- Department of Ophthalmology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China
Single-cell RNA sequencing (scRNA-seq) has facilitated a deeper comprehension of the molecular mechanisms behind eye diseases and has prompted the selection of precise therapeutic targets by examining the cellular and molecular intricacies at the single-cell level. This review delineates the pivotal role of scRNA-seq in elucidating the functions of innate immune cells within the context of ocular pathologies. Recent advancements in scRNA-seq have revealed that innate immune cells, both from the periphery and resident in the retina, are actively engaged in various stages of multiple eye diseases. Notably, resident microglia and infiltrating neutrophils exhibit swift responses during the initial phase of injury, while peripheral monocyte-derived macrophages exhibit transcriptomic profiles akin to those of activated microglia, suggesting their potential for long-term residence within the retina. The scRNA-seq analyses have underscored the cellular heterogeneity and gene expression alterations within innate immune cells, which, while sharing commonalities, exhibit disease-specific variations. These insights have not only broadened our understanding of the cellular and molecular mechanisms in eye diseases but also paved the way for the identification of candidate targets for targeted therapeutic interventions. The application of scRNA-seq technology has heralded a new era in the study of ocular pathologies, enabling a more detailed appreciation of the roles that innate immune cells play across a spectrum of eye diseases.
1 Introduction
The innate immune system serves as the first line of defense against pathogens and injuries. Innate immune cells such as microglia, monocyte-derived macrophages, and neutrophils are pivotal for the maintenance of retinal homeostasis and the formation of several inflammatory retinal disorders (1). The activation of innate immune cells is influenced by the ocular microenvironment, and traditional histological studies and inflammatory cytokine/chemokine examinations have confirmed their roles in eye diseases (2–6). Those innate immune cell-associated cytokines/chemokines can predict disease activity, some of which have become precise targets for treatment (7, 8). However, the function of each innate immune cell and their interactions remain complex and unclear. Dissecting their individual roles in pathogenesis is of great significance, as waves of innate immune response in the retina remain intricate under disease context (9–11).
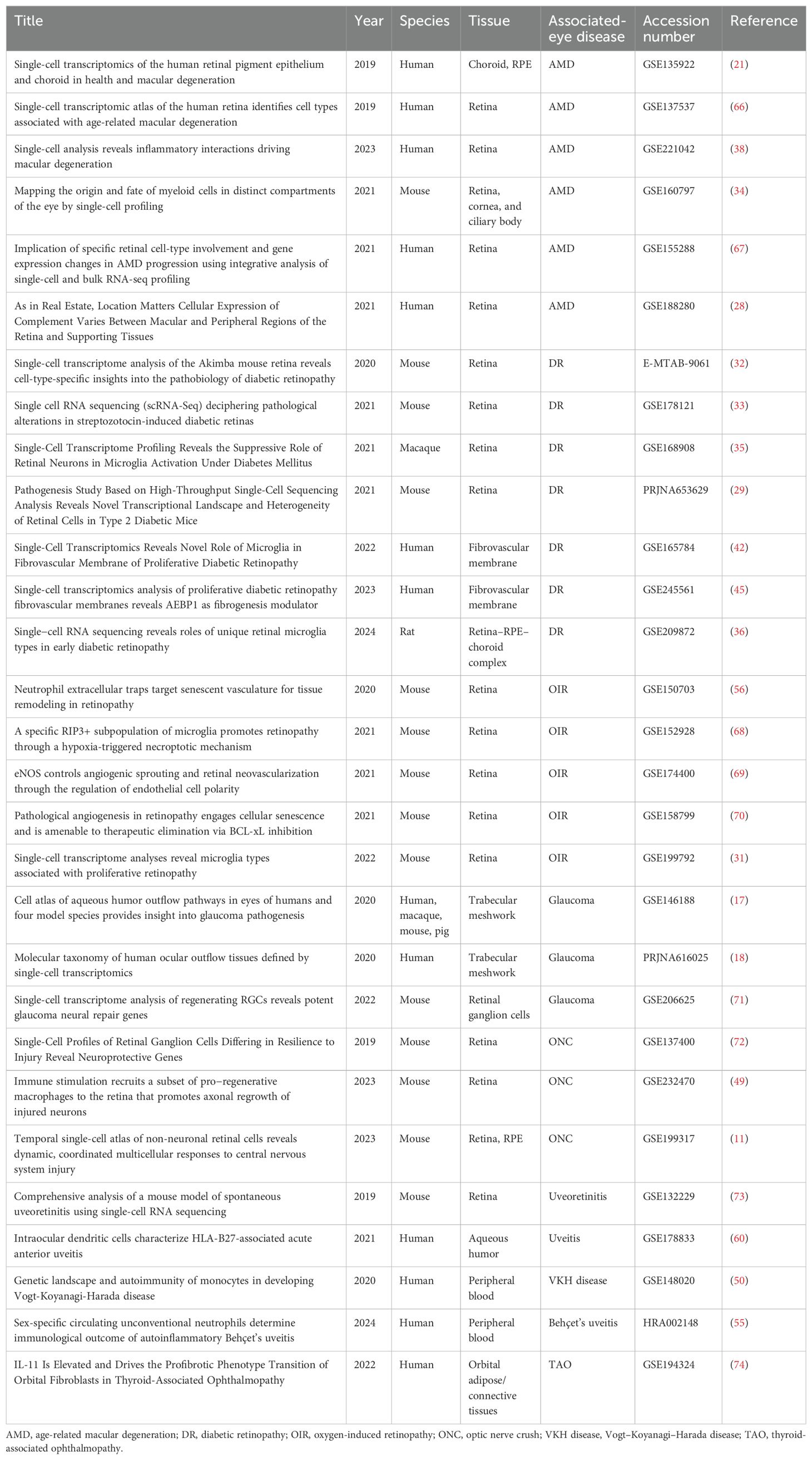
The development of single-cell RNA sequencing (scRNA-seq) has already contributed to profound discoveries. Compared with traditional bulk RNA sequencing (RNA-seq), scRNA-seq maps transcriptome to an individual cell, which has an edge on unveiling cellular composition differences among samples and assaying altered gene expression at the single-cell level, especially in studying complex tissues composed of many distinct cell types that are typically difficult to dissect experimentally like brain tissues and the retina (12, 13). The landmark study of scRNA-seq applied in ophthalmology began with murine retina profiled with Drop-seq barcoding strategy (14). Thereafter, a series of studies using scRNA-seq created a comprehensive transcriptome atlas of eye structures, including the cornea, aqueous humor, ciliary body, choroid, and retina (15–21). These available online public scRNA-seq data make it convenient for researchers to compare datasets to validate new outcomes (Table 1). In this review, we systematically summarized the roles of innate immune cells in cellular and molecular mechanisms behind several common eye diseases from the perspectives of scRNA-seq.
2 Microglia
Residing in the inner part of the neuronal retina, microglia, a specialized type of yolk sac-derived macrophage, can switch from a resting state to an activated state and work in surveillance and maintenance of homeostasis in the retina (22, 23).
Multiple types of proliferative retinopathy, such as proliferative diabetic retinopathy (PDR) and wet age-related macular degeneration (AMD), are relevant to the hyperactivation of microglia (24–27). ScRNA-seq performed on the retina and choroid from postmortem eyes found that microglia play an important role in early AMD (28). There are an increasing number of studies performing scRNA-seq on rodent models with retinal neovascularization, validating functions of microglia in proliferative retinopathy (29–34).
2.1 Microglia predominating in pathogenic cell interactions in DR
ScRNA-seq was performed on the retina of cynomolgus monkeys with non-proliferative diabetic retinopathy (NPDR) to assess the suppressive role of retinal microglia in diabetic retinopathy (DR) (35). Among the cell atlas, microglia exhibited the largest number of upregulated genes under diabetes, showing that hyperglycemia induced microglial activation and elevated inflammation levels. CellChat analysis found that microglia were the major source of TNF-α. Likewise, in a streptozotocin (STZ)-induced diabetic rat model, scRNA-seq data analysis found an inflammatory regulatory network centered on microglia in early DR (36). Here, M1 microglia were distinguished from M2 microglia by the different expression levels of CCR5, and M1 microglia were further divided into EGR2+ M1 microglia and EGR2− M1 microglia, the former of which showed a greater proinflammatory effect and participated in the regulation of retinal vascular permeability in the early stages of DR by interacting with endothelial cells (ECs).
In PDR, microglia also contribute to retinal vascular dysfunction. Ben et al. applied scRNA-seq in a diabetic mouse model and found two microglia clusters (37). The cluster more enriched in the DR group was mainly involved in inflammatory responses and exhibited immunoregulatory signatures. Abundant ligand–receptor-paired interactions were found between this microglia cluster and DR-enriched ECs. Notably, the CSF1/CSF1R signaling pathway was discovered between microglia and ECs, leading to inflammation-mediated vascular dysfunction in PDR. CSF1R signaling blockade can attenuate the inflammatory response and vascular dysfunction, which represents a promising therapeutic approach for DR treatment.
2.2 Microglia playing double-edged roles in pathological neovascularization
A novel topologically inspired machine learning pipeline based on scRNA-seq data has been developed, finding microglia with inflammasome activation signatures in AMD (38). Kuchroo et al. profiled lesions from postmortem human retinas with AMD and control retinas. During the early stage of AMD, a dry AMD-specific activated microglia cluster appeared with upregulation of APOE, TYROBP, and SPP1, while cell type-specific transcriptional changes exclusively in the subcluster of microglia in late-stage neovascular AMD displayed an inflammasome-related signature including IL1B, NOD2, and NFKB1. CellPhoneDB interaction analysis unveiled significant interactions between this neovascular-specific microglia cluster with pro-angiogenic astrocytes through interleukin-1β (IL-1β) mediating pathological neovascularization in AMD. Therefore, inhibiting microglia-derived IL-1β could provide therapeutic benefits in neovascular AMD. Interestingly, not only did microglia play pathological roles in neovascular AMD, but there also existed a cluster of microglia with antiangiogenic features. Luo et al. compared public scRNA-seq data of retinas from mice with oxygen-induced retinopathy (OIR) with neonatal mouse spine after spinal cord injury, finding a similar cluster of microglia with consistent antiangiogenic characteristics (39). Those microglia expressing antiangiogenic factor TSP-1 made up a limited quantity of microglia in the OIR retina and thus failed to play protective roles in retinal angiogenesis.
2.3 Microglia or macrophages: which contributing to the formation of FVMs?
DR can cause preventable vision impairment and blindness due to neovascularization and the formation of vitreous fibrovascular membranes (FVMs) leading to retinal hemorrhage and detachment (40). The cell components of FVMs were considered complex, including retinal glial cells, macrophages/monocytes, fibroblasts, and vascular ECs (41). Hu et al. performed scRNA-seq on surgically harvested PDR FVMs and generated a comprehensive cell atlas of FVM, among which the major cell population was found to be microglia/macrophages (42). Those myeloid cells with marker GPNMB were further identified as activated microglia, differentiated from retina-resident P2RY12+ microglia, and increased in PDR setting through pseudotime analysis, presenting both profibrotic and fibrogenic properties. Corano Scheri et al. queried the conclusion in this work but re-analyzed the online deposited raw data and considered monocyte-derived macrophages constituting the majority of immune cells (43). These contradictory results arise from the fact that monocyte-derived macrophages and microglia are indistinguishable by transcriptomic profiling, as microglia may rapidly respond to local insults by downregulating homeostatic microglial markers like P2RY12 and exhibited disease-associated markers such as the expression of macrophage marker APOE (44). Undoubtedly, monocyte-derived macrophages consisted of FVMs in this work, and scRNA-seq data of FVMs from Corano Scheri et al. also found a cluster of macrophages with marker SPP1 expressing proangiogenic genes and potentially contributing to the formation of FVMs (45). The discrepancy between the scRNA-seq data of FVMs in these two studies may be attributed to complex causes. For one thing, the heterogeneity of patients may affect the cell components of FVMs. Patients with different disease histories, for example, whether there existed vitreous hemorrhage or other systematic diseases, can exhibit varied immune cell diversity and responses. For another, different courses of DR may also contribute to this inconsistency. There is consensus that monocyte-derived macrophages activate in the early phase after injury while resident microglia adapt to pathological status in the long term (46). Therefore, further lineage tracing experiments are indispensable to determine the ontogeny of whether microglia or monocyte-derived macrophages dominate in the pathogenesis of FVM.
3 Monocyte
Apart from microglia, peripheral immune cells, such as monocytes, can also invade and respond to injuries in the retina. Recent evidence shows that the persistence of monocyte-derived macrophages in both the retina and subretinal space exacerbates pathogenic inflammation, participating in retina degeneration (47).
3.1 Monocyte-derived macrophages remaining in retina long term after retinal degeneration
In a mouse model of inducible photoreceptor degeneration, scRNA-seq revealed distinct populations of monocyte-derived macrophages after degeneration (48). Monocyte-derived cells adopted a microglia-like genomic phenotype and remained in the retina. Analysis of scRNA-seq data found a hybrid-like cluster of monocyte-derived macrophages with both pathological microglial and traditional monocytic markers 1 week and 20 days after retinal degeneration, which supported the hypothesis that monocyte-derived macrophages adopted a microglia-like phenotype and remained long-term in the retina suffering altered immune state.
3.2 Monocyte-derived macrophages participating in repairing period after optic nerve injury
Optic nerve crush (ONC) injury is characterized by an inflammatory cascade surrounding the optic nerve together with retinal ganglion cells. Two individual studies recently have both uncovered that peripheral monocyte-derived macrophages took part in the inflammatory resolution period after ONC as a supplement to activated resident microglia and border-associated macrophages (BAMs). Benhar et al. generated a scRNA-seq atlas at multiple time points after ONC injury, finding that retinal microglia and astrocytes activated in the early phase and recruited Ccr2+ monocytes from the circulation (11). Interactions between chemokines and their receptors were enriched between BAMs, neutrophils, and monocytes, including Ccl2, Ccl3, Ccl4, Ccl6, Ccl12, Cxcr2, and Ccr2. Those Ccr2+ monocytes gave rise to Ms4a7+ and Gpnmb+ macrophage subsets thereafter, characterized by active lipid metabolism and phagocytosis, which were mainly recruited by retinal astrocytes and microglia through Ccl2–Ccr2 signaling axis. Similarly, in the scRNA-seq data from Andries et al., microglia from the injured retina exhibited an elevated expression of the monocyte chemoattractant Ccl2, Ccl3, Ccl4, and Ccl12, recruiting monocyte-derived macrophages to ONC injury and presenting the expression of pro-regenerative secreted factors (49). Ligand–receptor interactions analysis using NicheNet identified the ligands Spp1, Thbs1, Vegfa, and Igf1 in one subset of monocyte-derived macrophages featured with pro-regenerative gene signatures, which are secreted proteins known to promote nerve survival. Therefore, monocyte-derived macrophages may function as a potential responder after ONC to promote axon regrowth and neuron regeneration via paracrine signaling.
3.3 Monocytes highly expressing inflammatory markers predicting autoimmune uveitis
Unlike other resident cell types in the retina, monocytes originate from the periphery, which can be isolated and enriched directly from peripheral blood mononuclear cells (PBMCs). ScRNA-seq was applied to encode the landscape of human monocytes from peripheral blood samples of patients with developing VKH disease (50). A disease-specific subset was identified as proinflammatory monocytes. Among differentially expressed genes (DEGs), ISG15 expression in monocytes from VKH patients was upregulated predominantly compared to normal controls. Markedly decreased ISG15 after 3 months of treatment was further functionally validated, suggesting that ISG15 level in circulating monocytes may reflect the treatment response in VKH disease. Similarly, scRNA-seq analyses of monocytes in PBMCs from Behçet’s disease revealed the accumulation of C1Q-high monocytes, the level of which can also aid in monitoring treatment efficacy (51).
4 Neutrophil
As first-line defenders against infection and injury, neutrophils can be rapidly recruited to the site of immune response (52). Evidence shows that neutrophils homing into the retina participate in the early stage of retinal diseases such as retinal detachment and AMD (10, 53).
4.1 Neutrophils acting as sentinel during rapid injury
Once an injury occurs, neutrophils are “first-line defense” cells arriving at the site of the retina. In an acute retinal arterial ischemia model of Macaca fascicularis, Li et al. used scRNA-seq and made bioinformatics analyses just 6 hours after ligation of the ophthalmic artery, in which neutrophils were a non-negligible part in immune cells after rapid ischemic injury (54). Likewise, neutrophils were found infiltrated to the injured retina very soon at the first stage after ONC injury (11, 49). The substantial increase in the number of neutrophils was transient, however; subsequently, macrophages dominated. Further analyses discovered that neutrophil infiltration mainly attributed to the upregulation of chemoattractants in retinal microglia, BAMs, and astrocytes after injury.
4.2 Imbalance of unconventional neutrophil subsets determining sex-specific outcome of Behçet’s uveitis
There exist male-biased incidence, severity, and poor prognosis of auto-inflammatory Behçet’s uveitis. Wang et al. recently applied scRNA-seq to circulating neutrophils from healthy individuals and Behçet’s uveitis to elucidate sex-specific heterogeneity in neutrophil composition under normal and auto-inflammatory conditions (55). In healthy individuals, male donors tend to have more inflammatory phenotype neutrophils, while neutrophil-mediated T-cell regulatory functions were relatively active in women. Loss of T-cell regulatory neutrophils was only observed in men under disease conditions, and dysregulation of two unconventional neutrophil (interferon-α responsive and T-cell regulatory) subsets in male patients led to male-biased severity. Interferon-α2a could directly promote unconventional neutrophil subsets to a more immune-regulatory phenotype via Annexin A1 signaling in women, specifically to maintain homeostasis. These findings prompted a promising female-specific sensitive therapy strategy to treat Behçet’s uveitis.
4.3 Neutrophil extracellular traps leading to vascular remodeling in retinal neovascularization
An unconventional role of neutrophils in vascular remodeling during late-stage sterile inflammation in OIR mice was found with the utility of scRNA-seq, where neutrophil extracellular traps (NETs) can remodel and clear pathological retinal vasculature through apoptotic elimination of senescent ECs (56). One cluster was identified as neutrophil specially enriched at P17 of OIR when vascular regression was initiated. Gene set variation analysis (GSVA) on scRNA-seq data suggested that neuronal and vascular units have distinct patterns of cellular senescence and secretory phenotypes in OIR, and further experiments confirmed that senescent vascular endothelium triggered the release of NETs. More recently, Dong et al. performed scRNA-seq on the retina–choroid complex from choroidal neovascularization (CNV) mice and found an enhanced Ccl4–Ccr5 interaction from neutrophils to natural killer (NK) cells in the CNV group (57). Their study demonstrated that NK cells interact with neutrophils, converting neutrophils from a pro-senescence state to an anti-senescence state by initiating NET formation, to eradicate senescent cells.
5 Discussion
Traditional methods and studies usually focus on single-cell type and molecular mechanisms in the context of diseases. Results from RNA-seq are robust. For example, DEGs from microglia by RNA-seq in different disease contexts contained both proinflammatory genes and negative regulator genes, of which a proinflammatory signature was predominant (58). The function of microglia was consequently regarded as promoting inflammation. However, scRNA-seq makes it possible to subdivide microglia into proinflammatory and anti-inflammatory subsets and dissect the protective roles of microglia (36). Therefore, the advent and maturation of the scRNA-seq technique have leveraged the utility of high-throughput sequencing to the single-cell level, thus making it possible to explore pathogenesis on the cellular and molecular levels in the field of ophthalmology (Figure 1A).
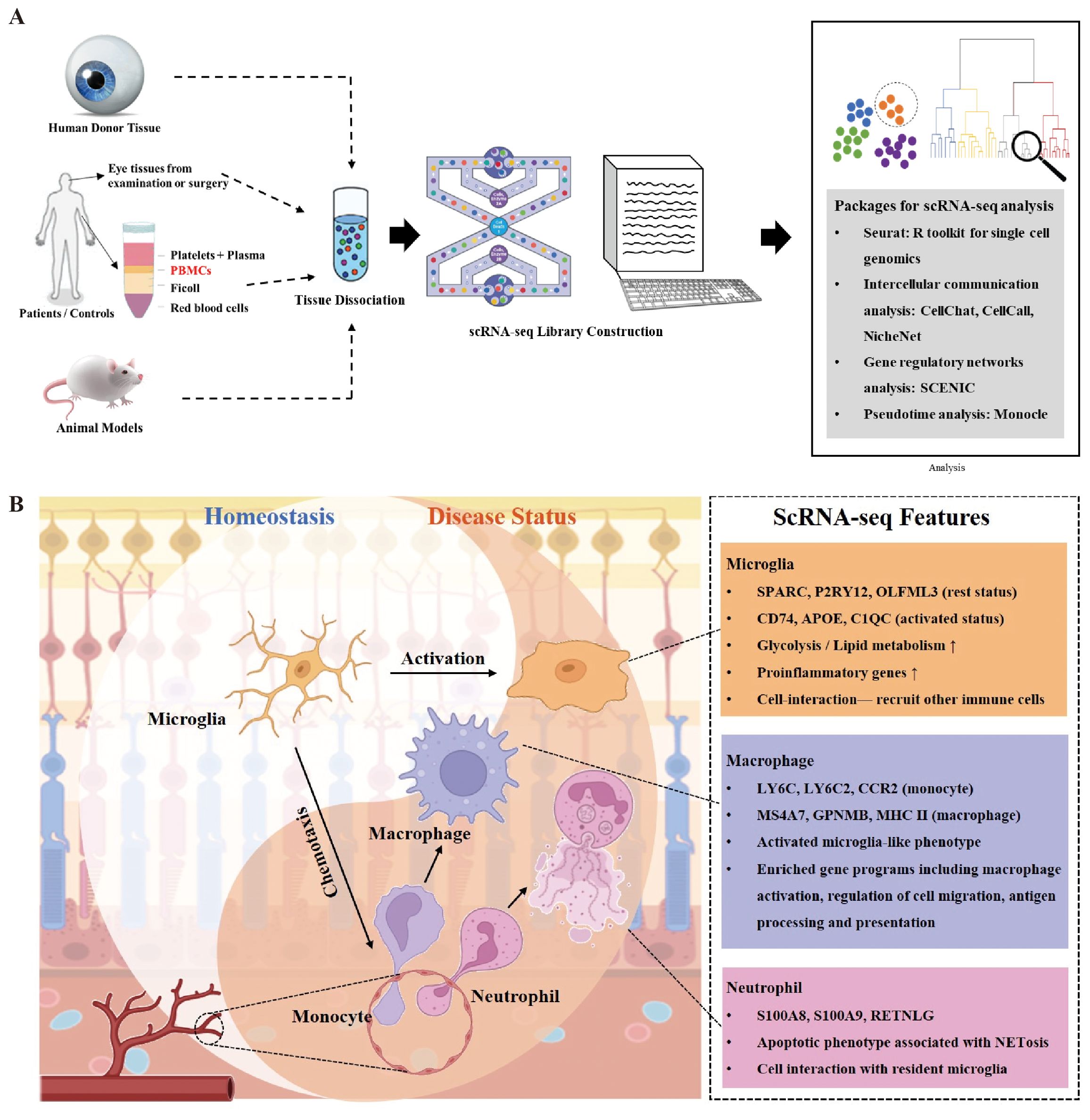
Figure 1. Overview of innate immune cell functions in retina in context of injuries and diseases. (A) There are generally four sources of ocular tissue that are used in most single-cell RNA sequencing (scRNA-seq) experiments in the field of ophthalmology, including human postmortem donors, ocular tissues obtained from examinations or surgeries (e.g., aqueous humor), peripheral blood, and animal models. After tissue dissociation, scRNA-seq library is built for further analysis, such as identification of cell clusters and investigation of gene expression using the Seurat package. Packages focusing on intercellular communication, gene regulatory networks, and pseudotime analysis are summarized. (B) Main cell types of innate immunity respond to retinal injuries and diseases including resident microglia, monocyte-derived macrophages, and neutrophils infiltrated from circulation. Rest microglia play important roles in defense of retinal homeostasis, which can turn to activated status and recruit other immune cells such as monocytes and neutrophils rapidly at the first stage of injury. Neutrophils as primary defense cells participate in cell interactions with both microglia and monocytes to amplify inflammation. Neutrophil extracellular traps (NETs) from neutrophils have multiple influences on retinal microenvironment. Monocyte-derived macrophages share similar transcriptomes with activated microglia and remain in retina after injury.
The ocular innate immune system consists of epithelial barriers, tissue-resident myeloid, and glial cells, which can initiate an inflammatory immune response and result in further recruitment of neutrophils, monocytes, and NK cells in response to infection or injury. Impaired innate immunity participates in the pathobiology of eye diseases (59). Though we mainly focus on microglia, monocytes, and neutrophils in this review, other cell types of the innate immune system are non-negligible. Recent studies applying scRNA-seq found NK cells play important roles in NET formation to inhibit pathological retinal neovascularization, while dendritic cells featured the most active intercellular interactions in HLA-B27-positive uveitis (57, 60). Comprehensive cell atlas in retinal diseases established by scRNA-seq has distinguished classical unremarkable innate immune cells as novel pathogenic cell types and uncovered new pathological mechanisms in innate immune cells throughout the stages of diseases in the retina (Figure 1B).
The role of the scRNA-seq technique can be generally divided into three aspects. 1) It acts in a monodrama. ScRNA-seq is an unbiased method to establish a comprehensive cell atlas. 2) It functions as a whistleblower to start the show, which means scRNA-seq can make disease-specific cell types, and DEGs stand out so as to explore new directions for pathogenesis. 3) It has a flexible role throughout the show, meaning it can create a comprehensive cell atlas, analyze DEGs and susceptible pathways, validate cell proportions and gene expression after intervention, explore interactions between cells, and so forth, once needed.
However, there do exist limitations. First, it is known that transcript expressions do not always correlate with protein levels, so it is necessary to validate levels of specific transcripts identified by scRNA-seq at the protein level. Second, analyses of scRNA-seq data may obtain biased and misleading results, as the expensive costs of this technique limit the number of samples in one single study. Substantial samples are needed in statistical analysis to reach a conclusion. Therefore, it is no wonder that different studies applying the scRNA-seq technique in the same pathological condition sometimes found contradictory results (43). A larger number of samples or integration with accessible public scRNA-seq data may reduce the possibility of drawing incorrect conclusions. Third, though scRNA-seq data can reveal the metabolic phenotype of disease-associated cell clusters at the transcriptional level, it is hard to elucidate the cause-and-effect relationship between metabolic phenotype and pathological phenotype. It is confusing whether the metabolic switch of cells causes diseases or whether metabolic changes are adaptations to disease conditions. The efficacy of interfering cells’ metabolic conditions to prevent disease progression remains hazy. ScRNA-seq data can provide clues, while solid evidence requires further experiments to confirm them. A recent study applying scRNA-seq identified activated retinal microglia with metabolic and inflammatory disturbances as the main source of IL-1β in STZ-induced DR mouse retina. Metabolomics analyses further confirmed that activated microglia strongly increased the level of purine metabolism and glycolysis, together with significant metabolism changes in triacylglycerol accumulation. Treatment with both glycolysis and triacylglycerol synthesis inhibitor attenuated IL-1β expression (61). With the rapid development of single-cell multi-omics technologies, integrating transcriptome, genome, epigenome, epitranscriptome, proteome, metabolome, and other omics and acquiring more precise information at the same time have become possible, which may solve these problems (62). A comprehensive single-cell multi-omics atlas of the human retina has been established with a combination of angle-nuclei RNA-seq and single-nuclei ATAC-seq (63). Single-cell multi-omics analyses have uncovered pathogenic genes in eye diseases (64, 65). Further studies should make efforts to integrate transcriptome with proteome or metabolome at the single-cell level to elucidate pathological cause-and-effect relationships behind eye diseases from genotype to phenotype. In addition, it is worth noting that the result of clusterization and annotation of cells may be subjective to some degree with different researchers. For example, it is difficult to distinguish microglia from macrophages, as they share many markers at the transcriptomic level (31, 43, 44). Therefore, it remains technically challenging due to the phenotypic overlap between homologous cells, and validation needs further experiments. More difficultly, identifying the small subset of cellular populations that drive diseases in scRNA-seq data remains a challenge. New methods like Cellular Analysis with Topology and Condensation Homology (CATCH) have been developed to solve this problem (38).
6 Conclusion
In the last few years, advancements in scRNA-seq have built a comprehensive cell atlas for multiple eye diseases. Roles of innate immune cells and their complex interactions in eye diseases have been refreshed thanks to the development of scRNA-seq. Emphasis on innate immune cells in eye diseases will be greater with a much deeper elucidation of accessible public scRNA-seq data.
Author contributions
CL: Conceptualization, Methodology, Writing – original draft. XM: Conceptualization, Methodology, Writing – review & editing. SY: Conceptualization, Methodology, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Key Research and Development Program of China (2017YFA0104103) and the National Natural Science Foundation of China (81870694, 82201239).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Murakami Y, Ishikawa K, Nakao S, Sonoda K-H. Innate immune response in retinal homeostasis and inflammatory disorders. Prog Retin Eye Res. (2020) 74:100778. doi: 10.1016/j.preteyeres.2019.100778
2. Streilein JW, Stein-Streilein J. Does innate immune privilege exist? J Leukoc Biol. (2000) 67:479–87. doi: 10.1002/jlb.67.4.479
3. Gupta N, Brown KE, Milam AH. Activated microglia in human retinitis pigmentosa, late-onset retinal degeneration, and age-related macular degeneration. Exp Eye Res. (2003) 76:463–71. doi: 10.1016/S0014-4835(02)00332-9
4. Stitt AW, Gardiner TA, Archer DB. Histological and ultrastructural investigation of retinal microaneurysm development in diabetic patients. Br J Ophthalmol. (1995) 79:362–7. doi: 10.1136/bjo.79.4.362
5. Yoshimura T, Sonoda K-h, Sugahara M, Mochizuki Y, Enaida H, Oshima Y, et al. Comprehensive analysis of inflammatory immune mediators in vitreoretinal diseases. PloS One. (2009) 4:e8158. doi: 10.1371/journal.pone.0008158
6. Oh H, Takagi H, Takagi C, Suzuma K, Otani A, Ishida K, et al. The potential angiogenic role of macrophages in the formation of choroidal neovascular membranes. Invest Ophthalmol Vis Sci. (1999) 40:1891–8.
7. Qiao H, Sonoda K-H, Ariyama A, Kuratomi Y, Kawano Y-I, Ishibashi T. CXCR2 Expression on neutrophils is upregulated during the relapsing phase of ocular Behcet disease. Curr Eye Res. (2005) 30:195–203. doi: 10.1080/02713680490904331
8. Chun DW, Heier JS, Topping TM, Duker JS, Bankert JM. A pilot study of multiple intravitreal injections of ranibizumab in patients with center-involving clinically significant diabetic macular edema. Ophthalmology. (2006) 113:1706–12. doi: 10.1016/j.ophtha.2006.04.033
9. Cruz-Herranz A, Oertel FC, Kim K, Cantó E, Timmons G, Sin JH, et al. Distinctive waves of innate immune response in the retina in experimental autoimmune encephalomyelitis. JCI Insight. (2021) 6(11):e149228. doi: 10.1172/jci.insight.149228
10. Maidana DE, Gonzalez-Buendia L, Pastor-Puente S, Naqvi A, Paschalis E, Kazlauskas A, et al. Peripheral monocytes and neutrophils promote photoreceptor cell death in an experimental retinal detachment model. Cell Death Dis. (2023) 14:834. doi: 10.1038/s41419-023-06350-6
11. Benhar I, Ding J, Yan W, Whitney IE, Jacobi A, Sud M, et al. Temporal single-cell atlas of non-neuronal retinal cells reveals dynamic, coordinated multicellular responses to central nervous system injury. Nat Immunol. (2023) 24:700–13. doi: 10.1038/s41590-023-01437-w
12. Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. (2015) 16:133–45. doi: 10.1038/nrg3833
13. Grun D, van Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell. (2015) 163:799–810. doi: 10.1016/j.cell.2015.10.039
14. Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. (2015) 161:1202–14. doi: 10.1016/j.cell.2015.05.002
15. Li DQ, Kim S, Li JM, Gao Q, Choi J, Bian F, et al. Single-cell transcriptomics identifies limbal stem cell population and cell types mapping its differentiation trajectory in limbal basal epithelium of human cornea. Ocul Surf. (2021) 20:20–32. doi: 10.1016/j.jtos.2020.12.004
16. Kaplan N, Wang J, Wray B, Patel P, Yang W, Peng H, et al. Single-cell RNA transcriptome helps define the limbal/corneal epithelial stem/early transit amplifying cells and how autophagy affects this population. Invest Ophthalmol Vis Sci. (2019) 60:3570–83. doi: 10.1167/iovs.19-27656
17. van Zyl T, Yan W, McAdams A, Peng Y-R, Shekhar K, Regev A, et al. Cell atlas of aqueous humor outflow pathways in eyes of humans and four model species provides insight into glaucoma pathogenesis. Proc Natl Acad Sci U S A. (2020) 117:10339–49. doi: 10.1073/pnas.2001250117
18. Patel G, Fury W, Yang H, Gomez-Caraballo M, Bai Y, Yang T, et al. Molecular taxonomy of human ocular outflow tissues defined by single-cell transcriptomics. Proc Natl Acad Sci U S A. (2020) 117:12856–67. doi: 10.1073/pnas.2001896117
19. Youkilis JC, Bassnett S. Single-cell RNA-sequencing analysis of the ciliary epithelium and contiguous tissues in the mouse eye. Exp Eye Res. (2021) 213:108811. doi: 10.1016/j.exer.2021.108811
20. Lukowski SW, Lo CY, Sharov AA, Nguyen Q, Fang L, Hung SS, et al. A single-cell transcriptome atlas of the adult human retina. EMBO J. (2019) 38:e100811. doi: 10.15252/embj.2018100811
21. Voigt AP, Mulfaul K, Mullin NK, Flamme-Wiese MJ, Giacalone JC, Stone EM, et al. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc Natl Acad Sci U S A. (2019) 116:24100–7. doi: 10.1073/pnas.1914143116
22. Fernandes A, Miller-Fleming L, Pais TF. Microglia and inflammation: conspiracy, controversy or control? Cell Mol Life Sci. (2014) 71:3969–85. doi: 10.1007/s00018-014-1670-8
23. Vecino E, Rodriguez FD, Ruzafa N, Pereiro X, Sharma SC. Glia-neuron interactions in the mammalian retina. Prog Retin Eye Res. (2016) 6(11):e149228. doi: 10.1016/j.preteyeres.2015.06.003
24. Blasiak J, Sobczuk P, Pawlowska E, Kaarniranta K. Interplay between aging and other factors of the pathogenesis of age-related macular degeneration. Ageing Res Rev. (2022) 81:101735. doi: 10.1016/j.arr.2022.101735
25. Xu Y, Cui K, Li J, Tang X, Lin J, Lu X, et al. Melatonin attenuates choroidal neovascularization by regulating macrophage/microglia polarization via inhibition of RhoA/ROCK signaling pathway. J Pineal Res. (2020) 69:e12660. doi: 10.1111/jpi.12660
26. Hata M, Hata M, Andriessen EM, Juneau R, Pilon F, Crespo-Garcia S, et al. Early-life peripheral infections reprogram retinal microglia and aggravate neovascular age-related macular degeneration in later life. J Clin Invest. (2023) 133(4):e159757. doi: 10.1172/JCI159757
27. Kinuthia UM, Wolf A, Langmann T. Microglia and inflammatory responses in diabetic retinopathy. Front Immunol. (2020) 11:564077. doi: 10.3389/fimmu.2020.564077
28. Zauhar R, Biber J, Jabri Y, Kim M, Hu J, Kaplan L, et al. As in real estate, location matters: cellular expression of complement varies between macular and peripheral regions of the retina and supporting tissues. Front Immunol. (2022) 13:895519. doi: 10.3389/fimmu.2022.895519
29. Niu T, Fang J, Shi X, Zhao M, Xing X, Wang Y, et al. Pathogenesis study based on high-throughput single-cell sequencing analysis reveals novel transcriptional landscape and heterogeneity of retinal cells in type 2 diabetic mice. Diabetes. (2021) 70:1185–97. doi: 10.2337/db20-0839
30. Mao P, Shen Y, Mao X, Liu K, Zhong J. The single-cell landscape of alternative transcription start sites of diabetic retina. Exp Eye Res. (2023) 233:109520. doi: 10.1016/j.exer.2023.109520
31. Liu Z, Shi H, Xu J, Yang Q, Ma Q, Mao X, et al. Single-cell transcriptome analyses reveal microglia types associated with proliferative retinopathy. JCI Insight. (2022) 7(23):e160940. doi: 10.1172/jci.insight.160940
32. Van Hove I, De Groef L, Boeckx B, Modave E, Hu T-T, Beets K, et al. Single-cell transcriptome analysis of the Akimba mouse retina reveals cell-type-specific insights into the pathobiology of diabetic retinopathy. Diabetologia. (2020) 63:2235–48. doi: 10.1007/s00125-020-05218-0
33. Sun L, Wang R, Hu G, Liu H, Lv K, Duan Y, et al. Single cell RNA sequencing (scRNA-Seq) deciphering pathological alterations in streptozotocin-induced diabetic retinas. Exp Eye Res. (2021) 210:108718. doi: 10.1016/j.exer.2021.108718
34. Wieghofer P, Hagemeyer N, Sankowski R, Schlecht A, Staszewski O, Amann L, et al. Mapping the origin and fate of myeloid cells in distinct compartments of the eye by single-cell profiling. EMBO J. (2021) 40:e105123. doi: 10.15252/embj.2020105123
35. Xiao Y, Hu X, Fan S, Zhong J, Mo X, Liu X, et al. Single-cell transcriptome profiling reveals the suppressive role of retinal neurons in microglia activation under diabetes mellitus. Front Cell Dev Biol. (2021) 9:680947. doi: 10.3389/fcell.2021.680947
36. Wang Y, Yang X, Zhang Y, Hong L, Xie Z, Jiang W, et al. Single-cell RNA sequencing reveals roles of unique retinal microglia types in early diabetic retinopathy. Diabetol Metab Syndr. (2024) 16:49. doi: 10.1186/s13098-024-01282-3
37. Ben S, Ma Y, Bai Y, Zhang Q, Zhao Y, Xia J, et al. Microglia-endothelial cross-talk regulates diabetes-induced retinal vascular dysfunction through remodeling inflammatory microenvironment. iScience. (2024) 27:109145. doi: 10.1016/j.isci.2024.109145
38. Kuchroo M, DiStasio M, Song E, Calapkulu E, Zhang L, Ige M, et al. Single-cell analysis reveals inflammatory interactions driving macular degeneration. Nat Commun. (2023) 14:2589. doi: 10.1038/s41467-023-37025-7
39. Luo Q, Jiang Z, Jiang J, Wan L, Li Y, Huang Y, et al. Tsp-1+ microglia attenuate retinal neovascularization by maintaining the expression of Smad3 in endothelial cells through exosomes with decreased miR-27a-5p. Theranostics. (2023) 13:3689–706. doi: 10.7150/thno.84236
40. Wong TY, Cheung CM, Larsen M, Sharma S, Simo R. Diabetic retinopathy. Nat Rev Dis Primers. (2016) 2:16012. doi: 10.1038/nrdp.2016.12
41. Snead DRJ, James S, Snead MP. Pathological changes in the vitreoretinal junction 1: epiretinal membrane formation. Eye (Lond). (2008) 22:1310–7. doi: 10.1038/eye.2008.36
42. Hu Z, Mao X, Chen M, Wu X, Zhu T, Liu Y, et al. Single-cell transcriptomics reveals novel role of microglia in fibrovascular membrane of proliferative diabetic retinopathy. Diabetes. (2022) 71:762–73. doi: 10.2337/db21-0551
43. Corano Scheri K, Fawzi AA. Comment on Hu et al. Single-Cell Transcriptomics Reveals Novel Role of Microglia in Fibrovascular Membrane of Proliferative Diabetic Retinopathy. Diabetes (2024) 73(7):e6–e8. doi: 10.2337/db24-0041
44. Mao X, Xu M, Liu Q, Yuan S. Response to Comment on Hu et al. Single-Cell Transcriptomics Reveals Novel Role of Microglia in Fibrovascular Membrane of Proliferative Diabetic Retinopathy. Diabetes. (2024) 73(7):e9-e10. doi: 10.2337/dbi24-0024
45. Corano Scheri K, Lavine JA, Tedeschi T, Thomson BR, Fawzi AA. Single-cell transcriptomics analysis of proliferative diabetic retinopathy fibrovascular membranes reveals AEBP1 as fibrogenesis modulator. JCI Insight. (2023) 8(23):e172062. doi: 10.1172/jci.insight.172062
46. Yu C, Roubeix C, Sennlaub F, Saban DR. Microglia versus monocytes: distinct roles in degenerative diseases of the retina. Trends Neurosci. (2020) 43:433–49. doi: 10.1016/j.tins.2020.03.012
47. Beguier F, Housset M, Roubeix C, Augustin S, Zagar Y, Nous C, et al. The 10q26 risk haplotype of age-related macular degeneration aggravates subretinal inflammation by impairing monocyte elimination. Immunity. (2020) 53(2):429–41.e8. doi: 10.1016/j.immuni.2020.07.021
48. Ronning KE, Karlen SJ, Burns ME. Structural and functional distinctions of co-resident microglia and monocyte-derived macrophages after retinal degeneration. J Neuroinflammation. (2022) 19:299. doi: 10.1186/s12974-022-02652-2
49. Andries L, Kancheva D, Masin L, Scheyltjens I, Van Hove H, De Vlaminck K, et al. Immune stimulation recruits a subset of pro-regenerative macrophages to the retina that promotes axonal regrowth of injured neurons. Acta Neuropathol Commun. (2023) 11:85. doi: 10.1186/s40478-023-01580-3
50. Hu Y, Hu Y, Xiao Y, Wen F, Zhang S, Liang D, et al. Genetic landscape and autoimmunity of monocytes in developing Vogt-Koyanagi-Harada disease. Proc Natl Acad Sci U S A. (2020) 117:25712–21. doi: 10.1073/pnas.2002476117
51. Zheng W, Wang X, Liu J, Yu X, Li L, Wang H, et al. Single-cell analyses highlight the proinflammatory contribution of C1q-high monocytes to Behçet’s disease. Proc Natl Acad Sci U S A. (2022) 119:e2204289119. doi: 10.1073/pnas.2204289119
52. Tu H, Ren H, Jiang J, Shao C, Shi Y, Li P. Dying to defend: neutrophil death pathways and their implications in immunity. Adv Sci (Weinh). (2024) 11:e2306457. doi: 10.1002/advs.202306457
53. Ghosh S, Padmanabhan A, Vaidya T, Watson AM, Bhutto IA, Hose S, et al. Neutrophils homing into the retina trigger pathology in early age-related macular degeneration. Commun Biol. (2019) 2:348. doi: 10.1038/s42003-019-0588-y
54. Li L, Zuo S, Liu Y, Yang L, Ge S, Ye F, et al. Single-cell transcriptomic sequencing reveals tissue architecture and deciphers pathological reprogramming during retinal ischemia in macaca fascicularis. Invest Ophthalmol Vis Sci. (2024) 65:27. doi: 10.1167/iovs.65.1.27
55. Wang Q, Ma J, Gong Y, Zhu L, Tang H, Ye X, et al. Sex-specific circulating unconventional neutrophils determine immunological outcome of auto-inflammatory Behçet’s uveitis. Cell Discovery. (2024) 10:47. doi: 10.1038/s41421-024-00671-2
56. Binet F, Cagnone G, Crespo-Garcia S, Hata M, Neault M, Dejda A, et al. Neutrophil extracellular traps target senescent vasculature for tissue remodeling in retinopathy. Science. (2020) 369(6506):eaay5356. doi: 10.1126/science.aay5356
57. Dong X, Song Y, Liu Y, Kou X, Yang T, Shi SX, et al. Natural killer cells promote neutrophil extracellular traps and restrain macular degeneration in mice. Sci Transl Med. (2024) 16:eadi6626. doi: 10.1126/scitranslmed.adi6626
58. Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat Neurosci. (2020) 23:194–208. doi: 10.1038/s41593-019-0566-1
59. Das S, Ahmad Z, Suryawanshi A, Kumar A. Innate immunity dysregulation in aging eye and therapeutic interventions. Ageing Res Rev. (2022) 82:101768. doi: 10.1016/j.arr.2022.101768
60. Kasper M, Heming M, Schafflick D, Li X, Lautwein T, Meyer Zu Horste M, et al. Intraocular dendritic cells characterize HLA-B27-associated acute anterior uveitis. Elife. (2021) 10:e67396. doi: 10.7554/eLife.67396
61. Lv K, Ying H, Hu G, Hu J, Jian Q, Zhang F. Integrated multi-omics reveals the activated retinal microglia with intracellular metabolic reprogramming contributes to inflammation in STZ-induced early diabetic retinopathy. Front Immunol. (2022) 13:942768. doi: 10.3389/fimmu.2022.942768
62. Baysoy A, Bai Z, Satija R, Fan R. The technological landscape and applications of single-cell multi-omics. Nat Rev Mol Cell Biol. (2023) 24:695–713. doi: 10.1038/s41580-023-00615-w
63. Liang Q, Cheng X, Wang J, Owen L, Shakoor A, Lillvis JL, et al. A multi-omics atlas of the human retina at single-cell resolution. Cell Genom. (2023) 3:100298. doi: 10.1016/j.xgen.2023.100298
64. Wang SK, Nair S, Li R, Kraft K, Pampari A, Patel A, et al. Single-cell multiome of the human retina and deep learning nominate causal variants in complex eye diseases. Cell Genom. (2022) 2(8):100164. doi: 10.1101/2022.03.09.483684
65. Lopez Soriano V, Dueñas Rey A, Mukherjee R, Coppieters F, Bauwens M, Willaert A, et al. Multi-omics analysis in human retina uncovers ultraconserved cis-regulatory elements at rare eye disease loci. Nat Commun. (2024) 15:1600. doi: 10.1038/s41467-024-45381-1
66. Menon M, Mohammadi S, Davila-Velderrain J, Goods BA, Cadwell TD, Xing Y, et al. Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat Commun. (2019) 10:4902. doi: 10.1038/s41467-019-12780-8
67. Lyu Y, Zauhar R, Dana N, Strang CE, Hu J, Wang K, et al. Implication of specific retinal cell-type involvement and gene expression changes in AMD progression using integrative analysis of single-cell and bulk RNA-seq profiling. Sci Rep. (2021) 11:15612. doi: 10.1038/s41598-021-95122-3
68. He C, Liu Y, Huang Z, Yang Z, Zhou T, Liu S, et al. A specific RIP3+ subpopulation of microglia promotes retinopathy through a hypoxia-triggered necroptotic mechanism. Proc Natl Acad Sci U.S.A. (2021) 118(11):e2023290118. doi: 10.1073/pnas.2023290118
69. Smith TL, Oubaha M, Cagnone G, Boscher C, Kim JS, El Bakkouri Y, et al. eNOS controls angiogenic sprouting and retinal neovascularization through the regulation of endothelial cell polarity. Cell Mol Life Sci. (2021) 79:37. doi: 10.1007/s00018-021-04042-y
70. Crespo-Garcia S, Tsuruda PR, Dejda A, Ryan RD, Fournier F, Chaney SY, et al. Pathological angiogenesis in retinopathy engages cellular senescence and is amenable to therapeutic elimination via BCL-xL inhibition. Cell Metab. (2021) 33(4):818–32.e7. doi: 10.1016/j.cmet.2021.01.011
71. Li L, Fang F, Feng X, Zhuang P, Huang H, Liu P, et al. Single-cell transcriptome analysis of regenerating RGCs reveals potent glaucoma neural repair genes. Neuron. (2022) 110(16):2646–63.e6. doi: 10.1016/j.neuron.2022.06.022
72. Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, et al. Single-cell profiles of retinal ganglion cells differing in resilience to injury reveal neuroprotective genes. Neuron. (2019) 104(6):1039–55.e12. doi: 10.1016/j.neuron.2019.11.006
73. Heng JS, Hackett SF, Stein-O’Brien GL, Winer BL, Williams J, Goff LA, et al. Comprehensive analysis of a mouse model of spontaneous uveoretinitis using single-cell RNA sequencing. Proc Natl Acad Sci U S A. (2019) 116:26734–44. doi: 10.1073/pnas.1915571116
Keywords: single-cell RNA sequencing, innate immunity, retina, microglia, monocyte, neutrophil, eye diseases
Citation: Lu C, Mao X and Yuan S (2024) Decoding physiological and pathological roles of innate immune cells in eye diseases: the perspectives from single-cell RNA sequencing. Front. Immunol. 15:1490719. doi: 10.3389/fimmu.2024.1490719
Received: 03 September 2024; Accepted: 09 October 2024;
Published: 31 October 2024.
Edited by:
Xiaoquan Rao, Case Western Reserve University, United StatesReviewed by:
Jiaqian Luo, Cornell University, United StatesMin Zhang, University of Kentucky, United States
Copyright © 2024 Lu, Mao and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Songtao Yuan, c29uZ3Rhb3l1YW5AbmptdS5lZHUuY24=
 Chen Lu
Chen Lu Xiying Mao
Xiying Mao Songtao Yuan*
Songtao Yuan*