- 1Department of Molecular Cardiovascular Pharmacology, Graduate School of Pharmaceutical Sciences, Chiba University, Chiba, Japan
- 2Laboratory for Immune Homeostasis, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
The thymus is a central organ that controls T cell development. Thymic epithelial cells (TECs) create a unique microenvironment essential for the differentiation of major histocompatibility complex (MHC)-restricted and self-tolerant T cells. TECs present a complex of self-peptides and MHC molecules (self-pMHCs) to immature T cells and regulate their survival and differentiation based on their affinity for self-pMHCs. The processing of self-peptides in TECs depends on bulk protein degradation systems, specifically autophagy and proteasomes. Studies using autophagy- and proteasome-deficient mouse models have demonstrated that these degradation systems in TECs are indispensable for maintaining immune homeostasis. Although autophagy and proteasomes are ubiquitous in nearly all eukaryotic cells, TECs exhibit unique characteristics in their autophagy and proteasome functions. Autophagy in TECs is constitutively active and independent of stress responses, while TEC proteasomes contain specialized catalytic subunits. This review summarizes the distinctive characteristics of autophagy and proteasomes in TECs and their roles in immune system regulation.
1 Introduction
The thymus is the central organ responsible for producing immunocompetent T cells. Within the thymus, thymic epithelial cells (TECs) create a specialized microenvironment for the generation of T cells that express major histocompatibility complex (MHC)-restricted and self-tolerant T-cell receptors (TCRs) (1, 2). TECs are categorized into two major subtypes based on their localization: cortical thymic epithelial cells (cTECs) and medullary thymic epithelial cells (mTECs) (3–5). Bone marrow-derived T cell progenitors enter the thymus through blood vessels at the corticomedullary junction of the thymic lobules. The CD4–CD8– double-negative (DN) immature thymocytes then migrate to the outer cortex, where they interact with cTECs and differentiate into CD4+CD8+ double-positive (DP) T cells that express a diverse repertoire of TCRs. These DP cells undergo positive selection through interactions with cTECs displaying self-peptide–MHC complexes (self-pMHCs) (6–8). During this process, cTECs present self-pMHCs to DP cells and facilitate their survival and differentiation into CD4 or CD8 single-positive T cells (CD4 or CD8 T cells) that express TCRs with moderate affinity for self-pMHCs. MHC class I (MHC-I) and MHC class II (MHC-II) molecules present self-peptides to CD8 and CD4 T cells, respectively. Positively selected T cells migrate into the medulla and interact with mTECs. mTECs express a wide variety of peripheral tissue-specific antigens, in part through the function of the transcriptional regulator Aire (9), and also by replicating peripheral tissue-specific gene expression programs, a function carried out by the recently identified subset of mimetic TECs (10). T cells bearing a TCR with a high affinity for self-pMHC on mTECs undergo apoptosis during negative selection (7, 9). Alternatively, strong TCR signaling facilitates translocation of self-reactive CD8 T cells from thymus to the periphery and their development into mature and self-tolerant clones (11). Interactions between T cells and mTECs also promote the differentiation of immunosuppressive regulatory T cells (Tregs) in the thymus (12, 13).
The generation of self-peptides in TECs depends on bulk protein degradation systems, such as autophagy and proteasomes, which are ubiquitously present in eukaryotic cells and regulate various biological processes, including cell survival, proliferation, differentiation, and death (14). Compared with dendritic cells, which rely on the capture of exogenous proteins by endocytosis for processing of peptides for MHC-II, TECs are less active in endocytosis to present exogenous peptides (15, 16). Therefore, it is reasonable that TECs utilize cytoplasmic protein degradation systems, i.e. autophagy and proteasomes, for processing of self-peptides from endogenous self-antigens. Interestingly, autophagy and proteasomes in TECs exhibit unique characteristics compared with those in other cell types. While autophagy activation in most cells depends on cellular stress, autophagy in TECs is constitutively active and appears to be independent of stress responses (17, 18). Additionally, proteasomes in TECs contain unique catalytic subunits (19, 20). These distinctive characteristics are crucial for the processing of self-peptides and the selection of T cells (14). This review focuses on the molecular processes and functions of autophagy and proteasomes in TECs.
1.1 General Mechanisms of Autophagy
Autophagy, originally identified as a process to accelerate material and energy recycling, involves the digestion of proteins and organelles via autophagy-specific double-membrane vesicles that fuse with lysosomes, forming autophagosomes. This non-selective bulk autophagy is known as macroautophagy (21–23). Under non-starvation conditions, the mechanistic/mammalian target of rapamycin (mTOR) complex (mTORC1) suppresses autophagy by inactivating Unc51-like kinase 1 (ULK1), a protein kinase implicated in autophagy initiation. Starvation reduces mTOR activity, leading to the activation of ULK1 (24, 25). Activated ULK1 then phosphorylates proteins involved in autophagy initiation, including beclin-1, a key protein that activates vacuolar protein sorting 34 (VPS34) (26). VPS34, a catalytic subunit of class III phosphatidylinositol 3-kinase (PI3K), plays a role in endocytosis, intracellular vesicular trafficking, and autophagosome formation (27).
Autophagy is not limited to starvation; it is also activated by various forms of cellular stress, such as mitochondrial damage, oxidative stress, and hypoxia. The selective autophagy of mitochondria, known as mitophagy, has gained considerable attention (22, 28). Mitochondrial damage, primarily caused by reactive oxygen species (ROS), causes mitophagy via phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1) and the E3 ubiquitin ligase, Parkin (29, 30).
1.2 Role of autophagy in TECs in T cell selection and development
Autophagy in TECs has been primarily observed in autophagy reporter transgenic mice that ubiquitously express a green fluorescent protein (GFP)-fused form of microtubule-associated protein light chain 3 (LC3), which is crucial for autophagosome formation. In TECs, constitutive activation of autophagy occurs even in the absence of starvation or infection, indicating that autophagy is triggered by stress-independent mechanisms (17). As TECs are less active in endocytosis to present exogenous peptides, autophagy is likely to highly active to process endogenously expressed self-peptides in TECs (15, 16). Studies of purified thymic stromal cells revealed that cTECs exhibited higher autophagic activity than mTECs. The role of autophagy in TECs was initially investigated using autophagy-deficient mice with a targeted disruption of Atg5, a gene essential for autophagosome formation (18). Transplantation of an Atg5-deficient thymus into athymic nude mice showed that autophagy deficiency did not affect overall T cell development. However, it impaired the shaping of the T cell repertoire and led to severe colitis and multi-organ inflammation in the recipient nude mice, indicating that autophagy in TECs is required for the selection of the T cell repertoire and the establishment of tolerance (18). While this study could not determine whether self-tolerance induction was ascribed to autophagy in mTECs or cTECs, subsequent studies suggested that autophagy supports the loading of antigens expressed by Aire+ mTECs onto MHC-II for the negative selection of CD4 T cells (31).
The role of autophagy in MHC class II-mediated CD4 T cell development has also been explored in mice lacking genes crucial for lysosomal-autophagosomal proteolysis and autophagosome formation. The lysosomal endopeptidase cathepsin L is highly expressed in cTECs and regulates the processing of MHC-II invariant chain (Ii) and generation of self-peptides for MHC-II-mediated CD4 T cell development (32, 33). Endosomal and lysosomal thymus-specific serine protease (TSSP) contributes to the diversification of the functional CD4 T cell repertoire in the thymus (34, 35). Lysosomal-associated membrane protein 2 (LAMP2) is a lysosomal protein that promotes the fusion of autophagic vacuoles with lysosomes during macroautophagy (36, 37). LAMP2 is highly expressed in cTECs. Although deletion of Lamp2 in thymic stromal cells did not affect the overall differentiation of cTECs and mTECs, it specifically impaired CD4 T cell development without affecting the CD8 lineage. Mechanistically, inactivation of autophagy in Lamp2-deficient cTECs caused defects in MHC-II processing, leading to a marked reduction in CD4 TCR repertoire diversity (38).
The role of VPS34, a catalytic subunit of class III PI3K, in TECs was elucidated by the TEC-specific disruption of Vps34 in mice. Loss of VPS34 in TECs resulted in almost complete inactivation of autophagy in both cTECs and mTECs and abolished the positive selection of CD4 T cells without affecting CD8 T cells. TCR sequencing revealed that deletion of Vps34 in TECs altered T cell repertoire properties and reduced clonal sharing. cTECs lacking Vps34 exhibited an increased abundance of invariant chain intermediates bound to surface MHC class II molecules, indicating altered antigen processing (39). Collectively, autophagy deficiency due to the deletion of Lamp2 or Vps34 in TECs demonstrates that autophagy is indispensable for the processing of self-pMHC class II complexes in TECs and the generation of a broad CD4 TCR repertoire (Figure 1).
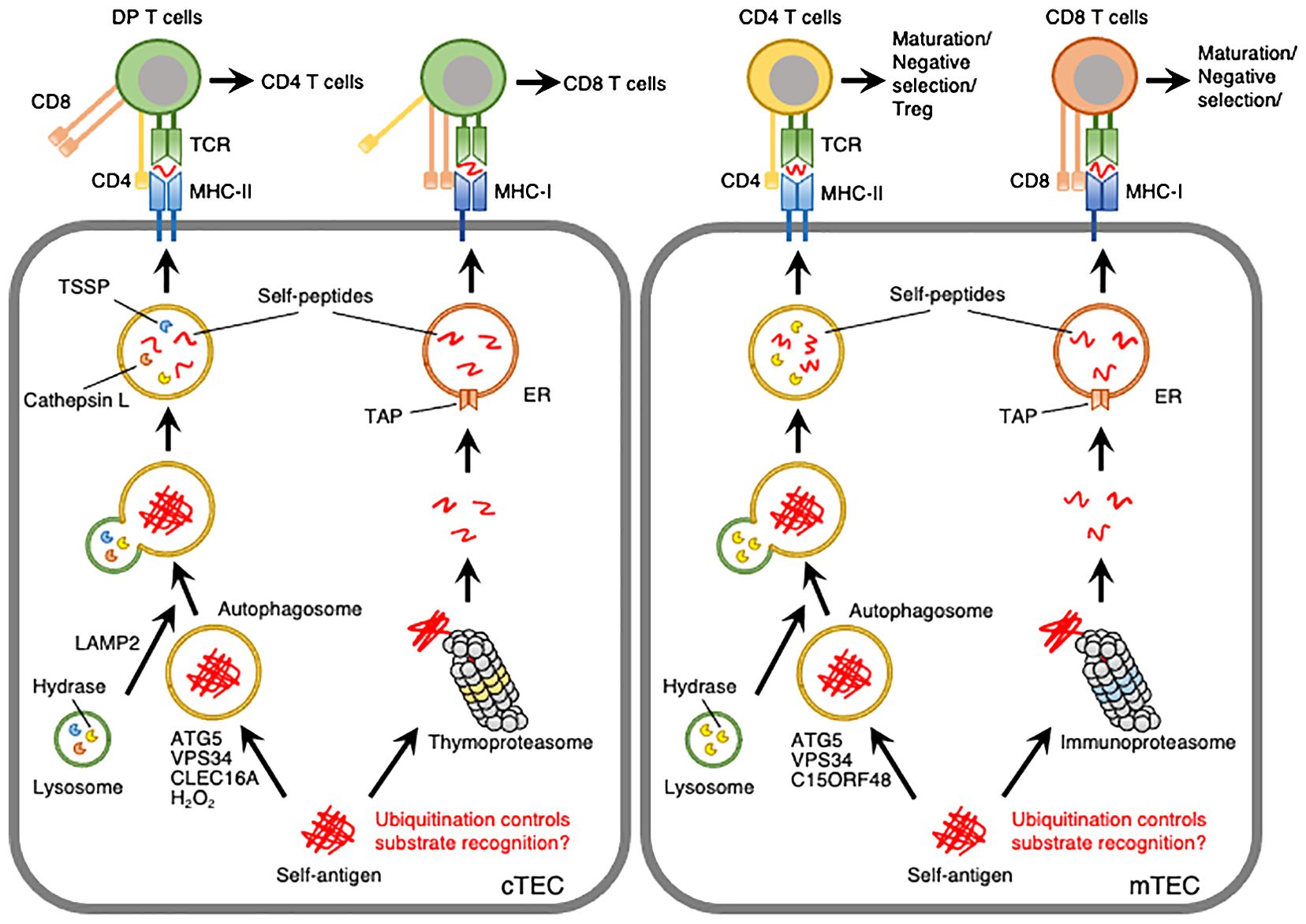
Figure 1. Model depicting autophagy- or proteasome-mediated processing of self-peptides in cTECs and mTECs. Self-antigens expressed in TECs are processed into self-peptides by autophagy or proteasomes and then loaded onto MHC-II or MHC-I molecules, respectively. ATG5 and VPS34 are required for autophagy in cTECs and mTECs. LAMP2, CLEC16A, and hydrogen peroxide (H2O2) are involved in autophagy in cTECs. C15ORF48 is closely associated with autophagy initiation in mTECs and, to a lesser extent, in cTECs. Thymoproteasomes and immunoproteasomes are expressed in cTECs and mTECs, respectively, with distinct catalytic subunits and substrate-processing activities. Post-translational modifications, such as ubiquitination, may be involved in substrate recognition by autophagy and proteasomes.
1.3 Molecular mechanisms of autophagy in TECs
While the importance of autophagy in TECs for T cell selection and development has been established in autophagy-deficient mouse models, the molecular mechanisms underlying autophagy in TECs remain unclear. Autophagy in TECs is constitutively active and should be dependent on specific mechanisms that may differ from those of typical autophagy (17, 18).
C-type lectin domain family 16A (CLEC16A) is a membrane-associated endosomal protein that functions as an E3 ubiquitin ligase and is implicated in various autoimmune disorders, such as multiple sclerosis, type 1 diabetes, and systemic lupus erythematosus (40). Genetic evidence has demonstrated the involvement of CLEC16A in autophagy in various cell types (41–43). A previous study showed that whole-body knockdown of Clec16a in non-obese diabetic (NOD) mice with type 1 diabetes reduced autophagic activity in cTECs and affected T cell selection in the thymus. Interestingly, while Atg5 disruption causes autoimmunity, Clec16a knockdown mice exhibit T cell hyporeactivity and suppression of autoimmune phenotypes, even though both impair autophagy activity in TECs (44). Although the exact mechanism behind the suppression of autoimmunity by Clec16a deficiency is unclear, CLEC16A-dependent autophagy in TECs may regulate T cell maturation without significantly impacting the T cell repertoire.
mTOR is a key regulator of various biological processes, including cell growth, differentiation, and autophagy (45). The role of mTOR in TECs was investigated using pharmacological inhibition of mTOR kinase activity and TEC-specific deletion of Mtor. The mTOR inhibitor rapamycin induces severe thymic atrophy and reduces the number of TECs. TEC-specific deletion of Mtor caused a significant reduction in mTECs, defects in thymocyte differentiation, and autoimmune phenotypes, such as multi-organ immune cell infiltration and autoreactive antibodies in sera. Mechanistically, deletion of Mtor in TECs led to upregulation of autophagy-mediated degradation of β-catenin and inactivation of Wnt/β-catenin signaling, suggesting that mTOR promotes TEC development and maturation by suppressing autophagy-mediated over-degradation of β-catenin (46). Since the role of mTOR in autophagy-dependent processing of self-peptides in TECs remains unclear, further studies on the role of mTOR in TECs are needed.
H2O2 is a relatively long-lived, cell-permeable ROS that acts as an inducer of autophagy (47). H2O2 activates AMP-activated protein kinase (AMPK) in conjunction with a decrease in the ATP: AMP ratio (48), leading to ULK1-mediated autophagy. Additionally, H2O2 enhances the oxidation of ATG4, thereby inactivating the LC3-delipidation activity and promoting autophagosome formation (49). H2O2 also initiates autophagy by altering the thiol redox state (50), inhibiting mTOR signaling (51), and inducing beclin-1 expression (52). cTECs exhibit low expression of the H2O2-quenching enzyme catalase, suggesting the involvement of H2O2 in autophagy within these cells (53). This involvement has been studied using transgenic mice stably expressing mitochondria-targeted human catalase (mCat Tg). mCat Tg mice showed a significant reduction in mitochondrial H2O2 levels and autophagy in cTECs without affecting mTECs. Furthermore, these mice exhibited defects in the clonal deletion of thymocytes and developed autoimmune phenotypes, such as increased tissue infiltration of lymphocytic cells and serum antinuclear antigen reactivity (54). These findings suggest that the production of H2O2, facilitated by the low expression of catalase, induces constitutive autophagy and regulates the clonal deletion of T cells in cTECs (Figure 1).
Recently, our studies have demonstrated that the mitochondrial protein chromosome 15 open reading frame 48 (C15ORF48) plays a crucial role in initiating autophagy in TECs (55). C15ORF48 functions as an accessory subunit of the electron transport chain complex IV, where it suppresses cytochrome c oxidase activity (56, 57). We identified C15ORF48 as an inducer of autophagy in human lung cancer cells. Notably, C15ORF48 promotes autophagy independently of mTOR phosphorylation, which is an indicator of starvation stress, or mitophagic activity. This suggests that C15ORF48 initiates autophagy independently of starvation stress or mitochondrial damage. C15ORF48 has been shown to lower the mitochondrial membrane potential and reduce intracellular ATP levels, thereby activating the pro-autophagic signaling pathway AMPK-ULK1. Single-cell RNA sequencing analysis of thymic cells revealed high expression levels of C15orf48 in both cTECs and mature mTECs. Importantly, whole-body C15orf48 knockout mice showed significant reduction in autophagy in mTECs and, to a lesser extent, in cTECs. Moreover, C15orf48-deficient mice exhibited autoimmune features, including the production of self-reactive antibodies in the serum, multi-organ infiltration of inflammatory cells, and glomerulitis-like IgG accumulation in the kidneys. Transplantation of C15orf48-deficient thymic stroma into athymic nude mice resulted in similar autoimmune phenotypes. Global T cell development remained largely unaffected by C15orf48-deficiency, except for a slight reduction in mature CD4 T cells. Thus, this study highlights the role of C15ORF48 in the induction of stress-independent autophagy in TECs, likely independent of mitophagy, and suggests it may shape the T cell repertoire by regulating T cell selection, particularly in mTECs (55) (Figure 1). Future studies are needed to explore the impact of C15orf48-deficiency on the T cell repertoire.
1.4 Thymoproteasomes in cTECs
Proteasomes are multicatalytic complexes that regulate protein degradation. In general, ubiquitination of target proteins is a hallmark of proteasome-mediated degradation (58). The cytosolic proteasome is a 26S protein complex consisting of a 20S enzymatic core particle flanked by two 19S regulatory particles. The 19S regulatory particles recognize ubiquitinated target proteins. The 20S core particle is a multicatalytic protease complex composed of 28 subunits arranged cylindrically into four heteroheptameric rings: α1-7, β1-7, β1-7, and α1-7. Within the proteasome, the β1, β2, and β5 subunits function as catalytic components (58). Peptides processed by proteasomes in the cytosol are translocated into the lumen of the endoplasmic reticulum (ER) by a transporter associated with antigen processing (TAP) and subsequently loaded onto MHC-I molecules (59). cTECs uniquely express the catalytic subunit β5t (Psmb11), which forms a specialized proteasome known as the thymoproteasome (19). The β5t subunit has altered proteolytic activity that leads to the preferential cleavage of proteins at hydrophilic peptide residues, reduced chymotrypsin-like activity, and reduced enzyme kinetics compared with β5 (60). The β5t-containing proteasome generates peptides with not only different amino acid residues but also different quantities from those produced by other proteasomes, and these quantitative and qualitative differences may lead to the presentation of unique MHC-I-associated peptides (61, 62). Targeted disruption of Psmb11 leads to defects in the maturation of CD8 T cells, alterations in the TCR repertoire, and impaired T cell responses (19, 63, 64). Since proteasomes in TECs are crucial for generating peptides presented by MHC-I molecules (65), these findings indicate that thymoproteasomes contribute to the generation of self-peptides necessary for the positive selection of CD8 T cells (Figure 1).
1.5 Immunoproteasomes in mTECs
mTECs express a different type of proteasome, the immunoproteasome, which contains the catalytic subunits β1i (Psmb9), β2i (Psmb10), and β5i (Psmb8) (20, 62). Psmb8, Psmb9, and Psmb10 triple knockout mice showed a reduction in CD8 T cells and decreased expression of MHC-I in the thymus. Mass spectrometry analysis of peptides associated with MHC-I revealed that immunoproteasome deficiency altered the MHC-I-associated peptide repertoire (66). The peptide-processing-independent role of the immunoproteasome in mTECs was shown in Psmb8 and Psmb10 double knockout (dKO) mice. These mice exhibited low expression of Psmb9, which rendered them almost complete deficiencies in immunoproteasomes. dKO mice exhibited accelerated thymic involution, characterized by a reduction in CD8 T cells and mTECs, as well as the induction of multiorgan autoimmune manifestations. Loss of Psmb8 and Psmb10 led to proteotoxic stress in mTECs, resulting in the exhaustion of postnatal mTEC progenitors (67). These findings suggest that the immunoproteasome plays a crucial role in generation of MHC-I-associated self-peptides and maintaining mTEC homeostasis, thereby supporting MHC-I-mediated CD8 T cell selection.
The thymus lacking thymoproteasomes and immunoproteasomes was examined in Psmb8, Psmb9, Psmb10, and Psmb11 quadruple knockout (4KO) mice. The 4KO mice showed a severe defect in generation of CD8 T cells without affecting that of CD4 T cells. Loss of thymoproteasomes and immunoproteasomes increased apoptosis-dependent negative selection of CD8 T cells in the thymus (68). These results suggest that the restricted expression of thymoproteasomes and immunoproteasomes in the thymus is likely to promote a switch in the self-peptides presented by cTECs and mTECs. The difference in self-peptides displayed in the cortex and the medulla may be crucial for eviction of positively selected thymocytes from subsequent negative selection and the development of CD8 T cells (62).
2 Discussion
Autophagy- or proteasome-deficient mouse models have demonstrated the indispensability of these proteolytic systems in TECs for the processing of self-peptides and the selection and differentiation of T cells. However, molecular details about how autophagy and proteasomes in TECs selectively degrade self-peptides have been largely unknown. Most likely, the reason of this limitation is the difficulty in isolation of enough amount of primary TECs from thymus for biochemical and cell biological analyses (16, 69). Ubiquitination functions as a signal for substrate recognition in autophagy as well as proteasome-mediated degradation (22, 58). Ubiquitinated proteins interact with the autophagy machinery through autophagy adaptors and promote autophagosome formation (70). Therefore, it is plausible that the ubiquitination of self-peptides regulates their processing by autophagy and proteasomes in TECs. However, whether self-peptides are ubiquitinated in TECs remains unclear. As CLEC16A can associate with ubiquitin ligases and deubiquitinases (71–73), CLEC16A may be involved in ubiquitination of self-peptides in TECs. A study on the post-translational modifications of self-peptides in TECs would be instrumental in understanding the mechanisms underlying autophagy-mediated selective processing of self-peptides.
Recent studies have revealed that mTECs express lineage-defining transcription factors and convert themselves into mimetic TECs that possess characteristics of various peripheral-tissue cells (10). It is obscure whether these mimetic TECs utilize constitutive autophagy and immunoproteasomes for processing of self-peptides as observed in mTECs. Given that Aire is involved in accumulation of mimetic TECs (10) and that C15orf48 is highly expressed in Aire+ mature mTECs (55), mimetic TECs may have at least C15ORF48-dependent constitutive autophagy for processing of self-peptides. This hypothesis should be examined by isolating mimetic cells from thymus.
In conclusion, considering the importance of autophagy and proteasomes in TECs for the development and selection of immunocompetent T cells, understanding the molecular mechanisms of these protein degradation systems is essential for gaining insights into the nature of the immune system and immune-associated disorders. Therefore, the relationship between autophagy and proteasomes in TECs and human autoimmune diseases warrants further investigation.
Author contributions
NY: Conceptualization, Writing – original draft, Writing – review & editing. YT: Writing – review & editing. TA: Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by grants-in-aid for Scientific Research (24K09792, 20H03441) (NY, TA) and Early-Career Scientists (24K18269) (YT) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology, the Suzuken Memorial Foundation (YT), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (YT), Takeda Science Foundation (NY), TERUMO Life Science Foundation (NY), the Research Foundation for Pharmaceutical Sciences (NY), and CREST from Japan Science and Technology Agency (JPMJCR2011) (TA).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gill J, Malin M, Sutherland J, Gray D, Hollander G, Boyd R. Thymic generation and regeneration. Immunol Rev. (2003) 195:28–50. doi: 10.1034/j.1600-065x.2003.00077.x
2. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol. (2014) 14:377–91. doi: 10.1038/nri3667
3. Abramson J, Anderson G. Thymic epithelial cells. Annu Rev Immunol. (2017) 35:85–118. doi: 10.1146/annurev-immunol-051116-052320
4. Anderson G, Takahama Y. Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. (2012) 33:256–63. doi: 10.1016/j.it.2012.03.005
5. Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol. (2006) 6:127–35. doi: 10.1038/nri1781
6. Anderson G, Lane PJ, Jenkinson EJ. Generating intrathymic microenvironments to establish T-cell tolerance. Nat Rev Immunol. (2007) 7:954–63. doi: 10.1038/nri2187
7. Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. (2009) 9:833–44. doi: 10.1038/nri2669
8. Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. (2006) 24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601
9. Mathis D, Benoist C. Aire. Annu Rev Immunol. (2009) 27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532
10. Michelson DA, Hase K, Kaisho T, Benoist C, Mathis D. Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells. Cell. (2022) 185:2542–58 e18. doi: 10.1016/j.cell.2022.05.018
11. Badr ME, Zhang Z, Tai X, Singer A. CD8 T cell tolerance results from eviction of immature autoreactive cells from the thymus. Science. (2023) 382:534–41. doi: 10.1126/science.adh4124
12. Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. (2012) 30:531–64. doi: 10.1146/annurev.immunol.25.022106.141623
13. Wing K, Sakaguchi S. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat Immunol. (2010) 11:7–13. doi: 10.1038/ni.1818
14. Sousa LG, Rodrigues PM, Alves NL. T-cell selection in the thymus: New routes toward the identification of the self-peptide ligandome presented by thymic epithelial cells. Eur J Immunol. (2023) 53:e2250202. doi: 10.1002/eji.202250202
15. Klein L, Roettinger B, Kyewski B. Sampling of complementing self-antigen pools by thymic stromal cells maximizes the scope of central T cell tolerance. Eur J Immunol. (2001) 31:2476–86. doi: 10.1002/1521-4141(200108)31:8<2476::aid-immu2476>3.0.co;2-t
16. Ohigashi I, Matsuda-Lennikov M, Takahama Y. Peptides for T cell selection in the thymus. Peptides. (2021) 146:170671. doi: 10.1016/j.peptides.2021.170671
17. Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. (2004) 15:1101–11. doi: 10.1091/mbc.e03-09-0704
18. Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. (2008) 455:396–400. doi: 10.1038/nature07208
19. Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. (2007) 316(5829):1349–53. doi: 10.1126/science.1141915
20. Murata S, Takahama Y, Kasahara M, Tanaka K. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol. (2018) 19:923–31. doi: 10.1038/s41590-018-0186-z
21. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. (2011) 147:728–41. doi: 10.1016/j.cell.2011.10.026
22. Vargas JNS, Hamasaki M, Kawabata T, Youle RJ, Yoshimori T. The mechanisms and roles of selective autophagy in mammals. Nat Rev Mol Cell Biol. (2023) 24:167–85. doi: 10.1038/s41580-022-00542-2
23. Wang L, Klionsky DJ, Shen HM. The emerging mechanisms and functions of microautophagy. Nat Rev Mol Cell Biol. (2023) 24:186–203. doi: 10.1038/s41580-022-00529-z
24. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. (2009) 20:1981–91. doi: 10.1091/mbc.e08-12-1248
25. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. (2011) 13:132–41. doi: 10.1038/ncb2152
26. Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. (2001) 2:330–5. doi: 10.1093/embo-reports/kve061
27. Backer JM. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem J. (2016) 473:2251–71. doi: 10.1042/bcj20160170
28. Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. (2011) 12(1):9–14. doi: 10.1038/nrm3028
29. Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. (2010) 189:211–21. doi: 10.1083/jcb.200910140
30. Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PloS Biol. (2010) 8:e1000298. doi: 10.1371/journal.pbio.1000298
31. Aichinger M, Wu C, Nedjic J, Klein L. Macroautophagy substrates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance. J Exp Med. (2013) 210:287–300. doi: 10.1084/jem.20122149
32. Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, et al. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science. (1998) 280:450–3. doi: 10.1126/science.280.5362.450
33. Honey K, Nakagawa T, Peters C, Rudensky A. Cathepsin L regulates CD4+ T cell selection independently of its effect on invariant chain: a role in the generation of positively selecting peptide ligands. J Exp Med. (2002) 195(10):1349–58. doi: 10.1084/jem.20011904
34. Gommeaux J, Gregoire C, Nguessan P, Richelme M, Malissen M, Guerder S, et al. Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur J Immunol. (2009) 39:956–64. doi: 10.1002/eji.200839175
35. Viret C, Lamare C, Guiraud M, Fazilleau N, Bour A, Malissen B, et al. Thymus-specific serine protease contributes to the diversification of the functional endogenous CD4 T cell receptor repertoire. J Exp Med. (2011) 208:3–11. doi: 10.1084/jem.20100027
36. Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. (2000) 406:902–6. doi: 10.1038/35022595
37. Eskelinen EL, Illert AL, Tanaka Y, Schwarzmann G, Blanz J, Von Figura K, et al. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. (2002) 13:3355–68. doi: 10.1091/mbc.e02-02-0114
38. Rodrigues PM, Sousa LG, Perrod C, Maceiras AR, Ferreirinha P, Pombinho R, et al. LAMP2 regulates autophagy in the thymic epithelium and thymic stroma-dependent CD4 T cell development. Autophagy. (2023) 19:426–39. doi: 10.1080/15548627.2022.2074105
39. Postoak JL, Song W, Yang G, Guo X, Xiao S, Saffold CE, et al. Thymic epithelial cells require lipid kinase Vps34 for CD4 but not CD8 T cell selection. J Exp Med. (2022) 219:e20212554. doi: 10.1084/jem.20212554
40. Pandey R, Bakay M, Hakonarson H. CLEC16A-an emerging master regulator of autoimmunity and neurodegeneration. Int J Mol Sci. (2023) 24:8224. doi: 10.3390/ijms24098224
41. Kim S, Naylor SA, DiAntonio A. Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function. Proc Natl Acad Sci U S A. (2012) 109:E1072–81. doi: 10.1073/pnas.1120320109
42. Soleimanpour SA, Gupta A, Bakay M, Ferrari AM, Groff DN, Fadista J, et al. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell. (2014) 157:1577–90. doi: 10.1016/j.cell.2014.05.016
43. Redmann V, Lamb CA, Hwang S, Orchard RC, Kim S, Razi M, et al. Clec16a is critical for autolysosome function and purkinje cell survival. Sci Rep. (2016) 6:23326. doi: 10.1038/srep23326
44. Schuster C, Gerold KD, Schober K, Probst L, Boerner K, Kim MJ, et al. The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity. (2015) 42:942–52. doi: 10.1016/j.immuni.2015.04.011
45. Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. (2020) 21:183–203. doi: 10.1038/s41580-019-0199-y
46. Liang Z, Zhang L, Su H, Luan R, Na N, Sun L, et al. MTOR signaling is essential for the development of thymic epithelial cells and the induction of central immune tolerance. Autophagy. (2018) 14:505–17. doi: 10.1080/15548627.2017.1376161
47. Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. (2014) 15(6):411–21. doi: 10.1038/nrm3801
48. Choi SL, Kim SJ, Lee KT, Kim J, Mu J, Birnbaum MJ, et al. The regulation of AMP-activated protein kinase by H(2)O(2). Biochem Biophys Res Commun. (2001) 287(1):92–7. doi: 10.1006/bbrc.2001.5544
49. Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. (2007) 26:1749–60. doi: 10.1038/sj.emboj.7601623
50. Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. (2015) 22:377–88. doi: 10.1038/cdd.2014.150
51. Zhao S, Li L, Wang S, Yu C, Xiao B, Lin L, et al. H2O2 treatment or serum deprivation induces autophagy and apoptosis in naked mole-rat skin fibroblasts by inhibiting the PI3K/Akt signaling pathway. Oncotarget. (2016) 7:84839–50. doi: 10.18632/oncotarget.13321
52. Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ. (2008) 15:171–82. doi: 10.1038/sj.cdd.4402233
53. Griffith AV, Venables T, Shi J, Farr A, van Remmen H, Szweda L, et al. Metabolic damage and premature thymus aging caused by stromal catalase deficiency. Cell Rep. (2015) 12:1071–9. doi: 10.1016/j.celrep.2015.07.008
54. Semwal MK, Hester AK, Xiao Y, Udeaja C, Cepeda S, Verschelde JS 2nd, et al. Redox status regulates autophagy in thymic stromal cells and promotes T cell tolerance. Proc Natl Acad Sci U S A. (2022) 119:e2204296119. doi: 10.1073/pnas.2204296119
55. Takakura Y, Machida M, Terada N, Katsumi Y, Kawamura S, Horie K, et al. Mitochondrial protein C15ORF48 is a stress-independent inducer of autophagy that regulates oxidative stress and autoimmunity. Nat Commun. (2024) 15:953. doi: 10.1038/s41467-024-45206-1
56. Floyd BJ, Wilkerson EM, Veling MT, Minogue CE, Xia C, Beebe ET, et al. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol Cell. (2016) 63:621–32. doi: 10.1016/j.molcel.2016.06.033
57. Lee CQE, Kerouanton B, Chothani S, Zhang S, Chen Y, Mantri CK, et al. Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity. Nat Commun. (2021) 12:2130. doi: 10.1038/s41467-021-22397-5
58. Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci. (2009) 85:12–36. doi: 10.2183/pjab.85.12
59. Suh WK, Cohen-Doyle MF, Fruh K, Wang K, Peterson PA, Williams DB. Interaction of MHC class I molecules with the transporter associated with antigen processing. Science. (1994) 264:1322–6. doi: 10.1126/science.8191286
60. Sasaki K, Takada K, Ohte Y, Kondo H, Sorimachi H, Tanaka K, et al. Thymoproteasomes produce unique peptide motifs for positive selection of CD8(+) T cells. Nat Commun. (2015) 6:7484. doi: 10.1038/ncomms8484
61. Kuckelkorn U, Stubler S, Textoris-Taube K, Kilian C, Niewienda A, Henklein P, et al. Proteolytic dynamics of human 20S thymoproteasome. J Biol Chem. (2019) 294:7740–54. doi: 10.1074/jbc.ra118.007347
62. Frantzeskakis M, Takahama Y, Ohigashi I. The role of proteasomes in the thymus. Front Immunol. (2021) 12:646209. doi: 10.3389/fimmu.2021.646209
63. Nitta T, Murata S, Sasaki K, Fujii H, Ripen AM, Ishimaru N, et al. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity. (2010) 32:29–40. doi: 10.1016/j.immuni.2009.10.009
64. Takada K, Van Laethem F, Xing Y, Akane K, Suzuki H, Murata S, et al. TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8(+) T cells. Nat Immunol. (2015) 16:1069–76. doi: 10.1038/ni.3237
65. Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. (1999) 17:739–79. doi: 10.1146/annurev.immunol.17.1.739
66. Kincaid EZ, Che JW, York I, Escobar H, Reyes-Vargas E, Delgado JC, et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol. (2011) 13:129–35. doi: 10.1038/ni.2203
67. St-Pierre C, Morgand E, Benhammadi M, Rouette A, Hardy MP, Gaboury L, et al. Immunoproteasomes control the homeostasis of medullary thymic epithelial cells by alleviating proteotoxic stress. Cell Rep. (2017) 21:2558–70. doi: 10.1016/j.celrep.2017.10.121
68. Kincaid EZ, Murata S, Tanaka K, Rock KL. Specialized proteasome subunits have an essential role in the thymic selection of CD8(+) T cells. Nat Immunol. (2016) 17:938–45. doi: 10.1038/ni.3480
69. Nakagawa Y, Ohigashi I, Nitta T, Sakata M, Tanaka K, Murata S, et al. Thymic nurse cells provide microenvironment for secondary T cell receptor alpha rearrangement in cortical thymocytes. Proc Natl Acad Sci U S A. (2012) 109:20572–7. doi: 10.1073/pnas.1213069109
70. Mizushima N. Ubiquitin in autophagy and non-protein ubiquitination. Nat Struct Mol Biol. (2024) 31:208–9. doi: 10.1038/s41594-024-01217-6
71. Pearson G, Chai B, Vozheiko T, Liu X, Kandarpa M, Piper RC, et al. Clec16a, nrdp1, and USP8 form a ubiquitin-dependent tripartite complex that regulates beta-cell mitophagy. Diabetes. (2018) 67:265–77.
72. Gingerich MA, Liu X, Chai B, Pearson GL, Vincent MP, Stromer T, et al. An intrinsically disordered protein region encoded by the human disease gene CLEC16A regulates mitophagy. Autophagy. (2023) 19:525–43. doi: 10.1080/15548627.2022.2080383
Keywords: thymus, thymic epithelial cells, T cells, autophagy, proteasomes
Citation: Yamaguchi N, Takakura Y and Akiyama T (2024) Autophagy and proteasomes in thymic epithelial cells: essential bulk protein degradation systems for immune homeostasis maintenance. Front. Immunol. 15:1488020. doi: 10.3389/fimmu.2024.1488020
Received: 29 August 2024; Accepted: 09 October 2024;
Published: 25 October 2024.
Edited by:
Miho Shinzawa, National Institutes of Health (NIH), United StatesReviewed by:
Magdalena Paolino, Karolinska Institutet (KI), SwedenCopyright © 2024 Yamaguchi, Takakura and Akiyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Noritaka Yamaguchi, eWFtYWd1Y2hpbm9yaXRha2FAY2hpYmEtdS5qcA==; Taishin Akiyama, dGFpc2hpbi5ha2l5YW1hQHJpa2VuLmpw
 Noritaka Yamaguchi
Noritaka Yamaguchi Yuki Takakura1,2
Yuki Takakura1,2 Taishin Akiyama
Taishin Akiyama