- 1Department of Gastroenterology, The Third Xiangya Hospital, Central South University, Changsha, Hunan, China
- 2Department of General Surgery, Zhongshan Hospital of Traditional Chinese Medicine Affiliated to Guangzhou University of Traditional Chinese Medicine, Zhongshan, Guangdong, China
- 3School of Medical Laboratory Science, Hebei North University, Zhangjiakou, Hebei, China
- 4Department of General Surgery, The First People's Hospital of Qingzhen City, Guiyang, Guizhou, China
- 5Department of Infectious Diseases, Affiliated Hospital of Zunyi Medical University, Zunyi, Guizhou, China
Background: Helicobacter pylori (H. pylori) may be present in the intestinal mucosa of patients with inflammatory bowel disease (IBD), which is a chronic inflammation of the gastrointestinal tract. The role of H. pylori in the pathogenesis of IBD remains unclear. In this study, bioinformatics techniques were used to investigate the correlation and co-pathogenic pathways between H. pylori and IBD.
Methods: The following matrix data were downloaded from the GEO database: H. pylori-associated gastritis, GSE233973 and GSE27411; and IBD, GSE3365 and GSE179285. Differential gene analysis was performed via the limma software package in the R environment. A protein−protein interaction (PPI) network of DEGs was constructed via the STRING database. Cytoscape software, through the CytoHubba plugin, filters the PPI subnetwork and identifies Hub genes. Validation of the Hub genes was performed in the validation set. Immune analysis was conducted via the CIBERSORT algorithm. Transcription factor interaction and small molecule drug analyses of the Hub genes were also performed.
Results: Using the GSE233973 and GSE3365 datasets, 151 differentially expressed genes (DEGs) were identified. GO enrichment analysis revealed involvement in leukocyte migration and chemotaxis, response to lipopolysaccharides, response to biostimulatory stimuli, and regulation of interleukin-8 (IL-8) production. Ten Hub genes (TLR4, IL10, CXCL8, IL1B, TLR2, CXCR2, CCL2, IL6, CCR1 and MMP-9) were identified via the PPI network and Cytoscape software. Enrichment analysis of the Hub genes focused on the lipopolysaccharide response, bacterial molecular response, biostimulatory response and leukocyte movement. Validation using the GSE27411 and GSE179285 datasets revealed that MMP-9 was significantly upregulated in both the H. pylori and IBD groups. The CIBERSORT algorithm revealed immune infiltration differences between the control and disease groups of IBD patients. Additionally, the CMap database identified the top 11 small molecule compounds across 10 cell types, including TPCA-1, AS-703026 and memantine, etc.
Conclusion: Our study revealed the co-pathogenic mechanism between H. pylori and IBD and identified 10 Hub genes related to cellular immune regulation and signal transduction. The expression of MMP-9 is significantly upregulated in both H. pylori infection and IBD. This study provides a new perspective for exploring the prevention and treatment of H. pylori infection and IBD.
1 Introduction
Helicobacter pylori (H. pylori) is a gram-negative, microaerophilic, spiral-shaped bacterium that infects approximately 50% of the global population and is classified as a Group I human carcinogen. The association between the gastric microbiome and H. pylori plays a significant role in the development and progression of gastric cancer (GC) (1, 2). H. pylori infection is closely associated with chronic gastritis, peptic ulcers, GC and mucosa-associated lymphoid tissue (MALT) lymphoma. H. pylori infection can cure chronic active gastritis and peptic ulcer disease, reduce the risk of peptic ulcer bleeding in aspirin users, and lower the risk of GC in infected individuals (3, 4). Research has shown that H. pylori infection increases the risk of developing colorectal cancer (CRC) (5, 6), and reduces the risk of inflammatory bowel disease (IBD) (7, 8). In conclusion, H. pylori may be related to IBD, and the specific mechanism needs to be further studied.
IBD is a chronic, recurrent inflammatory disease of the gastrointestinal tract that includes ulcerative colitis (UC) and Crohn’s disease (CD) (9). The main gastrointestinal symptoms of IBD include abdominal pain, diarrhea, bloody stools and weight loss (10). Currently, the aetiology and pathogenesis of IBD are not fully understood. Some studies have shown that genetic and environmental factors, gut microbiota imbalance, intestinal mucosal immune responses, and damage to the intestinal mucosal barrier play important roles in IBD (11–14). Multiple studies have shown that H. pylori infection is associated with IBD and that H. pylori may influence the immunoregulatory functions related to IBD (8, 15, 16). However, the specific molecular mechanism is still unclear.
In this study, we used bioinformatics methods to comprehensively analyze the relationship between H. pylori and IBD on the basis of sequencing data of the two diseases from databases. By analyzing the coexpression of DEGs via GO and KEGG analyses, identifying candidate Hub genes (core genes coexpressed between H. pylori and IBD), predicting small-molecule drugs and performing transcription factor analysis, we explored the potential pathogenic mechanisms common to the two diseases.
2 Methods
2.1 Matrix data download and analysis
In accordance with the previous literature on IBD selection strategies (17–19), through the search of H. pylori datasets in the Gene Expression Omnibus (GEO) database, we identified 4 transcriptome datasets, including H. pylori (GSE233973 and GSE27411) and IBD (GSE3365 and GSE179285), and collected basic clinical information (sample size, GEO_accession, Type, Sample_organism, Diagnosis, Age, Race, Gender and Country; for further details (Supplementary Table 1). First, we analyzed the data from the GSE3365 and GSE233973 datasets and discovered a large number of coexpressed DEGs. We used R language for the generation analysis of the GSE3365 and GSE233973 datasets and verified the Hub genes of the GSE27411 and GSE179285 datasets.
2.2 Screening of coexpressed DEGs
For the GSE3365 and GSE233973 datasets, the limma software package was used to identify DEGs in the data in the R environment, and the screening criteria were P.adj. value < 0.05 and |LogFC| > 0.585.
2.3 Functional enrichment analysis of DEGs
GO, KEGG and GSEA (20, 21) pathway enrichment analyses were performed via R packages such as org.Hs.eg.db, clusterProfiler, enrichplot and enrichment plot. GSEA is used to identify important functional items between H. pylori or IBD and control samples and to annotate gene pathways. p < 0.05 indicated that enrichment was statistically significant.
2.4 PPI network construction and Hub gene screening
A total of 151 DEGs were input into the STRING platform to delete isolated genes. Cytoscape software was used to screen the Hub genes, and the MCC algorithm was used to calculate the top 10 genes in the PPI network. Finally, the Hub genes were input into the GeneMANIA network tool to analyze the gene coexpression network (22).
2.5 Verification of H. pylori and IBD Hub genes
In the validation datasets GSE27411 and GSE179285, the ggpubr package in R software was used to verify the expression levels of Hub genes in H. pylori and IBD and to determine their diagnostic ability for H. pylori and IBD.
2.6 Immune infiltration analysis
With the CIBERSORT calculation method (23), the proportion of immune cells in each sample was calculated, as were the expression levels of genes in the 22 immune cells.
2.7 CMap analysis to identify small molecule drugs
Connectivity Map (CMap): A tool or method that uses gene expression signatures to link small molecules, genes and diseases. We input the Hub genes into CMap to identify small molecule compounds with potential therapeutic effects and screen for predicted small molecule drugs. The structure of small molecule drugs was screened by PubChem.
2.8 Transcription factor regulatory network and differential analysis
TRRUST v2 (TRRUST version 2) is a manually curated database of human and mouse transcriptional regulatory networks (24). The transcription factors that interact with the Hub genes were analyzed via the TRRUST v2 transcriptional regulatory interaction database. The Hub genes were input into the TRRUST database, a transcription factor regulatory network diagram was constructed, and the transcription factors were output. Differential analysis of the transcription factors was performed via the GSE3365 and GSE233973 datasets. In addition, the common databases used for data processing in this article are detailed in Supplementary Table 9.
3 Results
3.1 Identification of DEGs in H. pylori and IBD
Through the H. pylori dataset (GSE233973) from the GEO database, a total of 6,942 DEGs were identified, as shown in the volcano plot, including 3,368 upregulated genes and 3,754 downregulated genes (Figure 1A). The heatmap displays the top 50 most upregulated and downregulated genes (Figure 1C), with OLFM4 being the most upregulated gene. From the IBD dataset (GSE3365), a total of 591 DEGs were identified. As shown in the volcano plot, there were 311 upregulated genes and 280 downregulated genes (Figure 1B), the heatmap displays 50 differential genes (Figure 1D), with SERPINB2 being the most significantly upregulated gene. Finally, 151 overlapping genes were identified between the H. pylori and IBD datasets, including 126 upregulated genes and 25 downregulated genes (Figures 1E, F). A list of DEGs and common genes can be found in Supplementary Tables 2–4.
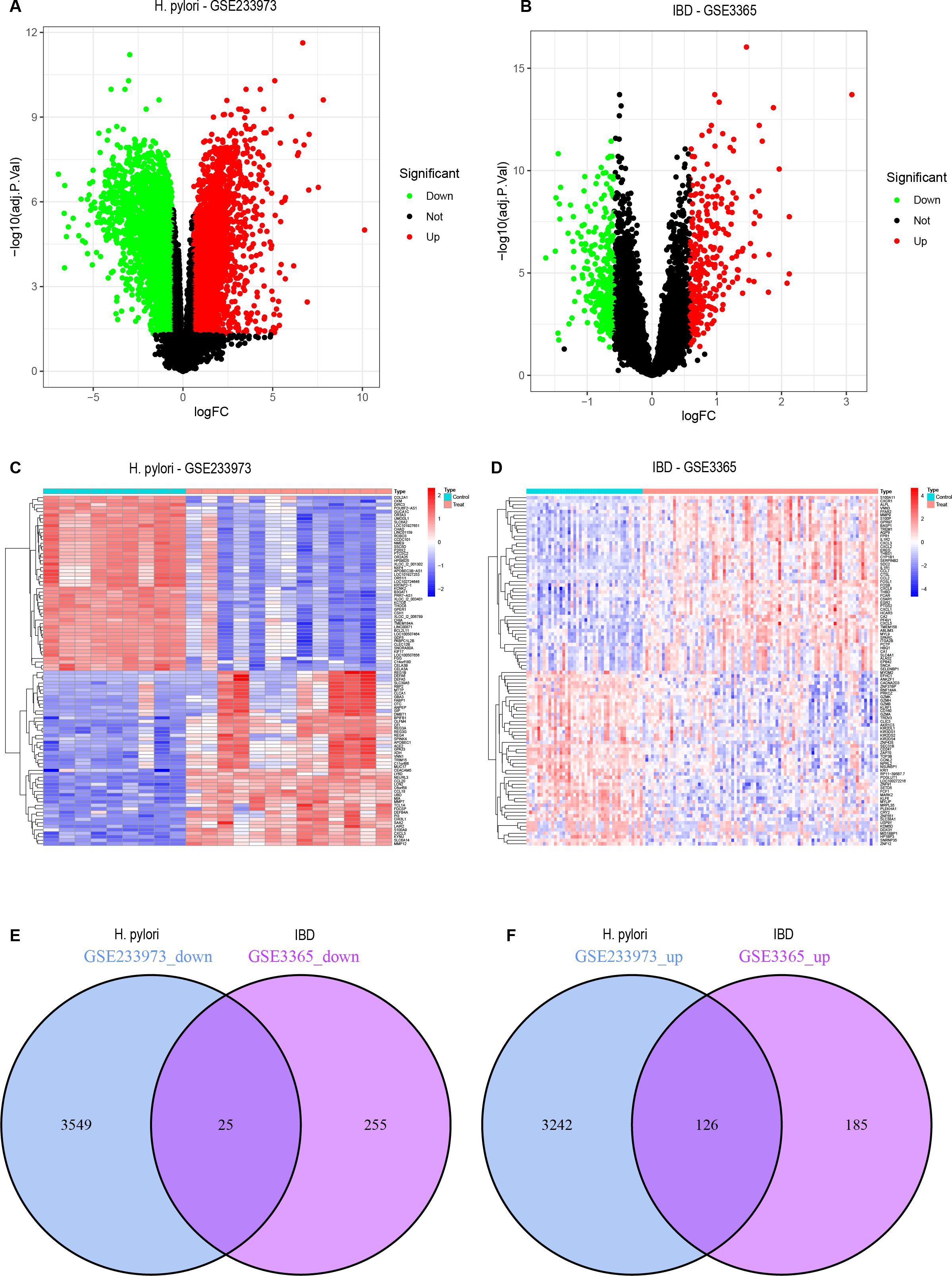
Figure 1. DEGs volcano plots, heatmaps and co-expressed genes. (A) Volcano plot representing DEGs from the GSE233973 dataset. (B) Volcano plot representing DEGs from the GSE3365 dataset. (C) Heatmap representing DEGs from the GSE233973 dataset. (D) Heatmap representing DEGs from the GSE3365 dataset. (E) Venn diagram of the coexpressed downregulated DEGs shared between the GSE233973 and GSE3365 datasets. (F) Venn diagram of the coexpressed upregulated DEGs shared between the GSE233973 and GSE3365 datasets. Red represents upregulated genes, green represents downregulated genes, and grey represents genes with no differential expression.
3.2 Functional enrichment analysis of DEGs
In the functional enrichment analysis, 151 DEGs were coexpressed by H. pylori and IBD. By GSEA analysis, in IBD (GSE3365), GO enrichment analysis revealed that these DEGs were enriched mainly in CELL_CHEMOTAXIS, CELLULAR_RESPONSE_TO_BIOTIC_STIMULUS, GRANULOCYTE_CHEMOTAXIS, GRANULOCYTE_MIGRATION and HEMOSTASIS. KEGG enrichment analysis suggested that these DEGs were enriched mainly in CHEMOKINE_SIGNALING_PATHWAY, COMPLEMENT_AND_COAGULATION_CASCADES, CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION, ECM_RECEPTOR_INTERACTION KEGG_FOCAL_ADHESION and other pathways (Figures 2A, B). In H. pylori (GSE233973), GO enrichment analysis revealed that these DEGs were enriched mainly in ACTIVATION_OF_IMMUNE_RESPONSE, ADAPTIVE_IMMUNE_RESPONSE_BASED_ON_SOMATIC_RECOMBINATION_OF_IMMUNE_RECEPT ORS_BUILT, ALPHA_BETA_T_CELL_ACTIVATION, and ANTIGEN_PROCESSING_AND_PRESENTATION on the equal path. KEGG enrichment analysis indicated that these DEGs were enriched mainly in ALLOGRAFT_REJECTION, ANTIGEN_PROCESSING_AND_PRESENTATION, AUTOIMMUNE_THYROID_DISEASE and CELL_ADHESION_MO LECULES_CAMS, CHEMOKINE_SIGNALING_PATHWAY and other pathways (Figures 2C, D). In addition, the enrichment analysis focused on four main aspects: reactions to lipopolysaccharides, leukocyte chemotaxis, cellular chemotaxis and leukocyte migration (Figures 2E, F).
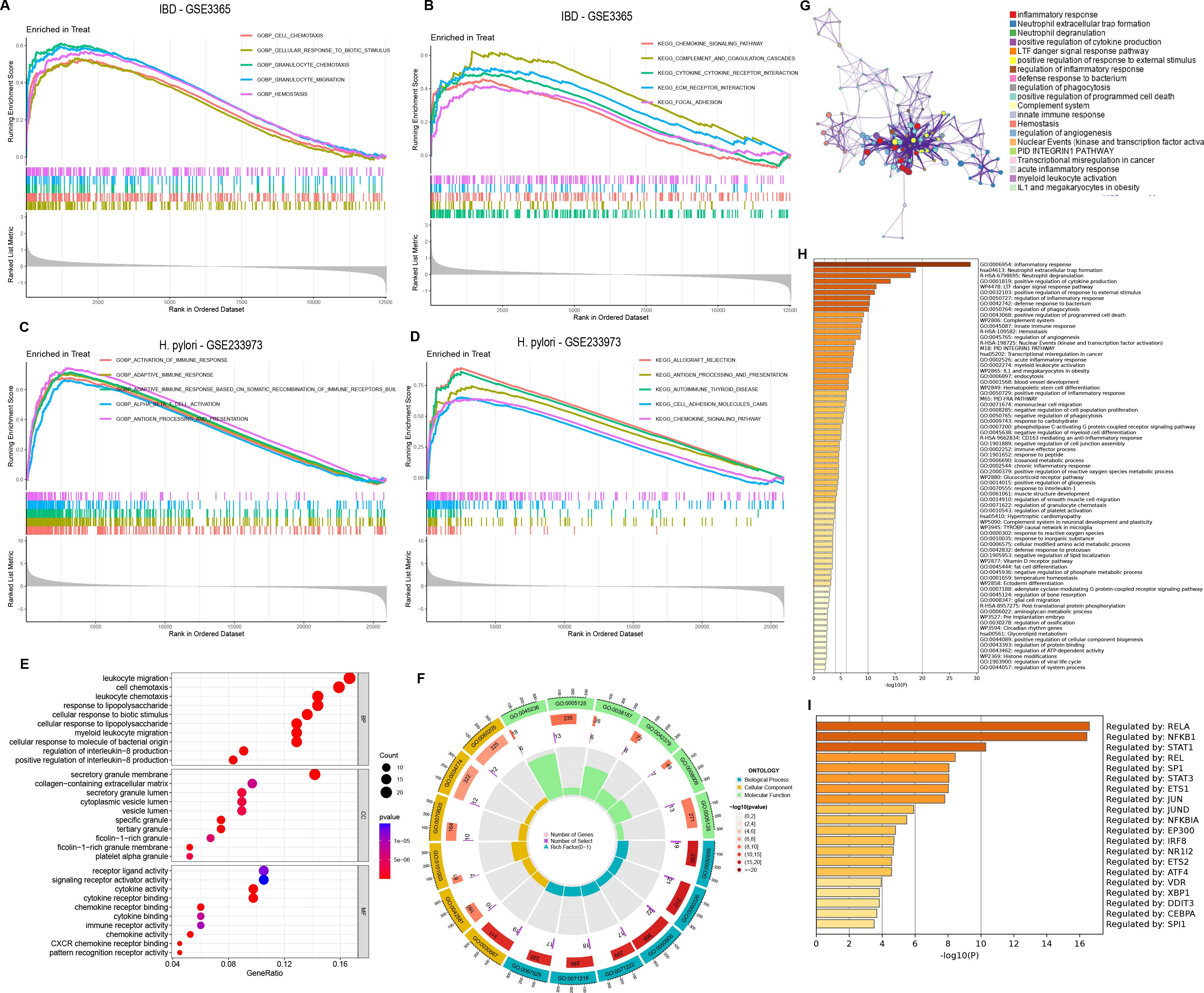
Figure 2. Functional enrichment and pathway enrichment analysis of coexpressed DEGs in H. pylori and IBD. (A–D) GSEA of coexpressed DEGs, with GO and KEGG enrichment analyses of the GSE3365 and GSE233973 datasets. Treat represents disease group. (E) GO analysis of coexpressed DEGs, including BP, biological process; CC, cellular component; MF, molecular function. (F) Circular plot of the GO analysis results. (G) PPI network analysis of coexpressed DEGs. (H) Enrichment analysis of 151 coexpressed DEGs via the Metascape online tool. (I) Transcription factor analysis of coexpressed DEGs.
Furthermore, to further elucidate the enriched pathways of the DEGs, the MetScape database played a significant role in disease enrichment analysis, interorgan correlation, and transcription factor interactions. We found that different genes in the MetScape database may exhibit different functional group distributions, with the positive regulation of the inflammatory response being the most prominent (Figure 2G). Moreover, enrichment analysis based on the MetScape database revealed a common role of inflammation in the pathogenesis of H. pylori infection and IBD (Figure 2H). Finally, transcription factor analysis of the DEGs revealed RELA, BFKB and STAT1 as the most commonly regulated genes (Figure 2I).
3.3 Extraction and enrichment analysis of H. pylori and IBD Hub genes
PPI analysis of the DEGs coexpressed in H. pylori and IBD was performed via the STRING database. To further categorize the DEGs into functional groups, we input the 151 DEGs into the STRING database and removed the independent genes to construct a PPI network diagram (Figure 3A). The analysis was conducted via the CytoHubba plugin in Cytoscape software, with the visualization results shown in Cytoscape (Figure 3B). On the basis of the visualization results, MCODE analysis yielded two subnetwork diagrams of DEGs, wherein the genes within each subnetwork exhibited strong interconnections (Figures 3C, D). The MCC and degree algorithms were used to analyze the PPI network, identifying the top 10 Hub genes: TLR4, IL10, CXCL8, IL1B, TLR2, CXCR2, CCL2, IL6, CCR1 and MMP-9. These 10 Hub genes were identified as candidate diagnostic markers, with IL6 being the most significant within the group (Figure 3E; Supplementary Table 5). GeneMANIA network analysis revealed the interaction network of the 10 Hub genes, with the outer circle indicating the interaction relationships with the Hub genes (Figure 3F).
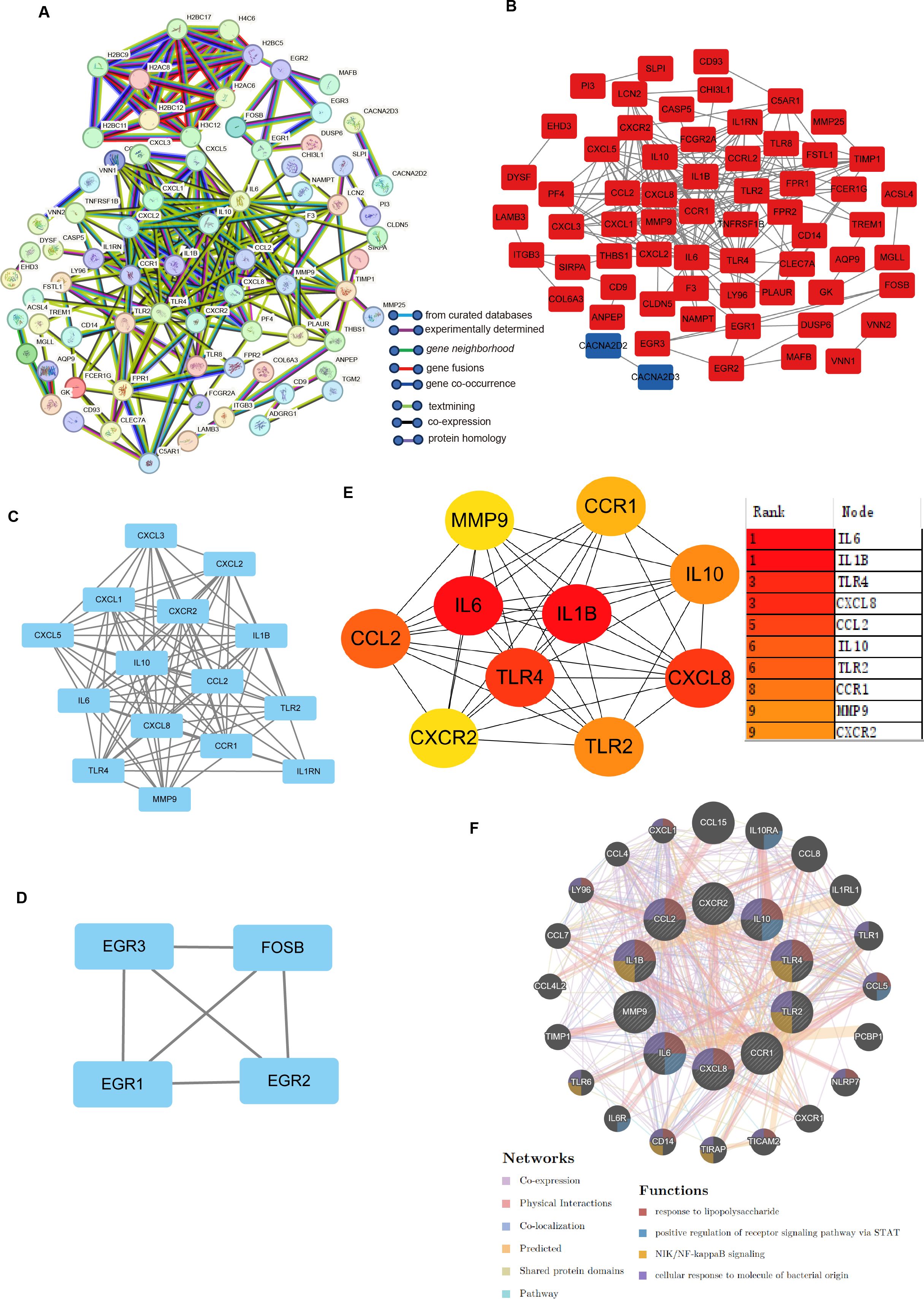
Figure 3. Identification of hub genes and functional interaction network diagram. (A) PPI network analysis of DEGs (STRING database). (B, E) The top 10 Hub genes identified by the degree and MCC algorithms via the CytoHubba plugin in Cytoscape. (C, D) Visualization of the PPI network and important modules and subnetworks. (F) GeneMANIA diagram showing the coexpression interactions between the 10 identified shared Hub genes and their neighbouring genes. The colour codes indicate the functions shared by the genes.
In the GO functional enrichment analysis, the 10 Hub genes associated with H. pylori and IBD were enriched in three key aspects: cellular response to lipopolysaccharide, cellular response to molecule of bacterial origin and cellular response to biotic stimulus (Figures 4A, B). Furthermore, KEGG analysis suggested that these Hub genes are mainly enriched in Malaria, Chagas disease, Lipid and atherosclerosis and Cytokine-cytokine receptor interaction, it is particularly noteworthy that the KEGG analysis indicated a significant association between these Hub genes and IBD (Figure 4C).
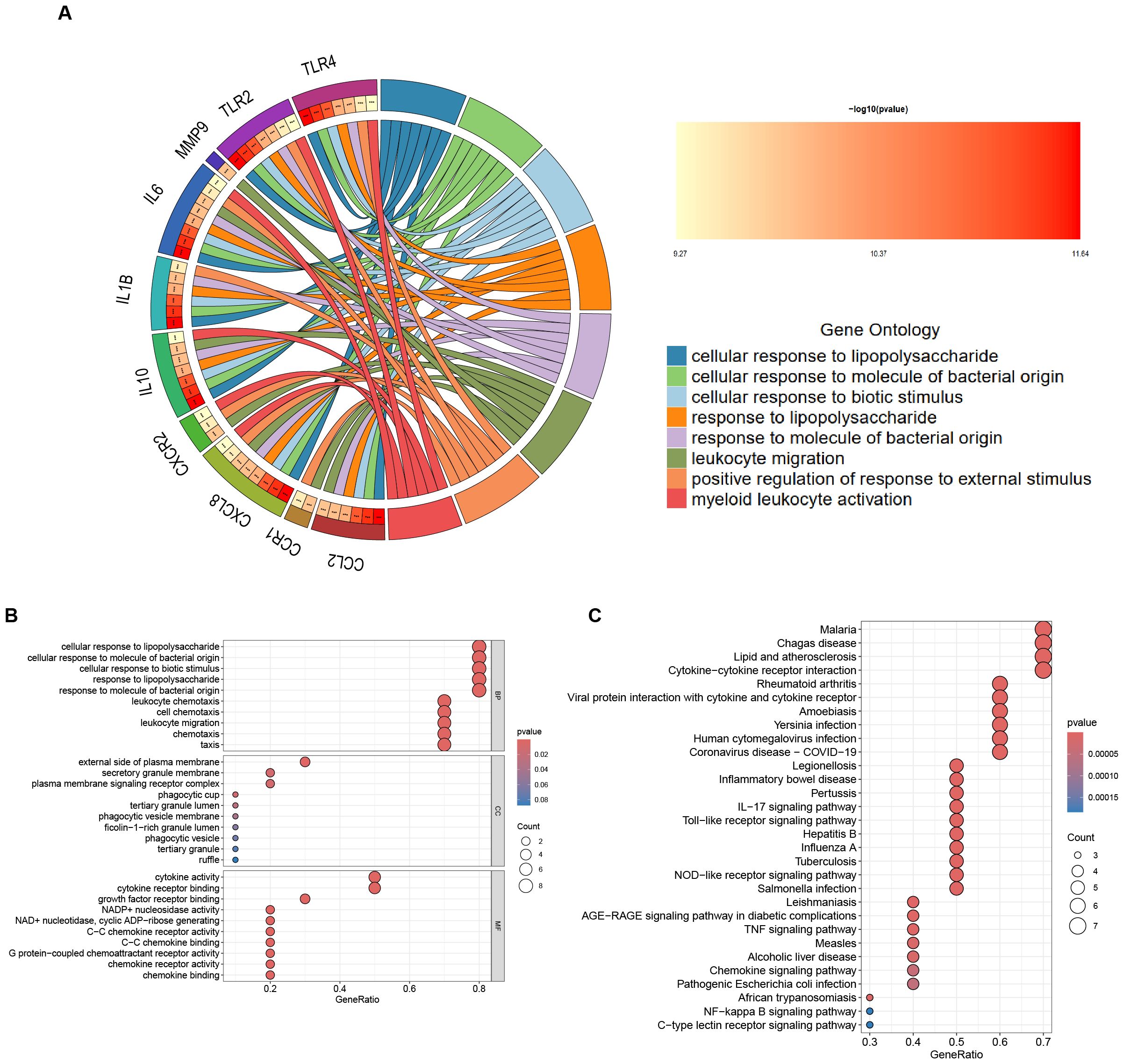
Figure 4. Enrichment Analysis of Hub Genes. (A) The circular diagram visualizing the results of the GO enrichment analysis is presented below: the left semicircle denotes Hub genes, which represent gene names, whereas the right semicircle indicates GO terms. The different colours correspond to different GO terms. Notably, the connections between the two semicircles illustrate the enrichment of genes within specific GO terms. ***p < 0.001. (B) GO enrichment analysis of the following Hub genes: BP, biological process; CC, cellular component; MF, molecular function. (C) KEGG enrichment analysis of the Hub genes.
3.4 Validation of Hub Genes in H. pylori and IBD
The Hub genes were validated in the H. pylori dataset (GSE27411), revealing upregulated expression of the Hub genes, with MMP-9 showing a significant increase in expression (Figure 5A). In the IBD dataset (GSE179285), the Hub genes were also found to be upregulated, with MMP-9, IL6, CCL2, TLR4, TLR2, IL1B and CXCR2 showing significant increases in expression (Figure 5B).
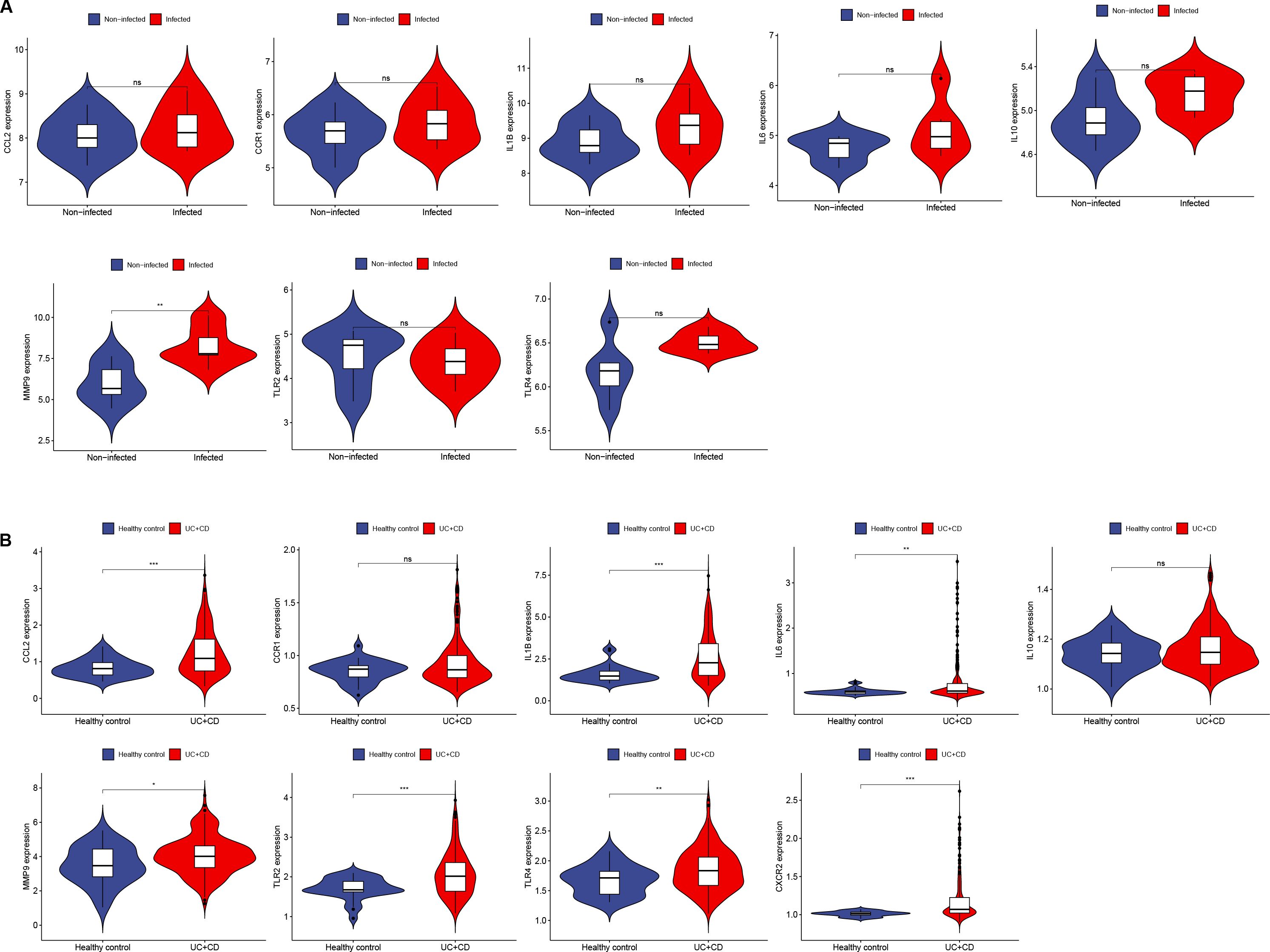
Figure 5. Expression of Hub genes in H. pylori and IBD. (A, B) Expression of 10 Hub genes in GSE27411 and GSE179285. (A) Blue represents noninfected individuals, and red represents H. pylori patients. (B) Blue represents normal individuals, and red represents UC+CD patients. *p < 0.05; **p < 0.01; ***p < 0.001; ns, no statistical significance.
3.5 Immune infiltration analysis
Enrichment analysis revealed that inflammatory responses and immune reactions play crucial roles in the development of both diseases. In the IBD datasets (GSE3365 and GSE179285), we investigated different patterns of immune infiltration on the basis of 22 types of immune cells via the CIBERSORT method (Figures 6A, C). The distribution of immune cells varied between the two IBD datasets, which may be attributed to differences in sample size and the clinical characteristics of the patients. In IBD, immune cell infiltration was increased in CD8+ T cells, monocytes, M0 macrophages, activated mast cells and neutrophils, whereas memory B cells, resting NK cells and resting mast cells were decreased (Figures 6B, D). Owing to the small sample size of the H. pylori datasets, no valid results were obtained. Moreover, we reviewed the relevant literature and reported that H. pylori is related to the aforementioned immune cells. Therefore, immune responses and inflammatory reactions may occur in both H. pylori-infected individuals and individuals with IBD.
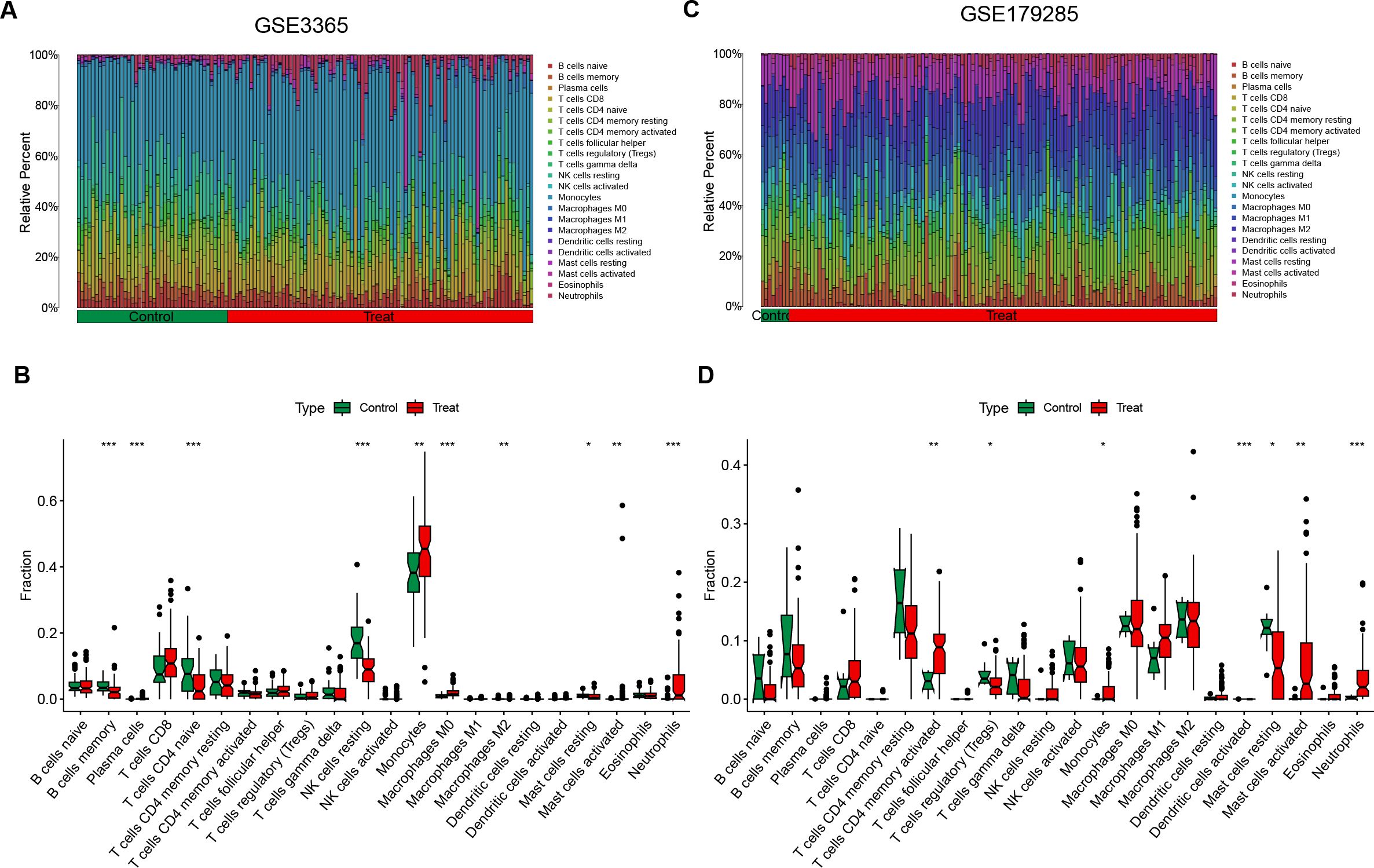
Figure 6. Immune cell infiltration analysis of DEGs in IBD. (A, C) These figures represent the extent of infiltration of various immune cells between the IBD disease group and the normal group. (B, D) Violin plots depicting the differences in immune-infiltrating cells between the IBD disease group and the normal group. The horizontal axis represents the names of the immune cells, and the vertical axis represents the content of the immune cells. Green represents the normal group, and red represents the disease group. Treat represents IBD disease group. *p < 0.05; **p < 0.01; ***p < 0.001; ns indicates no statistical significance.
3.6 Prediction of potential small-molecule drugs
To identify potential small-molecule drugs that target H. pylori and IBD, we utilized the CMap method to analyze the Hub genes. A total of 11 potentially effective drugs were predicted (score < −97) (Figure 7A; Supplementary Table 6), including TPCA-1, AS-703026, memantine, WT-171, vemurafenib, RHO-kinase-inhibitor-III, alpha-linolenic acid, PD-169316, TG-101348, dexamethasone and phentolamine. We further analyzed the interactions between these 11 predicted drugs and Hub genes via the Search Tool for Interactions of Chemicals (STITCH) database (25). The results revealed that alpha-linolenic acid could interact with IL6; dexamethasone could interact with IL10, IL6, IL1B, CCR1, CCL2 and TLR4; and phentolamine could interact with CCR1 (Figure 7B). Additionally, the unique chemical and 3D structures of these three drugs (alpha-linolenic acid, dexamethasone and phentolamine) were predicted via the PubChem (Figures 7C–H).
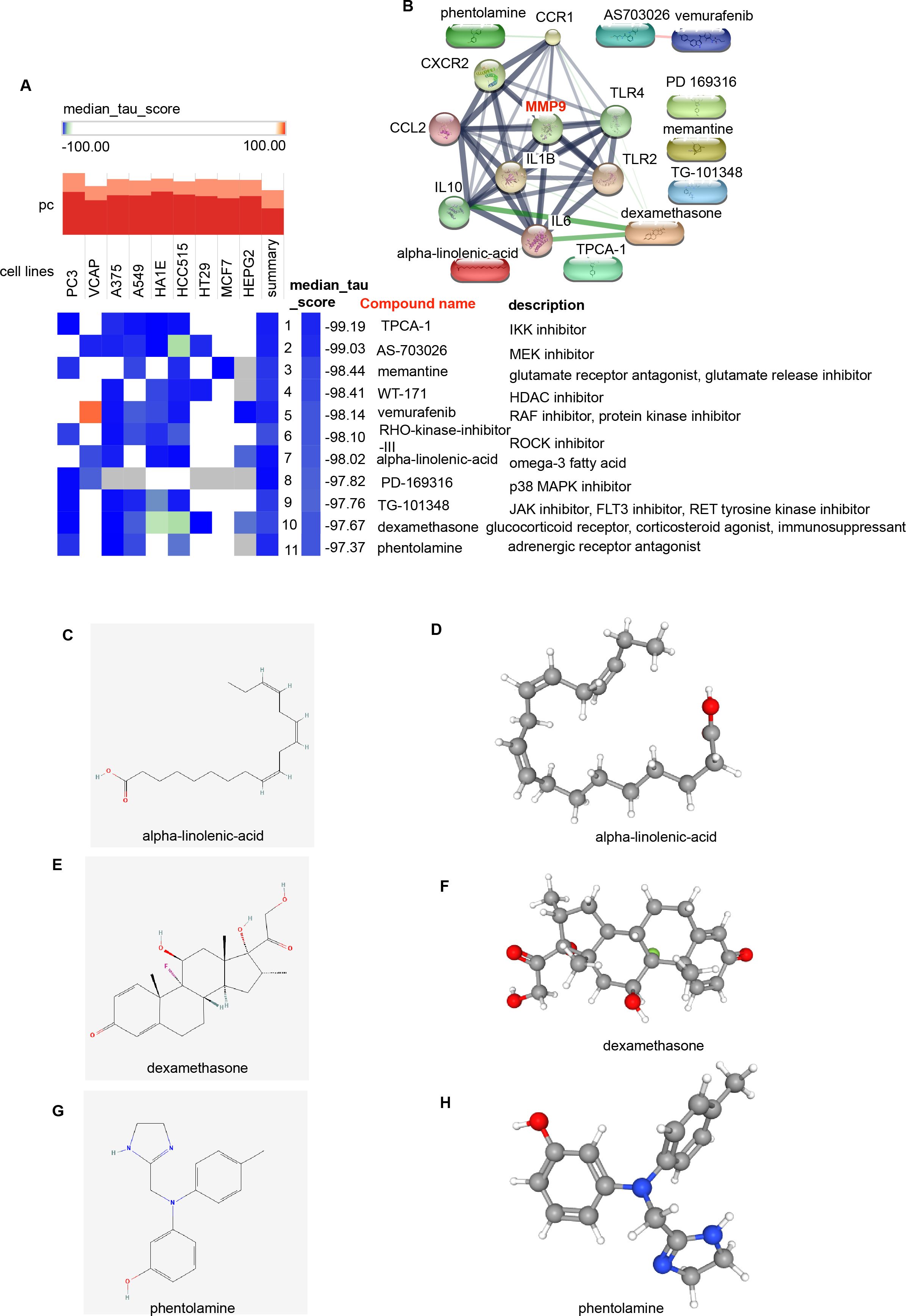
Figure 7. Screening of potential small-molecule compounds for the treatment of H. pylori and IBD via cMAP analysis. (A) Heatmap showing the top 11 compounds with significantly negative enrichment scores (score < -97) in 10 cell lines and a description of those top 11 compounds via cMAP analysis. We choose Perturbagen Type: compound. (B) PPI network diagram of the 11 compounds and Hub genes. (C, E, G) Chemical structures of the 3 compounds (alpha-linolenic acid, dexamethasone and phentolamine). (D, F, H) 3D structures of the 3 compounds. cMAP, Connectivity Map.
3.7 Expression of transcription factors in H. pylori and IBD
The expression of transcription factors in H. pylori and IBD patients was analyzed via TRUST to investigate the interactions of transcription factors with Hub genes. A transcription factor regulatory network was constructed to identify transcription factors that interact with Hub genes (Figure 8A; Supplementary Table 7). The top 9 transcription factors (those interacting with more than three Hub genes) were selected, including NFKB1, RELA, STAT1, STAT3, JUN, SP1, REL, CEBPA and CEBPB (Supplementary Table 8). Further validation of transcription factor expression was conducted in the GSE3365 and GSE233973 datasets. In the GSE3365 dataset, NFKB1, REL and RELA were significantly downregulated, and STAT1 was significantly upregulated (Figure 8B); in the GSE233973 dataset, CEBPB, NFKB1, STAT1 and STAT3 were significantly upregulated, and JUN was significantly downregulated (Figure 8C).
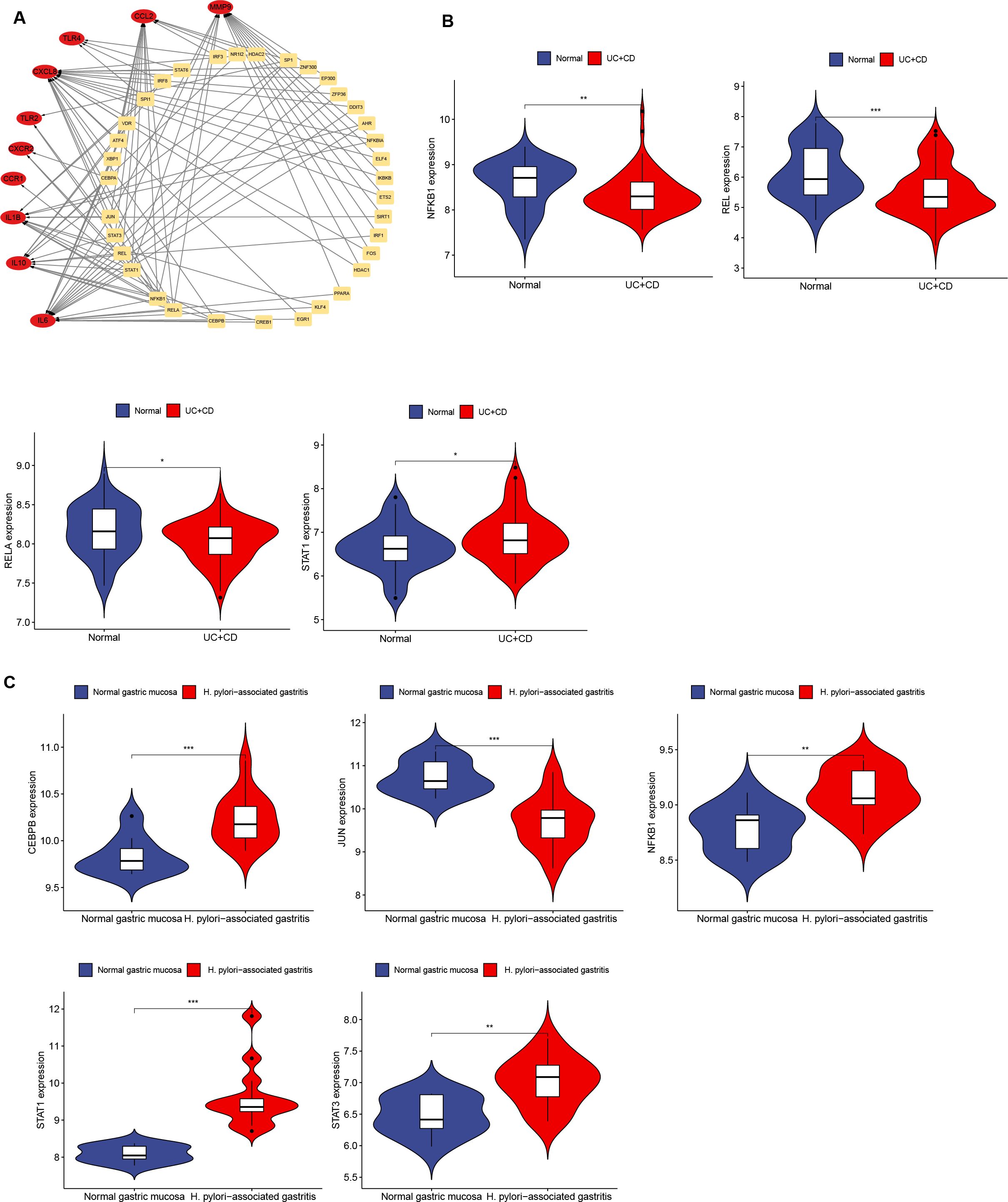
Figure 8. Network interaction between Hub genes and transcription factors and validation in H. pylori and IBD. (A) Network interaction diagram of transcription factors and Hub genes. (B, C) Expression of transcription factors in H. pylori-infected and IBD patients, with transcription factors showing significant differences. Blue represents the normal group, and red represents the disease group. *p < 0.05; **p < 0.01; ***p < 0.001.
4 Discussion
Studies have shown that IBD is an autoimmune disease and that H. pylori infection and colonization have a protective effect against IBD. The mechanisms involved include alterations in inflammatory responses and immune reactions, with improvements in living standards, decreases in H. pylori infection rates, and increases in H. pylori eradication rates, and the incidence of IBD has gradually increased in some regions (26, 27). Additionally, some animal experiments have confirmed a negative correlation between H. pylori infection and the onset of IBD (28, 29). Studies have shown that H. pylori activate Toll-like receptors (TLRs), triggering proinflammatory or anti-inflammatory effects, during the progression of H. pylori infection. Inflammatory factors (IL-6, IL-8, IL-18, IL-32, IL-1β, TNF-α, CCL2, CCL20, CXCL1 and CXCL10) and immune cells (dendritic cells, regulatory T cells, B cells and M2 macrophages) play key roles in the persistence, differentiation, malignant progression, immune infiltration, immune response and immune evasion associated with H. pylori-related diseases, involving various pathogenic mechanisms (30–36).
The pathogenesis of IBD is related to immune system impairment, where activation of the immune system promotes inflammatory responses, leading to tissue damage. Inflammatory factors play important regulatory roles in intestinal mucosal inflammation. Changes in the expression of IL-6, IL-12, IL-23, IL-32, TNF-α and IL-1β promote mucosal inflammation. Therapeutic antibodies and targeted drugs for these factors have been developed and are gradually being used in clinical IBD treatment (37–40). Furthermore, inflammation of the intestinal mucosa is exacerbated by the actions of immune cells, including neutrophils, T lymphocytes, B lymphocytes, macrophages, and dendritic cells (41–45). Therefore, inflammatory factors, immune cells and immune-related genes are key hubs linking H. pylori and IBD. This study identified the DEGs commonly expressed in both H. pylori-infected patients and IBD patients and explored potential targets to provide a theoretical basis for the management and treatment of patients with both H. pylori and IBD.
In this study, we utilized the H. pylori and IBD datasets to identify 151 overlapping DEGs. GO and KEGG enrichment analyses indicated that these DEGs were enriched primarily in response to chemokine signaling, the inflammatory response and the immune response. These findings suggest that these DEGs are involved in the regulation of immune and inflammatory responses. Therefore, disruption of the immune system and the inflammatory microenvironment may be major factors in the pathogenesis of H. pylori infection and IBD. Using the MCC and degree algorithms, 10 Hub genes were subsequently identified: TLR4, IL10, CXCL8, IL1B, TLR2, CXCR2, CCL2, IL6, CCR1 and MMP-9. GO and KEGG enrichment analyses were performed on these Hub genes, which focused mainly on responses to lipopolysaccharides, responses to molecules of bacterial origin, responses to biotic stimuli and leukocyte migration. These genes were identified as diagnostic markers for H. pylori and IBD. Validation using the H. pylori and IBD datasets revealed that the expression of Hub genes was upregulated in patients with H. pylori infection and IBD, with the expression of MMP-9 significantly upregulated.
Matrix metalloproteinases (MMPs) are a class of glycosylated endopeptidases that degrade extracellular matrix (ECM) molecules, including gelatinase A (MMP-2) and gelatinase B (MMP-9). These MMPs are involved in various inflammatory diseases, such as H. pylori-associated gastritis and gastric and duodenal ulcers. Initially, H. pylori was shown to activate MMP-9 expression in gastric epithelial cells, contributing to the development of H. pylori-related gastric diseases (46, 47). In clinical samples, MMP-9 mRNA expression was found to be significantly upregulated in H. pylori-positive patients, with a notable association with CagA status and peptic ulcer disease (PUD) (48). MMP-9 plays a critical role in gastric remodeling in children with H. pylori-related gastritis (49). Moreover, studies on single nucleotide polymorphisms (SNPs) of the MMP-9 gene revealed that the rs3918249-rs17576 CG haplotype increased the risk of H. pylori-positive PUD (50); SNPs in the MMP-9 gene were also associated with H. pylori-positive gastric ulcers (GU) (51). Mechanistically, H. pylori lipopolysaccharide (LPS)-induced gastric mucosal MMP-9 release is associated with the activation of the MAPK, ERK, and p38 pathways (52). H. pylori can promote the progression of H. pylori-related GC by upregulating semaphorin 5A (Sema5A) and regulating the ERK/MMP-9 signaling pathway (53). These studies are consistent with our findings, which demonstrate that MMP-9 is significantly upregulated in H. pylori infection in both the training and validation sets. These studies indicate that MMP-9 plays a crucial role in H. pylori-related gastric diseases, although the specific molecular mechanisms involved require further investigation.
In the intestinal mucosa of IBD patients, mice, and dogs, MMP-9 has been shown to promote the development of colitis (54–56). In children with UC, its expression increases with the severity of the disease (57). Fibroblasts in IBD continuously express IL-21, which enhances MMP-9 production (58). Some studies have suggested that in mouse models of acute and chronic dextran sulfate sodium (DSS)-induced colitis and acute 2,4,6-trinitrobenzene sulfonic acid-induced colitis, the parameters associated with chronic colitis in MMP-9 knockout mice are similar. Pharmacological inhibition of MMP-9 with bioactive peptides does not improve DSS-induced colitis, suggesting that the upregulation of MMP-9 may be a consequence of intestinal inflammation rather than a cause (59). Mechanistic studies have shown that MMP-9 increases intestinal tight junction (TJ) permeability via the upregulation of myosin light-chain kinase (MLCK) through the p38 kinase signaling pathway in both in vivo and in vitro models (60). Additionally, MMP-9 regulates intestinal epithelial tight junction barrier function through NF-κB pathway activation of MLCK (61). The long noncoding RNA (lncRNA) growth arrest-specific transcript 5 (GAS5) can regulate MMP-9 expression, mediating tissue damage in IBD patients. Inflammatory tissues show downregulation of GAS5 and upregulation of MMP-9 (62). Furthermore, a mutated CXCL8 protein (G31P) binds to CXCR1/2, rendering CXCL8 inactive, reducing MMP-9 expression, and alleviating colonic fibrosis (63). The upregulation of MMP-9 in IBD is consistent with our validation results, but MMP-9 affects the progression of IBD through multiple pathways. On the basis of the enrichment analysis results in this study, the current research is insufficient to fully explain the pathogenesis of IBD, and further studies are needed to provide a theoretical basis for clinical IBD treatment.
Therefore, MMP-9 may play a significant role in the pathogenesis and progression of both H. pylori infection and IBD, and modulating this enzyme could have important implications. The findings of this study provide key clues for further research into the connection between these two diseases and their potential therapeutic targets.
Moreover, MMP-9 is critically involved in a variety of diseases, particularly in cancer. In studies on lymphoma, EGR-1, an early growth response protein 1, is expressed by stromal cells within lymphoma tissues and inhibits the transcriptional activation of the MMP-9 gene in these stromal cells (64). MMP-9 inhibitors have been shown to enhance the functionality of TCR-T cells, which are suppressed by EBV-induced M2 macrophages. The combination of MMP-9 inhibitors with TCR-T-cell therapy has demonstrated effective treatment outcomes for EBV-positive solid tumours (65). Tumor necrosis factor alpha-induced protein 8 (TIPE) has been identified as a promoter of CRC metastasis through its regulatory effects on MMP-9 expression (66). Targeting mTORC1 can facilitate the progression from ductal carcinoma in situ (DCIS) to invasive ductal carcinoma (IDC) via the inhibition of MMP-9 (67). In oral squamous cell carcinoma (OSCC), MED1, a subunit of the Mediator complex, activates the transcription of MMP-9 while simultaneously inhibiting CD8+ T-cell antitumor immune responses (68).
Within the context of inflammation, injury repair mechanisms also highlight the significant role played by MMP-9. Recent studies have indicated that unbound bilirubin (UCB) and its derivative dimethyl ester bilirubin (BD1) effectively inhibit the neutrophil-mediated activity of MMP-9 while reducing proinflammatory cytokine gene expression and modulating MAPK pathway signaling to alleviate psoriasis-like skin inflammation in mice (69). Furthermore, the commensal bacterium Enterococcus faecalis activates the expression of MMP-9 in host intestinal tissue; pharmacological inhibition of this activation can prevent anastomotic leakage in rat models (70). In cases of ischemic stroke, MMP-9 inhibitors have been shown to specifically reduce the levels of MMP-9, inflammation, and activation while improving neurological function (71). Research on tissue damage and regeneration indicates that liver-specific inhibition of MMP-9 accelerates liver regeneration by 40% following partial hepatectomy; it also prevents proteolytic cleavage of vascular endothelial growth factor-mesenchymal stem cell-derived factor in hepatic tissue, thereby mitigating damage and enhancing regenerative capacity (72). In hydrogel studies, cinnamaldehyde (CA), a specific TRPA1 agonist, is incorporated into a dual-layer hybrid hydrogel (CA@HA-Gel) designed to respond to matrix MMP-9, which inhibits ferroptosis in wound endothelial cells and significantly promotes healing in diabetic wounds (73). In conclusion, MMP-9 plays a critical role in various biological processes, including inflammation, the injury response, tissue regeneration, tumor progression and immune regulation.
In the pathogenesis of H. pylori infection and IBD, immune cell infiltration plays an important role. Immune infiltration analysis revealed the relationships between immune cells and cellular molecules in H. pylori infection and IBD, which may affect the immune response and inflammatory processes in these diseases. We utilized CIBERSORT to analyze the immune cell infiltration patterns of DEGs in both diseases, and the immune infiltration results were similar. In IBD, immune cell infiltration is increased in CD8+ T cells, monocytes, M0 macrophages, activated mast cells, and neutrophils, whereas memory B cells, resting NK cells, and resting mast cells are decreased. Studies have shown that specific infiltration is dominated by intraganglionic B cells and monocytes in CD, whereas the myenteric plexus (MP) in UC is infiltrated by CD8+ T cells, leading to neuronal apoptosis. In a DSS-induced colitis model, monocyte, B cell, and CD8+ T-cell infiltration also occurs (74). Different clinical outcomes of CD are related to the exhaustion of CD8+ T cells. In active CD, activated CD39+ and CD39+PD-1+ CD8+ T-cell subsets are enriched at low frequencies, whereas during remission, exhaustion-related transcriptional characteristics are upregulated in these subsets (75). Monocytes and macrophages play important roles in regulating the immune response of the intestinal mucosa in IBD. Macrophages produce proinflammatory and anti-inflammatory cytokines, chemokines, toxic mediators, and macrophage extracellular traps (METs), regulating processes such as epithelial cell proliferation, angiogenesis, fibrosis, and the immune response. Additionally, several IBD susceptibility genes related to the maturation and function of macrophages, such as RUNX3, IL21R, GTF2I, and LILRB3, play key roles in the pathogenesis of IBD (76). Synbindin, a classical membrane tethering factor, regulates TLR4 signaling and suppresses the proinflammatory activation of macrophages in response to the microbiota during colitis (77). Moreover, IL-10 is a key regulator of monocyte IL-23 production, providing the basis for targeted therapies such as IL-23p19 and/or IL-1α/IL-1β in IBD patient subgroups (78–80). Our findings, which are consistent with those of other studies, show that the intestinal mucosa in IBD patients contains many activated memory CD4 T cells, M1 macrophages, M2 macrophages, activated mast cells, and neutrophils (81–84). Thus, immune cells play an essential role in regulating immune responses in the intestinal mucosa and provide strategies for the treatment of IBD.
In exploring the relationship between core genes and immune cell infiltration, we emphasize a comprehensive discussion and literature review concerning the association between MMP-9 and the aforementioned significant immune cells. In OSCC, MED1 activates MMP-9 transcription and inhibits the antitumor immune response of CD8+ T cells (68). MMP-9 suppresses the infiltration and cytotoxic activity of CD8+ T cells, thereby providing support for the existence of an inhibitory tumor immune microenvironment (TIME) and resistance to PD-1 inhibitors associated with gain-of-function mutations in CTNNB1 (85). In chronic hepatitis B (CHB), MMP-9 plays an important role in regulating the intrahepatic anti-HBV CD8 T-cell response by mediating the release of soluble CD100 (86). Patients with prostate cancer can increase the expression of MMP-9 in their peripheral blood NK cells and secrete factors that recruit and polarize monocytes (87). Following surgery for acute aortic dissection, platelets release MMP-9 to reprogram monocytes, thereby contributing to postoperative recovery (88). MMP-9 is a central protein related to the immune system and is associated with immune cell infiltration in patients with spinal tuberculosis (STB) (89). In glioblastoma patients, the release of MMP-9 by tumor-infiltrating neutrophils contributes to the efficacy of bevacizumab (90). Therefore, MMP-9 is associated not only with CD8+ T cells, monocytes, and neutrophils but also with other immune cells.
In the analysis of immune infiltration related to H. pylori infection, the small sample size of H. pylori infection data from the GEO datasets resulted in fewer immune infiltrates than did the results of CIBERSORT. In the future, we plan to conduct transcriptomic sequencing on a large number of H. pylori-infected gastric mucosa samples to address the limitations of small sample sizes and perform effective immune infiltration analysis. According to studies on H. pylori and immune cell infiltration, H. pylori infection induces the infiltration of T lymphocytes and enhances apoptosis through inflammatory responses (91–95). H. pylori can induce the expression of cystathionine γ-lyase (CTH) in macrophages, promoting persistent inflammation (96). H. pylori infection impacts the TIME, leading to a poorer prognosis in de novo gastric diffuse large B-cell lymphoma (gDLBCL) (97, 98). Immune cells play crucial roles in the pathogenesis of H. pylori-related diseases.
In addition, this study identified potential small-molecule compounds from the CMap database by screening DEGs in H. pylori and IBD and revealed that alpha-linolenic acid, dexamethasone, and phentolamine interact with several Hub genes. We further used the PubChem tool to predict the unique chemical and 3D structures of these three drugs. These small-molecule drugs may improve the pathological conditions of H. pylori infection and IBD, providing potential directions for the treatment of these diseases and their associated complications.
The Hub genes were closely related to small molecule drugs and transcription factors. As shown in Figure 7B, dexamethasone was closely associated with the Hub genes; however, there was no direct correlation between MMP-9 and dexamethasone, as depicted in Figure 7B, although indirect relationships were present. Dexamethasone has been shown to downregulate both MMP-9 expression and oxidative stress levels in mice suffering from eosinophilic meningitis induced by Angiostrongylus cantonensis infection (99). MMP-9 is correlated with the selected small-molecule drugs. Dexamethasone induces the secretion of MMP-9 both in vitro and in vivo, thereby enhancing the production of mature brain-derived neurotrophic factor (mBDNF), which is essential for synaptic plasticity in adults (100). As shown in Figure 8, several transcription factors identified through the selection of Hub genes revealed that in IBD, several transcription factors were significantly downregulated (NFKB1, REL and RELA), whereas STAT1 expression was upregulated. RELA may play an important role in coronary artery disease (CAD) by targeting MMP-9 (101). Additionally, MMP-9 may participate in the development of various diseases through the STAT1-related signaling pathway (89, 102–104). Dexamethasone plays a pivotal role in the clinical management of IBD. Through our bioinformatics exploration, we identified common Hub genes associated with H. pylori infection and IBD, particularly MMP-9, which was significantly upregulated under both conditions. On the basis of our literature review informed by bioinformatics, we hypothesize that the activation of this pathway increases the expression of transcription factors, activates MMP-9, and promotes inflammatory responses as well as immune cell activation. Furthermore, the small-molecule drugs we identified may modulate the expression of MMP-9 through various pathways, indicating a need for further investigation.
MMP-9 is a common diagnostic marker for both H. pylori and IBD, and although some relevant studies exist in these two diseases, the underlying mechanisms remain unclear. Due to funding limitations, further validation and in-depth exploration of these mechanisms were not conducted. In the future, our research group will further investigate the pathogenesis of these two diseases to provide a theoretical basis and direction for disease prevention and treatment.
5 Conclusion
In this study, we revealed that the inflammatory response and immune response may be the common pathogenic mechanisms of H. pylori infection and IBD. Additionally, we identified 10 Hub genes (TLR4, IL10, CXCL8, IL1B, TLR2, CXCR2, CCL2, IL6, CCR1 and MMP-9) with significant diagnostic relevance, among which MMP-9 was significantly upregulated in both the training and validation sets for these two diseases. MMP-9 may modulate target genes associated with inflammation and immune responses via transcription factor activation, and the small-molecule drugs identified could serve as a significant strategy for inhibiting MMP-9, thereby providing a scientific rationale for clinical interventions in H. pylori and IBD.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author/s.
Author contributions
MZ: Investigation, Methodology, Software, Validation, Visualization, Data curation, Writing – original draft. TL: Software, Validation, Visualization, Writing – review & editing. LL: Software, Validation, Writing – review & editing, Methodology. YZ: Methodology, Software, Writing – review & editing. QC: Methodology, Software, Writing – review & editing. FW: Methodology, Supervision, Writing – review & editing. YX: Methodology, Software, Writing – review & editing, Investigation, Validation, Visualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Independent Exploration and Innovation Project of Central South University (No.2024ZZTS0161); the National Natural Science Foundation of China (No.82270594); the National Demonstration Pilot Project for the Inheritance and Development of Traditional Chinese Medicine -Construction project between Guangzhou University of Chinese Medicine and Zhongshan Hospital of Traditional Chinese Medicine (No.GZYZS2024G39).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1492810/full#supplementary-material
References
1. Zeng R, Gou H, Lau HCH, Yu J. Stomach microbiota in gastric cancer development and clinical implications. Gut. (2024) 73(12):2062–73. doi: 10.1136/gutjnl-2024-332815
2. Guo Y, Zhang Y, Gerhard M, Gao JJ, Mejias-Luque R, Zhang L, et al. Effect of Helicobacter pylori on gastrointestinal microbiota: a population-based study in Linqu, a high-risk area of gastric cancer. Gut. (2020) 69(9):1598–607. doi: 10.1136/gutjnl-2019-319696
3. Chen YC, Malfertheiner P, Yu HT, Kuo CL, Chang YY, Meng FT, et al. Global prevalence of helicobacter pylori infection and incidence of gastric cancer between 1980 and 2022. Gastroenterology. (2024) 166(4):605–19. doi: 10.1053/j.gastro.2023.12.022
4. Pellicano R, Ianiro G, Fagoonee S, Settanni CR, Gasbarrini A. Review: Extragastric diseases and Helicobacter pylori. Helicobacter. (2020) 25 Suppl 1:e12741. doi: 10.1111/hel.v25.s1
5. Ralser A, Dietl A, Jarosch S, Engelsberger V, Wanisch A, Janssen KP, et al. Helicobacter pylori promotes colorectal carcinogenesis by deregulating intestinal immunity and inducing a mucus-degrading microbiota signature. Gut. (2023) 72:1258–70. doi: 10.1136/gutjnl-2022-328075
6. Engelsberger V, Gerhard M, Mejías-Luque R. Effects of Helicobacter pylori infection on intestinal microbiota, immunity and colorectal cancer risk. Front Cell Infect Microbiol. (2024) 14:1339750. doi: 10.3389/fcimb.2024.1339750
7. Nanda S. Infection: Helicobacter pylori may protect against IBD–a mechanistic insight. Nat Rev Gastroenterol Hepatol. (2011) 8(6):299. doi: 10.1038/nrgastro.2011.77
8. Castaño-Rodríguez N, Kaakoush NO, Lee WS, Mitchell HM. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut. (2017) 66:235–49. doi: 10.1136/gutjnl-2015-310545
9. Saez A, Herrero-Fernandez B, Gomez-Bris R, Sánchez-Martinez H, Gonzalez-Granado JM. Pathophysiology of inflammatory bowel disease: innate immune system. Int J Mol Sci. (2023) 24(2):1526. doi: 10.3390/ijms24021526
10. Ma C, Wu W, Lin R, Ge Y, Zhang C, Sun S, et al. Critical role of CD6highCD4+ T cells in driving th1/th17 cell immune responses and mucosal inflammation in IBD. J Crohns Colitis. (2019) 13:510–24. doi: 10.1093/ecco-jcc/jjy179
11. Huibregtse IL, van Lent AU, van Deventer SJ. Immunopathogenesis of IBD: insufficient suppressor function in the gut? Gut. (2007) 56(4):584–92. doi: 10.1136/gut.2006.103523
12. Lautenschlager SA, Barry MP, Rogler G, Biedermann L, Schreiner P, Siebenhüner AR. Lifestyle factors associated with inflammatory bowel disease: data from the Swiss IBD cohort study. BMC Gastroenterol. (2023) 23(1):71. doi: 10.1186/s12876-023-02692-9
13. Liu Z, Liu R, Gao H, Jung S, Gao X, Sun R, et al. Genetic architecture of the inflammatory bowel diseases across East Asian and European ancestries. Nat Genet. (2023) 55(5):796–806. doi: 10.1038/s41588-023-01384-0
14. Massironi S, Viganò C, Palermo A, Pirola L, Mulinacci G, Allocca M, et al. Inflammation and malnutrition in inflammatory bowel disease. Lancet Gastroenterol Hepatol. (2023) 8(6):579–90. doi: 10.1016/S2468-1253(23)00011-0
15. Sonnenberg A, Turner KO, Saboorian H, Singhal A, Genta RM. The occurrence of gastritis in microscopic colitis and inflammatory bowel disease. Clin Gastroenterol Hepatol. (2023) 21(5):1356–1358.e2. doi: 10.1016/j.cgh.2022.03.019
16. Lin KD, Chiu GF, Waljee AK, Owyang SY, El-Zaatari M, Bishu S, et al. Effects of anti-helicobacter pylori therapy on incidence of autoimmune diseases, including inflammatory bowel diseases. Clin Gastroenterol Hepatol. (2019) 17(10):1991–9. doi: 10.1016/j.cgh.2018.12.014
17. Sun HW, Zhang X, Shen CC. The shared circulating diagnostic biomarkers and molecular mechanisms of systemic lupus erythematosus and inflammatory bowel disease. Front Immunol. (2024) 15:1354348. doi: 10.3389/fimmu.2024.1354348
18. Zhang H, Mo Y, Wang L, Zhang H, Wu S, Sandai D, et al. Potential shared pathogenic mechanisms between endometriosis and inflammatory bowel disease indicate a strong initial effect of immune factors. Front Immunol. (2024) 15:1339647. doi: 10.3389/fimmu.2024.1339647
19. Wang Y, Xie D, Ma S, Shao N, Zhang X, Wang X. Exploring the common mechanism of vascular dementia and inflammatory bowel disease: a bioinformatics-based study. Front Immunol. (2024) 15:1347415. doi: 10.3389/fimmu.2024.1347415
20. Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. (2003) 34(3):267–73. doi: 10.1038/ng1180
21. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. (2005) 102(43):15545–50. doi: 10.1073/pnas.0506580102
22. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. (2010) 38:W214–20. doi: 10.1093/nar/gkq537
23. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. (2015) 12(5):453–7. doi: 10.1038/nmeth.3337
24. Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. (2018) 46:D380–d386. doi: 10.1093/nar/gkx1013
25. Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. (2016) 44:D380–4. doi: 10.1093/nar/gkv1277
26. Yu Y, Zhu S, Li P, Min L, Zhang S. Helicobacter pylori infection and inflammatory bowel disease: a crosstalk between upper and lower digestive tract. Cell Death Dis. (2018) 9(10):961. doi: 10.1038/s41419-018-0982-2
27. Arnold IC, Hitzler I, Müller A. The immunomodulatory properties of Helicobacter pylori confer protection against allergic and chronic inflammatory disorders. Front Cell Infect Microbiol. (2012) 2:10. doi: 10.3389/fcimb.2012.00010
28. Wu J, Lin K, Zeng J, Liu W, Yang F, Wang X, et al. Role of DC-SIGN in Helicobacter pylori infection of gastrointestinal cells. Front Biosci (Landmark Ed). (2014) 19(5):825–34. doi: 10.2741/4250
29. Higgins PD, Johnson LA, Luther J, Zhang M, Sauder KL, Blanco LP, et al. Prior Helicobacter pylori infection ameliorates Salmonella typhimurium-induced colitis: mucosal crosstalk between stomach and distal intestine. Inflammation Bowel Dis. (2011) 17(6):1398–408. doi: 10.1002/ibd.21489
30. Pachathundikandi SK, Tegtmeyer N, Backert S. Masking of typical TLR4 and TLR5 ligands modulates inflammation and resolution by Helicobacter pylori. Trends Microbiol. (2023) 31(9):903–15. doi: 10.1016/j.tim.2023.03.009
31. Meliț LE, Mărginean CO, Mărginean CD, Mărginean MO. The relationship between toll-like receptors and helicobacter pylori-related gastropathies: still a controversial topic. J Immunol Res. (2019) 2019:8197048. doi: 10.1155/2019/8197048
32. Pachathundikandi SK, Tegtmeyer N, Arnold IC, Lind J, Neddermann M, Falkeis-Veits C, et al. T4SS-dependent TLR5 activation by Helicobacter pylori infection. Nat Commun. (2019) 10(1):5717. doi: 10.1038/s41467-019-13506-6
33. Tegtmeyer N, Neddermann M, Lind J, Pachathundikandi SK, Sharafutdinov I, Gutiérrez-Escobar AJ, et al. Toll-like receptor 5 activation by the cagY repeat domains of helicobacter pylori. Cell Rep. (2020) 32(11):108159. doi: 10.1016/j.celrep.2020.108159
34. Kumar Pachathundikandi S, Brandt S, Madassery J, Backert S. Induction of TLR-2 and TLR-5 expression by Helicobacter pylori switches cagPAI-dependent signaling leading to the secretion of IL-8 and TNF-α. PloS One. (2011) 6(5):e19614. doi: 10.1371/journal.pone.0019614
35. Smith SM, Freeley M, Moynagh PN, Kelleher DP. Differential modulation of Helicobacter pylori lipopolysaccharide-mediated TLR2 signaling by individual Pellino proteins. Helicobacter. (2017) 22(1):10.1111/hel.12325. doi: 10.1111/hel.2017.22.issue-1
36. Neuper T, Frauenlob T, Sarajlic M, Posselt G, Wessler S, Horejs-Hoeck J. TLR2, TLR4 and TLR10 shape the cytokine and chemokine release of H. pylori-infected human DCs. Int J Mol Sci. (2020) 21(11):3897. doi: 10.3390/ijms21113897
37. Moschen AR, Tilg H, Raine T. IL-12, IL-23 and IL-17 in IBD: immunobiology and therapeutic targeting. Nat Rev Gastroenterol Hepatol. (2019) 16(3):185–96. doi: 10.1038/s41575-018-0084-8
38. Kotla NG, Rochev Y. IBD disease-modifying therapies: insights from emerging therapeutics. Trends Mol Med. (2023) 29(3):241–53. doi: 10.1016/j.molmed.2023.01.001
39. Levin AD, Wildenberg ME, van den Brink GR. Mechanism of action of anti-TNF therapy in inflammatory bowel disease. J Crohns Colitis. (2016) 10(8):989–97. doi: 10.1093/ecco-jcc/jjw053
40. Digby-Bell JL, Atreya R, Monteleone G, Powell N. Interrogating host immunity to predict treatment response in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. (2020) 17(1):9–20. doi: 10.1038/s41575-019-0228-5
41. Danne C, Skerniskyte J, Marteyn B, Sokol H. Neutrophils: from IBD to the gut microbiota. Nat Rev Gastroenterol Hepatol. (2024) 21(3):184–97. doi: 10.1038/s41575-023-00871-3
42. Hegarty LM, Jones GR, Bain CC. Macrophages in intestinal homeostasis and inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. (2023) 20(8):538–53. doi: 10.1038/s41575-023-00769-0
43. Frede A, Czarnewski P, Monasterio G, Tripathi KP, Bejarano DA, Ramirez Flores RO, et al. B cell expansion hinders the stroma-epithelium regenerative cross talk during mucosal healing. Immunity. (2022) 55(12):2336–2351.e12. doi: 10.1016/j.immuni.2022.11.002
44. Bauché D, Joyce-Shaikh B, Jain R, Grein J, Ku KS, Blumenschein WM, et al. LAG3(+) regulatory T cells restrain interleukin-23-producing CX3CR1(+) gut-resident macrophages during group 3 innate lymphoid cell-driven colitis. Immunity. (2018) 49(2):342–352.e5. doi: 10.1016/j.immuni.2018.07.007
45. Hedl M, Lahiri A, Ning K, Cho JH, Abraham C. Pattern recognition receptor signaling in human dendritic cells is enhanced by ICOS ligand and modulated by the Crohn’s disease ICOSLG risk allele. Immunity. (2014) 40(5):734–46. doi: 10.1016/j.immuni.2014.04.011
46. Mori N, Sato H, Hayashibara T, Senba M, Geleziunas R, Wada A, et al. Helicobacter pylori induces matrix metalloproteinase-9 through activation of nuclear factor kappaB. Gastroenterology. (2003) 124(4):983–92. doi: 10.1053/gast.2003.50152
47. Slomiany BL, Slomiany A. Role of protein kinase D2 phosphorylation on Tyr in modulation by ghrelin of Helicobacter pylori-induced up-regulation in gastric mucosal matrix metalloproteinase-9 (MMP-9) secretion. Inflammopharmacology. (2016) 24:119–26. doi: 10.1007/s10787-016-0267-2
48. Bagheri N, Sadeghiani M, Rahimian G, Mahsa M, Shafigh M, Rafieian-Kopaei M, et al. Correlation between expression of MMP-9 and MMP-3 in Helicobacter pylori infected patients with different gastroduodenal diseases. Arab J Gastroenterol. (2018) 19(4):148–54. doi: 10.1016/j.ajg.2018.11.001
49. Helmin-Basa A, Kubiszewska I, Wiese-Szadkowska M, Strzyżewska E, Skalska-Bugała A, Balcerowska S, et al. Expression of matrix metalloproteinases in the circulating immune cells in children with helicobacter pylori infection-correlation with clinical factors. Int J Mol Sci. (2023) 24(21):15660. doi: 10.3390/ijms242115660
50. Minyaylo O, Ponomarenko I, Reshetnikov E, Dvornyk V, Churnosov M. Functionally significant polymorphisms of the MMP-9 gene are associated with peptic ulcer disease in the Caucasian population of Central Russia. Sci Rep. (2021) 11(1):13515. doi: 10.1038/s41598-021-92527-y
51. Dvornyk V, Ponomarenko I, Minyaylo O, Reshetnikov E, Churnosov M. Association of the functionally significant polymorphisms of the MMP9 gene with H. pylori-positive gastric ulcer in the Caucasian population of Central Russia. PloS One. (2021) 16(9):e0257060. doi: 10.1371/journal.pone.0257060
52. Slomiany BL, Slomiany A. Helicobacter pylori-elicited induction in gastric mucosal matrix metalloproteinase-9 (MMP-9) release involves ERK-dependent cPLA2 activation and its recruitment to the membrane-localized Rac1/p38 complex. Inflammopharmacology. (2016) 24:87–95. doi: 10.1007/s10787-016-0261-8
53. Pan G, Wang X, Wang Y, Li R, Li G, He Y, et al. Helicobacter pylori promotes gastric cancer progression by upregulating semaphorin 5A expression via ERK/MMP9 signaling. Mol Ther Oncolytics. (2021) 22:256–64. doi: 10.1016/j.omto.2021.06.002
54. Munakata S, Tashiro Y, Nishida C, Sato A, Komiyama H, Shimazu H, et al. Inhibition of plasmin protects against colitis in mice by suppressing matrix metalloproteinase 9-mediated cytokine release from myeloid cells. Gastroenterology. (2015) 148(3):565–578.e4. doi: 10.1053/j.gastro.2014.12.001
55. Koelink PJ, Overbeek SA, Braber S, Morgan ME, Henricks PA, Abdul Roda M, et al. Collagen degradation and neutrophilic infiltration: a vicious circle in inflammatory bowel disease. Gut. (2014) 63(4):578–87. doi: 10.1136/gutjnl-2012-303252
56. Hanifeh M, Rajamäki MM, Syrjä P, Mäkitalo L, Kilpinen S, Spillmann T. Identification of matrix metalloproteinase-2 and -9 activities within the intestinal mucosa of dogs with chronic enteropathies. Acta Vet Scand. (2018) 60(1):16. doi: 10.1186/s13028-018-0371-y
57. Kofla-Dłubacz A, Matusiewicz M, Krzesiek E, Noga L, Iwańczak B. Metalloproteinase-3 and -9 as novel markers in the evaluation of ulcerative colitis activity in children. Adv Clin Exp Med. (2014) 23(1):103–10. doi: 10.17219/acem/37031
58. Monteleone G, Caruso R, Fina D, Peluso I, Gioia V, Stolfi C, et al. Control of matrix metalloproteinase production in human intestinal fibroblasts by interleukin 21. Gut. (2006) 55(12):1774–80. doi: 10.1136/gut.2006.093187
59. de Bruyn M, Breynaert C, Arijs I, De Hertogh G, Geboes K, Thijs G, et al. Inhibition of gelatinase B/MMP-9 does not attenuate colitis in murine models of inflammatory bowel disease. Nat Commun. (2017) 8:15384. doi: 10.1038/ncomms15384
60. Al-Sadi R, Youssef M, Rawat M, Guo S, Dokladny K, Haque M, et al. MMP-9-induced increase in intestinal epithelial tight permeability is mediated by p38 kinase signaling pathway activation of MLCK gene. Am J Physiol Gastrointest Liver Physiol. (2019) 316(2):G278–g290. doi: 10.1152/ajpgi.00126.2018
61. Al-Sadi R, Engers J, Haque M, King S, Al-Omari D, Ma TY. Matrix Metalloproteinase-9 (MMP-9) induced disruption of intestinal epithelial tight junction barrier is mediated by NF-κB activation. PloS One. (2021) 16(4):e0249544. doi: 10.1371/journal.pone.0249544
62. Lucafò M, Pugnetti L, Bramuzzo M, Curci D, Di Silvestre A, Marcuzzi A, et al. Long non-coding RNA GAS5 and intestinal MMP2 and MMP9 expression: A translational study in pediatric patients with IBD. Int J Mol Sci. (2019) 20(21):5280. doi: 10.3390/ijms20215280
63. Walana W, Ye Y, Li M, Wang J, Wang B, Cheng JW, et al. IL-8 antagonist, CXCL8(3-72)K11R/G31P coupled with probiotic exhibit variably enhanced therapeutic potential in ameliorating ulcerative colitis. BioMed Pharmacother. (2018) 103:253–61. doi: 10.1016/j.biopha.2018.04.008
64. Bouchard F, Bélanger SD, Biron-Pain K, St-Pierre Y. EGR-1 activation by EGF inhibits MMP-9 expression and lymphoma growth. Blood. (2010) 116(5):759–66. doi: 10.1182/blood-2009-12-257030
65. Chen Y, Ouyang D, Wang Y, Pan Q, Zhao J, Chen H, et al. EBV promotes TCR-T-cell therapy resistance by inducing CD163+M2 macrophage polarization and MMP9 secretion. J Immunother Cancer. (2024) 12(6):e008375. doi: 10.1136/jitc-2023-008375
66. Chen H, Ye Y, Yang Y, Zhong M, Gu L, Han Z, et al. TIPE-mediated up-regulation of MMP-9 promotes colorectal cancer invasion and metastasis through MKK-3/p38/NF-κB pro-oncogenic signaling pathway. Signal Transduct Target Ther. (2020) 5(1):163. doi: 10.1038/s41392-020-00276-7
67. Chen G, Ding XF, Pressley K, Bouamar H, Wang B, Zheng G, et al. Everolimus inhibits the progression of ductal carcinoma in situ to invasive breast cancer via downregulation of MMP9 expression. Clin Cancer Res. (2020) 26(6):1486–96. doi: 10.1158/1078-0432.CCR-19-2478
68. Li Z, Sun M, Yang R, Wang Z, Zhu Q, Zhang Y, et al. Mediator complex subunit 1 promotes oral squamous cell carcinoma progression by activating MMP9 transcription and suppressing CD8(+) T cell antitumor immunity. J Exp Clin Cancer Res. (2024) 43(1):270. doi: 10.1186/s13046-024-03191-9
69. Bharatha M, Nandana MB, Praveen R, Nayaka S, Velmurugan D, Vishwanath BS, et al. Unconjugated bilirubin and its derivative ameliorate IMQ-induced psoriasis-like skin inflammation in mice by inhibiting MMP9 and MAPK pathway. Int Immunopharmacol. (2024) 130:111679. doi: 10.1016/j.intimp.2024.111679
70. Shogan BD, Belogortseva N, Luong PM, Zaborin A, Lax S, Bethel C, et al. Collagen degradation and MMP9 activation by Enterococcus faecalis contribute to intestinal anastomotic leak. Sci Transl Med. (2015) 7(286):286ra68. doi: 10.1126/scitranslmed.3010658
71. Arkelius K, Wendt TS, Andersson H, Arnou A, Gottschalk M, Gonzales RJ, et al. LOX-1 and MMP-9 inhibition attenuates the detrimental effects of delayed rt-PA therapy and improves outcomes after acute ischemic stroke. Circ Res. (2024) 134:954–69. doi: 10.1161/CIRCRESAHA.123.323371
72. Wang X, Maretti-Mira AC, Wang L, DeLeve LD. Liver-selective MMP-9 inhibition in the rat eliminates ischemia-reperfusion injury and accelerates liver regeneration. Hepatology. (2019) 69(1):314–28. doi: 10.1002/hep.30169
73. Lin C, Hu Y, Lin Z, Du L, Hu Y, Ouyang L, et al. MMP-9 responsive hydrogel promotes diabetic wound healing by suppressing ferroptosis of endothelial cells. Bioact Mater. (2025) 43:240–54. doi: 10.1016/j.bioactmat.2024.09.006
74. Wiese JJ, Manna S, Kühl AA, Fascì A, Elezkurtaj S, Sonnenberg E, et al. Myenteric plexus immune cell infiltrations and neurotransmitter expression in crohn’s disease and ulcerative colitis. J Crohns Colitis. (2024) 18(1):121–33. doi: 10.1093/ecco-jcc/jjad122
75. Globig AM, Mayer LS, Heeg M, Andrieux G, Ku M, Otto-Mora P, et al. Exhaustion of CD39-expressing CD8(+) T cells in crohn’s disease is linked to clinical outcome. Gastroenterology. (2022) 163(4):965–981.e31. doi: 10.1053/j.gastro.2022.06.045
76. Lu H, Suo Z, Lin J, Cong Y, Liu Z. Monocyte-macrophages modulate intestinal homeostasis in inflammatory bowel disease. biomark Res. (2024) 12(1):76. doi: 10.1186/s40364-024-00612-x
77. Ai L, Ren Y, Zhu M, Lu S, Qian Y, Chen Z, et al. Synbindin restrains proinflammatory macrophage activation against microbiota and mucosal inflammation during colitis. Gut. (2021) 70(12):2261–72. doi: 10.1136/gutjnl-2020-321094
78. Aschenbrenner D, Quaranta M, Banerjee S, Ilott N, Jansen J, Steere B, et al. Deconvolution of monocyte responses in inflammatory bowel disease reveals an IL-1 cytokine network that regulates IL-23 in genetic and acquired IL-10 resistance. Gut. (2021) 70(6):1023–36. doi: 10.1136/gutjnl-2020-321731
79. Griffin H, Ceron-Gutierrez L, Gharahdaghi N, Ebrahimi S, Davies S, Loo PS, et al. Neutralizing autoantibodies against interleukin-10 in inflammatory bowel disease. N Engl J Med. (2024) 391(5):434–41. doi: 10.1056/NEJMoa2312302
80. Bernshtein B, Curato C, Ioannou M, Thaiss CA, Gross-Vered M, Kolesnikov M, et al. IL-23-producing IL-10Rα-deficient gut macrophages elicit an IL-22-driven proinflammatory epithelial cell response. Sci Immunol. (2019) 4(36):eaau6571. doi: 10.1126/sciimmunol.aau6571
81. Tang X, Hu W, You W, Fang T. Exploration of key ferroptosis-related genes and immune infiltration in Crohn’s disease using bioinformatics. Sci Rep. (2023) 13(1):12769. doi: 10.1038/s41598-023-40093-w
82. De Zuani M, Dal Secco C, Frossi B. Mast cells at the crossroads of microbiota and IBD. Eur J Immunol. (2018) 48(12):1929–37. doi: 10.1002/eji.201847504
83. Lai LJ, Shen J, Ran ZH. Natural killer T cells and ulcerative colitis. Cell Immunol. (2019) 335:1–5. doi: 10.1016/j.cellimm.2018.08.010
84. Kiss M, Czimmerer Z, Nagy L. The role of lipid-activated nuclear receptors in shaping macrophage and dendritic cell function: From physiology to pathology. J Allergy Clin Immunol. (2013) 132(2):264–86. doi: 10.1016/j.jaci.2013.05.044
85. Cai N, Cheng K, Ma Y, Liu S, Tao R, Li Y, et al. Targeting MMP9 in CTNNB1 mutant hepatocellular carcinoma restores CD8(+) T cell-mediated antitumor immunity and improves anti-PD-1 efficacy. Gut. (2024) 73(6):985–99. doi: 10.1136/gutjnl-2023-331342
86. Ding H, Hu H, Tian F, Liang H. A dual immune signature of CD8+ T cells and MMP9 improves the survival of patients with hepatocellular carcinoma. Biosci Rep. (2021) 41(3):BSR20204219. doi: 10.1042/BSR20204219
87. Gallazzi M, Baci D, Mortara L, Bosi A, Buono G, Naselli A, et al. Prostate cancer peripheral blood NK cells show enhanced CD9, CD49a, CXCR4, CXCL8, MMP-9 production and secrete monocyte-recruiting and polarizing factors. Front Immunol. (2020) 11:586126. doi: 10.3389/fimmu.2020.586126
88. Shen WY, Li H, Zha AH, Luo RY, Zhang YL, Luo C, et al. Platelets reprogram monocyte functions by secreting MMP-9 to benefit postoperative outcomes following acute aortic dissection. iScience. (2023) 26(6):106805. doi: 10.1016/j.isci.2023.106805
89. Zhou C, Liang T, Jiang J, Chen J, Chen T, Huang S, et al. MMP9 and STAT1 are biomarkers of the change in immune infiltration after anti-tuberculosis therapy, and the immune status can identify patients with spinal tuberculosis. Int Immunopharmacol. (2023) 116:109588. doi: 10.1016/j.intimp.2022.109588
90. Jiguet-Jiglaire C, Boissonneau S, Denicolai E, Hein V, Lasseur R, Garcia J, et al. Plasmatic MMP9 released from tumor-infiltrating neutrophils is predictive for bevacizumab efficacy in glioblastoma patients: an AVAglio ancillary study. Acta Neuropathol Commun. (2022) 10(1):1. doi: 10.1186/s40478-021-01305-4
91. Talayev V, Svetlova M, Zaichenko I, Voronina E, Babaykina O, Neumoina N, et al. CCR6(+) T helper cells and regulatory T cells in the blood and gastric mucosa during Helicobacter pylori infection. Helicobacter. (2024) 29(3):e13097. doi: 10.1111/hel.13097
92. Cook KW, Letley DP, Ingram RJ, Staples E, Skjoldmose H, Atherton JC, et al. CCL20/CCR6-mediated migration of regulatory T cells to the Helicobacter pylori-infected human gastric mucosa. Gut. (2014) 63(10):1550–9. doi: 10.1136/gutjnl-2013-306253
93. Tsai HF, Hsu PN. Interplay between Helicobacter pylori and immune cells in immune pathogenesis of gastric inflammation and mucosal pathology. Cell Mol Immunol. (2010) 7(4):255–9. doi: 10.1038/cmi.2010.2
94. Koch MRA, Gong R, Friedrich V, Engelsberger V, Kretschmer L, Wanisch A, et al. CagA-specific gastric CD8(+) tissue-resident T cells control helicobacter pylori during the early infection phase. Gastroenterology. (2023) 164(4):550–66. doi: 10.1053/j.gastro.2022.12.016
95. Xie J, Wen J, Chen C, Luo M, Hu B, Wu D, et al. Notch 1 is involved in CD4(+) T cell differentiation into th1 subtype during helicobacter pylori infection. Front Cell Infect Microbiol. (2020) 10:575271. doi: 10.3389/fcimb.2020.575271
96. Latour YL, Sierra JC, Finley JL, Asim M, Barry DP, Allaman MM, et al. Cystathionine γ-lyase exacerbates Helicobacter pylori immunopathogenesis by promoting macrophage metabolic remodeling and activation. JCI Insight. (2022) 7(12):e155338. doi: 10.1172/jci.insight.155338
97. Deng Y, Su W, Zhu J, Ji H, Zhou X, Geng J, et al. Helicobacter pylori infection disturbs the tumor immune microenvironment and is associated with a discrepant prognosis in gastric de novo diffuse large B-cell lymphoma. J Immunother Cancer. (2021) 9(10):e002947. doi: 10.1136/jitc-2021-002947
98. Mayerhoefer ME, Raderer M, Lamm W, Weber M, Kiesewetter B, Rohrbeck J, et al. CXCR4 PET/MRI for follow-up of gastric mucosa-associated lymphoid tissue lymphoma after first-line Helicobacter pylori eradication. Blood. (2022) 139(2):240–4. doi: 10.1182/blood.2021013239
99. Tsai HC, Chen YH. Dexamethasone downregulates the expressions of MMP-9 and oxidative stress in mice with eosinophilic meningitis caused by Angiostrongylus cantonensis infection. Parasitology. (2021) 148(2):187–97. doi: 10.1017/S0031182020001870
100. Martinelli S, Anderzhanova EA, Bajaj T, Wiechmann S, Dethloff F, Weckmann K, et al. Stress-primed secretory autophagy promotes extracellular BDNF maturation by enhancing MMP9 secretion. Nat Commun. (2021) 12(1):4643. doi: 10.1038/s41467-021-24810-5
101. Wang C, Li Q, Yang H, Gao C, Du Q, Zhang C, et al. MMP9, CXCR1, TLR6, and MPO participant in the progression of coronary artery disease. J Cell Physiol. (2020) 235(11):8283–92. doi: 10.1002/jcp.v235.11
102. Nosaka M, Ishida Y, Kimura A, Kuninaka Y, Inui M, Mukaida N, et al. Absence of IFN-γ accelerates thrombus resolution through enhanced MMP-9 and VEGF expression in mice. J Clin Invest. (2011) 121(7):2911–20. doi: 10.1172/JCI40782
103. Liu Z, Dou C, Jia Y, Li Q, Zheng X, Yao Y, et al. RIG-I suppresses the migration and invasion of hepatocellular carcinoma cells by regulating MMP9. Int J Oncol. (2015) 46(4):1710–20. doi: 10.3892/ijo.2015.2853
Keywords: bioinformatics analysis, Helicobacter pylori, inflammatory bowel disease, diagnostic biomarkers, molecular mechanism
Citation: Zhang M, Liu T, Luo L, Zhang Y, Chen Q, Wang F and Xie Y (2024) Common diagnostic biomarkers and molecular mechanisms of Helicobacter pylori infection and inflammatory bowel disease. Front. Immunol. 15:1492810. doi: 10.3389/fimmu.2024.1492810
Received: 08 September 2024; Accepted: 13 November 2024;
Published: 05 December 2024.
Edited by:
Narendra Prasad Singh, University of South Carolina, United StatesReviewed by:
Urmi Halder, University of South Carolina, United StatesShruthi Thada, University of South Carolina, United States
Copyright © 2024 Zhang, Liu, Luo, Zhang, Chen, Wang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuxin Xie, eXV4aW54aWUyMDI0QDE2My5jb20=; Fen Wang, d2Zlbi1qdWR5QGNzdS5lZHUuY24=
†These authors have contributed equally to this work
 Minglin Zhang
Minglin Zhang Tong Liu2†
Tong Liu2† Qijiao Chen
Qijiao Chen Yuxin Xie
Yuxin Xie