
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 20 January 2025
Sec. Molecular Innate Immunity
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1490478
This article is part of the Research TopicPattern Recognition Receptors: Balancing Inflammation and Immune HomeostasisView all 3 articles
Toll-like receptors (TLRs) play an important role in the recognition of viral particles and activation of the innate immune system, but their role in SARS-CoV-2 infection is still poorly characterized. In the present study, we investigated the role of Toll-like receptor 10 (TLR10) in modulating the immune response during SARS-CoV-2 infection. The results showed that overexpression of TLR10 in A549 lung epithelial cells, immunostimulated with SARS-CoV-2 proteins S and N mainly downregulated proinflammatory cytokines and interferons and affected gene expression in the cocultured THP-1 monocytes. Our results suggest that TLR10 could mediate the extent of SARS-CoV-2 infection by downregulating the release of inflammatory cytokines and chemokines such as CXCL10, IL6, IL8, and IFNβ. Modulation of TLR10 expression could have implications for the treatment of patients with severe COVID-19, in whom excessive inflammation leading to the development of acute respiratory distress syndrome (ARDS) is a key feature. However, further research is needed to fully understand the impact of modulating TLR10 expression on the antiviral response and the overall balance of the immune response during SARS-CoV-2 infection.
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a recently emerged virus that causes coronavirus disease 2019 (COVID-19). Since 2019, it has spread worldwide, affecting global health, social life and the world economy. SARS-CoV-2 is a β-coronavirus that belongs to the Coronaviridae family and is an enveloped positive-strand RNA (+ssRNA) virus. The virus consists of four main structural proteins, namely the spike glycoprotein (S), the small envelope glycoprotein (E), the membrane glycoprotein (M) and the nucleocapsid phosphoprotein (N), as well as several accessory proteins, such as the papain-like protease (P) (1).
Infection of the respiratory tract with SARS-CoV-2 leads to an extremely diverse clinical spectrum. It ranges from asymptomatic forms and mild symptoms such as fever, dry cough and shortness of breath to severe symptoms that require hospitalization. In the most severe cases of SARS-CoV-2 infection, acute overproduction and uncontrolled release of proinflammatory cytokines (hypercytokinemia, also known as “cytokine storm”) can trigger excessive systemic inflammation leading to acute respiratory distress syndrome (ARDS) and multiple organ failure (2). The severity of the disease has been associated with elevated levels of various proinflammatory cytokines and chemokines, such as interleukin (IL)1β, IL6, IL8, C-X-C motif chemokine ligand 10 (CXCL10), chemokine (C-C motif) ligand 20 (CCL20), tumor necrosis factor alpha (TNFα), and a delayed type I interferone (IFN) response (3–6).
Airway epithelial cells interface with the external environment and play a key role in recognizing airborne pathogens and activating the host immune system (7). Pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs), which recognize pathogen-associated molecular patterns (PAMPs) and trigger an immune response, are important components of the innate immune system. Members of the TLR family are the best-studied PRRs that can also recognize structural proteins of SARS-CoV-2 (8). TLRs on endosomes recognize intracellular components of viruses; for example, TLR7 and TLR8 can recognize viral single-stranded RNA (9, 10), TLR3 recognizes transient dsRNA intermediates formed during replication of the virus (11), while TLRs on the cell membrane recognize viral proteins. The S protein is thought to be recognized by both TLR2 (12) and TLR4 (13), while protein E is recognized by TLR2 (14). It has been shown that not only the viral infection but also various individual SARS-CoV-2 proteins are immunogenic both in vivo and in vitro (15) and trigger the expression of proinflammatory cytokines in immune (14, 15) and epithelial cells (12).
TLR10 is the only poorly characterized member of the TLR family. It belongs to the TLR2 subfamily and can homodimerize in the presence of its respective ligands or act in paired combination with other members of the subfamily (TLR1, TLR2, TLR6) (16). To our knowledge, the role of TLR10 in the recognition of SARS-CoV-2 has not yet been investigated, but it is known that TLR10 is expressed in lung epithelial cells (17). Our recent results confirmed that TLR10 is indeed present in A549 lung epithelial cells and showed that its overexpression by CRISPRa downregulates some of the proinflammatory mediators in cells immunostimulated with dsRNA, LPS or Pam3Cys (18). Studies also show that upregulation of TLR10 is possible by adding the biologically active form of vitamin D, calcitriol (1,25-dihydroxycholecalciferol), to the cell growth medium (19). However, the effects of other vitamins (e.g. vitamin C and B vitamins) associated with the prevention and management of COVID-19 (20) on TLR10 expression in cell cultures have not been determined. As several vitamins, especially D and C, have been associated with the prevention and treatment of diseases, including SARS-CoV-2 infections (e.g. 21, 22), we were interested in whether the addition of vitamins to the growth medium affects TLR10 expression; if so, one could speculate that the immunomodulatory effects of vitamins could be explained, at least in part, by their effect on the regulation of TLR10 expression.
The aim of this study was to investigate the potential immunoregulatory effects of TLR10 upregulation in the A549 cell line immunostimulated with SARS-CoV-2 S and N proteins. To our knowledge, this is one of the first studies to address the potential immunomodulatory effects of TLR10 in lung epithelial cells during immunostimulation with SARS-CoV-2 proteins in an in vitro system. Harnessing the immunomodulatory effects of TLR10 could have potential therapeutic applications for the treatment of patients with severe COVID-19 as well as for the management of other inflammatory diseases.
The human lung epithelial cell line A549 was purchased from the American Type Culture Collection (ATCC). Cells were grown in DMEM (Sigma Aldrich) supplemented with 10% FBS (Sigma–Aldrich), 2 mM L-glutamine (Thermo Fisher Scientific), and 0.01% penicillin-streptavidin (Sigma–Aldrich) at 37°C and 5% CO2.
The human mononuclear cell line THP-1 (ATCC) was grown in RPMI 1640 medium (Sigma–Aldrich) supplemented with 10% FBS (Sigma–Aldrich) and 0.01% penicillin-streptavidin (Sigma–Aldrich) at 37°C and 5% CO2.
Endogenous TLR10 was overexpressed in A549 cells with CRISPR/dCas9 as previously described (18). Briefly, cells were transfected using Lipofectamine 3000 (Thermo Fisher Scientific) at approximately 70% confluence with the vectors pCMV-dCas9:NLS: VPR (23), encoding dCas9-VPR, and pGGAselect (N0309AAVIAL, New England BioLabs), encoding sgRNAs for TLR10 overexpression, (A549-TLR10OE) or with an empty pGGAselect vector representing TLR10 control expression (A549C). To avoid cell apoptosis, cells were washed with PBS four hours after transfection, and the medium was changed. All experiments were performed 48 hours after transfection. Localization of TLR10 in A549C and A549-TLR10OE was determined using immunostaining (see Supplementary Section S4).
In addition, ascorbic acid (A4544, Merck), folic acid (F8758, Merck), calciferol (D1530, Merck) and riboflavin (R9504, Merck) were added to the growth medium of non-transfected A549 cells at two different concentrations (50 or 200 µM) to determine their effects on TLR10 expression after 24 and 48 hours of incubation.
Cells were immunostimulated with final concentrations of 500 ng/mL SARS-CoV-2 protein S (RP87668, Thermo Fisher Scientific) or SARS-CoV-2 protein N (RP87707, Thermo Fisher Scientific) and incubated for four and 24 hours (corresponding to “early” and “late” immune response, respectively) to simulate immune challenge of lung epithelial cells during SARS-CoV-2 infection. RNA was extracted from the cells and gene expression was quantified by real-time PCR. In addition, the growth media were collected for the quantification of cytokines.
Cells were plated on 12 mm Transwell™ plates with 0.4 μm pore polyester membrane inserts (3460, Corning). One day before transfection, A549 cells were seeded at a concentration of 1×105 cells/well in the upper chamber of the Transwell system (Figure 1). On the day of transfection, cells were transfected with pCMV-dCas9:NLS: VPR and pGGAselect as described above to obtain A549-TLR10OE and A549C cells. After four hours of incubation, the cells were washed with PBS and the medium was changed. After two days, THP-1 cells were seeded into the lower chamber at a concentration of 0.5×106 cells/well. A549-TLR10OE and A549C cells were stimulated with SARS-CoV-2 S or N protein at a final concentration of 500 ng/mL for four or 24 hours, respectively. The cell culture medium was collected and RNA was isolated from THP-1 cells as described above.
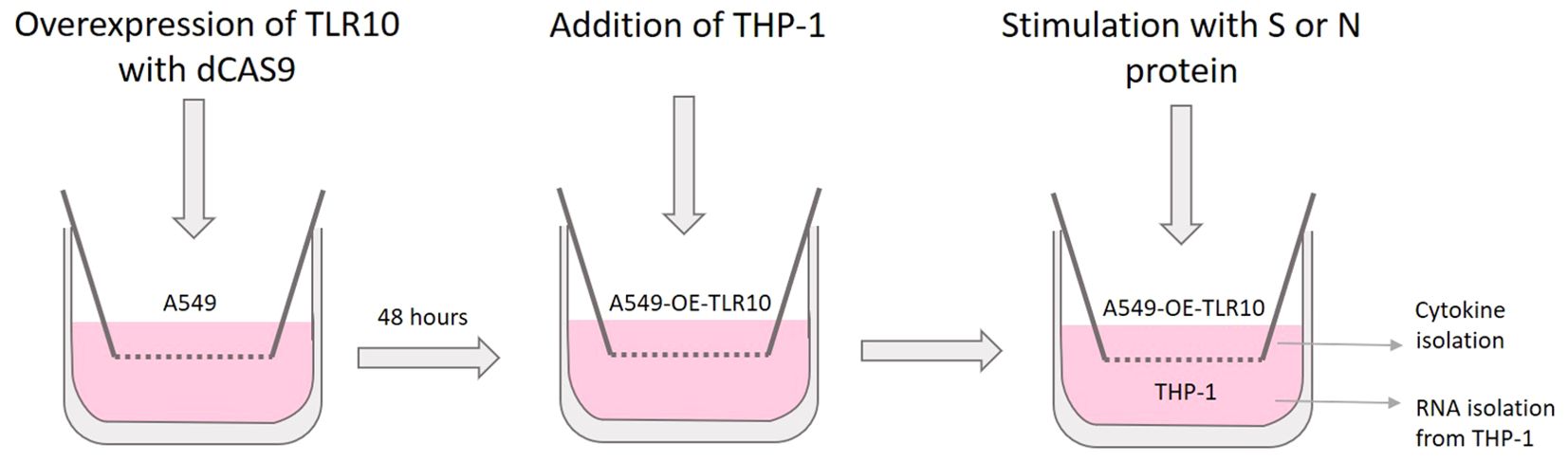
Figure 1. Schematic representation of the co-culture experiments involving A549 and THP-1 cells in a transwell system.
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The extracted RNA was diluted to a final concentration of approximately 100 ng/µL RNA and treated with DNase I (Thermo Fisher Scientific). The treated RNA was reverse transcribed into single-stranded cDNA using a High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) and stored at -20°C.
Digital PCR (dPCR) was performed using a QIAcuity instrument (QIAGEN). Reactions were prepared using a QIAcuity Probe PCR Kit (QIAGEN) in a volume of 40 μL. Gene expression experiments were performed in duplicate. The expression of the target gene TLR10 (Hs01935337_s1) and the reference gene TOP1 (Hs00243257) was measured using TaqMan Gene Expression Assays (Thermo Fisher Scientific). Reactions using TaqMan probes at a final concentration of 0.8 µM were loaded onto a 26k nanoparticle dPCR nanoplate (QIAcuity) and performed as follows: 2 min, 95°C, 40× (15 s at 95°C, 15 s at 60°C). QIAcuity analysis software (QIAGEN) was used to analyze the results. Poisson distribution was used to calculate the number of copies of the target molecule per positive partition. To normalize the data, the copies/unit RNA of the target gene were multiplied by the normalization factor (geometric mean of TOP1), and the final data were displayed as normalized copies/unit RNA.
The cDNA was mixed with PowerUp™ SYBR® Green Master Mix (Applied Biosystems) and aliquoted into the wells of a pre-made qPCR profiling array (RT2 Profiler PCR Array, GeneGlobe ID-PAHS-018Z, Qiagen) containing a panel of 84 genes associated with the TLR signaling pathway. Amplification results were normalized to the five housekeeping genes ACTB, B2M, GAPDH, HPRT1, and RPLP0. Fold changes in gene expression between immunostimulated A549C and immunostimulated A549-TLR10OE cells were calculated using the 2−ΔΔCT method (24).
Further expression analyses were performed for individual genes identified as differentially expressed during qPCR array screening and/or consistently associated with the SARS-CoV-2 immune response in the literature. Primers were designed for the genes of interest using the online tool PrimerBlast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi (accessed May 27, 2023) (Supplementary Table S1). All the reactions were performed in triplicate. Thermal cycling conditions were as follows: 10 min at 95°C, 40 cycles at 95°C for 15 s and at 60°C for 1 min. Primer efficiencies were determined for all the primer pairs using a six-log cDNA dilution range. All the determined primer pair efficiencies were in the range of 100 ± 10% and the R2 ≥ 0.99. Relative differential expression between immunostimulated A549C and immunostimulated A549-TLR10OE cells was calculated using the 2-ΔΔCt method (24). A gene was considered differentially expressed if it showed at least a two-fold change.
The cell culture medium of A549 and THP-1 cells was removed four and 24 hours after incubation with SARS-CoV-2 proteins S and N, respectively, and centrifuged at 1000 × g for 10 minutes at 4°C. The concentrations of IL8 (assay range: 15.6-1,000 pg/mL, BMS204-3INST, Thermo Fisher Scientific), IL1β (assay range: 7.8-500 pg/mL, BMS224INST, Thermo Fisher Scientific), CXCL10 (assay range: 3.1-200 pg/mL, BMS284INST, Thermo Fisher Scientific), IFNβ (assay range: 50-4000 pg/mL, 414101, Thermo Fisher Scientific), IL10 (assay range: 50-4000 pg/mL, BMS215INST, Thermo Fisher Scientific), CCL20 (assay range: 0.8-600 pg/mL, EHCCL20, Thermo Fisher Scientific) and TNFα (assay range: 7.8-500 pg/mL, BMS223INST, Thermo Fisher Scientific) were measured according to the manufacturer’s instructions.
Approximately 5×105 A549 cells were seeded in 6-well plates and transfected with plasmids to overexpress TLR10 as previously described. After two days, cells were lysed in ice-cold RIPA lysis buffer supplemented with protease and phosphatase inhibitor cocktails (4906845001, Roche). Protein concentration was determined using the Pierce BCA Protein Assay Kit (# 23227, Thermo Fisher Scientific) according to the manufacturer’s instructions. Approximately 15 μg of the cell lysate was transferred to a precast NuPAGE 4-12% gel (NP0321BOX, Thermo Fisher Scientific), electrophoresed at 180 V for two hours and immunoblotted onto a PVDF membrane. Membranes were stained with primary antibodies against TLR10 (1:1000) (PRS3275, Sigma–Aldrich) and β-actin (1:5000) (MA1-140, Thermo Fisher Scientific). Secondary HRP-conjugated anti-rabbit antibody (1:5000) (31460, Thermo Fisher Scientific) and HRP-conjugated anti-mouse antibody (1:5000) (31430, Thermo Fisher Scientific) were used for visualization, and the HRP signal recorded after two hours of incubation with TrueBlue peroxidase substrate (5510-0030, LGS Diagnostics).
Data were analyzed using Prism 8 version 10.2.2 software (GraphPad). Statistical significance was determined using a two-tailed, unpaired Student’s t-test (p ≤ 0.05 was considered statistically significant).
Overexpression of endogenous TLR10 in immunostimulated A549 cells significantly altered the expression of several cytokines and genes associated with TLR signaling. Furthermore, overexpression of TLR10 in A549 cells co-cultured with THP-1 cells altered the expression of genes in THP-1 cells.
Using the CRISPR/dCas9 system, we were able to increase the expression of TLR10 by approximately 24-fold (Figure 2A). The induction appears to translate to the protein level, which is visible in the Western blot (Figure 2B; Supplementary Figure S1). After overexpression, TLR10 was mainly localized at the cell membrane, but to a lesser extent also in the cytoplasm (possibly in endosomes) (Supplementary Figure S2). Supplementation of the growth medium with calcitriol (active form of vitamin D) significantly affected TLR10 expression, while supplementation with other vitamins (ascorbic acid – vitamin C, folic acid – vitamin B9, and riboflavin – vitamin B2) had no statistically significant effect on the expression of TLR10 at any of the concentrations used. The addition of the biologically active form of vitamin D to the growth medium at a high concentration (200 µM) increased the expression of TLR10 approximately four-fold after 24 hours of incubation and approximately 16-fold after 48 hours of incubation (Figure 2C).
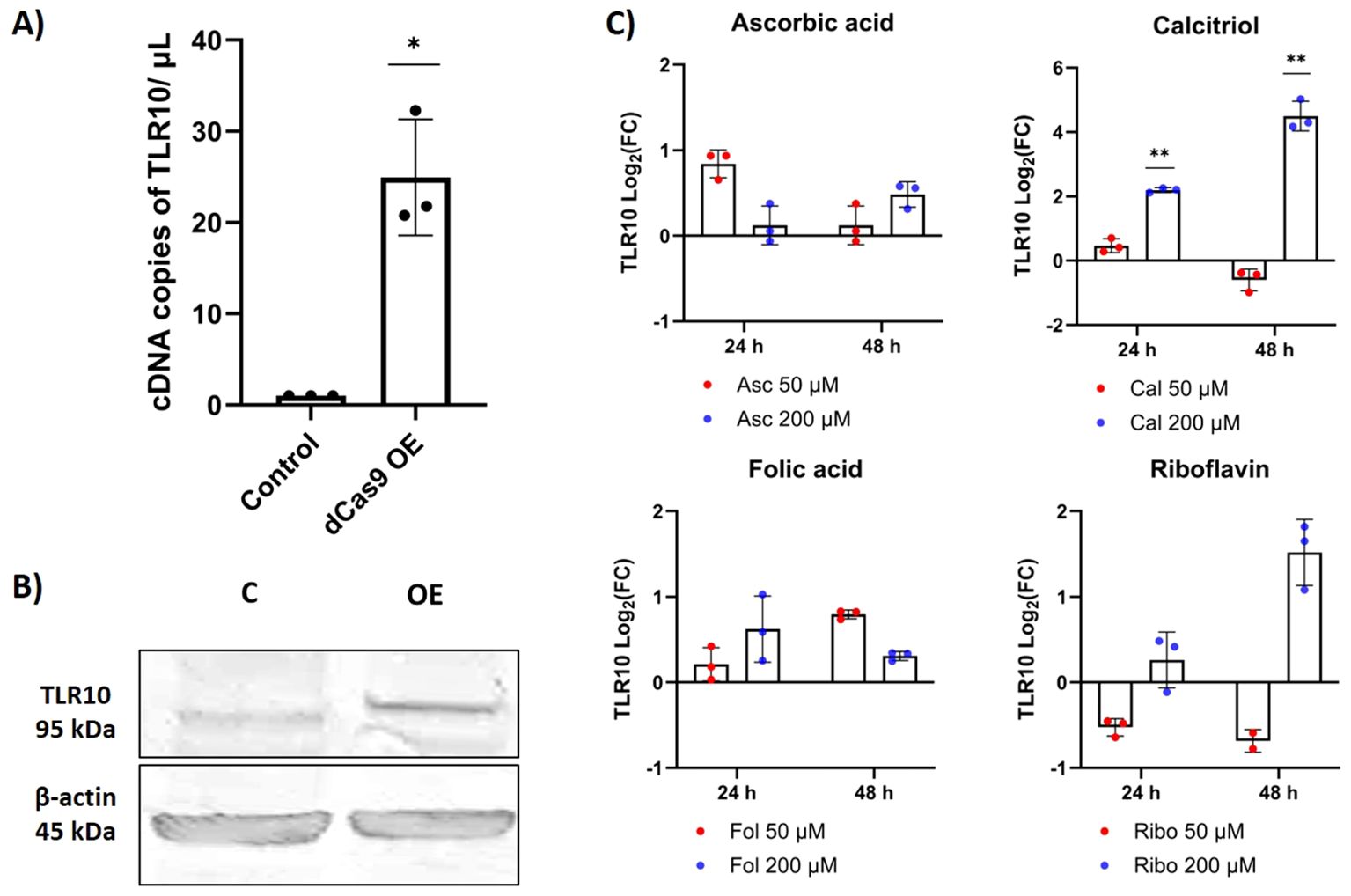
Figure 2. Differential expression of TLR10. (A) Differential expression of TLR10 induced by CRISPR/dCas9, evaluated by dPCR. (B) Western blot showing TLR10 protein 48 hours after transfection. (C) Differential expression of TLR10 after addition of different vitamins to the cell growth medium, evaluated by qPCR. Data are presented as mean with SEM of three independent experiments; *p ≤ 0.05, **p ≤ 0.01 by unpaired Student’s t-test.
Both S and N protein induced the expression of the proinflammatory cytokines TNFα and IL1β and the chemokines CXCL10 and IL8, while IFNβ was downregulated in A549 cells (p > 0.05). In general, N protein induced the relative expression of inflammatory mediators in A549 cells earlier than S protein as shown by the expression data for four- and 24-hour time points after immunostimulation (Figure 3). The highest differential expression in immunostimulated A549 cells was observed for IL8 and CXCL10, for both virulence factors, and for TNFα in the case of N protein challenge after four hours (Figure 3A). The induction of inflammatory mediators in THP-1 cells was weaker than in A549 cells. The S protein increased the expression of TNFα, IL8 and IL1β at both time points after stimulation, while N protein induced the expression of these markers only in the “early” phase of immune stimulation (after four hours) (Figure 3B). No statistically significant difference in IFNβ expression was observed after immunostimulation with the S or N protein.
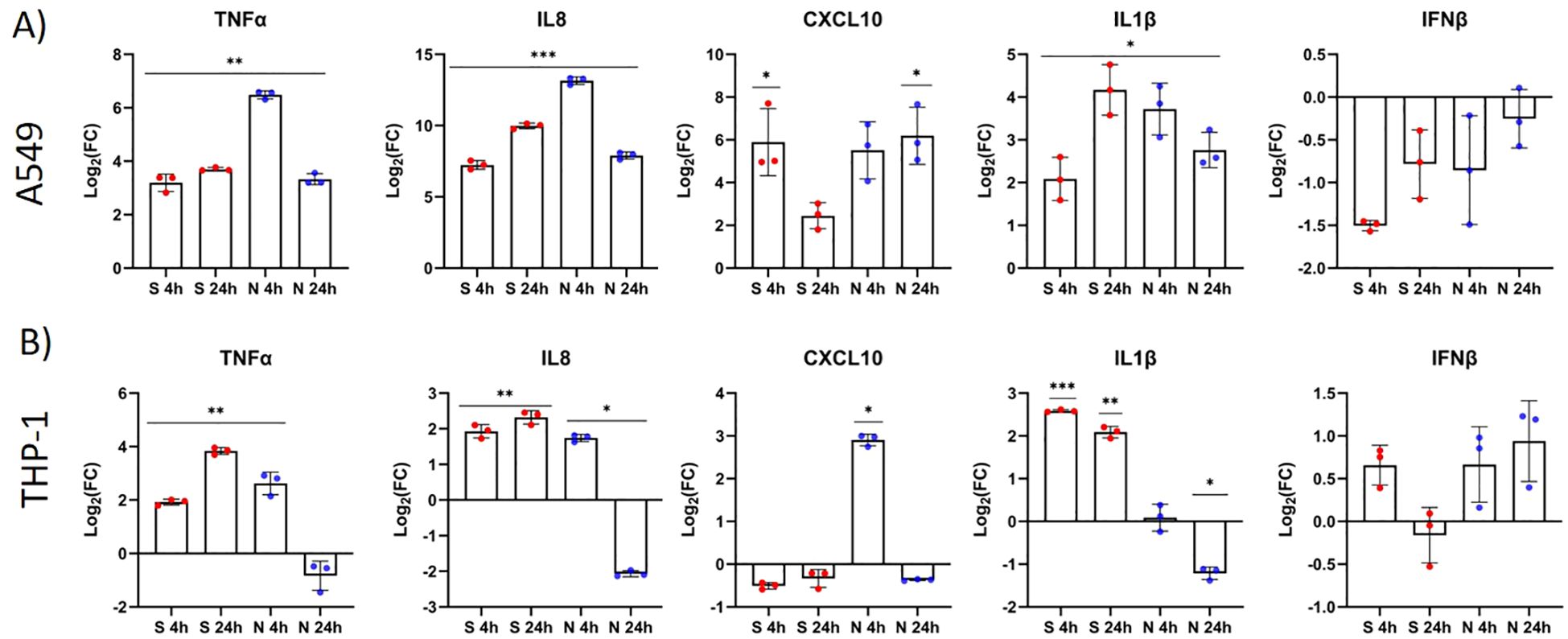
Figure 3. The S and N proteins of SARS-CoV-2 induce the differential expression of inflammatory markers (cytokines) in A549 and THP-1 cells. A549 cells (A) and THP-1 cells (B) were stimulated with the SARS-CoV-2 protein S (red) or N (blue) (both 500 ng/mL) for four and 24 hours. Gene expression of TNFα, IL8, CXCL10, IL1β and IFNβ was measured by RT–qPCR and compared between immunostimulated and untreated cells. Data are presented as mean with SEM of three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001 according to unpaired Student’s t tests.
Using a qPCR array focusing on genes associated with the TLR signaling pathway, we investigated the differential expression of 84 genes involved in this pathway. We compared the expression of the genes in immunostimulated cells overexpressing TLR10 (A549-TLR10OE) with immunostimulated cells with native TLR10 expression (A549C) (Supplementary Table S2; Figure 4A). Gene expression analysis revealed five upregulated (NFRKβ, TBK1, TLR10, NFKβIL1, IRAK1) and 13 downregulated (CSF3, IL6, CXCL10, IFNβ1, CSF2, IL1β, TNFα, CLE4E, IFNα, CCL2, NFKβIA, IL1A, TLR3) genes four hours after stimulation with the S protein (≥ 2-fold change and a p-value ≤ 0.05 was used as a threshold for differential expression). Prominent immune mediators such as IL6, CXCL10, IL1β, TNFα, IFNβ and the colony-stimulating factors CSF2 and CSF3 were downregulated, while genes encoding adaptor proteins, kinases and transcription factors such as IRAK1, TBK1, FOS and ELK were upregulated in response to TLR10 overexpression. Twenty-four hours after stimulation, the effect of TLR10 overexpression diminished, as only three genes were downregulated (IL6, CSF3 and IRAK2).
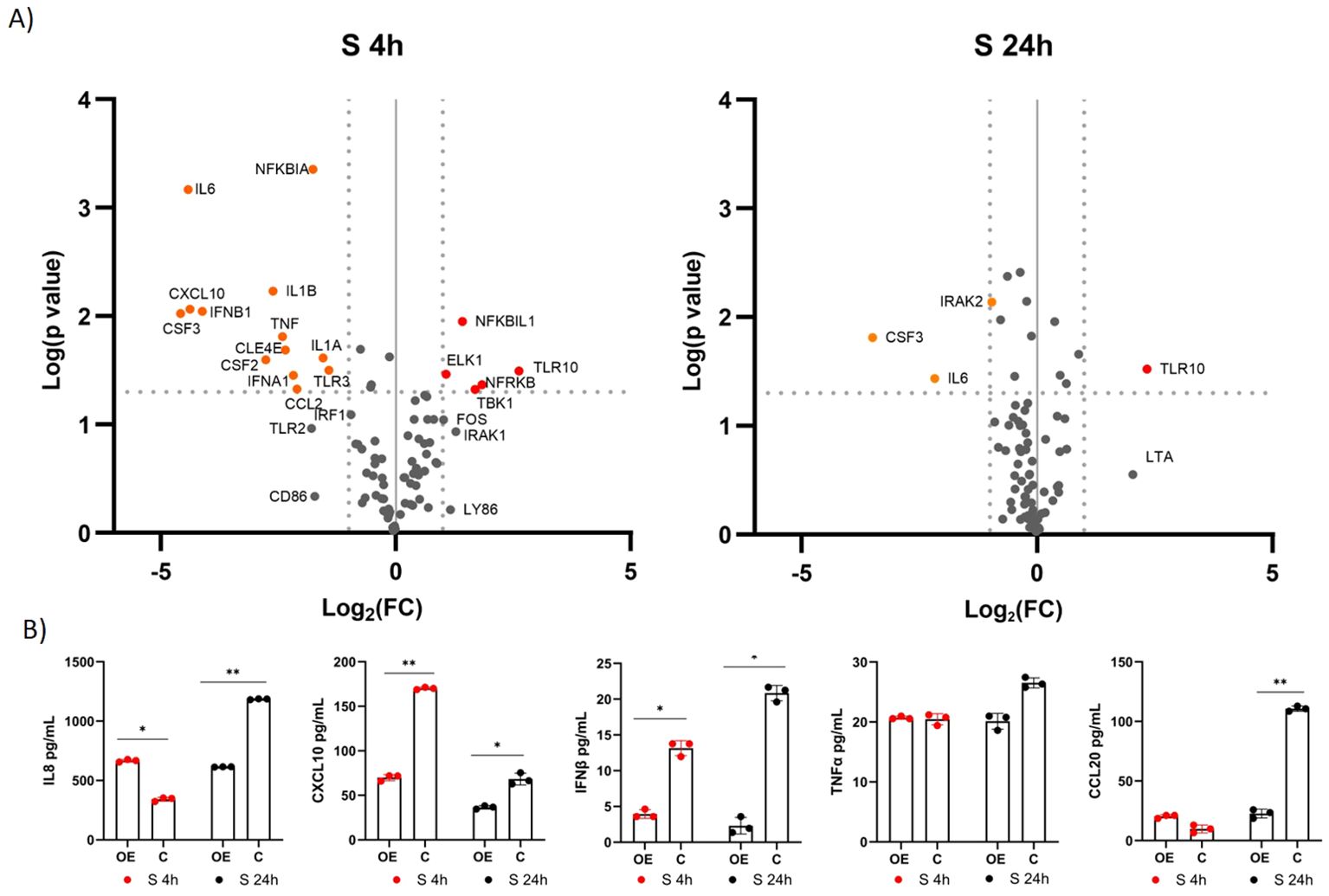
Figure 4. Differential expression of immune-associated genes in A549-TLR10OE cells challenged with the S protein. (A) a volcano plot of differential expression between immunostimulated A549-TLR10OE and immunostimulated A549C cells challenged for four and 24 hours as determined by qPCR arrays. Log2-fold changes are plotted against the p-values of the t-test. Significantly downregulated genes are marked as orange dots, while significantly upregulated genes are marked as red dots. Fold-change (≥ 2-fold, vertical lines) and statistically significant difference (p ≤ 0.05, horizontal line) were used as threshold for differential expression. (B) Cytokine secretion was measured by ELISA. Data are presented as mean with SEM of three independent experiments; *p ≤ 0.05, **p ≤ 0.01 by unpaired Student’s t-test. OE, overexpressed; C, control.
To further investigate the effects of TLR10 overexpression, protein concentrations of known inflammatory markers, namely, IL8, CXCL10, IFNβ, TNFα, CCL20, and IL1β, were measured by ELISA (Figure 4B). The concentrations of the cytokines in the medium were not entirely consistent with the qPCR results, e.g. TNFα levels were not significantly different between the treatment and the control, while IL8 levels were elevated in the sample overexpressing TLR10 at four hours after immunostimulation. Additionally, we were unable to detect IL1β in the growth medium, while the measured IFNβ levels were below the range value of the assay (50 pg/mL).
A similar response to immunostimulation with S protein was observed after immunostimulation with N protein (Supplementary Table S2; Figure 5A). After four hours, 12 genes were downregulated (CD180, CXCL10, CSF2, CD180, LTA, IFNβ1, TNFα, BTK, FOS, IL6, NFKBIA, TLR2), and five were upregulated (CD14, TLR10, TBK1, IRAK1, MAP2K3) in response to TLR10 overexpression. After 24 hours, the effect of TLR10 overexpression diminished, as only four genes were downregulated (IL1β, CLE4E, CD86, and CD80) and four were upregulated (CSF2, CSF3, and IFNβ1). Cytokine secretion was measured by ELISA for IL8, CXCL10, IFNβ, and TNFα, CCL20, and IL1β (Figure 5B). The concentrations of cytokines in the medium were not entirely consistent with the qPCR results, for example concentrations of TNFα were not significantly different between treatment and control, while CCL20 levels were elevated in the sample overexpressing TLR10 24 hours after immunostimulation. We were unable to detect IL1β in the growth medium, while the measured IFNβ levels were below the range value of the assay (50 pg/mL).
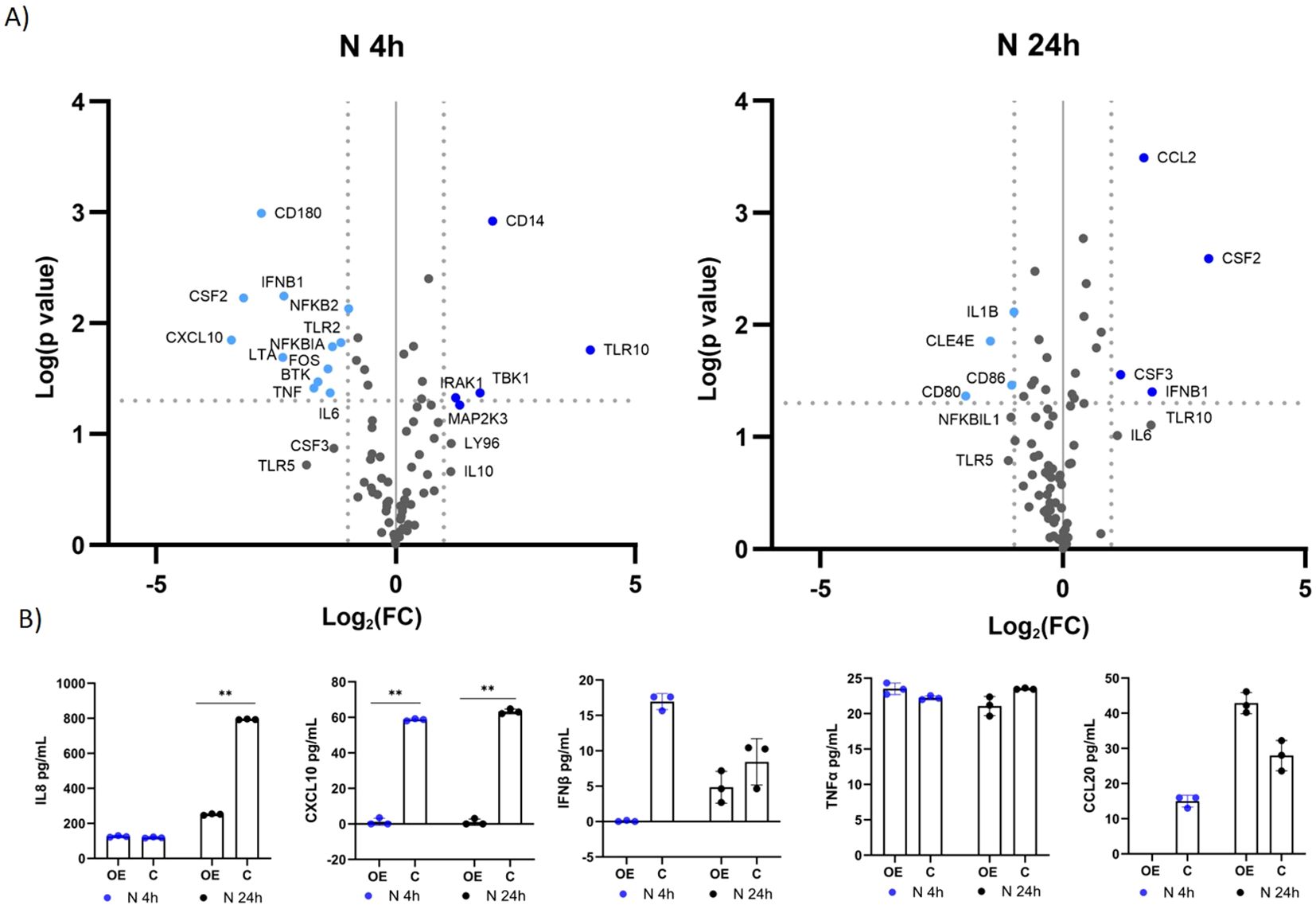
Figure 5. Differential expression of immune-associated genes in A549-TLR10OE challenged with the N protein. (A) a volcano plot of differential expression between immunostimulated A549-TLR10OE and immunostimulated A549C cells challenged for four and 24 hours as determined by qPCR arrays. Log2-fold changes are plotted against the p-values of the t-test. Significantly downregulated genes are marked as light blue dots, while significantly upregulated genes are marked as dark blue dots. Fold-change (≥ 2-fold, vertical lines) and statistically significant difference (p ≤ 0.05, horizontal line) were used as threshold for differential expression. (B) Cytokine secretion was measured by ELISA. Data are presented as mean with SEM of three independent experiments; **p ≤ 0.01 by unpaired Student’s t-test. OE, overexpressed; C, control.
In the co-culture experiment, we investigated whether overexpression of TLR10 in immunostimulated A549 cells affects gene expression in the monocytic human leukemia cell line THP-1. The results showed that overexpression of TLR10 in immunostimulated A549-TLR10OE altered expression of genes in co-cultured THP-1, by downregulating expression of various proinflammatory cytokines (TNFα, IL8, CXCL10, and IL1β) four and 24 hours after immunostimulation with N protein and after four hours in the case of S protein, while their expression was upregulated 24 hours after immunostimulation with S protein (Figures 6A, C). In response to TLR10 overexpression in immunostimulated A549 cells, granulocyte-macrophage colony-stimulating factors 2 (CSF2) and CSF3, which activate monocytes to produce inflammatory cytokines, were downregulated in A549-TLR10OE (Figures 4A, 5A) and in A549-TLR10OE co-cultured with THP-1.
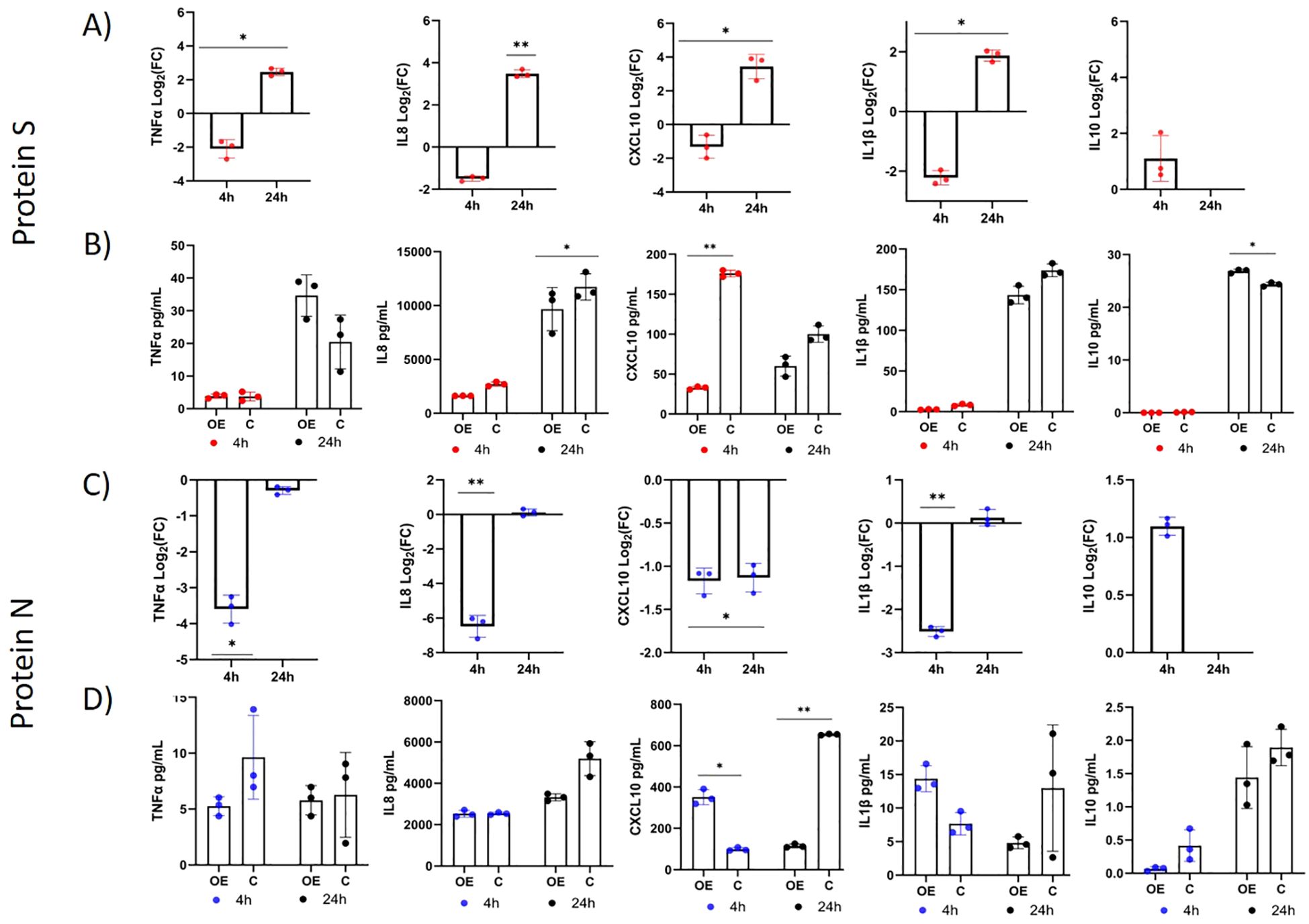
Figure 6. Differential expression of immune-associated genes in THP-1 cells co-cultured with immunostimulated A549-TLR10OE or immunostimulated A549C cells (A549 cells were challenged with the N or S protein for four and 24 h). (A, C) The differential expression of TNFα, IL8, CXCL10, IL10 and IL1β in THP-1 cells between coculture with immunostimulated A549-TLR10OE and immunostimulated A549C cells. (B, D) The protein concentrations of TNFα, CXCL10, IL10, IL1β, and IL8 in the co-culture medium measured by ELISA. Data are presented as mean with SEM of three independent; *p ≤ 0.05, **p ≤ 0.01 by unpaired Student’s t-test. OE, overexpressed; C, control.
The overexpression of TLR10 affected the concentrations of cytokines in the co-culture medium (Figures 6B, D). In the case of immunostimulation with S protein (Figure 6B), the concentrations of IL8 (concentrations in some samples exceeded recommended assay range value of 1000 pg/mL), IL1β and CXCL10 were lower in the case of TLR10 overexpression at both time- points after immunostimulation, while the concentrations of IL10 and TNFα were higher 24 hours after immunostimulation and undetectable four hours after stimulation. In the case of four-hour immunostimulation with N protein, the concentrations of the cytokines CXCL10 and IL1β were higher in the A549-TLR10OE co-culture medium than in the A549C co-culture medium. However, 24 hours after immunostimulation the levels of all the cytokines were lower in the A549-TLR10OE co-culture medium (significantly in case of CXCL10). The concentrations of TNFα and IL10 were lower in the A549-TLR10OE co-culture medium than in the A549C co-culture medium at both time points after immunostimulation with N protein (p > 0.05), but the measured levels of IL10 and TNFα (in some of the samples) were below the recommended range values for the assays (7.8 pg/mL for TNFα and 50 pg/mL for IL10).
Although studies have shown that some TLRs recognize SARS-CoV-2 and trigger an inflammatory response, the role of TLR10 in SARS-CoV-2 infection has not yet been investigated. TLR10 is considered an orphan receptor without a clear function, a specific ligand or a specific mode of action (16). Since mice lack a functional TLR10 receptor (25), studies on TLR10 have mainly been conducted in vitro and have yielded contradictory results. Depending on cell type, ligand, expression type and dimerization (homo or heterodimers), TLR10 has been associated with either a proinflammatory (26, 27) or an anti-inflammatory function (28, 29). TLR10 forms heterodimers with the TLR2 subfamily; TLR10-TLR1/TLR6 dimers have been shown to have proinflammatory effects (30), while TLR10-TLR2 dimers have been associated with anti-inflammatory effects (25). Recent studies have shown that TLR10 is involved in triggering the innate immune response against viruses by recognizing dsRNA (31) as well as viral proteins such as HIV-1 (32) and influenza virus proteins (33). A study by Zheng et al. (14) showed that TLR2 and MyD88 are required for the inflammatory response induced by β-coronaviruses (14). Therefore, we hypothesized that TLR10 may be involved in immune signaling in response to certain SARS-CoV-2 structural proteins, possibly through heterodimerization with TLR2.
To investigate the potential role of TLR10 in the inflammatory response induced by SARS-CoV-2 proteins in lung epithelial cells, we overexpressed endogenous TLR10 in A549 cells and immunostimulated the cells with SARS-CoV-2 S or N protein. In addition to CRISPR-induced activation (CRISPRa) we were able to successfully induce expression of endogenous TLR10 by supplementing the growth medium of challenged A549 cells with an active form of vitamin D. It is known that the active form of vitamin D interacts with the nuclear vitamin D receptor (VDR). VDR then forms a heterodimer with retinoid-X-receptor alpha (RXR-α) and the complex can bind to the vitamin D response elements located in the promoter regions of the target genes. Three potential VDR binding sites have been identified within the proximal promoter of TLR10, located within 250 base pairs upstream of the transcription start site (34). The interaction of vitamin D with TLR10 could potentially influence the body’s defense mechanisms against viral infections, including SARS-CoV-2. Further research is needed to fully understand the effects of this signaling pathway in the context of COVID-19 and other viral infections.
Studies on serum cytokines in patients with COVID-19 have shown that the concentrations of many cytokines and chemokines, such as IL6, TNFα, CSFs, monocyte chemoattractant proteins (MCPs) and CXCL10 (5, 35), are elevated in response to SARS-CoV-2 infection and correlate positively with the severity of the disease (36, 37). Therefore, downregulation of the aforementioned markers could alleviate the symptoms of COVID-19 and prevent hyperinflammation. First, we investigated whether the S or N protein triggers inflammatory signaling pathways and cytokine production in A549 cells. We found that the presence of S or N protein in the medium caused differential expression of inflammatory genes. Next, we investigated whether the expression level of TLR10 affects the expression of inflammatory mediators in immunostimulated A549 cells. We found that overexpression of TLR10 downregulates the expression of several proinflammatory cytokines. The genes that were significantly (p < 0.05) downregulated in both cases (after stimulation with the S or N protein) at least one time-point after immunostimulation (four or 24 hours after immunostimulation) were IL6, CXCL10, TNFα, IFNβ, CSF2, and NFKBIA, while the only upregulated gene in both cases was TBK1.
The vast majority of inflammatory cells that infiltrate the lungs after infection are monocytes and macrophages (38, 39). Angiotensin converting enzyme 2 (ACE2), through which SARS CoV-2 enters target cells, is only weakly expressed in monocytes (40), and the signal for their activation during SARS-CoV-2 infections is thought to originate from paracrine signaling of infected tissue. Several known molecules involved in monocyte migration are TNFα, CCL2, IL1β, IL8, and CSFs (41, 42). The co-culture experiments with immunostimulated A549 and monocytic THP-1 cells were performed to investigate the effects of TLR10 overexpression in the immunostimulated epithelial A549 cell line on TLR pathway-associated gene expression of immune cells (THP-1) in a co-culture cell model. Cell co-cultures contribute to a better understanding of complex biological processes, as a variety of signaling pathways and mechanisms require direct interactions between cells, including paracrine signaling through cytokines, growth factors, and transcriptional regulators that are activated or repressed in response to pathogens (43). Our results suggest that overexpression of TLR10 in A549 epithelial cells influences cytokine and chemokine expression in co-cultured THP-1 cells. The response of THP-1 cells to TLR10 overexpression in co-cultured A549 cells immunostimulated with S or N protein (Figure 6) was downregulation of several proinflammatory cytokines, consistent with previous reports (12). However, when the co-cultures were immunostimulated with S protein, we found that some of the cytokines (IL8, CXCL10, IFNβ, TNFα) in the THP-1 cells were initially downregulated in the “early” phase (4 hours) after immunostimulation and upregulated in the “late” phase (24 hours), but the concentrations of all measured cytokines (except of TNFα) were lower in A549-TLROE co-cultures than in A549C co-cultures 24 hours after immunostimulation (Figure 6B). Furthermore, S protein immunostimulation of A549-TLR10OE cells co-cultured with THP-1 led to a significant upregulation of the anti-inflammatory cytokine IL10 in THP-1 cells. When N protein immunostimulated A549-TLR10OE and THP-1 cells were co-cultured, the expression of inflammatory genes such as IL8, CXCL10, and IL1β was downregulated in THP-1 cells after 4 hours compared to A549C co-cultures (Figure 6C), but the cytokine concentrations in the medium were not downregulated until 24 hours. Such reduced correlations between mRNA expression and protein levels could be explained by transcriptional delays – the time when transcription starts and when protein is formed (44, 45), which is why transcriptional regulation may not be immediately visible at the protein level.
In the present study, we demonstrated that TLR10 is able to modulate the immune response to SARS-CoV-2 proteins S and N in A549 and THP-1 cells. In general, we observed a downregulation of inflammatory cytokines when TLR10 was overexpressed in immunostimulated A549 cells. However, some contradictory results (e.g. upregulation of some proinflammatory cytokines in immunostimulated A549-TLR10OE cells compared to immunostimulated A549C cells) and discrepancies between the transcriptional regulation of some genes and their protein levels were observed, especially in the »early« phase after immunostimulation, which may indicate that the mechanisms of TLR10 functions are complex and depend on virulence factor, incubation time, cell type, and TLR10 concentration. Furthermore, such discrepancies could be caused by the kinetics of CRISPRa regulation, as overexpression is not stable but decreases over time, as well as by possible epigenetic effects acting on RNA-mediated processes and preventing or delaying translation. In addition, we were unable to detect IL1β in A549 medium by ELISA or to optimize the dilutions of some of the samples so that their levels would fall within the recommended assay range (e.g. the measured IL10 concentrations in co-culture medium was below the recommended assay range value).
Our results suggest that TLR10 may mediate the extent of inflammation induced by SARS-CoV-2 by downregulating the release of inflammatory cytokines and chemokines such as CXCL10, IL6, IL8, and IFNβ. Whether the immunosuppressive effect of TLR10 on epithelial cells is desirable in patients with severe COVID-19 needs to be investigated in further experiments, as overexpression of TLR10 also leads to downregulation of IFNβ, a key component of the antiviral response that is already suppressed by the elusive mechanism of SARS-CoV-2 (46). Delayed type-I interferon signaling promotes the accumulation of proinflammatory cytokines and chemokines, which may worsen the prognosis of patients. However, we believe that the immunomodulatory effects of TLR10 may represent an opportunity for intervention approaches to rebalance the excessive immune response at a later stage of infection, when an excess of inflammatory cytokines can become harmful to the tissue (cytokine storm).
Further studies are needed to determine the TLR10 ligands, to explain the mode of action by which it exerts immunomodulatory functions, and to determine the expression kinetics of target genes when CRISPRa is used. The lung is an organ suitable for drug delivery in aerosolized form by inhalation. Recent advances in inhalation strategies (47) may enable the development of inhalable liquid or powder CRISPR-based formulations for therapies and, in perspective, lead to the development of novel treatment strategies that utilize the immunomodulatory functions of TLR10.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
ŠK: Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft. MN: Conceptualization, Formal analysis, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing. JO: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Slovenian Research and Innovation Agency (ARIS) through research programme P4-0220 and Slovenian young researchers programme (ŠK).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1490478/full#supplementary-material
1. Astuti I, Ysrafil. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab Syndrome: Clin Res Rev. (2020) 14:407–12. doi: 10.1016/j.dsx.2020.04.020
2. Wang J, Jiang M, Chen X, Montaner LJ. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol. (2020) 108:17–41. doi: 10.1002/JLB.3COVR0520-272R
3. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. (2020) 724:718–24. doi: 10.1126/science.abc6027
4. Liu X, Yin S, Chen Y, Wu Y, Zheng W, Dong H, et al. LPS-induced proinflammatory cytokine expression in human airway epithelial cells and macrophages via NF-κB, STAT3 or AP-1 activation. Mol Med Rep. (2018) 17:5484–91. doi: 10.3892/mmr.2018.8542
5. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
6. Coperchini F, Chiovato L, Croce L. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Frontiers in Immunology. (2021) 12:1–8. doi: 10.3389/fimmu.2021.668507
7. Bals R, Hiemstra PS. Innate immunity in the lung: How epithelial cells fight against respiratory pathogens. Eur Respir J. (2004) 23:327–33. doi: 10.1183/09031936.03.00098803
8. Onofrio L, Caraglia M, Facchini G, Margherita V, Placido S, Buonerba C. Toll-like receptors and COVID-19: A two-faced story with an exciting ending. Future Sci OA. (2020) 6:10–3. doi: 10.1073/pnas.1410293111
9. Poulas K, Farsalinos K, Zanidis C. Activation of TLR7 and innate immunity as an efficient method against COVID-19 pandemic: imiquimod as a potential therapy. Nat Rev Immunol. (2020) 13:875–87. doi: 10.1038/nri3547
10. Frank MG, Wallach T, Raden M, Hinkelmann L, Brehm M, Rabsch D, et al. Distinct SARS-CoV-2 RNA fragments activate Toll-like receptors 7 and 8 and induce cytokine release from human macrophages and microglia. Frontiers Immunol. (2023) 13:1–14. doi: 10.3389/fimmu.2022.1066456
11. Lee IH, Lee JW, Kong SW. A survey of genetic variants in SARS-CoV-2 interacting domains of ACE2, TMPRSS2 and TLR3/7/8 across populations. Infection, Genetics and Evolution. (2020) 85 (5):104507. doi: 10.1016/j.meegid.2020.104507
12. Khan S, Shafiei MS, Longoria C, Schoggins JW, Savani RC, Zaki H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. ELife. (2021) 10:1–26. doi: 10.7554/eLife.68563
13. Aboudounya MM, Heads RJ. COVID-19 and toll-like receptor 4 (TLR4): SARS-coV-2 may bind and activate TLR4 to increase ACE2 expression, facilitating entry and causing hyperinflammation. Mediators of Inflammation. (2021) 14:8874339. doi: 10.1155/2021/8874339
14. Zheng M, Karki R, Williams EP, Yang D, Fitzpatrick E, Vogel P, et al. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat Immunol. (2021) 22:829–38. doi: 10.1038/s41590-021-00937-x
15. Haystead T, Lee E, Cho K, Gullickson G, Hughes P, Krafsur G, et al. Investigation of SARS-CoV-2 individual proteins reveals the in vitro and in vivo immunogenicity of membrane protein. Sci Rep. (2023) 13:1–11. doi: 10.1038/s41598-023-49077-2
16. Fore F, Indriputri C, Mamutse J, Nugraha J. TLR10 and its unique anti-inflammatory properties and potential use as a target in therapeutics. Immune Network. (2020) 20:1–10. doi: 10.4110/in.2020.20.e21
17. Sha Q, Truong-Tran AQ, Plitt JR, Beck LA, Schleimer RP. Activation of airway epithelial cells by toll-like receptor agonists. Am J Respir Cell Mol Biol. (2004) 31:358–64. doi: 10.1165/rcmb.2003-0388OC
18. Knez Š, Narat M, Ogorevc J. Differential gene expression induced by different TLR agonists in A549 lung epithelial cells is modulated by CRISPR activation of TLR10. Biomolecules. (2023) 13(1):19. doi: 10.3390/biom13010019
19. Verma R, Kim JY. 1,25-dihydroxyvitamin D3 facilitates M2 polarization and upregulates TLR10 expression on human microglial cells. Neuroimmunomodulation. (2016) 23 (2):75–80.
20. Sinopoli A, Sciurti A, Isonne C, Santoro MM, Baccolini V. The efficacy of multivitamin, vitamin A, vitamin B, vitamin C, and vitamin D supplements in the prevention and management of COVID-19 and long-COVID: an updated systematic review and meta-analysis of randomized clinical trials. Nutrients. (2024) 16(9):1345. doi: 10.3390/nu16091345
21. Majidi N, Rabbani F, Gholami S, Gholamalizadeh M, BourBour F, Rastgoo S, et al. The effect of vitamin C on pathological parameters and survival duration of critically ill coronavirus disease 2019 patients: A randomized clinical trial. Front Immunol. (2021) 12:717816. doi: 10.3389/fimmu.2021.717816
22. Kümmel LS, Krumbein H, Fragkou PC, Hünerbein BL, Reiter R, Papathanasiou KA, et al. Vitamin D supplementation for the treatment of COVID-19: A systematic review and meta-analysis of randomized controlled trials. Front Immunol. (2022) 13:1023903. doi: 10.3389/fimmu.2022.1023903
23. Chavez A, Wiegand DJ, Ter-ovanesyan D, Braff JL, Davidsohn N. Highly-efficient Cas9-mediated transcriptional programming. Nature. (2015) 12 (4):326–8. doi: 10.1038/nmeth.3312
24. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
25. Guan Y, Ranoa DRE, Jiang S, Mutha SK, Li X, Baudry J, et al. Human TLRs 10 and 1 share common mechanisms of innate immune sensing but not signaling. J Immunol. (2010) 184:5094–103. doi: 10.4049/jimmunol.0901888
26. Regan T, Nally K, Carmody R, Houston A, Shanahan F, MacSharry J, et al. Identification of TLR10 as a key mediator of the inflammatory response to listeria monocytogenes in intestinal epithelial cells and macrophages. J Immunol. (2013) 191:6084–92. doi: 10.4049/jimmunol.1203245
27. Hasan U, Chaffois C, Gaillard C, Saulnier V, Merck E, Tancredi S, et al. Human TLR10 is a functional receptor, expressed by B cells. J Immunol. (2005) 174:2942–50. doi: 10.4049/jimmunol.174.5.2942
28. Oosting M, Cheng SC, Bolscher JM, Vestering-Stenger R, Plantinga TS, Verschueren IC, et al. Human TLR10 is an anti-inflammatory pattern-recognition receptor. Proceedings of the National Academy of Sciences of the United States of America (2014) 111 (42):E4478–84. doi: 10.1073/pnas.1410293111
29. Stappers MHT, Oosting M, Ioana M, Reimnitz P, Mouton JW, Netea MG, et al. Genetic variation in TLR10, an inhibitory toll-like receptor, influences susceptibility to complicated skin and skin structure infections. J Infect Dis. (2015) 212:1491–9. doi: 10.1093/infdis/jiv229
30. Hasan UA, Dollet S, Vlach J. Differential induction of gene promoter constructs by constitutively active human TLRs. Biochem Biophys Res Commun. (2004) 321:124–31. doi: 10.1016/j.bbrc.2004.06.134
31. Lee SMY, Yip TF, Yan S, Jin DY, Wei HL, Guo RT, et al. Recognition of double-stranded RNA and regulation of interferon pathway by toll-like receptor 10. Front Immunol. (2018) 9:516. doi: 10.3389/fimmu.2018.00516
32. Henrick BM, Yao XD, Zahoor MA, Abimiku A, Osawe S, Rosenthal KL. TLR10 senses HIV-1 proteins and significantly enhances HIV-1 infection. Front Immunol. (2019) 10:482. doi: 10.3389/fimmu.2019.00482
33. Lee SMY, Kok KH, Jaume M, Cheung TKW, Yip TF, Lai JCC, et al. Toll-like receptor 10 is involved in induction of innate immune responses to influenza virus infection. Proc Natl Acad Sci United States America. (2014) 111:3793–8. doi: 10.1073/pnas.1324266111
34. Verma R, Jung JH, Kim JY. 1,25-Dihydroxyvitamin D3 up-regulates TLR10 while down-regulating TLR2, 4, and 5 in human monocyte THP-1. J Steroid Biochem Mol Biol. (2014) 141:1–6. doi: 10.1016/j.jsbmb.2013.12.012
35. Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. (2020) 53:25–32. doi: 10.1016/j.cytogfr.2020.05.003
36. Mabrey FL, Morrell ED, Wurfel MM. TLRs in COVID-19: How they drive immunopathology and the rationale for modulation. Innate Immun. (2021) 27:503–13. doi: 10.1177/17534259211051364
37. Karki R, Sharma BR, Tuladhar S, Williams EP, Zalduondo L, Samir P, et al. Synergism of TNF-α and IFN-γ Triggers inflammatory cell death, tissue damage, and mortality in SARS-coV-2 infection and cytokine shock syndromes. Cell. (2021) 184:149–168.e17. doi: 10.1016/j.cell.2020.11.025
38. Soy M, Keser G, Atagündüz P, Tabak F, Atagündüz I, Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. (2020) 39:2085–94. doi: 10.1007/s10067-020-05190-5
39. Hou F, Xiao K, Tang L, Xie L. Diversity of macrophages in lung homeostasis and diseases. Front Immunol. (2021) 12:753940. doi: 10.3389/fimmu.2021.753940
40. Ropa J, Cooper S, Capitano ML, Van’t Hof W, Broxmeyer HE. Human hematopoietic stem, progenitor, and immune cells respond ex vivo to SARS-coV-2 spike protein. Stem Cell Rev Rep. (2021) 17:253–65. doi: 10.1007/s12015-020-10056-z
41. Paolini A, Borella R, Biasi S, Neroni A, Mattioli M, Lo D, et al. Cell death in coronavirus infections: uncovering its role during COVID-19. Cells. (2021) 10(7):1585. doi: 10.3390/cells10071585
42. Gschwandtner M, Derler R, Midwood KS. More than just attractive: how CCL2 influences myeloid cell behavior beyond chemotaxis. Front Immunol. (2019) 10:2759. doi: 10.3389/fimmu.2019.02759
43. Duell BL, Cripps AW, Schembri MA, Ulett GC. Epithelial cell coculture models for studying infectious diseases: Benefits and limitations. J Biomed Biotechnol. (2011) 852419:1–9. doi: 10.1155/2011/852419
44. Josić K, López JM, Ott W, Shiau LJ, Bennett MR. Stochastic delay accelerates signaling in gene networks. PloS Computational Biology. (2011) 7 (11).doi: 10.1371/journal.pcbi.1002264
45. Gedeon T, Bokes P. Delayed protein synthesis reduces the correlation between mRNA and protein fluctuations. Biophysical Journal. (2012) 103(3):377–85.
46. Li JY, Liao CH, Wang Q, Tan YJ, Luo R, Qiu Y, et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. (2020) 286:198074. doi: 10.1016/j.virusres.2020.198074
Keywords: SARS-CoV-2, COVID-19, Toll-like receptor 10, interferons, proinflammatory cytokines, innate immunity
Citation: Knez Š, Narat M and Ogorevc J (2025) TLR10 overexpression modulates immune response in A549 lung epithelial cells challenged with SARS-CoV-2 S and N proteins. Front. Immunol. 15:1490478. doi: 10.3389/fimmu.2024.1490478
Received: 03 September 2024; Accepted: 16 December 2024;
Published: 20 January 2025.
Edited by:
Liliana Oliveira, Universidade do Porto, PortugalReviewed by:
Karen Bohmwald, Autonomous University of Chile, ChileCopyright © 2025 Knez, Narat and Ogorevc. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jernej Ogorevc, amVybmVqLm9nb3JldmNAYmYudW5pLWxqLnNp
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.