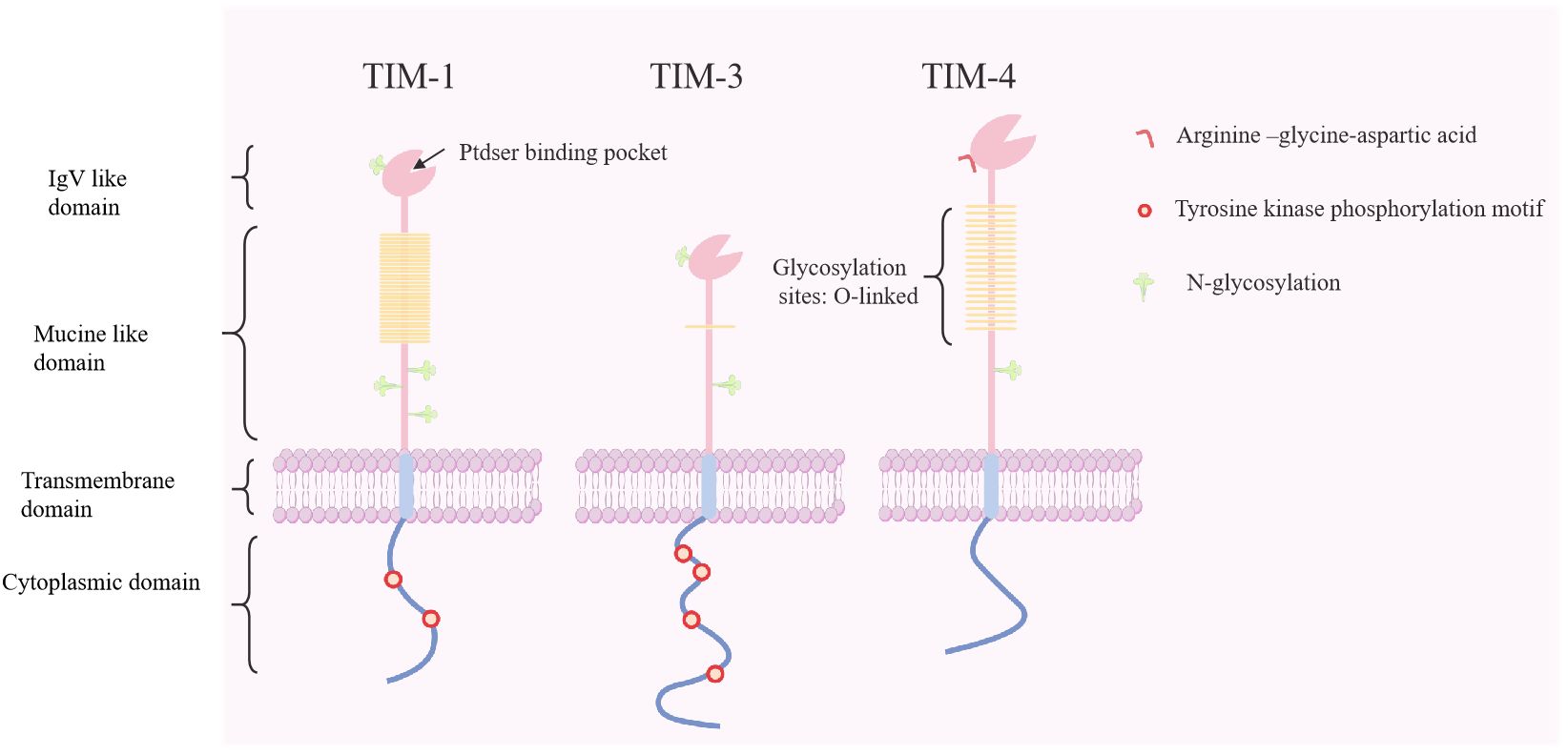
Figure 1. Molecular structure of human T cell immunoglobulin and mucin proteins (TIM-1, -3, and -4). The TIM genes encode type I membrane proteins that contain an Ig V-like domain, an O-linked glycosylated mucin domain, a transmembrane domain, and a cytoplasmic domain with tyrosine-phosphorylated motifs. TIM-4 contains RGD motifs that can interact with integrins and participate in intercellular adhesion. Ptdser, phosphatidylserine; RGD, arginine−glycine−aspartic acid.
2.1 Functional characteristics of TIM-1
Initially identified as the hepatitis A virus receptor (HAVCR1) and later as a human kidney injury molecule, TIM-1 is found on B cells, DCs, mast cells, and invariant natural killer T (iNKT) cells, playing a crucial role in immune activation (43–45). As a potential co-stimulatory molecule, it is well established that TIM-1 exerts immune effects by maintaining Breg suppression and stimulating effector T cell activity and homeostasis (8, 46). The diverse biological roles of TIM-1 open up new avenues for the treatment of autoimmune diseases, viral infections and tumors (47–49). Previous studies have suggested the potentially diverse roles of TIM-1 in inducing immune tolerance in transplantation.
2.2 Role of TIM-1 in transplantation
Recent studies have highlighted the pivotal role of TIM-1 in preventing and mitigating allograft rejection. The agonistic TIM-1-specific mAb 3B3 disrupts mouse allograft tolerance by interacting with effector T cells and Tregs (50). Additionally, TIM-1 not only serves as a surface marker but also as a crucial surface molecule that induces and maintains regulatory B cell (Breg) function in mice (51, 52). In a model of islet transplantation, anti-CD45RB and anti-TIM-1 (RMT1-10) antibodies increased interleukin (IL)-10 expression in TIM-1+ Bregs and antigen-specific transplantation tolerance (52, 53). This combined antibody therapy relies on TIM-1 expression, IL-10-producing Bregs, and Tregs (54). Altered IL-10 levels and accelerated allograft rejection have been observed in TIM-1 knockout and mutant mice (46). Recent findings indicate that the inhibitory function of ex vivo expansion of human B cells partly relies on TIM-1, which maintains long-term regulatory function and human allogeneic skin graft survival by positively regulating STAT3 phosphorylation (55). The TIM-1 signaling pathway is not only targeted after allogeneic transplantation, but also as a new therapeutic strategy to improve post-transplant complications (56).
2.3 Functional characteristics of TIM-3
TIM-3 serves as a suppressor molecule involved in T cell activation and is a marker of T cell depletion in tumors and chronic viral infections (57). Subsequently, TIM-3 was found to accelerate tumor progression and support maternal-fetal tolerance (58, 59). Galectin-9 (Gal-9), the first ligand identified for TIM-3, eliminates interferon γ-producing Th1 cells, thereby reducing the severity and mortality of experimental autoimmune encephalomyelitis (60) (Table 2). TIM-3 interacts with different ligands and mediates various immune responses, making it a promising target for immunotherapy.
2.4 Role of TIM-3 in transplantation
Initially considered as a marker for terminally differentiated effector T cells, TIM-3 has been found to influence Treg acquisition and function, providing new insights into the mechanisms of transplant rejection (94). The natural TIM-3 ligand Gal-9 limits Th1 activation, thereby protecting specific Treg responses and attenuating allograft rejection (95). When allograft rejection occurs, increased expression of TIM-3 on the recipient’s NK cells stimulates IFN-γ production through interaction with Gal-9 (96). Therefore, high serum levels of soluble TIM-3 and sGal-9 serve as prospective biomarkers for diagnosing and predicting renal transplant dysfunction (97, 98). Additionally, hepatocytic Gal-9 signaling via TIM-3+CD4+ T cells mitigate ischemia-reperfusion injury (IRI) during orthotopic liver transplantation in recipient mice (72). TIM-3+CD4+ and TIM-3+CD8+ T cells in allogeneic transplantation models exhibit a depleted dysfunctional phenotype owing to continuous stimulation by allogeneic antigens (99). This early induction and establishment of T cell dysfunction ultimately mediate and maintain the phenotypic and functional characteristics of self-tolerance or exhaustion (100). Moreover, inhibitory receptors such as TIM-3 and PD-1 ensure that Treg are depleted after graft rejection to prevent microbial and tumor unresponsiveness and to balance immunomodulatory functions (16). High pretransplant T-cell expression of PD-1 and Tim-3 co-suppressor receptors correlated positively with the incidence of posttransplant infection (101). Clinical studies have shown that elevated CEACAM1 levels are associated with a favorable outcome in orthotopic liver transplantation. Recent evidence confirms that T cell CEACAM1 - TIM-3 crosstalk inhibits Kupffer cell NF-ΚB phosphorylation, attenuates post-transplant liver injury and promotes T cell homeostasis (81). Overall, TIM-3 has shown potential applications in transplantation, but more thorough mechanisms of action need to be explored.
2.5 Functional characteristics of TIM-4
Traditionally known to be primarily expressed on the surface of APCs, including macrophages, mature DCs, B1 cells, and iNKT cells, recent studies have also identified TIM-4 expression in fibroblasts (13, 102). This diverse expression profile suggests potential multifaceted roles of TIM-4 in immune regulation and cellular interactions. Structurally, despite the lack of a cytoplasmic tail for intracellular signaling, the TIM-4 extracellular IgV domain contains arginine-glycine-aspartate (RGD) motifs, which predominantly facilitates APC-T cell adhesion (7, 103).
Initial studies have suggested that TIM-4 acts as a natural ligand for TIM-1, contributing to helper T cell proliferation and favoring Th2 immune responses (104). However, further investigations have revealed the nuanced effects of TIM-4 on T cell responses. Depending on the concentration of TIM-4 stimulation and the state of T cell activation, TIM-4 has contrasting effects on T cell proliferation (62, 105). These findings suggest that the influence of TIM-4 on T cells may involve receptors other than the known TIM-1 receptor, especially during the initial T cell surface expression.
As a phosphatidylserine receptor, TIM-4 contributes to the creation of an environment of immune tolerance by clearing apoptotic cells and debris, simultaneously suggesting potential risks associated with infection and tumorigenesis (91, 106). Overall, the function of TIM-4 as a potent co-stimulatory signal in APCs revealed its diverse and context-dependent biological activities. Its precise biological effects seem to be closely linked to the type of ligands it interacts with and the specific sites of T cell activation. Understanding the intricate interactions of TIM-4 with various receptors and their dual roles in immune tolerance and potential pathogenic processes remains an area of active research in immunology.
2.6 Role of TIM-4 in transplantation
Few studies have investigated TIM-4 in the context of transplantation. Researchers have focused on understanding TIM-4 expression in specific immune cells, particularly macrophages and DCs, as these cells play crucial roles in the modulation of TIM-4 to promote tolerance in human transplantation (107, 108).
Prior to 2010, studies exploring the direct relationship between TIM-4 and transplantation immunity were lacking. However, in 2010, Uchida et al. hypothesized that blocking the TIM-1–TIM-4 signaling pathway might alleviate hepatic IRI. The proposed intervention presented a novel approach aimed at extending the survival and success of transplanted organs (109). In the following year, Rong et al. provided initial evidence supporting this hypothesis by demonstrating that disrupting the TIM-1–TIM-4 pathway could inhibit CD4 T cell activation. This inhibition protected renal function and reduced local leukocyte recruitment and activation, offering a promising novel target for the treatment of acute kidney injury (61).
Subsequent studies further reinforced these initial findings, consistently showing that blocking TIM-4 signaling conferred protection against hepatic IRI. Notably, these studies highlight the significance of TIM-4-mediated phagocytosis, which is involved in activating the innate immune system and represents a crucial aspect of this process (110, 111). Indeed, these studies underscore the potential therapeutic implications of targeting TIM-4 in mitigating transplantation-related complications, and hold promise for developing novel strategies to enhance the success of organ transplantation.
Macrophages, particularly tissue-resident macrophages such as CD169+ macrophages, play a critical role in modulating immune responses and influencing transplant outcomes. For instance, genetic ablation of TIM-4 in CD169+ tissue-resident macrophages improve their survival. However, this alteration does not seem to affect the effective stimulation of Treg production or promote the prolonged survival of cardiac allografts (15).
Kupffer cells (KCs), the dominant macrophages in the liver, have been identified as critical mediators of tolerance following liver transplantation. KCs promote tolerance through mechanisms involving upregulation of FasL-induced apoptosis and cytokine secretion in T cells (112, 113). Disrupting TIM-4 signaling in KCs in combination with transforming growth factor (TGF)-Β treatment significantly induces the transformation of inducible Tregs and ameliorates acute rejection after liver transplantation. This effect occurs via inhibition of the IL-4–STAT6–Gata3 signaling pathway, thereby modulating immune responses and improving tolerance induction (114).
However, studies on mice with congenital TIM-4 deficiency have reported an autoimmune response due to nonspecific immune activation. This is because of defects in the ability to eliminate apoptotic cells, suggesting a crucial role for TIM-4 in maintaining immune homeostasis and preventing autoimmunity (115). Moreover, DCs, which are highly specialized APCs, are key players in the induction of inflammation and immune tolerance (116). In a skin transplantation model, disruption of TIM-4 co-stimulatory signaling on DCs enhanced the transfer of naïve CD4 cells to inducible Tregs, while limiting the transfer of IL-4/STAT-6 signaling. This modulation attenuates the Th2 response and effectively prolongs graft survival, highlighting the potential of targeting TIM-4 on DCs to modulate immune responses in transplantation scenarios (117).
Collectively, these findings emphasize the intricate role of TIM-4 in regulating immune responses involving macrophages, KCs, and DCs in transplantation scenarios, suggesting its potential as a target for therapeutic interventions to modulate immune tolerance and improve graft survival.
2.7 TIM proteins as phosphatidylserine receptors
Structurally, TIM proteins create a cavity with a distinctive “pocket” structure in the immunoglobulin variable region, securely binding to phosphatidylserine (93, 118) (Figure 2). During apoptosis, phosphatidylserine exposure to the plasma membrane triggers phagocytosis, which is essential for tissue homeostasis and immune regulation (119, 120). TIM-1 signaling by T and iNKT cells prevents recipient survival by inhibiting acute graft-versus-host disease after hematopoietic cell transplantation (20). TIM-1-expressing renal epithelial cells aid in phagocytosis of damaged cells, thereby limiting inflammation (121, 122). In addition to its role in phagocytosis, TIM-3 utilizes functional antibodies with phosphatidylserine to enhance T cell activation and anti-tumor activity (123). TIM-4, as a surface receptor, indirectly modulates inflammation and tumor progression through immune cell clearance (43, 106).
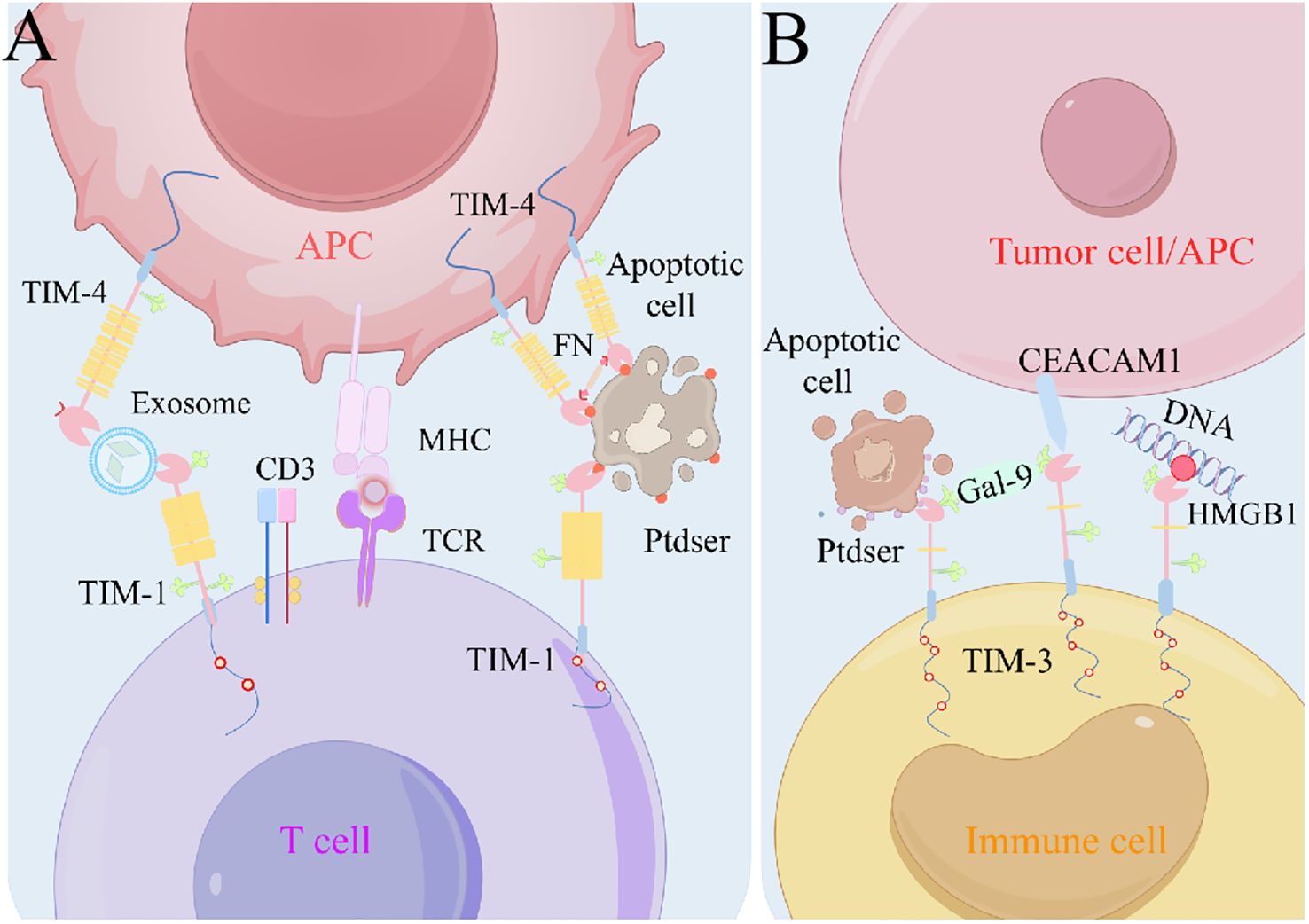
Figure 2. Models of TIM-ligand interactions. (A) TIM-1 can interact with Ptdser on the surface of apoptotic cells, or TIM-1 and TIM-4 interact via exosome bridging. TIM-4 is used as a bolus molecule to immobilize apoptotic cells near phagocytes to initiate efferocytosis. (B) Gal-9 can promote TIM-3 oligomerization and thus the interaction with other TIM-3 ligands, such as CEACAM1–TIM-3. Ptdser released from apoptotic cells can bind the FG-CC′ cleavage site of TIM-3. In addition, TIM-3 can bind HMGB1 and thus inhibit nucleic acid-mediated anti-tumor immunity. APC, antigen-presenting cell; CEACAM1, carcinoembryonic antigen cell adhesion molecule 1; Gal-9, galectin-9; HMGB1, high-mobility group protein B1; MHC, major histocompatibility complex; Ptdser, phosphatidylserine; TCR, T cell receptor; TIM, T cell immunoglobulin and mucin domain.
3 Expression and functions of miRNAs
3.1 Biogenesis of miRNAs
miRNAs are a class of small noncoding RNAs present in animals, plants, and some viruses that play a crucial regulatory role in transcription by either cleaving target mRNAs or inhibiting their translation (124). The gene sequences encoding miRNAs are arranged differently within the genome. Some miRNAs are organized as mono-cis-parallels with autonomous promoters, whereas others are arranged in multi-cis-parallels, sharing a common promoter and being transcribed into multiple miRNA clusters (125, 126). In certain cases, miRNA genes are located within the exons (Figure 3). RNA polymerase II is typically responsible for miRNA transcription. This process generates primary precursors known as pri-miRNAs, which adopt a typical hairpin structure and contain a 5′- and a 3′-polyadenylated tail. Subsequently, pri-miRNA undergoes precise cleavage in the nucleus by Drosha and DiGeorge Syndrome Critical Region 8(DGCR8), a nucleic acid endonuclease of the RNase III family, producing pre-miRNAs with stem-loop structures (127).
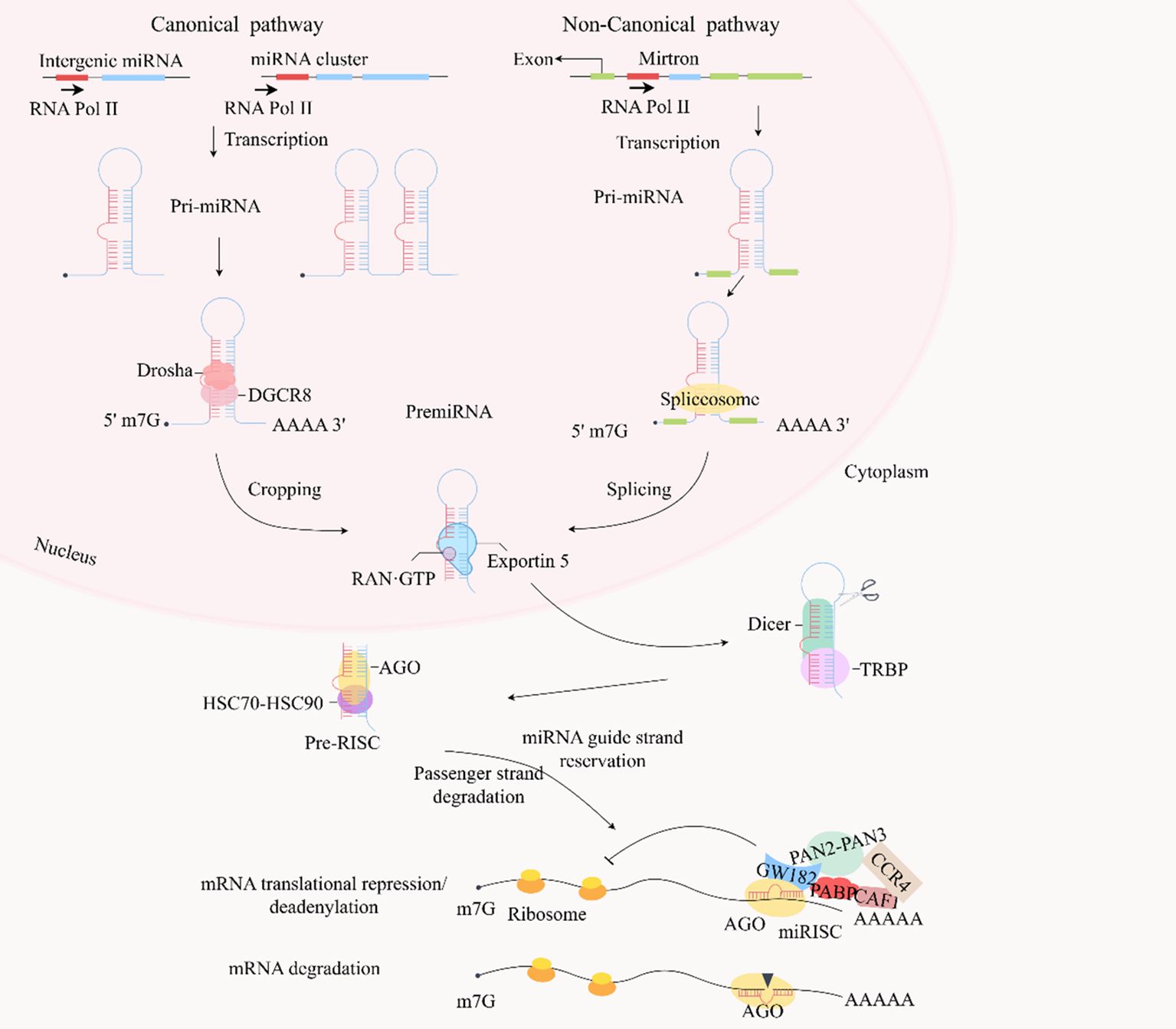
Figure 3. In the canonical pathway, typical miRNA genes are encoded by introns in the transcript, generating single or multiple cis-transcripts, but some miRNAs are encoded by exonic regions. miRNAs in the same cluster are co-transcribed and undergo additional post-transcriptional regulation. Most miRNAs generate primary transcription products (pri-miRNAs) in response to RNA polymerase II, which have the original hairpin structure of the embedded miRNA sequence. The primary precursor (pri-miRNA) is cleaved by the microprocessor complex (including Drosha and DGCR8) into a stem-loop structure of about 70 nucleotides called pre-miRNA. Drosha is an endonuclease responsible for processing and cropping the pri-miRNA, whereas DGCR8 is a protein that binds the pri-miRNA to Drosha. Furthermore, some pre-miRNAs are produced in the nucleus in very short introns (mirtrons) by splicing and debranching without Drosha/DGCR8 processing. The pre-miRNAs are then exported to the cytoplasm via Exportin 5 and RAN-GTP. Dicer in the cytoplasm cleaves the pre-miRNA by TRBP-assisted cleavage of the pre-miRNA, releasing a dsRNA of about 20 bp. The dsRNA is then loaded onto the AGO protein and the HSC70-HSC90 complex. The passenger strand is degraded, and the guide strand is retained in the AGO protein, ultimately forming a RISC. This RISC prevents the initiation of translation by inhibiting ribosome elongation and facilitates de-adenylation of poly(A) by recruiting GW182, PABP, CCR4-CAF1, and PAN2-PAN3 to promote mRNA attenuation. These mRNAs are cleaved and degraded when the RISC can target mRNAs that are nearly fully complementary. dsRNA, double-stranded RNA; miRNA, microRNA; RISC, RNA-induced silencing complex; TRBP, TAR RNA-binding protein.
The pre-miRNA, approximately 70 nucleotides in length, is formed by the Drosha enzyme and exported from the nucleus to the cytoplasm via Exportin 5. In the cytoplasm, it is further processed by Dicer/TAR RNA-binding protein (TRBP)/AGO into double-stranded RNA (dsRNA) consisting of a guide strand and a passenger strand. The guide strand, typically around 22 nucleotides long, enters the miRNA-induced silencing complex (RISC), leading to translational repression or degradation of the target mRNA, whereas the passenger strand is released and subsequently degraded (124, 128, 129). More recently, it was discovered that miRNA biogenesis can occur independently of the conventional Drosha–DGCR8 pathway. Some pre-miRNAs are produced in the nucleus in very short introns by splicing and debranching (130).
3.2 Clustered miRNAs
Approximately 25% of human miRNA genes are organized into clusters, wherein a single cluster contains two or more miRNA genes (131). Although multiple miRNA primary transcripts are generated from the same gene cluster, differential expression arises because of complex regulatory mechanisms. For instance, the 23a–27a–24-2 cluster, comprising three miRNAs, exhibits dysregulation in specific tumors and leukemias, where sometimes only one or two miRNAs are expressed (132). Conversely, some clustered miRNAs show coordinated expression, with a change in a single miRNA gene within the cluster, triggering a chain reaction that affects the other pri/mature miRNAs (133). Current research supports the idea that miRNAs within the same cluster often target overlapping sets of genes, implying enhanced specificity in targeting and increased interconnectedness within the regulatory network (134). miRNA clusters display homogeneity, multiplicity, and paradoxical functions with respect to the roles of individual miRNAs.
3.3 Modes of miRNA regulation
miRNAs serve as fundamental components in RISC, which comprises AGO proteins along with certain cofactors (127). Initially, it was believed that miRNAs exert post-transcriptional control over their targets by regulating processes such as translation elongation, protein degradation, and ribosomal release (135). In mammals, the seed sequence at the 5′ end of the miRNA (nucleotides 2-8) recognizes the 3′ or 5′ UTR of the target mRNA (126). Typically, this recognition involves incomplete base pairing, ultimately leading to cleavage and degradation of the target mRNA. In addition, miRNA-mediated target decay and deadenylation ultimately lead to reduced protein production and fine-tuned gene expression (136).
3.4 Role of miRNAs in transplantation
The use of miRNAs as noninvasive biomarkers has shown promising potential for the diagnosis, prognosis, and treatment of various aspects of organ transplantation, particularly liver transplantation (137).
3.4.1 Liver transplantation
Reperfusion injury is a major concern after liver transplantation and a leading cause of graft failure and rejection (138). Serum miR-122 levels have been proposed as independent markers of persistent liver injury and early liver allograft dysfunction (139). Hepatocyte-derived miR-122 triggers M1 polarization of KCs, exacerbating hepatic IRI by modulating specific pathways (140). The early elevation of serum levels of miRNAs, including miR-122, miR-146a, and miR-192, has shown potential as powerful markers for predicting graft injury and acute rejection after liver transplantation, often preceding changes in transaminase levels (141).
miR-155 plays a role in inflammation, immunity, and tumorigenesis in liver disease. Inhibition of miR-155 expression in KCs results in positive outcomes by activating anti-inflammatory pathways, enhancing the survival of liver allografts, and attenuating inflammatory injury and apoptosis after IRI (142, 143). MiRNAs such as miR-155 and miR-181a may also serve as potential noninvasive biomarkers. Pre-transplant miR-155 levels identified patients at low immunological risk, and the combination of miR-181a and miR-155 levels acted as an early and noninvasive biomarker for preventing acute T cell-mediated rejection (TCMR) and subclinical rejection (144).
These findings suggest that specific miRNAs hold promise as reliable and early markers for assessing graft injury, predicting rejection episodes, and monitoring complications, such as HCC recurrence after liver transplantation. Further research and validation studies could enhance their clinical utility for improving patient outcomes and graft survival.
3.4.2 Renal transplantation
The use of miRNAs as diagnostic and prognostic markers in renal transplantation has shown considerable potential for addressing various aspects of graft health, rejection, and long-term dysfunction.
Recent validation studies have highlighted that miRNAs, including miR-142-5p, miR-142-3p, miR-155 and miR-223, have high specificity in biopsy specimens and help predict TCMR in allogeneic kidney transplantation (145). Interestingly, Pierre’s group identified a variety of miRNAs that interact with the TIM gene, including miR-142-3p and miR-142-5p by analyzing miRNA profiles in kidney allograft samples. However, alloimmune injury pathways are often not unique or specific, and miRNAs such as miR-142-3p or miR-155-5p have been associated not only with IFTA but also with acute rejection or TCMR (146).
miR-21 is a crucial marker of chronic renal dysfunction after transplantation. Silencing miR-21 directly activates Notch2, inhibits the development of renal fibrosis and inflammation, and ultimately prevents chronic allograft dysfunction (147). Changes in miR-21 expression levels in plasma, urine, and graft tissue serve as diagnostic markers for identifying renal injury and dysfunction over time (148).
Moreover, miRNAs, including miR-19a, miR-886-5p, miR-126, miR-223, and miR-24, have been validated as independent predictors of HCC recurrence within the Milan criteria after liver transplantation, aiding the prognosis and management of HCC after transplantation (149).
Richard and colleagues conducted an analysis of microRNA expression in peripheral blood mononuclear cells (PBMC) from patients with chronic antibody-mediated rejection (CAMR) and those with stable graft function, revealing a significant upregulation of miR-142-5p in CAMR (150). This finding was validated and analyzed, indicating that miR-142-5p functions not only as a potential biomarker for CAMR but also plays a role in regulating the immune status of patients.
TCMR, treatable without causing graft failure but associated with chronic or progressive renal dysfunction, has been associated with specific miRNA profiles, aiding in the prediction and understanding of this type of rejection (151).
These findings underscore the potential of miRNAs as noninvasive and specific biomarkers for diagnosing rejection types, monitoring graft health, and predicting chronic dysfunction in renal transplantation. Continued research and validation are essential to refine their clinical utility and enhance their role in improving patient outcomes after transplantation.
3.4.3 Heart transplantation
The role of miRNAs in heart transplantation has emerged as a promising avenue for diagnosing graft rejection, understanding immune responses, and improving outcomes. Recent studies have shed light on the specific miRNAs associated with acute cellular rejection (ACR) and ABMR after heart transplantation.
Identified and validated in 2020, miR-181a-5p showed promise as a marker for ACR in heart transplantation (152). Its specificity and high negative predictive value render it a potential diagnostic tool. A 2021 study identified miR-139-5p, miR-151a-5p, and miR-186-5p as predictive markers for the subsequent development of rejection after heart transplantation (153).
T cell-derived exosomal miR-142-3p is elevated during cardiac allograft rejection, contributing to increased vascular permeability by downregulating the expression of the endothelial Rab11 family of interacting proteins 2 (RAB11FIP2) (154).
miR-146a and miR-155 are involved in the regulation of immune response and rejection mechanisms. Deletion of miR-146a in Tregs exerts tissue-protective effects and transiently prolongs cardiac survival in transplanted mice (155). miR-155 serves as a regulator of allograft rejection by affecting T cell proliferation and macrophage function (142, 156, 157).
Inhibition of miR-155 has shown promising results in suppressing macrophage maturation, downregulating T cell responses, and inducing graft immune tolerance. Using antagomiR-155 delivered through ultrasound-targeted microbubble destruction technology reduces the degree of ACR and improves allogeneic heart survival (158). Ultrasound-guided microbubble disruption technology, capable of delivering cationic microbubbles with miRNA155 silencers to target tissues, is considered a more desirable immunosuppressive therapy for ACR (159).
While these studies highlight the potential of miRNAs as diagnostic markers and therapeutic targets in heart transplantation, further research is necessary to validate these findings in larger cohorts and to standardize diagnostic approaches, considering the heterogeneity of treatment protocols across transplant centers. Developing miRNA-based interventions holds promise for improving rejection detection and for managing post-transplantation outcomes in heart transplantation.
4 Potential associations of TIM proteins with miRNAs
The relationship between miRNAs, specifically miR-155, and the TIM-3 pathway has been extensively studied in the context of various inflammatory and immune responses, including chronic infections and transplantation. However, the direct implications and specific roles of miR-155 and TIM-3 in allograft tolerance and transplantation immunity need to be further elucidated.
miR-155 is a crucial regulator of inflammation and immunity, affecting various immune cell activities such as macrophage polarization, differentiation of T helper cell subsets such as Th17 and Tregs, and cytokine production (160). miR-155 modulates the expression of suppressor of cytokine signaling 1 (SOCS1), a key negative regulator of the JAK–STAT pathway (161). This miRNA can influence macrophage phenotypes, including the M1/M2 balance, and affect the local inflammatory response in certain contexts, such as liver transplantation and hepatic IRI (32, 162, 163).
TIM-3, an inhibitory co-receptor expressed on immune cells, interacts with different ligands such as Gal-9 and plays a role in regulating immune responses (164, 165). Through its interactions, TIM-3 affects T cell polarization, cytokine production, DC maturation, and other immune activities (166, 167). The interplay between TIM-3 and miR-155 has been studied in inflammation and immune regulation, particularly in controlling adaptive and innate immune cell activation. However, direct evidence regarding their roles in allograft tolerance, specifically in transplantation immunity, is yet to be thoroughly investigated. Understanding the specific contributions of miR-155 and TIM-3 in allograft tolerance might offer potential therapeutic avenues for modulating immune responses and improving transplantation outcomes.
The interactions between other miRNAs (miR-142 and miR-330) and members of the TIM family (TIM-1 and TIM-3) have been studied in various contexts, shedding light on their roles in immune regulation, inflammatory responses, and tolerance induction in different physiological settings, including transplantation and maternal-fetal tolerance (23, 26, 59, 168). Studies have shown that miR-142-3p plays a role in modulating TIM-1 transcription, influencing endothelial cell permeability, and reducing systemic inflammatory responses during viral infections (169, 170). It reports that miR-142-3p are upregulated in biopsies from patients with microvascular inflammation typical of Antibody-mediated rejection (ABMR) (171). Elevated miR-142 levels have been observed in patients with cardiac and renal transplant rejection, indicating its potential as a biomarker for monitoring graft rejection. The regulatory function of miR-142 in targeting TGF-Β sensitivity and enhancing Treg development has been linked to promoting cardiac allograft tolerance by targeting Tgfbr1 (168). Contradictory findings have been reported regarding the effects of miR-142 knockdown in specific cells. While Treg-specific knockdown led to severe autoimmune disease, transient knockdown enhanced Treg survival and improved skin graft survival (172). After in situ liver transplantation, TIM-1 blockade not only inhibits macrophage recruitment and infiltration, but also enhances Th2/Treg differentiation and improves IRI (173). TIM-1 signaling, in turn, can maintain and induce baseline levels of Bregs and clear apoptotic cells during transplantation to produce IL-10, which promotes immune tolerance and survival (46). Even TIM-1+ Bregs affect Th differentiation, thereby inhibiting Th1/Th17 cells and promoting Th2 cells and Foxp3+ Tregs, which are dependent on IL-10 expression (174).
miR-330-5p protects against myocardial IRI and apoptosis by modulating TIM-3 transcription and translation, thereby reducing the expression of the inflammatory mediator NLRP3 (24). In a model of myocardial IRI, downregulation of miR-330 inhibited left ventricular remodeling via the TGF-Β1–Smad3 pathway (175). miR-330–TIM-3 interactions promote macrophage M2 polarization, inhibiting local inflammation and insulin resistance (26). TIM-3 activity in innate immune cells, facilitated by miR-330, contributes to trophoblast invasion and angiogenesis, essential for maintaining maternal-fetal tolerance (176).
5 Hypothetical insights from the mechanism process
Although there is no direct evidence in the literature suggesting that miRNA and TIM may play an emerging role in transplantation immunity. However, we seem to be able to propose a plausible hypothesis for such an interaction mechanism through the signaling axis they share.
The miRNA/TIM/TLR signaling axis: In a model of lung transplantation, miR-21 and miR-122 ameliorate graft dysfunction and ischemia-reperfusion injury by negatively regulating the TLR signaling (177). Activation of the TLR signaling pathway alters macrophage miR-21 expression, which influences macrophage polarization status and inflammatory responses (178). The interaction between the two acts as a feedback regulator that modulates the initiation and termination of inflammation, providing a fundamental argument for post-transplant immune regulation (179). Furthermore, in addition to TLRs themselves, miRNAs also regulate TLR-related signaling proteins that regulate related pathways. For example, in Kupffer’s disease, miR-146a/b can act as a negative regulator to control the TLR4 pathway to prevent liver transplant injury by down-regulating IRAK1 and TRAF6 (180). TIM-3 inhibits the production of inflammatory factors associated with the TLR pathway by suppressing NF-ΚB to create an immune-tolerant microenvironment (181). Interestingly, HMGB1 promotes TIM-1Breg cell expansion through TLR2/4 and mitogen-activated protein kinase (MAPK) signaling pathways, providing new evidence for immune tolerance (182). Surprisingly, miRNAs were able to attenuate inflammatory and oxidative responses through the HMGB1/TLR4/NF-ΚB axis (183). Although the relationship between miRNAs and TIM-targeted regulation has long been clear. However, data show that miRNAs bind to mRNAs encoding the 3’-UTR of TIM-3 (36). All these data are sufficient to suggest that the miRNA/TIM/TLR may become a new signaling axis for immune regulation before and after transplantation.
miRNA/TIM/PI3K/AKT signaling axis: The ability of miRNAs to make early prediction and intervention of post-transplantation acute kidney injury through PI3K/AKT signaling pathway was found by prediction (184). miR-21 accelerates wound healing and angiogenesis in grafted skin by activating PI3K/AKT and ERK1/2 signaling (185). Upregulation of miR-221 was able to target PTEN to activate PI3K/AKT to restore contractile function and ameliorate myocardial injury in transplanted myocardium (186). Binding of Gal-9 to Tim-3 can inhibit activation of the PI3K/AKT pathway and enhance the function of Treg cells, thereby attenuating acute GVHD and inducing immune tolerance (187). In the AML model, elevated TIM-3 promotes M2 macrophage polarization, leading to elevated PI3K and AKT levels to accelerate tumor immune escape (188). Through PI3K/AKT signaling, it has long been clear that miRNAs can promote tumor metastasis, immune escape and microenvironmental remodeling (189). MiRNAs have a novel mechanism to balance immune injury and tolerance in viral infection and anti-tumor with respect to TIM signaling capacity in T/NK cells (190). In summary, we believe that induction of immune tolerance and improvement of graft function in the transplant microenvironment are the main themes of this pathway.
TIM/miR/SOCS1 signaling axis: Recent literature suggests that miR-142 and miR-155 exhibit differential expression patterns in the miRNA profiles of kidney transplant samples, with both being upregulated in biopsies from patients exhibiting microvascular inflammation characteristic of rejection (146, 171). Furthermore, it has been demonstrated that miR-155 directly targets SOCS1, thereby promoting immune cell activation and enhancing the immune response (161, 191). Collectively, these findings indicate that modulation of the miR-155/SOCS1 axis may offer novel insights into the mechanisms underlying transplantation immunity. In a similar vein, the miR-142/SOCS1 axis may play a significant role in disease pathogenesis by influencing T cell differentiation and enhancing the secretion of specific cytokines, including IL-6 and IL-8 (192, 193). These effects can adversely impact transplanted organs and elevate the risk of graft rejection. As illustrated in Table 1, existing studies have validated the regulatory roles of miR-142 and miR-155 in the modulation of TIM-1 and TIM-3, respectively.
These findings suggest that intricate interactions between miRNAs and members of the TIM family modulate immune responses, regulate inflammatory processes, and influence tissue-specific responses. These interactions can have diverse effects on various immune cells, leading to implications in transplantation tolerance, inflammation modulation, and maternal-fetal immune regulation. Further studies are required to better understand the precise mechanisms and outcomes of miRNA–TIM interactions in transplantation settings and harness their potential for therapeutic interventions aimed at promoting immune tolerance and mitigating transplant rejection.
6 Discussion
The field of transplantation medicine has evolved substantially over the years, offering life-saving treatments for individuals with organ failure or tissue damage. Despite these advancements, post-transplantation complications remain a considerable challenge. Issues such as graft rejection, IRI, allograft dysfunction, and infections can jeopardize successful organ transplantation. Enhancing long-term graft function and survival outcomes requires a personalized treatment approach tailored to individual immune responses.
The TIM family of proteins is a focal point of transplantation research. Modulation of the TIM pathways using blocking antibodies or soluble proteins has shown promise in altering immune responses. These approaches aim to tilt the balance toward tolerance by providing co-inhibitory signals to T and B cells or suppressing innate immune cells. However, varying affinities and epitopes of TIM antibodies can lead to different T cell effects, resulting in immune cell dysfunction. Moreover, TIM proteins act as receptors for phosphatidylserine, contributing not only to the regulation of innate immunity but also to the control of adaptive immune responses, adding complexity to their roles in transplantation.
miRNAs are key regulators of gene expression and have shown promise in transplantation immunology. Analysis of circulating and tissue-specific miRNAs has suggested them as diagnostic and prognostic biomarkers, offering insights into efficacy and predicting transplantation outcomes. These miRNAs hold the potential as the specific markers for assessing immune responses and status of transplanted organs.
Importantly, a reciprocal regulatory relationship exists between the TIM proteins and miRNAs. TIM proteins can regulate miRNAs through various mechanisms; conversely, miRNAs can influence the expression of TIM proteins. This intricate interplay has been observed in various contexts, including tumorigenesis, viral infections, and metabolic disorders, such as insulin resistance in diabetes mellitus. Exploring and understanding this reciprocal regulation in the context of transplant immune tolerance can offer new avenues for clinical studies and potential therapeutic interventions.
In the realm of future transplantation research in miRNA and TIM, several promising avenues beckon our exploration. Initially, we should focus on the study of specific miRNAs, such as miR-21, miR-155, and miR-133a-5p. Utilizing databases and software like miRWalk and TargetScan, we can predict potential binding sites for these miRNAs. Concurrently, in the context of transplantation, it’s imperative to collect plasma, urine, or tissue samples from patients before and after transplantation or drug administration. These samples can undergo miRNA sequencing, followed by screening and validation of differentially expressed genes. To investigate downstream signaling molecule alterations, protein microarrays can be employed to identify differential proteins, which can then be verified using luciferase reporter genes for miRNA binding to the 3’UTR of genes.
Furthermore, the expression patterns of miRNAs may vary between different transplanted organs, indicating tissue-specific regulatory mechanisms. Hence, we should prioritize the study of post-transplantation immunomodulatory capacity on miRNA. This includes the regulation of immune cell function and response strength in adaptive immunity (T/B cells) and innate immunity (NK and macrophages). In terms of signaling pathways, our focus should be on influencing cell differentiation/activation/effector function, integrating transcriptomic, proteomic, and other multi-omics data with experimental validation for comprehensive analysis and screening.
Ultimately, leveraging the regulatory role of miRNAs, it’s crucial to devise novel therapeutic strategies for a safe and effective approach to the transplantation site. Nanoparticle delivery technology can be utilized to transport specific immunomodulatory genes to transplanted tissues, thereby inducing local immunosuppressive cytokine production and fostering immune tolerance. Additionally, considering the fragility of miRNAs, Ultrasound Targeted Microbubbles Destruction offers a non-invasive, targeted gene delivery technique that is safe, efficient, and specific.
In summary, the intersection between TIM proteins and miRNAs represents a promising area for further investigation of transplantation immune tolerance. Understanding the complex interplay between these molecules and their regulatory roles may lead to innovative therapeutic strategies aimed at promoting immune tolerance and improving long-term outcomes in transplant recipients.
Author contributions
JT: Writing – original draft. XS: Software, Writing – original draft. HQ: Data curation, Writing – original draft. QD: Writing – review & editing. LW: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82305289), Science and Technology Development Plan of Suzhou (SKY2023078), Zhangjiagang Science and Technology Bureau Healthcare Guidance Project (ZKYL2313), and Zhangjiagang Health Youth Science and Technology Project (ZJGQNKJ202208), and Science and Technology Development Plan of Suzhou(KJXW2021066).
Acknowledgments
This is a short text to acknowledge the contributions of Zhangjiagang TCM Hospital Affiliated to Nanjing University of Chinese Medicine and QD that aided the efforts of the authors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Duneton C, Winterberg PD, Ford ML. Activation and regulation of alloreactive T cell immunity in solid organ transplantation. Nat Rev Nephrol. (2022) 18:663–76. doi: 10.1038/s41581-022-00600-0
2. Griffith BP, Shah A, Awad MA. Current status and outcomes in heart transplantation: A narrative review. Rev Cardiovasc Med. (2022) 23:11. doi: 10.31083/j.rcm2301011
3. Roberts MB, Fishman JA. Immunosuppressive agents and infectious risk in transplantation: managing the “Net state of immunosuppression. Clin Infect Dis. (2021) 73:e1302–e17. doi: 10.1093/cid/ciaa1189
4. Jane-Wit D, Fang C, Goldstein DR. Innate immune mechanisms in transplant allograft vasculopathy. Curr Opin Organ Transplant. (2016) 21:253–7. doi: 10.1097/MOT.0000000000000314
5. Dreyer CH, Rasmussen M, Pedersen RH, Overgaard S, Ding M. Comparisons of efficacy between autograft and allograft on defect repair in vivo in normal and osteoporotic rats. BioMed Res Int. (2020) 2020:9358989. doi: 10.1155/2020/9358989
6. Loupy A, Lefaucheur C. Antibody-mediated rejection of solid-organ allografts. N Engl J Med. (2018) 379:1150–60. doi: 10.1056/NEJMra1802677
7. Rodriguez-Manzanet R, DeKruyff R, Kuchroo VK, Umetsu DT. The costimulatory role of tim molecules. Immunol Rev. (2009) 229:259–70. doi: 10.1111/j.1600-065X.2009.00772.x
8. Du P, Xiong R, Li X, Jiang J. Immune regulation and antitumor effect of tim-1. J Immunol Res. (2016) 2016:8605134. doi: 10.1155/2016/8605134
9. Freeman GJ, Casasnovas JM, Umetsu DT, DeKruyff RH. Tim genes: A family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol Rev. (2010) 235:172–89. doi: 10.1111/j.0105-2896.2010.00903.x
10. Tang R, Rangachari M, Kuchroo VK. Tim-3: A co-receptor with diverse roles in T cell exhaustion and tolerance. Semin Immunol. (2019) 42:101302. doi: 10.1016/j.smim.2019.101302
11. Banerjee H, Kane LP. Immune regulation by tim-3. F1000Res. (2018) 7:316. doi: 10.12688/f1000research.13446.1
12. Liu Y, Chen H, Chen Z, Qiu J, Pang H, Zhou Z. Novel roles of the tim family in immune regulation and autoimmune diseases. Front Immunol. (2021) 12:748787. doi: 10.3389/fimmu.2021.748787
13. Liu W, Xu L, Liang X, Liu X, Zhao Y, Ma C, et al. Tim-4 in health and disease: friend or foe? Front Immunol. (2020) 11:537. doi: 10.3389/fimmu.2020.00537
14. Ueno T, Habicht A, Clarkson MR, Albin MJ, Yamaura K, Boenisch O, et al. The emerging role of T cell ig mucin 1 in alloimmune responses in an experimental mouse transplant model. J Clin Invest. (2008) 118:742–51. doi: 10.1172/JCI32451
15. Thornley TB, Fang Z, Balasubramanian S, Larocca RA, Gong W, Gupta S, et al. Fragile tim-4-expressing tissue resident macrophages are migratory and immunoregulatory. J Clin Invest. (2014) 124:3443–54. doi: 10.1172/JCI73527
16. Gupta S, Thornley TB, Gao W, Larocca R, Turka LA, Kuchroo VK, et al. Allograft rejection is restrained by short-lived tim-3+Pd-1+Foxp3+ Tregs. J Clin Invest. (2012) 122:2395–404. doi: 10.1172/JCI45138
17. Hill M, Tran N. Mirna interplay: mechanisms and consequences in cancer. Dis Model Mech. (2021) 14:dmm047662. doi: 10.1242/dmm.047662
18. Harden JT, Krams SM. Micro-rnas in transplant tolerance. Curr Opin Organ Transplant. (2018) 23:66–72. doi: 10.1097/mot.0000000000000479
19. Hamdorf M, Kawakita S, Everly M. The potential of micrornas as novel biomarkers for transplant rejection. J Immunol Res. (2017) 2017:4072364. doi: 10.1155/2017/4072364
20. Iliopoulou BP, Hsu K, Perez-Cruz M, Tang SW, Pang WW, Erkers T, et al. Blockade of tim-1 on the donor graft ameliorates graft-versus-host disease following hematopoietic cell transplantation. Blood Adv. (2019) 3:3419–31. doi: 10.1182/bloodadvances.2019000286
21. Wei L, Peng Y, Shao N, Zhou P. Downregulation of tim-1 inhibits the proliferation, migration and invasion of glioblastoma cells via the mir-133a/tgfbr1 axis and the restriction of wnt/beta-catenin pathway. Cancer Cell Int. (2021) 21:347. doi: 10.1186/s12935-021-02036-1
22. Moghaddam Y, Andalib A, Mohammad-Ganji M, Homayouni V, Sharifi M, Ganjalikhani-Hakemi M. Evaluation of the effect of tim-3 suppression by mir-498 and its effect on apoptosis and proliferation rate of hl-60 cell line. Pathol Res Pract. (2018) 214:1482–8. doi: 10.1016/j.prp.2018.07.019
23. Kansakar U, Gambardella J, Varzideh F, Avvisato R, Jankauskas SS, Mone P, et al. Mir-142 targets tim-1 in human endothelial cells: potential implications for stroke, Covid-19, zika, ebola, dengue, and other viral infections. Int J Mol Sci. (2022) 23:10242. doi: 10.3390/ijms231810242
24. Zuo W, Tian R, Chen Q, Wang L, Gu Q, Zhao H, et al. Mir-330-5p inhibits nlrp3 inflammasome-mediated myocardial ischemia–reperfusion injury by targeting tim3. Cardiovasc Drugs Ther. (2020) 35:691–705. doi: 10.1007/s10557-020-07104-8
25. Fooladinezhad H, Khanahmad H, Ganjalikhani-Hakemi M, Doosti A. Negative regulation of tim-3 expression in aml cell line (Hl-60) using mir-330-5p. Br J Biomed Sci. (2016) 73:129–33. doi: 10.1080/09674845.2016.1194564
26. Sun J, Huang Q, Li S, Meng F, Li X, Gong X. Mir-330-5p/tim-3 axis regulates macrophage M2 polarization and insulin resistance in diabetes mice. Mol Immunol. (2018) 95:107–13. doi: 10.1016/j.molimm.2018.02.006
27. Emamdoost F, Khanahmad H, Ganjalikhani-hakemi M, Doosti A. The mir-125a-3p inhibits tim-3 expression in aml cell line hl-60 in vitro. Indian J Hematol Blood Transfusion. (2016) 33:342–7. doi: 10.1007/s12288-016-0733-4
28. Yang Z, Shan N, Deng Q, Wang Y, Hou Y, Mei J, et al. Extracellular vesicle-derived microrna-18b ameliorates preeclampsia by enhancing trophoblast proliferation and migration via notch2/tim3/mtorc1 axis. J Cell Mol Med. (2021) 25:4583–95. doi: 10.1111/jcmm.16234
29. Wang Y, Cheng Q, Liu J, Dong M. Leukemia stem cell-released microvesicles promote the survival and migration of myeloid leukemia cells and these effects can be inhibited by microrna34a overexpression. Stem Cells Int. (2016) 2016:1–8. doi: 10.1155/2016/9313425
30. Yan K, Fu Y, Zhu N, Wang Z, Hong J-L, Li Y, et al. Repression of lncrna neat1 enhances the antitumor activity of cd8+T cells against hepatocellular carcinoma via regulating mir-155/tim-3. Int J Biochem Cell Biol. (2019) 110:1–8. doi: 10.1016/j.biocel.2019.01.019
31. Zhang F, Zhao J, Sun D, Wei N. Mir-155 inhibits transformation of macrophages into foam cells via regulating ceh expression. Biomedicine Pharmacotherapy. (2018) 104:645–51. doi: 10.1016/j.biopha.2018.05.068
32. Jiang X, Zhou T, Xiao Y, Yu J, Dou S, Chen G, et al. Tim-3 promotes tumor-promoting M2 macrophage polarization by binding to stat1 and suppressing the stat1-mir-155 signaling axis. OncoImmunology. (2016) 5:1850–7. doi: 10.1080/2162402x.2016.1211219
33. Yan M, Zhang Y, Chang S. Chitosan nanoparticles loaded with tgf-Β1 inhibit cervical cancer cell progression through down-regulation of microrna-155 and activation of tim-3 pathway. J Biomed Nanotechnology. (2021) 17:1850–7. doi: 10.1166/jbn.2021.3146
34. Zhou J, Jiang Y, Zhang H, Chen L, Luo P, Li L, et al. Clinicopathological implications of tim3+ Tumor-infiltrating lymphocytes and the mir-455-5p/galectin-9 axis in skull base chordoma patients. Cancer Immunology Immunotherapy. (2019) 68:1157–69. doi: 10.1007/s00262-019-02349-1
35. Ai Y, Wu S, Gao H, Wei H, Tang Z, Li X, et al. Repression of Crnde Enhances the Anti-Tumor Activity of Cd8 + t Cells against Oral Squamous Cell Carcinoma through Regulating Mir-545-5p and Tim-3. J Cell Mol Med. (2021) 25:10857–68. doi: 10.1111/jcmm.16909
36. Zhang M, Gao D, Shi Y, Wang Y, Joshi R, Yu Q, et al. Mir-149-3p reverses cd8+T-cell exhaustion by reducing inhibitory receptors and promoting cytokine secretion in breast cancer cells. Open Biol. (2019) 9:190061. doi: 10.1098/rsob.190061
37. Hojati Z, Ganjalikhani-Hakemi M, Ameri M, Alimohammadi-Jelodar SF, Dehbashi M, Mohammad Ganji M, et al. Evaluation of silencing effect of mir-133a-5p mimic on tim-3 expression in aml (Hl-60) cell line. Indian J Clin Biochem. (2019) 35:359–66. doi: 10.1007/s12291-019-00834-z
38. Yu T, Ju Z, Luo M, Hu R, Teng Y, Xie L, et al. Elevated expression of mir-146a correlates with high levels of immune cell exhaustion markers and suppresses cellular immune function in chronic hiv-1-infected patients. Sci Rep. (2019) 9:18829. doi: 10.1038/s41598-019-55100-2
39. Xu C, Zhai J, Fu Y. Overexpression of nuclear enriched autosomal transcript 1 facilitates cell proliferation, migration invasion, and suppresses apoptosis in endometrial cancer by targeting microrna-202-3p/T cell immunoglobulin and mucin domain 4 axis. Cancer Biotherapy Radiopharmaceuticals. (2022) 37:815–23. doi: 10.1089/cbr.2020.3902
40. Kuchroo VK, Umetsu DT, DeKruyff RH, Freeman GJ. The tim gene family: emerging roles in immunity and disease. Nat Rev Immunol. (2003) 3:454–62. doi: 10.1038/nri1111
41. Kane LP. Tim family proteins and autoimmunity. Autoimmunity. (2007) 40:405–8. doi: 10.1080/08916930701464871
42. Meyers JH, Sabatos CA, Chakravarti S, Kuchroo VK. The tim gene family regulates autoimmune and allergic diseases. Trends Mol Med. (2005) 11:362–9. doi: 10.1016/j.molmed.2005.06.008
43. McGrath MM. Diverse roles of tim4 in immune activation: implications for alloimmunity. Curr Opin Organ Transplant. (2018) 23:44–50. doi: 10.1097/MOT.0000000000000487
44. Tami C, Silberstein E, Manangeeswaran M, Freeman GJ, Umetsu SE, DeKruyff RH, et al. Immunoglobulin a (Iga) is a natural ligand of hepatitis a virus cellular receptor 1 (Havcr1), and the association of iga with havcr1 enhances virus-receptor interactions. J Virol. (2007) 81:3437–46. doi: 10.1128/JVI.01585-06
45. Angiari S, Constantin G. Regulation of T cell trafficking by the T cell immunoglobulin and mucin domain 1 glycoprotein. Trends Mol Med. (2014) 20:675–84. doi: 10.1016/j.molmed.2014.10.003
46. Yeung MY, Ding Q, Brooks CR, Xiao S, Workman CJ, Vignali DA, et al. Tim-1 signaling is required for maintenance and induction of regulatory B cells. Am J Transplant. (2015) 15:942–53. doi: 10.1111/ajt.13087
47. Bod L, Kye Y-C, Shi J, Torlai Triglia E, Schnell A, Fessler J, et al. B-cell-specific checkpoint molecules that regulate anti-tumor immunity. Nature. (2023) 619:348–56. doi: 10.1038/s41586-023-06231-0
48. Wang J, Qiao L, Hou Z, Luo G, Ou JHJ. Tim-1 promotes hepatitis C virus cell attachment and infection. J Virol. (2017) 91:e01583-16. doi: 10.1128/jvi.01583-16
49. Liu Y, Chen Z, Qiu J, Chen H, Zhou Z. Altered tim-1 and il-10 expression in regulatory B cell subsets in type 1 diabetes. Front Immunol. (2021) 12:773896. doi: 10.3389/fimmu.2021.773896
50. Degauque N, Mariat C, Kenny J, Zhang D, Gao W, Vu MD, et al. Immunostimulatory tim-1–specific antibody deprograms tregs and prevents transplant tolerance in mice. J Clin Invest. (2008) 118:735–41. doi: 10.1172/jci32562
51. Cherukuri A, Rothstein DM. Regulatory and transitional B cells: potential biomarkers and therapeutic targets in organ transplantation. Curr Opin Organ Transplant. (2022) 27:385–91. doi: 10.1097/MOT.0000000000001010
52. Ding Q, Yeung M, Camirand G, Zeng Q, Akiba H, Yagita H, et al. Regulatory B cells are identified by expression of tim-1 and can be induced through tim-1 ligation to promote tolerance in mice. J Clin Invest. (2011) 121:3645–56. doi: 10.1172/jci46274
53. Li S, Li X, Yang M, Wei L, Wei L, Deng S, et al. Identification of the subsets of il-10-producing regulatory B cells in the course of tolerance induction and maintenance in islet allotransplantation. Transplant Proc. (2018) 50:3900–5. doi: 10.1016/j.transproceed.2018.04.065
54. Lee KM, Kim JI, Stott R, Soohoo J, O'Connor MR, Yeh H, et al. Anti-cd45rb/anti-tim-1-induced tolerance requires regulatory B cells. Am J Transplant. (2012) 12:2072–8. doi: 10.1111/j.1600-6143.2012.04055.x
55. Shankar S, Stolp J, Juvet SC, Beckett J, Macklin PS, Issa F, et al. Ex vivo-expanded human cd19+Tim-1+ Regulatory B cells suppress immune responses in vivo and are dependent upon the tim-1/stat3 axis. Nat Commun. (2022) 13:3121. doi: 10.1038/s41467-022-30613-z
56. Zheng Y, Wang L, Chen M, Liu L, Pei A, Zhang R, et al. Inhibition of T cell immunoglobulin and mucin-1 (Tim-1) protects against cerebral ischemia-reperfusion injury. Cell Communication Signaling. (2019) 17:112. doi: 10.1186/s12964-019-0417-4
57. Huang YH, Zhu C, Kondo Y, Anderson AC, Gandhi A, Russell A, et al. Ceacam1 regulates tim-3-mediated tolerance and exhaustion. Nature. (2015) 517:386–90. doi: 10.1038/nature13848
58. Zhao L, Cheng S, Fan L, Zhang B, Xu S. Tim-3: an update on immunotherapy. Int Immunopharmacol. (2021) 99:107933. doi: 10.1016/j.intimp.2021.107933
59. Li M, Sun F, Xu Y, Chen L, Chen C, Cui L, et al. Tim-3(+) decidual mphis induced th2 and treg bias in decidual cd4(+)T cells and promoted pregnancy maintenance via cd132. Cell Death Dis. (2022) 13:454. doi: 10.1038/s41419-022-04899-2
60. Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, et al. The tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. (2005) 6:1245–52. doi: 10.1038/ni1271
61. Rong S, Park JK, Kirsch T, Yagita H, Akiba H, Boenisch O, et al. The tim-1:Tim-4 pathway enhances renal ischemia-reperfusion injury. J Am Soc Nephrol. (2011) 22:484–95. doi: 10.1681/ASN.2010030321
62. Meyers JH, Chakravarti S, Schlesinger D, Illes Z, Waldner H, Umetsu SE, et al. Tim-4 is the ligand for tim-1, and the tim-1-tim-4 interaction regulates T cell proliferation. Nat Immunol. (2005) 6:455–64. doi: 10.1038/ni1185
63. Shim JA, Lee E-S, Choi B, Sohn S. The role of T cell immunoglobulin mucin domains 1 and 4 in a herpes simplex virus-induced behçet’s disease mouse model. Mediators Inflammation. (2013) 2013:1–13. doi: 10.1155/2013/903948
64. Guo Y-Y, Yin C-J, Zhao M, Guo L-T, Su R-F, Fu X-X, et al. Effect of rmt1-10 on the immunological characteristics of dendritic cells cultured in vitro and corneal european review for medical and pharmacological sciences. Eur Rev Med Pharmacol Sci. (2019) 23:9150–62. doi: 10.26355/eurrev_201911_19405
65. Feng B-S, Chen X, He S-H, Zheng P-Y, Foster J, Xing Z, et al. Disruption of T-cell immunoglobulin and mucin domain molecule (Tim)–1/tim4 interaction as a therapeutic strategy in a dendritic cell–induced peanut allergy model. J Allergy Clin Immunol. (2008) 122:55–61.e7. doi: 10.1016/j.jaci.2008.04.036
66. Corneillie L, Lemmens I, Montpellier C, Ferrie M, Weening K, Van Houtte F, et al. The phosphatidylserine receptor tim1 promotes infection of enveloped hepatitis E virus. Cell Mol Life Sci. (2023) 80:326. doi: 10.1007/s00018-023-04977-4
67. Jayasinghe MK, Gao C, Yap G, Yeo BZJ, Vu LT, Tay DJW, et al. Red blood cell-derived extracellular vesicles display endogenous antiviral effects and enhance the efficacy of antiviral oligonucleotide therapy. ACS Nano. (2023) 17:21639–61. doi: 10.1021/acsnano.3c06803
68. Thio CL-P, Lai AC-Y, Wang J-C, Chi P-Y, Chang Y-L, Ting Y-T, et al. Identification of a pd-L1+Tim-1+ Inkt subset that protects against fine particulate matter–induced airway inflammation. JCI Insight. (2022) 7:e164157. doi: 10.1172/jci.insight.164157
69. Angiari S, Donnarumma T, Rossi B, Dusi S, Pietronigro E, Zenaro E, et al. Tim-1 glycoprotein binds the adhesion receptor P-selectin and mediates T cell trafficking during inflammation and autoimmunity. Immunity. (2014) 40:542–53. doi: 10.1016/j.immuni.2014.03.004
70. Mori Y, Ajay AK, Chang JH, Mou S, Zhao H, Kishi S, et al. Kim-1 mediates fatty acid uptake by renal tubular cells to promote progressive diabetic kidney disease. Cell Metab. (2021) 33:1042–61.e7. doi: 10.1016/j.cmet.2021.04.004
71. Yamanishi Y, Kitaura J, Izawa K, Kaitani A, Komeno Y, Nakamura M, et al. Tim1 is an endogenous ligand for lmir5/cd300b: lmir5 deficiency ameliorates mouse kidney ischemia/reperfusion injury. J Exp Med. (2010) 207:1501–11. doi: 10.1084/jem.20090581
72. Liu Y, Ji H, Zhang Y, Shen X, Gao F, He X, et al. Recipient T cell tim-3 and hepatocyte galectin-9 signaling protects mouse liver transplants against ischemia-reperfusion injury. J Hepatol. (2015) 62:563–72. doi: 10.1016/j.jhep.2014.10.034
73. Li YH, Zhou WH, Tao Y, Wang SC, Jiang YL, Zhang D, et al. The galectin-9/tim-3 pathway is involved in the regulation of nk cell function at the maternal-fetal interface in early pregnancy. Cell Mol Immunol. (2016) 13:73–81. doi: 10.1038/cmi.2014.126
74. Yang R, Sun L, Li CF, Wang YH, Yao J, Li H, et al. Galectin-9 interacts with pd-1 and tim-3 to regulate T cell death and is a target for cancer immunotherapy. Nat Commun. (2021) 12:832. doi: 10.1038/s41467-021-21099-2
75. Ni X, Wu W, Sun X, Ma J, Yu Z, He X, et al. Interrogating glioma-M2 macrophage interactions identifies gal-9/tim-3 as a viable target against pten-null glioblastoma. Sci Adv. (2022) 27:eabl5165. doi: 10.14791/btrt.2022.10.Suppl
76. Folgiero V, Cifaldi L, Li Pira G, Goffredo BM, Vinti L, Locatelli F. Tim-3/gal-9 interaction induces ifngamma-dependent ido1 expression in acute myeloid leukemia blast cells. J Hematol Oncol. (2015) 8:36. doi: 10.1186/s13045-015-0134-4
77. Goncalves Silva I, Yasinska IM, Sakhnevych SS, Fiedler W, Wellbrock J, Bardelli M, et al. The tim-3-galectin-9 secretory pathway is involved in the immune escape of human acute myeloid leukemia cells. EBioMedicine. (2017) 22:44–57. doi: 10.1016/j.ebiom.2017.07.018
78. Dolina JS, Braciale TJ, Hahn YS. Liver-primed cd8+ T cells suppress antiviral adaptive immunity through galectin-9-independent T-cell immunoglobulin and mucin 3 engagement of high-mobility group box 1 in mice. Hepatology. (2014) 59:1351–65. doi: 10.1002/hep.26938
79. Yasinska IM, Goncalves Silva I, Sakhnevych SS, Ruegg L, Hussain R, Siligardi G, et al. High mobility group box 1 (Hmgb1) acts as an "Alarmin" to promote acute myeloid leukemia progression. Oncoimmunology. (2018) 7:e1438109. doi: 10.1080/2162402X.2018.1438109
80. Huang S, Liu D, Sun J, Zhang H, Zhang J, Wang Q, et al. Tim-3 regulates sepsis-induced immunosuppression by inhibiting the nf-kappab signaling pathway in cd4 T cells. Mol Ther. (2022) 30:1227–38. doi: 10.1016/j.ymthe.2021.12.013
81. Kojima H, Kadono K, Hirao H, Dery KJ, Torgerson T, Yao S, et al. T cell ceacam1-tim-3 crosstalk alleviates liver transplant injury in mice and humans. Gastroenterology. (2023) 165:1233–48.e9. doi: 10.1053/j.gastro.2023.07.004
82. Lake CM, Voss K, Bauman BM, Pohida K, Jiang T, Dveksler G, et al. Tim-3 drives temporal differences in restimulation-induced cell death sensitivity in effector cd8(+) T cells in conjunction with ceacam1. Cell Death Dis. (2021) 12:400. doi: 10.1038/s41419-021-03689-6
83. Wu M, Wu A, Zhang X, Li Y, Li B, Jin S, et al. Identification of a novel small-molecule inhibitor targeting tim-3 for cancer immunotherapy. Biochem Pharmacol. (2023) 212:115583. doi: 10.1016/j.bcp.2023.115583
84. Jiang W, Li F, Jiang Y, Li S, Liu X, Xu Y, et al. Tim-3 blockade elicits potent anti-multiple myeloma immunity of natural killer cells. Front Oncol. (2022) 12:739976. doi: 10.3389/fonc.2022.739976
85. Matsumoto H, Fujita Y, Onizawa M, Saito K, Sumichika Y, Yoshida S, et al. Increased ceacam1 expression on peripheral blood neutrophils in patients with rheumatoid arthritis. Front Immunol. (2022) 13:978435. doi: 10.3389/fimmu.2022.978435
86. Kristenson L, Badami C, Ljungberg A, Islamagic E, Tian Y, Xie G, et al. Deletion of the tmem30a gene enables leukemic cell evasion of nk cell cytotoxicity. Proc Natl Acad Sci U.S.A. (2024) 121:e2316447121. doi: 10.1073/pnas.2316447121
87. Ma S, Tian Y, Peng J, Chen C, Peng X, Zhao F, et al. Identification of a small-molecule tim-3 inhibitor to potentiate T cell–mediated antitumor immunotherapy in preclinical mouse models. Sci Transl Med. (2023) 15:6752. doi: 10.1126/scitranslmed.adg6752
88. Zhao P, Xu L, Wang P, Liang X, Qi J, Liu P, et al. Increased expression of human T-cell immunoglobulin- and mucin-domain-containing molecule-4 in peripheral blood mononuclear cells from patients with system lupus erythematosus. Cell Mol Immunol. (2010) 7:152–6. doi: 10.1038/cmi.2009.118
89. Ding Q, Mohib K, Kuchroo VK, Rothstein DM. Tim-4 identifies ifn-Γ–expressing proinflammatory B effector 1 cells that promote tumor and allograft rejection. J Immunol. (2017) 199:2585–95. doi: 10.4049/jimmunol.1602107
90. Hu T, Fan X, Ma L, Liu J, Chang Y, Yang P, et al. Tim4-tim1 interaction modulates th2 pattern inflammation through enhancing sirt1 expression. Int J Mol Med. (2017) 40:1504–10. doi: 10.3892/ijmm.2017.3150
91. Rhein BA, Brouillette RB, Schaack GA, Chiorini JA, Maury W. Characterization of human and murine T-cell immunoglobulin mucin domain 4 (Tim-4) igv domain residues critical for ebola virus entry. J Virol. (2016) 90:6097–111. doi: 10.1128/JVI.00100-16
92. Sims B, Farrow A, Williams S, Bansal A, Krendelchtchikov A, Gu L, et al. Role of tim-4 in exosome-dependent entry of hiv-1 into human immune cells. Int J Nanomedicine. (2017) 12:4823–33. doi: 10.2147/ijn.S132762
93. Kobayashi N, Karisola P, Pena-Cruz V, Dorfman DM, Jinushi M, Umetsu SE, et al. Tim-1 and tim-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. (2007) 27:927–40. doi: 10.1016/j.immuni.2007.11.011
94. Sanchez-Fueyo A, Tian J, Picarella D, Domenig C, Zheng XX, Sabatos CA, et al. Tim-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nat Immunol. (2003) 4:1093–101. doi: 10.1038/ni987
95. Chou F-C, Kuo C-C, Wang Y-L, Lin M-H, Yen BL, Chang D-M, et al. Overexpression of galectin-9 in islets prolongs grafts survival via downregulation of th1 responses. Cell Transplant. (2013) 22:2135–45. doi: 10.3727/096368912x657891
96. Gleason MK, Lenvik TR, McCullar V, Felices M, O'Brien MS, Cooley SA, et al. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood. (2012) 119:3064–72. doi: 10.1182/blood-2011-06-360321
97. Li YM, Shi YY, Li Y, Yan L, Tang JT, Bai YJ, et al. Soluble tim-3 and gal-9 are associated with renal allograft dysfunction in kidney transplant recipients: A cross-sectional study. Int Immunopharmacol. (2018) 55:330–5. doi: 10.1016/j.intimp.2018.01.008
98. Shahbaz SK, Barabadi M, Ahmadpour P, Pourrezagholi F, Nafar M, Foroughi F, et al. Sequential monitoring of tim-3 mrna expression in blood and urine samples of renal transplant recipients. Transplant Immunol. (2019) 54:9–16. doi: 10.1016/j.trim.2018.10.007
99. Schietinger A, Greenberg PD. Tolerance and exhaustion: defining mechanisms of T cell dysfunction. Trends Immunol. (2014) 35:51–60. doi: 10.1016/j.it.2013.10.001
100. Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting tim-3 and pd-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. (2010) 207:2187–94. doi: 10.1084/jem.20100643
101. Mysore KR, Ghobrial RM, Kannanganat S, Minze LJ, Graviss EA, Nguyen DT, et al. Longitudinal assessment of T cell inhibitory receptors in liver transplant recipients and their association with posttransplant infections. Am J Transplant. (2018) 18:351–63. doi: 10.1111/ajt.14546
102. Savill J, Gregory C. Apoptotic ps to phagocyte tim-4: eat me. Immunity. (2007) 27:830–2. doi: 10.1016/j.immuni.2007.12.002
103. Park D, Hochreiter-Hufford A, Ravichandran KS. The phosphatidylserine receptor tim-4 does not mediate direct signaling. Curr Biol. (2009) 19:346–51. doi: 10.1016/j.cub.2009.01.042
104. Foks AC, Engelbertsen D, Kuperwaser F, Alberts-Grill N, Gonen A, Witztum JL, et al. Blockade of tim-1 and tim-4 enhances atherosclerosis in low-density lipoprotein receptor–deficient mice. Arteriosclerosis Thrombosis Vasc Biol. (2016) 36:456–65. doi: 10.1161/atvbaha.115.306860
105. Rodriguez-Manzanet. R, Meyers. JH, Balasubramanian. S, Slavik. J, Kassam. N, Dardalhon. V, et al. Tim-4 expressed on antigen-presenting cells induces T cell expansion and survival. J Immunol. (2008) 180:4706–13. doi: 10.4049/jimmunol.180.7.4706
106. Chow A, SChad S, Green MD, Hellmann MD, Allaj V, Ceglia N, et al. Tim-4(+) cavity-resident macrophages impair anti-tumor cd8(+) T cell immunity. Cancer Cell. (2021) 39:973–88.e9. doi: 10.1016/j.ccell.2021.05.006
107. Locati M, Curtale G, Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev Pathol. (2020) 15:123–47. doi: 10.1146/annurev-pathmechdis-012418-012718
108. Ezzelarab M, Thomson AW. Tolerogenic dendritic cells and their role in transplantation. Semin Immunol. (2011) 23:252–63. doi: 10.1016/j.smim.2011.06.007
109. Uchida Y, Ke B, Freitas MC, Ji H, Zhao D, Benjamin ER, et al. The emerging role of T cell immunoglobulin mucin-1 in the mechanism of liver ischemia and reperfusion injury in the mouse. Hepatology. (2010) 51:1363–72. doi: 10.1002/hep.23442
110. Ni M, Zhang J, Sosa R, Zhang H, Wang H, Jin D, et al. T-cell immunoglobulin and mucin domain-containing protein-4 is critical for kupffer cell homeostatic function in the activation and resolution of liver ischemia reperfusion injury. Hepatology. (2021) 74:2118–32. doi: 10.1002/hep.31906
111. Zhang Y, Shen Q, Liu Y, Chen H, Zheng X, Xie S, et al. Hepatic ischemic preconditioning alleviates ischemia-reperfusion injury by decreasing tim4 expression. Int J Biol Sci. (2018) 14:1186–95. doi: 10.7150/ijbs.24898
112. Chen Y, Liu Z, Liang S, Luan X, Long F, Chen J, et al. Role of kupffer cells in the induction of tolerance of orthotopic liver transplantation in rats. Liver Transpl. (2008) 14:823–36. doi: 10.1002/lt.21450
113. Chen GS, Qi HZ. Effect of kupffer cells on immune tolerance in liver transplantation. Asian Pac J Trop Med. (2012) 5:970–2. doi: 10.1016/S1995-7645(12)60184-9
114. Wu H, Xu X, Li J, Gong J, Li M. Tim−4 blockade of kcs combined with exogenous tgf−Β Injection helps to reverse acute rejection and prolong the survival rate of mice receiving liver allografts. Int J Mol Med. (2018) 42:346–58. doi: 10.3892/ijmm.2018.3606
115. Rodriguez-Manzanet R, Sanjuan MA, Wu HY, Quintana FJ, Xiao S, Anderson AC, et al. T and B cell hyperactivity and autoimmunity associated with niche-specific defects in apoptotic body clearance in tim-4-deficient mice. Proc Natl Acad Sci U.S.A. (2010) 107:8706–11. doi: 10.1073/pnas.0910359107
116. Waisman A, Lukas D, Clausen BE, Yogev N. Dendritic cells as gatekeepers of tolerance. Semin Immunopathol. (2017) 39:153–63. doi: 10.1007/s00281-016-0583-z
117. Yeung MY, McGrath MM, Nakayama M, Shimizu T, Boenisch O, Magee CN, et al. Interruption of dendritic cell-mediated tim-4 signaling induces regulatory T cells and promotes skin allograft survival. J Immunol. (2013) 191:4447–55. doi: 10.4049/jimmunol.1300992
118. Kerr D, Tietjen GT, Gong Z, Tajkhorshid E, Adams EJ, Lee KYC. Sensitivity of peripheral membrane proteins to the membrane context: A case study of phosphatidylserine and the tim proteins. Biochim Biophys Acta Biomembranes. (2018) 1860:2126–33. doi: 10.1016/j.bbamem.2018.06.010
119. Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. (2010) 39:407–27. doi: 10.1146/annurev.biophys.093008.131234
120. Uribe-Querol E, Rosales C. Phagocytosis: our current understanding of a universal biological process. Front Immunol. (2020) 11:1066. doi: 10.3389/fimmu.2020.01066
121. Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. (2008) 118:1657–68. doi: 10.1172/JCI34487
122. Yang L, Brooks CR, Xiao S, Sabbisetti V, Yeung MY, Hsiao LL, et al. Kim-1-mediated phagocytosis reduces acute injury to the kidney. J Clin Invest. (2015) 125:1620–36. doi: 10.1172/JCI75417
123. Zhang D, Jiang F, Zaynagetdinov R, Huang H, Sood VD, Wang H, et al. Identification and characterization of M6903, an antagonistic anti-tim-3 monoclonal antibody. Oncoimmunology. (2020) 9:1744921. doi: 10.1080/2162402X.2020.1744921
124. Krol J, Loedige I, Filipowicz W. The widespread regulation of microrna biogenesis, function and decay. Nat Rev Genet. (2010) 11:597–610. doi: 10.1038/nrg2843
125. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of micrornas: biology, functions, therapeutics, and analysis methods. J Cell Physiol. (2019) 234:5451–65. doi: 10.1002/jcp.27486
126. Khan Z, Suthanthiran M, Muthukumar T. Micrornas and transplantation. Clin Lab Med. (2019) 39:125–43. doi: 10.1016/j.cll.2018.10.003
127. Li S, Le TN, Nguyen TD, Trinh TA, Nguyen TA. Bulges control pri-mirna processing in a position and strand-dependent manner. RNA Biol. (2021) 18:1716–26. doi: 10.1080/15476286.2020.1868139
128. Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of mirna regulation. Genomics Proteomics Bioinf. (2009) 7:147–54. doi: 10.1016/S1672-0229(08)60044-3
129. Harris A, Krams SM, Martinez OM. Micrornas as immune regulators: implications for transplantation. Am J Transplant. (2010) 10:713–9. doi: 10.1111/j.1600-6143.2010.03032.x
130. Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microrna-class regulatory rnas in drosophila. Cell. (2007) 130:89–100. doi: 10.1016/j.cell.2007.06.028
131. Kabekkodu SP, Shukla V, Varghese VK, DS J, Chakrabarty S, Satyamoorthy K. Clustered mirnas and their role in biological functions and diseases. Biol Rev Camb Philos Soc. (2018) 93:1955–86. doi: 10.1111/brv.12428
132. Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the micrornas in the mir-23a~27a~24-2 cluster and its implication in human diseases. Mol Cancer. (2010) 9:232. doi: 10.1186/1476-4598-9-232
133. Lataniotis L, Albrecht A, Kok FO, Monfries CAL, Benedetti L, Lawson ND, et al. Crispr/cas9 editing reveals novel mechanisms of clustered microrna regulation and function. Sci Rep. (2017) 7:8585. doi: 10.1038/s41598-017-09268-0
134. Wang Y, Luo J, Zhang H, Lu J. Micrornas in the same clusters evolve to coordinately regulate functionally related genes. Mol Biol Evol. (2016) 33:2232–47. doi: 10.1093/molbev/msw089
135. Catalanotto C, Cogoni C, Zardo G. Microrna in control of gene expression: an overview of nuclear functions. Int J Mol Sci. (2016) 17:1712. doi: 10.3390/ijms17101712
136. Westholm JO, Lai EC. Mirtrons: microrna biogenesis via splicing. Biochimie. (2011) 93:1897–904. doi: 10.1016/j.biochi.2011.06.017
137. Zhou M, Hara H, Dai Y, Mou L, Cooper DK, Wu C, et al. Circulating organ-specific micrornas serve as biomarkers in organ-specific diseases: implications for organ allo- and xeno-transplantation. Int J Mol Sci. (2016) 17:1232. doi: 10.3390/ijms17081232
138. Hu ZQ, Lu Y, Cui D, Ma CY, Shao S, Chen P, et al. Micrornas and long non-coding rnas in liver surgery: diagnostic and therapeutic merits. Hepatobiliary Pancreat Dis Int. (2020) 19:218–28. doi: 10.1016/j.hbpd.2020.04.002
139. Roderburg C, Benz F, Vargas Cardenas D, Koch A, Janssen J, Vucur M, et al. Elevated mir-122 serum levels are an independent marker of liver injury in inflammatory diseases. Liver Int. (2015) 35:1172–84. doi: 10.1111/liv.12627
140. Liu L, Xiao F, Sun J, Wang Q, Wang A, Zhang F, et al. Hepatocyte-derived extracellular vesicles mir-122-5p promotes hepatic ischemia reperfusion injury by regulating kupffer cell polarization. Int Immunopharmacol. (2023) 119:110060. doi: 10.1016/j.intimp.2023.110060
141. Farid WR, Verhoeven CJ, de Jonge J, Metselaar HJ, Kazemier G, van der Laan LJ. The ins and outs of micrornas as biomarkers in liver disease and transplantation. Transpl Int. (2014) 27:1222–32. doi: 10.1111/tri.12379
142. Li J, Gong J, Li P, Li M, Liu Y, Liang S, et al. Knockdown of microrna-155 in kupffer cells results in immunosuppressive effects and prolongs survival of mouse liver allografts. Transplantation. (2014) 97:626–35. doi: 10.1097/TP.0000000000000061
143. Xiao Q, Ye QF, Wang W, Fu BQ, Xia ZP, Liu ZZ, et al. Mild hypothermia pretreatment protects hepatocytes against ischemia reperfusion injury via down-regulating mir-122 and igf-1r/akt pathway. Cryobiology. (2017) 75:100–5. doi: 10.1016/j.cryobiol.2017.01.005
144. Millan O, Ruiz P, Orts L, Ferre P, Crespo G, Santana M, et al. Monitoring of mir-181a-5p and mir-155-5p plasmatic expression as prognostic biomarkers for acute and subclinical rejection in de novo adult liver transplant recipients. Front Immunol. (2019) 10:873. doi: 10.3389/fimmu.2019.00873
145. Soltaninejad E, Nicknam MH, Nafar M, Ahmadpoor P, Pourrezagholi F, Sharbafi MH, et al. Differential expression of micrornas in renal transplant patients with acute T-cell mediated rejection. Transpl Immunol. (2015) 33:1–6. doi: 10.1016/j.trim.2015.05.002
146. Mahtal N, Lenoir O, Tinel C, Anglicheau D, Tharaux P-L. Micrornas in kidney injury and disease. Nat Rev Nephrol. (2022) 18:643–62. doi: 10.1038/s41581-022-00608-6
147. Schauerte C, Hübner A, Rong S, Wang S, Shushakova N, Mengel M, et al. Antagonism of profibrotic microrna-21 improves outcome of murine chronic renal allograft dysfunction. Kidney Int. (2017) 92:646–56. doi: 10.1016/j.kint.2017.02.012
148. Chen Y-J, Hsu C-T, Tsai S-F, Chen C-H. Association between circulating micrornas (Mir-21-5p, mir-20a-5p, mir-29b-3p, mir-126-3p and mir-101-3p) and chronic allograft dysfunction in renal transplant recipients. Int J Mol Sci. (2022) 23:12253. doi: 10.3390/ijms232012253
149. Han ZB, Zhong L, Teng MJ, Fan JW, Tang HM, Wu JY, et al. Identification of recurrence-related micrornas in hepatocellular carcinoma following liver transplantation. Mol Oncol. (2012) 6:445–57. doi: 10.1016/j.molonc.2012.04.001
150. Mari B, Danger R, Paul C, Giral M, Lavault A, Foucher Y, et al. Expression of mir-142-5p in peripheral blood mononuclear cells from renal transplant patients with chronic antibody-mediated rejection. PLoS One. (2013) 8:e60702. doi: 10.1371/journal.pone.0060702
151. Kim MY, Brennan DC. Therapies for chronic allograft rejection. Front Pharmacol. (2021) 12:651222. doi: 10.3389/fphar.2021.651222
152. Constanso-Conde I, Hermida-Prieto M, Barge-Caballero E, Núñez L, Pombo-Otero J, Suárez-Fuentetaja N, et al. Circulating mir-181a-5p as a new biomarker for acute cellular rejection in heart transplantation. J Heart Lung Transplant. (2020) 39:1100–8. doi: 10.1016/j.healun.2020.05.018
153. Kennel PJ, Yahi A, Naka Y, Mancini DM, Marboe CC, Max K, et al. Longitudinal profiling of circulating mirna during cardiac allograft rejection: A proof-of-concept study. ESC Heart Failure. (2021) 8:1840–9. doi: 10.1002/ehf2.13238
154. Sukma Dewi I, Celik S, Karlsson A, Hollander Z, Lam K, McManus J-W, et al. Exosomal mir-142-3p is increased during cardiac allograft rejection and augments vascular permeability through down-regulation of endothelial rab11fip2 expression. Cardiovasc Res. (2017) 113:440–52. doi: 10.1093/cvr/cvw244
155. Lu J, Wang W, Li P, Wang X, Gao C, Zhang B, et al. Mir-146a regulates regulatory T cells to suppress heart transplant rejection in mice. Cell Death Discovery. (2021) 7:165. doi: 10.1038/s41420-021-00534-9
156. Van Aelst LNL, Summer G, Li S, Gupta SK, Heggermont W, De Vusser K, et al. Rna profiling in human and murine transplanted hearts: identification and validation of therapeutic targets for acute cardiac and renal allograft rejection. Am J Transplant. (2016) 16:99–110. doi: 10.1111/ajt.13421
157. Feng Z, Xia Y, Zhang M, Zheng J. Microrna-155 regulates T cell proliferation through targeting gsk3Β in cardiac allograft rejection in a murine transplantation model. Cell Immunol. (2013) 281:141–9. doi: 10.1016/j.cellimm.2013.04.001
158. He M, Jin Q, Deng C, Fu W, Xu J, Xu L, et al. Amplification of plasma micrornas for non-invasive early detection of acute rejection after heart transplantation with ultrasound-targeted microbubble destruction. Ultrasound Med Biol. (2023) 49:1647–57. doi: 10.1016/j.ultrasmedbio.2023.03.020
159. Yi L, Chen Y, Jin Q, Deng C, Wu Y, Li H, et al. Antagomir-155 attenuates acute cardiac rejection using ultrasound targeted microbubbles destruction. Advanced Healthcare Materials. (2020) 9:e2000189. doi: 10.1002/adhm.202000189
160. Hu J, Huang S, Liu X, Zhang Y, Wei S, Hu X, et al. Mir-155: an important role in inflammation response. J Immunol Res. (2022) 2022:1–13. doi: 10.1155/2022/7437281
161. Nie M, Liu J, Yang Q, Seok HY, Hu X, Deng ZL, et al. Microrna-155 facilitates skeletal muscle regeneration by balancing pro- and anti-inflammatory macrophages. Cell Death Dis. (2016) 7:e2261–e. doi: 10.1038/cddis.2016.165
162. Tang B, Wang Z, Qi G, Yuan S, Yu S, Li B, et al. Microrna-155 deficiency attenuates ischemia-reperfusion injury after liver transplantation in mice. Transplant Int. (2015) 28:751–60. doi: 10.1111/tri.12528
163. Song C, Wang G, Ma X, Mao P, Lu W, Zhang H, et al. The effect of mir-155–5p on M1 polarization of kupffer cells and immune response during liver transplantation through regulating the expression of kdm5d. Mol Immunol. (2023) 155:17–26. doi: 10.1016/j.molimm.2023.01.003
164. Wang Y, Feng T, Li H, Xiong Y, Tao Y. Gal-9/tim-3 signaling pathway activation suppresses the generation of th17 cells and promotes the induction of foxp3+ Regulatory T cells in renal ischemia-reperfusion injury. Mol Immunol. (2023) 156:136–47. doi: 10.1016/j.molimm.2023.03.008
165. Lamarthée B, Callemeyn J, Van Herck Y, Antoranz A, Anglicheau D, Boada P, et al. Transcriptional and spatial profiling of the kidney allograft unravels a central role for fcyriii+ Innate immune cells in rejection. Nat Commun. (2023) 14:4359. doi: 10.1038/s41467-023-39859-7
166. Li M, Peng X, Qian J, Sun F, Chen C, Wang S, et al. Galectin-9 regulates htr8/svneo function via jnk signaling. Reproduction. (2021) 161:1–10. doi: 10.1530/rep
167. Cai L, Zhou H, Fang Z, Yuan J, Niki T, Hirashima M, et al. Galectin-9 in combination with rapamycin induces cardiac allograft tolerance in mice. Transplantation. (2013) 96:379–86. doi: 10.1097/TP.0b013e31829b07b5
168. Anandagoda N, Roberts LB, Willis JCD, Sarathchandra P, Xiao F, Jackson I, et al. Dominant regulation of long-term allograft survival is mediated by microrna-142. Am J Transplant. (2020) 20:2715–27. doi: 10.1111/ajt.15907
169. Iwasaki K, Yamamoto T, Inanaga Y, Hiramitsu T, Miwa Y, Murotani K, et al. Mir-142-5p and mir-486-5p as biomarkers for early detection of chronic antibody-mediated rejection in kidney transplantation. Biomarkers. (2017) 22:45–54. doi: 10.1080/1354750X.2016.1204000
170. Mari B, Sukma Dewi I, Hollander Z, Lam KK, McManus J-W, Tebbutt SJ, et al. Association of serum mir-142-3p and mir-101-3p levels with acute cellular rejection after heart transplantation. PLoS One. (2017) 12:e0170842. doi: 10.1371/journal.pone.0170842
171. Tinel C, Lamarthée B, Callemeyn J, Van Loon E, Sauvaget V, Morin L, et al. Integrative omics analysis unravels microvascular inflammation-related pathways in kidney allograft biopsies. Front Immunol. (2021) 12:738795. doi: 10.3389/fimmu.2021.738795
172. Anandagoda N, Willis JCD, Hertweck A, Roberts LB, Jackson I, Gökmen MR, et al. Microrna-142–mediated repression of phosphodiesterase 3b critically regulates peripheral immune tolerance. J Clin Invest. (2019) 129:1257–71. doi: 10.1172/jci124725
173. Zhang Y, Ji H, Shen X, Cai J, Gao F, Koenig KM, et al. Targeting tim-1 on cd4 T cells depresses macrophage activation and overcomes ischemia-reperfusion injury in mouse orthotopic liver transplantation. Am J Transplant. (2013) 13:56–66. doi: 10.1111/j.1600-6143.2012.04316.x
174. Cherukuri A, Mohib K, Rothstein DM. Regulatory B cells: tim-1, transplant tolerance, and rejection. Immunol Rev. (2021) 299:31–44. doi: 10.1111/imr.12933
175. Liu ZY, Pan HW, Cao Y, Zheng J, Zhang Y, Tang Y, et al. Downregulated microrna-330 suppresses left ventricular remodeling via the tgf-beta1/smad3 signaling pathway by targeting sry in mice with myocardial ischemia-reperfusion injury. J Cell Physiol. (2019) 234:11440–50. doi: 10.1002/jcp.27800
176. Cui L, Sun F, Xu Y, Li M, Chen L, Chen C, et al. Tim-3 coordinates macrophage-trophoblast crosstalk via angiogenic growth factors to promote pregnancy maintenance. Int J Mol Sci. (2023) 24:1538. doi: 10.3390/ijms24021538
177. Lu Y, Wang Y-L, Liu Q, Zhou P, Mei P-Y, Li J-S, et al. Mirna-122 promotes ischemia-reperfusion injury after lung transplantation via the toll-like receptor signaling pathway. Curr Med Sci. (2021) 41:1231–8. doi: 10.1007/s11596-021-2487-y
178. Riechert G, Maucher D, Schmidt B, Schumann J. Mirna-mediated priming of macrophage M1 differentiation differs in gram-positive and gram-negative settings. Genes. (2022) 13:211. doi: 10.3390/genes13020211
179. Li Y, Shi X. Micrornas in the regulation of tlr and rig-I pathways. Cell Mol Immunol. (2012) 10:65–71. doi: 10.1038/cmi.2012.55
180. Jiang W, Ni Q, Tan L, Kong L, Lu Y, Xu X, et al. The microrna-146a/B attenuates acute small-for-size liver graft injury in rats. Liver Int. (2014) 35:914–24. doi: 10.1111/liv.12674
181. Wang S, Cao C, Piao H, Li Y, Tao Y, Zhang X, et al. Tim-3 protects decidual stromal cells from toll-like receptor-mediated apoptosis and inflammatory reactions and promotes th2 bias at the maternal-fetal interface. Sci Rep. (2015) 5:9013. doi: 10.1038/srep09013
182. Ye L, Zhang Q, Cheng Y, Chen X, Wang G, Shi M, et al. Tumor-derived exosomal hmgb1 fosters hepatocellular carcinoma immune evasion by promoting tim-1+ Regulatory B cell expansion. J ImmunoTherapy Cancer. (2018) 6:145. doi: 10.1186/s40425-018-0451-6
183. Xia H, Wang D, Guo X, Wu K, Huang F, Feng Y. Catalpol Protects against Spinal Cord Injury in Mice through Regulating Microrna-142-Mediated Hmgb1/Tlr4/Nf-Κb Signaling Pathway. Front Pharmacol. (2021) 11:630222. doi: 10.3389/fphar.2020.630222
184. Guo D, Fan Y, Yue J-R, Lin T. A regulatory mirna–mrna network is associated with transplantation response in acute kidney injury. Hum Genomics. (2021) 15:69. doi: 10.1186/s40246-021-00363-y
185. Wu D, Kang L, Tian J, Wu Y, Liu J, Li Z, et al. Exosomes derived from bone mesenchymal stem cells with the stimulation of fe3o4 nanoparticles and static magnetic field enhance wound healing through upregulated mir-21-5p. Int J Nanomedicine. (2020) 15:7979–93. doi: 10.2147/ijn.S275650
186. Hao C, Lu Z, Zhao Y, Chen Z, Shen C, Ma G, et al. Overexpression of gata4 enhances the antiapoptotic effect of exosomes secreted from cardiac colony-forming unit fibroblasts via mirna221-mediated targeting of the pten/pi3k/akt signaling pathway. Stem Cell Res Ther. (2020) 11:251. doi: 10.1186/s13287-020-01759-8
187. Pang N, Tudahong S, Zhu Y, He J, Han C, Chen G, et al. Galectin-9 alleviates acute graft-versus-host disease after haplo-hematopoietic stem cell transplantation by regulating regulatory T cell/effector T cell imbalance. Immunity Inflammation Dis. (2024) 12:e1177. doi: 10.1002/iid3.1177
188. Ning Q, Jian T, Cui S, Shi L, Jian X, He X, et al. Tim-3 facilitates immune escape in benzene-induced acute myeloid leukemia mouse model by promoting macrophage M2 polarization. Ecotoxicology Environ Saf. (2023) 266:115532. doi: 10.1016/j.ecoenv.2023.115532
189. Hong BS, Ryu HS, Kim N, Kim J, Lee E, Moon H, et al. Tumor suppressor mirna-204-5p regulates growth, metastasis, and immune microenvironment remodeling in breast cancer. Cancer Res. (2019) 79:1520–34. doi: 10.1158/0008-5472.Can-18-0891
190. Cheng YQ, Ren JP, Zhao J, Wang JM, Zhou Y, Li GY, et al. Microrna-155 regulates interferon-Γproduction in natural killer cells via tim-3 signaling in chronic hepatitis C virus infection. Immunology. (2015) 145:485–97. doi: 10.1111/imm.12463
191. Prieto I, Kavanagh M, Jimenez-Castilla L, Pardines M, Lazaro I, Herrero del Real I, et al. A mutual regulatory loop between mir-155 and socs1 influences renal inflammation and diabetic kidney disease. Mol Ther - Nucleic Acids. (2023) 34:102041. doi: 10.1016/j.omtn.2023.102041
192. Talebi F, Ghorbani S, Chan WF, Boghozian R, Masoumi F, Ghasemi S, et al. Microrna-142 regulates inflammation and T cell differentiation in an animal model of multiple sclerosis. J Neuroinflamm. (2017) 14:55. doi: 10.1186/s12974-017-0832-7
Keywords: T cell immunoglobulin and mucin domain, microRNAs, transplantation, allograft rejection, allograft tolerance
Citation: Tao J, Shen X, Qian H, Ding Q and Wang L (2024) TIM proteins and microRNAs: distinct impact and promising interactions on transplantation immunity. Front. Immunol. 15:1500228. doi: 10.3389/fimmu.2024.1500228
Received: 23 September 2024; Accepted: 06 November 2024;
Published: 22 November 2024.
Edited by:
Rita Maccario, San Matteo Hospital Foundation (IRCCS), ItalyReviewed by:
Baptiste Lamarthée, Université de Franche-Comté, FranceMarcello Maestri, University of Pavia, Italy
Copyright © 2024 Tao, Shen, Qian, Ding and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Ding, ZGluZ3FAdXBtYy5lZHU=; Lihong Wang, empnenl3bGhAbmp1Y20uZWR1LmNu
 Jialing Tao
Jialing Tao Xiaoxuan Shen2
Xiaoxuan Shen2 Qing Ding
Qing Ding Lihong Wang
Lihong Wang